Abstract
One of the most widely used microscopy techniques in biology and medicine is fluorescence microscopy, offering high specificity in labeling as well as maximal sensitivity. For live-cell imaging, the ideal fluorescence microscope should offer high spatial resolution, fast image acquisition, three-dimensional sectioning, and multicolor detection. However, most existing fluorescence microscopes have to compromise between these different requirements. Here, we present a multiplane, multicolor wide-field microscope that uses a dedicated beam splitter for recording volumetric data in eight focal planes and for three emission colors with frame rates of hundreds of volumes per second. We demonstrate the efficiency and performance of our system by three-dimensional imaging of multiply labeled fixed and living cells. The use of commercially available components makes our proposed microscope straightforward for implementation, thus promising for widely used applications.
Introduction
The ultimate goal of fluorescence microscopy is to record three-dimensional images of cellular structures with maximal spatial and temporal resolution and high signal/noise ratio, ideally in a robust and easy-to-use manner. Although super-resolution microscopy has pushed the limits of spatial resolution down to a few nanometers (1, 2, 3, 4, 5, 6, 7, 8), it could do so only by compromising on the simplicity of the utilized hardware or data analysis approaches, on image acquisition speed, and/or on the ability of imaging several structures or molecules with different emission colors at the same time, i.e., multiplexing (9, 10, 11). However, (three-dimensional) imaging speed and multiplexing are pivotal for following multiple cellular processes in vivo in three dimensions and with sufficient temporal resolution.
To achieve maximal acquisition speed of whole volume images, several multiplane wide-field imaging modalities have been recently developed. In 2013, Abrahamsson and co-workers introduced the first multiplexed 3D wide-field epifluorescence microscope using a chromatically corrected grating that splits the emission light into several detection channels, each corresponding to a different focal plane and to two emission colors (12). The authors of (13) successfully used such a grating-based multiplane microscope even for single-molecule localization microscopy, although the signal/noise ratio goes increasingly down with increasing number of simultaneously imaged planes. Later, the Abrahamsson group improved the chromatic aberration correction and optical efficiency of this microscope by incorporating two gratings and two aberration-compensating prisms, one grating-prism pair for one color (14). Although the development of these systems is a formidable technological achievement, their technological complexity (complex micro-optical phase gratings and chromatic-aberration-correcting multifaceted prisms) and demands on alignment has prevented its widespread application so far. Furthermore, up to now such a system was realized only for a maximum of two spectral channels.
As an alternative, Leutenegger and colleagues developed a glass-based beam-splitter device that allows for multiplane imaging similar to the diffracting grating approach by Abrahamsson, but in a much simpler manner and with tremendously reduced chromatic aberration. Although the first setup of this beam splitter still used an array of individual 50/50 beam splitters and mirrors, it was later simplified into a single glass piece that offers maximal simplicity in implementation, maximal mechanical stability, and nearly no chromatic aberration over the spectral range of 500–700 nm (15, 16, 17, 18).
In this work, we expand this multiplane beam-splitter system with an additional spectral beam splitter to achieve simultaneous three-color imaging in eight focal planes with a volumetric image acquisition rate of up to several 100 volumes per second in three spectral windows. Besides providing high imaging speed and color multiplexing, our system is straightforward to implement and to use, both in hardware and in data analysis.
Materials and methods
Setup
In the illumination path, three light sources, a 100 mW laser at 473 nm (CNI, Changchun, China), a 200 mW laser at 561 nm (CNI), and a 140 mW laser at 637 nm (Obis; Coherent, Santa Clara, CA) are incorporated for fluorescence excitation. All three beams are aligned to a common path, spatially filtered, and expanded. The collimated beam is guided into the illumination path of a conventional inverted epifluorescence microscope (IX-71; Olympus, Tokyo, Japan) equipped with a high-NA water immersion objective (UPLSAPO 60× 1.2 NA W; Olympus). A schematic of the illumination path is shown in Fig. S1.
