Highlights
-
•
LvBigPEN, a member of penaeidins from L. vannamei, was regulated by host AP-1 signaling pathway.
-
•
LvBigPEN could bind to both Gram-negative bacteria and Gram-positive bacteria.
-
•
LvBigPEN could destroy bacterial cells and bind to DNA.
-
•
LvBigPEN played an important role in defense against V. parahaemolyticus infection.
Keywords: Antimicrobial peptide, Penaeidins, V. parahaemolyticus, AP-1 pathway
Abstract
Penaeidins are members of an antimicrobial peptide (AMP) family that have broad anti-microbial activities only found in penaeid shrimps. The LvBigPEN, a member of penaeidins from shrimp Litopenaeus vannamei, has showed antiviral activity against white spot syndrome virus (WSSV) in our previous report. However, whether LvBigPEN possesses potential anti-bacterial activities is still unknown. Herein, we found that the LvBigPEN played an important role in restricting the infection of Vibrio parahaemolyticus, a natural and Gram-negative bacteria pathogen in shrimp. The transcription of LvBigPEN was strongly induced after V. parahaemolyticus challenge. RNA interference (RNAi) mediated knockdown of LvBigPEN showed that LvBigPEN had a potential antibacterial function against V. parahaemolyticus. Microorganism binding assays indicated that rLvBigPEN could bind to both Gram-negative bacteria and Gram-positive bacteria. Transmission electron microscopy (TEM) analysis showed its ability to destroy bacterial cells in vitro. Besides, in a gel retardation assay, rLvBigPEN could bind to plasmid DNA and bacteria (V. parahaemolyticus) genomic DNA in a concentration-dependent manner. Moreover, the AP-1 pathway could participate in the transcription of LvBigPEN by the dual luciferase reporter assays. Taken together, these results suggested that LvBigPEN possessed the antibacterial activity against V. parahaemolyticus and may be alternative agents for the prevention and treatment of diseases caused by V. parahaemolyticus.
1. Introduction
Pacific white shrimp (Litopenaeus vannamei) is one of the most important commercial marine species in the world [1]. However, the shift of shrimp farming modes, such as higher density and super-intensive culture in semi-closed or closed water, has led to a remarkable increase in outbreaks of various diseases associated with several protozoal, fungal, bacterial, and viral agents in recent years [2]. Vibrio parahaemolyticus is the common bacterial pathogen of shrimps. In particular, the bacteria V. parahaemolyticus carrying a virulence plasmid of PirA/B is thought to cause a serious disease to farmed Penaeid shrimp, usually referred to as “early mortality syndrome” (EMS), also known as “acute hepatopancreatic necrosis disease” (AHPND) [3]. Outbreak of diseases often occurs when the homeostasis between pathogens and host resistance is disrupted. Meanwhile, the abuses of antibiotics in shrimp culture with the attempt to control bacterial disease have caused serious threats to ecological environment and human health [4,5]. Therefore, there is an intense desire for environmentally friendly and effective prophylaxis for diseases caused by V. parahaemolyticus.
Host innate immune system plays a significant role in protecting organisms from pathogenic invasion, particularly in invertebrates that lacking the adaptive immunity [6]. Antimicrobial peptides (AMPs) are important components of the innate immune system. AMPs are originally defined as membrane-active molecules with small molecular mass (<10 kDa) that show antimicrobial activities [7]. In shrimps, several families of AMPs have been identified and characterized, including penaedins (PENs), crustins (CRUs), anti-lipopolysaccharide factors (ALFs), lysozymes (LYZs) and stylicins (STYs) [8,9]. Growing researches show that these AMPs have diversity activities against bacteria, fungi, as well as viruses.
Penaeidins are unique cationic molecules that consist of an N-terminal proline-rich region (PRR) and a C-terminal cysteine-rich region (CRR) within six conserved cysteine residues forming three disulfide bonds [10,11]. Previous study showed that they can be classified into four distinct subgroups: PEN2, PEN3, PEN4 and PEN5 (as PEN1 turned out to be the variant of PEN2) based on amino acid sequence comparisons and the position of specific amino acids [12]. Notably, usually more than one subgroup was found in a penaeid shrimp. For example, three penaeidins subgroups (PEN2, PEN3 and PEN4), were identified in L. vannamei and Litopenaeus setiferus [13], whilst two subgroups of penaeidins, PEN3 and PEN5, were found in Fenneropenaeus chinensis [14] and P. monodon [15]. In our previous study, we identified a new member of penaeidin, named LvBigPEN (accession no. MN149368), and uncovered that LvBigPEN could function as an antiviral effector against white spot syndrome virus (WSSV) though antagonizing viral envelope proteins to block its entry [16]. As members of AMPs, most penaeidins have been identified to have significant antibacterial activities [8], although their antibacterial mechanisms are still unknown. In the present study, we found that LvBigPEN has antibacterial activity with ability to destroy bacterial membrane structure and bind with bacterial DNA in vitro. RNA interference also showed that LvBigPEN provided a protect role against V. parahaemolyticus in vivo. Together, these results could help us better understand the function of penaeidin in shrimp innate immunity.
2. Materials and methods
2.1. Animals and bacteria
Healthy shrimp L. vannamei (average 5 g each) were purchased from the local shrimp farm in Zhanjiang, Guangdong Province, China, and cultured in recirculating water tank system filled with air-pumped sea water with 5 ‰ salinity at 27 °C, and fed to satiation three times/day on commercial diet. The Gram-negative bacteria used in our analysis included Vibrio parahaemolyticus (ATCC25922), Aeromonas hydrophila (ATCC35654), Pseudomonas aeruginosa (ATCC17802) and Escherichia coli (ATCC27853). The Gram-positive bacteria contained Staphylococcus aureus (ATCC29213), Enterococcus faecalis (ATCC29212), Micrococcus luteus (ATCC49732) and Bacillus subtilis (ATCC6633). All these bacteria were purchased from Guangdong Microbial Culture Collection Center and cultured in Luria broth (LB) medium overnight at 30 °C or 37 °C. Each bacterium was quantified by counting the microbial colony-forming units (CFU) per milliliter on LB agar plates. For bacterial challenge experiment, the final injection concentration of V. parahaemolyticus was adjusted to yield ∼1 × 105 CFU in 50 μl PBS [17].
2.2. Total RNA extraction and cDNA synthesis
Total RNA was extracted from different tissues of shrimp using the Eastep Super Total RNA Extraction Kit (Promega, Shanghai, China). The genomic DNA of shrimp tissues was extracted using a genomic DNA Extraction Kit (Omega, Guangzhou, China), according to the manufacturer's instructions. First strand cDNA synthesis was performed using a cDNA Synthesis Kit (Takara, Dalian, China), following the manufacturer's recommendations.
