Graphical abstract
Keywords: Euglenozoa, Metakinetoplastina, RNA editing, Mitochondrion, Maxicircle, guide RNA, ATPase 6
Abbreviations: U-indel editing, Uridine insertion/deletion editing
Abstract
The kinetoplastids are unicellular flagellates that derive their name from the ‘kinetoplast’, a region within their single mitochondrion harboring its organellar genome of high DNA content, called kinetoplast (k) DNA. Some protein products of this mitochondrial genome are encoded as cryptogenes; their transcripts require editing to generate an open reading frame. This happens through RNA editing, whereby small regulatory guide (g)RNAs direct the proper insertion and deletion of one or more uridines at each editing site within specific transcript regions. An accurate perspective of the kDNA expansion and evolution of their unique uridine insertion/deletion editing across kinetoplastids has been difficult to achieve. Here, we resolved the kDNA structure and editing patterns in the early-branching kinetoplastid Trypanoplasma borreli and compare them with those of the well-studied trypanosomatids. We find that its kDNA consists of circular molecules of about 42 kb that harbor the rRNA and protein-coding genes, and 17 different contigs of approximately 70 kb carrying an average of 23 putative gRNA loci per contig. These contigs may be linear molecules, as they contain repetitive termini. Our analysis uncovered a putative gRNA population with unique length and sequence parameters that is massive relative to the editing needs of this parasite. We validated or determined the sequence identity of four edited mRNAs, including one coding for ATP synthase 6 that was previously thought to be missing. We utilized computational methods to show that the T. borreli transcriptome includes a substantial number of transcripts with inconsistent editing patterns, apparently products of non-canonical editing. This species utilizes the most extensive uridine deletion compared to other studied kinetoplastids to enforce amino acid conservation of cryptogene products, although insertions still remain more frequent. Finally, in three tested mitochondrial transcriptomes of kinetoplastids, uridine deletions are more common in the raw mitochondrial reads than aligned to the fully edited, translationally competent mRNAs. We conclude that the organization of kDNA across known kinetoplastids represents variations on partitioned coding and repetitive regions of circular molecules encoding mRNAs and rRNAs, while gRNA loci are positioned on a highly unstable population of molecules that differ in relative abundance across strains. Likewise, while all kinetoplastids possess conserved machinery performing RNA editing of the uridine insertion/deletion type, its output parameters are species-specific.
1. Introduction
The kinetoplastids are a group of unicellular flagellates of the phylum Euglenozoa that possess many particularities related to their cell biology, biochemistry, and gene expression [1]. Its most well-known members belong to the family Trypanosomatidae (subclass Metakinetoplastina: order Trypanosomatida) [2], [3] and include parasites transmitted by insects to mammals that cause severe diseases such as sleeping sickness, Chagas disease, and leishmaniases [4]. Substantially less understood are members of this phylum belonging to other orders [2]. Trypanoplasma borreli is an iconic species of the family Trypanoplasmatidae (order Parabodonida). It is an obligate bloodstream parasite of marine and freshwater fish vectored by hematophagous leeches [5]. The outcome of fish infection is primarily determined by host immunity and the level of mutual host-parasite adaptation [6], [7]. Leech and fish-derived isolates are morphologically indistinguishable and have been cultured extensively in rich medium. Because of this, it would be difficult to say whether parasites derived from these different hosts metabolically differ.
Historically, perhaps the most arresting feature of kinetoplastids was the extreme abundance and unusual structure of DNA in their single, reticulated mitochondrion. While this so-called kinetoplast (k)DNA carries organellar rRNA genes and a subset of the suite of typical mitochondrion-encoded genes, the expression mechanism of some of their mRNAs is bizarre. They are encoded in the kDNA as cryptogenes and to become translatable, their transcripts require multiple targeted insertions and deletions of one or more uridines (Us) at numerous editing sites. The process is termed RNA editing of the uridine insertion/deletion type (U-indel editing). While we now know that mechanistically different types of RNA editing frequently occur in viruses and bacteria, as well as in single and multicellular eukaryotes [8], at the time of discovery the post-transcriptional insertion of four uridines into COII (coxII) mRNA of trypanosomes [9] was difficult to explain. Consequently, the range of explanations was wide [10]. To test the various hypotheses, it was not only important to dissect the RNA and protein machinery responsible for RNA editing in the most common model organisms Trypanosoma brucei and Leishmania tarentolae of the family Trypanosomatidae, but also to look for this process in the distantly related kinetoplastid protists. Of these, Trypanoplasma borreli was the most prominent candidate. The finding of U-indel editing in its mitochondrial transcripts [11], [12] indeed had important evolutionary implications [13]. The nuclear genome of this species has been sequenced [14], yet, very little progress occurred in our understanding of its peculiar organellar genome and transcriptome.
A subset of mitochondrial transcripts of all kinetoplastid flagellates examined so far is subject to the process of RNA editing. Being composed of either free supercoiled or catenated relaxed circles, the size but not the coding capacity of the kDNA is variable and species-specific [8], [15]. The editing of kDNA-derived transcripts is performed by several protein complexes with the assistance of small RNA molecules called guide (g) RNAs [1].
The kDNA of trypanosomatids has a very uniform arrangement, present as a single network of thousands of mutually catenated, gRNA-bearing minicircles, and dozens of maxicircles, on which a standard set of mitochondrial-encoded genes reside [16]. Hence, the mRNA substrates of editing and the gRNAs that provide information for the exact insertions and deletions of uridines are derived from distinct components of the kDNA [17], [18]. These molecules are packed into an electron-dense disk located close to the basal body of the flagellum [19]. Much less is known about kDNA structure outside the family Trypanosomatidae, with transmission electron microscopy evidence suggesting that in different lineages it evolved in a variety of complex and much less regular structures [20], [21]. In the case of T. borreli, its massive kDNA is dispersed throughout the mitochondrial lumen, although apparently in a condensed enough region to be easily detected by light microscopy [12]. In fact, in terms of the sheer amount of DNA, it is one of the most extensive organellar genomes known so far [22]. Yet, our knowledge about the kDNA and mitochondrial transcriptome of kinetoplastids outside of the trypanosomatids is fragmentary at best. It would, therefore, be highly informative to characterize its kDNA molecules, gRNAs and U-indel editing, only a few details of which are known. If the observed patterns, extent, variability, and progression of RNA editing in this fish pathogen were comparable with those traits in trypanosomatids, it would question the alleged superiority of their compactly packed and catenated kDNA disk structure [23].
In this work, we use the features of editing apparent from long read DNA sequence data and RNA-seq reads, as well as computational methods specifically designed for the dissection of U-indel editing to substantially improve our understanding of the T. borreli kDNA structure and U-indel editing patterns.
2. Materials & methods
2.1. Strain identity, parasite growth, nucleic acid purification, and library generation
Trypanoplasma borreli Tt-JH was isolated from a tench (Tinca tinca) in 1986 in the vicinity of Jindřichův Hradec (Czechia) and verified as in [24]. The parasites were cultivated as described previously [25]. The total DNA and RNA were isolated using Nucleospin DNA and RNA XS kits (Macherey-Nagel, Düren, Germany) from 5 × 108 cells. The library was prepared starting from 16 μg high molecular weight genomic DNA. The sample was sheared using a Megaruptor 1 to 25 kb (Diagenode, Seraing (Ougrée), Belgium). All sheared DNA was then used as input into library preparation, using SMRTbell Template Prep Kit 1.0 (Pacific Biosciences, Menlo Park, USA) and following the standard protocol. Prior to sequencing, the library was size selected on the Blue pippin (Sage Science, Beverly, USA) at >7 kb cut-off (S1 marker, 0.75 % gel cassette).
2.2. Read sequencing and preprocessing
Sequencing was performed on a Pacific Biosciences RSII, using SMRT® Cell 8Pac V3 cells. All SMRTcells had 240-minute movies, and stage start acquisitions. This yielded a total of 156,405 reads and 1.252 trillion bases read. Total cellular RNA was sequenced on an Illumina HiSeq 4000 with a strand-independent 100 bp paired-end protocol, yielding 25.2 million read pairs. The RNAseq data is accessible under SRR9331469 in the Sequence Read Archive [26]. For downstream processing with T-Aligner [27], [28], RNAseq reads were quality-checked with FastQC v. 0.11.9 [29] and quality-trimmed with trimmomatic v. 0.39 [30]. Trimmed reads were merged with the paired-end read merger PEAR v. 0.9.6 [31], and 69.3 % of read pairs were successfully merged. Merged and unmerged reads were combined into the single file used as input for all T-Aligner pipelines.
2.3. Kinetoplast DNA assembly
PacBio sequencing reads were used to assemble draft contigs with Flye v. 2.8.3 [32] and Canu v. 2.2 [33]. As the actual organization of the kinetoplast genome was not known, it was safest to include reads that would likely map to the nuclear genome as raw input to avoid the potential false filtering of mitochondrial reads. The kDNA maxicircle contig was extracted using previously published T. borreli mitochondrial rRNA and mRNA coding region sequences (GenBank accession numbers U11682 and U14181 [11], [12]) as a blastn (BLAST suite v. 2.5.0+ [34]) query against the assembled contigs database. The circular nature of the maxicircle contig was confirmed by the presence of uniquely mapped reads overlapping the junction point when PacBio reads were mapped to the circularly rotated assembled maxicircle contig with minimap2 v. 2.22 [35]. The junction point was located in the repeat supercluster area of the divergent region; therefore, the exact sequence of the junction point is ambiguous. This ambiguity is represented by the addition of ‘NNN’ on the scaffold submitted to GenBank under accession number OP278005. Tools were utilized exactly as described in [36] to acquire information conveyed in Fig. 1A.
Fig. 1.
