ABSTRACT
Glycolysis is an ancient, widespread, and highly conserved metabolic pathway that converts glucose into pyruvate. In the canonical pathway, the phosphofructokinase (PFK) reaction plays an important role in controlling flux through the pathway. Clostridium thermocellum has an atypical glycolysis and uses pyrophosphate (PPi) instead of ATP as the phosphate donor for the PFK reaction. The reduced thermodynamic driving force of the PPi-PFK reaction shifts the entire pathway closer to thermodynamic equilibrium, which has been predicted to limit product titers. Here, we replace the PPi-PFK reaction with an ATP-PFK reaction. We demonstrate that the local changes are consistent with thermodynamic predictions: the ratio of fructose 1,6-bisphosphate to fructose-6-phosphate increases, and the reverse flux through the reaction (determined by 13C labeling) decreases. The final titer and distribution of fermentation products, however, do not change, demonstrating that the thermodynamic constraints of the PPi-PFK reaction are not the sole factor limiting product titer.
IMPORTANCE The ability to control the distribution of thermodynamic driving force throughout a metabolic pathway is likely to be an important tool for metabolic engineering. The phosphofructokinase reaction is a key enzyme in Embden-Mayerhof-Parnas glycolysis and therefore improving the thermodynamic driving force of this reaction in C. thermocellum is believed to enable higher product titers. Here, we demonstrate switching from pyrophosphate to ATP does in fact increases the thermodynamic driving force of the phosphofructokinase reaction in vivo. This study also identifies and overcomes a physiological hurdle toward expressing an ATP-dependent phosphofructokinase in an organism that utilizes an atypical glycolytic pathway. As such, the method described here to enable expression of ATP-dependent phosphofructokinase in an organism with an atypical glycolytic pathway will be informative toward engineering the glycolytic pathways of other industrial organism candidates with atypical glycolytic pathways.
KEYWORDS: Clostridium, metabolomics, metabolic engineering, advanced biofuels, Clostridium thermocellum
INTRODUCTION
Clostridium thermocellum is a promising candidate for production of lignocellulosic biofuels due to its strong native ability to consume cellulose. Metabolic engineering has resulted in strains that produce ethanol at high yield (>80% of theoretical) (1–3). However, titers above 30 g/L have not been achieved (1, 2, 4), thus limiting commercial viability of the engineered strains (5).
C. thermocellum belongs to a group of organisms that specialize in cellulose fermentation. These organisms typically have a limited substrate range, preferring to grow on crystalline cellulose and its hydrolysis products (6). For many of them, Embden-Mayerhof-Parnas (EMP) glycolysis is the only pathway for substrate assimilation, although several variations are present (7). In these organisms, glycolysis appears to be closer to thermodynamic equilibrium (8, 9), in part due to the different cofactor usage for several of its reactions (8). In the case of C. thermocellum glycolysis, it was previously found that pyrophosphate (PPi) was the main cofactor used in the 6-phosphofructokinase (PFK) reaction instead of ATP; in addition PPi is also necessary for conversion of phosphoenolpyruvate to pyruvate via the pyruvate-phosphate dikinase (PPDK) reaction. It was also previously demonstrated that biosynthetic reactions were insufficient to account for the PPi needed to explain glycolytic flux, suggesting therefore that a nonbiosynthetic reaction was responsible for generating the majority of the PPi needed by glycolysis (10). A recent study attempted to identify the primary source of PPi, but the findings were inconclusive (11).
Among these several atypical aspects of C. thermocellum glycolysis (10), the use of PPi instead of ATP as a high-energy phosphate donor for the PFK reaction has a large effect on the overall thermodynamics of the pathway for several reasons. One is that the standard Gibbs free energy of the PPi-PFK reaction (EC 2.7.1.90, ΔrG′° = −4.6 ± 1.4 kJ/mol) is lower than that of the ATP-PFK reaction (EC 2.7.1.11, ΔrG′° = −17.8 ± 1.3 kJ/mol) (12). Another is that the ATP/ADP ratio is around 10 in many organisms, whereas the PPi/Pi ratio is around 0.1 (13), which serves to increase the thermodynamic driving force of the ATP-linked reaction, while decreasing the driving force of the PPi-linked reaction. The low thermodynamic driving force of the PPi-PFK reaction can explain both the high reversibility of glycolysis in this organism and the large size of the intracellular hexose phosphate pools (glucose-6-phosphate [G6P] and fructose-6-phopshate [F6P]) in this organism compared to organisms with canonical glycolysis, such as Escherichia coli and Thermoanaerobacterium saccharolyticum, where the hexose phosphate pools are much smaller (8). It may also explain the low ethanol tolerance of C. thermocellum.
Growth of C. thermocellum is inhibited in the presence of 5 g/L ethanol and completely inhibited at concentrations of 15 g/L (14). A comparison of changes to relative intracellular metabolite concentrations in both C. thermocellum and T. saccharolyticum in the presence of increasing ethanol concentrations revealed that concentrations of glycolytic intermediates began to accumulate in C. thermocellum cells at much lower ethanol concentrations than in T. saccharolyticum (15). Of note, the relative concentrations of the hexose phosphate (G6P and F6P) pool—both being immediately upstream of the PFK reaction—increased significantly as a result of ethanol addition in C. thermocellum, whereas in T. saccharolyticum the relative hexose phosphate pool sizes remained relatively similar between the control and ethanol addition experiments, indicating that in C. thermocellum, this reaction is close to thermodynamic equilibrium and the upstream metabolite pool sizes are largely controlled by mass action effects.
The free energy change of a reaction can also be determined by measuring the relative forward and reverse flux in a reaction (16). In brief, a more thermodynamically favorable reaction will exhibit greater forward flux than reverse flux. Isotope labeling experiments where cells are fed a mixture of naturally labeled and universally labeled substrates can be used to determine the relative forward and reverse fluxes of a reaction, allowing one to infer differences in free energy changes (16, 17). Experiments with 13C-labeled substrates have shown that glycolysis is much closer to thermodynamic equilibrium in C. thermocellum compared to organisms that use the canonical pathway (e.g., E. coli and T. saccharolyticum) (8).
Furthermore, a thermodynamic analysis of elementary flux modes of alternative glycolytic pathways identified the PPi-PFK reaction as one of three important genetic interventions for increasing the overall thermodynamic driving force of the cellobiose to ethanol pathway (18).
Finally, in experiments designed to increase product titer by growing C. thermocellum in the presence of high concentration of substrate, a significant fraction of the substrate is left unconsumed or converted to products upstream of glycolysis (i.e., glucose) (1, 4).
These converging lines of evidence are the basis for our hypothesis that the low thermodynamic driving force of the PPi-PFK reaction limits ethanol titer in C. thermocellum and that replacing that enzyme with an ATP-linked enzyme would increase ethanol titer by allowing for more complete substrate consumption and reducing inhibition by product accumulation. It should be noted that the C. thermocellum genome contains a putative ATP-pfk gene, Clo1313_0997 (19); however, enzymatic assays to determine the cofactor usage of the PFK reaction in C. thermocellum indicate that the reaction is exclusively PPi dependent, with no detectable ATP-dependent activity (10), suggesting that either this native ATP-Pfk was inactive or possessing activity below our limit of detection in cell extracts, or that our assay conditions were not suitable for measuring its activity. Due to this observation, and that the native ATP-Pfk is not fully characterized and understood, it was decided that expressing a heterologous ATP-pfk—in this study taken from T. saccharolyticum (20)—with detectable activity was necessary.
