Abstract
Background
As an “immune-privileged organ”, the liver has higher rates of both spontaneous tolerance and operational tolerance after being transplanted compared with other solid organs. Also, a large number of patients still need to take long-term immunosuppression regimens. Liver transplantation (LT) rejection involves varieties of pathophysiological processes and cell types, and a deeper understanding of LT immune response is urgently needed.
Methods
Homogenic and allogeneic rat LT models were established, and recipient tissue was collected on postoperative day 7. The degree of LT rejection was evaluated by liver pathological changes and liver function. Differentially expressed genes (DEGs) were detected by transcriptome sequencing and confirmed by reverse transcription-polymerase chain reaction. The functional properties of DEGs were characterized by the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome pathway analyses. The cells infiltrating the graft and recipient spleen and peripheral blood were evaluated by immunofluorescence and flow cytometry.
Result
A total of 1,465 DEGs were screened, including 1,177 up-regulated genes and 288 down-regulated genes. GO enrichment and KEGG pathway analysis indicated that DEGs were involved in several immunobiological processes, including T cell activation, Th1, Th2 and Th17 cell differentiation, cytokine-cytokine receptor interaction and other immune processes. Reactome results showed that PD-1 signaling was enriched. Further research confirmed that mRNA expression of multiple immune cell markers increased and markers of T cell exhaustion significantly changed. Flow cytometry showed that the proportion of Treg decreased, and that of PD-1+CD4+ T cells and PD-1+CD8+ T cells increased in the allogeneic group.
Conclusion
Using an omic approach, we revealed that the development of LT rejection involved multiple immune cells, activation of various immune pathways, and specific alterations of immune checkpoints, which would benefit risk assessment in the clinic and understanding of pathogenesis regarding LT tolerance. Further clinical validations are warranted for our findings.
Keywords: RNA-seq, liver transplantation, regulatory T cells, immune checkpoint, differentially expressed genes, GO, KEGG pathway
Introduction
Liver transplantation (LT) is the most effective way to treat end-stage liver disease (1). In the United Kingdom, 1- and 5-year survival rates were reported to be over 90% and 80%, respectively (2). However, patients with long-term application of immunosuppressive (IS) drugs also face the risks of serious adverse events, accounting for 58% of deaths after three years in LT recipients (3, 4). Rejection is one of the important reasons leading to the failure of liver transplantation. Understanding the mechanism of LT rejection and inducing immune tolerance can provide more specific strategies and improve postoperative quality of life.
LT immunity is an adaptive immune response involving activation of T and B lymphocytes (5). T cells can be divided into different subgroups according to their function. Regulatory T cells (Tregs), a specialized subset of CD4+ T cells expressing the key transcription factor Foxp3, play an important role in operational tolerance after solid organ transplantation (6, 7). Tregs could inhibit activation and proliferation of multiple immune cells through inhibitory costimulatory molecules, such as T cell immunoglobulin domain and mucin domain 3 (TIM-3), programmed cell death protein 1 (PD-1), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) (8–11). Studies have also shown that costimulatory pathways blockade could limit the activation of T cells reversely (12), which may alter the immune response against allograft and attenuate rejection, prolonging graft survival time (13). Currently, the potential for adoptive cell therapy with Tregs to promote transplant tolerance is being actively explored (14). Although most results of these trials are optimistic, many experimental and clinical unanswered questions are slowing the progression of this new therapeutic alternative (15). In addition, cytokines with pro-inflammatory and regulatory properties are also considered as potential therapeutic targets for inhibiting or enhancing the immune response of recipients (16–18). These results suggest that the content of cytokines, proliferation, and differentiation of T cell subsets, and inhibition of costimulatory molecules are essential for the formation of immune tolerance.
To date, several reports have focused on the effects of different factors on gene expression patterns in regenerating rat livers (19–21), post-transplant tumor recurrence (22), before and after human liver perfusion (23–25), and even the microenvironment of steatotic liver graft (26) using the next-generation RNA sequencing (RNA-seq) or at the single‐cell level. However, to our knowledge, our research is the first to reveal differential expression profiles between immune tolerance and rejection models of rat liver transplantation by RNA-seq, complemented with additional validation using quantitative real-time PCR, immunofluorescence, and flow cytometry. We found that LT rejection involves the participation of various immune cells, the activation of immune process, and the changes of multiple immune checkpoints. These results provide a strong theoretical basis for the potential clinical risks, related immune process, and pathogenesis of LT, and promote us to design rational drugs for the treatment of liver dysfunction caused by LT rejection.
Materials and methods
Animals
Kamada’s two-cuff method was used to establish a rat LT model (27). Lewis and Brown Norway (BN) rats, each weighing 210-240 g, were utilized as liver donors and recipients to establish allogeneic rejection models. Lewis-rats were used to construct homogenic tolerance models. All operations were carried out in accordance with the guidelines of the Ethics Committee of Shanghai General Hospital, Affiliated Hospital of Shanghai Jiao Tong University School of Medicine (Ethical code: 2019SQ147).
Histopathological examination and blood sample testing
Liver tissue was collected from the recipient rats for histopathological examination. The rejection activity index (RAI) was independently determined by three pathologists using the Banff Schema (International Panel, 1997) (28). Serum samples were obtained from the recipients for evaluation of liver function on postoperative day (POD) 7. The levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), γ-glutamyl transpeptidase (GGT, γ-GT), and total bilirubin (TBIL) were examined with an automated chemical analyzer (Hitachi 7600-10; Hitachi High-Technologies, Japan). The ELISA kits of necrosis factor-β (TNF-β), interferon-γ (IFN-γ), and interleukin-2 (IL-2), IL-4 were purchased from MULTISCIENCE (Shanghai, China).
RNA isolation, library construction and sequencing
Liver tissues of recipient rats were collected on POD 7. Total RNA was isolated from graft liver tissue using TRIzol reagent (ThermoFisher, Waltham, MA, USA) according to the manufacturer’s guideline, and the purity and concentration of RNA were detected by agarose electrophoresis or a standard Agilent 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA, USA). Briefly, mRNA was captured by magnetic oligo(dT) beads and fragmented, and the first-strand cDNA was generated using random hexamers. After the library was constructed, PCR amplification was used to enrich fragments, and 450bp was optimal size. Then, the total concentration and effective concentration of the library were detected using an Agilent 2100 Bioanalyzer. The standardized cDNA libraries were sequenced on an Illumina HiSeq2500 sequencer by the way of paired-end. The data were stored in the form of FASTQ. The raw sequencing data were filtered and evaluated for quality. The clean reads were mapped to the reference genome (Rattus_norvegicus.Rnor_6.0.dna.toplevel.fa) using BWT algorithm of HISAT2 (29). After the statistical analysis, the software HTSeq-count (30) was used to screen the differentially expressed genes (DEGs) by the following criteria: fold change >2 or < −2, false discovery rate (FDR) < 0.05.
Bioinformatic analysis
The Gene Ontology (GO) (31, 32) is a free available public resource that describes the role of genes in biological systems. The GO terms were comprised of the following three divisions: biological process (BP), cellular component (CC) and molecular function (MF). Adj. P<0.05 was regarded as statistically significant (33).