T. borreli kDNA includes a maxicircle and gRNA-containing elements that can be assembled into ScaI-flanked contigs. A. The maxicircle scheme. The outer track defines approximate boundaries of major structural compartments of a maxicircle: CR, coding region; 5P, ND5-promixal repeat compartment of the divergent region; 12P, 12S-proximal repeat compartment of the divergent region composed of tandem repeat arrays; AO - assembly overlap point, the place of molecule circularization in assembly (see Section 2.3). The numbers represent position in kilobases. The next track in from the circle’s exterior is a 24-mer repeat histogram, showing the frequency of the 24-mer per maxicircle, kmers with highest observed frequency are blue, others are green. The height of the track is normalized on the highest observed frequency. The next track in (violet band) is the location of tandem repeats detected with the ‘mreps’ tool. The next track in (green) is GC-content in fraction form from 0 to 1, represented by the height of the track at each position. The inner track of the circle shows the regions of sequence identity by connecting them with colored ribbons (blue – sequence identity of 95–100 %, green – 85–95 %, yellow – 80–85 %), and the inverted repeats detected with ‘einverted’ tool (dark pink/violet arcs connecting the ends of each repeat). The source of tools used to generate tracks are described in Section 2.3. B. Scheme of a ScaI-flanked contig (not to scale). The ellipsis shown on each terminus indicates that there are additional ScaI-containing repeats of variable number on the end beyond those shown. C. Contig 1 5′ ScaI repeat region. The portion shown is to scale and is the innermost section that is covered by an entire PacBio read. The CSB3 sequence is shown; red nucleotides indicate those that differ from what is considered consensus among trypanosomes. D. Map of unique gRNA-coding regions of Contigs 1, 2, and 3 (top to bottom). The top track shows proximal motifs and gRNA loci (rectangular blocks) for one strand, the bottom track shows the same for the opposite strand. The loci are color coded based on the mRNA that they primarily align to. A6, blue; COI, pink; CYb, orange; RPS12, yellow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Other components of the T. borreli kDNA were identified by scanning the Flye-assembled contigs database with blastn for ScaI-containing repeats (GenBank: U14184 and U14185) and for two previously known gRNAs and their flanking regions (GenBank: U47932 and U47933) [12], [37]. This resulted in the acquisition of gRNA-containing contigs 1–14 that are flanked by ScaI-containing repeats. The sequence of gRNA-containing contig 6 was manually extracted from its initial contig, which also contained nuclear chromosomal DNA. The last three gRNA-containing contigs were identified by searching incomplete sequences found in the Flye assembly in the set of Canu-assembled contigs (complete flanked contigs 15–17). Typically, the length of repeat regions was higher than most PacBio reads used for assembly. All contigs were trimmed leaving the most proximal several kilobases of flanking repeats before employing gRNA-identifying procedures. The contig GenBank accession numbers are OP242806-OP242822.
Maxicircle and ScaI-flanked contig coverage was estimated by PacBio read mapping with minimap2 with further processing of bam files with SAMtools ‘view’ and ‘depth’ [38]. Quantile coverage was calculated using a custom python script. For assessment of coverage in a different strain, we used paired-end DNA sequencing reads of T. borreli K-100 (ATCC 50432, GenBank accession number ERR316180) and the same processing protocol, but mapped with BWA-MEM [39].
2.4. T-Aligner version update
We used the newest T-Aligner v. 4.0.5f (https://github.com/jalgard/T-Aligner). This version contains a dynamic open reading frame (ORF) search depth (the number of paths traced in its read graph now depends on coverage; this mode addresses the 3′ coverage bias of most complex cryptogene products), multithreading at the ORF tracing step, and an improved gRNA finder tool with a flexible scoring model. The gRNA finder now scans potential gRNA-containing sequences for a fixed length seed match that exceeds a score threshold (default scoring: +2 match, +0 G:U, −2 mismatch); every seed is extended step by step in both directions, each time choosing the best-scoring extension; and the step size is variable (default: 4). The best-scoring extended match that is greater than threshold value is reported for each region. Scoring will also request the presence of an anchor region at the 5′ end of the gRNA, which is usually necessary for gRNA:pre-mRNA interactions.
2.5. Maxicircle annotation and mitochondrial transcriptome assembly
The expression profile of the maxicircle was obtained by aligning trimmed merged and unmerged RNA reads on the assembled maxicircle with BWA-MEM and T-Aligner’s ‘alignlib’ tool, separately extracting only a portion of the edited reads (reads with 5 or more edited sites are considered edited) in a separate ‘.bam’ file specifically for the generation of locus maps showing edited versus not-edited reads. SAMtools and BEDTools [38], [40] were used to convert T-Aligner’s ‘.taf’ files to sorted ‘.bam’ files, which were visualized using a custom R script.
The set of typical kDNA gene sequences (especially the previously obtained well-curated sets available for Leptomonas pyrrhocoris and Trypanosoma cruzi [28], [41]) were manually aligned to the maxicircle sequence to locate maxicircle genes. T-Aligner’s ‘alignlib’ module was used to align RNA sequencing reads and detect edited domains. For the four cryptogenes, approximate boundaries were flanked with ∼ 50–100 bp of adjacent sequence and used as references for T-Aligner’s ‘findorfs’ module. Edited mRNA sequences were reconstructed with ‘findorfs’ using ‘extension’ ORF tracing mode and ‘aln_mismatch_max’ option set to 0. For CYb and COI, ‘aln_min_segment’ was set to 20 and ‘orf_search_depth’ was set to 3 and −1 respectively (the negative value turns on the dynamic ORF search depth mode in which T-Aligner dynamically increases the number of possible paths when a sufficient coverage threshold is met). The set of mRNAs assembled with T-Aligner was then subjected to blastp searches against reference proteins of Leptomonas pyrrhocoris, Trypanosoma cruzi, T. lewisi, and T. vivax [28], [41], [42], [43] to detect the best canonically edited mRNA candidates. Typical of transcriptome analysis, the overlapping of edited reads contributing to the ORF along a cryptogene locus provides evidence of an entire mRNA, but it is still a reconstruction rather than a single fully sequenced product [28]. The number of total reads supporting each full length edited cryptogene ORF is as follows: A6, 1409; COI, 82,241; CYb, 40,406; RPS12, 2917. The sequences of T. borreli A6, COI, CYb, and RPS12 mRNAs were submitted to GenBank under accession numbers OP242802-OP242805.
The maxicircle divergent region was annotated as described previously, using the same scripts and protocols for repeat annotation and data visualization [36]. The visualization scheme of the maxicircle is generated with the pipeline described in [36].
2.6. Identification of putative gRNA loci in ScaI-flanked gRNA-containing contigs
Putative gRNA coding loci were detected on ScaI-flanked contigs with ‘findgrna’ tool from T-Aligner’s suite (https://github.com/jalgard/T-Aligner) with ‘—seed_length 25 –seed_score 27 –gu 14 –mm 3 –anchor 2 –length 27 –score 32′ strict (at most 3 mismatches in total, at most 14 G:U pairs in total, minimal alignment length of 27) or ‘—seed_length 20 –seed_score 22 –gu 16 –mm 4 –anchor 2 –length 24 –score 27′ relaxed (at most 4 mismatches and 16 G:U pairs, minimal length of 24) search settings. Both forward and reverse strands of each ScaI-flanked contig were examined. Alignments above threshold called raw hits were considered as possible gRNA coding loci and filtered further for the presence of the RYAGGCTTT motif sequence between 20 and 50 bp downstream of the hit region [37].
Reconstruction of an editing cascade model for each canonical edited mRNA was performed by inspection of the filtered output of ‘findorfs’ and assembling the best gRNA:mRNA pairs on the map with manual refinement of alignment boundaries.
2.7. Editing stringency assessment
At each potential editing site, the percentage of editing events that contribute to the identified translatable product for each gene was determined as described in detail elsewhere [27], [41].
3. Results and discussion
3.1. New evidence modifies our understanding of the T. borreli kDNA
Based on the limited available data, previous studies suggested that the genetic arrangement of T. borreli kDNA differs from that of other studied kinetoplastids. We generated T. borreli DNA and transcriptomic read datasets suitable to mine for kinetoplast-derived fragments. We used PacBio technology to sequence and assemble complete kDNA molecules. Moreover, a paired-end Illumina poly(A)-enriched library was used to characterize the T. borreli mitochondrial gene expression.
An early attempt to characterize the mRNA and rRNA-containing kDNA molecule in T. borreli estimated its size to be unusually large for a kDNA, approximately 80–90 kb, albeit with a modest coding region of approximately 6 kb [12]. However, another group utilizing a different technology estimated it to be approximately 37 kb, similar to that of well-studied trypanosomatids [11]. To resolve this disparity, we assembled this molecule using long sequencing reads well suited for this purpose [44], [45], [46]. The circular molecule was found to be approximately 43 kb, similar to the 37 kb size previously estimated with Southern blotting analysis [11].
The T. borreli maxicircle is partitioned into two major regions: the coding region and the divergent region, similar to maxicircles of trypanosomatid species [36] (Fig. 1A). The divergent region consists of organizational domains, also previously characterized in the trypanosomatid maxicircles. Specifically, these are two repeat-containing units termed 5P and 12P that typically flank the coding region [36]. However, this synteny is not conserved in T. borreli. Instead, large tandem repeats containing sequence analogous to 5P flank the coding region from both sides (Fig. 1A). A supercluster of 17 imperfect copies of repeat blocks that can be classified as 12P-like (by virtue of its repeat pattern, not the sequence of the repeat units) lies between the 5P domains (Fig. 1A). Its larger size is the reason why the size of the T. borreli maxicircle slightly exceeds that of other studied maxicircles. Notably, the T. borreli maxicircle possesses abundant inverted repeats, including, uniquely, some positioned in the coding region (violet arcs in Fig. 1A).
Previous characterization of the gRNA-encoding kDNA concluded that it (most likely) consisted of enormous circular molecules of an estimated size of 170–200 kb [12], [37]. This was a surprising finding, as in trypanosomatids one or several gRNAs are encoded on small circular molecules [47], [48]. The same historical studies further demonstrated that the putative 170–200 kb molecules possessed regions of repetitive sequence, in which the ScaI restriction endonuclease recognition site appeared at regular intervals of about 1 kb [12], [37]. We attempted to confirm the existence of very large, circular gRNA-containing kDNA molecules using the same PacBio read set used to assemble the T. borreli maxicircle. We anticipated that the result would be either the previously proposed 170–200 kb circles, or molecules similar to the trypanosomatid minicircles. The usage of previously determined ScaI-containing sequences and two gRNA-containing sequences to probe our assembled contigs for parts of the presumed large circles resulted in the detection of a total of 17 contigs in a range of sizes averaging ∼ 70 kb. Each of them was flanked by a series of ScaI-containing repeats positioned in an inverse orientation (Fig. 1B). Various numbers of the ScaI repeats flanked a unique inner sequence. As noted previously [12], each ScaI-containing repeat unit also harbors a sequence very similar to the minicircle conserved sequence block 3 (CSB3) of other trypanosomatids (Fig. 1C). The CSB3 12-mer is invariably present in minicircles and was proposed to be their origin of replication [49], [50]. Relative to the CSB3 orientation, the ScaI repeats are oriented inward rather than outward on each contig. For further analysis, we trimmed the terminal repeats leaving only a few copies per end.
