Highlights
-
•
In situ hybridisation of Ca. Branchiomonas cysticola shows that the bacterium is associated with inflammation and necrosis in which histologically detectable epitheliocysts were not visible.
-
•
New multiplex real-time PCR assays and RNA scope in situ hybrdizaton methods for a list of gill pathogens represent a rapid and sensitive diagnostic tool.
-
•
The results from multiplex real-time PCR and in situ hybridisation corresponded well.
Keywords: Respiratory disease, Poxvirus, Bacterial gill infection, in situ hybridisation, Ca. Piscichlamydia salmonis
Abstract
Gill diseases may cause high mortalities in farmed Atlantic salmon. In seawater reared fish co-infections involving the epitheliocystis associated bacterium Ca. Branchiomonas cysticola, the microsporidian Desmozoon lepeophtherii, the causative agent of amoebic gill disease Paramoeba perurans and salmon gill poxvirus are common and histopathological lesions may be complex. Here, we report detection of these agents utilising multiplex real-time PCR and link the presence of agents to histopathologically visible gill lesions by in situ hybridisation (ISH) utilising RNAscope®. We show that Ca. Branchiomonas cysticola infections may remain undetected if diagnostic investigations are restricted to histopathology alone. Further, positive in situ labelling of Ca. Branchiomonas cysticola was observed within epitheliocysts, but also in small foci within areas of inflammation and necrosis in which histologically detectable epitheliocysts were not visible. In situ labelling of D. lepeophtherii corresponded well with tissue distribution patterns previously associated with this microsporidian. Salmon gill poxvirus was associated with apoptotic gill epithelial cells, while Ca. Piscichlamydia salmonis could not be associated with pathological changes. The multiplex real-time PCRs utilised were rapid and sensitive diagnostic tools and the results corresponded well with ISH. This study shows that the agents involved in complex gill disease can be linked to lesions using ISH and suggests that Ca. B. cysticola plays a crucial role in the development of gill disease in the farming of salmon in Norway.
1. Introduction
In farmed Atlantic salmon (Salmo salar L), hereafter termed salmon, there are few specific gill disease entities, in the sense that clear causal relationships between individual agents, clinical signs of disease and pathological manifestation are rare. Amoebic gill disease (AGD) caused by Paramoeba perurans [1] and salmon gill poxvirus disease (SGPVD) caused by salmon gill poxvirus (SGPV) [2,3] are, however, exceptions [3, 4]. Challenge experiments for AGD [5] and SGPVD [3], demonstrate a clear link between clinical disease, histopathology and detection of the respective aetiological agents. Not surprisingly, transcriptome analysis of infected fish suggests that expression of multiple host genes is modified following infection [6], [7], [8]. Such single agent infections are, however, more the exception rather than the rule in commercial settings. ‘Complex gill disease ‘or ‘complex gill disorder’ (CGD) are terms now used to describe gill disease manifestation in which the histopathological pattern is complex [9,10] and overlaps with ‘multifactorial gill disease’, where multiple distinguishable gill diseases are present. The list of agents infecting gills of salmon is long and the pathogenicity of individual agents is poorly understood. The microsporidian Desmozoon lepeophtherii not only causes severe lesions in the gills, but is also associated with peritonitis, intestinal infection and pathological changes [11]. While some epitheliocystis-associated bacteria e.g. Ca. Piscichlamydia salmonis [12] and Ca. Sygnamydia salmonis [13] do not appear to be associated with severe gill disease, diagnostic histopathological observations and experimental study [14] indicates that the epitheliocystis-associated bacterium Ca. Branchiomonas cysticola [15], may well play a significant role in development of severe gill inflammation and necrosis [9]. Members of the genus Tenacibaculum may also cause severe skin and gill infections with extensive necrosis of gill filaments and lamellae [16]. SGPV [2] can lead to severe gill lesions in salmon, with apoptosis of gill epithelial cells and in some cases, high, acute mortality.
In 2019, we performed an extensive investigation of histopathological lesions in CGD affected salmon [9]. In that study, necrosis and inflammation was clearly associated with Ca. B. cysticola infection, while ballooning degenerative cells containing light brown pigmented material were only observed when D. lepeophtherii was present. The aim of the present study was to investigate these issues in greater depth using in situ hybridization (ISH) to link the presence of specific agents to pathological features in affected gills. Further, the study involved validation of two novel diagnostic multiplex real-time PCRs for detection of P. perurans, SGPV, Ca. B. cysticola, D. lepeophtherii and a salmon reference gene.
2. Material and methods
2.1. Fish samples
Gills from sea-farmed salmon with suspected gill disease were studied. Gill samples from five farms included in a previous study [9], and gills from a further four diagnostic cases were selected for histopathological investigation and ISH for Ca B. cysticola, D. lepeoptherii and SGPV. Single-plex PCR analyses for detection of P. perurans, Ca B. cysticola, D. lepeoptherii and Ca. P. salmonis had been used in the previous study [9]. For the new diagnostic cases, new multiplex PCRs targeting all four agents and salmonids was developed.
2.2. Histopathology, probe design and ISH staining
All formalin-fixed paraffin-embedded (FFPE) gill samples were serially sectioned and stained with hematoxylin and eosin (H&E). To study the distribution of the infectious agents, ISH for Ca. B. cysticola, D. lepeoptherii and SGPV was performed on a subset of serial sections. In gills displaying autolysis, where a specific agent was not detected by PCR, or in the one case where there was a lack of individual labelling of sample containers to link real-time PCR and histology, ISH was not performed, except for few sections serving as negative controls (Table 1). An RNAscope ISH protocol was applied using RNAscope® 2.5 HD Red Chromogenic Reagent Kit (Advanced Cell Diagnostics Inc) as described previously [3]. In this method, paired double-Z oligonucleotide probes (RNAscope specifically targeting unique and conserved sequence regions of Ca. B. cysticola (Acc. No. JN968376, RNAscope Cat NO. 812001), D. lepeoptherii (Acc. No. KR187184, RNAscope Cat NO. 812011) and SGPV (Acc. No KT159937, RNAscope Cat. No. 540201) were used. Briefly, FFPE sections were first deparaffinised in xylene and dehydrated through a series of alcohol washes. The dehydrated sections were treated with hydrogen peroxide at room temperature for 10 min to block endogenous peroxidases. The sections were then boiled in target retrieval buffer for 15 min and incubated with protease at 40°C for 15 min. The slides were then hybridized with the probes on the individual serial sections specified earlier at 40°C for 2 hours and then run through a sequence of signal amplification (40°C for 15 or 30 min) and washing steps. Finally, the hybridization signal was detected using the chromogenic Fast Red substrate. All slides were counterstained using hematoxylin stain for 2 min.
