Abstract
Glycans that are abundantly displayed on vertebrate cell surface and secreted molecules are often capped with terminal sialic acids (Sias). These diverse 9-carbon-backbone monosaccharides are involved in numerous intrinsic biological processes. They also interact with commensals and pathogens, while undergoing dynamic changes in time and space, often influenced by environmental conditions. However, most of this sialoglycan complexity and variation remains poorly characterized by conventional techniques, which often tend to destroy or overlook crucial aspects of Sia diversity and/or fail to elucidate native structures in biological systems, i.e. in the intact sialome. To date, in situ detection and analysis of sialoglycans has largely relied on the use of plant lectins, sialidases, or antibodies, whose preferences (with certain exceptions) are limited and/or uncertain. We took advantage of naturally evolved microbial molecules (bacterial adhesins, toxin subunits, and viral hemagglutinin-esterases) that recognize sialoglycans with defined specificity to delineate 9 classes of sialoglycan recognizing probes (SGRPs: SGRP1–SGRP9) that can be used to explore mammalian sialome changes in a simple and systematic manner, using techniques common in most laboratories. SGRP candidates with specificity defined by sialoglycan microarray studies were engineered as tagged probes, each with a corresponding nonbinding mutant probe as a simple and reliable negative control. The optimized panel of SGRPs can be used in methods commonly available in most bioscience labs, such as ELISA, western blot, flow cytometry, and histochemistry. To demonstrate the utility of this approach, we provide examples of sialoglycome differences in tissues from C57BL/6 wild-type mice and human-like Cmah−/− mice.
Keywords: Adhesin, acetylation, bacterial toxin, Sialoglycan-recognizing probes, Sialoglycans
Introduction
All cells in nature are covered with a dense and complex array of sugar chains (Varki and Kornfeld 2022). In vertebrates, the outermost ends of the branches on this glycan forest are often capped with monosaccharides called sialic acids (Sias), which have enormous intrinsic complexity (Chen and Varki 2010; Schauer and Kamerling 2018). Most current methods to study this important and dynamic aspect of the glycome are too specialized for an average scientist to employ, and many aspects of the “sialome” (Altheide et al. 2006) are thus poorly studied. Given their ubiquitous presence and terminal position, Sias have been exploited as primary, transient, or co-receptors by a diverse range of commensal or pathogenic microorganisms (Severi et al. 2007; Matrosovich et al. 2015). These interactions are typically mediated by microbial proteins that have evolved high binding specificity toward sialoglycans because of the ongoing evolutionary arms race between microbes and hosts and can differentiate their target Sias by types, modifications, substitutions, and/or linkage to underlying glycans.
Here, we develop Sia-specific probes from such microbial proteins, harnessing their Sia specificity, and if found insufficient‚ assessed other available probes to generate a simple and reliable toolkit that can be used to easily monitor dynamic changes of Sias in normal and abnormal states. This set of sialoglycan-recognizing probes (SGRPs) can confirm whether a biological sample has any sialic acids or not; if so, the type of common Sia variations (particularly O-acetylation), linkage to underlying glycans, and the presence of N-acetyl or N-glycolyl groups. We assessed a number of Sia-specific binding proteins for specificity toward different mammalian Sia types and/or linkage to underlying glycans. Specifically, 9 types of SGRPs were defined from bacterial serine-rich repeat (SRR) adhesins, bacterial B5 toxins, viral hemagglutinins (HAs), and hemagglutinin-esterases (HEs) and compared with previously known invertebrate and plant lectins, selected Siglecs, and polyclonal and monoclonal antibodies. Upon identification of the best SGRP for each class of sialoglycans, a mutant inactive probe was also developed as an internal control of each probe’s specificity. To ensure minimal loss of sensitive Sia modifications/substitutions, experimental conditions were optimized, and specificity of each probe was tested with positive and negative controls, such as pretreatment with specific sialidases or esterases, or mild periodate oxidation of the Sia side chain. The binding specificities of SGRPs were confirmed using a sialoglycan microarray presenting a diverse array of more than 100 mammalian sialoglycans and demonstrated by examples of laboratory methods of ELISA, western blotting, and fluorescence detection by flow cytometry and histological analysis. An example of application of SGRPs is provided, showing sialoglycome changes in mice with human-like loss of the CMAH enzyme.
We of course realize that the absence of reactivity to a given probe is not an evidence of absence of a particular sialoglycan, and reactivity only indicates that the cognate glycan component is very likely to be present. Moreover, given the vast diversity of terminal sialoglycans in nature, we could not address all possibilities. Furthermore, our array does not represent all possible sialoglycans in nature (our estimate is >100,000 possibilities for terminal structures (Sasmal et al., BioRxiv 2021 May 28.446191)] and does not include branched structures nor some motifs found on other arrays (Fukui et al. 2002; Kletter et al. 2013; Arthur et al. 2014; Muthana and Gildersleeve 2014; Stencel-Baerenwald et al. 2014; Klamer et al. 2017). Thus, these probes are not meant to replace more rigorous chemical and structural analysis by experts. Rather they are developed for a nonexpert to discover interesting sialome patterns and changes in various biological systems, which are then worth exploring further.
Results and discussion
Defining distinct classes of SGRPs
We sought to define a set of SGRPs for detection of the most common mammalian sialoglycan variants, with nonbinding mutants as controls. This approach simplifies in-situ detection for all major types of Sias (SGRP1), typical N-acyl modifications at C-5 of Sias (SGRP2, SGRP5), linkages to underlying glycans (SGRP3, SGRP6, SGRP8), and occurrence of O-acetyl groups (SGRP4, SGRP7, SGRP9). The SGRP numbering system (see Table 1) for these probes make them easier to remember. To define a given probe, we used a superscript and added NB for the nonbinding control. Thus, for example, the Yersinia enterocolitica toxin B subunit (YenB) that recognizes all Neu5Ac and Neu5Gc forms and linkages is designated as SGRP1YenB and the nonbinding variant as SGRP1YenBNB. The current set of SGRPs does not include probes specific for 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid (Kdn), which, although naturally found in mammals, occurs in limited amounts and primarily in the free form (Kawanishi et al. 2021).
Table 1.
Sialoglycan recognition probes and their specificities.
| Class of SialoGlycan Recognition Probe | Sialoglycans classes | Identified molecule | Pfam IDs | Sialoglycan preferences | SGRPs Nomenclature | |
|---|---|---|---|---|---|---|
| Binding | Non-binding | |||||
| SGRP1 | All Sialic Acids (Sias) | YenB | Pertussis_S2S3 (PF02918) | All Sias | SGRP1YenB | SGRP1YenBNB |
| SGRP2 | 5-N-acetylneuraminic acid (Neu5Ac) | PltB | Pertussis_S2S3 (PF02918) | All Neu5Ac glycans | SGRP2PltB | SGRP2PltBNB |
| SGRP3 | α-2-3 Linked Sialic Acids (α-2-3 Sias) | HsaBR | GspA_SrpA_N (PF20164) | All α2–3-linked Sias | SGRP3Hsa | SGRP3HsaNB |
| SGRP4 | 4-O-Acetylated Sialic Acids (4-OAc Sias) | MHV | Hema_esterase (PF03996) | 4-OAcetylated Neu5Ac | SGRP4MHV | SGRP4MHVNB |
| SGRP5 | 5-N- glycolylneuraminic acid (Neu5Gc) | IgY | Ig (PF00047) | All Neu5Gc glycans | SGRP5IgY | SGRP5IgYNB |
| SGRP6 | α-2-6 Linked Sialic Acids (α-2-6 Sias) | SNA | Ricin_B_lectin (PF00652) | All α2–6 Linked Sia | SGRP6SNA | N/A |
| SGRP7 | 7-O-Acetylated Sialic Acids (7-OAc Sias) | BCoV | Hema_esterase (PF03996) | 7–9-di O-Acetylated Sia | SGRP7BCoV | SGRP7BCoVNB |
| SGRP8 | α-2-8 Linked Di-Sialic Acid linkage | TeT | Toxin_R_bind_C (PF07951) | Disialic linkages | SGRP8TeT | SGRP8TeTNB |
| SGRP9 | 9-O-Acetylated Sialic Acids (9-OAc Sias) | PToV | Hema_esterase (PF03996) | All 9-O-Acetylated Sia | SGRP9PToV | SGRP9PToVNB |
Table representing the classes of sialoglycan recognition probes (SGRPs), their presumed binding specificities, the most appropriate molecules as observed by assessment of sialoglycan binding by various glycomic methods, their experimentally confirmed sialoglycan preferences and defining SGRP nomenclature suggesting class of probe along with source of probe as mentioned in superscripts. The names of probes ending with NB represent the nonbinding variant of SGRPs.
Search for naturally evolved microbial molecules with defined specificity toward specific aspects of sialoglycans
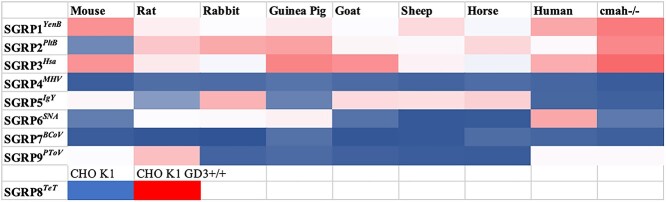
SGRP candidates were defined by sialoglycan microarray studies, with a corresponding nonbinding mutant as a negative control. The essential criterion for a protein to qualify as an SGRP is that it must show specificity toward the preferred Sia modification/linkage. Detailed analyses and specificities and their practicability toward probing mammalian sialoglycans are discussed in subsequent sections. As mentioned in Table 1, we could not identify any microbial candidate having a better or broader specificity for SGRP5 (N-glycolyl-Sias) than our previously described affinity-purified Neu5Gc chicken polyclonal IgY, or for SGRP6 Sambucus nigra agglutinin (SNA), the conventionally used lectin probe for α2–6-linked Sias.
SGRP1YenB detects all mammalian sialoglycans on the microarray
While α2–3Sia-binding MAL (Maackia amurensis lectin) and α2–6Sia-binding SNA together recognize the majority of Sia linkages, their preferences cannot be generalized for all types of Sias. Previously, Wheat germ agglutinin (WGA) and Limax flavus agglutinin (LFA) have been reported as having broad-spectrum Sia specificity, but their preferences toward Neu5Ac limit their utility as probes to detect all types of Sias (Bhavanandan and Katlic 1979; Miller et al. 1982; Cummings et al. 2015). For an SGRP that binds all types of Sia, we first considered YpeB (Yersinia pestis Toxin B subunit that recognizes both Neu5Ac- and Neu5Gc-terminated glycans [detailed sialoglycan preferences were recently reported (Khan et al. 2022), see also Fig. S1, see online supplementary material for a color version of this figure]. However, despite its ability to recognize most major classes of mammalian sialoglycans in glycan array and serum ELISAs, YpeB did not bind 4-OAc-Sias (Fig. S1, see online supplementary material for a color version of this figure). We investigated additional B subunits of AB5 bacterial toxins and selected YenB (Y. enterocolitica toxin B subunit) based on homology with YpeB and Salmonella Typhimurium ArtB, and broad host specificity (Sasmal et al., BioRxiv 2021 May 28.446191). Using His6-tagged YenB (Fig. 1, also reported in Sasmal et al., bioRxiv 2021 May 28.446191), we confirmed the recognition of both Neu5Ac and Neu5Gc including 9-O- and 4-O-acetylated Sias, a clear advantage over WGA, LFA, and our initial candidate YpeB (Fig. S2, see online supplementary material for a color version of this figure). Notably, YenB did not show binding to nonsialylated glycans in the same assay. To biotinylate YenB without affecting Sia-binding, we attempted to clone with additional tags for biotinylation (SNAP, ACP, and Avi) but the modifications resulted in poor protein quality and yield, leading to reduced binding in glycan arrays. A direct N-hydroxysuccinimide (NHS)-biotin conjugation of YenB was therefore optimized to obtain the final biotinylated probe (SGRP1YenB) that showed no change in binding in comparison to nonbiotinylated YenB. It was previously established that a serine residue contributes critically to Neu5Ac binding, while a tyrosine residue interacts with the extra OH group at the C5-acyl chain of Neu5Gc and is thus critical for its binding (Byres et al. 2008). As reported elsewhere (Sasmal et al., BioRxiv 2021 May 28.446191), we aligned the YenB sequence with those of Escherichia coli SubB and S. Typhimurium ArtB and predicted the conserved serine (S31) and tyrosine (Y100) in YenB. Mutating these critical sites for Sia recognition (YenB, S31A;Y100F) produced SGRP1YenBNB as an internal control for Sia-binding SGRP1YenB (Fig. S3a and b, see online supplementary material for a color version of this figure). The final binding and nonbinding specificities for SGRP1YenB were tested on a sialoglycan microarray with nearly 130 mammalian sialoglycan types, suggesting all-inclusive Sia specificity in SGRP1, and a complete lack of Sia-binding by SGRP1YenBNB (Fig. S4, see online supplementary material for a color version of this figure). Since there are no other molecules known to possess YenB-like Sia specificities, the pair of SGRP1YenB and SGRP1YenBNB are currently the most appropriate probe to detect all mammalian Sia types. The utility of SGRP1YenB as a tool in situ Sia detection through ELISAs, western blotting, IHC, and flow cytometry is described and discussed below. We of course realize that all possible sialoglycans and nonsialylated glycans have not been tested here (please see Conclusions and Perspectives section).
Fig. 1.
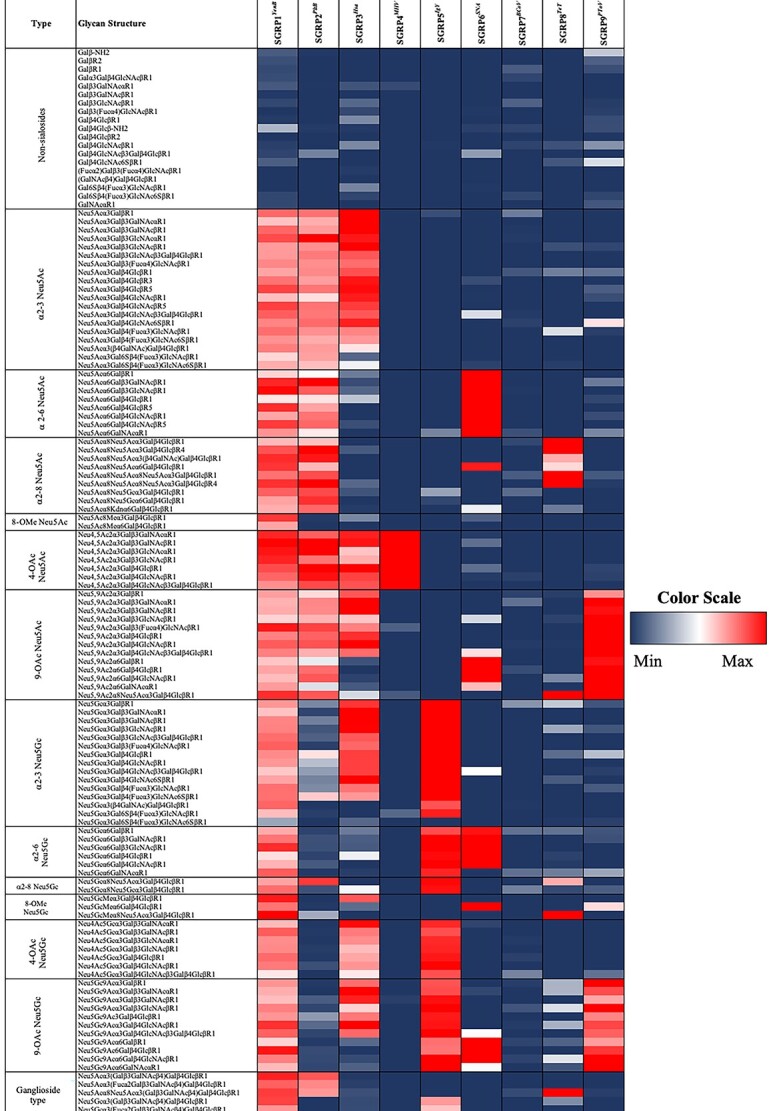
Sialoglycan microarray binding studies of proposed SGRPs. Heatmap analysis of SGRPs (see Table I for details of nomenclature) binding to mammalian (sialo) glycans; SGRP1YenB (30 μg/ml), SGRP2PltB (30 μg/ml), SGRP3Hsa (30 μg/ml), SGRP4MHV (30 μg/ml), SGRP5IgY (20 μg/ml), SGRP6SNA (20 μg/ml), SGRP7BCoV (60 μg/ml), SGRP8TeT (30 μg/ml), and SGRP9PToV (30 μg/ml) in microarray experiments. SGRPs binding efficiencies are displayed in red for highest binding (saturated or 100%), blue for minimum or no binding (0%), and intermediate binding represented by colors ranging between blue and red. Ranks; red (100%, or maximum), blue (0%, or minimum). Markedly reduced or absent binding was seen with the no-binding control probes studied simultaneously (see Fig. S4, see online supplementary material for a color version of this figure).
