Abstract
The ring-infected erythrocyte surface antigen (RESA) is a dense-granule protein of Plasmodium falciparum which binds to the cytoskeletal structure of the erythrocyte after parasite invasion. It is currently under trial as a vaccine candidate. In an effort to characterize further the antibody responses to this antigen, we have panned two independent libraries of random peptides expressed on the surface of filamentous phage with a monoclonal antibody (MAb 18/2) against RESA. One library consisted of a potentially constrained 17-mer peptide fused with the gpVIII phage coat protein, and the other displayed an unconstrained 15-mer as a fusion with the minor phage coat protein gpIII. Several rounds of biopanning resulted in enrichment from both libraries clones that interacted specifically with MAb 18/2 in protein-blotting and enzyme-linked immunosorbent assay experiments. Nucleotide sequencing of the random oligonucleotide insert revealed a common predominant motif: (S/T)AVDD. Several other clones had related but degenerate motifs. Thus, a monoclonal antibody against a malarial antigen can select common mimotopes from different random peptide libraries. We envisage many uses for this technology in malaria research.
Libraries of random peptides expressed on the surface of phage provide vast pools of diverse molecular structures from which peptides with binding affinities toward desired molecules can be selected (39). Screening such libraries has emerged as a powerful tool in the identification of small peptides that mimic structural and functional features of larger molecules, e.g., identification of epitopes or mimotopes (a peptide that mimics an epitope but has a different primary amino acid sequence) on antigens (28). In addition, small peptides with affinity for molecules involved in biological interactions (such as receptors) can be isolated and assessed as possible functional modulators. We have used phage peptide technology to obtain mimotopes for an important malarial antigen, the ring-infected erythrocyte surface antigen (RESA).
RESA is a protein produced by the most pathogenic of the human malaria parasites, Plasmodium falciparum. Although some culture-adapted parasite isolates do not express RESA, it is found in all field isolates of P. falciparum, suggesting that it facilitates survival of the parasite in vivo (31). RESA is produced in the final stages of schizont development and stored in dense granules within the developing merozoite (3). Following rupture and merozoite reinvasion, RESA is secreted into the newly formed parasitophorous vacuole and then transported, by an unknown mechanism, to the erythrocyte membrane skeleton (16). RESA interacts with erythrocyte spectrin and stabilizes the erythrocyte membrane (17, 22).
The spectrin-binding region of the RESA polypeptide has been mapped to the region between amino acids 670 and 770 (20). RESA also contains a region that has homology to the conserved J region of the Escherichia coli molecular chaperone, DnaJ (11). This region may be responsible for the proposed chaperone-like activity of RESA (17, 21). These functional regions of the RESA molecule are flanked by two regions of repetitive acidic amino acid sequence, the so-called 5′ and 3′ repeat regions. These acidic repeats represent immunodominant epitopes (19) and are recognized by sera of people who are naturally exposed to malaria (32). Indeed, a number of studies examining malaria endemicity and other seroepidemiological parameters have relied on synthetic peptides corresponding to the linear repeat sequences of RESA (29, 33, 34). The function of the repetitive sequences of RESA is not clear. Many malaria antigens have extensive regions of their amino acid sequence composed of repetitive sequences, some of which are probably the targets of the protective immune response (6). Other repeats, including some that are recognized as dominant epitopes by the host immune system, may function as molecular “smoke screens,” decreasing the ability of the host to mount an effective immune response (4, 26).
Although RESA is not exposed at the surface of the infected erythrocyte (3) and is not essential for growth in vitro (12), evidence from several studies has indicated that antibodies against RESA can inhibit the invasion of merozoites into the host erythrocyte (1, 38). Moreover, immunization of Aotus monkeys with recombinant RESA offers some protection from malaria challenge (14). This has led to the idea that antibodies to the RESA molecule might cross-react with another malarial protein that plays an important role in invasion or development of the intraerythrocytic parasite. A diacidic motif found within both the 5′ and 3′ repeat regions of RESA is also found within the repeat regions of the falciparum interspersed repeat antigen, the FC27 S-antigen, Pf332, Pf11.1, and erythrocyte band 3 (6, 25, 38). A human monoclonal antibody (MAb 33G2) has been shown to cross-react with Pf322 and RESA (27). Indeed, anti-Pf322 antibodies that cross-react with the acidic repeat regions at the C terminus of RESA were found to inhibit the growth of parasites even when the parasite strain did not express RESA (38). These studies suggest that antibodies recognizing the repeat regions of RESA may be important antimalarial agents due to their promiscuous binding activity and to the presence of diacidic motifs in many parasite antigens.
In this study, we used phage peptide technology to obtain information about the binding specificity of an anti-RESA monoclonal antibody, MAb 18/2. MAb 18/2 was raised against a C-terminal recombinant fragment of RESA (residues 893 to 1073) containing the 3′ repeat sequences (5). It also cross-reacts with 5′ repeat sequences of RESA which are also rich in acidic amino acids (5). MAb 18/2 has been used extensively as a research tool to reveal much about the role of RESA in the newly infected erythrocyte (17, 22). It was used to pan two independent random peptide libraries displayed on the surface of filamentous phage. Peptides that bind to the antigen-binding site of MAb 18/2 might be expected to mimic the structure of the acidic repeat regions of RESA. It was anticipated that these peptides would provide information about the fine structure of epitopes on RESA and might prove useful in understanding the nature of the epitopes of cross-reactive antigens that may help parasites evade the protective immune response.
MATERIALS AND METHODS
Parasites.
The P. falciparum cloned line FAC-8 was continuously cultured as described by Trager and Jensen (37).
Phage library preparation.
The 15-mer phage peptide library was kindly provided by George Smith, University of Missouri, Columbia, Mo. (35), and the 17-mer library was provided by Jamie Scott, Simon Fraser University, British Columbia, Canada (10). Phage were amplified, based on a procedure by Smith and Scott (36), by infecting a log-phase culture of E. coli K91 and shaking overnight at 37°C in Luria-Bertani medium containing 2% tetracycline. The supernatant was twice clarified by pelleting the cells at 10,000 × g for 15 min, and 0.2 volume of PEG solution (20% polyethylene glycol 8000, 2.5 M NaCl) was added. The sample was incubated at 4°C for at least 2 h before being centrifuged at 16,000 × g for 50 min to precipitate the phage. The phage pellet was resuspended in 1 ml of TBS (50 mM Tris, 150 mM NaCl [pH 7.5]) and stored at 4°C with 0.02% NaN3.
Panning the phage libraries.
An adaptation of the technique used by Parmley and Smith (30) was used to screen the phage peptide libraries on MAb 18/2. The wells of a 96-well enzyme-linked immunosorbent assay (ELISA) plate (Maxisorp; Nunc International) were coated with MAb 18/2 (0.5 μg) in 100 μl of binding buffer (0.1 M NaHCO3) overnight at 4°C in a humidified container. The wells were blocked for 2 h at 4°C with 400 μl of blocking solution (0.5% bovine serum albumin [BSA], 0.1 M NaHCO3 [pH 8.6]). Phage (approximately 1011 particles) in 100 μl of probing solution (0.5% BSA in TBS) were added to the wells and left for 50 min at room temperature. After incubation, the wells were washed vigorously twice in the first round and four times in subsequent rounds of panning with TBS-T (0.5% Tween 20 in TBS) to remove nonbinding phage. Phage bound to the antibody were eluted with 100 μl of elution solution (0.1 M glycine [pH 2.2]) for 15 min at room temperature and neutralized with 7 μl of 2 M Tris. The titer of eluted phage was estimated, and an aliquot of the eluted fraction was used to infect E. coli K91 cells for amplification.
