Abstract
NLRs are important intracellular pattern recognition receptors and play an essential role in innate immunity for vertebrates. However, research on sturgeon NLRs is still scarce. In this study, the NLRC3-like gene from Siberian sturgeon (Acipenser baerii) (AbNLRC3-like) was cloned and characterized. The AbNLRC3-like full-length cDNA was composed of 3310 bp with a 166 bp of 5′-UTR, 2838 bp open reading frame (ORF) and 306 bp 3′-UTR, encoding 945 amino acids. Structural analysis of AbNLRC3-like showed the typical NLRs structure, including a central NACHT domain and a C-terminal 12 LRR motifs. Quantitative real-time PCR (qRT-PCR) analysis revealed that AbNLRC3-like was widely distributed in all tested tissues with a relatively high expression level in the mid-kidney, head-kidney and spleen. After Streptococcus iniae infection, the mRNA expression of AbNLRC3-like was significantly up-regulated at the pre-mortality period and the recovering period in valvula intestine, but it was significantly down-regulated around the mortality period in the duodenum and spleen. In splenic leukocyte, the mRNA expression of AbNLRC3-like was significantly induced by LPS, PGN and Poly(I:C). These results suggested that AbNLRC3-like may play a critical role in the immune response of Siberian sturgeon during pathogen challenge. To our best knowledge, this is the first report of the NLRC3 gene in sturgeon.
Keywords: Siberian sturgeon, NLRC3, Streptococcus iniae, PAMPs, Immune response
1. Introduction
The innate immune system is the host's first line of defense and plays a crucial role in resisting pathogen invasion. NOD-like receptors (NLRs) are important intracellular pattern recognition receptors in innate immunity, which can recognize pathogen-associated molecular patterns (PAMPs), including LPS, PGN and Poly(I:C) [1], [2], [3]. The identified twenty-three NLRs in human were divided into four main subfamilies: NLRA, NLRB, NLRC and NLRP by the differences in N-terminal domains [4]. Interestingly, the NLRs found in teleost fish are far more than that in mammals, for instance nearly 400 NLR proteins are identified in the zebrafish genome, and research has mainly focused on the NLRC subfamily [5,6]. To date, mammals NOD1, NOD2, NLRC3, NLRC5 and NLRX1 homologous genes were found in fish.
NLRC3 is a non-inflammasome-forming member of the NLR family in mammalian who was first identified as an inhibitor of T cell activation [7]. Subsequent studies confirmed that NLRC3 inhibited inflammation and immune response by negatively regulating the activation of NF-κB and STING-dependent pathway to reduce the production of type I interferon [7,8]. In addition, NLRC3 can inhibit the activation of PI3K-mTOR signal axis [9]. In fish, NLRC3 was reported involved in innate immunity against a variety of pathogen infections in at least 11 species, including zebrafish (Danio rerio) [5,10,11], channel catfish (Ictalurus punctatus) [12], [13], [14], goldfish (Carassius auratus L.) [15], Nile tilapia (Oreochromis niloticus) [16], turbot (Scophthalmus maximus L.) [17], Asian seabass (Lates calcarifer) [18], miiuy croaker (Miichthys miiuy) [19], blunt snout bream (Megalobrama amblycephala) [20], Japanese flounder (Paralichthys olivaceus) [21], rainbow trout (Oncorhynchus mykiss) [22] and Atlantic salmon (Salmo salar) [23]. These studies suggest that fish NLRC3 can be involved in the immune response in vivo and in vitro. However, its role in fish immunology response mechanism is still unclear.
