ABSTRACT
Controlled gene expression is crucial for engineering bacteria for basic and applied research. Inducible systems enable tight regulation of expression, wherein a small-molecule inducer causes the transcription factor to activate or repress transcriptional initiation. The T7 expression system is one of the most widely used inducible systems, particularly for high overexpression of proteins. However, it is well known that the highly active T7 RNA polymerase (RNAP) has several drawbacks, including toxicity to the host and substantial leaky expression in the absence of an inducer. Much work has been done to address these issues; current solutions require special strains or additional plasmids, making the system more complicated and less accessible. Here, we challenge the assumption that the T7 expression system is the best choice for obtaining high protein titers. We hypothesized that expression from strong inducible promoters expressed from high-copy plasmids could compete with expression levels obtained from T7 RNAP but that such promoters would possess improved control of transcription. Employing inducible systems from a toolbox we developed previously, we demonstrate that our plasmids consistently give higher outputs and greater fold changes over basal expression than the T7 system across rich and minimal media. In addition, we show that they outperformed the T7 system when we used an engineered metabolic pathway to produce lycopene.
IMPORTANCE Genetic systems for protein overexpression are required tools in microbiological and biochemical research. Ideally, these systems include standardized genetic parts with predictable behavior, enabling the construction of stable expression systems in the host organism. Modularity of a genetic system is advantageous, so that the expression system can be easily moved into a host that best suits the needs of a given experiment. The T7 expression system lacks both predictability and stability and requires special host strains to function. Despite these limitations, it remains one of the most popular systems for protein overproduction. This study directly compared the T7 system to four inducible systems from our broad-host-range plasmid toolbox and demonstrated these alternative expression systems have distinct advantages over the T7. The systems are entirely plasmid-based and not constrained to a specific bacterial host, expanding the options for high-level protein expression across strains.
KEYWORDS: T7 system, inducible expression, plasmid toolbox, protein overexpression
INTRODUCTION
Escherichia coli has been a workhorse in the field of microbiology for decades, serving as both a model organism and intracellular workbench for molecular biology studies (1–4). A variety of systems exist for heterologous protein expression in these cellular factories, with the T7 expression system among the most popular (5–7). This system involves a chromosomally encoded bacteriophage T7 RNA polymerase (T7 RNAP), most often generated with the λDE3 lysogen. The T7 promoter regulates expression of the target gene and is usually contained on a plasmid. The T7 RNAP recognizes its promoter sequence with stringent specificity and is very efficient, generating high polymerase flux to maximize target protein production (7, 8).
For greater control of gene expression, the native T7 RNAP promoter can be replaced with the lacUV5 inducible promoter, while an inducible variant of the T7 promoter, T7lac, is often used to drive expression of the target gene (6, 7, 9). Even with these parts in place, T7 systems are notoriously leaky. Due to the high activity of the T7 RNAP, even low-level basal expression of the polymerase leads to high expression of the target gene. This basal expression of T7 RNAP decreases the stability of protein production strains, mainly when the target proteins affect cell fitness (10–12). Additionally, the high processivity of T7 RNAP can come at a large fitness cost to the host due to competition for cellular resources (5, 7, 13, 14). Numerous strategies have been used to increase the stringency of T7 RNAP repression, but toxicity and leakiness remain concerns (15–23). The conventional T7 system also lacks tunability, meaning inducer concentration is not correlated to protein output levels in a dose-dependent manner (22). Moreover, induction often results in a mixed population of cells that express the target protein at different levels. Uniformity of expression can vary depending on the available carbon source and the presence of toxicity-escape mutants, which can lower or abolish protein production (21, 22, 24–27). These problems necessitate the use of freshly transformed cells due to the propensity for chromosomal mutations of the host strain that diminish levels of T7 RNAP (19, 28).
Still, systems for high-level overexpression of recombinant proteins from single genes or multigene pathways are extremely valuable. The regulated coexpression of multiple genes is necessary for building biosynthetic pathways and can reduce the occurrence of nonfunctional protein aggregates (2). The Duet plasmids (Novagen) were developed to meet this need, as they enable the coexpression of up to eight genes on four compatible vectors (29). In the Duet system, λDE3 lysogens are the required host, and T7lac promoters regulate all target genes, so many of the limitations discussed above apply. While plasmid-borne T7 RNAP can also be used, the aforementioned problems are often exacerbated (30–32). Significantly, use of the T7 promoter to regulate all inserted genes only allows for a rough measure of tunability in the form of relative copy number, making fine control of gene expression virtually impossible (33).
More recently, an impressive work by Meyer et al. greatly expanded the tools available for coexpression with the development of the Marionette system. In this system, E. coli strains house 12 evolved transcriptional regulators on the chromosome and cognate output promoters on a single plasmid (34). The researchers demonstrated that many of the promoters have large dynamic ranges, and several could be used together to construct a biosynthetic pathway where each gene could be independently tuned. However, the system was restricted to the Marionette strains, which include all regulatory elements integrated into the chromosome and lack the additional level of tunability that comes from varying copy number with different plasmid backbones (35).
We recently developed a broad-host-range plasmid toolbox for tunable gene expression and tested it across nine species of Proteobacteria (36). Having attained very high levels of expression in many of these species, we wondered if some of the inducible systems were capable of competing with the expression levels obtained from T7 RNAP in E. coli. We hypothesized that expression from our toolbox promoter-regulator pairs would enable high protein overexpression and improved control of transcription. Our plasmid system's ease of assembly helped us efficiently construct a collection of 28 plasmid variants to test four toolbox inducible systems on a set of origin and marker backbones. We were interested in demonstrating a dynamic range of expression using high- and low-copy plasmids, tuning transcription via titrated inducer or a combination of both strategies.
For the work presented here, we added four E. coli origins of replication to our toolbox, and we report characterization data for four inducible expression systems to demonstrate their large dynamic range and utility in gene coexpression experiments in E. coli. We also assessed whether these plasmids could outperform canonical T7 promoter plasmids in strength, stability, and utility.
RESULTS
Plasmid design, tunability, and context dependence.
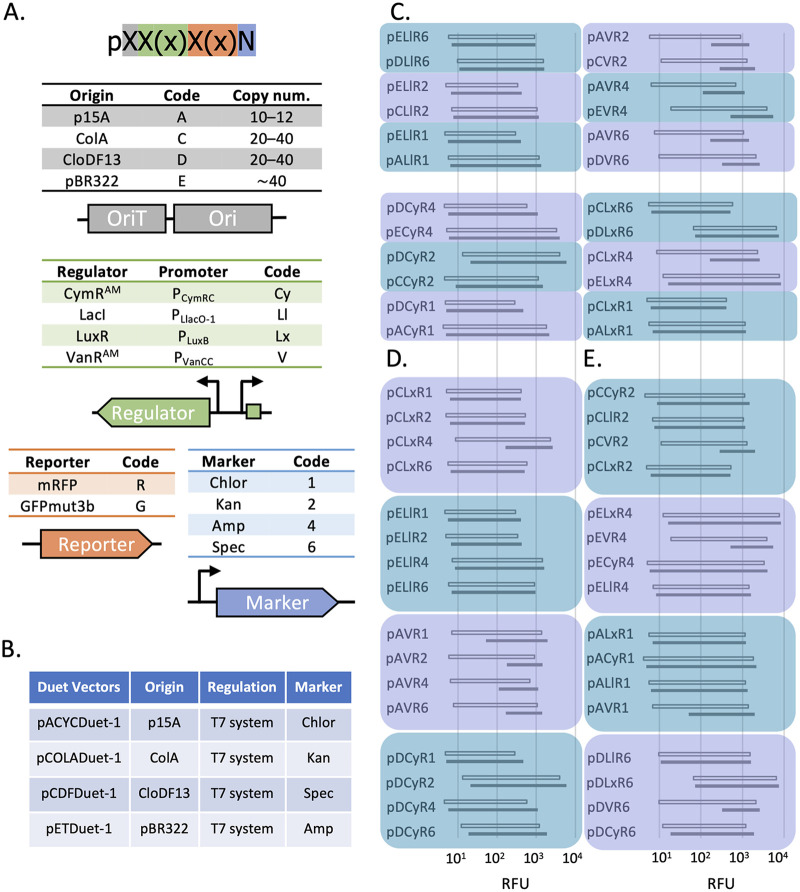
Plasmids were constructed with four interchangeable parts with common linker sequences, so that combinatorial assembly by ligation-independent cloning could be quickly performed (36). Building on our previously described broad-host-range plasmid toolbox, we developed a protein overexpression toolbox with the genetic part variants shown in Fig. 1A. Plasmids were named to represent each incorporated part, based on the specified codes. For reference, genetic parts included in the Duet plasmids are listed in Fig. 1B. With four antibiotic markers, four promoter-regulator pairs, and the addition of four enteric bacteria-specific origins of replication to our toolbox, we constructed 28 plasmid variants and characterized their expression in E. coli. Cells were induced during early log phase, and expression was measured at late-exponential and stationary phases of growth (see Materials and Methods). The expression data in Fig. 1C to E are grouped in shaded boxes, such that only one genetic part is varied within each box, to assess how this single variable changed the expression levels. Our a priori assumption was that changing the marker would have little influence on the expression levels, while changing origin and regulator would have greater influence.
