Summary
The mammalian genome encodes thousands of long non-coding RNAs (lncRNAs) many of which are developmentally regulated and differentially expressed across tissues, suggesting their potential roles in cellular differentiation. Despite this expression pattern, little is known about how lncRNAs influence lineage commitment at the molecular level. Here, we demonstrate that perturbation of an ES cell/early embryonic lncRNA, Pluripotency-associated transcript 4 (Platr4), directly influences the specification of cardiac mesoderm lineage differentiation. We show that Platr4 acts as a molecular scaffold or chaperone interacting with the Hippo signaling pathway molecules Yap and Tead4 to regulate the expression of a downstream target gene, Ctgf, which is crucial to the cardiac lineage program. Importantly, Platr4 knockout mice exhibit myocardial atrophy and valve mucinous degeneration, which are both associated with reduced cardiac output and sudden heart failure. Together, our findings provide evidence that Platr4 is required in cardiac lineage specification and adult heart function in mice.
Keywords: Platr4, lncRNA, Ctgf, Tead4, mesendoderm lineage, cardiac function, Hippo signaling pathway
Graphical Abstract
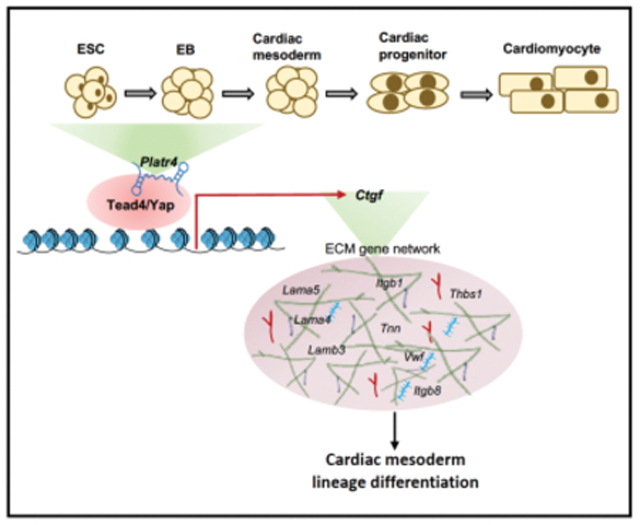
eTOC Blurb
Hazra et al. characterized the detailed molecular mechanism by which an ESC-specific and early embryonic lncRNA, Platr4 functions. They demonstrated that Platr4 interacts with the transcription factor Tead4 and its coactivator Yap to modulate a downstream target gene, Ctgf, which ultimately affects the cardiac mesoderm lineage program and adult heart function.
Introduction
Recent genomic and transcriptomic analyses1–3 have identified that a large part of mammalian genomes encode for long non-coding RNAs (lncRNAs), which are greater than 200 nucleotides in length4, 5. LncRNAs are the product of RNA polymerase II, are capped and often polyadenylated and spliced, but do not code for proteins6. They have been shown to have diverse properties, including cell/tissue specific expression, binding to transcription factors or other regulatory molecules including miRNAs, and regulating transcription or RNA splicing7–10. Hundreds of lncRNAs have been identified in embryonic stem cells (ESCs)8, 11, and a few well-studied examples1, 8, 12 suggest that many of these lncRNAs have critical functions in ESC biology, though in many cases their regulatory roles are not fully understood. ESCs have the capacity to self-renew and differentiate into all three germ layers. Studies in ESCs have shown that lncRNAs are present in the regulatory biological networks that contribute to stemness, self-renewal, and cell fate specification8, 11. Several lncRNAs have been shown to be involved in aspects of differentiation. For example, Eomesodermin (Eomes) expressing the mesendoderm progenitor lncRNA Meteor interacts with Eomes and epigenetically regulates mesendoderm specification and cardiac differentiation13, 14. In addition, Braveheart (Bvht), a heart-specific lncRNA, functions upstream of mesoderm posterior 1 (Mesp1) and regulates a cardiovascular gene network by interacting with SUZ12, a core component of the PRC2 complex15. Despite these tissue/developmental-specific roles of lncRNAs, their function in ESCs during lineage specification and differentiation has yet to be investigated. Interestingly, cardiac-specific lncRNAs have been shown to be involved in cardiac function and diseases16.
Connective tissue growth factor (Ctgf), also known as cellular communication network 2 (Ccn2) is a candidate extracellular matrix (ECM) protein, essential for normal embryonic development and the maintenance of normal cardiac function17–19. Ctgf is a secreted protein expressed in endothelial cells20. Endothelial cells undergo endothelial-to-mesenchymal transition (EndoMT), which is essential for heart valve development and is regulated by the Tgfβ pathway; this pathway can induce both epithelial-to-mesenchymal transition (EMT) and EndoMT21, 22. It has also been demonstrated that Ctgf is critical for inducing the differentiation of many cell types; however, a role in cardiomyocyte specification has not yet been established2, 23, 24. Ctgf is a direct downstream target of Yes-associated protein (Yap) and its cofactors, transcriptional enhanced associate domain (Tead) family members25.
Yap, a major transcriptional co-activator of the Hippo signaling pathway, is critical for early cell fate determination during embryogenesis and is required for the proper differentiation of ESCs26. Yap does not have its own DNA binding domain: it usually interacts with DNA binding transcription factors, such as Teads, to regulate gene transcription27. In mammalian genomes, the Tead gene encodes for four homologues (Tead1-4) that have the same domain structure, though they are expressed in distinct patterns depending on tissue and developmental stage28, indicating their unique functions in biological processes. For example, in mouse ESCs, depletion of Yap and Tead1 or Tead4 downregulates the pluripotency markers Oct4 and Sox2, with the concomitant expression of differentiation markers, such as Brachyury (T), alpha-fetoprotein, and Gata429. Moreover, the Yap/Tead4 complex with p300 activates anti-apoptotic genes and maintains ESC differentiation30. Furthermore, during blastocyst development modulation of Tead4–Yap expression is crucial in cell fate determination of trophectoderm (TE) from the inner cell mass (ICM)26. In human induced pluripotent stem cells, a YAP complex with TEAD3 promotes cardiac mesoderm differentiation into the cardiovascular lineage31, suggesting that in certain contexts, YAP may play an important role in the differentiated state. Taken together, all of these findings suggest that YAP/TEAD complexes and their functional transcription outputs in different contexts are critical for ESC maintenance and differentiation. Interestingly, the recently identified primate-specific lncRNA BANCR, mainly expressed in fetal cardiomyocytes and a downstream target of the YAP/TEAD4 signaling pathway, is critical for cardiomyocyte migration and ventricular enlargement of the primate heart32.
Here, we investigated an ESC-specific lncRNA Platr4 (Pluripotency associated transcript 4), which interacts in a complex containing the Hippo signaling molecules Yap and Tead4 to modulate their downstream target gene, Ctgf, in cardiovascular lineage commitment. Our analysis determine that Platr4 regulates genes at the transcriptional level and that the loss of Platr4 in ESCs results in a significant depletion of Ctgf at both the RNA and protein levels. Moreover, overexpression of Ctgf rescued the Platr4 knockout (KO) phenotype, demonstrating that Platr4 functions in trans and upstream of Ctgf. We further showed that depletion of Platr4 in ESCs affects the downstream differentiation program by modulating the expression level of primitive streak (Gsc, Mixl1), mesoderm (Gata4, Tbx5), and cardiogenic endothelial (Sox17, foxa2, Cdh5, Pecam1, Flk-1, Vwf) markers. In addition, a directed differentiation technique confirmed a ciritcal role of Platr4 in mesoderm specification, especially in the cardiac mesoderm lineage program.
Interestingly, our study further verified that depletion of Platr4 reduced the expression of genes associated with major (collagens, laminin, fibronectin) and minor (matrix metalloproteinases, tenascin-C, thrombospondins) extracellular matrix (ECM) components. ECM components and their interactions are essential for mesoderm specification, especially in endocardial cell specification, which are critical for heart valve development, maturation, and function33, 34. Importantly, deletion of Platr4 in a KO mouse model resulted in a pathophysiological condition of myocardial atrophy, fibrosis, valve defects with fibrocartilaginous metaplasia, and mucinous degeneration, as well as disrupted cardiomyocyte contractility, an overall decrease in cardiac output and sudden death. Together, our findings identify a mechanism of action of a lncRNA with the Hippo signaling pathway molecules Yap and Tead4 regulating the cardiogenic mesoderm lineage differentiation program and adult heart valve development and function.
Results
Platr4 is an ESC-specific and early embryonic lncRNA
Platr4 lncRNA was initially identified in a differential RNA sequencing (RNA-seq) screen, comparing mouse ESCs and their differentiation into neural progenitor cells11. Rapid amplification of cDNA ends (RACE) cloning followed by Sanger sequencing indicated Platr4 to be a 1,034 nucleotide-long polyadenylated RNA encoded from two exons, which is consistent with the findings of our RNA-seq and Northern blot analyses (Figures 1A, 1B and S1A). The exact chromosomal location is chr3:41443239-41447627. Examination of the Platr4 sequence with the Coding Potential Calculator (CPC)35, Coding-Potential Assessment Tool (CPAT)36, and PhyloCSF37 indicated that the Platr4 transcript does not have protein-coding capacity (Figures S1B and S1C). Publicly available ENcylopedia of DNA Elements (ENCODE) RNA-seq datasets of adult mouse tissues showed that Platr4 is expressed in ESCs, but not in neural progenitor cells nor in any adult tissues (Figure 1C). Cellular fractionation of ESCs followed by quantitative RT-PCR (qRT-PCR) demonstrated that Platr4 is predominantly enriched in the nucleus (Figure 1D). Consistent with the nuclear fractionation, single-molecule RNA fluorescent in situ hybridization (smRNA-FISH) identified that Platr4 is predominantly localized in nuclei (Figure 1E). In addition, we performed smRNA-FISH on whole mount and paraffin sections of embryos, ranging from embryonic day 3.0 (E3.0) to 12.5 (E12.5). We found prominent expression of Platr4 in both the trophectoderm and the inner cell mass of the E4.0 embryos (blastocysts) (Figure 1F) and very low expression at E3.0 embryos (morula), consistent with the qRT-PCR data in preimplantation embryos (Figure S1D). We also detected prominent Platr4 expression in the ectoderm, primitive streak, and epiblast at E6.5, very low at E8.5, but not at E10.5 or E12.5 (Figure S1E).
Figure 1: Platr4 is an ESC-specific and early embryonic lncRNA.
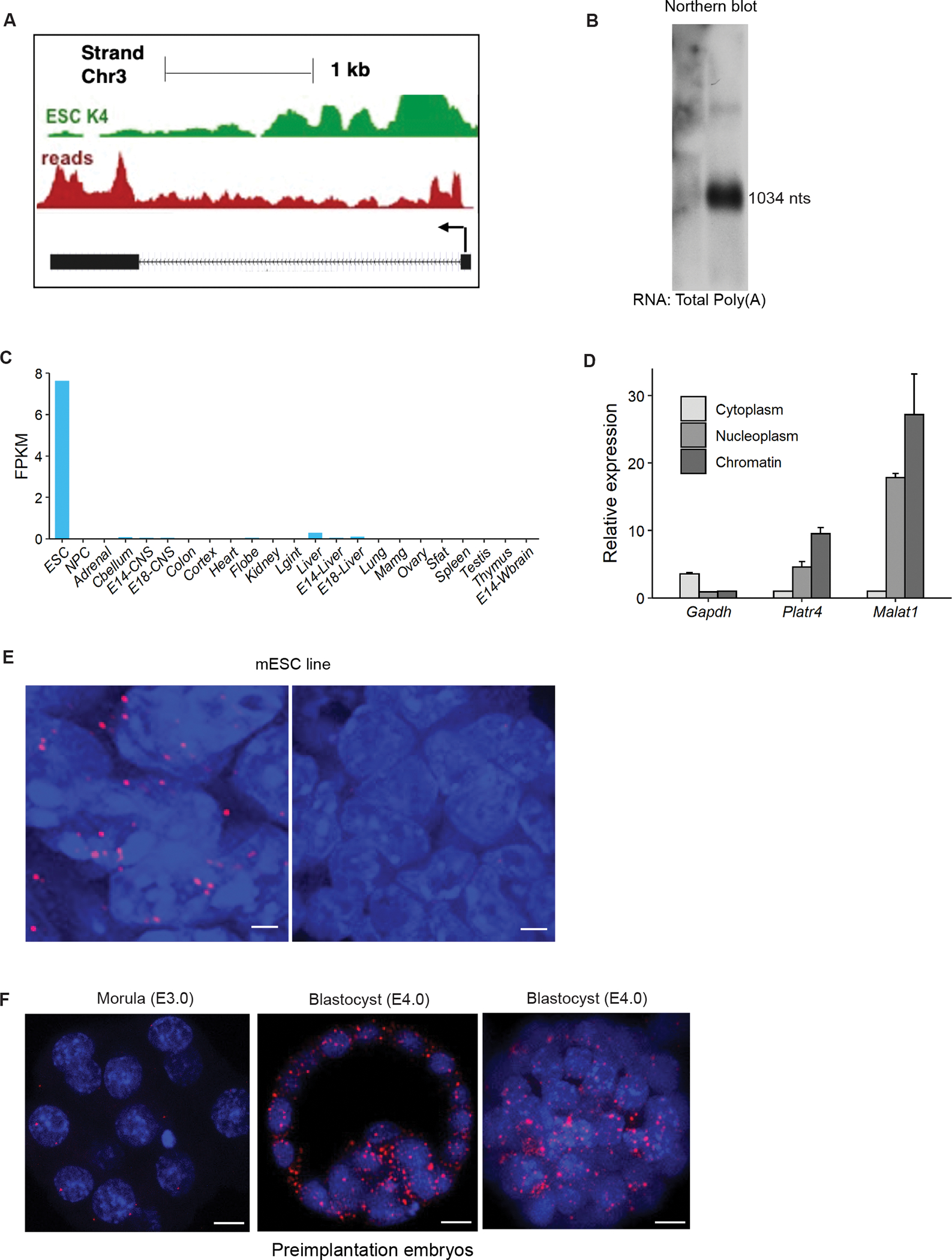
(A) A UCSC genome browser view of the Platr4 locus. Figure also showed ChIP-seq tracks of H3K4me3 and RNA-seq reads of ESCs. (B) Northern blot data showed that Platr4 is 1,034 nucleotides in length. (C) Fragments per kilobase per million mapped reads (FPKM) values for Platr4 in various mouse tissue types from ENCODE data sets. (D) Subcellular fractionation followed by qRT-PCR showed the localization of Platr4 transcripts. Gapdh and Malat1 were used as cytoplasmic and nuclear markers for quality control, respectively. Data are presented as mean values ± SEM (N=3 independent experiments). (E) Single-molecule RNA-FISH images indicate localization of Platr4 RNA transcripts (red dots) within nuclei in control ESCs. No detectable Platr4 transcripts are observed in Platr4-KO ESCs. Scale bars, 20 μm. (F) Single-molecule RNA-FISH in pre-implanted embryos (morula, E3.0 and blastocysts, E4.0). Images are shown at both single z-plane and maximum intensity of Z-projection. Scale bars, 12 μm.
Platr4 is essential for ESC differentiation, but not for maintenance of pluripotency
To dissect the molecular function of Platr4, we knocked out Platr4 in two mouse ESC lines (V6.5 and AB2.2) using the CRISPR/Cas9 approach. We designed two single guide RNAs (sgRNAs) spanning the promoter region (+300 bp/−200 bp) relative to the transcription start site of Platr4 to create a genomic deletion verified by genomic PCR in both V6.5 and AB2.2 lines and by Sanger sequencing (Figures 2A, S2A and S2B). We performed nucleofection of the transiently expressing Cas9 and sgRNAs to enhance Cas9 specificity and reduce off-target activity38. A sgRNA targeting Renilla luciferase was used as a non-targeting control. Luciferase control vs. Platr4-KO cells were single-cell sorted 48 hours after nucleofection. We detected a near-complete loss (99%) of the Platr4 transcript in ESCs, verified by Northern blot, qRT-PCR, and smRNA-FISH (Figures 2B, 1E and S2C). Interestingly, the loss of Platr4 in ESCs did not affect colony morphology (Figure 2C), cell cycle kinetics and proliferation (Figure S2D), nor expression of master pluripotency factors, such as Pou5f1 (Oct4), Nanog, and Sox2 (Figures 2C and S2E). Thus, Platr4 is likely not essential for maintaining ESC pluripotency and self-renewal.
