Abstract
Protein‐based pharmaceuticals are increasingly important, but their inherent instability necessitates a “cold chain” requiring costly refrigeration during production, shipment, and storage. Drying can overcome this problem, but most proteins need the addition of stabilizers, and some cannot be successfully formulated. Thus, there is a need for new, more effective protective molecules. Cytosolically, abundant heat‐soluble proteins from tardigrades are both fundamentally interesting and a promising source of inspiration; these disordered, monodisperse polymers form hydrogels whose structure may protect client proteins during drying. We used attenuated total reflectance Fourier transform infrared spectroscopy, differential scanning calorimetry, and small‐amplitude oscillatory shear rheometry to characterize gelation. A 5% (wt/vol) gel has a strength comparable with human skin, and melts cooperatively and reversibly near body temperature with an enthalpy comparable with globular proteins. We suggest that the dilute protein forms α‐helical coiled coils and increasing their concentration drives gelation via intermolecular β‐sheet formation.
Keywords: calorimetry, desiccation tolerance, Fourier transform infrared spectroscopy, gels, principle component analysis, protein disorder, protein–protein interactions, rheometry, tardigrades
1. INTRODUCTION
Proteins are only marginally stable. 1 The advent of protein‐based pharmaceuticals, diagnostics, and industrial enzymes has brought this problem to the fore. Anhydrobiosis, the ability to survive desiccation, is found in every Kingdom of life. 2 , 3 This state is achieved via several mechanisms, but primarily by preventing the loss of function upon rehydration. 4 Similarly, the chemical and pharmaceutical industries exploit a variety of compounds from trehalose to bovine serum albumin to stabilize dried proteins. The discovery of desiccation‐tolerance proteins in tardigrades, 5 amazing animals that survive a variety of stresses, 6 sparked interest in new methods for stabilizing proteins. 7 , 8 , 9 , 10 Here, we analyze the structure and physical properties of a particular tardigrade desiccation‐tolerance protein as a function of temperature and concentration to devise a hypothesis about its ability to form gels and protect dry client proteins. 1
The low sequence similarity of tardigrade cytosolic abundant heat soluble (CAHS) proteins to other proteins 5 , 7 hints at novel mechanisms to expand the toolbox of protective molecules. 11 CAHS proteins are intrinsically disordered, 5 , 7 confer desiccation tolerance to other organisms, stabilize enzymes, 8 , 10 , 12 and are biocompatible, 13 suggesting a potential avenue for eliminating the cold chain.
The protein used here, CAHS D, phase separates and fibrillates when expressed in higher eukaryotic cells and can form gels in buffer. 14 , 15 , 16 Predictions based on primary structure reveal three regions, N‐terminal and C‐terminal domains (residues 1–90 and 196–227) and a large linker (91–195; Figure 1). The linker is predicted to comprise amphiphilic α‐helices; 5 , 15 nuclear magnetic resonance spectroscopy (NMR) data show their locations. 15 The terminal domains are more disordered but have β‐sheet propensity (Figure 1). 18
FIGURE 1.
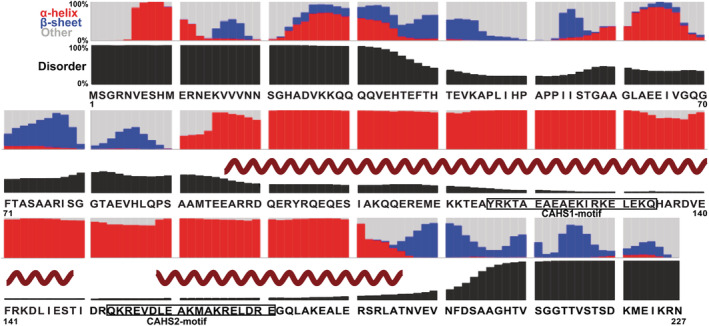
Primary structure analysis of CAHS D; 17 , 18 Helix from NMR data; 15 CAHS1 and 2 motifs from analysis of conserved repeats. 5 CAHS, cytosolic abundant heat soluble protein
We used rheology (from the Greek for flow) to study the physical properties of soft matter. More specifically, the technique provides information about how a substance responds to an applied force in terms of its storage modulus and the loss modulus. The storage modulus reveals how energy is stored, which in hydrogels is related to crosslink density. 19 The loss modulus reveals how energy is dispersed. If the storage modulus exceeds the loss modulus, the sample behaves more like a solid, if the loss modulus is higher the sample behaves like a liquid. These factors and their response to the frequency of the applied force and to temperatures provide data on the samples microstructure. 20
The secondary structure of a gel can be inferred through attenuated total reflectance Fourier transform infrared (ATR‐FTIR) spectroscopy, which is one of the few techniques that works on dry, gelled, and solution samples, The amide 1 region (1,600–1,700 cm−1) corresponds to the amide‐carboxyl hydrogen bond strength throughout a protein's mainchain. The angle with which the amide and carboxyl bond has a large effect on the bond strength. Different secondary structures have canonical bond angles, which affect FTIR, in terms of the frequency and linewidth of peaks within the Amide 1 region. The presence of multiple secondary structures with similar dihedral angles necessitates spectral deconvolution and requires careful analysis to attribute‐specific subpeaks to appropriate structures.