FIG 1.
Plasmid toolbox genetic parts, nomenclature, and induction results. (A) The plasmid toolbox included several options of each of four genetic parts, and plasmids were constructed according to a previously described combinatorial assembly strategy. Plasmid names were based on the codes provided, in the following order: origin, regulator, reporter, and marker. (B) Genetic parts of the Duet vectors (Novagen) included in this work. For induction experiments, mrfp or gfpmut3b was cloned into the first multiple-cloning site downstream of the T7lac promoter. (C, D, and E) Expression range of 28 plasmids in E. coli. Induction data are shown for all plasmid variants included in this study and are grouped to best show the effect of a single genetic part on overall expression, with plasmids grouped with only the origin part changed (C), with only the marker changed (D), and with only the promoter-regulator changed (E). For each plasmid, expression data are shown from the late-exponential (open bar) and stationary-phase (filled bar) time points. Bars represent the induction range of mRFP for each plasmid, with fluorescence in the absence of inducer plotted at the left end of each bar and induced expression plotted at the right end. Inducer concentrations are listed in Table S3 in the supplemental material. All measurements were in E. coli MG1655 grown in rich medium and are the averages of three technical replicates.
Pairwise comparisons between plasmids that differed only in their origin showed that expression levels changed greatly between some plasmids (Fig. 1C). In E. coli, the pACYC backbone has a low plasmid copy number, pCOLA and pCDF have medium plasmid copy numbers, and the pET backbone has a high plasmid copy number (2, 29) (Fig. 1A). Plasmid copy number is often directly related to gene expression because more DNA templates are available for transcription, though this relationship is not always maintained (35, 37–39). Our expression data mostly followed copy number trends, with all expression systems on a pACYC backbone giving a maximal expression around 103 relative fluorescent units (RFU), while systems on pCDF and pET backbones were more likely to approach 104 RFU. LacI-regulated systems on a pET backbone were notable exceptions; while pELx, pECy, and pEV had outputs near 104 RFU by late-exponential phase, pELl was at or under 103 RFU after an overnight induction (Fig. 1D and E). Indeed, expression from LacI/PLlacO-1 was consistently lower than the other promoter-regulator pairs tested across origin and marker combinations, suggesting that this system has a lower output than the others, rather than its plasmid context.
In Fig. 1D, expression levels after changing only the marker were compared within four different origin-regulator pairs, and in Fig. 1E, inducible systems were compared on the same origin-marker backbone. At the late-exponential-phase measurement, leakiness and expression levels were remarkably similar for pCLx, pELl, and pAV, regardless of the marker. Expression from pDCy was comparatively less consistent and was influenced by the marker to a greater extent (Fig. 1D). Similarly, in the late-exponential phase, plasmids with the pCDF backbone varied with the promoter-regulator more than other origins (Fig. 1E). Another trend was the high level of uninduced expression of monomeric red fluorescent protein (mRFP) in VanRAM-regulated systems after overnight growth. This was true across plasmid backbones, suggesting a characteristic intrinsic to VanRAM/PVanCC in E. coli. While the original description of the evolved VanRAM showed leakiness in M9 medium (34), our results showed high uninduced expression in rich medium as well. All other systems maintained a low basal expression level, except for pCLxR4, which became leaky at stationary phase.
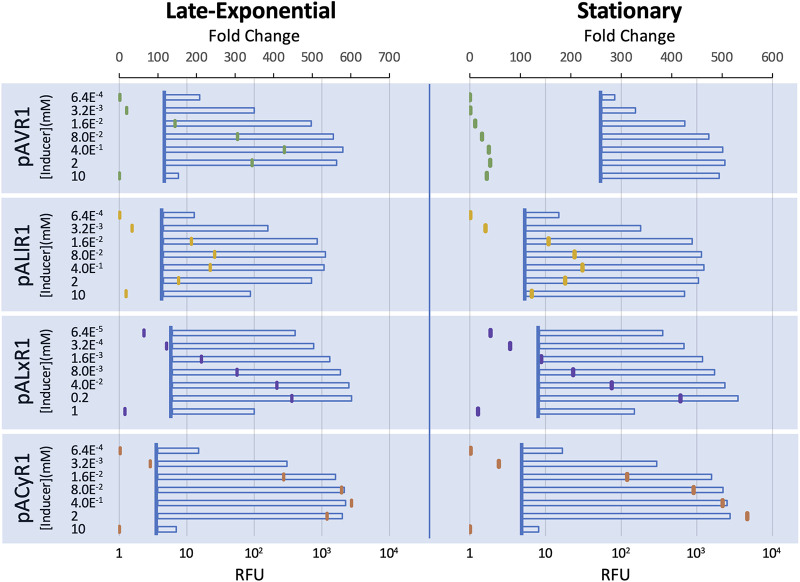
We next assessed titrating the expression level by changing the inducer concentration. The pA-R1 plasmids with each promoter-regulator pair were screened with various inducer levels, and the fluorescence was measured at late-exponential and stationary phases (Fig. 2, see also Fig. S2 in the supplemental material). Both the LacI- and VanRAM-regulated systems exhibit a bell curve of expression levels across titrated inducer concentrations, particularly at the late-exponential time point. Across inducer concentrations, the VanRAM/PVanCC system was inducible from 19- to nearly 430-fold at 6 h postinduction, though much of this range was lost by stationary phase due to increased leakiness in the absence of inducer. After an overnight induction, pAVR1 had a maximal fold change of 40 with 2 mM vanillate. At the late-exponential measurement, the LacI-regulated system had a smaller range of induction across inducer concentrations than did VanRAM/PVanCC. However, LacI/PLlacO-1 maintained a relatively low level of basal expression through stationary phase and remained inducible over 200-fold at both 0.4 mM and 80 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Importantly, LacI/PLlacO-1 was sensitive to inducer such that it exhibited a 30-fold change at 3.2 μM IPTG and a >150-fold change at 16 μM IPTG at both time points measured, suggesting a space for further titrations to tune expression while minimizing inducer cost.
FIG 2.
Expression across titrated inducer concentrations. Expression of mRFP in E. coli MG1655 strains containing plasmids pACyR1, pALxR1, pALlR1, and pAVR1 were induced with titrated inducer and measured at the late-exponential and stationary phase of growth. Graphs show average RFU induction data (horizontal bars) and calculated fold change (colored notches) within each data cluster. Vertical lines at each cluster represent the average fluorescence of uninduced samples. Inducer was serially diluted 5-fold from 10 mM (cumate, IPTG, Van) or 1 mM (OC6) across seven concentrations for each plasmid. All measurements in rich medium and are the averages of three technical replicates.
Across the inducer concentrations tested, the LuxR-regulated system was tunable at concentrations titrated from 200 μM 3-oxohexanoyl-homoserine lactone (OC6), with fold change values following a nearly log-linear response. At stationary phase, LuxR/PLuxB was also inducible to 40-fold at the lowest concentration of inducer tested (64 nM OC6), suggesting that this system is highly sensitive and that further inducer titrations may continue the log-linear trend seen across most of this data set. Though CymRAM/PCymRC had the highest maximal fold change of the entire data set at both time points measured, this system exhibited the most binary response. Though expression reached 50- to 80-fold with 3.2 μM cumate at both time points, fold changes <3 or >300 were found at other inducer concentrations. The considerable distance between outputs for induction at 3.2 μM and 16 μM cumate may indicate space for more fine-tuned expression.
Expression stability over time.
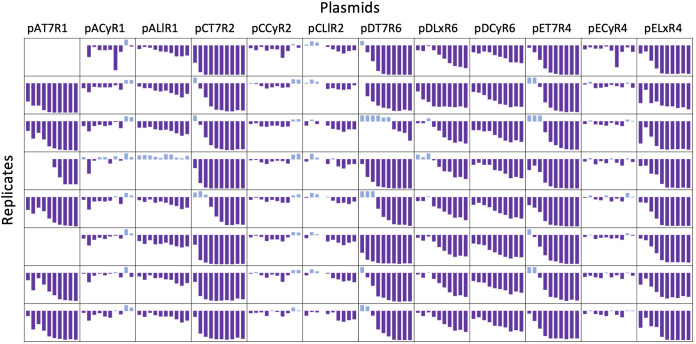
We were interested in testing the stability of our plasmids to ascertain if expression remained consistent throughout several passages. If this were true, our toolbox systems would demonstrate a distinct advantage over T7 expression systems in BL21(DE3) cells by maintaining stable and predictable high-level expression. Our experiment measured mRFP expression from eight toolbox plasmids and four Duet plasmids in minimal medium over 12 daily passages under the pressure of continuous induction. Eight replicates were included for each of the 12 plasmids to monitor changes in mRFP expression (Fig. 3). On day 11, one replicate from each sample was struck on a plate spread with inducer and photographed under blue light to visualize colonies with red fluorescence (see Fig. S3).