Kyoto Encyclopedia of Genes and Genomes (KEGG) is an integrated database resource, which aims to link genomic information with higher order functional information by computerizing current knowledge on cellular processes and by standardizing gene annotations (34).
The Reactome Knowledgebase systematically links proteins to their molecular functions, providing a resource that serves both as an archive of biological processes and as a tool for discovering unexpected functional relationships (35).
The GO annotation, KEGG pathways, and Reactome pathways were used to visualization and pathway enrichment analysis of DEGs, with a corrected P-value < 0.05 defined as significantly enriched.
Quantitative real-time polymerase chain reaction
RNA was extracted as previously described. The extracted RNA was analyzed by Nano Drop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) to ensure RNA quality and purity. After quantified, RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix (Perfect Real Time; Takara). RT‐PCR was performed using SYBR® Premix Ex Taq™ (Tli RNaseH Plus; Takara, Japan) in a Light Cycler Real‐time PCR System (Roche) (36). The corresponding primer sequences are provided in Supplement Table 1 .
Immunofluorescence assay
Paraffin sections were de-paraffinized, rehydrated and antigen retrieval according to standard protocols. Then, slices were incubated with anti-CD3 (1:200, Abcam, ab16669), anti-CD4 (1:200, Abcam, ab237722), anti-CD8 (1:200, Abcam, ab33786), and anti-Foxp3 (1:100, Abcam, ab215206). Fluorophore-conjugated secondary antibodies were incubated for one hour at room temperature (1:200, Abcam). The slides were imaged with fluorescence microscopes (Leica, Barcelona, Spain).
Flow cytometry
Spleens of recipient rats were collected, grounded, and filtered to obtain cell suspension, which was centrifuged at 1600 rpm at 4°C for 5 mins. The precipitation was resuspended by pre-cooled phosphate buffer solution. Double volume of lymphocyte separation solution purchased from Sigma-Aldrich was added and centrifuged at 2000 RPM for 25 min to obtain cell suspension. CD4+ T cells and CD8+ T cells were isolated by LS column (Miltenyi Biotec, Germany, #130-042-401) with the method of antibiotic microbeads (Miltenyi Biotec, Germany, # 130-090-319, # 130-090-318) as previously described (37, 38). Sorted cells were incubated directly with diluted fluorochrome-conjugated monoclonal antibodies as shown below: anti-CD4 (Invitrogen, #11-0040-82, FITC), anti-CD8a (Invitrogen, #11-0084-82, FITC), anti-PD-1 (Proteintech, #65211, Coralite647) and anti-Foxp3 (eBioscience, #12-5773-82, PE).
Statistical analysis
Statistical analyses were performed by GraphPad Prism V7.0. Results are shown as representative images or as mean ± standard deviation (SD) of at least three independent experiments. Differences between LT tolerance and rejection groups were estimated by the Student’s t-test. For all tests, statistical significance was considered at a P value < 0.05.
Results
The allogeneic LT group has a higher degree of rejection than the homogeneic group
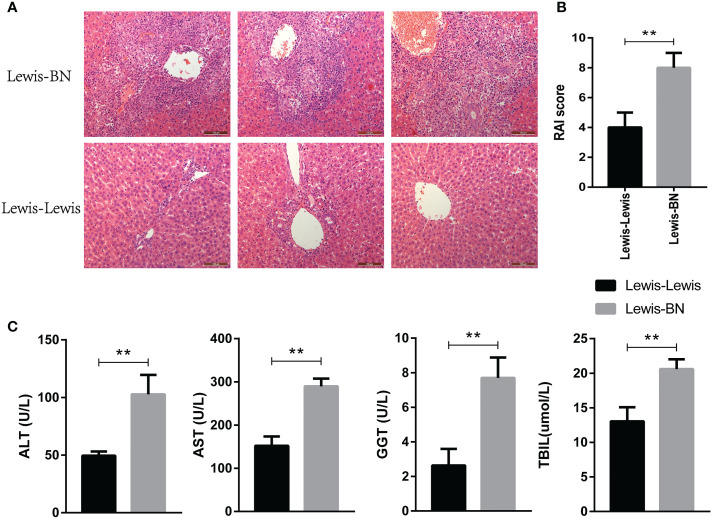
The homogeneic and allogeneic rat LT models were constructed and the rejection degree was evaluated on POD 7. Histopathological changes of liver were graded according to the Banff model (28). As shown in Figure 1A , rejection was relentless in the allogeneic LT model, as evidenced by marked infiltration of inflammatory cells into most portal areas, damage of bile duct epithelial cells and increased venous endothelial inflammation. The RAI score was shown in Figure 1B (t=4.899, P=0.008). In addition, the recipients in the homogeneic LT group shown better liver function indexes compared with allogeneic group (ALT: t=5.349 P=0.0059; AST: t=8.479 P=0.0011; GGT: t=5.77 P=0.0045; TBIL: t=5.279 P=0.0062). These results indicated that the allogeneic LT group has a higher degree of rejection than the homogeneic group.
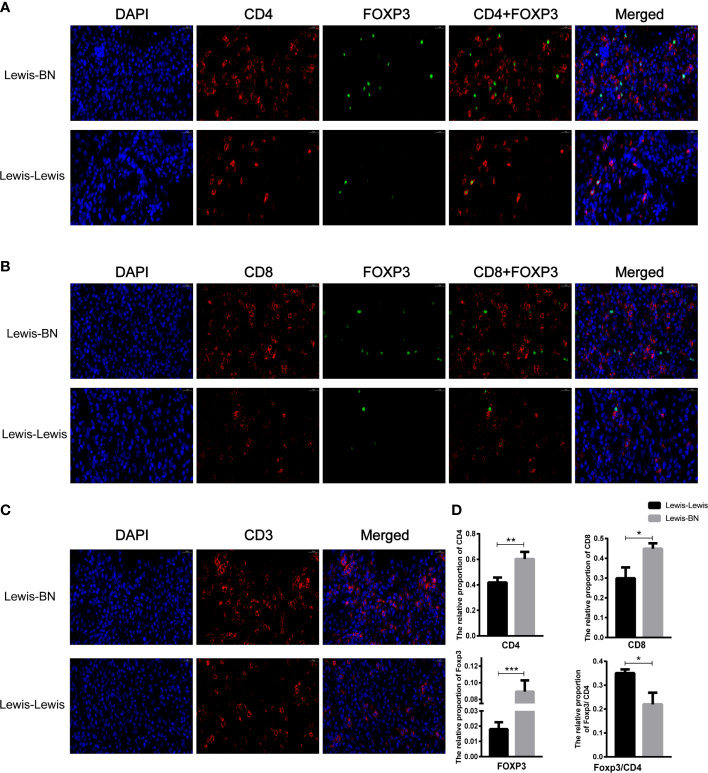
Figure 1.