2. RESULTS AND DISCUSSION
2.1. Bulk properties
CAHS D gels are clear (Figure 2a), solidify rapidly below the gelation point and are 95%–97% water by mass. Gelation is reversible; repeated heating and cooling do not affect the properties of the gel if evaporation is prevented (Figure S1). The gels are self‐healing; small amounts of heat or sustained pressure are sufficient to restore the integrity of a torn gel.
FIGURE 2.
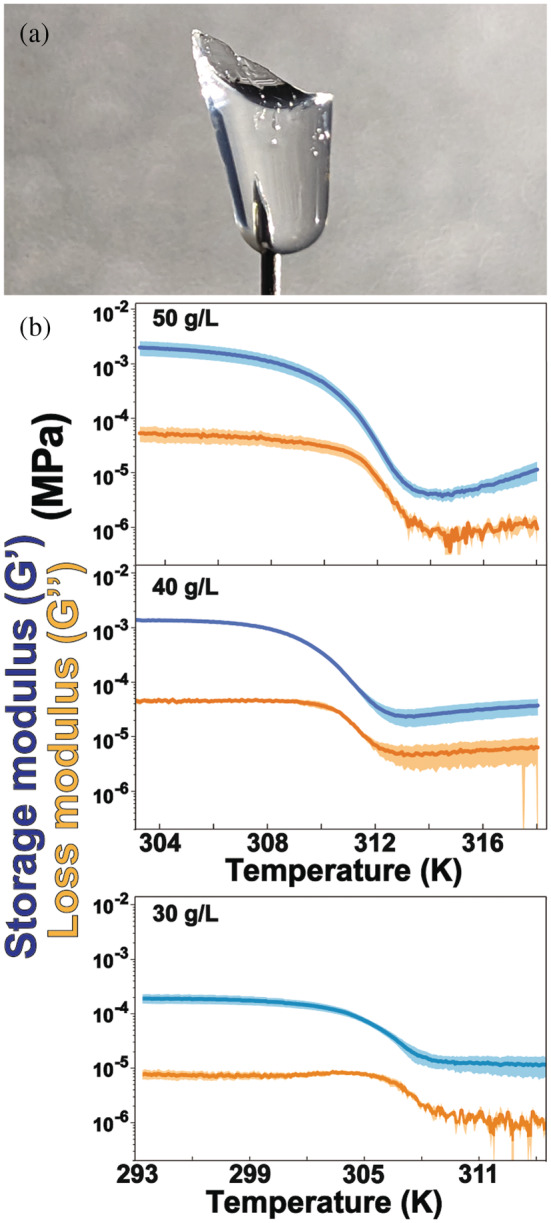
(a) Image of 50 g/L CAHS D gel on a needle. (b) Heating‐induced dissociation was monitored by measuring storage‐moduli (blue) and loss‐moduli (orange), which are shown with their SDs of the mean (shaded regions). The lower concentration gels (30 g/L) have lower liquification points; therefore, data collection began at a lower temperature. Conditions: 30–50 g/L CAHS D gel, 13.7 mM NaCl, 0.27 mM KCl, 0.8 mM Na2HPO4, 0.2 mM KH2PO4, and pH 7.4. CAHS, cytosolic abundant heat soluble protein
Phase separation does not occur under our conditions (Figure S2) yet the primary structure criteria suggest that CAHS D is a random coil, and the number of aromatic‐, positive, negative, polar, and nonpolar residues falls within the bounds of a homotypic weakly phase‐separating protein. 21 However, the formidable linker length (104 residues) and low valence increase the probability of gelation without phase separation, 22 an observation supported by SAXS data indicating a sharp transition between gel and liquid phases. 15 Phase separation inside cells suggests a requirement for macromolecular crowding. 23 , 24
We used small‐amplitude oscillatory shear rheometry to characterize the stiffness, strength, and other properties 25 , 26 of CAHS D gels as a function of concentration and temperature (Figure 2b, Table 1). At lower temperatures, the storage modulus, G', a measure of stiffness, rises steeply from 10−4 to 10−3 MPa between 30 and 40 g/L protein with a more modest increase between 40 and 50 g/L. G' is independent of frequency from 0.1 to 100 Hz, while the dynamic viscosity decreases linearly (Figure S3). This independence indicates a strong gel, 25 meaning it retains the shape of its container (Figure 2a).
TABLE 1.