FIG 3.
Stability over extended passages. Sparkline plots of percent change in fluorescence compared to baseline measurements for all replicates across four Duet vectors and eight toolbox plasmids. Plots are bound on the y axis by −100% and 20%. Data were excluded from graphing when growth of replicate was below a threshold OD of 0.2. For replicates that grew over the threshold after the first day of measurements, the first fluorescence measurement taken when cultures were above an OD of 0.2 was used as the baseline measurement. All plotted fluorescence was normalized to growth, and inducer concentrations are listed in Table S3.
The percent change in growth-normalized fluorescence on each day was compared to the benchmark measurement (day 2) and plotted for each replicate (Fig. 3). The T7 systems were very unstable, consistent with previous studies (30). Among the Duet vectors, there were no apparent differences in system behavior among the different origin and marker backbones. The T7 expression systems consistently lost protein production capacity across all plasmid genetic backgrounds, and most replicates had lost over 80% of their fluorescence signal by day 7. The loss of fluorescence occurred most rapidly in pCOLADuet-1, with five of the eight replicates exhibiting over a 75% decrease compared to benchmark measurements by day 4. Though pETDuet-1 possesses the origin with the highest copy number, the percent change throughout the experiment was not notably different from the others. In fact, at least one replicate from all other Duet vectors lost almost 60% fluorescence after just 2 to 3 passages, whereas this degree of reduction was not seen in pETDuet-1 until day 5. This suggested that copy number alone does not determine the stability of an expression vector, and toxicity-escape mutations cannot necessarily be avoided by using a low-copy plasmid.
Among the toolbox expression systems, those tested on pACYC and pCOLA backbones best maintained expression over the 12-day experiment (Fig. 3). Expression levels from pACyR1 and pCCyR2 were particularly stable, and by day 12, only three of 16 replicates from these two plasmids had decreased expression compared to benchmark measurements, with the greatest decrease at just 12%. Though mRFP expression from LacI/PLlacO-1 on pA and pC backbones generally decreased throughout the experiment compared to benchmark measurements, percent decreases were 20 and 30% on average by day 12 for pCLlR2 and pALlR1, respectively. Conversely, the expression declines from pDLxR6 and pDCyR6 were only slightly less pronounced than that from the T7 system on the same backbone, though pCDFDuet-1 lost fluorescence sooner and was less consistent across replicates. A notable anomaly among our toolbox plasmids was pELxR4. Fluorescence from each replicate in pETDuet-1 and pELxR4 was less than half the benchmark measurements by day 7, and most had decreased over 90% by day 12. CymRAM/PCymRC on the same backbone, pECyR4, lost an average of 6% fluorescence by day 12, compared to 93% from pELxR4. Again, expression from pETDuet-1 was less consistent across replicates, likely a result of accumulated mutations that affected protein production in different ways.
To measure changes in promoter activity at the cellular level, we applied flow cytometry to the day 12 samples (Fig. S4). In the Duet vectors, cell counts peaked at different fluorescence intensities and the spread of peaks on the x axis representing relative fluorescence varied considerably (Table S4). With our toolbox plasmids, the peaks were more narrow and uniform, though there was increased variability among replicates in pDLxR6 and pELxR4, as expected based on the population measurements. For most of the Duet vector samples, the geometric mean of fluorescence intensity was an order of magnitude lower than those of the toolbox plasmids, and three of the four Duet vectors had replicates with a coefficient of variation an order of magnitude higher than any replicate from the toolbox plasmids (Table S4). This highlights the poor stability and lack of predictability in T7 expression and an overall decline in protein production. The fully plasmid-based inducible systems in this toolbox on the same backbones consistently generated higher protein levels after many subculturings and had reduced cell-to-cell variation.
Growth medium-dependent properties.
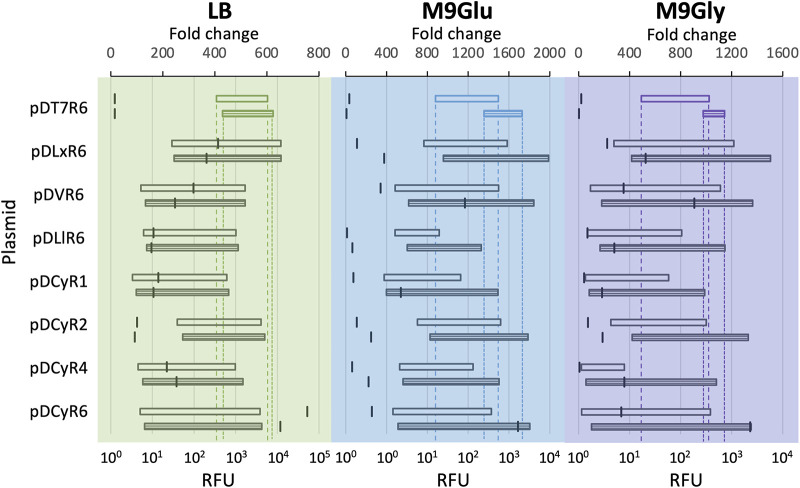
We next took the pD toolbox plasmid variants and pCDFDuet-1 with mrfp cloned into the first multiple cloning site and compared the expression levels in E. coli BL21(DE3) with different growth media: Luria broth (LB), M9 supplemented with glucose (M9Glu), and M9 supplemented with glycerol (M9Gly). While we expected overall expression to be higher in LB, we were interested in comparing expression levels between M9 with glucose and M9 with glycerol, as glycerol is less expensive than glucose and is being used as an alternative carbon source in metabolic engineering experiments (40, 41). In both rich and minimal media, all toolbox plasmids exhibited lower leaky expression than pCDFDuet-1. At the same time, many maintained output levels that were as high or higher when induced (Fig. 4). Output levels from the T7lac promoter across all media tested were less than 15-fold above basal expression by the stationary-phase time point, and in all cases, these low fold changes could be attributed to high uninduced expression, highlighting a lack of controlled induction.
FIG 4.
Comparison between T7 and toolbox regulated promoters in E. coli BL21(DE3). Expression levels of mRFP regulated by the T7 system and toolbox systems regulated by LuxR, CymRAM, VanRAM, and LacI were measured from a set of eight pCDF plasmids in LB and minimal media with two different carbon sources. Charts show average RFU induction data (horizontal bars) and calculated fold change (horizontal notches) with measurements taken at late-exponential (open bar) and stationary (horizontally-striped bar) phases of growth. Horizontal bars represent the induction range of mRFP for each toolbox plasmid, with fluorescence in the absence of inducer plotted at the left end of each bar and induced expression plotted at the right end. The induction ranges from pCDFDuet-1 (pDT7R6) extend through the charts with dotted lines.
In LB, the highest mRFP expression over basal levels came from pDCyR6, with a fold change of over 650 at both time points taken. Here, leaky expression from pDCyR6 remained 2 orders of magnitude below that of the T7 promoter by stationary phase and was among the lowest of all expression vectors tested. Expression from pDCyR6 also had the biggest fold change of the pD plasmid variants by stationary phase in M9Glu and M9Gly, with similarly low uninduced expression. In this way, pDCyR6 provides a versatile option for tightly regulated plasmid-based expression in different culturing conditions. While induction profiles showed that pDCyR6 and pDCyR2 had similar maximal RFU outputs across media types, pDCyR2 had lower fold changes due to leaky expression, consistent with our previous screening data (Fig. 1D and E).
While very high levels of inducible expression are often desirable, some experiments require low expression to match physiological levels or avoid overburdening the host (39). Of the four CymRAM-regulated systems tested here, pDCyR1 consistently had the lowest expression across the three media types by stationary phase and remained very tightly off after overnight growth. The pDLlR6 plasmid had the lowest induced expression from the three remaining systems while still tightly off in the absence of an inducer, consistent with data showing that LacI/PLlacO-1 generally had lower induced expression levels (Fig. 1D and E).
The pDLxR6 plasmid had the highest overall RFU output, with expression 4-fold and 12-fold higher than the T7 promoter in M9Glu and M9Gly, respectively, after an overnight induction and growth and over 350-fold in rich media at both time points. This was consistent with our previous data, ranking pDLxR6, pELxR4, and pDCyR2 among the highest-expressing plasmids in the toolbox. Though pDLxR6 and pDCyR2 had higher leaky expression than the same systems on different backbones, these plasmids are still useful where high output is necessary and expression in the absence of inducer is less of a concern. Though the LuxR construct was leaky at both time points, its expression was still an order of magnitude lower than uninduced T7 expression on the same pCDF backbone in both rich and minimal media.
Multiplasmid strains.