SGRP2PltB recognizes all Neu5Ac-terminated glycans on the microarray
Neu5Ac is the most abundant Sia type and occurs in all Sia-expressing organisms. Certain mammals (including humans) lack a functional CMAH enzyme to convert cytidine 5′-monophosphate N-acetylneuraminic acid (CMP-Neu5Ac) into CMP-Neu5Gc and express excess Neu5Ac as a primary Sia type. Despite such wide distribution and roles in human physiology/immunity, there has so far been no direct probe to selectively detect all forms and linkages of Neu5Ac in situ. Among commonly known probes for sialoglycans, WGA shows relative specificity toward Neu5Ac-glycans, but it neither recognizes all sialoglycans terminating with Neu5Ac nor is it exclusive to Neu5Ac, showing a dual preference for Neu5Ac and GlcNAc (Bhavanandan and Katlic 1979) (Fig. S2a, see online supplementary material for a color version of this figure). Previously, we identified Neu5Ac-specific binding in PltB, the B subunit of Typhoid toxin which preferentially bound human erythrocytes and tissues rich in Neu5Ac over the corresponding Neu5Gc-rich samples from Chimpanzees (Deng et al. 2014b). Similar Neu5Ac-specific patterns of PltB binding were further studied recently by others who also reported its binding O-acetylated Neu5Ac in both α2–3 and α2–6-linked sialoglycans (Nguyen et al. 2020). In another paper (Sasmal et al., BioRxiv 2021 May 28.446191), we report on the binding of PltB to a range of naturally occurring Neu5Ac but not Neu5Gc-glycans, underlining PltB’s appropriateness as a Neu5Ac-recognizing probe. Identifying the poor expression and purification quality of the His6-tagged Neu5Ac-binding domain of PltB with an additional ACP tag (NEB), the final biotinylated probe version of PltB (SGRP2PltB) was derived by NHS- biotinylation of PltB. This biotinylated-PltB shows comparable specificity toward Neu5Ac glycans as the unmodified protein (Fig. 1). The Neu5Ac-binding domain of PltB was characterized previously and a serine residue was reported to be critical for Neu5Ac-binding by PltB (Deng et al. 2014b). Thus, PltBS35ANB, an internal nonbinding mutant control of SGRP2, was also produced, biotinylated, and included in all studies (Fig. S3c and d, see online supplementary material for a color version of this figure).
The sialoglycan microarray with mammalian sialoglycan types showed the Neu5Ac-specific binding pattern of SGRP2PltB, while SGRP2PltBNB completely lacks binding (Fig. S4, see online supplementary material for a color version of this figure). In agreement with previous data SGRPPltB (Deng et al. 2014b), high binding preferences toward Neu5Ac were seen regardless of substitutions, modifications, or linkages to underlying glycans. SGRP2PltB demonstrates comparable binding for α2–3, α2–6, and α2–8-linked Neu5Ac and distinguishes them from α2–3, α2–6, or α2–8-linked Neu5Gc (Fig. 1). Notably, SGRP2PltB also exhibited recognition for Neu4,5Ac2 and Neu5,9Ac2. Taken together, SGRP2PltB showed definite superiority over the plant lectin WGA as a Neu5Ac-binding probe (Fig. S2a, see online supplementary material for a color version of this figure).
SGRP3Hsa selectively recognizes all α2–3-linked Sias on the microarray
Lectins from M. amurensis seeds (Kawaguchi et al. 1974) have been the gold standard for detection for Sias α2–3-linked to penultimate Gal. Initially designated as “strongly mitogenic M. amurensis leukoagglutinin (MAL) and ‘strongly hemagglutinating M. amurensis hemagglutinin” (MAH), they are also known as MAL-I and MAL-II, respectively. While both lectins require at least a Siaα2–3Gal disaccharide structure to bind, MAL-I shows stronger binding to Siaα2–3Galβ1-4GlcNAc/Glc (a trisaccharide common in N-glycans) and MAH (MAL-II) is selective toward Siaα2–3Galβ1-3GalNAcs (Kooner et al. 2021), typically found in O-linked glycans (Geisler and Jarvis 2011). Among Siaα2–3-linked structures tested, MAH prefers 9-O-acetyl Sias with Neu5Ac over Neu5Gc and Ser/Thr-linked O-glycan structures (Knibbs et al. 1991; Konami et al. 1994; Brinkman-Van der Linden et al. 2002). However, MAL-I can also recognize 3-O-sulfated Gal terminated oligosaccharides, i.e. it does not show exclusivity toward sialylated sequences (Bai et al. 2001). Instead, MAL-I displays widespread affinity toward α2–3-linked Neu5Ac and Neu5Gc, but with selective inclination toward Asn-linked over Ser/Thr-linked glycans. Attempts to improve Maackia lectins (Kaku et al. 1993) did not yield a general-purpose Siaα2–3-recognizing probe.
We looked into Sia-binding properties of previously reported Siglec-like-domain-containing ligand-binding regions (BRs) of Streptococcus gordonii SRR adhesins (Bensing et al. 2004; Deng et al. 2014a; Bensing et al. 2016). We selected GspB-BR of S. gordonii strain M99, Hsa-BR of S. gordonii Stain DL1 (Challis), and UB10712-BR of S. gordonii strain UB10712 and investigated their suitability for a comprehensive Siaα2–3-linkage identifying probe. All 3 BRs were expressed as SNAPf-His6 fusions in the pGEX-3X vector in a bacterial expression system along with their nonbinding variants HsaBR (R340E), GspBBR (R484E), and UB10712BR (R338E) (Fig. S3, see online supplementary material for a color version of this figure).
The pairs of HsaBR/HsaBRNB, GspBBR/GspBBRNB, and UB10712BR/ UB10712BRNB were biotinylated using SNAP-benzyl guanine chemistry and tested with the sialoglycan microarray. Despite structural similarities, the 3 biotinylated BRs displayed uniquely different ligand binding profiles, including differential recognition of sialyl Lewis antigens and sulfated glycans. While GspBBR selectively binds sialyl-T antigen (Neu5Acα2–3Galβ1-3GalNAc) and related structures, HsaBR displays broader specificity covering NeuAcα2–3Galβ1-4GlcNAc and sialyl-T antigen (Fig. 1 and Fig. S5a, see online supplementary material for a color version of this figure) (Bensing et al. 2004; Takamatsu et al. 2005; Deng et al. 2014a). In comparison to HsaBR, GspBBR imparts lesser specificity toward α2–3 than α2–6 Sia linkages (Bensing et al. 2004) and falls short of the binding range of HsaBR, which not only includes trisaccharide and oligosaccharide but also disaccharide Sias. 9-O-Acetylation on Sia did not block GspBBR or HsaBR binding, but sulfation enhanced HsaBR binding (Deng et al. 2014a). Sia binding preferences of UB10712BR remained comparable to α2–3-linked Sia specificities of HsaBR and GspBBR. UB10712BR bound to a range of α2–3-sialyl linkages including sialyl Lewis X, 3′-sialyllactosamine and their sulfated forms but preferred Neu5Ac over Neu5Gc sequences (Fig. S5a, see online supplementary material for a color version of this figure) (Bensing et al. 2016). Confirming the Sia-specificity of these BRs, the NB variants did not show any binding of Sia linkages/modifications on the array (Figs S4 and S5, see online supplementary material for a color version of these figures). The data obtained with biotinylated BRs also agree with previously published reports on GST-fusion BRs (Bensing et al. 2004, 2016; Takamatsu et al. 2005; Deng et al. 2014a), and the comparable binding abilities of these biotinylated probes with the original BRs confirmed that biotinylation did not affect the Sia specificity of these proteins.
We asked if HsaBR could be a replacement for MAL and MAH, conventional lectins for this class of probes (SGRP3). In an experiment to compare Siaα2–3-binding preferences of Biotin-HsaBR, Biotin-MAL, and Biotin-MAH, HsaBR showed significantly pronounced Siaα2–3-binding ability regardless of Sia modifications (O-acetylation, O-sulfation) or glycan structures (disaccharides, trisaccharides, oligosaccharides) (Fig. S6a, see online supplementary material for a color version of this figure). Taken together with the glycan array data, we chose biotinylated HsaBR as our SGRP 3 probe (SGRP3Hsa) and its nonbinding variant HsaBR (R340E) as the nonbinding control (SGRP3HsaNB).
SGRP4MHV as a probe for 4-OAc-Sias
A major contributor to mammalian sialoglycan diversity is O-acetylation, substituting the sialic acid hydroxyl groups at C4, C7, C8, and/or C9 (Klein and Roussel 1998; Kamerling and Gerwig 2006). The presence or absence of these O-acetyl moieties can block or promote the binding of cellular and microbial lectins, and their regulations through sialate-O-acetylesterases (SOAEs) and sialate-O-transferases (SOATs) act as the molecular switches to control several cellular functions and interactions (Cariappa et al. 2009; Pillai 2013). In contrast to O-acetylation at C7 or C9-OH, 4-O-acetylated Sia and its structural and functional significance have yet to be explored in detail, and 4-OAc-Sias have been difficult to study even by chemical methods, due to variable expression or absence in many animal species, dynamic occurrence in pathological conditions, resistance to conventional sialidases, lability to acidic conditions (Manzi et al. 1990), and masking of Sias from detection by some lectins. Equine erythrocytes, α2-macroglobulins, and sera were used to study 4-OAc-“HD3”-reactive antibodies due to their high 4-O-acetyl-N-acetylneuraminic acid (Neu4,5Ac2) content (30–50 percent of total Sias) (Hanaoka et al. 1989). Guinea pigs are another common source of mammalian 4-OAc-Sias. Neu4,5Ac2 comprises a considerable share in serum (30% of all Sias) and liver (10% of all Sias) Sias in guinea pig, besides traces of 4-O-acetyl-N-glycolylneuraminic acid (Neu5Gc4Ac) (Iwersen et al. 1998).
In humans, 4-OAc has been reported in tumor-associated antigens of colon cancers, melanomas, and gastric cancers using “HD” antigen-specific antibodies, for example a chicken antibody specific to Neu5Gc4Ac-lactosylceramide (4-OAc-HD3) recognized Neu5Gc4Ac in GM3 ganglioside fractions of human colon cancer tissues (Miyoshi et al. 1986). Similar 4-OAc-HD3-reactive HD antibodies have also been reported in sera of patients with malignancies and liver diseases (Higashihara et al. 1991). Two chicken MAbs HU/Ch2–7 and HU/Ch6–1 reacted with Neu5Gc4Ac (Asaoka et al. 1992). Despite frequent reports on heterogenous 4-OAc in HD antigens in human cancers, there has not been a reliable conventional probe for in situ detection of this entire class of sialoglycans.
Previously, a Sia-binding lectin with specificity for O-acetyl Sia was purified from the hemolymph of the California coastal crab Cancer antennarius, which was more precise than other known lectins from horseshoe crab (Limulus Polyphemus) and slug (L. flavus) but also showed affinity toward 9-O-acetyl in addition to 4-O-acetyl Sias (Ravindranath et al. 1985). A lectin from Tritrichomonas foetus, a parasitic protozoan that causes abortion in cows, was reported to react preferentially with Neu4,5Ac2 over de-O-acetylated Sias, and agglutinated equine erythrocytes containing Neu4,5Ac2 efficiently, but its preferences were also not exclusive for 4-O-acetylated Sias (Babál et al. 1999).
In general, O-acetylation of Sias can be a major receptor determinant for some viruses. Among the 5 viruses shown to initiate infections via O-Ac-Sias were the influenza C viruses, human coronavirus OC43, Bovine Coronaviruses BCoV, and porcine encephalomyelitis virus (PToV), but none of them exhibited binding of Neu4,5Ac2 (Herrler et al. 1985; Rogers et al. 1986; Vlasak et al. 1987; Vlasak et al. 1988; Schultze et al. 1991a). Infectious salmon anemia virus (ISAV), the causative agent of infections in Atlantic salmons, showed specificity for, and hydrolysis of 4-OAc-Sias (Hellebø et al. 2004). ISAV preferentially de-O-acetylated free and glycosidically bound Neu4,5Ac2 and showed lower and no hydrolysis for free and bound Neu5,9Ac2, respectively. ISAV exhibited hydrolysis of both Neu4,5Ac2 and Neu5Gc4Ac at comparable efficacy, which was a significant advantage over known 4-OAc-Sia-binding molecules but its affinity for free Neu5,9Ac2 although lower than influenza C virus (Hellebø et al. 2004), restricted possibilities to derive a comprehensive probe for 4-OAc-Sias.
Murine coronavirus mouse hepatitis virus (MHV-stain S) expresses a hemagglutinin-esterase that exhibits comparable sialate-4-O-acetylesterase enzymatic activity to that of ISAV (Regl et al. 1999). Unlike comparable esterases from other sources, MHV-S HE protein specifically de-O-acetylates Neu4,5Ac2, but not Neu5,9Ac2, and converts glycosidically bound Neu4,5Ac2-rich glycoproteins from horse and guinea pigs to Neu5Ac (Regl et al. 1999). MHV-S hydrolyzes acetyl esters from free as well as glycosidically linked Neu4,5Ac2. Interestingly, MHV-A59 and several other MHV strains do not express a HE (Luytjes et al. 1988; Shieh et al. 1989; Yokomori et al. 1991). Previously, the MHV-S HE ectodomain, released from HE-Fc by thrombin-cleavage, was reported to exhibit proper sialate-4-O-acetylesterase activity when assayed for substrate specificity with a synthetic di-O-acetylated Sia (4,9-di-O-acetyl-N-acetylneuraminic acid α-methylglycoside, Neu4,5,9Ac3αMe) (Langereis et al. 2012).
Our collaborators had previously expressed the esterase-inactive MHV-S-HE ectodomain as a fusion protein with a C-terminal Fc domain of human IgG1 and investigated Neu4,5Ac2 distribution in human and mouse tissues (Langereis et al. 2015). In another study, we further modified the virolectin by fusing MHV-S-HE ectodomain-Fc to a hexahistidine (His6) sequence and detected higher expression of 4-OAc-Sias in horse and guinea pig respiratory tract tissues than mouse, where it was mostly localized in the gastrointestinal tract. 4-OAc-Sias were also found in a small number of cells within the duck, dog, and ferret respiratory tissues screened‚ but not so far in the tissues of humans or pigs (Wasik et al. 2017).
To derive a stable 4-OAc-Sia-binding probe, we expressed the MHV-S-HE esterase inactive ectodomain (S45A), and the nonbinding mutant MHV-S-HE (F212A), as fusion proteins with the C-terminal Fc domain of human IgG1, along with Avi-tag for permanent biotinylation (Fig. S3, see online supplementary material for a color version of this figure). Sia-binding and nonbinding proteins MHV-S-HE protein probes were biotinylated and tested for 4-OAc-Sia specificity on sialoglycan microarray. As anticipated, biotinylated Sia-binding MHV-S-HE (S45A) exhibited very specific recognition of 4-OAc-Sias, while the nonbinding mutant MHS-S HE (F212A) did not show any binding with any sialylated or nonsialylated glycan on the microarray, confirming its suitability as a nonbinding control (Fig. 1; Fig. S4, see online supplementary material for a color version of this figure). Biotinylated MHV-S-HE (S45A) binds exclusively to Neu4,5Ac2α3Galβ3GalNAcαR1, Neu4, 5Ac2α3Galβ3GalNAcβR1, Neu4,5Ac2α3Galβ3GlcNAcαR1, Neu4,5Ac2α3Galβ3GlcNAcβR1, Neu4,5Ac2α3Galβ4GlcβR1, Neu4,5Ac2α3Galβ4GlcNAcβR1, and Neu4,5Ac2α3Galβ4GlcNAcβ3Galβ4GlcβR1 (Fig. 1). Despite such high specificity and avidity for Neu4,5Ac2, MHV-S-HE (S45A) did not show any binding of 4-OAc-Neu5Gc-glycans (Fig. 1). Currently, the sialoglycan microarray does not include α2–6-linked 4-OAc-Sias (Neu5Ac or Neu5Gc), hence the binding of MHV-S-HE α2–6-linked 4-OAc-Sias is not discussed here.
Considering the rarity of Neu5Gc4Ac and limited knowledge about 4-OAc-Sia-recognizing proteins, MHV-S HE-derived fusion proteins provide the most useful probes for in situ detection of 4-OAc-Sias, represented mostly by Neu4,5Ac2. Given its exclusive preference for 4-OAc-Sias, the biotinylated MHV-S-HE esterase inactive ectodomain appears to be the best probe for this class of sialoglycans (SGRP4MHV and SGRP4MHVNB) and we demonstrate its utility through ELISA, western blotting, FACS, and histochemistry. We expect that SGRP4MHV will facilitate research on functional significance of 4-OAc-Sias, help in finding more comprehensive 4-OAc-Sia-binding proteins, and allow us to seek an improved version of SGRP4 with recognition of both Neu4,5Ac2 and Neu5Gc4Ac.
