Abstract
Elice16Indures® Plant Conditioner combines the effects of a number of herbs to increase the yield of dicotyledonous plants in the field. This crop enhancer can also be used in organic farming applying low doses with ULV spraying by drone. Reducing the ecological footprint is the basis for sustainable crop production. By using the crop enhancer, a better crop can be achieved with less impact on the environment. EU Member States attach great importance to rapeseed (Brassica napus). Due to its versatility, it is one of the supported plants. Plant conditioner applied in different phenological phases (BBCH 51 and BBCH 67) of winter oilseed rape at a dose of 240 g/ha of.
By using Elice16Indures, the value of the vegetation index and the yield can be increased.
RNA-seq data set of field Elice16Indures-treated and non-treated (control) rapeseed plants are presented. For RNA-seq experiments, two samples were taken from leaf tissues in the phenological phase of BBCH 69 from control and treated plots, 2 days after treatment.
Illumina NextSeq 550 sequence reads were uploaded to the NCBI SRA database after preprocessing. Combined read sets were de novo assembled and functional annotation with the output transcripts were performed. The entire dataset of identified coding sequences (transcripts) was deposited in the NCBI TSA database. The SRA and TSA datasets are under the BioProject access PRJNA838472. The data series reported in this study may open up new opportunities to increase the efficiency of organic rapeseed production.
Keywords: Biofarming, Yield enhancer, Rapeseed, RNA-seq, Brassica napus, Transcriptome, Illumina sequencing
Specifications Table
| Subject | Plant Science: Plant Physiology |
| Specific subject area | RNA-seq profiling of control and Elice16Indures conditioner-treated rapeseed plants was performed using field leaf tissue samples. |
| Type of data | Table Database record Figure |
| How the data were acquired | Leaf samples (approximately 40–50 mg of plant tissue) were collected from plots sprayed with Elice16Indures plant conditioner and from control plots, two days after treatment. Three biological replicates from both treatments were used and samples were mixed per treatment and sequenced using the Illumina NextSeq550 platform. The raw reads were filtered, cleaned and trimmed for de novo transcriptome assembly. Further analysis of the pairwise analysis of differentially expressed genes (DEGs) were performed. Top 50 DEGs were visualized on a heat map. Functional annotation was performed using BLAST to determine the gene ontology (GO) and the biochemical functions were assigned to the coding sequences. The most specific genes were plotted on a bar graph. |
| Data format | Raw Analyzed Filtered |
| Description of data collection | RNA-seq data set of field control and Elice16Indures-treated rapeseed plants are presented. For RNA-seq experiments, samples were taken from leaf tissues in the phenological phase of BBCH 69 from control and treated (240 g/ha) plots on 26/4/2020, 2 days after treatment. |
| Data source location | • EduCoMat Ltd • Keszthely • Hungary |
| Data accessibility | The BioProject and raw reads are available in National Center for Biotechnology Information database under the accessions: Repository name: Brassica napus raw sequence reads Data identification number: PRJNA838472 Direct link to datasets: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA838472 Repository name: Eukaryotic sample from Brassica napus Data identification number: SRR19224059 Direct link to datasets: https://www.ncbi.nlm.nih.gov/sra/?term=SRR19224059 Repository name: Eukaryotic sample from Brassica napus_treated Data identification number: SRR19224058 Direct link to datasets: https://www.ncbi.nlm.nih.gov/sra/?term=SRR19224058 Repository name: Brassica napus Raw sequence reads Transcriptome Shotgun Assembly Data identification number: GJYZ00000000 |
| Direct link to datasets: https://www.ncbi.nlm.nih.gov/Traces/wgs/?val=GJYZ01 (under “Contigs” tab) Repository name: Count Table, Annotation Table, Differential Expressed Genes (DEG) by NOIseq program, Most specific upregulated genes by Fisher exact test, Most specific downregulated genes by Fisher exact test (in an excel file on separate worksheets) Data identification number (DOI):10.17632/hp4hjpjyfh.1 Direct link to datasets: https://data.mendeley.com/datasets/hp4hjpjyfh/1 |
Value of the Data
-
•
The widespread use of environmentally friendly technologies is a prerequisite for sustainable agricultural production. Reducing the ecological footprint is a common task for all farmers. The use of Elice16Indures, which can also be used in organic farming, has a number of cultivation benefits as the product stimulates biochemical processes as response to abiotic and biotic stresses. Datasets from treated rapeseed plants may provide information on the yield-enhancing effect and underlying genetic background of the investigated material.
-
•
Rapeseed is a versatile crop supported in most government of countries around the world. However, in order to improve the quantitative and qualitative parameters, it also requires high levels of chemicals. Chemical-free production is essential in organic farming, so the use of Elice16Indures offers a promising alternative to economical rapeseed production. The published dataset will also facilitate the development of future yield-enhancing products.