Phage titer determinations.
Phage were subjected to titer determination by the procedure described by Scott and Smith (35). Phage were subjected to serial 10-fold dilutions with 100 μl of water in a 96-well microtiter plate (Maxisorp; Nunc International). To each of the phage dilutions, 100 μl of log-phase E. coli K91 cells was added, and the mixture was incubated at room temperature for 20 min to allow the phage to infect the E. coli cells. A 20-μl aliquot of each dilution was spread onto agar plates containing 4% tetracycline and incubated overnight at 37°C. Phage infection of bacteria confers resistance to tetracycline, and such colonies were counted and expressed as CFU per milliliter.
Western blotting.
A total of 1010 phage particles from selected clones were boiled for 3 min in the presence of sodium dodecyl sulfate (SDS) sample buffer (10% glycerol, 63 mM Tris [pH 6.8], 2% SDS, 0.0025% bromophenol blue) with or without 10% β-mercaptoethanol and applied to 10 to 20% Tricine gels (Novex, San Diego, Calif.). Separated proteins were then transferred to a polyvinylidene difluoride transfer membrane (PVDF-Plus; Millipore, Bedford, Mass.), and the membrane was blocked overnight in 5% Blotto (5% skim milk powder) and probed with MAb 18/2. Horseradish peroxidase (HRP)-conjugated anti-mouse immunoglobulin G (IgG) was used as a secondary antibody, and binding was detected by enhanced chemiluminescence (ECL) (Pierce Chemical Co. Rockford, Ill.).
ELISAs.
Phage enzyme-linked immunosorbent assays (ELISAs) were performed, using a procedure similar to that described by Harlow and Lane (24), by coating 96-well plates (Nunc) with MAb 18/2, polyclonal rabbit antisera raised against rat chaperone 60, anti-cpn60 (a kind gift from N. Hoogenraad, Department of Biochemistry, La Trobe University, Bundoora, Australia), or 4D6, a mouse monoclonal antibody reactive with apical membrane antigen (AMA-1) from Plasmodium chabaudi (5 μg/ml) (6a). Phage, diluted in probing solution (0.5% BSA in TBS), were added to the wells and incubated for 40 min at room temperature. The wells were washed five times with TBS-T, and bound phage were detected with an anti-M13–HRP antibody (Pharmacia Biotech, Quarry Bay, Hong Kong) by using o-phenylenediamine as an enzyme substrate. For competition experiments, 1010 phage particles were added to the MAb 18/2-coated wells (0.5 μg/well) in the presence of increasing amounts of recombinant RESA (RESA-322 [17]) or synthetic peptide. In ELISAs involving the detection of synthetic peptides, 96-well plates were coated with 1 μg of synthetic peptide. MAb 18/2, diluted in probing solution, was added to the wells as the primary antibody and detected with an anti-mouse IgG-HRP antibody (Amersham Australia Pty Ltd.) as described above. For competition experiments, increasing amounts of synthetic peptide were added along with the primary antibody. ELISAs involving pooled human sera from Papua New Guinea were performed in essentially that same manner as described above. An anti-human IgG-HRP antibody (Amersham Australia Pty Ltd.) was used to detect the human sera.
Nucleotide sequencing.
PCR was used to amplify the region of the phage genome encoding the peptide sequence with phage DNA released from E. coli K91 cells by boiling. The relevant primers were as follows: 15-mer library (gpIII), 5′ primer (GAT AAA CCG ATA CAA TTA AAG) and 3′ primer (CAC AGA CAA CCC TCA TAG); 17-mer library (gpVIII), 5′ primer (CTG AAG AGA GTC AAA AGC) and 3′ primer (CAA TTT CTT AAT GGA AAC). The amplified fragments were sequenced at least twice by automated dye terminator cycle sequencing (Amersham Australia Pty Ltd.). Sequences were analyzed with DNASIS V2.1 (Hitachi Software Engineering Co., Ltd.) computer software.
Peptide synthesis.
Peptides 15(1) (CFDYAPYVSAVDDIC), 17(3) (GLKNCTVQPWDATDVCD), 17(3j) (GAQLDCTVKTPDVWDCN), and 8-mer [(EENVEHDA)4] were synthesized by Auspep Pty Ltd. Peptides RESA 0 (SNDQKYSIEDSLTIK), 11-mer [(DDEHVEEPTVA)2], BSA-11-mer [(DDEHVEEPTVA)3], and 4-mer [(EENV)8] were synthesized by the Joint Protein Structure Laboratory (Walter and Eliza Hall Institute of Medical Research and the Ludwig Institute for Cancer Research, Melbourne, Australia).
Reduction and alkylation of peptides.
Peptides 17(3), 15(1), and 17(3j) (20 μl; 1 mg/ml) were each combined with an equal volume of 40 mM dithiothreitol and incubated at 70°C for 5 min to reduce disulfide bonds. A 10-μl volume of 0.2 M iodoacetamide was then added, and the mixture was incubated for 60 min to alkylate each reduced peptide. The peptides were subsequently immobilized on the wells of an ELISA plate (1 μg/well) and probed with MAb 18/2 as described previously.
Indirect-immunofluorescence assay.
Indirect-immunofluorescence assays were carried out essentially as described previously (9) with MAb 18/2 as a primary antibody and a fluorescein isothiocyanate-labelled anti-mouse secondary antibody (Sigma Chemical Co., St. Louis, Mo.).
Peptide inhibition of antibody binding to parasite-produced RESA.
P. falciparum FAC-8 was grown to approximately 10% ring stage parasitemia. The erythrocytes were isolated by centrifugation and incubated with 2 volumes of 0.2% saponin (Sigma Chemical Co.) at 37°C for 20 min to release the hemoglobin. The lysed cells were washed in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4 [pH 8]) until all the hemoglobin was removed from the cells. A 25-μl volume of packed cells, diluted in 150 μl of probing solution, was incubated with 0.1 μg of MAb 18/2 in the presence or absence of peptide for 40 min at room temperature. The cells were washed four times with TBS, and the amount of bound MAb 18/2 was assessed by using an alkaline phosphatase-coupled anti-mouse IgG (Sigma Chemical Co.) followed by adding 200 μl of p-nitrophenyl phosphate alkaline phosphatase substrate as specified by the manufacturer (Sigma Chemical Co.). The cells were pelleted, and the supernatant was analyzed spectrophotometrically at 405 nm.
RESULTS
Panning and ELISA screening.
