Abstract
After nearly 20 years of research, it is now established that mutations within the estrogen receptor (ER) gene, ESR1, frequently occur in metastatic breast cancer and influence response to hormone therapy. Though early studies presented differing results, sensitive sequencing techniques now show that ESR1 mutations occur at a frequency between 20 and 40% depending on the assay method. Recent studies have focused on several “hot spot mutations,” a cluster of mutations found in the hormone-binding domain of the ESR1 gene. Throughout the course of treatment, tumor evolution can occur, and ESR1 mutations emerge and become enriched in the metastatic setting. Sensitive techniques to continually monitor mutant burden in vivo are needed to effectively treat patients with mutant ESR1. The full impact of these mutations on tumor response to different therapies remains to be determined. However, recent studies indicate that mutant-bearing tumors may be less responsive to specific hormonal therapies, and suggest that aromatase inhibitor (AI) therapy may select for the emergence of ESR1 mutations. Additionally, different mutations may respond discretely to targeted therapies. The need for more preclinical mechanistic studies on ESR1 mutations and the development of better agents to target these mutations are urgently needed. In the future, sequential monitoring of ESR1 mutational status will likely direct personalized therapeutic regimens appropriate to each tumor’s unique mutational landscape.
Keywords: ESR1 Mutations, Breast Cancer, Selective Estrogen Receptor Downregulator (SERDs), Palbociclib, Fulvestrant
Endocrine Resistance
Hormone receptor-positive disease is the most common presentation of breast cancer. The antiestrogens tamoxifen (Tam) and fulvestrant (Ful), along with aromatase inhibitors (AIs), are the most frequently prescribed hormonal agents targeting the ER, with excellent palliation in the metastatic setting and long-term delay of first recurrence with adjuvant therapy. However, despite improvements in the efficacy of targeted endocrine therapies following the introduction of the AIs and extended durations of Tam/AI therapy, the development of endocrine resistance remains a major cause of first recurrence and mortality in ER-positive patients. It is unlikely that complete loss of ER signaling is the driving force for the majority of endocrine resistance, as both metastatic tumors and breast cancer cell lines with acquired Tam resistance frequently retain ER expression [1–3] and can remain responsive to second or third line hormonal therapy.
ER-positive tumors do not consistently respond to hormonal therapy and have considerable heterogeneity in response to therapeutic agents. Notably, tumors can be refractory to one type of treatment (for example Tam or a nonsteroidal AI) but sensitive to another type (steroidal AI) [4]. Thus, ER-expressing tumors are not a homogenous group, even within the well-defined luminal A or B molecular subtypes. Unfortunately, these molecular subtype classifications do not adequately predict the heterogeneous response of ER-positive tumors. Thus, optimal selection of endocrine agents will require a better understanding of the mechanisms associated with the evolution of hormone resistance and the adaptation of individual tumors to the selective pressure of therapeutics. As mechanisms of resistance are elucidated, selection of patients for specific hormonal agents may be based on a combination of personalized genomics and adaptive pathway biomarkers which predict response, rather than clinical criteria alone.
The recent ESO-ESMO consensus guidelines for advanced breast cancer define acquired endocrine resistance as a relapse after the first 2 years of adjuvant endocrine treatment, a relapse within 12 months of completing adjuvant endocrine treatment, or progressive disease greater than 6 months after initiating endocrine therapy for metastatic breast cancer (MBC) [5, 6]. Unfortunately, patients with acquired resistance have a low probability of responding to further endocrine treatments for extended periods. Thus, MBC remains incurable, with median 5-year survival rates of less than 25%. Undoubtedly, endocrine resistance is common and an important clinical problem in MBC. The main identified mechanisms of endocrine resistance are related to the upregulation of escape survival pathways, such as the HER2 growth factor receptor family, and the PI3K/Akt/mammalian target of rapamycin (mTOR) pathways. However, in the past few years, there have been a dramatic change in the understanding of endocrine resistance and a general acceptance of the “rediscovery” that ESR1 mutations and alterations (amplifications and translocations) are a central mechanism of resistance [7, 8]. This review will focus on ESR1 alterations as a leading mechanism of intrinsic and acquired resistance in ER-positive advanced breast cancer.
Preclinical Models of Resistance
One of the first preclinical models of hormone resistance was developed by Craig Jordan and colleagues by growing MCF-7 breast cancer cells in athymic nude mice with estrogen supplementation [9]. Withdrawal of estrogen and long-term treatment of these xenograft tumors with Tam resulted in the eventual outgrowth of Tam-stimulated tumors [10]. These Tam-stimulated tumors could be serially retransplanted and displayed a stable resistant phenotype. One of several transplanted tumor lines was found to contain a single D351Y ESR1 mutation whose transcriptional activity was indeed stimulated by Tam treatment [11]. Subsequent studies demonstrated the structural basis for the switch to estrogen-like action of Tam by substitutions at this critical residue [12]. Although this specific ESR1 mutation has not been found in clinical samples, these studies underscore that temporal evolution of ESR1 mutations might occur under the selective pressure of antiestrogen treatment. However, as the majority of Tam-stimulated lines contained wild-type (WT) ER, ESR1 mutations are not thought to be the major mechanism for the agonist activity of Tam in this model.
A number of breast cancer sublines with acquired Tam resistance have been developed, the majority of which exhibit increases in growth factor receptor and subsequent downstream signaling, and in some cases, increased ER levels [13–16]. From these studies, investigators concluded that mechanisms other than alterations in ER must be important for the development of Tam-resistant growth. However, to our knowledge, reports of the presence of ESR1 mutations in Tam-resistant sublines is limited. Based on recent findings in detecting frequent ESR1 mutations in metastatic tumors arising after multiple lines of endocrine therapy (discussed below), deep genomic sequencing of ESR1 in sublines with acquired resistance is likely warranted.
Sublines mimicking AI resistance have also been generated via several techniques: long-term adaptation to estrogen withdrawal [17], transfection of ER-positive cells with aromatase enzyme and maintenance in vitro [18], or grown in vivo as tumor xenografts in mice [19, 20]. Similar to that seen in Tam-resistant lines, AI resistance was accompanied by activation of growth factor receptors (EGFR and IGF-1R), the PI3K/Akt/MAPK, and mTOR pathways. Enhanced estrogen hypersensitivity was inherent in many of the lines maintained in the absence of estrogen and demonstrates that cells adapt to estrogen deprivation during AI treatment by activation of alternate signaling pathways leading to ligand-independent activation of ER [21]. These preclinical studies were some of the first to demonstrate that growth factor signaling is an important contributor to the development of endocrine resistance, and suggest that the use of signal transduction inhibitors will provide a promising alternative therapeutic strategy. However, we now know that breast cancer cells are clearly able to continue evolving and harness additional bypass pathways for growth and cell survival in the presence of signal transduction inhibitor monotherapy, and therefore, the efficacy of these inhibitors is also limited by rapid acquired resistance. The co-targeting of these adaptive pathways, along with ER signaling antagonism, is a current clinical approach under investigation [22]. Indeed, since many of the growth factor receptor pathways ultimately activate the downstream mTOR complex, mTOR has proven a promising and efficacious clinical target to restore hormone sensitivity in AI-resistant MBC [23–25].
Genomic Characterization of Breast Cancer-Derived Xenograft Models Reveals Multiple ESR1 Alterations
The genomic landscape of primary breast cancers has recently been thoroughly explored using next generation sequencing (NGS) [26, 27]. However, the genomic basis of metastatic and endocrine-resistant breast cancer remains poorly understood. The molecular differences between primary and metastatic tumors have been a controversial area, but some studies have demonstrated that a subset of primary tumors have inherent expression signatures found in metastatic tumors, and maintain these distinct features during tumor progression [28, 29]. However, the prevailing model of metastasis holds that most primary tumor cells have low metastatic potential and that rare cells within the primary tumor acquire metastatic capacities through somatic mutation events which are selected for during tumor dissemination [30, 31].
Li et al. took a pioneering approach to examine ER-positive, endocrine-resistant tumors by establishing patient-derived xenograft (PDX) models, which appear to predominantly maintain the hormone response profile of the originating patient tumor [32]. Using deep whole-genome NGS techniques, these ER-positive PDXs were found to contain ESR1 hormone-binding domain (HBD) mutations (Y537S and E380D), ESR1 amplification, and ESR1 translocations (ESR1/YAP1). Most of the ESR1 alterations were present in the original patient material; however, one mutation (E380D) arose during serial passage of the PDX models, showing that tumors can continue to evolve when maintained as xenografts in mice. Two additional ESR1 translocations, ESR1/AKAP12 and ESR1/POLH, were reported in The Cancer Genome Atlas (TCGA) analysis of primary tumors [33]. A recurrent ESR1/CCDC170 fusion associated with the luminal B molecular subtype has been reported to also engage growth factor receptor signaling which reduces endocrine sensitivity [34]. Although at present these gain-of-function ESR1 translocations are relatively infrequent (< 5%), they could play a significant role in the emergence of aggressive subpopulations in progressing tumors.
