Abstract
Background
Compared with traditional drugs, nanomaterial drugs have the benefits of improving the solubility, bioavailability, and absorption rate of insoluble drugs. Nanoporous complexes can increase the efficiency with which drugs can penetrate the blood-brain barrier and reach target organs. Ginsenoside Rg1 is an effective drug that promotes angiogenesis. Ginsenoside Rg1 composite nanoparticles were employed to induce the expression of several key epigenetic enzymes and then activate the VEGF and Notch pathways after the onset of ischemic brain lesions.
Methods
We constructed nanoparticles to fully encapsulate the therapeutic drug (ginsenoside Rg1), which can be transferred into brain tissue via the receptor-mediated transfer of drug-encapsulated nanoparticles. Evaluation of the therapeutic effect of ginsenoside Rg1 complex nanovesicles (CNV) was performed by in vitro and in vivo experiments. Real-time polymerase chain reaction (RT- PCR), Western blot, immunohistochemistry staining (IHC), and Co-immunoprecipitation (co-IP) were employed to screen for epigenetic enzymes with an up-regulated expression post ginsenoside Rg1-CNV intervention. RNA sequencing, shRNA knockdown, and chromatin Immunoprecipitation (ChIP) sequencing were performed to detect the target genes of ginsenoside Rg1-CNV that regulate angiogenesis. Then, bioinformatic analysis was performed to investigate the mechanism of action of epigenetic modifying enzymes in regulating target genes.
Results
The average of the synthesized ginsenoside Rg1-CNV was 203.78±6.83 nm, the polydispersion index was 0.135±0.007, and the Zeta potential was 23.13±1.65 mV. Through in vivo and in vitro experiments, we found that it promotes the proliferation, migration, and tubular formation of brain microvascular endothelial cells (BMECs). Meanwhile, the intervention of ginsenoside Rg1-CNV promoted the demethylation of H3K27me3 within the promoter region of VEGF-A and Jagged1 genes and reduced the H3K27me3 modification within this region.
Conclusion
The ginsenoside Rg1 nanoparticles may be an available blood-brain barrier penetrating agent for ischemic stroke.
Keywords: ginsenoside Rg1, nanoparticles, angiogenesis, blood-brain barrier, epigenetic, ischemic stroke
Introduction
Stroke involves multiple diseases that seriously endanger human health. According to its etiology, a stroke can be divided into ischemic and hemorrhagic types, of which ischemic stroke accounts for about 60%–80% of stroke patients.1 Ischemic stroke represents the common form of cerebrovascular disease. It is the leading cause of permanent disability or death.2 Currently, intravenous thrombolysis with tissue-type plasminogen activator (trPA) and endovascular thrombectomy (EVT) remains the mainstay of acute stroke therapy within the initial 4.5 h after stroke onset. Additionally, neuroprotectants could mitigate the ischemia/reperfusion injury, including edaravone and fingolimod,3 but their clinical effects cannot achieve the desired condition, primarily due to the rigid blood-brain barrier (BBB) that limits drug delivery to the ischemic regions.4 On the other hand, a large number of neurons cause irreversible degeneration and necrosis in ischemic regions, which results from ischemia/reperfusion injury.5 The damaged brain tissue could be restored by angiogenesis, which provides oxygen and nutrients to improve recovery in the penumbra area,6 and allows macrophages to clear necrotic nerve cells or tissues.7 Besides, new vascular endothelial cells can release a variety of cytokines to nourish nerve cells and induce neurogenesis, and communication between nerves could also be repaired.8
Vascular endothelial growth factor and Notch signaling pathway have a crucial role in brain angiogenesis. VEGF can promote vascular permeability, extracellular matrix degeneration, vascular endothelial cell migration, proliferation, and angiogenesis.9 In rats, VEGF signaling was highly activated within minutes after ischemic stroke, and VEGF overexpression was still observed in glial cells up to three weeks later.10,11 A VEGF injection within 24 h after onset significantly stimulated brain angiogenesis and narrowed the penumbra area.12 Neuronal survival and neuronal morphological complexity can be maintained by inducing VEGF secretion.13 Both VEGF and its modified related materials have been shown to have neurotrophic and neuroprotective effects on neuronal cells within cerebral ischemia as well.14–16 Additionally, Notch pathway activation is a vital regulatory factor in angiogenesis and the ultimate formation of functional vessels.17 As one of the Notch ligands, Jagged1 can stimulate the endothelial front-end cells to generate new vascular buds and branch blood vessels.18 The blood supply to the infarct area is extremely deficient after an ischemic stroke, and new blood vessels are urgently required to repair the necrotic area and nourish nerve cells. Determining which drugs specifically activate the expression of Jagged1 in endodermal cells to promote angiogenesis has gained exponentially increasing attention from researchers.
Although the activation of VEGF and Jagged1 expression has been shown to promote angiogenesis after ischemia/reperfusion injury, research on their activation mechanism only focuses on the upregulation of the promoter region of the transcription factor binding gene. The deep molecular regulation mechanism, especially the epigenetic regulation mechanism, remains to be poorly understood. Histone modification is the most common and widespread epigenetic regulation, which primarily includes posttranslational modifications such as acetylation, methylation, phosphorylation, ubiquitination, and sumoylation.19 VEGF and Jagged1 are two important regulatory genes in the process of angiogenesis. Which epigenetic enzymes modify VEGF and Jagged1? Can changing this epigenetic modification interfere with the process of angiogenesis after ischemic stroke? These questions merit further investigation in future research.
Ginsenoside Rg1 is a sterol compound, triterpenoid saponin. It is the primary active component of the traditional Chinese medicine ginseng. It has a wide variety of therapeutic values for central nervous system (CNS) diseases for the neuron protective effects. The different types of ginsenosides have similar basic structures, all of them contain steroidal nuclei consisting of 30 carbon atoms arranged in four rings, which are divided into two groups according to the number of hydroxyl groups: the ginsenodiol type contains the most ginsenosides, such as ginsenosides Rb1, Rb2, Rb3, Rc, Rd, Rg3, Rh2 and the glycosidic group PD; the ginsenotriol type contains the ginsenosides Re, Rg1, Rg2, Rh1 and the glycosidic group PT. Among them, ginsenoside Rg1 at different concentrations had no toxic effects on normal cells of human or experimental animals.20 Prior studies have demonstrated that ginsenoside Rg1 can induce the upregulation of HIF-1 expression in human umbilical vein endothelial cells (HUVEC) and promote angiogenesis.21 Meanwhile, ginsenoside Rg1 can also induce glucocorticoid receptor (GR) and fibroblast growth factor receptor in HUVEC to promote angiogenesis.22 However, several reports have investigated the epigenetic regulatory mechanisms and specific target molecules of ginsenoside Rg1. The studies also showed that it is unable to cross the blood-brain barrier to achieve good therapeutic effects.23,24 Brain microvascular endothelial cells (BMECs) are an important component of the blood-brain barrier. Unlike endothelial cells Unlike endothelial cells, BMECs have tighter junctions, which prevent most molecules from entering the brain. Unlike endothelial cells, BMECs have tighter connections, which prevent most molecules from entering the brain tissue, even many small molecules.25,26 In contrast, brain-targeted drug delivery systems are able to cross the blood-brain barrier and therefore have an important role to play. The brain-targeted drug delivery system is able to cross the blood-brain barrier and is therefore of great research value. Transferrin receptors are highly expressed in cerebrovascular endothelial tissue, and the monoclonal antibody OX26 The monoclonal antibody OX26 has a high affinity for the transferrin receptor and can be used to target the brain tissue. The monoclonal antibody OX26 has a high affinity for the transferrin receptor and can target the brain tissue. OX26 monoclonal antibody coupled with various polymeric materials can achieve brain targeting of various small molecule drugs. Targeted delivery of a variety of small molecule drugs.23,24,27,28 Ginsenoside Rg1 is unable to cross the blood-brain barrier, based on the special physiological structure and function of the brain. Based on the special physiological structure and function of the brain, it is important to prepare biocompatible and safe biodegradable drug carriers for the treatment of brain diseases. Our previous studies have shown that the ginsenoside Rg1 nanoparticle complex can significantly increase the efficiency of drug penetration into the blood-brain barrier to target organs in model rats, promoting the recovery of cerebral infarction.29 Therefore, the construction of novel complex nanovesicles and the preparation of ginsenoside Rg1-loaded complex nanovesicles (Rg1-CNV) are expected to further improve the bioavailability of the effective active components of Traditional Chinese medicine and have an increased therapeutic effect on diseases.
This study first investigated the epigenetic regulation of angiogenesis after ischemic stroke. Ginsenoside Rg1-CNV induced the expression of several key epigenetic enzymes and then specifically activated the VEGF and Notch pathways after ischemic stroke. The aim is to elucidate the specific modes of action of epigenetic regulation in VEGF and Notch pathways and to provide a new molecular regulatory research basis and candidate drugs for the treatment of ischemic stroke.
Materials and Methods
Reagents
Ginsenoside Rg1 was obtained from Shanghai Yuan Ye Biological Technology Co., Ltd(B21057). The endothelial cell medium (ECM) was acquired from Science ll (Carlsbad, CA, USA). Green streptomycin and Pancreatic enzymes were purchased from Hyclone (SH30042.01). CD31 antibody, VWF antibody, Alexa Fluor 488 donkey anti-rabbit IgG (H+L), and Alexa Fluor 594 donkey anti-mouse IgG(H+L) were purchased from Abcam (Cambridge, UK). BSA, DMSO, triphenyl tetrazolium chloride (TTC), and dilution buffer were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). PBS was purchased from Nanjing Sheng Xing Co. Paraformaldehyde, TritonX-100, and Toluidine blue were obtained from Aladdin (Los Angeles, CA, USA). MTT was purchased from Beyotime Co. (Shanghai, China). SYBR Green I, an RT-PCR kit, and RNAiso Plus were purchased from Takara (Kyoto, Japan).
Preparation of Ginsenoside Nanoparticles
Blank nanoparticles and ginsenoside nanoparticles were prepared. Amphiphilic polyglutamate and phenylalanine ethyl ester copolymer were prepared according to the Akaga method. About 20 mg of copolymer and 5mg of ginsenoside Rg1 were dissolved and mixed in DMSO, then slowly added into the aqueous phase and constantly stirred. The amphiphilic polymer could load the hydrophobic molecule ginsenoside Rg1 to form nanoparticles. Nanoparticles were enriched by centrifugation centrifugation (12,000 rpm/10 min) for enrichment, and unloaded drug ginsenoside Rg1’s during centrifugation remained in the supernatant was removed. 10 mg of Rg1-NP was coupled with 1 mL of OX26 antibody (100 μg/mL) in EDCI The uncoupled OX26 monoclonal antibody in the solution was further removed by centrifugation (12,000 rpm/10 min).
Characterization of Ginsenoside Nanoparticles
The average particle size, polydispersity index and zeta potential of the prepared ginsenoside Rg1 nanoparticles were determined by laser nanoparticle sizer. The temperature was determined at 25 °C, and each sample was repeated three times.