One factor that complicates a simple replacement of PPi-pfk with ATP-pfk in C. thermocellum is that the PPi-dependent phosphofructokinase enzyme in C. thermocellum is believed to have an additional role in the nonoxidative pentose phosphate pathway, where it catalyzes the interconversion of sedoheptulose-7-phosphate (S7P) to sedoheptulose-1,7-bisphosphate (SBP) (19, 21, 22) to allow for interconversion of hexoses and pentoses. Deleting the PPi-Pfk protein from C. thermocellum would therefore disrupt not only glycolysis but also the pentose phosphate pathway. In the direction of hexose to pentose conversion, SBP must be converted to S7P (21, 22). The high thermodynamic driving force of the ATP-Pfk enzyme effectively prevents the reverse reaction, and it therefore appears necessary to introduce transaldolase activity to allow replacement of the PPi-pfk gene with and ATP-pfk gene.
In this study, we investigated the effect of changing the phosphate donor of the PFK reaction from PPi to ATP on the thermodynamic driving force of glycolysis in C. thermocellum by deleting the native PPi-pfk gene (Clo1313_1876) and expressing a heterologous ATP-pfk gene from T. saccharolyticum (Tsac_1362).
RESULTS
Decoupling glycolysis from the pentose phosphate pathway.
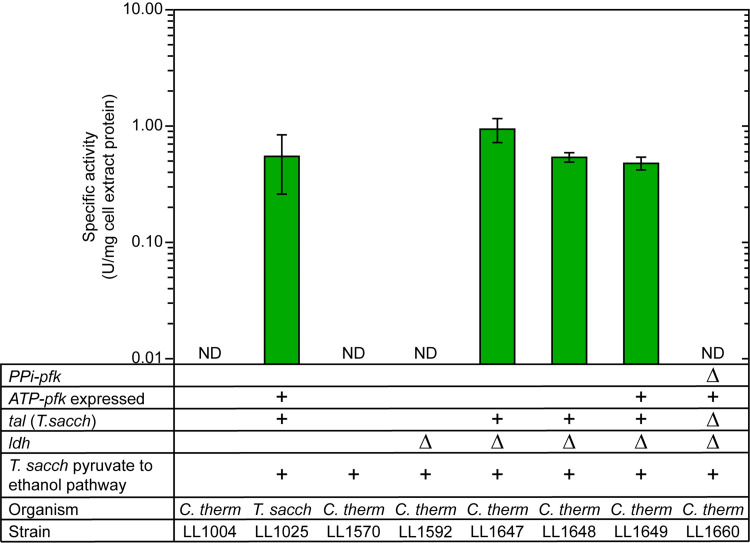
Strain LL1570 (1) (Table 1) was chosen as the reference strain for this study since it incorporates previous engineering with the T. saccharolyticum pyruvate to ethanol pathway—expression of T. saccharolyticum adhA, nfnAB, adhEG544D, pforA, and ferredoxin (1, 23)—to improve ethanol yield and titer; this made it the logical starting point for metabolic engineering efforts to further improve ethanol titer. In addition, since lactate is a minor but nonetheless undesired byproduct of C. thermocellum fermentation (24), the ldh gene was deleted from strain LL1570 to create strain LL1592 to redirect carbon flux from lactate to ethanol production. Following the ldh deletion, the first step in replacing PPi-linked PFK activity with ATP-linked activity was to decouple glycolysis from the nonoxidative pentose phosphate pathway (PPP) by eliminating the need for SBP activity. To this end, the transaldolase gene from T. saccharolyticum, Tsac_0327, and its promoter were integrated immediately downstream of the PPi-pfk gene. Enzyme assays confirmed that the resulting strain, LL1647, had gained transaldolase activity levels that were comparable to those in T. saccharolyticum cell extracts (Fig. 1).
TABLE 1.
Strains used in this study
| Strain | Organism | Description | Accession no. | Source or reference(s) |
|---|---|---|---|---|
| LL1004 | C. thermocellum | DSM 1313 | CP002416 | DSMZ |
| LL1025 | T. saccharolyticum | Strain JW/YS-485L | CP003184 | 42 |
| LL1570 | C. thermocellum | Engineered C. thermocellum with T. saccharolyticum pyruvate to ethanol pathway (T. saccharolyticum adhA, nfnAB, adhEG544D, pforA, ferredoxin), deletion of C. thermocellum pfors | SRP144049 | 1, 23 |
| LL1592 | C. thermocellum | LL1570 Δldh | SRP181986 | This study |
| L1647 | C. thermocellum | LL1592 PPi-pfk::pTsac_0327-Tsac_0327 | SRP222666 | This study |
| LL1648 | C. thermocellum | LL1647 Clo1313_0718::Tsac_1362 | SRP222662 | This study |
| LL1649 | C. thermocellum | LL1648 ΔClo1313-0717-0718::pTsac_1362 | SRP222669 | This study |
| LL1660 | C. thermocellum | LL1649 ΔClo1313_1876 Δtal | SRP246483 | This study |
| LL1661 | C. thermocellum | Sister colony #1 of LL1660 | SRP246561 | This study |
| LL1662 | C. thermocellum | Sister colony #2 of LL1660 | SRP246510 | This study |
| LL1663 | C. thermocellum | Sister colony #3 of LL1660 | SRP246511 | This study |
| T7 express | E. coli | fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10–TetS)2 [dcm] R(zgb-210::Tn10–TetS) endA1 Δ(mcrC-mrr)114::IS10 | NEBa |
New England Biolabs, Ipswich, MA.
FIG 1.
Transaldolase (TAL) specific activities of strains used in this study. “ND” indicates that transaldolase activity was not detected, i.e., it was below our limit of detection (<0.01 U/mg cell extract protein). Cth, C. thermocellum; Tsac, T. saccharolyticum. The “+” signs indicate the presence of a genetic feature, and the “Δ” sign indicates that a gene is deleted. Strain LL1004 is WT C. thermocellum and is a negative control for TAL activity. Strain LL1025 is WT T. saccharolyticum and is a positive control for TAL activity. Error bars represent 1 standard deviation (n ≥ 3).
Toxicity of heterologous expression of ATP-pfk.
Since C. thermocellum only grows on C6 sugars and sugar polymers and exclusively uses glycolysis for substrate assimilation, the PFK reaction is essential for growth. Thus, the next step in replacing PPi-pfk with ATP-pfk was heterologous expression of an ATP-pfk gene. Initial attempts to express the T. saccharolyticum ATP-pfk used the strong constitutive Clo1313_2638 promoter (25) to drive ATP-pfk expression. It was observed that the expression plasmid failed to give transformants despite positive controls obtaining colony counts (see Table S1 in the supplemental material) comparable to previously reported values (26). A no-promoter control ATP-pfk expression plasmid (pLL1383) could be transformed into wild-type C. thermocellum at an efficiency similar to that of an empty vector, indicating that the lack of transformants with the promoter-containing ATP-pfk expression plasmid was linked to expression and thus activity of the ATP-pfk gene, and not due to the sequence of the ATP-pfk gene.