Table 1.
Overview of gill material included in the study and results of the performed PCR and in situ hybridisation. The degree of staining is scored semi-quantitatively from 0 with no staining to 2, suggesting moderate staining and 3 as extensive staining. The distribution is evaluated as focal (f) or multifocal (mf). * indicates samples that have been run on multiplex PCR.
| FARM and fish ID | Ca. Branchiomonas cysticola | Desmozoon lepeoptherii | Salmon gill poxvirus | Paramoeba perurans | Ca.Piscichlamydia salmonis | ||||
|---|---|---|---|---|---|---|---|---|---|
| ISH (0-3) | PCR | PCR | ISH (0-3) | ||||||
| 1-1 | no ct | n.a | 24 | 1 f | n.a | n.a | no ct | no ct | n.a |
| 1-2 # | no ct | 0 | 24,6 | 1 f | n.a | n.a | no ct | no ct | n.a |
| 2-1 | 22,1 | 1,5 mf | no ct | n.a | no ct | n.a | no ct | no ct | n.a |
| 2-2 | 22,1 | 1,5 mf | no ct | n.a | no ct | n.a | no ct | no ct | n.a |
| 2-3 | 21,9 | 1,5 mf | no ct | n.a | 29,9 | n.a | no ct | no ct | n.a |
| 2-4 # | 21 | 1,5 mf | no ct | 0 | no ct | n.a | no ct | no ct | n.a |
| 3-1 | 32,9 | n.a | 34,9 | n.a | n.a | n.a | no ct | 19,8 | 1 f |
| 3-2 # | 35,0 | n.a | 33,3 | n.a | n.a | n.a | no ct | 19,7 | 1 f |
| 4-1 # | 17,2 | 2 mf | 19,5 | 2 mf | n.a | n.a | no ct | no ct | n.a |
| 5-1* | no ct | - | no ct | - | no ct | - | no ct | n.a | n.a |
| 5-2* | 21,6 | - | no ct | - | no ct | - | no ct | n.a | n.a |
| 5-3* | 24,6 | - | 32,9 | - | 24,7 | - | no ct | n.a | n.a |
| 6-1 # | 18,5 | 2,5 mf | 20 | 2,5 mf | n.a | n.a | 25,1 | no ct | n.a |
| 6-2 | 21,5 | 1 f | 23,5 | 1 f | n.a | n.a | 20,9 | no ct | n.a |
| 6-3 | 29,2 | 0,5 f | 26,0 | 0,5 f | n.a | n.a | 32,3 | no ct | n.a |
| 7-1* | 24,3 | 1,5 mf | 27,9 | 1 mf | 30,9 | n.a | no ct | n.a | n.a |
| 7-2* | 20,1 | 2 mf | 35 | 0 | 19,5 | 2f | no ct | n.a | n.a |
| 7-3* # | 23,3 | 1,5mf | no ct | n.a | 21 | 1,5f | no ct | n.a | n.a |
| 7-4* | 24 | 1,5mf | no ct | n.a | 22,2 | 1,5f | no ct | n.a | n.a |
| 7-5* | 23,2 | - | no ct | - | 24,4 | - | no ct | n.a | n.a |
| 8-1* | 17,5 | autolysis | 19,5 | autolysis | 24,2 | autolysis | no ct | n.a | n.a |
| 8-2* | 29 | 0 | 30,2 | 0 | 29,4 | 0 | no ct | n.a | n.a |
| 8-3* | 27,5 | n.a | 29,8 | n.a | no ct | not run | no ct | n.a | n.a |
| 8-4* | 21,4 | 2 mf | 24,8 | 1,5 mf | 25,4 | 1 f | no ct | n.a | n.a |
| 8-5* | 23,4 | 2 mf | 28,1 | 1,5 f | 25,6 | f f | no ct | n.a | n.a |
| 8-6* | 17,1 | autolysis | 21,4 | autolysis | 24,2 | autolysis | 28,4 | n.a | n.a |
| 8-7* # | 21,1 | 2mf | 25,3 | 1,5 mf | 26,2 | 0,5 f | no ct | n.a | n.a |
| 9-1* | 17,1 | 2 mf | 26,5 | 1 mf | 22,9 | 0 | 29,3 | n.a | n.a |
| 9-2* | 17,3 | 2,5 mf | 24,9 | 1 f | 23,6 | 0,5 f | 29,6 | n.a | n.a |
| 9-3* # | 17 | 2,5 mf | 22,9 | 2 mf | 31,5 | 0 | 28,5 | n.a | n.a |
The presence of Ca. Piscichlamydia salmonis, was investigated in paraffin sections from farm 3. In short, sections were incubated with dig-labelled anti-sense probes provided by Exigon (5’DIG-AATCGACTTAGGCAGTCTCGT-3’DIG). The sections were hybridized overnight at 54°C and immunodetection was performed using PO conjugated anti-digoxigenin fab fragments (Roche Diagnostics). The signal was amplified using a TSA Biotin system-kit (PerkinElmer) with streptavidin horseradish peroxidase and visualization was performed using ACE as a substrate.
The degree of staining was scored semi-quantitatively from 0 (no staining), 1 (little staining), 2 (moderate staining) and 3 (extensive staining). The distribution was evaluated as focal (f) or multifocal (mf). Images were taken from representative areas with sufficient technical quality to further explain and illustrate histopathological features.