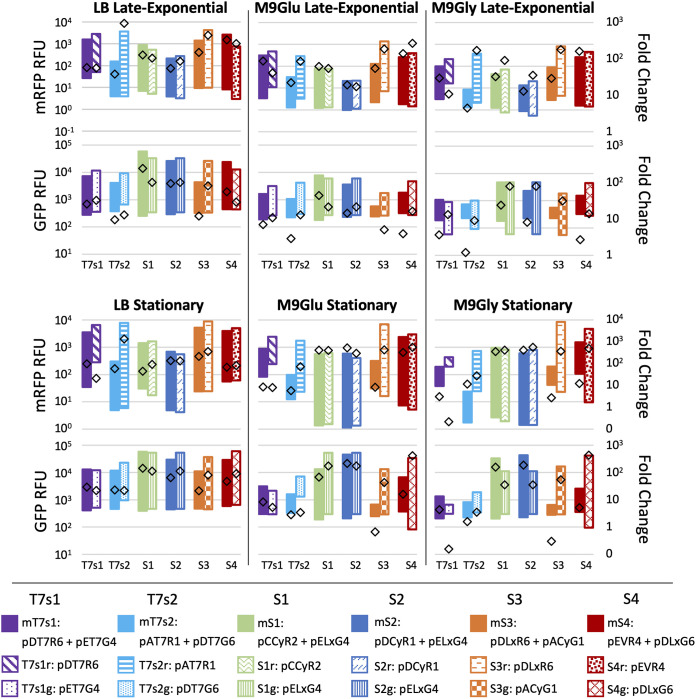
We next measured expression in strains that possessed two plasmids to determine whether this increased burden affected expression. The main feature of the Duet vectors is expression of several target genes from multiple plasmids with compatible origins, though because PT7lac regulates all genes, expression is tunable only through variation in copy number (29). In our plasmid system, target genes are controlled by different inducible systems on compatible plasmids, enabling more fine-tuned and temporal control for expressing different genes or operons. Though plasmid-based expression of multiple genes is employed frequently in metabolic engineering, direct comparisons to the widely used T7 system are lacking (15, 42, 43). Accordingly, E. coli BL21(DE3) strains were constructed that possessed Duet and toolbox plasmids both as single-plasmid and multiplasmid systems. Strains were organized into six groups, with multiplasmid strains designated mT7s1 mT7s2, and mS1-4 and single-plasmid strains designated with the group name plus “r” or “g,” as listed in Table 1. The single-plasmid strains in this experiment were also used for the experiments shown in Fig. 1, 3, and 4; however, the single-plasmid strain labels in Table 1 are used here for clarity. Across groups, induction profiles from toolbox multiplasmid strains where both plasmids were induced simultaneously were compared to the Duet multiplasmid strains in the presence and absence of IPTG, and within groups expression levels from multiplasmid strains were compared to those of single-plasmid strains (Fig. 5).
TABLE 1.
| Group | Strain | Plasmid(s) |
|---|---|---|
| T7s1 | mT7s1 | pDT7R6 + pET7G4 |
| T7s1r | pDT7R6 | |
| T7s1g | pET7G4 | |
| T7s2 | mT7s2 | pAT7R1 + pDT7G6 |
| T7s2r | pAT7R1 | |
| T7s2g | pDT7G6 | |
| S1 | mS1 | pCCyR2 + pELxG4 |
| S1r | pCCyR2 | |
| S1g | pELxG4 | |
| S2 | mS2 | pDCyR1 + pELxG4 |
| S2r | pDCyR1 | |
| S2g | pELxG4 | |
| S3 | mS3 | pDLxR6 + pACyG1 |
| S3r | pDLxR6 | |
| S3g | pACyG1 | |
| S4 | mS4 | pEVR4 + pDLxG6 |
| S4r | pEVR4 | |
| S4g | pDLxG6 |
Each strain group includes a multiplasmid strain and two single-plasmid strains, each possessing one of the two plasmids included in the multiplasmid strain.
FIG 5.
Expression data from induction experiments of single- and multiplasmid strains of E. coli BL21(DE3). Each strain group on the x axis shows induction data from the multiplasmid strain (solid bars) and single-plasmid strain (patterned bars), with mRFP readings at the top and GFP readings at the bottom of each time point pairing. Expression of GFP and mRFP from multiplasmid strains was induced simultaneously by both cognate inducers, and expression from single-plasmid strains was induced by the single cognate inducer. Vertical bars represent the induction range of mRFP or GFP for each strain, with fluorescence in the absence of inducer plotted at the bottom of each bar and induced expression plotted at the top. Fold change for each strain is represented by diamonds. Strains were tested in three medium types (LB, M9Glu, and M9Gly), and data were recorded at two time points. Inducer concentrations are listed in Table S3. All data are averages of triplicates.
First, we compared the effect of genetic context in the six multiplasmid strains (Table 1). Expression from the T7 promoters in mT7s1 and mT7s2 was consistent with expected behavior based on plasmid copy number. Green fluorescent protein (GFP) expression from pETDuet-1 in mT7s1 was consistently higher than expression from all other systems in the multiplasmid Duet strains, and mRFP expression from the low-copy pACYCDuet-1 plasmid in mT7s2 was the lowest. Among the toolbox plasmids, mRFP expression from the CymRAM-regulated systems in mS1 and mS2 were very similar by the stationary-phase time point, and GFP expression from pELxG4 in both mS1 and mS2 was consistently higher than GFP expression from pDLxG6 in mS4. These trends remained consistent across both time points in both rich and minimal media, supporting the idea that copy number itself can effectively be a rough mechanism for tuning expression of target genes on different plasmids.
Among the toolbox strains, mS1 and mS2 both possessed pELxG4, and mRFP was expressed from CymRAM-regulated systems on different plasmid backbones (Fig. 5). Though the pCDFDuet-1 and pCOLADuet-1 backbones had similar copy numbers, mRFP expression from pCCyR2 in mS1 was consistently higher than that from pDCyR1 in mS2 in the presence of only cumate and both cognate inducers (Fig. S5; Fig. 5). Strain mS3 possessed pDLxR6, which is among the highest-expressing toolbox plasmids in single-plasmid strains (Fig. 1, 3, and 5). Surprisingly, mRFP expression from pDLxR6 in mS3 was the lowest of the four toolbox multiplasmid strains by stationary phase in minimal medium conditions. Independent induction of pDLxR6 in mS3 showed similar results. These induction profiles exemplified how changing plasmid pairings and culturing media influence independent and dual expression in multiplasmid systems in unexpected ways.
We then compared expression between multi- and single-plasmid strains. Expression levels from multiplasmid strains were generally expected to be lower than from single-plasmid strains containing the same plasmids individually, due to an increased plasmid burden. Expression from the single-plasmid Duet strains was consistently higher and more leaky than expression of the same reporter in mT7s1 and mT7s2, except for T7s1g, which had slightly lower induced expression levels than GFP expression from mT7s1 at stationary phase. Expression from T7s2r was markedly higher than mRFP expression from mT7s2, which had the lowest expression level of all Duet multiplasmid strains. This could have been due to the phenomenon of T7 RNAP sequestration (44, 45), where the amount of T7 RNAP available for transcription is split between each copy of different plasmids and expression from the lower-copy plasmid, in this case pAT7R1, can be disproportionately reduced by the presence of higher-copy plasmids, here pDT7G6. Differences between single- and multiplasmid systems were most extreme for groups S3 and S4 in minimal media. By stationary phase, fold changes were 2 orders of magnitude higher for both S3r and S3g compared to mRFP and GFP expression from mS3, respectively; with the exception of S4r in M9Glu, fold changes from S4r and S4g were an order of magnitude higher than that from mS4. Conversely, mRFP expression from mS1 and mS2 was not notably different than that from S1r and S2r single-plasmid strains, and fold changes were similar across media types. Interestingly, the GFP expression from mS1 was higher than from S1g across all measurements except for M9Glu after overnight growth. By stationary phase, GFP expression from mS2 was higher than that from S2g.
In comparison, strains with two toolbox plasmids had a much higher dynamic range than multiplasmid Duet strains in most cases, especially in minimal media. With IPTG induction of mT7s1 and mT7s2 in minimal media, neither mRFP nor GFP were inducible over 90-fold at late-exponential phase, and the fold change dropped to a maximum of 12 after an overnight induction, mostly due to leaky expression. In contrast, at least one multiplasmid toolbox strain expressed both GFP and mRFP over 60-fold in LB and over 185-fold in M9Glu and M9Gly by the stationary-phase time point. Moreover, the overall expression of each reporter in at least one toolbox strain was higher than T7lac-driven reporters after an overnight induction across all media conditions.
These data suggest that toolbox inducible expression systems can effectively be utilized in a multiplasmid system and outperform widely used T7 systems in several key ways. First, toolbox plasmids allow independent expression of target genes with minimal cross talk, a feature inherently unattainable with multiplasmid Duet systems. Second, expression in the absence of inducer was similarly low or lower in the toolbox systems than in Duet systems. Based on our previous data (Fig. 4), this was expected and held true in multiplasmid systems. Finally, toolbox systems had induced expression levels that were higher than T7 systems in almost every case, even when both inducers were present. Though the toxicity of the bacteriophage expression system is often seen as a necessary evil to obtain very high expression levels, our data suggest the same expressions can be achieved in a more controllable manner and without the associated issues.
Lycopene production comparison.
