Abstract
Acacia mangium is an important wood for commercial products especially pulp and medium-density fibreboard. However, it is susceptible to Ceratocystis fimbriata infection, leading to Ceratocystis wilt. Therefore, the present work aimed to (i) establish the diversity of endophytic fungi in different plant parts of A. mangium,and (ii) evaluate the antifungal potentials of the isolated and identified endophytic fungi against C. fimbriata. Endophytic fungal identification was conducted by PCR amplification and sequencing of the internal transcribed spacer 1 (ITS1) and ITS4 regions of nuclear ribosomal DNA. A total of 66 endophytic fungi were successfully isolated from different parts of A. mangium; leaf (21), stem (13), petiole (12), root (9), flower (6), and fruit (5). The endophytic fungal isolates belonged to Ascomycota (95.5%) and Zygomycota (4.5%). For Ascomycota 13 genera were identified: Trichoderma (28.6%), Nigrospora (28.6%), Pestalotiopsis (12.7%), Lasiodiplodia (9.5%), Aspergillus (6.3%), Sordariomycetes (3%), and Neopestalotiopsis, Pseudopestalotiopsis, Eutiarosporella, Curvularia, Fusarium, Penicillium, and Hypoxylon each with a single isolate. For Zygomycota, only Blakeslea sp. (5%) was isolated. Against C. fimbriata, Trichoderma koningiopsis (AC 1S) from stem, Nigrospora oryzae (AC 7L) from leaf, Nigrospora sphaerica (AC 3F) from the flower, Lasiodiplodia sp. (AC 2 U) from fruit, Nigrospora sphaerica (AC 4P) from petiole, and Trichoderma sp. (AC 9R) from root exhibited strong inhibition for C. fimbriata between 58.33 to 69.23%. Thus, it can be concluded that certain endophytic fungi of A. mangium have the potential to be harnessed as anti-Ceratocystis agent in future biotechnological applications.
Keywords: Acacia mangium, endophytic fungi, Ceratocystis fimbriata , Ceratocystis wilt, antagonism
Introduction
Acacia mangium Willd., a fast-growing and flowering leguminous tree native to Indonesia, Papua New Guinea, and Australia, has been introduced and cultivated into humid tropical lowland regions of Asia, South America, and Africa (Pinyopusarerk et al., 1993). In 1966, forest plantation of A. mangium began in Sabah, Malaysia, pioneered by D.I. Nicholson, an Australian forester. Commercial cultivation of A. mangium began in 1976 (Udarbe and Hepburn, 1986; Pinyopusarerk et al., 1993). The species was considered promising due to its stellar performance, superior growth, and multiple uses especially for pulp and medium-density fibreboard (Potter et al., 2006). Furthermore, pharmacological studies have also shown that the leaves of A. mangium exhibit antibacterial (Sarah Shafiei et al., 2017), antifungal (Mihara et al., 2005), antifilarial, and antihelmintic (Chaki et al., 2015) properties.
Despite its various commercial applications, A. mangium is susceptible to the infection of the ascomycetous pathogen, Ceratocystis fimbriata, which infects the wounds of A. mangium trees in plantations, and causes the Ceratocystis wilt disease (Kile, 1993; Roux and Wingfield, 2009; Tarigan et al., 2011; Brawner et al., 2015). Wounded tree caused by humans, other mammals including monkeys, elephants, squirrels or boring insects, and others factor such as wind, are likely to increase the disease spreading and tree mortality as the wound become the entrance for this Ceratocystis species to invade (Nasution et al., 2019). In Malaysia at the year of 2011, a severe case which was the first report of this disease infected approximately 40% of A. mangium trees in plantation at Tawau, Sabah. Later, this disease spreads to other regions on A. mangium plantation in Sabah such as Pitas, Kota Belud and Sipitang, where the incidence of this disease were about in range of 6–60% (Mandy and Wickneswari, 2014; Farid et al., 2018). Johor, Pahang and Sarawak were also reported faced the same disease problems to the A. mangium plantation in respective state. 50% out of 1,500 trees that were accessed in a 2-year-old Acacia mangium plantation in Johor have been infected by this disease (Farid et al., 2018). The main reason of this disease spreading and uncontrollable was due to lack of knowledge, researches and studies on how to overcome or prevent this disease to happen towards Acacia mangium trees (Lee, 2018).
There are no specific methods or guidelines established on how to handle this disease in Malaysia yet up to now. But there were several actions that commonly are used by the plantation managers to prevent the infection of this disease. As Ceratocystis species penetrate and invade the trees by wounds, this problems can be prevent by avoid the occurance of wound itself (Kile, 1993; Harrington, 2013; Nasution et al., 2019). Silviculture practice should be done in correct way and cautions. The timing of doing work for silviculture is also important to reduce the risk of disease development (Pilotti et al., 2016; Farid et al., 2018). Problems involved with wildlife in plantation areas also are count on in management such as establishment of wildlife management plan to overcome the conflicts occurred (Farid et al., 2018). Chemical control is one of application they used to delay the symptoms of the disease development and help the infected trees to live longer for at least 2 years (Blaedow, 2009; Nasution et al., 2019). Although the use of chemical fungicides are more preferred due to their rapid action, they are often associated with high production and application costs, human health hazards, restriction by domestic and international regulatory limits, trade bans, residual effects, environmental pollution, resistance development in pests, and potential elimination of beneficial natural enemies of the targeted pests (Yazid et al., 2020). Therefore, biological control is seen as a safer and cheaper alternative. Biological control is the use of living organisms (including microorganisms) to eliminate or reduce the density of pests / pathogens to safe levels (Wyckhuys et al., 2013). Often, indigenous organisms or microorganisms are utilised as biological control agent to minimise the risk of introducing foreign species that might grow uncontrollably and in turn become invasive. One such example of indigenous organisms or microorganisms is endophyte. The research is about using a microorganism (endophyte) to fight the pathogen (Ceratocystis fimbriata) which is one of biological control.
Like many other plant species, A. mangium is also associated with endophytes. Endophytes are usually bacteria or fungi that endosynbiotically live within a plant host without causing disease. These endophytes function to enhance the plant host growth and nutrient acquisition improve the plant host’s ability to tolerate abiotic stresses or decrease biotic stresses by enhancing the plant host’s resistance to infections (Farahat, 2020). Recently, an endophytic actinomycete of the genus Fodinicola was isolated from the roots of A. mangium, and has shown potential activity as a beneficial plant-growth promoter and specialised secondary metabolite producer (Phạm et al., 2020).
Despite endophytic fungi being regarded as new sources of novel bioactive compounds (Daouk et al., 1995; Cui et al., 2015), biological activities, and biotechnological developments, their true potential in controlling A. mangium diseases caused by C. fimbriata remains underexplored and underreported. Moreover, the leaf and root parts of A. mangium have been found to provide the habitats for various endophytic fungi (Mihara et al., 2005; Sarah Shafiei et al., 2017; Phạm et al., 2020). Nevertheless, besides leaf and root, other plant parts of the species should also be explored for endophytic fungi which might offer novel species or strains that possess valuable bioactive compounds useful in controlling the Ceratocystis wilt disease. Therefore, the objectives of the present work were (i) to establish the diversity of endophytic fungi in different plant parts of A. mangium, and (ii) to evaluate the antifungal potentials of the isolated and identified endophytic fungi against C. fimbriata.
Materials and methods
Plant materials
Ten seedlings of Acacia mangium (≈30–50 cm in height) and 2 A. mangium trees (≈30 cm in diameter at breast height) free from disease and insect infestation were randomly sampled, and identified at Serdang, Selangor (coordinate E 101○ 42.6333 N 2○ 59.1833). The root, stem, petiole, and leaf from healthy A. mangium seedlings were sampled in three replicates, respectively. In addition, three replicates of flower and fruit were also sampled from mature trees, respectively. Each plant part was cut into five 0.5 cm2 segments using a blade. These plant parts were washed thoroughly under running tap water to remove adherent debris on the surface.
Isolation of endophytic fungi
Plant part segments were surface-sterilised following the protocol suggested by Nuangmek et al. (2021). Briefly, the plant part segments were washed thoroughly under running tap water, immersed in 70% ethanol (Cerilliant Corporation, United States) for 1 min, soaked in 4% NaOCl (Malay-Sino Chemical, Malaysia) for 1 min, rinsed thrice in sterile distilled water, and blot-dried using a sterile filter paper. Next, the surface-sterilised plant part segments were excised 1–2 mm from the edge, and explant-plated onto a Potato Dextrose Agar (PDA; Merck Milipore, Germany). The PDA plates were incubated at 27°C for 7 d. Single hyphae growing out from the cultivated plant part segments were sub-cultured onto fresh PDA. Pure cultures were grouped according to the six types of plant parts (root, stem, petiole, leaf, flower, and fruit). Isolates were group based on colour and morphology on PDA (Yoo and Eom, 2012). Cultures were maintained on PDA for 5 d before sub-cultured into Potato Dextrose Broth (PDB; Neogen®, United States) while shaken at 150 rpm at 26°C for 3–6 d. Following incubation, the culture supernatant was filtered through Whatman filter paper (Cytiva™ Sigma-Aldrich Chemie GmbH, Germany) before being used for genomic DNA extraction.
