Abstract
Objective
Deep learning algorithms were used to develop a model for predicting the staging and grading of renal clear cell carcinoma to inform clinicians’ treatment plans.
Methods
Clinical and pathological information was collected from 878 patients diagnosed with renal clear cell carcinoma in the Department of Urology, Peking University First Hospital. The patients were randomly assigned to the test set (n = 702) or the verification set (n = 176). Pathological staging and grading of renal clear cell carcinoma were predicted by preoperative clinical variables using deep learning algorithms. Receiver operating characteristic curves were used to evaluate the predictive accuracy as measured by the area under the receiver operating characteristic curve (AUC).
Results
For tumor pathological staging, AUC values of 0.933, 0.947, and 0.948 were obtained using the BiLSTM, CNN-BiLSTM, and CNN-BiGRU models, respectively. For tumor pathological grading, the AUC values were 0.754, 0.720, and 0.770, respectively.
Conclusions
The proposed model for predicting renal clear cell carcinoma allows for accurate projection of the staging and grading of renal clear cell carcinoma and helps clinicians optimize individual treatment plans.
Keywords: Renal clear cell carcinoma, deep learning, staging, grading, predictive model, treatment optimization
Introduction
Renal cell carcinoma, which accounts for 2% to 3% of malignant tumors in adults, is the most lethal malignant tumor in the urinary system. Its incidence is second only to prostate cancer and bladder cancer. Renal clear cell carcinoma is the main type of kidney cancer. It accounts for 82% to 90% of tumors in the kidney.1,2 The clinical staging of kidney cancer mainly relies on the combination of imaging and clinical data, which often has some deviation from the pathological staging of kidney cancer. The pathological staging of kidney cancer is the gold standard for diagnosis; however, the surgical strategy is mainly based on clinical staging before treatment. Therefore, better prediction of the pathological staging before surgery can better guide doctors in formulating treatment plans. Patients’ prognostic status after surgery is positively correlated with the renal function classification.3 Accurate preoperative pathological grading of renal clear cell carcinoma can also help clinicians choose better treatment plans.
Applications based on deep learning algorithms have developed rapidly in recent years, and their use in the medical field has continued to increase each year.4,5 Multiple studies have shown that deep learning algorithms outperform traditional statistical prediction models.6,7 While traditional multivariate prediction models have limited accuracy, deep learning models can make better use of extracted features, providing improved performance over the traditional models.8 In this study, multiple deep learning algorithms were used to construct a predictive model for the pathological staging and grading of renal clear cell carcinoma to assist clinicians in developing personalized treatment plans.
Methods
Patient selection
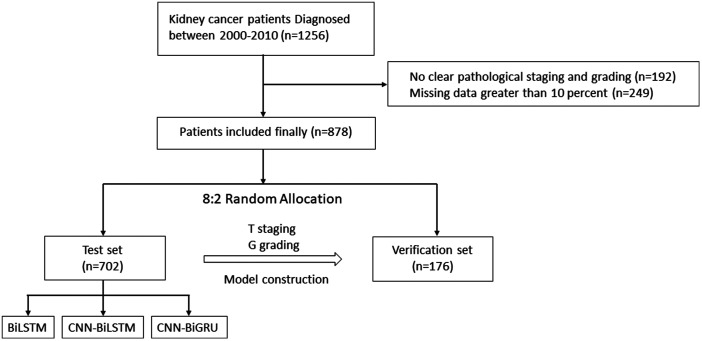
Clinical and pathological information was collected from 878 patients diagnosed with renal clear cell carcinoma in the Department of Urology, Peking University First Hospital from 23 January 2000 to 29 December 29. For patients with <10% missing clinical data, Lagrange interpolation was used to fill in any missing values.9 We preprocessed the input features before feeding them into the model. The non-numeric parts of the input variables were processed using one-hot encoding, and the numeric parts of the input variables were normalized. The inclusion criteria were confirmation of the diagnosis of renal clear cell carcinoma by postoperative pathology as well as complete clinical information, including sex, age, symptoms, chronic medical history, and selected preoperative tests. The exclusion criteria were the absence of clear postoperative pathological staging information and >10% missing clinical data. Patients who met the inclusion criteria were randomly divided into a test set (n = 702) and a verification set (n = 176) at a ratio of 8:2. A deep learning algorithm was used to construct a prediction model for the pathological staging of renal clear cell carcinoma through the test set, and the accuracy of the model was verified through the verification set. The detailed study design is shown in Figure 1. The prediction model constructed for this study was equivalent to a multivariate prediction model. The output of the prediction model was the specific T stage or G grade of the tumor. This study was approved by our ethics committee (Biomedical Research Ethics Committee of Peking University First Hospital, Beijing, China; Approval No. 2015[977]), and the requirement for informed consent was waived because the study did not involve any interventions.