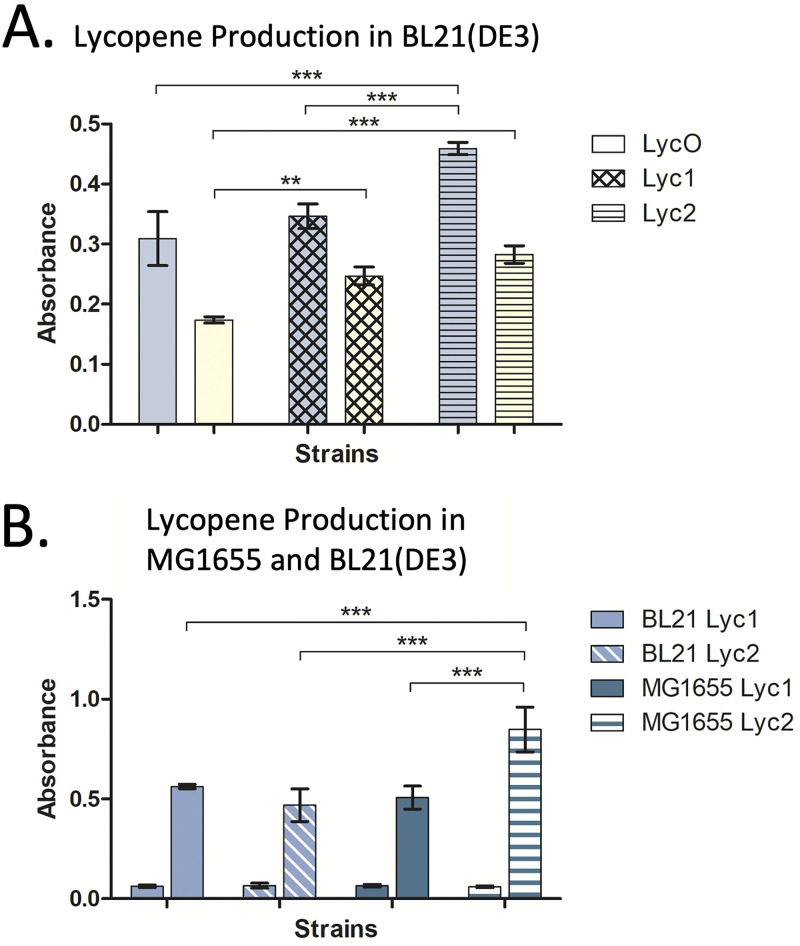
To demonstrate that our toolbox plasmids can also compete with the T7 promoter in metabolic engineering experiments, we recloned a previously described pathway for lycopene production and compared it against the original plasmids (46). In the previous two-plasmid system, the IUP genes ChK, IPK, and idi were expressed from the pro4 constitutive promoter (pSEVA228-pro4IUPi), while the lycopene genes crtl, crtB, ipi, and ggpps were expressed from a low-copy plasmid with the T7 promoter (p5T7-LYCipi-ggpps). To compare our toolbox plasmids, we cloned the IUP and lycopene genes under the control of CymRAM/PCymRC and LuxR/PLuxB, respectively. For consistency, the same markers were used, and the p15A and RK2 origins were chosen that closely matched the low plasmid copy numbers used in the original work (29, 46–48).
We first measured lycopene production in BL21(DE3) cells with three different plasmid pathways: (i) the original p5T7-LYCipi-ggpps and pSEVA228-pro4IUPi system, designated LycO, (ii) pALxLyc6 and pSEVA228-pro4IUPi, designated Lyc1, and (iii) pALxLyc6 and pRCyIUP2, designated Lyc2. The results showed that the lycopene production from Lyc2 was significantly higher than Lyc1 and LycO in minimal medium (Fig. 6A). In rich medium, Lyc1 and Lyc2 produced significantly more lycopene than LycO as well.
FIG 6.
Lycopene production in two strains of E. coli. Strains MG1655 and BL21(DE3) were tested for lycopene production via a two-plasmid system incorporating a pathway developed by Stephanopoulos et al. (A) Lycopene pathways were screened in BL21(DE3) on three different plasmid backbones, including p5T7-LYCipi-ggpps and pSEVA228-pro4IUPi (LycO), pALxLyc6 and pSEVA228-pro4IUPi (Lyc1), and pALxLyc6 and pRCyIUP2 (Lyc2). Induced lycopene production is shown in two medium conditions: minimal (blue bars) and rich (yellow bars). (B) Lyc1 and Lyc2 were compared in E. coli MG1655 and BL21(DE3) in minimal media. For each pair of bars, the first bar represents uninduced expression and the second shows induced expression. Lycopene was quantified through absorbance readings taken at 475 nm. Data shown are averages of three replicates, with standard deviations displayed. Statistical significance was determined with a two-way analysis of variance and Bonferroni’s multiple-comparison test. **, P < 0.01; ***, P < 0.001.
Because our toolbox plasmids are not constrained to BL21(DE3) strains, we also tested the E. coli K-strain MG1655. The strains were screened in parallel with Lyc1 and Lyc2 systems in BL21(DE3) (Fig. 6B). Surprisingly, we found that the highest lycopene production came from the Lyc2 system in MG1655, with production 13-fold over basal levels, compared to 6-fold from the same system in BL21(DE3). Induced lycopene production from Lyc2 in MG1655 was significantly higher than production from Lyc1 in both MG1655 and BL21(DE3) and from Lyc2 in BL21(DE3). Output from the Lyc1 systems in the two E. coli strains was not significantly different.
Overall, lycopene was notably lower in BL21(DE3) (Fig. 6A) than in the same strain in the assay comparing BL21(DE3) to MG1655 (Fig. 6B). However, this may have been due to a difference in protocols during the subculturing of the strains, rather than physiological differences between the two strains. In the assay across BL21(DE3) strains, the cells were subcultured twice prior to induction, similar to the fluorescence assays (see Materials and Methods). While this method has been shown to increase output in other studies (34, 36), it had the effect of decreasing lycopene in this experiment.
DISCUSSION
In this study, we expanded the application of our previously described plasmid toolbox to include both a large dynamic range and high overexpression in E. coli. Utilizing our combinatorial assembly method (36), we efficiently constructed plasmids with four promoter-regulator pairs, four antibiotic markers, and four enteric bacteria-specific origins of replication, generating a collection of 28 variants. After validating that LuxR/PLuxB, CymRAM/PCymRC, LacI/PLlacO-1, and VanRAM/PVanCC were functional on these vector backbones in E. coli, we assessed the dynamic range of expression compared to T7 systems in the same genetic background and found that our toolbox promoter-regulator pairs outperformed the T7 system in most cases. We showed that our toolbox systems were tunable, capable of independent expression in multiplasmid systems, and produced higher titers of the end product in a reengineered metabolic pathway. These findings challenge the use of the T7 system as the default when protein overexpression is desired.
Protein overexpression is necessary to generate high titers of value-added end products (46, 49, 50), to confirm recombinant protein function (51, 52), or to generate sufficient levels of a target protein for functional or structural studies (53–55). The T7 system is often chosen due to the high processivity of the RNAP and specificity in recognizing its promoter (5, 7). The T7 RNAP has been lauded for its high activity based on the premise that more mRNA would result in more protein, but this relationship quickly breaks down when host resources are exhausted, leading to growth inhibition and low protein yields (7, 19, 56–58). The system exhibits substantial leaky expression, as only small amounts of T7 RNAP can lead to high target gene expression in the absence of an inducer. In fact, omitting the inducer entirely and treating the T7 RNAP as constitutively expressed has been used effectively to generate membrane and secretory proteins (59). However, high uninduced expression can cause difficulty in obtaining transformants, for example, when the metabolic burden for protein production is too high or when the heterologous protein is toxic to the host (6, 30).
In protein overexpression experiments, it is often necessary to manage the level of T7 RNAP activity to balance protein production and host growth (60). Because expression of and from T7 RNAP is not easily tunable by titrating inducer concentration, alternative solutions have been utilized including the addition of T7 lysozyme to inhibit the activity of T7 RNAP (18–20, 61) and the construction or identification of mutants that are better equipped to handle the metabolic stress (21, 22, 26, 27, 60). T7 lysozyme is usually included on a separate plasmid, and while it can decrease leaky expression by up to 10 times, it also decreases protein expression following induction and results in growth inhibition as the lysozyme, a bifunctional enzyme, can also cut a specific bond in the cell wall of E. coli (11, 18). Moreover, additional plasmids complicate the system, limiting options in multiplasmid experiments and increasing any necessary fine-tuning for optimal protein production (18, 29).
T7 expression systems are notoriously unstable and frequently mutate to reduce the burden of protein overexpression on the host cell. This instability is often the result of mutation rather than plasmid loss (30), and toxicity-escape mutations have been found within the lacUV5 promoter regulating T7 RNAP expression (21), within the regulatory region of the T7 RNAP gene (62), and within LacI (27). These mutations often dampen the production or activity of T7 RNAP or lower the affinity of T7 RNAP to its promoter (26–28, 30, 60). Mutant hosts have emerged from overproduction experiments, themselves becoming popular protein-producing strains and also helping to inform targeted mutations that offer increased control and reduced toxicity (21, 23, 63). The obvious drawback to these proposed solutions is the necessity for specific strains, e.g., the Walker strains, C44(DE3) and C45(DE3), and in some cases additional inducers and plasmids (21–23). Additionally, these mutations are unpredictable and often have undesirable effects, including the total loss of target gene expression (28, 64, 65).