DNA extraction and PCR amplification
A total of 100 mg of fungal mycelia harvested from PDB was used for fungal genomic DNA extraction. Fungal genomic DNA was extracted as previously described by Landum et al. (2016), in accordance with the manufacturer’s instructions, using the FAVORGEN Fungi/ Yeast Genomic DNA Extraction Mini Kit (Taiwan). The nuclear ribosomal DNA internal transcribed spacer (ITS) of the fungal isolates were amplified using the forward primer, ITS-F (5’-CTT GGT CAT TTA GAG GAA GTA A-3′) and the reverse primer, ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3′; White et al., 1990). The final reaction volume was 25 μl, containing 12.5 μl of 2X PCRBio Tag Mix Red (PCR Biosystems, UK), 0.4 μM of forward and reverse primers, and 10 mg of genomic DNA template. For negative control, the DNA was replaced with distilled water to verify the absence of contamination. The PCR was carried out using MyCycler™ (Bio-Rad, USA), programmed for 5 min at 95°C; 30 cycles for 30 s at 95°C, 30 s at 54.8°C, and 1 min at 72°C; and a final 10 min extension at 72°C. The PCR products were separated using 1% agarose gel in 1X TAE buffer (90 mM Tris-acetate and 2 nM EDTA, pH 8.0), stained with ethidium bromide (0.5 μg/ml), and visualised using FluorChem TM (Alpha Innotech, USA). The PCR products were sequenced by Apical Scientific Sdn. Bhd. (Malaysia). The sequences were deposited in NCBI GenBank, and compared with those already deposited in there via BLAST searches.
Sequence and phylogenetic analyses
The resulting DNA sequences were aligned using MUSCLE software embedded in MEGA software version 10.0.5 (Kumar et al., 2018), and manually trimmed and edited to obtain the complete sequences. Homology searches were carried out using the BLAST program against the NCBI GenBank database.1 The Maximum Likelihood tree was constructed using MEGA software version 10.0.5 with all positions containing gaps and missing data were included for analysis. Clade supports were calculated based on 1,000 bootstrap replications. A total of 64 sequences of close relatives were downloaded from the NCBI GenBank, and combined with sequences of the 66 endophytic fungi isolated in the present work for phylogenetic tree construction. Two wood decay macrofungi namely Schizophyllum commune (phylum Basidiomycota, family Schizophyllaceae) and Phellinus gabonensis (phylum Basidiomycota, family Hymenochaetaceae) were included as out-group.
Antagonism assay
Endophytic fungal isolates were cultivated on PDA plates at 26°C for 7 days. The antagonistic activity was evaluated through the dual culture assay against C. fimbriata. The pathogenic C. fimbriata (FRIM1162) isolate used in this study was isolated from a infected Acacia mangium (Syazwan et al., 2021) and maintained at 27°C on PDA media at the Mycology & Pathology Unit, Forest Research Institute Malaysia (FRIM). Briefly, a fungal disc of 5 mm in diameter was taken from C. fimbriata, and placed 3 cm from the margin of the PDA plate (9 cm in diameter). Next, a 5 mm disc of the endophytic fungus was placed 3 cm from the margin of the PDA plate, and directly opposite of the C. fimbriata disc. Inoculated PDA plates were incubated at room temperature for 7 days. PDA plates inoculated with C. fimbriata in the absence of endophytic fungus served as negative controls. The assay was performed in triplicates. Observations were carried out for 6 days, after which the mycelial radial growth of test pathogen (C. fimbriata) on a control plate (rl) and in the presence of the antagonistic fungus (r2) were measured, and the percentage inhibition (I%) in mycelial growth was calculated as: I% = [(r1 – r2) / r1] × 100 (Hajieghrari et al., 2008). The I% data were analysed statistically with ANOVA using the SAS statistical software. To examine the significance between endophytic fungal isolates, Fisher’s LSD was performed at p ≤ 0.05.
Results
Identification of endophytic fungi
A total of 66 endophytic fungal isolates were successfully isolated from different parts of healthy A. mangium (Table 1); 21 from leaf, 12 from petiole, 13 from stem, nine from root, six from flower, and five from fruit. Correspondingly, 66 isolates were successfully amplified using primers ITS1 and ITS4. The endophytic fungal isolates mostly belonged to Ascomycota (95.5%) followed by Zygomycota (4.5%) based on the BLAST searches analysis (Table 2). For Ascomycota, 13 genera were identified; Trichoderma (28.6%), Nigrospora (28.6%), Pestalotiopsis (12.7%), Lasiodiplodia (9.5%), Aspergillus (6.3%), Sordariomycetes (3%), and genera that were represented by a single isolate were Neopestalotiopsis, Pseudopestalotiopsis, Eutiarosporella, Curvularia, Fusarium, Penicillium, and Hypoxylon. Only Blakeslea sp. (4.5%) of Zygomycota was identified in the present work (Table 1). All the fungal ITS rDNA sequences exhibited high similarity with existing sequences in the NCBI database (Table 1).
Table 1.
Endophytic fungi isolated from different plant part of healthy Acacia mangium.
| Plant part | Individual nnumber | Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fruit | 1 | 2 | 1 | 1 | 5 | ||||||||||
| Flower | 1 | 2 | 1 | 1 | 1 | 6 | |||||||||
| Leaf | 4 | 2 | 1 | 4 | 1 | 1 | 8 | 21 | |||||||
| Petiole | 1 | 1 | 1 | 1 | 4 | 4 | 12 | ||||||||
| Stem | 2 | 1 | 1 | 1 | 4 | 3 | 1 | 13 | |||||||
| Root | 2 | 3 | 2 | 1 | 1 | 9 | |||||||||
| Total | 8 | 1 | 1 | 6 | 1 | 4 | 1 | 18 | 1 | 1 | 18 | 2 | 1 | 3 | 66 |
| Pestalotiopsis | Pseudopestalotiopsis | Neopestalotiopsis | Lasiodiplodia | Eutiarosporella | Aspergillus | Penicillium | Trichoderma | Fusarium | Curvularia | Nigrospora | Sordariomycetes | Hypoxylon | Blakeslea | ||
Table 2.
Percentage of identity matches of 66 fungal isolates from different plant parts of Acacia mangium based on ITS sequences using BLAST analyses, and their percentage of inhibition against Ceratocystis fimbriata.