Sturgeon is an ancient freshwater fish that has survived for hundreds of millions of years. The adaptation of the immune system in the evolution of sturgeon is worthy of attention. In aquaculture, sturgeons show a relatively strong disease resistance, but enteritis is still a typical infection symptom [24], [25], [26]. The previous study constructed streptococcal enteritis in Siberian sturgeon (Acipenser baerii) by low lethal doses of Streptococcus iniae (S. iniae) [27]. Then, the valvula intestinal transcriptome data hinted that NLRC may be involved in the development of streptococcal enteritis (Data no published). However, the immune response of A. baerii against pathogen invasion remains mostly unknown. In this study, the AbNLRC3-like was cloned and characterized. The tissue distribution and expression analysis in streptococcal enteritis were investigated. Moreover, the expression responses of splenic leukocytes after PAMPs stimulation were detected. The present results contributed to understanding the immune strategy of fish against infective diseases.
2. Materials and methods
2.1. Fish
Siberian sturgeons were collected from a local fish farm in Pengzhou (Sichuan, China) and maintained in aerated water tanks at 24 ± 0.5 ℃ for two weeks before experiments. Fish were fed twice daily with commercial feed in the amount of 1% of their body weight. Approximately one third of the water was replaced twice daily. Three Siberian sturgeon (0.75 ± 0.14 kg) were used for tissue distribution experiment, ninety-six Siberian sturgeon (68.88 ± 14.70 g) were used for immune response in S. iniae infection, and six Siberian sturgeon (1.63 ± 0.18 kg) were used for in vitro stimulation experiment. All animal procedures were approved by the Animal Care and Use Committee of Sichuan Agricultural University.
2.2. RNA extraction and cDNA library construction
Total RNA of each sample was extracted with Trizol Reagent (Takara, Japan) according to the manufacturer's protocol. After checking the quality by electrophoresis on 1% agarose gel and NanoDrop 2000c spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), the RNA was transcribed into cDNA by PrimeScript® RT Reagent Kit with gDNA Eraser (Takara, Japan).
2.3. Amplification of AbNLRC3-like cDNA
The cDNA from the Siberian sturgeon spleen was used as a template. According to the Siberian sturgeon Transcriptome Shotgun Assembly project accession GIPE00000000 design the primes. Then, RACE PCR was applied to obtain the full cDNA length of AbNLRC3-like using SMART™ RACE cDNA Application Kit (Takara, Japan). All the PCR products were purified and ligated into the pMD19-T vector (Takara, Japan). The positive clones were sequenced by Sangon Biotech (Shanghai, China). The sequenced fragments were further assembled with Seqman 7.1 software to obtain the full-length cDNA of AbNLRC3-like. All primers used for gene clone were listed in Table S1.
2.4. Bioinformatics analysis of AbNLRC3-like
The ORF of AbNLRC3-like was deduced by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The protein domains of AbNLRC3-like were predicted using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl.heidelberg.de/). The physicochemical features of proteins, including molecular weight, amino acid composition, theoretical isoelectric point, were predicted using ProParam software in ExPasy server [28]. The signal peptide was predicted by SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP) [29]. Sequence identities of amino acids were calculated using Clustalx 1.83 software. Multiple sequence alignment and phylogenetic tree were constructed by MEGA X software using the Neighbor-joining method, with the bootstrap set as 1000.
2.5. Expression profiles of AbNLRC3-like mRNA
Three healthy Siberian sturgeons were used for tissue expression detection. 96 healthy Siberian sturgeons were used for tissue expression response after S. iniae infection at 1 d, 3 d, 5 d, 7 d and 10 d. Six healthy Siberian sturgeon were used for in vitro splenocyte culture, and samples were collected at 6 h, 12 h, 24 h, 48 h and 72 h after stimulation with LPS (25 μg/mL), PGN (10 μg/mL) and Poly(I:C) (50 μg/mL). Extraction of total RNA and then preparation of cDNA were described above. The primers for quantitative PCR (qPCR) are listed in Table S1, and according the SYBR® Premix Ex Taq™ II(Takara, Japan) manufacturer's instructions to Real-time quantitative PCR. In these experiments, β-actin was amplified as the reference gene.
