Abstract
Most cases of Escherichia coli meningitis develop as a result of hematogenous spread, but it is not clear how circulating E. coli crosses the blood-brain barrier. A TnphoA mutant of E. coli K1 RS218 was shown to be significantly less invasive than its parent strain in bovine and human brain microvascular endothelial cells (BMEC), which constitute the blood-brain barrier. More importantly, traversal of the blood-brain barrier was significantly less with this mutant than with the parent strain in newborn rats with experimental hematogenous meningitis. A DNA segment containing the TnphoA insertion site was cloned from RS218, and the cloned DNA complemented the TnphoA mutant in invasion of BMEC. Nucleotide sequence revealed a near identity to that of a hypothetical yijP gene (also called f577) in the E. coli K-12 genome. Sequence analysis indicated that the E. coli K1 yijP gene likely encodes a 66.6-kDa membrane protein. Deletion and complementation experiments indicated that the yijP gene was involved in E. coli K1 invasion of BMEC, i.e., the invasive ability of E. coli K1 was significantly reduced after yijP was deleted and was restored by complementation with a plasmid containing the yijP open reading frame. This is the first demonstration that the yijP gene locus plays a role in the pathogenesis of E. coli K1 meningitis.
Escherichia coli is the most common gram-negative microorganism causing meningitis in the neonatal period (19, 26). Most cases of E. coli meningitis occur as a result of hematogenous spread (14), but it is unclear how circulating E. coli crosses the blood-brain barrier. The entry of circulating E. coli into the central nervous system (CNS) is most likely to occur at sites of the blood-brain barrier, which is composed of brain microvascular endothelial cells (BMEC).
To study the mechanism of E. coli translocation from blood to the CNS, we have developed both in vitro and in vivo models of the blood-brain barrier. The in vitro model of the blood-brain barrier was established by isolation and cultivation of BMEC (12, 22, 23). The resulting BMEC exhibited transendothelial electrical resistance of 100 to 600 Ω/cm2 (15, 20), a unique property of the brain microvascular endothelial monolayer compared to systemic vascular endothelium. The in vivo model of E. coli meningitis was established by induction of hematogenous meningitis in 5-day-old rats (14). In this experimental meningitis model, bacteria are injected via subcutaneous or intracardiac injection, resulting in bacteremia and subsequent entry into the CNS. Using these models of the blood-brain barrier, we have shown that invasion of BMEC is a requirement for E. coli K1 penetration of the blood-brain barrier in vivo (12). In an attempt to identify E. coli K1 structures contributing to invasion of BMEC, we initially searched for any homology with the known proteins involved in invasion of eukaryotic cells by meningitis causing bacteria. We identified that E. coli outer membrane protein A (OmpA) has a sequence homology with Neisseria Opa proteins (18), which have been shown to be involved in invasion of eukaryotic cells (30). We have subsequently shown that E. coli OmpA contributes to invasion of BMEC (18). Our other approach to identify E. coli structures contributing to invasion of BMEC was by use of the transposon TnphoA. These investigations have identified two ibe loci, ibeA (previously named ibe10 [12]) and ibeB (11), which are located at different sites in E. coli K1 chromosome.
In our previous TnphoA mutagenesis of E. coli K1 RS218, which is a cerebrospinal fluid (CSF) isolate from a neonate with meningitis, we identified another mutant named 23A-20, which possessed OmpA, IbeA, and IbeB but exhibited significantly less invasion into BMEC in vitro compared to the parent strain (12). In the present study we identified, by cloning and characterizing the TnphoA insertion site of the mutant 23A-20, that the yijP locus contributes to E. coli traversal across the blood-brain barrier.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains, plasmid vectors, and their relevant characteristics are described in Table 1. Strain 23A-20 is a TnphoA insertion mutant of E. coli K1 E44 as described previously (12). Plasmids constructed in this study are illustrated in Fig. 1 or described in the text. E. coli strains were cultured at 37°C in Luria broth (LB; 1% tryptone, 0.5% yeast extract, 0.5% NaCl), brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.), or a synthetic medium (SM) based on M9 (21) supplemented with thiamine and nicotinamide (10 μg of each per ml) and pyruvate (10 mM) as a carbon source. Columbia agar with 5% sheep blood (Remel, Lenexa, Kans.) was used. When necessary, the medium was supplemented with ampicillin (Ap; 100 μg/ml), kanamycin (Km; 40 μg/ml), chloramphenicol (Cm; 25 μg/ml), or rifampin (Rif; 50 μg/ml).