The microstructures of the ginsenoside nanoparticles were observed by phosphomolybdic acid staining and transmission electron microscopy. After the sample was processed, it was placed in the H-7000 transmission electron microscope (a product of Hitachi Company in Japan). The structure was observed and photographed below. Nanoparticles were stored at 4, 25, and 37 °C with inert gas in the dark, respectively. To investigate the physical stability of the nanoparticles, the average particle size and polydispersion index (PDI) of the samples were measured under three different storage conditions.
Encapsulation Efficacy
The encapsulation efficiency of the ginsenoside nanocapsules was investigated using ginsenoside Rg1 as the detection index. Ginseng saponinRg1 standard product (purity above 98%, Shanghai Yuenye Biotechnology Co., LTD.), HPLC system (ShimadZU 20AT, Japan), chromatographic column, Kromasil ODS C18 Column (5 µm, 250 mm×4.6 mm), protection column, and Agilent ODS C18 Column (5 µm, 12.5 mm x 4.6 mm) were used. The mobile phase comprised acetonitrile-0.1% phosphoric acid solution = 40–60 (v/v), the flow rate was 1.0 mL/min, the column temperature was 35 °C, the detection wavelength was 203 nm, and the injection volume was 20 μL.
The standard ginsenoside Rg1 of 10.0 mg was accurately measured and dissolved in chromatographic methanol as the reserve solution for later use. The blank chromatographic methanol diluted the reserve solution to 100.0, 50.0, 10.0, 5.0, 2.0, and 1.0 μg/mL, respectively. The peak area (Y) of ginsenoside Rg1 and the mass of ginsenoside Rg1 were taken as ordinates. The standard curve with concentration (X) acted as the abscissa, and the equation of the standard curve and the correlation coefficient (r) were computed.
We applied low, medium, and high concentrations standards of 1.0 and 10.0 within the standard curve using reserved mother liquor and 100.0 μg/mL as the quality control sample (QC). According to the above chromatographic method of sampling analysis, each concentration sample was repeated three independent times, the peak areas of ginsenoside Rg1 in the sample were substituted into the standard curve, and the ratio of ginsenoside Rg1 concentration under various peak areas to the concentration of the prepared ginsenoside Rg1 standard solution was calculated. The methodological accuracy (%) of ginsenoside Rg1 in the HPLC system at low, medium, and high concentrations was calculated.
Low, medium, and high concentration standard samples of 1.0, 10.0, and 100.0 μg/mL were prepared according to the above sample treatment methods. The QC samples were injected for analysis, and each concentration sample was repeated three times. In a 45 °C water bath with nitrogen, 0.01 mL ginsenoside Rg1 standard solution was accurately absorbed at a concentration of 100 µg/mL. The sample containing standard ginsenoside Rg1 was prepared according to the sample analysis method above. The amount of ginsenoside Rg1 in the prepared sample was then calculated (μg). The ginsenoside Rg1 content (μg) was calculated as a ratio to the control ginsenoside Rg1 content. The average recovery and standard deviation were obtained.
The dialysis tubes were cut into dialysis bags (molecular weight cutoff, 10 KD) of a certain length and then rinsed with distilled water for nanoparticle dialysis. Rg1 nanoparticles were then added to it. The ginsenoside nanoparticles were demulsified with chromatographic methanol, and the content of ginsenoside Rg1 (W1) in the nanoparticles was accurately determined by HPLC, and then added into the dialysis bag. The content of free ginsenoside Rg1 in the dialysate was calculated (W2) after 48 h. The ginsenoside nanoparticles were stored at room temperature and at 4 °C for ten days. The pH values of the buffers were 5.0, 6.0, and 7.0. The encapsulation rate changes of ginsenoside nanoparticles under various storage conditions were measured successively. Nanoparticles are stored under seal at 4°C for short-term storage and freeze-dried and sealed for long-term storage.
Drug Release Behavior in vitro
100 mL of PBS buffer was pipetted into the beakers of different treatment groups, and 1 mL of the prepared ginsenoside nanoparticles was precisely pipetted into the dialysis bag, which was placed in the buffer and stirred magnetically in a constant temperature water bath at 37°C. The PBS buffer was pipetted after 1h, 2h, 4h, 8h, 12h, 24h, 36h, 48h, 72h of dialysis, and supplemented with equal volumes of Then the dialysate was aspirated from the outside of the dialysis bag and analyzed by HPLC. The content of free ginsenoside Rg1 in the dialysate was calculated accurately by HPLC.
Establishment and Protocols of a Rat Model
All animal experiments were provided free access to the Guiding Principles for the Care and Use of Laboratory Animals according to the Regulations of the People’s Republic of China for the Administration of Laboratory Animals. These studies were approved by the Ethics Committee of Jinling Hospital on Animal Resources (approval number: 2018GKJDWLS-03-198).
A total of 80 Sprague Dawley (SD) rats were bought from Qinglong Mountain Animal Breeding Farm (Nanjing, China). Twenty were selected as the blank control group, and the other 60 were divided into three groups. The model control group was given normal saline; the ginsenoside Rg1 group was given 40 mg/kg-1•d-1 ginsenoside Rg1; and the last group was given 40 mg/kg-1•d-1 ginsenoside Rg1-CNV by gavage as the ginsenoside Rg1-CNV group. After 14 days of continuous feeding, all rats were sacrificed and collected for the detection of relevant experimental indexes. Ischemic stroke was modeled by thread embolism.30 The rats were anesthetized with pentobarbital sodium. The common carotid artery (CCA), internal carotid artery (ICA), and external carotid artery (ECA) were carefully separated. The thread plug was inserted into the ECA and ICA, then continued through the beginning of the middle cerebral artery (MCA) and reached the fine anterior cerebral artery. Two hours after the blood flow obstruction of the right cerebral, MCA was realized. The plug was pulled out, and the wound was closed.
Evaluation of Infarct Volume by TTC Staining
TTC staining was used to evaluate infarct volume. The fresh brain tissue was cut into 1–2 mm thick slices and placed into 2% TTC at 37 °C to keep the staining uniform. The ischemic damage area of the brain slice was photographed and analyzed using Photoshop software.31 Pink penumbra brain tissue was positioned between the core, viable brain tissue was stained with a red color, and brain infarction tissue was pale-colored.32
Histology
Fresh tissues were fixed with 4% paraformaldehyde for more than 24 h. For histochemistry, the cerebral hemispheres were cut in the coronal plane, and the brain tissue of different groups was acquired from the identified ischemic cerebral cortex and paraffin-embedded section.33 Sections of 5 μm were stained with hematoxylin and eosin (HE). For Nissl’s staining, the tissue of the target site was flattened, paraffin-embedded, and sectioned, with a thickness of 4 μm. After dewaxing, the slices were placed at 60 °C and stained with 0.5% toluidine blue for 40 min. The Nissi bodies appeared to have a dark blue color.34 For immunohistochemistry, after BMEC culture for 24 h, paraformaldehyde (4% pure) was added and fixed for 30 min. After sealing with 5% BSA, it was removed, and 100μL H3K27me3 primary antibodies diluted at 1:200 were added to each well overnight at 4 °C. After three washes with PBS, 100 μL secondary antibodies were added to each well and incubated at 37 °C for 30 min. They have washed with PBS again and then removed. 50–100 μL fresh DAB was then added to each section. After complete color development, distilled water was washed, hematoxylin was redyed, hematoxylin was absorbed, water was washed, and photographs were taken by the microscope.
Cell Culture and Identification
Cell culture: After isolating and extracting rat brain microvascular endothelial cells (BMECs), PBS was added and rinsed. After 1 mL of trypsin digestion for 1–3 min, the digested cells were centrifuged at 1500 rpm for 10 min. After the supernatant was dumped, 3 mL of the medium was introduced to resuspend the cells, which were subcultured at 1:3. Cell identification: 4% paraformaldehyde was added to the cells and fixed at room temperature for 10 min. CD31 and VWF antibodies (1:100) were diluted in PBS-1%BSA. The primary antibody was added and incubated overnight at 4 °C. The fluorescent secondary antibodies were diluted in PBS-1% BSA solution at a ratio of 1:400, and were added and incubated at room temperature in the dark for 1 h. Next, 100 ng/mL DAPI was added and incubated at room temperature for 10 min. Finally, photographs were taken directly under a fluorescence microscope.
Proliferation, Migration, and Tubular Formation of BEMCs
The control group and Rg1-CNV groups were set up to evaluate the impact of ginsenoside Rg1-CNV on the proliferation, migration, and tubular formation of BMECs. Proliferation: PBS and Rg1-CNV (final concentration of 10uM) were added. Each group was repeated three times and cultured in an incubator for 6h. Following the instructions of the MTT kit, the cell proliferation of the two groups was analyzed. Migration: In each group, PBS and Rg1-CNV (10 µM) were added to the upper chamber three times. They were then placed into an incubator for 9 h. The upper chamber liquid was blotted, and about 400 μL paraformaldehyde and 400 μL crystal violet dye were added at room temperature for 10 min. The migrating cells were observed with a microscope (Nikon Ts2R, Japan) and counted. Tubular formation: PBS and Rg1-CNV (final concentration of 10 µM) were added. Each group was repeated once and placed in an incubator for 9 h. The lumen formation was analyzed using the ImageJ software Angiogenesis Analyzer Plugin.
Real-Time Polymerase Chain Reaction (RT- PCR)
The rats were divided into four groups: Sham, MCAO+ NS, MCAO+ Rg1 (40mg/kg/d, normal Rg1), and MCAO+ Rg1-CNV (40mg/kg/d). An appropriate amount of tissue was extracted, and 1 mL Trizol was added. They were then placed at room temperature for 2 min. 200 µL of chloroform was added to extract RNA. Isopropyl was used to precipitate. After being dissolved in DEPC, the concentration was detected by Nanodrop. The reverse transcription experiment was conducted following the instructions of the reverse transcription kit (TAKARA, RR047). Super pure water 4ul SYBR Green qPCR Master Mix 10 µL (Takra) upstream, primer 0.5 μL (10 μM), and downstream primer 0.5 μL (10 μM) were used; the total volume was 20 μL. A two-step method was applied for the quantification of PCR. The conditions were: 96 °C for 30s and 56 °C for 15s, for 40 cycles. The gene expression level was calculated using three measurements, with GADPH as an internal reference. Table 1 lists the primer sequences.
Table 1.