SGRP5IgY recognizes all Neu5Gc-terminated glycans on the microarray
Humans are genetically defective in synthesizing the common mammalian Sia Neu5Gc but can metabolically incorporate small amounts of this Sia from dietary sources into glycoproteins and glycolipids of human tumors, fetuses, and some normal tissues (Tangvoranuntakul et al. 2003). A mutually nonexclusive hypothesis has been put forward to suggest endogenous Neu5Gc production by tumor cells (Bousquet et al. 2018), but supporting data is very incomplete. Regardless, Neu5Gc has been observed in breast, ovarian, prostate, colon, and lung cancers. There is also a need for sensitive and specific detection of Neu5Gc in human tissues and biotherapeutic products. Previously, a number of different monoclonal antibodies against Neu5Gc have been reported which recognized Neu5Gc only in the context of particular underlying sequences and generally lack the ability to detect Neu5Gc on other structurally related or unrelated glycans (Ohashi et al. 1983; Higashi et al. 1984; Higashi et al. 1985; Hirabayashi et al. 1987; Higashi et al. 1988; Miyake et al. 1988; Tai et al. 1988). Most microbial Neu5Gc-binding proteins binding Neu5Gc-glycans are also not completely specific for Neu5Gc, as there is always some cross-reactivity with few Neu5Ac glycans (Ohashi et al. 1983; Higashi et al. 1984; Higashi et al. 1988).
To explore Neu5Gc-specific probes from microbial sources, we considered our previously reported Neu5Gc-binding preferences of subtilase cytotoxin (SubAB), an AB5 toxin secreted by some strains of Shiga toxigenic E. coli (STEC) (Byres et al. 2008). The B5 subunits of this toxin (SubB) exhibited strong preference for Neu5Gc-terminating glycans. SubB showed 20-fold less binding to Neu5Ac and over 30-fold less if the Neu5Gc linkage was changed from α2–3 to α2–6. Using molecular modeling and site directed mutations, Day and colleagues reduced the α2–3 to α2–6-linkage preference while maintaining or enhancing the selectivity of SubB for Neu5Gc over Neu5Ac (Day et al. 2017; Wang et al. 2018). This SubB analog, SubB2M (SubBΔS106/ΔT107 mutant), did display further improved specificity toward Neu5Gc, bound to α2–6-linked-Neu5Gc, and could discriminate Neu5Gc- over Neu5Ac-glycoconjugates in glycan microarrays, surface plasmon resonance and ELISA assays. SubB2M also showed promising diagnostic properties in detecting presumed Neu5Gc-glycans in serum of patients with all stages of ovarian cancer (Shewell et al. 2018) and breast cancer (Shewell et al. 2022). While the SubB2M as reported has a clear and strong preference for Neu5Gc over Neu5Ac glycans the preference is not absolute. The polyclonal chicken IgY also has the advantage of recognizing all 40+ Neu5Gc-bearing glycans, with no cross reactivity at all with Neu5Ac-glycans. Since the goal was to have as broad-spectrum a probe for Neu5Gc-glycans as possible, we chose to go with the biotinylated-IgY, using IgY from nonimmunized chickens as a negative control (Fig. S7, see online supplementary material for a color version of this figure).
Considering all results from the Neu5Gc-binding molecules discussed or tested above, there is no contemporary probe better than the affinity-purified chicken polyclonal anti-Neu5Gc-IgY to detect a broader range of Neu5Gc-glycans. Hence, we selected chicken polyclonal-specific anti-Neu5Gc-IgY as SGRP5 and biotinylated this along with its nonbinding control IgY (from nonimmunized chickens) as the final set of SGRP5IgY (Fig. S4, see online supplementary material for a color version of this figure). The utility of chicken Neu5Gc-IgY as SGRP5IgY in general lab-used methods including ELISA, western blotting, FACS, and histochemistry is discussed in an earlier article, and the results obtained here confirmed its Neu5Gc-specificity as previously reported (Diaz et al. 2009; Samraj et al. 2015). In a long run, it is necessary to identify a monoclonal IgY that can detect all forms of Neu5Gc-sialoglycans without any cross-reactivity with Neu5Gc.
SGRP6SNA recognizes all α2–6-Sias on the array
Unlike bacteria, some viruses that cause upper respiratory infections such as human influenza A, B viruses, and human coronavirus OC43 exhibit preferential affinity toward α2–6Sias, which are abundant in the upper airway epithelial brush border in humans (Nicholls et al. 2007; Jia et al. 2020). Influenza viral haemagglutinins (HA) are the major glycoproteins that allow the recognition of cells in the upper respiratory tract or erythrocytes by binding to α2–6Sias, making them potential candidates for α2–6Sias-binding probes. We investigated a range of viral HAs in the form of HA-Fc fusion proteins (soluble HA fused to human IgG1 Fc) for their sialoglycan-binding specificity using the glycan microarray (Fig. S8, see online supplementary material for a color version of this figure). Among the tested HA-Fc fusion proteins, Cali09 HA-Fc derived from California/04/2009 H1N1 showed selective binding to α2–6-linked Sias, most prominently with Neu5Acα6Galβ4GlcNAcβR5, followed by Neu5Acα6Galβ4GlcNAcβR1 and Neu5,9Ac2α6Galβ4GlcN AcβR1. Despite strong preferences for α2–6Sias, none showed binding with a full range of α2–6Sias, especially α2–6Neu5Gc glycans. The failure of Cali09-HA-Fc to recognize a number of α2–6-linked sialosides, and its binding to a few α2–3-linked sialosides, questioned its suitability as an exclusive probe for α2–6-linked Sias (Fig. S8, see online supplementary material for a color version of this figure). Aichi68-HA-Fc derived from the hemagglutinin of Aichi/2/1968 H3N2 strain also lacked robustness and specificity showing indiscriminate binding toward a number of α2–6 and α2–3Sias (Fig. S8, see online supplementary material for a color version of this figure). Surprisingly one candidate, PR8 HA-Fc derived from Influenza strain A/Puerto Rico/8/1934 (PR8 H1N1), even showed prominent binding of α2–3-sialylated glycans instead of α2–6Sias (Fig. S8, see online supplementary material for a color version of this figure). Among other tested viral haemagglutinins, SC18 HA-Fc derived from influenza A H1N1 (A/SouthCarolina/1/18) exhibited selective binding to a few α2–6Sias such as Neu5Acα6Galβ4GlcNAcβR5 and Neu5Acα6Galβ4GlcNAcβR1, while another virolectin Mem-HA-Fc from influenza A H1N1 (A/Memphis/1/1987) bound to α2–3Sias and α2–6Sias without any strong preference for either linkage (Fig. S8, see online supplementary material for a color version of this figure). Taken together, it can be inferred that the viral haemagglutinins tested exhibited promising specificity toward α2–6Sias but lacked the robustness and binding dynamics required for a probe. Our results and observations appear true for several other viral hemagglutinins not included here (Lehmann et al. 2006).
As an alternative α2–6Sia-specific microbial probe, we reviewed the α2–6Sia-binding properties of a recombinant lectin (PSL) from the mushroom Polyporus squamosus that was reported for its high affinity binding with Neu5Acα6Galβ4GlcNAc (Tateno et al. 2004; Kadirvelraj et al. 2011). However, PSL showed a high order preference toward α2–6 over α2–3Sias but bound exclusively to Neu5Acα6Gal on N-linked glycoproteins, so we did not experimentally investigate this molecule further as an α2–6Sia-specific probe.
As an α 2–6Sia-binding alternative from mammals, we considered the well-characterized vertebrate sialic acid-dependent adhesion molecule CD22/Siglec-2 a member of the immunoglobulin superfamily expressed by B lymphocytes that binds specifically to Neu5Acα6Galβ4GlcNAc (Powell et al. 1993, 1995; Kelm et al. 1994; Sjoberg et al. 1994; Brinkman-van der Linden et al. 2000). hCD22-Fc (human CD22 fused with the Fc region of human IgG1) showed high affinity for a few α2–6Sias on microarray (Fig. S9, see online supplementary material for a color version of this figure), but lacked the avidity required for a probe, particularly for α2–6-linked Neu5Gc. With a possibility to characterize 2 SGRP6 candidates: one specific for α2–6Neu5Ac and the other for α2–6Neu5Gc, we tried exploiting the evolutionary derived strong preference of mouse siglec-2 (mCD22) for α2–6Neu5Gc. mCD22-hFc showed exclusive binding toward α2–6Sias but preferred Neu5Gc in general (Fig. S9, see online supplementary material for a color version of this figure). Interestingly, 9-O-acetylation completely aborted hCD22’s binding to α2–6Neu5Ac (Fig. S9, see online supplementary material for a color version of this figure) but did not affect mCD22 binding to α2–6Sias. Nevertheless, the relatively poor avidity and inability of hCD22 to recognize O-Ac-substitution in α2–6Neu5Ac and the nonspecificity of mCD22 binding α2–3 sialyl-LNnT glycan (Neu5Gcα3Galβ4GlcNAcβ3Galβ4Glc) limited their competency as α2–6Sia-binding probes, even in combination (Fig. S9, see online supplementary material for a color version of this figure).
Among other molecules, we also analyzed the adhesin of nontypeable Haemophilus influenzae which was reported for high affinity toward α2–6-linked Neu5Ac (Atack et al. 2018). According to this study, HMW2 bound with high affinity to α2–6-linked Neu5Ac such as Neu5Acα6Galβ4GlcNAc (α2–6-sialyllactosamine) and could discriminate it from α2–3Sias (<120-fold lesser binding with α2–3 Sialyllactosamine). HMW2 indeed showed appreciable preference for the α2–6 over the α2–3-linkage but was not able to recognize α2–6Neu5Gc, disqualifying it from consideration as a comprehensive probe for α2–6Sias. In the absence of an evolutionarily derived microbial protein as a dynamic probe for α2–6Sias, it appears that SNA (S. nigra or elderberry bark lectin) is still the best available probe for α2–6Sias.
SNA exhibits high preference for the terminal Neu5Acα6Gal /GalNAc sequences in both N-linked and O-linked glycans (Shibuya et al. 1987). Due to its ability to discriminate Siaα6Gal/GalNAc from Siaα2–3Gal/GalNAc and ubiquitous binding to Neu5Ac/Gc with/without O-Ac substitutions, SNA has been the standard probe for studying α2–6Sias. Using Vector lab’s Biotin-SNA (Catalog number B1305, lot number Z1002), we investigated SNA’s Sia-binding efficiency in sialoglycan microarray and observed very high preference toward α2–6Sias for both Neu5Ac and Neu5Gc and no measurable binding to α2–3Sias (Fig. 1). Interestingly, the O-acetyl substitutions that were a major concern for hCD22, mCD22, viral hemagglutinins, and HMW2 did not hinder SNA’s binding to α2–6Sias. Apart from its binding to 6-O-sulfated Galβ4GlcNAc (Yamashita et al. 1992), SNA displayed an exclusive preference for α2–6Sias in disaccharides, trisaccharides, and oligosaccharides in Asn or Ser/Thr-linked glycans. So far, there appears to be no better molecule than SNA to probe α2–6Sias, hence we selected Biotin-SNA as SGRP6 (SGRP6SNA) for this study. SGRP6SNA is the exception to the set of SGRPs in not having an internal control of specificity. We characterized SGRP6SNA as α2–6Sia-probe without any nonbinding variant in routine lab assay methods.
SGRP7BCoV recognizes 7,9-diOAc-Sias
Except for some claims in microorganisms (Lewis et al. 2004; Gurung et al. 2013) and on human lymphocytes (Wipfler et al. 2011), 7-OAc-Sias are not commonly reported in natural glycans because of their instability. During biosynthesis, the primary attachment site of O-acetyl groups was exclusively to the C-7 hydroxyl of Sias, and the ester group migrates from C-7 to C-9 (Vandamme-Feldhaus and Schauer 1998). A hypothesis was proposed that SOAT enzyme would effectively be a 7-O-acetyltransferase incorporating O-acetyl groups primarily at C-7 of sialic acids, followed by their migration to the primary hydroxyl group at C-9 and transfer of an additional O-acetyl residue to C-7, resulting in di- and tri-O-acetylated species (Schauer 1987). This suggests that in nature, 7-OAc will be represented as 7,9-diOAc or 7,8,9-triOAc, but largely as 7,9-diOAc in mammalian sialoglycans. Considering that there is no stable 7-OAc, there may not be a true 7-OAc-Sia-binding protein. We defined specificities of SGRP7 to 7,9-diOAc-Sias, and 7/9di,9-diOAc has traditionally been studied in bovine submaxillary mucin, where the amount of 7-OAc- and 7,9-diOAc-Sias combined is almost twice of 9-mono-O-Ac-Sias (Langereis et al. 2015).
We also reviewed the sialoglycan specificity of other lectins and microbial proteins, reported to bind to 7,9-diOAc-Sias. Lectins from C. antennarius, Achatina fulica, and T. foetus showed higher preferences for 9-OAc and somewhat to 4-OAc but negligible to poor affinities for 7,9-diOAc-Sias (Stewart et al. 1978; Ravindranath et al. 1985; Babál et al. 1999). Viruses exhibit prominent binding to O-Ac-Sias but most show either 9-OAc preference (Influenza C viruses, human coronavirus OC43, porcine encephalomyelitis virus) or 4-OAc preference (ISAV, Puffinosis virus, mouse hepatitis virus strains-S, DVIM, JHM) binding over 7,9-diOAc (Herrler et al. 1985; Rogers et al. 1986; Vlasak et al. 1987, 1988; Schultze et al. 1991a; Klausegger et al. 1999; Hellebø et al. 2004; Langereis et al. 2010, 2015). The hemagglutinin esterases from bovine toroviruses (BTOV-B150, BToV-Breda), bovine coronaviruses (strains; Mebus and Lun), and equine coronavirus (ECoV-NC99) preferentially cleave 7,9-diOAc substrates over mono 4- or 9-OAc-substrates (Langereis et al. 2009; Langereis et al. 2015). Bovine viruses exhibit selective binding toward 7,9-diOAc, particularly BCoV-Mebus having relatively pronounced preferences toward 7,9-diOAc variants of both Neu5Gc and Neu5Ac, but lower preferences for 9-OAcs (Langereis et al. 2015). It is interesting that BCoV esterase selectively removes all 9-OAc residues in 7,9-diOAc-Sias in BSM, leaving 7-OAc attached. The residual 7-OAc residues do not attract binding of BCoV anymore, demonstrating that Sia 9-O-acetylation is a strict requirement and that mono 7-OAc-Sias do not serve as ligands (Langereis et al. 2015). The S protein, another hemagglutinin in BCoV, showed high specificity exclusively for 9-OAc-Sias with no preference for 7-OAc and this raises possibility that HE is the only receptor-binding protein for 7,9-di-OAc in BCoV-Mebus (Schultze et al. 1991b; Schultze and Herrler 1992).
Since BCoV-Mebus-HE is best characterized for 7,9-diOAc-Sia specificities, we expressed its esterase inactive mutant (BCoV-Mebus-HE S40A) and corresponding nonbinding mutant (BCoV-Mebus-HE F211A), each fused to human IgG-Fc followed by a His6-tag and an Avi-tag. Both proteins were biotinylated with an Avi-tag and tested for preferences on the sialoglycan microarray (Fig. 1, Fig. S4, see online supplementary material for a color version of this figure). While the nonbinding mutant does not show binding with any sialylated or nonsialylated glycans in the array, the Sia-binding molecule BCoV-Mebus-HE S40A exhibited some affinity for 9-OAc-Sias (Fig. 1), agreeing with its Sia-binding pattern on sialoglycan microarray reported previously (Langereis et al. 2015). However, BCoV-Mebus-HE, S40A did not exhibit any binding to 4-OAc, most 9-OAc-Sias, and nonacetylated sialyloligosaccharides. The sialoglycan microarray did not contain 7/9di-Sias, so it is not known whether the protein binds that form.
SGRP8TeT binds to a major class of terminal α2–8-linked disialosides
Oligosaccharide sequences in gangliosides and a few glycoproteins that terminate in Sias are not always mono-sialylated. In addition to α2–3 or α2–6-linkage with penultimate Gal, Sia can be linked with another Sia predominantly by α2–8-linkage forming α2–8-linked di, αoligo, and polysialic acid chains. α2–8-linked disialyl structures are critical component of neuronal gangliosides and these disialyl gangliosides, especially GD3 and GD2, have been utilized as tumor markers for melanomas, gliomas, and neuroblastomas (Miyoshi et al. 1986; Hanaoka et al. 1989; Higashihara et al. 1991; Asaoka et al. 1992). Due to their utility in cancers either as biomarker for in vivo immunolocalization or in phase I and II trials to target disseminated neuroblastoma, a range of anti-disialoganglioside monoclonal antibodies have also been generated. For example, R24 is a mouse IgG3 monoclonal antibody (MAb) that reacts with the ganglioside GD3‚ expressed by cells of neuroectodermal origin (Kemminer et al. 2001). Similarly, a list of anti-GD2-specific MAbs; 3F8 (Heiner et al. 1987), BW704 (Manzke et al. 2001), 14G2a, 15G2a (Castel et al. 2010), AI-201, AI-287, AI-410. AI-425 (Kawashima et al. 1988), and anti-GD3 antibodies such as MAbs AI-245 and AI-267 (Kawashima et al. 1988), Ch14.18 and Ch14.18/CHO have been characterized for their preferential binding to disialyl gangliosides. Furthermore, MAbs JONES, D1.1, and 27A showed affinity for 9-OAc-GD3 but not 9-OAc-GD2, while Mab 8A2 binds both (Sjoberg et al. 1992). Despite the large number of GD3 and GD2 MAbs, there has not been a single one that could serve as a comprehensive probe for the broader range of disialylated glycans.