TABLE 1.
Bacterial strains and plasmid vectors used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| RS218 | O18:K1:H7 | 1 |
| E44 | Rifr of RS218 | 11 |
| 23A-20 | E44(yijP::TnphoA) | This study |
| D12 | E44(ΔyijP) | This study |
| DH5α | F′ recA1 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 recA1 deoR Δ(lacZYA-argF) U169 (θ 80lacZM15) | Gibco-BRL |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc:: Mu (λpir) pro endA hsdA hsdR supF | 25 |
| Vectors | ||
| pUC13 | Apr; lacZ′ | 27 |
| pCVD442 | Apr; sacB oriV oriT | 10 |
| pCVD433 | Tcr (pACYC184 with a MluI linker) | 9 |
| pIB307 | Cmr (temperature-sensitive pSC101 derivative) | 6 |
| pK194 | Kmr; lacZ′ (pACYC184 derivative) | 13 |
FIG. 1.
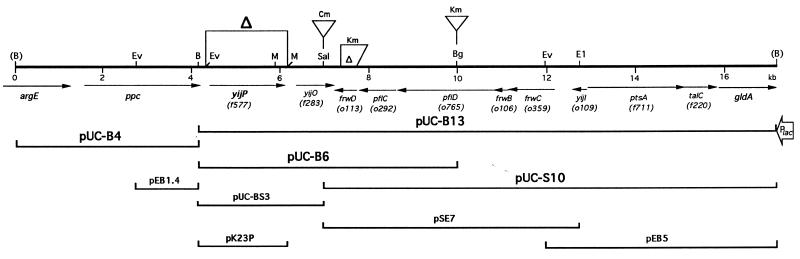
Gene organization of the cloned DNA and the physical maps of plasmid derivatives. All of these DNA fragments were cloned on pUC13, except pK23P, which was cloned on pK194. Restriction sites: B, BamHI; (B), BamHI from the vector; Bg, BglII; E1, EcoRI; Ev, EcoRV; M, MluI; Sal, SalI. Only the sites involved in plasmid construction are shown. The orientation of the lacZ promoter in the vector is indicated. Deletion (Δ) and antibiotic cassette insertion sites in the E44 chromosome are also depicted. The functions of the ORFs with alternative names are unknown.
Tissue cultures and invasion assays.
BMEC were prepared from bovine and human brains (22, 23), and invasion assays were performed as previously described (11). The primary bovine BMEC were used, and the human BMEC were immortalized by transfection with simian virus 40 large T antigen (24). In invasion assays, bacteria were added to confluent monolayers of BMECs with a multiplicity of infection of 100. The monolayers were incubated for 1.5 h at 37°C to allow invasion to occur. The number of intracellular bacteria was determined on blood agar after extracellular bacteria were killed by incubation of the monolayers with experimental medium containing gentamicin (100 μg/ml). Results were expressed either as the percent invasion ([number of intracellular bacteria recovered/number of bacteria inoculated] × 100) or as the relative invasion (invasion as a percentage of the parent E. coli K1 strain).
Neonatal rat model of hematogenous E. coli K1 meningitis.
The TnphoA insertion mutant 23A-20 was examined for its ability to enter the CNS by using our newborn rat model of experimental hematogenous E. coli meningitis as described previously (14). Briefly, at 5 days of age, all members of each litter were randomly divided into two groups to receive, via intracardiac injection, 6.2 × 106 CFU of the parent strain E44 or 1.1 × 107 CFU of the mutant strain 23A-20. Our pilot experiments revealed that these bacterial inocula for strains E44 and 23A-20 produced bacteremia of 105 to 108 CFU/ml of blood in almost 100% of the infected animals within 2 h of inoculation. This level of bacteremia has been shown to be sufficient for allowing circulating E. coli to enter the CNS (13). At 1 to 2 h after bacterial inoculation, blood and CSF specimens were collected for quantitative cultures. Blood and CSF specimens obtained from animals infected with 23A-20 were cultured in BHI with Km.