Figure 2 : Platr4 is essential for ESC differentiation, but not for pluripotency maintenance.
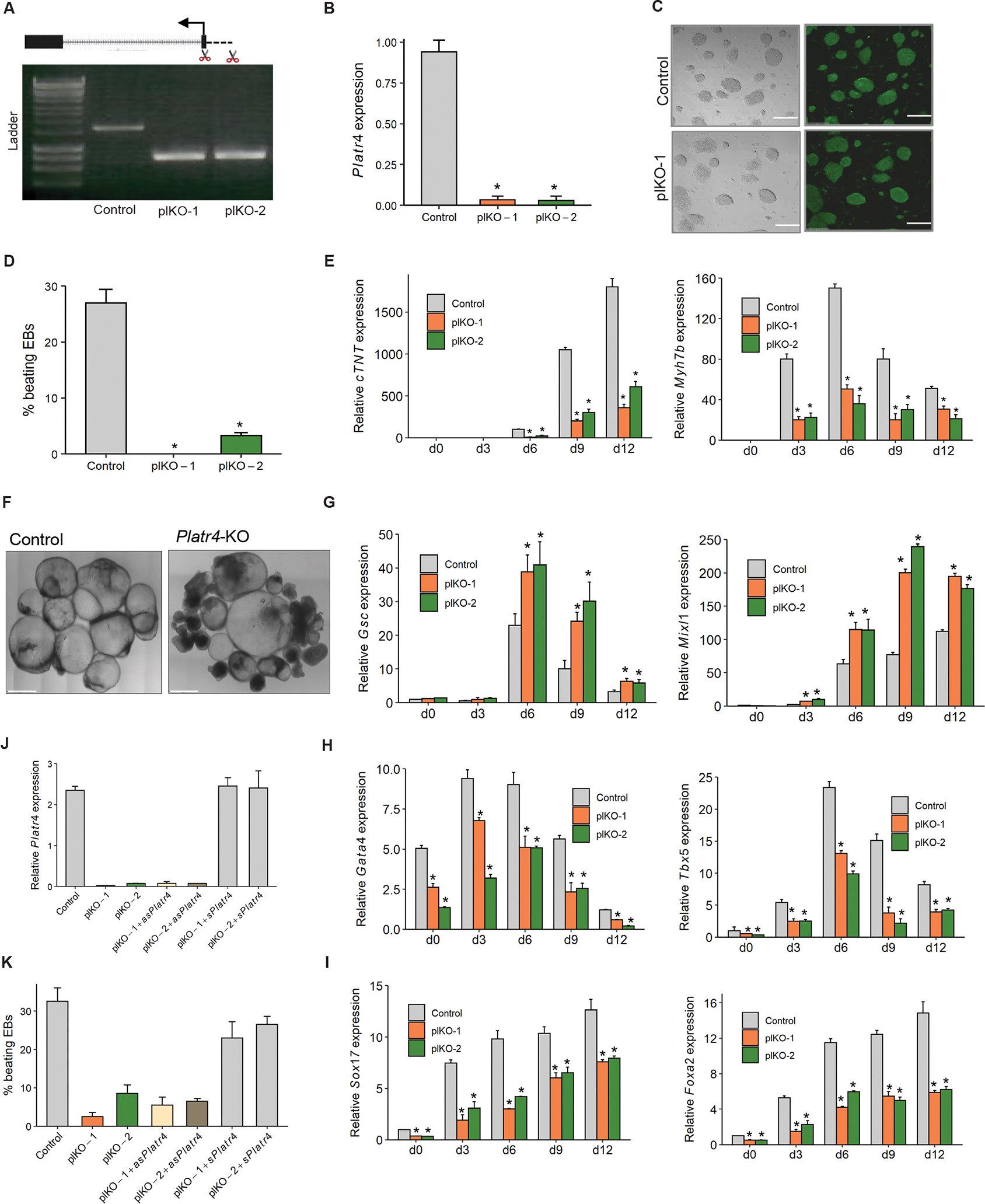
(A) CRISPR/Cas9-mediated deletion of Platr4-KO (plKO) clones in ESCs. A pair of sgRNAs near the transcription start site of Platr4 were used to establish a ~600 bp genomic deletion. plKO clones were verified by genomic PCR (clones 1, 2), and a Renilla Luciferase sgRNA was used as a negative control (control). (B) Deletion of Platr4 transcripts was further verified by qRT-PCR. (C) Platr4-KO (plKO) in ESCs did not affect colony morphology or immunostaining of Pou5f1. Left panel shows bright field image. Scale bar, 150 μm. (D) Percentage loss of spontaneously beating EBs in Platr4-KO clones compared to control at day 12 of differentiation. N=100 EBs were counted, and experiments were performed in triplicate. (E) The relative level of cardiac Troponin T (cTnT) and myosin heavy chain 7b (myh7b) were measured by qRT-PCR using control vs. Platr4-KO (plKO) EBs at different time points upon differentiation. (F) Control vs. Platr4-KO EBs at day 12. Platr4-KO EBs show a smaller size and darker cavity compared to control EBs. Scale bar, 200 μm (G, H, I) qRT-PCR analysis of expression levels of primitive streak (Gsc, Mixl1) and mesoderm (Gata4, Tbx5) markers and endocardial/endothelial markers (Sox17, Foxa2) in Platr4-KO EBs compared to control. (J) Ectopic expression of Platr4 in Platr4-KO ESCs measured by qRT-PCR. ‘asPlatr4’ and sPlatr4 stand for antisense and sense Platr4. Luc. control stands for luciferase control (K) Ectopic expression of Platr4 in Platr4-deleted EBs can rescue the contracting EBs. ‘asPlatr4’ and sPlatr4 stand for antisense and sense Platr4. Data (Figures B, D, E, G, H, I, J and K) are presented as mean values ± SEM. N=3 independent experiments. *p < 0.05 (Student’s t-test).
ESCs have the capacity to differentiate into derivatives of the three germ layers: endoderm, mesoderm, and ectoderm. Given its expression in early embryos, we chose to study the impact of the Platr4 loss on lineage commitment and differentiation. We next induced control vs. Platr4-KO ESCs to differentiate by withdrawing leukemia inhibitory factor (LIF) and allowing cells to aggregate into embryoid bodies (EBs) by the hanging drop method. The EBs form cell types corresponding to all three germ layers and are a routinely used model system to assess the early differentiation potential of ESCs39. We further confirmed that Platr4 lncRNA is expressed in ESCs and is significantly downregulated upon their differentiation (Figure S2F). During the differentiation process, we first observed abnormalities in the spontaneous contraction of EBs derived from Platr4-depleted cells. In control cells, 27% of EBs showed beating by day 12, compared to 0% (clone-1) and 3% (clone-2) in the Platr4-KO derived EBs (Figure 2D). Consistent with this observation, we performed qRT-PCR at different time points using Platr4-depleted EBs and found decreased expression levels of cardiac Troponin T (cTnT) and myosin heavy chain 7b (myh7b), two important proteins involved in cardiomyocyte contraction (Figure 2E). In addition, morphological abnormalities, i.e., smaller size and darker cavities, were observed in Platr4-depleted EBs compared to control EBs (Figure 2F), consistent with the observation that bright cavity EBs have greater differentiation capacity than dark cavity EBs40. We next differentiated EBs in the presence of retinoic acid, since it enhances the efficiency of ESCs’ conversion to a neural phenotype41, and found that the neuroectoderm markers Sox1, Nestin were unaltered in Platr4-depleted EBs (Figure S2G). Although Platr4-depleted EBs significantly upregulated the levels of primitive streak markers (Goosecoid, Mixl1), but the expression levels of mesoderm markers (Gata4, Tbx5) and endocardial/endothelial markers (Sox17, Foxa2) were reduced (Figures 2G, 2H and 2I)42. Notably, Sox17 is not only an endoderm-specific marker, it is also expressed in vascular endothelial cells of the endocardium43. Single cell RNA-seq studies showed that Sox17 mutants transcriptomically misregulate the proliferation of endocardial and myocardial cells44 and affect the differentiation of mouse ESCs into cardiomyocytes in vitro45. In addition, Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential46. Foxa2 is selectively expressed in cardiovascular cells of both ventricles, as well as in epicardial cells, which play an important role in the formation of the cardiac valve47. Together, these data demonstrate that Platr4 likely functions in the specification of the cardiac mesoderm lineage.
To exclude the possibility that the phenotypes observed in the Platr4-KO cells were caused by disturbing chromatin structure rather than the specific loss of the Platr4 transcript, we generated single-cell ectopic expression rescue clones of Platr4 in Platr4-depleted ESCs. Notably, transient rescue of Platr4 significantly increased the percentage of beating EBs (Figures 2J and 2K), indicating that the Platr4 transcript plays an important role in these processes in situ and likely exerts its effect in trans.
Platr4 regulates the expression of ECM genes
To examine whether Platr4 functions in cis or in trans, we examined the expression levels of neighboring genes (Sclt1, Jade1, and D3Ertd751e) within a 100 kb window and determined an equivalent level of expression in Platr4-KO and control ESCs (Figure S3A), consistent with Platr4 exerting its function in trans (Figures 2J and 2K). LncRNAs play an important role in the molecular circuitry of the undifferentiated ESC state to control pluripotency and lineage differentiation 8. Therefore, to explore the transcriptional control of Platr4 in the ESC state and to understand its function during differentiation, we analyzed global gene expression profiles using poly(A)+ RNA-seq on control vs. Platr4-KO ESCs. To this end, a large number of differentially expressed genes (DEGs) (scaled expression) were identified between Platr4-KO and control ESCs (Figures 3A, 3B and S3B, Table S1). The top ten significantly downregulated genes were validated by qRT-PCR (Figure 3C). A Database for Annotation, Visualization and Integrated Discovery (DAVID) gene ontology (GO) term pathway analysis in control vs. Platr4-KO ESCs showed that DEGs were significantly enriched in eight significant pathways, including the ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathways, all of which play vital roles in stem cell differentiation during mammalian development48, 49 (Figure 3D). Previous studies reported that ECM components (fibronectin, collagens, and laminins) promote ESC differentiation towards mesendoderm/mesoderm/endoderm lineages50–52. Interestingly, we found that deletion of Platr4 in ESCs reduced the expression of genes encoding ECM constituents (Figures 3E and S3F). We further performed an RNA-seq experiment using control vs. Platr4-KO EBs and assessed DEGs (log fold change), which were analyzed on different days (D0–D12) (Figure S3C and table S1). A DAVID GO term analysis showed that the EB DEGs—like the ESC DEGs—were significantly enriched in eight pathways, including the ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathways, which are crucial for ESC differentiation. (Figure S3D).
Figure 3: Platr4 regulates the expression of ECM genes.
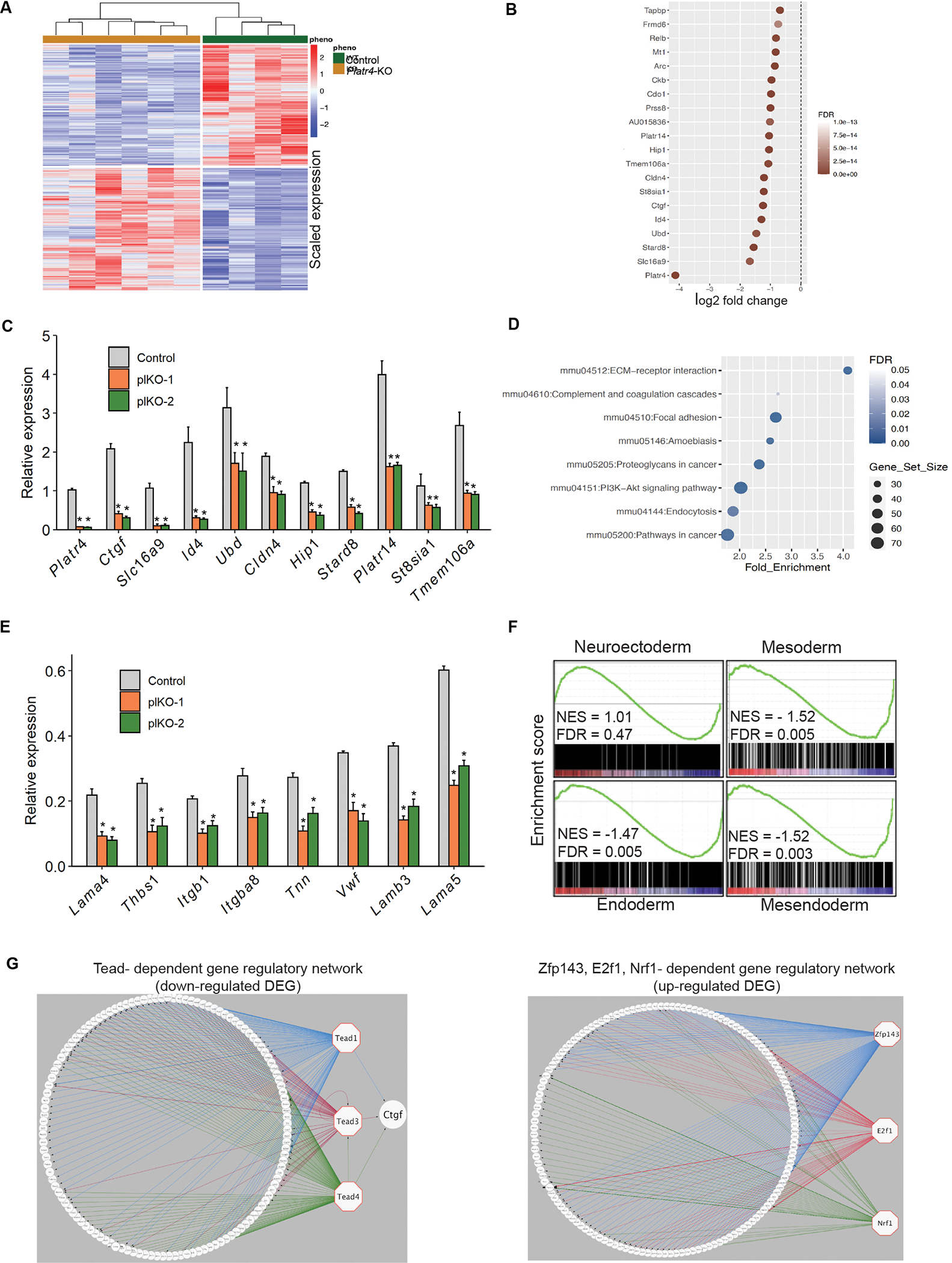
(A) Heatmap of differently expressed genes (DEGs) (scaled expression) in control and Platr4-KO (plKO) ESCs. (B) Top 20 downregulated genes upon deletion of Platr4 in ESCs. (C) The top ten downregulated genes were quantified by qRT-PCR using control vs. downregulated genes in Platr4-KO (plKO) ESC clones. Data are presented as mean values ± SEM. (N=3 independent experiments). (D) Analysis of significant GO terms in downregulated DEGs that were dysregulated upon deletion of Platr4 in ESCs. (E) The genes encoded for ECM components were quantified by qRT-PCR using control vs. Platr4-KO (plKO) ESC clones. Data are presented as mean values ± SEM. (N= 3 independent experiments). (F) Gene set enrichment analysis profiles of DEGs. The gene lists are shown in Table S1. (G) iRegulon analyses detected the enriched transcription factor motif in downregulated (left panel) and upregulated (right panel) DEGs visualized by Cytoscape software. Normalized Enrichment Score (NES) >3.
Next, a customized GSEA analysis showed that the downregulated DEGs were significantly associated with the mesendoderm lineage, though expression of neuroectoderm lineage genes were unaltered (Figure 3F and table S1), consistent with our previous observation that Platr4 is critical to mesoderm lineage specification (Figures 2G and 2H).
Platr4 exerts its functions in trans and we hypothesized that it may interact with transcription factors to regulate the expression level of target genes in the ESC state. To assess this hypothesis, we performed an iRegulon (Cytoscape) analysis53 to predict the potential transcription factors from the co-expressed gene set of Platr4 lncRNA. We used a normalized enrichment score >3 for the significant enrichment of target transcription factors and generated iRegulon interactome maps with transcription factors using upregulated and downregulated DEGs. Zfp143, E2f, and Tead family transcription factors were enriched with Platr4 co-expressed genes in upregulated and downregulated DEGs (Figures 3G, S3E and Table S1).
