Abstract
All current smooth muscle cell (SMC) Cre mice similarly recombine floxed alleles in vascular and visceral SMCs. Here, we present an Itga8-CreERT2 knock-in mouse and compare its activity with a Myh11-CreERT2 mouse. Both Cre drivers demonstrate equivalent recombination in vascular SMCs. However, Myh11-CreERT2 mice, but not Itga8-CreERT2 mice, display high activity in visceral SMC-containing tissues such as intestine, show early tamoxifen-independent activity, and produce high levels of CreERT2 protein. Whereas Myh11-CreERT2-mediated knockout of serum response factor (Srf) causes a lethal intestinal phenotype precluding analysis of the vasculature, loss of Srf with Itga8-CreERT2 (SrfItga8) yields viable mice with no evidence of intestinal pathology. Male and female SrfItga8 mice exhibit vascular contractile incompetence, and angiotensin II causes elevated blood pressure in wild type, but not SrfItga8, male mice. These findings establish the Itga8-CreERT2 mouse as an alternative to existing SMC Cre mice for unfettered phenotyping of vascular SMCs following selective gene loss.
Keywords: mouse, Cre recombinase, knockout, smooth muscle, serum response factor, integrin alpha 8
Editor summary:
Warthi and colleagues generate an alpha 8 integrin-cre driver that enables gene targeting specifically in vascular smooth muscle cells, and show in a proof-of-principle study that using the Itga8-CreERT2 knock-in mouse for selective ablation of the Srf gene causes vascular defects but not a lethal visceral myopathy observed in a smooth muscle cell-specific Myh11-CreERT2-driven Srf loss, attesting to an increased vascular muscle cell-specificity of the Itga8-CreERT locus.
Conditional gene disruption requires two components derived from bacteriophage P1: a Cyclization recombination (Cre) gene encoding a 343 amino acid (38 kDa) tyrosine site-specific recombinase and 34-base pair locus of crossing (x) over of P1 (loxP) sequences having dyad symmetry, flanking (or floxing) the region of DNA to be excised.1 The first application of the Cre/loxP system for conditional knockout of a gene utilized an Lck promoter-driven Cre transgene to inactivate Polb in T cells.2 Soon after, temporal inactivation of a gene was introduced with the fusion of Cre to a tamoxifen-responsive ligand binding domain of the estrogen receptor.3 These pioneering studies underscore two considerations in the design of conditional knockout experiments: judicious placement of loxP sites around a sequence to be excised and selection of a well-characterized, cell-restricted Cre driver. The spatial control of Cre expression is of crucial importance for gene disruption in smooth muscle cells (SMCs) which enfold and support blood and lymphatic vessels, the airways of the respiratory system, and all hollow organs of the abdominal cavity.
SMCs have distinct embryological origins4, 5 and contractile properties,6, 7 but they express a common set of cell-restricted genes that maintain normal SMC homeostasis.8 Several of these genes have been harnessed for the development of SMC Cre driver mice, each of which has advantages and disadvantages.9 A shared characteristic of all current SMC Cre driver mice is the comparable recombination activity in both vascular and visceral SMC lineages.9 SMC-wide Cre activity can lead to severe, often lethal, visceral myopathies that impede efforts to understand the biology of key genes within vascular SMCs (VSMCs).10-14 Further, whole-body knockout of some SMC-restricted genes results in embryonic or postnatal death, portending lethality even with an SMC conditional knockout approach.15-18 Various gene editing strategies will accelerate the development of new floxed alleles for conditional knockout studies in SMCs,19-22 and many gene products will likely be essential for visceral SMC homeostasis. Thus, a more VSMC-restricted Cre driver mouse is urgently needed to avoid the confounding effects of visceral myopathies observed with existing SMC Cre strains.
Alpha 8 integrin (Itga8) was first cloned from an embryonic chicken cDNA library and the 160 kilodalton protein was shown to interact with the beta 1 integrin subunit.23 Expression studies demonstrated ITGA8 protein in VSMCs and mesangial cells (an SMC-like cell) of the adult rat kidney.24, 25 A subtractive hybridization screen in VSMCs reported a partial cDNA to rat Itga8 that was used to show prominent Itga8 mRNA in the adult rat aorta.26 Subsequent work demonstrated the down-regulation of ITGA8 with vascular injury27 and its role in the maintenance of VSMC differentiation.28 A more recent study found abundant Itga8 mRNA in VSMCs, with minimal expression in visceral SMC-containing tissues of mouse, rat, and human origin. This prompted a suggestion that Itga8 might represent an ideal locus for targeting with an inducible Cre to establish a new mouse model for VSMC-restricted knockout studies.29 Here, the generation and characterization of an Itga8-CreERT2 mouse are reported. Comparative studies indicate important advantages of the Itga8-CreERT2 mouse over existing SMC Cre drivers for the unambiguous determination of gene function in VSMCs.
Results
Itga8-CreERT2 mice expressed normal levels of ITGA8
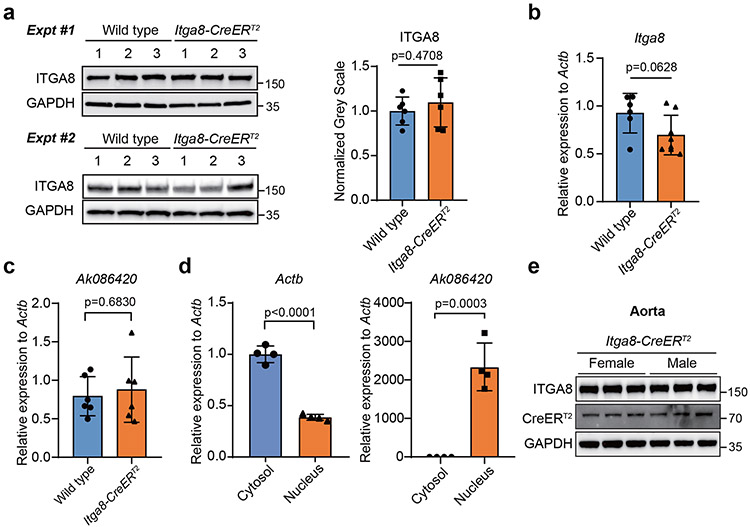
Compared to most traditional SMC markers, Itga8 mRNA is preferentially expressed in VSMCs (Supplementary Fig. 1). Accordingly, the first exon of Itga8 was targeted with a CreERT2 cassette in embryonic stem cells to establish a new inducible CreERT2 mouse (Extended Data Fig. 1). Mice homozygous for Itga8-CreERT2 displayed renal agenesis and postnatal death (Supplementary Fig. 2), a phenotype consistent with a previous Itga8 loss-of-function study.30 Because ITGA8 protein expression was not reported in heterozygous knockout mice,30 it was important to determine whether one functional Itga8 allele is sufficient for normal levels of ITGA8 protein. Heterozygous Itga8-CreERT2 aorta showed indistinguishable levels of ITGA8 protein and a slight reduction of Itga8 mRNA versus wild type aorta (Extended Data Fig. 2a,b). There was little change in the expression of a 5’ overlapping nuclear long noncoding RNA (Supplementary Fig. 1 and Extended Data Fig. 2c,d). Male and female Itga8-CreERT2 mice exhibited similar levels of ITGA8 and CreERT2 protein in the aorta (Extended Data Fig. 2e) and they bred well with no overt signs of pathology. These results demonstrated wild type levels of ITGA8 protein with only one functional Itga8 allele, thus supporting accurate phenotyping of Itga8-CreERT2-mediated gene knockouts without the confounding effects of attenuated ITGA8 protein.