2.3. Quantitative RT-PCR
To explore the expression of LvBigPEN in response to bacterial infection, the treated groups were injected with 50 μl V. parahaemolyticus suspension (1 × 105 CFU) at the second abdominal segment of each shrimp, while the control groups were injected with phosphate buffer saline (PBS) solution. Hemocytes of challenged shrimps were collected at 0, 4, 8, 12, 24, 36, 48, 72 h (h) post injection, and the samples at each time point were pooled from 15 shrimps. Total RNA and quantitative RT-PCR (qRT-PCR) were performed as described previously [18]. Expression levels of LvBigPEN were calculated using the Livak (2−ΔΔCT) method after normalization to L. vannamei EF-1α (GU136229). Primer sequences were listed in Table 1.
Table 1.
Primers used in this paper.
| Primers | Sequences (5′−3′) |
|---|---|
| Quantitative PCR | |
| LvBigPEN-F | ACCACAGACCCCAAGTCCTA |
| LvBigPEN-R | AGTTCCGGCAGATTTCGGTT |
| c-Fos-F | CCATTACAGCTGTGGCTACGAGT |
| c-Fos-R | GGTCTGTTCGATGTTCCTCAAG |
| c-Jun-F | GACGCCCTCCCAGTTCTTCTT |
| c-Jun-R | CTGGTGGAGATGGCATCCTG |
| V. parahemolyticus - | |
| 16s-F | GGTGTAGCGGTGAAATGCGTAG |
| V. parahemolyticus - | |
| 16s-R | CCACAACCTCCAAGTAGACATCG |
| EF-1α-F | TATGCTCCTTTTGGACGTTTTGC |
| EF-1α-R | CCTTTTCTGCGGCCTTGGTAG |
| Protein expression | |
| LvBigPEN-F | GAGGGGCCGCCTGGAGTGCTGCGTCCTC |
| LvBigPEN-R | ACAGCAGGAGTTCCAGCGCTTGCAG |
| RNA interference | |
| T7-LvBigPEN-F | GGATCCTAATACGACTCACTATAGGGCCTATTGCTCGCCCTCA |
| T7-LvBigPEN-R | GGATCCTAATACGACTCACTATAGGCGGCAGATTTCGGTTTCC |
| T7-c-Fos-F | GGATCCTAATACGACTCACTATAGGCCCACTTCGTCCTCGTCTTCG |
| T7-c-Fos-R | GGATCCTAATACGACTCACTATAGGAGGTCCTCCTCGTGCTCCAT |
| T7-c-Jun-F | GGATCCTAATACGACTCACTATAGGACCATCCTCAACAGCAACACG |
| T7-c-Jun-R | GGATCCTAATACGACTCACTATAGGCGCTCCTGGCACTCCATATC |
| T7-GFP-F | GGATCCTAATACGACTCACTATAGGATGGTGAGCAAGGGCGAGGAG |
| T7-GFP-R | GGATCCTAATACGACTCACTATAGGTTACTTGTACAGCTCGTCCATGCC |
| Dual-luciferase | |
| pGL3-LvBigPEN-F | GGGGTACCCACATACACACACATATATATATGTGTCTGTC |
| pGL3-LvBigPEN-R | CCGCTCGAG GTCTGTCCTCTTCGTGCTGATCAAG |
2.4. Recombinant proteins expression and purification
The coding sequences of LvBigPEN (without N-terminal signal peptide and stop code) was amplified by PCR using corresponding primers (Table 1) and subcloned into pET-32a (+) plasmid (Merck Millipore, Darmstadt, Germany). After confirming that construction was correct by sequencing, the recombinant plasmid was transferred into E. coli Rosetta (DE3) cells (TransGen Biotech, Beijing, China). Then, positive clones harboring the desired fragment were selected for inducing expression. After 4 h of induction with 0.1 mM IPTG at 30 °C, cells were pelleted by centrifugation and sonicated for 30 min on ice water. The supernatant from the sonicated proteins was purified by using Ni-NTA agarose (Qiagen, Düsseldorf, Germany) according to the manufacturer's instructions. The recombinant Trx-His-tag (rTrx-His-tag) was induced as a control and purified in the same way. The purified recombinant LvBigPEN (rLvBigPEN) and rTrx-His-tag proteins were checked by Coomassie staining or western blotting. The concentration of the purified proteins was determined using a BCA protein assay kit (Beyotime Biotechnology, Shanghai, China).
2.5. SDS-PAGE and western blotting
The SDS-PAGE and western blotting were used to identify the purified recombinant protein rLvBigPEN. After by SDS-PAGE and western blotting, the proteins were separated on 12.5% SDS PAGE gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany). And then blocking with 5% nonfat milk diluted in TBST buffer (150 mM NaCl, 3 mM EDTA, 0.1% Tween-20, 50 mM Tris–HCl, pH = 8.0) for 1 h, and the membrane was incubated with 1:1000 mouse anti-6 × His (Sigma, St Louis, MO, USA) for 2 h at 25 °C. The PVDF membranes were washed for three times with TBST and then incubated with 1:2000 goat anti-mouse IgG (H+L) HRP secondary antibody (Promega, Shanghai, China) for 1 h. Membranes were developed using an enhanced chemiluminescent (ECL) blotting substrate (Thermo Fisher Scientific, Waltham, MA, USA), and the chemiluminescent signal was detected using the 5200 Chemiluminescence Imaging System (Tanon, Shanghai, China).
2.6. DsRNA-mediated RNA interference (RNAi)
The dsRNAs, including target to LvBigPEN and GFP (as a control), were generated by in vitro transcription with the T7 RiboMAX Express RNAi System kit (Promega, Shanghai, China) using the primers shown in Table 1. The quality of dsRNA was checked by 1.5% gel electrophoresis, and the quantity was determined by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNAi assay was performed as described previously [19]. The dsRNAs of LvBigPEN and GFP in length were 378 and 504 bp, respectively. The shrimps of experimental group were treated with the injection of LvBigPEN dsRNA (10 μg each shrimp in 50 μl PBS), while the control groups were injected with GFP dsRNA or PBS, respectively. Gill and stomach tissues were sampled from 9 shrimps in each group at 48 h post injection, and tissues from 3 shrimps pooled together, then qRT-PCR was used to investigate the RNA interference efficiency.