The long-term, estrogen-deprived MCF-7 model undergoes widespread genomic changes during adaptation to estrogen withdrawal, including amplification of the ESR1 locus with subsequent increases in ER protein levels [35]. Whether ESR1 gene amplification occurs in breast tumors, though, has been a controversial question. Holst et al. reported frequent ESR1 amplification in 21% of breast tumors using fluorescent in situ hybridization (FISH). Amplification was associated with a significantly longer survival in Tam-treated patients [36]. This finding has been confirmed in another retrospective patient study [37]. In contrast, another study found that amplification was associated with poor disease-free and overall survival in Tam-treated patients [38], but it was found in only 1% of tumors using a variety of array hybridization platforms [39]. These discrepancies may be related to different technical protocols. Moelans et al. recently employed an RNase FISH protocol with multiplex ligation-dependent probe amplification to demonstrate that the FISH signals being interpreted as ESR1 amplification were sensitive to RNase treatment, indicating that FISH was detecting accumulation of ESR1 transcripts in cells expressing high levels of ER RNA, rather than gene amplification [40]. Thus, although it is apparent that ESR1 gene amplification can occur in tumors and be propagated in PDX models [32], amplification in breast tumors may be a relatively infrequent event and, thus, not likely an attractive clinical target.
ESR1 Mutations in Primary Breast Tumors: a Controversy Resolved
The general consensus for almost 20 years was that ESR1 mutations in primary disease were either not present or were very rare [41]. Recently, TCGA reported no ESR1 mutations in 390 primary tumors, apparently confirming this consensus opinion [33]. However, in 2000, our group reported a somatic K303R ESR1 mutation in one third of premalignant breast hyperplasias and, subsequently, found it in almost 50% of primary breast tumors [42, 43]. K303R ESR1 mutation studies are listed in Table 1. Consistent with TCGA data, several earlier reports failed to detect the K303R ESR1 mutation in primary tumors using standard fluorescent sequencing technologies [47, 48]. We subsequently reported that the discrepancy was due to poor base incorporation in both the forward and reverse strands of ESR1 using dye-labeled terminator sequencing techniques employed by these earlier investigators [43]. Conway et al. have also reported this problem with detection of the ESR1 K303R mutation in invasive breast tumors, albeit the reported frequency of the mutation was low (6%) in their studies [44]. It has also recently been shown that the K303R mutation frequency (29%) was higher in tumors from women with a family history of breast cancer [46] and that mutation-positive cases were more likely to have a first-degree family history of breast cancer [45]. We assert that it is time to challenge the long-standing dogma concerning ESR1 mutations in primary cancer, and conduct a reevaluation of the contradictory data that exists in the literature, rather than dismiss these data as outliers [49]. It is especially important to reconsider sequence detection methods, as the K303R mutation cannot be resolved with traditional core sequencing or NGS. Preliminary data using a sensitive droplet digital polymerase chain reaction (ddPCR) method confirm that the K303R mutation is indeed present at a low frequency, but often resides in a small subpopulation within primary tumors (Gu and Fuqua, unpublished).
Table 1.
Summary of relevant K303R-positive patient cohorts
| Study | Frequency | Method | Location detected | Reference |
|---|---|---|---|---|
| Fuqua et al. 2000 | 34% (20/59) | PCR amplification | Typical hyperplasia | [42] |
| Herynk et al. 2007 | 50% (133/267) | SNaPshot™ | Invasive breast cancer | [43] |
| Conway et al. 2004, 2006 | 5.7% (37/653) | SSCP | Invasive breast cancer | [44, 45] |
| Abbasi et al. 2013 | 10.7% (16/150) | SSCP | Invasive breast cancer | [46] |
Overexpression of the K303R ESR1 mutation in ER-positive breast cancer cells conferred hypersensitivity to estrogen-stimulated growth [42], consistent with the hypersensitive growth of long-term estrogen-deprived MCF-7 breast cancer cells. K303R-expressing cells also displayed reduced sensitivity to Tam treatment, but only when growth factor signaling was engaged [50], demonstrating that enhanced growth factor receptor-ER crosstalk is one mechanism of relative endocrine resistance associated with expression of this ESR1 mutation. Expression of the K303R mutation also resulted in significantly increased hormone-independent activity and conferred resistance to AIs through dynamic interactions with the IGF-1R signaling pathway [51–53]. The K303R mutation altered genomic transcriptional output with enhanced expression of components both upstream and downstream of the IGF-1R receptor signaling network [54]. This mutation is a classical gain-of-function mutation with the lysine to arginine substitution rendering the receptor an enhanced substrate for phosphorylation by several kinase cascades [53, 55]. Finally, the mutation enhanced bidirectional communication with and response to signals from the microenvironment [56], an important, though underexplored, area of ER crosstalk. The influence of ESR1 mutations on paracrine signaling is an exciting potential area for mutation-specific therapeutic intervention.
The K303R mutation was associated with recurrence-free survival in univariate analyses of tumors from 267 untreated breast cancer patients, but it was not an independent prognostic factor for outcomes in multivariate analyses [43]. Its presence was also associated with biologic measures of poor outcome, including larger tumor size, older age, and axillary lymph node positivity. Since the mutation is present in untreated patients, and not the consequence of treatment selection in primary tumors, it will be important to determine whether the mutation occurs spontaneously, is driven by exogenous carcinogens or hormone exposure, or is the result of endogenous DNA damage. We originally proposed that the K303R ESR1 mutation may confer a proliferative advantage in premalignant lesions due to its hypersensitivity, especially in postmenopausal women with lower levels of circulating estrogen. Similarly, cells with this hyperproliferative mutation could provide a favorable environment to facilitate the accumulation of additional mutational events that drive tumor progression. Studies examining this mutation in metastatic tumors are underway.
“Rediscovery” of ESR1 Mutations in the HBD
A number of years ago, we originally hypothesized that maintenance of ER expression, along with selection of specific ESR1 mutations, was a key event in breast cancer progression, most likely due to the selective pressure of hormonal treatment in metastatic patients [7]. Our argument was based not only on identifying two key ESR1 mutational hot spots [42, 57] but also on the fact that these mutations could provide biologic functions that would be selected for in emerging populations of metastatic deposits, e.g., estrogen hypersensitivity and/or independence.
In 1997, 30 tumors from MBC patients were screened for ESR1 mutations using single-strand polymorphism conformation analyses coupled with Sanger sequencing techniques [57]. Sequencing was performed from polymerase chain reaction (PCR)-amplified DNA primed with ESR1-specific oligonucleotide primers, similar to the reduced exome sequencing performed in the contemporary laboratory setting. Three missense mutations were detected (S47T, K531E, and Y537N), corresponding to a 10% mutation frequency rate in metastatic tumors. The frequency of specific HBD ESR1 mutations detected in individual sequencing studies is shown in Table 2. S47T and K531E were found to exhibit transcriptional activity similar to WT ER, and were not studied further. The Y537N mutation displayed potent, estradiol-independent activity in ER-negative breast cancer cells but was virtually unaffected by estradiol, Tam, or fulvestrant treatment. At that time, it was speculated that if present in other metastatic tumors, the mutation might contribute to breast cancer progression, and its constitutive activity might present as endocrine-resistant disease. It is important to note that the patient with the Y537N substitution presented with advanced stage IV breast cancer, and it was an ER, progesterone receptor (PR)-negative metastatic bone tumor that was sequenced after treatment with diethylstilbestrol (DES) therapy. Since estrogen treatment had little effect on the high constitutive activity of the Y537N mutation, and the patient sample was clinically ER, PR-negative, it is unlikely that a mutant subclonal population was selected for by DES treatment. Unfortunately, after this single report, sensitive ESR1 gene-specific PCR amplification and sequencing of metastatic breast tumors was not validated by other laboratories, and surprisingly, the therapeutic implications of ESR1 mutations in breast tumors were underappreciated until now.
Table 2.