Other proposed solutions include utilizing different inducible systems, constitutive promoters, or negative feedback loops to control T7 RNAP (16, 30, 44), splitting T7 RNAP to alleviate stress and toxicity (66), or inhibiting the T7 RNAP through other means (32, 66–69). Though effective, splitting T7 RNAP is labor-intensive, and reconfiguring the T7 system to change its regulation necessitates reengineering and may still result in toxicity intrinsic to T7 RNAP (30, 58).
In the work presented here, we demonstrated that our plasmid toolbox systems are capable of high, tightly controlled expression. The combinatorial strategy we have developed for plasmid assembly with choice of origin, marker, inducible system, and target gene is straightforward and efficient (36). Our plasmids offer a dynamic range of expression levels, and most systems are tunable through titrated inducer concentration (Fig. 2) and are stable over several passages (Fig. 3), all without the need for special mutant strains or auxiliary plasmids. Crucially, toolbox expression systems can achieve higher expression levels than T7 systems on the same genetic backbone across rich and minimal media (Fig. 4). Overexpression of fluorescent proteins can result in inclusion bodies, and the proportion of soluble to insoluble protein is influenced by multiple factors that include the expression vector, culturing conditions, host genetic background, and expressed protein (40, 70). Fluorescent protein aggregation has a mild to moderate effect on specific fluorescence, or the fluorescence per microgram of protein, which varies according to the specific protein being expressed (71, 72). While our fluorescence data were obtained from whole-cell measurements, we believe that loss of fluorescence signal due to inclusion bodies was unlikely to have significantly influenced our results for two reasons. First, expression trends were consistent across rich and minimal media culturing conditions, different reporter proteins, and the two E. coli strains used in this study. Second, variability due to genetic context was minimized with use of the same plasmid backbone and host for comparisons between toolbox and T7 expression systems.
Our plasmids also provided independent control over different target proteins in coexpression experiments. While the Duet vectors are important additions to the plasmids available for coexpression, their combined usage has been associated with unexpected problems. When both high- and low-copy Duet vectors are used in a metabolic pathway, T7 RNAP sequestration can occur where expression from genes on the lower-copy plasmid are less than expected (45). This was evident in our experiments when we compared induced expression from strain mT7s2 to expression from a pACYCDuet-1 single-plasmid strain, T7s2r (Fig. 5). This differential partitioning phenomenon is inherent to the orthogonality of T7 RNAP to its promoter and can occur when multiple T7 promoters are used in the same system (44). Because the T7lac promoter regulates all target genes in the Duet vectors, independent and tunable expression is not possible. Our toolbox promoter-regulator pairs can effectively replace the T7 system in a synthetic multiplasmid pathway for synthesizing lycopene. We argue that the modularity and tunability of our plasmid toolbox offer advantages over those currently available for protein overproduction in E. coli and have the potential to be expanded to other hosts.
Conventional T7 systems, with a chromosomally integrated T7 RNAP and cognate expression plasmid, have been used in nonmodel hosts but are often inefficient or completely nonfunctional, likely due to phage polymerase-associated toxicity (73). In developing successful phage-derived expression systems outside of E. coli, researchers have turned to part mining T7-like expression systems (73–75) or using similar strategies to those discussed above for controlling the activity of T7 RNAP. Notably, the UBER and HITES systems have been used effectively in Gram-negative and Gram-positive hosts. Still, they require that T7 RNAP basal expression be repressed through elaborate feedback loops or antisense RNA (31, 44). Even still, toxicity remains a concern and continues to restrict use of the T7 expression system in nonmodel bacteria (76). In a previous study, we constructed and tested broad-host-range vectors containing 12 inducible systems, including the four utilized here, across nine members of the Proteobacteria. We demonstrated that LuxR/PLuxB, CymRAM/PCymRC, LacI/PLlacO-1, and VanRAM/PVanCC have a range of induction levels in these species and can be used successfully in two-plasmid systems (36). Where E. coli BL21(DE3) is not an ideal host for protein overproduction or structure/function studies, our expression systems can be moved and utilized in other bacteria quickly and effectively.
Overall, we achieved high expression from our toolbox plasmids in both E. coli BL21(DE3) and MG1655 in rich and minimal media conditions, demonstrating that our inducible systems can accommodate versatile protein production strategies. We also demonstrated the evolutionary stability of our expression systems compared to T7 systems through an extended passage experiment under the pressure of constant induction. Finally, we reengineered a previously constructed metabolic pathway that utilized the T7 system to incorporate our toolbox promoter-regulators and showed that the reengineered pathway produced a higher titer of the end product, lycopene. We argue that the benefits of orthogonality in the T7 system is outweighed by its toxicity and lack of control over expression and that our toolbox plasmids offer an alternative with distinct advantages. These expression plasmids further expand the broad-host-range toolbox for investigating, coordinating, and optimizing gene expression in E. coli.
MATERIALS AND METHODS
Plasmid construction and transformation.
Plasmids were assembled using NEB HiFi Assembly with PCR-amplified genetic parts using a protocol established in previous work from this lab (36). A list of regulatory parts and their sources are available in Table S2 in the supplemental material, and schematics of genetic parts are in Fig. S1. Plasmids available at Addgene are listed in Table S1.
All recipient E. coli strains were transformed via electroporation using the following protocol: 5 mL cultures were started from isolated colonies and incubated with shaking overnight. The following day, the cells were subcultured 1:50 in 5 mL of fresh medium until cells reached exponential growth or an optical density (OD) of approximately 0.5. The total culture volume was then spun down in microcentrifuge tubes at 5,000 × g for 2 min. Culture supernatants were aspirated, and cell pellets were resuspended in 1 mL of 300 mM sucrose at room temperature and centrifuged again for an additional 2 min at 5,000 rpm. The wash was repeated, and then cells were resuspended in a final volume of 1/10 of the initial culture volume. Suspensions of 50 μL were electroporated in a 1-mm electroporation cuvette, and cells were electroporated at 1.8 kV. Cells were recovered in 1 mL of LB and incubated for 1 h at 37°C.
Both plasmids were electroporated together for the multiple plasmid systems tested in Fig. 5. Strains utilized in the lycopene production experiments in Fig. 6 could not be efficiently transformed simultaneously, and the transformations were done sequentially. In this way, wild-type E. coli MG1655 or BL21(DE3) cells were transformed with the first plasmid following the protocol outlined above, and from the transformation plate, a single colony was grown and the same process was followed to transform the second plasmid.
Fluorescence assays.
As displayed in Fig. 1 to 5, fluorescence measurements were taken as follows. For each set of plasmids housed in E. coli MG1655 to be screened, glycerol stocks were struck onto fresh plates. Isolated colonies were used to inoculate 1 mL of medium in a deep-well plate and incubated on a plate shaker overnight. The following day, the cultures were diluted to an OD of 0.1 in 1 mL of fresh medium and antibiotics in a deep-well plate. At exponential phase the cultures were diluted into 100 μL of fresh medium in a 96-well plate (Costar, black, clear-bottom) to an OD of 0.07 and incubated with shaking for 0.5 h. At this point, 100 μL of medium with antibiotic and 2× inducer was added to wells to induce samples, and 100 μL of medium with antibiotic only was added to uninduced control wells. The plate was incubated on a plate shaker, and fluorescence and OD measurements were taken at 1, 2, 4, 6, and 24 h postinduction in a plate reader (Molecular Devices SpectraMax M3). All experiments were performed in three technical replicates, and both wild-type and empty vector controls (cells transformed with a plasmid lacking mrfp) were included on each plate as negative controls. The same protocol was followed for measurements in E. coli BL21(DE3), except all cultures were started from freshly transformed cells.
Calculations and data analysis were performed using Microsoft Excel. Each screening data set was first organized into time point OD and RFU measurements, and the OD was adjusted to a 1-cm pathlength by dividing by a factor of 0.56 or 0.28 for the measurement of 200 or 100 μL of culture, respectively (36). As noted in figure descriptions, raw fluorescence data were either used directly or normalized to optical density readings. Fold change values were calculated by subtracting the uninduced fluorescence from the induced fluorescence and dividing this value by the uninduced fluorescence.
Stability screen experiments.
(i) Cultures. Cultures were started from freshly transformed BL21(DE3) cells in 1 mL M9 glucose (0.4%) with the appropriate antibiotics in a deep-well plate. Eight technical replicates were included for each of the 12 strains under study. The deep-well plate was grown with shaking overnight at 37°C, and the following day, the cultures were diluted 1:1,000 into fresh medium in a deep-well plate, marking day 1 of the screen. This plate was grown with shaking overnight, and the following day, the culture was diluted 1:100 into two deep-well plates: one with glucose-supplemented minimal medium and relevant antibiotics only and one with relevant inducers added to the medium. This process was repeated, diluting 1:100 of overnight cultures from the induced and uninduced plates into fresh medium with and without inducer, respectively, for 12 total days.