| No. | Endophytic isolate ID | Plant part | Inhibition activities (%) (mean ± standard error) | GenBank Accession number | ITS region | ||||
|---|---|---|---|---|---|---|---|---|---|
| Match identity (%) | E-value | Identification in GenBank | BLAST match in GenBank | Phylum, Class, Family | |||||
| 1 | AC 1R | Root | 55 ± 0.58 | MW254902 | 99.28 | 0 | Blakeslea trispora | HQ248186 | Zygomycota, Zygomycetes, Choanephoraceae |
| 2 | AC 2R | Root | 0 ± 0.00 | MW254903 | 99.63 | 0 | Trichoderma gamsii | KX009501 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 3 | AC 3R | Root | 0 ± 0.00 | MW254904 | 100 | 0 | Aspergillus aculeatinus | MK281555 | Ascomycota, Eurotiomycetes, Trichocomaceae |
| 4 | AC 4R | Root | 44 ± 2.08 | MW254905 | 99.38 | 0 | Nigrospora sphaerica | MH368102 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 5 | AC 5R | Root | 20 ± 0.00 | MW254913 | 99.21 | 0 | Aspergillus niger | MN474007 | Ascomycota, Eurotiomycetes, Trichocomaceae |
| 6 | AC 6R | Root | 8.88 ± 0.66 | MW254916 | 99.63 | 0 | Trichoderma spirale | MN227543 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 7 | AC 7R | Root | 14.28 ± 0.43 | MW254942 | 99.17 | 0 | Sordariomycetes sp. | JQ759985 | Ascomycota, Sordariomycetes, |
| 8 | AC 8R | Root | 25 ± 2.89 | MW254956 | 99.58 | 0 | Nigrospora oryzae | MN382281 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 9 | AC 9R | Root | 58.33 ± 5.02×1015 | MW254964 | 99.81 | 0 | Trichoderma sp. | MK870905 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 10 | AC 1S | Stem | 58.33 ± 5.02 ×1015 | MW254907 | 99.81 | 0 | Trichoderma koningiopsis | KY807125 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 11 | AC 2S | Stem | 33.33 ± 0.29 | MW254909 | 99.79 | 0 | Nigrospora sphaerica | KJ572188 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 12 | AC 3S | Stem | 0 ± 0.00 | MW254914 | 99.63 | 0 | Pestalotiopsis vismiae | KP747709 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 13 | AC 4S | Stem | 0 ± 0.00 | MW254920 | 99.81 | 0 | Pestalotiopsis sp. | KY413701 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 14 | AC 5S | Stem | 45.45 ± 5.02 ×1015 | MW254924 | 99.45 | 0 | Trichoderma sp. | MK870688 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 15 | AC 6S | Stem | 20 ± 3.61 | MW254925 | 99.15 | 0 | Lasiodiplodia theobromae | GQ502461 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 16 | AC 7S | Stem | 45 ± 0.00 | MW254931 | 99.25 | 0 | Trichoderma gamsii | KX009501 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 17 | AC 8S | Stem | 40 ± 0.00 | MW254937 | 99.59 | 0 | Nigrospora oryzae | MN38228 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 18 | AC 9S | Stem | 0 ± 0.00 | MW254940 | 99.63 | 0 | Trichoderma ovalisporum | FJ442652 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 19 | AC 10S | Stem | 0 ± 0.00 | MW254944 | 99.8 | 0 | Aspergillus niger | MN559950 | Ascomycota, Eurotiomycetes, Trichocomaceae |
| 20 | AC 11S | Stem | 45.45 ± 2.60 | MW254951 | 100 | 0 | Sordariomycetes sp. | KC178665 | Ascomycota, Sordariomycetes, |
| 21 | AC 12S | Stem | 14.28 ± 0.30 | MW254954 | 97.98 | 0 | Eutiarosporella sp. | KX464132 | Ascomycota, Dothideomycetes, Botryosphaeriales |
| 22 | AC 13S | Stem | 0 ± 0.00 | MW254959 | 100 | 0 | Nigrospora sp. | MT556677 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 23 | AC 1 l | Leaf | 55.55 ± 5.02 ×1015 | MW254906 | 99.63 | 0 | Trichoderma gamsii | KM103313 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 24 | AC 2 l | Leaf | 37.5 ± 1.44 | MW254908 | 99.38 | 0 | Nigrospora sphaerica | MN625838 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 25 | AC 3 l | Leaf | 45.45 ± 0.75 | MW254910 | 99.81 | 0 | Trichoderma gamsii | KX009501 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 26 | AC 4 l | Leaf | 16.67 ± 9.53 | MW254918 | 100 | 0 | Curvularia pandanicola | MH275056 | Ascomycota, Dothideomycetes, Pleosporaceae |
| 27 | AC 5 l | Leaf | 16.67 ± 0.00 | MW254919 | 99.63 | 0 | Pestalotiopsis microspora | MT597837 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 28 | AC 6 l | Leaf | 45.45 ± 1.16 | MW254921 | 99.81 | 0 | Pestalotiopsis microspora | EU137910 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 29 | AC 7 l | Leaf | 58.3 ± 5.02 ×1015 | MW254922 | 98.77 | 0 | Nigrospora oryzae | MN382281 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 30 | AC 8 l | Leaf | 28.57 ± 2.51 ×1015 | MW254923 | 99.58 | 0 | Fusarium chlamydosporum | MT448890 | Ascomycota, Sordariomycetes, Nectriaceae |
| 31 | AC 9 l | Leaf | 0 ± 0.00 | MW254926 | 99.38 | 0 | Nigrospora sphaerica | MN566004 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 32 | AC 10 l | Leaf | 30 ± 5.77 | MW254934 | 99.57 | 0 | Lasiodiplodia theobromae | KF293981 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 33 | AC 11 l | Leaf | 12.5 ± 6.93 | MW254936 | 99.63 | 0 | Trichoderma koningiopsis | JQ617301 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 34 | AC 12 l | Leaf | 0 ± 0.00 | MW254938 | 99.44 | 0 | Pestalotiopsis neglecta | MN006391 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 35 | AC 13 l | Leaf | 22.22 ± 0.00 | MW254939 | 99.62 | 0 | Trichoderma gamsii | KX009501 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 36 | AC 14 l | Leaf | 0 ± 0.00 | MW254943 | 99.58 | 0 | Nigrospora oryzae | JX966549 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 37 | AC 15 l | Leaf | 33.33 ± 1.59 | MW254945 | 99.79 | 0 | Nigrospora sp. | MT561433 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 38 | AC 16 l | Leaf | 45.45 ± 5.02 ×1015 | MW254946 | 99.58 | 0 | Lasiodiplodia theobromae | MK696043 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 39 | AC 17 l | Leaf | 40 ± 5.77 | MW254948 | 99.43 | 0 | Pestalotiopsis vismiae | KP747709 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 40 | AC 18 l | Leaf | 25 ± 0.00 | MW254949 | 99.59 | 0 | Nigrospora sphaerica | MT043797 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 41 | AC 19 l | Leaf | 0 ± 0.00 | MW254950 | 99.8 | 0 | Aspergillus aculeatus | KJ605160 | Ascomycota, Eurotiomycetes, Trichocomaceae |
| 42 | AC 20 l | Leaf | 40 ± 5.77 | MW254962 | 99.59 | 0 | Nigrospora sphaerica | MH368102 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 43 | AC 21 l | Leaf | 0 ± 0.00 | MW254963 | 99.58 | 0 | Nigrospora sphaerica | MT561433 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 44 | AC 1P | Petiole | 0 ± 0.00 | MW254917 | 99.81 | 0 | Trichoderma crissum | MK911703 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 45 | AC 2P | Petiole | 0 ± 0.00 | MW254932 | 99.79 | 0 | Nigrospora sphaerica | MT561433 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 46 | AC 3P | Petiole | 50 ± 4.91 | MW254933 | 97.68 | 0 | Nigrospora sphaerica | MN795570 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 47 | AC 4P | Petiole | 58.33 ± 5.02 ×1015 | MW254935 | 99.38 | 0 | Nigrospora sphaerica | KJ572188 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 48 | AC 5P | Petiole | 45.45 ± 5.02 ×1015 | MW254947 | 99.79 | 0 | Nigrospora sphaerica | MT561433 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 49 | AC 6P | Petiole | 45 ± 5.77 | MW254952 | 99.8 | 0 | Penicillium rolfsii | MK120600 | Ascomycota, Eurotiomycetes, Trichocomaceae |