Validation of differences in liver transplantation rejection between Lewis-Lewis group and Lewis-BN group on POD 7. (A, B) Hepatic morphologic changes and RAI scores of recipient rats. (C) Peripheral blood of each group was taken for analysis and determination of serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), γ-glutamyl transpeptidase (GGT) and total bilirubin (TBIL) by automatic biochemical analyzer (N = 3). *P<0.05, **P < 0.01, ***P < 0.001.
RNA-seq profiling showed allograft rejection involved significant immune dysregulation
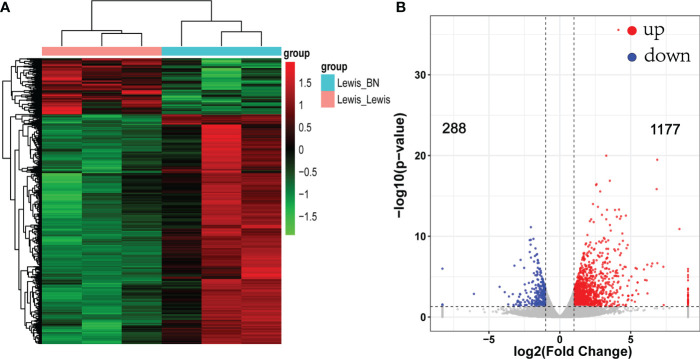
To reveal the molecular mechanism related to graft rejection, gene expression changes between the two groups were identified by RNA-seq. A total of 845,870,684 reads were obtained. After filtering and quality control, an average of 92.7% reads are available. The allograft models were taken as the experimental group. Using criteria of fold-change > 2 and FDR < 0.05 to define DEGs. Compared with the control group, a total of 1,465 DEGs were identified, of which 1,177 up-regulated genes and 288 down-regulated genes ( Figure 2 ). Among them, the top five down-regulated genes were SLC24A2, Pex5l, Kif28p, Brinp3, and Nudt10. However, the genes related to various immune components significantly upregulated in the allogeneic group with increased transplantation rejection response. The markers of T cell (CD2,CD3, CD28), B cell (CD19,CD79A), T-cells/NK-cells/T-cell activation and migration (ICOS, LCK, GZMA/B, CCR6, CCR7) and cellular infiltrates, such as dendritic cell markers CD80 and CD86 were significantly up-regulated. In addition to the markers mentioned above, multiple immune pathways, including the Th1 (STAT1, TBX21,IFNG, CXCL9/CXCL10/CXCL11), Th2 (CCR5, CCL11) and T cell exhaustion (LAG3, TIGIT, HAVCR2) were also up-regulated. The top five genes with the highest expression up-regulation related to immune regulation were CRTAM, IFNG, TIGIT, IL21, CD3E. Changes in the expression of chemokine receptor families and markers of immune cells suggest that multiple immune pathways are involved in the activation and participation of liver transplantation rejection.
Figure 2.
Heat map and volcano map of differentially expressed genes (DEGs). (A) Each column represents a sample. Red indicates up-regulated genes; green indicates down-regulated genes. (B) Volcano map of DEGs. fold change >2 or < −2, FDR < 0.05.
DEGs functional annotation and enrichment analysis
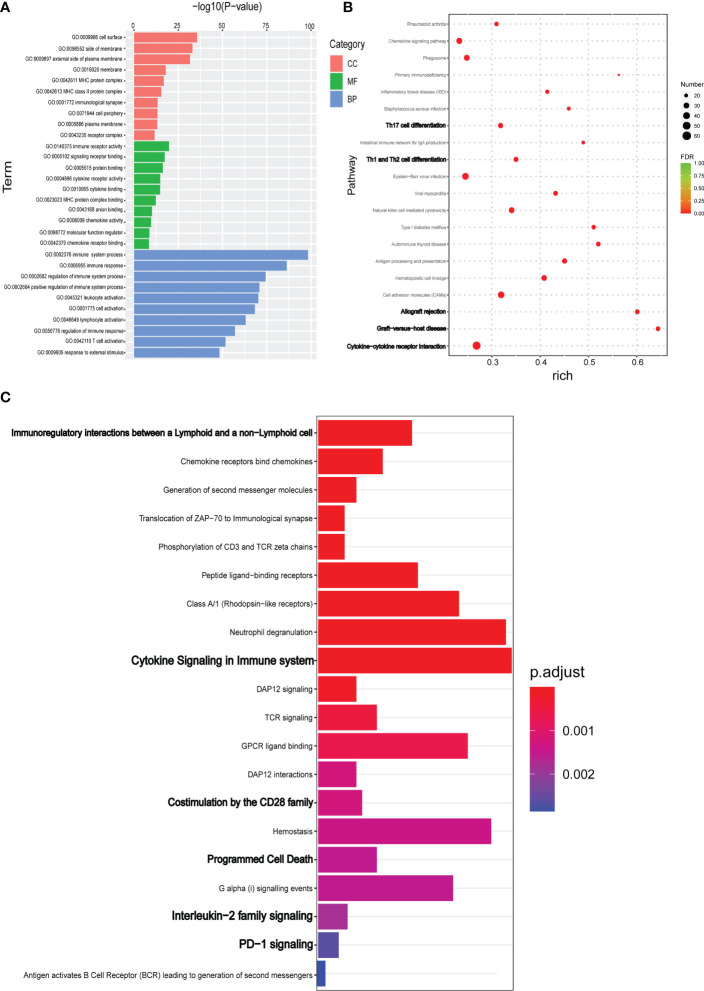
DEGs were annotated using the GO database to examine the biological functions and pathways. As shown in the Figure 3A , DEGs were mainly involved in the biological processes such as immune response, regulation of the immune system process and T cell activation, etc., which were consistent with the above speculation. The most enriched MF terms include immune receptor activity, signaling receptor binding, protein binding, cytokine receptor activity, and chemokine binding. In addition to other cell components such as cell surface, side of membrane, and external side of plasma membrane, MHC and MHC class II protein complexes are also enriched ( Figure 3A ).
Figure 3.
DEGs identified by RNA-seq were evaluated for functional enrichment using multiple gene annotation databases. (A) TopGO was used for GO enrichment analysis. GO enrichment analysis results of DEGs were classified according to molecular function (MF), biological process (BP) and cell component (CC). The top 10 GO term items with minimum p-value and most significant enrichment in each GO category were selected. (B) KEGG pathway enrichment analysis of DEGs. According to KEGG enrichment results, the top 20 KEGG pathways with the smallest FDR value, namely the most significant enrichment, were selected. (C) For the results of Reactome enrichment analysis of DEGs, the top 20 Reactome results with minimum p-value were selected.
KEGG database was used to functionally annotate the observed gene expression changes to identify potential pathways. KEGG pathway analysis indicated that DEGs were enriched in cytokine-to-cytokine receptor interaction, graft-versus-host disease, allograft rejection, cell adhesion molecules, Th1/Th2/Th17 cell differentiation, NK cell–mediated cytotoxicity, and antigen processing and presentation ( Figure 3B ). In addition to the classic KEGG pathway analysis, the Reactome database was also used to further analyze possible pathways involved in DEGs. As shown in the Figure 3C , the main enrichment pathway of DEGs were immunoregulatory interactions between a Lymphoid and a non−Lymphoid cell, Chemokine receptors bind chemokines, Cytokine Signaling in immune system, TCR signaling, Programmed Cell Death, and PD−1 signaling ( Figure 3C ).