2.6. Statistical analysis
All data were expressed as mean ± SEM. Statistical analysis was analyzed using SPSS Statistics package 19.0 (SPSS Inc., Chicago, Illinois). A paired sample t-test was applied to check the significance of the data and differences were consideredas statistically significant at p < 0.05. GraphPad Prism 8 (Graphpad Inc., China) was used for drawing.
3. Results and discussion
NLRs are important pattern-recognition receptors that play an essential role in innate immunity. However, to date, there is little known about NLRs in Siberian sturgeon. In this study, NLRC3-like was identified and reported from the Siberian sturgeon for the first time, and its tissue distribution expression profiles and the expression pattern in S. iniae infection and PAMPs stimulation were analyzed.
3.1. Bioinformatics analysis of AbNLRC3-like
The AbNLRC3-like full-length cDNA is 3310 bp, including the 5′ non-coding region of 166 bp, the 3′ non-coding region of 306 bp and the open reading frame of 2838 bp encoding 945 amino acid residues. AbNLRC3-like contained an in-frame stop condon upstream of the start condon (ATG) and a polyadenylation signal (AATAA motif) before the Poly A tail (Fig. S1), suggesting that the complete ORF of the gene had been obtained. By using SMART, the results show that AbNLRC3-like has a NACHT domain and 12 LRR motifs. Then the physical and chemical characteristics of AbNLRC3-like were analyzed using the ProtParam program. The relative molecular weight of the AbNLRC3-like protein is 103.66 kDa and the isoelectric point (pI) value is 5.72. There is no signal peptide or transmembrane domain in AbNLRC3-like protein, and this result is consistent with the research in turbot [17], Asian seabass [18], miiuy croaker [19] and blunt snout bream [20]. These results suggest AbNLRC3-like locates in the cytosol [12,30].
3.2. Comparative structural and phylogenetic analysis of AbNLRC3-like
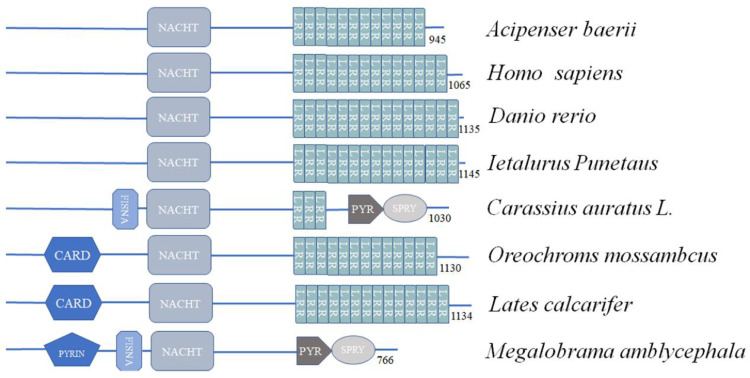
The AbNLRC3-like has a NACHT domain and C-terminal LRR motifs, which is similar to NLRC3 of human, zebrafish, goldfish [15], and channel catfish [14], but the N-terminal effector domain is absence in AbNLRC3-like (Fig. 1). Turbot NLRC3 possesses CARD effector domain [17], but the N-terminal of blunt snout bream NLRC3 is Pyrin domain [20]. Studies in mammals have shown that NLRC3 is a pro-inflammatory signal inhibitor, which can inhibit the activation of NF-κB. NLRC3 seems to function by interacting with TRAF6, which is a key protein for NF-κB transcription and activation in the nucleus. The NLRC3-TRAF6 complex passes through the 2 TRAF-binding sites in the NLR sequence ((P/S/A/T)-X-(Q/E)-E, where “P/S/A/T” indicates proline, serine, alanine or threonine, “X” indicates any amino acid, and “Q/E” indicates glutamine or glutamic acid). There are two TRAF-binding sites in the NACHT domain of AbNLRC3-like, site 1: PDQE, site 2: SAEE. It hinted AbNLRC3-like may bind TRAF to active NF-κB transcription. Meanwhile, there are differences in the number of LRR motifs that may play important roles in the ligand recognition and binding of NLRs. The difference in the number of LRR motifs may be related to the specificity of ligand recognition. AbNLRC3-like has 12 LRR motifs, while Nile tilapia NLRC3 has 13 LRR motifs, human and turbot NLRC3 have 14 LRR motifs, zebrafish and Asian seabass NLRC3 have 15 LRR motifs [17,20]. These NLRC3 can participate in the response of pathogens infected and PAMPs stimulation, the difference in the specificity of fish NLR ligands whether is caused by the difference in the number of LRR motif still remains to be further studied.