In bacterial challenge experiments, after 48 h LvBigPEN dsRNA injection, shrimps were injected again with 1 × 105 CFU V. parahaemolyticus (n = 35 in each group) or PBS as a control. The gills from each group (9 shrimps) were sampled for qRT-PCR to detect the relative bacteria content of V. parahaemolyticus. The cumulative mortality of each group was recorded every 4 h. The Mantel–Cox (log-rank χ2 test) method was subjected to analyze differences between groups using GraphPad Prism software (Graphpad, San Diego, CA, USA).
In parallel, rescue experiments were performed to monitor the effect of rLvBigPEN on V. parahaemolyticus proliferation levels in vivo or cumulative mortality after knockdown of LvBigPEN in shrimp. After 48 h LvBigPEN dsRNA injection, 10 μg rLvBigPEN was first incubated with 1 × 105 CFU V. parahaemolyticus (n = 35 in each group) for 1 h, and then the mixture was inoculated into the experimental shrimp by injection. The rTrx-His-tag protein was used as a control. Likewise, the bacteria content and cumulative mortality were analyzed as described above.
2.7. Dual-luciferase reporter assays
The transcription factor binding sites in promoter region of LvBigPEN were predicted by JASPAR database (http://jaspardev.genereg.net/), and found it contained an AP-1 binding motif located at −215 to −201 (GTTTACGTAATTAAA). The promoter region of LvBigPEN was cloned using the specific primers (Table 1) and then linked into pGL3-Basic (Promega, Shanghai, China) to generate reporter plasmid pGL3-BigPEN. The L. vannamei c-Fos and c-Jun expression vectors (pAc-Lvc-Fos-V5 and pAc-Lvc-Jun-V5) were obtained from our previous studies [20].
Since no permanent shrimp cell line was available, Drosophila Schneider 2 (S2) cell line (ATCC CRL 1963) was used instead in order to detect the effects of L. vannamei AP-1 (Lvc-Fos, Lvc-Jun) on the promoter activity of LvBigPEN. S2 cells were cultured at 28 °C in Schneider's Insect Medium (Sigma-Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum (Gibco, Grand Island, NY). For dual-luciferase reporter assays, S2 cells were plated into a 96-well plate, and 12 h later, the cells of each well were transfected with 0.05 μg of firefly luciferase reporter gene plasmids, 0.001 μg pRL-TK renilla luciferase plasmid (Promega, Shanghai, China), and 0.05 μg protein expression plasmids or empty pAc5.1A plasmids (as controls) using the Fugene HD Transfection Reagent (Promega, Shanghai, China) according to the user's manual. Forty-eight hours post-transfection, the dual-luciferase reporter assays were performed in order to calculate the relative ratios of firefly and renilla luciferase activities using the Dual-Glo Luciferase Assay System kit (Promega, Shanghai, China), according to the manufacturer's instructions. All experiments were repeated six times.
2.8. Antibacterial activity assay of rLvBigPEN
The minimum inhibitory concentration (MIC) was determined by a liquid growth inhibition assay as previously described with slight modifications [21]. Briefly, the bacteria were inoculated into LB broth, cultured at 37 °C until the logarithmic growth phase, and finally diluted with Poor Broth (1% (w/v) tryptone, 0.5% (w/v) NaCl, pH 7.5) to 1 × 105 CFU mL−1. Serial of 2-fold dilution of rLvBigPEN ranging from 0 to 50 μM were made in the PBS buffer. Then, 50 μL of rLvBigPEN of different concentrations and 50 μL bacterial solution were added to a 96-well plate, mixed well and incubated at 28 °C for 16 h. Bacterial growth was evaluated by the culture absorbance at 600 nm measured by the ELX800 Universal Microplate Reader (Bio-Tek, Winooski, VT, USA). Bacteria incubated with rTrx-His-tag were used as controls. Wells without bacteria were used as negative controls. If the absorbance value of the assay well is consistent with the negative control, the minimum antimicrobial peptide concentration of its antipode is determined as the minimum antimicrobial concentration. All tests were performed in duplicate and replicated at least three times.
2.9. Detection of rLvBigPEN binding to bacteria
To determine the antibacterial function of rLvBigPEN, a bacterial binding activity assay was performed on four Gram-negative bacteria and four Gram-positive bacteria following a previously reported method with a little modification [22]. Briefly, bacteria were cultured to log phase, and washed with PBS buffer, then resuspended and adjusted to a final concentration of 1 × 108 CFU/ mL. A 200 μl bacterial was mixed with 50 μg rLvBigPEN, and incubated with shaking for 1 h at 25 °C. The rTrx-His-tag and PBS were used as negative control and blank control, respectively. After incubation, the mixtures were centrifuged at 5000 rpm for 10 min, and the bacterial segment was wash with PBS six times. Then, 300 μl of 7% SDS was added to the segment for 10 min at 25 °C, and the mixture was centrifuged at 12,000 rpm for 3 min for protein elution. Finally, the precipitates were washed with PBS six times and centrifuged at 12,000 rpm for 10 min per wash. The precipitates were prepared for SDS-PAGE and western blotting.
2.10. DNA gel retardation assay
For plasmid DNA, plasmid pAc5.1a was extracted purified using Plasmid Mini Kit (Omega, Guangzhou China). The steps were carried out according to a previously reported method with a little modification [23]. The plasmid DNA (500 ng) was mixed with increasing amounts of rLvBigPEN (0–500 ng) in 20 μl of 10 mM Tris, 1 mM EDTA buffer, pH 8.0, and the mixtures were incubated at room temperature for 30 min. The rTrx-His-tag was used as the controls. After incubation, 1% agarose electrophoresis was detected and analyzed and photographed.
For bacterial DNA, V. parahaemolyticus was cultured overnight and harvested, and then genomic DNA was isolated by using the Tissue DNA Kit (Omega, Guangzhou, China) according to the protocol. The purity and integrity of the V. parahaemolyticus DNA were tested by spectrophotometry (A260/A280) and 1% agarose gel electrophoresis. The V. parahaemolyticus DNA (500 ng) was mixed with increasing amounts of rLvBigPEN (0–500 ng) in 20 μl of 10 mM Tris, 1 mM EDTA buffer, pH 8.0, and the mixtures were incubated at 25 °C for 30 min, then subjected to electrophoresis on a 1% agarose gel.
2.11. Transmission electron microscopy (TEM)
The effect of rLvBigPEN treatment on V. parahaemolyticus was observed using transmission electron microscopy (TEM). The same treatment method was used to obtain the suspension of bacteria as described above [22]. The bacteria were cultured to the appropriate concentration and incubated at 28 °C with an equal volume of rLvBigPEN (final concentration of 50 μM) for 1 h and 2 h. PBS or rTrx-His-tag were used as controls. The bacterial pellets were fixed with 2.5% glutaraldehyde in 1 × PBS overnight at 4 °C and washed three times with 1 × PBS. After washing with distilled water three times, the samples were counterstained with 2% sodium phosphotungstate for 1 min and then observed under a transmission electron microscopy (JEM-100CXII, JEOL, Tokyo, Japan).