Properties of CAHS D gels (13.7 mM NaCl, 0.27 M KCl, 0.8 mM Na2HPO4, 0.2 mM KH2PO4, and pH 7.4)
| CAHS D (g/L) | Transition temperature, T m (K) | (kcal/Mol) | G'(MPa) | ||
|---|---|---|---|---|---|
| FTIRPC1 | DSC | Rheology | DSC | Rheology | |
| 30 | 300 ± 100 | ND a | 303.3 ± 0.9 | ND a | 2.0 ± 0.5 × 10−4 |
| 40 | 308.3 ± 0.3 | 308.2 ± 0.1 | 309.4 ± 0.2 | 150 ± 10 | 1.49 ± 0.06 × 10−3 |
| 50 | 310 ± 2 | 309.91 ± 0.06 | 309.86 ± 0.07 | 130 ± 10 | 2.0 ± 0.6 × 10−3 |
Note: FTIR data are from fitting a two‐state reaction using the integrated Gibbs‐Helmholtz equation with ΔC p o' set to zero. Uncertainties from FTIR are 95% confidence intervals. Uncertainties from DSC and rheology are the SDs of the mean from triplicate analysis.
Abbreviations: CAHS, cytosolic abundant heat soluble; DSC, differential scanning calorimetry; FTIR, Fourier transform infrared.
ND, Sample did not provide usable data.
We then studied the heat‐induced transition. The onset of liquification is the temperature where the storage modulus begins to decrease. 27 For our gels, the storage modulus always exceeds the loss modulus. This observation indicates the persistence of gel structure even after liquification, 26 but at concentrations ≤20 g/L the gel is too weak to give reliable data (<0.5 × 10−5 MPa; Figure S4). In summary, the protein retains at least some structure at all temperatures examined. To put the rheology data into perspective, the stiffness of a 50 g/L (5% wt/vol) CAHS D gel near room temperature is comparable with that of a 4.5% wt/vol collagen gel, which is about twice the collagen concentration in a serving of Jell‐O brand gelatin. However, the tardigrade protein gel has a higher liquification point. 28 , 29
Like globular proteins, physical protein gels can be stabilized by both hydrogen bonds and hydrophobic interactions. Trifluoroethanol strengthens the gel indicating melting in our samples occurs by the breaking (dissociation) of intramolecular and intermolecular hydrogen bonds, 16 , 30 and is corroborated by the enthalpy change (~120 kcal/mol) from the differential scanning calorimetry (DSC)‐measured endotherm (Table 1) which is similar to the enthalpy change for an average globular protein of similar melting temperature and size. 31 The observation of a single feature in the thermogram (Figure S5) with a mid‐point temperature matching the FTIR data (Figure 3e) indicates that the structural rearrangement and melting are coupled, cooperative, and consistent with a two‐state reaction. 1
FIGURE 3.
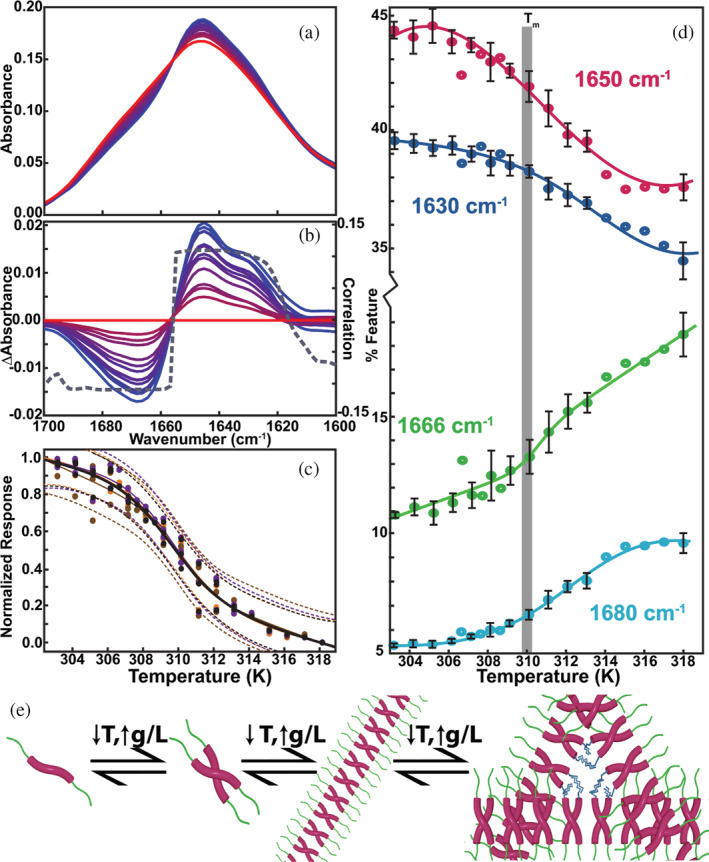
(a) Spectra of a 50 g/L CAHS D from 303 K (blue) to 318 K (red). (b) Difference spectra relative to the 318 K spectrum and loading plot of the first principal component (PC1, gray curve). (c) Melting curves with two‐state fits constructed using absorbance data (orange, 1,502 cm−1; brown, 1,622 cm−1; purple, 1,644 cm−1) and PC1 (black) with 95% confidence intervals (dashes). (d) Calculated percentage of various features from iterative peak fitting of Voigt profiles. Conditions: 13.7 mM NaCl, 0.27 M KCl, 0.8 mM Na2HPO4, 0.2 mM KH2PO4, and pH 7.4 (e) Presumed model of gelation, from monomer to oligomers that form gelation‐competent fibrils. CAHS, cytosolic abundant heat soluble protein.