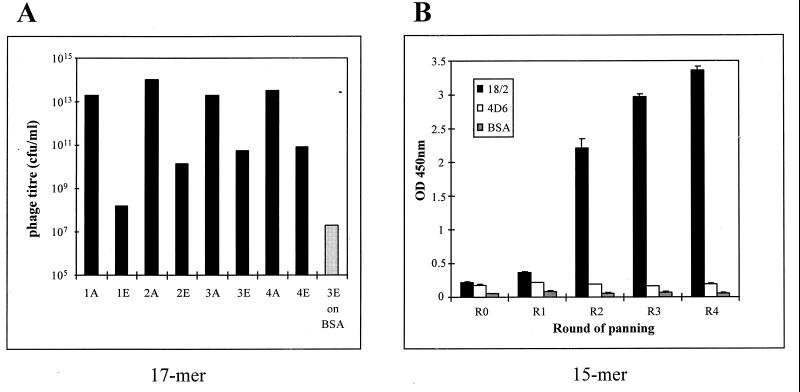
To identify peptides that can mimic structural features of the malarial protein RESA, we used the anti-RESA MAb 18/2 to pan two independent phage-displayed random peptide libraries. One of the libraries contains phage expressing random 15-residue peptides fused to the minor coat protein gpIII and present at 5 copies per phage particle (35), and the other library contains phage expressing approximately 250 copies of a 17-residue peptide library (X15CX), with an invariant penultimate cysteine residue, as a fusion to the gpVIII coat protein (10). The latter library has a diversity of 1.2 × 108 clones. Four rounds of panning resulted in an approximately 1,000-fold enrichment of eluted phage (Fig. 1A). Phage pools after two rounds of panning exhibited enhanced binding to MAb 18/2, whereas no binding to BSA or the irrelevant antibody MAb 4D6 was seen with pooled phage from any of the four rounds of panning, indicating enrichment of phage with specific binding characteristics during the panning procedure (Fig. 1B).
FIG. 1.
Enrichment of phage during panning. (A) Titers of phage applied to (bars A) and eluted from (bars E) plastic wells at each round of panning. The phage collected after three rounds of panning (3E) were used to pan on a control protein, BSA. (B) Phage ELISA of pools of clones after each round of panning. Pooled phage were amplified, and 1010 phage were added to each well of a microtiter plate precoated with MAb 18/2, anti-AMA-1 (4D6), or BSA. Phage were detected with an anti-M13-HRP antibody. OD 450nm, optical density at 450 nm.
Amino acid sequences of peptide inserts in phage that bind to MAb 18/2.
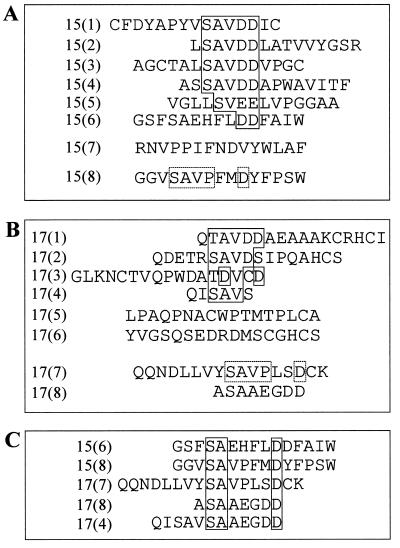
Although the precise epitope of MAb 18/2 has not been determined, the antibody recognizes both the 5′ and 3′ repeat regions of the RESA antigen. After two or four rounds of panning, individual phage clones were propagated and the region of DNA corresponding to the random peptide was sequenced. Peptides isolated from the 15-mer library after two rounds of panning were fairly diverse. After four rounds of panning, this diversity decreased. After four rounds of panning, most of the isolated sequences contained a common SAVDD motif within different amino acid sequence contexts (Fig. 2A). There was some degeneracy in the motif, with one peptide [15(5)] having SVEE and another [15(6)] having only the diacidic (DD) residues in common. Although there was no exact identity between the peptides isolated and RESA sequences (19), the majority of the peptides contained a diacidic sequence immediately preceded by a hydrophobic residue, which is a motif found in both the 5′ and 3′ RESA repeats. Acidic residues were significantly enriched in the selected peptides (Fig. 2). This is significant, since Couet et al. have estimated that the expected occurrence of acidic residues in the 15-mer library is only 5% (15). In contrast, the proportion of acidic residues in the isolated peptides was 14%, almost threefold higher than expected, perhaps reflecting the acidic nature of the epitope on RESA recognized by MAb 18/2. One peptide isolated from the 15-mer library, 15(7), did not appear to have any homology to the consensus motif (Fig. 2A).
FIG. 2.
Deduced amino acid sequences of individual clones obtained after two to four rounds of panning of the phage libraries on MAb 18/2. (A) Sequences of inserts from phage isolated from the 15-mer library. (B) Sequences of inserts from phage isolated from the 17-mer library. Regions of similarity within the peptides are boxed. (C) Alignment of selected clones from the two libraries revealing a secondary motif (boxed) within the amino acid sequences. The AAEEGDD sequence in the last two clones is derived from the gene III protein immediately following the insert.
The peptides isolated from the 17-mer library were more diverse in sequence but were again rich in acidic amino acids. The presence of a common motif was less obvious; however, the preference for serine, threonine, valine, and aspartic acid suggests a loose motif with structural similarity to SAVDD. Interestingly, one of the peptides [17(1)] contained the motif TAVDD (Fig. 2B), which is clearly related to the SAVDD motif identified from the 15-mer library. Hence, we propose the consensus motif (S/T)AVDD. Further comparison of the clones from the two different libraries identified peptides 15(8) and 17(7) as sharing a different motif, SAVPXXD, where X is any amino acid (Fig. 2C). These two peptides, when grouped with other selected peptides, allowed the assignment of a second consensus sequence, SAXXXXD, which was present in five peptides within different amino acid contexts. The binding of a representative peptide, 15(6) (GSFSAEHFLDDFAIW), to MAb 18/2 has been confirmed (Fig. 3 and 4B). Interestingly, sequences of two of these five peptides [peptides 17(8) and 17(4) (Fig. 2C)] align with this second consensus motif only if the phage protein backbone of gpVIII to which the peptides are fused is considered to contribute to the binding. This may explain the surprising observation that a phage displaying a peptide insert of only two residues [alanine and serine; peptide 17(8)] was selected from the library. We also isolated peptides from these libraries [i.e., peptides 15(7), 17(5), and 17(6)] that did not conform to any of these consensus patterns.
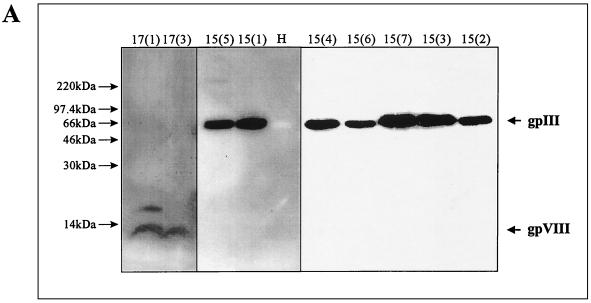
FIG. 3.
Interaction of isolated phage clones with MAb 18/2. (A) Analysis by Western blotting. Selected phage clones from each of the two libraries and wild-type M13 phage as a control were separated by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide), transferred to PVDF membranes, and probed with MAb 18/2. The positions of peptides 15(5), 15(1), 15(4), 15(6), 15(7), 15(3), and 15(2) fused to gpIII and peptides 17(1) and 17(3) fused to gpVIII were visualized by ECL. No binding was observed to gpIII on wild-type M13 phage (H). (B) Effect of reduction and alkylation on binding of mimotopes to MAb 18/2. Peptides 15(1), 17(3), and 17(3j), either cyclized or reduced and alkylated (R+A), were immobilized on the wells of an ELISA plate and probed with MAb 18/2. Binding was detected by HRP-conjugated anti-mouse antibody. OD 450nm, optical density at 450 nm.