Neither CreERT2 showed recombination activity in myeloid cells
Over 340 floxed genes have been targeted with various SMC Cre driver mice, 90% of which were excised with several versions of Tagln-Cre or Myh11-Cre (Supplementary Table 1). Previous work with Tagln-Cre (aka Sm22-Cre) reported recombination in myeloid cell lineages.31 For this reason, circulating cells (cleared of red blood cells) from tamoxifen-treated male and female Itga8-CreERT2 mice carrying the mT/mG reporter were isolated and compared to similarly prepared cells from tamoxifen-treated Myh11-CreERT2-mT/mG male mice.32 As expected, fluorescence-activated cell sorting revealed GFP positive circulating cells from an Sm22-Cre mouse33; however, essentially no GFP fluorescence was observed in circulating cells from Itga8-CreERT2 or Myh11-CreERT2 mice (Extended Data Fig. 3a). Moreover, analysis of bone marrow aspirates showed no activity of Itga8-CreERT2 or Myh11-CreERT2 in megakaryocytes or surrounding stromal cells (Extended Data Fig. 3b). Given the broad activity of Sm22-driven transgenes in cells of the vessel wall,9 the balance of experiments were conducted in Itga8-CreERT2 and Myh11-CreERT2 mice only.
Itga8-CreERT2 showed preferential recombination in VSMCs
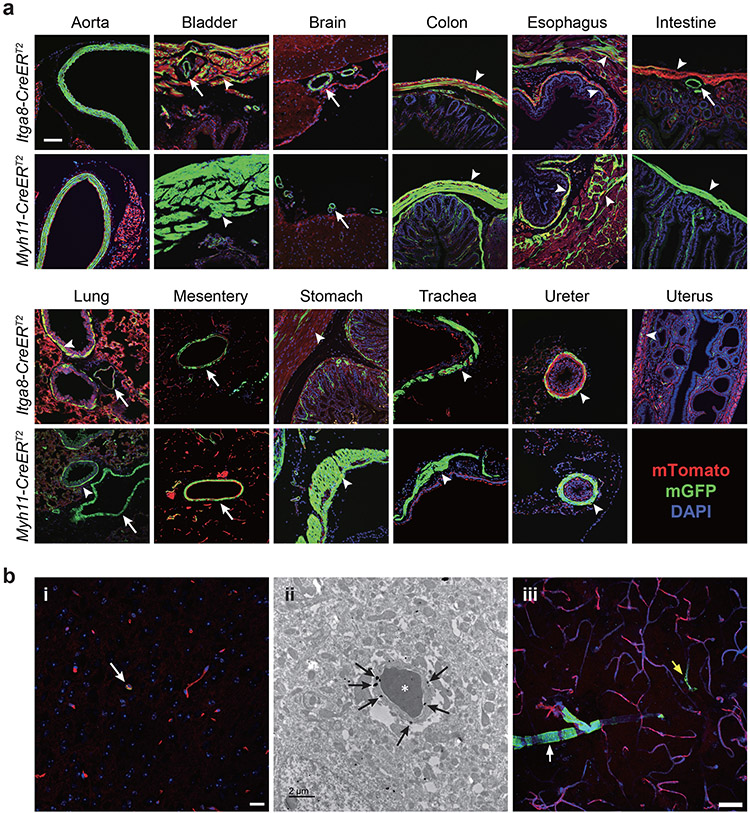
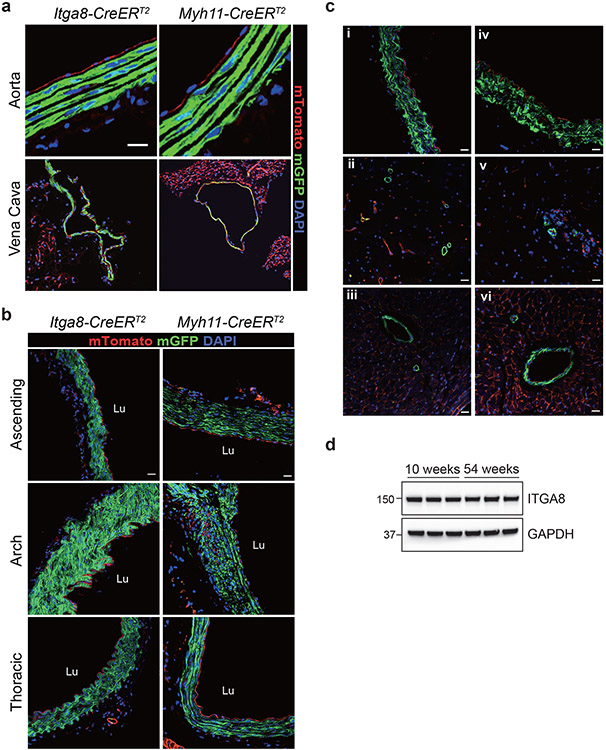
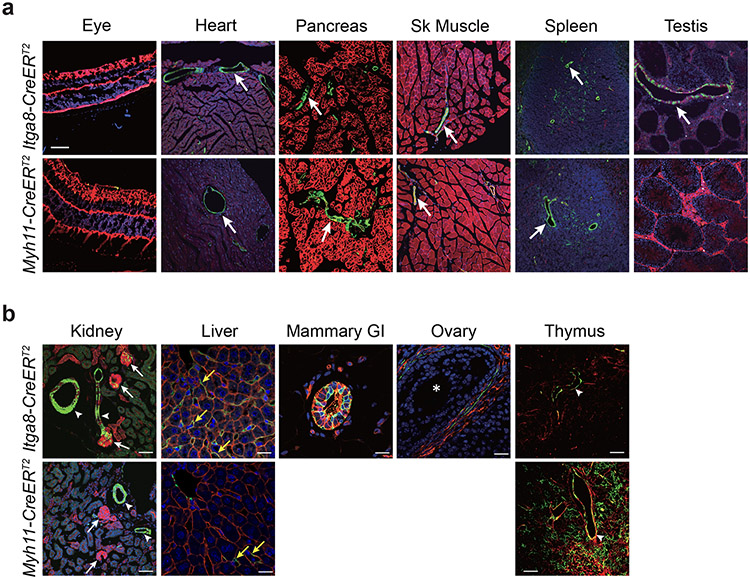
Confocal immunofluorescence microscopy was used to compare recombination activities across multiple tissues in age-matched Itga8-CreERT2 and Myh11-CreERT2 mice. Both strains demonstrated recombination activity in VSMCs of the aorta, mesentery, and microvasculature of the brain, as well as vessels in visceral organs such as intestine and bladder (Fig. 1a). Higher magnification, z-stacked images further confirmed recombination in VSMCs of the aorta and vena cava with no activity in endothelial or adventitial cells (Extended Data Fig. 4a,b). The activity of each CreERT2 driver was seen in VSMCs of the ascending aorta (mix of secondary heart field- and neural crest-derived VSMCs), aortic arch (neural crest-derived VSMCs), and thoracic/descending aorta (paraxial mesoderm-derived VSMCs) (Extended Data Fig. 4b). Confocal microscopy and immunogold electron microscopy demonstrated sporadic Itga8-CreERT2 activity in pericytes of the brain (Fig. 1b). Importantly, whereas Myh11-CreERT2 mice showed high recombination activity in visceral SMCs of bladder, colon, esophagus, intestine, stomach, and ureter, Itga8-CreERT2 mice displayed lower activity in these tissues, particularly intestine (Fig. 1a). Low Itga8-CreERT2 activity was also seen in the uterus (Fig. 1a); no comparison could be made in the uterus with Myh11-CreERT2 because this transgene integrated on the Y chromosome.32 Comparable activity of Itga8-CreERT2 was observed in both male and female mice, whether derived from male or female Itga8-CreERT2 parents, up to the age of 54 weeks (Extended Data Fig. 4c), consistent with equivalent levels of ITGA8 protein in young versus old mice (Extended Data Fig. 4d). Neither Itga8-CreERT2 nor Myh11-CreERT2 mice displayed obvious activity outside of VSMCs in tissues of the eye, heart, pancreas, skeletal muscle, spleen, and germ cells of the testis and ovary (Extended Data Fig. 5a,b). However, Myh11-CreERT2 mice exhibited pronounced activity in interstitial cells of the thymus, and Itga8-CreERT2 mice showed activity in glomerular cells of the kidney (Extended Data Fig. 5b). Itga8-CreERT2 activity was also detected in sinusoids of the liver, thecal cells surrounding mature follicles of the ovary, and in myoepithelial cells of the mammary gland (Extended Data Fig. 5b). Collectively, these results demonstrate some similarities, but important differences, in the recombination activities of Itga8-CreERT2 versus Myh11-CreERT2.