We did not obtain evidence that the ScaI-flanked contigs were parts of large circular molecules of several hundred kilobases. For circular molecules of the size previously determined, multiple ScaI-flanked contigs would have to be assembled into each circle, and we would expect similar DNA read coverage across the large circle. Taken separately, average and median coverages of each contig were largely very similar, indicating an even coverage across each contig. Yet, the coverage of different contigs spanned a fivefold range (Table 1). The simplest explanation of this observation is that the various contigs belong to separate molecules, the circular or linear nature of which we cannot confidently determine. Neither the Flye nor the Canu assemblers marked any contigs as circular in the output files. All contigs ended with tandem long imperfect repeats oriented in opposing directions, such the contig ends cannot be overlapped. This suggests (but does not prove) a linear status, particularly since the homology of the regions between ScaI sites is over 99 %.
Table 1.
Features of the 17 ScaI-flanked contigs containing putative gRNAs of T. borreli and their DNA read coverage in two strains. Columns contain contig ID, contig length, number of high-confidence gRNA hits per contig and the read coverage (average and median) for two strains: Tt-JH and K-100. For Tt-JH PacBio reads were used to estimate the coverage, for K-100 paired-end Illumina reads. The ‘Ratio’ column is the ratio of the median to the average read coverage.
| Contig | Length, bp | gRNAs | Tt-JH strain |
K-100 strain |
||||
|---|---|---|---|---|---|---|---|---|
| Average | Median | Ratio | Average | Median | Ratio | |||
| 1 | 72,623 | 27 | 538 | 542 | 1.01 | 608 | 0 | 0.00 |
| 2 | 67,545 | 18 | 408 | 405 | 0.99 | 3492 | 3073 | 0.88 |
| 3 | 69,164 | 20 | 360 | 369 | 1.02 | 526 | 0 | 0.00 |
| 4 | 71,326 | 23 | 256 | 265 | 1.03 | 449 | 0 | 0.00 |
| 5 | 74,611 | 23 | 244 | 237 | 0.97 | 1350 | 713 | 0.53 |
| 6 | 67,281 | 25 | 240 | 232 | 0.97 | 749 | 0 | 0.00 |
| 7 | 59,764 | 24 | 193 | 200 | 1.04 | 1071 | 403 | 0.38 |
| 8 | 62,950 | 26 | 190 | 191 | 1.01 | 2811 | 2449 | 0.87 |
| 9 | 81,573 | 25 | 240 | 182 | 0.76 | 602 | 0 | 0.00 |
| 10 | 67,594 | 23 | 141 | 123 | 0.87 | 839 | 298 | 0.36 |
| 11 | 73,206 | 26 | 832 | 126 | 0.15 | 1283 | 339 | 0.26 |
| 12 | 47,387 | 20 | 115 | 103 | 0.89 | 1313 | 756 | 0.58 |
| 13 | 67,800 | 27 | 66 | 58 | 0.88 | 590 | 0 | 0.00 |
| 14 | 61,768 | 23 | 97 | 92 | 0.95 | 530 | 0 | 0.00 |
| 15 | 84,266 | 30 | 144 | 127 | 0.88 | 566 | 0 | 0.00 |
| 16 | 70,495 | 30 | 180 | 131 | 0.73 | 1092 | 405 | 0.37 |
| 17 | 71,295 | 30 | 617 | 637 | 1.03 | 532 | 0 | 0.00 |
The relative abundance of various minicircle classes in trypanosomatids is typically malleable, differing greatly among isolates [27], [51], [52], [53]. To examine this possibility in T. borreli, we mapped the DNA reads of another T. borreli strain, K-100, onto our 17 assembled gRNA-containing contigs. The coverage for only 2 of the 17 contigs was robust, while there was basically no coverage for the other contigs (Table 1). Technically, for many contigs, some K-100 reads did map (there is a number is the ‘Average’ column), yet K-100 median coverage was calculated to be zero. In these cases, K-100 reads hits to only one or a few specific positions on each contig. If only a narrow region of high similarity exists between K-100 reads and a Tt-JH contig, a homologous contig in K-100 is unlikely. The lack of many clear Tt-JH gRNA-containing contig homologues among K-100 assembled contigs is consistent with the losses and gains of gRNA-containing molecules among strains. This is a common feature of the kDNA minicircle population of various trypanosomatids. Taken together, our findings do not suggest that particularly large circular molecules encode gRNAs in T. borreli.
3.2. Analysis of expression and editing of the T. borreli maxicircle mRNAs fills a gap in evolutionary knowledge of these processes
While the putative open reading frames (ORFs) of the T. borreli maxicircle were identified nearly thirty years ago, a complete maxicircle transcriptome remains unavailable. We are filling this gap with this study. Firstly, we profiled RNA read coverage on the maxicircle coding region to pinpoint transcription unit boundaries and subsequently determined their relative expression, distinguishing between the edited and unedited mapped reads (Fig. 2). We did this by removing Us from all reads and from the maxicircle, so that reads both with and without U insertions and deletions would map to their maxicircle origin. Edited reads were defined as those having 5 or more instances of U insertions and/or deletions relative to the maxicircle sequence. Thus, the definition of an “edited” read is independent of whether it came from a fully edited, translatable mRNA or one in the process of being edited, as this can be difficult or impossible to unambiguously determine.
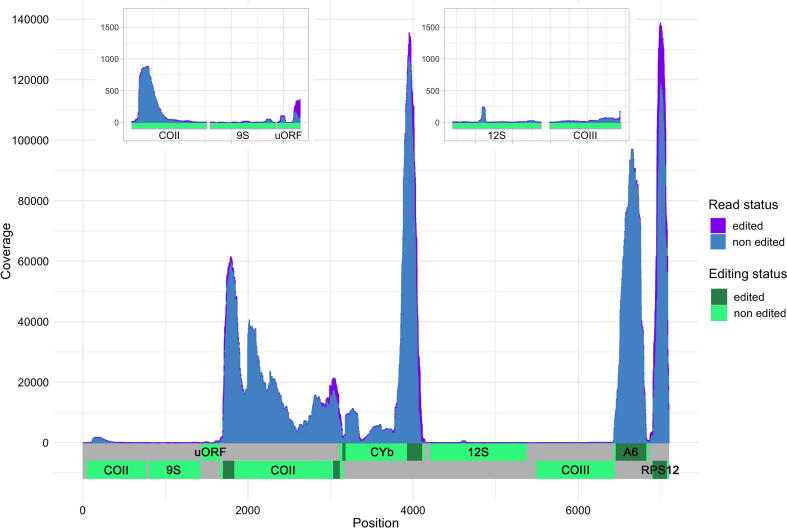
Fig. 2.
Expression of the maxicircle coding region genes and cryptogenes is variable. Expression coverage profile of the approximately 6 kb coding region of the T borreli maxicircle. The Y axis shows total per base read coverage with Illumina paired-end poly(A)-enriched reads. Inset are two low-coverage regions with a linear, lower amplitude Y axis scale to visualize relative coverage of low-coverage regions. The fraction of edited reads (defined as those with five or more U insertions and/or deletions relative the maxicircle sequence to which it maps) is shown in violet. Reads with four or fewer of these differences relative to the maxicircle are considered non edited (differences could be reasonably attributed to sequencing errors or incorrect trimming) and are shown in blue. Genes are schematically placed on the respective strands; edited domains of the genes are highlighted. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Very few reads mapped to 9S and 12S rRNA genes, likely due to the poly(A) enrichment of the sequenced cDNA library, although in a previous study high levels of rRNAs were detected despite such enrichment [28]. The small region previously identified as “G” in the originally published maxicircle coding region fragment [11] is either not transcribed or its RNA product is very unstable. Another small region between 9S and COI denoted as ‘unknown ORF’ (uORF) in Fig. 2 (‘RF’ in [11]) is irregularly covered by a low number of RNA-seq reads. It is plausible that the expression patterns of T. borreli may be different in different strains or hosts, or for freshly isolated parasites as compared to those with a long culture history.
Next, we verified and expanded the products of editing of T. borreli maxicircle transcripts. One mystery we aimed to solve was whether a cryptogene for the protein A6 (ATPase subunit 6) was present in its kDNA. The A6 gene was found in all previously sequenced trypanosomatids, as well as in Perkinsela sp. and Bodo saltans, and the representatives of Prokinetoplastina [54], [55], [56], [57], [58]. However, no evidence for either an A6 gene or cryptogene on the T. borreli maxicircle has been previously noted [12]. Utilizing T-Aligner software [27], we reconstructed a translatable edited A6 mRNA of the same size as that of other kinetoplastid A6 mRNAs. Reads comprising the A6 ORF are derived from the genome locus between COIII and RPS12 encoded in the opposite orientation, previously denoted as ‘GRII’ in [12]. Its transcript is edited throughout its entire length. We also reconstructed ORFs for the RPS12 mRNA edited throughout its length, and mRNAs for CYb and COI that are edited at only parts of their transcripts. A small portion of the reads mapped to the uORF display evidence of editing, yet no mRNA could be assembled from these reads. For ORFs reconstructed with T-Aligner, there is no DNA read assembly to confirm their sequences; thus, there is an inherently higher ambiguity for these products than for properly encoded mRNAs. However, the fact that translations of the T-Aligner reconstructions share similar approximate termini, length, and translated protein sequences with their kinetoplastid homologues speak in favor of their accuracy (Fig. S1). In conclusion, four of the seven potentially protein-coding transcripts derived from the T. borreli maxicircle require editing to generate translatable mRNAs. For all four cryptogenes, the start codon is generated by editing; likewise, generation of the stop codon requires editing for all cryptogenes, except CYb.
While there was some coverage variability of CYb and COI, we note that the observed variability is retained when only non-edited reads are mapped in a traditional manner with BWA-MEM. The coverage variability is thus unlikely to be an artifact of T-less mapping with T-Aligner. Additionally, since similar coverage differences exist in previous analyses of kinetoplastid mitochondrial genomes [27], [28], [41], we speculate that it has something to do with the stability of these molecules, perhaps even during library processing. If this is the case, these variabilities are present in libraries prepared by three independent research groups. It is also possible that oligo(A) regions within mitochondrial mRNAs may be sufficient to capture decay intermediates in the oligo(dT) affinity step that could (in theory) be mapped to the loci.