Primers in RT-PCR
| Name | Sequence | Product Size |
|---|---|---|
| JMJD3-F | TGCTTGGATGAAGGGCTCAG | 286 |
| JMJD3-R | CAGATAGGGAGGTGGTGGGA | |
| UTX-F | GGCCTCAGACCTAGACCTCA | 244 |
| UTX-R | GAGGGAAAAAGGCATCACGC | |
| GAPDH-F | TGATGGGTGTGAACCACGAG | 152 |
| GAPDH-R | AGTGATGGCATGGACTGTGG |
Western Blot
Tissue from rat cerebral ischemia was collected into an EP tube, then 1000 μL of RIPA lysate was added and lysed on ice for 60 min. Centrifugation was performed at 12,000 rpm at 4 °C for 5 min. The protein content was determined according to the instructions of the Beyotime BCA assay kit. They were then dissolved for 15 min at 50 °C in a sample buffer containing sodium dodecyl sulphate (SDS) and stored at −20 °C. On a 4% polyacrylamide concentrated gel, SDS–polyacrylamide gel electrophoresis (PAGE) was performed. After the sample entered the separation gel, electrophoresis was applied at 120 V for 90 min. The proteins were transferred onto PVDF membranes (Millipore, ISEQ00010) with a Bio-Rad Mini Protean III apparatus (Bio-Rad, CA, USA). The film was soaked from bottom to top with TBS, moved to a plate containing a blocking solution (5% skimmed milk powder TBST solution), and then closed by shaking on a decolorization shaking table at room temperature for 1 h. The primary antibody was diluted with a TBST solution containing 1% BSA at 1:1000 and incubated at 4 °C overnight. Horseradish peroxidase-labelled goat anti-rabbit IgG and goat anti-mouse IgG were diluted with TBST at 1:5000 and photographed by a Tanon5200 chemiluminescence imager. The primary and secondary antibodies are listed in Table 2.
Table 2.
The Primary and Secondary Antibodies in WB
| Names | Dilution |
|---|---|
| Anti-JMJD3 antibody | 1:10,000 |
| Anti-UTX antibody | 1:1000 |
| Anti-GAPDH antibody | 1:5000 |
| Anti-H3K27me3 antibody | 1:500 |
| Anti-H3 antibody | 1:1000 |
| Goat Anti-Rabbit IgG | 1:5000 |
| Goat Anti-Mouse IgG | 1:5000 |
RNA Sequencing Analysis
RNA-seq was employed to analyze the brain tissues isolated from each group. Double-end sequencing was performed for qualified RNAs. The sequencing sample library was completed following the experimental instructions, which were constructed by a Qubit® 2.0 Fluorometer for concentration and Agilent2100 for library size. Cluster generation and primer hybridization of the first sequence were applied on the cBot supporting the Illumina sequencer. Sequencing reagents were prepared according to the Illumina User Guide, and flow cells with clusters were placed on the machine. A paired-end procedure was used for two-terminal sequencing. The sequencing process was controlled by Illumina’s Data Collection software, and real-time data analysis was performed.
Bioinformatics Analysis
The mRNAs identified in rat samples were included in the analysis. Hisat2 (version 2.0.4) was used to conduct the genome aligement, and Stringtie (version 1.3.0) to analyze the genome-level expression quantification. Differently expressed genes (DEGs) were detected using the edgeR package. Statistical significance was defined as an adjusted P value<0.05, and the biological significance was defined as a difference of fold-change ≥2, which was exhibited as a Valcano spot. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the Python programming language to achieve the enrichment analysis algorithm. The Matplotlib package was used for visualization.
Co-Immunoprecipitation (Co-IP)
In in vivo and in vitro experiments, after the brain tissues or BMECs were frozen and ground, RIPA lysate was added to ice lysate cells, and the supernatant was collected. Protein concentration was measured by the BCA method and adjusted to 2–3 mg/mL. The volume of each IP sample was 500 µL, and each sample was retained for 30 µL as input. Protein A/G Bead (50 µL 50% Bead concentration is required for each IP sample) was prepared and then added to each 500 μL protein sample solution at 4 °C. This was followed by magnetic bead-coupled antibodies to precipitate the bait protein. SDS-PAGE was used to separate the protein complex, and Western blot was used to detect the target protein.
Chromatin Immunoprecipptation (ChIP) Sequencing
The BMECs were cultured in a 10 cm petri dish, and 10 µM Rg1-CNV was added when the cells reached 70% fusion degree. After 9 h of culture, cell crosslinking and ultrasonic fragmentation were completed. Each IP experiment required a 450 μL Dilution Buffer and a 2.25 μL protease inhibitor. Test samples included positive control anti-Histone H3 (4.0 μg antibody per tube), negative normal rabbit IgG (5.0 μg antibody per tube), and H3K27me3 antibody (5.0 μg antibody per tube). 450 μL diluent (containing 2.2 μL of protease inhibitor) was added and gently mixed. 5 μL was removed and frozen as input. Different antibodies and 20 μL protein A magnetic beads were added to pull down the target protein and the DNA it binds, followed by high and low-salt elution to remove impurities. The elution protein/DNA complex and the anticrosslinked protein/DNA complex were separate DNA. The purified DNA was directly used for sequencing.
Construction of a Plasmid
The shuttle plasmid K5342 and packaging plasmid were used for the establishment of control low expression, control shRNA, JMJD3 low expression, UTX low expression, and JMJD3+UTX low expression. The plasmids were from Shanghai Genepharma Bio-Tech Company (Shanghai, China). JMJD3 and UTX in each group were detected at 48 h after transfection by qPCR.
The recombinant viral plasmid encoding lentivirus particles and its three auxiliary packaging plasmids were prepared, respectively, for high-purity endotoxin-free extraction. 293T cells were co-transfected with the transfection reagent RNAi Mate. After 6 h of transfection, the cells were replaced with a complete medium, and after 72 h of culture, a cell supernatant rich in lentivirus particles was collected. A concentrated solution of lentivirus was obtained with a high titer, which was used to infect the target cells. Resistance genes and fluorescent proteins on lentiviral vectors can be used to screen target cells after infection.
Statistics Analysis
Statistics analysis was performed using GrraphPad Prism 8.0 software. A T-test was used to evaluate the difference in measurement data between the two groups. A one-way ANOVA analysis of variance test and a Bonferroni-corrected posttest were used for comparing more than two groups. P<0.05 was considered statistically significant. All data were presented as mean ±SD.
Results
Preparation and Characterization of Ginsenoside Nanoparticles
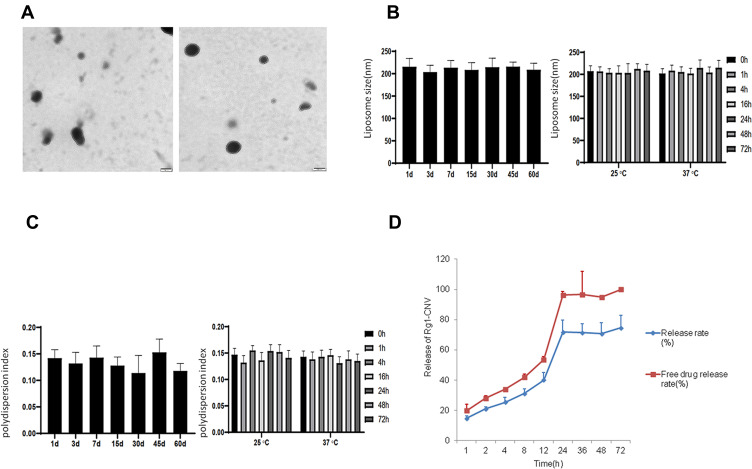
According to the results of the laser nanoparticle size analyzer, the average particle size of ginsenoside nanoparticles was 203.78±6.83 nm, the polydispersion index was 0.135±0.007, and the Zeta potential was 23.13±1.65 mV (Figure 1A). When the nano-liposomes were stored at 4 °C for 60 d, the average particle size and polydispersion index did not change significantly at 25 and 37 °C for 72 h (Figures 1B and C). Under the selected chromatographic analysis conditions, the drug ginsenoside Rg1 was well separated from impurities by comparison. The results demonstrate that there was no interference peak at the corresponding retention time of ginsenoside. Its retention time was about 17.015 min. In the extraction method and chromatographic conditions, ginsenoside Rg1 was in the concentration range of 1.0–100.0 µg/mL, and the linear relationship was good using the linear regression method to calculate the slope, intercept and correlation coefficient, and standard curve square. The process and correlation coefficients are as follows: Ginsenoside Rg1: Y= 11,912 X -- 5285.2 R =0.9999. The mean and standard deviation of the peak area of ginsenoside Rg1 in the samples at various concentrations were calculated, and the coefficient of variation (RSD) of the peak area of high-performance liquid chromatography at low, medium, and high concentrations was calculated. The precision of the HPLC method was obtained (%). Under the given extraction method and detection conditions, the average recovery of ginsenoside Rg1 in the shell of nanoparticles was 98.54%, and RSD was 3.82%.
Figure 1.
Preparation and characterization of ginsenoside nanoparticles. (A) Ginsenoside nanoparticles in electron microscopy. (B) The change of particle size of nanoparticles at different storage temperatures. (C) The change of polydispersion index of nanoparticles at different storage temperatures. (D) Release of Rg1-CNV(n=3).
As the formula = (W1-W2)/W1*100%, the encapsulation ratio of ginsenosides in nanoparticles was calculated. According to the detection, the encapsulation rate of ginsenoside Rg1 was 62.83±2.84%. In the calculation of ginsenoside leakage rate, the formula = (encapsulation rate measured on the first day - encapsulation rate measured on the 10th day)/encapsulation rate measured on the first day *100%, the leakage rate of ginsenoside nanoparticles was 5.98±0.58% at 4 °C and 13.64±1.84% at room temperature. The known formula is the release rate RE=W2/W1*100%. According to the results of detection, ginsenoside Rg1 was under pH5.0. The release rate of ginsenoside Rg1 was 1.54±0.28% at pH6.0 and 0.93±0.11% at pH7.0. Under these conditions, the release rate of ginsenoside Rg1 was 0.55±0.071%. The release rate of ginsenoside Rg1 decreased gradually, indicating that the stability of the nanoparticles decreased under low pH conditions. The content of free ginsenoside Rg1 in the dialysate was accurately calculated (Figure 1D). The results indicated that the release rate of ginsenoside Rg1 increased gradually with the extension of release time, but the release remained stable after 24h.