Fig. 1. Comparative recombination activity of Itga8-CreERT2 versus Myh11-CreERT2 in adult tissues.
a, Paired tissues from 10-week-old mice carrying indicated CreERT2 and mT/mG reporter. Green fluorescent protein (GFP), reflecting CreERT2-mediated excision of tdTomato cassette in the mT/mG reporter, is primarily confined to SMCs. DAPI indicates nuclear staining. White arrows indicate blood vessels; white arrowheads indicate visceral smooth muscle cells. Scale bar represents 100μm for aorta and uterus and 50μm for all other panels. b, Minimal Itga8-CreERT2 activity in pericytes of brain shown by mT/mG-derived GFP fluorescence (arrow in panel i), immunogold (arrows) electron microscopy (ii), and 2D maximum projection confocal microscopy (iii). An arteriole (white arrow) and capillary (yellow arrow) are highlighted in panel iii. Asterisk in panel ii denotes a red blood cell. Scale bars are 20μm, 2μm, and 50μm for panels i, ii, and iii, respectively. Data are representative of at least two independent male or female mice analyzed over the course of five years in two independent labs.
Tamoxifen-independent activity and CreERT2 protein expression
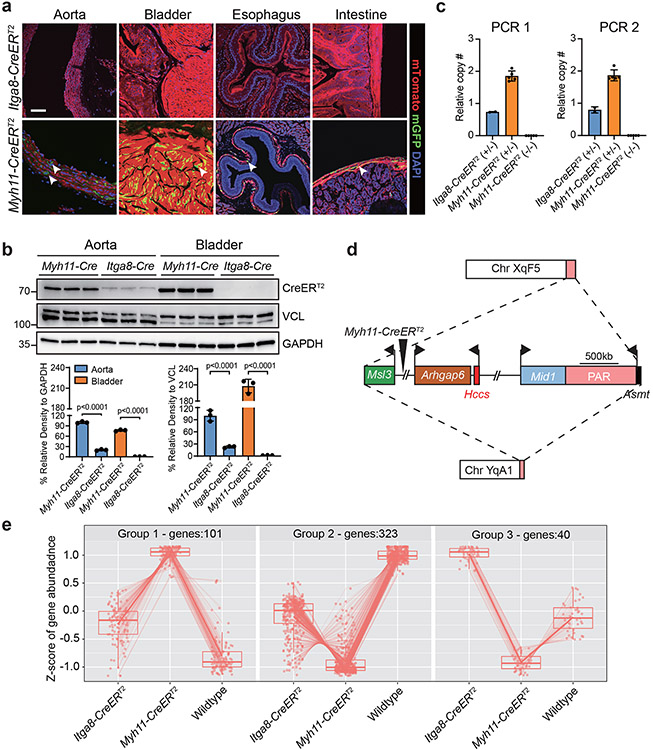
There is evidence for tamoxifen-independent (or leaky) recombination in mice with the CreERT2 cassette.34 Accordingly, individually-housed Itga8-CreERT2 or Myh11-CreERT2 male mice carrying the mT/mG reporter were analyzed for recombination in the absence of tamoxifen. Whereas no detectable recombination of the mT/mG reporter was noted in young Itga8-CreERT2 mice, clear evidence of tamoxifen-independent recombination was visible in similarly-aged tissues of Myh11-CreERT2 mice (Fig. 2a). There was no leaky activity in Itga8-CreERT2 mice up to 6 months of age suggesting high fidelity activity of this CreERT2 strain; however, low-level leaky recombination was seen in the aorta of 54-week-old Itga8-CreERT2 mice, although the extent of leakiness was much less than that of age-matched Myh11-CreERT2 mice (Extended Data Fig. 6). To begin probing the basis for early-onset leaky Myh11-CreERT2 activity, CreERT2 protein levels were analyzed in the aorta and bladder of age-matched young male mice. Western blotting showed five-fold greater CreERT2 protein in the aorta of Myh11-CreERT2 mice than Itga8-CreERT2 mice (Fig. 2b); higher levels of CreERT2 were seen in the bladder of Myh11-CreERT2 mice (Fig. 2b). In contrast, CreERT2 expression was undetectable in the bladder of Itga8-CreERT2 mice by Western analysis (Fig. 2b), although activity was evident with more sensitive confocal imaging (Fig. 1a). The level of Myh11 mRNA expression is much higher than Itga8 mRNA,35 suggesting promoter strength may underlie differences in CreERT2 protein levels between the two strains. To further explore potential explanations for the differences in CreERT2 protein levels, Sanger sequencing of both CreERT2 drivers was carried out and the results disclosed a ~20% difference in nucleotide sequence (Supplementary Fig. 3). Close inspection of nucleotide differences revealed Myh11-CreERT2 to be codon optimized for more efficient translation of the bacterial Cre protein in mammals (Supplementary Fig. 3).36 Moreover, quantitative PCR of genomic DNA from the Myh11-CreERT2 strain showed two copies of the transgene (Fig. 2c). The latter finding prompted efforts to physically map the Myh11-CreERT2 transgene using nanopore long read sequencing.37 Interestingly, X chromosome-specific sequence near the pseudoautosomal region flanked the Myh11-CreERT2 transgene, suggesting initial integration on the X chromosome (Fig. 2d).