Analysis and validation of RNA-seq data
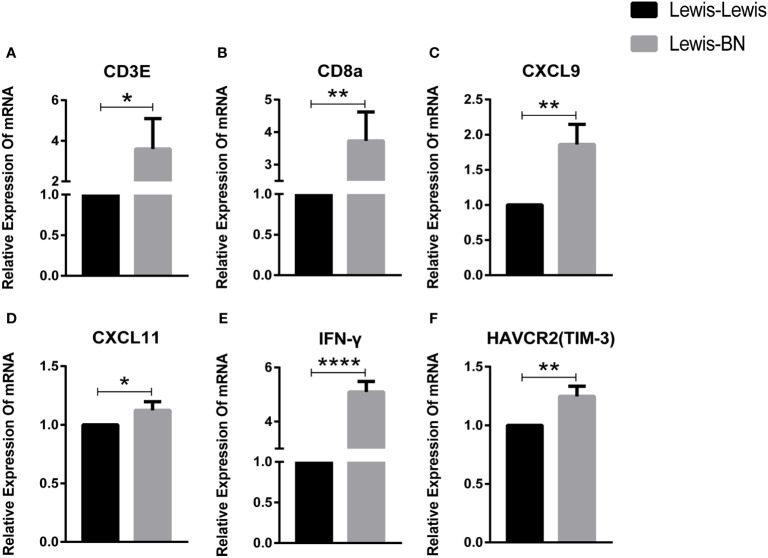
To further identify key genes involved in the progress of LT rejection, we selected ten genes associated with T cell activation or immunosuppression. The expression of T cell markers, CD3e and CD8a, increased (CD3e: P=0.0397; CD8a: P=0.006), while the expression difference in CD4 was not significant. The main Th1 cytokines, IFN-γ, and Th1 pathway, CXCL9 and CXCL11, were significantly increased (IFN-γ: P<0.0001; CXCL9: P=0.0065; CXCL11: P=0.442), while FOXP3, the Treg marker and Th17 cytokine, IL-17a, showed no significant changes. In addition, the marker of T cell exhaustion TIM-3 showed significant differences (P=0.0077), but PDCD1 mRNA did not differ between the two groups ( Figure 4 ).
Figure 4.
Fold-changes of immune mediators in the liver of homogenic and allogeneic rat liver transplantation recipients were measured by quantitative real-time PCR. *P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The cells infiltrating graft liver were increased in the rejection group
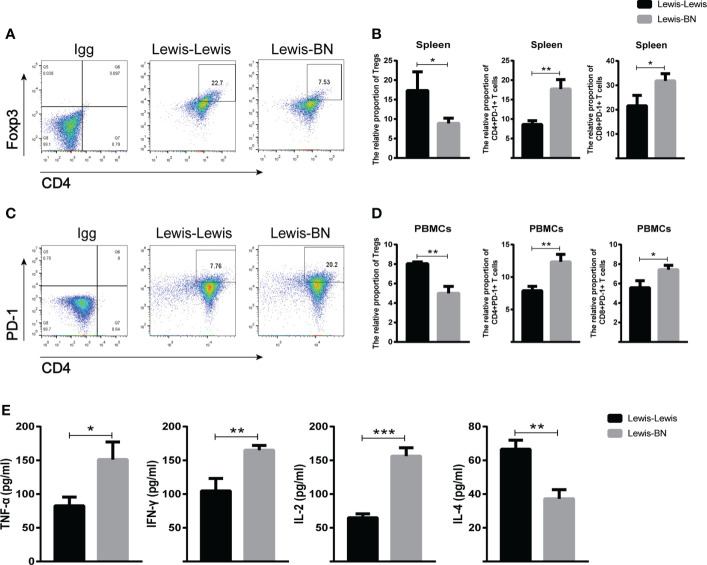
We further explored the cells infiltrating graft to confirm the reliability of the expression profiles generated by the RNA-Seq and DEGs analysis. Compared with the control group, the content of CD4 and CD8 T cell was up-regulated in the allograft (CD4+ T cell: t=4.641, P=0.097; CD8+ T cell: t=4.201, P==0.0137), and the expression of Foxp3 was also increased (t=8.665, P<0.001). However, the ratio of Foxp3/CD4 showed an opposite trend in allograft (t=4.433, P=0.0114) ( Figure 5 ). We also investigated the proportion of Foxp3+ T cells and the expression of PD-1 in the recipient spleen and PBMCs. The expression level of Foxp3 on T cells showed lower expression on CD4+ T cells compared with the control group (Spleen: Tregs, t=2.942, P=0.0423, PBMCs: Tregs, t=7.166, P=0.002). The expression of PD-1 on CD4+ and CD8+ T cells was up-regulated with the aggravation of rejection (Spleen: PD-1+CD4+ T cell, t=6.106, P=0.036; PD-1+CD8+ T cell, t=3.425, P=0.02676; PBMCs: PD-1+CD4+ T cell, t=5.75, P=0.0045; PD-1+CD8+ T cell, t=3.818, P=0.0188) ( Figures 6A–D ). The above results were consistent with the previous results of DEGs analysis. The proportion of CD4+ T and CD8+ T cells in the transplant rejection group was increased, and the expression of immune checkpoint PD-1 was also up-regulated. However, the proportion of Tregs was higher in the control group.
Figure 5.
Comparison of the content of cells infiltrating graft between two groups. (A-D) The cells infiltrating the graft were examined by immunofluorescence staining. *P<0.05, **P < 0.01, ***P < 0.001.
Figure 6.
(A–D) Spleen and peripheral blood of recipient rats in each group was collected on POD 7. The content of CD4+Foxp3+, CD4+PD-1, CD8+PD-1 T cells were detected by flow cytometry. (E) The serum levels of TNF-β, IFN-γ, IL -2 and IL-4 in each group was detected on POD 7. *P<0.05, **P < 0.01, ***P < 0.001.
Comparison of cytokine content in peripheral blood
Serum cytokines secreted by T cells play an important role in regulating various immune responses, including LT (39). Therefore, we measured the levels of TNF-α, IFN-γ, IL-2, and IL-4 by ELISA kit. The results indicated that TNF-α, IFN-γ and IL-2 increased in allogeneic transplant recipients (TNF-α: t=4.072, P=0.0152; IFN-γ: t=5.31, P=0.006; IL-2: t=11.62, P=0.0003) while IL-4 decreased (t=6.641, P=0.0027) ( Figure 6E ). Together, these results further suggest that multiple cytokines secreted by Th1 and Th2 cells are involved in the immune rejection of LT.
Discussion
Compared with other solid organs, a small portion of stable LT recipients showed sustained graft tolerance after complete withdrawal from IS, known as operational tolerance (40). However, the majority of patients still require a lifetime medication regimen. Therefore, the processes and mechanisms that control the outcome of LT remain to be thoroughly understood.