To reduce toxicity of ATP-pfk expression, we engineered the ribosome binding site (RBS) to create plasmids with different predicted translation initiation rates (27) for the T. saccharolyticum ATP-pfk gene (Table 2). Lower predicted translation initiation rates allowed us to observe transformants in wild-type C. thermocellum (see Table S1); however, when these transformants were assayed for PFK activity, ATP-PFK activities were still below our limit of detection (<0.01 U/mg cell extract protein; see Table S1).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Accession no. | Source or reference |
|---|---|---|---|
| pDGO143 | C. thermocellum expression vector | KX259110 | 26 |
| pLL1381 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; native RBS with predicted translation initiation rate of ~95,000 arbitrary units | ON809502 | This study |
| pLL1382 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; modified RBS with predicted translation initiation rate of ~500 arbitrary units | ON809503 | This study |
| pLL1383 | pDGO143 with T. saccharolyticum ATP-pfk; no promoter driving ATP-pfk expression | ON809504 | This study |
| pLL1384 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; modified RBS with predicted translation initiation rate of ~4,000 arbitrary units | ON809505 | This study |
| pLL1385 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; modified RBS with predicted translation initiation rate of ~8,000 arbitrary units | ON809506 | This study |
| pLL1386 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; modified RBS with predicted translation initiation rate of ~14,000 arbitrary units | ON809507 | This study |
| pLL1387 | pDGO143 with C. thermocellum Clo1313_2638 promoter driving T. saccharolyticum ATP-pfk; modified RBS with predicted translation initiation rate of ~27,000 arbitrary units | ON809508 | This study |
| pLL1388 | Integration vector; introduces T. saccharolyticum transaldolase and its native promoter immediately downstream of C. thermocellum PPi-pfk gene | ON809509 | This study |
| pLL1389 | Integration vector; introduces T. saccharolyticum ATP-phosphofructokinase downstream of Clo1313_0718 (putative ADP-glucose synthase), with a spacer sequence between Clo1313_0718 and the ATP-pfk | ON809510 | This study |
| pLL1390 | Replacement vector; deletes Clo1313_0717-0718 and the spacer sequence, and replaces it with the T. saccharolyticum ATP-pfk promoter sequence | ON809511 | This study |
| pLL1391 | Deletion vector; deletes C. thermocellum PPi-pfk. In strain LL1647 and its derivatives, this plasmid also deletes the previously introduced T. saccharolyticum transaldolase | ON809512 | This study |
| pLL1392 | Deletion vector; deletes C. thermocellum PPi-pfk only in strain LL1647 and its derivatives | ON809513 | This study |
Simultaneous expression of ATP-pfk and deletion of PPi-pfk.
The apparent toxicity of a heterologous ATP-Pfk in C. thermocellum needed to be addressed. One possible explanation included the creation of a futile cycling reaction between an ATP-Pfk and PPi-Pfk, with net result being conversion of ATP to PPi. However, since C. thermocellum does not have a cytosolic pyrophosphatase activity that would hydrolyze the PPi from this cycling reaction—which would result in ATP wastage—we considered alternatives.
A second and more likely explanation was that expression of ATP-Pfk in C. thermocellum shifted cofactor usage at the PFK reaction away from PPi to ATP, thus reducing the glycolytic demand for PPi as a phosphoryl donor. Given that PPi in C. thermocellum comes from both biosynthesis and primarily from a nonbiosynthetic reaction, the reduced demand for PPi from the PFK reaction would result in accumulation of PPi in the cell, as the nonbiosynthetic reaction continues to generate PPi. The effect of PPi accumulation would be inhibition of biosynthetic reactions, leading to arrest in cell growth (28). This therefore suggested that expression of ATP-pfk is conditional on disruption of the nonbiosynthetic PPi mechanism, so that PPi accumulation could be mitigated.
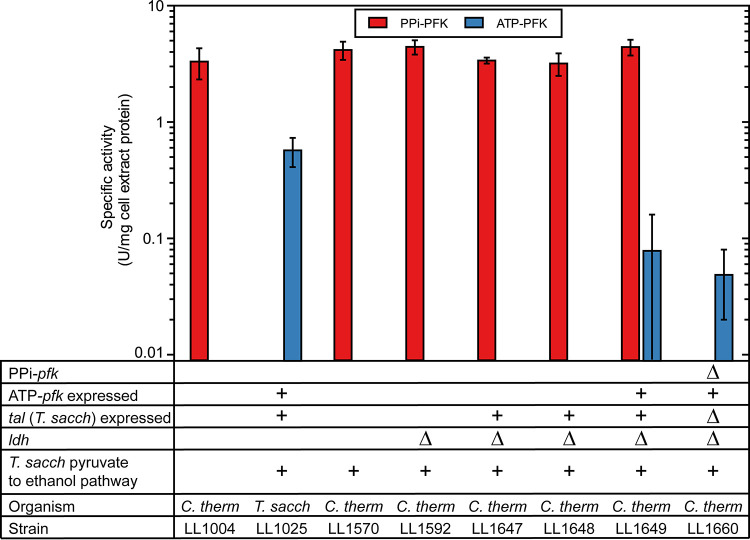
Since our initial approach of sequentially expressing a heterologous ATP-pfk gene failed, we devised a two-step strategy to allow for simultaneous activation of ATP-pfk expression and disruption of PPi generation. It was previously reported that there were several candidates for PPi generation in C. thermocellum, of which two were believed to be the likely sources: the membrane-bound pyrophosphatase encoded by the gene Clo1313_0823, and the ADP-glucose synthase enzyme complex encoded by the two-gene operon, Clo1313_0717-0718 (10, 11). Since it has previously been reported that both the membrane-bound pyrophosphatase (Clo1313_0823) gene (20) and the ppdk gene (10) could be deleted from C. thermocellum, we hypothesized that the APD-glucose synthase enzyme complex (used for glycogen cycling) was the likely candidate for PPi generation. In the first step, the T. saccharolyticum ATP-pfk was integrated immediately downstream of the ADP-glucose synthase operon (Clo1313_0717-0718) (see Fig. S1). To avoid unintended ATP-PFK activity due to unwanted translation of this ATP-pfk gene, a spacer sequence was included between the ADP-glucose synthase operon and the T. saccharolyticum ATP-pfk coding sequence, with the predicted effect of significantly reducing the translation rate of the ATP-pfk gene. In the second step, the ADP-glucose synthase operon, as well as the insertion sequence, was replaced by the T. saccharolyticum ATP-pfk promoter sequence, thus simultaneously removing ADP-glucose synthase (the hypothesized PPi-generating mechanism), and expressing ATP-pfk (see Fig. S1) to ensure that PFK activity was complemented prior to deleting the PPi-pfk. Gain of ATP-PFK activity in the resulting strain (LL1649) was confirmed by detection of low ATP-PFK activity (~0.08 U/mg cell extract protein) in cell extracts of strain LL1649 (Fig. 2). While the ATP-PFK activity observed in strain LL1649 was much lower than that in T. saccharolyticum cell extracts (~0.57 U/mg cell extract protein), it was still greater than that of non-ATP-PFK-expressing strains, where ATP-PFK was below the limit of detection (<0.01 U/mg cell extract protein). Analysis of relative gene expression levels revealed that ATP-pfk expression in strain LL1649 was comparable to that in T. saccharolyticum (see Fig. S2). Proteomic analyses of strains LL1649 and wild-type T. saccharolyticum subsequently confirmed that the relative protein abundance of ATP-Pfk protein was ~10-fold lower in strain LL1649 than in T. saccharolyticum (see Fig. S3), consistent with the observed difference in enzyme activity (Fig. 2). The simplest way to reconcile these observations is that ATP-Pfk expression is limited at the level of translation.
FIG 2.
PFK activities for strains used in this study, with PPi (red) or ATP (blue) as the cofactor. Where there is no column plot, PFK activity was below the limit of detection (<0.01 U/mg cell extract protein). The “+” signs indicate the presence of a genetic feature, and the “Δ” sign indicates that a gene is deleted. Strain LL1004 is WT C. therm(ocellum) and is a negative control for ATP-PFK activity, and a positive control for PPi-PFK activity. Strain LL1025 is WT T. sacch(arolyticum), and is a positive control for ATP-PFK activity and a negative control for PPi-PFK activity. Error bars represent 1 standard deviation (n ≥ 3).