FTIR spectra were acquired at several temperatures and protein concentrations to understand changes in structure associated with melting. The amide 1‐regions and amide 2‐regions (1,500–1,700 cm−1) were processed, and their components of maximal variance calculated. 32 The wavelength dependence of the first principal component (PC1) is consistent with the difference spectra acquired between 303 and 318 K. PC1 embodies the main eigenvector, which explains 65% of the variance (Figure S6). The isosbestic point at 1,655 cm−1 is consistent with two‐state melting (Figure 3a,b). The midpoint temperatures (T m) from fitting PC1 scores and intensities at specific wavenumbers to a two‐state reaction with linear pretransition and posttransition baselines 33 are within uncertainty of one another (Figure 3c). The T m values were also derived from fitting secondary structure transitions (Figure 3d). The values are within the uncertainty of those from the PC1 scores (Figure 3c). The T m is like that from both DSC (Figure S6) and the liquification temperature from rheology (Figure 3c, Table 1). The estimated standard‐state enthalpy of liquification, , from FTIR and rheology data is positive, and consistent with DSC (Table 1), despite the paucity of points in the transition.
CAHS D forms clear, strong, reversible, self‐healing gels that set on cooling, but unlike gelatin and sodium alginate, CAHS D gels set without heat or cosolute‐induced denaturation. 34 From these characteristics, we suggest that CAHS D forms a flexible fine‐stranded hydrogel. 35 The 10‐fold increase in G' from 30 to 40 g/L (Figure 1b) follows from the rearrangement in structure seen in the FTIR data. This increase is mirrored in the increased T m, where large changes are seen between 20 and 40 g/L and plateauing near body temperature at 50 g/L (Table 1).
2.2. Structural properties
Secondary structure was assessed by fitting Voigt profiles 36 to the amide 1 region for spectra acquired from 303 to 318 K at a variety of concentrations (Figure S7). The centers of the fits were identified from maxima in second derivative spectra (Figure S7). We fitted four distinct peaks at 1,630, 1,650, 1,666, and 1,680 cm−1, which can be assigned based on context. The 1,630 and 1,680 cm−1 are classic β‐sheets while the 1,666 cm−1 feature is assigned to turns. The large line width of the ~1,650 cm−1 peak suggests overlap of random coil and α‐helix features (Figure S8). 37
As temperature increases, the content of low‐frequency features (~1,630 and ~1,650 cm−1) decreases and the content of high frequency features (~1,680 and ~1,666 cm−1) increases (Figure 3). The temperature loading plot for PC1 suggests that at low temperatures the 1,650 cm−1 feature is dominated by α‐helix content, as the amide 2 region displays a sharp peak at 1,550 cm−1 (Figure S9). 36 , 38 The 1,550–1,600 cm−1 region is associated with the random coil component and is negative, indicating that coil content increases with increasing temperature, despite the net decrease in area, which means that as temperature increases there is a shift from α‐helix to random coils independent of the decrease in area.
The combination of features that increase on cooling‐induced gelation, 1,650 and 1,630 cm−1, are associated with coiled coils. 39 The decreasing 1,666 and 1,680 cm−1 features are associated with turns and antiparallel β‐sheets, respectively. 40 The high temperature baseline for the 1,666 cm−1 is positive (Figure 3d), suggesting that as the temperature increases past melting other features transition into turns.
The concentration dependence of the FTIR data acquired below T m provides complementary information about structure changes in the gel (Figure S8). Increasing the CAHS D concentration from 20 to 50 g/L decreases the high‐frequency (1,680 cm−1) and increases the low‐frequency (1,630 cm−1) features (Figure S10). The central features (1,650 and 1,666 cm−1) are nearly unchanged (Figure S10). The 1,630 cm−1 peak suggests a 3‐to‐4 strand β‐sheet, 40 and the 1,650 cm−1 feature is assigned to α‐helix. 37 Thus, coil‐like features are temperature‐, but not concentration‐ dependent, while β‐sheets content depends on both concentration and temperature.
Taken together, the NMR, rheological, and FTIR data suggest that upon cooling at high CAHS D concentrations, random coils change to coiled coils, forming the backbone of the gel. The concentration dependence of coiled coil formation is consistent with NMR observations. 15 Primary structure analysis 18 suggests the termini 15 have a propensity to form β‐strands (Figure 1). The low temperature, concentration dependent FTIR data suggest that gel formation involves intermolecular β‐sheet formation, probably via the termini. These data suggest that the α‐helical coiled coils comprise the backbone of a gelation‐competent oligomer with low‐valency sticky ends (Figure 3e).
3. CONCLUSION
Gelation is driven by the transition from sticky intramolecular β‐structures, identified as the high‐frequency FTIR features at the termini, to intermolecular β‐sheets. These low‐frequency spectral features are broad, 41 suggesting a distribution of nonspecific interactions between terminal β‐sheets, which allow formation of junctions in a variety of geometries.