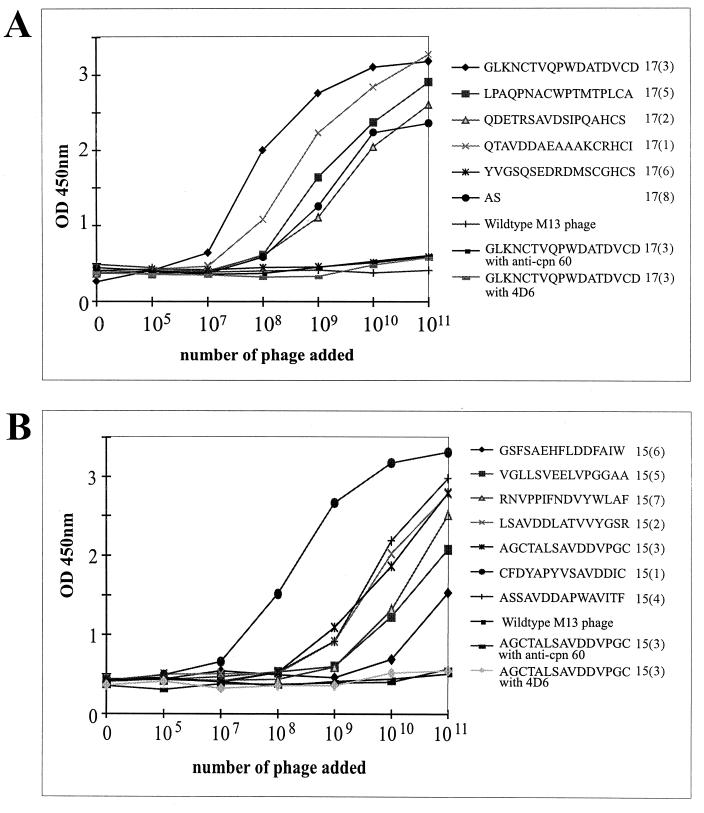
FIG. 4.
Analysis of specificity and affinity of binding of phage-displayed peptides to MAb 18/2. Increasing numbers of phage of representative clones from the 17-mer library (A), the 15-mer library (B), and wild-type M13 phage were applied to wells of an ELISA plate coated with MAb 18/2. The binding of peptides 15(3) and 17(3) to two other antibodies, anti-chaperone 60 and anti-AMA-1 (4D6), were also examined. This data is representative of an experiment that was performed on three separate occasions. OD 450nm, optical density at 450 nm.
Binding specificity of selected phage to MAb 18/2.
When the proteins of the selected phage were separated under nonreducing conditions by SDS-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride (PVDF) membranes, MAb 18/2 recognized the peptide fused gene product; however, there was no binding of MAb 18/2 to wild-type M13 proteins which did not display any foreign peptides (Fig. 3A).
A number of the isolated peptides from both libraries contained two cysteine residues. This was particularly noted for peptides isolated from the 17-mer library, where there was an invariant cysteine engineered into the sequence. This suggested that an intramolecular disulfide bond might contribute to the conformation of some of the peptide structures. To further investigate this possibility, phage clones from the cysteine-containing 15-mer library were separated under reducing conditions. These reduced polypeptides were still recognized by MAb 18/2 on a Western blot (Fig. 3A), suggesting that a disulfide bond between the two cysteine residues present in some peptides is not essential for the interaction with the monoclonal antibody antigen-binding site. MAb 18/2 was also able to bind to both peptides 15(1) and 17(3) when produced as cyclized peptides; however, we have shown that when these peptides are reduced and alkylated, the binding of 15(1) to MAb 18/2 is unaffected whereas the binding of 17(3) is significantly reduced. Nevertheless, there is still substantial binding of 17(3) to MAb 18/2 (Fig. 3B). Mass spectrometry has confirmed that more than 95% of the peptides were correctly reduced and alkylated (data not shown).
Phage isolated from both libraries also bound to MAb 18/2-coated ELISA plates (Fig. 4). In contrast, phage lacking a peptide (wild-type M13 phage) did not bind to MAb 18/2, and one phage clone [17(6)] with the insert sequence YVGSQSEDRDMSCGHCS, which lacks the consensus motif, also did not bind (Fig. 4A). Phage from both libraries, which showed high-affinity binding to MAb 18/2, did not bind to an irrelevant monoclonal antibody, MAb 4D6, or to antisera to the irrelevant protein, Cpn60 (Fig. 4). Taken together, these data indicate that the majority of the isolated peptide sequences bind specifically to the antigen-binding site of MAb 18/2 and display a range of apparent binding affinities and hence mimic epitopes on RESA.
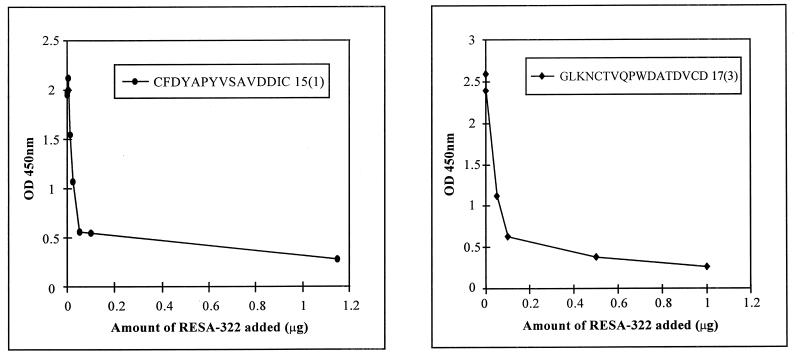
RESA protein inhibits binding of MAb 18/2 to selected phage.
If the peptide sequences mimic epitopes on RESA, RESA would be expected to compete with phage displaying these sequences for binding to MAb 18/2. Figure 5 demonstrates that a recombinant bacterially expressed form of RESA (amino acids 322 to 1073), which includes both repeat regions, reduces the binding of selected phage clones to immobilized MAb 18/2 in a dose-dependent manner.
FIG. 5.
Inhibition by RESA of the binding of phage clones to MAb 18/2. MAb 18/2 was immobilized on the wells of an ELISA plate, and a constant amount of phage from the 15-mer library (A) or the 17-mer library (B) was added to each well in the presence of increasing amounts of RESA-322. Binding of phage was assessed with an anti-M13–HRP antibody. OD 450nm, optical density at 450 nm.
Synthetic peptides are RESA mimotopes.