Figure 1.
Flowchart of the specific patient screening process in the present study.
Study variables
The following data of the selected patients were modeled by 22 variables: preoperative clinical characteristics, including sex, age, body mass index, symptoms (e.g., low back pain, abdominal pain, and/or hematuria), and chronic medical history (e.g., history of hypertension, diabetes, and/or coronary heart disease); preoperative laboratory data, including the creatinine, albumin, sodium, calcium, phosphorus, magnesium, triglyceride, cholesterol, and uric acid concentrations and the white blood cell, lymphocyte, neutrophil, eosinophil, and basophil counts; and tumor information, including the maximum tumor diameter and tumor location (right and/or left kidney).
Deep learning analysis
Deep learning models are mainly based on convolutional neural networks (CNNs) and recurrent neural networks (RNNs). RNNs include both long short-term memory (LSTM) and gated recurrent unit (GRU) models.10,11 BiRNN, which can encode both front-to-back and back-to-back information, is a combination of an anterior RNN and a posterior RNN. The RNNs in BiRNN can be replaced with LSTM and GRU, forming BiLSTM and BiGRU, respectively.
The GRU consists of an update gate and a reset gate. The update gate controls the entry of information from the previous moment to the current state, and the reset gate controls the entry of information from the previous state.12 LSTM is a common neural network architecture for sequence modeling and consists of a forget gate, an input gate, and an output gate. The forget gate determines the information to be retained, the input gate determines the information to be added, and the output gate determines the next hidden state.13 Gates are used to process past and updated information and pass it on, allowing the continuous extraction of the features over time, and models are constructed by processing the features (i.e., patient information variables).14
A CNN is highly advantageous for extracting local features, and an RNN is advantageous for extracting features in the time dimension. The CNN and RNN algorithms can be combined to construct CNN-BiLSTM and CNN-BiGRU models, which can be used to address the limitations of the individual models and build a more reasonable prediction model.15
The hyperparameters of the deep learning models using the Adam optimization algorithm were as follows: learning rate = 0.001, Adam = True, momentum = 0.9, weight decay = 1e-8, and batch size = 16.
Performance verification
The area under the receiver operating characteristic curve (AUC) was used to evaluate the performance of each predictive model.16,17 Accuracy, precision, and the F1-score were also used to evaluate the performance of the model.18
Statistical analysis
Continuous variables are expressed as mean ± standard deviation. Student’s t test was conducted to analyze differences between groups. Python 3.8.1 for Windows was used for deep learning analysis. Other analyses were performed with R statistical software version 3.4.1, where P < 0.05 was considered statistically significant.
Results
Patient baseline characteristics
This study involved 878 eligible patients with renal clear cell carcinoma. The patients were randomly assigned to the test set (n = 702) or the verification set (n = 176). The patients’ mean age was 56.28 ± 12.84 years, and the proportions of men and women were 69.2% and 30.8%, respectively. The proportions of pathological T-stage Ta-T1a, T1b, T2, T3, and T4 tumors were 44.6%, 30.4%, 9.3%, 14.4%, and 1.3%, respectively, and the proportions of G-grade G1, G2, G3, and G4 tumors were 23.9%, 63.2%, 12.9%, and 0.0%, respectively. G1, G2, G3, and G4 accounted for 23.9%, 63.2%, 12.9%, and 0.0%, respectively. The clinical and pathological characteristics of all patients with renal clear cell carcinoma in this study are shown in Table 1.
Table 1.
Clinical and pathological characteristics of patients with renal clear cell carcinoma in the present study.