Interestingly, the abundance of cryptogene transcripts is much higher than that of the correctly encoded gene transcripts. For cryptogenes, the portion of reads with U insertions or deletions in each site over the total reads mapped on the site is quite low (Fig. 2, portion of edited reads is colored violet on the coverage plot). Among all edited regions of the coding repertoire, the 3′ end of RPS12 and the 3′ edited domains of CYb and COI include the greatest proportion of those reads that are edited. We did note some low abundance expression peaks in loci characterized as unedited that include mapped reads with U insertions and deletions, such as one in 12S. These reads likely originate from other (probably non-maxicircle) genomic loci that share sequence similarity with these maxicircle loci. This ‘read multimapping’ effect is commonly observed in low-complexity genomic regions such as the AT-rich region of 12S, and in our case, 2 non-T mismatches per read alignment are permitted.
3.3. Models of reconstructed cryptogene editing cascades and locations of putative gRNA genes
Five gRNA were identified and characterized in 1996 from a gRNA library (one gRNA for RPS12 and CYb each, and three gRNAs for COI) [37]. These molecules were shorter than gRNAs of the better studied L. tarentolae. The RPS12 and CYb gRNA genes were determined to exist on the same ‘Component I’ DNA that encoded the ScaI-containing repeats. Little effort has been made ever since to either expand the number or precisely characterize the position of T. borreli gRNA genes on the DNA molecules that encode them. With our updated, extended and better-supported data on the translatable products of U-indel editing in T. borreli, it was possible to characterize its putative gRNA population.
To search for specific genomic gRNA loci, we performed alignments between the edited regions of the four edited ORFs and the 17 ScaI-flanked contigs. The gRNA:mRNA interactions are often imprecise. Such imprecision results from the fact that the local alignments are short, and because G:U base pairing and mismatches may be tolerated by the editing machinery [59], [60]. Complicating matters further, typical gRNA length and the number of hypothetically permitted G:U pairs and mismatches vary among species [27], [41]. As the mRNA:gRNA pairing parameters are unknown for T. borreli, we performed searches with strict and relaxed settings (Section 2.6). This method is similar to previous gRNA loci searches [27], [52], [61], where initial parameters were chosen using prior knowledge of at least a few gRNA:mRNA alignments. We allowed the anchor length, defined in the alignment tool as having exact Watson-Crick pairing with the mRNA, to be as low as 2 base pairings. This is because pairings of the 5 formerly sequenced T. borreli gRNAs with their cognate mRNAs suggest that G:U pairs and even mismatches are permitted in their anchor regions. Strict settings were initially selected based on the composition of known gRNAs and then relaxed by reducing seed score, to allow adjacent mismatches. While some putative gRNAs identified under a reduced seed score could be false positives, a verification approach to separate actual gRNA loci from false positives relies on conserved sequence motifs that are often found near the “true” gRNAs [62], [63]. A nucleotide motif was identified in two of the five originally identified gRNAs for which genomic sequence context had been determined [37]. We used the sequence context of identified gRNAs to further refine the identity and position of the motif: the sequence ‘RYAGGCTTT’ located 20–40 bp downstream of the sequence ‘hit’ and upstream of the gRNA loci. The presence of this motif was used to cull the larger library to a “high-confidence” set of 420 putative gRNA loci on the ScaI-flanked contigs (Table S1). The high-confidence set appeared to have fewer G:U pairs than the set generated with relaxed settings, and although median values for length and number of mismatches did not seem to vary, the overall distribution around these metrics did (Fig. S2). All 17 contigs are approximately equally covered with putative gRNA loci of the high-confidence category positioned on both strands (Fig. 1D). No pattern of gRNA gene placement on contigs could be discerned. On average, 23 high-confidence putative gRNA loci populate the central non-repetitive region of the ScaI-flanked contig.
Before proceeding, the likelihood of this sequence being a legitimate proximal motif of gRNA loci was assessed by using SEA and ACE tools from MEME suite in an enrichment analysis. We compared the presence of ‘RYAGGCTTT’ in the 50 bp downstream of each mRNA hit on the ScaI-flanked contigs to its presence in a set of 6,000 randomly extracted 50 bp sequences from ScaI-flanked contigs. Both algorithms detected highly significant (‘expected value’ in the e-23 or e-28 range) 3 × to 4 × enrichment of the motif, which confirms that a substantial portion of the contig:mRNA alignments is associated with it. While sequencing of small RNA may unambiguously prove that ‘RYAGGCTTT’ is a gRNA-associated sequence motif, the presented analysis is very suggestive that this is the case.
We compared the general properties (gRNA length based on the alignment, number of G:Us per base, number of mismatches per base and the anchor region characteristics) of the T. borreli motif-filtered putative gRNA set with the gRNA populations of other species – L. tarentolae, L. pyrrhocoris, and T. brucei [27], [64], [65]. Our high-confidence set of putative T. borreli gRNAs has a simple, unimodal distribution of these parameters with median values of:i/ 28 bp length; ii/ 0.11 as a proportion of per-base G:Us, and iii/ 0.13 as a proportion of per-base mismatches. These parameters have their median values of 43, 0.2, and 0.06 for L. tarentolae, 42, 0.4, and 0.03 for T. brucei, and 32, 0.2, and 0.1 for L. pyrrhocoris. We conclude that T. borreli has relatively short gRNAs, its RNA editing mechanism permits high mismatch in the mRNA:gRNA alignment, and that relative to other species, G:U pairing is infrequently utilized. However, the putative T. borreli gRNA anchor regions frequently contain G:U pairs and even mismatches, which are rare in the gRNAs of other studied flagellates. The anchor regions are the gRNA 5′ ends that initially bind them to their cognate mRNA region, allowing the rest of the gRNA to direct editing of the upstream mRNA region. Complete editing is accomplished by the sequential utilization of gRNAs in the 3′ to 5′ direction along the whole transcript during the editing process.
We next determined whether editing in T. borreli could be entirely accomplished with our identified high-confidence putative gRNA set. For each mature edited mRNA we manually generated gRNA cascade models, i.e. the putative pattern of gRNA usage from 3′ to 5′. Putative gRNAs that were the longest and scored highest in alignment by our algorithm were placed in the cascades first, followed by slightly lower scoring and shorter putative gRNAs. The gRNA cascade model of the A6 transcript is shown in Fig. 3 and models for COI, CYb, and RPS12 are shown in Fig. S3.
Fig. 3.
A redundant gRNA cascade model can be assembled for the edited regions of A6. Lines connecting gRNA to edited RNA bases indicate canonical pairing, ‘:’ indicates a G:U pair, and ‘#’ a mismatch. Red-orange ‘T’s in the DNA are those deleted to generate the edited product. Red-orange ‘U’s in the edited RNA indicate those inserted by editing. Dashes within DNA and RNA sequences are present to facilitate spacing for alignment of the DNA and RNA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
There seems to be substantially redundant gRNA coverage for the edited mRNAs of T. borreli, since editing of most regions can be properly guided by two or more overlapping, albeit different gRNAs. Further, during gRNA cascade assembly, once an RNA region was already well covered, we decided to ignore additional alignments with lower scoring putative gRNAs for Figs. 3 and S3. While gRNA redundancy is evident in other species [27], the degree of redundancy is very high in T. borreli. For instance, 18 gRNAs are involved in L. pyrrhocoris RPS12 editing, with redundancy of coverage for each nucleotide reaching 3 × to 4 × in some regions by visual inspection [27]. In comparison, utilizing all T. borreli high-confidence putative gRNAs, 237 and 110 gRNAs align with edited A6 and RPS12 mRNAs, respectively, resulting in 7 × to 12 × redundant coverage across the genes by visual inspection (note that the L. pyrrhocoris gRNAs are typically longer than those of T. borreli, thus each pairing covers more of the mRNA).
To quantify this phenomenon, the sum of lengths for all gRNAs aligning to the edited areas of each mRNA was divided by the edited region length. This ‘redundancy score‘ should increase as coverage redundancy increases. The T. borreli gRNA redundancy scores for A6 and RPS12 were 9.7 and for the edited sections of CO1 and CYb they were 10.1. In contrast, the average T.brucei gRNA redundancy score for extensively edited mRNAs was collectively 8.7 when the gRNA set from [66] was used. Not surprisingly, the scores using L. tarentolae gRNAs [64] and L. pyrrhocoris gRNAs [27], is 2.9 and 3.6, respectively, for the extensively edited mRNAs. However, this measure is limited in its ability to convey a true picture of the qualities of gRNA coverage redundancy, as the degree of gRNA:mRNA alignment redundancy varies greatly across an mRNA.
We wanted to verify that the observed T. borreli gRNA redundancy was not due to an overly permissive gRNA identification scheme. To test this, for published minicircle datasets in [64] (L. tarentolae), [52], [66] (T. brucei), and [27] (L. pyrrhocoris), initial gRNA sets were obtained using the same strict and relaxed settings applied to the T. borreli ScaI-flanked contigs (for the ‘find_grna’ tool), followed by filtering based on the appropriate species-specific motif. Our methodology produced gRNA datasets for these organisms (all with small RNA-validated gRNA sets) that aligned to tested mRNAs with a coverage redundancy like that which was previously determined (Fig. S4 shows T. brucei and L. tarentolae findings).
Many gRNAs that are part of T. borreli editing cascades have single nucleotide mismatches to their mRNAs. Mismatch regions in gRNA:mRNA alignments with no redundant gRNA possessing the proper match at that site appear in all three well-characterized alignments of kinetoplastid mitochondrial edited mRNAs. Therefore, it is our assumption that the secondary structure of the RNA portion of the editing site tolerates the existing mismatches. All putative gRNAs in the cascades should be considered equally likely to guide editing at any one site, in the absence of any other evidence. However, as suggested previously [27], the degree of tolerance for mismatches appears to be species specific.
In a characterization of L. pyrrhocoris gRNAs [27], nearly all gRNAs identified in the analysis of assembled minicircles were validated by small RNA-seq. We demonstrated that by using the full repertoire of identified gRNAs, we could explain by mRNA:gRNA pairings the directing of ∼80–85 % of total editing events observed in the transcriptome (pairings include numbers of Us inserted or deleted that contribute to a canonically edited sequence and mRNA non-canonical insertions and deletions). We likewise performed this analysis with the T. borreli high-confidence gRNAs set, finding that 90 % and 87 % of editing events observed among RPS12 and A6 cryptogenes reads, respectively, can be explained using these gRNAs. Such high percentages implicitly suggest that we identified a nearly complete gRNA repertoire for T. borreli.