Molecular cloning.
Standard molecular techniques (21) were used unless otherwise specified. MluI-digested genomic DNA from the mutant 23A-20 was cloned in the same site of pCVD433 as described (9). Kmr transformants were selected, and a plasmid containing an extra 2-kb insert was purified and partially sequenced by using the 5′ primer Tnp5 and 3′ primer Tnp3, which are complementary to the ends of TnphoA (12). Plasmid DNA was prepared by using the Miniprep Spin Column (Qiagen, Valencia, Calif.). Nucleotide sequencing was carried out with the Applied Biosystems automatic sequencer (Foster City, Calif.).
The genomic library of E. coli K1 RS218 was constructed by using the LambdaGEM-12 system (Promega, Madison, Wis.) according to the manufacturer’s instructions. The library was screened with 32P-labeled PCR DNA containing the TnphoA insertion site. The recombinant λ DNAs were isolated from the positive plaques and analyzed by restriction digestion. The cloned DNA was subcloned in pUC13 as illustrated in Fig. 1, except for pK23P, which contains only the yijP open reading frame (ORF) in the vector pK194. This plasmid was constructed as follows: a 325-bp PCR fragment was generated between two MluI sites at the 3′ end of yijP (Fig. 1), in which the second MluI was converted to PstI through primer design. This MluI-PstI PCR DNA was used to replace the 1.2-kb MluI-PstI fragments in pUC-BS3 to produce the pUC23P. A sequencing reaction was performed to verify the 3′ end of yijP in pUC23P. The 2-kb BamHI-PstI fragment containing yijP ORF was then transferred to pK194 at the same sites to produce pK23P.
Complementation analysis.
The recombinant plasmids were introduced into E44 or its yijP mutant derivatives by electroporation as previously described (29). Transformants were tested for their invasion in BMEC. The assays were repeated at least three times, each time in triplicate wells. Results were expressed as relative invasion by comparing to the invasion of the parent strain E44 containing the vector.
Construction of yijP deletion mutant in E. coli.
The EcoRV-to-MluI fragments of 1.85 kb containing the entire yijP ORF was deleted from pUC-BS3 (termed pBS-Δibe) (Fig. 1). The 1.1-kb BamHI-HindIII fragment containing the yijP-deleted DNA was cleaved from pBS-Δibe and inserted into the HindIII-BamHI sites in pEB1.4 (Fig. 1). This recombinant 2.55-kb yijP-deleted DNA was cleaved with HindIII-EcoRI and cloned in the suicide plasmid pCVD442 at the SmaI site (termed pCVD-Δibe). The yijP gene was then deleted from E44 chromosome by the method described earlier (10). Briefly, pCVD-Δibe was transferred into E44 by conjugation with SM10λpir as the host strain. Apr Rifr transconjugants were selected and grown in LB without antibiotics at 32°C for 6 h and then diluted and plated on LB containing no NaCl and 5% sucrose at 30°C. The sucrose-resistant Aps Rifr colonies were selected and tested for the deletion of yijP by PCR as described previously (29) by using the TaqPlus Long PCR System (Stratagene, La Jolla, Calif.). E44 DNA, pCVD-Δibe, and an Apr Rifr transconjugant colony were used as control templates in such PCR reactions. The primers used were: PPC-C (5′-CAGAGTCTATTCAGCTAC) located at the end of the ppc gene, F283-R (5′-AGATAGCTGACGTCGTGA) located at the beginning of f283, and 23A3R (5′-CAGGCTGAAATACTGGCT) and 23A-5 (5′-GCCAGCCTGATTAAAGAC), both of which were located in the middle of yijP.
Construction of antibiotic cassette insertion mutants.