| Variables | Characteristic | Training group | Test group | P value |
|---|---|---|---|---|
| Sex | Male | 125 (73.1) | 483 (68.3) | 0.92 |
| Female | 46 (26.9) | 224 (31.7) | ||
| Symptoms | Present | 36 (21.1) | 174 (24.6) | 0.57 |
| Absent | 135 (78.9) | 533 (75.4) | ||
| History of chronic disease | Present | 76 (44.4) | 314 (44.4) | 0.48 |
| Absent | 95 (55.6) | 393 (55.6) | ||
| Tumor location | Left | 79 (46.2) | 356 (50.4) | 0.81 |
| Right | 91 (53.2) | 344 (48.7) | ||
| Bilateral | 1 (0.6) | 6 (0.8) | ||
| Pathological T stage | Ta-T1a | 88 (51.5) | 304 (43.0) | 0.51 |
| T1b | 42 (24.6) | 225 (31.8) | ||
| T2 | 14 (8.2) | 68 (9.6) | ||
| T3 | 23 (13.5) | 103 (14.6) | ||
| T4 | 4 (2.3) | 7 (1.0) | ||
| Pathological G grade | G1 | 41 (24.0) | 169 (23.9) | 0.31 |
| G2 | 108 (63.2) | 447 (63.2) | ||
| G3 | 22 (12.9) | 91 (12.9) | ||
| G4 | 0 (0.0) | 0 (0.0) | ||
| Age, years | 56.28 ± 12.84 | 55.4 ± 14.67 | 0.18 | |
| BMI, kg/m2 | 25.22 ± 6.84 | 25.03 ± 3.47 | 0.13 | |
| Creatinine level, µmol/L | 84.89 ± 33.59 | 84.49 ± 13.72 | 0.47 | |
| Albumin level, g/L | 41.75 ± 6.58 | 43.56 ± 22.91 | 0.98 | |
| Blood sodium level, mmol/L | 139.71 ± 12.02 | 140.28 ± 3.48 | 0.89 | |
| Blood calcium level, mmol/L | 2.27 ± 0.21 | 2.23 ± 0.29 | 0.57 | |
| Blood phosphorus level, mmol/L | 1.13 ± 0.21 | 1.35 ± 0.17 | 0.46 | |
| Blood magnesium level, mmol/L | 0.92 ± 0.10 | 1.73 ± 10.82 | 041 | |
| Triglycerides, mmol/L | 1.54 ± 1.06 | 1.50 ± 0.89 | 0.21 | |
| Cholesterol level, mmol/L | 4.41 ± 0.97 | 4.40 ± 0.81 | 0.57 | |
| Uric acid level, µmol/L | 328.84 ± 92.80 | 325.01 ± 73.12 | 0.98 | |
| Hemoglobin, g/L | 130.95 ± 39.73 | 128.81 ± 42.44 | 0.44 | |
| Platelets, ×109/L | 203.89 ± 92.16 | 200.25 ± 86.87 | 0.11 | |
| Leukocytes, ×109/L | 6.08 ± 2.59 | 5.84 ± 2.62 | 0.19 | |
| Lymphocytes, ×109/L | 1.67 ± 0.75 | 1.61 ± 0.96 | 0.83 | |
| Neutrophils, ×109/L | 3.72 ± 2.08 | 3.58 ± 1.98 | 0.55 | |
| Eosinophils, ×109/L | 0.15 ± 0.13 | 0.14 ± 0.12 | 0.24 | |
| Basophils, ×109/L | 0.03 ± 0.02 | 0.03 ± 0.12 | 0.19 | |
| Maximum tumor diameter, cm | 4.97 ± 2.30 | 4.75 ± 2.11 | 0.27 | |
Data are presented as n (%) or mean ± standard deviation.
Performance of different models
A deep learning algorithm was used to construct a prediction model for pathological staging and grading of renal cancer. The variables included in the model were sex; age; body mass index; symptoms (e.g., low back pain, abdominal pain, and/or hematuria); chronic disease (e.g., hypertension, diabetes, and/or coronary heart disease); preoperative creatinine, albumin, serum sodium, blood calcium, blood phosphorus, blood magnesium, triglyceride, cholesterol, hemoglobin, and uric acid concentrations; preoperative white blood cell, lymphocyte, neutrophil, eosinophil, and basophil counts; and tumor information, including the maximum tumor diameter and tumor location (left and/or right kidney). There were no significant differences in the patients’ characteristics between the test set and verification set.