2.12. Statistical analysis
All data were presented as means ± SD. Student's t-test was used to calculate the comparisons between groups of numerical data. For cumulative mortality, data were subjected to statistical analysis using GraphPad Prism software (Graphpad, San Diego, CA, USA) to generate the Kaplan–Meier plot (log-rank χ2 test).
3. Results
3.1. Sequence analysis and multiple sequence alignment of LvBigPEN
In our previous study, we identified a member of penaeidins, named LvBigPEN (accession no. MN149368), which belonged to a new sub-group of penaeidins [16]. The full-length of LvBigPEN transcript was 1581 bp long with an ORF of 810 bp, which encoded a peptide of 269 residues with a predicted molecular weight of 29.22 kDa and a theoretical pI of 11.24 (Fig. 1A). The LvBigPEN contained a 20 residues signal peptide in N-terminal, an RPT domain and a conserved PEN domain with six conservative cysteines in C-terminal (Fig. 1A,B).
Fig. 1.
The full-length cDNA sequence and deduced amino acid sequences of LvBigPEN. Nucleotides and amino acids were numbered on the left of the sequences. The ORF of the nucleotide sequence was shown in upper-case letters, while the 5′ and 3′ -UTR sequences were shown in lowercase. Amino acid sequence was represented with one-letter codes above the nucleotide sequence. The signal peptide was shown in red line, the RPT and PEN domain of LvBigPEN were shown in blue and yellow line. The conserved cysteine residues were shaded with yellow, and the poly A signal (aataa) was box with black line. (B) The schematic representation of the LvBigPEN protein. The signal peptide is shown in red box. The RPT and PEN domain of LvBigPEN were shown in blue and yellow box, respectively.
3.2. Critical role of LvBigPEN in defense against V. parahaemolyticus infection
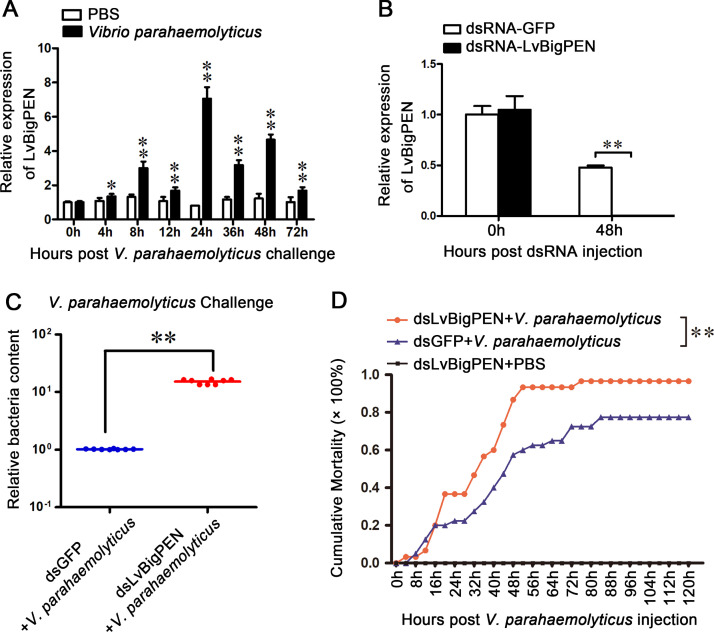
To investigate whether LvBigPEN can respond to bacterial infection, qRT-PCR was used to detect the expression levels of LvBigPEN at different time points after V. parahaemolyticus challenge. In hemocytes, the expression levels of LvBigPEN in infected shrimp but not the control shrimp were dramatically up-regulated at 8 h with a 2.99-fold increase, and then remained at a high level during 24–48 h with 7.06-, 3.18-, and 4.66-fold at 24 h, 36 h and 48 h, respectively (Fig. 2A). The result indicated that LvBigPEN could play an important role against V. parahaemolyticus infection. To determine the function of LvBigPEN in response to V. parahaemolyticus infection, we suppressed LvBigPEN expression in vivo via the RNA interference (RNAi) strategy. The results showed that the mRNA level of LvBigPEN was effectively suppressed by the specific dsRNA, which was downregulated to 0.01-fold of the GFP dsRNA injection groups (control) (Fig. 2B). After knockdown of LvBigPEN, shrimp were infected with V. parahaemolyticus by intramuscular injection. The result showed that the relative bacteria content of the dsRNA-LvBigPEN group were significantly higher than those of the control group, with a 14.94-fold increase being apparent (Fig. 2C). In addition, the cumulative mortality of shrimp in the LvBigPEN-knockdown group was significantly higher than that in the GFP-knockdown group (P < 0.01) (Fig. 2D). All of the results above manifested that LvBigPEN played a crucial role in the immune defense against V. parahaemolyticus infection.
Fig. 2.
Silencing of LvBigPEN during V. parahaemolyticus infected shrimp. (A) Expression profiles of LvBigPEN in hemocytes from V. parahaemolyticus challenged shrimp. Quantitative RT-PCR was performed in triplicate for each sample. The statistical significance was calculated using Student's t-test (**P < 0.01 and *P < 0.05). (B) Quantitative RT-PCR analysis of the silencing efficiencies of LvBigPEN in Gills. The internal control was LvEF-1α. Samples were taken at 48 h post-injection and analyzed by quantitative RT-PCR using gene-specific primers for LvBigPEN or GFP. Differences were analyzed using Student's t-test (**P < 0.01). (C) The relative bacterial loads in gills from each group (8 shrimps) were detected by qRT-PCR at 24 h post-infection. Differences between the experimental and control groups were analyzed using Student's t-test (**P < 0.01). (D) Shrimp cumulative mortality following treatment with dsRNAs and infection with V. parahaemolyticus. Cumulative mortality was recorded every 8 h. Differences in cumulative mortality levels between treatments were analyzed by Kaplan–Meier plot (log-rank χ2 test) (**P < 0.01).