Fig. 2. Distinguishing features of the Myh11-CreERT2 mouse.
a, Tamoxifen-independent recombination activity in 10-week-old male Myh11-CreERT2 tissues. Arrowheads point to SMCs with leaky activity. Images of aorta and bladder replicated in two additional animals from two independent labs; images of esophagus and intestine replicated in one independent experiment. b, Western blot of CreERT2 protein from 8-week-old male mice from each CreERT2 driver (n=3 mice per genotype). Units of measure for molecular weight markers here and in all subsequent blots is in kilodaltons. p values determined by one-way ANOVA with Tukey’s Student’s t-test. c, qPCR of Myh11-CreERT2 transgene from adult spleen (n=5 wild type and Myh11-CreERT2 mice; n=2 Itga8-CreERT2 mice). Two primer pairs (PCR 1 and PCR 2) used that amplify non-overlapping sequences in ERT2. d, Long read sequence mapping of approximate site-of-integration for Myh11-CreERT2 transgene (large triangle). Dashed lines represent initial translocation of the Myh11-CreERT2 transgene from X chromosome (top) carrying X-linked genes surrounding the Myh11-CreERT2 transgene (middle) to Y chromosome (bottom). Bent arrows indicate transcription start sites of each gene. Scale bar is an approximation. PAR, pseudoautosomal region. e, Bulk RNA-seq summary of differentially expressed genes in the absence of tamoxifen in 8-week-old male mice (n=3 mice per genotype). Group 1 represents significantly elevated genes in Myh11-CreERT2 aorta versus Itga8-CreERT2 and wild type; Group 2 represents significantly elevated genes in wild type aorta; Group 3 represents significantly elevated genes in Itga8-CreERT2 aorta. A list of differentially expressed genes is provided in Supplementary Table 2. Values for box whisker plots can be found in the source data file.
The higher expression of CreERT2 protein in Myh11-CreERT2 mice, and the unexpected presence of a segment of X chromosome sequence around the Y-integrated transgene,32 suggested there could be perturbations in the vascular transcriptome of this CreERT2 driver strain. To investigate this idea, bulk RNA-seq studies were conducted and the results showed an altered transcriptome in the aorta of Myh11-CreERT2 and, to a lesser extent, Itga8-CreERT2 mice (Fig. 2e). Specifically, 101 upregulated genes (Group 1) and 323 down-regulated genes (Group 2) of Myh11-CreERT2 deviated further from wild type than Itga8-CreERT2 (Fig. 2e). Interestingly, 40 genes (Group 3) upregulated in Itga8-CreERT2 aorta were down-regulated in Myh11-CreERT2 as compared to wild type aorta (Fig. 2e). Of note, there were no down-regulated genes unique to Itga8-CreERT2. Consistent with there being two copies of the Myh11-CreERT2 transgene (Fig. 2c), two X-linked genes (Arhgap6 and Hccs), found near the Myh11-CreERT2 insertion site (Fig. 2d), were expressed two-fold higher as compared to wild type and Itga8-CreERT2 aorta (Supplementary Table 2). Taken together, these results show several distinctive characteristics in the Myh11-CreERT2 driver strain including early, unscheduled recombination, high-level CreERT2 protein expression, and a more divergent baseline transcriptome likely driven, at least in part, by an unusual translocation event.
Similar recombination efficiencies between each CreERT2
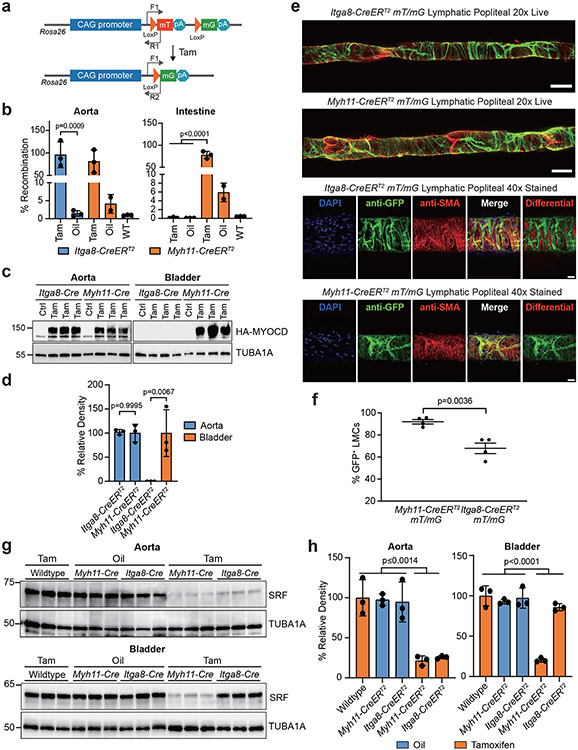
The differential expression of CreERT2 protein suggested there could be differences in recombination efficiency between the two CreERT2 driver strains. To address this possibility, Myh11-CreERT2 and Itga8-CreERT2 mice were crossed with several independently floxed mice to ascertain relative recombination rates. Quantitative PCR before and after tamoxifen administration showed comparable recombination of the mT/mG reporter in the aorta of both CreERT2 strains (Fig. 3a,b). As expected, there was high-level recombination in the intestine of Myh11-CreERT2 mice, but negligible amounts in Itga8-CreERT2 mice (Fig. 3a,b). Similar findings were observed in the bladder of a new tamoxifen-inducible Myocardin mouse model (Fig. 3c,d). Next, the activity of each CreERT2 strain was quantitated in popliteal lymphatic vessels. Results revealed ~90% and ~70% recombination efficiency in lymphatic vessels of the mT/mG reporter in Myh11-CreERT2 and Itga8-CreERT2 mice, respectively (Fig. 3e,f). Recombination efficiencies were somewhat lower in popliteal artery and vein, but the Myh11-CreERT2 activity appeared higher than Itga8-CreERT2 (Extended Data Fig. 7). Finally, each CreERT2 strain was crossed to a floxed Srf mouse33 treated with oil or tamoxifen. Tamoxifen alone had no effect on the expression of SRF in wild type aorta (Extended Data Fig. 8a,b). Normalized SRF protein was similarly knocked down in the aorta of each CreERT2 strain; however, only Myh11-CreERT2 displayed significant knockdown of SRF in the bladder (Fig. 3g,h). Overall, despite lower expression of CreERT2 protein in Itga8-CreERT2 mice, excision of floxed sequences approximated that seen in Myh11-CreERT2 mice.
Fig. 3. Recombination efficiency of Itga8-CreERT2 versus Myh11-CreERT2.
a, Schematic of mT/mG reporter at the Rosa26 locus and position of primers (arrows) for measuring recombination. b, qPCR of genomic DNA following Oil or Tamoxifen (Tam) administration in each strain of CreERT2 (n=3 mice per condition and genotype, save Oil-treated Myh11-CreERT2 condition with n=2). Note leaky activity in Oil-treated Myh11-CreERT2. c, Similar study design as in panel b only an inducible Myocd transgenic mouse was bred to each CreERT2 driver for Oil (Ctrl) or tamoxifen (Tam) administration and Western blotting for the presence of the HA-tagged MYOCD protein. d, Quantitative data of Western blots in panel c; (n=3 independent mice per genotype). e, Representative 2D maximum projections of popliteal lymphatic vessels isolated from each CreERT2 mouse (n=4 mice per genotype). Top panel, stitched maximum projections of live GFP and tdTomato fluorescence imaged with confocal microscopy at 20x. Note presence of GFP negative valves. Bottom panel, popliteal lymphatic vessels fixed and stained with DAPI, anti-GFP and anti-smooth muscle alpha actin (SMA) and imaged with confocal microscopy at 40x. The scale bars are 100μm for stitched images and 25μm for immunostained images. f, Quantitation of GFP+ fluorescence in lymphatic muscle cells (LMCs; n=4 mice per genotype). g, Western blot of SRF protein in aorta and bladders of each indicated genotype with Oil or Tam administration (n=3 mice per genotype). h, Quantitative data of panel g; percent relative density to the TUBA1A loading control (n=3 independent mice per condition). Error bars represented by mean ± standard deviation. p values determined by one-way ANOVA and Tukey’s t-test for panels b and h and unpaired two-tailed Student’s t-test for panels d and f.