Evaluation of the Therapeutic Effect of Ginsenoside Rg1-CNV
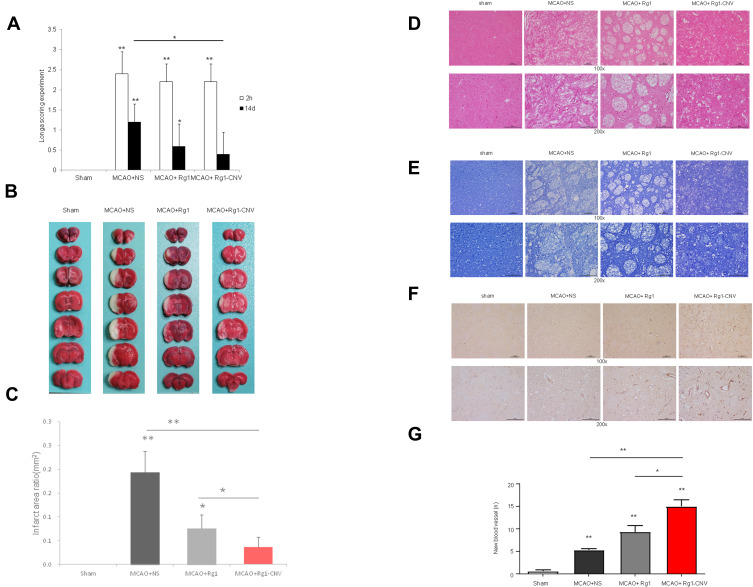
The neurological function of the Sham group was 0, and there no neurological defects were observed. At the 2 h observation point, the neurological function score of the MCAO+NS group was significantly higher than that of the Sham group. The scores of the MCAO+ Rg1 group and MCAO+ Rg1-CNV group were slightly lower than those of the MCAO group. At 14 d after modeling, compared with the Sham group, the neurological function scores of the MCAO+NS group still significantly increased, while the scores of the MCAO+ Rg1 group and the MCAO+ Rg1-CNV group gradually decreased, especially since no significant difference was observed between the MCAO+ Rg1-CNV group and the Sham group (Figure 2A). In brain TCC staining (Figure 2B), the white areas of the brain represent infarcts. The infarct area was the largest in the model group and the smallest in the MCAO+ Rg1-CNV group (Figure 2C). The HE staining results suggested that the number of neurons in the MCAO+NS group was significantly reduced and the tissue edema was obvious compared with the sham group; the number of neurons in the MCAO+ Rg1 group was slightly increased compared with the MCAO group; the number of neurons in the MCAO+ Rg1-CNV group was significantly increased compared with the MCAO group (Figure 2D). Nissl’s staining provided us with results that were basically consistent with HE staining (Figure 2E). After MCAO modeling, the neovascularization in brain tissue increased, while Rg1 injection promoted the increase of neovascularization, and Rg1- CNV injection further promoted the increase of neovascularization (Figures 2F and G).
Figure 2.
Evaluation of the therapeutic effect of ginsenoside Rg1-CNV. (A) Longa scoring experiment N=5. (B) Brain TCC staining N=5. (C) Infarct area ratio N=5. (D) Histochemistry N=3. (E) Nissl’s staining N=3. (F) Immunocytochemistry of CD31 markers N=3. (G) The number of new blood vessel N=3. * P<0.05, ** P<0.01.
Epigenetic Modification Enzymes Regulated by Ginsenoside Rg1-CNV
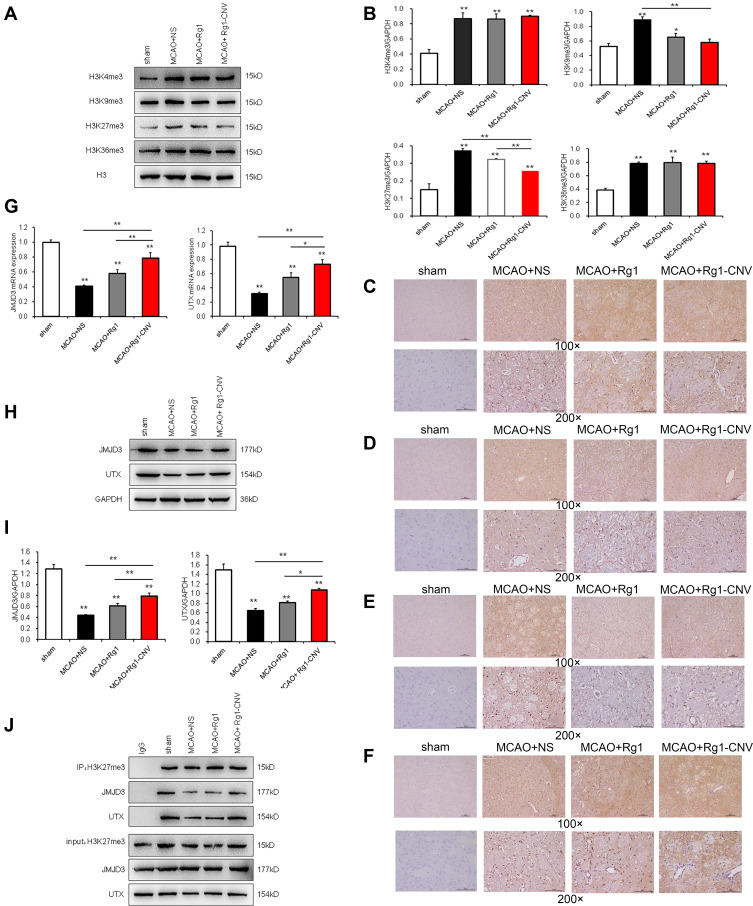
H3K4Me3, H3K9Me3, H3K27Me3, and H3K36Me3 methylation modifications in the brain tissue of rats after MCAO modeling were increased (Figure 3A and B). When Rg1 was added to the modeling, H3K4Me3, H3K9Me3, and H3K36Me3 modifications showed no significant changes, while H3K27Me3 decreased (Figure 3E). When the Rg1-CNV was added, H3K4Me3 and H3K36Me3 modifications were not significantly changed (Figures 3C and F), while H3K9Me3 and H3K27Me3 were decreased (Figures.3D and E). Compared with the Rg1 group, intracellular H3K27Me3 was decreased when Rg1-CNV was injected. No significant changes were observed in H3K4Me3, H3K9Me3, and H3K36Me3 (Figures 3C, D and F).
Figure 3.
Epigenetic modification enzymes regulated by ginsenoside Rg1-CNV. (A) Gray scale of epigenetic modification enzymes. (B) Protein expression of epigenetic modification enzymes. (C) Immunocytochemistry of H3K4me3 in every groups. (D) Immunocytochemistry of H3K9me3 in every groups. (E) Immunocytochemistry of H3K27me3 in every groups. (F) Immunocytochemistry of H3K36me3 in every groups. (G) The mRNA expression of JMJD3 and UTX. (H) Gray scale analysis of JMJD3 and UTX. (I) Protein expression of JMJD3 and UTX. (J) Co-IP detects the binding of H3K27me3 with JMJD3 and UTX* P<0.05, ** P<0.01.
Compared with the Sham group, the mRNA expression of UTX and JMJD3 in rat brain tissue was down-regulated after MCAO modeling. UTX and JMJD3 expression was up-regulated after Rg1 was added. The expression of UTX and JMJD3 in the Rg1-CNV group was significantly higher than in the MCAO+NS group (Figure 3G). Compared with the MCAO+Rg1 group, the expression of UTX and JMJD3 in the MCAO+Rg1-CNV group significantly increased. As shown in the protein trends of UTX and JMJD3, this was consistent with the mRNA expression (Figures 3H and I). Next, we applied IP detection to detect H3K27me3. The experiment was successful, and IgG served as a negative control. JMJD3 and UTX were detected in all IP samples, indicating that they had a binding effect on H3K27me3. In the rat MCAO model, the binding effect of UTX and JMJD3 to H3K27me3 was reduced, while the binding ability was slightly increased when Rg1 was injected. Meanwhile, when Rg1-CNV was added, the binding of UTX and JMJD3 to H3K27me3 significantly increased (Figure 3J).
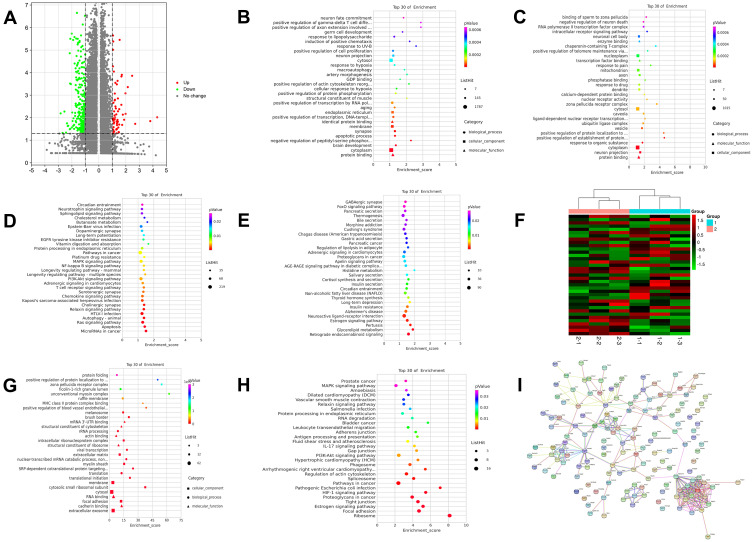
According to the quality statistics in Table 3, the sequencing results of all samples were excellent, and the base distribution was balanced. Bioinformatics analysis revealed that there were 7671 downregulated differentially expressed genes (DEGs) and 4186 up-regulated DEGs in the Rg1-CNV treatment group compared to the MCAO control group (Figure 4A). Further GO enrichment analysis showed that the 7671 downregulated DEGs were enriched in biological processes, such as brain development, apoptotic process, and positive regulation of cell proliferation; cellular components, such as cytoplasm, synapse, and endoplasmic reticulum; and molecular function, such as protein binding, identical protein binding and GDP binding (Figure 4B). The 4186 up-regulated DEGs were discovered in biological processes such as responses to organic substances, positive regulation of protein establishment, and intracellular receptor signaling pathway; cellular components such as cytoplasm, neuron projection, and cytosol; and molecular function such as protein binding, ligend-dependent nuclear receptor transcription, and enzyme binding (Figure 4C).
Table 3.
Pretreatment of Data
| Sample ID | Raw Reads | Clean Reads | Clean Ratio | rRNA Trimed | rRNA Ratio | No rRNA Pair |
|---|---|---|---|---|---|---|
| X11 | 41,811,144 | 38,485,949 | 92.05% | 38,410,682 | 0.20% | 35,895,868 |
| X12 | 39,463,104 | 36,073,146 | 91.41% | 35,906,999 | 0.46% | 33,353,052 |
| X13 | 40,628,153 | 35,826,155 | 88.18% | 34,251,233 | 0.41% | 32,251,069 |
| X21 | 42,526,391 | 37,914,283 | 89.15% | 35,169,283 | 0.37% | 33,618,527 |
| X22 | 41,734,082 | 38,104,821 | 91.30% | 37,970,934 | 0.35% | 35,371,026 |
| X23 | 61,479,024 | 59,462,534 | 96.72% | 59,145,093 | 0.53% | 58,151,914 |
Notes: Clean ratio = (Clean reads/Raw reads)%; rRNA ratio = [(Clean reads - rRNA trimed)/ Clean reads]%; X11-X13 was MCAO control group; X21-x23 was Rg1-CNV treatment group.
Figure 4.