3.3. rLvBigPEN restricted V. parahaemolyticus infection in vivo
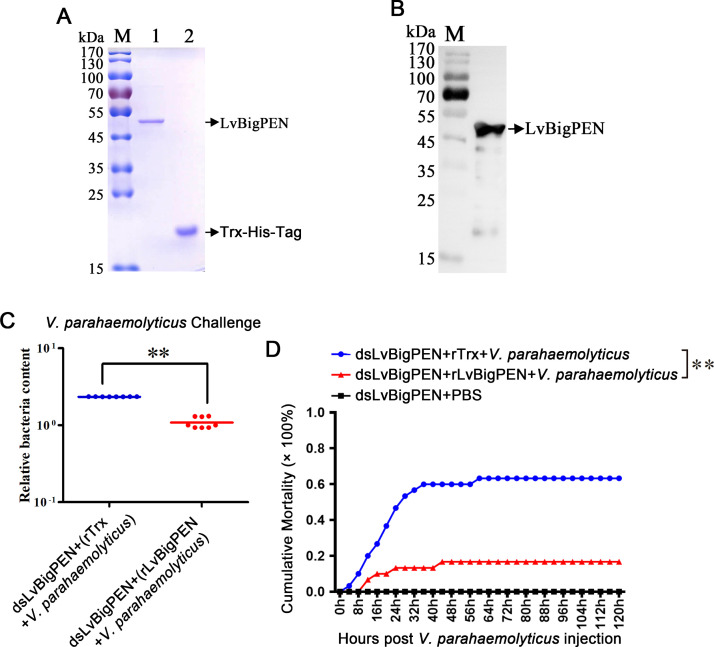
To study the biological activities of LvBigPEN toward V. parahaemolyticus in vitro, the rLvBigPEN and rTrx-His-tag were expressed and purified (Fig. 3A). The results from western blotting of purified rLvBigPEN revealed one band, indicating that no contamination occurred (Fig. 3B). The concentrations of rLvBigPEN and rTrx-His-tag were all adjusted to 1.0 μg/μl by BCA method.
Fig. 3.
rLvBigPEN facilitates V. parahaemolyticus clearance in vivo. (A) SDS-PAGE analysis of the recombinant LvBigPEN. Line 1, purified recombinant LvBigPEN protein (black arrow); Line 2, rTrx-His-tag protein (black arrow). (B) Purified rLvBigPEN protein were analyzed by western blotting with anti-6 × His-antibody (black arrow). (C) The relative bacterial contents in gills from each group (8 shrimps) were detected by qRT-PCR. Forty-eight hours post-dsRNA injection, the shrimp were injected with V. parahaemolyticus premixed with purified rLvBigPEN. Injection with a similar amount of a mixture of V. parahaemolyticus with rTrx-His-tag protein were used as controls. Differences between the experimental and control groups were analyzed using Student's t-test (**P < 0.01). (D) Shrimp cumulative mortality following treatment with dsRNAs, and 48 h post-dsRNA injection, the shrimps were injected with V. parahaemolyticus premixed with purified rLvBigPEN or rTrx-His-tag. Cumulative mortality was recorded every 8 h. Differences in cumulative mortality levels between treatments were analyzed by Kaplan–Meier plot (log-rank χ2 test) (**P < 0.01).
To further confirm the anti-V. parahaemolyticus function of LvBigPEN in vivo, RNAi experiments coupled with rLvBigPEN rescue was performed. After knockdown of LvBigPEN, the shrimp were infected with V. parahaemolyticus mixed with rLvBigPEN or rTrx-His-tag (as a control) by intramuscular injection. The result showed that the relative bacteria content of the rLvBigPEN rescue group were significantly lower than those of the rTrx-His-tag control group, with a 2.16-fold decrease being apparent (Fig. 3C). In addition, the cumulative mortality of shrimp in the rescue group was significantly lower than that in the control group (P < 0.01) (Fig. 3D). These results strongly indicated that rLvBigPEN could restrict V. parahaemolyticus infection in shrimp.
3.4. Antimicrobial activity of rLvBigPEN in vitro
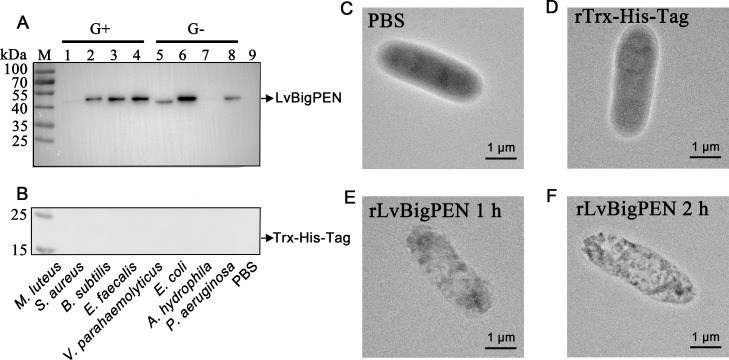
To further study the antibacterial mechanism of rLvBigPEN, microorganism binding assays were carried out by western blotting. The results showed that rLvBigPEN could bind to several Gram-negative bacteria (V. parahaemolyticus, E. coli and P. aeruginosa), and some Gram-positive bacteria (S. aureus, B. subtilis and E. faecalis) (Fig. 4A). In the control group, rTrx-His-tag could not bind to either Gram-negative bacteria or Gram-positive bacteria (Fig. 4B). These results indicated that rLvBigPEN might be involved in binding activities to bacterial cells.
Fig. 4.
Anti-bacterial activity of rLvBigPEN protein. (A-B) Binding activity of rLvBigPEN (A) or rTrx-His-tag (B) to different bacteria. Western blotting assay showed the results of bacteria incubated with rLvBigPEN or rTrx-His-tag. Line 1–4 represent gram-positive bacteria: Micrococcus luteus, Staphylococcus aureus, Bacillus subtilis and Enterococcus faecalis treated by recombinant proteins; Line 5–8 represent gram-negative bacteria: Vibrio Parahaemolyticus, Escherichia coli, Aeromonas hydrophila and Pseudomonas aeruginosa treated by recombinant proteins; Line 9 represents PBS treated by recombinant protein. (C-F) TEM images of V. parahaemolyticus treated with rLvBigPEN. The V. parahaemolyticus was incubated with rLvBigPEN at 28 °C for 1 h (E) and 2 h (F) and the images showed the morphology and structure of V. parahaemolyticus by TEM examination. The PBS (C) and rTrx-His-tag (D) used as controls. Scale bar: 1 μm.
To test the antimicrobial activity of rLvBigPEN, liquid growth inhibition assays were performed. The minimal inhibitory concentrations against Gram-positive or Gram-negative bacteria for rLvBigPEN were listed in Table 2. Based on the MIC values, rLvBigPEN showed superior inhibitory abilities against Gram-negative bacteria, including V. parahaemolyticus, A. hydrophila, P. aeruginosa and E. coli with MIC values of 12.5, 25, 25 and 25 μM, respectively. The MIC values for Gram-positive bacteria, including E. faecalis, S. aureus and M. luteus were 25, 25 and 25 μM, respectively. However, rLvBigPEN showed a little inhibitory ability against Gram-positive bacteria of B. subtilis, and the MIC value was 50 μM (Table 2).