| 50 | AC 7P | Petiole | 20 ± 5.77 | MW254957 | 100 | 0 | Trichoderma longibrachiatum | FJ462745 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 51 | AC 8P | Petiole | 30 ± 5.77 | MW254958 | 99.37 | 0 | Neopestalotiopsis cubana | LC521857 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 52 | AC 9P | Petiole | 45.45 ± 5.02 ×1015 | MW254961 | 99.79 | 0 | Pestalotiopsis sp. | JN116590 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 53 | AC 10P | Petiole | 45.45 ± 5.02 ×1015 | MW254965 | 99.57 | 0 | Lasiodiplodia theobromae | MT075441 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 54 | AC 11P | Petiole | 14.28 ± 0 | MW254966 | 70.1 | 4.00E-20 | Trichoderma sp. | GU973813 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 55 | AC 12P | Petiole | 20 ± 5.77 | MW254967 | 99.44 | 0 | Trichoderma koningiopsis | MT102395 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 56 | AC 1F | Flower | 8.88 ± 0.00 | MW254911 | 99.28 | 0 | Blakeslea trispora | HQ248186 | Zygomycota, Zygomycetes, Choanephoraceae |
| 57 | AC 2F | Flower | 8.88 ± 5.14 | MW254915 | 94.87 | 0 | Hypoxylon monticulosum | KY610404 | Ascomycota, Sordariomycetes, Hypoxylaceae |
| 58 | AC 3F | Flower | 58.33 ± 5.02 ×1015 | MW254927 | 100 | 0 | Nigrospora sphaerica | MT561433 | Ascomycota, Sordariomycetes, Trichosphaeriales |
| 59 | AC 4F | Flower | 40 ± 0.00 | MW254941 | 99.62 | 0 | Trichoderma longibrachiatum | MH745146 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 60 | AC 5F | Flower | 45.45 ± 2.60 | MW254953 | 99.59 | 0 | Pseudopestalotiopsis theae | KX401429 | Ascomycota, Sordariomycetes, Pestalotiopsidaceae |
| 61 | AC 6F | Flower | 45.45 ± 5.02 ×1015 | MW254955 | 100 | 0 | Trichoderma koningiopsis | JQ278013 | Ascomycota, Sordariomycetes, Hypocreaceae |
| 62 | AC 1 U | Fruit | 8.88 ± 0.00 | MW254912 | 100 | 0 | Pestalotiopsis microspore | EU137910 | Ascomycota, Sordariomycetes, Sporocadaceae |
| 63 | AC 2 U | Fruit | 69.23 ± 0.00 | MW254928 | 99.79 | 0 | Lasiodiplodia theobromae | MK696044 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 64 | AC 3 U | Fruit | 25 ± 2.89 | MW254929 | 98.39 | 0 | Blakeslea trispora | HQ248186 | Zygomycota, Zygomycetes, Choanephoraceae |
| 65 | AC 4 U | Fruit | 45 ± 2.89 | MW254930 | 99.79 | 0 | Lasiodiplodia venezuelensis | MH865369 | Ascomycota, Dothideomycetes, Botryosphaeriaceae |
| 66 | AC 5 U | Fruit | 16.16 ± 0.00 | MW254960 | 100 | 0 | Trichoderma harzianum | MF537642 | Ascomycota, Sordariomycetes, Hypocreaceae |
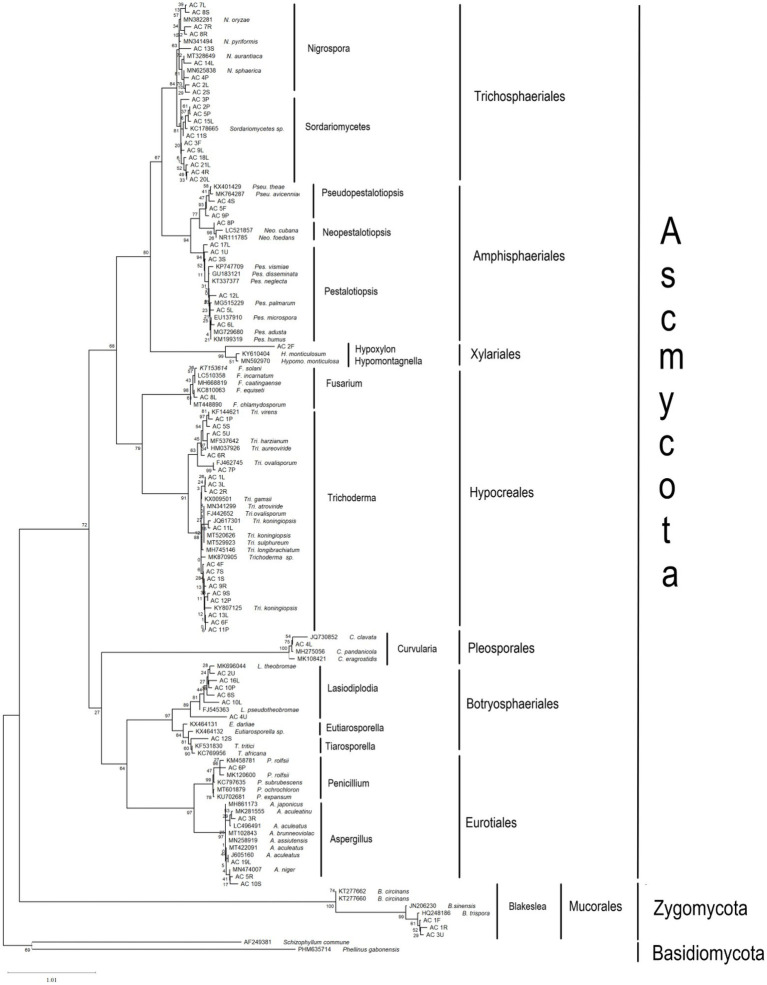
The ITS sequences obtained in the present work were deposited in the NCBI GenBank (MW254902 - MW254967) for future reference. A total of 66 sequences of close relatives were downloaded from the NCBI GenBank, and combined with sequences of the 66 endophytic fungi for phylogenetic tree construction (Figure 1). Nine different orders were observed, of which six belonged to Ascomycota (Amphisphaeriales, Brotryosphaerialase, Eurotiales, Hypocreales, Pleosporales and Trichosphaeriales), one belonged to Zygomycota, and two belonged to Basidiomycota (out-group). Most of the endophytic fungal isolates clustered under the order Trichosphaeriales (20 isolates) belonged to genus Nigrospora, and under the order Hypocreales (19 isolates) belonged to genera Fusarium and Trichoderma. Tables 3 and 4 summarises these results.
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on rDNA ITS sequences of endophytic fungal isolates and fungal ITS sequences from the GenBank. ML tree was constructed using the Kimura 2-parameter (K2) model and gamma distributed (+G) model. All positions containing gaps and missing data were included for analysis. Clade supports were calculated based on 1,000 bootstrap.
Table 3.
Endophytic fungal orders from the phylum Ascomycota.
| No. | ID | GenBank Accession no. | Plant part | Amphisphaeriales | |
|---|---|---|---|---|---|
| 1 | AC 3S | MW254914 | Stem | Pestalotiopsis vismiae | Pestalotiopsis clade (94% bootstrap) |
| 2 | AC 4S | MW254920 | Stem | Pestalotiopsis sp. | |
| 3 | AC 9P | MW254961 | Petiole | Pestalotiopsis sp. | |
| 4 | AC 5L | MW254919 | Leaf | Pestalotiopsis microspora | |
| 5 | AC 6L | MW254921 | Leaf | Pestalotiopsis microspora | |
| 6 | AC 12L | MW254938 | Leaf | Pestalotiopsis neglecta | |
| 7 | AC 1U | MW254912 | Fruit | Pestalotiopsis microspora | |
| 8 | AC 5F | MW254953 | Flower | Pseudopestalotiopsis theae | |
| 9 | AC 17L | MW254948 | Leaf | Pestalotiopsis vismiae | Pseudopestalotiopsis clade (77% bootstrap) |
| 10 | AC 8P | MW254958 | Petiole | Neopestalotiopsis cubana Brotryosphaerialase | Neopestalotiopsis clade (77% bootstrap) |
| 11 | AC 6S | MW254925 | Stem | Lasiodiplodia theobromae | Lasiodiplodia clade (97% bootstrap) |
| 12 | AC 10P | MW254965 | Petiole | Lasiodiplodia theobromae | |
| 13 | AC 10L | MW254934 | Leaf | Lasiodiplodia theobromae | |
| 14 | AC 16L | MW254946 | Leaf | Lasiodiplodia theobromae | |
| 15 | AC 2U | MW254928 | Fruit | Lasiodiplodia theobromae | |
| 16 | AC 4U | MW254930 | Fruit | Lasiodiplodia venezuelensis | |
| 17 | AC 12S | MW254954 | Stem | Eutiarosporella sp. Eurotiales | Eutiarosporella clade (97% bootstrap) |
| 18 | AC 10S | MW254944 | Stem | Aspergillus niger | Aspergillus clade (97% bootstrap) |
| 19 | AC 3R | MW254904 | Root | Aspergillus aculeatinus | |
| 20 | AC 5R | MW254913 | Root | Aspergillus niger | |
| 21 | AC 19L | MW254950 | Leaf | Aspergillus aculeatus | |
| 22 | AC 6P | MW254952 | Petiole | Penicillium rolfsii Hypocreales | Penicillium clade (97% bootstrap) |
| 23 | AC 2R | MW254903 | Root | Trichoderma gamsii | Trichoderma clade (95% bootstrap) |
| 24 | AC 6R | MW254916 | Root | Trichoderma spirale | |
| 25 | AC 9R | MW254964 | Root | Trichoderma sp. | |
| 26 | AC 1S | MW254907 | Stem | Trichoderma koningiopsis | |
| 27 | AC 5S | MW254924 | Stem | Trichoderma sp. | |
| 28 | AC 7S | MW254931 | Stem | Trichoderma gamsii | |
| 29 | AC 9S | MW254940 | Stem | Trichoderma ovalisporum | |
| 30 | AC 1P | MW254917 | Petiole | Trichoderma crissum | |
| 31 | AC 7P | MW254957 | Petiole | Trichoderma longibrachiatum | |
| 32 | AC 11P | MW254966 | Petiole | Trichoderma sp. | |
| 33 | AC 12P | MW254967 | Petiole | Trichoderma koningiopsis | |
| 34 | AC 1L | MW254906 | Leaf | Trichoderma gamsii | |
| 35 | AC 3L | MW254910 | Leaf | Trichoderma gamsii | |
| 36 | AC 11L | MW254936 | Leaf | Trichoderma koningiopsis | |
| 37 | AC 13L | MW254939 | Leaf | Trichoderma gamsii | |
| 38 | AC 4F | MW254941 | Flower | Trichoderma longibrachiatum | |
| 39 | AC 5U | MW254960 | Fruit | Trichoderma harzianum | |
| 40 | AC 6F | MW254955 | Fruit | Trichoderma koningiopsis | |
| 41 | AC 8L | MW254923 | Leaf | Fusarium chlamydosporum Pleosporales | Fusarium clade (95% bootstrap) |
| 42 | AC 4L | MW254918 | Leaf | Curvularia pandanicola | Curvularia clade (95% bootstrap) |
| Hypocreales | |||||
| 43 | AC 4R | MW254905 | Root | Nigrospora sphaerica | Nigrospora and Sordariomycete polytomy clade (95% bootstrap) |
| 44 | AC 8R | MW254956 | Root | Nigrospora oryzae | |
| 45 | AC 2S | MW254909 | Stem | Nigrospora sphaerica | |
| 46 | AC 8S | MW254937 | Stem | Nigrospora oryzae | |
| 47 | AC 13S | MW254959 | Stem | Nigrospora sp. | |
| 48 | AC 2P | MW254932 | Petiole | Nigrospora sphaerica | |
| 49 | AC 3P | MW254933 | Petiole | Nigrospora sphaerica | |
| 50 | AC 4P | MW254935 | Petiole | Nigrospora sphaerica | |
| 51 | AC 5P | MW254947 | Petiole | Nigrospora sphaerica | |
| 52 | AC 2L | MW254908 | Leaf | Nigrospora sphaerica | |
| 53 | AC 7L | MW254922 | Leaf | Nigrospora oryzae | |
| 54 | AC 9L | MW254926 | Leaf | Nigrospora sphaerica | |
| 55 | AC 14L | MW254943 | Leaf | Nigrospora oryzae | |
| 56 | AC 15L | MW254945 | Leaf | Nigrospora sp. | |
| 57 | AC 18L | MW254949 | Leaf | Nigrospora sphaerica | |
| 58 | AC 20L | MW254962 | Leaf | Nigrospora sphaerica | |
| 59 | AC 21L | MW254963 | Leaf | Nigrospora sphaerica | |
| 60 | AC 3F | MW254927 | Flower | Nigrospora sphaerica | |
| 61 | AC 7R | MW254942 | Root | Sordariomycetes sp. | |
| 62 | AC 11S | MW254951 | Stem | Sordariomycetes sp. Hypocreales | |
| 63 | AC 2F | MW254915 | Flower | Hypoxylon monticulosum | Hypoxylon clade (99% bootstrap) |
Table 4.