We speculate that the surrounding gel protects client proteins from irreversible denaturation by forming a “molecular shield,” 42 which persists and is enhanced by drying. 10 The gel's stability up to body temperature, strength near that of human skin, 43 self‐healing properties, biocompatibility, 13 and ability to protect enzyme activity 8 make CAHS D a promising candidate for a tunable delivery system for stabilizing protein‐based drugs.
4. MATERIALS AND METHODS
4.1. Purification and sample preparation
The Escherichia coli codon‐optimized structural gene for CAHS D with an N‐terminal histidine tag was synthesized (Integrated DNA Technologies) and cloned into a pET28b expression vector and the resulting vector transformed into BL21star (DE3) E. coli cells. A single bacterial colony was used to inoculate 150 ml of Lennox broth (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) supplemented with 60 g/L kanamycin. The culture was shaken at 37°C overnight (New Brunswick Scientific Innova I26, 225 rpm). Flasks (1 L) of Lennox Broth were inoculated with 15 ml of overnight culture and shaken at 37°C until the optical density at 600 nm reached 0.6. IPTG (1 mM final concentration) was added to induce expression.
After 3 h, the cells were pelleted at 1,000 gfor 30 min at 10°C. The pellets were resuspended in 10 ml of a solution comprising 20 mM Tris, 50 mM NaCl, pH 7.4, then stored overnight at 20°C. Cells were thawed in a 37°C water bath and then lysed via heat shock by boiling in a beaker of water for 15 min. Lysates were cooled and the cell debris pelleted by centrifugation at 20,000 gfor 30 min at 10°C. The lysate was mixed 1:1 with HUa buffer (3 M urea, 20 mM Na2HPO4, 500 mM NaCl, 10 mM imidazole, and pH 7.4) and filtered (45 μm).
CAHS D was purified using a HisTrap HP column (Cytiva) by gradient elution from 0% to 30% HUb (3 M urea, 20 mM Na2HPO4, 500 mM NaCl, 150 mM imidazole, and pH 7.4) over 30 column volumes using an AKTA Start (Cytiva) FPLC. Fractions were visualized on a Criterion™ TGX™ Precast gel (Bio‐Rad, Hercules, CA) with Coomassie blue staining. Fractions showing one band indicative of CAHS D were loaded into SnakeSkin™ Dialysis Tubing 10 k MWCO (Thermo Fisher, Waltham, MA) and dialyzed against TEV buffer (5 mM Tris and 0.5 mM ethylenediaminetetraacetic acid), until the concentration of urea was <2 M. Dithiothreitol was added to a final concentration of 1 mM immediately prior to adding 1 mg of TEV protease. The samples were then placed on an orbital shaker for 16 h. The samples were then diluted 1:1 with HUa and filtered (0.45 μm) before being loaded on a HisTrap column to remove the protease and remaining tagged protein. The flowthrough was collected and dialyzed twice against phosphate buffer (13.7 mM NaCl, 0.27 KCl, 0.8 mM Na2HPO4, 0.2 mM KH2PO4, and pH 7.4), and then against distilled and deionized water (17.4 Ω) until the calculated concentration of all non‐protein components was <1 nM. Purified CAHS D was flash frozen in an ethanol‐dry ice bath and lyophilized.
Samples were prepared by dissolving lyophilized CAHS D in buffer and alternating heat treatment in a 42°C heat block and brief treatment at 100°C before vortexing and pulse centrifugation on a tabletop centrifuge to pellet undissolved protein. Samples were then melted and aliquoted with a positive displacement pipette. CAHS D concentration was quantified with the Pierce Coomassie Plus assay using a bovine serum albumin standard (Thermo Fisher) in 96‐well plates. 44 Aliquots were then diluted to the desired concentrations.
4.2. Attenuated total reflectance Fourier transform infrared
FTIR spectra were recorded on a dry‐air purged BioTools Prota‐3 S spectrophotometer equipped with a HgCdTe detector and a ZnSe‐diamond attenuated ATR crystal. Spectra were acquired from 805 to 5,500 cm−1 at 4 cm−1 resolution, using 100 scans. Background, buffer (13.7 mM NaCl, 0.27 KCl, 0.8 mM Na2HPO4, 0.2 mM KH2PO4, and pH 7.4), and vapor spectra were acquired at the appropriate temperatures using a Peltier temperature controller (Pike).
Empty, path‐, buffer‐, and sample‐ spectra were acquired and preprocessed for each sample using Prota3s (BioTools) software. The background spectrum was subtracted from both the buffer‐ and sample‐ spectra to produce absorbance spectra. The buffer absorbance spectrum was then subtracted from the sample absorbance spectrum in the same proportion for all samples. To ensure complete buffer subtraction, the procedure was performed such that the region at 3,750 cm−1 is non‐negative and the region from 2,000 to 1,800 cm−1 is flat.