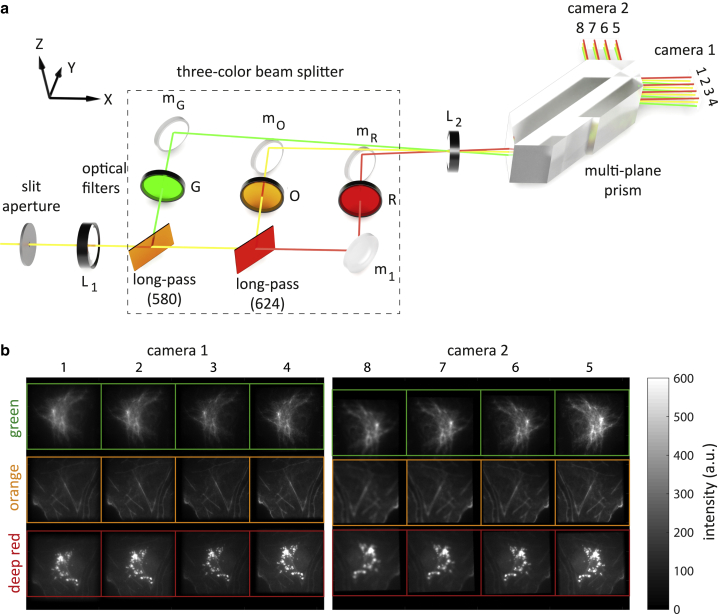
The fluorescence detection path comprises a color-splitting unit and a multiplane prism exhibited in Fig. 1 a. After passing the first lens L1, a custom-built three-color beam splitter splits the fluorescence light into three spectral channels along the vertical direction: a green channel (G, 450–580 nm), an orange channel (O, 580–624 nm), and a deep red channel (R, 650–700 nm). To spatially separate these color channels on the camera chip, each color is reflected under a slightly different vertical angle; the middle color channel, i.e., the orange beam, propagates in the plane, and the green and deep red beams are tilted by angles ±θ. The angle θ is determined by the required vertical offset Δy of the images on the camera and the focal length f2 of the final tube lens as θ ≥ arcsin(Δy/f2). In our case, we have Δy = 3.5 mm and f2 = 200 mm, resulting in θ ≈ 1°. Note that the length of the optical path is the same for all three color channels. The mirrors mG, mO, and mR align the beams of the three channels to a common vertical plane. Here, the orange and green beams pass over the mirrors mO and mR, respectively. A detailed schematic of the spectral beam-splitting unit with intercomponent distances is provided in Fig. S2.
Figure 1.
(a) Detection path of the three-color multiplane fluorescence microscope. An adjustable slit aperture crops the light at the microscope’s side-port, thus defining the field of view. Lenses L1 and L2 comprise a 4f optics with magnification 1.33 that relays the fluorescence light to the sCMOS cameras 1 and 2. Between the lenses, the light sequentially passes two dichroic mirrors. The resulting three color channels of green (λ < 580 nm), orange (580 < λ < 624 nm), and red (λ > 624 nm) are further filtered with emission band-pass filters G (513/17 nm), O (593/40 nm), and R (692/40 nm), respectively. An off-axis tilt of the dichroic mirrors deflects the propagation direction of the green and deep red color away from the horizontal plane. Next, the multiplane prism generates eight images with uniformly increasing optical path length, corresponding to eight distinct focal planes inside the sample. (b) Raw eight-plane three-color images of a fixed fluorescently labeled COS-7 cell. Different focal planes are shown along the horizontal direction, different colors along the vertical direction. The different colors correspond to labeled vimentin, actin, and mitochondria with the corresponding peak emission wavelengths at 520, 593, and 692 nm, respectively. The numbers above the images show the axial order of the image planes (one is closest to sample surface, eight is farthest).