Fig. 1.
Schematic representation of the domain architecture of NLRC3 in different animals, obtained by using SMART. Abbreviations: CARD, caspase associated recruitment domain; PYRIN, pyrin domain; FISNA, fish-specific NACHT associated domain; NACHT: nucleotide-binding/oligomerisation domain; LRR, leucine-rich repeats; PRY-SPRY, PRY-SPRY or B30.2 domain.
The deduced amino acid sequence of AbNLRC3-like has less than 30% identity with the currently known NLRC3 in teleost (Table S2), except for the chondrostean Acipenser ruthenus (XP. 034,772,582.1) with the highest identity as 99%. The consistency of NLRC3 NACHT domain and LRR domain between Siberian sturgeon and other species is less than 35% (Table S3, Table S4). The results of multiple sequence alignment of the NLRC3 motif NACHT reported in the literature show that there are three conserved motifs in NACHT, namely Walker A (GVAGIGKT), Walker B (LLILDGLDEFR) and Sensor 1 (TSRP) (Fig. 2). In the Walker A motif, the lysine residue directly interacts with the phosphate part of ATP, and the Walker B motif is the region of ATPase responsible for ATP hydrolysis and oligomerization. Sensor 1 is responsible for the interaction with the γ phosphate of ATP [31]. To further verify the evolutionary relationship between AbNLRC3 and other known vertebrate NLRCs, MEGA X was used to carry out multiple sequence alignments, and then the phylogenetic tree was constructed by the Neighbour-joining method. AbNLRC3-like firstly clustered with A. ruthenus NLR, and then clustered with NLRC3 of other known teleost fishes in the evolutionary tree (Fig. 3).
Fig. 2.
Multiple alignments of the deduced amino acid sequence of the NACHT domain of NLRC3 from different species. Dashes (-) indicate gaps, black shadow indicates identical residues, and gray shadow indicates similar residues in the aligned amino acid sequences. Walker A, Walker B and Sensor 1 motif is boxed.
Fig. 3.
Phylogenetic tree analysis of the deduced amino acid sequences of NLR.
3.3. Tissue distribution of AbNLRC3-like
The qRT-PCR was used to detect the tissue distribution of AbNLRC3-like transcripts. The results showed AbNLRC3-like transcripts was ubiquitously expressed in all 12 tissues selected, including duodenum (Du), liver (Li), valvula intestine (Vi), pyloric caeca (Pc), rectum (Re), esophagus (Es), stomach (St), Gill (Gi), spleen (Sp), skin (Sk), head-kidney (Hk) and middle-kidney (Mk). Similar results were found in other fish species, including channel catfish [14], rainbow trout [22] and goldfish [15], indicating that NLRC3 may play a crucial role in pathogen identification in multiple tissues(Fig. 4). Interestingly, our results show that AbNLRC3-like mRNA is expressed highest in the mid-kidney and head kidney but is lower in the intestine (Fig. 4). Similarly, the channel catfish NLRC3 and turbot NLRC3a also have a high expression in the head kidney [12]. In contrast, goldfish NLRC3-like and Asian seabass NLRC3 highly expressed in the intestine [15,18]. These results indicate that fish NLRC3 is highly expressed in immune or mucosal immune-related organs, suggesting that NLRC3 may play an important role in fish immunity.
Fig. 4.