Endophytic fungal order from the phylum Zygomycota.
Antagonism assay
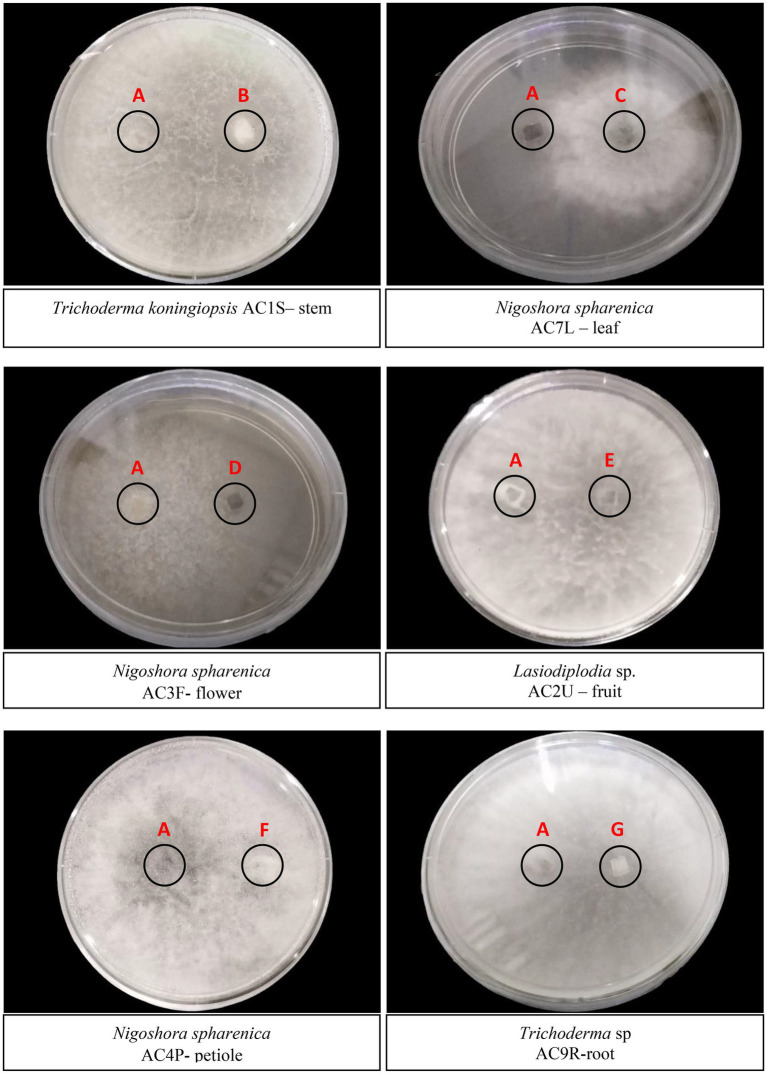
All 66 endophytic fungal isolates were tested in the antagonism assay against C. fimbriata. After 5 days of incubation, six fungal isolates namely Trichoderma koningiopsis (AC 1S) stem, Nigrospora oryzae (AC 7L) leaf, Nigrospora sphaerica (AC 3F) flower, Lasiodiplodia sp. (AC 2 U) fruit, Nigrospora sphaerica (AC 4P) petiole, and Trichoderma sp. (AC 9R) root were observed to exhibit stronger inhibition where the mycelia of the antagonists had breached into C. fimbriata colony (Figure 2). Of these, four fungal isolates namely T. koningiopsis (AC 1S) stem, Lasiodiplodia sp. (AC 2 U) fruit, N. sphaerica (AC 4P) petiole, and Trichoderma sp. (AC 9R) root colonised almost 99% of the culture plate. Although N. sphaerica (AC 7 l) leaf and N. sphaerica (AC 3F) flower did not colonise the entire culture plate, there was no growth of C. fimbriata observed.
Figure 2.
Dual culture plate assay between six endophytic fungal isolates against the pathogen C. fimbriata (A). (B) Trichoderma koningiopsis AC1S – stem; (C) Nigoshora spharenica AC7L – leaf; (D) Nigoshora spharenica AC3F - flower; (E) Lasiodiplodia sp. AC2U – fruit; (F) Nigoshora spharenica AC4P-petiole; (G) Trichoderma sp. AC9R-root. The plates were cultivated for 5 days at 27°C. Radial growths were measured and interaction were observed.
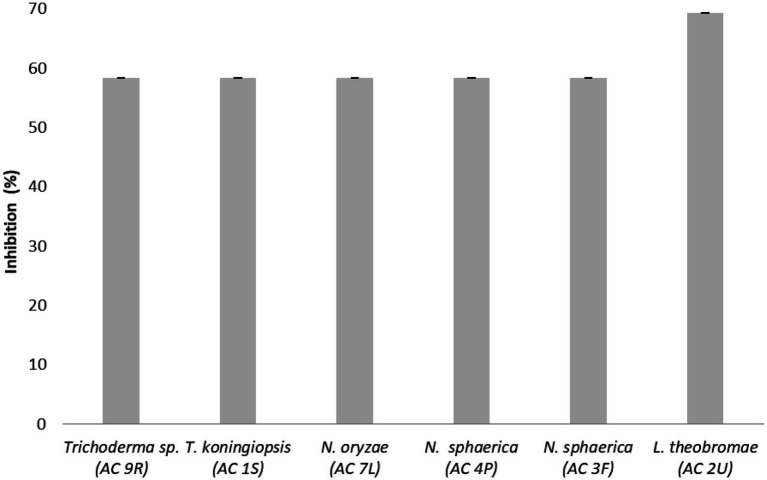
The inhibition percentages (I%) of endophytic fungi against the pathogen C. fimbriata in dual culture assay are shown in Figure 3. Lasiodiplodia sp. (AC 2 U) isolated from fruit recorded the highest I% (69.23%), followed by Trichoderma sp. (AC 9R) isolated from root, Nigrospora sphaerica (AC 4P) isolated from petiole, Nigrospora sphaerica (AC 3F) isolated from flower, Trichoderma koningiopsis (AC 1S) isolated from stem, and Nigrospora oryzae (AC 7L) isolated from leaf with value 58.33%, respectively.
Figure 3.
Inhibition percentages (I%) of endophytic fungi against the pathogen Ceratocystis fimbriata in dual culture assay. Data are mean ± standard error (SE) of triplicates.