A major advantage of RNA-seq is its ability to identify potential novel biomarkers (41). The high genomic match between rat and human makes it a powerful model for elucidating the immune mechanisms of LT (42). There have been numbers of reports on liver regeneration pathways, ischemia-reperfusion at different time points, and different perfusion methods of rats and humans (19, 22, 24, 25, 43). Single-cell techniques have also been reported to assess the plasticity and phenotype of immune cells in the microenvironment of liver grafts (26). However, mRNA changes in rats at 7 days after homo/allogeneic LT have not been reported. And this is the first comprehensive transcriptomic analysis of rat graft liver on POD 7. DEGs identified were enriched in multiple immune processes, regulation of immune response, lymphocyte activation, and cell activation. KEGG results showed that Th1, Th2, and Th17 cell differentiation, NK-cell-mediated cytotoxicity, antigen presentation, and other immune mechanisms participated in the progression of transplant rejection. In addition to the involvement of the immune system, Reactome database analysis showed that PD-1 signaling pathway and cytokine signaling were also involved.
The rejection of allogeneic transplantation is mainly mediated by the recognition of non-self donor alloantigens (44–46), which leads to T cell activation and proliferation. MHC class I molecules present intracellular epitopes to CD8+ cytotoxic T cells, while MHC class II molecules are mainly responsible for presenting epitopes from extracellular substances to CD4+ helper T cells (47). Activated T cells work through a variety of effector mechanisms, including T cell-mediated direct damage to bile ducts, endothelium, and liver cells, as well as indirect effects through cytokine production and tissue destructive inflammatory cell recruitment (44, 48, 49). These effects explain the histological appearance of typical acute T cell-mediated rejection (48). CD8+ T cells could differentiate into cytotoxic T-cells able to exert direct cell damage on the allograft (44), and are also the main effector lymphocytes responsible for mediating tissue damage.
The outcome of liver is determined by the balance of effector and regulatory immune cell activities (50). Tregs are thought to be involved in inducing LT immune tolerance (45, 51–55). At present, several clinical trials on Treg treatment are under way (56), but there are still great challenges regarding the efficacy and safety of Treg treatment before it is truly implemented in routine clinical application. We analyzed the infiltrating cells in the liver and functional T cell subsets in the spleen and peripheral blood and found that the proportion of CD4+ T cells and CD8+ T cells, as well as the expression of Foxp3, were significantly increased in the rejection group. However, Foxp3/CD4 ratio decreased, suggesting that the content of Treg cells increased in the tolerant group. In addition, our data further indicated the important role of Treg cells in inducing immune tolerance in liver transplantation.
Most scholars believe that PD-1 plays a crucial role in inducing and maintaining the tolerance of peripheral transplant (57–59), besides, it is also involved in T cell exhaustion. overexpression of PD-1 on CD8+ T cells can induce cancer cells to escape from anti-tumor immune response and promote transplant tolerance (60–62). Similar to PD-1, TIGIT, a novel immune checkpoint, which is mainly expressed on NKs, CD8+ T cells, CD4+ T cells, and Treg cells (63), is well known for its important role in tumor immunity and autoimmune diseases (64–66). In transplantation immunity, TIGIT can regulate the severity of graft-versus-host disease (GVHD) by affecting the function of Treg cells and the number of donor antigen-reactive T cells (67–69). However, our results were not consistent with prevailing thinking. Reactome analysis indicated that DEGs were enriched in the activation of PD-1 signaling pathway. The expression of PD-1 on CD4+ T and CD8+ T cells in spleen and peripheral blood showed an increasing trend. And the mRNA expression of LAG3, HAVCR2(TIM-3), and TIGIT were significantly up-regulated in the rejection group, while PDCD1 was not. Currently, there are few studies on the simple application of immune checkpoint inhibitors (ICIs) in LT patients. Most patients suffer from malignant tumors or liver cancer before LT (70–73). Therefore, accurate conclusions cannot be drawn about the role of immune checkpoints in liver transplantation rejection. The mechanism of immune checkpoints such as PD-1, LAG3, and TIM-3 in liver transplantation remains to be further studied.
The activation and differentiation of T cells depend on the selection of costimulatory molecules and the composition of cytokines in the environment (74). On the one hand, cytokines secreted by Th1 cells, such as IFN-γ, TNF-α, and IL-2, can adversely affect the graft by recruiting and activating effector T cells (16, 17, 75). On the other hand, immunomodulatory cytokines secreted by Th2 cells, such as IL-4 and IL-10, can induce tolerance to liver allografts (16, 39, 76). Our results showed that TNF-α, IFN-γ, and IL-2 increased in allograft recipients, while IL-4 decreased. It is consistent with the above conclusion. Besides, the content of cytokine can also significantly affect the expression of PD-1/PD-L1 ( 77, 78). PD-1 expression was found to be elevated in spleen and peripheral blood, we speculated that the content of cytokine may also play a part in it. To sum up, the most important thing is to regulate the balance of various cytokines and use their advantages and disadvantages to achieve the maintenance of homeostasis.
There are some limitations to this study. First of all, the sample size is small. Although the current results show a significant difference in gene expression between the homogeneic group and the allogeneic rejection group, future studies still need a larger cohort and other strains of rats to evaluate its accuracy. Secondly, our study only proposed the hypothesis of possible immune-based treatments for LT, which needs to be confirmed in future proof-of-concept studies and clinical trials.
In summary, the current genomic and cellular profiling study of rat liver transplantation firstly provides a comprehensive molecular fingerprint of the immune alterations between the allogeneic group and the homogeneic group. Our results reveal a broad spectrum of immune system regulation including significant changes in T lymphocytes and a variety of cytokines, as well as maladjustment of immune checkpoints in liver transplantation rejection, providing a novel and promising insight into the understanding and induction of self-tolerance in LT.
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE210164.
Ethics statement
The animal study was reviewed and approved by The Ethics Committee of Shanghai General Hospital, Affiliated Hospital of Shanghai Jiao Tong University School of Medicine. Ethical code: 2019SQ147.
Author contributions
CW participated in research-design; LJ and LS participated in the performance of the research; SF was responsible for the design. XJ provided guidance on the research. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (grant number 81670595 and 81970568).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.947437/full#supplementary-material
Abbreviations
LT, Liver transplantation; IS, Immunosuppressive drugs; Th, Helper T cells; Tregs, Regulatory T cells; TIM-3, T cell immunoglobulin domain and mucin domain 3; PD-1, Programmed cell death protein 1; TIGIT, T cell immunoreceptor with Ig and ITIM domains; RNA-seq, RNA sequencing; TNF-β, Tumor necrosis factor-β; IFN-γ, Interferon-γ; IL-2, Interleukin-2; PBMCs, Peripheral blood Mononuclear Cells; RAI, Rejection activity index; POD, Postoperative day; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; GGT, γ-GT, γ-glutamyl transpeptidase; TBIL, Total bilirubin; DEGs, Differentially expressed genes; FDR, False discovery rate; (GO, Gene Ontology; BP, Biological processes; MF, Molecular factor; CC, Cell components; KEGG, Kyoto Encyclopedia of Genes and Genomes; GVHD, graft-versus-host disease; ICIs, Immune checkpoint inhibitors.