There is one caveat regarding the high gRNA coverage redundancy in T. borreli putative gRNA editing cascade models. For editing to proceed with its currently accepted processive mechanism, a “leaving” gRNA must have directed editing of a sufficient length of mRNA to serve as a platform for the anchor region binding of the subsequent gRNA. In some positions within edited domains, such as the g25/g26 gRNA binding regions of A6 (Fig. 3), the putative leaving mRNA does not sufficiently overlap with the subsequent upstream gRNA to allow for a platform for its anchor region. A lower scoring gRNAs not included in the cascades may serve to direct editing in these gaps. It is also possible that some gRNAs may be encoded on the maxicircle, or on mitochondrial DNA molecules that lack ScaI-containing repeats. Our search would not detect these gRNAs. However, as the minimum gRNA anchor region length for T. borreli is unknown, it did not seem useful to speculate further whether we had identified all its necessary gRNAs for editing. Rather, we conclude that the high-confidence putative gRNA set appears to provide a higher degree of guiding redundancy than those of better-studied kinetoplastids.
Our putative gRNA analysis bolstered previous findings on the origins and evolution of gRNAs and their utilization in RNA editing. The origins of the gRNA sequences and their utilization in editing is a fascinating mystery. One hypothesis is that gRNAs evolved from duplicated and “repackaged” elements of ancient, correctly encoded precursors of the current cryptogenes. For various reasons, that hypothesis is questioned [18]. We also note a lack of evidence of upstream or downstream “parent mRNA” anywhere near putative gRNA loci on the ScaI-flanked contigs that might be expected from the myriad duplication events that would be required to result in the current gRNA loci in its seemingly random arrangement. This finding is shared in all examined maxicircle populations to date. We also investigated the associations between gRNA position and their respective loci on ScaI-flanked contigs in the cascade model. There was no strong overall correlation, but other patterns were observed that warrant further study. We developed linkage plot diagrams to illustrate this (an example is shown in Fig. 4). The presented scheme connects the randomly selected gRNA loci of ScaI-flanked contigs 2 and 12 to gRNAs in the cascade models of all four cryptogenes. Firstly, we noticed a frequent inclusion of putative gRNAs of the same locus in multiple gRNA models, suggesting that a single gRNA is capable of directing correct editing events in different cryptogenes (Fig. 4; red lines). Among 420 high-confidence putative gRNAs, we found 265 molecules (63 % of all identified gRNAs) likely participating in single-locus editing, 124 (30 %) that could potentially participate in editing of two loci, and 26 (6 %) that could direct editing of three loci. Conversely, the scheme illustrates that editing of a given mRNA position can be directed by any of several gRNAs encoded on different ScaI-flanked contigs, resulting in a redundant coverage (Fig. 4; dark violet lines). Finally, a few putative gRNAs capable of canonically directing editing of a single mRNA (Fig. 4; dark gold line) also align with regions of mRNAs that are not normally edited (Fig. 4; pale gold line). There is growing evidence that minicircles are constantly transcribed, often across their full length [64]. This finding in T. borreli of apparent shared, multi-locus, accidental or evolving gRNA use demonstrates how probable it would be for unrelated sequence to “fix” into a functional gRNA sequence once the transcription of a particular sequence, originally unrelated to mitochondrial mRNAs, confers advantage.
Fig. 4.
A linkage plot diagram reveals the inherent potential for flexibility of gRNA populations in directing editing. Randomly-selected ScaI-flanked contigs 2 and 12 and the four T. borreli edited mRNAs were plotted to show alignments between the mRNAs and putative gRNA loci on the contigs. Alignments are plotted in grey. An example alignment where one position on the contig mapped to multiple edited mRNA loci is shown in dark violet. An example where several positions on the two contigs map to a single edited site on an mRNA is shown in red. Gold linkages indicate a single contig locus that aligns with a region of edited mRNA (A6, dark gold), and a region on another mRNA that is not edited in the canonical, translatable product (CYb, light gold). The mRNA sequences on the scheme are shown proportionally 500-fold larger than those of the contigs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. T. borreli transcriptome reveals extensive species-specific differences in U-indel editing
Our reconstructions of the four fully edited ORFs allowed us to assess similarities and differences in the quantitative parameters of the U-indel editing. A prominent feature of this process is the apparently stochastic nature of editing, reflected by the high frequency of mRNAs with a sequence incompatible with a single “canonically” edited sequence [67]. Once the sequence of a fully edited and translatable mRNA is determined, it is possible to compare all high throughput sequencing reads with both edited and pre-edited transcripts derived from a kDNA maxicircle. Many reads contain editing events such as Us inserted or deleted at sites other than the standard positions, or they contain an incompatible number of inserted and/or deleted Us. In T. brucei, PCR-based whole-mRNA sequencing approaches revealed that such “non-canonical“ editing events are typically located in the transcript region that is being actively edited at the time of collection [61], [68]. However, the degree to which this is the case in other kinetoplastid species is poorly understood. Previously, we developed a way to visualize editing events captured in sequencing reads in a matrix format [27], [28]. We generated the same matrices for the four extensively edited transcripts of T. borreli (Fig. S5). By and large, the overall editing picture is the same as observed in the dot matrix plots of T. cruzi and L. pyrrhocoris [27], [41]. In the edited regions, editing also occasionally occurs in positions where it disrupts the ORF (examples indicated by red arrowheads in Fig. S5). Interestingly, these dot matrices reveal off-target editing events in positions that are also shown to be covered by gRNAs annealing to multiple mRNAs in linkage plot diagrams (e.g., Fig. 4). A few examples of likely off-target editing events in regions not normally edited are indicated by blue boxes in Fig. S5, although many more are also evident within these dot matrixes. These editing states are detected in just a few reads. Clearly, the T. borreli U-indel RNA editing mechanism generates both canonical and non-canonical editing events.
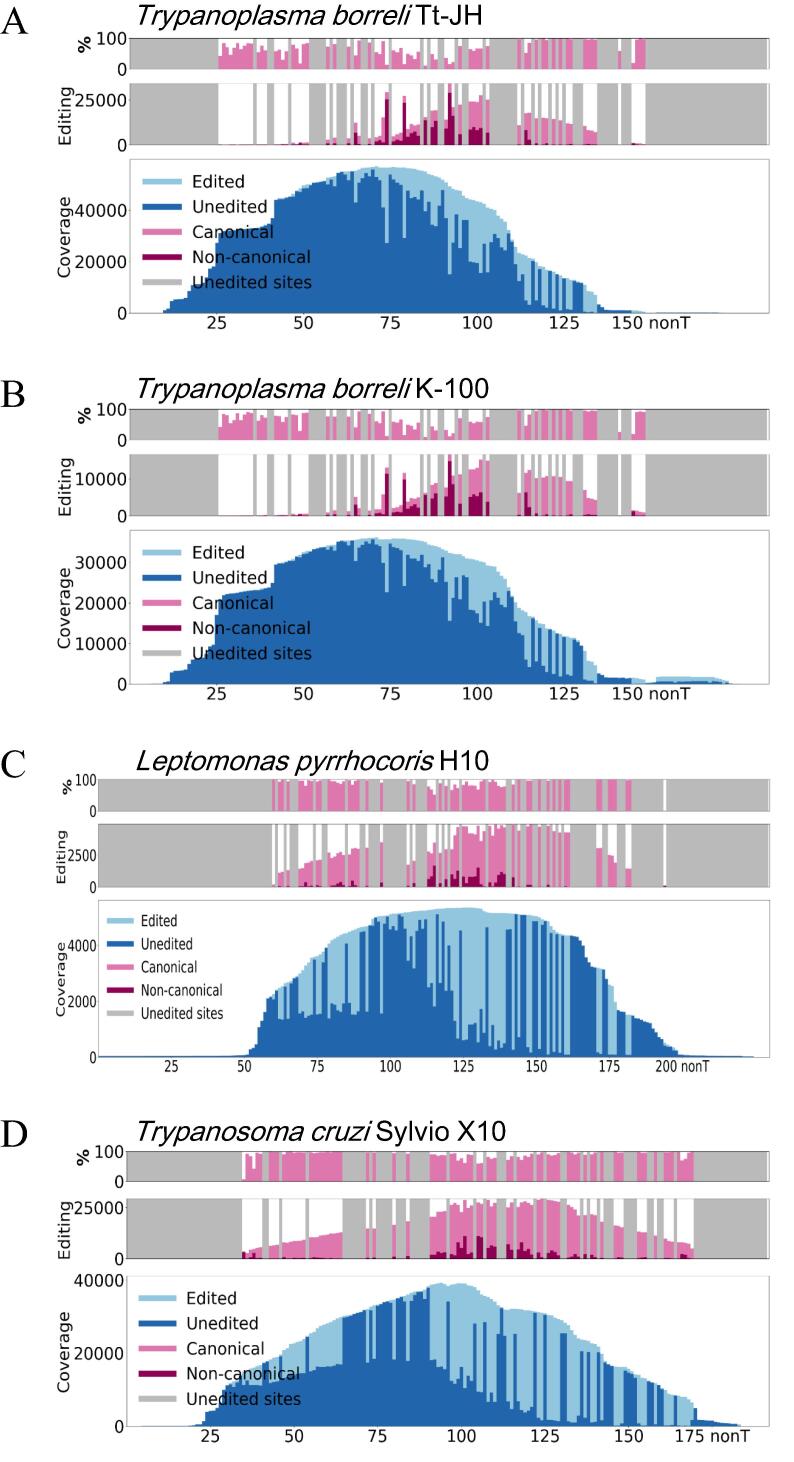
We initially hypothesized that since the number and length of edited domains in T. borreli is lower as compared with trypanosomatids investigated in this respect, editing will be more straightforward, resulting in fewer non-canonical editing events. However, our finding of a rich and complex gRNA repertoire in this fish parasite (Fig. 3, Fig. 4 and S3; Table S1) suggests that this view is overly simplistic. To measure the relative proportions of canonical and non-canonical editing events in sequence read populations at each potential editing site, we have previously developed a dedicated bioinformatics tool, which allowed us to compare “productive editing” in L. pyrrhocoris and T. cruzi and determine that the degree of non-canonical editing events is higher in T. cruzi, where it moreover varies significantly among its strains [41]. At the time, we speculated that a higher incidence of non-canonical editing events may reflect that Trypanosoma spp. have a higher proportion of maxicircle cryptogenes relative to L. pyrrhocoris. However, the productive editing plots of the T. borreli cryptogenes do not support this explanation, as exemplified by the RPS12 productive editing plots for two available strains of T. borreli (Fig. 5A-B). A decrease in the ratio of canonical to non-canonical editing events is observed particularly at the sites in the center of RPS12 mRNA, a situation reminiscent to that described previously in L. pyrrhocoris and T. cruzi [41]. However, the ratio of non-canonical editing events is even higher in T. borreli RPS12 than that in T. cruzi and L. pyrrhocoris RPS12, with their more complex edited transcriptomes (Fig. 5C-D). We also documented frequent non-canonical editing events in the edited domains of the other three T. borreli cryptogenes, particularly at specific sites (Fig. S6).