Summary of reported frequencies of ligand-binding domain mutations
| Study | Amino acid change | Frequency | Source |
|---|---|---|---|
| Zhang et al. 1997 [57] | Y537N | 3.3% (1/30) | Metastasis |
| Li et al. 2013 [32] | Y537S | 13.6% (3/22) | Metastasis |
| E380Q | 4.5% (1/22) | Metastasis | |
| Merenbakh-Lamin et al. 2013 [58] | D538G | 38% (5/13) | Metastasis |
| Robinson et al. 2013 [59] | L536Q | 9% (1/11) | Metastasis |
| Y537S | 27% (3/11) | Metastasis | |
| D538G | 18% (2/11) | Metastasis | |
| Toy et al. 2013 [60] | Y537S | 14% (5/36) | Metastasis |
| D538G | 8% (3/36) | Metastasis | |
| S463P | 3% (1/36) | Metastasis | |
| L536R | 3% (1/36) | Metastasis | |
| V534E | 3% (1/36) | Metastasis | |
| Y537N | 3% (1/36) | Metastasis | |
| De Mattos-Arruda et al. 2014 [61] | E380Q | 2% (7/287) | Primary tumor |
| E380Q | 68% (106/157) | Metastasis | |
| E380Q | 46% (339/737) | CTC | |
| E380Q | 19% (158/823) | CTC | |
| E380Q | 58% (160/275) | CTC | |
| E380Q | 53% (534/1009) | CTC | |
| Jeselsohn et al. 2014 [62] | E380Q | 0.74% (1/134) | Metastasis |
| Y537N | 1.15% (2/134) | Metastasis | |
| Y537S | 1.15% (2/134) | Metastasis | |
| Y537C | 1.15% (2/134) | Metastasis | |
| D538G | 1.72% (3/134) | Metastasis | |
| 344insC | 1.15% (2/134) | Metastasis | |
| Guttery et al. 2015 [63] | E380Q | 2% (1/48) | cfDNA |
| Y537S | 2% (1/48) | cfDNA | |
| D538G | 2% (1/48) | cfDNA | |
| Niu et al. 2015 [64] | Y537S | 5% (11/217) | Metastasis |
| D538G | 4% (9/217) | Metastasis | |
| Y537C | 2% (4/217) | Metastasis | |
| Y537N | 1% (2/217) | Metastasis | |
| V533 M | 0.5% (1/217) | Metastasis | |
| L536P | 0.5% (1/217) | Metastasis | |
| Y537P | 0.5% (1/217) | Metastasis | |
| D538P | 0.5% (1/217) | Metastasis | |
| Sefrioui et al. 2015 [65] | Y537S | 29% (2/7) | Metastasis |
| D538G | 43% (3/7) | Metastasis | |
| Y537N | 29% (2/7) | Metastasis | |
| Schiavon et al. 2015 [66] | D538G | 11% (15/128) | cfDNA |
| Y537S | 2% (3/128) | cfDNA | |
| Y537N | 3% (4/128) | cfDNA | |
| Y537C | 2% (2/128) | cfDNA | |
| L536R | 2% (2/128) | cfDNA | |
| Takeshita et al. 2015 [67] | Y537S | 45% (5/11) | Metastasis |
| Y537C | 27% (3/11) | Metastasis | |
| Y537N | 36% (4/11) | Metastasis | |
| D538G | 36% (4/11) | Metastasis | |
| Wang et al. 2015 [68] | D538G | 13% (2/15) | Metastasis |
| Y537C | 7% (1/15) | Metastasis | |
| Chandarlapaty et al. 2016 [69] | Y537S |
13.3% 7.8% (72/541) |
cfDNA |
| D538G | 21.1% (114/541) | cfDNA | |
| Clatot et al. 2016 [70] | Y537S | 14.6% (21/144) | cfDNA |
| Y537N | 11.1% (16/144) | cfDNA | |
| Y537C | 1.4% (2/144) | cfDNA | |
| D538G | 16.7% (24/144) | cfDNA | |
| Gyanchandani et al. 2016 [71] | D538G | 18.8% (3/16) | cfDNA |
| Y537S | 12.5% (2/16) | cfDNA | |
| Y537N | 12.5% (2/16) | cfDNA | |
| Hrebien et al. 2016 [72] | D538G | 21.1% (15/71) | cfDNA |
| Ma et al. 2016 [73] | E380Q | 6.3% (1/16) | Primary tumor/metastasis |
| S576L | 6.3% (1/16) | Primary tumor/metastasis | |
| Spoerke et al. 2016 [74] | Y537N | 4% (2/47) (of mets) | Metastasis |
| Y537N | 4% (2/47) | cfDNA | |
| L536Q | 2% (1/47) | Metastasis | |
| L536Q | 2% (1/47) | cfDNA | |
| Y537C | 6% (3/47) | Metastasis | |
| Y537C | 6% (3/47) | cfDNA | |
| D538G | 9% (4/47) | Metastasis | |
| D538G | 11% (5/47) | cfDNA | |
| P535H | 2% (1/47) | Metastasis | |
| P535H | 2% (1/47) | cfDNA | |
| E380Q | 13% (6/47) | Metastasis | |
| E380Q | 15% (7/47) | cfDNA | |
| Y537S | 2% (1/47) | Metastasis | |
| Y537S | 2% (1/47) | cfDNA | |
| L536H | 2% (1/47) | cfDNA | |
| S463P | 2% (1/47) | cfDNA | |
| Takeshita et al. 2016 [75] | Y537S | 14% (6/42) | cfDNA |
| D538G | 5% (2/42) | cfDNA | |
| Y537N | 12% (5/42) | cfDNA | |
| Wang et al. 2016 [76] | Y537C | 3% (1/29) | cfDNA |
| Y537S | 4% (1/24) | Metastasis | |
| Y537S | 7% (2/29) | cfDNA | |
| D538G | 7% (3/43) | Primary | |
| D538G | 11.4% (4/35) | Metastasis | |
| D538G | 21% (6/29) | cfDNA | |
| Chu et al. 2017 [77] | Y537S | 26.3% (5/19) | Metastasis |
| D538G | 52.6% (10/19) | Metastasis | |
| Y537N | 10.5% (2/19) | Metastasis | |
| Fribbens et al. 2017 [78] | D538G | 14.2% (51/360) | cfDNA |
| Y537N | 3.9% (14/360) | cfDNA | |
| Y537S | 6.4% (23/360) | cfDNA | |
| E380Q | 6.1% (22/360) | cfDNA | |
| S463P | 1.1% (4/360) | cfDNA | |
| Y537C | 1.4% (5/360) | cfDNA | |
| L536R | 0.3% (1/360) | cfDNA | |
| Page et al. 2017 [79] | Y537N | 3% (1/39) | cfDNA |
| Y537S | 5% (2/39) | cfDNA | |
| E380Q | 8% (3/39) | cfDNA | |
| Shaw et al. 2017 [80]a | E380Q | 50% (2/4) | CTC |
| Y537C | 25% (1/4) | CTC | |
| D538G | 25% (1/4) | CTC | |
| Yanagawa et al. 2017 [1] | Y537C | 2% (1/46) | Metastasis |
| Y537N | 2% (1/46) | Metastasis | |
| D538G | 2% (1/46) | Metastasis | |
| Y537S | 5%(2/38) | cfDNA | |
| G557R | 2% (1/46) | Metastasis | |
| D538G | 8% (3/38) | cfDNA | |
| S463P | 3% (1/36) | cfDNA | |
| L536H | 3% (1/36) | cfDNA |
aStudy looked at 112 women with MBC, only 5 were analyzed for CTCs, 4 being ER+
Sixteen years later, two laboratories published correlating results using NGS of metastatic tumors, validating the presence of estrogen-independent ESR1 mutations occurring within the HBD of ER [59, 60]. Both reports found highly recurrent mutations surrounding the site of the original mutation detected in 1997 (L536S, Y537S, D538G), demonstrating that this location is most likely a genomic “hot spot” for activating HBD ESR1 mutations. Robinson et al. [59] sequenced 11 metastatic patients and showed that 54% harbored ESR1 mutations. Toy et al. [60] sequenced two cohorts of patients and reported frequencies of 25–50 and 11% for HBD ESR1 mutations, respectively. In a retrospective study of 217 ER-positive patients, Niu et al. used NGS to determine a 12.1% frequency of HBD ESR1 mutations [64]. All specimens in this study were collected from MBC treated with at least one line of AIs, most of which were heavily pretreated with other hormonal agents. Most patients also carried three or more additional genomic alterations. Three patients achieved stable disease with a combination of exemestane and everolimus, though notably, all of these patients harbored only the D538G mutation. Sufficiently powered clinical studies are necessary to determine whether discrete mutations may respond differently to targeted therapies.