Among mammalian proteins, myelin-associated glycoprotein (MAG) shows high binding for specific gangliosides in order of GQ1bα > GT1aα, GD1α > GD1a, GT1b > > GM3, GM4, but not for GM1, GD1b, GD3, and GQ1b. Despite high affinity for disia and oligosias, MAG’s binding to gangliosides is not exclusive for di-Sia linkages (Ito et al. 1999; Vinson et al. 2001). Siglec-7 is also reported to react mainly with disialyl structures such as disialyl Lewis α, disialyl galactosyl globoside, and ganglioside GD3, although it also lacks the probe such as binding dynamics for this class (Hashimoto et al. 2019). Among lectins reported to bind to disialylated oligosaccharides, C. antennarius lectin binds with GD3 (Ravindranaths et al. 1988), Agrocybe cylindracea lectin binds with GD1a and GD1b while not clearly differentiating other structures containing NeuAcα3Galβ3GalNAc (Yagi et al. 1997). WGA however preferred GT1 and GD1 over other gangliosides and exhibits very high affinity for monosialylated glycans and GlcNAc also, hence cannot be used as a di-Sia linkage-specific probe (MONSIGNY et al. 1980).
Infectious microbes possess promising affinity for disialyl oligosaccharides in accordance with the glycan environment of their anchorage surfaces. Porcine sapelovirus binds specifically to GD1α (Kim et al. 2016) and Helicobacter pylori recognizes GD2, GD1α, and GD1β by its sialic acid-binding adhesin (SabA) (Benktander et al. 2018). In agreement with high affinity for single ganglioside as observed in Vibrio cholerae (GM1), the majority of gut infecting bacteria and viruses exhibit binding with a specific sialoglycan structure, and their affinity cannot be generalized for range of disialyloligosaccharide sequences.
Tetanus (TeT) and botulinum neurotoxins (BoT) are produced by anaerobic bacteria Clostridium tetani and Clostridium botulinum, respectively, and share significant structural and functional similarity (Iwamori et al. 2008). Not only do BoT (serotypes A to G) and TeT exhibit affinities for similar di and oligosialogangliosides but also share significant sequence homology. In both BoT and TeT, the 150 Kda single chain can be cleaved in to a 50-Kda N-terminal light chain and 100-Kda C-terminal heavy chain. The HC fragment plays the primary role in receptor binding and can be further cleaved into a 50-Kda N-terminal fragment (HN) and a 50-Kda C-terminal fragment (HC). HC domain or HCR is characterized by the amino acid sequence homology among BoTs and TeT, which suggests that a conserved amino acid motif in this domain may define a common carbohydrate recognition site. Since both BoT and TeT possess comparable binding specificity for gangliosides, we preferred TeT to investigate its potential for a comprehensive disialyl linkage recognizing probe. Selection of TeT over BoT was also based on detailed reports on TeT binding to gangliosides (Halpern and Loftus 1993; Louch et al. 2002; Chen et al. 2008; Chen et al. 2009), and the feasibility to select a single serotype of TeT‚ (where BoT has multiple serotypes). An optimal receptor-binding domain (residues 1105–1315) from TeT-HCR was expressed as SNAPf-His6 fusion in the pGEX-3X vector in a bacterial expression system along with its mutants TeT-HCR R1226L, TeT-HCR W1289A, and TeT-HCR R1226L/W1289A (Figs S3 and S10, see online supplementary material for a color version of these figures). Expressed proteins were biotinylated by SNAP-Biotin chemistry as described in experimental procedures and tested for their sialoglycan affinity in microarray (Fig. 1, Figs S4 and S10, see online supplementary material for a color version of these figures). TeT has been shown to specifically bind gangliosides of the G1b series, GD1b or GT1b. In accordance with a published report (Chen et al. 2009), TeT-HCR bound with a range of sialogangliosides including monosialylated GM1α, fucosyl-GM1, and oligosialylated GD1α, GD3, GT3 (Fig. S10, see online supplementary material for a color version of this figure). The high affinity of TeT-HCR for GM1 and fucosyl-GM1 questioned its candidacy as di-Sia linkage-specific probe. Single mutant, TeT-HCR R1226L, and double mutant, TeT-HCR R1226L/W1289A, did not bind with any glycans on array, suggesting the significance of arginine residue at 1226 position in sialoglycan identification (Figs S4 and S10, see online supplementary material for a color version of this figure). Another single mutant TeT-HCR W1289A showed appreciable enhancement in affinity toward GD3, GT3 while loss in binding toward monosialylated and asialylated glycans on array (Fig. S10, see online supplementary material for a color version of this figure). With respect to TeT-HCR (native protein), the tryptophan mutant TeT-HCR W1289A had pronounced specificity toward disialogangliosides structures particularly Neu5Acα8Neu5Acα3Galβ4GlcβR1, Neu5Acα8Neu5Acα3Galβ4GlcβR4, Neu5Acα8Neu5Acα8Neu5Acα3Galβ4GlcβR4, and Neu5,9Ac2α8Neu5Acα3Galb β4GlcβR1 (Fig. S10, see online supplementary material for a color version of this figure). However, we noticed that TeT-HCR W1289A exhibited some minor binding with monosialogangliosides on the array but based on its high affinity for the range of α2–8-linked disialyl oligosaccharides and lack of other available options (MAbs, plant lectins, siglecs, viral, bacterial proteins) to derive a more potent probe for this class, we selected biotinylated TeT-HCR W1289A as SGRP8 (SGRP8TeT), and the double mutant, Biotinylated TeT-HCR R1226L/W1289A (SGRP8TeTNB) as a nonbinding control of SGRP8TeT due to its complete loss of binding for any glycans on array. We excluded the potential interaction of SGRP8Tet toward polysialic acid by testing SGRP8 candidates in an ELISA experiment containing several gangliosides and colominic acid (Fig. S10, see online supplementary material for a color version of this figure). The broad specificity of several bacterial toxins including TeT-HCR toward GM1, GD3, GT3, and GQ series of glycans could be related with evolutionary preference to bind with most available ligands in their neural niche. This nonexclusive binding with multiple ligands by toxins might have advantage for the microbes but reduces our possibility to derive a molecule with probe such as robustness and dynamic specificity. Here, we show that site-specific modifications could improve the binding range in otherwise “nonspecific” probe candidates and characterize SGRP8TeT as α2–8-disiaoligosaccharide-binding probe through general laboratory methods later in this report.
SGRP9PToV is a competent probe for all 9-OAc-Sias on the microarray
Of all O-Ac-Sia modifications in nature, 9-O-acetylation is the most common in cells and tissues of humans and other animals. Their distribution is highly variable and implicated in embryonic development, host–pathogen interactions, and immunity. Although there have been monoclonal antibodies against O-acetylated gangliosides, these MAbs remained highly specific in recognition of O-acetyl esters only when presented by specific underlying sugar chain (Varki 1992). Few Sia-specific lectins recognizing 9-OAc modifications have been reported, for example a lectin from the marine crab C. antennarius showed affinity for 9-OAc and was used to demonstrate the presence of tumor-associated 9-OAc-GD3 on human melanoma cells (Ravindranath et al. 1985). Cancer antennarius lectin also showed significant affinity for Neu4,5Ac2 and did not show affinity for a broad range of 9-OAc. Achatinin-H a lectin from the hemolymph of African land snail A. fulica did not bind to Neu4,5Ac2 but failed to exhibit probe-like preference for a number of 9-OAc-Sias (Mandal and Basu 1987). Another lectin from the protozoan, T. foetus, showed promising affinities for 9-OAc-Sias but also bound to de-O-acetylated Sias with relatively high affinity, confirming its unsuitability as a probe (Babál et al. 1999).
9-OAc-Sia-binding-specific influenza C virus hemagglutinin esterase (Inf-CHE) was previously utilized as whole virions or recombinant protein for assessing a wide spectrum of sialoglycoconjugates such as mucins, serum glycoproteins, or gangliosides containing naturally or synthetically O-acetylated sialic acids (Muchmore and Varki 1987). The influenza C virus hemagglutinin-esterase is a membrane-bound glycoprotein that binds specifically to 9-OAc-Sias (hemagglutinin activity) and then hydrolyzes the O-acetyl group (receptor-destroying activity). Inf-CHE can specifically cleave O-acetyl groups from Neu5,9Ac2 but not from 7-O-acetyl-N-acetylneuraminic acid (Neu5,7Ac2), and very slowly from Neu4,5Ac2 (Zimmer et al. 1992). Previously, we demonstrated that inactivation of Inf-CHE esterase by treatment with the serine esterase inhibitor diisopropyl fluorophosphate (DFP) resulted in stable and irreversible binding with 9-OAc-Sias (Muchmore and Varki 1987). However, using the whole virion as a probe was complicated due to variations in purity and stability of preparations, poor reproducibility, high nonspecific background, and lack of linearity in response. To avoid these limitations, we replaced Inf-C virions with a recombinant soluble chimeric molecule composed of the extracellular domain of Inf-CHE fused to the Fc region of human IgG1 (CHE-Fc). DFP inactivation of CHE-Fc stabilized the hemagglutinin activity and yielded the probe CHE-FcD that was more specific for 9-OAc-Sias than the whole Inf-C virion (Klein et al. 1994; Martin et al. 2003). CHE-FcD provided a better alternative but did not qualify as a standard probe for 9-OAc-Sias due to selective preference for Neu5,9Ac2 over Neu9Ac5Gc glycans, nonspecific binding to Neu5,7Ac2, and hazards related to use of DFP (Fig. S11, see online supplementary material for a color version of this figure) (Hellebø et al. 2004).
To derive a better probe for 9-OAc-Sias, we reviewed other O-Ac-esterases from Inf-C such as mammalian coronaviruses: bovine (bovine coronaviruses; BCoV strains- Mebus, LUN, Breda, B150), birds (puffinosis virus), human (human coronaviruses, strains-OC43, HKU1), equine (ECoV-NC99), murine (mouse hepatitis virus strains-S, DVIM, JHM), and porcine (porcine torovirus strains; PToV, strains- Markelo, P4), reported previously (Klausegger et al. 1999; Langereis et al. 2010, 2015). Based on these reports, which also includes previous studies on OAc-Sia specificity of nidovirus HEs, we selected Porcine Torovirus P4 strain (PToV) hemagglutinin esterase to investigate as a probe, expressing the PToV HE ectodomain in insect Hi-five cells as fusion proteins with a C-terminal Fc domain of human IgG1 (PToV-HE-Fc). Instead of DFP, we used site-directed mutagenesis (S46A) to inactivate the esterase activity in PToV-HE-Fc fusion protein. PToV-HE-Fc showed high selectivity toward 9-OAc-Sias and demonstrated applicability to revealing the 9-OAc-Sia in human and animal tissues and cell lines (Langereis et al. 2015; Wasik et al. 2017).
We expressed 9-OAc-Sia-binding protein (PToV-HE-Fc, S46A) and nonbinding protein (PToV-HE-Fc, F271A) fused to human IgG-Fc followed by a His6 tag and an Avi-tag (Fig. S3, see online supplementary material for a color version of this figure). Both binding and nonbinding proteins were biotinylated using the Avi-tag and investigated for their affinities in a sialoglycan microarray. The esterase-inactive probe, PToV-HE-Fc (S46A), bound exclusively with 9-OAc-Sias including Neu5Ac and Neu5Gc, while hemagglutinin inactive nonbinding PToV-HE-Fc (F271A) did not bind any sialylated or nonsialylated glycans (Fig. 1, Fig. S4, see online supplementary material for a color version of this figure). PToV-HE-Fc (S46A) showed binding toward 9-OAc Neu5Ac and Neu5Gc α2–3, α2–6 and α2–8-linked to their penultimate sugars. In a similar microarray, CHE-FcD showed efficient binding with a number of Neu5,9Ac2-glycans but did not show similar affinity for Neu5Gc9Ac and remained a weaker binder to 9-OAc-Sias in general (Fig. S11, see online supplementary material for a color version of this figure). PToV-HE-Fc (S46A) exhibited significantly stronger binding toward a wide range of 9-OAc-Sia-glycans in comparison with CHE-FcD. Taken together, the efficacy, exclusivity, and reproducibility of PToV-HE-Fc (S46A), we selected biotinylated PToV-HE-Fc (S46A) as comprehensive probe (SGRP9PToV) for 9-OAc-Sia and biotinylated nonbinding PToV-HE-Fc (F271A) as SGRP9PToVNB, the internal control of SGRP9PToV’s specificity. We characterize the utility of PToV-HE-Fc (S46A) as SGRP9PToV in general laboratory methods in sections below.
Use of the panel of SGRPs in common methods
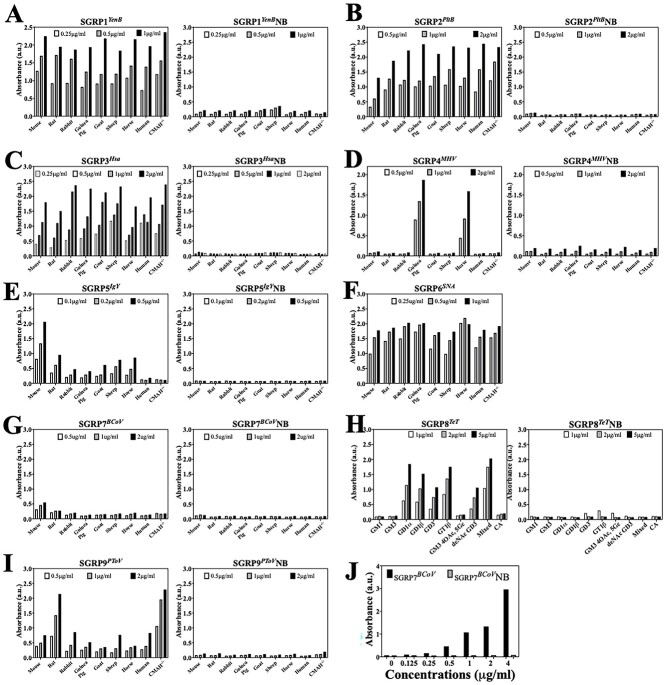
Although detailed sialoglycan microarray analysis provides insights into the Sia specificity of SGRPs, it is essential for SGRPs to exhibit equally precise affinities toward their ligands in ensemble of glycans represented in biological samples. In particular, the multi-antennary sialoglycans and structurally overlapping sialoglycans in biological samples may have different interactions with probes than observed with individual sialoglycans in the glycan arrays. Qualitative Sia binding and ligand specificity of SGRPs were therefore tested by ELISA experiments with blood sera from 9 animal species, each signifying diverse sialoglycan composition in terms of the presence or absence of N-glycolyl at C5 of Sia, O-acetylation, types of glycosidic bonds to penultimate sugars, and overall oligosaccharide sequence. For example, mouse serum is high in Neu5Gc-Sias in comparison with Cmah−/− mouse that remain exclusive for Neu5Ac-Sias and have relatively higher O-acetylation. Horse and guinea pig sera were included for their high representation of 4-OAc-Sias which was absent in other sera, as observed in HPLC analysis of Sias (Table S1). Similarly, we included erythrocytes from 9 animal species, expecting to derive a relatable pattern of SGRPs binding with sera and RBCs. HPLC analysis of surface sialome of erythrocytes (Table S2) suggests high variance in Sia diversity among animals which would be interesting to detect with SGRPs. We emphasized minimal loss of heat and pH labile O-Acs during assays and modified procedures accordingly.
SGRP1YenB exhibits Sia-specific binding to all mammalian sera
As results summarized in Fig. 2 suggest, SGRP1YenB exhibits comparable affinity to all sera, indicating its broad range of Sia specificities. In accordance with the glycan array (Fig. 1), SGRP1YenB exhibits strong binding toward sera whether it is Neu5Gc rich (mouse, goat) or Neu5Ac abundant (human, rat, and Cmah−/− mouse). Furthermore, high proportions of Sia modifications such as 9-OAc substitutions (rat, rabbit, and Cmah−/− mouse) and, more prominently, 4-OAc substitutions (guinea pig and horse serum) did not inhibit SGRP1YenB’s binding to serum sialoglycans (Fig. 2a). To exclude the potential contribution of serum protein interactions with SGRP1YenB from our observations, we performed binding assays with sera after mild periodate oxidation of sera followed by borohydride reduction, generaing side-chain truncated-Sias with a terminal hydroxyl at the 7th carbon. SGRP1YenB showed significantly reduced binding to periodate oxidized sera that was proportional to the loss of desired Sia ligands after the truncation (Fig. S12a, see online supplementary material for a color version of this figure). The question remained as to whether the residual binding of SGRP1YenB to sera, observed even after periodate treatment was due to the interaction with nonsialylated structures in serum, or O-acetyl substitutions protecting Sias from complete oxidation. When O-acetyl esters were removed by mild base treatment before periodate oxidation, the residual SGRP1YenB binding was completely abolished, thereby excluding the role of protein–protein interaction in SGRP1YenB’s binding to mammalian sera (Fig. S12a, see online supplementary material for a color version of this figure). In the same experiment, base treatment without periodate oxidation did not significantly influence SGRP1YenB binding to sera, confirming the probe’s unbiased preference for Sias as seen in glycan array also (Fig. 1). Significantly, the nonbinding variant SGRP1YenB did not show any binding with any serum at all tested concentrations (Fig. 2a).
Fig. 2.