Bioinformatics analysis and ChIP-seq. (A) Volcano plot. (B) Top 30 of GO enrichment analysis of gene expression all down. (C) Top 30 of GO enrichment analysis of gene expression all up. (D) Top 30 of KEGG enrichment analysis of gene expression all down. (E) Top 30 of KEGG enrichment analysis of gene expression all up. EdgeR was used to analyze the differential genes among samples.35 After the p-value was obtained, multiple hypothesis tests were conducted to correct, and the threshold of p-value was determined by controlling FDR (False Discovery Rate) 36,37. Statistical significance was defined as adjusted p-value <0.05, and the biological significance was defined as a difference of fold-change≥2, which were exhibited as Volcano spot. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes. (F) Heat map of histone H3K27trime binding had obvious changes in genes. Group 1 was MCAO model group, and group 2 was MCAO+ Rg1-CNV, treatment group. (G) Top of 30 GO enrichment. (H) Top of 30 KEGG enrichment. (I) String hires image.
In addition, KEGG enrichment analysis showed that the 7671 downregulated DEGs were associated with various pathways such as the Ras signaling pathway, chemokine signaling pathway, and MAPK signaling pathway (Figure 4D). The 4186 up-regulated DEGs demonstrated neuroactive ligand-receptor interaction, retrograde endocannabinoid signaling, and histidine metabolism (Figure 4E). The results of Chip-Seq detected obvious changes in the Histone H3K27trime binding gene (Figure 4F). A total od 152 proteins were bound by histone H3K27me3, among which 34 were down-regulated, 18 were up-regulated, and 100 were not significantly changed in the MCAO+ Rg1-CNV treatment group (Figures 4G and H). The sequencing results demonstrated that the histone H3K27 of VEGFA, EPO, JAG2, and other genes related to angiogenesis had triple methylation modification (Figure 4I).
Effects of Ginsenoside Rg1-CNV on Proliferation, Migration, and Tubular Formation of BMECs
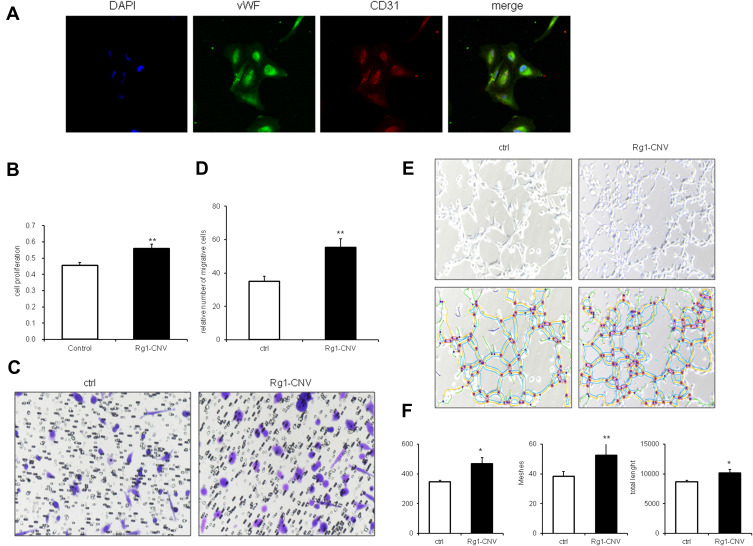
To further verify the impact of ginsenoside Rg1-CNV on the angiogenesis of ischemic brain tissue, we successfully isolated BMECs in vitro and administered ginsenoside Rg1-CNV intervention after culture and identification to observe the effects of ginsenoside Rg1-CNV on the proliferation, migration, and canalization of BMECs. In Figure 5A, as endothelial-specific markers, both vWF and CD31 were positive in the isolated cells, indicating that the isolated cells were brain microvascular endothelial cells (BMECs).
Figure 5.
Effects of ginsenoside Rg1-CNV on proliferation, migration, and tubular formation of BMECs. (A) Identification of rat brain microvascular endothelial cells by immunofluorescence images (400×). (B) Proliferation of BMECs on data statistic. (C) Migration of BMECs in microscope. (D) Migration of BMECs on data statistic. (E) The upper row is tubular formation of BMECs in white-light picture, the lower row is software analysis diagram of tubular formation by Image J v1.8.0. (F) BEMCs tubular formation data statistic. Meshes analyze the number of mesh formed in lumen, Nodes refers to the number of points in lumen, and total length refers to the length of lumen. * P<0.05, ** P<0.01.
As shown in Figure 5B, compared with normal cultured cells, the introduction of Rg1-CNV promoted cell proliferation. The number of migrated cells in the Rg1-CNV group was greater than that in the control group, indicating that Rg1-CNV promoted cell migration (Figures 5C and D). As shown in Figures 5E and F compared with the control group, the mesh, dot, and lumen length in the Rg1-CNV group increased, indicating that Rg1-CNV accelerated the formation of lumen.
Ginsenoside Rg1-CNV Up-Regulates JMJD3 and UTX to Promote the Demethylation of H3K27me3 in the Promoter Region of VEGF-A and Jagged1 Genes
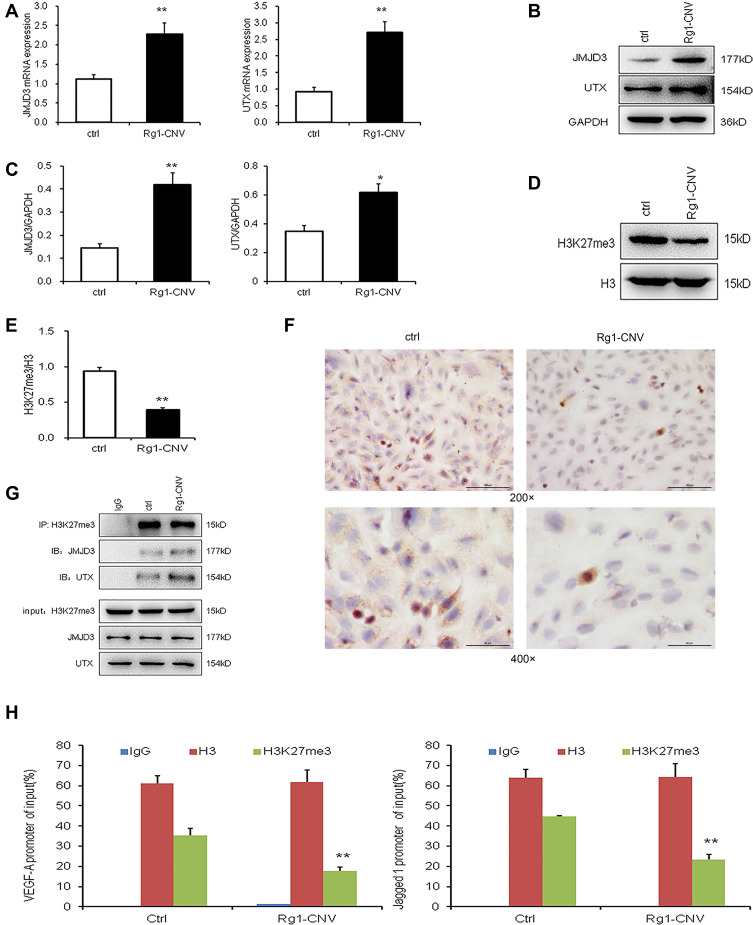
Rg1-CNV increased the mRNA expression of histone demethylase UTX and JMJD3 in cells compared to the control group (Figure 6A). As shown in Figures 6B and C, Rg1-CNV promoted the intracellular expression of histone demethylases UTX and JMJD3. As for the epigenetic enzymes screened by the previous in vivo experiment, the BEMCs were differently treated by Rg1-CNV, and we found that Rg1-CNV inhibits intracellular histone H3K27me3 methylation (Figures 6D and E), and the protein of H3K27me3 in Rg1-CNV group was lower than that in the control group (Figure 6E), which was consistent with microscopic observations (Figure 6F). Next, the H3K27me3 antibody was used for IP detection. JMJD3 and UTX were detected in all IP samples, indicating that UTX and JMJD3 had a binding effect on H3K27me3. When Rg1-CNV was added, the binding effect of UTX and JMJD3 on H3K27me3 increased (Figure 6G). As shown in Figure 6H, the intervention of ginsenoside Rg1-CNV promoted the demethylation of H3K27me3 in the promoter region of VEGF-A and Jagged1 genes, and also reduced the H3K27me3 modification in this region.
Figure 6.
Ginsenoside Rg1-CNV up-regulates JMJD3 and UTX to promote the demethylation of H3K27me3 in the promoter region of VEGF-A and Jagged1 genes. (A) mRNA expression of JMJD3 and UTX. (B) Western blot analysis of JMJD3 and UTX. (C) Protein expression of JMJD3 and UTX. (D) Western blot analysis of H3K27me3. (E) Protein expression of H3K27me3. (F) Comparison of H3K27me3 expression in each group by Immunohistochemical staining. (G) co-IP detection results of BMECs cells after different treatment. (H) ChIP detection results of ChIP detection results of H3K27me3 demethylation in the promoter regions of VEGF-A and Jagged1 genes (IgG was used as the negative control, H3 was used as the positive control). * P<0.05,** P<0.01.
Rescue
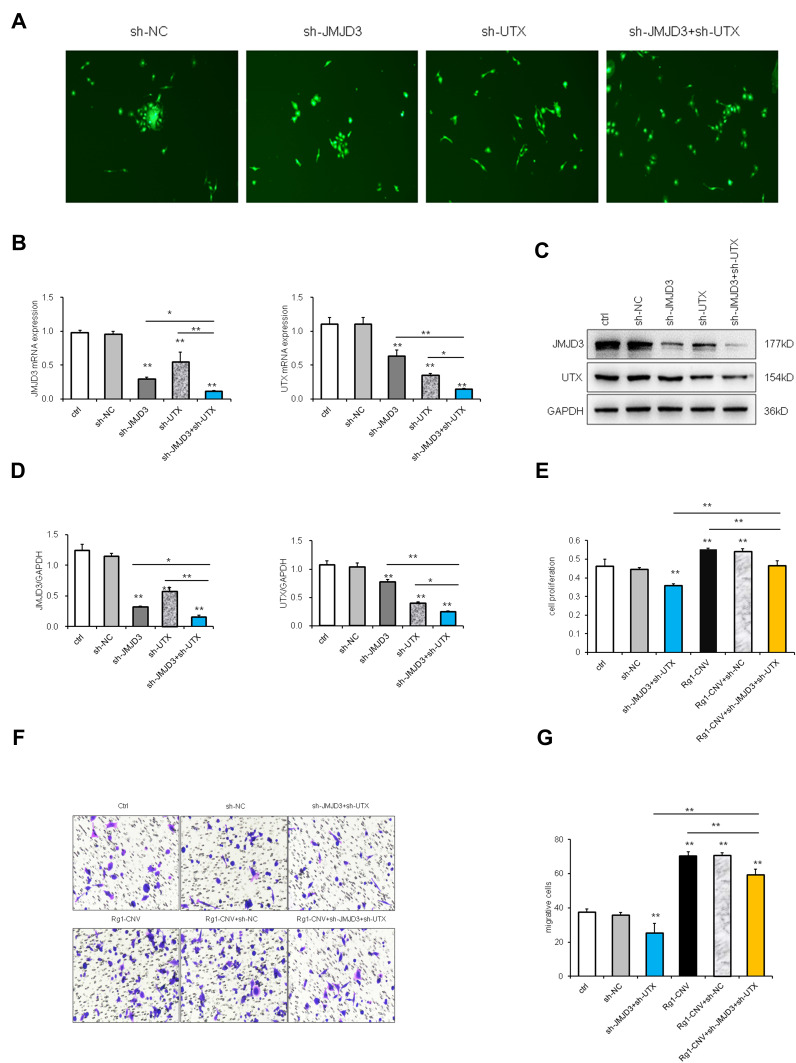
The rescue experiment was designed to investigate the function and target genes of epigenetic modification enzymes induced by ginsenoside Rg1-CNV in angiogenesis. The shuttle plasmid K5342 and packaging plasmid were applied to establish control low expression, control shRNA, JMJD3 low expression, UTX low expression, and JMJD3+UTX low expression groups. The fluorescence expression of BMECs cells was observed under the microscope, indicating that the lentivirus was successfully transfected and intracellular expression was initiated (Figure 7A).