Newer studies posit that ESR1 mutations are indeed present in primary tumors, but only occur at very low frequencies [81]. ESR1 mutations have been reported in low frequencies in primary tumors: detected at < 1% in a recent analysis of 772 primary TCGA tumors [82] and reported at 2.4% in TCGA primary tumors. ESR1 mutations have also been detected using NGS in one primary ovarian cancer tumor [83] and four cases of endometrial cancer in the TCGA database [59]. Thus, understanding ESR1 mutational effects could have implications beyond just breast cancer. These studies also demonstrate that ESR1 mutations are detectable in primary tumors; however, due to their low frequencies, NGS may not be adequately sensitive for diagnostic purposes.
The more sensitive ddPCR can detect rare point mutations in small tumor subpopulations. Our lab identified ESR1 mutations in primary breast tumors at a frequency of 2–12% using ddPCR [81]. Sequencing using ddPCR has also been used to estimate ESR1 mutation frequencies in metastatic samples. The BOLERO-2 trial [69] enrolled women to evaluate the efficacy of adding the mTOR inhibitor everolimus to the steroidal AI exemestane to treat advanced MBC that had progressed on nonsteroidal AIs. In a retrospective study utilizing cell-free DNA (cfDNA) from 541 evaluable patients, ddPCR detected ESR1 mutations in 28.8% of patients. Importantly, compared to patients with WT ESR1, these mutations were associated with shorter overall survival. Additionally, while both the D538G and WT patients’ progression-free survival increased with the addition of everolimus, Y537S mutation-bearing patients received no added benefit with combined therapy. While this difference may be due to the lack of statistical power, it could also indicate that the HBD mutations promote different biological phenotypes. In 2015, Takeshita et al. [67] also applied ddPCR technology to a study of 55 metastatic breast cancer samples. They found a comparable overall frequency of 20% of the most frequent ESR1 HBD mutations (Y537N, Y537S, D538G, and Y537C). They also noted a 2.5% frequency of mutations in primary breast tumors. They reported that half of the ESR1 mutations found in MBC were not present in the corresponding primary tumor, and a few patients harbored double or triple mutations. These studies confirm that ESR1 mutations are readily detectable using ddPCR and occur in both primary and MBC and that the mutational profile can be polyclonal.
From these studies, comprehensively listed in Table 2, it is clear that ESR1 mutations occur at low frequencies in primary breast tumors, but at higher frequencies in MBC. These studies also indicate that ESR1 mutations can be associated with shorter progression-free survival, and may need to be treated based on precise mutation type. However, in order to treat ESR1 mutations specifically and effectively, the mechanisms by which ESR1 mutations impact cancer progression must first be unraveled.
Biologic Mechanisms Associated with ESR1 Mutations
Fewer than 20% of MBC tumors lose ER expression, and ER initiates invasion and promotes metastasis [7]. In light of recently published studies demonstrating the high frequency of ESR1 mutations in MBC, we must understand how mutant ERs promote metastasis. Early studies on the K303R mutation revealed altered ER phosphorylation status [51]. This increased ER phosphorylation at several residues promotes interaction with growth factor signaling and enhanced binding of coactivators [51]. It is not known if a similar mechanism may present in the hot spot HBD mutations. Studies to date have focused on understanding the conformational changes created by mutations in the HBD.
The Y537 residue is important for ER dimerization and activation in response to ligand binding [84], which is required for ER to bind DNA and initiate gene transcription [54]. Structural analyses have suggested that both the Y537S and D538G mutations lead to a novel hydrogen bond with the mutated amino acid [58, 60, 85]. This changes the receptor to an agonist conformation even in the absence of ligand, resulting in increased coactivator recruitment, decreased recruitment of Hsp90 chaperone protein, and in one study, increased phosphorylation at the S118 ER residue [60]. MCF-7 cells transfected with mutant ER showed increased hormone-independent transcriptional activation, proliferation, xenograft growth in vivo, and a decreased response to selective estrogen receptor downregulators (SERDs) [58, 60, 85]. Of note, one of these studies observed an additional hydrogen bond in the backbone of the D538G mutated receptor and an apparently smaller overall conformational change than that seen in the Y537S mutation [60]. As the BOLERO-2 study noted different responses to treatment between these two mutations [69], this structural alteration could be therapeutically important. However, in order to fully understand how specific individual mutations affect patient response, it will be essential to detect and monitor them clinically.
Monitoring ESR1 Mutations In Vivo: a Moving Target
To answer the imperative question of how to best treat patients with ESR1 mutations, a reliable method to sequentially monitor the evolution and emergence of mutants during treatment and tumor progression must be established. Since ESR1 mutations are rarely detected in primary tumors, we must be able to ascertain when they appear during tumor progression, and monitor their emergence during treatment and metastasis to assess how to change therapeutic strategies.
Circulating tumor cells (CTCs) can be used to monitor changes in the genomic landscape of tumors during disease progression. Yu et al. [86] isolated CTCs from patient’s plasma and created stable in vitro cultures where they then characterize a number of ESR1 mutations. Though no ESR1 mutations were observed in primary tumors, they were detected in metastatic samples and, interestingly, in tumor cells grown under low estrogen conditions in vitro. CTCs from patients with active metastatic disease who had ER-positive primary tumors retained ER expression. The authors also tested in vitro response to a number of single and combination targeted therapies. They found that mutant-harboring CTC lines were relatively resistant to selective estrogen receptor modulators (SERMs), SERDs, and the mTOR inhibitor everolimus, but were uniquely sensitive to Hsp90 inhibition. Though it is sometimes difficult to isolate enough CTCs from patients to perform sequencing, this study demonstrates that CTCs can be cultured in vitro to test therapeutic sensitivity.
Isolating cfDNA from the plasma of breast cancer patients is an attractive alternative to isolating CTCs. Several groups have compared cfDNA to CTC DNA and have found that cfDNA faithfully recaptures the genomic landscape of both CTCs and the original tumor [61, 80]. Despite great intra- and intertumoral heterogeneity, ESR1 mutations are generally enriched or appear in serial biopsies in patients progressing over time, with little to no mutations detected in the primary tumor. However, when the mutation was present in the primary tumor, it was maintained in the metastasis [85].
As summarized in a recent review [87], utilizing serial plasma cfDNA samples as “liquid biopsies” to monitor ESR1 mutation progression is an exciting diagnostic option. Liquid biopsies can be obtained at primary diagnosis and through the course of treatment and progression to MBC, where tissue biopsies are often difficult or impossible to obtain. Subsequent liquid biopsies postsurgery, postadjuvant treatment, and during metastatic progression could then be analyzed by ddPCR for the emergence and enrichment of ESR1 mutations. This clinical scenario allows for the detection of ESR1 mutant subclones during their emergence in disease progression, different lines of treatment, and metastatic onset. This clinical paradigm will likely elucidate how ESR1 mutations’ impact tumor response to therapy and, importantly, determine how ESR1 mutations impact tumor evolution.
Evolution of ESR1 Mutations During Treatment Course
ESR1 mutant cells could either evolve during tumor progression due to the selective pressure of treatment or represent an initial small subset of cells that evade treatment, as represented in Fig. 1. Indeed, compared to other molecular alterations, ESR1 mutations appear to be a bona fide acquired mutation with the high discordant frequency between primary and metastatic tumors [62, 88]. In Fumagalli et al., ESR1 mutations were found in 10.8% of patients who had received at least 5 years of hormone therapy. They noted that of the many alternations (p35, PI3K, etc.) identified, only ESR1 exhibited discordance between primary and metastatic samples, indicating a unique role for ESR1 mutations in promoting tumor progression.
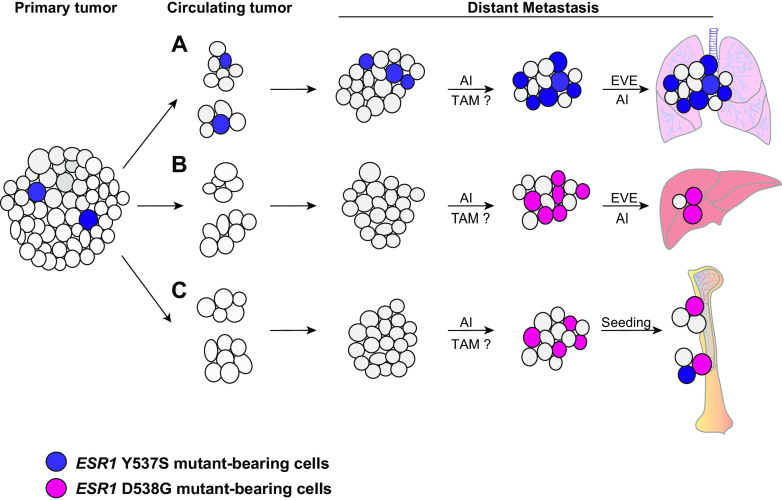
Fig. 1.