2.3. Multiplex real-time PCR
A real-time PCR assay to detect SGPV was designed to target the major capsid protein gene [2] (Table 2, accession: NC_027707). A modified version of the real-time PCR developed by Dowes et al [17]. was used to detect P. perurans. Following alignment of available P. perurans sequences in NCBI covering the amplicon we chose to a degenerate the reverse primer in this assay as shown in Table 2. A real-time PCR assay that targets the salmonid host elongation factor 1 alfa (Acc. No NC_027326) gene was designed with the purpose of acting as a DNA extraction control. P.perurans, D. lepeophtherii (ref) and the salmonid reference gene were combined in one multiplex PCR and Ca. Branchiomonas cysticola [18], SGPV and the salmonid reference gene were combined in another multiplex PCR assay.
Table 2.
Primer and probe sequences with consentrations.
| Organism | Oligo name | Sequence 5′>3′ | C, nM |
|---|---|---|---|
| Paramoeba perurans | NP1 | AAAAGACCATGCGATTCGTAAAGT | 300 |
| NP2_D | ATTCTTTTCGGAGARTGGAAATT | 900 | |
| NPP_FAM | 6FAM-ATCATGATTCACCATATGTT-MGBNFQ | 200 | |
| Desmozoon lepeophtherii | Nuc-F | CGGACAGGGAGCATGGTATAG | 600 |
| Nuc-R | GGTCCAGGTTGGGTCTTGAG | 600 | |
| Nuc P_VIC | VIC-TTGGCGAAGAATGAAA-MGBNFQ | 200 | |
| Ca Branchiomonas cysticola | BPf2 | AATACATCGGAACGTGTCTAGTG | 500 |
| BPr2 | GCCATCAGCCGCTCATGTG | 500 | |
| BPp2_FAM | 6FAM-CTCGGTCCCAGGCTTTCCTCTCCCA-BHQ1 | 250 | |
| Salmon gill poxvirus | poxForw2 | AAGCGTTGATCTCCTTGTCG | 400 |
| poxRev2 | CGAATACATGAAGGCCACGA | 400 | |
| poxProbe2_HEX | HEX-AACAGTTGTCGGGTGCTCGGATTGTT-BHQ1 | 200 | |
| Salmonids | ssEF1a3F | GTGTGGAGACTGGAACCCTG | 75 |
| ssEF1a3R | GTTGAAGCCSACGTTGTCAC | 75 | |
| ssEF1a3pr | CY5-TCGTCACCTTCGCCCCCGCT-BHQ2 | 37,5 |
In silico specificity was analysed with a multiplex version of ePCR (an offline version of primerblast) [[18], [19], [20]]. The script ePCR multiplex.py(https://github.com/karinlag/ePCR_multiplex/blob/master/ePCR_multiplex.py) generated all possible primer combinations. PCRs were performed with Brilliant Multiplex QPCR Master Mix (Agilent) on an Aria MX instrument (Agilent) utilising a PCR program of 1 × 95°C for 10 min and 45 × 95°C x 10 seconds 60°C x. Primer and probe sequences and concentrations are shown in Table 2. Amplification efficiency and linearity were obtained by analysing a dilution series for each target in each multiplex PCR assay.
3. Results
Gill samples from salmon farmed at nine sea locations along the coast of Norway (Fig. 1) were studied. Samples from farms 1–4 and 6 were collected during a field study carried out in 2012–2013 [9], while samples from farms 5 and 7–9 represented diagnostic cases involving gill disease in Norwegian farmed salmon in the period 2018–2020 (Table 1). The cases are presented below in order of increasing frequency of multiple agents.
Fig. 1.
Location of farms included in this study along the coast of Norway.
Farm 1: Fish (n=2) from farm 1, situated in Western Norway (Fig. 1), were sampled in March 2013.
D. lepeophtherii was present at Ct values of 24.0 and 24.6 (Table 1) and was the only agent detected in the gills of these fish.
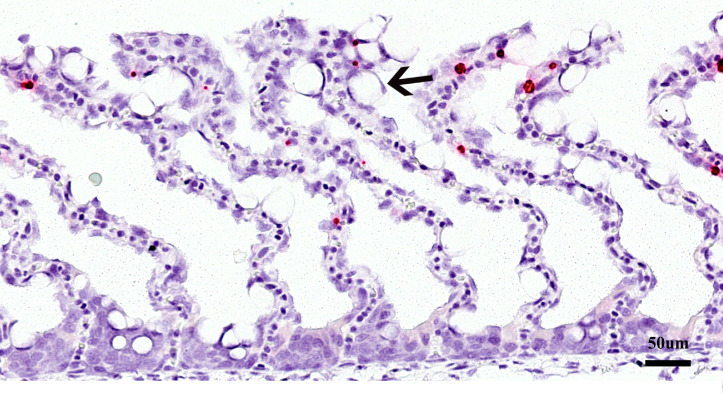
Histopathology revealed moderate mucous cell hyperplasia (Fig. 2), sparse clubbing (thickening of the marginal part of lamellae) and in one of the two fish a few lamellae with telangiectasia were observed. ISH staining revealed only few small foci of D. lepeophtherii, and any association with specific gill pathology was not obvious (Fig. 2).
Fig. 2.
Section of Atlantic salmon gill from farm 1 (fish 1-2) and analysed with in situ hybridization for Desmozoon lepeophtherii. Note thin lamellae with some mucus cell hyperplasia (arrows). Few spots (red) of labelling for D. lepeophtherii.
Farm 2: Fish (n=4) from farm 2, situated in the north of Norway (Fig. 1) weighed about 1 kg were sampled in November 2012. High levels of morbidity, reduced appetite and growth were reported.
Ca. B. cysticola was the only agent detected by PCR with the exception of a single fish positive for SGPV (Ct value of 29.9). Ca. B. cysticola Ct values ranged between 21 and 22.1 (Table 1).
Histopathology revealed moderate to extensive hyperplasia of epithelial cells, moderate hyperplasia of mucus cells, moderate numbers of sub-epithelial inflammatory cells, as well as cell-debris (Fig. 3a–c) and sparse pustule-like lesions. Focal haemorrhage and thrombosis of lamellar vessels were observed. Surprisingly, while no epitheliocysts could be seen in H&E stained sections, ISH revealed positive staining of small, rounded foci of Ca. B. cysticola (Fig. 3b) within areas of inflammation (Fig. 3c),and upon closer investigation of the corresponding area in the H&E stained section some basophilic, dense, rounded structures were seen.
Fig. 3.