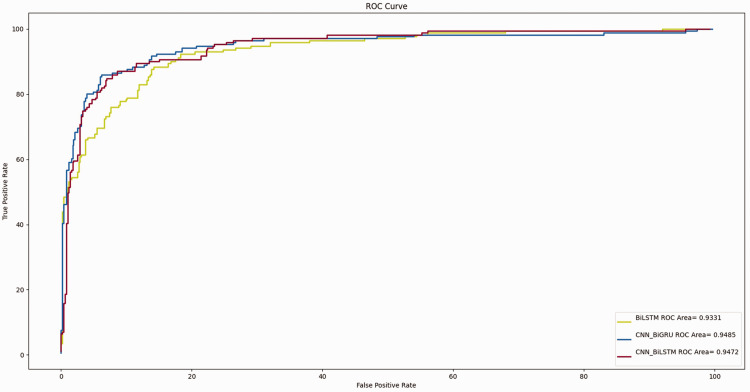
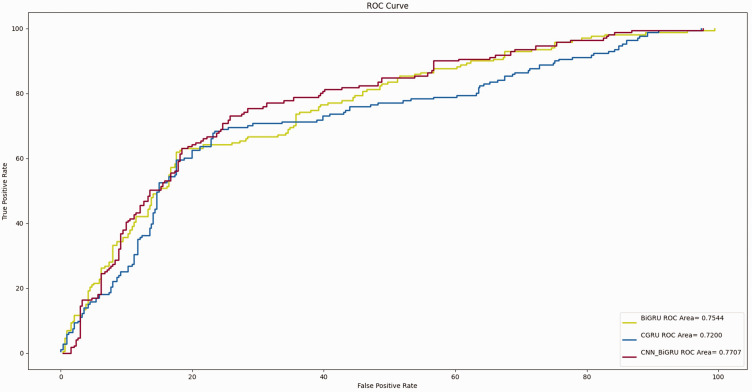
The mean AUCs of the T-stage prediction models constructed by the BiLSTM, CNN-BiLSTM, and CNN-BiGRU algorithms were 0.933, 0.947, and 0.948, respectively (Figure 2). The mean AUCs of the G-grade prediction models constructed by the BiGRU, CGRU, and CNN-BiGRU algorithms were 0.754, 0.722, and 0.771, respectively (Figure 3). The T-stage prediction model constructed by CNN-BiGRU in the internal verification set had better prediction results than the G-grade prediction model constructed by the CNN-BiGRU algorithm.
Figure 2.
Receiver operating characteristic curve of the T stage based on deep learning.
Figure 3.
Receiver operating characteristic curve of the G grade based on deep learning.
The performance of the models was also judged by the accuracy, precision, and F1-score. The T-stage prediction models constructed by the BiLSTM, CNN-BiLSTM, and CNN-BiGRU algorithms all had good stability; the G-grade prediction models constructed by the BiGRU, CGRU, and CNN-BiGRU algorithms also all had good stability (Tables 2 and 3).
Table 2.
Efficacy of deep learning algorithm based on T-stage detection of renal clear cell carcinoma.
| Algorithms | Accuracy | Precision | AUC | F1-score |
|---|---|---|---|---|
| BiLSTM | 78.1% (95% CI, 76.47%–79.72%) | 79.2% (95% CI, 77.4%–81.0%) | 0.933 (95% CI, 0.922–0.936) | 77.8 |
| CNN-BiLSTM | 76.9% (95% CI, 76.4%–77.4%) | 77.0% (95% CI, 75.3%–78.7%) | 0.947 (95% CI, 0.924–0.969) | 81.3 |
| CNN-BiGRU | 79.0% (95% CI, 77.7%–80.2%) | 82.0% (95% CI, 80.7%–83.2%) | 0.948 (95% CI, 0.923–0.972) | 83.6 |
CI, confidence interval; AUC, area under the receiver operating characteristic curve.
Table 3.
Efficacy of deep learning algorithm based on G-grade detection of renal clear cell carcinoma.
| Algorithms | Accuracy | Precision | AUC | F1-score |
|---|---|---|---|---|
| BiGRU | 62.6% (95% CI, 60.1%–65.0%) | 62.2% (95% CI, 61.2%–63.1%) | 0.754 (95% CI, 0.740–0.759) | 61.2 |
| CGRU | 63.2% (95% CI, 61.7%–64.6%) | 63.5% (95% CI, 62.7%–64.2%) | 0.72 (95% CI, 0.69–0.744) | 63.2 |
| CNN-BiGRU | 63.7% (95% CI, 63.0%–64.3%) | 67.9% (95% CI, 65.4%–70.3%) | 0.77 (95% CI, 0.745–0.794) | 64.3 |
CI, confidence interval; AUC, area under the receiver operating characteristic curve.