Table 2.
Antimicrobial activities of purified LvBigPEN protein.
| Microorganisms | MIC (μM) |
|---|---|
| Gram-negative bacteria | |
| Vibrio parahaemolyticus | 12.5 |
| Aeromonas hydrophila | 25 |
| Pseudomonas aeruginosa | 25 |
| E. coli | 25 |
| Gram-positive bacteria | |
| Enterococcus faecalis | 25 |
| staphylococcus aureus | 25 |
| Micrococcus luteus | 25 |
| Bacillus subtilis | 50 |
Minimal inhibitory concentration is defined as the lowest protein concentration harvesting visible growth inhibition function, compared with the negative control.
3.5. Ultrastructural changes in bacteria using TEM
To explore the action of LvBigPEN against V. parahaemolyticus, we investigated the potential effect of rLvBigPEN on bacterial microstructure. The morphological and structural changes of V. parahaemolyticus cells before and after incubation with rLvBigPEN were observed under TEM. TEM clearly showed differences in morphology between the untreated and rLvBigPEN treated bacteria. The image of bacterial cells from the PBS treated group and rTrx-His-tag treated group revealed that cells had a normal shape and inner membrane with an undamaged, complete architecture, and the outer membrane was round and smooth (Fig. 4C,D). In rLvBigPEN treated group, the cell cavity became small at 1 h (Fig. 4E), and bacterial cell was severely damaged at 2 h (Fig. 4F). The result showed that rLvBigPEN could execute its inhibitory activity against V. parahaemolyticus possibly damaging bacterial microstructure.
3.6. The binding activity of rLvBigPEN to DNA
Since some antibacterial peptides have been reported to inhibit DNA synthesis by directly binding DNA [23], the DNA binding ability of LvBigPEN was evaluated in a gel retardation assay. The rTrx-His-Tag (as a control) and increased concentrations of rLvBigPEN were mixed with a fixed amount (500 ng) of pAc5.1a plasmid DNA, and then the complexes were electrophoresed on a 1% agarose gel. Compared with controls, the plasmid DNA incubated with peptide (rLvBigPEN) was decrease in brightness (Fig. 5A). At a peptide/DNA weight ratio of 0.4, a fraction of the plasmid DNA was still able to migrate into the gel, whereas, at a weight ratio of 0.6, complete retardation of DNA was observed, showing that DNA was aggregated by rLvBigPEN (Fig. 5B). After 500 ng V. parahaemolyticus genomic DNA was incubated with rTrx-His-Tag or gradient concentrations of rLvBigPEN, the result showed that the V. parahaemolyticus genomic DNA was decrease in brightness in a rLvBigPEN concentration dependent manner (Fig. 5C). At the peptide/DNA weight ratio of 0.6-0.8, a fraction of the bacterial DNA was still able to migrate into the gel, whereas, at a higher weight ratio, complete retardation of the bacterial DNA was observed, showing that the bacterial DNA was aggregated by rLvBigPEN (Fig. 5D). All the results showed that rLvBigPEN exhibited a binding capability toward plasmid or V. parahaemolyticus genomic DNA.
Fig. 5.
Gel retardation analysis of the binding of rLvBigPEN to DNA. (A) Binding activity of rLvBigPEN to plasmid DNA. The pAc5.1A plasmid DNA (500 ng) was treated with rTrx-His-tag (Line 1), and gradient concentrations of rLvBigPEN (0, 100, 200, 300, 400, 500 ng) (Line 2–7). (B) The panel shows the unshifted fraction of plasmid DNA versus the same peptide/plasmid weight ratio. (C) Binding activity of rLvBigPEN to V. parahaemolyticus DNA. The V. parahaemolyticus DNA (500 ng) was treated with rTrx-His-tag (Line 1), and gradient concentrations of rLvBigPEN (0, 100, 200, 300, 400, 500 ng) (Line 2–7). (D) The panel shows the unshifted fraction of V. parahaemolyticus DNA versus the same peptide/plasmid weight ratio.
3.7. LvBigPEN was regulated by AP-1 pathway
In shrimp L. vannamei, the c-Fos and c-Jun (AP-1) are the downstream transcription factors of JNK-MAPK signaling pathway [20,24]. The AP-1 family proteins have showed to play a key role in the synthesis of immune effector molecules, such as antimicrobial peptides (AMPs) in response to infection [24]. To explore whether c-Fos and c-Jun were able to regulate the expression of LvBigPEN in vitro, a dual-luciferase reporter assay was performed in S2 cells. The promoter sequence and putative AP-1 binding site of LvBigPEN were showed in Fig. 6A. The result showed that expression of c-Fos and c-Jun significantly improved the transcriptional activity of pGL3-LvBigPEN (Fig. 6B). To address whether the expression of LvBigPEN was regulated by c-Fos and c-Jun in vivo, an RNAi experiment was performed. We observed that the mRNA levels of LvBigPEN were significantly decreased in gills from c-Fos- or c-Jun-silenced shrimps post V. parahaemolyticus infection, confirming that expression of LvBigPEN was regulated by AP-1 (c-Fos and c-Jun) (Fig. 6C). By qRT–PCR, we confirmed that c-Fos and c-Jun were effectively suppressed by corresponding dsRNAs (P < 0.01) (Fig. 6D,E). Taken together, the results suggested that AP-1 transcription factors (c-Fos and c-Jun) participated in the transcriptional expression of LvBigPEN in response to V. parahaemolyticus infection.
Fig. 6.
Regulation of LvBigPEN promoter activity by shrimp AP-1. (A) The promoter sequence of LvBigPEN. The putative AP-1 binding site and the start codon ATG were boxed. (B) Dual-luciferase reporter assays were performed to analyze the effects of the overexpression of Lvc-Fos and Lvc-Jun on the promoter activities of LvBigPEN in Drosophila S2 cells. All data are representative of three independent experiments. The statistical significance was calculated using Student's t-test (**P < 0.01). Expression levels of Lvc-Fos and Lvc-Jun proteins were verified by western-blotting. (C) The mRNA levels of LvBigPEN in the gills of Lvc-Fos- and Lvc-Jun-silenced shrimp 48 h post V. parahaemolyticus infection, the expression values were normalized to LvEF-1a. The statistical significance was calculated using Student's t-test, **P < 0.01. (D-E) Effective knockdown for Lvc-Fos (D) and Lvc-Jun (E) in gills by dsRNA was confirmed by qRT-PCR. Differences were analyzed using Student's t-test (**P < 0.01).