Serial sections of Atlantic salmon gill from farm 2 (fish 2-4) stained with (a, c) haematoxylin and eosin and (b) in situ hybridization for Ca. Branchiomonas cysticola. (a) Some lamellae are thickened due to the presence of sub-epithelial inflammatory cells, cellular debris /necrosis (arrows) and some epithelial hyperplasia. The boxed area is magnified in Figure c. (b) Note small foci of labelling for Ca. B. cysticola, not observed in the H&E section. Lamellar fusion is present. (c) Note inflammatory cells (arrows) and debris (asterisk).
Farm 3: Fish (n=2) from farm 3, situated in Southern Norway (Fig. 1), weighed between 1.4 and4 kg and were sampled in September 2012. There was low mortality on the day of sampling, but gill problems had been reported from previous recent samplings.
Ca. P. salmonis was present at Ct values of 19.7 and 19.8, respectively, (Table 1). Only low levels of Ca. B. cysticola and D. lepeoptherii (Ct values > 32) were detected.
Histopathology revealed a degree of lamellar clubbing, but otherwise very few lesions and few epitheliocysts were observed in the two gills examined. ISH staining for Ca. P. salmonis revealed rounded foci and small structures consistent with bacterial cells in larger numbers (Fig. 4a) than estimated by histopathological examination, (Fig. 4b,c).
Fig. 4.
Sections of Atlantic salmon gill from farm 3 (fish 3-2) stained with in situ hybridization for Ca. Piscichlamydia salmonis (brown). (a) The upper box is magnified in b, while the lower box is magnified in c. (a-c) thin lamellae with no apparent host reaction to the bacteria. Small spots of staining of few bacteria (arrows) that were not observed in H&E stained sections. Note also sparse mucus cell hyperplasia (arrowheads)
Farm 4: Fish from farm 4 (n=1), situated in mid-Norway (Fig. 1), weighed around 1 2 kg and were sampled in October 2012. Some increased mortality and reduced appetite were reported at the farm.
Concurrent infection with both B. cysticola and D. lepeoptherii was identified in this fish, at Ct values of 17.2 and 19.5, respectively (Table 1).
Histopathological examination revealed extensive lamellar marginal adhesion (Fig. 5a,b), sparse epithelial- and mucus cell- hyperplasia, as well as some thrombosis and telangiectasia in lamellar vessels. Foci of epitheliocysts, a few Trichodina- and costia-like parasites (Fig. 5a,b) were also seen. Sparse ballooning, degenerative cells, containing a light brown granular pigment, were observed in some lamellae.
Fig. 5.
Sections of Atlantic salmon gill from farm 4 (fish 4-1) stained with (a) haematoxylin and eosin, (b) in situ hybridization for Desmozoon lepeoptherii. (a-b) lamellar adhesion, Ichthyobodo-like structure (arrow) and Trichodina in the lower left of both images. Note also some separation of the epithelial layer from the basal membrane indicating oedema (asterisk).
ISH revealed clear labelling of Ca. B. cysticola within epitheliocysts observed in the H&E sections. ISH for D. lepeophtherii revealed small foci, evenly distributed throughout the lamellae and filament (Fig. 5b). An association with any specific gill pathology and presence of D. lepeophtherii was not obvious.
Farm 5: The fish (n=3) from farm 5, situated in south-western Norway (Fig. 1), weighed around 1 kg and were sampled in July 2020. Moribund fish with loss of appetite, but no increased mortality, were reported.
Of the three fish sampled from farm 5, the gills of one were PCR negative for all four agents tested, one tested positive for Ca. B. cysticola alone (Ct value 21.6) and the third tested positive for Ca. B. cysticola, D. lepeoptherii and SGPV with Ct values of 24.6, 32.9 and 24.7, respectively (Table 1).
Correlation of histopathological assessment, agent load and ISH was not possible in this case due to inappropriate identification of individual sample containers. Histopathology revealed sparse lamellar clubbing in one fish, while foci of epitheliocysts, few haemorrhages and the sparse presence of sub-epithelial inflammatory cells were observed in a few lamellae of one of the remaining two fish. ISH staining revealed clear small foci of Ca. B. cysticola (Fig. 6)
Fig. 6.
Sections of Atlantic salmon gill from farm 5 tested with in situ hybridization for Ca. Branchiomonas cysticola. Note many small foci with labelling and some thickening and lamellar adhesion. Some mucus cells are present (arrow). Epithelicysts were not observed in H&E stained sections.
Farm 6: The fish from farm 6 (n=3), situated in south-western region Norway (Fig. 1), weighed about 1 2 kg and were sampled in November 2012. Fish at this farm displayed reduced appetite.
In all three fish, mixed infections with Ca. B. cysticola (Ct values 18.5–29.2), D. lepeophtherii, (Ct values 20–26.0), and P. perurans (Ct values 20.9–32.3), were identified (Table 1).