Platr4 interacts with Tead4 and Yap signaling molecules in the Hippo signaling pathway
The iRegulon analysis identified that Platr4 might exert its function by regulating Tead family transcription factors, since they have the highest enrichment score (Figure S3E). First, we found that the RNA and protein expression levels of Tead1, 2, 3 and 4 were equivalent in control and Platr4-KO ESCs (Figures 4A and S4A). Tead transcription factors exert their function by a transcriptional coactivator known as Yap54. Therefore, we also measured Yap RNA and protein levels in control and Platr4-KO ESCs (Figures 4A and S4A) and found that Yap expression was also equivalent in both control and KO ESCs. Next, we performed a small interfering RNA (siRNA) knockdown (KD) analysis of each Tead family member and Yap and found that they did not affect the Platr4 expression level, although the immediate downstream targets of Yap and Tead (Ctgf, Vgll3, and Gli2)55 were significantly downregulated (Figures 4B, 4C, S4B, and S4C). Since we found significant enrichment of Tead family members in downregulated Platr4 DEGs (Figure 3G), we performed RNA immunoprecipitation using antibodies against Tead1, 2, 3, 4 and Yap protein to assess their specific interactions with Platr4. Interestingly, we found that Platr4 interacted specifically with Tead4 and Yap (Figures 4D and 4E), but not with Tead1, Tead2 or Tead3 (Figure S4D). Next, using in vitro biotin-RNA pull-down, we further confirmed this specific interaction of Tead4 and Yap with Platr4 in ESCs, but not with Tead1 or Tead2 (Figure 4F). To identify the Tead4 binding region(s), we used in vitro transcription to generate four non-overlapping biotinylated Platr4 fragments (P1–P4, 200 nucleotides each) spanning full-length Platr4. All four fragments bound to Tead4 and Yap proteins (Figure 4G), suggesting that the Tead4 binding sites may be distributed diffusely on Platr4 lncRNA. In contrast, Tead1, Tead2 or Tead3 did not interact with any region of Platr4 (Figure S4D), validating the specific interaction of the Platr4/Tead4/Yap binding. To further map the Tead4 binding sites on Platr4, we performed a UV cross-linking immunoprecipitation and qPCR (CLIP-qPCR) assay56 using ten pairs of primers with overlapping 200 bp amplicons, which allowed detection of the protected Platr4 RNA segments bound by Tead4 and mapping of Tead4 binding sites on Platr4 at 200 nucleotide intervals. We detected at least one major peak in each fragment (Figure 4H), suggesting that Platr4 contains multiple Tead4 binding sites.
Figure 4: Platr4 interacts with Tead4 and Yap signaling molecules in the Hippo signaling pathway.
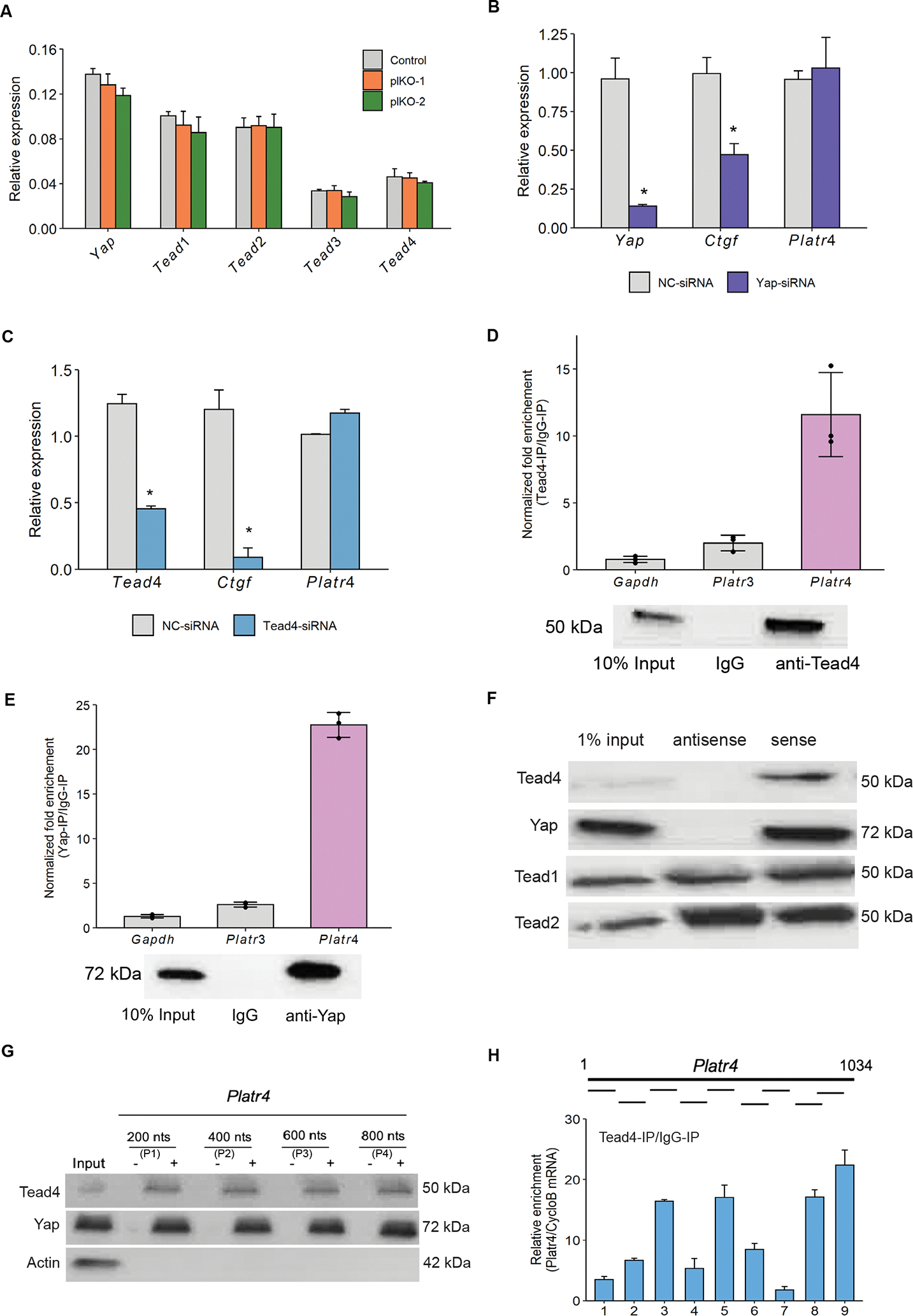
(A) The relative expression levels of Yap, Tead1, Tead2, Tead3 and Tead4 in control vs. Platr4-KO (plKO) clones, measured by qRT-PCR. (B & C) qRT-PCR analysis of the Platr4 level in ESCs using Yap-siRNA, Tead4-siRNA and non-targeting (NC)-siRNA. (D & E) RNA immune precipitation assay confirmed the interaction of Platr4 with Tead4 and Yap using Tead4 and Yap antibodies. Fold enrichment of Platr4 over IgG signal is shown. Gapdh and Platr3 (lncRNA) transcripts were used as controls (upper panel). Western blot analysis of Tead4 and Yap was performed to confirm enrichment (lower panel). (F) Biotin-RNA pull-downs using the full-length Platr4 transcript in ESC nuclear lysate showed specific binding to Tead4 and Yap, but not with Tead1 or Tead2. (G) Biotin-RNA pull-downs using unlabeled (−) and biotin-labeled (+) Platr4 fragments (P1–P4) in ESC nuclear lysate and immunoblot was performed using Tead4 and Yap antibodies. Actin antibody used as a control. (H) CLIP-qPCR analysis showed Platr4 RNA segments bound by Tead4 and detected by qPCR using 9 pairs of primers. Data (Figures A, B, C, and H) are presented as mean values ± SEM. N=3 independent experiments. *p < 0.05 (Student’s t-test).
Platr4 functions upstream of connective tissue growth factor (Ctgf)
It has been established that Ctgf is a target gene regulated by Tead4 and Yap25. Interestingly, we found that Ctgf was one of the top five significantly downregulated genes in the RNA-seq data (Figures 3B and 3C). Furthermore, we found that both the RNA and protein levels of Ctgf were significantly reduced in Platr4-KO vs. control ESCs (Figures 5A and 5B). Moreover, ectopic expression of Platr4 in Platr4-KO ESCs resulted in a corresponding increase in the mRNA level of Ctgf (Figure 5C), but not the level of Vgll3 or Gli2 (Figures S5A and S5B). These results suggest that Ctgf is a direct downstream target of Platr4 and that it functions in trans. Next, we performed CRISPR/Cas9 KD using sgRNAs targeting Ctgf in puromycin-resistant ESCs for downstream functional assays. Both the RNA and protein expression levels of Ctgf in Ctgf-KD ESCs were verified (Figures 5D and S5C). Control and Ctgf-KD ESCs were induced to differentiate via withdrawal of LIF by allowing cells to aggregate into EBs by the hanging drop method. We found that reducing Ctgf in EBs at day 12 phenocopied the Platr4-KO EBs and that there was a significant reduction in spontaneous contracting EBs: 24% of control EBs showed an observable beat at day 12 compared to just 12% of Ctgf-KD EBs (Figure 5E). Notably, ectopic expression of the full-length Ctgf gene in Platr4-KO ESCs increased the percentage of contracting EBs in Platr4-depleted cells (Figure 5F). Together, these data suggest that Ctgf is a potential regulator of cardiomyocyte differentiation and a critical downstream target of Platr4. In addition, to examine the effect of Tead4 on Ctgf expression in ESCs, we performed either ectopic overexpression (Figures 5G and S5D) or siRNA-mediated KD (Figure 4C) of Tead4. The results verified that modulating the Tead4 expression level influences the expression level of Ctgf in ESCs, suggesting that the Platr4/Tead4 ribonucleic protein complex is essential for regulating Ctgf. We further performed chromatin immunoprecipitation (ChIP) using a Tead4 antibody, and multiple qPCR primer pairs showed that Tead4 has a high occupancy over the Ctgf promoter, as suggested in other studies57, 58. Notably, the occupancy was impaired in Platr4-KO ESCs and was able to be restored upon ectopic expression of Platr4 in Platr4-KO ESCs (Figure 5H). Together, these results provide compelling evidence that the interaction of Tead4 protein with Platr4 lncRNA is required for Tead4 to bind to regulatory motifs in the Ctgf gene in ESCs. This is the first demonstration that the interaction between a lncRNA and a transcription factor is necessary to modulate a downstream target gene and regulate cardiac lineage differentiation.
Figure 5: Platr4 functions upstream of connective tissue growth factor (Ctgf).
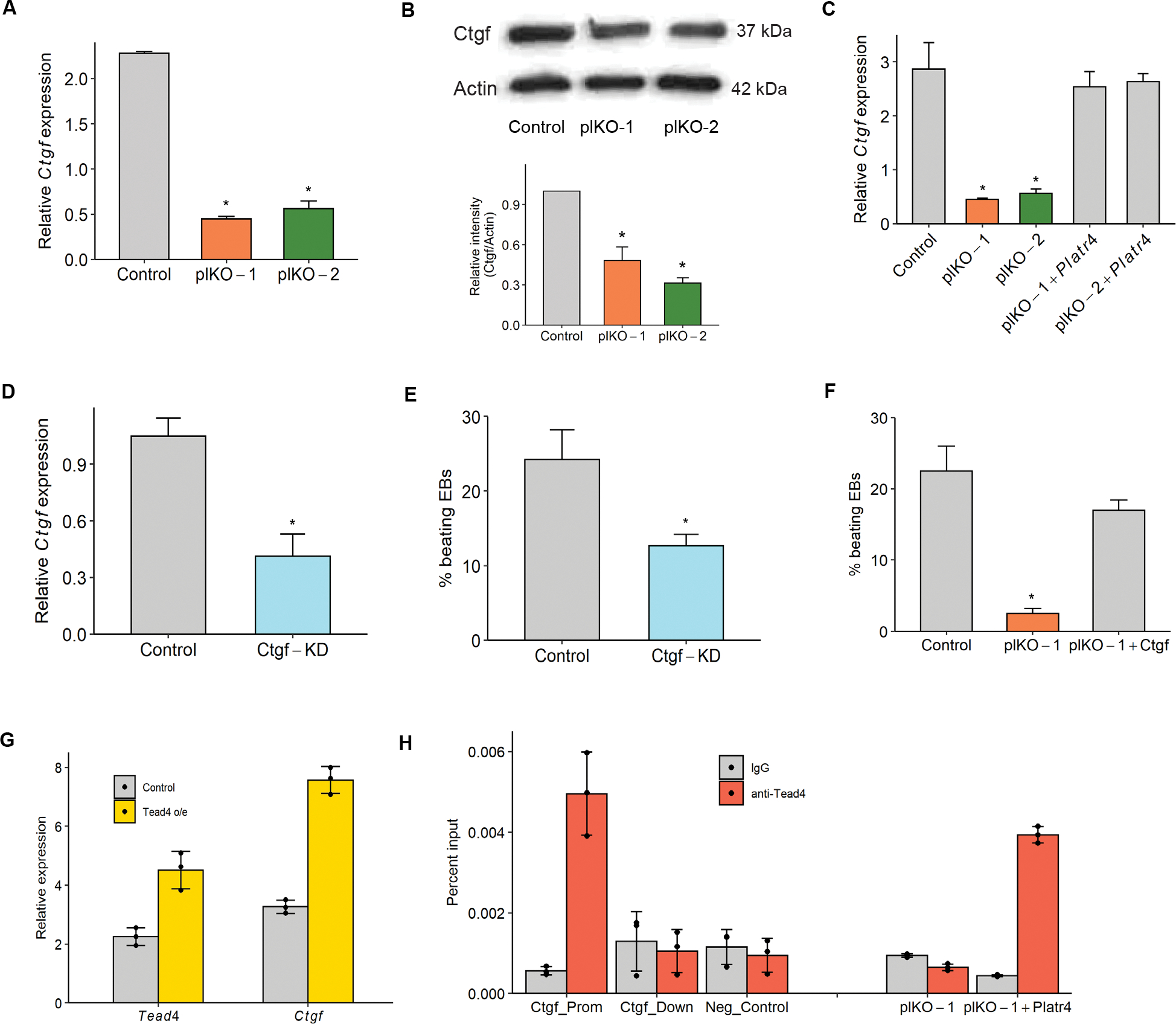
(A) Relative expression of Ctgf in control vs. Platr4-KO (plKO) ESC clones. (B) Western blot analysis of Ctgf level (top and bottom panel) in control and Platr4-KO (plKO) ESC clones (N=2 independent experiments). (C) Ctgf level is rescued upon ectopic expression of Platr4 in Platr4-KO (plKO) cells as determined by qRT-PCR. (D) CRISPR/Cas9-mediated knockdown of Ctgf in ESCs, verified by both qRT-PCR (N=2 independent experiment). KD stands for knockdown. (E) Reduced percentage of contracting EBs upon downregulation of Ctgf compared to control. (F) Ectopic expression of Ctgf in Platr4-KO (plKO) cells rescues the percentage of contracting EBs in Platr4-KO ESCs upon differentiation. (G) qRT-PCR analysis of increased Ctgf expression in ectopic overexpression of Tead4. (H) ChIP-qPCR analysis displayed Tead4 occupancy over the Platr4 targeting region of the Ctgf DNA locus in control, Platr4-KO (plKO), and ectopic Platr4 expression in Platr4-KO ESCs. Ctgf-prom and Ctgf-down stand for promoter and downstream gene, respectively. Gapdh was used as a negative control. All experiments were performed in triplicate, otherwise mentioned in the respective figures. Data are presented as mean values ± SEM. *p < 0.05 (Student’s t-test).