(ii) Fluorescence measurements. On each day, 200 μL of overnight culture was transferred from both deep-well plates into 96-well plates (Costar, black, clear-bottom) to read fluorescence in a plate reader (Molecular Devices SpectraMax M3). On day 11, 50 μL of culture was taken from induced cultures and struck onto LB plates with the appropriate antibiotic and relevant inducer. After an overnight incubation at 37°C, pictures were taken of each plate under blue light to visualize mRFP fluorescence of colonies.
(iii) Visualization. The day 12 plate of induced cultures was used for cytometry analysis. After overnight growth, 500 μL was transferred to a deep-well plate and spun down in a plate spinner until cells were pelleted in each well. The supernatant of minimal medium was aspirated, and the pellet was resuspended in 500 μL of a 4% formalin cell fixing solution and incubated for 10 min. The plate was then spun down again to pellet the cells, the fixing solution was removed using suction, and the cells were resuspended in 500 μL of phosphate-buffered saline.
(iv) Flow cytometry. Measurements were taken using a green laser (488 nm) with the standard 670 LP filter, and 10,000 events were used for analysis. Samples were left ungated to allow for the detection of multiple peaks. Data were analyzed using FlowJo (version 10.8.1) and are displayed as histograms. The geometric mean and coefficient of variation are shown for each replicate in Table S4.
Lycopene experiments.
Cultures were started from freshly transformed BL21(DE3) and MG1655 cells and grown overnight at 37°C in 5 mL of LB medium with the appropriate antibiotics. Overnight cultures were inoculated at 1% (vol/vol) into 3 mL of M9 medium supplemented with 0.32% glucose, 0.5% Casamino Acids, and ATCC trace mineral supplement and the appropriate antibiotics and grown in glass tubes with rubber stoppers at 37°C until cultures reached exponential phase. At this point, the cognate inducer(s) and 25 mM isoprenol were added to the cultures, and the glass vials were sealed with a Teflon-coated stopper and crimp sealed to prevent the evaporation of isoprenol. Inducer concentrations per strain were as follows: 0.1 mM IPTG for LycO, 10 μM OC6 for Lyc1, 10 μM OC6 and 100 μM cumate for Lyc2. Cultures were then grown overnight, and lycopene extraction was performed the following morning. A 500 μL volume of each culture was spun down in 1.5 mL microcentrifuge tubes at 16,000 × g for 1 min. The supernatant was decanted, and the pellet was resuspended in 1 mL of a solution of 50% ethanol and 50% acetone to extract lycopene. The tubes were then vortexed for 15 min and centrifuged again for 1 min at 16,000 × g to remove particulates. The extraction took place in a darkened room, as lycopene is light sensitive. A 200 μL aliquot was then transferred to a microplate, and absorbance was recorded at 475 nm.
ACKNOWLEDGMENTS
Funding was provided by the University of Florida Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences. L.A.S. was supported by a GSPA fellowship from the University of Florida.
Footnotes
Supplemental material is available online only.
Contributor Information
Christopher R. Reisch, Email: creisch@ufl.edu.
Martha Vives, Unversidad de los Andes.
REFERENCES
- 1.Hewitt L, McDonnell JM. 2004. Screening and optimizing protein production in E. coli. Methods Mol Biol 278:1–16. 10.1385/1-59259-809-9:001. [DOI] [PubMed] [Google Scholar]
- 2.Tolia NH, Joshua-Tor L. 2006. Strategies for protein coexpression in Escherichia coli. Nat Methods 3:55–64. 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- 3.Gold L. 1990. Expression of heterologous proteins in Escherichia coli. Methods Enzymol 185:11–14. 10.1016/0076-6879(90)85004-8. [DOI] [PubMed] [Google Scholar]
- 4.Adams BL. 2016. The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth Biol 5:1328–1330. 10.1021/acssynbio.6b00256. [DOI] [PubMed] [Google Scholar]
- 5.Studier W, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185:60–89. 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 6.Dubendorf JW, Studier FW. 1991. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J Mol Biol 219:45–59. 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 7.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlin M, Mcgrath J, Waskell L. 1970. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature 228:227–231. 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg AH, Lade BN, Chui DS, Lin SW, Dunn JJ, Studier FW. 1987. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56:125–135. 10.1016/0378-1119(87)90165-X. [DOI] [PubMed] [Google Scholar]
- 10.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Terpe K. 2006. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72:211–222. 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 12.Pan SH, Malcolm BA. 2000. Reduced background expression and improved plasmid stability with pET vectors in BL21(DE3). Biotechniques 29:1234–1238. 10.2144/00296st03. [DOI] [PubMed] [Google Scholar]
- 13.Dong H, Nilsson L, Kurland CG. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J Bacteriol 177:1497–1504. 10.1128/jb.177.6.1497-1504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bervoets I, Van Brempt M, Van Nerom K, Van Hove B, Maertens J, De Mey M, Charlier D. 2018. A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res 46:2133–2144. 10.1093/nar/gky010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW. 2006. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 40:355–364. 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- 16.Wycuff DR, Matthews KS. 2000. Generation of an AraC-araBAD promoter-regulated T7 expression system. Anal Biochem 277:67–73. 10.1006/abio.1999.4385. [DOI] [PubMed] [Google Scholar]
- 17.de Boer HA, Comstock LJ, Vasser M. 1983. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci USA 80:21–25. 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier FW. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol 219:37–44. 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 19.Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier J-W. 2008. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci USA 105:14371–14376. 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spehr V, Frahm D, Meyer TF. 2000. Improvement of the T7 expression system by the use of T7 lysozyme. Gene 257:259–267. 10.1016/S0378-1119(00)00400-5. [DOI] [PubMed] [Google Scholar]
- 21.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 22.Angius F, Ilioaia O, Amrani A, Suisse A, Rosset L, Legrand A, Abou-Hamdan A, Uzan M, Zito F, Miroux B. 2018. A novel regulation mechanism of the T7 RNA polymerase based expression system improves overproduction and folding of membrane proteins. Sci Rep 8:8572. 10.1038/s41598-018-26668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SK, Lee DH, Kim OC, Kim JF, Yoon SH. 2017. Tunable control of an Escherichia coli expression system for the overproduction of membrane proteins by titrated expression of a mutant lac repressor. ACS Synth Biol 6:1766–1773. 10.1021/acssynbio.7b00102. [DOI] [PubMed] [Google Scholar]
- 24.Anilionyte O, Liang H, Ma X, Yang L, Zhou K. 2018. Short, auto-inducible promoters for well-controlled protein expression in Escherichia coli. Appl Microbiol Biotechnol 102:7007–7015. 10.1007/s00253-018-9141-z. [DOI] [PubMed] [Google Scholar]
- 25.Blazeck J, Alper HS. 2013. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J 8:46–58. 10.1002/biot.201200120. [DOI] [PubMed] [Google Scholar]
- 26.Schlegel S, Genevaux P, de Gier JW. 2015. De-convoluting the genetic adaptations of E. coli C41(DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep 10:1758–1766. 10.1016/j.celrep.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Kwon SK, Kim SK, Lee DH, Kim JF. 2015. Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci Rep 5:16076. 10.1038/srep16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vethanayagam JG, Flower AM. 2005. Decreased gene expression from T7 promoters may be due to impaired production of active T7 RNA polymerase. Microb Cell Fact 4:3. 10.1186/1475-2859-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Held D, Yaeger K, Novy R. 2003. New coexpression vectors for expanded compatibilities in E. coli. Innovations. [Google Scholar]
- 30.Tan SI, Ng IS. 2020. New insight into plasmid-driven T7 RNA polymerase in Escherichia coli and use as a genetic amplifier for a biosensor. ACS Synth Biol 9:613–622. 10.1021/acssynbio.9b00466. [DOI] [PubMed] [Google Scholar]
- 31.Liang X, Li C, Wang W, Li Q. 2018. Integrating T7 RNA polymerase and its cognate transcriptional units for a host-independent and stable expression system in single plasmid. ACS Synth Biol 7:1424–1435. 10.1021/acssynbio.8b00055. [DOI] [PubMed] [Google Scholar]
- 32.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. 2012. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res 40:8773–8781. 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosano GL, Morales ES, Ceccarelli EA. 2019. New tools for recombinant protein production in Escherichia coli: a 5-year update. Protein Sci 28:1412–1422. 10.1002/pro.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, Voigt CA. 2019. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat Chem Biol 15:196–204. 10.1038/s41589-018-0168-3. [DOI] [PubMed] [Google Scholar]