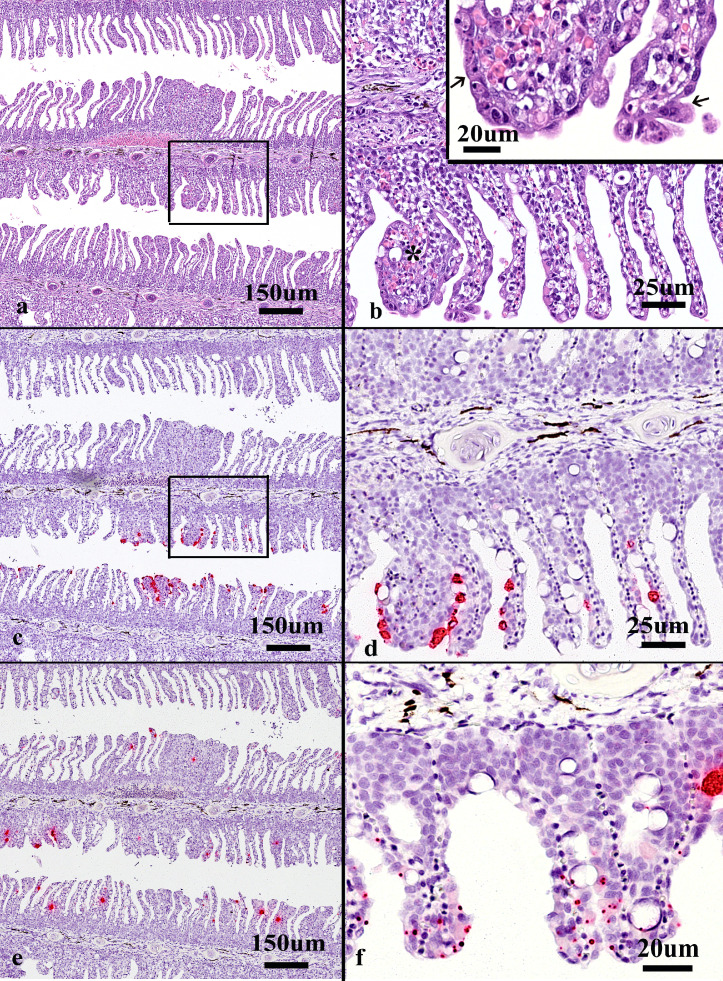
Histopathology revealed few findings in the fish with lowest agent load (fish 6-3. Table 1), however, in the gills of the two other fish, characteristic segmental AGD-like lesions were seen at low magnification, (Figs. 7a–c, 8). At higher magnification, the lesions comprised extensive epithelial cell hyperplasia, but also areas with some rounded, fragmented debris, few ballooning degenerative cells containing sparse light brown granular pigment (Fig. 8a,b), mucus cell hyperplasia and the notable presence of amoebae (Fig. 8a). Few inflammatory cells were seen within filaments and filament sinusoids. In some areas in the gills of a single fish, many clear epitheliocysts were visible (Figs. 7a, c, 8a,d).
Fig. 7.
Serial sections of Atlantic salmon gill from farm 6 (fish 6-1) stained with (a) haematoxylin and eosin (b) in situ hybridization for Desmozoon lepeoptherii and (c-d) Ca. Branchiomonas cysticola. (a) segmental hyperplasia (arrowheads) characteristic for amoebic gill disease. An amoeba is indicated by a vertical arrow and is magnified in the insert (arrow). Epitheliocysts are present (horizontal arrow). Part of this area is magnified in Figure 8. (b) note widespread labelling of D. lepeoptherii. (c) Ca. B. cysticola are clearly labelled in the form of large epitheliocysts. (d) marginal lamellar adhesion and multiple round Ca. B. cysticola-labelled bodies, some apparently located on or close to the surface of epithelial cells. The bacteria could not be seen in H&E sections.
Fig. 8.
Serial sections of Atlantic salmon gill from farm 6 (fish 6-1) stained with (a-b) haematoxylin and eosin, in situ hybridization for (c) Desmozoon lepeoptherii and for (d) Ca. Branchimonas cysticola. The images show details of the same gill shown in Figure 7. (a) lower arrow indicates an amoeba, also magnified in the insert (arrow). The arrowhead indicates an area with several epitheliocysts. (b) The magnified area shows “vacuoles”, some containing light brown pigmented granular material (asterisk) and a comparable distribution of staining is seen in c. (c) Extensive staining in area of segmental hyperplasia. (d) labelling of epitheliocysts in addition to staining of small rounded foci.
ISH revealed the presence of Ca. B. cysticola within epitheliocysts, and in areas of lamellar adhesion, where no individual bacterial cells could be seen in H&E preparations (Fig. 7d). ISH results for D. lepeophtherii varied among the three fish, with almost no labelling in the fish with the highest Ct value and most labelling in the fish with low Ct value. In the latter fish, labelling was observed evenly throughout the gill, but was more pronounced in areas of epithelial hyperplasia (Figs. 7b, 8c) and with a similar pattern and distribution as the rounded, fragmented debris seen in the H&E sections (Fig. 8b,c).
Farm 7: Fish from farm 7 (n=4), situated in Northern Norway (Fig. 1), weighed around 2 kg, were moribund and were sampled in October 2018. They were submitted for analysis following mechanical delousing treatment with subsequently high mortality. Different levels of gill lesions were observed at autopsy.
Concurrent infection with Ca. B. cysticola (Ct values 20.1–24.3), SGPV (Ct values 19.5–30.9) and D. lepeophtherii (Ct values > 28.0 or negative) were identified by PCR (Table 1).
Histopathology revealed moderate epithelial hyperplasia, multifocal lamellar adhesion and fusion, the extensive presence of subepithelial inflammatory and necrotic cells, multifocal haemorrhages, thrombosis (Fig. 9a–f) and a few apoptotic epithelial cells in two of the four gills (Fig. 9b). ISH revealed labelling of SGPV in apoptotic cells (Fig. 9c,d) and Ca. B. cysticola within epitheliocysts, and in small foci which were not observed in the H&E sections (Fig. 9e,f).
Fig. 9.
Serial sections of Atlantic salmon gill from farm 7 (fish 7-3) stained with (a-b) haematoxylin and eosin, in situ hybridization for (c-d) salmon gill poxvirus and for (e-f) Ca. Branchiomonas cysticola. The boxed area in a and c is magnified in b and d, respectively. (a) Note the thickened lamellae due to epithelial hyperplasia and sub-epithelial debris. (b) Asterisk indicates area of haemorrhage, cellular debris and inflammatory cells. Apoptotic-like epithelial cell magnified in the insert (arrows). (c-d) extensive labelling of swollen epithelial cells for SGPV. (e-f) presence of Ca. B. cysticola in areas of lamellar adhesion and inflammation, where the bacteria could not be observed in H&E sections.
Farm 8: Fish from farm 8 (n=7), situated in Western Norway (Fig. 1), weighed around 2.5 kg and were sampled in September 2020. There was increased mortality on the farm and gill disease was suspected to be partly responsible.
Concurrent infections with Ca. B. cysticola (Ct values 17.1–29.0), D. lepeoptherii, (Ct values 19.5–30.2), SGPV (Ct values 24.2–40.2) and P. perurans, with one positive fish at a Ct value of 28.4, were found (Table 1).