Platr4 is required for cardiac mesoderm lineage differentiation
Next, a directed differentiation technique was applied to recapitulate aspects of normal early cardiac development. Differentiation of ESCs into cardiomyocytes from the mesoderm lineage are assessed by the initial expression of T and Eomes15, 59. Therefore, to study Platr4 function in cardiac cell fate from the mesodermal lineage, we employed a directed in vitro cardiomyocyte differentiation assay that permits isolation of cell populations at well-defined stages (ESCs, EBs, precardiac mesoderm, cardiac progenitors, and cardiomyocytes)15 (Figure 6A). Using this assay, we found that Platr4-KO EBs were smaller in size and had an irregular shape at day 4 (precardiac mesoderm) compared to controls (Figure 6B), although no morphological changes were observed at day 2, a time point that preceded the addition of cardiac growth factors. Despite the relatively elevated levels of T and Eomes in the precardiac mesoderm and cardiac progenitor populations in Platr4-KO cells compared to control cells (Figure 6C), the expression of core cardiac transcription factors at day 4 (precardiac mesoderm), day 6 (cardiac progenitors), and day 12 (cardiomyocytes) was significantly downregulated in Platr4-KO cells (Figures 6D, 6E & 6F). Interestingly, the cell surface markers Flk1 and PdgfRα, which are enriched in cardiac progenitor populations, were also downregulated in Platr4-KO cells (Figure 6G). Flk1 and PdgfRα represent early cardiac mesoderm markers60. These data further support the idea that loss of Platr4 affects the cardiac lineage. Notably, CRISPR/Cas9-mediated depletion of Ctgf showed similar effects to those of Platr4 deletion (Figures S6A and S6B). Interestingly, we also found that deletion of Ctgf in ESCs decreased the expression of genes encoding ECM constituents (Figure S6C).
Figure 6: Platr4 is required for cardiac mesoderm lineage differentiation.
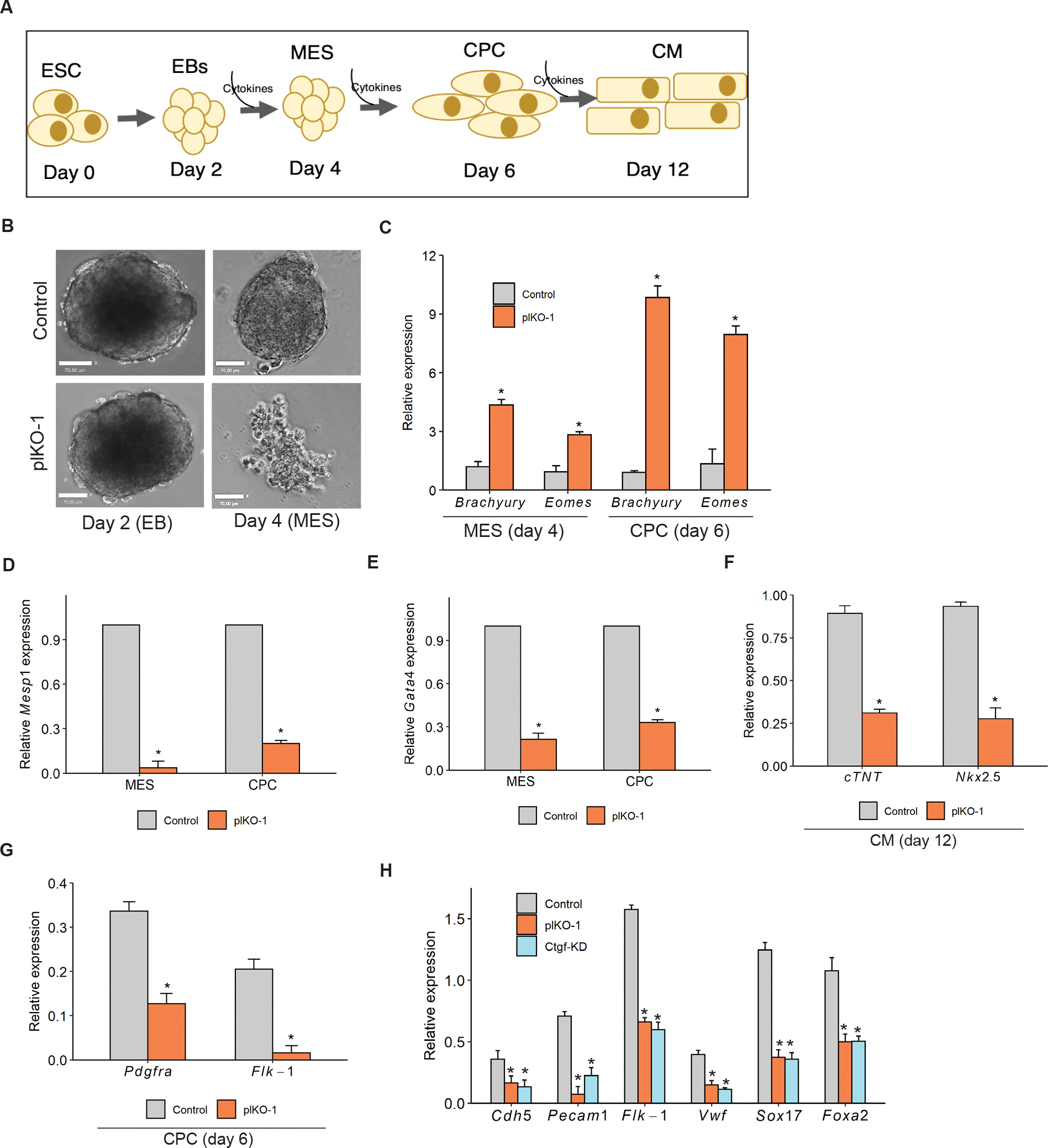
(A) ESCs were differentiated into cardiomyocytes (CM) and progressed through cardiac mesoderm (MES) and cardiac progenitor cell (CPC) stages using serum-free media with the sequential addition of cytokines. (B) Platr4-KO (plKO) EBs showed a smaller size at the MES stage compared to controls at day 4. Scale bar, 50 μm. (C) The relative expression level of Brachyury and Eomes were elevated compared to control at the noted days. (D, E & F) Relative expression of core cardiac transcription factors displayed significant downregulation in Platr4-KO (plKO) compared to control cells at different stages of differentiation. (G) The relative level of cell surface markers (Pdgfrα and Flk-1) was reduced in Platr4-depleted CPCs. (H) qRT-PCR measurement of the reduced level of expression of cardiac-endothelial markers in Platr4-KO (plKO) and Ctgf-knockdown (Ctgf-KD) ESCs. All experiments were performed in triplicate. Data (Figures C, D, E, F, G and H) are presented as mean values ± SEM. *p < 0.05 (Student’s t-test).
Since we found that KO of Platr4 resulted in reduced beating of EBs and endothelial cells are known to play a crucial role in establishing heart valve structures during embryonic development61, we differentiated ESCs to cardiogenic endothelial cells62. Our data identified that relative expression of the cardiac endothelial cell markers (Cdh5, Pecam1, Flk-1, Vwf, Sox17, Foxa2)63, was significantly reduced in both Platr4-KO ESC clone and Ctgf-KD ESCs compared to control cells (Figure 6H). Importantly, cardiac endothelial cells play an important role in cardiomyocyte differentiation from ESCs64. Together, our results further support an essential role of Platr4 in mesoderm specification, especially in the cardiac lineage differentiation program.
Platr4-KO mice exhibit fibrosis, valve degeneration, and reduced cardiomyocyte contractility
To assess the in vivo function of Platr4 we generated a Platr4-KO mouse in the C57BL/6J strain using two sgRNAs targeting the transcription start site and first exon of Platr4, as described earlier (Figure 2A); three KO founders were genotyped (Figure S7A) and used to establish three independent lines. The Platr4-KO alleles were backcrossed to the C57BL/6 background for ten generations to yield a pure C57BL/6 genetic background. Heterozygotes intercrossed with wild-type (WT) mice generated WT and Platr4-KO littermates (Figures S7B and S7C). All homozygous KO mice used for breeding were fertile and showed no gross physical abnormalities compared to WT mice. Interestingly, we observed the sudden death of young adult and mature adult mice (40%, N = 4/20) in the KO cohort but not in the WT cohort. When we examined hematoxylin & eosin (H&E)-stained heart tissue sections from the young adult mice that had died suddenly (N=3/cohorts) vs. those that had not died suddenly (N=10/cohorts) (Figure S7D), we found that 60% of the KO mice showed valve defects with fibrocartilaginous metaplasia (Figures 7A and S7E), fibro-osseous metaplasia, and mucinous degeneration compared to WT (Figures 7B and S7F). In addition, we found perivascular and myocardial mineralization (60% of KO mice, N=10/group) (Figure 7C) and myocardial atrophy and fibrosis (Figure S7G).
Figure 7: Platr4-KO mice exhibit fibrosis, valve degeneration, and reduced cardiomyocyte contractility.
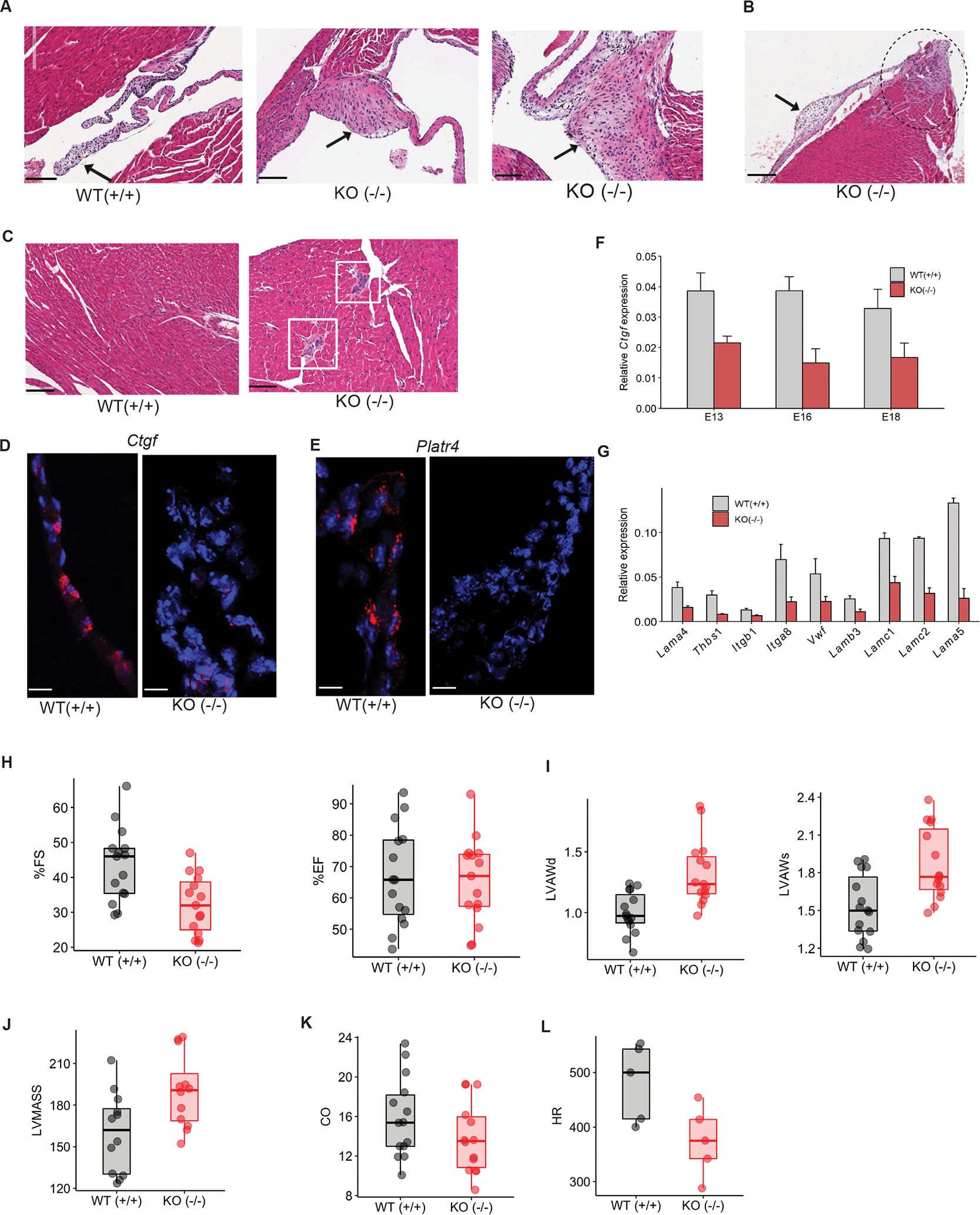
(A) H&E staining of the adult heart from WT (+/+) mice showed normal valve tissue, and Platr4-KO (−/−) mice showed valve defects with fibrocartilaginous metaplasia. Scale bar, 100 μm. (B) H&E staining of the adult Platr4-KO (−/−) heart showed fibrocartilaginous metaplasia and myxomatous degeneration of the valves. Scale bar, 100 μm. (C) H&E staining of KO (−/−) mice showed mineralization, but the WT heart was normal. Scale bar, 100 μm. (D, E) Single-molecule RNA-FISH indicates expression of Ctgf and Platr4 RNA transcripts (red dots) in the control adult heart valve [WT (+/+)] but not in the Platr4-KO [KO (−/−)] adult heart valve. Scale bars, 50 μm. (F) Relative expression of Ctgf in the control vs. Platr4-KO (−/−) embryonic hearts at different stages of development. (G) qRT-PCR analysis of the relative expression level of genes encoding ECM components in the WT (+/+) vs. Platr4-KO (−/−) adult heart. (H) Echocardiography analysis for the %FS and %EF of WT (+/+) and Platr4-KO (−/−) mice. ‘FS’and ‘EF’stands for Fractional Shortening and Ejection Fraction respectively. (I) Echocardiography analysis for LVAW (Left Ventricle Anterior Wall) in diastole (d) or systole (s) in WT (+/+) and Platr4-KO (−/−) mice. (J, K, L) Echocardiography analysis of LVMASS (Left Ventricular Mass), cardiac output (CO), heart rate (HR) (N=5) in WT (+/+) and Platr4-KO (−/−) mice. Data are presented as mean values ± SEM (N=15/group). *p < 0.05 (two-tailed Student’s t-test).
Interestingly, using smRNA-FISH, we detected Platr4 and Ctgf expression in the valve area of the adult heart tissue sections in the WT mice but not in those of the Platr4-KO mice (Figures 7D and 7E), suggesting that Platr4 is expressed in ESCs and the early developing embryo, and with specific expression in the adult mouse heart valve. In addition, we found reduced Ctgf RNA and protein levels in the embryonic and adult Platr4-KO mouse heart (Figures 7F, S7H and S7I). In addition, we detected the expression level of key cardiac transcription factors (Gata4, Nkx2-5, Mef2c, Tbx5)65 reduced in adult Platr4-KO mouse heart (Figure S7J). Notably, we also found that the relative expression of ECM genes was also reduced in the Platr4-KO compared to that of WT adult heart (Figure 7G). To further assess heart defects, we performed an echocardiography analysis on WT vs. KO mice. Echocardiography can visualize cardiovascular structures and measure cardiac function in mice due to its advanced spatial-temporal resolution. Our findings showed a significant (30%) decrease in the percentage of fractional shortening, without altering the percentage of the ejection fraction in KO compared to WT mice (Figure 7H). This observation supports our in vitro cardiomyocyte dysfunction data, since the percentage of fractional shortening indicates changes in left ventricle chamber size and myocyte contractility66. We further demonstrated increased ventricular wall thickness and ventricular mass in KO vs. WT mice, indicating ventricular hypertrophy (Figures 7I and 7J). Interestingly, a significant reduction of cardiac output and heart rate in KO compared to WT mice (Figures 7K and 7L) may explain our observation of heart failure and sudden death, since cardiac output is an important parameter for measuring cardiac dysfunction67. Thus, the genetic loss of Platr4 impacts cardiac valve development and function in vivo. therefore, our data support a role for Platr4 function in cardiac mesoderm lineage specification and heart valve function.
Discussion
The dynamic regulation of transcriptional programs is vital to lineage specification during mammalian development, and disruption or faulty regulation of these processes can lead to various developmental disorders. Bvht was the first lncRNA identified in mice to display a crucial role in cardiac commitment; however, its loss had no phenotype in a KO mouse model, and a human orthologue has thus far not been identified15, 68. More recent studies have shown the epigenetic regulation of lncRNAs in mesendoderm lineage commitment and their role in embryonic heart development14, 69. Recently, the muscle-specific lncRNA Charme has been reported to control skeletal and cardiac myogenesis in in vitro and in vivo systems70. In addition, the chromatin-associated lncRNA Myheart was shown to be highly expressed in the adult mammalian heart, and it has been shown to repress cardiomyopathy specifically in the mouse heart71.