4. Discussion
AMPs are found in evolutionarily diverse organisms ranging from prokaryotes, invertebrates to vertebrates [25]. In invertebrates, AMPs, as the first-line of defense to resist invading pathogens, are crucial components of nonspecific innate immunity [26]. Penaeidins are a type of AMPs only found in penaeid shrimps, which have different biological activities and play an important role in the immune defense to infection [27]. This family is highly cationic, consisting of a highly conserved leader peptide followed by an N-terminal proline-rich domain (PRD) and a C-terminal cysteine-rich domain (CRD) [10]. In our previous study, we uncovered that LvBigPEN possessed antiviral activity against WSSV, however, whether LvBigPEN play a role in defense against bacterial infection is still unknown. In this study, RNAi knockdown of LvBigPEN resulted in higher mortality and bacteria contents, meanwhile rLvBigPEN was able to effectively rescue silencing of LvBigPEN-mediated mortality and bacteria contents. Besides, liquid growth inhibition assays demonstrated that rLvBigPEN have a wide range of antibacterial activity multiple Gram-positive or Gram-negative bacteria. Taken together, the antibacterial function of LvBigPEN was determined in vivo and in vitro.
Although several members of penaeidins were showed to possess antibacterial activity [28,29], the accurate antibacterial mechanism is largely uncovered. Commonly, AMPs execute antibacterial function thought diverse mechanisms including destroying bacterial structure, promoting agglutination and phagocytose, interfering bacterial proliferation [8,30]. Indeed, a previous study of a penaeidin gene from Marsupenaeus japonicas revealed that MjPen-II could bind to bacteria via interacting with polysaccharides, thus promoting bacterial agglutination [29]. In this study, the antibacterial mechanism of rLvBigPEN was also explored. LvBigPEN could directly bind to several bacteria, and was able to destroy the structure of V. parahaemolyticus might though binding to superficial membrane. Besides, rLvBigPEN was observed to interact with DNA of V. parahaemolyticus, which could interfere bacterial proliferation. All these results were well consistent with the protective role of LvBigPEN in vivo. The primary structure of LvBigPEN protein was consisted of several cationic amino acid residues, which could endow it can bind with the negatively charged bacterial membrane and penetrate it, as well as interacting with negatively charged DNA. Similarly, the antimicrobial peptide, Indolicidin that contain cationic amino acid residues, was shown to bind to DNA and inhibit DNA synthesis and induce filamentation [23]. Thus, the possible mode of LvBigPEN against V. parahaemolyticus was that of its ability to bind directly to this bacterium, after attacking and penetrating to the membrane, and binding to bacterial genomic DNA.
AMPs function as effectors that are induced by many innate signaling pathways in response to infection. The expressional level of LvBigPEN was strongly up-regulated after bacterial infection. In previous study, our results showed that both Dorsal and Relish (NF-κB), the downstream transcription factors of Toll and IMD pathways, respectively, could be involved in the regulation of the LvBigPEN after WSSV infection in vivo [16]. In present study, we showed that AP-1 transcription factors (c-Fos and c-Jun) play a role in regulating the transcriptional expression of LvBigPEN in response to V. parahaemolyticus infection. The c-Fos and c-Jun, belonging to the activator protein-1 (AP-1) family, are the transcription factors of the IMD-MAPK branch [31, 32]. Lvc-Fos interacted with Lvc-Jun, and they could function as transcription factors to activate antimicrobial peptides (AMPs) of both Drosophila and shrimps [20]. In shrimp L. vannamei, the upstream-regulatory region of LvPEN4 contains many putative transcription-factor-binding sites, including STATx, AP-1, Dorsal, and GATA [33]. In shrimp P. monodon, the promoter sequences of PEN536 and PEN411 contain several transcription-factor-binding motifs, such as TATA box, GATA, dorsal, and AP-1 [34]. Therefore, penaeidins seem to be regulated by many innate signaling pathways, such as Toll pathway, IMD pathway, and others, in response to pathogen invasion.
In conclusion, we characterized that LvBigPEN played an important function in defense against bacterial infection, in addition to its antiviral activity. Based on our results observed in this study, it can be speculated that bacterial infection could induce the activation of some signaling pathways such as NF-κB and MAPK pathways to stimulate the expression of LvBigPEN, which exhibited antimicrobial activity against V. parahaemolyticus though binding to bacterial components including superficial membrane and DNA, thus destroying bacterial structure and/or interfering bacterial proliferation.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgement
This research was supported by National Key Research and Development Program of China (2018YFD0900600/2018YFD0900500), National Natural Science Foundation of China (32022085/31930113); Independent Research and Development Projects of Maoming Laboratory (2021ZZ007); Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (SML2021SP301) and Key-Area Research and Development Program of Guangdong Province (2018B020204001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Jianguo He, Email: lsshjg@mail.sysu.edu.cn.
Chaozheng Li, Email: lichaozh@mail2.sysu.edu.cn.