Figure 7.
Rescue experiment (1). (A) BMECs fluorescence diagram (100×). (B) mRNA expression of JMJD3 and UTX. (C) Western blot analysis of JMJD3 and UTX. (D) Protein expression of JMJD3 and UTX. (E) Proliferation of BEMCs on data statistic. (F) Migration of BEMCs in microscope. (G) Migration of BEMCs on data statistic. * P<0.05. ** P<0.01.
As shown in Figure 7B, the intracellular UTX and JMJD3 did not change significantly when the cells were infected with the sh-NC lentivirus. The intracellular UTX expression and JMJD3 expression were significantly down-regulated when the cells were infected with the sh-UTX lentivirus. The intracellular JMJD3 expression and UTX expression were significantly down-regulated when the cells were infected with the sh-JMJD3 virus. When sh-UTX and sh-JMJD3 lentivirus were co-infected, the expression of UTX and JMJD3 in cells was significantly down-regulated. Western blot analysis revealed that the protein trends of UTX and JMJD3 were consistent with their trends in mRNA expression (Figures 7C and D). When the expression of UTX and JMJD3 was down-regulated, cell proliferation was down-regulated accordingly, while Rg1-CNV promoted cell proliferation. Compared with the Rg1-CNV group, the expression of UTX and JMJD3 was simultaneously down-regulated, and cell proliferation was down-regulated (Figure 7E). Cell migration was down-regulated in the sh-UTX and sh-JMJD3 group, whereas the Rg1-CNV treatment increased the number of migration cells, indicating that UTX and JMJD3 promoted cell migration (Figures 7F and G).
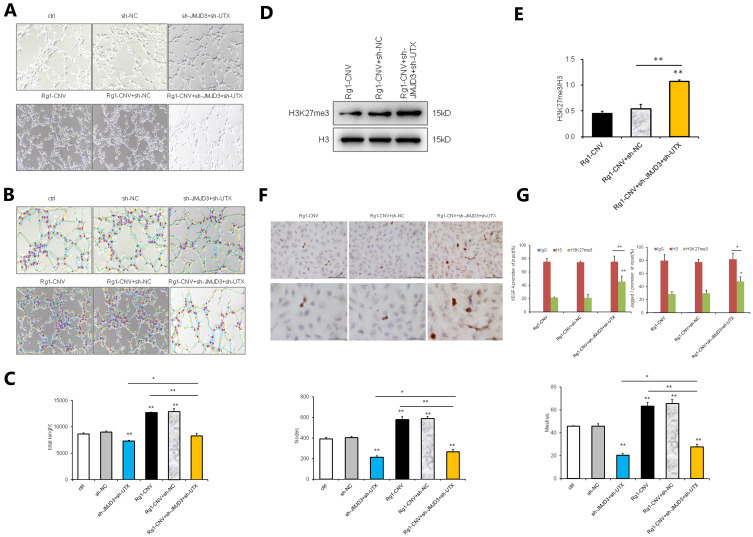
When the expression of JMJD3 and UTX was down-regulated, the lumen formation of the Rg1-CNV group increased. In contrast, when the expression of JMJD3 and UTX was down-regulated, the lumen formation of the Rg1-CNV group decreased (Figures 8A-C), demonstrating that Rg1-CNV promoted cell lumen development.
Figure 8.
Rescue experiment (2). (A) Tubular formation of BEMCs in white-light picture. (B) Tubular formation of BEMCs analysis diagram by software ImageJ v1.8.0. (C) Tubular formation of BEMCs analysis on data statistic. Meshes analysis of the number of holes formed in lumen, Nodes is the number of points in lumen, and Total Length is the length of lumen. (D) Western blot analysis of H3K27me3. (E) Protein expression of H3K27me3. (F) Comparison of H3K27me3 expression in each group by Immunohistochemical staining. (G) ChIP detection results of H3K27me3 demethylation in the promoter regions of VEGF-A and Jagged1 genes (IgG was used as the negative control, and H3 was used as the positive control). * P<0.05. ** P<0.01.
Intracellular H3K27me3 did not change significantly when cells were transfected with sh-NC lentivirus. However, when cells were transfected with sh-UTX and sh-JMJD3 lentivirus, their expression was up-regulated (Figures 8D and E). The H3K27me3 protein expression in the Rg1-CNV group was consistent with that in the Rg1-CNV + sh-NC group, the methylation in the Rg1-CNV + sh-JMJD3 + sh-UTX group was increased, and the H3K27me3 protein modification level was up-regulated (Figure 8F). As shown in Figure 8G, H3K27me3 in the histone of VEGF and the Jagged1 promoter region was modified. When the expression of JMJD3 and UTX was down-regulated by sh-JMJD3 and sh-UTX, the expression of H3K27me3 demethylation was higher than that in the Rg1-CNV group. These results demonstrate that ginsenoside Rg1-CNV intervention promoted H3K27me3 demethylation to reduce H3K27me3 methylation in the promoter regions of VEGF-A and Jagged1 genes.
Discussion
The primary role of ginsenoside Rg1 in angiogenesis has been heavily investigated in recent research.21,22 However, the epigenetic regulatory mechanism and specific target molecules of ginsenoside Rg1 have rarely been reported in the existing literature. Our previous study found that ginsenoside Rg1 can significantly induce the expression of histone H3K27-specific demethylases JMJD3 and UTX. Additionally, the H3K27 methylation level in brain microvascular endothelial cells (BMECs) of ischemic stroke model rats was significantly down-regulated post ginsenoside Rg1 intervention. Meanwhile, VEGF and Jagged1 expressions were significantly up-regulated. Due to the important role of VEGF and Notch signaling pathways in angiogenesis, it is of great significance to investigate and up-regulate their activation levels to improve the therapeutic effect they have. Additionally, the bioavailability of oral administration is relatively low, primarily due to the destruction of the acidic environment of gastric juice, the degradation of enzymes in coliform flora, and the role of the liver as the first gateway. Determining how active substances inherent in traditional Chinese medicine may efficiently pass the blood-brain barrier and act on target tissues is an urgent problem that merits further investigation in future research.
Compared with traditional drugs, nanomaterial drugs have the advantage of improving the solubility, bioavailability, and absorption rate of insoluble drugs.38–40 Our nanoparticles were prepared using an amphiphilic copolymer of polyglutamic acid and ethyl phenylalanine. Amphiphilic polymeric carrier materials have one end that is hydrophilic at one end and lipophilic at the other end. The lipophilic property can make the carrier material encapsulate some fat-soluble substances, and the hydrophilic nature can make the wrapped substances have a good hydration layer. This structure also has certain spatial advantages, which can improve the stability of the wrapped substance and avoid adhesion to molecules such as serum albumin during in vivo transport, resulting in a decrease in efficacy. Polyglutamic acid (PGA) is obtained by microbial fermentation and is degraded to a single glutamic acid, which is an essential amino acid for the human body41–43 and has a high water solubility. It can form a good hydration layer in the solution and improve the water solubility of some hydrophobic molecules. PGA polymeric materials are non-immunogenic and have good biodegradability and biocompatibility, and the metabolites are not toxic to humans. PGA can be amidated and coupled with the hydrophobic molecule L-phenylalanine ethyl ester to produce microspheres, tissue engineering materials, etc. PGAs are widely used in the pharmaceutical industry for the preparation of amphiphilic block copolymers. PGAs are commonly used for nanoparticles, which are modified with hydrophilic PEG chains to avoid the uptake of mononuclear macrophages in the body cycle.44 The nanoparticle surface is modified with hydrophilic PEG chains to avoid uptake by the mononuclear macrophage system in the body cycle44 and prolong its retention time in vivo. PGA can reduce the adsorption of enzymes on the nanoparticle surface and increase drug delivery system and drug stability. Our previous study showed that ginsenoside Rg1 nanoparticles can greatly increase the efficiency of drug penetration through the blood-brain barrier to target organs in model rats, and promote the recovery of cerebral infarction.31 Meanwhile, in this study, the ginsenoside Rg1-CNV we used was further modified based on the previous nanomaterial; the results obtained indicate that this improved the drug-carrying capacity and stability. The diameter of the delivery system for intracerebral drugs is usually around 200 nm, and can be more efficiently taken up by receptor-mediated endocytosis, which can achieve a good effect on the brain.45,46 The average of our synthesized ginsenoside Rg1-CNV was 203.78±6.83nm, therefore, the enrichment effect on brain tissue has a size advantage. The PDI is described the molecular weight distribution of polymers, and a lower PDI indicates a uniform molecular weight distribution. In general, PDI<0.3 was thought to be a good polymer dispersion. Our PDI was 0.135±0.007, indicating that our ginsenoside Rg1-CNV had good polymer dispersion.47 The zeta potential can be thought of as a dispersion system’s stability. A higher potential causes particles to repel each other, resulting in a more stable dispersion system.48 In our study, the Zeta potential was 23.13±1.65mV, indicating that the ginsenoside Rg1-CNV could maintain stability in aqueous phase despite strong mutual electrostatic repulsion. The results in Figure 1D show the release of ginsenoside Rg1-CNV in a slower and sustainable manner. Drugs should be released from a drug carrier system in a sustained rather than rapid manner. Slow-release formulations, as opposed to rapid release formulations, can maintain a longer effective concentration of medicine in blood and reduce drug dose frequency. The ginsenoside Rg1 could be loaded into nanoparticles, preventing degradation and increasing drug stability. Consequently, we predict that CNV nanoparticles can assist ginsenoside Rg1 to traverse the BBB more efficiently to target tissues. Obviously, we have a deeper exploration of the mechanism of therapeutic effect of ginsenoside Rg1-CNV in this study.