Here, we report that the depletion of the nuclear enriched lncRNA Platr4 in ESCs significantly represses downstream mesendoderm determinants, cardiac endothelial markers, and especially core cardiac transcription factors, without affecting pluripotency master transcription factors, such as Pou5f1 (Oct4), Nanog and Sox2. Our results also showed that deleting Platr4 in mouse ESCs reduced the level of cardiac endothelial markers and core cardiac transcription factors in mesoderm-directed differentiated EBs. Consistent with this observation, we found that Platr4-KO mice exhibited valve defects and myocardial atrophy cardiac dysfunction and that overall heart function was impaired. Together, our findings reveal that Platr4 is required in the undifferentiated ESC network to regulate the downstream mesoderm differentiation.
The important effectors of the Hippo signaling pathway, Yap and its interacting partner Tead, play key roles in many diverse biological processes, including progenitor proliferation, stem cell identity, and lineage fate specification72. The mammalian Tead family consists of four members with a similar domain architecture, Tead1-4, and each Tead has cell- tissue-specific expression28, 73, 74. Although all Tead isoforms share a common protein structure, their gene regulation mechanism is context-specific due to their cell- and tissue-specific expression (Stein et a., 2015). Yap and Tead4 play an essential role in trophectoderm lineage determination26, 75, 76. Among the four Tead members, Tead1 and Tead4 are highly expressed in cardiac and skeletal muscle28, 77, 78. A recent study in human ESCs showed that Yap/Tead4 cooperate with Wnt/β-catenin to induce mesendodermal differentiation genes and regulate embryonic cardiac mesoderm formation79. In addition, two studies have shown that the Hippo pathway and Yap regulate cardiomyocyte function and heart size80, 81. Tead4 alone is expressed in developing skeletal muscle in mouse embryos, and it interacts with Myod1 and Myog transcription factors in the C2C12 mouse myoblast cell line28, 82. Furthermore, cardiac muscle-specific transgenic overexpression of Tead4 has been shown to induce cardiac contractile dysfunction83. In addition, many studies have reported crosstalk between Tead4 and lncRNAs in various cancers and in muscle development, suggesting its critical role in different biological contexts84–86. Here, we showed that the direct interaction between Yap, Tead4, and the lncRNA Platr4 is required to modulate their shared downstream target gene Ctgf in mouse ESCs, suggesting that Platr4 may function as a molecular scaffold or chaperone, bringing Yap and Tead4 to the Ctgf loci. Furthermore, our ChIP-qPCR result indicated high occupancy of Tead4 on the Ctgf promoter, supporting its functional role in partnering with Platr4 to regulate the Ctgf gene. In addition, both Tead4 gain-of-function and loss-of-function resulted in altered Ctgf expression (upregulation and downregulation, respectively), indicating a transcriptional regulatory role of Tead4 in this context. Thus, our results for the first time showed that the interaction of Tead4/Yap with Platr4 lncRNA is required for the transcriptional regulation of Ctgf, thereby impacting cardiac lineage differentiation and optimal heart function. Future studies will examine the potential role of Platr4 in regulating other target genes, as well as in regulating Tead4 gene expression programs.
We identified the involvement of Ctgf in cardiomyocyte fate determination and function, as Ctgf is a downstream target of Platr4 lncRNA. Ctgf is essential for embryonic development, and global Ctgf-KO mice display skeletal anomalies and perinatal death87. An abnormal Ctgf level is associated with multiple disorders, including fibrosis and cancer in many organs and tissues88. Interestingly, like Platr4, Ctgf is also expressed in both the inner cell mass and trophectoderm lineage in mouse blastocysts and is highly expressed in the mesoderm lineage at E6.589. Furthermore, it has been shown that Ctgf is highly expressed in the mouse embryonic heart at E12.5–E13.0 and at E14.5, and then in adult hearts, Ctgf is restricted to the aorta and to the pulmonary valves90. Interestingly, we also found a restricted Platr4 expression pattern in the adult mouse heart valve. Homozygous deletion of Platr4 in mice showed reduced Ctgf expression in both embryonic and adult hearts and resulted in various pathological conditions, including valve mucinous degeneration. In addition, a significant pathological condition of heart disease is cardiac fibrosis, caused by the excessive deposition of ECM components in the heart. Ctgf regulates the fibrotic process as a matrix protein and is a known biomarker for cardiac fibrosis91. It has also been shown to be required for initiating the fibrosis process in several tissues and organs, including the heart92. We also observed cardiac fibrosis in the Platr4-KO mice. Moreover, plasma Ctgf concentration is used as a biomarker for cardiac dysfunction in chronic heart failure patients93. Here, we report for the first time that depleting Ctgf in ESCs impacts cardiomyocyte contractility during lineage differentiation, consistent with the finding that Ctgf protein is involved in cardiomyocyte differentiation71.
In addition, ECM components are also critical regulators of ESC differentiation in various lineages and of somatic cell reprogramming at different developmental stages in vitro94, 95. Furthermore, ECM components, both major (collagens) and minor (fibronectin, laminins), make up almost 90% of the heart and play diverse functional roles within heart valves96. Interestingly, CTGF is an ECM-associated protein and a direct downstream modulator of the TGF-β signaling pathway during heart valve development97–99. We found that depletion of Platr4 affects ECM components and downregulation of the TGF-β gene. Moreover, Ctgf regulates EMT genes to differentiate fibroblasts into myofibroblasts100 and EMT initiation is the first step of endothelial cells to form the heart valve101. Failure to undergo EMT or disruption of signals from endothelial cells to valvular cushion cells results in a variety of valve defects102. In addition, ECM organization in mature valves is conserved across species, and structural and compositional alterations in the ECM of the valve cause valve disease to develop103. Therefore, our data suggest that Platr4 may be involved in cardiac lineage differentiation and healthy heart valve function through a highly organized network of ECM components.
Cardiovascular disease, especially valve disease, is the leading cause of death worldwide, resulting in >25,000 deaths annually in the United States104. Therefore, understanding the molecular pathways of cardiovascular and endocardial lineage differentiation is crucial for understanding the fundamental facts of heart valve development and applying that knowledge to regenerative medicine. Our findings highlight a new mechanism in the Hippo signaling pathway, which controls mesodermal lineage differentiation, specifically cardiac lineage differentiation, and adult heart valve function. These findings could lay the groundwork for new approaches to studying cardiac diseases in vitro and have potential for developing regenerative stem cell therapies. Based on synteny, Platr4 has three potential human orthologues—RP11-130C6.1, RP11-184M15.1, and RP11-184M15.2, located in chromosome 4 (q28.1-28.3)—which will be the subject of future study. Clinical case studies have reported that deletion of chromosome 4 (q28.1–31.3) is associated with cardiac defects, including arrhythmia, ventricular or atrial septal defects, thickening (hypertrophy) of the heart muscle (myocardium), and structural heart defects105–107. Thus, we anticipate that one or more of these potential human lncRNA transcripts may be the human orthologue of Platr4. These transcripts may therefore be ideal candidate(s) for exploring their functional role in heart valve development and function and may act as potential biomarkers for valvular heart disease.
Limitations of this study:
In this study we identified that Platr4 interacts with the Hippo signaling molecules, Yap and Tead4 to regulate a downstream target gene, Ctgf, in the cardiac mesoderm lineage differentiation process. Although we identified Ctgf as a top target of Platr4, we cannot rule out the possibility that Platr4 also regulates other genes that were identified in our differential RNA-seq analysis comparing wild-type to KO ESCs. Therefore, future studies are needed to assess the possibility that Platr4 may regulate other target genes, in association with different protein complexes, to modulate additional aspects of the cardiac differentiation program. Also of future interest is how the early loss of expression of Platr4 impacts the late onset of disease. The possibility of an early redundancy mechanism that is lost or incapable of carrying out full rescue in the adult will be an interesting avenue of future exploration.
STAR★METHODS
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, David L. Spector (spector@cshl.edu).
Materials availability
All cell lines and knockout mice generated in this study will be made available upon request to the lead contact and will require a completed materials transfer agreement and reasonable compensation by requestor for its processing and shipping.
Data and code availability
No custom codes were generated in this study.
Raw RNA-Sequencing data generated in this study have been deposited at NCBI SRA website and are publicly available as of the date of 09/18/2022: PRJNA880813 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA880813/) and PRJNA881397 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881397/)
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Oct-3/4 Antibody | Santa Cruz | Cat # sc-5279 |
| Anti-mouse Alexa Fluor 488 | Thermofischer | Catalog # A28175 |
| Anti-TEAD4 antibody - ChIP Grade | Abcam | Cat # ab58310 |
| Anti-TEAD4 antibody | Abcam | Cat # ab97460 |
| Anti-TEAD1 antibody | BD Biosciences | Cat # 610923 |
| Anti-TEAD1 antibody | Cell Signaling Technology | Cat #12292 |
| Anti-TEAD2 antibody | Biorbyt | Cat # orb382464 |
| Anti-TEAD3 antibody | Novus Biologicals | Cat # NBP1-83949 |
| CTGF/CCN2 antibody | Novus Biologicals | Cat # NB100-724SS |
| Monoclonal anti-β-Actin | Millipore-Sigma | Cat # A5441 |
| Anti-Yap1-antobody | Novus Biologicals | Cat # NB110-58358 |
| Bacterial and virus strains | ||
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | Cat # 48138 |
| pSpCas9(BB)-2A-Puro (PX459) V2.0 | Addgene | Cat # 62988 |
| Chemicals, peptides, and recombinant proteins | ||
| Pregnant mare’s serum gonadotropin (PMSG) | ProSpec | Cat # HOR-272 |
| Human chorionic gonadotropin (hCG) | Sigma Aldrich | Cat # C1063 |
| KSOM Mouse Embryo Media | Millipore Sigma | Cat # MR-121-D |
| EmbryoMax® M2 Medium | Millipore Sigma | Cat # MR-015-D |
| Mineral oil | Ovoil, Vitrolife | Cat # 10029 |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | Cat # 19200 |
| Acidic Tyrode’s solution | Sigma-Aldrich | Cat # MR-004-D, |
| Phosphate-buffered saline (PBS) | Thermo Fisher | Cat # 144444 |
| Bovine serum albumin (BSA) | Thermo Fisher | Cat # 15260037 |
| Molecular Grade Water | G-Biosciences | Cat # 786-73C |
| Triton X-100 | Sigma-Aldrich | Cat # X100 |
| SUPERase•In RNase Inhibitor | Thermo Fisher | Cat # AM2696 |
| Agarose | Millipore Sigma | Cat # A0701-25G |
| 20x saline-sodium citrate (SSC) buffer | G- Biosciences | Cat # R020 |
| RNAscope Multiplex Fluorescent V2 Assay | Advanced Cell Diagnostics | Cat # 323120 |
| Mouse Platr4 probe set | RNAscope Target Probe C1 | Cat # 300031 |
| Opal 570 | Akoya Biosciences | Cat # FP1488001KT |
| RNase A | Thermo Fisher | Cat # EN0531 |
| RNase H | Epicenter | Cat # H39500 |
| Leukemia inhibitory factor (LIF) | Millipore | Cat # ESG1107 |
| Iscove’s Modification of DMEM | Corning | Cat # 15-016-CV |
| B27 Supplement Gibco | Thermo Fischer | Cat # 0080085SA |
| Trizol Reagent | Thermo Fischer | Cat # 15596018 |
| Ultrahyb Ultrasensative hybridization buffer | Thermo Fischer | Cat # AM8670 |
| NorthernMax 10X Running Buffer | Thermo Fischer | Cat # AM8671 |
| NorthernMax Denaturing Buffer (10X) | Thermo Fischer | Cat # AM8676 |
| Proteinase K Solution | Viagen Biotech | Cat # 501-PK |
| Poly(A)Purist™ MAG Kit | Thermo Fischer | Cat # AM1922 |
| Retinoic Acid | Sigma | Cat # R2625-50MG |
| L-Ascorbic acid | Sigma | Cat # A7506-25g |
| PowerUp™ SYBR® Green Master Mix | Thermo Fisher | Cat # A25743 |
| MEM NEAA | Thermo Fisher | Cat # 11140-050 |
| Sodium Pyruvate | Sigma | Ca t# P2256-25G |
| Pierces™ IP Lysis Buffer | Thermo Fisher | Cat # 87787 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher | Cat # 89900 |
| N2 Supplement 100x | Thermo Fisher | Cat # 17502048 |
| ON-TARGETplus Mouse Tead4 (21679) siRNA - SMARTpool | Dharmacon | Cat # L-057322-01-0005 |
| ON-TARGETplus Mouse Tead2 (21677) siRNA - SMARTpool | Dharmacon | Cat # L-060552-01-0005 |
| ON-TARGETplus Mouse Tead3 (21678) siRNA - SMARTpool | Dharmacon | Cat # L-044127-01-0005 |
| ON-TARGETplus Mouse Yap1 siRNA | Dharmacon | Cat # L-046247-01-0005 |
| ON-TARGETplus Mouse Tead2 (21677) siRNA | Dharmacon | Cat # L-060552-01-0005 |
| ON-TARGETplus Non-targeting Pool | Dharmacon | Cat # D-001810-10-05 |
| SuperScript II Reverse Transcriptase | Life Technologies | Cat # 18064014 |
| Penicillin-Streptomycin 10,000U/mL | Life Technologies | Cat # 15140122 |
| BSA fraction V (7.5%) | Thermo fischer | Cat # 15260037 |
| dPBS | Life Technologies | Cat # 14190-250 |
| TaqMan Reverse Transcription Reagents | Thermo fischer | Cat # 4304134 |
| Chromatin IP DNA purification kit | Active motif | Cat # 58002 |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 10-well, 50 μl #4561094 | Biorad | Cat # 4561094 |
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat # RS-122-2001 |
| Retinoic acid | Sigma | Cat # R2625-100MG |
| 1-Thioglycerol | Sigma | Cat # M6145-25ML |
| DR4 MEF Feeder Cells, P3, Irradiated | Applied Stem cell | Cat # ASF-1014 |
| EmbryoMax® PMEF P3 Strain CF-1, Irradiated | Millipore | Cat # R-4169444.2 |
| Poly(A)Purist™ MAG Kit | Thermo Fisher | Cat # AM1922 |
| Proteinase K Solution | Viagen Biotech | Cat # 501-PK |
| Iscove’s Modification of DMEM | Corning | Cat # 15-016-CV |
| Ham’s F-12 Medium | Corning | Cat # 10-080-CV |
| StemPro-34 | GIBCO | Cat # 10639011 |
| human VEGF | R&D system | Cat # 293-VE |
| human Activin A | R&D system | Cat # 338-AC |
| human BMP4 | R&D system | Cat # 314-BP |
| human basic FGF | R&D system | Cat # 233-FB |
| Deposited data | ||
| Raw RNAseq data of control vs Platr4-KO mouse ES cells | This paper | SRA data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA880813/ |
| Raw RNAseq data of control vs Platr4-KO differentiated mouse ES cells | This paper | SRA data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881397/ |
| Unprocessed western blot images | This paper | Mendeley Data: https://data.mendeley.com/datasets/2ddgwynm42 |
| Experimental models: Cell lines | ||
| Mouse ES cell line (V6.5) | Eggan et al., 2001116 | CSHL Gene Targeting Facility |
| Mouse ES cell line (AB2.2) | ATCC | Cat # SCRC-1023 |
| Knocout ES cell lines | This Paper | NA |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratory, Farmington, CT USA | Cat # 000664 |
| Platr4-KO mouse | This paper | NA |
| Oligonucleotides | ||
| 5’ RACE outer: ACCACACCCCTAACAGTACCAC | This paper | NA |
| 5’ RACE inner: TGTACGCTGGATAGAACTGCAT | This paper | NA |
| 3’ RACE outer: CCATCATGGGCTTTGCTAGT | This paper | NA |
| 3’ RACE inner: CCCACGAGGGTCAACAGTAG | This paper | NA |
| Platr4-primer-F: GCCCTGGAGAAGTCACAGAG | This paper | NA |
| Platr4-primer-R: ATGGGGAAGAAGGAAGAGGA | This paper | NA |
| gPCR_Platr4-F1: GGGCTCAAAGAAGGTGACTG | This paper | NA |
| gPCR_Platr4-R1: GGCCTCAAATTCAGAGACCA | This paper | NA |
| gPCR_Platr4-F2: GGCCAATTGTTCCACAAGAAG | This paper | NA |
| gPCR_Platr4-R2: AGAGACCACCTCCCTCTGCT | This paper | NA |
| Recombinant DNA | ||
| Tead4 (NM_011567) Mouse Tagged ORF Clone | Origene | Cat # MR219506 |
| Ctgf (BC006783) Mouse Untagged Clone | Origene | Cat #: MC200503 |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| R | R | https://www.r-project.org/ |
| UCSC genome browser | UCSC | https://genome.ucsc.edu/ |
| ensembl genome browser | EMBL-EBI | http://useast.ensembl.org/index.html |
| ZEN microscope software | ZEISS | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| The sgRNAs design software | http://crispr.mit.edu/ | |
| Other | ||
| Microscope, inverted | Nikon | TS100 |
| Stereoscope dissection microscope | Leica | MZ12.5 |
| Microscope, confocal | Zeiss | LSM 780 |
Microscopy data reported in this paper will be shared by the lead contact upon request
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal ethics
All animal procedures were approved by the Cold Spring Harbor Laboratory (CSHL) Institutional Animal Care and Use Committee. Animals were housed under controlled conditions (12-h light, 12-h dark cycle, 19–22 C) with ad libitum access to food and water and pathogen-free conditions in the CSHL Animal Shared Resource, fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Embryonic, young adult and mature adult mice were used for the experiments.