After successfully achieving chromosomal expression of ATP-pfk, we proceeded to delete the native PPi-pfk gene. Two plasmids were used; one to delete only the PPi-pfk coding sequence (pLL1392), and the other to delete both the PPi-pfk coding sequence, as well as the T. saccharolyticum transaldolase expression cassette that had originally been introduced (pLL1391). We expected that deletion of the transaldolase would not be possible due to its anticipated essential role (see Introduction); however, the deletion attempt using plasmid pLL1391 worked, resulting in a strain that had neither PPi-pfk nor transaldolase (strain LL1660). The deletions were confirmed both by loss of PPi-PFK (Fig. 2) and TAL (Fig. 1) activities in strain LL1660.
Investigating changes to thermodynamic driving force of the PFK reaction.
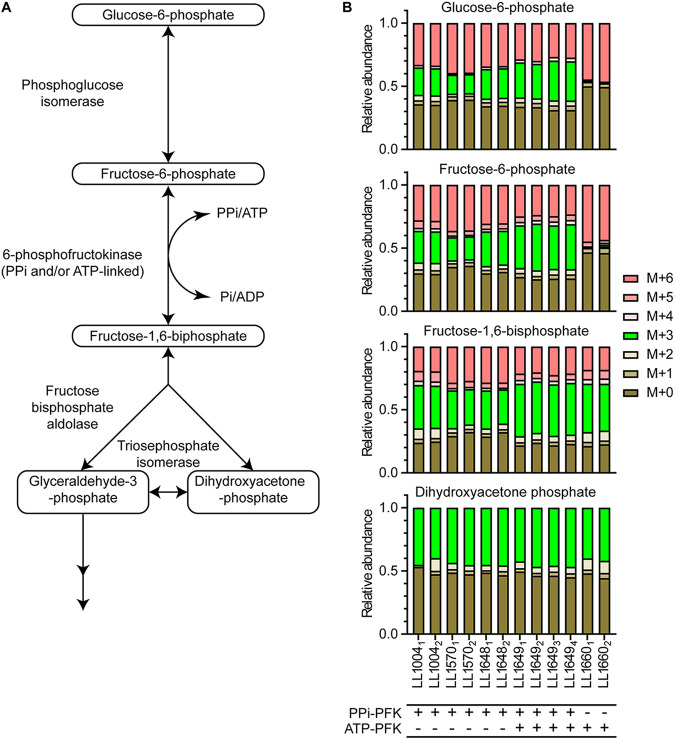
To determine whether replacing the C. thermocellum PPi-pfk with a heterologous ATP-pfk leads to greater thermodynamic driving force in glycolysis, C. thermocellum strains were cultured on a 50:50 ratio of naturally and uniformly 13C-labeled cellobiose ([U-13C12]cellobiose). As previously described (8), the forward PFK reaction will produce a 50:50 mixture of unlabeled and fully labeled fructose-1,6-bisphosphate (FBP), in turn producing dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P) in a similar 50:50 ratio of unlabeled to labeled. However, reverse flux of the fructose bisphosphate aldolase has a 50% chance to produce M+3-labeled FBP (FBPM+3; i.e., with three 13C carbon atoms) from this mixed pool of G3P and DHAP; the FBPM+3 in turn can be converted to M+3-labeled fructose-6-phosphate (F6PM+3) by reverse PFK flux and then to M+3 glucose-6-phosphate (G6P) by reverse phosphoglucose isomerase flux (Fig. 3A). The relative abundance of the M+3 isotopomer in the F6P pool therefore gives an indication of the reverse flux through the PFK reaction. A smaller ratio of F6PM+3 versus FBPM+3 would therefore represent a less reversible PFK reaction, which in turn would suggest a more thermodynamically favorable forward PFK reaction (9).
FIG 3.
(A) Metabolic diagram of glycolysis around the PFK reaction and the enzymes that catalyze the reactions. (B) 13C-labeling patterns for key glycolytic metabolites (glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, and dihydroxyacetone phosphate) for cells fed a 50:50 mixture of uniformly labeled and naturally labeled glucose. The “M+” notation indicates the number of 13C-labeled carbon atoms (i.e., M+6 indicates six labeled carbon atoms). The subscript after the strain ID number indicates biological replicate number. The cofactor specificity of the PFK reaction is indicated below the strain name: “+” indicates the presence of a reaction, and “−” indicates the absence of a reaction.
It was observed that the relative abundance of the M+3 fractions for both G6P and F6P were more or less similar among wild-type C. thermocellum and engineered C. thermocellum strains that still contained the PPi-pfk gene (Fig. 3B); the ratios observed for the M+3 F6P and G6P were similar to previously reported results (8). Strains with both PPi-pfk and ATP-pfk genes (LL1649) showed an M+3 labeling pattern similar to that of the wild type. Deletion of the PPi-pfk to create strain a strain with only ATP-PFK activity (LL1660) substantially reduced the M+3 G6P and F6P fractions, suggesting that switching the cofactor usage of the PFK reaction from PPi to ATP decreased the reversibility of the PFK reaction in C. thermocellum.
The M+3-labeled fraction is not observed in glucose-1-phosphate (G1P) (see Fig. S4). Since phosphoglucomutase is present in C. thermocellum (29), and the reaction is close to equilibrium under standard conditions (ΔG′° = 0.8 kJ/mol [12]), the lack of reversibility of this reaction suggests that it is actively regulated.
In comparing metabolite pool sizes across the different samples, we further observe that expression and replacement of the PPi-pfk gene with an ATP-pfk gene (strain LL1660) resulted in a significant decrease in the F6P pool size, and a corresponding increase in the FBP pool size (Fig. 4), resulting in a much greater FBP/F6P ratio than strains that still contained the more reversible PPi-PFK pathway, making the FBP/F6P pool ratios of this C. thermocellum strain more similar to that of organisms with a canonical glycolysis pathway, such as E. coli (30) and T. saccharolyticum (8). Other significant changes to metabolite pool sizes include a transient increase in pyruvate concentrations when both PPi-pfk and ATP-pfk were present (strain LL1649) and a decrease in the ATP pool once all PFK activity was ATP-linked (strain LL1660).
FIG 4.

(A) Comparison of log2-transformed peak area values for glycolytic metabolites. (B) FBP/F6P ratios for the different samples. Subscripts after the strain ID number indicates replicate number. The cofactor specificity of the PFK reaction is indicated below the strain name: the “+” indicates the presence of a reaction, and “−” indicates the absence of a reaction.
Effect on fermentation.
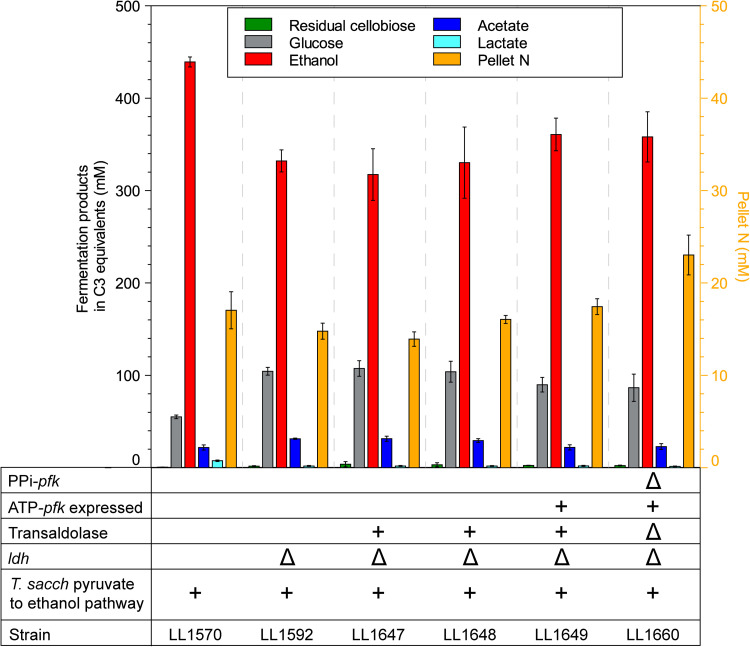
The engineered strains in this study were cultured on 60 g/L cellobiose in defined medium to determine whether replacing PPi-pfk with ATP-pfk affected the quantity or distribution of fermentation products. Consumption of cellobiose and production of glucose, ethanol, and acetate (the primary fermentation products in the parent strain) were largely unaffected. A slight increase in biomass production (measured by pellet nitrogen) was observed upon deletion of the PPi-pfk gene (strain LL1660) (Fig. 5).