To evaluate the therapeutic effect of ginsenoside Rg1-CNV, rat models of ischemic stroke were established, and model rats were treated with oral administration. The differences in pathological morphology, the volume of infarcted brain tissue, the density of neovascularization, and behavioral signs of nerve defect between the control group and treatment group were all monitored. Our results reveal that ginsenoside Rg1-CNV has a significant therapeutic effect on the ischemic stroke of rat models, which verifies the results reported by a prior study on the protective effect of ginsenoside Rg1 on cerebral ischemia injury.49 On one hand, after the culture and identification of BMECs in vitro, ginsenoside Rg1-CNV was administered to promote the proliferation, migration, and tubular formation of BMECs. This suggests that ginsenoside Rg1-CNV has an angiogenic effect on BMECs in ischemic injury. On the other hand, in the process of verifying the target genes regulated by ginsenoside Rg1-CNV and the corresponding epigenetic inheritance regulatory mechanism, we found that the intervention of ginsenoside Rg1-CNV promoted the demethylation of H3K27me3 in the promoter region of VEGF-A and Jagged1 genes, and reduced the H3K27me3 modification in the promoter region of these genes. VEGF-A is the primary form of VEGF, and is a highly specific pro-vascular endothelial cell growth factor.50 VEGF can significantly promote vascular endothelial cell proliferation, migration, and chemotaxis in bone, lungs, kidneys, brain, and tumors.51–53 Numerous reports are available on the expression and activation of VEGF after cerebral ischemia injury. For example, in the experimental rat model, the VEGF signal is highly activated within a few minutes after the occurrence of an ischemic stroke, and the overexpression of VEGF can still be observed in glial cells after three weeks.54,55 Jagged1 is one of the ligands of Notch. The Notch signaling pathway is another important regulatory signal of angiogenesis that is initiated by the binding of the Notch receptor and ligand. Notch molecules are activated after ligand and receptor binding in the pathway, releasing NICD and initiating the transcription of Hes1 and other downstream target genes of Notch to play a regulatory role.56 We conclude that ginsenoside Rg1-CNV may potentially have a vital function in boosting angiogenesis based on the above two parts of the research.
To better understand how ginsenoside Rg1-CNV promotes angiogenesis, we searched for epigenetic enzymes regulated by ginsenoside Rg1-CNV at the mRNA and protein levels and knocked, and then used a rescue experiment to confirm the function of the target epigenetic enzymes in angiogenesis. Interestingly, we found that the binding of UTX and JMJD3 to H3K27me3 was enhanced post Rg1-CNV intervention. UTX (KDM6B) and JMJD3 (KDM6A) are both members of the Jumonji family. They mainly demethylate H3K27, whose methylation levels are altered by Polycomb-group proteins.57,58 H3K27 methylation and demethylation modifications have a critical role in a variety of biological processes, including X chromosome inactivation, genomic imprinting, stem cell maintenance, inflammatory response, and carcinogenesis.58,59 When we knocked down the expression of UTX and JMJD3 in the rescue experiment, the cell proliferation, migration, and tubular formation were previously down-regulated compared to the Rg1-CNV group. Meanwhile, the expression of H3K27me3 was higher than the group with a normal expression of UTX and JMJD3. A prior study also demonstrated that H3K27 trimethylation levels in glomerular podocyte development can directly regulate podocyte differentiation by influencing the Notch signaling pathway.60 Therefore, we speculate that UTX and JMJD3 may influence H3K27me3 demethylation in angiogenesis. In other words, the expression changes of UTX and JMJD3 indicate that ginsenoside Rg1-CNV may specifically induce the demethylation of histone H3K27. Chemosynthesis plays a vital role in regulating the expression of some genes in angiogenesis.
GO enrichment analysis showed that the DEGs regulated by ginsenoside Rg1-CNV concentrated on the positive regulation of cell proliferation, intracellular receptor signaling pathway, enzyme binding, etc. KEGG enrichment analysis showed that regulated DEGs were associated with various pathways. This suggests that the candidate target genes regulated by ginsenoside Rg1-CNV may be expressed via epigenetic enzymes, thus resulting in the activation of angiogenesis pathways. The results of ChIP-seq confirmed that the histone H3K27 of VEGFA, EPO, JAG2, and other genes related to angiogenesis had triple methylation modification. Moreover, in the rescue experiment, the downregulation of UTX and JMJD3 promoted H3K27me3 demethylation of VEGF-A and Jagged1 gene promoter regions. In other words, Rg1-CNV enhances UTX and JMJD3 binding to H3K27me3 and reduces H3K27me3 demethylation in VEGF-A and Jagged1 promoter regions. Therefore, the expression of VEGF and Jagged1 is significantly up-regulated. Thus, our findings suggest that ginsenoside Rg1-CNV nanoparticles have an ideal contribution to the treatment of ischemic stroke, and that their application in clinical treatment has promising theoretical feasibility in the near future.
Conclusion
In this study, our prepared ginsenoside Rg1-CNV has good physical stability and stable drug release rate. The results of a series of in vivo experiments demonstrated that ginsenoside Rg1-CNV can significantly promote angiogenesis in rats with ischemic stroke. Meanwhile, we explored the mechanism of ginsenoside Rg1-CNV promoting angiogenesis through in vitro experiments and bioinformatics analysis. The ginsenoside Rg1-CNV is a BBB penetrating agent by which the epigenetic enzymes UTX and JMJD3 are induced to demethylate histone H3K27me3 in the VEGF-A and Jagged1 promoter regions, thus activating angiogenesis. Our ginsenoside nanoparticles may be a promising strategy for the treatment of ischemic stroke in the near future.
Funding Statement
This work was supported by the National Natural Science Foundation of China(81803943).
Abbreviations
BBB, blood-brain barrier; CNV, complex nanovesicles; PDI, polydispersion index; H3K4Me3, histone H3 lysine 4 tri-methylation; H3K9Me3, histone H3 lysine 9 tri-methylation; H3K27Me3, histone H3 lysine 27 tri-methylation; H3K36Me3, histone H3 lysine 36 tri-methylation; DEGs, differentially expressed genes; BEMCs, brain microvascular endothelial cells;VEGF, vascular endothelial growth factor; SD, Sprague Dawley; CCA, common carotid artery; ICA, internal carotid artery; ECA, external carotid artery; MCA, middle cerebral artery; RNA-seq, RNA sequencing; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; co-IP, Co-immunoprecipitation; ChIP-seq, Chromatin Immunoprecipptation sequencing.
Data Sharing Statement
All data generated or analyzed during this study are present in this article. Research data are not shared.
Ethics Approval and Consent to Participate
All animal procedures were performed according to the national regulations and approved by the Animal Ethics Committee of Jinling Hospital, Jiangsu Province, China (2018GKJDWLS-03-198).
Consent for Publication
All authors agree to be published.
Disclosure
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Woodruff TM, Thundyil J, Tang SC, Sobey CG, Taylor SM, Arumugam TV. Pathophysiology, treatment, and animal and cellular models of human ischemic stroke. Mol Neurodegener. 2011;6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papanagiotou P, Ntaios G. Endovascular Thrombectomy in Acute Ischemic Stroke. Circ Cardiovasc Interv. 2018;11(1):e005362. doi: 10.1161/CIRCINTERVENTIONS.117.005362 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi S, Fukuma S, Ikenoue T, Fukuhara S, Kobayashi S. Effect of Edaravone on Neurological Symptoms in Real-World Patients With Acute Ischemic Stroke. Stroke. 2019;50(7):1805–1811. doi: 10.1161/STROKEAHA.118.024351 [DOI] [PubMed] [Google Scholar]
- 4.He W, Mei Q, Li J, et al. Preferential Targeting Cerebral Ischemic Lesions with Cancer Cell-Inspired Nanovehicle for Ischemic Stroke Treatment. Nano Lett. 2021;21(7):3033–3043. doi: 10.1021/acs.nanolett.1c00231 [DOI] [PubMed] [Google Scholar]
- 5.Chouchani ET, Pell VR, Gaude E, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann CJ, Harms U, Rex A, et al. Vascular signal transducer and activator of transcription-3 promotes angiogenesis and neuroplasticity long-term after stroke. Circulation. 2015;131(20):1772–1782. doi: 10.1161/CIRCLATIONNAHA.114.013003 [DOI] [PubMed] [Google Scholar]
- 7.Perego C, Fumagalli S, Zanier ER, et al. Macrophages are essential for maintaining a M2 protective response early after ischemic brain injury. Neurobiol Dis. 2016;96:284–293. doi: 10.1016/j.nbd.2016.09.017 [DOI] [PubMed] [Google Scholar]
- 8.Greenberg DA, Jin K. From angiogenesis to neuropathology. Nature. 2005;438(7070):954–959. doi: 10.1038/nature04481 [DOI] [PubMed] [Google Scholar]
- 9.Ferrara N. Role of vascular endothelial growth factor in regulation of physiological angiogenesis. Am J Physiol Cell Physiol. 2001;280(6):C1358–66. doi: 10.1152/ajpcell.2001.280.6.C1358 [DOI] [PubMed] [Google Scholar]
- 10.Abe K, Setoguchi Y, Hayashi T, Itoyama Y. Dissociative expression of adenoviral-mediated E. coli LacZ gene between ischemic and reperfused rat brains. Neurosci Lett. 1997;226(1):53–56. doi: 10.1016/s0304-3940(97)00243-7 [DOI] [PubMed] [Google Scholar]
- 11.Zhang ZG, Zhang L, Jiang L, et al. VEGF enhances angiogenesis and promotes blood-brain barrier leakage in the ischemic brain. J Clin Invest. 2000;106(7):829–838. doi: 10.1172/JCI9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Jin K, Xie L, et al. Greenberg. VEGF-induced neuroprotection, neurogenesis, and angiogenesis after focal cerebral ischemia. J Clin Invest. 2003;111(12):1843–1851. doi: 10.1172/JCI200317977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CC, Wang LC, Su YT, et al. Synthetic α5β1 integrin ligand PHSRN is proangiogenic and neuroprotective in cerebral ischemic stroke. Biomaterials. 2018;185:142–154. doi: 10.1016/j.biomaterials.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Fu X, Yu L, et al. Correction to: preconditioning with VEGF Enhances Angiogenic and Neuroprotective Effects of Bone Marrow Mononuclear Cell Transplantation in a Rat Model of Chronic Cerebral Hypoperfusion. Mol Neurobiol. 2020;57:3989–3990. doi: 10.1007/s12035-020-01898-2 [DOI] [PubMed] [Google Scholar]