Specific binding of SGRPs to sialoglycans in mammalian sera. The figure presents SGRPs binding of sialoglycans in biological samples, here represented by mammalian sera from mouse, rat, rabbit, Guinea pig, goat, sheep, horse, human, and Cmah−/− mouse. Panels A–I (except panel H) show binding efficiency of SGRPs and their nonbinding controls toward sera in concentration-dependent manner, as marked in individual panels. Panel H shows binding of SGRP8TeT and SGRP8TeTNB of purified gangliosides at concentrations mentioned in panel. Panel J demonstrates the 7,9-diOAc-Sia-specific binding of SGRP7BCoV with bovine submaxillary mucin, while its mutant SGRP7BCoVNB shows no affinity toward BSM. Binding of biotinylated SGRPs was detected and developed using Avidin-HRP (1:1,500). ELISA for each SGRP pair was performed under the same experimental conditions and validity of binding was confirmed by background signal wells treated only with secondary antibody (Avidin-HRP), not shown here.
SGRP2PltB exhibits Neu5Ac-specific binding to mammalian sera
SGRP2PltB binds to Neu5Ac and its derivatives that constitute a major fraction of Sia population in mammalian sera. Accordingly, SGRP2PltB exhibited binding to all serum types tested in a concentration dependent manner (Fig. 2b). However, SGRP2PltB exhibits an interesting pattern of binding toward Neu5Gc rich mouse serum vs. Neu5Ac-rich sera, especially human and Cmah−/− mouse sera. The nonbinding mutant SGRP2PltBNB failed to bind serum from any animal source. Binding assays with mild base and periodate-treated sera confirmed that SGRP2PltB’s binding toward serum was exclusively to Sia and was devoid of nonspecific interactions (Fig. S12b, see online supplementary material for a color version of this figure). The SGRP2PltB also exhibited residual binding after periodate oxidation that was further reduced with prior base treatments, but base treatment alone did not noticeably influence the SGRP2PltB binding to sera.
SGRP3Hsa exhibits α2–3Sia-specific binding to mammalian sera
SGRP3Hsa is specific toward Sias linked to underlying glycans with α2–3-linkage that coexist with α2–6Sias and α2–8Sias in cells, tissues, and other biological samples. SGRP3Hsa bound all sera in a concentration dependent manner. In a different experiment to test Sia-specific binding of SGRP3Hsa, the probe exhibited sera binding directly proportionate to their Sia constituents (Fig. S12c, see online supplementary material for a color version of this figure), excluding nonspecific interaction with nonsialylated structures in serum. Collectively, the results demonstrate SGRP3Hsa’s indiscriminate binding to α2–3Sia in serum, irrespective of O-acetylation, Neu5Ac, or Neu5Gc, a finding that agrees with observations from sialoglycan microarray (Fig. 1). SGRP3HsaNB, the nonbinding version of SGRP3Hsa showed no binding with any serum at tested concentrations (Fig. 2c).
SGRP4MHV exhibits 4-OAc-Sia-specific binding to mammalian sera
SGRP4MHV represents a probe for an exclusive class of 4-O-acetylated sialoglycans, which show a high abundance in blood components and tissues of certain animals. In our collection of sera, 4-OAc is represented by guinea pig and horse (38.7% and 32.4% of total Sia content, respectively, Table S1) and accordingly SGRP4MHV exhibited strong binding to these sera (Fig. 2d). The negligible binding of SGRP4MHV to other sera suggests that its specific binding to guinea pig and horse serum was due to 4-OAc-Sias exclusively found in these samples. Importantly, SGRP4MHV exhibited no binding to serum rich in 9-OAc-Sias (Cmah−/−, rat, and rabbit). confirming its ability to detect 4-OAc-Sias over 9-OAc-Sias. In similar experiments, SGRP4MHVNB did not bind to any serum, confirming its utility as a nonbinding control. Periodate treatment did not have much effect on SGRP4MHV’s binding toward guinea pig and horse serum, but base treatment followed by periodate oxidation completely blocked Sia detection (Fig. S12d, see online supplementary material for a color version of this figure). These results, along with sialoglycan glycan microarray confirm that even in complex biological samples such as serum, SGRP4MHV binds only to 4-OAc-Sias.
SGRP5IgY exhibits Neu5Gc-specific binding to mammalian sera
While 5-N-glycolyl-Sias (Neu5Gc) are a major component of Sia diversity in mammals, Neu5Gc is absent in humans due to loss of functional Cmah, and also in Cmah −/− mice. The serum binding assay (Fig. 2e) showed that SGRP5IgY had very strong affinity for the WT mouse serum and also bound to all sera except human and Cmah −/− mouse. Significantly, SGRP5IgY showed a reverse binding pattern to that observed with SGRP2PltB (Fig. 2b), suggesting an approach to double-check the specificity of such complimentary probes. SGRP5IgYNB did not bind any serum tested in similar experiments, as anticipated. A binding assay with base- and periodate-treated sera supported the specificity of SGRP5IgY, as removal of OAc esters did not affect its binding to serum, while removal of Sias abolished binding (Fig. S12e, see online supplementary material for a color version of this figure).
SGRP6SNA exhibits α2–6-specific binding to mammalian sera
SGRP6SNA exhibits strong and general binding with all mammalian sera in binding experiments, suggesting its robustness as α2–6Sia-recognition probe. As observed in glycan array (Fig. 1), SGRP6SNA does not discriminate between α2–6Neu5Ac and α2–6Neu5Gc, with or without OAc esters. Similarly, the serum binding data (Fig. 2f) shows the binding of SGRP6SNA to mouse, rabbit, guinea pig, human, or Cmah−/− mouse sera which contain Neu5Gc, Neu5Ac and Neu5,9Ac2, Neu4,5Ac2, Neu5Ac, Neu5Ac, and Neu5,9Ac2, respectively. An experiment with periodate oxidized serum confirmed that SGRP6SNA binds to sera in a Sia-specific manner and does not bind serum proteins (Fig. S12f, see online supplementary material for a color version of this figure).
SGRP7BCoV exhibits 7,9-diOAc-Sia-specific binding to bovine submaxillary mucin
SGRP7BCoV exhibited insignificant binding to tested mammalian sera, likely due to very strong preference toward 7,9-diOAc-Sias, absent in these sera (Fig. 2g). The probe does possess inconsistent and minor binding toward 9-OAc-Sias but those were not detected in the sera sialoglycans. To confirm SGRP7BCoV’s recognition of 7, 9-diOAc-Sias, we studied binding with immobilized BSM, enriched in di- and tri-OAc-sialoglycans. As anticipated, SGRP7BCoV showed dose-dependent binding patterns to BSM, while the nonbinding mutant SGRP7BCoVNB showed no detectable signals (Fig. 2j). We confirmed its specificity toward OAc-Sias in base and mild periodate-oxidized BSM. The data (Fig. S12g, see online supplementary material for a color version of this figure) shows that SGRP7BCoV binds BSM in OAc-dependent manner, and depletion of OAc esters resulted in dramatic reduction in SGRP7BCoV’s binding, comparable to loss after complete depletion of Sias.
SGRP8TeT exhibits DiSia linkage-specific binding to gangliosides
SGRP8TeT exhibits very specific binding toward di-Sia linkages, the sialoglycan moieties mostly represented by gangliosides in biological system. As a result, SGRP8TeT showed no detectable binding when tested against sera in ELISA experiment (Fig. S13a, see online supplementary material for a color version of this figure). In an ELISA experiment with immobilized purified gangliosides, SGRP8TeT showed dose-dependent binding toward di-Sia oligosaccharide (GD1, GD3, GT1) discriminating from the mono-Sia oligosaccharides (GM1, GM3) and polySia oligosaccharides conformations (Colominic acid) (Fig. 2h). The observd results from ganglioside binding experiments are in complete agreement with the sialoglycan microarrays performed with SGRP8TeT and confirm the advantage of the probe over native TeT-HCR (Fig. 1, Fig. S10a and b, see online supplementary material for a color version of this figure). The nonbinding mutant SGRP8TeTNB showed no detectable signals, signifying its utility as a control for SGRP8TeT’s specificity (Fig. 2h).
SGRP9PToV exhibits 9-OAc-Sia-specific binding to mammalian sera
SGRP9PToV binds to 9-OAc-Sias that are represented by all mammalian sera included for binding assays (Table S1). In observed results, SGRP9PToV demonstrated remarkably high affinity toward rat and Cmah−/− mouse serum, suggesting their high proportions of Neu5,9Ac2 (Fig. 2i). However, relatively lower binding to rabbit serum was interesting as rabbit serum contained the highest fraction of Neu5,9Ac2 among the tested sera, but the basis of such variance is not known. It is clear from the serum-binding data of SGRPs that they can demonstrate the presence or absence of the preferred ligand, but that may not always measure the proportions relative to other Sia types within the serum. No detectable signals from similar binding assays with SGRP9PToVNB confirm that SGRP9PToV binding was specific. As observed in different ELISA experiments with OAc-depleted sera, SGRP9PToV’s binding to mammalian sera including rat and Cmah−/− mouse showed a linear and reproducible binding exclusive to base labile OAc esters (Fig. S12h, see online supplementary material for a color version of this figure). The data showed that depletion of non-O-acetylated Sias does not influence SGRP9PToV binding significantly while removal of OAc esters from Sias or complete diminution of Sia population abolished its binding to sera (Fig. S12h, see online supplementary material for a color version of this figure). These results together with sialoglycan microarray data (Fig. 1) confirm that SGRP9PToV exhibits very strong preferences toward 9-OAc-Sias and does not interact with nonsialylated structures or proteins in complex biological samples.
Western blot analysis of SGRPs specificity toward sialoglycans in mammalian sera
To confirm that SGRPs are applicable to routine methods of glycoprotein analysis, we assessed SGRP’s qualitative detection of sialoglycoproteins by western blots. The gel electrophoresis and western blotting protocol were modified to minimize the loss of sensitive OAc-Sias (Varki and Diaz 1984; Higa et al. 1989). SGRP1YenB bound to all 9 sera included in the experiment, which is consistent with the serum binding ELISA experiments (Fig. 3a). SGRP2PltB also bound to sera in accordance with their Neu5Ac contents, with a noticeable difference between its affinity for WT and Cmah−/− mouse sera (Fig. 3c). SGRP3Hsa and SGRP6SNA showed binding to all sera, a pattern also observed in serum binding ELISAs (Fig. 3e and k). SGRP4MHV exclusively bound to guinea pig and horse sera (Fig. 3g), while SGRP9PToV exhibited high binding toward rat, rabbit, and Cmah−/− mouse sera (Fig. 3n), corresponding to OAc-Sia contents of these sera (Table S1). Interestingly, SGRP7BCOV showed a weak nonspecific binding with multiple sera in western blots (Fig. 3) and also showed a comparable weak nonspecificity in ELISA, which is likely due to its specificity toward 7,9-diOAc-Sias. We speculate that changes in pH and temperature during gel electrophoresis or blotting may result in migration of OAc esters, resulting in binding in western blotting, but we did not investigate this in detail. Blots, interrogated with nonbinding variants of SGRPs (Fig. 3) did not show any binding to sera, suggesting Sia-specific binding of SGRPs to sera. As SGRP8TeT did not bind sera in ELISA, it was excluded from western blot analysis of serum.
Fig. 3.
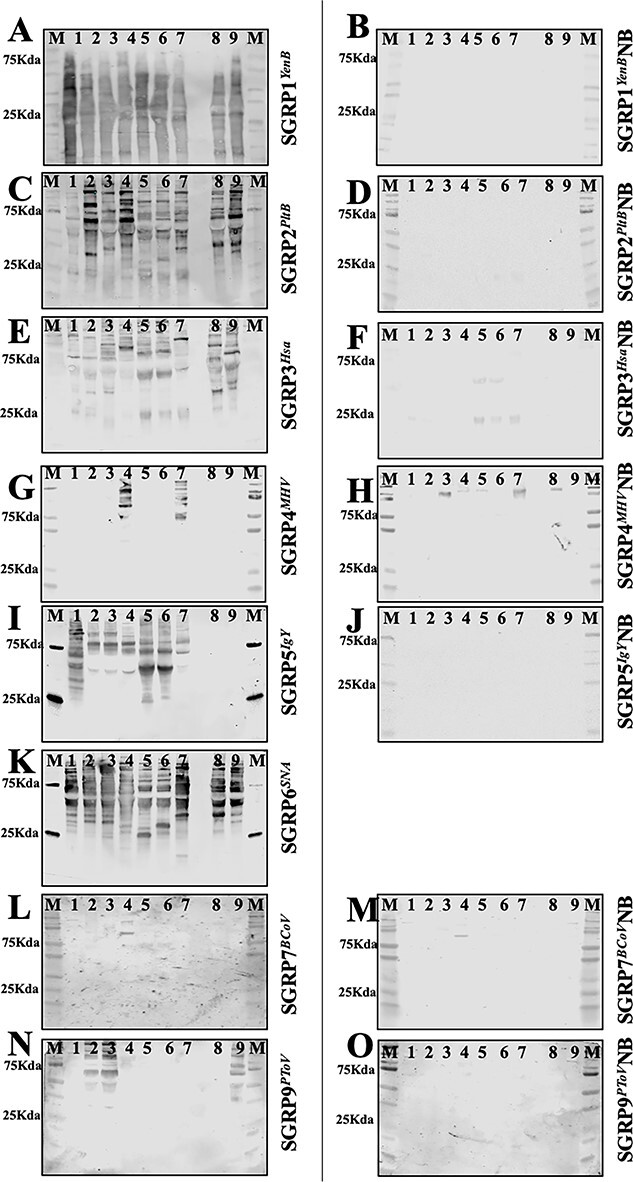
Specific recognition of sialoglycoproteins in mammalian sera by SGRPs in western blots. Mammalian sera from mouse (1), rat (2), rabbit (3), Guinea pig (4), goat (5), sheep (6), horse (7), human (8), and Cmah−/− mouse (9) were subjected to SDS-PAGE under conditions that protect labile O-acetyl groups immunoblotted and probed with SGRPs. M in each blot represents the molecular weight marker. Panels a and b show blots interrogated with SGRP1YenB and SGRP1YenBNB (both at 1 μg/ml); panels c and d show blots interrogated with 1-μg/ml SGRP2PltB and SGRP2PltBNB; panels e and f represent SGRP3Hsa and SGRP3HsaNB (both at 1 μg/ml); panels g and h show blots probed with 1 μg/ml of SGRP4MHV and SGRP4MHVNB; panels i and j show blot probed with 0.33-μg/ml SGRP5IgY and SGRP5IgYNB; panels k is blot interrogated by 1-μg/ml SGRP6SNA; panels l and m show blots probed by 1-μg/ml SGRP7BCoV and SGRP7BCoVNB, and panels n and o represent blots interrogated with 2-μg/ml SGRP9PToV and SGRP9PToVNB. Binding of biotinylated SGRPs was detectable and developed using streptavidin IRDye 680 (1:10,000). Analysis was done on a LiCor odyssey infrared imager. Both blots for each SGRP set were detected at the same time using the same conditions.
SGRPs binding to frozen tissue sections: testing utility for in situ detection of Sias
The panel shows the typical binding patterns observed with each of the probes (red color indicates binding) (Fig. 4). The nonbinding control shows no binding to any of the sections, as expected. SGRP1YenB detects mucins and blood vessels in many organs, and this is best demonstrated in the sections of kidney, where glomeruli are highlighted (Fig. 4a). The glomeruli as well as the capillary blood vessels between the tubules are prominent in the wild-type mouse, while in the Cmah−/− mouse, the capillaries between tubules are not as prominent (Fig. 4b). The nonbinding control shows no reactivity to any of the sections, as expected (Fig. 4c). SGRP2PltB detects mucins and blood vessels in many organs, and this is best demonstrated in the sections of kidney, where the glomeruli are highlighted, and also the capillaries in between the tubules are visible (Fig. 4d and e). The nonbinding control shows no binding to any of the sections, as expected (Fig. 4f). SGRP3Hsa detects mucins and blood vessels in many organs, and this is best demonstrated in the sections of kidney, where the glomeruli are prominently highlighted (Fig. 4g). The binding to the kidney glomeruli in Cmah−/− mouse is slightly more prominent (Fig. 4h). The nonbinding control shows no binding to any of the sections, as expected (Fig. 4i). SGRP4MHV showed selective detection of mucins, and this is best demonstrated here in the mucin contained within the acini of pancreas. Staining was faint in wild type and was much more prominent in the Cmah null animal (Fig. 4j and k). Islets of the pancreas (the endocrine portion) and the nonbinding control show no binding (Fig. 4l). SGRP5IgY detects blood vessels in many organs and demonstrated here by the detection of the capillaries within the sections from the pancreas in wild type, but not in Cmah−/−mice (Fig. 4m and n). The nonbinding control shows no binding to any of the sections (Fig. 4o). SGRP6SNA detects mucins and blood vessels in many organs. Here, we observe that SGRP6SNA detects blood vessels in the pancreas of the Cmah−/−mouse much better than it does in the wild-type animal (Fig. 4p and q). The tissue sections were incubated with secondary antibody Cy3-SA and served as control for SGRP6SNA specificity, and show no binding to any of the sections, as expected (Fig. 4r). SGRP7BCoV detects blood vessels in some organs but also detected white matter in the brain of the Cmah−/−mouse extremely well (Fig. 4t). The nonbinding control shows no binding to any of the sections (Fig. 4u). SGRP8TeT detected large areas of the nonnuclear neuropil in the brain from both the wild type and in the Cmah−/−mouse. The white matter in the brain from the Cmah−/−mouse was even more prominent, and the nonbinding control showed no binding to any of the sections (Fig. 4v, w, and x). SGRP9PToV detected mucins and also red blood cells in many of the organs. However, in sections of liver from the Cmah−/−mouse, there was an abnormally high expression in the sinusoidal endothelial cells around the central vein, indicating right heart failure in the Cmah−/−mouse (the portal triads have two sources of blood supply and are thus the better perfused areas in the liver) (Fig. 4y and z). The nonbinding control showed no binding to any of the sections, as expected (Fig. 4z1).