References
- 1. Li X, Li S, Wu B, Xu Q, Teng D, Yang T, et al. Landscape of immune cells heterogeneity in liver transplantation by single-cell RNA sequencing analysis. Front Immunol (2022) 13:90019. doi: 10.3389/fimmu.2022.890019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burke NT, Maurice JB, Nasralla D, Potts J, Westbrook R. Recent advances in liver transplantation. Frontline Gastroenterol (2022) 13(1):57–63. doi: 10.1136/flgastro-2020-101425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shaked A, DesMarais MR, Kopetskie H, Feng S, Punch JD, Levitsky J, et al. Outcomes of immunosuppression minimization and withdrawal early after liver transplantation. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surg (2019) 19(5):1397–409. doi: 10.1111/ajt.15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pruthi J, Medkiff KA, Esrason KT, Donovan JA, Yoshida EM, Erb SR, et al. Analysis of causes of death in liver transplant recipients who survived more than 3 years. Liver Transpl (2001) 7(9):811–5. doi: 10.1053/jlts.2001.27084 [DOI] [PubMed] [Google Scholar]
- 5. Ding M, He Y, Zhang S, Guo W. Recent advances in costimulatory blockade to induce immune tolerance in liver transplantation. Front Immunol (2021) 12:537079–. doi: 10.3389/fimmu.2021.537079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ni X, Wang Q, Gu J, Lu L. Clinical and basic research progress on treg-induced immune tolerance in liver transplantation. Front Immunol (2021) 12:535012. doi: 10.3389/fimmu.2021.535012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng G, Song X, Fujimoto S, Piccirillo CA, Nagai Y, Greene MI. Foxp3 post-translational modifications and treg suppressive activity. Front Immunol (2019) 10:2486. doi: 10.3389/fimmu.2019.02486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung CS, Yang KY, Li X, Chan VW, Ku M, Waldmann H, et al. Single-cell transcriptomics reveal that PD-1 mediates immune tolerance by regulating proliferation of regulatory T cells. Genome Med (2018) 10(1):71. doi: 10.1186/s13073-018-0581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hossain MS, Kunter GM, El-Najjar VF, Jaye DL, Al-Kadhimi Z, Taofeek OK, et al. PD-1 and CTLA-4 up regulation on donor T cells is insufficient to prevent GvHD in allo-HSCT recipients. PloS One (2017) 12(9):e0184254-e. doi: 10.1371/journal.pone.0184254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Li N, Li F, Zhou Z, Sang J, Chen Y, et al. Immune checkpoint proteins PD-1 and TIM-3 are both highly expressed in liver tissues and correlate with their gene polymorphisms in patients with HBV-related hepatocellular carcinoma. Med (Baltimore). (2016) 95(52):e5749. doi: 10.1097/MD.0000000000005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng R, Cassady K, Li X, Yao S, Zhang M, Racine J, et al. B7H1/CD80 interaction augments PD-1-dependent T cell apoptosis and ameliorates graft-versus-host disease. J Immunol (2015) 194(2):560–74. doi: 10.4049/jimmunol.1402157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hartigan CR, Sun H, Ford ML. Memory T-cell exhaustion and tolerance in transplantation. Immunol Rev (2019) 292(1):225–42. doi: 10.1111/imr.12824 [DOI] [PubMed] [Google Scholar]
- 13. Kinnear G, Jones ND, Wood KJ. Costimulation blockade: current perspectives and implications for therapy. Transplantation (2013) 95(4):527–35. doi: 10.1097/TP.0b013e31826d4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimozawa K, Contreras-Ruiz L, Sousa S, Zhang R, Bhatia U, Crisalli KC, et al. Ex vivo generation of regulatory T cells from liver transplant recipients using costimulation blockade. Am J Transpl (2022) 22(2):504–18. doi: 10.1111/ajt.16842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orozco G, Gupta M, Gedaly R, Marti F. Untangling the knots of regulatory T cell therapy in solid organ transplantation. Front Immunol (2022) 13:883855. doi: 10.3389/fimmu.2022.883855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang H, Wang Z, Zhang J, Gui Z, Han Z, Tao J, et al. Combined immunotherapy with belatacept and BTLA overexpression attenuates acute rejection following kidney transplantation. Front Immunol (2021) 12:618737. doi: 10.3389/fimmu.2021.618737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou JY, Alvarez CA, Cobb BA. Integration of IL-2 and IL-4 signals coordinates divergent regulatory T cell responses and drives therapeutic efficacy. Elife (2021) 10:e57417. doi: 10.7554/eLife.57417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lei H, Reinke P, Volk H-D, Lv Y, Wu R. Mechanisms of immune tolerance in liver transplantation-crosstalk between alloreactive T cells and liver cells with therapeutic prospects. Front Immunol (2019) 10:2667–. doi: 10.3389/fimmu.2019.02667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Colak D, Al-Harazi O, Mustafa OM, Meng F, Assiri AM, Dhar DK, et al. RNA-Seq transcriptome profiling in three liver regeneration models in rats: comparative analysis of partial hepatectomy, ALLPS, and PVL. Sci Rep (2020) 10(1):5213. doi: 10.1038/s41598-020-61826-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim A, Wu X, Allende DS, Nagy LE. Gene deconvolution reveals aberrant liver regeneration and immune cell infiltration in alcohol-associated hepatitis. Hepatology (2021) 74(2):987–1002. doi: 10.1002/hep.31759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J, Xu L, Wang P, Zheng X, Hu Y, Luo J, et al. RNA-Seq used to explore circRNA expression and identify key circRNAs during the DNA synthesis phase of mice liver regeneration. DNA Cell Biol (2020) 39(11):2059–76. doi: 10.1089/dna.2020.5750 [DOI] [PubMed] [Google Scholar]
- 22. Lee NP, Wu H, Ng KTP, Luo R, Lam TW, Lo CM, et al. Transcriptome analysis of acute phase liver graft injury in liver transplantation. Biomedicines (2018) 6(2):41. doi: 10.3390/biomedicines6020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mori N, Kida YS. Expression of genes involved in drug metabolism differs between perfusable 3D liver tissue and conventional 2D-cultured hepatocellular carcinoma cells. FEBS Open Bio. (2020) 10(10):1985–2002. doi: 10.1002/2211-5463.12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang P, Ming Y, Cheng K, Niu Y, Ye Q. Gene expression profiling in ischemic postconditioning to alleviate mouse liver Ischemia/Reperfusion injury. Int J Med Sci (2019) 16(2):343–54. doi: 10.7150/ijms.29393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoyer DP, Swoboda S, Treckmann JW, Benkö T, Paul A, Brocke-Ahmadinejad N, et al. Transcriptomic profiles of human livers undergoing rewarming machine perfusion before transplantation-first insights. Funct Integr Genomics (2021) 21(3-4):367–76. doi: 10.1007/s10142-021-00781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X, Lu D, Wang R, Lian Z, Lin Z, Zhuo J, et al. Single-cell profiling reveals distinct immune phenotypes that contribute to ischaemia-reperfusion injury after steatotic liver transplantation. Cell Prolif. (2021) 54(10):e13116. doi: 10.1111/cpr.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamada N, Calne RY. Orthotopic liver transplantation in the rat. technique using cuff for portal vein anastomosis and biliary drainage. Transplantation (1979) 28(1):47–50. doi: 10.1097/00007890-197907000-00011 [DOI] [PubMed] [Google Scholar]