ESCs culture:
Mouse ESCs were cultured using standard procedures in knockout DMEM medium containing 15% FBS, 0.1 mM 2-mercaptoethanol, 1000 units/ml leukemia inhibitory factor, nonessential amino acids, and sodium pyruvate with irradiated MEF feeders (Global Stem) on gelatin-coated plates.
Generation of knockout ESC lines:
Two single guide RNAs (sgRNAs) targeting the transcription start site (TSS) of Platr4 were placed into the pSpCas9(BB)-2A-GFP (PX458) vector (Addgene #48138). The sgRNAs were designed using http://crispr.mit.edu/. ESCs were transfected with plasmids using 4D-Nucleofector X Unit, program code CG104. Transfected ESCs were sorted 48 hours post-transfection and deposited as single cells into 96-well plates using a FACS Aria (SORP) Cell Sorter (BD Biosciences). Each cell clone was cultured and examined by genomic PCR, qRT-PCR, and Sanger sequencing to select a homozygous knockout clone. Cells were transfected with a sgRNA targeting Renilla luciferase, used as a negative control. sgRNA sequence and primers are provided in Key Resource Table.
Knockout mouse generation:
Platr4 knockout mice were generated in the Gene Targeting and Transgenic Core facility at the University of Rochester Medical Center108. Briefly, two sgRNAs (Key Resource Table) near the TSS were used to delete the promoter of Platr4 lncRNA. Guide RNA was in vitro transcribed by the T7 promoter using a MEGAshortscriptT7 Transcription Kit (Ambion, AM1354). In vitro transcribed RNAs were purified using a MEGAclear Transcription Clean-Up Kit (Ambion, AM1908). Fertilized embryos were collected from oviducts of superovulated females. sgRNAs (50 ng/μl) and Cas9 mRNA (100 ng/μl) were co-injected into mouse zygotes with well-recognized pronuclei and then transferred into the uterus of pseudopregnant ICR females. Founder mice were tail-snipped for genotyping and sequencing. After confirming genomic deletion, each founder mouse was successfully bred to the F1 generation using C57BL/6J mice for germline transmission. Heterozygous mice were backcrossed for ten generations to clean genetic background and generate homozygous mice.
METHOD DETAILS
ESC differentiation into embryoid bodies:
Feeder cells were depleted from ESCs by one hour of soaking and were resuspended to 20,000 cells/ml in a complete growth medium without leukemia inhibitory factor (LIF). The suspension was plated as 20 ul drops on the lid of the Petri dish, which were then inverted and incubated for two days to induce aggregation by the hanging drop method. Aggregated ESCs formed the embryoid body (EB). EBs were then collected and plated on standard gelatin-coated 6-well cell culture plates in a complete growth medium without LIF for 12 days to analyze the percentage of spontaneous beating EBs. Furthermore, EBs were further collected at different time points (day 0, 3, 5, 7, 9 and 12) to interpret gene expression and for hematoxylin & eosin (H&E) staining. To differentiate into the neuroectoderm lineage, EBs were grown in a complete growth medium containing 1 uM all-trans retinoic acid lacking LIF.
Cardiomyocyte differentiation:
ESCs were directed to differentiate into cardiomyocytes as described previously15. Briefly, cells were depleted from the feeder layer with a standard technique and aggregated into EBs using the hanging drop method. Next, EBs were dissociated and cultured at a density of 100,000 cells/ml for two days in serum-free media (3 parts Iscove’s Modification of DMEM (IMDM) (Cellgro): 1 part Ham’s F12 (Gibco), 0.05% bovine serum albumin (BSA), 2 mM GlutaMax (Gibco), B27 supplement (Gibco), N2 supplement (Gibco) supplemented with 50 mg/ml ascorbic acid, and 4.5×10−4 M monothioglycerol (Sigma)). Around 48 hours later, EBs were dissociated and re-aggregated in the presence of hVEGF (5 ng/mL), Activin A (5 ng/ml), and hBMP4 (0.25 ng/ml) (all from R&D Systems). EBs were further dissociated and replated at 500,000 cells/well in a 24 well plate in StemPro-34 (Gibco) supplemented with 5 ng/mL hVEGF, 10 ng/mL human basic FGF, and 25 ng/mL FGF10 (all from R&D Systems).
5’/3’ Rapid Amplification of cDNA Ends (RACE):
5’ and 3’ RACE was performed using the Ambion FirstChoice RLM-RACE kit according to the manufacturer’s protocol. Briefly, fragments were amplified by nested PCR using AmpliTaq Polymerase, PCR products were cloned using a pGEM-T Easy kit (Promega), and clones were sequenced using standard Sanger sequencing. See Key Resource Table for primer sequences.
Northern blot analysis:
A total of 5 μg of polyA+ enriched RNA (Dynabeads mRNA purification kit, Invitrogen) was resolved on a 1% denaturing polyacrylamide gel, transferred to a Hybond XL membrane (GE Healthcare Life Sciences), and crosslinked (Stratalinker 1800 UV, Stratagene). The membrane was prehybridized with ULTRAhyb buffer (Ambion) for two hours, then hybridized with the Platr4-specific radiolabeled DNA probe overnight at 42°C. Platr4-specific DNA probes were labeled with [γ−32P] ATP in a random primed labeling reaction using the Prime-It RmT Random Primer Labeling Kit (Stratagene). The next day, the hybridized blot was washed three times in 2xSSC/0.1%SDS and once in 0.1xSSC/0.1%SDS. The signal was quantified using a Fujifilm Life Science FLA-5100 imaging system and X-ray film.
Western blot analysis:
Cells were washed with PBS and lysed in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40 substitute, 1% sodium deoxycholate, and 0.1% SDS) supplemented with a 1X Roche protease inhibitor cocktail on ice for 30 minutes with occasional vortex. Lysate was then centrifuged at 13000 × g for 30 minutes. The supernatant was collected, and protein concentration was measured by BCA protein assay. Proteins were separated by SDS-PAGE on 4%–20% discontinuous polyacrylamide gels. The proteins were transferred onto a nitrocellulose membrane. The membrane was blocked in 5% non-fat dry milk for one hour at room temperature and incubated with primary antibodies (see below) at 4°C overnight. The following day membranes were washed three times for 5 minutes each in PBS-Tween (0.05%) and incubated with appropriate secondary antibody for one hour at room temperature. Then the membrane was washed three times, and the signal was visualized using west pico chemiluminescent substrate (Thermo Fisher Scientific). The primary antibodies were as follows: anti-Tead4 (Abcam, ab97460), anti-Tead1 (BD Biosciences, 610923), anti-Tead2 (Biorbyt, orb382464), anti-Tead3 (Novus Biologicals, NBP1-83949), and anti-Ctgf (Novus Biologicals, NB100-724SS); anti-Actin (Sigma, A5441) was used as the loading control.
RNA isolation and quantitative real-time PCR (qRT-PCR) assays:
Total RNA was extracted from cells or tissues using TRIzol according to the manufacturer’s instructions. Then, 1 μg of total RNA was used to synthesize cDNA using the TaqMan Reverse Transcription Reagent kit (Thermofischer). Next, 30 ng of cDNA was used to perform the qRT-PCR reaction using SYBR green PCR master mix on an ABI QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). The housekeeping genes Gapdh, CycloB, and Pabpc were used as internal controls to normalize the gene of interest. Each experiment had two samples, and each assay was run three times. Primer sequences are listed in Supplemental Table S2.
Cell fractionation:
Cell fractionation was performed as previously described109. Briefly, 5 million cells were resuspended in ice-cold lysis buffer containing 10 mM Tris pH 7.5, 150 mM NaCl, and 0.15% NP-40 substitute. The cytoplasmic fraction was separated from the nuclei by overlaying the cell suspension on a sucrose buffer containing 10 mM Tris pH7.4, 150 mM NaCl, and 24% sucrose and was centrifuged at 3500 × g for 10 minutes. The remaining nuclei pellet was rinsed with ice-cold PBS-ethylenediaminetetraacetic acid (EDTA) once and resuspended in urea buffer (1 M urea, 0.3 M NaCl, 7.5 mM MgCl2, 0.2 mM EDTA, and 1% NP-40 substitute) on ice for two minutes. The lysate was then centrifuged at 13000 × g for two minutes to separate the chromatin pellet from the supernatant comprising the nucleoplasm fraction. The cytoplasm fraction, nucleoplasm fraction, and chromatin pellet were used for RNA extraction using the TRIzol reagent according to the manufacturer’s protocol.
Cloning:
To overexpress specific genes, an insert containing the gene of interest was cloned into pBApo-EF1alpha Puro (TaKaRa) and pCMV6-entry (Origene) plasmids, following the manufacturer’s protocol. The plasmids were transformed into NEB Stable Competent E. coli using the heat shock method. Four to five colonies per plate were chosen and sequenced using standard Sanger sequencing.
Knockdown using small interfering RNA (siRNA):
The siRNA transfection was performed in ESCs using 4D-Nucleofector X Unit, program code CG104. ON-TARGET-plus SMARTpool siRNAs for Tead1, Tead2, Tead4, and non-targeting siRNA were purchased from Dharmacon Inc. The experiments were performed at least in triplicate.
RNA sequencing and analysis:
Total RNA was extracted from both ESCs and EBs using TRIzol according to the manufacturer’s instructions. RNA quality was assayed by running an RNA 6000 Nanochip on a 2100 Bioanalyzer (Agilent). Each RNA sample had an RNA integrity number (RIN) of 9 or above. Then, 500 ng of total RNA was used to prepare poly(A)+ enriched RNA-seq libraries using the Illumina TruSeq sample prep kit following the manufacturer’s protocol. The libraries were multiplexed and sequenced single-end 75 bp on the NextSeq500 platform (Illumina). Reads were then mapped to the mouse mm10 genome using RNA STAR110, and reads per gene record were counted using HTSeq-counts111 using the GENCODE M20 annotation. The list of differentially expressed genes was generated using DESeq2112, and a false discovery rate (FDR)-adjusted P-value of < 0.05 was set as the threshold for statistical significance. KEGG pathway and gene ontology term enrichment analyses were carried out using the R/Bioconductor packages GAGE113 and Pathview114.
iRegulon analysis:
iRegulon v1.3 (build: 2015-02-12) analysis was performed as described previously53. The master regulator transcription factors were identified using a differential gene set as the input for the motif search. The thresholds set for this motif enrichment analysis were as follows: the minimum normalized enrichment score (NES) > 3 and FDR on motif similarity < 0.001. Motif collection was set to 10 K (9713 PWMs), and putative regulatory search regions of 10 kb centered around the TSS (7 species) and 500 bp upstream of the TSS for the ranking option. Targeted transcription factors were obtained based on the HOMER, TRANSFAC, yeTFaSCo databases included in the iRegulon analysis.
RNA immunoprecipitation (RIP):
RIP was performed as previously described115. Briefly, 10 million ESCs were harvested in Tryple E, washed with cold PBS, resuspended in 2 ml of PBS, 2 ml of nuclear isolation buffer (1.28 M sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100), and 6 ml of nuclease-free water. The cells were incubated on ice for 20 minutes with intermittent vortex. The lysate was pelleted by centrifugation at 2500 × g for 15 minutes to harvest nuclei. Nuclei pellets were resuspended in 1 ml RIP buffer (150 mM KCl, 25 mM Tris pH 7.5, 5 mM EDTA, 0.5 mM Dithiothreitol (DTT), 0.5% NP-40, 100 U/ml SUPERase-IN, and 1X Roche protease inhibitor cocktail) and sonicated for 15 minutes using a Pico BioRuptor (30 seconds ON/OFF) (Diagenode) at 4°C. The lysate was then centrifuged at 16000 × g for 10 minutes. The supernatant was collected and incubated with 4 μg of Tead4 (Abcam #ab58310), Tead1 (Cell Signaling Technology #12292), Tead2 (Biorbyt #orb382464), Tead3 (Novus Biologicals #NBP1-8394) antibodies and IgG control for overnight at 4°C with gentle shaking. The next day, 40 μl of protein G and A beads (Thermo Fisher Scientific) for mouse and rabbit antibodies, respectively, were added to the immune-complex reactions and incubated for two hours at 4°C with gentle shaking. Beads were washed three times with RIP buffer and once with PBS. Then, the beads were collected for Western blot analysis and RNA extraction to perform qRT-PCR. The primer sequences are listed in Supplemental Table S2.
Chromatin immunoprecipitation (ChIP) coupled with quantitative PCR (ChIP-qPCR):
ChIP was performed according to the manufacturer’s protocol (Active Motif) with minor modification. A total of 15 million cells were crosslinked with 1% formaldehyde for 10 minutes at room temperature. The reaction was then quenched by adding glycine at a final concentration of 0.125 M at room temperature for 5 minutes. Cells were centrifuged at 2500 rpm for 10 minutes at 4°C, and the supernatant was discarded. The cell pellet was resuspended in lysis buffer and incubated on ice for 30 minutes, followed by a centrifugation step at 5000 rpm for 10 minutes at 4°C to pellet the nuclei. Next, the nuclei pellets were resuspended in shearing buffer and sonicated for 20 minutes using a Pico BioRuptor (30 seconds ON/OFF) at 4°C. The sonicated sample was centrifuged at a maximum speed for 15 minutes at 4°C, and the supernatant was transferred to a new Eppendorf tube for immunoprecipitation. Anti-Tead4 (Abcam #ab58310) antibodies were used for ChIP assay. ChIP-enriched DNA was quantified by qPCR on an ABI QuantStudio 6 Flex Real-Time PCR System (Applied Biosystem). The ChIP-qPCR primers can be found in Supplemental Table S2.
Embryo collection and in vitro culture:
Embryos were collected at embryonic day 0.5 (E0.5) from swollen ampullae treated with hyaluronidase (Sigma) at 37°C for 2–3 minutes to remove cumulus cells. Embryos were then washed three times in PBS with BSA (Sigma, 6 mg/ml) and cultured for three days in 15 μl drops of Potassium Simplex Optimized Medium (KSOM-AA, Millipore) covered with mineral oil (Ovoil, Vitrolife) in a humidified chamber at 37°C with 5% CO2. Post-implantation embryos were collected at different embryonic days (E6.5, E8.5, E10.5, and E12.5), fixed in 4% paraformaldehyde (PFA) overnight at 4°C, washed with PBS, and used for whole-mount RNA fluorescence in situ hybridization (FISH).
Single-molecule RNA FISH:
ESCs:
Single-molecule RNA FISH was performed according to the manufacturer’s protocol for the Affymetrix View ISH Cell Assay Kit (Thermo Fisher Scientific, QVT0050) using custom Type-6 primary probes targeting Platr4. ESCs were seeded onto acid-cleaned #1.5 glass coverslips (Electron Microscopy Sciences, 72230-01) for 24 hours to reach 80% confluence, then fixed in freshly prepared 4% PFA (Electron Microscopy Sciences, 19200). Fixed cells were permeabilized and protease digested before hybridization. The hybridization and signal amplification steps were performed according to the manufacturer’s instructions, and nuclei were counterstained with DAPI. Coverslips were mounted in ProLong Gold Antifade mounting medium (Thermo Fisher Scientific, P36930) for imaging on Zeiss LSM 710/780 confocal microscope systems.