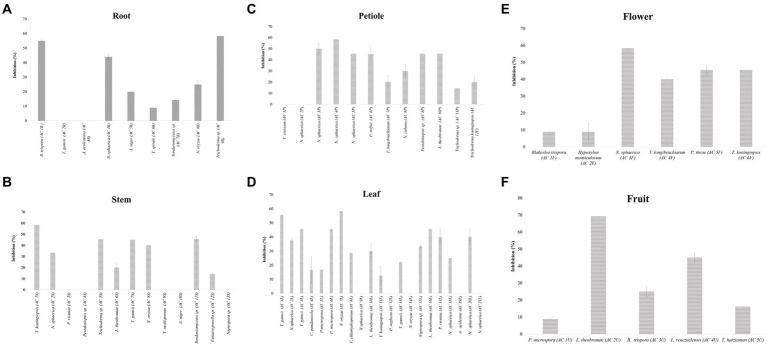
Thirteen endophytic fungi from various plant parts of A. mangium showed no inhibition against C. fimbriata (Figure 4) namely A. aculeatinus (AC 3R) isolated from root, A. aculeatus (AC 19L) isolated from leaf, A. niger (AC 10S) isolated from stem, N. oryzae (AC 14L) isolated from leaf, Nigrospora sp. (AC 13S) isolated from stem, N. sphaerica (AC 9L) isolated from leaf, N. sphaerica (AC 2P) isolated from petiole, P. neglecta (AC 12L) isolated from leaf, Pestalotiopsis sp. (AC 4S) isolated from stem, P. vismiae (AC 3S) isolated from stem, T. crissum (AC 1P) isolated from petiole, T. gamsii (AC 2R) isolated from root, and T. ovalisporum (AC 9S) isolated from stem.
Figure 4.
The inhibition percentages (I%) of endophytic fungi isolated from root (A), stem (B), petiole (C), leaf (D), flower (E), and fruit (F) against the pathogen Ceratocystis fimbriata. Data are mean ± standard error (SE) of triplicates. Means followed by the same letter in each group are not significantly different at α = 0.05 according to DuncanLSD.
Diversity of endophytic fungi
Endophytic fungi are ubiquitous, and every plant species examined to date have been found colonised by them (Arnold et al., 2001). A single plant species may harbour hundreds of endophytes which may inhabit all available tissues, including leaves, petioles, stems, twigs, barks, xylems, roots, fruits, flowers, and seeds (Chapela and Boddy, 1988; Fisher et al., 1993; Saikkonen et al., 1998; Jena and Tayung, 2013). In the present work, endophytic fungi were isolated from different plant parts of A. mangium with the highest number of isolates found in leaf and dominated by the genera Trichoderma and Nigrospora. Trichoderma spp. were present in all plant parts, while Nigrospora spp. were present in all but fruit. In total, 66 endophytic fungal isolates were obtained from different plant parts of A. mangium.
Trichoderma and Nigrospora have also been reported as endophytes in other plants such as Rauvolfia serpentine, Prosopis cineraria, and Piper nigrum (Gehlot et al., 2008; Dutta et al., 2014; Sopialena et al., 2018). Trichoderma is also found in many ecosystems, and can reduce the severity of plant diseases by inhibiting the plant pathogens in the soil through their highly potent antagonistic and mycoparasitic activities (Hermosa et al., 2012). Moreover, as revealed by research in recent decades, some Trichoderma strains can interact directly with roots, thus increasing plant growth potential, resistance to disease, and tolerance to abiotic stresses (Mastouri et al., 2010; Hermosa et al., 2012; Brotman et al., 2013). Nigrospora is also a beneficial member of the foliar endophytic community due to its mutualistic existence with their host plants, and having a potential for biological control strategies (Zakaria et al., 2016). Other than Nigrospora, Pestalotiopsis also is a beneficial member of the foliar endophytic community due to its ability to switch its nutritional mode, thus able to stay as an endophyte or switch to saprophyte when necessary (Douanla-Meli et al., 2013; Hamzah et al., 2018). Besides Trichoderma, Nigrospora, and Pestalotiopsis, other fungal genera such as Lasiodiplodia, Sordariomycetes, and Aspergillus have also been reported as predominant endophytic fungi in other plants species (Li et al., 2012; del Castillo et al., 2016), and have an antagonism ability (Chen et al., 2010). Fusarium too is a common endophytic fungal genus found in trees (Zakaria et al., 2010). Although it is widely available in most tropical plants investigated in past studies (Warman and Aitken, 2018), we recorded a low isolation frequency of Fusarium. Our finding also revealed lesser-known fungal genera, namely Eutiarosporella, Curvularia, Glomerella, and Hypoxylon in A. mangium.
In the present work, ITS sequences identified 63 endophytic fungal isolates from the phylum Ascomycota, and three from Zygomycota. The phylum Ascomycota has been reported to be the most common endophytic fungal phylum when isolated using standard isolation protocols (Koukol et al., 2012; Hamzah et al., 2018). Fungi from the phylum Zygomycota have been reported to be culture-method dependent (Crozier et al., 2006; Hamzah et al., 2018), which might explain the small isolate number reported in the present work. Comparative studies also show that only a small fraction of microorganisms in nature can be cultured using conventional microbiological techniques (Amann et al., 1995). There are many factors that can affect the microbial viability under laboratory conditions, for example the lack of knowledge about their nutritional requirements.
Antagonism activities against Ceratocystis fimbriata
Fungal antagonism can manifest in many ways such as nutrition competition, niche exclusion, mycoparasitism, and the production of extracellular metabolites (Siameto et al., 2010). These metabolites, especially antibiotics and lytic enzymes, have been widely applied in various fields like crop-pathogen controls. Endophytic microorganisms isolated from plants can produce various novel bioactive metabolites (Ramasamy et al., 2010). The bioactive metabolites produced by plants, microorganisms, and organisms are useful for the discovery and development of new drugs.
In the present work, Lasiodiplodia sp., T. koningiopsis, N. sphaerica and Trichoderma sp. successfully inhibited the pathogen C. fimbriata in the dual culture assay. The ability to out-grow the pathogen in vitro suggested that these fungi competed for the space and nutrient with the pathogen. In theory, biological agents with antifungal properties are known to secrete certain enzymes which break down their competitors’ cell wall, thus restricting their growth (Sharon et al., 2001). The antagonism displayed by Lasiodiplodia sp. was more aggressive as compared to other endophytic fungi (Figure 3). This could be attributed to the production of lytic enzymes by Lasiodiplodia sp. (Anitha and Rabeeth, 2010). The antagonism displayed by Lasiodiplodia sp., T. koningiopsis, N. sphaerica and Trichoderma sp. could also be explained by their secretion of secondary metabolites into the growth medium, as well as nutrient depletion in the growth medium (Robinson et al., 2014). The antagonism displayed might also be influenced by the antibiotics or hydrolytic enzymes they produced (Kamala and Indira, 2011). The difference in antagonism magnitude observed in the present work could also be dependent on specific fungal species (Kai et al., 2007). Previously, Lasiodiplodia sp. from the flower of Viscum coloratum also exhibited antimicrobial activity which could be due to the presence of cyclo-(Trp-Ala), ICA, indole-3-carbaldehyde, mullein, and 2-phenylethano in their extract (Qian et al., 2014). Lasiodiplodia sp. isolated from the twig of Aegle marmelos has also been shown to have in vitro fibrinolytic activities (Meshram and Saxena, 2016). Another plant parts such as bark and leaf of Terminalia sp. has also been isolated with Lasiodiplodia sp. which not only exhibited antimicrobial and antioxidant activities, but also aided the plant to withstand stressful environmental conditions (Patil et al., 2014).
Conclusion
Diversity of endophytic fungi were successfully isolated from different parts of A. mangium, with Trichoderma spp. being the most prevalent, and were isolated from all six plant parts. Against C. fimbriata, the crude extracts from Trichoderma spp., N. sphaerica, and Lasiodiplodia sp. exhibited strong inhibition in the dual culture assay. Thus, it can be concluded that certain endophytic fungi of A. mangium have the potential to be harnessed as anti-Ceratocystis agent in future biotechnological applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
RT designed the study, collected, identified plant materials, and edited the manuscript. RT and RZ conducted the experiments, drafted, and revised the manuscript. Data analysis performed by RT, MA, MM, NS, WAW-M-A, and AH. MH assisted in DNA extraction. RT, MA, MM, NS, and AH supervised. RT acquired funding. All authors contributed to the article and approved the submitted version.