Histopathology revealed autolysis in the gills of two fish (Table 1) and these were excluded from further histopathological assessment. In the gills of the fish with the lowest agent loads, many lamellae affected by telangiectasia were seen. Additionally, some sub-epithelial haemorrhage, inflammatory cell presence and moderate mucus cell hyperplasia were observed. In the three remaining fish, there was a degree of epithelial lifting (separation of the epithelium from the basal membrane), and epithelial cell hyperplasia with some rounded, fragmented debris and lamellar adhesion. Further, telangiectasia, lamellar haemorrhage, and mucus cell hyperplasia were noted. Epitheliocysts were present. ISH revealed labelling of Ca. B. cysticola within epitheliocysts and in small foci where no bacterial cells could be seen in the H&E stained sections (Fig. 10).
Fig. 10.
Serial sections of Atlantic salmon gill from farm 8 (fish 8-7) stained by in situ hybridization for (a) Ca. Branchiomonas cysticola and (b) salmon gill poxvirus. (a) extensive staining, both in large epitheliocysts and in small foci. Thick lamellae. (b) staining of cells in the marginal part of lamellae.
Farm 9: The fish from farm 9, situated in South Western Norway (Fig. 1), weighed around 1.3 kg and were sampled in October 2018. The fish were moribund and moderately high mortality was recorded. Bleeding from gills and gill disease was suspected by the fish health personnel. PCR detected concurrent infections with B. cysticola (Ct values 17.0–17.3), D. lepeoptherii (Ct values 22.9–26.5), P. perurans (Ct values 28.5–29.6) and SGPV (Ct values 22.9–31.5) (Table 1).
Histopathology revealed extensive epithelial hyperplasia, lamellar adhesion and fusion and structures consistent with amoebae. In some proliferative lesions, there were many cells containing light brown pigmented material (Fig. 11a) labelling positive for D. lepeoptherii (Fig. 11b). The sparse presence of sub-epithelial inflammatory cells was observed in association with visible epitheliocysts. ISH revealed foci of Ca. B. cysticola. The result of ISH for D. lepeoptherii varied among the three fish, with little labelling in the fish with lowest agent load and most in fish with higher load (Table 1).
Fig. 11.
Serial sections of Atlantic salmon gill from farm 9 (fish 9-3) stained with (a, c) haematoxylin and eosin, in situ hybridization for (b) Desmozoon lepeoptherii. (a) lamellae with moderate epithelial hyperplasia and abundant rounded, fragmented debris (in box) and granulated pigmented material (arrowheads). (b) shows extensive staining for D. lepeoptherii. The boxed areas in a-b is magnified in c. (c) demonstrating that the rounded, fragmented debris (arrow) have similar shape and distribution as staining for D. lepeophtherii.
The two multiplex PCR assays were analysed for non-specific PCR products in silico with all possible primer combinations using ePCR, but no non-specific amplicons were identified. Amplification efficiency (E) and linearity (R2) were determined from multiplex PCR analysis of a dilution curve with the relevant species. Three independent experiments were performed with three replicates in each (Table 3). The asymmetrical limit of detection (LODasym) is defined as a LOD in the presence of a high concentration of non-target DNA. For the two multiplexes,100 000 salmon haploid genome equivalents were used and the LODasym is reported in Table 3.
Table 3.
Key validation data for the two multiplex real-time PCR assays. nd = not determined.
| Species | Multiplex | Average E ± SD | Average R2 | LODasym |
|---|---|---|---|---|
| Paramoeba perurans | 1 | 95.1 ± 2.3 | 0.998 | 15 |
| Desmozoon lepeophtherii | 1 | 101.6 ± 2.5 | 0.996 | 5 |
| Ca. Branchiomonas cysticola | 2 | 87.7 ± 3.2 | 0.998 | 5 |
| Salmon gill poxvirus | 2 | 90.5 ± 3.1 | 0.996 | 7 |
| Salmo salar | 1 | 93.4 ± 1.6 | 0.997 | nd |
| Salmo salar | 2 | 89.3 ± 2.2 | 0.995 | nd |
4. Discussion
Our previous, extensive investigation of the histopathological findings in CGD affected salmon [9] aimed to establish a common understanding of CGD related histopathology by focusing on a wide range of gill lesions. In that study, PCR results and histopathological observations suggested that necrosis and inflammation were associated with the presence of Ca. B. cysticola, while ballooning, degenerative cells containing pigmented material were observed only in association with D. lepeophtherii infections. As well-established experimental models for most agents infecting gills are still lacking, the present study again focused on natural disease occurrences, but utilised new diagnostic approaches. Most of the fish in the present study suffered from CGD, as a wide range of histopathological lesions were present. ISH-analyses strongly indicate that features characteristic of CGD, including necrosis in hyperplastic lesions and the presence of sub-epithelial inflammatory cells and debris, appeared to be associated with Ca. B. cysticola infections, even in the absence of visible epitheliocysts. ISH-labelled Ca. B. cysticola was identified in many cases as widespread small foci, initially undetectable in H&E sections. However, closer investigation of H&E sections corresponding to areas identified as positive by ISH, allowed subsequent identification of small, rounded or irregular, basophilic structures that could putatively represent small or recently formed foci of bacteria.. Whether these small bacterial foci are intra- or extracellular is unclear. Hence, ISH labelling served as a guide for histopathological focus on specific lesions and structures in H&E sections that would most likely have otherwise been overlooked.
Intra-epithelial Ca. B. cysticola, in the form of epitheliocysts, are probably largely protected from host immune defences. However, some degree of inflammation was observed, indicating stages of the infection which may mobilise some inflammatory response. This is consistent with other studies involving betaproteobacterial infections of epithelial cells of the human respiratory tract [21]. Inflammation corresponds well with diagnostic experiences involving histopathological investigations of gill disorders, where sub-epithelial inflammatory cell infiltration and cell-debris are often observed, particularly in cases involving Ca. B. cysticola. Contradictory observations were, however, reported by Rodger and co-authors [22], who found no marked inflammatory cell response in relation to the presence of epitheliocysts in field cases of gill disorders in Atlantic salmon.