Itga8-CreERT2-mediated Srf knockout evades lethal visceral myopathy
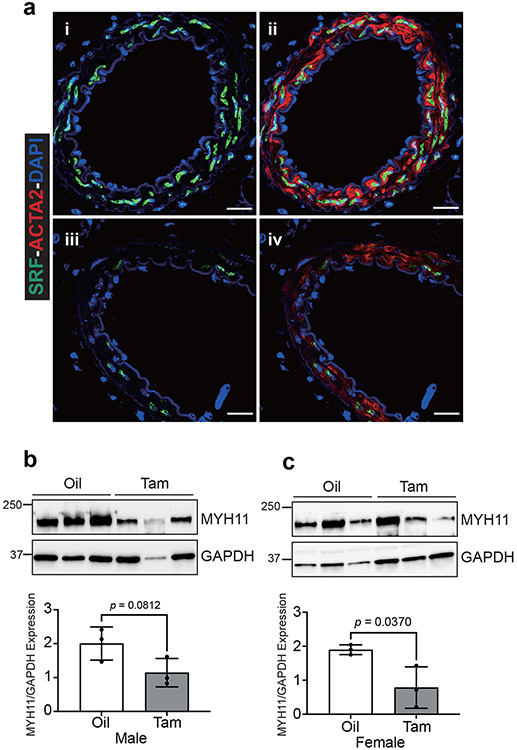
Srf loss-of-function studies using a Sm22-CreERT2 driver showed a fatal gastrointestinal phenotype.10, 11 Consistent with these findings, tamoxifen-treated homozygous floxed Srf mice carrying Myh11-CreERT2 (SrfMyh11) displayed lethality beginning three weeks following the first tamoxifen injection. Mutant mice showed a pale, distended intestine and a pronounced reduction in SRF expression within a thinned muscular layer of the small intestine (Extended Data Fig. 8c,d). In contrast, inducible knockout of Srf with Itga8-CreERT2 (SrfItga8) resulted in viable mice with no intestinal phenotype up to eight weeks post-tamoxifen administration (Extended Data Fig. 8c). Confocal immunofluorescence microscopy demonstrated a clear reduction of SRF and ACTA2 in the carotid artery of SrfItga8 mice, but not in bladder or intestinal SMCs (Extended Data Fig. 9a). Quantitative measures disclosed a ~75% decrease in SRF positive VSMCs in the carotid artery with little change in the intestine (Extended Data Fig. 9b,c). Western blot studies indicated a similar diminution of SRF in homozygous floxed (HoF) SrfItga8 knockout aorta, but no significant change in the bladder (Extended Data Fig. 9d-g). There was little change in SRF expression in the aorta of heterozygous floxed (HeF) SrfItga8 mice, suggesting compensatory expression from the remaining wild type allele (Extended Data Fig. 9d,f).
Because Itga8-CreERT2 activity was seen in the glomerulus (Extended Data Fig. 5b), conditional loss of Srf with this CreERT2 driver could have triggered a kidney phenotype. We therefore analyzed homozygous and heterozygous SrfItga8 mice for pathological changes in the kidney. Results revealed similar body weights, a normal histological appearance, and no indication of proteinuria in SrfItga8 mice (Supplementary Fig. 4). Taken together, these results support Itga8-CreERT2 as a suitable alternative to Myh11-CreERT2 in focused studies of Srf loss-of-function in VSMCs, without the confounding effects of an otherwise lethal visceral myopathy.
Vascular dysfunction in Itga8-CreERT2-mediated Srf knockout mice
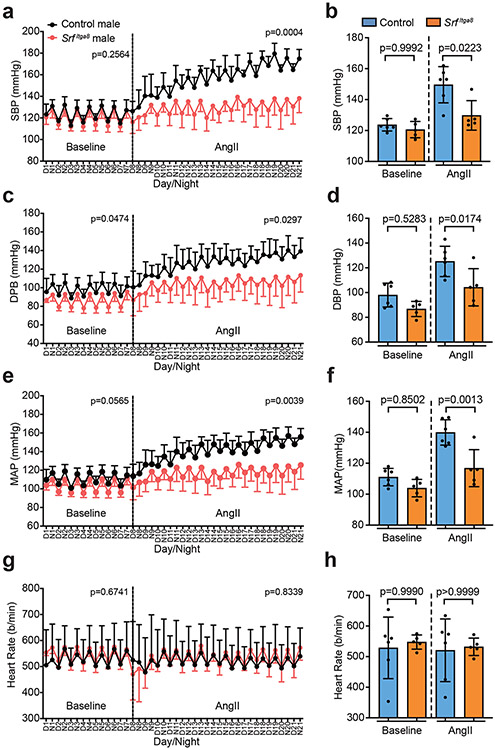
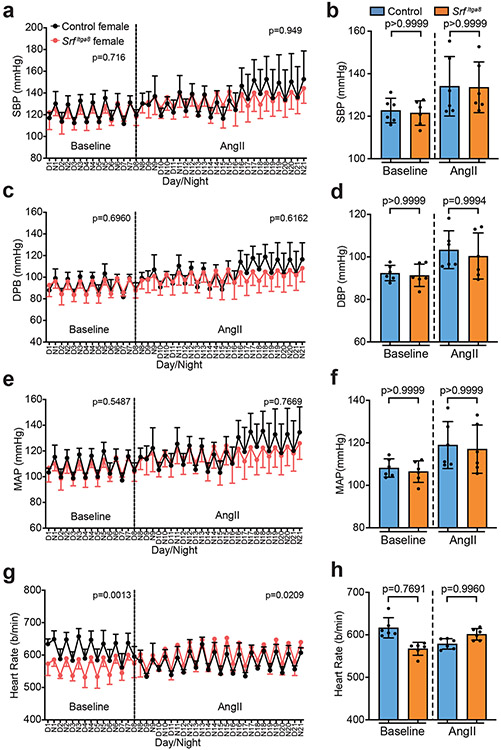
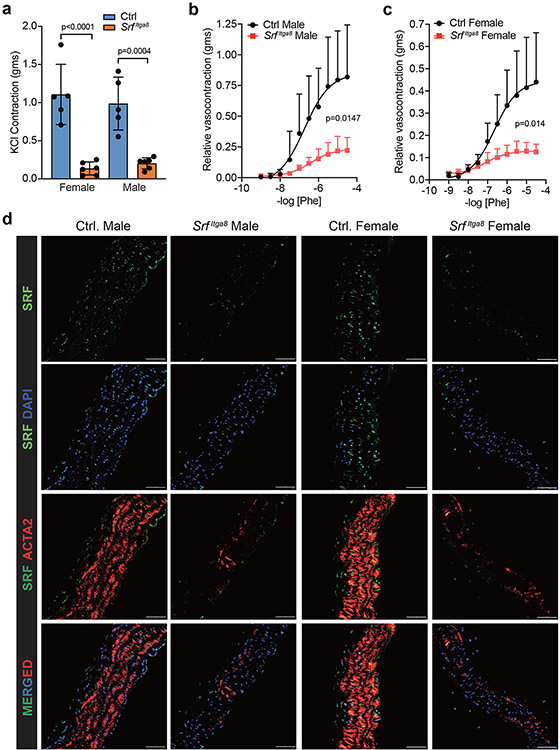
To formally establish an unambiguous phenotype associated with Srf gene loss in VSMCs, blood pressure and vascular contractile competence were assessed in male and female mice following Itga8-CreERT2-mediated knockout of Srf (SrfItga8). Telemetry measures in male mice demonstrated no change in baseline blood pressure. However, following angiotensin II infusion, significant increases in systolic, diastolic, and mean arterial pressures were observed in control mice, but not in SrfItga8 mice (Fig. 4). Consistent with a previous report,38 age-matched female mice failed to respond to angiotensin II and showed no changes in blood pressure, regardless of Srf status (Fig. 5). On the other hand, the aorta of male and female mice displayed attenuated contraction in response to KCl (Fig. 6a) and phenylephrine (Fig. 6b,c). Immunofluorescence confocal microscopy of the aorta from SrfItga8 mice indicated attenuated expression of SRF and ACTA2 in VSMCs (Fig. 6d). Similarly reduced SRF and contractile protein expression was seen in mesenteric arteries (Extended Data Fig. 10). Finally, bulk RNA-seq studies established a decrease in VSMC contractile genes in the aorta of SrfItga8 mice (Fig. 7a,b). Analysis of the top 250 down-regulated genes disclosed the SRF binding CArG box as the most significantly enriched cis regulatory element (Fig. 7c). Further assessment of the down-regulated gene set showed molecular functions related to the cytoskeleton and contractile machinery (Fig. 7d). Interestingly, the upregulated gene set in SrfItga8 aorta was most related to metabolism (Fig. 7e). Collectively, these results demonstrate an unambiguous viable mouse model of VSMC contractile incompetence following Itga8-CreERT2-mediated inactivation of Srf.