Fig. 5.
Canonical and noncanonical editing events in T. borreli show similarities and differences to patterns in other species. A-B. Presented is editing of RPS12 in two T. borreli strains, C. L. pyrrhocoris, and D. T. cruzi strain Sylvio at every non “T” position from 5′ to 3′ along the X axis. For each species/strain, the bottom plot depicts read coverage across the transcript, with the portion of reads edited at each location along the transcript shown in a lighter blue tone on top of the non-edited reads shown in darker blue. The middle plot is a bar plot with an X axis consistent with the bottom coverage plot. At each editing site the bar distinguishes the total number of reads in which an editing event at that site is observed and breaks down the number by canonical and noncanonical editing events. A, C, and G positions along the transcript that are not locations of editing for canonical edited sequence are blocked out in grey and not analyzed. The top plot shows a similar bar graph, but the Y axis represents percentage of editing that is canonical rather than absolute numbers of reads possessing editing at each site. C and D are reproduced from [41] for comparison purposes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Two other relatively unexplored editing parameters are the degree to which sequence conservation at the protein level is imparted through the U-indel editing mechanism, and the relative degree to which the same editing consists of insertion versus deletion events. Hence, it remains to be established whether or not this ratio is conserved across kinetoplastid species. The edited RPS12 and A6 mRNAs are convenient models with which to approach these questions. Four conserved regions of 8 to 12 amino acids interspersed with much more variable sequences of similar lengths can be found in multiple sequence alignments of predicted kinetoplastid RPS12 proteins. A similar pattern, albeit less pronounced, occurs in the protein product of A6. Indeed, alignments for both proteins rely heavily on these conserved domains. Fig. 6 shows portions of multiple alignments of selected RPS12 and A6 conserved regions from T. borreli and two distantly related trypanosomatids, and their corresponding DNA and edited mRNA sequences. Interestingly, T. borreli and (to a lesser extent) T. cruzi tend to use deletions to enforce the maintenance of conserved regions within RPS12, while L. pyrrhocoris rarely utilizes deletion editing. For the maintenance of conserved regions and length of the neighboring divergent regions of A6, T. borreli again seems to capitalize primarily on the deletion mechanism. The tendency of these flagellates to utilize the full capacity of U-indel editing in different ways in these regions suggests that kinetoplastids have become highly dependent on the editing mechanism to sustain amino acid sequence conservation of their mitochondrial genes.
Fig. 6.
Portions of RPS12 (top) and A6 (bottom) sequence alignments showing how deletion editing is differentially utilized between T. borreli and two other species in several specific regions of conserved amino acid sequence of the translated product. DNA, RNA, and amino acid (PEP) sequences are shown. Deleted ‘T’s in the DNA are in red–orange, inserted ‘u’s in the RNA are displayed in blue. In-sequence dashes are used for spacing for alignment. Tbor, T. borreli; Tcru, T. cruzi; Lpyr, L. pyrrhocoris. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Noting the pronounced usage of U deletions by T. borreli, we asked whether this phenomenon happens to be a feature of the selected regions, or whether the differences in U-insertion/deletion ratio are species-specific and consistent across entire transcriptomes. Hence, we explored this by two metrics. Firstly, we calculated the observed insertion/deletion event ratios for all reads mapped on the maxicircle, regardless of whether they were consistent with the final mature transcript from which they originated. These ratios were 2.3 for T. borreli, 3.7 for T. cruzi, and 4.2 for L. pyrrhocoris. The fact that they are all greater than 1 corroborates the general notion that insertion is the predominant form of U-indel editing. Secondly, we examined these same ratios within the assembled repertoire of mature edited sequences and found them to be 3.3 for T. borreli, 5.1 for T. cruzi, and 9.1 for L. pyrrhocoris. Two trends emerge from these metrics. Firstly, in all examined species the per-read ratios are lower than per-mRNA ratios. This means that the deletion events are less likely than insertion events to be incorporated into a translatable sequence. Conversely, we find them more frequently in the population of events categorized as non-canonical (Fig. 5). Secondly, the degree to which deletion and insertion are utilized by U-indel editing is a flexible parameter that is species-specific. Overall, we have identified several quantitative mechanism-linked parameters of editing that differ depending on the species examined. These parameters may be a suitable focus of future studies incorporating many more species to trace the evolution of U-indel editing across kinetoplastid protists.
4. Conclusions
Provocative early findings of coding and non-coding transcriptomes of the iconic T. borreli were long overdue for a follow-up that makes use of advanced sequencing and computational tools. Our characterization of the kDNA maxicircle specifies its length to be 42 kb, which is slightly larger than an early estimate of 37 kb [11], but substantially smaller than the other one [12]. However, we note that the maxicircle size reported in this and another previous study [11] utilized DNA from the strain Tt-JH isolated from a tench, whereas the 80 kb maxicircle-size estimate was based on the strain Tg-JH from a leech vector [12]. As the T. borreli maxicircle has repeat superclusters amenable to duplication, we cannot rule out that both early estimations of maxicircle size were accurate, since strain differences and decades in culture may play a significant role, as was shown in other kinetoplastids [18], [69].
The early finding of an unusually large molecule carrying gRNAs in T. borreli is partially supported by our findings. Although significantly smaller than the original estimates of up to 200 kb, the gRNA-containing contigs of ∼70 kb (Table 1) documented here are substantially larger than trypanosomatid minicircles, the size of which is usually around 1 kb and never exceeds 10 kb [53], [70]. However, we could not establish, even with exceptionally high read coverage, whether these ScaI-flanked contigs are linear or circular. Earlier claims of their circularity were based on ambiguous electrophoretic mobility experiments and on electron microscopy, with the latter described but not presented [12]. Naturally, linear chromosomes would require a very different replication mechanism compared to the circular molecules observed in other kinetoplastids. The replication mechanism of kDNA of trypanosomatids is extremely complex, with at least 6 DNA polymerases functionally implicated in T. brucei [16]. Due to the extreme amount of mitochondrial DNA in Trypanoplasma, exceeding by far that of the largest trypanosomatid kinetoplasts [22], we assume that replication machinery of linear gRNA-containing molecules would be comparably or even more complex, and would possibly be very different from that in Trypanosomatidae. However, too much experimental data is missing to speculate further.
Characterization of the T. borreli kDNA presented herein further supports the view of this organellar DNA being highly varied among kinetoplastids, both in sheer amount, structure, composition, and extent of RNA editing. Moreover, novel maxicircle features have been found, such as the presence of inverted repeats in its coding regions that lack a fixed pattern, and supercluster repeats with dodecameric and octameric structure. Similarly, the ScaI-flanked contigs with their telomeric-like repeats and the number of gRNA genes they carry are also a novel trait rather than just a variation of gRNA-carrying molecules. However, other major features remain conserved, namely the gRNA-containing molecules contain CSB3 and are present as a fluid repertoire only partially shared between strains, and conserved sequence motifs are often located a fixed distance from putative gRNA genes.
Our analysis of maxicircle transcription is based on a single sequence library, but this seems to be sufficient for characterizing the translatable products of all the T. borreli cryptogenes, especially when compared with the limited libraries composed of individual clones used to define fully edited mRNAs of T. brucei and L. tarentolae prior to the advent of high-throughput sequencing [71], [72], [73]. Our data further confirmed earlier observations from several trypanosomatid species that the maxicircle loci of highest abundance are those with products requiring editing [27], [41]. Consequently, we postulate that all organisms possessing U-indel editing have adjusted overall mRNA abundances to compensate for the extensive and apparently widespread inefficiency of the editing machinery in achieving translatable product from pre-edited transcript.
The T. borreli maxicircle genome and transcriptome analysis also confirmed the lack of expression of sequences resembling subunits of the NADH:ubiquinone oxidoreductase (mitochondrial respiratory complex I) that are normally found in most eukaryotic mitochondrial genomes. This finding is notable, as the related trypanosomes typically have at least eight complex I subunits encoded in their maxicircle. In addition to these mitochondrion-encoded subunits, trypanosomes also encode 4 core complex I subunits, and identified homologues of approximately half (∼15) of the mammalian complex I accessory factors in their nuclear genome [74]. In T. borreli none of the core nuclear-encoded subunits and only one accessory factor homologue can be identified by standard bioinformatics methods. This accessory subunit, acyl carrier protein (ACP, encoded by Tb927.3.860 in T. brucei) is also known to be a mitoribosome assembly factor [75], [76], and this role is likely what explains its presence in the T. borreli genome. We document here that T. borreli entirely lacks complex I of its mitochondrial respiratory chain entirely. The role and importance of the complex in kinetoplastids is currently uncertain [74] and its absence in T. borreli may shed light on these evolving questions.
An obvious limitation of this study is the lack of small RNA sequencing to confirm our gRNA discovery-by-alignment findings. However, several factors suggest that these results may be strong standing alone. Firstly, the five previously sequenced T. borreli gRNAs [37] fit very well with the gRNA length, G:U use, and mismatch parameter distributions obtained utilizing our alignment algorithm. Secondly, when we ran our ‘grnafind’ tool on the available L. tarentolae dataset [64] using settings similar to those applied for T. borreli, the algorithm properly recovered the set of well-annotated L. tarentolae genes and known L. tarentolae gRNA parameters with minimal noise. Thus, even with relaxed search settings the algorithm appears to be reliable. Thirdly, as in L. pyrrhocoris and L. tarentolae, T. borreli gRNAs can be partitioned into those proximal to a strongly conserved sequence motif and those proximal to poorly conserved motifs or possessing no motif at all. The experimentally confirmed gRNAs of L. pyrrhocoris and L. tarentolae are proximal to highly conserved motifs [27], [64] and, indeed, all editing cascades described herein (Figs. 3 and S4) utilize only gRNAs proximal to highly conserved motifs. Yet even following the exclusion of putative gRNAs without well-conserved proximal motifs, the level of redundancy across edited mRNAs remains particularly extensive in T. borreli. It is plausible that having more gRNA loci, allowed by the huge amount of kDNA in this flagellate [22], represents an evolutionary force driving similarly high ratios of the non-canonical to canonical editing events. However, sequencing the T. borreli gRNA population in the future would still be valuable: T. borreli gRNAs were reported to uniquely possess non-encoded oligo(U) sequence on both their 5′ and 3′ termini [37]. As gRNAs of other species invariably carry only non-encoded 3′ oligo(U) extensions [77], exploring this difference could lead to insights regarding gRNA processing.