FIG 5.
Fermentation product profiles for the C. thermocellum strains in this study. Note that concentrations for the fermentation products are reported in C3 sugar equivalents. The “+” sign indicates introduction of a genetic feature, and the “Δ” sign indicates deletion of a genetic feature. Error bars represent 1 standard deviation (n ≥ 3).
Resequencing analyses.
Whole-genome resequencing confirmed the presence of the various modifications in the strains created in this study (see File S3 in the supplemental material). In particular, it confirmed that ADP-glucose synthase was deleted and replaced by the T. saccharolyticum ATP-pfk promoter in strain LL1649 (as intended) and that this feature was preserved in strain LL1660 when both PPi-pfk and transaldolase were deleted.
We observed a partial genome duplication in strain LL1660 and three of its sister colonies (LL1661, LL1662, and LL1663) (see Fig. S5). The evidence for the gene duplication is that read depth is approximately doubled along the length of the region compared to the parent strain. The presence of a repeated insertion element at each end of the duplicated region suggests that the duplication was mediated by homologous recombination. Included in the duplicated region are the CDSs Clo1313_0790 to Clo1313_1248. This region includes several genes that may play a role in energy metabolism and cofactor cycling, including a putative native ATP-pfk gene (Clo1313_0997), the pta-ack pathway (Clo1313_1185-1186), the membrane-bound pyrophosphatase (Clo1313_0823), and the pyruvate phosphate dikinase gene (Clo1313_0949); however, the effect of the gene duplication on these activities was not investigated since its extent meant there was an impractical number of avenues of investigation to tackle in one study. One practical implication of this chromosome duplication event is to make it significantly more difficult to perform subsequent chromosomal modifications in this region due to the presence of two copies of each gene and potential for additional unintended recombination events.
DISCUSSION
The absence of transaldolase in C. thermocellum has led to speculation that this organism uses a variant of the PPP, where SBP-to-S7P conversion is necessary for the efficient generation of C5 compounds, and that this reaction is mediated by the PPi-Pfk enzyme (7, 21, 22). This was the motivation for introducing a transaldolase gene from T. saccharolyticum before deleting the PPi-pfk gene. However, the creation of a strain lacking both transaldolase and PPi-pfk suggests that our previous hypothesis about the pathway for C6-to-C5 interconversion was wrong. The pathway for interconversion of pentose and hexose sugars in this strain is not known. One possibility is that C. thermocellum has alternate means for interconverting hexoses and pentoses; one such pathway is the L-type pentose phosphate pathway (31), where octulose phosphates play an intermediary role for conversion between hexose phosphates and pentose phosphates. Further study will be needed to determine whether such a pathway operates in C. thermocellum.
Why was introducing the ATP-pfk gene so difficult? Initially, we suspected that simultaneous presence of both ATP-linked and PPi-linked PFK activity would be toxic. Since PPi-PFK activity is reversible, it presents the opportunity for a futile cycle where ATP-PFK converts F6P to FBP and PPi-PFK converts FBP to F6P, resulting in a net conversion of ATP + Pi → ADP + PPi. Indeed, in organisms that use fructose bisphosphatase (FBP; EC 3.1.3.11) for gluconeogenesis, the activities of FBP and PFK are regulated to ensure that only one is active (32). However, the successful generation of a strain where both activities are present (LL1649) eliminates that hypothesis. Furthermore, in that strain, ATP levels are similar to that of the wild-type strain (LL1004) (Fig. 4), indicating that if any futile cycling is happening, the flux is low enough that it does not significantly affect the adenylate pool. In other organisms, such as E. coli, PPi-pfk genes have been heterologously induced and expressed—for the purposes of purifying said PPi-Pfk proteins—alongside the native putative ATP-pfk (22, 33–36), further casting doubt on the futile cycling hypothesis.
One of the most significant metabolic impacts of the expression of ATP-pfk was an increase in the FBP pool. FBP is known to regulate many enzymes. In C. thermocellum cell extract, the addition of FBP has been shown to effectively abolish glycolytic flux (37). However, this would not explain the need to disrupt glycogen cycling in order for ATP-pfk expression to succeed.
The most likely explanation is that the introduction of ATP-pfk caused an increase in PPi levels, and eliminating the PPi generated by glycogen cycling (by deleting the ADP-glucose synthase reaction) was necessary to eliminate excess PPi. Although many organisms have a cytosolic pyrophosphatase to eliminate excess PPi, C. thermocellum does not (20). PPi levels are known to regulate the activity of both the pyruvate phosphate dikinase (Ppdk) and malic enzymes (38). The stimulatory role of decreasing PPi levels is also consistent with cell extract experiments (F6P stimulates flux, but FBP inhibits it) (37). Although many factors are still unknown regarding the role of PPi in C. thermocellum metabolism, the strains developed here provide a new and exciting avenue for testing hypotheses.
A surprising finding of this work was the extremely localized result of replacing the PPi-pfk gene with ATP-pfk gene. Our current understanding of C. thermocellum metabolism is that it operates close to thermodynamic equilibrium (8). Therefore, we would expect the introduction of a reaction with a high thermodynamic driving force (such as ATP-PFK) to deplete upstream metabolite pools and expand downstream ones. We found that this was, in fact, the case—at least for the metabolites immediately adjacent to the PFK reaction. However, this effect extended upstream only to G6P, and there was almost no effect on DHAP levels. This suggests active regulation of other reactions in glycolysis, or that the metabolic pathways downstream of PFK cannot carry the additional flux. The localized effect of this change further explains why the titer and distribution of fermentation products was largely unaffected.
MATERIALS AND METHODS
Strain and plasmid construction.
Figure 6 shows how the relationships among the strains used in this study; Table 1 provides full details for all strains used in this study. Plasmids used in this study are listed in Table 2; all plasmids were constructed via isothermal assembly (39), using a commercial kit from New England Biolabs (NEBuilder HiFi DNA Assembly Master Mix, catalog number E2621). Purification of DNA (plasmid or PCR products) was done with commercially available kits from Zymo Research and New England Biolabs. Transformation of C. thermocellum was performed as previously described (40); plasmid DNA that was to be transformed into C. thermocellum was purified from Escherichia coli BL21 derivative strains (New England Biolabs catalog number C2566) to ensure proper methylation of the plasmid DNA (41).
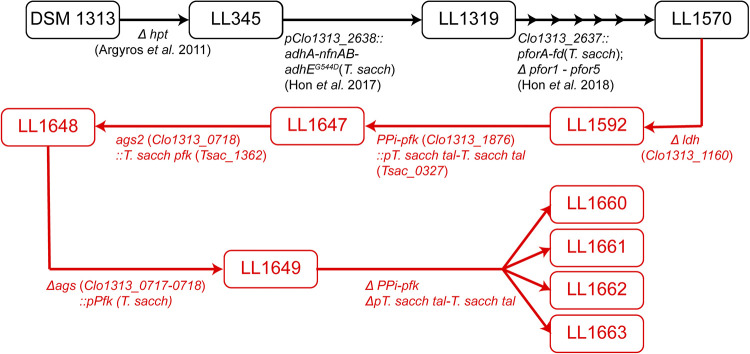
FIG 6.