Normal expression profile of AbNLRC3-like mRNA in different tissues of healthy Siberian sturgeon. Data are shown as relative expression ± SEM in 12 different tissues: Sk-Skin, Du-duodenum, Es-Esophagus, Re-Rectum, Mk-Middle kidney, St-Stomach, Vi-Valvula intestine, Hk-Head-kidney, Gi-Gill, Pc-Pyloric caeca, Li-Liver, Sp-Spleen. Different letters indicate significant differences at p < 0.05.
3.4. Expression of AbNLRC3-like in S. iniae infected
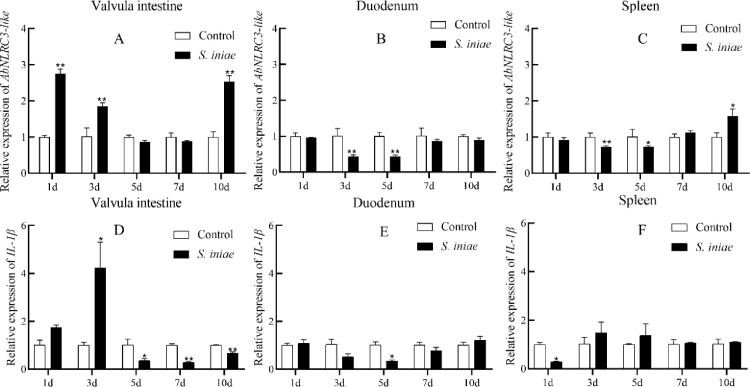
As a pattern recognition receptor, activation of NLRC3-like indicates that the host body is initiating resistance to foreign invasion pathogens. Our previous studies have shown that S. iniae infection in Siberian sturgeons can be divided into pre-mortality period (1–3 days post-infection, dpi), mortality period (4–6 dpi) and post-mortality period (7–10 dpi) [27]. The expression patterns of AbNLRC3-like in the three tissues tested showed significant differences. At 1, 3, 10 dpi, the expression of AbNLRC3-like mRNA in the valvular intestine showed a significant up-regulation trend (Fig.5). After a similar Vibrio alginolyticus infection, Asian seabass NLRC3 mRNA was significantly up-regulated at the early intestinal time point [18]. The expression of AbNLRC3-like mRNA in the duodenum and spleen was significantly down-regulated at pre-mortality and mortality period (3–5 dpi) (Fig. S4). IL-1β was a key cytokine of the NLR signaling pathway. However, our results indicate that the response pattern of IL-1β mRNA is different from that of NLRC3-like mRNA in vivo experiment (Fig. 5). A similar study showed that Nile tilapia was infected by Streptococcus agalactiae andNLRC3 mRNA in the intestine was significantly down-regulated at 3 dpi and up-regulated at 6 dpi and 9 dpi, while NLRC3 mRNA expression in the spleen was significantly down-regulated in 1–9 d [16]. After infection with different pathogens, channel catfish NLRC3 gene expression showed tissue specific profiles, channel catfish NLRC3 mRNA was significantly down-regulated after Edwardsiella ictaluri infection in intestine [13].The expression levels of channel catfish NLRC3 mRNA was significantly down-regulated at 12 h-72 h after Aeromonas hydrophila infection in spleen [12]. However, the expression of channel catfish NLRC3 mRNA was significantly up-regulated at 24 h-72 h after Edwardsiella tarda (E. tarda) infection in liver and only up-regulated at one time point in intestine and spleen [12]. After A. hydrophila infected blunt snout bream, NLRC3-like mRNA in head kidney, liver, intestine and body kidney time to peak at 12 h [20]. After E. tarda infection, Japanese flounder NLRC3 mRNAwas down-regulated at 4 h and then up-regulated at 8, 12, 24 h post infection (hpi) of spleen, head kidney and gill [21]. This study and other studies have shown that the NLRC3 response has tissue specificity. It plays an essential role in detecting multiple pathogens.
Fig. 5.