To interrupt the f283 gene, the 1.2-kb AccI-Bst31 fragment containing the Cmr gene was cleaved from pACYC184 and inserted at the SalI site in pUC-B6. The DNA was subcloned in pCVD442 and then transferred to E44 by conjugation, selecting for Cmr Rifr and Aps transconjugants. To completely delete the ORF o113, a 1-kb DNA fragment was amplified from upstream of o113, and another from downstream of o113 by using the Pfu DNA polymerase (Stratagene). These two fragments were cloned in pBluescript II KS(+) (Stratagene) and then transferred to the temperature-sensitive plasmid pIB307. A 1.2-kb Kmr cassette was isolated from pUC-4K (Pharmacia) and inserted in the middle of the o113-deleted DNA to obtain pIB-o113K. The plasmid was transformed into E44 by electroporation, selecting for Kmr Cms colonies at 40°C. To inactivate the ORF o765, the 6-kb EcoRI fragment from pUC-S10 containing o765 was subcloned in pACYC184 at the same site selecting for Tcr Cms. The 1.2-kb Kmr cassette from pUC-4K was inserted at the BglII site in the o765 ORF. This 7.2-kb recombinant DNA was subcloned in pIB307 and transformed into E44 as described above. All these insertion and/or deletion mutants were verified by PCR by using primers flanking the antibiotic-cassette-inserted sites.
Characterization of E. coli mutants.
E. coli RS218 (and E44) possessed both S and type 1 fimbriae, and the expression of S and type 1 fimbriae was tested by hemagglutination as described previously (18). K1 capsule was detected on an antiserum agar plate as described previously (14). E. coli mutant strains were examined by the Vitek Gram-Negative Identification+ Card and the automatic Vitek system for antimicrobial susceptibilities (bioMérieux Vitek, Inc., Hazelwood, Mo.). E. coli proteins were fractionated by differential centrifugation and examined on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels as previously described (18, 29).
Nucleotide sequence accession number.
The sequence of yijP from E. coli K1 RS218 has been submitted to the GenBank database under accession number AF112861.
RESULTS
Isolation and characterization of the mutant strain 23A-20 and its invasion in BMEC.
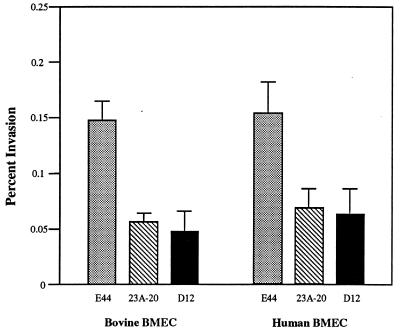
We have identified a TnphoA insertion mutant strain 23A-20 which exhibited significantly less invasive ability in bovine BMEC compared to the parent strain E44 (12). Southern hybridization with EcoRV- or MluI-cleaved genomic DNA of 23A-20 indicated that there was only one copy of TnphoA inserted in the chromosome (not shown). The mutant 23A-20 was found to be positive in the K1 capsule, S fimbriae, and type 1 fimbriae. Repeated in vitro invasion assays in both bovine and human BMECs showed that the mutant 23A-20 was significantly less invasive than E44 in both BMEC cell lines (Fig. 2).
FIG. 2.
Invasion frequencies of the E. coli K1 parent strain E44 and two yijP mutants in BMEC. Values are means of at least three independent assays; bars indicate the standard error of mean.
When E44 and 23A-20 were mixed 1:1 for invasion assays, the ratio of intracellular bacteria recovered were about 2.4:1 as determined by the percentage of Kmr colonies in the mixture of recovered intracellular 23A-20 (Kmr Rifr) and E44 (Rifr). The result was consistent with that of individual invasion assays (Fig. 2).
To test whether 23A-20 was indeed less invasive in vivo, the mutant 23A-20 and the parent strain E44 were administered via intracardiac injection to 5-day-old rats. Table 2 showed that 23A-20 was able to induce bacteremia in rats to a degree similar to that of E44. The rate of meningitis, defined as positive CSF cultures, induced by 23A-20 was, however, significantly less (P = 0.019) than that by E44. These data showed that the mutant strain 23A-20 was significantly less invasive in BMEC both in vitro and in vivo, indicating that the TnphoA inserted region may be important for E. coli K1 invasion of BMEC.
TABLE 2.
Comparison of bacterial counts in blood and number of animals with positive CSF culture between two groups of animals receiving E. coli K1 E44 or its TnphoA mutant 23A-20
| Strain (n = 24) | Bacteremia (mean log CFU/ml of blood ± SD) | No. of animals with positive CSF (%) |
|---|---|---|
| E44 | 7.53 ± 0.40 | 18 (75)a |
| 23A-20 | 7.80 ± 0.67 | 10 (42)a |
P = 0.019 by Fisher’s exact test.