Diagram of strain lineage. Strains shown in black are previously reported in literature. Strains in red were created in this study. Modifications that occurred between strains are listed below their respective arrows. Multiple arrows indicate several sequential modifications.
Media preparation and culture conditions.
All reagents used in this study were of molecular grade, and obtained from Sigma-Aldrich or Fisher Scientific, unless otherwise noted.
C. thermocellum and T. saccharolyticum strains were grown at 55°C under anaerobic conditions, either in tubes when grown in anaerobic chambers (Coy Laboratory Products, Grass Lakes, MI), with previously described environmental conditions (23), or in sealed serum bottles prepared as previously described (23).
Complex medium CTFUD was prepared as previously described (40); this medium was used for culturing C. thermocellum cells that were to be used in transformations or for preparing genomic DNA for strain resequencing. Defined medium MTC-5 was prepared as previously described (23) and was used for all other purposes. T. saccharolyticum cells (used to generate cell extracts of this bacterium for use in enzyme assays) were grown in MTC-6 medium (42).
Determination of transformation efficiencies.
Transformation efficiencies of the ATP-pfk expression plasmids was determined as previously described (26) by plating serial dilutions of recoveries and then counting the number of CFU where the dilution allowed for clear differentiation of C. thermocellum colonies. Transformation efficiency was determined from two independent transformations of the plasmids into wild-type C. thermocellum.
(i) Enzyme assays. Cells to be used for enzyme assays were grown, harvested, and lysed to obtain cell extract as previously described (23). Protein concentrations were determined using the Bradford protein dye assay (Bio-Rad, catalog number 1856210), with bovine serum albumin used as a protein standard. All enzyme assays were performed at 55°C under anaerobic conditions.
(ii) Transaldolase assay. Transaldolase activity was assayed by measuring the formation of glyceraldehyde-3-phosphate as previously described (43). The assay reaction was slightly modified from previously described (43), and contained 100 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 0.3 mM NADH, 1 mM erythrose-4-phosphate, 1 mM fructose-6-phosphate, 4 U/mL triosephosphate isomerase, 4 u/mL α-glycerophosphate dehydrogenase, and cell extract. The assay reaction was started with the addition of fructose-6-phosphate.
(iii) 6-Phosphofructokinase assay. The PFK activity was assayed by formation of fructose-1,6-bisphosphate and modified from a previously described method (10). The assay reaction contained 100 mM Tris-HCl, 5 mM MgCl2, 0.15 mM NADH, 1 mM fructose-6-phosphate, 4 U/mL fructose bisphosphate aldolase, 4 U/mL triosephosphate isomerase, 4 U/mL α-glycerophosphate dehydrogenase, cell extract, and 2 mM either PPi or ATP. The assay reaction was started by the addition of the phosphate donor (PPi or ATP).
Intracellular metabolite measurements.
(i) 13C labeling of C. thermocellum intracellular metabolites. C. thermocellum was grown on MTC-5 medium, with the main carbon source consisting of 3 mM naturally labeled cellobiose (i.e., unlabeled except for the naturally occurring abundance of 13C carbon atoms), and 3 mM uniformly labeled 13C-cellobiose ([UL-13C12]cellobiose), sourced from Omicron Biomedicals, catalog number CEL-002. Then, 8-mL cultures were inoculated with 8 or 0.8 μL of inoculum volume and grown at 55°C under anaerobic conditions. Freezer stocks were prepared by growing cells on MTC-5 with 5 g/L naturally labeled cellobiose to mid-exponential phase (optical density at 600 nm [OD600] of ~0.6 at the time the stocks were made).
Metabolite samples were harvested when the cells were growing at mid-exponential phase (OD600 ~0.5); to ensure that total metabolites extracted were similar across different cultures, the volume of culture to be harvested in milliliters was determined by dividing 2.5 by the OD600 value at the time of harvesting, i.e., ~5 mL of culture was harvested when the OD600 of the culture was ~0.5. Intracellular metabolites were collected as previously described (8) by vacuum filtering a culture through a 0.45-μm hydrophilic nylon filter to separate cells from medium. The filters were then placed in 1.6 mL of cold (−80°C) metabolite extraction solvent (40% methanol, 40% acetonitrile, 20% water) cell-side down. Cells were washed off the filter using the extraction solvent, and then metabolites were separated from cellular debris by centrifugation (8, 44). Metabolite samples were kept at −80°C when not used.
(ii) Analyses of C. thermocellum metabolite samples via LC-MS. Metabolite samples were analyzed by liquid chromatography-mass spectrometry (LC-MS) as described previously (8). Samples were dried under N2 gas and resuspended in solvent A (97:3 H2O-methanol with 10 mM tributylamine adjusted to pH 8.2 by the addition of acetic acid to an ~10 mM final concentration). Solvent B was 100% methanol, and the following gradient was used for chromatographic separation: 0 to 2.5 min, 5% B; 2.5 to 17 min, linear gradient from 5 to 95% B; 17 to 19.5 min, 95% B; 19.5 to 20 min, linear gradient from 95 to 5% B; and 20 to 25 min, 5% B. Separation was achieved on a 2.1 × 100-mm Acquity ultrahigh-pressure liquid chromatography ethylene bridge hybrid (UHPLC BEH) C18 column with a 1.7-μm particle size (Waters) at 25°C on a Vanquish UPLC coupled to a Q Exactive mass spectrometer (Thermo Scientific) by an electrospray ionization (ESI) source operating in negative mode. Mass spectrometry parameters were full MS-SIM (single ion-monitoring) scanning from 70 to 1,000 m/z, a resolution of 70,000 full width at half maximum (FWHM), a maximum injection time (IT) of 40 ms, and an automatic control gain (ACG) target of 1e6. Data were analyzed using the MAVEN software suite (45, 46), and compounds were identified by monoisotopic mass and retention time matching to pure standards. File S1 in the supplemental material contains data for 13C labeling of intracellular metabolites, as well as the relative metabolite abundance.
Fermentation product analyses.
Fermentation product analyses was done as previously described (1), with the modification of using 60 g/L cellobiose as the main carbon source instead of 50 g/L to ensure that ethanol titer was not limited by substrate concentration. Fermentations were performed in anaerobic sealed serum bottles with the headspace purged with 100% nitrogen and were cultured at 55°C for 168 h (7 days) before they were sampled for analyses.
Fermentation products were quantified by high performance liquid chromatography as previously described (47). Headspace composition in serum bottles was determined as previously described. Pellet nitrogen (used as a proxy for cell biomass) was measured as previously described (48).
Gene expression analyses.
Measuring the expression of the T. saccharolyticum ATP-pfk and C. thermocellum PPi-pfk was done via reverse transcriptase quantitative PCR (RT-qPCR) as previously described (26). The primers used for qPCR are reported in Table 3. ATP-pfk expression in each strain was normalized against the expression of the recA reference gene (49) to allow for comparison across different strains and species.
TABLE 3.
Primers used for RT-qPCR
| Primer | Target (species, gene, primer orientation) | Sequence (5′–3′) |
|---|---|---|
| XSH0367 | C. thermocellum, PPi-pfk, forward | ATGCATATTCGGACAATCC |
| XSH0905 | C. thermocellum, PPi-pfk, reverse | TGAGCTGCTCCGTAAACTGC |
| XSH0901 | T. saccharolyticum, ATP-pfk, forward | TAGAGACACGGCAACGTCAC |
| XSH0902 | T. saccharolyticum, ATP-pfk, reverse | ATAATCTCTGCTCCTCCAGC |
| XSH0198 | C. thermocellum, recA, forward | TTTACGGCCAGGGTATTTCA |
| XSH0199 | C. thermocellum, recA, reverse | GCCAATCTTCTGACCGTTGT |
| XSH0200 | T. saccharolyticum, recA, forward | GAAGCCTTAGTGCGAAGTGG |
| XSH0201 | T. saccharolyticum, recA, reverse | GAAGTCCAACATGTGCATCG |
Proteomics analyses.