- 35.Brewster RC, Weinert FM, Garcia HG, Song D, Rydenfelt M, Phillips R. 2014. The transcription factor titration effect dictates level of gene expression. Cell 156:1312–1323. 10.1016/j.cell.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster LA, Reisch CR. 2021. A plasmid toolbox for controlled gene expression across the Proteobacteria. Nucleic Acids Res 49:7189–7202. 10.1093/nar/gkab496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook TB, Rand JM, Nurani W, Courtney DK, Liu SA, Pfleger BF. 2018. Genetic tools for reliable gene expression and recombineering in Pseudomonas putida. J Ind Microbiol Biotechnol 45:517–527. 10.1007/s10295-017-2001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. 2006. A bottom-up approach to gene regulation. Nature 439:856–860. 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 39.Jones KL, Kim SW, Keasling JD. 2000. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab Eng 2:328–338. 10.1006/mben.2000.0161. [DOI] [PubMed] [Google Scholar]
- 40.Lozano Terol G, Gallego-Jara J, Sola Martínez RA, Martínez Vivancos A, Cánovas Díaz M, de Diego Puente T. 2021. Impact of the expression system on recombinant protein production in Escherichia coli BL21. Front Microbiol 12:682001. 10.3389/fmicb.2021.682001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clomburg JM, Gonzalez R. 2013. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol 31:20–28. 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Balzer S, Kucharova V, Megerle J, Lale R, Brautaset T, Valla S. 2013. A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli. Microb Cell Fact 12:26. 10.1186/1475-2859-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gawin A, Valla S, Brautaset T. 2017. The XylS/Pm regulator/promoter system and its use in fundamental studies of bacterial gene expression, recombinant protein production and metabolic engineering. Microb Biotechnol 10:702–718. 10.1111/1751-7915.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kushwaha M, Salis HM. 2015. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat Commun 6:7832. 10.1038/ncomms8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhamankar H, Tarasova Y, Martin CH, Prather KLJ. 2014. Engineering E. coli for the biosynthesis of 3-hydroxy-γ-butyrolactone (3HBL) and 3,4-dihydroxybutyric acid (3,4-DHBA) as value-added chemicals from glucose as a sole carbon source. Metab Eng 25:72–81. 10.1016/j.ymben.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Chatzivasileiou AO, Ward V, Edgar SM, Stephanopoulos G. 2019. Two-step pathway for isoprenoid synthesis. Proc Natl Acad Sci USA 116:506–511. 10.1073/pnas.1812935116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76:1648–1652. 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kues U, Stahl U. 1989. Replication of plasmids in gram-negative bacteria. Microbiol Rev 53:491–516. 10.1128/mr.53.4.491-516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Jain KK, Bhardwaj KN, Chakraborty S, Kuhad RC. 2015. Multiple genes in a single host: cost-effective production of bacterial laccase (cotA), pectate lyase (pel), and endoxylanase (xyl) by simultaneous expression and cloning in single vector in E. Coli. PLoS One 10:e0144379. 10.1371/journal.pone.0144379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Weng H, Zhou Z, Du G, Kang Z. 2019. Engineering of multiple modular pathways for high-yield production of 5-aminolevulinic acid in Escherichia coli. Bioresour Technol 274:353–360. 10.1016/j.biortech.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, Begley TP. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem Biol 10:1195–1204. 10.1016/j.chembiol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Buck B, Zamoon J, Kirby TL, DeSilva TM, Karim C, Thomas D, Veglia G. 2003. Overexpression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expr Purif 30:253–261. 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- 53.Grisshammer R. 2006. Understanding recombinant expression of membrane proteins. Curr Opin Biotechnol 17:337–340. 10.1016/j.copbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Wagner S, Baars L, Ytterberg AJ, Klussmeier A, Wagner CS, Nord O, Nygren P-A, van Wijk KJ, de Gier J-W. 2007. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteomics 6:1527–1550. 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Wagner S, Bader ML, Drew D, de Gier JW. 2006. Rationalizing membrane protein overexpression. Trends Biotechnol 24:364–371. 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Iost I, Guillerez J, Dreyfus M. 1992. Bacteriophage T7 RNA polymerase travels far ahead of ribosomes in vivo. J Bacteriol 174:619–622. 10.1128/jb.174.2.619-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shis DL, Bennett MR. 2014. Synthetic biology: the many facets of T7 RNA polymerase. Mol Syst Biol 10:745. 10.15252/msb.20145492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Rinas U. 2020. Recombinant protein production associated growth inhibition results mainly from transcription and not from translation. Microb Cell Fact 19:83. 10.1186/s12934-020-01343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Kuipers G, Niemiec Ł, Baumgarten T, Slotboom DJ, de Gier JW, Hjelm A. 2015. High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG. Microb Cell Fact 14:142. 10.1186/s12934-015-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li ZJ, Zhang ZX, Xu Y, Shi TQ, Ye C, Sun XM, Huang H. 2022. CRISPR-based construction of a BL21(DE3)-derived variant strain library to rapidly improve recombinant protein production. ACS Synth Biol 11:343–352. 10.1021/acssynbio.1c00463. [DOI] [PubMed] [Google Scholar]
- 61.Kuipers G, Karyolaimos A, Zhang Z, Ismail N, Trinco G, Vikström D, Slotboom DJ, de Gier JW. 2017. The tunable pReX expression vector enables optimizing the T7-based production of membrane and secretory proteins in. Microb Cell Fact 16:226. 10.1186/s12934-017-0840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alfasi S, Sevastsyanovich Y, Zaffaroni L, Griffiths L, Hall R, Cole J. 2011. Use of GFP fusions for the isolation of Escherichia coli strains for improved production of different target recombinant proteins. J Biotechnol 156:11–21. 10.1016/j.jbiotec.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Sun XM, Zhang ZX, Wang LR, Wang JG, Liang Y, Yang HF, Tao RS, Jiang Y, Yang JJ, Yang S. 2021. Downregulation of T7 RNA polymerase transcription enhances pET-based recombinant protein production in Escherichia coli BL21(DE3) by suppressing autolysis. Biotechnol Bioeng 118:153–163. 10.1002/bit.27558. [DOI] [PubMed] [Google Scholar]
- 64.Randall LL, Topping TB, Smith VF, Diamond DL, Hardy SJS. 1998. SecB: a chaperone from Escherichia coli. Methods Enzymol 290:444–459. 10.1016/s0076-6879(98)90037-4. [DOI] [PubMed] [Google Scholar]
- 65.Kuderová A, Nanak E, Truksa M, Brzobohatý B. 1999. Use of rifampicin in T7 RNA polymerase-driven expression of a plant enzyme: rifampicin improves yield and assembly. Protein Expr Purif 16:405–409. 10.1006/prep.1999.1079. [DOI] [PubMed] [Google Scholar]
- 66.Shis DL, Bennett MR. 2013. Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc Natl Acad Sci USA 110:5028–5033. 10.1073/pnas.1220157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Quijano JF, Kim J, Yeung E, Murray RM. 2021. Synthetic logic circuits using RNA aptamer against T7 RNA polymerase. Biotechnol J 17:e2000449. 10.1002/biot.202000449. [DOI] [PubMed] [Google Scholar]
- 68.Han T, Chen Q, Liu H. 2017. Engineered photoactivatable genetic switches based on the bacterium phage T7 RNA polymerase. ACS Synth Biol 6:357–366. 10.1021/acssynbio.6b00248. [DOI] [PubMed] [Google Scholar]
- 69.Ohuchi S, Mori Y, Nakamura Y. 2012. Evolution of an inhibitory RNA aptamer against T7 RNA polymerase. FEBS Open Bio 2:203–207. 10.1016/j.fob.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramón A, Señorale-Pose M, Marín M. 2014. Inclusion bodies: not that bad. Front Microbiol 5:2010–2015. 10.3389/fmicb.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.García-Fruitós E, González-Montalbán N, Morell M, Vera A, Ferraz RM, Arís A, Ventura S, Villaverde A. 2005. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Fact 4:27. 10.1186/1475-2859-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raghunathan G, Munussami G, Moon H, Paik H, An SSA, Kim YS, Kang S, Lee SG. 2014. A variant of green fluorescent protein exclusively deposited to active intracellular inclusion bodies. Microb Cell Fact 13:68. 10.1186/1475-2859-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao H, Zhang HM, Chen X, Li T, Wu Q, Ouyang Q, Chen GQ. 2017. Novel T7-like expression systems used for Halomonas. Metab Eng 39:128–140. 10.1016/j.ymben.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Liang T, Sun J, Ju S, Su S, Yang L, Wu J. 2021. Construction of T7-like expression system in Pseudomonas putida KT2440 to enhance the heterologous expression level. Front Chem 9:664967. 10.3389/fchem.2021.664967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Troeschel SC, Thies S, Link O, Real CI, Knops K, Wilhelm S, Rosenau F, Jaeger KE. 2012. Novel broad host range shuttle vectors for expression in Escherichia coli, Bacillus subtilis and Pseudomonas putida. J Biotechnol 161:71–79. 10.1016/j.jbiotec.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 76.Lammens EM, Nikel PI, Lavigne R. 2020. Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nat Commun 11:5294. 10.1038/s41467-020-19124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00939-22-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)