Our final important finding relates to the propensity of kinetoplastids to utilize the U-indel editing in its insertion rather than deletion mode. This parameter of editing is directly linked to the mechanism by which it is executed, as these different enzymatic processes are executed by only partially overlapping catalytic complexes [59], [78], [79], [80]. It is worth mentioning that our present analysis of differing ratios for insertions relative to deletions is far from perfect, the main reasons behind that being the narrow across-species analysis and the limited and hard-to-normalize confidence attributable to any particular insertion or deletion event. Attribution of confidence in editing events is largely due to differences in read coverage. For each transcript, coverage irregularity potentially skews the deletion events more than insertion events, as there are fewer of them. Still, our analysis convincingly demonstrated the high rate of U-deletions in T. borreli.
An initial motivation for this work was to determine protein-coding genes, gRNAs, and the editing patterns in the highly dispersed and consequently less organized kDNA of T. borreli. The documented linear gRNA-carrying DNA molecules are consistent with the diffuse kDNA structure observed by electron microscopy [22]. Moreover, the high redundancy of gRNAs, a relatively small number of sequences requiring editing, a very high fraction of non-canonical editing events, and an enhanced use of the U-deletion mechanism suggest that editing may be less “controlled” or less “efficient” in this early-branching bodonid than in the extensively studied, likely more derived, trypanosomatids [41], [61], [63], [68].
CRediT authorship contribution statement
Evgeny S. Gerasimov: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization, Funding acquisition. Dmitry A. Afonin: Formal analysis, Investigation, Data curation, Writing – review & editing, Visualization. Oksana A. Korzhavina: Formal analysis, Investigation, Data curation, Writing – review & editing, Visualization. Julius Lukeš: Validation, Writing – review & editing, Funding acquisition. Ross Low: Investigation, Data curation. Neil Hall: Resources, Writing – review & editing, Supervision. Kevin Tyler: Resources, Writing – review & editing. Vyacheslav Yurchenko: Conceptualization, Data curation, Writing – review & editing, Visualization, Funding acquisition, Project administration. Sara L. Zimmer: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by the Grant Agency of Czech Republic (GAČR 22-01026S) to V.Y. and J.L. and the Russian Science Foundation (RSF 19-74-10008) to E.G., D.A., and O.K. Next-generation sequencing and library construction were done via the BBSRC National Capability in Genomics and Single Cell (BB/CCG1720/1) at the Earlham Institute, by members of the Genomics Pipelines Group. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.11.023.
Contributor Information
Julius Lukeš, Email: jula@paru.cas.cz.
Ross Low, Email: Ross.Low@earlham.ac.uk.
Neil Hall, Email: neil.hall@earlham.ac.uk.
Kevin Tyler, Email: K.Tyler@uea.ac.uk.
Vyacheslav Yurchenko, Email: vyacheslav.yurchenko@osu.cz.
Sara L. Zimmer, Email: szimmer3@d.umn.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Maslov D.A., Opperdoes F.R., Kostygov A.Y., Hashimi H., Lukeš J., Yurchenko V. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology. 2019;146(1):1–27. doi: 10.1017/S0031182018000951. [DOI] [PubMed] [Google Scholar]
- 2.Kostygov A.Y., Karnkowska A., Votýpka J., Tashyreva D., Maciszewski K., Yurchenko V., et al. Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses. Open Biol. 2021;11 doi: 10.1098/rsob.200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukeš J., Butenko A., Hashimi H., Maslov D.A., Votýpka J., Yurchenko V. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol. 2018;34(6):466–480. doi: 10.1016/j.pt.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Stuart K., Brun R., Croft S., Fairlamb A., Gürtler R.E., McKerrow J., et al. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest. 2008;118(4):1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lom J. In: Biology of the trypanosomes and trypanoplasms of fish. Lumsden W.H.R., Evans D.A., editors. Academic Press; London: 1979. Biology of the trypanosomes and trypanoplasms of fish; pp. 269–337. [Google Scholar]
- 6.Losev A., Grybchuk-Ieremenko A., Kostygov A.Y., Lukeš J., Yurchenko V. Host specificity, pathogenicity, and mixed infections of trypanoplasms from freshwater fishes. Parasitol Res. 2015;114(3):1071–1078. doi: 10.1007/s00436-014-4277-y. [DOI] [PubMed] [Google Scholar]
- 7.Saeij J.P., de Vries B.J., Wiegertjes G.F. The immune response of carp to Trypanoplasma borreli: kinetics of immune gene expression and polyclonal lymphocyte activation. Dev Comp Immunol. 2003;27(10):859–874. doi: 10.1016/s0145-305x(03)00083-1. [DOI] [PubMed] [Google Scholar]
- 8.Lukeš J., Kaur B., Speijer D. RNA editing in mitochondria and plastids: weird and widespread. Trends Genet. 2021;37(2):99–102. doi: 10.1016/j.tig.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Benne R., Van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 10.Gray M.W. Evolutionary origin of RNA editing. Biochemistry. 2012;51(26):5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- 11.Lukeš J., Arts G.J., van den Burg J., de Haan A., Opperdoes F., Sloof P., et al. Novel pattern of editing regions in mitochondrial transcripts of the cryptobiid Trypanoplasma borreli. EMBO J. 1994;13(21):5086–5098. doi: 10.1002/j.1460-2075.1994.tb06838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslov D.A., Simpson L. RNA editing and mitochondrial genomic organization in the cryptobiid kinetoplastid protozoan Trypanoplasma borreli. Mol Cell Biol. 1994;14(12):8174–8182. doi: 10.1128/mcb.14.12.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslov D.A., Avila H.A., Lake J.A., Simpson L. Evolution of RNA editing in kinetoplastid protozoa. Nature. 1994;368(6469):345–348. doi: 10.1038/368345a0. [DOI] [PubMed] [Google Scholar]
- 14.Carrington M., Doro E., Forlenza M., Wiegertjes G.F., Kelly S. Transcriptome sequence of the bloodstream form of Trypanoplasma borreli, a hematozoic parasite of fish transmitted by leeches. Genome Announc. 2017;5(9):e01712–e1806. doi: 10.1128/genomeA.01712-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Záhonová K., Lax G., Leonard G., Sinha S., Richards T., Lukeš J., et al. Single-cell genomics unveils a canonical origin of the diverse mitochondrial genomes of euglenozoan. BMC Biol. 2021;19:103. doi: 10.1186/s12915-021-01035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen R.E., Englund P.T. Network news: the replication of kinetoplast DNA. Annu Rev Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 17.Stuart K., Panigrahi A.K. RNA editing: complexity and complications. Mol Microbiol. 2002;45(3):591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- 18.Simpson L., Thiemann O.H., Savill N.J., Alfonzo J.D., Maslov D.A. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci U S A. 2000;97(13):6986–6993. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4(6):623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 20.Lukeš J., Jirků M., Avliyakulov N., Benada O. Pankinetoplast DNA structure in a primitive bodonid flagellate, Cryptobia helicis. EMBO J. 1998;17(3):838–846. doi: 10.1093/emboj/17.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukeš J., Guilbride D.L., Votýpka J., Zíková A., Benne R., Englund P.T. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 2002;1(4):495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukeš J., Wheeler R., Jírsová D., David V., Archibald J.M. Massive mitochondrial DNA content in diplonemid and kinetoplastid protists. IUBMB Life. 2018;70(12):1267–1274. doi: 10.1002/iub.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poinar G., Jr. Early Cretaceous trypanosomatids associated with fossil sand fly larvae in Burmese amber. Mem Inst Oswaldo Cruz. 2007;102(5):635–637. doi: 10.1590/s0074-02762007005000070. [DOI] [PubMed] [Google Scholar]
- 24.Yurchenko V., Lukeš J., Xu X., Maslov D.A. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae) J Eukaryot Microbiol. 2006;53(2):103–111. doi: 10.1111/j.1550-7408.2005.00078.x. [DOI] [PubMed] [Google Scholar]
- 25.Pecková H., Lom J. Growth, morphology and division of flagellates of the genus Trypanoplasma (Protozoa, Kinetoplastida) in vitro. Parasitol Res. 1990;76(7):553–558. doi: 10.1007/BF00932559. [DOI] [PubMed] [Google Scholar]
- 26.Katz K, Shutov O, Lapoint R, Kimelman M, Brister JR, O'Sullivan C (2022) The Sequence Read Archive: a decade more of explosive growth. Nucleic Acids Res 50(D1): D387-D390. [DOI] [PMC free article] [PubMed]
- 27.Gerasimov E.S., Gasparyan A.A., Afonin D.A., Zimmer S.L., Kraeva N., Lukeš J., et al. Complete minicircle genome of Leptomonas pyrrhocoris reveals sources of its non-canonical mitochondrial RNA editing events. Nucl Acids Res. 2021;49(6):3354–3370. doi: 10.1093/nar/gkab114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerasimov E.S., Gasparyan A.A., Kaurov I., Tichý B., Logacheva M.D., Kolesnikov A.A., et al. Trypanosomatid mitochondrial RNA editing: dramatically complex transcript repertoires revealed with a dedicated mapping tool. Nucl Acids Res. 2018;46(2):765–781. doi: 10.1093/nar/gkx1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S (2019) FastQC: a quality control tool for high throughput sequence data. Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 2022 August 27.