- 15.Choi NY, Kim JY, Hwang M, et al. Atorvastatin rejuvenates neural stem cells injured by oxygen-glucose deprivation and induces neuronal differentiation through activating the PI3K/Akt and ERK pathways. Mol Neurobiol. 2019;56(4):2964–2977. [DOI] [PubMed] [Google Scholar]
- 16.Wang DP, Jin KY, Zhao P, et al. Neuroprotective Effects of VEGF-A Nanofiber Membrane and FAAH Inhibitor URB597 Against Oxygen-Glucose Deprivation-Induced Ischemic Neuronal Injury. Int J Nanomedicine. 2021;16:3661–3678. doi: 10.2147/IJN.S307335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Kilic E, Kilic U, et al. VEGF overexpression induces post-ischaemic neuroprotection, but facilitates haemodynamic steal phenomena. Brain. 2005;128(Pt 1):52–63. doi: 10.1093/brain/awh325 [DOI] [PubMed] [Google Scholar]
- 18.Benedito R, Roca C, Sorensen I, et al. The Notch Ligands Dll4 and Jagged1 Have Opposing Effects on Angiogenesis. Cell. 2009;137(6):1124–1135. doi: 10.1016/j.cell.2009.03.025 [DOI] [PubMed] [Google Scholar]
- 19.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YS, Seo EK, Gyllenhaal C, et al. Panax ginseng: a role in cancer therapy? Integr Cancer Ther. 2003;2(1):13–33. [DOI] [PubMed] [Google Scholar]
- 21.Leung KW, Ng HM, Tang MK, Wong CC, Wong RN, Wong AS. Ginsenoside-Rg1 mediates a hypoxia-independent upregulation of hypoxia-inducible factor-1alpha to promote angiogenesis. Angiogenesis. 2011;14(4):515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung LW, Leung KW, Wong CK, Wong RN, Wong AS. Ginsenoside-Rg1 induces angiogenesis via non-genomic crosstalk of glucocorticoid receptor and fibroblast growth factor receptor-1. Cardiovasc Res. 2011;89(2):419–425. doi: 10.1093/cvr/cvq300 [DOI] [PubMed] [Google Scholar]
- 23.Yue PJ, He L, Qiu SW, et al. OX26/CTX-conjugated PEGylated liposome as a dual-targeting gene delivery system for brain glioma. Mol Cancer. 2014;13(191):1476–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thom G, Burrell M, Haqqani AS, et al. Enhanced Delivery of Galanin Conjugates to the Brain through Bioengineering of the Anti-Transferrin Receptor Antibody OX26. Mol Pharm. 2018;15(4):1420–1431. doi: 10.1021/acs.molpharmaceut.7b00937 [DOI] [PubMed] [Google Scholar]
- 25.Shen J, Zhao Z, Shang W, et al. Ginsenoside Rg1 nanoparticle penetrating the blood-brain barrier to improve the cerebral function of diabetic rats complicated with cerebral infarction. Int J Nanomedicine. 2017;12:6477–6486. doi: 10.2147/IJN.S139602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramalho MJ, Sevin E, Gosselet F, et al. Receptor-mediated PLGA nanoparticles for glioblastoma multiforme treatment. Int J Pharm. 2018;545(1–2):84–92. doi: 10.1016/j.ijpharm.2018.04.062 [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Zhao H, Shu L, et al. Preparation and evaluation of Baicalin-loaded cationic solid lipid nanoparticles conjugated with OX26 for improved delivery across the BBB. Drug Dev Ind Pharm. 2015;41(3):353–361. doi: 10.3109/03639045.2013.861478 [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Zhang L, He Q, et al. Effect of Baicalin-loaded PEGylated cationic solid lipid nanoparticles modified by OX26 antibody on regulating the levels of baicalin and amino acids during cerebral ischemia-reperfusion in rats. Int J Pharm. 2015;489(1–2):131–138. doi: 10.1016/j.ijpharm.2015.04.049 [DOI] [PubMed] [Google Scholar]
- 29.Shen J, Zhao Z, Shang W, et al. Ginsenoside Rg1 nanoparticle penetrating the blood-brain barrier to improve the cerebral function of diabetic rats complicated with cerebral infarction. Int J Nanomedicine. 2017;12:6477–6486. doi: 10.2147/IJN.S139602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer CJ, Ischemic stroke: experimental models and reality. Acta Neuropathol. 2017;133(2):245–261. doi: 10.1007/s00401-017-1667-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Shuaib A, Li Q. Quantification of infarct size on focal cerebral ischemia model of rats using a simple and economical method. J Neurosci Methods. 1998;84(1–2)::9–16. doi: 10.1016/s0165-0270(98)00067-3 [DOI] [PubMed] [Google Scholar]
- 32.Gill R, Sibson NR, Hatfield RH, et al. A Comparison of the Early Development of Ischaemic Damage following Permanent Middle Cerebral Artery Occlusion in Rats as Assessed Using Magnetic Resonance Imaging and Histology. J Cereb Blood Flow Metab. 1995;15(1):1–11. doi: 10.1038/jcbfm.1995.1 [DOI] [PubMed] [Google Scholar]
- 33.Zhao L, Xu J, Wang Q, et al. Protective effect of rhGLP-1 (7–36) on brain ischemia/reperfusion damage in diabetic rats. Brain Res. 2015;30:153–159. doi: 10.1016/j.brainres.2015.01.014 [DOI] [PubMed] [Google Scholar]
- 34.Ye XC, Hao Q, Ma WJ, et al. Dectin-1/Syk signaling triggers neuroinflammation after ischemic stroke in mice. J Neuroinflammation. 2020;17(1):17. doi: 10.1186/s12974-019-1693-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1)::139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korthauer K, Kimes PK, Duvallet C, et al. A practical guide to methods controlling false discoveries in computational biology. Genome Biol. 2019;20(1):118. doi: 10.1186/s13059-019-1716-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge Y, Sealfon SC, Speed TP. Some step-down procedures controlling the false discovery rate under dependence. Stat Sin. 2008;18(3):881–904. [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Gu W, Chen J, Chen W, Xu ZP. Co-delivery of siRNAs and anti-cancer drugs using layered double hydroxide nanoparticles. Biomaterials. 2014;35(10):3331–3339. doi: 10.1016/j.biomaterials.2013.12.095 [DOI] [PubMed] [Google Scholar]
- 39.Peng C, Xu J, Yu M, et al. Tuning the In Vivo Transport of Anticancer Drugs Using Renal-Clearable Gold Nanoparticles. Angew Chem Int Ed Engl. 2019;58(25):8479–8483. doi: 10.1002/anie.201903256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin BT, Guo J, Presas E, Donovan MD, Alonso MJ, O’Driscoll CM. Pharmacokinetic, pharmacodynamic and biodistribution following oral administration of nanocarriers containing peptide and protein drugs. Adv Drug Deliv Rev. 2016;106(Pt B):367–380. doi: 10.1016/j.addr.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 41.van den Hoven JM, Van Tomme SR, Metselaar JM, Nuijen B, Beijnen JH, Storm G. Liposomal drug formulations in the treatment of rheumatoid arthritis. Mol Pharm. 2011;8(4):1002–1015. doi: 10.1021/mp2000742 [DOI] [PubMed] [Google Scholar]
- 42.Thomas TP, Goonewardena SN, Majoros IJ, et al. Folate-targeted nanoparticles show efficacy in the treatment of inflammatory arthritis. Arthritis Rheum. 2011;63(9):2671–2680. doi: 10.1002/art.30459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajaj I, Singhal R. Poly (glutamic acid)--an emerging biopolymer of commercial interest. Bioresour Technol. 2011;102(10):5551–5561. doi: 10.1016/j.biortech.2011.02.047 [DOI] [PubMed] [Google Scholar]
- 44.Abellan-Pose R, Rodriguez-Evora M, Vicente S, et al. Biodistribution of radiolabeled polyglutamic acid and PEG-polyglutamic acid nanocapsules. Eur J Pharm Biopharm. 2017;112:155–163. doi: 10.1016/j.ejpb.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 45.Ramalho MJ, Sevin E, Gosselet F, et al. Receptor-mediated PLGA nanoparticles for glioblastoma multiforme treatment. Int J Pharm. 2018;545(1–2):84–92. [DOI] [PubMed] [Google Scholar]
- 46.Henna TK, Raphey VR, Sankar R, et al. Carbon nanostructures: the drug and the delivery system for brain disorders. Int J Pharm. 2020;587:119701. doi: 10.1016/j.ijpharm.2020.119701 [DOI] [PubMed] [Google Scholar]
- 47.Arfors KE, Rutili G, Svensjo E. Microvascular transport of macromolecules in normal and inflammatory conditions. Acta Physiol Scand Suppl. 1979;463:93–103. [PubMed] [Google Scholar]
- 48.Mishra PR, Al Shaal L, Muller RH, et al. Production and characterization of Hesperetin nanosuspensions for dermal delivery. Int J Pharm. 2009;371(1–2):182–189. doi: 10.1016/j.ijpharm.2008.12.030 [DOI] [PubMed] [Google Scholar]
- 49.Lin M, Sun W, Gong W, et al. Ginsenoside Rg1 protects against transient focal cerebral ischemic injury and suppresses its systemic metabolic changes in cerabral injury rats. Acta Pharm Sin B. 2015;5(3):277–284. doi: 10.1016/j.apsb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrara N, Gerber H-P, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9(6):669–676. doi: 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- 51.Zhao T, Zhao W, Meng W, et al. VEGF-C/VEGFR-3 pathway promotes myocyte hypertrophy and survival in the infarcted myocardium. Am J Transl Res. 2015;7(4):697–709. [PMC free article] [PubMed] [Google Scholar]
- 52.Quittet MS, Touzani O, Sindji L, et al. Effects of mesenchymal stem cell therapy, in association with pharmacologically active microcarriers releasing VEGF, in an ischaemic stroke model in the rat. Acta Biomater. 2015;15:77–88. doi: 10.1016/j.actbio.2014.12.017 [DOI] [PubMed] [Google Scholar]
- 53.Iacovelli R, Sternberg CN, Porta C, et al. Inhibition of the VEGF/VEGFR Pathway Improves Survival in Advanced Kidney Cancer: a Systematic Review and Meta-Analysis. Curr Drug Targets. 2015;16(2)::164–170. doi: 10.2174/1389450115666141120120145 [DOI] [PubMed] [Google Scholar]
- 54.Abe K, Setoguchi Y, Hayashi Y, Itoyama Y. Dissociative expression of adenoviral-mediated E-coli LacZ gene between ischemic and reperfused rat brains. Neurosci Lett. 1997;226(1):53–56. doi: 10.1016/S0304-3940(97)00243-7 [DOI] [PubMed] [Google Scholar]
- 55.Zhang ZG, Zhang L, Jiang Q, et al. VEGF enhances angiogenesis and promotes blood-brain barrier leakage in the ischemic brain. J Clin Invest. 2000;106(7):829–838. doi: 10.1172/JCI9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- 57.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 58.Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–U710. doi: 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- 59.Martinez-Garcia E, Licht JD. Deregulation of H3K27 methylation in cancer. Nat Genet. 2010;42(2):100–101. doi: 10.1038/ng0210-100 [DOI] [PubMed] [Google Scholar]
- 60.Majumder S, Thieme K, Batchu SN, et al. Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J Clin Invest. 2018;128(1):483–499. doi: 10.1172/JCI95946 [DOI] [PMC free article] [PubMed] [Google Scholar]