Observed patterns of clonal evolution in mutant-bearing tumors. A primary tumor contains few to no ESR1 mutant-bearing cells. However, it can seed clusters of circulating cells that eventually become mutant-expressing metastases through several different evolutionary patterns. As demonstrated in panel A, circulating cell clusters may bear the same mutation, here Y537S, as the primary tumor. When the circulating cell clusters colonize a distal organ and become a metastatic tumor, they may continue to express the mutation, even increasing in frequency. Treatment with an AI (and possibly a SERD such as Tam, though clinical evidence is lacking) in the metastatic setting may actually cause an increase in mutant frequency. As seen in the BOLERO-2 trial [69], combinatorial treatment in AI + Eve was not effective in Y537S-bearing tumors. In B, the primary tumor seeds circulating cell clusters which lack ESR1 mutations, even in the metastatic tumor. However, hormonal treatment may provide selective pressure for the emergence of ESR1 tumors in the metastatic setting. In this case the mutation is D538G, which does respond to AI + Eve treatment. In C, hormonal therapy again selects for expression of the D538G mutation, and the process of seeding for micrometastases selects for emergence of a second mutation.
It has long been appreciated that there is clonal heterogeneity within tumors. Miller et al. sought to define how this clonal heterogeneity could be remodeled by sequencing 22 primary tumors before and after neoadjuvant AI therapy [89]. They found that in three cases, AI therapy selected for the emergence and enrichment of ESR1 mutations. This small study underscores the concept that therapy can influence clonal evolution, and enrich for ESR1 mutations. More studies with greater power are needed to address the outstanding question: does the choice of endocrine agent influence clonal expansion of ESR1 mutations?
Although a proof-of-principle study using cfDNA to predict relapse in early breast cancer patients failed to note ESR1 mutations as having significant predictive value [90], another study successfully used cfDNA to track ESR1 mutant evolution [66]. They noted an enrichment in ESR1 clones only in MBC patients treated with AI exclusively in the metastatic setting. This evolutionary pattern was also observed in the BOLERO-2 trial [69] and in a preclinical study of ESR1 mutations in PDX tumors or in vitro cell lines [32]. This observation could have implications for the treatment of MBC patients, both those who present with ESR1 mutations in the primary setting, and to prevent the expansion of ESR1 mutations in the metastatic setting. These studies are likely the “tip of the iceberg,” as noted in Gu and Fuqua [87], and further underscore the importance of therapeutic monitoring.
ESR1 Mutations’ Response to Treatment
Preclinical studies have predicted that tumors bearing ESR1 HBD mutations will exhibit different responses to treatment compared to those with WT ER. In a retrospective study using ddPCR sequencing of cfDNA from patients with MBC, Takeshita et al. noted that the frequency of ER mutations changes over time and with treatment [75]. They observed that ESR1 mutations increased over time and with AI therapy, supporting the hypothesis that these clones may be adapting to AI therapeutic conditions. The presence of ESR1 mutations was correlated with shorter time to treatment failure; 83% of MBC patients with ESR1 mutations exhibited a poor response to and shorter duration of effective endocrine control.
The BOLERO-2 trial [69, 91] saw an important change in ESR1 mutation prevalence with respect to the timing of AI therapy. A threefold increase in ESR1 mutation frequency was seen in patients treated with an AI for the first time in the metastatic setting (33 vs. 11%), compared to those who were treated with an AI in the adjuvant setting. This confirmed what an earlier smaller study [90] had observed: mutations were rarely seen in patients after adjuvant AI, implying that treatment was enriching ESR1 mutations in the metastatic setting [66]. A similar observation was also made in the phase II FERGI trial, which enrolled women with MBC who had failed previous AI therapy. The FERGI trial added a pan-PI3K inhibitor to fulvestrant treatment and concluded that tumors bearing ESR1 mutations were rarely found before AI treatment but were prevalent in MBC that had progressed on AI therapy [74].
These critical studies highlight four points: (1) the need for mutation tracking through cfDNA in plasma, (2) the importance of the timing of AI treatment in the adjuvant vs. metastatic setting, and (3) the possibility that AI treatment alone in the metastatic setting may provide a positive selection for hormone-independent ESR1 mutations, although it has recently been reported that Tam-only-treated MBC can harbor mutations [92], and (4) the potential importance of fulvestrant for the effective treatment of MBC. It is of paramount importance to unravel these questions, ideally through long-term follow-up of patients with ESR1 mutations. These studies will likely reinforce the current standard of care for AI adjuvant therapy after primary detection but might indicate against the current practice of treating MBC patients with steroidal AI therapy alone. It also predicts the importance of alternate antiestrogen (fulvestrant and novel targeted agents) therapies in the treatment of MBC with ESR1 mutations.
SERM/SERDs in the Treatment of Tumors with Mutant ESR1
If AI treatment provides a selection pressure for mutant-bearing clonal outgrowth in patients, the next option for therapy is likely a more effective SERM and/or SERD. Various studies have predicted that ESR1 mutations will still respond to SERMs or SERDs, though perhaps at a decreased sensitivity based on in vitro, xenograft, and PDX preclinical models [32, 58, 60, 85]. A study using MCF-7 cells with CRISPR-Cas9 knock-in of the Y537S ESR1 mutation found that cells were partially resistant to Tam and fulvestrant [93]. Tam has never been shown to be more effective with higher dosing [94]; however, fulvestrant may [95, 96]. Fulvestrant at 500 mg is more effective in preventing progression in ER mutant-bearing tumors [95]. This indicates that high-dose fulvestrant may be a promising therapeutic option. It also highlights the need for the development of novel, more potent SERMs/SERDs.
The quest for better SERDs has recently taken into consideration how these novel SERDs might specifically address the clinical consequences of ESR1 mutations. Oral SERDs are an especially attractive alternative to fulvestrant, whose dose is limited due to the amount of drug feasible to inject intramuscularly. The oral SERD GDC-0810 was effective in in vitro and xenograft models. The authors specifically included an elegant CRISPR-Cas9 knock-in model of mutant ER Y537S [97] to test GDC-0810’s ability to inhibit mutant cell growth. GDC-0810 was able to competitively bind mutant ER. A phase I clinical trial to evaluate this promising drug is currently recruiting patients (NCT01823835). A second new oral SERD, termed AZD9496, significantly inhibited PDX growth expressing the D538G mutation [98]. In an inducible mutant-expressing cell line, AZD9496 was also able to downregulate mutant ER and block PR induction better than Tam or fulvestrant. AZD9496 is currently in a phase I trial (NCT02248090). Finally, combining an oral SERD with targeted therapy (PI3K pathway inhibitor or CDK4/6 inhibitor) showed increased inhibitory effects compared to monotherapy alone [98], underscoring the potential utility of combinatorial therapy in mutant patient populations.
Combination Therapy to Treat Tumors with ESR1 Mutations
An effective strategy to treat MBC patients with ESR1 mutations is to combine selected targeted therapy with hormone therapy. With studies showing ESR1 mutants’ decreased sensitivity to SERDs and SERMs, the need for alternative targets for therapy is immediate. Harrod et al. combined fasudil with a CDK7 inhibitor in an MCF-7 Y537S knock-in preclinical model [93]. This combination reversed initial resistance to single agent therapy, promoted complete growth suppression, and reduced phosphorylation of ER at S118. Using RNA-Seq, they found that mutant ER had a unique gene set compared to WT ER, including elevated CDK7 expression. They hypothesized that mutant-bearing tumors might be more sensitive to targeted therapy aimed at suppressing the activity of these super-induced genes. CDK7 inhibitors have not yet been tested in MBC.
The PALOMA-1 clinical trial combined the AI letrozole with the CDK4/6 inhibitor palbociclib and found a significant increase in progression-free survival with the combined treatment compared to letrozole alone. In a smaller study of 16 patients with ESR1 mutation-positive MCB treated with letrozole plus palbociclib, no difference in progression-free survival or overall survival was observed compared to WT tumors [71]. Unfortunately, this treatment regimen did not prevent an enrichment of ESR1 mutations during treatment. Wardell et al. utilized a novel SERM-SERD hybrid (SSH) in combination with palbociclib in in vitro and animal PDX models of ESR1-expressing resistant cells [99]. They found that both a SERD-palbociclib and an SSH-palbociclib combination effectively inhibited growth in these preclinical models. Clinical testing of novel SERM/SERD/SSHs in combination with palbociclib is currently underway (single dose SERD GDC-9010 plus palbociclib phase II NCT01823835; SSH bazedoxifene plus palbociclib phase II/I NCT02448771). Palbociclib is also being evaluated clinically in combination with hormonal therapy (letrazole or tamoxifen plus palbociclib phase II NCT03065621; tamoxifen plus palbociclib as first line metastatic therapy phase II NCT02668666; fulvestrant plus palbociclib as second line therapy after progression on AI plus palbociclib phase II NCT02738866).