Spectra were processed using an Orange Datamining workflow and smoothed using a Savitsky–Golay filter with a window of 5 to remove fine noise and 4‐component principal component analysis denoising. 45 , 46 Smoothed bands were processed using a positive rubber band baseline‐correction and vector‐normalization. Difference spectra were generated by subtracting the smoothed highest temperature spectrum from the remaining spectra for a given concentration. The smoothed spectra were fit with Voigt profiles using non‐linear least‐squares regression. 36 Peaks were assigned to secondary structures and their areas normalized against the total peak area (excluding contribution from aromatics and amide 2) and multiplied by 100 to obtain the percent of each type (Figure S8).
Second derivative spectra were calculated using a Savitsky–Golay filter with a window size of 11 points and a second‐order polynomial. 47 The principal components of the smoothed spectra were calculated and the loading plots generated to assess the wavelength dependence of maximal variance. The PCA scoring at each temperature was then fit to a two‐state transition, derived from Equations (6) to (9) in reference 33 using MATLAB to estimate the melting temperature, T m and van't Hoff enthalpy of melting,
4.3. Rheology
Small angle oscillatory shear rheological data were acquired using a TA Instruments strain‐controlled ARES‐G2 rheometer with a 25‐mm parallel plate geometry. Samples were warmed in a 60°C oven for 5 min and the stage heated to 45°C. Samples (450 μl) were applied as the stage slowly rotated to ensure even application, without bubbles. The shear gap was set to ensure a flat edge and excess sample removed. A layer of silicon oil (10 MPa·sec, Sigma 146153) was placed around the edge to prevent evaporation. 48 A strain sweep was conducted to identify the best strain for our experiments, 0.1%. The sample was cured for 2 h below the gel point to find the plateau modulus. The temperature was then ramped at 0.5°C/min, with frequency scans from 1 to 100 Hz at 30°C and in the region where tan(δ) reached a maximum.
4.4. Differential scanning calorimetry
CAHS D gels were prepared as described above. Samples (20 μl) weighing ~19 mg were sealed in Tzero Hermetic Aluminum pans and loaded into a TA Instruments DSC 250 equipped with a TA Instruments Refrigerated Cooling System 90. An identical pan containing 18 μl of buffer was used as a reference. The sample cell was subject to a N2(g) (50 ml/min) purge. To eliminate differing thermal histories, samples were heated at 10°C/min to 50°C, then cooled at 10°C/min to 10°C, and held there for 10 min. 49 , 50 Samples were then heated at 1.5°C/min to 50°C and kept there for 2 min. Thermograms were analyzed using Trios V5.1.0.56403 software, with the minimum of the denaturation endothermic peak reported as the transition (melting) temperature (T m) and the modified standard‐state enthalpy of melting ΔH m o' as the area of the endotherm. 49 , 51 Samples were then cooled at 10°C/min to 10°C and heated at 1.5°C/min to ~10°C above their melting temperature to assess reversibility. The resulting T m values were within 0.5°C and ΔH m o' values were within 5% of those measured from the first scan.
4.5. Fluorescence assay
A 500 μL sample of CAHS D (10 g/L) was prepared as described in Section 4.1. Atto 488 NHS ester (1 μL of 10 mM) was added and allowed to react for 30 min at room temperature. Samples were aliquoted into each well of a glass‐bottomed plate and diluted to a final volume of 100 μL. Fluorescence was assessed on a Nikon Ti‐E stand equipped with a Yokogawa CSU‐W1 spinning disc confocal unit, a Plan‐Apochromat 100x/1.49 NA oil immersion objective, a Zyla sCMOS camera (Andor), and Nikon NIS‐Elements software version 4.60, and a 488 nm laser.
AUTHOR CONTRIBUTIONS
Jonathan E. Eicher: Conceptualization (equal); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); software (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (equal). Julia A. Brom: Data curation (supporting); formal analysis (supporting); investigation (supporting); validation (supporting); visualization (supporting). Joanna M. Atkin: Conceptualization (supporting); supervision (supporting). Shikun Wang: Validation (supporting). Sergei S. Sheiko: Conceptualization (supporting); funding acquisition (supporting); resources (supporting). Gary J. Pielak: Conceptualization (equal); funding acquisition (lead); project administration (lead); resources (lead); supervision (lead); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Appendix S1. Supporting Information
ACKNOWLEDGEMENTS
We thank Rafael F. Irgolič for automating FTIR data acquisition, Rina Dukor and Juanita Sanchez for FTIR support, Luke Hatfield for Figure 3e, Foad Vashahi for advice on rheology, Wilton T. Snead for assistance with microscopy, and Elizabeth Pielak for comments on the article. This work was supported by NIH grant R01GM127291 (G.J.P.) and NSF grants CHE‐2203505 (G.J.P.), DMR 1921835 (S.S.S.), and DMR 2004048 (S.S.S.).