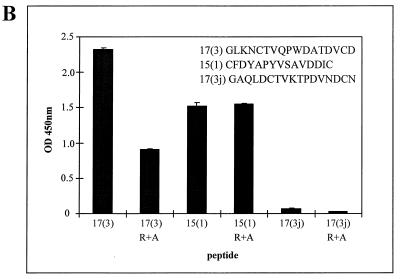
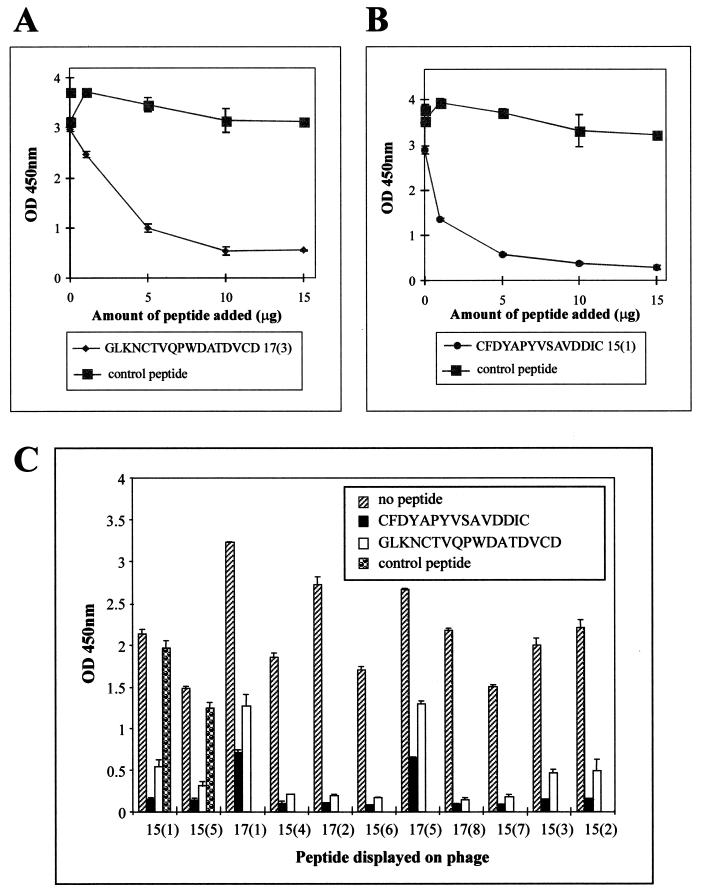
To confirm that the peptide sequences are true mimics of structural features of RESA, a representative peptide from each library was synthesized [17(3) (GLKNCTVQPWDATDVCD) and 15(1) (CFDYAPYVSAVDDIC)] and their binding specificities for MAb 18/2 were determined. Both peptides inhibited the binding to MAb 18/2 of phage displaying the corresponding peptide (Fig. 6A and B). This demonstrates that the phage scaffold is not necessary for the binding of the mimotopes to MAb 18/2. A peptide with the same amino acid composition as peptide 17(3) but with a randomized sequence [peptide 17(3j) (GAQLDCTVKTPDVWDCN)] had no effect on binding (Fig. 6A and B). Thus the primary sequence, and not simply the acidic nature of the peptide, appears to be critical for efficient binding to the antigen-binding site of MAb 18/2. Interestingly, the two synthetic peptides were capable of inhibiting the binding of most of the other phage-displayed mimotopes to MAb 18/2 despite the marked differences in amino acid sequences (Fig. 6C). It would appear, therefore, that despite the wide divergence of mimotope sequences identified in this study, they all have a similar fine specificity (i.e., they bind to spatially overlapping sites within the antigen-binding site).
FIG. 6.
Inhibition by synthetic peptides of the interaction of phage clones with MAb 18/2. (A and B) MAb 18/2 was immobilized on the wells of an ELISA plate and probed with a constant amount (1010 phage) of phage clone 17(3) in the presence of increasing amounts of synthetic 17(3) peptide or the control peptide 17(3j) (A) or phage clone 15(1) in the presence of increasing amounts of synthetic 15(1) peptide or the control peptide 17(3j) (B). (C) MAb 18/2 was immobilized on the wells of an ELISA plate and probed with a constant amount (1010 phage) of 11 different phage clones in the presence of no peptide, 5 μg of a synthetic version of peptide 17(3), 5 μg of peptide 15(1), or 5 μg of the control peptide 17(3j). OD 450nm, optical density at 450 nm.
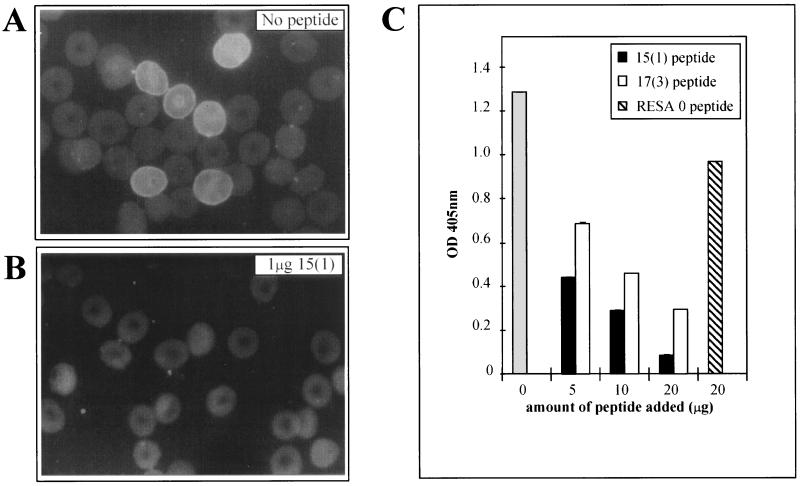
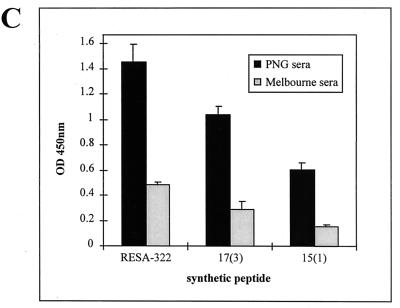
MAb 18/2 specifically recognizes RESA associated with the membrane skeleton of erythrocytes infected with ring stage parasites. We performed immunofluorescence experiments with MAb 18/2 on parasites growing in vitro (Fig. 7). The two synthetic peptides, 17(3) (GLKNCTVQPWDATDVCD) and 15(1) (CFDYAPYVSAVDDIC), inhibited the binding of MAb 18/2 to RESA as shown by a decrease in the signal in the immunofluorescence assay (Fig. 7). An irrelevant peptide, SNDQKYSIEDSLTIK (RESA 0), consisting of a randomized stretch of amino acids from RESA was unable to inhibit fluorescence (Fig. 7). Confirmation that both 15(1) and 17(3) peptides can block the binding of MAb 18/2 to RESA in situ was obtained by measuring the amount of antibody bound in the presence of mimotope in a modified ELISA format (see Materials and Methods). Both peptides blocked binding of the antibody to RESA, whereas an irrelevant peptide had little effect (Fig. 7C). Thus, binding of MAb 18/2 to authentic RESA (i.e., as it is presented by intraerythrocytic parasites) is inhibited by these phage-derived mimotopes. This emphasizes the structural similarity between these peptides and immunodominant regions of RESA. To explore the relationship between the mimotopes isolated in this study and the RESA repeat regions, we first examined the binding of MAb 18/2 to peptides representing the major repeats of RESA. MAb 18/2 recognized all three peptides, i.e., 4-mer, BSA-conjugated 8-mer, and BSA-conjugated 11-mer, as well as 17(3) and 15(1) mimotopes (see Materials and Methods for peptide sequences), although the 8-mer peptide appeared less reactive than the others (Fig. 8A). This confirms that MAb 18/2 reacts with sequences in both the 5′ and 3′ repeats of RESA. Interestingly, all three peptides representing the RESA repeats were able to block the binding of MAb 18/2 to both 17(3) and 15(1) mimotopes (Fig. 8B). These results strengthen the proposal that mimotopes selected on MAb 18/2 accurately represent authentic antigenic sites on RESA. To obtain support for the assertion that these mimotopes are relevant to the natural immune response induced by RESA, we examined the ability of sera from Papua New Guinea to recognize the phage clones. In ELISAs, pooled sera from malaria-infected individuals from a region of Papua New Guinea where malaria is endemic (8) showed a fourfold-higher response to peptide 17(3) and a threefold higher response to peptide 15(1) than the background signal observed with pooled sera from a group of non-infected control individuals in Melbourne (Fig. 8C).