Identification of D. lepeophtherii in histological preparations is challenging using common histochemical stains, as this small, widely distributed parasite occurs typically as individual organisms or in small clusters. The lesions associated with D. lepeophtherii in this study correspond well with previous reports also utilising in situ methodology. In contrast to the digoxygenin labelled probes used previously by Weli and colleagues [11], the RNA scope based labelling used in the current study, although specific to the target organisms, did appear to “spread” to the surrounding tissues, thus making histopathological interpretation of the surrounding tissues difficult. This phenomenon combined with the high intensity of staining observed suggests that the method could be further optimised.
A previous study carried out by Steinum and colleagues indicated that Ca. P. salmonis may contribute, possibly in a minor role, to multifactorial gill disease [23]. The lack of association between observable Ca. P. salmonis cysts, ISH non-cyst labelled Ca. P. salmonis and histopathological change in the present study combined with identification of Ca. B. cysticola as the by far most common and epitheliocystis-related agent in the investigated material [18], when taken together, may imply a less important role for Ca. P. salmonis in gill disease.
SGPV has been shown in previous studies to be associated with apoptotic gill epithelial cells [2,3,6] with virus replication taking place in these cells. This is again supported by the current work. In previous studies, infection with SGPV was suspected to be a precursor in some cases of CGD, as the virus compromises the gill epithelial barrier and paves the way for secondary infections [24]. This theory is also supported by another previous study demonstrating that host immune genes are downregulated during and after SGPV infection [6,8]. In the current work, a full history of the fish, covering general health and gill health status during the production cycle was not available, and assessment of primary and secondary gill infections was therefore not possible. This is particularly challenging in cases of multifactorial aetiology, as aetiological agents and their associated pathology may not necessarily be simultaneously present.
There is no doubt that PCR was more sensitive than ISH for detection of D. lepeophtherii, Ca. B. cysticola and SGPV, as in some cases, when the agent load was low, as assessed by PCR, ISH was negative. A plausible explanation for this, is the uneven distribution of infectious agents in the gills, especially in the case of Ca. B. cysticola. Two multiplex assays were validated both by in silico specificity analysis and by PCR analysis of known samples. Specificity in a multiplex PCR depends not only on each primer pair, but any combination of primers can lead to a non-specific PCR product if a matching template is present in the sample. All possible primer combinations were assessed but no significant non-specific amplicons were identified. Amplification efficiency in a multiplex context was determined to range between 89.3 and 101.6 % which were well within our method performance parameter acceptance values of 80–110%. Linearity measured as R2 over a dilution series were determined to be > 0.995 for all assays and found acceptable relative to the method performance parameter acceptance value for R2 = 0.96 (for a qualitative method). LODasym were determined to be 15 target copies per analysis or less and found to be acceptable. In conclusion, the two multiplex assays were found to have passed the validation tests. Studies on CGD, have been mainly descriptive, focusing on histopathological features of the disease. The increasingly available “omics” field of analyses (transcriptomics, proteomics, metabolomics) now offer novel opportunities for investigation of compromised gills. However, as CGD lesions are commonly found unevenly distributed, there are challenges associated with restricting ‘omic’ analyses to areas of morphological change. In a recent work, duplex in situ hybridisation [8] was utilised, allowing two targets, characterising the host response, to be detected simultaneously. These types of integrated analysis, and further efforts to establish experimental models for infectious gill diseases should be the focus of future studies on CGD, in order to get a broader understanding of the infection dynamics, disease development and host responses. In a broad perspective, there is reason to believe that disease diagnostics of farmed fish would benefit from a more holistic approach allowing a more in depth understanding of pathophysiological processes.
Declaration of Competing Interest
Name of the manuscript: “Multi-agent in situ hybridization confirms Ca. Branchiomonas cysticola as a major contributor in complex gill disease in Atlantic salmon”
The authors whose names are listed immediately below certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript. Author names:
Mona Cecilie Gjessing, Bjørn Spilsberg Terje Marken Steinum MaritAmundsen , Lars Austbø, Haakon Hansen, Duncan Colquhoun, Anne Berit Olsen
Acknowledgments
The authors would like to thank Lisa Furnesvik and Hanne Nilsen at NVI for submitting paraffin embedded tissue from case 7–9 and The Research Council of Norway for financing part of the work (project 267491).
References
- 1.Young N.D., Crosbie P.B., Adams M.B., Nowak B.F., Morrison R.N. Neoparamoeba perurans n. sp., an agent of amoebic gill disease of Atlantic salmon (Salmo salar) Int. J. Parasitol. 2007;37(13):1469–1481. doi: 10.1016/j.ijpara.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Gjessing M.C., Yutin N., Tengs T., Senkevich T., Koonin E., Ronning H.P., Alarcon M., Ylving S., Lie K.I., Saure B., Tran L., Moss B., Dale O.B. Salmon gill poxvirus, the deepest representative of the chordopoxvirinae. J. Virol. 2015;89(18):9348–9367. doi: 10.1128/JVI.01174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoen E., Tartor H., Amundsen M., Dale O.B., Sveinsson K., Ronning H.P., Gronneberg E., Dahle M.K., Gjessing M.C. First record of experimentally induced salmon gill poxvirus disease (SGPVD) in Atlantic salmon (Salmo salar L.) Vet. Res. 2020;51(1):63. doi: 10.1186/s13567-020-00787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouton A., Crosbie P., Cadoret K., Nowak B. First record of amoebic gill disease caused by neoparamoeba perurans in South Africa. J. Fish Dis. 2014;37(4):407–409. doi: 10.1111/jfd.12133. [DOI] [PubMed] [Google Scholar]