Conclusions
Though the frequency and relevance of ESR1 mutations in breast cancer has been debated for nearly 20 years, recent studies clearly demonstrate that ESR1 mutations are present in primary and metastatic tumors, are enriched in the metastatic setting, and affect progression-free survival and response to hormone therapy. The frequency of ESR1 HBD mutations in primary tumors remains low and sometimes undetectable with traditional sequencing techniques. In MBC patients, ESR1 HBD frequencies range from 12 to 40%. Though some sequencing methods are very sensitive for detection of subclonal ESR1 mutations, the ddPCR technology of plasma DNA is efficient for detection of ESR1 mutations even at low frequencies. Thus, ESR1 mutation status can be monitored real-time throughout treatment and disease progression. Mutations are a bona fide, and probably the dominant, mechanism of acquired hormone resistance in patients; their influence on response to therapy thus warrants deep exploration.
If ongoing clinical trials confirm what preliminary studies have observed, ESR1 HBD mutations may warrant new clinical approaches to the management of MBC. These mutations appear to be enriched during treatment with an AI alone in MBC. This observation leads to three clinical possibilities which should be studied: (1) adjuvant treatment of primary tumors could include Tam or fulvestrant treatment sequencing to prevent enrichment of ESR1 mutations, (2) continual monitoring of ESR1 mutations during AI monotherapy, and (3) MBC patients bearing ESR1 mutations should not be treated with a steroidal AI alone. The unmet clinical need for better SERM/SERD/SSHs is clear, and more potent SERDs and SSHs are currently entering clinical trials. Another option for treating ESR1 mutations currently in clinical trials is the combination of hormonal therapy with targeted therapy, most promisingly fulvestrant plus palbociclib.
The long duration of recurrence risk in women with ER-positive breast cancer is recognized, and how best to precisely determine who should receive prolonged endocrine therapy would be of great value. The apparent heterogeneity of ER-positive breast cancer and the mechanisms of resistance dictate a paradigm shift toward translational research using liquid biopsy specimens from patients that progress during endocrine therapy in the metastatic setting. Though there is still much to be discovered about the role of ESR1 mutations in breast cancer, there is enough evidence to conclude that the detection of ESR1 mutations should now be considered an ancillary diagnostic test in patients with disease progression during AI treatment. Our continued understanding of the mechanisms behind ESR1 mutations’ effects on tumor progression and metastasis will be imperative to best arm women for the fight against their breast cancer.
Tam tamoxifen, AIs aromatase inhibitors, ER estrogen receptor, ESO-ESMO European School of Oncology-European School of Medical Oncology, HER2 human epidermal growth factor receptor 2, ESR1 estrogen receptor gene, WT wild-type, NGS next generation sequencing, PDX patient-derived xenograft, HBD hormone-binding domain, TCGA The Cancer Genome Atlas, FISH fluorescent in situ hybridization, ddPCR droplet digital polymerase chain reaction, PR progesterone receptor, DES diethylstilbestrol, cfDNA cell-free deoxyribonucleic acid, MBC metastatic breast cancer, CTCs circulating tumor cells, SERM selective estrogen receptor modulator, SERD selective estrogen receptor degrader, SSH SERM/SERD hybrid, IC50 half maximal inhibitory concentration
Funding Information
The study received funding from the National Institutes of Health NCI RO1 CA207270, Breast Cancer Research Foundation 16-056, National Institutes of Health NCI R01CA072038, and Cancer Prevention Research Institute of Texas RP150440.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Hull DF, 3rd, Clark GM, Osborne CK, Chamness GC, Knight WA, 3rd, McGuire WL. Multiple estrogen receptor assays in human breast cancer. Cancer Res. 1983;43(1):413–416. [PubMed] [Google Scholar]
- 2.Gutierrez MC, Detre S, Johnston S, Mohsin SK, Shou J, Allred DC, Schiff R, Osborne CK, Dowsett M. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol. 2005;23(11):2469–2476. doi: 10.1200/JCO.2005.01.172. [DOI] [PubMed] [Google Scholar]
- 3.Herman ME, Katzenellenbogen BS. Response-specific antiestrogen resistance in a newly characterized MCF-7 human breast cancer cell line resulting from long-term exposure to trans-hydroxytamoxifen. J Steroid Biochem Mol Biol. 1996;59(2):121–134. doi: 10.1016/S0960-0760(96)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Gluck S. Extending the clinical benefit of endocrine therapy for women with hormone receptor-positive metastatic breast cancer: differentiating mechanisms of action. Clin Breast Cancer. 2014;14(2):75–84. doi: 10.1016/j.clbc.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso F, Costa A, Norton L, Senkus E, Aapro M, Andre F, Barrios CH, Bergh J, Biganzoli L, Blackwell KL, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2) Breast. 2014;23(5):489–502. doi: 10.1016/j.breast.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso F, Costa A, Norton L, Senkus E, Aapro M, Andre F, Barrios CH, Bergh J, Biganzoli L, Blackwell KL, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2)dagger. Ann Oncol. 2014;25(10):1871–1888. doi: 10.1093/annonc/mdu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua SA. The role of estrogen receptors in breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2001;6(4):407–417. doi: 10.1023/A:1014782813943. [DOI] [PubMed] [Google Scholar]
- 8.Giguere V. Estrogen receptor mutations in breast cancer—an anticipated “rediscovery?”. Mol Endocrinol. 2014;28(4):427–428. doi: 10.1210/me.2014-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottardis MM, Robinson SP, Jordan VC. Estradiol-stimulated growth of MCF-7 tumors implanted in athymic mice: a model to study the tumoristatic action of tamoxifen. J Steroid Biochem. 1988;30(1–6):311–314. doi: 10.1016/0022-4731(88)90113-6. [DOI] [PubMed] [Google Scholar]
- 10.Gottardis MM, Jordan VC. Development of tamoxifen-stimulated growth of MCF-7 tumors in athymic mice after long-term antiestrogen administration. Cancer Res. 1988;48(18):5183–5187. [PubMed] [Google Scholar]
- 11.Wolf DM, Jordan VC. The estrogen receptor from a tamoxifen stimulated MCF-7 tumor variant contains a point mutation in the ligand binding domain. Breast Cancer Res Treat. 1994;31(1):129–138. doi: 10.1007/BF00689683. [DOI] [PubMed] [Google Scholar]
- 12.Levenson AS, MacGregor Schafer JI, Bentrem DJ, Pease KM, Jordan VC. Control of the estrogen-like actions of the tamoxifen-estrogen receptor complex by the surface amino acid at position 351. J Steroid Biochem Mol Biol. 2001;76(1–5):61–70. doi: 10.1016/S0960-0760(00)00143-6. [DOI] [PubMed] [Google Scholar]
- 13.Knowlden JM, Hutcheson IR, Barrow D, Gee JM, Nicholson RI. Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: a supporting role to the epidermal growth factor receptor. Endocrinology. 2005;146(11):4609–4618. doi: 10.1210/en.2005-0247. [DOI] [PubMed] [Google Scholar]
- 14.Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, Barrow D, Wakeling AE, Nicholson RI. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144(3):1032–1044. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 15.Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66(24):11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 16.Shaw LE, Sadler AJ, Pugazhendhi D, Darbre PD. Changes in oestrogen receptor-alpha and -beta during progression to acquired resistance to tamoxifen and fulvestrant (Faslodex, ICI 182,780) in MCF7 human breast cancer cells. J Steroid Biochem Mol Biol. 2006;99(1):19–32. doi: 10.1016/j.jsbmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Santen RJ, Song RX, Zhang Z, Kumar R, Jeng MH, Masamura S, Lawrence J, Jr, MacMahon LP, Yue W, Berstein L. Adaptive hypersensitivity to estrogen: mechanisms and clinical relevance to aromatase inhibitor therapy in breast cancer treatment. J Steroid Biochem Mol Biol. 2005;95(1–5):155–165. doi: 10.1016/j.jsbmb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Masri S, Hong Y, Wang X, Phung S, Yuan YC, Wu X. New experimental models for aromatase inhibitor resistance. J Steroid Biochem Mol Biol. 2007;106(1–5):8–15. doi: 10.1016/j.jsbmb.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodie A, Jelovac D, Macedo L, Sabnis G, Tilghman S, Goloubeva O. Therapeutic observations in MCF-7 aromatase xenografts. Clin Cancer Res. 2005;11(2 Pt 2):884s–888s. [PubMed] [Google Scholar]