FIG. 7.
Inhibition of MAb 18/2 binding to parasite-expressed RESA by mimotopes. (A and B) Immunofluorescence experiments were carried out with MAb 18/2 to visualize RESA on infected erythrocyte membranes (A). This fluorescence was greatly reduced by the presence of 1 μg of 15-mer peptide (B). (C) Infected cultures were washed and incubated with MAb 18/2 in the absence of peptide (shaded bar) in the presence of various concentrations of 15(1) peptide or 17(3) peptide, or in the presence of 20 μg of a control peptide RESA 0. After addition of an alkaline phosphatase-coupled anti-mouse IgG, MAb 18/2 binding was determined by assaying the supernatants spectrophotometrically at 405 nm (OD 405nm).
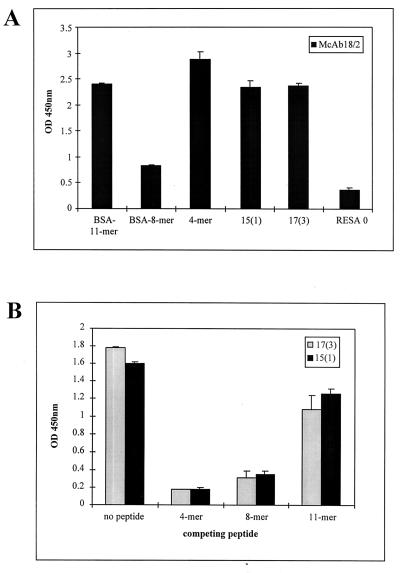
FIG. 8.
Binding of MAb 18/2 to mimotopes and synthetic peptides corresponding to the RESA repeats. (A) BSA-conjugated peptides 11-mer (DDEHVEEPTVA)3 and 8-mer (EENVEHDA)4 (see Materials and Methods for details), 4-mer, 15(1), 17(3), and RESA 0 were immobilized on the wells of an ELISA plate and probed with MAb 18/2. Bound antibody was detected with an HRP-coupled anti-mouse IgG. (B) Inhibition by synthetic peptides of the interaction of peptides 17(3) and 15(1) with MAb 18/2. Immobilized peptides 17(3) and 15(1) were probed with MAb 18/2 in the presence of 10 μg of the 4-mer, 8-mer, and 11-mer peptides. Bound antibody was detected with an HRP-coupled anti-mouse IgG. (C) Binding of pooled human sera to RESA and two synthetic mimotopes, 17(3) and 15(1). Recombinant RESA-322 and peptides were immobilized on the wells of an ELISA plate and probed with pooled human sera from individuals from Papua New Guinea (PNG) and Melbourne. Bound antibody was detected with HRP-coupled anti-human IgG. OD 450nm, optical density at 450 nm.
DISCUSSION
In this study, we have examined the utility of random peptide libraries displayed on the surface of bacteriophage as repositories of molecular structures, some of which will mimic structural and perhaps functional regions of important malaria antigens. We chose to pan two random peptide libraries with MAb 18/2, which recognizes epitopes within the acidic repeats of the malarial antigen RESA. MAb 18/2 was raised against a C-terminal fragment of RESA fragment. This region of RESA contains the 3′ repeats, which consist of 29 copies of a 4-amino-acid sequence (EENV) plus 5 copies of an extended (8-amino-acid) version of this repeat (EENVEHDA). The exact epitope recognized by MAb 18/2 has not been mapped; however, it is likely to include the hydrophobic, diacidic motif which is present in the 5′ and 3′ repeats of RESA (19) recognized by MAb 18/2.
Panning of two independent random peptide libraries with MAb 18/2 identified a set of peptides that bound to the antigen-binding site of this antibody. Although the selected peptides had no extensive identity to sequences within RESA, they had characteristics that might be expected of RESA mimotopes. It is not clear why there was no selection of peptides that closely resembled the 4-mer sequence, EENV, or the 8-mer sequence, EENVEHDA. It is possible that these sequences are not represented in the two libraries used in this study or, alternatively, are not tolerated by the phage or the bacteria in which the phage are propagated due to adverse effects or production of unfavorable conformations of the phage fusion protein and hence would not be accessible for selection during the panning procedure. Isolation of mimotopes—peptides that are not identical to the original epitope but are still able to bind to the antigen-binding site of an antibody due to sufficient structural similarities—from random peptide libraries has been reported by others (18, 23). Indeed, it appears to be uncommon to isolate an exact match for an epitope from a random peptide library. In this study, the overall negative charge of the isolated peptides was high, suggesting that the peptides mimic the high glutamate and aspartic acid content of RESA repeats. The consensus motif identified from the 15-mer library, SAVDD, had a closely related counterpart, TAVDD, in the set of peptides enriched from the 17-mer library. The consensus motif (S/T)AVDD most closely resembles the sequences TVADD, TVADE, and TVAEE, which are found within the 5′ repeat region of RESA, perhaps suggesting that MAb 18/2 may have a higher affinity for epitopes within the 11-mer repeats. This result is slightly surprising, since although MAb 18/2 reacts with both 3′ and 5′ repeats, the antibody was raised against a recombinant protein containing the 4-mer and 8-mer repeats only. A deeper understanding of this interaction was obtained by competition experiments between the mimotopes and synthetic peptides representing the three dominant repeat motifs in RESA, 4-mer (EENV)8, 8-mer (EENVEHDA)4, and 11-mer (DDEHVEEPTVA).
We have shown that MAb 18/2 recognized all three synthetic peptides from the 5′ and 3′ repeats of RESA and that all three RESA peptides were able to inhibit the binding of MAb 18/2 to the mimotopes (Fig. 8A and B), demonstrating the shared properties of the mimotopes and the RESA-derived sequences. It was interesting that the synthetic 8-mer gave the lowest binding to MAb 18/2, whereas it was the 11-mer that was the least effective at blocking the interaction between MAb 18/2 and the mimotopes, perhaps reflecting the local conformational differences between peptides attached to a solid substrate and those same peptides in solution.
Pf332, a megadalton protein consisting primarily of 11-amino-acid repeat sequences (2, 27), contains three sequences (SVTEE, SVTDE, and SVTED) that resemble the consensus (S/T)AVDD motif identified in this study. The human monoclonal antibody (MAb 33G2) is an efficient inhibitor of parasite invasion in vitro (38) and has been shown to recognize both RESA and Pf332. Although it is not known whether MAb 18/2 or the antisera generated against the mimotopes identified here are inhibitory, one study has revealed a correlation between antibodies against the RESA 4-mer repeated epitope and protection against malaria infection (7) while another study has demonstrated that antibodies to RESA repeats inhibit malaria invasion (13). Ahlborg et al. have also shown that antibodies reactive with RESA repeats inhibit the growth of malaria parasites (1). Interpretation of these results is unclear because of the network of cross-reactions that exist between P. falciparum antigens containing hydrophobic-diacidic motifs. Peptides isolated by panning phage display libraries may help define more clearly the functional cross-reactivities between antibodies to different malarial antigens.