Next, all three colors are passed through a tube lens (L2) and a commercially available multiplane prism (patent EP3049859A1; Scoptonic imaging technologies, Torun, Poland). This prism is geometrically designed in such a way that it splits an input beam through multiple reflections into two sets of four neighboring output beams with increasing optical pathlength (15,19). The focal lengths of lens L1 and L2 are chosen in such a way that the final magnification increases by a factor of 1.33. After the multibeam splitter, the two sets of four images are recorded by two sCMOS cameras (ORCA-Flash 4.0V2; Hamamatsu, Hamamatsu, Japan), with the second camera shifted, as compared to the first camera, by an additional distance 4d/n ∼9.16 mm away from the prism (here, n = 1.46 is the refractive index of the prism material). In this way, all eight images recorded by the two cameras correspond to eight equidistantly placed focal planes in sample space, with an optical path difference of Δl = d/n between them. An example of a recorded set of 8 × 3 images (eight focal plane times three colors) is shown in Fig. S3. The field aperture prevents lateral overlap of neighboring images on the camera chips.
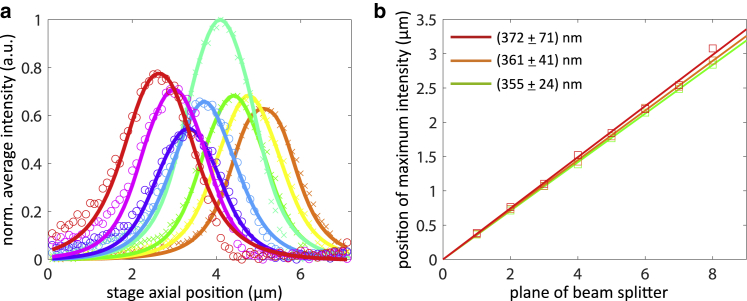
To determine the precise distances between focal planes and their relative light-collection efficiencies, a stack of z-scan images over an axial range of 7 μm in steps of 100 nm of immobilized tetra-spectral fluorescent beads was recorded (TetraSpeck carboxylate microspheres, 0.1 μm, T7279; Thermo Fisher Scientific, Waltham, MA). Fig. S4 shows one such image of fluorescent beads. Fluorescence excitation was done using the 473 and 561 nm lasers. The average bead brightness in each plane over the z-scan can be well-fitted with a Gaussian function (see Fig. 2). From these 24 fits, the axial shift of each plane and the relative collection efficiency of each plane can be determined. Notice that the latter is not identical for all focal planes because of slight imperfections of the multiplane beam-splitter prism.
Figure 2.
Brightness and interplane distance calibration. (a) Normalized average intensity of image planes in the green channel as a function of axial scan position (scan step equals to 100 nm). Solid lines represent Gaussian fits, and crosses (camera 1) and open circles (camera 2) are measured data. (b) Linear fits of z-scan positions of intensity maxima as a function of image for each color channel. These fits give an interplane distance of 355 ± 24 nm (mean ± standard deviation of seven sequential inter-plane distance measurements) for the green channel, 361 ± 41 nm for the orange channel, and 372 ± 71 nm for the deep red channel.
As can be seen from Fig. 2 b, the maxima of the Gaussian fits for each image plane follow a clear linear dependence in plane number, from which the uniform interplane distance for each color channel can be determined. These distances are slightly different for the three color channels (less than 5%) because of chromatic effects. The color-dependent differences of interplane distance accumulate to maximally 140 nm over the full axial depth range of ∼2.5 μm over the entire spectral range from 520 to 685 nm. This is consistent with analytical color aberration values (∼220 nm) over a range from 500 to 700 nm obtained by ray tracing analysis of the multiplane beam-splitter prism (see Supporting material in (15)).