Eicher JE, Brom JA, Wang S, Sheiko SS, Atkin JM, Pielak GJ. Secondary structure and stability of a gel‐forming tardigrade desiccation‐tolerance protein. Protein Science. 2022;31(12):e4495. 10.1002/pro.4495
Review Editor: Jean Baum
Funding information National Institute of General Medical Sciences, Grant/Award Number: R01GM127291; National Science Foundation, Grant/Award Numbers: CHE 2203505, DMR 1921835, DMR 2004048
DATA AVAILABILITY STATEMENT
Data are available upon reasonable request to the corresponding author.
REFERENCES
- 1. Creighton TE. The biophysical chemistry of nucleic acids and proteins. Eastbourne, East Sussex: Helvetian Press, 2010. [Google Scholar]
- 2. Hand SC, Menze MA, Toner M, Boswell L, Moore D. LEA proteins during water stress: Not just for plants anymore. Annu Rev Physiol. 2011;73:115–134. [DOI] [PubMed] [Google Scholar]
- 3. Boothby TC, Pielak GJ. Intrinsically disordered proteins and desiccation tolerance: Elucidating functional and mechanistic underpinnings of anhydrobiosis. Bioessays. 2017;39:1700119. [DOI] [PubMed] [Google Scholar]
- 4. Barrett J. Metabolic responses to anabiosis in the fourth stage juveniles of Ditylenchus dipsaci (Nematoda). Proc R Soc B: Biol Sci. 1982;216:159–177. [Google Scholar]
- 5. Yamaguchi A, Tanaka S, Yamaguchi S, et al. Two novel heat‐soluble protein families abundantly expressed in an anhydrobiotic tardigrade. PLoS One. 2012;7:e44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schill RO. Water bears: The biology of tardigrades. Germany: Springer International Publishing, 2018. [Google Scholar]
- 7. Boothby TC, Tapia H, Brozena AH, et al. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell. 2017;65:975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piszkiewicz S, Gunn KH, Warmuth O, et al. Protecting activity of desiccated enzymes. Protein Sci. 2019;28:941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brom JA, Pielak GJ. Desiccation‐tolerance and globular proteins adsorb similar amounts of water. Protein Sci. 2022;31:e4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crilly CJ, Brom JA, Warmuth O, Esterly HJ, Pielak GJ. Protection by desiccation‐tolerance proteins probed at the residue level. Protein Sci. 2022;31:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piszkiewicz S, Pielak G. Protecting enzymes from stress‐induced inactivation. Biochemistry. 2019;58:3825–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crilly CJ, Eicher JE, Warmuth O, Atkin JM, Pielak GJ. Water's variable role in protein stability uncovered by liquid‐observed vapor exchange NMR. Biochemistry. 2021;60:3041–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esterly HJ, Crilly CJ, Piszkiewicz S, Shovlin DJ, Pielak GJ, Christian BE. Toxicity and immunogenicity of a tardigrade cytosolic abundant heat soluble protein in mice. Front Pharmacol. 2020;11:565969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yagi‐Utsumi M, Aoki K, Watanabe H, et al. Desiccation‐induced fibrous condensation of CAHS protein from an anhydrobiotic tardigrade. Sci Rep. 2021;11:21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malki A, Teulon J‐M, Camacho‐Zarco AR, et al. Intrinsically disordered tardigrade proteins self‐assemble into fibrous gels in response to environmental stress. Angew Chem Int Ed. 2022;61:e202109961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanaka A, Nakano T, Watanabe K, et al. Stress‐dependent cell stiffening by tardigrade tolerance proteins that reversibly form a filamentous network and gel. PLoS Biol. 2022;20:e3001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S, Li W, Liu S, Xu J. RaptorX‐property: A web server for protein structure property prediction. Nucleic Acids Res. 2016;44:W430–W435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urban G, Magnan CN, Baldi P. SSpro/ACCpro 6: Almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, deep learning and structural similarity. Bioinformatics. 2022;38:2064–2065. [DOI] [PubMed] [Google Scholar]
- 19. Lv X, Liu C, Shao Z, Sun S. Tuning physical crosslinks in hybrid hydrogels for network structure analysis and mechanical reinforcement. Polymers (Basel). 2019;11(2):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao Y, Mezzenga R. Design principles of food gels. Nature Food. 2020;1:106–118. [DOI] [PubMed] [Google Scholar]
- 21. Saar KL, Morgunov AS, Qi R, et al. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc Natl Acad Sci U S A. 2021;118:e2019053118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harmon TS, Holehouse AS, Rosen MK, Pappu RV. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife. 2017;6:e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. André AAM, Spruijt E. Liquid‐liquid phase separation in crowded environments. Int J Mol Sci. 2020;21:5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Speer SL, Stewart CJ, Sapir L, Harries D, Pielak GJ. Macromolecular crowding is more than hard‐core repulsions. Annu Rev Biophys. 2022;51:267–300. [DOI] [PubMed] [Google Scholar]