References
- 1.Stentiford G.D., Neil D.M., Peeler E.J., Shields J.D., Small H.J., Flegel T.W., Vlak J.M., Jones B., Morado F., Moss S., Lotz J., Bartholomay L., Behringer D.C., Hauton C., Lightner D.V. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012;110(2):141–157. doi: 10.1016/j.jip.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Li C., Weng S., He J. WSSV-host interaction: host response and immune evasion. Fish Shellfish Immunol. 2019;84:558–571. doi: 10.1016/j.fsi.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 3.De Schryver P., Defoirdt T., Sorgeloos P. Early mortality syndrome outbreaks: a microbial management issue in shrimp farming? PLoS Pathog. 2014;10(4) doi: 10.1371/journal.ppat.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabello F.C., Godfrey H.P., Buschmann A.H., Dolz H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016;16(7):E127–E133. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 5.Su H., Liu S., Hu X., Xu X., Xu W., Xu Y., Li Z., Wen G., Liu Y., Cao Y. Occurrence and temporal variation of antibiotic resistance genes (ARGs) in shrimp aquaculture: aRGs dissemination from farming source to reared organisms. Sci. Total Environ. 2017;607-608:357–366. doi: 10.1016/j.scitotenv.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann J.A., Kafatos F.C., Janeway C.A., Ezekowitz R.A. Phylogenetic perspectives in innate immunity. Science. 1999;284(5418):1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 7.Kai-Larsen Y., Gudmundsson G.H., Agerberth B. A review of the innate immune defence of the human foetus and newborn, with the emphasis on antimicrobial peptides. Acta Paediatr. 2014;103(10):1000–1008. doi: 10.1111/apa.12700. [DOI] [PubMed] [Google Scholar]
- 8.Tassanakajon A., Amparyup P., Somboonwiwat K., Supungul P. Cationic antimicrobial peptides in penaeid shrimp. Mar. Biotechnol. 2010;12(5):487–505. doi: 10.1007/s10126-010-9288-9. (NY) [DOI] [PubMed] [Google Scholar]
- 9.Rolland J.L., Abdelouahab M., Dupont J., Lefevre F., Bachere E., Romestand B. Stylicins, a new family of antimicrobial peptides from the Pacific blue shrimp Litopenaeus stylirostris. Mol. Immunol. 2010;47(6):1269–1277. doi: 10.1016/j.molimm.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Gueguen Y., Garnier J., Robert L., Lefranc M.P., Mougenot I., de Lorgeril J., Janech M., Gross P.S., Warr G.W., Cuthbertson B., Barracco M.A., Bulet P., Aumelas A., Yang Y., Bo D., Xiang J., Tassanakajon A., Piquemal D., Bachere E. PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev. Comp. Immunol. 2006;30(3):283–288. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Vaseeharan B., Shanthi S., Chen J.C., Espineira M. Molecular cloning, sequence analysis and expression of Fein-Penaeidin from the haemocytes of Indian white shrimp Fenneropenaeus indicus. Results Immunol. 2012;2:35–43. doi: 10.1016/j.rinim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuthbertson B.J., Shepard E.F., Chapman R.W., Gross P.S. Diversity of the penaeidin antimicrobial peptides in two shrimp species. Immunogenetics. 2002;54(6):442–445. doi: 10.1007/s00251-002-0487-z. [DOI] [PubMed] [Google Scholar]
- 13.Gross P.S., Bartlett T.C., Browdy C.L., Chapman R.W., Warr G.W. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific White Shrimp, Litopenaeus vannamei, and the Atlantic White Shrimp, L. setiferus. Dev. Comp. Immunol. 2001;25(7):565–577. doi: 10.1016/s0145-305x(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 14.Kang C.J., Xue J.F., Liu N., Zhao X.F., Wang J.X. Characterization and expression of a new subfamily member of penaeidin antimicrobial peptides (penaeidin 5) from Fenneropenaeus chinensis. Mol. Immunol. 2007;44(7):1535–1543. doi: 10.1016/j.molimm.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Chiou T.T., Lu J.K., Wu J.L., Chen T.T., Ko C.F., Chen J.C. Expression and characterisation of tiger shrimp Penaeus monodon penaeidin (mo-penaeidin) in various tissues, during early embryonic development and moulting stages. Dev. Comp. Immunol. 2007;31(2):132–142. doi: 10.1016/j.dci.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Xiao B., Fu Q., Niu S., Zhu P., He J., Li C. Penaeidins restrict white spot syndrome virus infection by antagonizing the envelope proteins to block viral entry. Emerg. Microbes Infect. 2020;9(1):390–412. doi: 10.1080/22221751.2020.1729068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Yin B., Wang S., Fu Q., Xiao B., Lu K., He J., Li C. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PLoS Pathog. 2018;14(9) doi: 10.1371/journal.ppat.1007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Wang S., Lu K., Yin B., Xiao B., Li S., He J., Li C. An invertebrate STING from shrimp activates an innate immune defense against bacterial infection. FEBS Lett. 2017;591(7):1010–1017. doi: 10.1002/1873-3468.12607. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Wang S., Chen Y., Lu K., Yin B., Li S., He J., Li C. Identification of two p53 isoforms from Litopenaeus vannamei and their interaction with NF-kappaB to induce distinct immune response. Sci. Rep. 2017;7:45821. doi: 10.1038/srep45821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C., Li H., Wang S., Song X., Zhang Z., Qian Z., Zuo H., Xu X., Weng S., He J. The c-Fos and c-Jun from Litopenaeus vannamei play opposite roles in Vibrio parahaemolyticus and white spot syndrome virus infection. Dev. Comp. Immunol. 2015;52(1):26–36. doi: 10.1016/j.dci.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Imjongjirak C., Amparyup P., Tassanakajon A. Two novel antimicrobial peptides, arasin-likeSp and GRPSp, from the mud crab Scylla paramamosain, exhibit the activity against some crustacean pathogenic bacteria. Fish Shellfish Immunol. 2011;30(2):706–712. doi: 10.1016/j.fsi.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H.Q., Cheng W.Z., Zheng L.B., Wang P.P., Liu Q.H., Li Z., Li T.J., Wei Y.M., Mao Y., Yu X.Y. Identification of a group D anti-lipopolysaccharide factor (ALF) from kuruma prawn (Marsupenaeus japonicus) with antibacterial activity against Vibrio parahaemolyticus. Fish Shellfish Immun. 2020;102:368–380. doi: 10.1016/j.fsi.2020.04.039. [DOI] [PubMed] [Google Scholar]
- 23.Hsu C.H., Chen C., Jou M.L., Lee A.Y., Lin Y.C., Yu Y.P., Huang W.T., Wu S.H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005;33(13):4053–4064. doi: 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallio J., Leinonen A., Ulvila J., Valanne S., Ezekowitz R.A., Ramet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7(5–6):811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Bulet P., Stocklin R., Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahlapuu M., Hakansson J., Ringstad L., Bjorn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016;6 doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Destoumieux D., Munoz M., Bulet P., Bachere E. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda) Cell. Mol. Life Sci. 2000;57(8–9):1260–1271. doi: 10.1007/PL00000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B., Zhang C., Qin X., Shi L., Zhao M. Identification and function of penaeidin 3 and penaeidin 5 in Fenneropenaeus merguiensis. Fish Shellfish Immunol. 2019;89:623–631. doi: 10.1016/j.fsi.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 29.An M.Y., Gao J., Zhao X.F., Wang J.X. A new subfamily of penaeidin with an additional serine-rich region from kuruma shrimp (Marsupenaeus japonicus) contributes to antimicrobial and phagocytic activities. Dev. Comp. Immunol. 2016;59:186–198. doi: 10.1016/j.dci.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 2020;19(5):311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 31.Karin M., Liu Z., Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9(2):240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 32.Li C., Wang S., He J. The two NF-kappaB pathways regulating bacterial and WSSV infection of shrimp. Front. Immunol. 2019;10:1785. doi: 10.3389/fimmu.2019.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Leary N.A., Gross P.S. Genomic structure and transcriptional regulation of the penaeidin gene family from Litopenaeus vannamei. Gene. 2006;371(1):75–83. doi: 10.1016/j.gene.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Ho S.H., Song Y.L. Cloning of penaeidin gene promoter in tiger shrimp (Penaeus monodon) Fish Shellfish Immunol. 2009;27(1):73–77. doi: 10.1016/j.fsi.2009.05.001. [DOI] [PubMed] [Google Scholar]