Fig. 4. Chronic blood pressure measurements in Itga8-CreERT2 mediated Srf knockout male mice.
Effects of Srf knockout (SrfItga8) on systolic blood pressure (a, b), diastolic blood pressure (c, d), mean arterial pressure (e, f) and heart rate (g, h) in male mice as measured by radio-telemetry over 3 weeks. Control mice here and in Figures 5 and 6 are oil-treated, homozygous floxed Srf mice carrying Itga8-CreERT2. n=6 oil control and n=5 SrfItga8 mice. Error bars represented by mean ± standard deviation. Each biological replicate represents the mean over days 1-7 (baseline) or days 8-21 (AngII). p values determined by two-way ANOVA for temporal study (a,c,e,g) and one-way ANOVA and Tukey’s t-test for baseline and AngII (b,d,f,h).
Fig. 5. Chronic blood pressure measurements in Itga8-CreERT2 mediated Srf knockout female mice.
Effects of SrfItga8 on systolic blood pressure (a, b), diastolic blood pressure (c, d), mean arterial pressure (e, f) and heart rate (g, h) in female mice as measured by telemetry over 3 weeks. n=6 mice per condition. Error bars represented by mean ± standard deviation. Each biological replicate represents the mean over days 1-7 (baseline) or days 8-21 (AngII). p values determined by two-way ANOVA for temporal study (a,c,e,g) and one-way ANOVA and Tukey’s t-test for baseline and AngII (b,d,f,h).
Fig. 6. Vascular contractile activity in Itga8-CreERT2 mediated Srf knockout mice.
a, KCl-stimulated contractile activity in female (n=5) and male (n=6) aorta under control or SrfItga8 condition. The same groups of Male (b) and female (c) mouse aortas were treated with varying doses of Phenylephrine [Phe]. d, Confocal immunofluorescence microscopy of indicated proteins in control (Oil) and SrfItga8 male and female aortic segments from the same mice used in KCl and Phe treatments. Images are representative of two independent mice. The scale bars are 50μm. Error bars represented by mean ± standard deviation. p values determined by one-way ANOVA and Tukey’s t-test (a) or two-way ANOVA (b,c).
Fig. 7. Bulk RNA-seq of aortas from Itga8-CreERT2 mediated Srf knockout mice.
a, Down-regulated expression of SMC genes in aorta of heterozygous (n=2) versus homozygous (n=3) SrfItga8 knockout mice. b, Volcano plot of differentially expressed genes in SrfItga8 knockout aorta. c, oPOSSUM 3.0 analysis of over-represented transcription factor binding sites in promoter/intron of top 250 down-regulated genes in SrfItga8 knockout aorta. Enrichment scores for gene ontology terms (Molecular Function) related to downregulated (d) or upregulated (e) genes in SrfItga8 knockout aorta.
Discussion
All current SMC Cre driver mice recombine floxed DNA similarly in both vascular and visceral SMC lineages. Consequently, the inactivation of a critical gene in SMCs of the intestine, for example, may result in a lethal phenotype and the inability to accurately define gene function in VSMCs. Here, preferential activity of Itga8-CreERT2 was demonstrated in VSMCs with relatively less activity in visceral SMCs of the gastrointestinal and urinary tracts. Such restricted CreERT2 activity resulted in viable mice upon inactivation of Srf with no gross or histological evidence of pathology in the intestine. These findings are consistent with mT/mG reporter data and quantitative experiments showing similar levels of SRF protein in intestinal SMCs of wild type and SrfItga8 mice. In sharp contrast, Srf loss-of-function with the Myh11-CreERT2 driver provoked a lethal intestinal phenotype; a similar finding was reported previously with suppression of SRF-dependent pro-survival microRNAs and resultant intestinal SMC apoptosis.39 The latter phenotypes and the intestinal pseudo-obstruction seen with conditional knockout of Srf using an inducible Sm22-Cre driver10, 11 have hampered efforts to study Srf loss-of-function in VSMCs, particularly in models that require extended time for disease to be evident. Importantly, despite much lower levels of CreERT2 protein as compared to the Myh11-CreERT2 driver, Itga8-CreERT2 efficiently recombined the floxed Srf locus revealing an unequivocal VSMC phenotype of contractile incompetence. Based on the results of this study, the Itga8-CreERT2 mouse would be expected to circumvent other lethal visceral myopathies following gene loss with existing SMC Cre drivers.
Although short-term (14-15 days) studies of conditional loss of Srf in VSMCs with Sm22-CreERT2 have been reported, yielding insight into mechanosensitivity and blood flow40 as well as arterial stiffness,41 phenotyping occurred at a time when dilation of the intestine, defective peristalsis, and cachexia begin to be manifest.10, 11 The latter changes likely elicited systemic inflammation that could impact vascular function. As shown here, angiotensin II-induced increases in systemic blood pressure were abolished in male mice lacking normal levels of SRF indicating the phenotype was solely due to an intrinsic defect in VSMCs with no contribution from distal pathological events. Thus, future Srf loss-of-function studies can now be conducted in VSMCs without the confounding effects of early onset gastrointestinal defects or other visceral pathologies. Further, we anticipate that the Itga8-CreERT2 mouse will find utility as more floxed alleles emerge for studies in acute vascular disease models of intimal hyperplasia and chronic models such as arteriovenous fistula failure, atherosclerosis, hypertension, transplant arteriopathy, and arterial calcification.