The spectral separation of the three-color beam splitter is not perfect. The green emission is recorded with 14.68% in the orange channel and with 0.97% in the deep red channel. Similarly, orange emission is recorded with 13.38% in the deep red channel (see Fig. 1 b). Finally, for determining the transmission efficiency of the three-color beam-splitting system, we measured the intensity of 20 tetra-spectral beads in the orange channel with and without the three-color beam-splitter and long-pass filters in place. In this way, we found a transmission efficiency of ∼89%.
Image analysis
All image processing was done with custom-written MATLAB (The MathWorks, Natick, MA) routines. To generate a z-stack for each color channel, neighboring images (see colored rows in Fig. S4) are cropped, weighted by the corresponding collection efficiency obtained from the calibration measurements, and ordered by increasing axial position. All images are then co-registered using image cross correlation. Next, the abovementioned spectral bleedthrough is corrected by subtracting their intensities with corresponding spectral weighting factors from each color channel.
For reconstructing the 3D information of our volumetric image stacks, we applied 20 iterations of a 3D Lucy-Richardson algorithm (20,21) (MATLAB function deconvlucy) using precalculated point spread functions. These 3D point spread functions are calculated using the wave-optical theory of Wolf and Richards (22,23). Before applying the Richardson-Lucy deconvolution, the bottom and top image planes are mirrored and padded to each side of the image stack to reduce the boundary artifacts (24). Finally, all three color stacks are cropped to the common axial range. The cropped volume is nearly 95% of the initially acquired volume, which demonstrates the small axial chromatic aberration of our imaging system.
Cell culture, transfection, and immunostaining
COS-7 cell line CRL-1651 was purchased from American Type Culture Collection (Manassas, VA). All cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, 1 mM sodium pyruvate, and 100 U/mL penicillin-streptomycin in a humidified 5% CO2 atmosphere at 37°C. Cells were tested negative for mycoplasma contamination.
For fixed cell samples, cells were grown on glass coverslips for 24 h, fixed in 3.6% formaldehyde in phosphate-buffered saline (PBS) for 15 min, and washed with PBS. Then, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min and blocked with 3% bovine serum albumin (BSA) in PBS for 30 min at room temperature. Next, cells were incubated with rabbit monoclonal anti-vimentin antibody (SP20, 1:500; Invitrogen, Carlsbad, CA) in 3% BSA in PBS for 1 h. After washing with PBS, cells were incubated with Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (ab6717, 1:1000; Abcam, Cambridge, UK) together with 250 nM MitoTracker Deep Red FM (M22426; Invitrogen) and 100 nM phalloidin-Atto 550 (Atto-Tec) in 3% BSA in PBS for 1 h. After staining, the cells were washed with PBS and rinsed with distilled water. The samples were mounted on glass slides using Fluoroshield (F6182; Sigma-Aldrich, St. Louis, MO).
For live-cell measurements, cells were grown on glass coverslips for 24 h and then transfected with both mCherry-lamin B1-10 and GFP-connexin-43 using ViaFect (Promega, Madison, WI). The mCherry-lamin B1-10 was a gift from Michael Davidson (Addgene plasmid #55069). GFP-tagged rat connexin-43 was generated earlier (25). Staining of mitochondria and live-cell imaging were performed ∼12 h after transfection. For this purpose, 500 mM MitoTracker Deep Red FM was added directly to culture medium for 5 min at 37°C. Then, coverslips were mounted into self-made microscope imaging chambers, and cells were supplied with fresh culture medium. During imaging, cells were kept in a humidified 5% CO2 atmosphere at 37°C.