Expression profile of AbNLRC3-like (A, B and C) and IL-1β (D, E and F) mRNA in various tissues at different time-points in response to bacterial challenge with S. iniae. Quantification of AbNLRC3-like levels was normalized to β-actin. The “*” denotes significant differences (p < 0.05) relative to the control group. “**” means p < 0.01.
3.5. Expression of AbNLRC3-like mRNA in splenic leukocyte following LPS, PGN and Poly(I:C) stimulation
The spleen is one of the critical immune organs of fish. It is composed of many monocytes/macrophages, dendritic cells, and lymphocytes. At the same time, NLRC3 is mainly expressed in immune response cells [7]. To further investigate the function of AbNLRC3-like, their gene expressions in primary splenic leukocyte following LPS, PGN and Poly(I:C) stimulation were analyzed. After LPS and PGN stimulation of leucocyte from the spleen, NLRC3-like mRNA expression was significantly up-regulated from 6 h to 72 h after infection, while the expression of NLRC3-like mRNA after Poly(I:C) stimulation was significantly up-regulated at both 6 h and 12 h, and significantly down-regulated at 48 h and 72 h. Interestingly, the expression of AbNLRC3-like mRNA reached its peak at 6 h after PGN and Poly(I:C) stimulation, while peaked at 12 h after LPS stimulation (Fig. 6). Similar peak differences appear in rainbow trout, both LPS and Poly(I:C) can induce a significant up-regulation of rainbow trout NLRC3 mRNA expression. The difference is that the expression of rainbow trout NLRC3 mRNA reached a significant at 4 h after LPS stimulation, while Poly(I:C) did not reach it until 8 h [22]. In vitro studies of Asian seabass kidney cell line stimulated by LPS, PGN and Poly(I:C) showed that NLRC3 mRNA expression was significantly up-regulated at some time points [18]. In peripheral blood leucocytes, the expression of Japanese flounder NLRC3 mRNA was up-regulated in 2 h after LPS stimulation, reached the maximum expression level at 8 h, subsequent gradually decreased to the basal level[21]. After LPS and PGN stimulated splenic leukocyte, IL-1β mRNA expression was significantly up-regulated at 6 h, 12 h and 24 h, but had no significant change at 48 h and 72 h. After Poly(I:C) stimulation, IL-1β mRNA was significantly up-regulated at 6 h and 12 h, and significantly down-regulated at 24 h, 48 h and 72 h, which was similar with the trend of NLRC3-like mRNA expression profiles after Poly(I:C) stimulation. These findings indicate that piscine NLRC3 may be a critical intracellular pattern recognition receptor involved in recognizing and responding to multiple pathogens.
Fig. 6.
Expression profile of AbNLRC3-like (A, B and C)and IL-1β (D, E and F)mRNA in s Splenic leukocyte at different time-points in response to LPS, PGN or Poly(I:C). Quantification of AbNLRC3-like levels was normalized to β-actin. The “*” denotes significant differences (p < 0.05) relative to the control group. “**” means p < 0.01.
In short, we characterized a new AbNLRC3-like gene and provided new information about AbNLRC3-like in the innate immunity of sturgeon. Furthermore, the expressions of AbNLRC3-like mRNA after S. iniae infection in vivo and PAMPs stimulation in vitro were detected. It confirmed that the AbNLRC3-like gene might play an essential role in the Siberian sturgeon response against pathogens. Further studies can verify the NLRC3-like formation of inflammasome or NODosome and specific role of NLRC3-like in immune response through gene knockdown. These studies not only contribute to enrich the immunological background of fish NLR, but also provide a new target for the prevention and control of fish diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Opening Fund of Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River (No. NJTCCJSYSYS03) and Sichuan Innovation Team Project of Agricultural Industry Technology System (No. SCCXTD-15).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fsirep.2021.100042.
Contributor Information
Jun Wang, Email: wangjunzl@126.com.
Defang Chen, Email: chendf_sicau@126.com.