- 5.Crosbie P.B., Bridle A.R., Cadoret K., Nowak B.F. In vitro cultured Neoparamoeba perurans causes amoebic gill disease in Atlantic salmon and fulfils Koch's postulates. Int. J. Parasitol. 2012;42(5):511–515. doi: 10.1016/j.ijpara.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Gjessing M.C., Krasnov A., Timmerhaus G., Brun S., Afanasyev S., Dale O.B., Dahle M.K. The Atlantic salmon gill transcriptome response in a natural outbreak of salmon gill pox virus infection reveals new biomarkers of gill pathology and suppression of mucosal defense. Front. Immunol. 2020;11:2154. doi: 10.3389/fimmu.2020.02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boison S.A., Gjerde B., Hillestad B., Makvandi-Nejad S., Moghadam H.K. Genomic and transcriptomic analysis of amoebic gill disease resistance in Atlantic salmon (Salmo salar L.) Front. Genet. 2019;10:68. doi: 10.3389/fgene.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amundsen M.M., Tartor H., Andersen K., Sveinsson K., Thoen E., Gjessing M.C., Dahle M.K. Mucosal and systemic immune responses to salmon gill poxvirus infection in atlantic salmon are modulated upon hydrocortisone injection. Front. Immunol. 2021;12(2170) doi: 10.3389/fimmu.2021.689302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjessing M.C., Steinum T., Olsen A.B., Lie K.I., Tavornpanich S., Colquhoun D.J., Gjevre A.G. Histopathological investigation of complex gill disease in sea farmed Atlantic salmon. PLoS One. 2019;14(10) doi: 10.1371/journal.pone.0222926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguera P., Olsen A.B., Hoare J., Lie K.I., Lopez M.M., Poppe T., Rodger H.D. Complex gill disorder (CGD): a histopathology workshop report. Bull. Eur. Assoc. Fish Pathol. 2019;4(39):5. [Google Scholar]
- 11.Weli S.C., Dale O.B., Hansen H., Gjessing M.C., Ronneberg L.B., Falk K. A case study of Desmozoon lepeophtherii infection in farmed Atlantic salmon associated with gill disease, peritonitis, intestinal infection, stunted growth, and increased mortality. Parasit. Vectors. 2017;10(1):370. doi: 10.1186/s13071-017-2303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draghi A., 2nd, Popov V.L., Kahl M.M., Stanton J.B., Brown C.C., Tsongalis G.J., West A.B., Frasca S., Jr. Characterization of "Candidatus piscichlamydia salmonis" (order Chlamydiales), a chlamydia-like bacterium associated with epitheliocystis in farmed Atlantic salmon (Salmo salar) J. Clin. Microbiol. 2004;42(11):5286–5297. doi: 10.1128/JCM.42.11.5286-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nylund S., Steigen A., Karlsbakk E., Plarre H., Andersen L., Karlsen M., Watanabe K., Nylund A. Characterization of 'Candidatus Syngnamydia salmonis' (Chlamydiales, Simkaniaceae), a bacterium associated with epitheliocystis in Atlantic salmon (Salmo salar L.) Arch. Microbiol. 2015;197(1):17–25. doi: 10.1007/s00203-014-1038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiik-Nielsen J., Gjessing M., Solheim H.T., Litlabo A., Gjevre A.G., Kristoffersen A.B., Powell M.D., Colquhoun D.J. Ca. Branchiomonas cysticola, Ca. Piscichlamydia salmonis and Salmon Gill Pox Virus transmit horizontally in Atlantic salmon held in fresh water. J. Fish Dis. 2017;40(10):1387–1394. doi: 10.1111/jfd.12613. [DOI] [PubMed] [Google Scholar]
- 15.Toenshoff E.R., Kvellestad A., Mitchell S.O., Steinum T., Falk K., Colquhoun D.J., Horn M. A novel betaproteobacterial agent of gill epitheliocystis in seawater farmed Atlantic salmon (Salmo salar) PLoS One. 2012;7(3):e32696. doi: 10.1371/journal.pone.0032696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avendano-Herrera R., Toranzo A.E., Magarinos B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis. Aquat. Organ. 2006;71(3):255–266. doi: 10.3354/dao071255. [DOI] [PubMed] [Google Scholar]
- 17.Downes J.K., Henshilwood K., Collins E.M., Ryan A., O'Connor I., Rodger H.D., MacCarthy E., Ruane N.M. A longitudinal study of amoebic gill disease on a marine Atlantic salmon farm utilising a real-time PCR assay for the detection of Neoparamoeba perurans. Aquac. Environ. Interact. 2015;7(3):239–251. [Google Scholar]
- 18.Mitchell S.O., Steinum T.M., Toenshoff E.R., Kvellestad A., Falk K., Horn M., Colquhoun D.J. Candidatus Branchiomonas cysticola' is a common agent of epitheliocysts in seawater-farmed Atlantic salmon Salmo Salar in Norway and Ireland. Dis. Aquat. Organ. 2013;103(1):35–43. doi: 10.3354/dao02563. [DOI] [PubMed] [Google Scholar]
- 19.Schuler G.D. Sequence mapping by electronic PCR. Genome Res. 1997;7(5):541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nylund S., Nylund A., Watanabe K., Arnesen C.E., Karlsbakk E. Paranucleospora theridion n. gen., n. sp. (Microsporidia, Enterocytozoonidae) with a Life Cycle in the Salmon Louse (Lepeophtheirus salmonis, Copepoda) and Atlantic Salmon (Salmo salar) J. Eukaryot. Microbiol. 2010;57(2):95–114. doi: 10.1111/j.1550-7408.2009.00451.x. [DOI] [PubMed] [Google Scholar]
- 21.Chiu C.H., Ostry A., Speert D.P. Invasion of murine respiratory epithelial cells in vivo by Burkholderia cepacia. J. Med. Microbiol. 2001;50(7):594–601. doi: 10.1099/0022-1317-50-7-594. [DOI] [PubMed] [Google Scholar]
- 22.Rodger H.D., Murphy K., Mitchell S.O., Henry L. Gill disease in marine farmed Atlantic salmon at four farms in Ireland. Vet. Rec. 2011;168(25):668. doi: 10.1136/vr.d3020. [DOI] [PubMed] [Google Scholar]
- 23.Steinum T., Kvellestad A., Colquhoun D.J., Heum M., Mohammad S., Grontvedt R.N., Falk K. Microbial and pathological findings in farmed Atlantic salmon Salmo salar with proliferative gill inflammation. Dis. Aquat. Organ. 2010;91(3):201–211. doi: 10.3354/dao02266. [DOI] [PubMed] [Google Scholar]
- 24.Gjessing M.C., Thoen E., Tengs T., Skotheim S.A., Dale O.B. Salmon gill poxvirus, a recently characterized infectious agent of multifactorial gill disease in freshwater- and seawater-reared Atlantic salmon. J. Fish Dis. 2017;40(10):1253–1265. doi: 10.1111/jfd.12608. [DOI] [PubMed] [Google Scholar]