Results
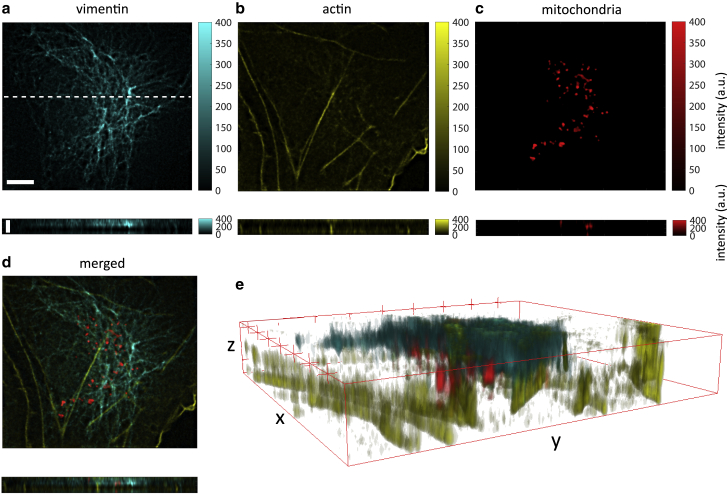
We first validated the volumetric three-color imaging capability of our system by imaging of fixed COS-7 cells with three different fluorescently labeled intracellular structures: vimentin, actin, and mitochondria. Fig. 3 shows transverse slices as well as a lateral views of the imaged 40 × 40 × 2.5 μm volume, with vimentin shown in cyan, actin in yellow, and mitochondria in red. Fig. 3 e renders a 3D view of the acquired data (generated by the 3D Viewer plugin in ImageJ). An animated 3D render of the imaged volume in different views is presented in Video S1.
Figure 3.
Instant 3D three-color imaging of a fixed COS-7 cell. (a) Vimentin (stained with FITC-conjugated goat anti-rabbit IgG) imaged through the green channel in an xy section (upper image) and an xz section (lower image) along the white dashed line highlighted in the transverse image. (b and c) The same views as (a) for actin (stained with phalloidin-Atto 550) and mitochondria (stained with MitoTracker Deep Red) imaged through the orange and deep red channels, respectively. (d) Overlay of the three components. (e) 3D render of the image data. Scale bars in upper and lower panels in (a) represent lengths of 5 and 2 μm, respectively.
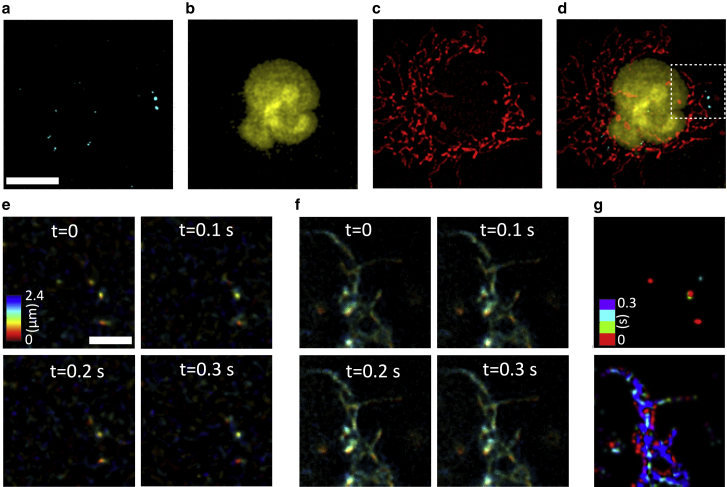
Fig. 4, a–d correspondingly show the connexin (in cyan), nuclei (in yellow), mitochondria (in red), and a merged image of all the organelles in a living COS-7 cell. Four consecutive time points of connexin and mitochondria highlighted in the dashed square in Fig. 4 d are represented in Fig. 4, e and f, respectively, in which the colors code the height of the samples from the surface. The acquisition time per each three-color volumetric data is 100 ms. The temporal evolution of the same organelles is represented in Fig. 4 g (upper panel for connexin and lower panel for mitochondria), in which now the time is color coded. One can observe both connexin and mitochondria’s lateral and axial mobility. See also Videos S2 and S3 accordingly for the connexin and mitochondria dynamics (height color coded) and Video S4 for the maximal intensity projection video of the merged color channels with connexin (yellow), nuclei (cyan), and mitochondria (red). Because of the highly dynamic behavior of connexin, the median intensity over frames is subtracted from each image to enhance the signal/noise ratio before deconvolution.