Appendix. Supplementary materials
References
- 1.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 4.Lim R.R., Wieser M.E., Ganga R.R., Barathi V.A., Lakshminarayanan R., Mohan R.R., Hainsworth D.P., Chaurasia S.S. NOD-like Receptors in the Eye: uncovering Its Role in Diabetic Retinopathy. Int. J. Mol. Sci. 2020;21(3) doi: 10.3390/ijms21030899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laing K.J., Purcell M.K., Winton J.R., Hansen J.D. A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol. Biol. 2008;8:42. doi: 10.1186/1471-2148-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang M., Xiong F., Wu X., Hu Y. The expanding and function of NLRC3 or NLRC3-like in teleost fish: recent advances and novel insights. Dev. Comp. Immunol. 2021;114 doi: 10.1016/j.dci.2020.103859. [DOI] [PubMed] [Google Scholar]
- 7.Conti B.J., Davis B.K., Zhang J., O'Connor, Jr W., Williams K.L., Ting J.P. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J. Biol. Chem. 2005;280(18):18375–18385. doi: 10.1074/jbc.M413169200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L., Mo J., Swanson K.V., Wen H., Petrucelli A., Gregory S.M., Zhang Z., Schneider M., Jiang Y., Fitzgerald K.A., Ouyang S., Liu Z.J., Damania B., Shu H.B., Duncan J.A., Ting J.P. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40(3):329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen I.C. Non-inflammasome forming NLRs in inflammation and tumorigenesis. Front. Immunol. 2014;5:169. doi: 10.3389/fimmu.2014.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T., Yan B., Lou L., Lin X., Yu T., Wu S., Lu Q., Liu W., Huang Z., Zhang M., Zhang W., Wen Z. Nlrc3-like is required for microglia maintenance in zebrafish. J. Genet. Genomics. 2019;46(6):291–299. doi: 10.1016/j.jgg.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Fang H., Wu X., Hu Y., Song Y., Zhang J., Chang M. NLRC3-like 1 inhibits NOD1-RIPK2 pathway via targeting RIPK2. Dev. Comp. Immunol. 2020;112 doi: 10.1016/j.dci.2020.103769. [DOI] [PubMed] [Google Scholar]
- 12.Li M., Wang Q., Lu Y., Chen S., Li Q., Sha Z. Expression profiles of NODs in channel catfish (Ictalurus punctatus) after infection with Edwardsiella tarda, Aeromonas hydrophila, Streptococcus iniae and channel catfish hemorrhage reovirus. Fish Shellfish Immunol. 2012;33(4):1033–1041. doi: 10.1016/j.fsi.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 13.Rajendran K.V., Zhang J., Liu S., Kucuktas H., Wang X., Liu H., Sha Z., Terhune J., Peatman E., Liu Z. Pathogen recognition receptors in channel catfish: I. Identification, phylogeny and expression of NOD-like receptors. Dev. Comp. Immunol. 2012;37(1):77–86. doi: 10.1016/j.dci.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Sha Z., Abernathy J.W., Wang S., Li P., Kucuktas H., Liu H., Peatman E., Liu Z. NOD-like subfamily of the nucleotide-binding domain and leucine-rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunol. 2009;33(9):991–999. doi: 10.1016/j.dci.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Xie J., Belosevic M. Characterization and functional assessment of the NLRC3-like molecule of the goldfish (Carassius auratus L.) Dev. Comp. Immunol. 2018;79:1–10. doi: 10.1016/j.dci.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Gao F., Pang J., Lu M., Yang X., Zhu H., Ke X., Liu Z., Cao J. Wang, Molecular characterization, expression and functional analysis of NOD1, NOD2 and NLRC3 in Nile tilapia (Oreochromis niloticus) Fish Shellfish Immunol. 2018;73:207–219. doi: 10.1016/j.fsi.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Hou Z., Ye Z., Zhang D., Gao C., Su B., Song L., Tan F., Song H., Wang Y., Li C. Characterization and expression profiling of NOD-like receptor C3 (NLRC3) in mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 2017;66:231–239. doi: 10.1016/j.fsi.