Fig. 4.
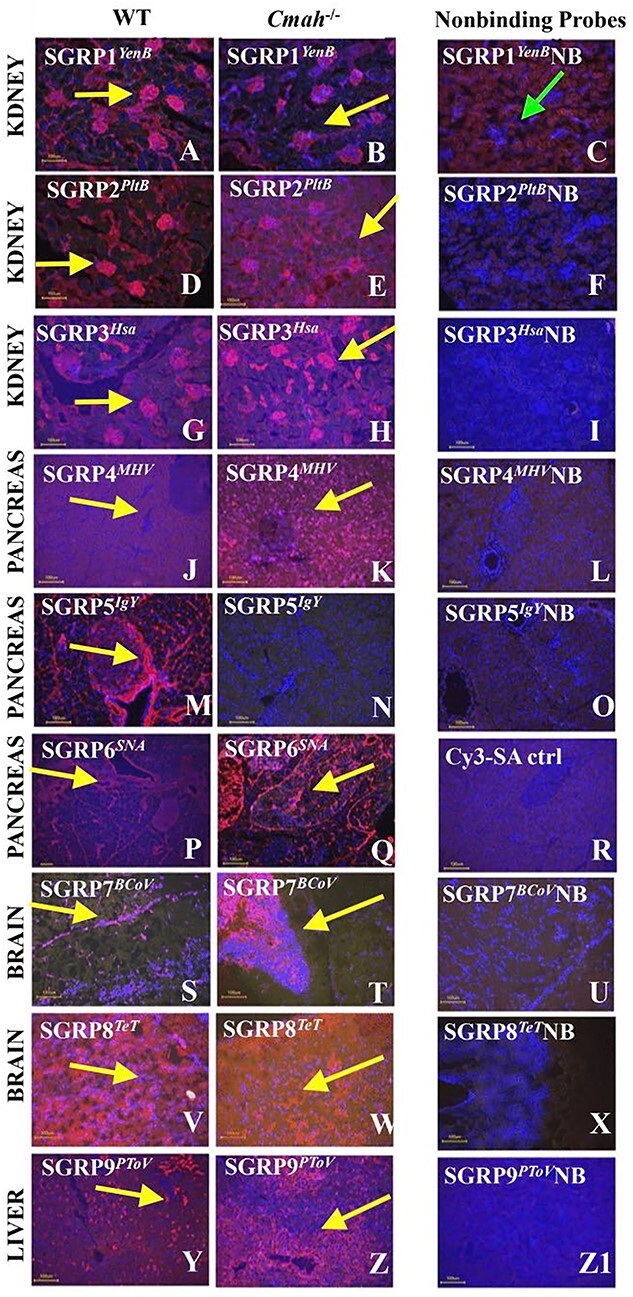
Histological analysis of frozen mouse tissues by SGRPs. Examples of binding patterns (red-Cy3 streptavidin) observed with each SGRP probe (on different organs of the wild-type and the cmah null mouse strain). Panels a, b, d, e, g, and h show the detection of kidney glomeruli, (yellow arrows) by SGRP1YenB and SGRP2PltB (each used at 5 μg/ml) and SGRP3Hsa, used at 3 μg/ml. In contrast, the nonbinding probe does not bind to glomeruli, as shown in panels c, f, and i. Pancreas exocrine mucin producing glands and intervening fibrous stroma are recognized by SGRP4MHV (used at 5 μg/ml), SGRP5IgY (used at 1 μg/ml) and SGRP6SNA (used at 2 μg/ml) indicated by arrows in panels g, h, j, k, and m. However, with SGRP5IgY as expected, there is no binding to Cmah null tissues; (panel n), the nonbinding controls also do not bind, as shown in panels l and o. Panel r represents secondary only control for SNA, with no binding. Brain meninges and white matter are recognized by SGRP7BCov (used at 10 μg/ml) and SGRP8TeT (used at 5 μg/ml), demonstrated with arrows on panels s, t, v, and w. The nonbinding controls do not bind shown in panels u and x. Liver macrophages are recognized in the wild-type animal by SGRPPToV, and interestingly, in the Cmah null animal, SGRPPToV binds to hepatocyte membranes closest to the central vein, indicating a different pathology in this genetically altered animal strain. The nonbinding control shows no binding (panel ZI). Nuclei are designated blue color by Hoechst stain.
SGRPs binding to mammalian erythrocytes in flow cytometry
We performed flow cytometry experiments with mammalian RBCs to demonstrate in situ detection of extracellular Sias by SGRPs (Fig. 5 and Fig. S14, see online supplementary material for a color version of this figure). As shown in Fig. 5a, SGRP1YenB bound to RBCs from all 9 animal sources tested, while its nonbinding version SGRP1YenBNB did not show any binding (Fig. 5a). In general, the total fluorescence intensity for nonbinder probes remained comparable to autofluorescence (RBCs without any treatment) and antibody control (RBCs detected with SA-PE only). SGRP2PltB bound with all RBCs except wild-type mouse. The most robust bindings of SGRP2PltB were detected with Cmah−/−mouse, guinea pig, rabbit, and rat followed by goat and human RBCs (Fig. 5b). SGRP2PltBNB did not show any noticeable binding with RBCs from any source. SGRP3Hsa bound with all types of RBCs with maximum binding detected on guinea pig, goat, and rat RBCs and minimum with rabbit, indicating variable abundance of α2–3Sias on these erythrocytes (Fig. 5c). SGRP3HsaNB did not significantly bind to any RBC type. SGRP4MHV showed no noticeable binding to any RBC type included in the experiment, and the binding as detected from SGRP4MHV and SGRP4MHVNB remained comparable, suggesting absence of 4OAc-Sias on the extracellular surface of these RBCs (Fig. 5d). SGRP5IgY bound with all RBCs except from Cmah−/− mouse as anticipated (Fig. 5e). SGRP5IgY also did not bind strongly to guinea pig and rat RBCs, suggesting less abundant Neu5Gc in these RBCs in comparison to mouse, horse, and rabbit. SGRP5IgYNB showed negligible detection, as expected from a nonbinding control probe. A stronger SGRP6SNA binding was detected with human, guinea pig, and rabbit in comparison to goat, sheep, and horse (Fig. 5f). Taking together with SGRP3Hsa’s binding to RBCs, the relative abundance of α2–3Sias over α2–6Sias can be inferred for certain mammalian RBCs such as goat, sheep, and guinea pig. In absence of nonbinding mutant of SGRP6SNA, the results are provided with autofluorescence and fluorescence antibody controls. The 7,9-diOAc-Sia-specific binder SGRP7BCoV showed no appreciable binding in flow cytometry experiments, suggesting undetectable amounts of its ligand in tested RBCs (Fig. 5g). The resultant binding of SGRP7BCoV and SGRP7BCOVNB remains comparable with RBCs; consistent with the probe’s response in the serum ELISA experiments, suggesting lack of SGRP7BCoV binding in absence of its exclusive ligand 7,9-diOAc-Sia. α2–8-linked disialyl-oligosaccharides-specific probe SGRP8TeT did not show any binding with RBCs tested (Fig. S13b, see online supplementary material for a color version of this figure), and hence, a set of GD3 transfected CHOK1 cells were used to investigate efficiency of SGRP8TeT in flow cytometry experiment. As shown in Fig. 5i, SGRP8TeT exhibited appreciable binding to GD3 expressing CHOK1 cells (CHOK1-GD3+/+) compared with normal CHOK1 cells. The nonbinding SGRP8TeTNB showed relatively minor binding with cells that was not affected by expression of GD3 gangliosides. As presented in Fig. 5h, SGRP9PToV showed remarkable binding with rat RBCs among all tested erythrocytes and its nonbinder probe SGRP9PToVNB remained completely undetectable. This experiment demonstrates the utility of SGRPs in the characterization of the extracellular sialome of cells, for example, showing that rat RBCs possess a dense Sia population that contains varying amounts of α2–3Sias, Neu5Ac, and 9-OAc-Sias than α2–6-Sias, Neu5Gc, and 4-OAc-Sias.
Fig. 5.
SGRPs recognize specific sialoglycans on mammalian erythrocytes. Comprehensive heat map representation of SGRP’s binding to mammalian erythrocytes and CHO-K1 cell line (SGRP8Tet); colors red and blue represent maximum and minimum binding efficiencies, respectively. The heat map represents the comprehensive binding pattern of SGRPs (5 μg/ml) to mammalian erythrocytes and CHO-K1 cell line (SGRP8Tet). SGRPs binding efficiencies are shown in red for highest binding (saturated or 100%), blue for minimum or no binding (0%), and intermediate binding represented by cells displaying colors between blue and red. As consistent with sialoglycan microarray data, no-binding control probes demonstrated significantly reduced or no binding in the same experiment.
Conclusions and perspectives
Here, we exploited naturally evolved Sia-recognizing microbial proteins to develop an approach to design and derive sialoglycan-detecting probes. We developed and characterized a set of practicable probes recognizing the most predominant classes of mammalian Sia types. The study also reviewed and re-examined the current standing of traditional probes or probe-like molecules that have been reported against known diversity of mammalian Sia types. We reviewed, assessed, and experimentally analyzed several probes to compare their suitability for use as updated SGRPs. In this process, we successfully identified an alternative for Maackia lectins in the form of SGRP3Hsa. This is important because these plant lectins can also recognize 3-O-sulfated glycans.
In most instances, we found that probes from microbes that infect mammals were superior to the cross-reacting plant lectins. A striking exception was utility of conventional α2–6Sia-binding S. nigra lectin elderberry which appeared to be better than several microbial and mammalian proteins tested or reviewed in this study. Among developed probes; SGRP2PltB, SGRP4MHV, SGRP8TeT, and SGRP9PToV represent novel probes for their class of Sia molecules, while SGRP1YenB and SGRP3Hsa showed substantial improvement over previously known probes. SGRP5IgY was also included in the final set, and SGRP6SNA remained unique as we could not identify better alternative from microbial sources. Neu5Gc-specific microbial proteins showed utility but could not match the specificity and robustness of anti-Neu5Gc IgY that was finally selected as SGRP5IgY. An exception to SGRPs set, SGRP7BCoV is characterized as an imperfect probe that is not completely selective for 7-OAcSias and that must be updated with a more precise SGRP7 if one is identified in the future.
There are efforts ongoing by other groups, both in academic and industry to provide state-of-the-art sialoglycan detecting probes, and we attempted to review most of such studies and theoretically compared SGRPs with those molecules. While we appreciate the alternate approaches developed and adopted by other groups (Yabe et al. 2007; Imamura et al. 2011; Dabelsteen et al. 2020; Büll et al. 2021; Nason et al. 2021), we remained focused on naturally evolved molecules to find probes toward predominant class of mammalian sialoglycan. This study advances the merits of proposed SGRPs and their utilities for a biological science investigator. To have a consolidated assumption about the “best” available probe for any given sialoglycan class, a real-time comparison is required which is outside the scope of this study. Also not addressed here are sulfate esters, which can substitute for sialic acids in some glycans.
In conclusion, this study provides a comprehensive set of probes for many (but not all) mammalian Sia types, designed and developed for the nonsialic acid experts. Several tests to demonstrate SGRPs specificity confirm their utility as practicable probes. We expect SGRPs to be adapted by both experts and nonexperts as tools of Sia recognition in their studies. In closing, we re-emphasize that all possible terminal glycan sequences cannot be studied at this time for cross-reactivity with the SGRPs. Also, this SGRP set is not meant to give definite proof of the presence of the sialoglycans in question. Rather, binding of each probe strongly suggests that a particular component (e.g. 9-O-acetyl-Sias with SGRP9) is present in the sample being probed. Absence of reactivity is not evidence of absence, and presence of reactivity only indicates that the component is very likely to be present. Moreover, given the vast diversity of sialic acids in nature, we cannot address all possibilities. For example, it is possible that a 9-O-lactyl group (known to be present in nature but very poorly studied, and not available synthetically at this time) might cross-react. On the other hand, the presence of an 8-O-methyl group (also known to be present in nature but again very poorly studied, and not easily available synthetically at this time) might disrupt the recognition of a 9-O-acetyl group by SGRP9. Thus, these probes are not meant to substitute for more rigorous chemical and structural analysis by experts. Rather, they are meant for the nonexpert to find interesting sialome patterns in various biological systems that are then worth pursuing further. The class of probes such as SGRP1, SGRP2, SGRP4, SGRP8 and SGRP9 described in this report are novel for their target sialoglycan types. The study is based on glycomic methods to identify probe candidates and demonstrate their applications through very generic laboratory methods that can be used even by nonglycobiologists. This approach is interesting as plenty of glycan profiling methods in trends are only useful for investigators with some background in glycobiology.
Materials and methods
Sialoglycan microarray with SGRPs
The sialoglycan microarray method was adapted and modified from the literature published earlier (Deng et al. 2014b). Briefly, defined sialosides with amine linker were chemoenzymatically synthesized and then quantitated utilizing an improved DMB-HPLC method (Ji et al. 2021), and 100 μM of sialoglycan solution (in 300-mM Na-phosphate buffer, pH 8.4) was printed in quadruplets on NHS-functionalized glass slides (PolyAn 3D-NHS; catalog# PO-10400401) using an ArrayIt SpotBot®Extreme instrument. The slides were blocked (0.05-M ethanolamine solution in 0.1 M Tris–HCl, pH 9.0), washed with warm Milli-Q water, and dried. The slides were rehydrated with 400 μl of ovalbumin (1% w/v, PBS) for 1 h in a humid chamber with gentle shaking, followed by incubating the SGRPs/proteins for 2 h at room temperature (gentle shaking). The slides were then washed with PBS-Tween (0.1% v/v). Following washes, the wells were then treated with Cy3-conjugated anti-His (Rockland Antibodies & Assays; Cat# 200-304-382) secondary antibody, for 1 h, followed by washing and scanning with a Genepix 4000B scanner (Molecular Devices Corp., Union City, CA) at a wavelength of 532 nm. Data analysis was performed using the Genepix Pro 7.3 software (Molecular Devices Corp., Union City, CA). The raw data analysis and sorting using the numerical codes were performed on Microsoft Excel. Every siaologlycan microarray experiment was repeated at least 3 times for reproducibility, and typical results are provided in the manuscript.
Serum binding assay with SGRPs
A saturable amount of sera (equivalent to 50-μg proteins), reconstituted in PBST (0.1% Tween 20), were added to individual wells in ELISA plates (Corning, 9018) and kept overnight at 4 °C for adherence. Unattached sera were removed, and wells were briefly rinsed with PBST before addition of 200 μl, 0.5% cold fish gelatin (Sigma- G7765-1 L, diluted in PBST) for blocking. Lectins, SGRPs, or other probes were reconstituted in diluting buffer (PBST with 0.05% cold fish gelatin) at desired concentrations and added to serum-coated wells for 1 h (or otherwise specified) at room temperature. Specifically, for MAL-I (VECTOR LAB, B1315) and MAL-II (B1265), 10-mM CaCl2 and 10-mM MnCl2 were added to diluting buffer to maximize lectin binding. Post-incubation, wells were vigorously washed with 250-μl PBST, 5 times and then incubated with Avidin-HRP (Biolegend, 405,103) for biotinylated probes at 1:2,000 dilution for 45 min at room temperature. Wells were again washed vigorously with 250-μl PBST 6–7 times, and 100 μl of TMB (BD OptEIA-55,214) was added to each well as substrate for HRP’s enzymatic activities and incubated for color development. The reactions were stopped by adding 40 μl of 2 N H2SO4 after 30 min, or if the wells specified as negative controls begin to develop color. Plates were read at 450 nm and readings were processed to observe binding patterns of tested molecules (Perkin Elmer, Enspire Alpha). For every experiment, wells treated with avidin-HRP only served as negative control of binding, along with nonbinding mutants of probes, whenever applicable.
For serum binding experiments with nonbiotinylated proteins, Fc-tagged or His6-conjugated proteins were probed. His6 detection was determined by anti-His antibody (mouse, Gene Script, #A00186, 1:4,000) followed by anti-mouse-HRP (CTS, #7076S, 1:1,500). In certain probes discussed in this article, for example LFA (EY Lab, BA1501–1, CHE-FcD etc.), the preconjugated combination of probe and secondary antibody was adopted for stronger signals, as mentioned in corresponding figure legends. Every serum binding ELISA experiment was repeated at least 3 times for reproducibility, and typical results are provided with the manuscript.