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30(5):614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 33.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37(23):4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerasimov E.S., Zamyatnina K.A., Matveeva N.S., Rudenskaya Y.A., Kraeva N., Kolesnikov A.A., et al. Common structural patterns in the maxicircle divergent region of Trypanosomatidae. Pathogens. 2020;9(2):100. doi: 10.3390/pathogens9020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuhira S., Simpson L. Guide RNAs and guide RNA genes in the cryptobiid kinetoplastid protozoan, Trypanoplasma borreli. RNA. 1996;2(11):1153–1160. [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez-Gonzalez R.H., Bonnal R., Caccamo M., Maclean D. Bio-SAMtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol Med. 2012;7(1):6. doi: 10.1186/1751-0473-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan AR (2014) BEDTools: the swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1-11.12.34. [DOI] [PMC free article] [PubMed]
- 41.Gerasimov E.S., Ramirez-Barrios R., Yurchenko V., Zimmer S.L. Trypanosoma cruzi strain and starvation-driven mitochondrial RNA editing and transcriptome variability. RNA. 2022;28(7):993–1012. doi: 10.1261/rna.079088.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R.H., Lai D.H., Zheng L.L., Wu J., Lukes J., Hide G., et al. Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen. Parasit Vectors. 2015;8:665. doi: 10.1186/s13071-015-1281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greif G., Rodriguez M., Reyna-Bello A., Robello C., Alvarez-Valin F. Kinetoplast adaptations in American strains from Trypanosoma vivax. Mutat Res. 2015;773:69–82. doi: 10.1016/j.mrfmmm.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Callejas-Hernández F., Herreros-Cabello A., Del Moral-Salmoral J., Fresno M., Gironès N. The complete mitochondrial DNA of Trypanosoma cruzi: maxicircles and minicircles. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.672448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufer A., Stark D., Ellis J. Evolutionary insight into the Trypanosomatidae using alignment-free phylogenomics of the kinetoplast. Pathogens. 2019;8(3):157. doi: 10.3390/pathogens8030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kay C., Williams T.A., Gibson W. Mitochondrial DNAs provide insight into trypanosome phylogeny and molecular evolution. BMC Evol Biol. 2020;20(1):161. doi: 10.1186/s12862-020-01701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yurchenko V., Kolesnikov A.A. Minicircular kinetoplast DNA of Trypanosomatidae. Mol Biol (Mosk) 2001;35(1):1–10. [PubMed] [Google Scholar]
- 48.Simpson L. The genomic organization of guide RNA genes in kinetoplastid protozoa: several conundrums and their solutions. Mol Biochem Parasitol. 1997;86(2):133–141. doi: 10.1016/s0166-6851(97)00037-6. [DOI] [PubMed] [Google Scholar]
- 49.Ray D.S. Conserved sequence blocks in kinetoplast minicircles from diverse species of trypanosomes. Mol Cell Biol. 1989;9(3):1365–1367. doi: 10.1128/mcb.9.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yurchenko V., Kolesnikov A.A., Lukeš J. Phylogenetic analysis of Trypanosomatina (Protozoa: Kinetoplastida) based on minicircle conserved regions. Folia Parasitol. 2000;47(1):1–5. doi: 10.14411/fp.2000.001. [DOI] [PubMed] [Google Scholar]
- 51.Camacho E., Rastrojo A., Sanchiz A., Gonzalez-de la Fuente S., Aguado B., Requena J.M. Leishmania mitochondrial genomes: maxicircle structure and heterogeneity of minicircles. Genes. 2019;10(10):758. doi: 10.3390/genes10100758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper S., Wadsworth E.S., Ochsenreiter T., Ivens A., Savill N.J., Schnaufer A. Assembly and annotation of the mitochondrial minicircle genome of a differentiation-competent strain of Trypanosoma brucei. Nucl Acids Res. 2019;47(21):11304–11325. doi: 10.1093/nar/gkz928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S.J., Zhang X., Lukeš J., Li B.Q., Wang J.F., Qu L.H., et al. Novel organization of mitochondrial minicircles and guide RNAs in the zoonotic pathogen Trypanosoma lewisi. Nucl Acids Res. 2020;48(17):9747–9761. doi: 10.1093/nar/gkaa700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.David V., Flegontov P., Gerasimov E., Tanifuji G., Hashimi H., Logacheva M.D., et al. Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, an endosymbiotic kinetoplastid. mBio. 2015;6(6):e01498–e10515. doi: 10.1128/mBio.01498-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blom D., de Haan A., van den Berg M., Sloof P., Jirků M., Lukeš J., et al. RNA editing in the free-living bodonid Bodo saltans. Nucl Acids Res. 1998;26(5):1205–1213. doi: 10.1093/nar/26.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolesnikov A.A., Merzlyak E.M., Bessolitsyna E.A., Fedyakov A.V., Schönian G. Reduction of the edited domain of the mitochondrial A6 gene for ATPase subunit 6 in Trypanosomatidae. Mol Biol (Mosk) 2003;37(4):637–642. [PubMed] [Google Scholar]
- 57.Gastineau R., Lemieux C., Turmel M., Davidovich N.A., Davidovich O.I., Mouget J.L., et al. Mitogenome sequence of a Black Sea isolate of the kinetoplastid Bodo saltans. Mitochondrial DNA B Resour. 2018;3(2):968–969. doi: 10.1080/23802359.2018.1507654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tikhonenkov D.V., Gawryluk R.M.R., Mylnikov A.P., Keeling P.J. First finding of free-living representatives of Prokinetoplastina and their nuclear and mitochondrial genomes. Sci Rep. 2021;11(1):2946. doi: 10.1038/s41598-021-82369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aphasizheva I., Alfonzo J., Carnes J., Cestari I., Cruz-Reyes J., Goringer H.U., et al. Lexis and grammar of mitochondrial RNA processing in trypanosomes. Trends Parasitol. 2020;36(4):337–355. doi: 10.1016/j.pt.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aphasizhev R., Aphasizheva I. Mitochondrial RNA editing in trypanosomes: small RNAs in control. Biochimie. 2014;100:125–131. doi: 10.1016/j.biochi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson R.M., Bruno A.E., Bard J.E., Buck M.J., Read L.K. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA. 2016;22(5):677–695. doi: 10.1261/rna.055160.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rusman F., Floridia-Yapur N., Tomasini N., Diosque P. Guide RNA repertoires in the main lineages of Trypanosoma cruzi: high diversity and variable redundancy among strains. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.663416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirby L.E., Koslowsky D. Cell-line specific RNA editing patterns in Trypanosoma brucei suggest a unique mechanism to generate protein variation in a system intolerant to genetic mutations. Nucl Acids Res. 2020;48(3):1479–1493. doi: 10.1093/nar/gkz1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simpson L., Douglass S.M., Lake J.A., Pellegrini M., Li F. Comparison of the mitochondrial genomes and steady state transcriptomes of two strains of the trypanosomatid parasite, Leishmania tarentolae. PLoS Negl Trop Dis. 2015;9(7):e0003841. doi: 10.1371/journal.pntd.0003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koslowsky D., Sun Y., Hindenach J., Theisen T., Lucas J. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucl Acids Res. 2014;42(3):1873–1886. doi: 10.1093/nar/gkt973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper S., Wadsworth E.S., Schnaufer A., Savill N.J. Organization of minicircle cassettes and guide RNA genes in Trypanosoma brucei. RNA. 2022;28(7):972–992. doi: 10.1261/rna.079022.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmer S.L., Simpson R.M., Read L.K. High throughput sequencing revolution reveals conserved fundamentals of U-indel editing. Wiley Interdiscip Rev RNA. 2018;9(5):e1487. doi: 10.1002/wrna.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tylec B.L., Simpson R.M., Kirby L.E., Chen R., Sun Y., Koslowsky D.J., et al. Intrinsic and regulated properties of minimally edited trypanosome mRNAs. Nucleic Acids Res. 2019;47(7):3640–3657. doi: 10.1093/nar/gkz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thiemann O.H., Maslov D.A., Simpson L. Disruption of RNA editing in Leishmania tarentolae by the loss of minicircle-encoded guide RNA genes. EMBO J. 1994;13(23):5689–5700. doi: 10.1002/j.1460-2075.1994.tb06907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yurchenko V., Hobza R., Benada O., Lukeš J. Trypanosoma avium: large minicircles in the kinetoplast DNA. Exp Parasitol. 1999;92(3):215–218. doi: 10.1006/expr.1999.4418. [DOI] [PubMed] [Google Scholar]
- 71.Maslov D.A., Sturm N.R., Niner B.M., Gruszynski E.S., Peris M., Simpson L. An intergenic G-rich region in Leishmania tarentolae kinetoplast maxicircle DNA is a pan-edited cryptogene encoding ribosomal protein S12. Mol Cell Biol. 1992;12(1):56–67. doi: 10.1128/mcb.12.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sturm N.R., Maslov D.A., Blum B., Simpson L. Generation of unexpected editing patterns in Leishmania tarentolae mitochondrial mRNAs: misediting produced by misguiding. Cell. 1992;70(3):469–476. doi: 10.1016/0092-8674(92)90171-8. [DOI] [PubMed] [Google Scholar]
- 73.Souza A.E., Myler P.J., Stuart K. Maxicircle CR1 transcripts of Trypanosoma brucei are edited and developmentally regulated and encode a putative iron-sulfur protein homologous to an NADH dehydrogenase subunit. Mol Cell Biol. 1992;12(5):2100–2107. doi: 10.1128/mcb.12.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duarte M., Tomás A.M. The mitochondrial complex I of trypanosomatids–an overview of current knowledge. J Bioenerg Biomembr. 2014;46(4):299–311. doi: 10.1007/s10863-014-9556-x. [DOI] [PubMed] [Google Scholar]
- 75.Saurer M., Ramrath D.J.F., Niemann M., Calderaro S., Prange C., Mattei S., et al. Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Science. 2019;365(6458):1144–1149. doi: 10.1126/science.aaw5570. [DOI] [PubMed] [Google Scholar]
- 76.Jaskolowski M., Ramrath D.J.F., Bieri P., Niemann M., Mattei S., Calderaro S., et al. Structural insights into the mechanism of mitoribosomal large subunit biogenesis. Mol Cell. 2020;79(4):629–644. doi: 10.1016/j.molcel.2020.06.030. [DOI] [PubMed] [Google Scholar]
- 77.Aphasizheva I., Aphasizhev R. Mitochondrial RNA quality control in trypanosomes. Wiley Interdiscip Rev RNA. 2021;12(3):e1638. doi: 10.1002/wrna.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28(1):122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simpson R.M., Bruno A.E., Chen R., Lott K., Tylec B.L., Bard J.E., et al. Trypanosome RNA Editing Mediator Complex proteins have distinct functions in gRNA utilization. Nucl Acids Res. 2017;45(13):7965–7983. doi: 10.1093/nar/gkx458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hashimi H., Zimmer S.L., Ammerman M.L., Read L.K., Lukeš J. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 2013;29(2):91–99. doi: 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.