Cloning of the TnphoA insertion region.
The TnphoA fragment in 23A-20 was cloned in pCVD433 in E. coli DH5α by selecting for the Kmr phenotype. A selected plasmid clone was purified, and DNA flanking the TnphoA insertion site was sequenced. A sequence of ca. 800 nucleotides was obtained and was used to search against databases by using the BLAST program (2). The sequence was found to be almost identical to an ORF named yijP (also called f577 [4, 5]), which is located at 90 min of the E. coli K-12 chromosome and encodes a hypothetical protein. The TnphoA insertion site was identified to be in the middle of yijP.
A 700-bp PCR DNA containing the TnphoA insertion site was synthesized and used to probe the genomic library of RS218. Several positive plaques were obtained, and a recombinant λ clone containing a 17-kb insert was selected for further study (Fig. 1). The cloned DNA was mapped with enzymes BamHI, BglII, EcoRI, HindIII, KpnI, SalI, and SmaI. Most of these restriction sites appeared to be identical to those in E. coli K-12 DNA, except that a SalI site in yijP and a KpnI site in o765 were missing in E. coli K1 RS218 compared with the E. coli K-12 genome (Fig. 1) (5). These results indicated that the cloned 17-kb segment from E. coli K1 RS218 was very similar to that in E. coli K-12. A sequencing reaction was performed from either end of this 17-kb segment, respectively. Database searching confirmed the presence of the argE gene and the gldA gene at either end of the cloned DNA (Fig. 1).
Complementation of 23A-20 for invasion of BMEC.
Two BamHI fragments from the recombinant λ DNA were first subcloned in pUC13 at the same site and named pUC-B4 and pUC-B13 (Fig. 1). All other deletions were later generated from these two plasmids. The direction of the yijP gene was opposite the lac promoter on the vector in pUC-B13; the plasmid containing the insert on the other direction was unstable in E. coli.
Most of the plasmids shown in Fig. 1 were transferred into the mutant 23A-20 or E44. Transformants were tested for invasion in BMEC together with E44(pUC13) and 23A-20(pUC13) as controls. As shown in Table 3, pUC-B6 and pUC-B13 were able to complement the mutant 23A-20 in our invasion assays to the level observed with the parent strain E44(pUC13). Of interest, pUC-B6 and pUC-B13 increased the BMEC invasion of the parent strain containing the vector by approximately 300%. All other constructs did not complement 23A-20, except that pUC-S10 appeared to increase the invasion of the mutant 23A-20 compared to 23A-20(pUC13), but the degree of complementation varied with relative invasion, ranging from 50 to 175%. These results indicated that the ORF f577 played a role in E. coli K1 invasion of BMEC, but the possible involvement of f283, o113, and o292 in E. coli invasion of BMEC was not excluded. Plasmid pUC-BS3 affected the growth of its host E. coli strain more than other plasmids, and complementation of 23A-20 with pUC-BS3 was not successful.
TABLE 3.
Relative invasion of E. coli K1 containing various plasmid constructs
| Strain | Plasmid | % Relative invasion (mean ± SD) |
|---|---|---|
| E44 | pUC13 | 100 |
| pUC-B13 | 355 ± 86 | |
| pUC-B6 | 273 ± 73 | |
| 23A-20 | pUC13 | 23 ± 6.2 |
| pUC-B13 | 156 ± 37 | |
| pUC-B6 | 135 ± 33 | |
| pUC-S10 | 109 ± 49 | |
| pSE7 | 6.5 ± 4.9 | |
| pEB5 | 30 ± 16 |
Nucleotide sequence of yijP.
The nucleotide sequence of yijP in pUC-B6 was determined. Compared to its counterpart in E. coli K-12 (GenBank accession number U00006), 40 of 1,731 nucleotides differed between these two ORFs (97.7% identities). However, only 5 of 577 deduced amino acid residues were different between them. These differences were located near the middle of the deduced YijP protein (Asn377 [K-12 strain] versus Lys [K1 strain], Asn403 versus Asp, Asp412 versus Glu, Ala450 versus Thr, and Asn451 versus Asp).