Western blotting
Commercial animal sera (rat, equine, goat, guinea pig, rabbit, sheep; Sigma, St.Louis, MO), wt mouse and human serum (Valley Biomedical, Winchester, VA) and in house Cmah−/− mouse serum were chromatographed on 12% CosmoPAGE Bis-Tris precast gels (Nacalai, USA, NU00212). The gels were run in CosmoPAGE MOPS running buffer pH 7 (Nacalai, USA, NU01004), with ice packs to keep running buffer temperature below 23 °C. The proteins were transferred to Immobilon FL (EMD Millipore Corp IPFL00010) membrane in transfer buffer containing 10% methanol and 0.2% SDS. Membranes were blocked overnight at 4 °C with 0.5% Cold water fish skin gelatin in PBST (PBS pH 7.4 with 0.1% Tween 20), with gentle agitation. The membranes were incubated at room temperature for 2 h with 1 μg/ml of all probes, except SGRP5IgY, which was used at 0.33 μg/ml, diluted in blocking buffer without Tween. Also, SGRP4MHV and SGRP9PToV were used at 2 μg/ml. Amounts of serum proteins immunoblotted for each SGRP were adjusted to highlight their ligand preferences. Specifically, 25-μg protein per well was loaded for SGRP1YenB, SGRP2PltB, and their nonbinding variants, 50-μg protein per well was loaded for SGRP3Hsa, SGRP4MHV, SGRP7BCoV, SGRP9PToV, and their nonbinding mutants, while 12.5-μg protein per well was loaded for SGRP5IgY, SGRP5IgYNB, and SGRP6SNA (Vector lab, B1305, lot number Z1002). Secondary antibody incubation was with Streptavidin IRDye 680 (Licor, 925–68,079) at 1:10,000 dilution in PBST at room temperature. Signal was read with Odyssey infrared imager (Licor Biosciences, Lincoln, NE). Interrogating SGRPs and corresponding nonbinding SGRPs were used at the same concentration and all images were obtained at the same time using the same imager settings.
Histology procedure
Frozen sections of various organs from the wild type and Cmah−/− mouse were air dried for 30 min, rehydrated in Tris-Buffered Saline pH 7.5, and overlaid with 0.5% Fish gelatin, which was then tipped off for blocking the endogenous biotin using the Avidin Biotin blocking kit from Vector labs (SP-2001). Sections were then fixed for 30 min in 10% Neutral buffered formalin (VWR-89370-094), washed and overlaid with either the binding or nonbinding probes, each used 5 μg/μl. Separate slides (Globe, 1358 W) were then overlaid with each of the Biotinylated probes and incubated in a humid chamber at room temperature for 30 min. Washing was then performed in TBS, followed by incubation with Cy3 Streptavidin (Red, Jackson Laboratory, 016-160-084) for 30 min. After washing again, nuclei were counterstained with Hoechst (Blue, Molecular Probes, H3570) for 2 min, washed and cover-slipped using an aqueous mounting medium (Vector lab H5501). Digital images were captured using the Keyence microscope and organized using Adobe Photoshop.
Flow cytometry
SGRPs were used at 5 μg/ml (or otherwise, mentioned in figure legends) for their RBC binding properties. For this experiment, blood from several animal species was resuspended in PBS (4°C) to make a 2% volume/volume dilution, and 200 μl of this blood suspension was supplemented with 200 μl of 10 μg/ml probe dilution in PBS (4°C) to make a final blood suspension and probe dilution of 1% and 5 μg/ml respectively (unless mentioned otherwise)· 400 μl of this blood suspension was incubated on ice for 1 h with intermittent gentle mixing to avoid settlement of RBCs. After incubation, 1 ml ice cold PBS was added to each tube and centrifuged at 1,000 RPM, 3 min at 4°C. The washing was repeated 2 more times, and then the pellet was gently resuspended in 300 μl of 1 μg/ml dilution of Streptavidin Cy3 (Jackson 016-170-084) and incubated for 45 min on ice. For autofluorescence control of RBCs, blood suspension was never treated either by probe or fluorescence antibody, while for fluorescent secondary antibody control, mock treated blood was incubated with the equivalent streptavidin-Cy3 dilution in similar conditions. After incubation, 1-ml ice cold PBS was added to each tube and the washes were repeated for a total of 3 times.The final pellet was gently resuspended in 500 μl of PBS and fluorescence was analyzed on FL2 channel on the FACS Caliber flow cytometer. The settings for acquisition of data and analysis remained the same for all probes and their nonbinding mutants in an experiment. Binding efficiencies of SGRPs and their nonbinding probes were plotted in red for highest binding (saturated or 100%), blue for minimum or no binding (0%), and intermediate binding represented by cells displaying colors between blue and red.
Data availability
All data are contained within the article and supporting information.
Funding
This work was supported by the National Institutes of Health Grants U01CA199792 and R01GM32373 (to AV), and additional generous financial support from BioLegend, San Diego.
Conflicts of interest statement: The authors declare no competing financial interests.
Supplementary Material
Contributor Information
Saurabh Srivastava, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Andrea Verhagen, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Aniruddha Sasmal, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Brian R Wasik, College of Veterinary Medicine, Cornell University, Ithaca, NY, USA.
Sandra Diaz, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Hai Yu, Department of Chemistry, University of California at Davis, Davis, CA, USA.
Barbara A Bensing, Department of Medicine, University of California, San Francisco, CA, USA; VA Medical Center, San Francisco, CA, USA.
Naazneen Khan, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Zahra Khedri, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Patrick Secrest, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Paul Sullam, Department of Medicine, University of California, San Francisco, CA, USA; VA Medical Center, San Francisco, CA, USA.
Nissi Varki, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
Xi Chen, Department of Chemistry, University of California at Davis, Davis, CA, USA.
Colin R Parrish, College of Veterinary Medicine, Cornell University, Ithaca, NY, USA.
Ajit Varki, Department of Cellular and Molecular Medicine, School of Medicine, University of California at San Diego, San Diego, CA, USA; Glycobiology Research and Training Center, University of California at San Diego, San Diego, CA, USA.
References
- Altheide TK, Hayakawa T, Mikkelsen TS, Diaz S, Varki N, Varki A. System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: Evidence for two modes of rapid evolution. J Biol Chem. 2006:281(35):25689–25702. [DOI] [PubMed] [Google Scholar]
- Arthur CM, Cummings RD, Stowell SR. Using glycan microarrays to understand immunity. Curr Opin Chem Biol. 2014:18:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka H, Nishinaka S, Wakamiya N, Matsuda H, Murata M. Two chicken monoclonal antibodies specific for heterophil Hanganutziu-Deicher antigens. Immunol Lett. 1992:32(1):91–96. [DOI] [PubMed] [Google Scholar]
- Atack JM, Day CJ, Poole J, Brockman KL, Bakaletz LO, Barenkamp SJ, Jennings MP. The HMW2 adhesin of non-typeable Haemophilus influenzae is a human-adapted lectin that mediates high-affinity binding to 2-6 linked N-acetylneuraminic acid glycans. Biochem Biophys Res Commun. 2018:503(2):1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babál P, Pindak FF, Russell LC, Gardner WA Jr. Sialic acid-specific lectin from Tritrichomonas foetus. Biochim Biophys Acta. 1999:1428(1):106–116. [DOI] [PubMed] [Google Scholar]
- Bai X, Brown JR, Varki A, Esko JD. Enhanced 3-O-sulfation of galactose in Asn-linked glycans and Maackia amurensis lectin binding in a new Chinese hamster ovary cell line. Glycobiology. 2001:11(8):621–632. [DOI] [PubMed] [Google Scholar]
- Benktander J, Barone A, Johansson MM, Teneberg S. Helicobacter pylori SabA binding gangliosides of human stomach. Virulence. 2018:9(1):738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Lopez JA, Sullam PM. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect Immun. 2004:72(11):6528–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Khedri Z, Deng L, Yu H, Prakobphol A, Fisher SJ, Chen X, Iverson TM, Varki A, Sullam PM. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology. 2016:26(11):1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavanandan VP, Katlic AW. The interaction of wheat germ agglutinin with sialoglycoproteins. The role of sialic acid. J Biol Chem. 1979:254(10):4000–4008. [PubMed] [Google Scholar]
- Bousquet PA, Sandvik JA, Jeppesen Edin NF, Krengel U. Hypothesis: Hypoxia induces de novo synthesis of NeuGc gangliosides in humans through CMAH domain substitute. Biochem Biophys Res Commun. 2018:495(1):1562–1566. [DOI] [PubMed] [Google Scholar]
- Brinkman-van der Linden ECM, Sjoberg ER, Juneja LR, Crocker PR, Varki N, Varki A. Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J Biol Chem. 2000:275(12):8633–8640. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Sonnenburg JL, Varki A. Effects of sialic acid substitutions on recognition by Sambucus nigra agglutinin and Maackia amurensis hemagglutinin. Anal Biochem. 2002:303(1):98–104. [DOI] [PubMed] [Google Scholar]
- Büll C, Nason R, Sun L, van Coillie J, Madriz Sørensen D, Moons SJ, Yang Z, Arbitman S, Fernandes SM, Furukawa S, et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc Natl Acad Sci U S A. 2021:118(17):e2026102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, Löfling JC, Smith DF, Wilce MCJ, Talbot UM, Chong DC, Yu H, Huang S, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008:456(7222):648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariappa A, Takematsu H, Liu H, Diaz S, Haider K, Boboila C, Kalloo G, Connole M, Shi HN, Varki N, et al. B cell antigen receptor signal strength and peripheral B cell development are regulated by a 9-O-acetyl sialic acid esterase. J Exp Med. 2009:206(1):125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel V, Segura V, Cañete A. Treatment of high-risk neuroblastoma with anti-GD2 antibodies. Clin Transl Oncol. 2010:12:788–793. [DOI] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010:5(2):163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Baldwin MR, Barbieri JT. Molecular basis for tetanus toxin coreceptor interactions. Biochemistry. 2008:47(27):7179–7186. [DOI] [PubMed] [Google Scholar]
- Chen C, Fu Z, Kim JJP, Barbieri JT, Baldwin MR. Gangliosides as high affinity receptors for tetanus neurotoxin. J Biol Chem. 2009:284(39):26569–26577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD, Darvill AG, Etzler ME, Hahn Ajit Varki MG, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al. Glycan-recognizing probes as tools. In: Varki A, et al., editors. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015 [PubMed] [Google Scholar]
- Dabelsteen S, Pallesen EMH, Marinova IN, Nielsen MI, Adamopoulou M, Rømer TB, Levann A, Andersen MM, Ye Z, Thein D, et al. Essential functions of Glycans in human epithelia dissected by a CRISPR-Cas9-engineered human Organotypic skin model. Dev Cell. 2020:54(5):669–684.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CJ, Paton AW, Higgins MA, Shewell LK, Jen FEC, Schulz BL, Herdman BP, Paton JC, Jennings MP. Structure aided design of a Neu5Gc specific lectin. Sci Rep. 2017:7(1):1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 2014a:10(12):e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galán JE, Varki A. Host adaptation of a bacterial toxin from the human pathogen salmonella Typhi. Cell. 2014b:159(6):1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz SL, Padler-Karavani V, Ghaderi D, Hurtado-Ziola N, Yu H, Chen X, Brinkman-van der Linden ECM, Varki A, Varki NM. Sensitive and specific detection of the non-human sialic acid N-glycolylneuraminic acid in human tissues and biotherapeutic products. PLoS One. 2009:4(1):e4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002:20(10):1011–1017. [DOI] [PubMed] [Google Scholar]
- Geisler C, Jarvis DL. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology. 2011:21(8):988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung MK, Ræder ILU, Altermark B, Smalås AO. Characterization of the sialic acid synthase from Aliivibrio salmonicida suggests a novel pathway for bacterial synthesis of 7-O-acetylated sialic acids. Glycobiology. 2013:23(7):806–819. [DOI] [PubMed] [Google Scholar]
- Halpern JL, Loftus A. Characterization of the receptor-binding domain of tetanus toxin. J Biol Chem. 1993:268(15):11188–11192. [PubMed] [Google Scholar]
- Hanaoka K, Pritchett TJ, Takasaki S, Kochibe N, Sabesan S, Paulson JC, Kobata A. 4-O-acetyl-N-acetylneuraminic acid in the N-linked carbohydrate structures of equine and Guinea pig alpha 2-macroglobulins, potent inhibitors of influenza virus infection. J Biol Chem. 1989:264(17):9842–9849. [PubMed] [Google Scholar]
- Hashimoto N, Ito S, Tsuchida A, Bhuiyan RH, Okajima T, Yamamoto A, Furukawa K, Ohmi Y, Furukawa K. The ceramide moiety of disialoganglioside (GD3) is essential for GD3 recognition by the sialic acid-binding lectin SIGLEC7 on the cell surface. J Biol Chem. 2019:294(28):10833–10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiner JP, Miraldi F, Kallick S, Makley J, Neely J, Smith-Mensah WH, Cheung NK. Localization of GD2-specific monoclonal antibody 3F8 in human osteosarcoma. Cancer Res. 1987:47(20):5377–5381. [PubMed] [Google Scholar]
- Hellebø A, Vilas U, Falk K, Vlasak R. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J Virol. 2004:78(6):3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G, Rott R, Klenk HD, Müller HP, Shukla AK, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985:4(6):1503–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa HH, Manzi A, Varki A. O-acetylation and de-O-acetylation of sialic acids. Purification, characterization, and properties of a glycosylated rat liver esterase specific for 9-O-acetylated sialic acids. J Biol Chem. 1989:264(32):19435–19442. [PubMed] [Google Scholar]
- Higashi H, Nishi Y, Fukui Y, Ikuta K, Ueda S, Kato S, Fujita M, Nakano Y, Taguchi T, Sakai S. Tumor-associated expression of glycosphingolipid Hanganutziu-Deicher antigen in human cancers. Gan. 1984:75(11):1025–1029. [PubMed] [Google Scholar]
- Higashi H, Hirabayashi Y, Fukui Y, Naiki M, Matsumoto M, Ueda S, Kato S. Characterization of N-glycolylneuraminic acid-containing gangliosides as tumor-associated Hanganutziu-Deicher antigen in human colon cancer. Cancer Res. 1985:45(8):3796–3802. [PubMed] [Google Scholar]
- Higashi H, Sasabe T, Fukui Y, Maru M, Kato S. Detection of gangliosides as N-glycolylneuraminic acid-specific tumor-associated Hanganutziu-Deicher antigen in human retinoblastoma cells. Jpn J Cancer Res. 1988:79(8):952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashihara T, Takeshima T, Anzai M, Tomioka M, Matsumoto K, Nishida K, Kitamura Y, Okinaga K, Naiki M. Survey of Hanganutziu and Deicher antibodies in operated patients. Int Arch Allergy Appl Immunol. 1991:95(2–3):231–235. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kasakura H, Matsumoto M, Higashi H, Kato S, Kasai N, Naiki M. Specific expression of unusual GM2 ganglioside with Hanganutziu-Deicher antigen activity on human colon cancers. Jpn J Cancer Res. 1987:78(3):251–260. [PubMed] [Google Scholar]
- Imamura K, Takeuchi H, Yabe R, Tateno H, Hirabayashi J. Engineering of the glycan-binding specificity of Agrocybe cylindracea galectin towards α(2,3)-linked sialic acid by saturation mutagenesis. J Biochem. 2011:150(5):545–552. [DOI] [PubMed] [Google Scholar]
- Ito H, Ishida H, Waki H, Ando S, Kiso M. Total synthesis of a cholinergic neuron-specific ganglioside GT1a alpha: a high affinity ligand for myelin-associated glycoprotein (MAG). Glycoconj J. 1999:16(10):585–598. [DOI] [PubMed] [Google Scholar]
- Iwamori M, Takamizawa K, Momoeda M, Iwamori Y, Taketani Y. Gangliosides in human, cow and goat milk, and their abilities as to neutralization of cholera toxin and botulinum type a neurotoxin. Glycoconj J. 2008:25(7):675–683. [DOI] [PubMed] [Google Scholar]
- Iwersen M, Vandamme-Feldhaus V, Schauer R. Enzymatic 4-O-acetylation of N-acetylneuraminic acid in Guinea-pig liver. Glycoconj J. 1998:15(9):895–904. [DOI] [PubMed] [Google Scholar]
- Ji Y, Sasmal A, Li W, Oh L, Srivastava S, Hargett AA, Wasik BR, Yu H, Diaz S, Choudhury B, et al. Reversible O-acetyl migration within the sialic acid side chain and its influence on protein recognition. ACS Chem Biol. 2021:16(10):1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Byrd-Leotis L, Matsumoto Y, Gao C, Wein AN, Lobby JL, Kohlmeier JE, Steinhauer DA, Cummings RD. The human lung Glycome reveals novel glycan ligands for influenza a virus. Sci Rep. 2020:10(1):5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadirvelraj R, Grant OC, Goldstein IJ, Winter HC, Tateno H, Fadda E, Woods RJ. Structure and binding analysis of Polyporus squamosus lectin in complex with the Neu5Ac{alpha}2-6Gal{beta}1-4GlcNAc human-type influenza receptor. Glycobiology. 2011:21(7):973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Mori Y, Goldstein IJ, Shibuya N. Monomeric, monovalent derivative of Maackia amurensis leukoagglutinin. Preparation and application to the study of cell surface glycoconjugates by flow cytometry. J Biol Chem. 1993:268(18):13237–13241. [PubMed] [Google Scholar]