Two of the peptide inserts selected from the 17-mer library by panning on MAb 18/2 contained large deletions. One insert consisted of only 2 amino acids, and the other contained 6 residues instead of the expected 17. Deviant sequences have previously been observed in this 17-mer library (10); however, it was surprising that a clone containing a peptide insert of only 2 amino acids bound to MAb 18/2 while phage containing no insert exhibited no binding activity. Presumably, the adjacent sequence of the phage protein gpVIII combines with the AS insert to constitute a mimotope of MAb 18/2. The involvement of the phage environment in binding of phage-displayed peptides to an antibody is a phenomenon that has been proposed previously (18). Interestingly, both deleted peptides end in a serine residue, which, when considered along with the next 6 adjacent gpVIII residues (AAEGDD), conforms to a second consensus motif, SAXXXXD, found in MAb 18/2-binding clones isolated from both libraries (Fig. 2C).
Of the two libraries we used, the first consists of 15 completely random amino acids expressed at the N terminus of the phage protein gpIII and the second has 15 random amino acids followed by a fixed cysteine residue, which is immediately followed by another random amino acid. This latter library can be considered to be “semiconstrained,” since single cysteine residues are not favored and panning tends to select for a second cysteine elsewhere within the peptide (10, 40). We did observe a number of second cysteine residues within the peptides isolated from the 17-mer library (Fig. 2B). It was also noted that two peptides containing two cysteine residues were isolated from the 15-mer library, although this library had no particular selection pressure in favor of disulfide bonds. Although disulfide bound formation between the two cysteine residues probably does occur for at least some of the isolated peptide inserts (40), the disulfide bridges in the selected peptides do not appear to be essential for the binding to MAb 18/2 since they were still able to bind to MAb 18/2 after elimination of the disulfide bond by reduction and alkylation. The disulfide bond in mimotope 17(3) does appear to play a role in maintaining the integrity of the peptide so that it can bind efficiently to MAb 18/2, since reduction and alkylation leads to a decrease in MAb 18/2 binding (Fig. 3B). Disulfide bond formation may play a role in stabilization of the peptides during their route through the bacterial periplasm or possibly in allowing the peptide to be efficiently packaged on the phage surface and extruded as completed phage particles.
Our results demonstrate that small peptides can mimic features of repeat regions of large P. falciparum proteins. The peptide mimotopes inhibit the binding of MAb 18/2 to a recombinant fragment of RESA and also to authentic RESA within the malaria parasite. Such peptide mimotopes and those corresponding to other malarial antigens may prove useful markers for monitoring the seroepidemiology in communities where malaria is endemic. Numerous studies examining malaria endemicity and other seroepidemiological parameters have relied on synthetic peptides corresponding to the linear repeat sequences of RESA (29, 33, 34). It seems likely that mimotopes isolated with anti-RESA antibodies would more closely mimic the presentation of RESA within the parasite, since any effects of conformation within the repeat regions may not be faithfully represented in short synthetic peptides based solely on primary sequence of the antigen. The observation that sera from individuals from a malaria-endemic area recognize synthetic mimotopes (Fig. 8C) supports this proposal. Furthermore small peptides have obvious advantages over parasite extracts or recombinant antigens in terms of stability and cost-effectiveness, and it may be possible to identify a small set of peptides that could represent many variants of a particular highly variable antigen.
We believe that random peptide libraries displayed on phage can be used to aid in the search for peptides with improved diagnostic and therapeutic potential as well as peptides for probing the molecular interactions between the malaria parasite and its host.
ACKNOWLEDGMENTS
This work was supported by the Australian Research Council and the National Health and Medical Research Council of Australia and La Trobe University Grants Scheme.
We thank Sue Mullins for excellent technical support.
REFERENCES
- 1.Ahlborg N, Iqbal J, Bjork L, Stahl S, Perlmann P, Berzins K. Plasmodium falciparum: differential parasite growth inhibition mediated by antibodies to the antigens Pf332 and Pf155/RESA. Exp Parasitol. 1996;82:155–163. doi: 10.1006/expr.1996.0020. [DOI] [PubMed] [Google Scholar]
- 2.Ahlborg N, Wahlin-Flyg B, Iqbal J, Perlmann P, Berzins K. Epitope specificity and capacity to inhibit parasite growth in vitro of human antibodies to repeat sequences of the Plasmodium falciparum antigen Ag332. Parasite Immunol. 1993;15:391–400. doi: 10.1111/j.1365-3024.1993.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 3.Aikawa M, Torii M, Sjolander A, Berzins K, Perlmann P, Miller L. Pf155/RESA antigen is localized in dense granules of Plasmodium falciparum merozoites. Exp Parasitol. 1990;71:326–329. doi: 10.1016/0014-4894(90)90037-d. [DOI] [PubMed] [Google Scholar]
- 4.al-Yaman F, Genton B, Reeder J, Mokela D, Anders R, Alpers M. Humoral response to defined Plasmodium falciparum antigens in cerebral and uncomplicated malaria and their relationship to parasite genotype. Am J Trop Med Hyg. 1997;56:430–435. doi: 10.4269/ajtmh.1997.56.430. [DOI] [PubMed] [Google Scholar]
- 5.Anders R, Barzaga N, Shi P-T, Scanlon D, Brown L, Thomas L, Brown G, Stahl H-D, Coppel R, Kemp D. Repetitive sequences in malaria antigens. UCLA Symp Mol Cell Biol New Ser. 1987;42:333–342. [Google Scholar]
- 6.Anders R, Murray L, Thomas L, Davern K, Brown G, Kemp D. Structure and function of candidate vaccine antigens in Plasmodium falciparum. Biochem Soc Symp. 1987;53:103–114. [PubMed] [Google Scholar]
- 6a.Anders, R. Unpublished data.