The sequence downstream of E. coli K1 yijP was not determined. According to the E. coli K-12 sequence, there are several genes of unknown function present downstream of yijP (Fig. 1). The deduced f283 product belonged to the AraC/XylS family of transcriptional regulators (5). The five-gene cluster, o113-o359, appeared to form an operon. Their predicted products were 23 to 47% similar to E. coli pyruvate formate catabolic enzymes (5).
Deletion and insertion mutagenesis in the E44 chromosome.
To further clarify the function of yijP and its downstream genes, an isogenic E44(ΔyijP) mutant was generated, and one of the colonies, named D12, was chosen for further study. PCR reactions with three pairs of primers indicated that yijP was as expected deleted in the D12 strain (not shown).
The growth characteristics of E44 and the ΔyijP mutant were the same in all of the media used (see Materials and Methods). Mutation in the ppc gene, which is upstream of yijP, was reported to render E. coli cells unable to grow on several carbon sources such as pyruvate and glutamate (3). However, the ΔyijP mutant was found to be able to grow on these carbon sources. The doubling time of the ΔyijP mutant in SM supplemented with pyruvate (10 mM) as the sole carbon source was about 70 min, identical to that observed with E44. Like E44, the ΔyijP mutant expressed both S fimbriae and type 1 fimbriae and possessed the K1 capsule. The fractionated protein profiles were identical between E44 and the ΔyijP mutant in the Coomassie blue-stained SDS-polyacrylamide gel (not shown), suggesting that YijP was a minor protein. The biochemical reactions were identical between E44 and the ΔyijP mutant on the Vitek Gram-Negative Identification+ Card. The antimicrobial susceptibility profile of these strains was also identical in the automatic Vitek system. Viability of D12 and 23A-20 cells was unchanged in water containing 0.5% Triton X-100 for 2 h, which was much longer than the exposure time to Triton X-100 (∼10 min) used in the in vitro BMEC invasion assay.
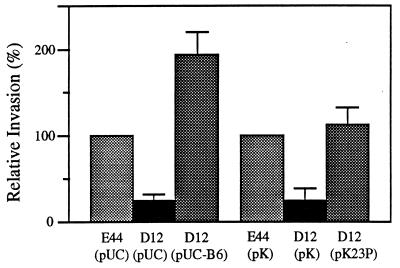
Results of invasion assays showed that deletion of yijP in E44 reduced the BMEC invasion frequency to the level of the TnphoA mutant 23A-20 (Fig. 2; P < 0.05 compared to the parent strain in both bovine and human BMECs). Plasmid pUC-B6 was able to complement D12 in invasion of BMEC compared to E44(pUC13) (Fig. 3). Furthermore, a low-copy-number plasmid pK194 containing only the yijP ORF was constructed (pK23P; Fig. 1), and this plasmid fully complemented the ΔyijP mutant in invasion of BMEC (Fig. 3).
FIG. 3.
Complementation of the ΔyijP mutant with the plasmids pUC-B6 and pK23P in the invasion of BMEC. Values are means of four separated experiments; bars indicate the standard error of the mean.
To study the possible involvement of the genes downstream of yijP in E. coli invasion of BMEC, f283, o113, and o765 were selectively interrupted by antibiotic cassette insertion (Fig. 1). The invasion frequency of these mutants in BMEC was found to be identical to that of E44, indicating that the downstream genes of yijP were probably not involved in E. coli K1 invasion of BMEC.
DISCUSSION
The most distressing aspect of bacterial meningitis is the limited improvement in the mortality and morbidity attributable to advances in antimicrobial chemotherapy and supportive care. A major contributing factor is our incomplete understanding of the pathogenesis associated with this disease. For example, at present it is unclear how circulating bacteria cross the blood-brain barrier. Our investigations of this issue with E. coli as a paradigm have shown that successful traversal of E. coli across the blood-brain barrier requires two independent steps of bacterium-BMEC interactions, i.e., E. coli binding to BMEC and the invasion of BMEC. We have previously shown that S fimbriae contribute to E. coli binding to BMEC (23); however, binding to BMEC via S fimbriae was not accompanied by invasion of BMEC. In contrast, we have shown that OmpA, IbeA, and IbeB contribute to E. coli K1 invasion of BMEC (11, 12, 18). The ibeA gene was found to be unique in CSF isolates of E. coli K1, while ompA and ibeB have homologues present in the E. coli K-12 genome. OmpA is functionally similar between E. coli K1 and K-12 strains, as shown by successful complementation of the noninvasive ompA deletion mutant of E. coli K1 to invade BMEC by the E. coli K-12 ompA gene (18). In addition, we have identified specific receptors for both OmpA (17) and IbeA (16) present only on BMEC, suggesting that OmpA and IbeA contribute to BMEC invasion via ligand-receptor interactions. IbeB displays the characteristics of outer membrane proteins (11), suggesting that the IbeB-mediated invasion of BMEC may also occur via ligand-receptor interactions. In addition, we have previously shown in the experimental hematogenous E. coli meningitis model that the K1 capsule is a critical determinant for E. coli to cross the blood-brain barrier as live bacteria (14).