- 25. Clark AH, Ross‐Murphy SB. Structural and mechanical properties of biopolymer gels. Berlin Heidelberg: Springer, 1987. [Google Scholar]
- 26. Lopes da Silva J, Rao M, Rheological behavior of food gels . Rheology of fluid and semisolid foods food engineering series. Boston, MA: Springer, 2007;p. 339–401. [Google Scholar]
- 27. Magami SM, Williams RL. Gelation via cationic chelation/crosslinking of acrylic‐acid‐based polymers. Polym Int. 2019;68:1980–1991. [Google Scholar]
- 28. Joly‐Duhamel C, Hellio D, Ajdari DM. All gelatin networks: 2. The master curve for elasticity. Langmuir. 2002;18:7158–7166. [Google Scholar]
- 29. Ahmed J. Rheological properties of gelatin and advances in measurement. In: Ahmed J, Ptaszek P, Basu S, editors. Advances in food rheology and its applications. Sawston: Woodhead Publishing, 2017; p. 377–404. [Google Scholar]
- 30. Belopolskaya TV, Tsereteli GI, Grunina N, Vaveliouk O. DSC study of the postdenaturated structures in biopolymer–water systems. J Therm Anal Calorim. 2012;62:75–88. [Google Scholar]
- 31. Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: A calorimetric study. J Mol Biol. 1974;86:665–684. [DOI] [PubMed] [Google Scholar]
- 32. Huang Y, Cavinato AG, Tang J, Swanson BG, Lin M, Rasco BA. Characterization of sol–gel transitions of food hydrocolloids with near infra‐red spectroscopy. LWT‐Food Sci Technol. 2007;40:1018–1026. [Google Scholar]
- 33. Cohen DS, Pielak GJ. Stability of yeast iso‐1‐cytochrome c as a function of pH and temperature. Protein Sci. 1994;3:1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larsen BE, Bjørnstad J, Pettersen EO, Tønnesen HH, Melvik JE. Rheological characterization of an injectable alginate gel system. BMC Biotechnol. 2015;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Foegeding EA. Food biophysics of protein gels: A challenge of nano and macroscopic proportions. Food Biophysics. 2006;1:41–50. [Google Scholar]
- 36. Sadat A, Joye IJ. Peak fitting applied to Fourier transform infrared and raman spectroscopic analysis of proteins. Appl Sci. 2020;10:5918. [Google Scholar]
- 37. Barth A, Zscherp C. What vibrations tell about proteins. Q Rev Biophys. 2002;35:369–430. [DOI] [PubMed] [Google Scholar]
- 38. De Meutter J, Goormaghtigh E. Amino acid side chain contribution to protein FTIR spectra: Impact on secondary structure evaluation. Eur Biophys J. 2021;50:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heimburg T, Schünemann J, Weber K, Geisler N. FTIR‐spectroscopy of multistranded coiled coil proteins. Biochemistry. 1999;38:12727–12734. [DOI] [PubMed] [Google Scholar]
- 40. Lomont JP, Ostrander JS, Ho J‐J, Petti MK, Zanni MT. Not all β‐sheets are the same: Amyloid infrared spectra, transition dipole strengths, and couplings investigated by 2D ir spectroscopy. J Phys Chem B. 2017;121:8935–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S, Ma J, Xu J. AUCpreD: Proteome‐level protein disorder prediction by AUC‐maximized deep convolutional neural fields. Bioinformatics. 2016;32:i672–i679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wise MJ, Tunnacliffe A. Popp the question: What do LEA proteins do? Trends Plant Sci. 2004;9:13–17. [DOI] [PubMed] [Google Scholar]
- 43. Holt B, Tripathi A, Morgan J. Viscoelastic response of human skin to low magnitude physiologically relevant shear. J Biomech. 2008;41:2689–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. [DOI] [PubMed] [Google Scholar]
- 45. Koziol P, Raczkowska MK, Skibinska J, et al. Comparison of spectral and spatial denoising techniques in the context of high definition FT‐IR imaging hyperspectral data. Sci Rep. 2018;8:14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Toplak M, Read ST, Sandt C, Borondics F. Quasar: Easy machine learning for biospectroscopy. Cell. 2021;10:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gautam R, Vanga S, Ariese F, Umapathy S. Review of multidimensional data processing approaches for raman and infrared spectroscopy. EPJ Tech Instrum. 2015;2:8. [Google Scholar]
- 48. Curtis D, Holder A, Badiei N, et al. Validation of optimal Fourier rheometry for rapidly gelling materials and its application in the study of collagen gelation. J Non‐Newtonian Fluid Mech. 2015;75:253–259. [Google Scholar]
- 49. Djabourov M, Nishinari K, Ross‐Murphy SB, Techniques for the characterization of physical gels . Physical gels from biological and synthetic polymers. Cambridge, England: Cambridge University Press, 2013;p. 18–63. [Google Scholar]
- 50. Qiao C, Wang X, Zhang J, Yao J. Influence of salts in the Hofmeister series on the physical gelation behavior of gelatin in aqueous solutions. Food Hydrocoll. 2021;110:106150. [Google Scholar]
- 51. Gornall JL, Terentjev EM. Helix‐coil transition of gelatin: Helical morphology and stability. Soft Matter. 2008;4:544–549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information
Data Availability Statement
Data are available upon reasonable request to the corresponding author.