- 7.Astagneau P, Chougnet C, Lepers J, Andriamangatiana-Rason M, Deloron P. Antibodies to the 4-mer repeat of the ring-infected erythrocyte surface antigen (Pf155/RESA) protect against Plasmodium falciparum malaria. Int J Epidemiol. 1994;23:169–175. doi: 10.1093/ije/23.1.169. [DOI] [PubMed] [Google Scholar]
- 8.Beck H, Felger I, Genton B, Alexander N, al-Yaman F, Anders R, Alpers M. Humoral and cell-mediated immunity to the Plasmodium falciparum ring-infected erythrocyte surface antigen in an adult population exposed to highly endemic malaria. Infect Immun. 1995;63:596–600. doi: 10.1128/iai.63.2.596-600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianco A, Crewther P, Coppel R, Stahl H, Kemp D, Anders R, Brown G. Patterns of antigen expression in asexual blood stages and gametocytes of Plasmodium falciparum. Am J Trop Med Hyg. 1988;38:258–267. doi: 10.4269/ajtmh.1988.38.258. [DOI] [PubMed] [Google Scholar]
- 10.Bonnycastle L, Mehroke J, Rashed M, Gong X, Scott J. Probing the basis of antibody reactivity with a panel of constrained peptide libraries displayed by filamentous phage. J Mol Biol. 1996;258:747–762. doi: 10.1006/jmbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Sander C, Valencia A, Bukau B. A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem Sci. 1992;17:129. doi: 10.1016/0968-0004(92)90319-5. [DOI] [PubMed] [Google Scholar]
- 12.Cappai R, van Schravendijk M, Anders R, Peterson M, Thomas L, Cowman A, Kemp D. Expression of the RESA gene in Plasmodium falciparum isolate FCR3 is prevented by a subtelomeric deletion. Mol Cell Biol. 1989;9:3584–3587. doi: 10.1128/mcb.9.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlsson J, Udomsangpetch R, Wahlin B, Ahlborg N, Berzins K, Perlmann P. Plasmodium falciparum: differential parasite reactivity of rabbit antibodies to repeated sequences in the antigen Pf155/RESA. Exp Parasitol. 1990;71:314–325. doi: 10.1016/0014-4894(90)90036-c. [DOI] [PubMed] [Google Scholar]
- 14.Collins W, Anders R, Pappaioanou M, Campbell G, Brown G, Kemp D, Coppel R, Skinner J, Andrysiak P, Favaloro J, Corcoran L, Broderson J, Mitchell G, Campell C. Immunization of Aotus monkeys with recombinant proteins of an erythrocyte surface antigen of Plasmodium falciparum. Nature. 1986;323:259–262. doi: 10.1038/323259a0. [DOI] [PubMed] [Google Scholar]
- 15.Couet J, Li S, Okamoto T, Ikezu T, Lisanti M. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 16.Culvenor J, Day K, Anders R. Plasmodium falciparum ring-infected erythrocyte surface antigen is released from merozoite dense granules after erythrocyte invasion. Infect Immun. 1991;59:1183–1187. doi: 10.1128/iai.59.3.1183-1187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Da Silva E, Foley M, Dluzewski A, Murray L, Anders R, Tilley L. The Plasmodium falciparum protein RESA interacts with the erythrocyte cytoskeleton and modifies erythrocyte thermal stability. Mol Biochem Parasitol. 1994;66:59–69. doi: 10.1016/0166-6851(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 18.Demangel C, Lafaye P, Mazie J. Reproducing the immune response against the Plasmodium vivax merozoite surface protein 1 with mimotopes selected from a phage-displayed peptide library. Mol Immunol. 1996;33:909–916. doi: 10.1016/s0161-5890(96)00058-2. [DOI] [PubMed] [Google Scholar]
- 19.Favaloro J, Coppel R, Corcoran L, Foote S, Brown G, Anders R, Kemp D. Structure of the RESA gene of Plasmodium falciparum. Nucleic Acids Res. 1986;14:8265–8277. doi: 10.1093/nar/14.21.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foley M, Corcoran L, Tilley L, Anders R. Plasmodium falciparum—mapping the membrane-binding domain in the ring-infected erythrocyte surface antigen. Exp Parasitol. 1994;79:340–350. doi: 10.1006/expr.1994.1096. [DOI] [PubMed] [Google Scholar]
- 21.Foley M, Tilley L. Home improvements—malaria and the red blood cell. Parasitol Today. 1995;11:436–439. doi: 10.1016/0169-4758(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 22.Foley M, Tilley L, Sawyer W, Anders R. The ring-infected erythrocyte surface antigen of Plasmodium falciparum associates with spectrin in the erythrocyte membrane. Mol Biochem Parasitol. 1991;46:137–147. doi: 10.1016/0166-6851(91)90207-m. [DOI] [PubMed] [Google Scholar]
- 23.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Nicosia A. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 25.Holmquist G, Udomsangpetch R, Berzins K, Wigzell H, Perlmann P. Plasmodium chabaudi antigen Pch105, Plasmodium falciparum antigen Pf155, and erythrocyte band 3 share cross-reactive epitopes. Infect Immun. 1988;56:1545–1550. doi: 10.1128/iai.56.6.1545-1550.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp D, Coppel R, Anders R. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 27.Mattei D, Berzins K, Wahlgren M, Udomsangpetch R, Perlmann P, Griesser H, Scherf A, Muller-Hill B, Bonnefoy S, Guillotte M, Langsley G, Pereira Da Silva L, Mercereau-Puijalon O. Cross-reactive antigenic determinants present on different Plasmodium falciparum blood-stage antigens. Parasite Immunol. 1989;11:15–30. doi: 10.1111/j.1365-3024.1989.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 28.Mennuni C, Santini C, Lazzaro D, Dotta F, Farilla L, Fierabracci A, Bottazzo G, Di Mario U, Cortese R, Luzzago A. Identification of a novel type 1 diabetes-specific epitope by screening phage libraries with sera from pre-diabetic patients. J Mol Biol. 1997;268:599–606. doi: 10.1006/jmbi.1997.0946. [DOI] [PubMed] [Google Scholar]
- 29.Modiano D, Chiucchiuini A, Petrarca V, Sirima B, Luoni G, Perlmann H, Esposito F, Coluzzi M. Humoral response to Plasmodium falciparum Pf155/ring-infected erythrocyte surface antigen and Pf332 in three sympatric ethnic groups of Burkina Faso. Am J Trop Med Hyg. 1998;58:220–224. doi: 10.4269/ajtmh.1998.58.220. [DOI] [PubMed] [Google Scholar]
- 30.Parmley S, Smith G. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- 31.Perlmann H, Berzins K, Wahlin B, Udomsangpetch R, Ruangjirachuporn W, Wahlgren M, Perlmann P. Absence of antigenic diversity in Pf155, a major parasite antigen in membranes of erythrocytes infected with Plasmodium falciparum. J Clin Microbiol. 1987;25:2347–2354. doi: 10.1128/jcm.25.12.2347-2354.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlmann H, Perlmann P, Berzins K, Wahlin B, Troye-Blomberg M, Hagstedt M, Andersson I, Hogh B, Petersen E, Bjorkman A. Dissection of the human antibody response to the malaria antigen Pf155/RESA into epitope specific components. Immunol Rev. 1989;112:115–132. doi: 10.1111/j.1600-065x.1989.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 33.Riley E, Allen S, Troye-Blomberg M, Bennett S, Perlmann H, Andersson G, Smedman L, Perlmann P, Greenwood B. Association between immune recognition of the malaria vaccine candidate antigen Pf155/RESA and resistance to clinical disease: a prospective study in a malaria-endemic region of west Africa. Trans R Soc Trop Med Hyg. 1991;85:436–443. doi: 10.1016/0035-9203(91)90207-f. [DOI] [PubMed] [Google Scholar]
- 34.Roy A, Sharma V, Chauhan V. The use of peptide ELISA in determining malaria endemicity. J Immunol Methods. 1994;167:139–143. doi: 10.1016/0022-1759(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 35.Scott J, Smith G. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 36.Smith G, Scott J. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 37.Trager W, Jensen J. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 38.Wahlin B, Sjolander A, Ahlborg N, Udomsangpetch R, Scherf A, Mattei D, Berzins K, Perlmann P. Involvement of Pf155/RESA and cross-reactive antigens in Plasmodium falciparum merozoite invasion in vitro. Infect Immun. 1992;60:443–449. doi: 10.1128/iai.60.2.443-449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrighton N, Farrell F, Chang R, Kashyap A, Barbone F, Mulcahy L, Johnson D, Barrett R, Jolliffe L, Dower W. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–464. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 40.Zhong G, Smith G, Berry J, Brunham R. Conformational mimicry of a chlamydial neutralization epitope on filamentous phage. J Biol Chem. 1994;269:24183–24188. [PubMed] [Google Scholar]