Funding
The present work was financially supported by Universiti Putra Malaysia under the Putra Grant Scheme (GP-IPM/2017/9565600).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors gratefully acknowledge the Laboratory of Wood Deterioration and Protection, Department of Natural Resource Industry, Faculty of Forestry and Environment, Universiti Putra Malaysia for the research facilities.
Footnotes
References
- Amann R. I., Ludwig W., Schleifer K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. doi: 10.1128/mr.59.1.143-169.1995, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitha A., Rabeeth M. (2010). Degradation of fungal cell walls of phytopathogenic fungi by lytic enzyme of Streptomyces griseus. African J. Plant Sci. 4, 61–66. [Google Scholar]
- Arnold A. E., Maynard Z., Gilbert G. S. (2001). Fungal endophytes in dicotyledonous neotropical trees: patterns of abundance and diversity. Mycol. Res. 105, 1502–1507. doi: 10.1017/S0953756201004956 [DOI] [Google Scholar]
- Blaedow R. A. (2009). Use of the systemic fungicide propiconazole for oak wilt management: an assessment of uncharacterized host-pathogen-fungicide interactions. University of Minnesota.
- Brawner J., Japarudin Y., Lapammu M., Rauf R., Boden D., Wingfield M. J. (2015). Evaluating the inheritance of Ceratocystis acaciivora symptom expression in a diverse Acacia mangium breeding population. South. For. 77, 83–90. doi: 10.2989/20702620.2015.1007412 [DOI] [Google Scholar]
- Brotman Y., Landau U., Cuadros-Inostroza Á., Takayuki T., Fernie A. R., Chet I., et al. (2013). Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:1003221. doi: 10.1371/annotation/8b818c15-3fe0-4e56-9be2-e44fd1ed3fae [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki S., Ghosh B., Bandyopadyhay S., Mookerjee M., Das S., Dastidar S. G. (2015). Detection of various phytochemical compounds from seeds of a. auriculiformis for possibilities of obtaining potent antimicrobial agents. Int. J. Biol. Pharm. Res. 6, 120–128. [Google Scholar]
- Chapela I. H., Boddy L. (1988). Fungal colonization of attached beech branches: II. Spatial and temporal organization of communities arising from latent invaders in bark and functional sapwood, under different moisture regimes. New Phytol. 110, 47–57. doi: 10.1111/j.1469-8137.1988.tb00236.x [DOI] [Google Scholar]
- Chen X. M., Dong H. L., Hu K. X., Sun Z. R., Chen J., Guo S. X. (2010). Diversity and antimicrobial and plant-growth-promoting activities of endophytic fungi in Dendrobium loddigesii Rolfe. J. Plant Growth Regul. 29, 328–337. doi: 10.1007/s00344-010-9139-y [DOI] [Google Scholar]
- Crozier J., Thomas S. E., Aime M. C., Evans H. C., Holmes K. A. (2006). Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 55, 783–791. doi: 10.1111/j.1365-3059.2006.01446.x [DOI] [Google Scholar]
- Cui J., Guo T., Ren Z., Zhang N., Wang M. (2015). Diversity and antioxidant activity of culturable endophytic fungi from alpine plants of Rhodiola crenulata, R. angusta, and R. sachalinensis. PLoS One 10:0118204. doi: 10.1371/journal.pone.0118204, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daouk R. K., Dagher S. M., Sattout E. J. (1995). Antifungal activity of the essential oil of Origanum syriacum L. J. Food Prot. 58, 1147–1149. doi: 10.4315/0362-028X-58.10.1147, PMID: [DOI] [PubMed] [Google Scholar]
- Del Castillo D. S., Parra D., Noceda C., Pérez-Martínez S. (2016). Co-occurrence of pathogenic and non-pathogenic fusarium decemcellulare and Lasiodiplodia theobromae isolates in cushion galls disease of cacao (Theobroma cacao L.). J. Plant Pro. Res. 56, 129–138. doi: 10.1515/jppr-2016-0020 [DOI] [Google Scholar]
- Douanla-Meli C., Langer E., Mouafo F. T. (2013). Fungal endophyte diversity and community patterns in healthy and yellowing leaves of citrus Limon. Fungal Ecol. 6, 212–222. doi: 10.1016/j.funeco.2013.01.004 [DOI] [Google Scholar]
- Dutta D., Puzari K. C., Gogoi R., Dutta P. (2014). Endophytes: exploitation as a tool in plant protection. Braz. Arch. Biol. Technol. 57, 621–629. doi: 10.1590/S1516-8913201402043 [DOI] [Google Scholar]
- Farahat M. (2020). Alleviation of salinity stress in wheat by ACC deaminase-producing bacillus aryabhattai EWR29 with multifarious plant growth-promoting attributes. Plant Arch. 20, 417–429. [Google Scholar]
- Farid A. M., Syazwan S. A., Azrul W. W. M., Patahayah M., Salleh S. M., Ong S. P. (2018). Ceratocystis fimbriata: A white listed invasive alien species (IAS) causing wilt disease on acacia Mangium plantation.
- Fisher P. J., Petrini O. S. B. C., Sutton B. C. (1993). A comparative study of fungal endophytes in leaves, xylem and bark of eucalyptus in Australia and England. Sydowia 45, 338–345. [Google Scholar]
- Gehlot P., Bohra N. K., Purohit D. K. (2008). Endophytic mycoflora of inner bark of Prosopis cineraria—a key stone tree species of Indian desert. Am. Eurasian J. Botany 1, 1–4. [Google Scholar]
- Hajieghrari B., Torabi-Giglou M., Mohammadi M. R., Davari M. (2008). Biological potential of some Iranian Trichoderma isolates in the control of soil borne plant pathogenic fungi. Afr. J. Biotechnol. 7, 967–972. [Google Scholar]
- Hamzah T. N. T., Lee S. Y., Hidayat A., Terhem R., Faridah-Hanum I., Mohamed R. (2018). Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Front. Microbiol. 9:1707. doi: 10.3389/fmicb.2018.01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington T. C. (2013). “Ceratocystis diseases”, in Infectious forest diseases. ed. P. Gonthier (Wallingford: CABI; ). 230–255. doi: 10.1079/9781780640402.0230 [DOI] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. doi: 10.1099/mic.0.052274-0, PMID: [DOI] [PubMed] [Google Scholar]
- Jena S. K., Tayung K. (2013). Endophytic fungal communities associated with two ethno-medicinal plants of Similipal biosphere reserve, India and their antimicrobial prospective. J. Appl. Pharm. Sci. 3, S7–S12. doi: 10.7324/JAPS.2013.34.S2 [DOI] [Google Scholar]
- Kai N., Effimert U., Berg G., Piechulla B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0, PMID: [DOI] [PubMed] [Google Scholar]
- Kamala T., Indira S. (2011). Evaluation of indigenous Trichoderma isolates from Manipur as biocontrol agent against Pythium aphanidermatum on common beans. 3 Biotech 1, 217–225. doi: 10.1007/s13205-011-0027-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile G. A. (1993). “Plant diseases caused by species of Ceratocystis sensu stricto and Chalara” in Ceratocystis and Ophiostoma: taxonomy, ecology, and pathogenicity. eds. Wingfield M., Seifert K., Webber J. (Minnesota USA: The American Phytopathological Society St. Paul; ), 173–183. [Google Scholar]
- Koukol O., Kolarík M., Kolárová Z., Baldrian P. (2012). Diversity of foliar endophytes in wind fallen Picea abies trees. Fungal Divers. 54, 69–77. doi: 10.1007/s13225-011-0112-2 [DOI] [Google Scholar]
- Kumar C. G., Mongolla P., Pombala S. (2018). Lasiosan, a new exopolysaccharide from Lasiodiplodia sp. strain B2 (MTCC6000): structural characterization and biological evaluation. Process Biochem. 72, 162–169. doi: 10.1016/j.procbio.2018.06.014 [DOI] [Google Scholar]
- Landum M. C., do Rosário Félix M., Alho J., Garcia R., Cabrita M. J., Rei F., et al. (2016). Antagonistic activity of fungi of Olea europaea L. against Colletotrichum acutatum. Microbiol. Res. 183, 100–108. doi: 10.1016/j.micres.2015.12.001, PMID: [DOI] [PubMed] [Google Scholar]
- Lee S. S. (2018). Observations on the successes and failures of acacia plantations in Sabah and Sarawak and the way forward. J. Trop. For. Sci. 30, 468–475. doi: 10.26525/jtfs2018.30.5.468475 [DOI] [Google Scholar]
- Li H. Y., Shen M., Zhou Z. P., Li T., Wei Y. L., Lin L. B. (2012). Diversity and cold adaptation of endophytic fungi from five dominant plant species collected from the Baima Snow Mountain, Southwest China. Fungal Divers. 54, 79–86. doi: 10.1007/s13225-012-0153-1 [DOI] [Google Scholar]
- Mandy M, Wickneswari R. (2014). Incidences and severity of vascular wilt in Acacia mangium plantations in Sabah, Malaysia. The 2014 UKM FST Postgraduate Colloquium. AIP Publishing LLC. 784–789.