A limitation of some Cre driver strains is the recombination of a floxed allele in the absence of tamoxifen.34 Here, the Itga8-CreERT2 mouse showed high fidelity recombination up to six months of age. In contrast, tamoxifen-independent Myh11-CreERT2 activity was evident in multiple tissues, including aorta, as early as 10 weeks of age. Such unscheduled recombination was more apparent in aged mice. A similar finding of tamoxifen-independent Myh11-CreERT2 activity was reported in SMCs of the stomach.42 These findings should alert investigators as to the best controls for inducible knockout studies. For example, in the absence of haploinsufficiency and no leaky CreERT2 activity, a stringent control would be tamoxifen-administered mice carrying CreERT2 and a heterozygous floxed allele. If, however, haploinsufficiency is suspected and/or the CreERT2 driver is leaky, then a less confounding control would be tamoxifen-treated mice carrying CreERT2 without loxP sites.43 Of note, there has been a suggestion that “pseudo loxP sites” in the genome could be subject to Cre-mediated recombination with unknown consequences.44 Clearly, careful analysis of a CreERT2 driver and the expression profile of floxed alleles is essential when contemplating a conditional knockout experiment.45
What is the basis for the precocious leaky activity seen in the Myh11-CreERT2 driver? One possibility could be the comparatively greater amounts of CreERT2 protein observed in this strain. Indeed, in vitro studies have correlated tamoxifen-independent activity with higher levels of CreERT2 protein.46 An overabundance of CreERT2 protein may favor dissociation from the HSP90 chaperone, which anchors CreERT2 in the cytoplasm thereby preventing CreERT2 entry into the nucleus in the absence of ligand.47 The finding of two copies of the Myh11-CreERT2 transgene and, perhaps, stronger Myh11 promoter activity over Itga8, likely contribute to the high-level CreERT2 protein observed in Myh11-CreERT2 mice. Theoretically, only four molecules of Cre are needed per cell to catalyze recombination between two loxP sites.48 Thus, the weak expression of CreERT2 demonstrated here in the Itga8-CreERT2 mouse has distinct advantages over the Myh11-CreERT2 strain by effecting comparable recombination efficiencies while displaying low-level leakiness observed only in mice greater than six months of age.
Another advantage of the Itga8-CreERT2 mouse is the ability to perform sex studies which enables compliance with the National Institutes of Health directive of addressing sex as a biological variable in biomedical research.49 The present Myh11-CreERT2 mouse carries the CreERT2 allele on the Y chromosome, precluding studies in females.9 A previous study reported translocation of Myh11-CreERT2 to the X chromosome and the generation of female mice that displayed some Myh11-CreERT2 activity.50 It should be pointed out, however, that the site of integration on the X chromosome was not mapped, and high doses of tamoxifen were needed to elevate recombination efficiency in hemizygous females.50 Here, using nanopore long-read sequencing,37 the Myh11-CreERT2 transgene was physically mapped in a male mouse. Results suggest that the transgene initially integrated at the terminal end of the X chromosome near the pseudoautosomal region where high rates of recombination occur during meiosis.51 The Myh11-CreERT2 transgene then translocated to the Y chromosome carrying at least two X-linked genes, which are more abundantly expressed in Myh11-CreERT2 mice than wild type or Itga8-CreERT2 mice. The precise insertion site on this “neo Y chromosome” remains unclear, but it must be within an interval between the pseudoautosomal region and the pseudoautosomal boundary (where identical X and Y sequences diverge) to explain the ~3% of Myh11-CreERT2 t(Y;X) mice observed in this lab over the last five years. This peculiar site of integration suggests several substrains of the original Myh11-CreERT2 32 driver may exist due to stochastic recombination and probable variation in breakpoints. Future engineering of Cre (or other recombinase) mice should use a more targeted approach for single copy integration, either within a gene of interest or a safe harbor locus on an autosome. Given early unscheduled recombination with Myh11-CreERT2, the unusual translocation events associated with its site of integration, and the high level activity in visceral SMC tissues, there is an opportunity to revisit previous conclusions drawn from the >100 alleles targeted in SMCs with this CreERT2 driver (Supplementary Table 1).
Despite the Itga8-CreERT2 mouse enabling sex studies in a more VSMC-restrictive manner without leaky activity up to at least six months of age, some limitations need comment. First, as loss of both functional Itga8 alleles is lethal,30 breeding the Itga8-CreERT2 mouse to homozygosity is pointless. Indeed, breeding any unmapped transgenic line to homozygosity is ill-advised since randomly integrated transgenes may disrupt critical coding or noncoding genes or essential regulatory elements.52 Second, Itga8 appears to be down-regulated under conditions of SMC de-differentiation,27 so the Itga8-CreERT2 driver may not be optimal for studies requiring inducible activity following disease onset, although there is evidence for the up-regulation of ITGA8 protein late in intimal disease.53 Further work in various vascular disease models will resolve this issue. Third, the activity profile of Itga8-CreERT2 during embryogenesis was not assessed, so it is unclear whether this CreERT2 driver will be suitable for developmental studies. Fourth, low Itga8-CreERT2 activity in uterine SMCs suggests this CreERT2 driver may not be useful for studying effects of conditional SMC gene loss in the female reproductive tract. Fifth, whether Itga8-CreERT2 is active in other SMC types such as those in arrector pili of skin and ciliary body of the eye remains to be determined. Finally, as with all current SMC Cre drivers, Itga8-CreERT2 exhibits activity in non-SMC types, most notably glomerular cells of the kidney. Thus, when using the Itga8-CreERT2 mouse for conditional knockouts, it may be necessary to assess kidney function through histological and biochemical assays.54 In this report, SrfItga8 mice exhibited normal glomerular and tubular structures with no evidence of proteinuria. Whether a kidney phenotype is manifest under stress conditions awaits further study. Further, since there is some level of Itga8-CreERT2 activity in visceral SMCs (Fig. 1a), it will be important to determine whether gene-specific phenotypes exist in these tissues under certain experimental conditions.
In addition to glomerular cells, Itga8-CreERT2 activity was observed in sinusoidal cells of the liver where endothelial cells and fibroblast-like stellate cells reside; only occasional liver sinusoidal activity was seen with Myh11-CreERT2. Given a previous report of ITGA8 in stellate cells,55 the lack of Itga8-CreERT2 activity in vascular endothelial cells as reported here, and the virtual absence of Itga8 mRNA expression in various endothelial cell populations,35 the activity of Itga8-CreERT2 in liver sinusoids likely occurs in stellate cells. Of note, no obvious Itga8-CreERT2 activity was observed in cardiac fibroblasts or adventitial fibroblasts of blood vessels. Whether activity emerges in these and other fibroblast populations under stress conditions is unknown at this time. While no Itga8-CreERT2 activity was detected in resting endothelial cells, activity may arise under conditions of endothelial-mesenchymal transition or in unique endothelial cell beds as reported for Myh11-CreERT2.56 Itga8-CreERT2 activity was also demonstrated in myoepithelial cells of the mammary gland and thecal cells encapsulating mature follicles of the ovary. We do not anticipate gene loss in the aforementioned non-SMC types to impact a VSMC phenotype. Nevertheless, investigators should be mindful of the full expression profile of a floxed gene to probe potential competing phenotypes outside the vascular or lymphatic vessel wall when using the Itga8-CreERT2 mouse. Continued characterization of Itga8-CreERT2 under various experimental conditions will provide further insight into its activity profile.