Whole-mount embryos:
Single-molecule RNA FISH was performed on formalin-fixed whole-mount embryos using RNAscope® Fluorescent Multiplex Reagent Kit 320850 (ACD #320850). Pretreatment of the tissue sections, hybridization, and signal amplification were all performed according to the manufacturer’s instructions. Mounted embryos were imaged on a Zeiss LSM710 or LSM780 spinning disk confocal microscope.
Immunofluorescence:
ESCs:
ESCs were cultured onto acid-cleaned #1.5 glass coverslips for 24 hours to reach 80–90% confluence. The following day, the cells were fixed in freshly prepared 4% PFA for 20 minutes at room temperature, washed three times with PBS, and permeabilized in PBS with 0.2% Triton-X and 0.1% Tween-20 for 30 minutes on ice. Permeabilized cells were blocked in PBS with 1% BSA for one hour at room temperature, then the cells were incubated with anti-Oct4 antibody (mouse monoclonal, Santa Cruz Biotechnology, sc-5279, dilution 1:100) overnight at 4°C. The next day, coverslips were washed three times in blocking reagent and stained with anti-mouse secondary antibody (Alexafluor 488). After washing three times, the nuclei were counterstained with DAPI and the coverslips were imaged on a Zeiss LSM710 or LSM780 spinning disk confocal microscope.
Hematoxylin and Eosin (H&E staining:
Heart tissue and EBs were fixed in 4% PFA overnight, washed three times in PBS, and dehydrated by a series of ethanol and xylene washes before embedding in paraffin. Paraffin-embedded tissue and EBs were sectioned and stained with H&E. Slides were scanned using an Aperio slide scanner (Leica Biosystems). A pathologist analyzed the sections to assess the histologic grade.
RNA pull-down:
This experiment was performed as previously described15. Briefly, 10 million cells were used to prepare the nuclear pellet, and the pellet was resuspended in RIP buffer (150 mM KCl, 25 mM Tris pH 7.5, 0.5 mM DTT, 0.5% NP40, 1 mM ABSEF, protease inhibitor cocktail (Roche), and 20 U/ml RNaseOut (Invitrogen)) and sonicated for 10 cycles using a Pico BioRuptor (30 seconds ON/OFF) at 4°C. The sonicated lysate was then centrifuged for 10 minutes at 13000 rpm. Then, 50 pmol of in vitro transcribed biotinylated RNA was added to the supernatant and incubated for one hour at room temperature, followed by the addition of 60 μl streptavidin Dynabeads (Invitrogen) and incubation for one hour at room temperature. The lysate was then washed four times with RIP buffer, and the beads were denatured in SDS buffer to release protein. Proteins were analyzed by Western blotting.
Genotyping:
Tail DNA was prepared using standard methods and analyzed by PCR. Knockout primer pairs were designed outside the deleted region, and wild-type primer pairs were created inside the deleted area (Supplemental Table S2). Knockout PCR bands from each founder were confirmed by Sanger sequencing.
Echocardiography:
All cardiac ultrasound imaging was performed using a Vevo3100 scanner (VisualSonics) and a MX550D transducer. In preparation for the scan and on the same days as imaging, mice were anesthetized with 2–3% isoflurane. The hair over the chest region was entirely removed by shaving and Nair (depilation cream). For the actual scan, mice were again anesthetized with 2–3% isoflurane, and their paws were lightly taped onto the copper plates of the heated imaging platform. A small quantity of Aquasonic gel was applied to the paws before taping to ensure a full ECG trace. Both long and short axis heart views were acquired in B and M mode and analyzed using the LV analysis software tool (VisualSonics). Each mouse was kept warm using a heated platform and warm Aquasonic ultrasound gel while scanning. The gel was placed on the mid-abdomen to the upper ventral side of the mice. Clips were taken of the long and short axis of each mouse’s heart in both B and M mode. After the scanning was completed, Vevo LAB software was used to analyze the images and perform the Cardiac Package of LV Trace on the long and short axis.
Quantification and statistical analysis:
All figures were assembled and statistical tests were performed using Microsoft Excel and R. Box plots depict the median and the 25th and 75th percentiles; whiskers correspond to minimum and maximum non-outlier values in Figures 7H, I, J, K and L. The P-value was calculated by two-tailed paired Student’s t-test. Significance was defined as P < 0.05. All data are presented as mean ± SEM. The number of samples (N) are indicated in each respective figure legend.
Supplementary Material
Table S1, Platr4 regulates the expression of ECM genes (Related to Figure 3)
Table S2, Primer sequences (Related to Figures 1–7 and Figures S1–S7)
Highlights.
Platr4 is an ESC-specific and early embryonic lncRNA
Platr4 is required for cardiogenic mesoderm differentiation
Platr4 functions upstream of Ctgf to regulate cardiac lineage differentiation
Platr4-knockout mice exhibit cardiac dysfunction
Acknowledgements
We thank all members of the Spector laboratory for important suggestions throughout the course of this research. We thank Dr. Tse-Luen (Erika) Wee, Director of the Cold Spring Harbor Laboratory (CSHL) Microscopy Shared Resource for assistance with imaging. We thank to Dr. Qing Gao, Director of the CSHL Histology Shared Resource for assistance with embedding and sectioning of mouse heart samples. We would also like to thank the staff of the Flow Cytometry, Animal, and Next-Gen Sequencing Shared Resources for services and technical expertise (NCI 2P3OCA45508). This research was supported by NIGMS R35GM131833 (to DLS) and by NIH R01 HD085904 (to MMS).
Footnotes
Declaration of interests
The authors declare no competing interests.
Inclusion and Diversity
We support inclusive, diverse and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Derrien T et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789. 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aban CE et al. (2021). Downregulation of E-cadherin in pluripotent stem cells triggers partial EMT. Sci Rep 11, 2048. 10.1038/s41598-021-81735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL & Chang HY (2012). Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166. 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC & Chang HY (2011). Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914. 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulitsky I & Bartel DP (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28, 503–510. 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao RW, Wang Y & Chen LL (2019). Cellular functions of long noncoding RNAs. Nat Cell Biol 21, 542–551. 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 10.Statello L, Guo CJ, Chen LL & Huarte M (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96–118. 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann JH et al. (2015). Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res 25, 1336–1346. 10.1101/gr.189027.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer TR et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11, 14. 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexanian M et al. (2017). A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun 8, 1806. 10.1038/s41467-017-01804-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo X et al. (2018). A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Cell Stem Cell 22, 893–908 e896. 10.1016/j.stem.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Klattenhoff CA et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobuss L, Bar C & Thum T (2019). Long Non-coding RNAs: At the Heart of Cardiac Dysfunction? Front Physiol 10, 30. 10.3389/fphys.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerritsen KG et al. (2016). Plasma CTGF is independently related to an increased risk of cardiovascular events and mortality in patients with atherosclerotic disease: the SMART study. Growth Factors 34, 149–158. 10.1080/08977194.2016.1210142. [DOI] [PubMed] [Google Scholar]
- 18.Leeuwis JW et al. (2010). Connective tissue growth factor is associated with a stable atherosclerotic plaque phenotype and is involved in plaque stabilization after stroke. Stroke 41, 2979–2981. 10.1161/STROKEAHA.110.589036. [DOI] [PubMed] [Google Scholar]
- 19.Rickard AJ et al. (2009). Deletion of mineralocorticoid receptors from macrophages protects against deoxycorticosterone/salt-induced cardiac fibrosis and increased blood pressure. Hypertension 54, 537–543. 10.1161/HYPERTENSIONAHA.109.131110. [DOI] [PubMed] [Google Scholar]
- 20.Guney MA et al. (2011). Connective tissue growth factor acts within both endothelial cells and beta cells to promote proliferation of developing beta cells. Proc Natl Acad Sci U S A 108, 15242–15247. 10.1073/pnas.1100072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bischoff J (2019). Endothelial-to-Mesenchymal Transition. Circ Res 124, 1163–1165. 10.1161/CIRCRESAHA.119.314813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abreu JG, Ketpura NI, Reversade B & De Robertis EM (2002). Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol 4, 599–604. 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croci S et al. (2004). Inhibition of connective tissue growth factor (CTGF/CCN2) expression decreases the survival and myogenic differentiation of human rhabdomyosarcoma cells. Cancer Res 64, 1730–1736. 10.1158/0008-5472.can-3502-02. [DOI] [PubMed] [Google Scholar]
- 24.Luo Q et al. (2004). Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem 279, 55958–55968. 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao B et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22, 1962–1971. 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishioka N et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16, 398–410. 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Pocaterra A, Romani P & Dupont S (2020). YAP/TAZ functions and their regulation at a glance. J Cell Sci 133. 10.1242/jcs.230425. [DOI] [PubMed] [Google Scholar]
- 28.Jacquemin P, Hwang JJ, Martial JA, Dolle P & Davidson I (1996). A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem 271, 21775–21785. 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- 29.Lian X et al. (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848–1857. 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBlanc L et al. (2018). Yap1 safeguards mouse embryonic stem cells from excessive apoptosis during differentiation. Elife 7. 10.7554/eLife.40167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Z et al. (2020). YAP/TEAD3 signal mediates cardiac lineage commitment of human-induced pluripotent stem cells. J Cell Physiol 235, 2753–2760. 10.1002/jcp.29179. [DOI] [PubMed] [Google Scholar]
- 32.Wilson KD et al. (2020). Endogenous Retrovirus-Derived lncRNA BANCR Promotes Cardiomyocyte Migration in Humans and Non-human Primates. Dev Cell 54, 694–709 e699. 10.1016/j.devcel.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva AC, Pereira C, Fonseca A, Pinto-do OP & Nascimento DS (2020). Bearing My Heart: The Role of Extracellular Matrix on Cardiac Development, Homeostasis, and Injury Response. Front Cell Dev Biol 8, 621644. 10.3389/fcell.2020.621644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segers VFM, Brutsaert DL & De Keulenaer GW (2018). Cardiac Remodeling: Endothelial Cells Have More to Say Than Just NO. Front Physiol 9, 382. 10.3389/fphys.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong L et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35, W345–349. 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L et al. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41, e74. 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin MF, Jungreis I & Kellis M (2011). PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282. 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slaymaker IM et al. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desbaillets I, Ziegler U, Groscurth P & Gassmann M (2000). Embryoid bodies: an in vitro model of mouse embryogenesis. Exp Physiol 85, 645–651. [PubMed] [Google Scholar]
- 40.Kim JM et al. (2011). Assessment of differentiation aspects by the morphological classification of embryoid bodies derived from human embryonic stem cells. Stem Cells Dev 20, 1925–1935. 10.1089/scd.2010.0476. [DOI] [PubMed] [Google Scholar]
- 41.Bain G, Kitchens D, Yao M, Huettner JE & Gottlieb DI (1995). Embryonic stem cells express neuronal properties in vitro. Dev Biol 168, 342–357. 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 42.Holtzinger A et al. (2015). New markers for tracking endoderm induction and hepatocyte differentiation from human pluripotent stem cells. Development 142, 4253–4265. 10.1242/dev.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misfeldt AM et al. (2009). Endocardial cells are a distinct endothelial lineage derived from Flk1+ multipotent cardiovascular progenitors. Dev Biol 333, 78–89. 10.1016/j.ydbio.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 44.Saba R et al. (2019). Endocardium differentiation through Sox17 expression in endocardium precursor cells regulates heart development in mice. Sci Rep 9, 11953. 10.1038/s41598-019-48321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saint-Jean L et al. (2019). Myocardial differentiation is dependent upon endocardial signaling during early cardiogenesis in vitro. Development 146. 10.1242/dev.172619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardot E et al. (2017). Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential. Nat Commun 8, 14428. 10.1038/ncomms14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lockhart MM, van den Hoff M & Wessels A The Role of the Epicardium in the Formation of the Cardiac Valves in the Mouse, in Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. (eds. Nakanishi T et al. ) 161–167 (Tokyo; 2016). [PubMed] [Google Scholar]
- 48.Chatzizacharias NA, Kouraklis GP & Theocharis SE (2010). The role of focal adhesion kinase in early development. Histol Histopathol 25, 1039–1055. 10.14670/HH-25.1039. [DOI] [PubMed] [Google Scholar]
- 49.Riley JK et al. (2005). The PI3K/Akt pathway is present and functional in the preimplantation mouse embryo. Dev Biol 284, 377–386. 10.1016/j.ydbio.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 50.Schenke-Layland K et al. (2007). Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells 25, 1529–1538. 10.1634/stemcells.2006-0729. [DOI] [PubMed] [Google Scholar]
- 51.Rozario T & DeSimone DW (2010). The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol 341, 126–140. 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor-Weiner H, Schwarzbauer JE & Engler AJ (2013). Defined extracellular matrix components are necessary for definitive endoderm induction. Stem Cells 31, 2084–2094. 10.1002/stem.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janky R et al. (2014). iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10, e1003731. 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev A, Kaneko KJ, Shu H, Zhao Y & DePamphilis ML (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15, 1229–1241. 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y et al. (2016). The TEAD Family and Its Oncogenic Role in Promoting Tumorigenesis. Int J Mol Sci 17. 10.3390/ijms17010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon JH & Gorospe M (2016). Cross-Linking Immunoprecipitation and qPCR (CLIP-qPCR) Analysis to Map Interactions Between Long Noncoding RNAs and RNA-Binding Proteins. Methods Mol Biol 1402, 11–17. 10.1007/978-1-4939-3378-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benhaddou A et al. (2012). Transcription factor TEAD4 regulates expression of myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death Differ 19, 220–231. 10.1038/cdd.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang W et al. (2018). miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis 9, 92. 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murry CE & Keller G (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680. 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y et al. (2016). Mesp1 Marked Cardiac Progenitor Cells Repair Infarcted Mouse Hearts. Sci Rep 6, 31457. 10.1038/srep31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao G, Kotick JD & Lincoln J (2012). Heart valve development, maintenance, and disease: the role of endothelial cells. Curr Top Dev Biol 100, 203–232. 10.1016/B978-0-12-387786-4.00006-3. [DOI] [PubMed] [Google Scholar]
- 62.Palpant NJ et al. (2017). Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat Protoc 12, 15–31. 10.1038/nprot.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pratumvinit B, Reesukumal K, Janebodin K, Ieronimakis N & Reyes M (2013). Isolation, characterization, and transplantation of cardiac endothelial cells. Biomed Res Int 2013, 359412. 10.1155/2013/359412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen K et al. (2010). Endothelial cells regulate cardiomyocyte development from embryonic stem cells. J Cell Biochem 111, 29–39. 10.1002/jcb.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akerberg BN et al. (2019). A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun 10, 4907. 10.1038/s41467-019-12812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gardin JM, Siri FM, Kitsis RN, Edwards JG & Leinwand LA (1995). Echocardiographic assessment of left ventricular mass and systolic function in mice. Circ Res 76, 907–914. 10.1161/01.res.76.5.907. [DOI] [PubMed] [Google Scholar]