Figure 4.
Instant 3D three-color live-cell imaging of a COS-7 cell. Maximal intensity projections of (a) connexin (stained with GFP) imaged through the green channel, (b) nuclei (stained with mCherry) imaged through the orange channel, (c) mitochondria (stained with MitoTracker Deep Red) imaged through the deep red channel, and (d) the merged image are shown. (e) Time series of connexin dynamics indicated in the white dashed square in (d) in four frames in which colors encode the height of the sample. (f) The same time points as (e) but showing the mitochondria dynamics. (g) Temporal evolution of the connexin (upper panel) and mitochondria mobility (lower panel) in 0.3 s (see color bar). Scale bars in (a) represent 10 μm and in (e) represent 3 μm.
Discussion
We presented an easy-to-realize multiplane three-color wide-field microscope capable of recording images with a frame rate of several hundred three-color volume images per second (determined solely by the image acquisition speed of the used sCMOS cameras). We described in detail the setup, its calibration, and image processing routines. Our system uses sCMOS cameras more efficiently than other existing multiplane single-color systems (12,15,19) because it employs nearly three-fourths of all pixels available in both sCMOS cameras. Our microscope uses only commercially available components (most prominently optical dichroic and band-pass filters and a commercial multiplane prism). We demonstrated the performance of this microscope by imaging multicolor labeled fixed and live cells.
It should also be mentioned that the magnification determines the interplane distance between planes and thus the axial range of volume imaging of our system. Therefore, this range can be extended (or reduced) by appropriately changing the image magnification, i.e., the overall magnification of the telescope lenses, the tube lens, and the microscope objective. In addition to the magnification, one should also select a proper numerical aperture to fulfill the sampling rate along the axial direction. The adequate axial sampling implies that the interplane spacing should not be more than half the axial resolution.
A potential future extension of our microscope could be its combination with super-resolution optical fluctuation imaging, extending the work of (26) to three-color imaging. The multicolor challenge, together with the challenge of photobleaching, could be alleviated by implementing exchangeable fluorophores (27) into multiplane imaging. The exploitation of such fluorophores has allowed other super-resolution microscopy methods to image large 3D volumes (28). Another important extension could be its combination with structured illumination microscopy by using appropriate structured illumination in the excitation path. This would not only increase the lateral resolution of our system but also significantly improves the optical sectioning, which is, at the moment, only achieved indirectly via Richardson-Lucy deconvolution.
Author contributions
S.M. and I.G. designed the microscopic setup. S.M. carried out all the experiments. S.M. and I.G. analyzed the data. E.B. prepared the biological samples. S.M. and J.E. wrote the article.
Acknowledgments
We thank Alexey Chizhik for his help in sketching Fig. 1 a.
S.M. and J.E. acknowledge financial support by the Deutsche Forschungsgemeinschaft (German Research Council) via the Collaborative Research Center SFB 889 “Cellular mechanisms of sensory processing,” project B08. J.E. thanks also for financial support by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy - EXC 2067/1- 390729940.
Declaration of interests
The authors declare no conflicts of interest.
Editor: Yuval Ebenstein.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.bpr.2021.100001.