-
•
The presented RNA-seq data contribute to the understanding of cellular plant mechanisms in rapeseed. Gene expression data from Elice16Indures treatments contribute to an environmentally friendly increase in yield averages. These datasets can help with transcriptomic studies that underpin the next generation of organic farming.
1. Data Description
Multi-purpose rapeseed is grown in large quantities worldwide, and its yield is an important value measure. Rapeseed is particularly sensitive to wintering conditions and the amount and distribution of precipitation. If negative effects occur during the growing season, it reacts with yield depression. Farmers are looking for new technological solutions to get the most out of their crops to create favorable growth conditions and reduce their negative reactions to stressors. Therefore, there is a growing demand for plant conditioning products with yield-enhancing effects [1], [2]–3]. QuantSeq 3′ mRNA sequencing [4,5] is a satisfactory method to gene expression profiling presented in response to the use of Elice16Indures, a plant conditioning, general immunostimulant and yield-enhancing product (https://gynki.hu/en/rimph-botanicals/products/) [6]. For RNA-seq experiments, samples were taken from leaf tissues in the phenological phase of BBCH 69 from control and treated plots on 26/4/2020, 2 days after treatment. Illumina NextSeq 550 reads were deposited in NCBI Sequence Read Archive (SRA), data can be found under the following access numbers: SRR19224059 (Brassica napus control); SRR19224058 (Brassica napus treated with Elice16Indures). Using these SRA datasets, de novo assembly and identification of coding sequences were performed and deposited in NCBI Transcriptome Shotgun Assemblies (TSA) database under the accession GJYZ00000000. DEGs of control and treated Brassica napus samples were determined by bowtie2 alignment fitting SRA reads to the TSA contigs. Transcript abundances by mRNA quantification are indicated in the count table which was deposited in Mendeley data, on worksheet 1 (https://data.mendeley.com/datasets/hp4hjpjyfh/1).
Using the NOIseq program [7,8] up- and downregulated genes as a response to the treatment were determined. A total of 583 upregulated (M > 0) and 781 downregulated (M < 0) contigs were obtained. The top 50 were plotted on a heat map (Fig. 1). DEGs were visualized and placed in Mendeley Data https://data.mendeley.com/datasets/hp4hjpjyfh/1, on worksheet 2. The table of functional annotation for pairwise DEGs is deposited in Mendeley https://data.mendeley.com/datasets/hp4hjpjyfh/1, on worksheet 3.
Fig. 1.
Representation of gene expression levels of top 50 contigs of DEGs under the conditions of control and Elice16Indures treated Brassica napus.
The most specific up- and downregulated genes determined by Fisher exact test [9] were deposited in Mendeley Data https://data.mendeley.com/datasets/hp4hjpjyfh/1, on worksheets 4 and 5 are presented on Figs. 2 and 3. The Fig. 1 shows the heatmap of top 50 contigs for which the change in gene expression is most pronounced during Elice16Indures treatment.
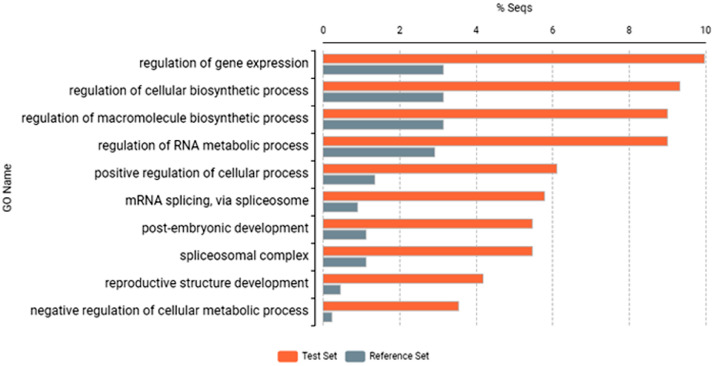
Fig. 2.
Most specific upregulated, annotated genes by Fisher's exact test. The sample marks are as follows, Reference Set: control, Test Set: Elice16Indures treated Brassica napus.
Fig. 3.
Most specific downregulated, annotated genes by Fisher's exact test. The sample marks are as follows, Reference Set: control, Test Set: Elice16Indures treated Brassica napus.
2. Experimental Design, Materials and Methods
Brassica napus cv. GK Mécses plants were grown under field conditions. Plant conditioner was applied in different phenological phases (BBCH 51 and BBCH 67) [10] of winter oilseed rape at a dose of 240 g/ha and leaf samples were collected on 26/4/2020 in Tata, Hungary. Leaf samples (40–50 mg/sample) were taken from the control and treated plots at the same time, in 3-3 repetitions. The samples were stored in liquid nitrogen and then shipped for sequencing [11]. The three replicates of each sample were pooled and sequenced by third party, Xenovea Ltd. Experimental design is summarized in Fig. 4.