Cells for proteomic analyses were cultured on defined MTC-5 or MTC-6 medium, depending on bacterial species, and harvested at mid-exponential phase (n = 3). Cells were pelleted, washed, and then processed for LC-MS/MS-based proteomic analyses as previously described (50). For each sample, 3-μg portions of tryptic peptides were loaded, separated, and analyzed by one-dimensional LC-MS/MS using a Vanquish uHPLC plumbed directly in-line with a Q Exactive Plus mass spectrometer (Thermo Scientific) operating in data-dependent acquisition. Tandem mass spectra were searched against the relevant C. thermocellum and T. saccharolyticum proteome databases using Proteome Discoverer v.2.3 and informatically postprocessed, as previously described (51).
Protein abundance raw and normalized data are provided in File S2 in the supplemental material.
Sequencing and resequencing analyses.
Routine Sanger sequencing of plasmids and PCR products was performed by Genewiz, Inc. (NJ, USA), with a minimum 2-fold coverage of sequences. Whole-genome resequencing was performed by the Department of Energy Joint Genome Institute using the Illumina MiSeq sequencing platform, with minimum 100-fold coverage. Strains were analyzed with the software CLC Genomics Workbench (Qiagen) using strain DSM1313 as the reference genome (GenBank accession NC_017304.1); reads were filtered against strain LL1570 (accession number SRP144049) (1) to exclude inherited mutations. A summary of the identified mutations is provided in File S3 in the supplemental material.
Data availability.
Whole-genome resequencing data for the strains in this study were deposited in the NCBI Sequence read archive under the accession numbers listed in Table 1. Plasmid sequence accession numbers are listed in Table 2. Raw LC/MS data for metabolite samples were deposited in Zenodo (10.5281/zenodo.7032172).
ACKNOWLEDGMENTS
Funding was provided by The Center for Bioenergy Innovation, a U.S. Department of Energy (DOE) Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Whole-genome resequencing was performed by the DOE Joint Genome Institute, a DOE Office of Science User Facility, and is supported by the Office of Science of the U.S. DOE under contract DE-AC02-05CH11231.
L.R.L. is a founder of Enchi Corporation, which has a financial interest in engineering C. thermocellum. S.H. is an employee of Enchi corporation.
S.H. and D.G.O. designed the experiments. S.H. performed strain and plasmid construction, biochemical assays, gene expression measurements, fermentations and subsequent analyses, generating metabolite samples in labeling experiments, and analyses of Sanger sequencing and whole-genome resequencing. T.J. and D.M.S. performed the analyses of C. thermocellum metabolite samples. R.J.G. and R.L.H. performed proteomic analyses of C. thermocellum and T. saccharolyticum. M.I.M. prepared whole-genome resequencing samples. S.H., D.G.O., T.J., and R.J.G. wrote the manuscript. D.A.-N., D.G.O., and L.R.L. revised the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Daniel G. Olson, Email: daniel.g.olson@dartmouth.edu.
Haruyuki Atomi, Kyoto University.
REFERENCES
- 1.Hon S, Holwerda EK, Worthen RS, Maloney MI, Tian L, Cui J, Lin PP, Lynd LR, Olson DG. 2018. Expressing the Thermoanaerobacterium saccharolyticum pforA in engineered Clostridium thermocellum improves ethanol production. Biotechnol Biofuels 11:242. 10.1186/s13068-018-1245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian L, Papanek B, Olson DG, Rydzak T, Holwerda EK, Zheng T, Zhou J, Maloney M, Jiang N, Giannone R, Hettich R, Guss A, Lynd L. 2016. Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol Biofuels 9:116. 10.1186/s13068-016-0528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazzoli R, Olson DG. 2020. Clostridium thermocellum: a microbial platform for high-value chemical production from lignocellulose. Adv Appl Microbiol 113:111–161. 10.1016/bs.aambs.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Holwerda EK, Olson DG, Ruppertsberger N, Stevenson DM, Murphy SJ-L, Maloney MI, Lanahan AA, Amador-Noguez D, Lynd LR. 2020. Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production. Biotechnol Biofuels 13:40. 10.1186/s13068-020-01680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dien BS, Cotta MA, Jeffries TW. 2003. Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol 63:258–266. 10.1007/s00253-003-1444-y. [DOI] [PubMed] [Google Scholar]
- 6.Lynd LR, Weimer PJ, Van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577. 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taillefer M, Sparling R. 2016. Glycolysis as the central core of fermentation. Adv Biochem Eng Biotechnol 156:55–77. 10.1007/10_2015_5003. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson TB, Korosh TC, Stevenson DM, Foster C, Maranas CD, Olson DG, Lynd LR, Amador-Noguez D. 2020. In vivo thermodynamic analysis of glycolysis in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum using 13C and 2H tracers. mSystems 5:e00736-19. 10.1128/mSystems.00736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JO, Tanner LB, Wei MH, Khana DB, Jacobson TB, Zhang Z, Rubin SA, Li SH-J, Higgins MB, Stevenson DM, Amador-Noguez D, Rabinowitz JD. 2019. Near-equilibrium glycolysis supports metabolic homeostasis and energy yield. Nat Chem Biol 15:1001–1008. 10.1038/s41589-019-0364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. 2013. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol 79:3000–3008. 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuil T, Hon S, Yayo J, Foster C, Ravagnan G, Maranas CD, Lynd LR, Olson DG, van Maris AJA. 2022. Functional analysis of H+-pumping membrane-bound pyrophosphatase, ADP-glucose synthase, and pyruvate phosphate dikinase as pyrophosphate sources in Clostridium thermocellum. Appl Environ Microbiol 88:e01857-21. 10.1128/aem.01857-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flamholz A, Noor E, Bar-Even A, Milo R. 2012. eQuilibrator: the biochemical thermodynamics calculator. Nucleic Acids Res 40(Database issue):D770–D775. 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noor E, Bar-Even A, Flamholz A, Reznik E, Liebermeister W, Milo R. 2014. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput Biol 10:e1003483. 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian L, Cervenka ND, Low AM, Olson DG, Lynd LR. 2019. A mutation in the AdhE alcohol dehydrogenase of Clostridium thermocellum increases tolerance to several primary alcohols, including isobutanol, n-butanol, and ethanol. Sci Rep 9:1–7. 10.1038/s41598-018-37979-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian L, Perot SJ, Stevenson D, Jacobson T, Lanahan AA, Amador-Noguez D, Olson DG, Lynd LR. 2017. Metabolome analysis reveals a role for glyceraldehyde-3-phosphate dehydrogenase in the inhibition of C. thermocellum by ethanol. Biotechnol Biofuels 10:276. 10.1186/s13068-017-0961-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JO, Rubin SA, Xu Y-F, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD. 2016. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol 12:482–489. 10.1038/nchembio.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. 2008. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3:1299–1311. 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dash S, Olson DG, Joshua Chan SH, Amador-Noguez D, Lynd LR, Maranas CD. 2019. Thermodynamic analysis of the pathway for ethanol production from cellobiose in Clostridium thermocellum. Metab Eng 55:161–169. 10.1016/j.ymben.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. 2012. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol 12:214. 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holwerda EK, Zhou J, Hon S, Stevenson DM, Amador-Noguez D, Lynd LR, van Dijken JP. 2020. Metabolic fluxes of nitrogen and pyrophosphate in chemostat cultures of Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. Appl Environ Microbiol 86:e01795-20. 