In the present study, we identified a TnphoA mutant 23A-20 that is less capable of invasion into BMEC in vitro. More importantly, this mutant was significantly less invasive into the CNS in vivo than the parent strain in the newborn rat model of hematogenous meningitis. These data suggested that a genetic locus in the mutant 23A-20 affected by TnphoA insertion was most likely to confer the ability of E. coli K1 to invade BMEC both in vitro and in vivo. Both the parent strain R218 (or E44) and its mutant 23A-20 possess the K1 capsule, OmpA, IbeA, and IbeB, as well as type 1 and S fimbriae. These findings suggested that a less-invasive property of 23A-20 was unlikely resulted from a polar effect of TnphoA on other known invasion genes. This concept was supported by our demonstration that (i) the yijP-deleted mutant of RS218 (strain D12) was less invasive in BMEC and that (ii) plasmids pUC13 and pK194 containing the yijP gene were able to complement the yijP mutants in the invasion of BMEC to the level of the parent strain (e.g., pUC-B6 and pK23P; Table 2 and Fig. 3). In contrast, other plasmids without the yijP gene were not able to restore the ability of 23A-20 to invade BMEC. In addition, selective interruption of other genes downstream of yijP by antibiotic cassette insertion did not reduce the invasion frequency of the parent strain. These findings indicated that yijP is an important component for E. coli K1 invasion of BMEC.
The nucleotide sequence and the deduced amino acid sequence of yijP were found to be almost identical to those of yijP in the E. coli K-12 genome, whose function is currently unknown. Analysis of the deduced amino acid sequence of YijP showed that it has some features of outer membrane protein, including a signal peptide-like sequence and five or six transmembrane segments at its N terminus (28). Comparison of the outer membrane protein profiles revealed that the parent strain and the ΔyijP mutant had identical patterns, suggesting that YijP is a minor protein. It is unclear how YijP contributes to E. coli K1 invasion of BMEC. We have previously shown that E. coli OmpA, a highly conserved outer membrane protein, contributes to E. coli K1 invasion of BMEC, and its N-terminal portion is involved in invasion (18). We have recently shown that IbeA interacts with a novel BMEC protein for E. coli K1 invasion of BMEC (16). Studies are in progress to examine whether the invasion phenotype is a result of direct interaction of YijP with BMEC and also to determine which domain(s) of YijP is involved in BMEC invasion.
During the preparation of the manuscript, a YijP homologue, termed L7028 (8) or ecf3 (7), was reported in the 92-kb virulence plasmid pO157 in E. coli O157:H7. The product of this E. coli O157 gene shares 82% identities with YijP, and it is predicted to have a similar secondary structure, i.e., a signal peptide-like sequence and five or six transmembrane segments in its N-terminal domain (28). pO157 is known to carry a number of virulence genes, including genes for hemolytic activity and adherence to intestinal cells (8). It would be interesting to find out whether this YijP homologue of E. coli O157 and E. coli K-12 YijP are able to complement the invasion phenotype in K1 E. coli (ΔyijP).
In summary, we identified the gene locus yijP which contributes to E. coli K1 RS218 to invade into the CNS. This is the first demonstration that YijP plays a role in E. coli meningitis. Taken together, our findings indicate that several E. coli determinants, including OmpA, IbeA, IbeB, and YijP, contribute to E. coli K1 crossing of the blood-brain barrier. Studies are in progress to determine how these different E. coli structures contribute to invasion of BMEC.