- 20.Jelovac D, Macedo L, Handratta V, Long BJ, Goloubeva OG, Ingle JN, Brodie AM. Effects of exemestane and tamoxifen in a postmenopausal breast cancer model. Clin Cancer Res. 2004;10(21):7375–7381. doi: 10.1158/1078-0432.CCR-04-0565. [DOI] [PubMed] [Google Scholar]
- 21.Santen RJ, Song RX, Zhang Z, Kumar R, Jeng MH, Masamura A, Lawrence J, Jr, Berstein L, Yue W. Long-term estradiol deprivation in breast cancer cells up-regulates growth factor signaling and enhances estrogen sensitivity. Endocr Relat Cancer. 2005;12(Suppl 1):S61–S73. doi: 10.1677/erc.1.01018. [DOI] [PubMed] [Google Scholar]
- 22.Johnston SR, Martin LA, Head J, Smith I, Dowsett M. Aromatase inhibitors: combinations with fulvestrant or signal transduction inhibitors as a strategy to overcome endocrine resistance. J Steroid Biochem Mol Biol. 2005;95(1–5):173–181. doi: 10.1016/j.jsbmb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Piccart M, Hortobagyi GN, Campone M, Pritchard KI, Lebrun F, Ito Y, Noguchi S, Perez A, Rugo HS, Deleu I, et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2dagger. Ann Oncol. 2014;25(12):2357–2362. doi: 10.1093/annonc/mdu456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, Abadie-Lacourtoisie S, Eymard JC, Debled M, Spaeth D, et al. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J Clin Oncol. 2012;30(22):2718–2724. doi: 10.1200/JCO.2011.39.0708. [DOI] [PubMed] [Google Scholar]
- 25.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 29.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van’t Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 30.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197(4306):893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 31.Poste G, Fidler IJ. The pathogenesis of cancer metastasis. Nature. 1980;283(5743):139–146. doi: 10.1038/283139a0. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veeraraghavan J, Tan Y, Cao XX, Kim JA, Wang X, Chamness GC, Maiti SN, Cooper LJ, Edwards DP, Contreras A, et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilar H, Sole X, Bonifaci N, Serra-Musach J, Islam A, Lopez-Bigas N, Mendez-Pertuz M, Beijersbergen RL, Lazaro C, Urruticoechea A, et al. Biological reprogramming in acquired resistance to endocrine therapy of breast cancer. Oncogene. 2010;29(45):6071–6083. doi: 10.1038/onc.2010.333. [DOI] [PubMed] [Google Scholar]
- 36.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, Lebeau A, Terracciano L, Al-Kuraya K, Janicke F, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39(5):655–660. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 37.Tomita S, Zhang Z, Nakano M, Ibusuki M, Kawazoe T, Yamamoto Y, Iwase H. Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer Sci. 2009;100(6):1012–1017. doi: 10.1111/j.1349-7006.2009.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen KV, Ejlertsen B, Muller S, Moller S, Rasmussen BB, Balslev E, Laenkholm AV, Christiansen P, Mouridsen HT. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res Treat. 2011;127(2):345–355. doi: 10.1007/s10549-010-0984-y. [DOI] [PubMed] [Google Scholar]
- 39.Brown LA, Hoog J, Chin SF, Tao Y, Zayed AA, Chin K, Teschendorff AE, Quackenbush JF, Marioni JC, Leung S, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40(7):806–807. doi: 10.1038/ng0708-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moelans CB, Holst F, Hellwinkel O, Simon R, van Diest PJ. ESR1 amplification in breast cancer by optimized RNase FISH: frequent but low-level and heterogeneous. PLoS One. 2013;8(12):e84189. doi: 10.1371/journal.pone.0084189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roodi N, Bailey LR, Kao WY, Verrier CS, Yee CJ, Dupont WD, Parl FF. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. J Natl Cancer Inst. 1995;87(6):446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- 42.Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O'Connell P, et al. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60(15):4026–4029. [PubMed] [Google Scholar]
- 43.Herynk MH, Parra I, Cui Y, Beyer A, Wu MF, Hilsenbeck SG, Fuqua SA. Association between the estrogen receptor alpha A908G mutation and outcomes in invasive breast cancer. Clin Cancer Res. 2007;13(11):3235–3243. doi: 10.1158/1078-0432.CCR-06-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway K, Parrish E, Edmiston SN, Tolbert D, Tse CK, Geradts J, Livasy CA, Singh H, Newman B, Millikan RC. The estrogen receptor-alpha A908G (K303R) mutation occurs at a low frequency in invasive breast tumors: results from a population-based study. Breast Cancer Res. 2005;7(6):R871–R880. doi: 10.1186/bcr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway K, Parrish E, Edmiston SN, Tolbert D, Tse CK, Moorman P, Newman B, Millikan RC. Risk factors for breast cancer characterized by the estrogen receptor alpha A908G (K303R) mutation. Breast Cancer Res: BCR. 2007;9(3):R36. doi: 10.1186/bcr1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbasi S, Rasouli M, Nouri M, Kalbasi S. Association of estrogen receptor-alpha A908G (K303R) mutation with breast cancer risk. Int J Clin Exp Med. 2013;6(1):39–49. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Yamashita H, Toyama T, Omoto Y, Sugiura H, Hara Y, Haruki N, Kobayashi S, Iwase H. Estrogen receptor alpha mutation (A-to-G transition at nucleotide 908) is not found in different types of breast lesions from Japanese women. Breast Cancer. 2003;10(1):70–73. doi: 10.1007/BF02967628. [DOI] [PubMed] [Google Scholar]
- 48.Tebbit CL, Bentley RC, Olson JA, Jr, Marks JR. Estrogen receptor alpha (ESR1) mutant A908G is not a common feature in benign and malignant proliferations of the breast. Genes Chromosomes Cancer. 2004;40(1):51–54. doi: 10.1002/gcc.20017. [DOI] [PubMed] [Google Scholar]
- 49.Oesterreich S, Davidson NE. The search for ESR1 mutations in breast cancer. Nat Genet. 2013;45(12):1415–1416. doi: 10.1038/ng.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giordano C, Cui Y, Barone I, Ando S, Mancini MA, Berno V, Fuqua SA. Growth factor-induced resistance to tamoxifen is associated with a mutation of estrogen receptor alpha and its phosphorylation at serine 305. Breast Cancer Res Treat. 2010;119(1):71–85. doi: 10.1007/s10549-009-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barone I, Brusco L, Fuqua SA. Estrogen receptor mutations and changes in downstream gene expression and signaling. Clin Cancer Res. 2010;16(10):2702–2708. doi: 10.1158/1078-0432.CCR-09-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barone I, Cui Y, Herynk MH, Corona-Rodriguez A, Giordano C, Selever J, Beyer A, Ando S, Fuqua SA. Expression of the K303R estrogen receptor-alpha breast cancer mutation induces resistance to an aromatase inhibitor via addiction to the PI3K/Akt kinase pathway. Cancer Res. 2009;69(11):4724–4732. doi: 10.1158/0008-5472.CAN-08-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barone I, Iacopetta D, Covington KR, Cui Y, Tsimelzon A, Beyer A, Ando S, Fuqua SA. Phosphorylation of the mutant K303R estrogen receptor alpha at serine 305 affects aromatase inhibitor sensitivity. Oncogene. 2010;29(16):2404–2414. doi: 10.1038/onc.2009.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herynk MH, Hopp T, Cui Y, Niu A, Corona-Rodriguez A, Fuqua SA. A hypersensitive estrogen receptor alpha mutation that alters dynamic protein interactions. Breast Cancer Res Treat. 2010;122(2):381–393. doi: 10.1007/s10549-009-0580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64(24):9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- 56.Barone I, Catalano S, Gelsomino L, Marsico S, Giordano C, Panza S, Bonofiglio D, Bossi G, Covington KR, Fuqua SA, et al. Leptin mediates tumor-stromal interactions that promote the invasive growth of breast cancer cells. Cancer Res. 2012;72(6):1416–1427. doi: 10.1158/0008-5472.CAN-11-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57(7):1244–1249. [PubMed] [Google Scholar]