- 28. Banff schema for grading liver allograft rejection: an international consensus document. Hepatology (1997) 25(3):658–63. doi: 10.1002/hep.510250328 [DOI] [PubMed] [Google Scholar]
- 29. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods (2015) 12(4):357–60. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (2015) 31(2):166–9. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat Genet (2000) 25(1):25–9. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res (2019) 47(D1):D330–d8. doi: 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gaudet P, Logie C, Lovering RC, Kuiper M, Lægreid A, Thomas PD. Gene ontology representation for transcription factor functions. Biochim Biophys Acta (BBA) - Gene Regul Mech (2021) 1864(11):194752. doi: 10.1016/j.bbagrm.2021.194752 [DOI] [PubMed] [Google Scholar]
- 34. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res (2000) 28(1):27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res (2020) 48(D1):D498–d503. doi: 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L, Wang T, Li S, Chen Z, Wu J, Cao W, et al. TDO2 promotes the EMT of hepatocellular carcinoma through kyn-AhR pathway. Front Oncol (2020) 10:562823. doi: 10.3389/fonc.2020.562823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lalfer M, Chappert P, Carpentier M, Urbain D, Davoust JM, Gross DA. Foxp3(+) regulatory and conventional CD4(+) T cells display similarly high frequencies of alloantigen-reactive cells. Front Immunol (2019) 10:521. doi: 10.3389/fimmu.2019.00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao W, Lu J, Li L, Qiu C, Qin X, Wang T, et al. Activation of the aryl hydrocarbon receptor ameliorates acute rejection of rat liver transplantation by regulating treg proliferation and PD-1 expression. Transplantation (2022). doi: 10.1097/tp.0000000000004205 [DOI] [PubMed] [Google Scholar]
- 39. Lu X, Cui J, Cui L, Luo Q, Cao Q, Yuan W, et al. The effects of human umbilical cord-derived mesenchymal stem cell transplantation on endometrial receptivity are associated with Th1/Th2 balance change and uNK cell expression of uterine in autoimmune premature ovarian failure mice. Stem Cell Res Ther (2019) 10(1):214–. doi: 10.1186/s13287-019-1313-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orlando G, Soker S, Wood K. Operational tolerance after liver transplantation. J Hepatol (2009) 50(6):1247–57. doi: 10.1016/j.jhep.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 41. Verma A, Muthukumar T, Yang H, Lubetzky M, Cassidy MF, Lee JR, et al. Urinary cell transcriptomics and acute rejection in human kidney allografts. JCI Insight (2020) 5(4):e131552. doi: 10.1172/jci.insight.131552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X, MacParland SA, Perciani CT. Immunological determinants of liver transplant outcomes uncovered by the rat model. Transplantation (2021) 105(9):1944–56. doi: 10.1097/TP.0000000000003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meng J, Zhao Z, Xi Z, Xia Q. Liver-specific Mettl3 ablation delays liver regeneration in mice. Genes Dis (2022) 9(3):697–704. doi: 10.1016/j.gendis.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ronca V, Wootton G, Milani C, Cain O. The immunological basis of liver allograft rejection. Front Immunol (2020) 11:2155. doi: 10.3389/fimmu.2020.02155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du X, Chang S, Guo W, Zhang S, Chen ZK. Progress in liver transplant tolerance and tolerance-inducing cellular therapies. Front Immunol (2020) 11:1326. doi: 10.3389/fimmu.2020.01326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anggelia MR, Cheng HY, Chuang WY, Hsieh YH, Wang AYL, Lin CH, et al. Unraveling the crucial roles of FoxP3+ regulatory T cells in vascularized composite allograft tolerance induction and maintenance. Transplantation (2021) 105(6):1238–49. doi: 10.1097/TP.0000000000003509 [DOI] [PubMed] [Google Scholar]
- 47. Demetris AJ, Bellamy CO, Gandhi CR, Prost S, Nakanuma Y, Stolz DB. Functional immune anatomy of the liver-as an allograft. Am J Transpl (2016) 16(6):1653–80. doi: 10.1111/ajt.13749 [DOI] [PubMed] [Google Scholar]
- 48. Dogan N, Hüsing-Kabar A, Schmidt HH, Cicinnati VR, Beckebaum S, Kabar I. Acute allograft rejection in liver transplant recipients: Incidence, risk factors, treatment success, and impact on graft failure. J Int Med Res (2018) 46(9):3979–90. doi: 10.1177/0300060518785543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang YC, Wu TJ, Wu TH, Lee CF, Chou HS, Chan KM, et al. The risk factors to predict acute rejection in liver transplantation. Transplant Proc (2012) 44(2):526–8. doi: 10.1016/j.transproceed.2012.01.041 [DOI] [PubMed] [Google Scholar]
- 50. Aghabi YO, Yasin A, Kennedy JI, Davies SP, Butler AE, Stamataki Z. Targeting enclysis in liver autoimmunity, transplantation, viral infection and cancer. Front Immunol (2021) 12:662134. doi: 10.3389/fimmu.2021.662134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terry LV, Oo YH. The next frontier of regulatory T cells: Promising immunotherapy for autoimmune diseases and organ transplantations. Front Immunol (2020) 11:565518. doi: 10.3389/fimmu.2020.565518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. López-Abente J, Martínez-Bonet M, Bernaldo-de-Quirós E, Camino M, Gil N, Panadero E, et al. Basiliximab impairs regulatory T cell (TREG) function and could affect the short-term graft acceptance in children with heart transplantation. Sci Rep (2021) 11(1):827. doi: 10.1038/s41598-020-80567-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu KY, Tong S, Wu CY, Ding XC, Chen JL, Ming Y, et al. Nlrp3 inflammasome inhibitor MCC950 ameliorates obliterative bronchiolitis by inhibiting Th1/Th17 response and promoting treg response after orthotopic tracheal transplantation in mice. Transplantation (2020) 104(6):e151-e63. doi: 10.1097/TP.0000000000003208 [DOI] [PubMed] [Google Scholar]