Discussion
Given that kidney cancer is a common tumor of the urological system, a quality treatment plan is vital for patients with kidney cancer, and the tumor stage is of great significance as an important reference for the treatment plan.19,20 In the present study, a pathological staging system for renal clear cell carcinoma was constructed by deep learning. The distinction between T1a and T1b in the model construction is important for patients to decide whether to undergo radical nephrectomy or partial nephrectomy, and the distinction among T2, T3, and T4 staging is important for patients to make clinical decisions regarding whether to undergo lymphatic dissection, subtractive nephrectomy, or systemic drug treatment. Fuhrman’s classification can also be used to predict the prognosis of patients. Based on the results of the proposed models, clinicians can optimize individual treatment plans to achieve better survival benefits and prolong a patient’s survival time.
The previous prediction models for kidney cancer staging and prognosis were mainly constructed by a nomogram. Li et al.21 constructed a prediction model for staging of kidney clear cell carcinoma using a gene set as the variable, and the model showed accuracy of 81.15% and an AUC of 0.86. Zheng et al.22 constructed a nomogram to predict preoperative renal clear cell carcinoma grading from 20 computed tomography parameter variables, and the model showed an accuracy AUC of 0.876 and 95% confidence interval of 0.812-0.939. In the present study, we established the first deep learning algorithm for construction of a prediction model for the preoperative staging of renal clear cell carcinoma, and the AUCs of the CNN-BiGRU model for tumor T staging and G grading were 0.948 and 0.77, respectively, with high accuracy.
Deep learning algorithms are a new type of algorithm that have been developed in recent years and offer greater advantages in processing data and extracting features than traditional single and multifactor regression analysis for constructing line graphs.14 Schulz et al.23 constructed a multimodal deep learning model for prediction of the kidney cancer prognosis using a deep learning algorithm. The multimodal deep learning model showed great performance in predicting the prognosis of patients with clear cell renal cell carcinoma, with a mean C-index of 0.7791 and a mean accuracy of 83.43%. The stability of the model was greatly improved over that of the traditional approach using a nomogram. The prediction model of renal clear cell carcinoma staging and grading constructed by the deep learning algorithm in this study has greater advantages than the traditional nomogram and is of greater reference value for practical clinical application.
Deep learning algorithms are now used in several fields, and they have been increasingly used in medicine in recent years.24,25 The predictive model of renal clear cell carcinoma staging constructed by deep learning algorithms can assist in the interpretation of puncture results to a certain extent, especially for patients with contraindications to puncture, as well as to avoid invasive tests and reduce the financial burden.26,27
It should be noted that because the data used to construct this model were limited to a single center, the application of the model is somewhat limited. In the future, multicenter collaboration is needed to expand the sample size and perform further external and clinical validation, thereby improving the stability of the model. For deep learning algorithms, adding imaging data to the variables can make better use of the patient’s clinical information. As a result, accurate models can be better constructed. However, a retrospective study is presented in this paper, and a large amount of patient imaging data are missing; imaging parameters were not included in this study. Our team is prospectively building a multicenter cohort for the kidney cancer staging prediction model, expanding the variables that have an impact on predicting outcomes (including imaging data such as computed tomography and magnetic resonance imaging) to further improve the accuracy of the model. In addition, the construction of the deep learning model resembles a black-box model, and the computer principles designed in a given interval are difficult to represent. We plan to further develop a visualization program to facilitate the application by clinicians and patients.
Conclusions
In this study, we developed a model for the prediction of renal clear cell carcinoma staging and grading related to deep learning algorithms based on our central database. The model constructed in our study will be useful in the treatment of patients with renal clear cell carcinoma.
Author contributions: Gao Wen-zhi wrote the initial draft of the manuscript and incorporated suggestions from the other authors into the final version. Guo Yuexian and Li Xuesong revised the initial draft of the manuscript and provided references. Tian Tai, Fu Zhixin, Gong Yanqing, and Liang Huanyu revised the initial draft of the manuscript and provided background information.
The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt (pending publication) of the following financial support for the research, authorship, and/or publication of this article: This study was funded by grants from the Clinical Medical Talents Training Program funded by the Hebei Provincial Government in 2021 (Item 96).