- 58.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D, et al. D538G mutation in estrogen receptor-alpha: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73(23):6856–6864. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 59.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, Ferrer-Lozano J, Perez-Fidalgo JA, Cristofanilli M, Gomez H, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guttery DS, Page K, Hills A, Woodley L, Marchese SD, Rghebi B, Hastings RK, Luo J, Pringle JH, Stebbing J, et al. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin Chem. 2015;61(7):974–982. doi: 10.1373/clinchem.2015.238717. [DOI] [PubMed] [Google Scholar]
- 64.Niu J, Andres G, Kramer K, Kundranda MN, Alvarez RH, Klimant E, Parikh AR, Tan B, Staren ED, Markman M. Incidence and clinical significance of ESR1 mutations in heavily pretreated metastatic breast cancer patients. Onco Targets Ther. 2015;8:3323–3328. doi: 10.2147/OTT.S92443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sefrioui D, Perdrix A, Sarafan-Vasseur N, Dolfus C, Dujon A, Picquenot JM, Delacour J, Cornic M, Bohers E, Leheurteur M, et al. Short report: Monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int J Cancer. 2015;137(10):2513–2519. doi: 10.1002/ijc.29612. [DOI] [PubMed] [Google Scholar]
- 66.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I, Lopez-Knowles E, Ribas R, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, Omoto Y, Iwase H. Droplet digital polymerase chain reaction assay for screening of ESR1 mutations in 325 breast cancer specimens. Transl Res. 2015;166(6):540–553. doi: 10.1016/j.trsl.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Wang T, Liu JH, Zhang J, Wang L, Chen C, Dai PG. A multiplex allele-specific real-time PCR assay for screening of ESR1 mutations in metastatic breast cancer. Exp Mol Pathol. 2015;98(2):152–157. doi: 10.1016/j.yexmp.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Voi M, Gnant M, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clatot F, Perdrix A, Augusto L, Beaussire L, Delacour J, Calbrix C, Sefrioui D, Viailly PJ, Bubenheim M, Moldovan C, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016;7(46):74448–74459. doi: 10.18632/oncotarget.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gyanchandani R, Kota KJ, Jonnalagadda AR, Minteer T, Knapick BA, Oesterreich S, Brufsky AM, Lee AV, Puhalla SL (2016) Detection of ESR1 mutations in circulating cell-free DNA from patients with metastatic breast cancer treated with palbociclib and letrozole. Oncotarget [DOI] [PMC free article] [PubMed]
- 72.Hrebien S, O’Leary B, Beaney M, Schiavon G, Fribbens C, Bhambra A, Johnson R, Garcia-Murillas I, Turner N. Reproducibility of digital PCR assays for circulating tumor DNA analysis in advanced breast cancer. PLoS One. 2016;11(10):e0165023. doi: 10.1371/journal.pone.0165023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma CX, Luo J, Naughton M, Ademuyiwa F, Suresh R, Griffith M, Griffith OL, Skidmore ZL, Spies NC, Ramu A, et al. A phase I trial of BKM120 (buparlisib) in combination with fulvestrant in postmenopausal women with estrogen receptor-positive metastatic breast cancer. Clin Cancer Res. 2016;22(7):1583–1591. doi: 10.1158/1078-0432.CCR-15-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, Omoto Y, Iwase H. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget. 2016;7(22):32504–32518. doi: 10.18632/oncotarget.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P, Bahreini A, Gyanchandani R, Lucas PC, Hartmaier RJ, Watters RJ, Jonnalagadda AR, Trejo Bittar HE, Berg A, Hamilton RL, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res. 2016;22(5):1130–1137. doi: 10.1158/1078-0432.CCR-15-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu D, Paoletti C, Gersch C, VanDenBerg DA, Zabransky DJ, Cochran RL, Wong HY, Toro PV, Cidado J, Croessmann S, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res. 2016;22(4):993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34(25):2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 79.Page K, Guttery DS, Fernandez-Garcia D, Hills A, Hastings RK, Luo J, Goddard K, Shahin V, Woodley-Barker L, Rosales BM, et al. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem. 2017;63(2):532–541. doi: 10.1373/clinchem.2016.261834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, Goddard KS, Hastings RK, Luo J, Ogle O, et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res. 2017;23(1):88–96. doi: 10.1158/1078-0432.CCR-16-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gelsomino L, Gu G, Rechoum Y, Beyer AR, Pejerrey SM, Tsimelzon A, Wang T, Huffman K, Ludlow A, Ando S, et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Treat. 2016;157(2):253–265. doi: 10.1007/s10549-016-3829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, Massard C, Levy C, Arnedos M, Lacroix-Triki M, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13(12):e1002201. doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McIntyre JB, Rambau PF, Chan A, Yap S, Morris D, Nelson GS, Kobel M. Molecular alterations in indolent, aggressive and recurrent ovarian low-grade serous carcinoma. Histopathology. 2017;70(3):347–358. doi: 10.1111/his.13071. [DOI] [PubMed] [Google Scholar]
- 84.Arnold SF, Notides AC. An antiestrogen: a phosphotyrosyl peptide that blocks dimerization of the human estrogen receptor. Proc Natl Acad Sci. 1995;92(16):7475–7479. doi: 10.1073/pnas.92.16.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu G, Fuqua SA. ESR1 mutations in breast cancer: proof-of-concept challenges clinical action. Clin Cancer Res. 2016;22(5):1034–1036. doi: 10.1158/1078-0432.CCR-15-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fumagalli D, Wilson TR, Salgado R, Lu X, Yu J, O’Brien C, Walter K, Huw LY, Criscitiello C, Laios I, et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann Oncol. 2016;27(10):1860–1866. doi: 10.1093/annonc/mdw286. [DOI] [PubMed] [Google Scholar]
- 89.Miller CA, Gindin Y, Lu C, Griffith OL, Griffith M, Shen D, Hoog J, Li T, Larson DE, Watson M, et al. Aromatase inhibition remodels the clonal architecture of estrogen-receptor-positive breast cancers. Nat Commun. 2016;7:12498. doi: 10.1038/ncomms12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 91.Fuqua SA, Rechoum Y, Gu G. ESR1 mutations in cell-free DNA of breast cancer: predictive “tip of the iceberg”. JAMA Oncol. 2016;2(10):1315–1316. doi: 10.1001/jamaoncol.2016.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartels S, Christgen M, Luft A, Persing S, Jodecke K, Lehmann U, Kreipe H (2017) Estrogen receptor (ESR1) mutation in bone metastases from breast cancer. Mod Pathol [DOI] [PubMed]
- 93.Harrod A, Fulton J, Nguyen VT, Periyasamy M, Ramos-Garcia L, Lai CF, Metodieva G, de Giorgio A, Williams RL, Santos DB et al (2016) Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer. Oncogene [DOI] [PMC free article] [PubMed]
- 94.Tormey DC, Lippman ME, Edwards BK, Cassidy JG. Evaluation of tamoxifen doses with and without fluoxymesterone in advanced breast cancer. Ann Intern Med. 1983;98(2):139–144. doi: 10.7326/0003-4819-98-2-139. [DOI] [PubMed] [Google Scholar]
- 95.Paoletti C, Larios JM, Muniz MC, Aung K, Cannell EM, Darga EP, Kidwell KM, Thomas DG, Tokudome N, Brown ME, et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol Oncol. 2016;10(7):1078–1085. doi: 10.1016/j.molonc.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bardia A, Iafrate JA, Sundaresan T, Younger J, Nardi V. Metastatic breast cancer with ESR1 mutation: clinical management considerations from the Molecular and Precision Medicine (MAP) Tumor Board at Massachusetts General Hospital. Oncologist. 2016;21(9):1035–1040. doi: 10.1634/theoncologist.2016-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Joseph JD, Darimont B, Zhou W, Arrazate A, Young A, Ingalla E, Walter K, Blake RA, Nonomiya J, Guan Z et al (2016) The selective estrogen receptor downregulator GDC-0810 is efficacious in diverse models of ER+ breast cancer. eLife 5 [DOI] [PMC free article] [PubMed]
- 98.Weir HM, Bradbury RH, Lawson M, Rabow AA, Buttar D, Callis RJ, Curwen JO, de Almeida C, Ballard P, Hulse M, et al. AZD9496: an oral estrogen receptor inhibitor that blocks the growth of ER-positive and ESR1-mutant breast tumors in preclinical models. Cancer Res. 2016;76(11):3307–3318. doi: 10.1158/0008-5472.CAN-15-2357. [DOI] [PubMed] [Google Scholar]
- 99.Wardell SE, Ellis MJ, Alley HM, Eisele K, VanArsdale T, Dann SG, Arndt KT, Primeau T, Griffin E, Shao J, et al. Efficacy of SERD/SERM hybrid-CDK4/6 inhibitor combinations in models of endocrine therapy-resistant breast cancer. Clin Cancer Res. 2015;21(22):5121–5130. doi: 10.1158/1078-0432.CCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]