- Mastouri F., Björkman T., Harman G. E. (2010). Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology 100, 1213–1221. doi: 10.1094/PHYTO-03-10-0091, PMID: [DOI] [PubMed] [Google Scholar]
- Meshram V., Saxena S. (2016). Potential fibrinolytic activity of an endophytic Lasiodiplodia pseudotheobromae species. 3. Biotech 6:114. doi: 10.1007/s13205-016-0428-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara R., Barry K. M., Mohammed C. L., Mitsunaga T. (2005). Comparison of antifungal and antioxidant activities of Acacia mangium and A. auriculiformis heartwood extracts. J. Chem. Ecol. 31, 789–804. doi: 10.1007/s10886-005-3544-x, PMID: [DOI] [PubMed] [Google Scholar]
- Nasution A., Glen M., Beadle C., Mohammed C. (2019). Ceratocystis wilt and canker–a disease that compromises the growing of commercial acacia-based plantations in the tropics. Aust. For. 82, 80–93. doi: 10.1080/00049158.2019.1595347 [DOI] [Google Scholar]
- Nuangmek W., Aiduang W., Kumla J., Lumyong S., Suwannarach N. (2021). Evaluation of a newly identified endophytic fungus, Trichoderma phayaoense for plant growth promotion and biological control of gummy stem blight and wilt of muskmelon. Front. Microbiol. 12:634772. doi: 10.3389/fmicb.2021.634772, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil M. P., Patil R. H., Patil S. G., Maheshwari V. L. (2014). Endophytic mycoflora of Indian medicinal plant, Terminalia arjuna and their biological activities. Int. J. Biotech. Well. Indus. 3, 53–61. doi: 10.6000/1927-3037.2014.03.02.3 [DOI] [Google Scholar]
- Phạm H. T. T., Suwannapan W., Koomsiri W., Inahashi Y., Také A., Matsumoto A., et al. (2020). Fodinicola acaciae sp. nov., an endophytic actinomycete isolated from the roots of Acacia mangium Willd. And its genome analysis. Microorganisms 8:467. doi: 10.3390/microorganisms8040467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotti M., Di Lernia G., Modesti V., Lumia V., Brunetti A. (2016). Outcome of Ceratocystis platani inoculations in Platanus× acerifolia in relation to season and inoculum dose. For. Biogeo. For. 9, 608–617. doi: 10.3832/ifor1594-008 [DOI] [Google Scholar]
- Pinyopusarerk K., Liang S. B., Gunn B. V. (1993). “Taxonomy, distribution, biology and use as an exotic” in Acacia mangium, growing and utilization. FAO, Bangkok, Thailand, and Winrock international, MPTS monograph series no. eds. Awang K., Taylor D., vol. 3, 1–20.
- Potter K., Rimbawanto A., Beadle C. (2006). Heart rot and root rot in tropical acacia plantations. Proceedings of a workshop held in Yogyakarta, Indonesia, 7–9 February 2006. Canberra, ACIAR Proceedings No. 124.
- Qian C. D., Fu Y. H., Jiang F. S., Xu X. H., Cheng D. Q., Ding B., et al. (2014). Lasiodiplodia sp. ME4-2, an endophytic fungus from the floral parts of Viscum coloratum, produces indole-3-carboxylic acid and other aromatic metabolites. BMC Microbiol. 14:297. doi: 10.1186/s12866-014-0297-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy K., Lim S. M., Bakar H. A., Ismail N., Ismail M. S., Ali M. F., et al. (2010). Antimicrobial and cytotoxic activities of Malaysia endophytes. Phytother. Res. 24, 640–643. doi: 10.1002/ptr.2891, PMID: [DOI] [PubMed] [Google Scholar]
- Robinson J. G., Nedergaard B. S., Rogers W. J., Fialkow J., Neutel J. M., Ramstad D., et al. (2014). Effect of evolocumab or ezetimibe added to moderate-or high-intensity statin therapy on LDL-C lowering in patients with hypercholesterolemia: the LAPLACE-2 randomized clinical trial. J. Am. Med. Assoc. 311, 1870–1883. doi: 10.1001/jama.2014.4030 [DOI] [PubMed] [Google Scholar]
- Roux J., Wingfield M. J. (2009). Ceratocystis species: emerging pathogens of non-native plantation eucalyptus and acacia species. South. For. 71, 115–120. doi: 10.2989/SF.2009.71.2.5.820 [DOI] [Google Scholar]
- Saikkonen K., Faeth S. H., Helander M., Sullivan T. J. (1998). Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 29, 319–343. doi: 10.1146/annurev.ecolsys.29.1.319 [DOI] [Google Scholar]
- Sarah Shafiei S. N., Ahmad K., Ikhsan N. F. M., Ismail S. I., Sijam K. (2017). Antibacterial activity of acacia spp. leaves extracts against Xanthomonas oryzae pv. Oryzae and screening for active phytochemical contents. J. Agric. Vet. Sci. 10, 49–60. [Google Scholar]
- Sharon E., Bar-Eyal M., Chet I., Herrera-Estrella A., Kleifeld O., Spiegel Y. (2001). Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Phytopathology 40, 2016–2020. doi: 10.1016/j.soilbio.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Siameto E. N., Okoth S., Amugune N. O., Chege N. C. (2010). Antagonism of Trichoderma harzianum isolates on soil borne plant pathogenic fungi from Embu District, Kenya. J. Yeast Fungal Res. 1, 47–54. [Google Scholar]
- Sopialena S., Suyadi S., Sahil M., Nurdiana J. (2018). The diversity of endophytic fungi associated with Piper nigrum in the tropical areas: a recent study from Kutai Kartanegara, Indonesia. Bio. J. Biol. Diversity 19, 2028–2034. doi: 10.13057/biodiv/d190607 [DOI] [Google Scholar]
- Syazwan S. A., Mohd-Farid A., Wan-Muhd-Azrul W.-A., Syahmi H. M., Zaki A. M., Ong S. P., et al. (2021). Survey, identification, and pathogenicity of Ceratocystis fimbriata Complex associated with wilt disease on Acacia mangium in Malaysia. Forests 12:1782. doi: 10.3390/f12121782 [DOI] [Google Scholar]
- Tarigan M., Roux J., Van Wyck M., Tjahjono B., Wingfield M. J. (2011). A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. S. Afr. J. Bot. 77, 292–304. doi: 10.1016/j.sajb.2010.08.006 [DOI] [Google Scholar]
- Udarbe M. P., Hepburn A. J. (1986). “Development of Acacia mangium as plantation species in Sabah” in Australian acacias in developing countries. Proceedings of international workshop held at the forestry training Centre, Gympie. ed. Turnbull J. W., vol. 16 (Queensland, Australia: Canberra, ACIAR Proceedings; ), 157–159. [Google Scholar]
- Warman N. M., Aitken E. A. (2018). The movement of fusarium oxysporum f. sp. cubense (sub-tropical race 4) in susceptible cultivars of banana. Frontiers. Plant Sci. 9:1748. doi: 10.3389/fpls.2018.01748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocol. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wyckhuys K. A. G., Lu Y., Morales H., Vazquez L. L., Legaspi J. C., Eliopoulos P. A., et al. (2013). Current status and potential of conservation biological control for agriculture in the developing world. Biol. Control 65, 152–167. doi: 10.1016/j.biocontrol.2012.11.010 [DOI] [Google Scholar]
- Yazid S. N. E., Selamat J., Ismail S. I., Magan N., Samsudin N. I. P. (2020). Phytopathogenic organisms and mycotoxigenic fungi: why do we control one and neglect the other? A biological control perspective in Malaysia. Compr. Rev. Food Sci. Food Saf. 19, 643–669. doi: 10.1111/1541-4337.12541, PMID: [DOI] [PubMed] [Google Scholar]
- Yoo J. J., Eom A. H. (2012). Molecular identification of endophytic fungi isolated from needle leaves of conifers in Bohyeon Mountain. Korea. Mycobiol. 40, 231–235. doi: 10.5941/MYCO.2012.40.4.231, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria L., Jamil M. I. M., Anuar I. S. M. (2016). Molecular characterisation of endophytic fungi from roots of wild banana (Musa acuminata). Trop. Life Sci. Res. 27, 153–162. PMID: [PMC free article] [PubMed] [Google Scholar]
- Zakaria L., Yaakop A. S., Salleh B., Zakaria M. (2010). Endophytic fungi from paddy. Trop. Life Sci. Res. 21, 101–107. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.