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
ORCID iD
Gao Wen-zhi https://orcid.org/0000-0002-5944-017X
References
- 1.Humphrey PA, Moch H, Cubilla AL, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs—part B: prostate and bladder tumours. Eur Urol 2016; 70: 106–119. [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005; 353: 2477–2490. [DOI] [PubMed] [Google Scholar]
- 3.Campbell SC, Clark PE, Chang SS, et al. Renal mass and localized renal cancer: evaluation, management, and follow-up: AUA guideline: part I. J Urol 2021; 206: 199–208. [DOI] [PubMed] [Google Scholar]
- 4.Jang HJ, Cho KO. Applications of deep learning for the analysis of medical data. Arch Pharm Res 2019; 42: 492–504. [DOI] [PubMed] [Google Scholar]
- 5.Kriegeskorte N, Golan T. Neural network models and deep learning. Curr Biol 2019; 29: R231–R236. [DOI] [PubMed] [Google Scholar]
- 6.Lucas M, Jansen I, Van Leeuwen TG, et al. Deep learning–based recurrence prediction in patients with non–muscle-invasive bladder cancer. Eur Urol Focus 2022; 8: 165–172. [DOI] [PubMed] [Google Scholar]
- 7.Feng JZ, Wang Y, Peng J, et al. Comparison between logistic regression and machine learning algorithms on survival prediction of traumatic brain injuries. J Crit Care 2019; 54: 110–116. [DOI] [PubMed] [Google Scholar]
- 8.Kwon JM, Lee Y, Lee Y, et al. An algorithm based on deep learning for predicting in-hospital cardiac arrest. J Am Heart Assoc 2018; 7: e008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kai Z, Jinchun S, Ke N, et al. Lagrange interpolation learning particle swarm optimization. PLoS One 2016; 11: e0154191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Li Y, Liang W. CNN-RNN based intelligent recommendation for online medical pre-diagnosis support. IEEE/ACM Trans Comput Biol Bioinform 2021; 18: 912–921. [DOI] [PubMed] [Google Scholar]
- 11.Yu SZ. Explicit duration recurrent networks. IEEE Trans Neural Netw Learn Syst 2022; 33: 3120–3130. doi:10.1109/tnnls.2021.3051019. [DOI] [PubMed] [Google Scholar]
- 12.Zheng W, Chen G. An accurate GRU-based power time-series prediction approach with selective state updating and stochastic optimization. IEEE Trans Cybern 2021; 1–13. doi:10.1109/tcyb.2021.3121312. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9: 1735–1780. [DOI] [PubMed] [Google Scholar]
- 14.Elhaj-Abdou MEM, El-Dib H, El-Helw A, et al. Deep_CNN_LSTM_GO: protein function prediction from amino-acid sequences. Comput Biol Chem 2021; 95: 107584. [DOI] [PubMed] [Google Scholar]
- 15.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- 16.Obuchowski NA, Bullen JA. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys Med Biol 2018; 63: 07TR1. [DOI] [PubMed] [Google Scholar]
- 17.Aggarwal R, Ranganathan P. Understanding diagnostic tests – part 3: receiver operating characteristic curves. Perspect Clin Res 2018; 9: 145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Kim DH, Jeong SN, et al. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent 2018; 77: 106–111. [DOI] [PubMed] [Google Scholar]
- 19.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014; 349: g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury N, Drake CG. Kidney cancer: an overview of current therapeutic approaches. Urol Clin North Am 2020; 47: 419–431. [DOI] [PubMed] [Google Scholar]
- 21.Li F, Yang M, Li Y, et al. An improved clear cell renal cell carcinoma stage prediction model based on gene sets. BMC Bioinformatics 2020; 21: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z, Chen Z, Xie Y, et al. Development and validation of a CT-based nomogram for preoperative prediction of clear cell renal cell carcinoma grades. Eur Radiol 2021; 31: 6078–6086. [DOI] [PubMed] [Google Scholar]
- 23.Schulz S, Woerl AC, Jungmann F, et al. Multimodal deep learning for prognosis prediction in renal cancer. Front Oncol 2021; 11: 788740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anwar SM, Majid M, Qayyum A, et al. Medical image analysis using convolutional neural networks: a review. J Med Syst 2018; 42: 226. [DOI] [PubMed] [Google Scholar]
- 25.Dhaka VS, Meena SV, Rani G, et al. A survey of deep convolutional neural networks applied for prediction of plant leaf diseases. Sensors (Basel) 2021; 21: 4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalifeh A, Autorino R, Eyraud R, et al. Three-year oncologic and renal functional outcomes after robot-assisted partial nephrectomy. Eur Urol 2013; 64: 744–750. [DOI] [PubMed] [Google Scholar]
- 27.Capitanio U, Terrone C, Antonelli A, et al. Nephron-sparing techniques independently decrease the risk of cardiovascular events relative to radical nephrectomy in patients with a T1a–T1b renal mass and normal preoperative renal function. Eur Urol 2015; 67: 683–689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.