- 67.Hoffman M et al. (2019). Myocardial Strain and Cardiac Output are Preferable Measurements for Cardiac Dysfunction and Can Predict Mortality in Septic Mice. J Am Heart Assoc 8, e012260. 10.1161/JAHA.119.012260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han X et al. (2018). Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J Mol Cell Biol 10, 175–178. 10.1093/jmcb/mjy003. [DOI] [PubMed] [Google Scholar]
- 69.Grote P et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24, 206–214. 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ballarino M et al. (2018). Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice. EMBO J 37. 10.15252/embj.201899697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han P et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Currey L, Thor S & Piper M (2021). TEAD family transcription factors in development and disease. Development 148. 10.1242/dev.196675. [DOI] [PubMed] [Google Scholar]
- 73.Anbanandam A et al. (2006). Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A 103, 17225–17230. 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacquemin P, Martial JA & Davidson I (1997). Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J Biol Chem 272, 12928–12937. 10.1074/jbc.272.20.12928. [DOI] [PubMed] [Google Scholar]
- 75.Yagi R et al. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836. 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 76.Nishioka N et al. (2008). Tead4 is required for specification of trophectoderm in preimplantation mouse embryos. Mech Dev 125, 270–283. 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Wang J et al. (2018). Effect of TEAD4 on multilineage differentiation of muscle-derived stem cells. Am J Transl Res 10, 998–1011. [PMC free article] [PubMed] [Google Scholar]
- 78.Joshi S et al. (2017). TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet 13, e1006600. 10.1371/journal.pgen.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Estaras C, Hsu HT, Huang L & Jones KA (2017). YAP repression of the WNT3 gene controls hESC differentiation along the cardiac mesoderm lineage. Genes Dev 31, 2250–2263. 10.1101/gad.307512.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J, Liu S, Heallen T & Martin JF (2018). The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol 15, 672–684. 10.1038/s41569-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 81.von Gise A et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A 109, 2394–2399. 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blais A et al. (2005). An initial blueprint for myogenic differentiation. Genes Dev 19, 553–569. 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen HH et al. (2004). Transcription enhancer factor-1-related factor-transgenic mice develop cardiac conduction defects associated with altered connexin phosphorylation. Circulation 110, 2980–2987. 10.1161/01.CIR.0000146902.84099.26. [DOI] [PubMed] [Google Scholar]
- 84.Chen M et al. (2020). Structural and Functional Overview of TEAD4 in Cancer Biology. Onco Targets Ther 13, 9865–9874. 10.2147/OTT.S266649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan BS et al. (2019). LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38, 5612–5626. 10.1038/s41388-019-0812-8. [DOI] [PubMed] [Google Scholar]
- 86.Li Z et al. (2016). Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front Physiol 7, 687. 10.3389/fphys.2016.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambi AG et al. (2012). The skeletal site-specific role of connective tissue growth factor in prenatal osteogenesis. Dev Dyn 241, 1944–1959. 10.1002/dvdy.23888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramazani Y et al. (2018). Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol 68–69, 44–66. 10.1016/j.matbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 89.Surveyor GA & Brigstock DR (1999). Immunohistochemical localization of connective tissue growth factor (CTGF) in the mouse embryo between days 7.5 and 14.5 of gestation. Growth Factors 17, 115–124. 10.3109/08977199909103520. [DOI] [PubMed] [Google Scholar]
- 90.Lopes LB et al. (2009). Cell permeant peptide analogues of the small heat shock protein, HSP20, reduce TGF-beta1-induced CTGF expression in keloid fibroblasts. J Invest Dermatol 129, 590–598. 10.1038/jid.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daniels A, van Bilsen M, Goldschmeding R, van der Vusse GJ & van Nieuwenhoven FA (2009). Connective tissue growth factor and cardiac fibrosis. Acta Physiol (Oxf) 195, 321–338. 10.1111/j.1748-1716.2008.01936.x. [DOI] [PubMed] [Google Scholar]
- 92.Shi-Wen X, Leask A & Abraham D (2008). Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev 19, 133–144. 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 93.Koitabashi N et al. (2008). Plasma connective tissue growth factor is a novel potential biomarker of cardiac dysfunction in patients with chronic heart failure. Eur J Heart Fail 10, 373–379. 10.1016/j.ejheart.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 94.Goh SK, Olsen P & Banerjee I (2013). Extracellular matrix aggregates from differentiating embryoid bodies as a scaffold to support ESC proliferation and differentiation. PLoS One 8, e61856. 10.1371/journal.pone.0061856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sart S, Ma T & Li Y (2014). Extracellular matrices decellularized from embryonic stem cells maintained their structure and signaling specificity. Tissue Eng Part A 20, 54–66. 10.1089/ten.TEA.2012.0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kodigepalli KM et al. (2020). Biology and Biomechanics of the Heart Valve Extracellular Matrix. J Cardiovasc Dev Dis 7. 10.3390/jcdd7040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen B et al. (2000). Mice mutant for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet 24, 296–299. 10.1038/73528. [DOI] [PubMed] [Google Scholar]
- 98.Qi X et al. (2007). Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development. Dev Biol 311, 136–146. 10.1016/j.ydbio.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 99.Song R et al. (2015). BMP-2 and TGF-beta1 mediate biglycan-induced pro-osteogenic reprogramming in aortic valve interstitial cells. J Mol Med (Berl) 93, 403–412. 10.1007/s00109-014-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee SW et al. (2013). Snail as a potential target molecule in cardiac fibrosis: paracrine action of endothelial cells on fibroblasts through snail and CTGF axis. Mol Ther 21, 1767–1777. 10.1038/mt.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.von Gise A & Pu WT (2012). Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 110, 1628–1645. 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barnett JV & Desgrosellier JS (2003). Early events in valvulogenesis: a signaling perspective. Birth Defects Res C Embryo Today 69, 58–72. 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- 103.Chen JH & Simmons CA (2011). Cell-matrix interactions in the pathobiology of calcific aortic valve disease: critical roles for matricellular, matricrine, and matrix mechanics cues. Circ Res 108, 1510–1524. 10.1161/CIRCRESAHA.110.234237. [DOI] [PubMed] [Google Scholar]
- 104.Bergmann O et al. (2009). Evidence for cardiomyocyte renewal in humans. Science 324, 98–102. 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frappaz D, Bourgeois J, Berthier JC, Laurent C & Bethenod M (1983). [Syndrome of terminal deletion of the long arm of chromosome 4. Apropos of a personal case with a review of the literature]. Pediatrie 38, 261–270. [PubMed] [Google Scholar]
- 106.Copelli S, del Rey G, Heinrich J & Coco R (1995). Brief clinical report: interstitial deletion of the long arm of chromosome 4, del(4)(q28-->q31.3). Am J Med Genet 55, 77–79. 10.1002/ajmg.1320550120. [DOI] [PubMed] [Google Scholar]
- 107.Ockey CH, Feldman GV, Macaulay ME & Delaney MJ (1967). A large deletion of the long arm of chromosome No. 4 in a child with limb abnormalities. Arch Dis Child 42, 428–434. 10.1136/adc.42.224.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han Y, Slivano OJ, Christie CK, Cheng AW & Miano JM (2015). CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice--brief report. Arterioscler Thromb Vasc Biol 35, 312–315. 10.1161/ATVBAHA.114.305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Conrad T & Orom UA (2017). Cellular Fractionation and Isolation of Chromatin-Associated RNA. Methods Mol Biol 1468, 1–9. 10.1007/978-1-4939-4035-6_1. [DOI] [PubMed] [Google Scholar]
- 110.Dobin A et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anders S, Pyl PT & Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Love MI, Huber W & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo W, Friedman MS, Shedden K, Hankenson KD & Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo W & Brouwer C (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y et al. (2016). Characterization of Long Noncoding RNA-Associated Proteins by RNA-Immunoprecipitation. Methods Mol Biol 1402, 19–26. 10.1007/978-1-4939-3378-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eggan K et al. (2001). Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A 98, 6209–6214. 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Platr4 regulates the expression of ECM genes (Related to Figure 3)
Table S2, Primer sequences (Related to Figures 1–7 and Figures S1–S7)
Data Availability Statement
No custom codes were generated in this study.
Raw RNA-Sequencing data generated in this study have been deposited at NCBI SRA website and are publicly available as of the date of 09/18/2022: PRJNA880813 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA880813/) and PRJNA881397 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881397/)
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Oct-3/4 Antibody | Santa Cruz | Cat # sc-5279 |
| Anti-mouse Alexa Fluor 488 | Thermofischer | Catalog # A28175 |
| Anti-TEAD4 antibody - ChIP Grade | Abcam | Cat # ab58310 |
| Anti-TEAD4 antibody | Abcam | Cat # ab97460 |
| Anti-TEAD1 antibody | BD Biosciences | Cat # 610923 |
| Anti-TEAD1 antibody | Cell Signaling Technology | Cat #12292 |
| Anti-TEAD2 antibody | Biorbyt | Cat # orb382464 |
| Anti-TEAD3 antibody | Novus Biologicals | Cat # NBP1-83949 |
| CTGF/CCN2 antibody | Novus Biologicals | Cat # NB100-724SS |
| Monoclonal anti-β-Actin | Millipore-Sigma | Cat # A5441 |
| Anti-Yap1-antobody | Novus Biologicals | Cat # NB110-58358 |
| Bacterial and virus strains | ||
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | Cat # 48138 |
| pSpCas9(BB)-2A-Puro (PX459) V2.0 | Addgene | Cat # 62988 |
| Chemicals, peptides, and recombinant proteins | ||
| Pregnant mare’s serum gonadotropin (PMSG) | ProSpec | Cat # HOR-272 |
| Human chorionic gonadotropin (hCG) | Sigma Aldrich | Cat # C1063 |
| KSOM Mouse Embryo Media | Millipore Sigma | Cat # MR-121-D |
| EmbryoMax® M2 Medium | Millipore Sigma | Cat # MR-015-D |
| Mineral oil | Ovoil, Vitrolife | Cat # 10029 |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | Cat # 19200 |
| Acidic Tyrode’s solution | Sigma-Aldrich | Cat # MR-004-D, |
| Phosphate-buffered saline (PBS) | Thermo Fisher | Cat # 144444 |
| Bovine serum albumin (BSA) | Thermo Fisher | Cat # 15260037 |
| Molecular Grade Water | G-Biosciences | Cat # 786-73C |
| Triton X-100 | Sigma-Aldrich | Cat # X100 |
| SUPERase•In RNase Inhibitor | Thermo Fisher | Cat # AM2696 |
| Agarose | Millipore Sigma | Cat # A0701-25G |
| 20x saline-sodium citrate (SSC) buffer | G- Biosciences | Cat # R020 |
| RNAscope Multiplex Fluorescent V2 Assay | Advanced Cell Diagnostics | Cat # 323120 |
| Mouse Platr4 probe set | RNAscope Target Probe C1 | Cat # 300031 |
| Opal 570 | Akoya Biosciences | Cat # FP1488001KT |
| RNase A | Thermo Fisher | Cat # EN0531 |
| RNase H | Epicenter | Cat # H39500 |
| Leukemia inhibitory factor (LIF) | Millipore | Cat # ESG1107 |
| Iscove’s Modification of DMEM | Corning | Cat # 15-016-CV |
| B27 Supplement Gibco | Thermo Fischer | Cat # 0080085SA |
| Trizol Reagent | Thermo Fischer | Cat # 15596018 |
| Ultrahyb Ultrasensative hybridization buffer | Thermo Fischer | Cat # AM8670 |
| NorthernMax 10X Running Buffer | Thermo Fischer | Cat # AM8671 |
| NorthernMax Denaturing Buffer (10X) | Thermo Fischer | Cat # AM8676 |
| Proteinase K Solution | Viagen Biotech | Cat # 501-PK |
| Poly(A)Purist™ MAG Kit | Thermo Fischer | Cat # AM1922 |
| Retinoic Acid | Sigma | Cat # R2625-50MG |
| L-Ascorbic acid | Sigma | Cat # A7506-25g |
| PowerUp™ SYBR® Green Master Mix | Thermo Fisher | Cat # A25743 |
| MEM NEAA | Thermo Fisher | Cat # 11140-050 |
| Sodium Pyruvate | Sigma | Ca t# P2256-25G |
| Pierces™ IP Lysis Buffer | Thermo Fisher | Cat # 87787 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher | Cat # 89900 |
| N2 Supplement 100x | Thermo Fisher | Cat # 17502048 |
| ON-TARGETplus Mouse Tead4 (21679) siRNA - SMARTpool | Dharmacon | Cat # L-057322-01-0005 |
| ON-TARGETplus Mouse Tead2 (21677) siRNA - SMARTpool | Dharmacon | Cat # L-060552-01-0005 |
| ON-TARGETplus Mouse Tead3 (21678) siRNA - SMARTpool | Dharmacon | Cat # L-044127-01-0005 |
| ON-TARGETplus Mouse Yap1 siRNA | Dharmacon | Cat # L-046247-01-0005 |
| ON-TARGETplus Mouse Tead2 (21677) siRNA | Dharmacon | Cat # L-060552-01-0005 |
| ON-TARGETplus Non-targeting Pool | Dharmacon | Cat # D-001810-10-05 |
| SuperScript II Reverse Transcriptase | Life Technologies | Cat # 18064014 |
| Penicillin-Streptomycin 10,000U/mL | Life Technologies | Cat # 15140122 |
| BSA fraction V (7.5%) | Thermo fischer | Cat # 15260037 |
| dPBS | Life Technologies | Cat # 14190-250 |
| TaqMan Reverse Transcription Reagents | Thermo fischer | Cat # 4304134 |
| Chromatin IP DNA purification kit | Active motif | Cat # 58002 |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 10-well, 50 μl #4561094 | Biorad | Cat # 4561094 |
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat # RS-122-2001 |
| Retinoic acid | Sigma | Cat # R2625-100MG |
| 1-Thioglycerol | Sigma | Cat # M6145-25ML |
| DR4 MEF Feeder Cells, P3, Irradiated | Applied Stem cell | Cat # ASF-1014 |
| EmbryoMax® PMEF P3 Strain CF-1, Irradiated | Millipore | Cat # R-4169444.2 |
| Poly(A)Purist™ MAG Kit | Thermo Fisher | Cat # AM1922 |
| Proteinase K Solution | Viagen Biotech | Cat # 501-PK |
| Iscove’s Modification of DMEM | Corning | Cat # 15-016-CV |
| Ham’s F-12 Medium | Corning | Cat # 10-080-CV |
| StemPro-34 | GIBCO | Cat # 10639011 |
| human VEGF | R&D system | Cat # 293-VE |
| human Activin A | R&D system | Cat # 338-AC |
| human BMP4 | R&D system | Cat # 314-BP |
| human basic FGF | R&D system | Cat # 233-FB |
| Deposited data | ||
| Raw RNAseq data of control vs Platr4-KO mouse ES cells | This paper | SRA data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA880813/ |
| Raw RNAseq data of control vs Platr4-KO differentiated mouse ES cells | This paper | SRA data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881397/ |
| Unprocessed western blot images | This paper | Mendeley Data: https://data.mendeley.com/datasets/2ddgwynm42 |
| Experimental models: Cell lines | ||
| Mouse ES cell line (V6.5) | Eggan et al., 2001116 | CSHL Gene Targeting Facility |
| Mouse ES cell line (AB2.2) | ATCC | Cat # SCRC-1023 |
| Knocout ES cell lines | This Paper | NA |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratory, Farmington, CT USA | Cat # 000664 |
| Platr4-KO mouse | This paper | NA |
| Oligonucleotides | ||
| 5’ RACE outer: ACCACACCCCTAACAGTACCAC | This paper | NA |
| 5’ RACE inner: TGTACGCTGGATAGAACTGCAT | This paper | NA |
| 3’ RACE outer: CCATCATGGGCTTTGCTAGT | This paper | NA |
| 3’ RACE inner: CCCACGAGGGTCAACAGTAG | This paper | NA |
| Platr4-primer-F: GCCCTGGAGAAGTCACAGAG | This paper | NA |
| Platr4-primer-R: ATGGGGAAGAAGGAAGAGGA | This paper | NA |
| gPCR_Platr4-F1: GGGCTCAAAGAAGGTGACTG | This paper | NA |
| gPCR_Platr4-R1: GGCCTCAAATTCAGAGACCA | This paper | NA |
| gPCR_Platr4-F2: GGCCAATTGTTCCACAAGAAG | This paper | NA |
| gPCR_Platr4-R2: AGAGACCACCTCCCTCTGCT | This paper | NA |
| Recombinant DNA | ||
| Tead4 (NM_011567) Mouse Tagged ORF Clone | Origene | Cat # MR219506 |
| Ctgf (BC006783) Mouse Untagged Clone | Origene | Cat #: MC200503 |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| R | R | https://www.r-project.org/ |
| UCSC genome browser | UCSC | https://genome.ucsc.edu/ |
| ensembl genome browser | EMBL-EBI | http://useast.ensembl.org/index.html |
| ZEN microscope software | ZEISS | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| The sgRNAs design software | http://crispr.mit.edu/ | |
| Other | ||
| Microscope, inverted | Nikon | TS100 |
| Stereoscope dissection microscope | Leica | MZ12.5 |
| Microscope, confocal | Zeiss | LSM 780 |
Microscopy data reported in this paper will be shared by the lead contact upon request
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.