- Kamerling JP, Gerwig GJ. Structural analysis of naturally occurring sialic acids. Methods Mol Biol. 2006:347:69–91. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Matsumoto I, Osawa T. Studies on hemagglutinins from Maackia amurensis seeds. J Biol Chem. 1974:249(9):2786–2792. [PubMed] [Google Scholar]
- Kawanishi K, Saha S, Diaz S, Vaill M, Sasmal A, Siddiqui SS, Choudhury B, Sharma K, Chen X, Schoenhofen IC, et al. Evolutionary conservation of human ketodeoxynonulosonic acid production is independent of sialoglycan biosynthesis. J Clin Invest. 2021:131(5):137681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima I, Tada N, Ikegami S, Nakamura S, Ueda R, Tai T. Mouse monoclonal antibodies detecting disialogangliosides on mouse and human T lymphomas. Int J Cancer. 1988:41(2):267–274. [DOI] [PubMed] [Google Scholar]
- Kelm S, Pelz A, Schauer R, Filbin MT, Tang S, Bellard, Schnaar RL, Mahoney JA, Hartnell A, Bradfield P, et al. Sialoadhesin, myelin-associated glycoprotein and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr Biol. 1994:4(11):965–972. [DOI] [PubMed] [Google Scholar]
- Kemminer SE, Conradt HS, Nimtz M, Sagi D, Peter-Katalinic J, Diekmann O, Drmic I, Muthing J. Production and molecular characterization of clinical phase i anti-melanoma mouse IgG3 monoclonal antibody R24. Biotechnol Prog. 2001:17(5):809–821. [DOI] [PubMed] [Google Scholar]
- Khan N, Sasmal A, Khedri Z, Secrest P, Verhagen A, Srivastava S, Varki N, Chen X, Yu H, Beddoe T, et al. Sialoglycan-binding patterns of bacterial AB5 toxin B subunits correlate with host range and toxicity, indicating evolution independent of a subunits. J Biol Chem. 2022:298(5):101900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Son KY, Koo KM, Kim JY, Alfajaro MM, Park JG, Hosmillo M, Soliman M, Baek YB, Cho EH, et al. Porcine Sapelovirus uses α2,3-linked sialic acid on GD1a ganglioside as a receptor. J Virol. 2016:90(8):4067–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamer Z, Staal B, Prudden AR, Liu L, Smith DF, Boons GJ, Haab B. Mining high-complexity motifs in Glycans: a new language to uncover the fine specificities of lectins and Glycosidases. Anal Chem. 2017:89(22):12342–12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausegger A, Strobl B, Regl G, Kaser A, Luytjes W, Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999:73(5):3737–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Roussel P. O-acetylation of sialic acids. Biochimie. 1998:80(1):49–57. [DOI] [PubMed] [Google Scholar]
- Klein A, Krishna M, Varki NM, Varki A. 9-O-acetylated sialic acids have widespread but selective expression: analysis using a chimeric dual-function probe derived from influenza C hemagglutinin-esterase. Proc Natl Acad Sci U S A. 1994:91(16):7782–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletter D, Singh S, Bern M, Haab BB. Global comparisons of lectin-glycan interactions using a database of analyzed glycan array data. Mol Cell Proteomics. 2013:12(4):1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibbs RN, Goldstein IJ, Ratcliffe RM, Shibuya N. Characterization of the carbohydrate binding specificity of the leukoagglutinating lectin from Maackia amurensis. Comparison with other sialic acid-specific lectins. J Biol Chem. 1991:266(1):83–88. [PubMed] [Google Scholar]
- Konami Y, Yamamoto K, Osawa T, Irimura T. Strong affinity of Maackia amurensis hemagglutinin (MAH) for sialic acid-containing Ser/Thr-linked carbohydrate chains of N-terminal octapeptides from human glycophorin a. FEBS Lett. 1994:342(3):334–338. [DOI] [PubMed] [Google Scholar]
- Kooner AS, Diaz S, Yu H, Santra A, Varki A, Chen X. Chemoenzymatic synthesis of Sialosides containing 7-N- or 7,9-Di-N-acetyl sialic acid as stable O-acetyl analogues for probing sialic acid-binding proteins. J Org Chem. 2021:86(21):14381–14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis MA, Zeng Q, Gerwig GJ, Frey B, Itzstein, Kamerling JP, de Groot RJ, Huizinga EG. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc Natl Acad Sci U S A. 2009:106(37):15897–15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis MA, van Vliet ALW, Boot W, de Groot RJ. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J Virol. 2010:84(17):8970–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis MA, Zeng Q, Heesters B, Huizinga EG, de Groot RJ. The murine coronavirus hemagglutinin-esterase receptor-binding site: A major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012:8(1):e1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis MA, Bakkers MJG, Deng L, Padler-Karavani V, Vervoort SJ, Hulswit RJG, van Vliet ALW, Gerwig GJ, de Poot SAH, Boot W, et al. Complexity and diversity of the mammalian Sialome revealed by Nidovirus Virolectins. Cell Rep. 2015:11(12):1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann F, Tiralongo E, Tiralongo J. Sialic acid-specific lectins: Occurrence, specificity and function. Cell Mol Life Sci. 2006:63(12):1331–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AL, Nizet V, Varki A. Discovery and characterization of sialic acid O-acetylation in group B streptococcus. Proc Natl Acad Sci U S A. 2004:101(30):11123–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louch HA, Buczko ES, Woody MA, Venable RM, Vann WF. Identification of a binding site for ganglioside on the receptor binding domain of tetanus toxin. Biochemistry. 2002:41(46):13644–13652. [DOI] [PubMed] [Google Scholar]
- Luytjes W, Bredenbeek PJ, Noten AFH, Horzinek MC, Spaan WJM. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988:166(2):415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal C, Basu S. An unique specificity of a sialic acid binding lectin AchatininH, from the hemolymph of Achatina fulica snail. Biochem Biophys Res Commun. 1987:148(2):795–801. [DOI] [PubMed] [Google Scholar]
- Manzi AE, Sjoberg ER, Diaz S, Varki A. Biosynthesis and turnover of O-acetyl and N-acetyl groups in the gangliosides of human melanoma cells. J Biol Chem. 1990:265(22):13091–13103. [PubMed] [Google Scholar]
- Manzke O, Russello O, Leenen C, Diehl V, Bohlen H, Berthold F. Immunotherapeutic strategies in neuroblastoma: Antitumoral activity of deglycosylated ricin a conjugated anti-GD2 antibodies and anti-CD3xanti-GD2 bispecific antibodies. Med Pediatr Oncol. 2001:36(1):185–189. [DOI] [PubMed] [Google Scholar]
- Martin LT, Verhagen A, Varki A. Recombinant influenza C hemagglutinin-esterase as a probe for sialic acid 9-O-acetylation. Methods Enzymol. 2003:363:489–498. [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Herrler G, Klenk HD. Sialic acid receptors of viruses. Top Curr Chem. 2015:367:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Collawn JF, Fish WW. Purification and macromolecular properties of a sialic acid-specific lectin from the slug Limax flavus. J Biol Chem. 1982:257(13):7574–7580. [PubMed] [Google Scholar]
- Miyake M, Ito M, Hitomi S, Ikeda S, Taki T, Kurata M, Hino A, Miyake N, Kannagi R. Generation of two murine monoclonal antibodies that can discriminate N-glycolyl and N-acetyl neuraminic acid residues of GM2 gangliosides. Cancer Res. 1988:48(21):6154–6160. [PubMed] [Google Scholar]
- Miyoshi I, Higashi H, Hirabayashi Y, Kato S, Naiki M. Detection of 4-O-acetyl-N-glycolylneuraminyl lactosylceramide as one of tumor-associated antigens in human colon cancer tissues by specific antibody. Mol Immunol. 1986:23(6):631–638. [DOI] [PubMed] [Google Scholar]
- Monsigny M, Roche AC, Sene C, Maget-Dana R, Delmotte F. Sugar-lectin interactions: how does wheat-germ agglutinin bind sialoglycoconjugates. Eur J Biochem. 1980:104(1):147–153. [DOI] [PubMed] [Google Scholar]
- Muchmore EA, Varki A. Selective inactivation of influenza C esterase: a probe for detecting 9-O-acetylated sialic acids. Science. 1987:236(4806):1293–1295. [DOI] [PubMed] [Google Scholar]
- Muthana SM, Gildersleeve JC. Glycan microarrays: powerful tools for biomarker discovery. Cancer Biomark. 2014:14(1):29–41. [DOI] [PubMed] [Google Scholar]
- Nason R, Büll C, Konstantinidi A, Sun L, Ye Z, Halim A, du W, Sørensen DM, Durbesson F, Furukawa S, et al. Display of the human mucinome with defined O-glycans by gene engineered cells. Nat Commun. 2021:12(1):4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Lee S, Yang YA, Ahn C, Sim JH, Kei TG, Barnard KN, Yu H, Millano SK, Chen X, et al. The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog. 2020:16(2):e1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JSM. Sialic acid receptor detection in the human respiratory tract: Evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007:8(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Sasabe T, Nishida T, Nishi Y, Higashi H. Hanganutziu-Deicher heterophile antigen in human retinoblastoma cells. Am J Ophthalmol. 1983:96(3):321–325. [DOI] [PubMed] [Google Scholar]
- Pillai S. Rethinking mechanisms of autoimmune pathogenesis. J Autoimmun. 2013:45:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LD, Sgroi D, Sjoberg ER, Stamenkovic I, Varki A. Natural ligands of the B cell adhesion molecule CD22beta carry N-linked oligosaccharides with alpha-2,6-linked sialic acids that are required for recognition. J Biol Chem. 1993:268(10):7019–7027. [PubMed] [Google Scholar]
- Powell LD, Jain RK, Matta KL, Sabesan S, Varki A. Characterization of sialyloligosaccharide binding by recombinant soluble and native cell-associated CD22. Evidence for a minimal structural recognition motif and the potential importance of multisite binding. J Biol Chem. 1995:270(13):7523–7532. [DOI] [PubMed] [Google Scholar]
- Ravindranath MH, Higa HH, Cooper EL, Paulson JC. Purification and characterization of an O-acetylsialic acid-specific lectin from a marine crab Cancer antennarius. J Biol Chem. 1985:260(15):8850–8856. [PubMed] [Google Scholar]
- Ravindranaths MH, Paulson JC, Irie RF. Human melanoma antigen O-acetylated ganglioside GD3 is recognized by Cancer antennarius lectin. J Biol Chem. 1988:263(4):2079–2086. [PubMed] [Google Scholar]
- Regl G, Kaser A, Iwersen M, Schmid H, Kohla G, Strobl B, Vilas U, Schauer R, Vlasak R. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J Virol. 1999:73(6):4721–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, Herrler G, Paulson JC, Klenk HD. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J Biol Chem. 1986:261(13):5947–5951. [PubMed] [Google Scholar]
- Samraj AN, Pearce OMT, Läubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A. 2015:112(2):542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. Sialic acids: metabolism of O-acetyl groups. Methods Enzymol. 1987:138:611–626. [DOI] [PubMed] [Google Scholar]
- Schauer R, Kamerling JP. Exploration of the sialic acid world. Adv Carbohydr Chem Biochem. 2018:75:1–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J Gen Virol. 1992:73(Pt 4):901–906. [DOI] [PubMed] [Google Scholar]
- Schultze B, Gross HJ, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. 1991a:65(11):6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Wahn K, Klenk HD, Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991b:180(1):221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007:153(Pt 9):2817–2822. [DOI] [PubMed] [Google Scholar]
- Shewell LK, Wang JJ, Paton JC, Paton AW, Day CJ, Jennings MP. Detection of N-glycolylneuraminic acid biomarkers in sera from patients with ovarian cancer using an engineered N-glycolylneuraminic acid-specific lectin SubB2M. Biochem Biophys Res Commun. 2018:507(1–4):173–177. [DOI] [PubMed] [Google Scholar]
- Shewell LK, Day CJ, Kutasovic JR, Abrahams JL, Wang J, Poole J, Niland C, Ferguson K, Saunus JM, Lakhani SR, et al. N-glycolylneuraminic acid serum biomarker levels are elevated in breast cancer patients at all stages of disease. BMC Cancer. 2022:22(1):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2-6)gal/GalNAc sequence. J Biol Chem. 1987:262(4):1596–1601. [PubMed] [Google Scholar]
- Shieh CK, Lee HJ, Yokomori K, la Monica N, Makino S, Lai MM. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989:63(9):3729–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoberg ER, Manzi AE, Khoo KH, Dell A, Varki A. Structural and immunological characterization of O-acetylated GD2. Evidence that GD2 is an acceptor for ganglioside O-acetyltransferase in human melanoma cells. J Biol Chem. 1992:267(23):16200–16211. [PubMed] [Google Scholar]
- Sjoberg ER, Powell LD, Klein A, Varki A. Natural ligands of the B cell adhesion molecule CD22 beta can be masked by 9-O-acetylation of sialic acids. J Cell Biol. 1994:126(2):549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald JE, Reiss K, Reiter DM, Stehle T, Dermody TS. The sweet spot: defining virus-sialic acid interactions. Nat Rev Microbiol. 2014:12(11):739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AD, Batty J, Harkiss GD. Genetic variation in plasma thyroxine levels and minimal metabolic rates of the mouse, Mus musculus. Genet Res. 1978:31(3):303–306. [DOI] [PubMed] [Google Scholar]
- Tai T, Kawashima I, Furukawa K, Lloyd KO. Monoclonal antibody R24 distinguishes between different N-acetyl- and N-glycolylneuraminic acid derivatives of ganglioside GD3. Arch Biochem Biophys. 1988:260(1):51–55. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, López JA, Griffiss JML, Sullam PM. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol Microbiol. 2005:58(2):380–392. [DOI] [PubMed] [Google Scholar]
- Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, Muchmore E. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci U S A. 2003:100(21):12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Winter HC, Goldstein IJ. Cloning, expression in Escherichia coli and characterization of the recombinant Neu5Acalpha2,6Galbeta1,4GlcNAc-specific high-affinity lectin and its mutants from the mushroom Polyporus squamosus. Biochem J. 2004: 382(Pt 2):667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme-Feldhaus V, Schauer R. Characterization of the enzymatic 7-O-acetylation of sialic acids and evidence for enzymatic O-acetyl migration from C-7 to C-9 in bovine submandibular gland. J Biochem. 1998:124(1):111–121. [DOI] [PubMed] [Google Scholar]
- Varki A. Diversity in the sialic acids. Glycobiology. 1992:2(1):25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Diaz S. The release and purification of sialic acids from glycoconjugates: Methods to minimize the loss and migration of O-acetyl groups. Anal Biochem. 1984:137(1):236–247. [DOI] [PubMed] [Google Scholar]
- Varki A, Kornfeld S. Historical background and overview. In: Varki A, et al., editors. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022 [Google Scholar]
- Vinson M, Strijbos PJLM, Rowles A, Facci L, Moore SE, Simmons DL, Walsh FS. Myelin-associated glycoprotein interacts with ganglioside GT1b. A mechanism for neurite outgrowth inhibition. J Biol Chem. 2001:276(23):20280–20285. [DOI] [PubMed] [Google Scholar]
- Vlasak R, Krystal M, Nacht M, Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology. 1987:160(2):419–425. [DOI] [PubMed] [Google Scholar]
- Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci U S A. 1988:85(12):4526–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shewell LK, Paton AW, Paton JC, Day CJ, Jennings MP. Specificity and utility of SubB2M, a new N-glycolylneuraminic acid lectin. Biochem Biophys Res Commun. 2018:500(3):765–771. [DOI] [PubMed] [Google Scholar]
- Wasik BR, Barnard KN, Ossiboff RJ, Khedri Z, Feng KH, Yu H, Chen X, Perez DR, Varki A, Parrish CR. Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere. 2017:2(5):e00379–e00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipfler D, Srinivasan GV, Sadick H, Kniep B, Arming S, Willhauck-Fleckenstein M, Vlasak R, Schauer R, Schwartz-Albiez R. Differentially regulated expression of 9-O-acetyl GD3 (CD60b) and 7-O-acetyl-GD3 (CD60c) during differentiation and maturation of human T and B lymphocytes. Glycobiology. 2011:21(9):1161–1172. [DOI] [PubMed] [Google Scholar]
- Yabe R, Suzuki R, Kuno A, Fujimoto Z, Jigami Y, Hirabayashi J. Tailoring a novel sialic acid-binding lectin from a ricin-B chain-like galactose-binding protein by natural evolution-mimicry. J Biochem. 2007:141(3):389–399. [DOI] [PubMed] [Google Scholar]
- Yagi F, Miyamoto M, Abe T, Minami Y, Tadera K, Goldstein IJ. Purification and carbohydrate-binding specificity of Agrocybe cylindracea lectin. Glycoconj J. 1997:14(2):281–288. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Umetsu K, Suzuki T, Ohkura T. Purification and characterization of a Neu5Ac alpha 2-->6Gal beta 1-->4GlcNAc and HSO3(−)-->6Gal beta 1-->GlcNAc specific lectin in tuberous roots of Trichosanthes japonica. Biochemistry. 1992:31(46):11647–11650. [DOI] [PubMed] [Google Scholar]
- Yokomori K, Banner LR, Lai MM. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991:183(2):647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer G, Reuter G, Schauer R. Use of influenza C virus for detection of 9-O-acetylated sialic acids on immobilized glycoconjugates by esterase activity. Eur J Biochem. 1992:204(1):209–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information.