Supporting material
References
- 1.Betzig E., Patterson G.H., et al. Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharonov A., Hochstrasser R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilemann M., van de Linde S., et al. Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 6.Chizhik A.I., Rother J., et al. Enderlein J. Metal-induced energy transfer for live cell nanoscopy. Nat. Photonics. 2014;8:124–127. [Google Scholar]
- 7.Chen F., Tillberg P.W., Boyden E.S. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gwosch K.C., Pape J.K., et al. Hell S.W. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods. 2020;17:217–224. doi: 10.1038/s41592-019-0688-0. [DOI] [PubMed] [Google Scholar]
- 9.Liyanage M., Coleman A., et al. Ried T. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 10.Tsurui H., Nishimura H., et al. Shirai T. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J. Histochem. Cytochem. 2000;48:653–662. doi: 10.1177/002215540004800509. [DOI] [PubMed] [Google Scholar]
- 11.Valm A.M., Cohen S., et al. Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahamsson S., Chen J., et al. Gustafsson M.G. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods. 2013;10:60–63. doi: 10.1038/nmeth.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajj B., Wisniewski J., et al. Dahan M. Whole-cell, multicolor superresolution imaging using volumetric multifocus microscopy. Proc. Natl. Acad. Sci. USA. 2014;111:17480–17485. doi: 10.1073/pnas.1412396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrahamsson S., Ilic R., et al. Bargmann C.I. Multifocus microscopy with precise color multi-phase diffractive optics applied in functional neuronal imaging. Biomed. Opt. Express. 2016;7:855–869. doi: 10.1364/BOE.7.000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descloux A., Grußmayer K.S., et al. Lasser T. Combined multi-plane phase retrieval and super-resolution optical fluctuation imaging for 4D cell microscopy. Nat. Photonics. 2018;12:165–172. [Google Scholar]
- 16.Descloux A., Müller M., et al. Huser T. High-speed multiplane structured illumination microscopy of living cells using an image-splitting prism. Nanophotonics. 2019;9:143–148. [Google Scholar]
- 17.Grußmayer K., Lukes T., et al. Radenovic A. Self-blinking dyes unlock high-order and multiplane super-resolution optical fluctuation imaging. ACS Nano. 2020;14:9156–9165. doi: 10.1021/acsnano.0c04602. [DOI] [PubMed] [Google Scholar]
- 18.Louis B., Camacho R., et al. Rocha S. Fast-tracking of single emitters in large volumes with nanometer precision. Opt. Express. 2020;28:28656–28671. doi: 10.1364/OE.401557. [DOI] [PubMed] [Google Scholar]
- 19.Geissbuehler S., Sharipov A., et al. Leutenegger M. Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nat. Commun. 2014;5:5830. doi: 10.1038/ncomms6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson W.H. Bayesian-based iterative method of image restoration. J. Opt. Soc. Am. 1972;62:55–59. [Google Scholar]
- 21.Lucy L.B. An iterative technique for the rectification of observed distributions. Astron. J. 1974;79:745–754. [Google Scholar]
- 22.Wolf E. Electromagnetic diffraction in optical systems I. An integral representation of the image field. Proc. R. Soc. Lond. A Math. Phys. Sci. 1959;253:349–357. [Google Scholar]
- 23.Richards B., Wolf E. Electromagnetic diffraction in optical systems II. Structure of the image field in an aplanatic system. Proc. R. Soc. Lond. A Math. Phys. Sci. 1959;253:358–379. [Google Scholar]
- 24.Liu R., Jia J. 2008 15th IEEE International Conference on Image Processing. IEEE; 2008. Reducing boundary artifacts in image deconvolution; pp. 505–508. [Google Scholar]
- 25.Butkevich E., Hülsmann S., et al. Majoul I. Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr. Biol. 2004;14:650–658. doi: 10.1016/j.cub.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 26.Grußmayer K.S., Geissbuehler S., et al. Lasser T. Spectral cross-cumulants for multicolor super-resolved SOFI imaging. Nat. Commun. 2020;11:3023. doi: 10.1038/s41467-020-16841-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glogger M., Spahn C., et al. Heilemann M. Multi-color, bleaching-resistant super-resolution optical fluctuation imaging with oligonucleotide-based exchangeable fluorophores. Angew. Chem. Int. Ed. Engl. 2021;133:6380–6383. doi: 10.1002/anie.202013166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spahn C., Hurter F., et al. Heilemann M. Protein-specific, multicolor and 3D STED imaging in cells with DNA-labeled antibodies. Angew. Chem. Int. Ed. Engl. 2019;58:18835–18838. doi: 10.1002/anie.201910115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.