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Paria A., Deepika A., Sreedharan K., Makesh M., Chaudhari A., Purushothaman C.S., Thirunavukkarasu A.R., Rajendran K.V. Identification of Nod like receptor C3 (NLRC3) in Asian seabass, Lates calcarifer: characterisation, ontogeny and expression analysis after experimental infection and ligand stimulation. Fish Shellfish Immunol. 2016;55:602–612. doi: 10.1016/j.fsi.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Li J., Kong L., Gao Y., Wu C., Xu T. Characterization of NLR-A subfamily members in miiuy croaker and comparative genomics revealed NLRX1 underwent duplication and lose in actinopterygii. Fish Shellfish Immunol. 2015;47(1):397–406. doi: 10.1016/j.fsi.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Zhou F., Zhan Q., Ding Z., Su L., Fan J., Cui L., Chen N., Wang W., Liu H. A NLRC3-like gene from blunt snout bream (Megalobrama amblycephala): molecular characterization, expression and association with resistance to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2017;63:213–219. doi: 10.1016/j.fsi.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Li S., Chen X., Hao G., Geng X., Zhan W., Sun J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immunol. 2016;50:79–90. doi: 10.1016/j.fsi.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Álvarez C.A., Ramírez-Cepeda F., Santana P., Torres E., Cortés J., Guzmán F., Schmitt P., Mercado L. Insights into the diversity of NOD-like receptors: identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol. Immunol. 2017;87:102–113. doi: 10.1016/j.molimm.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Pontigo J.P., Yañez A., Sanchez P., Vargas-Chacoff L. Characterization and expression analysis of Nod-like receptor 3 (NLRC3) against infection with Piscirickettsia salmonis in Atlantic salmon. Dev. Comp. Immunol. 2021;114 doi: 10.1016/j.dci.2020.103865. [DOI] [PubMed] [Google Scholar]
- 24.Deng M., Yu Z., Geng Y., Wang K., Chen D., Huang X., Ouyang P., Chen Z., Zhong Z., Lai W. Outbreaks of Streptococcosis associated with Streptococcus iniae in Siberian sturgeon (Acipenser baerii) in China. Aquac. Res. 2017;48(3):909–919. [Google Scholar]
- 25.Shchelkunov I.S., Shchelkunova T.I., Shchelkunov A.I., Kolbassova Y.P., Didenko L.V., Bykovsky A.P. First detection of a viral agent causing disease in farmed sturgeon in Russia. Dis. Aquat. Organ. 2009;86(3):193–203. doi: 10.3354/dao02124. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L., Geng Y., Lei X., Yu Z., Huang X., Chen D., Ouyang P., Cao S., Han R. Isolation and identification of Streptococcus iniae from sturgeon and its pathological lesions of infection. Acta Agriculturae Zhejiangensis. 2018;30(2):203–210. (in Chinese) [Google Scholar]
- 27.Chen D., Peng S., Chen D., Yang F., Liu J., Wang J., Liu Q., Huang X., Ouyang P., Wang K., Li Z., Geng Y. Low lethal doses of Streptococcus iniae caused enteritis in Siberian sturgeon (Acipenser baerii) Fish Shellfish Immunol. 2020;104:654–662. doi: 10.1016/j.fsi.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., Grosdidier A., Hernandez C., Ioannidis V., Kuznetsov D., Liechti R., Moretti S., Mostaguir K., Redaschi N., Rossier G., Xenarios I., Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:597–603. doi: 10.1093/nar/gks400. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen H. Predicting secretory proteins with SignalP. Methods Mol. Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 30.Motta V., Soares F., Sun T., Philpott D.J. NOD-like receptors: versatile cytosolic sentinels. Physiol. Rev. 2015;95(1):149–178. doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- 31.Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.