- 54. Pathak S, Meyer EH. Tregs and mixed chimerism as approaches for tolerance induction in islet transplantation. Front Immunol (2020) 11:612737. doi: 10.3389/fimmu.2020.612737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin-Moreno PL, Tripathi S, Chandraker A. Regulatory T cells and kidney transplantation. Clin J Am Soc Nephrol. (2018) 13(11):1760–4. doi: 10.2215/CJN.01750218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu J, Liu Z, Li C, Wei Q, Zheng S, Saeb-Parsy K, et al. Regulatory T cell therapy following liver transplantation. Liver Transpl (2021) 27(2):264–80. doi: 10.1002/lt.25948 [DOI] [PubMed] [Google Scholar]
- 57. Morita M, Fujino M, Jiang G, Kitazawa Y, Xie L, Azuma M, et al. PD-1/B7-H1 interaction contribute to the spontaneous acceptance of mouse liver allograft. Am J Transpl (2010) 10(1):40–6. doi: 10.1111/j.1600-6143.2009.02859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tanaka K, Albin MJ, Yuan X, Yamaura K, Habicht A, Murayama T, et al. PDL1 is required for peripheral transplantation tolerance and protection from chronic allograft rejection. J Immunol (2007) 179(8):5204–10. doi: 10.4049/jimmunol.179.8.5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu B, Yang XB, Sang XT. Liver graft rejection following immune checkpoint inhibitors treatment: a review. Med Oncol (2019) 36(11):94. doi: 10.1007/s12032-019-1316-7 [DOI] [PubMed] [Google Scholar]
- 60. Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol (2019) 234(6):8509–21. doi: 10.1002/jcp.27782 [DOI] [PubMed] [Google Scholar]
- 61. Liu Y, Liang X, Dong W, Fang Y, Lv J, Zhang T, et al. Tumor-repopulating cells induce PD-1 expression in CD8(+) T cells by transferring kynurenine and AhR activation. Cancer Cell (2018) 33(3):480–94.e7. doi: 10.1016/j.ccell.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 62. Amobi-McCloud A, Muthuswamy R, Battaglia S, Yu H, Liu T, Wang J, et al. IDO1 expression in ovarian cancer induces PD-1 in T cells via aryl hydrocarbon receptor activation. Front Immunol (2021) 12:678999–. doi: 10.3389/fimmu.2021.678999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu L, You X, Han S, Sun Y, Zhang J, Zhang Y. CD155/TIGIT, a novel immune checkpoint in human cancers (Review). Oncol Rep (2021) 45(3):835–45. doi: 10.3892/or.2021.7943 [DOI] [PubMed] [Google Scholar]
- 64. Guillerey C, Harjunpää H, Carrié N, Kassem S, Teo T, Miles K, et al. TIGIT immune checkpoint blockade restores CD8(+) T-cell immunity against multiple myeloma. Blood (2018) 132(16):1689–94. doi: 10.1182/blood-2018-01-825265 [DOI] [PubMed] [Google Scholar]
- 65. Ostroumov D, Duong S, Wingerath J, Woller N, Manns MP, Timrott K, et al. Transcriptome profiling identifies TIGIT as a marker of T-cell exhaustion in liver cancer. Hepatology (2021) 73(4):1399–418. doi: 10.1002/hep.31466 [DOI] [PubMed] [Google Scholar]
- 66. Sato K, Yamashita-Kanemaru Y, Abe F, Murata R, Nakamura-Shinya Y, Kanemaru K, et al. DNAM-1 regulates Foxp3 expression in regulatory T cells by interfering with TIGIT under inflammatory conditions. Proc Natl Acad Sci U.S.A. (2021) 118(21): e2021309118. doi: 10.1073/pnas.2021309118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zeng Q, Yuan X, Cao J, Zhao X, Wang Y, Liu B, et al. Mycophenolate mofetil enhances the effects of tacrolimus on the inhibitory function of regulatory T cells in patients after liver transplantation via PD-1 and TIGIT receptors. Immunopharmacol Immunotoxicol (2021) 43(2):239–46. doi: 10.1080/08923973.2021.1891247 [DOI] [PubMed] [Google Scholar]
- 68. van der List ACJ, Litjens NHR, Klepper M, Betjes MGH. Expression of senescence marker TIGIT identifies polyfunctional donor-reactive CD4+ T cells preferentially lost after kidney transplantation. Front Immunol (2021) 12:656846. doi: 10.3389/fimmu.2021.656846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang D, Hu W, Xie J, Zhang Y, Zhou B, Liu X, et al. TIGIT-fc alleviates acute graft-versus-host disease by suppressing CTL activation via promoting the generation of immunoregulatory dendritic cells. Biochim Biophys Acta Mol Basis Dis (2018) 1864(9 Pt B):3085–98. doi: 10.1016/j.bbadis.2018.06.022 [DOI] [PubMed] [Google Scholar]
- 70. Nordness MF, Hamel S, Godfrey CM, Shi C, Johnson DB, Goff LW, et al. Fatal hepatic necrosis after nivolumab as a bridge to liver transplant for HCC: Are checkpoint inhibitors safe for the pretransplant patient? Am J Transpl (2020) 20(3):879–83. doi: 10.1111/ajt.15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tabrizian P, Florman SS, Schwartz ME. PD-1 inhibitor as bridge therapy to liver transplantation? Am J Transpl (2021) 21(5):1979–80. doi: 10.1111/ajt.16448 [DOI] [PubMed] [Google Scholar]
- 72. Luo Y, Teng F, Fu H, Ding GS. Immunotherapy in liver transplantation for hepatocellular carcinoma: Pros and cons. World J Gastrointest Oncol (2022) 14(1):163–80. doi: 10.4251/wjgo.v14.i1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Munker S, De Toni EN. Use of checkpoint inhibitors in liver transplant recipients. United Eur Gastroenterol J (2018) 6(7):970–3. doi: 10.1177/2050640618774631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Toomer KH, Malek TR. Cytokine signaling in the development and homeostasis of regulatory T cells. Cold Spring Harb Perspect Biol (2018) 10(3):a028597. doi: 10.1101/cshperspect.a028597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang XX, Bian RJ, Wang J, Zhang QY. Relationship between cytokine gene polymorphisms and acute rejection following liver transplantation. Genet Mol Res (2016) 15(2). doi: 10.4238/gmr.15027599 [DOI] [PubMed] [Google Scholar]
- 76. Chen L, Zheng L, He W, Qiu M, Gao L, Liu J, et al. Cotransfection with IL-10 and TGF-β1 into immature dendritic cells enhances immune tolerance in a rat liver transplantation model. Am J Physiol Gastrointest Liver Physiol (2014) 306(7):G575–81. doi: 10.1152/ajpgi.00283.2013 [DOI] [PubMed] [Google Scholar]
- 77. Wang Y, Xi Y, Han F, Liu Y, Li N, Ren Z, et al. Vascularized composite allograft rejection is delayed by infusion of IFN-γ-conditioned BMSCs through upregulating PD-L1. Cell Tissue Res (2019) 376(2):211–20. doi: 10.1007/s00441-018-2967-y [DOI] [PubMed] [Google Scholar]
- 78. Qian J, Wang C, Wang B, Yang J, Wang Y, Luo F, et al. The IFN-γ/PD-L1 axis between T cells and tumor microenvironment: hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflamm (2018) 15(1):290. doi: 10.1186/s12974-018-1330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE210164.