10.1128/AEM.01795-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Susskind BM, Warren LG, Reeves RE. 1982. A pathway for the interconversion of hexose and pentose in the parasitic amoeba Entamoeba histolytica. Biochem J 204:191–196. 10.1042/bj2040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koendjbiharie JG, Hon S, Pabst M, Hooftman R, Stevenson DM, Cui J, Amador-Noguez D, Lynd LR, Olson DG, van Kranenburg R. 2020. The pentose phosphate pathway of cellulolytic clostridia relies on 6-phosphofructokinase instead of transaldolase. J Biol Chem 295:1867–1878. 10.1074/jbc.RA119.011239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon S, Olson DG, Holwerda EK, Lanahan AA, Murphy SJ, Maloney MI, Zheng T, Papanek BA, Guss AM, Lynd LR. 2017. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab Eng 42:175–184. 10.1016/j.ymben.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Biswas R, Prabhu S, Lynd LR, Guss AM. 2014. Increase in ethanol yield via elimination of lactate production in an ethanol-tolerant mutant of Clostridium thermocellum. PLoS One 9:e86389–7. 10.1371/journal.pone.0086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson DG, Maloney M, Lanahan AA, Hon S, Hauser LJ, Lynd LR. 2015. Identifying promoters for gene expression in Clostridium thermocellum. Metab Eng Commun 2:23–29. 10.1016/j.meteno.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hon S, Lanahan AA, Tian L, Giannone RJ, Hettich RL, Olson DG, Lynd LR. 2016. Development of a plasmid-based expression system in Clostridium thermocellum and its use to screen heterologous expression of bifunctional alcohol dehydrogenases (adhEs). Metab Eng Commun 3:120–129. 10.1016/j.meteno.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espah Borujeni A, Salis HM. 2016. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism. J Am Chem Soc 138:7016–7023. 10.1021/jacs.6b01453. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Brevet A, Fromant M, Leveque F, Schmitter J-M, Blanquet S, Plateau P. 1990. Pyrophosphatase is essential for growth of Escherichia coli. J Bacteriol 172:5686–5689. 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang YHP. 2010. A highly active phosphoglucomutase from Clostridium thermocellum: cloning, purification, characterization and enhanced thermostability. J Appl Microbiol 108:39–46. 10.1111/j.1365-2672.2009.04396.x. [DOI] [PubMed] [Google Scholar]
- 30.Bennett BD, Kimball EH, Gao M, Osterhout R, van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5:593–599. 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JF, MacLeod JK. 2006. The metabolic significance of octulose phosphates in the photosynthetic carbon reduction cycle in spinach. Photosynth Res 90:125–148. 10.1007/s11120-006-9113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Link H, Kochanowski K, Sauer U. 2013. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat Biotechnol 31:357–361. 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- 33.Ding YHR, Ronimus RS, Morgan HW. 2000. Sequencing, cloning, and high-level expression of the pfp gene, encoding a PPi-dependent phosphofructokinase from the extremely thermophilic eubacterium Dictyoglomus thermophilum. J Bacteriol 182:4661–4666. 10.1128/JB.182.16.4661-4666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding YR, Ronimus RS, Morgan HW. 2001. Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes. J Bacteriol 183:791–794. 10.1128/JB.183.2.791-794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reshetnikov AS, Rozova ON, Khmelenina VN, Mustakhimov II, Beschastny AP, Murrell JC, Trotsenko YA. 2008. Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Methylococcus capsulatus Bath. FEMS Microbiol Lett 288:202–210. 10.1111/j.1574-6968.2008.01366.x. [DOI] [PubMed] [Google Scholar]
- 36.Rozova ON, Khmelenina VN, Trotsenko YA. 2012. Characterization of recombinant PPi-dependent 6-phosphofructokinases from Methylosinus trichosporium OB3b and Methylobacterium nodulans ORS 2060. Biochemistry (Mosc) 77:288–295. 10.1134/S0006297912030078. [DOI] [PubMed] [Google Scholar]
- 37.Cui J, Stevenson D, Korosh T, Amador-Noguez D, Olson DG, Lynd LR. 2020. Developing a cell-free extract reaction (CFER) system in Clostridium thermocellum to identify metabolic limitations to ethanol production. Front Energy Res 8. https://www.frontiersin.org/articles/10.3389/fenrg.2020.00072/full. [Google Scholar]
- 38.Taillefer M, Rydzak T, Levin DB, Oresnik IJ, Sparling R. 2015. Reassessment of the transhydrogenase ‘malate shunt’ in Clostridium thermocellum ATCC 27405 through kinetic characterization of malic enzyme and malate dehydrogenase. Appl Environ Microbiol 81:2423–2432. 10.1128/AEM.03360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson DG, Lynd LR. 2012. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol 510:317–330. 10.1016/B978-0-12-415931-0.00017-3. [DOI] [PubMed] [Google Scholar]
- 41.Guss AM, Olson DG, Caiazza NC, Lynd LR. 2012. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels 5:30. 10.1186/1754-6834-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Olson DG, Lanahan AA, Tian L, Murphy SJ-L, Lo J, Lynd LR. 2015. Physiological roles of pyruvate ferredoxin oxidoreductase and pyruvate formate-lyase in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biotechnol Biofuels 8:138. 10.1186/s13068-015-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprenger GA, Schörken U, Sprenger G, Sahm H. 1995. Transaldolase B of Escherichia coli K-12: cloning of its gene, talB, and characterization of the enzyme from recombinant strains. J Bacteriol 177:5930–5936. 10.1128/jb.177.20.5930-5936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson DG, Hörl M, Fuhrer T, Cui J, Zhou J, Maloney MI, Amador-Noguez D, Tian L, Sauer U, Lynd LR. 2017. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng 39:169–180. 10.1016/j.ymben.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinforma 37:14.11.1–14.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melamud E, Vastag L, Rabinowitz JD. 2010. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82:9818–9826. 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holwerda EK, Thorne PG, Olson DG, Amador-Noguez D, Engle NL, Tschaplinski TJ, van Dijken JP, Lynd LR. 2014. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol Biofuels 7:155. 10.1186/s13068-014-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holwerda EK, Ellis LD, Lynd LR. 2013. Development and evaluation of methods to infer biosynthesis and substrate consumption in cultures of cellulolytic microorganisms. Biotechnol Bioeng 110:2380–2388. 10.1002/bit.24915. [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Walker C, Dien B, Giannone RJ, Slininger P, Thompson SR, Trinh CT. 2021. Exploring proteomes of robust Yarrowia lipolytica isolates cultivated in biomass hydrolysate reveals key processes impacting mixed sugar utilization, lipid accumulation, and degradation. mSystems 6:e00443-21. 10.1128/mSystems.00443-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Presley GN, Werner AZ, Katahira R, Garcia DC, Haugen SJ, Ramirez KJ, Giannone RJ, Beckham GT, Michener JK. 2021. Pathway discovery and engineering for cleavage of a β-1 lignin-derived biaryl compound. Metab Eng 65:1–10. 10.1016/j.ymben.2021.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 to S5. Download aem.01258-22-s0001.pdf, PDF file, 1.0 MB (1MB, pdf)
Data Set S1. Download aem.01258-22-s0002.xlsx, XLSX file, 0.06 MB (62.8KB, xlsx)
Data Set S2. Download aem.01258-22-s0003.xlsx, XLSX file, 0.8 MB (847.5KB, xlsx)
Data Set S3. Download aem.01258-22-s0004.xlsx, XLSX file, 0.02 MB (21.4KB, xlsx)
Data Availability Statement
Whole-genome resequencing data for the strains in this study were deposited in the NCBI Sequence read archive under the accession numbers listed in Table 1. Plasmid sequence accession numbers are listed in Table 2. Raw LC/MS data for metabolite samples were deposited in Zenodo (10.5281/zenodo.7032172).