ACKNOWLEDGMENTS
This study was supported by NIH grants R01-NS 26310 (to K.S.K.), R29-AI40635 (to S.-H.H.), and a CHLA Research Institute career development fellowship (to Y.W.).
We thank D. Gally of University of Newcastle for providing pIB307 and the Clinical Microbiology Laboratory of Childrens Hospital Los Angeles for testing E. coli strains with the Vitek system.
REFERENCES
- 1.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Six widespread bacterial clones among Escherichia coli K-1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashworth J M, Kornberg H L F.R.S. The anaplerotic fixation of carbon dioxide by Escherichia coli. Proc R Soc Lond Ser B. 1966;165:179–188. doi: 10.1098/rspb.1966.0063. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Burland V, Plunkett III G, Sofia H J, Daniels D L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol Microb. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 7.Boerlin P, Chen S, Colbourne J K, Johnson R, De Grandis S, Gyles C. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect Immun. 1998;66:2553–2561. doi: 10.1128/iai.66.6.2553-2561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Calderwood S B, Donohue-Rolfe A, Keusch G T, Kaper J B. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade Hep-2 cells. Infect Immun. 1990;58:1565–1571. doi: 10.1128/iai.58.6.1565-1571.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Kaper J B. Construction of an eac deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S H, Chen Y H, Fu Q, Stins M, Wang Y, Wass C, Kim K S. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67:2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S H, Wass C A, Fu Q, Prasadarao N A, Stins M F, Kim K S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jobling M G, Holmes R K. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZα and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990;18:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K S, Itabashi H, Gemski P, Sadoff J, Warren R L, Cross A S. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest. 1992;90:897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nizet V, Kim K S, Stins M, Jonas M, Chi E Y, Nguyen D, Rubens C E. Invasion of brain microvascular endothelial cells by group B streptococci. Infect Immun. 1997;65:5074–5081. doi: 10.1128/iai.65.12.5074-5081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasadarao N V, Wass C A, Huang S H, Kim K S. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect Immun. 1999;67:1131–1138. doi: 10.1128/iai.67.3.1131-1138.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasadarao N V, Wass C A, Kim K S. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrie. Infect Immun. 1996;64:154–160. doi: 10.1128/iai.64.1.154-160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S H, Kim K S. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quagliarello V, Scheld W M. Bacterial meningitis: pathogenesis, pathophysiology, and progress. N Engl J Med. 1992;327:864–872. doi: 10.1056/NEJM199209173271208. [DOI] [PubMed] [Google Scholar]
- 20.Ring A, Weiser J N, Tuomanen E I. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J Clin Invest. 1998;102:347–360. doi: 10.1172/JCI2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Stins M F, Gilles F, Kim K S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol. 1997;76:81–90. doi: 10.1016/s0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 23.Stins M F, Prasadarao N V, Ibric L, Wass C A, Luckett P, Kim K S. Binding of S-fimbriated Escherichia coli to brain microvascular endothelial cells. Am J Pathol. 1994;145:1228–1236. [PMC free article] [PubMed] [Google Scholar]
- 24.Stins M F, Prasadarao N V, Zhou J, Arditi M, Kim K S. Transfection of bovine brain microvascular endothelial cells with SV40-large T antigen: development of an immortalized cell line. In Vitro. 1997;33:243–247. doi: 10.1007/s11626-997-0042-1. [DOI] [PubMed] [Google Scholar]
- 25.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unhanand M, Musatafa M M, McCracken G H, Nelson J D. Gram-negative enteric bacillary meningitis: a twenty-one year experience. J Pediatr. 1993;122:15–21. doi: 10.1016/s0022-3476(05)83480-8. [DOI] [PubMed] [Google Scholar]
- 27.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 28.Wang, Y., and K. S. Kim. Unpublished results.
- 29.Wang Y, Rawlings M, Gibson D T, Labbé D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 30.Weel J F L, Hopman C T P, van Putten J P M. In situ expression and localization of Neisseria gonorrhoeae opacity protein infected epithelial cells: apparent role of Opa proteins in cellular invasion. J Exp Med. 1991;173:1395–1405. doi: 10.1084/jem.173.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]