Fig. 4.
Timeline of Elice16Indures treatment and sample collection of field rapeseed experiment.
3. Sequencing and Bioinformatics
NGS libraries were prepared using QuantSeq 3′mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen GmbH, Wien, 510 Austria). Diluted samples were sequenced using the NextSeq 500/550 High Output v2 Kit on the NextSeq550 platform (Illumina, San Diego, CA, USA) to generate 1 × 86 bp single-ended reads.
Reads were processed using Trimmomatic software [12], which allows removal of adapters and contaminating sequences. De novo transcriptome was generated using Trinity and Bowtie2 programs [13,14].
To estimate transcript abundancies, sample reads were aligned to the de novo transcriptome assembled from combined read sets. The read counts are detailed in the CountTable.
Functional annotation and Gene Ontology (GO) analyzes were performed with the OmicsBox. BioBam program package [15]. With the help of GO annotation, we determined the molecular function, the intracellular location and the role of genes in biochemical processes [15].
Differential gene expressions were calculated based on CountTable. This numerical analysis was performed in a pairwise comparison using OmicsBox.BioBam. The used application is based on the NoiSeq program implementing simulation of technical replicates and evaluating the significance of individual genes between two experimental conditions.
Ethics Statements
The manuscript adheres in all to Ethics in publishing standards.
CRediT authorship contribution statement
Kincső Decsi: Writing – original draft, Visualization, Validation. Géza Hegedűs: Software, Investigation. Barbara Kutasy: Validation, Visualization. Eszter Virág: Conceptualization, Validation, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgments
The work was funded by the KFI_16-1-2017-0457 - Development and production of a plant-based pesticide-plant conditioner for use in organic farming - project of the Hungarian Government. We express our thanks to József Péter Pallos, executive director, RIMPH Ltd. for the project financiering, to Márta Kiniczky, for producing of the investigated product and to Zsófia Thomas-Nyári, for the project supervision and administration. We are grateful for the Plant-Art Research Ltd, Hungary for the performance of field experiments.
Data Availability
References
- 1.Szczepanek M., Siwik-Ziomek A. P and K accumulation by rapeseed as affected by biostimulant under different NPK and S fertilization doses. Agronomy. 2019;9(9):477. doi: 10.3390/agronomy9090477. [DOI] [Google Scholar]
- 2.Siwik-Ziomek A., Szczepanek M. Soil extracellular enzyme activities and uptake of N by oilseed rape depending on fertilization and seaweed biostimulant application. Agronomy. 2019;9(9):480. doi: 10.3390/agronomy9090480. [DOI] [Google Scholar]
- 3.Gavelienė V., et al. Effect of biostimulants on cold resistance and productivity formation in winter rapeseed and winter wheat. Ir. J. Agric. Food Res. 2018;57:71–83. https://www.jstor.org/stable/26555013 [Google Scholar]
- 4.Moll P., et al. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat. Methods. 2014;11(12):i–iii. doi: 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- 5.Corley S.M., et al. QuantSeq. 3′ sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci. Rep. 2019;9(1):1–15. doi: 10.1038/s41598-019-55434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decsi K., et al. RNA-seq datasets of field soybean cultures conditioned by Elice16Indures® biostimulator. Data Brief. 2022 doi: 10.1016/j.dib.2022.108182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarazona S., et al. NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet J. 2011;17(B):18–19. doi: 10.14806/ej.17.B.265. [DOI] [Google Scholar]
- 8.J. Gutierrez, October 24, 2019. Reanalyzing the A. galli transcriptomic response to an anthelmintic drug with OmicsBox. Michaela M. Martis et al. in 2017, doi: 10.1371/journal.pone.0185182. [DOI]
- 9.Al-Shahrour F., Díaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 10.Zadoks J.C., Chang T.T., Konzak C.F. A decimal code for the growth stages of cereals. Weed Res. 1974;14(6):415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x. [DOI] [Google Scholar]
- 11.Hegedűs G., et al. Transcriptome datasets of β-Aminobutyric acid (BABA)-primed mono-and dicotyledonous plants, Hordeum vulgare and Arabidopsis thaliana. Data Brief. 2022 doi: 10.1016/j.dib.2022.107983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabherr M.G., et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29(7):644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. Bioinformatics and S. Valencia, OmicsBox–Bioinformatics made easy (Version 1.4.11). BioBam Bioinformatics. March 3, 2019. https://www.biobam.com/omicsbox/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.