An ongoing question in vascular biology is whether SMC Cre drivers are active in pericytes, an SMC-like cell that is essential for maintaining capillary integrity and function.57 A previous report found Myh11-CreERT2 activity in pericytes of the retina, cornea, and hindlimb skeletal muscle.58 In contrast, only an occasional pericyte of the brain showed evidence of Itga8-CreERT2 activity, a finding consistent with an analysis of Itga8 mRNA expression in single-cell RNA sequencing of the mouse brain.35 Thus, interrogating Srf loss-of-function phenotypes in pericytes using the Itga8-CreERT2 driver may be uninformative. Recently, however, an elegant study using Pdgfrb-CreERT2 to disrupt Srf in pericytes found defective migration of these cells during sprouting angiogenesis in the newborn retina resulting in faulty vascular remodeling, aberrant blood flow patterns, and the formation of arteriovenous shunts.59 Table 1 summarizes notable distinctions between Itga8-CreERT2 and Myh11-CreERT2 mice.
Table 1.
Contrasting characteristics of two SMC Cre drivers
| Itga8-CreERT2 | Myh11-CreERT2 |
|---|---|
| Single-copy knock-in on Chr 2 | Two-copy transgene on Chr Y |
| Chromosome stability | Exhibits t(Y;X) in ~ 3% of pups |
| Male and female studies | Male studies only |
| Lightly codon-optimized Cre | Heavily codon-optimized Cre |
| Low-level CreERT2 expression | High-level CreERT2 expression |
| More stable transcriptome | Altered transcriptome |
| Low-level leaky Cre in old mice | High-level leaky Cre in mice |
| Active preferentially in VSMCs | Equally active in all SMC lineages |
| Evades visceral myopathies | Causes many visceral myopathies |
| Active in mesangial, stellate, thecal, and myoepithelial cells of kidney, liver, ovary, and mammary gland, respectively | Active in corneal endothelial cells,56 interstitial cells of the thymus, and in some stellate cells of liver |
| Low-level pericyte activity | Active in some pericytes58 |
Little is known about the transcriptional/post-transcriptional control of Itga8. In vitro luciferase and transgenic mouse assays failed to show the importance of a conserved SRF-binding site located four kilobases upstream of the start site of Itga8 transcription.29 However, the Myocardin (MYOCD) coactivator stimulated Itga8 expression in an SRF-independent manner.29 The latter finding distinguishes Itga8 from most other SMC markers that have functional SRF-binding sites in the promoter or first intron.60 Elucidating the transcriptional control of Itga8 and other VSMC-restricted genes will inform the molecular basis for preferential expression in VSMCs and the further development or refinement of Cre driver mice for VSMC conditional knockout studies.
In summary, the Itga8-CreERT2 mouse displays preferential activity in VSMCs, an attribute that has yet to be shown in any existing SMC-restricted Cre driver. As such, the Itga8-CreERT2 mouse represents a solution to the problem of competing, sometimes lethal, phenotypes arising from the inactivation of critical genes in visceral SMCs such as the intestine.
Methods
Mouse models
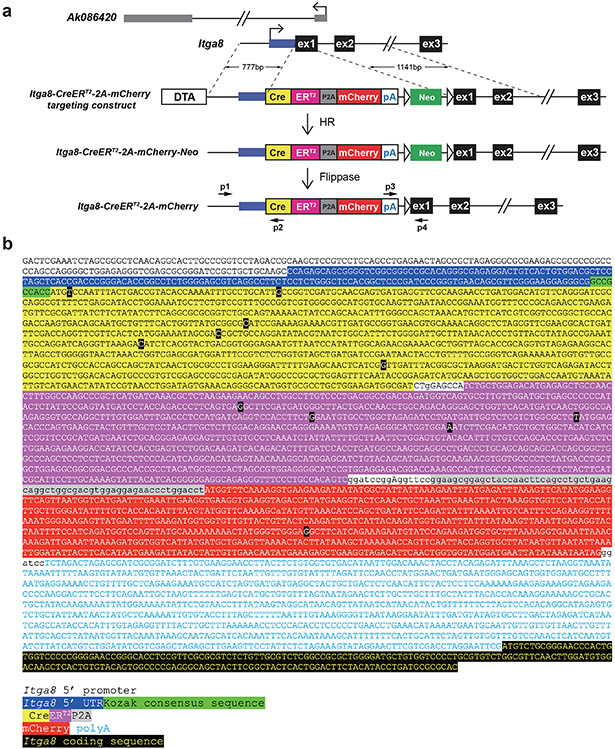
All mouse studies were approved by local Institutional Animal Care and Use Committees at the University of Rochester during the period of 2016-2019 (approval #101587); Medical College of Georgia from 2019-present (approval number #2019-1000); and the University of Missouri (approval #27320). An Institutional Biosafety Committee at Augusta University approved the use of Tamoxifen for inducible activation of relevant Cre driver mice (approval #1942). The ethical treatment of mice was in accordance with ARRIVE guidelines. All mice were maintained in micro-isolator cages containing water and pelleted food (Teklad Global Irradiated 18% Protein Rodent Diet 2918) ad libitum. Rooms were temperature and humidity controlled under a 12-hour light (600-1800 hours), 12-hour dark (1800-600 hours) cycle. The Myh11-CreERT2 mouse (stock 019079) and membrane tdTomato/membrane GFP (mT/mG) reporter (stock 007676) were obtained from the Jackson laboratory. The Sm22-Cre and floxed Srf strains have been described previously.33 To generate the Itga8-CreERT2 allele, a DNA fragment containing CreERT2-P2A-mCherry coding sequence and SV40 polyadenylation signal (CreERT2-P2A-mCherry-pA) was synthesized by GENEWIZ. A 0.7-kb 5’-homology arm comprising promoter and untranslated exon 1 of Itga8 and a 1.1-kb 3’-homology arm containing the coding sequence of exons one and two, intron one, and a portion of intron two of Itga8, were PCR-amplified from C57BL/6J genomic DNA (primers listed in Supplementary Table 4). The homology arms were cloned, along with an FRT flanked pGK-Neo, pGK-DTA, and CreERT2-P2A-mCherry-pA cassette, into pBluescript SKII (+) to generate the Itga8-CreERT2 knock-in construct (Extended Data Fig. 1). The targeting construct was linearized with AscI and electroporated into C57BL/6J-129S6 hybrid G4 embryonic stem cells (ESCs).61 The targeted Itga8-CreERT2 ESC clones were confirmed by long and accurate PCR genotyping using external primers to identify the 1.0 kb 5’ and 1.3 kb 3’ targeted DNA fragments (primers listed in Supplementary Table 4). Validated ESCs were microinjected into C57BL/6J blastocysts to generate mouse chimeras. By crossing the chimeras with FLPeR mice in C57BL/6J background (Jackson Laboratory, 009086), the Neo cassette was removed to produce the Itga8-CreERT2 mouse line. Sanger sequencing both strands verified the final knockin allele. Itga8-CreERT2 activity was assessed in early (N2-N3) and later (>N10) generation mice of age 8-12 weeks (unless specified otherwise) and were maintained strictly on a C57BL/6J background with continuous refreshing of the breeder colony every fifth generation to mitigate effects of genetic drift and inadvertent generation of a substrain. Older Itga8-CreERT2 (24 and 54 weeks) and Myh11-CreERT2 (54 weeks) mice, each carrying the mTmG reporter, were used to evaluate tamoxifen-independent activity during aging. The inducible Myocd mouse (iMyocd) was generated in the same ESC line as above with an HA-tagged Myocd cDNA cloned downstream of a CAG-driven stop-floxed cassette in the Rosa26 locus. A list of mouse models used in this report is provided in Supplementary Table 3.
Tamoxifen administration
Cre mouse lines were crossed with mT/mG fluorescent reporter mice, a floxed Srf mouse, or an inducible Myocd mouse. Inducible Cre activity was achieved by intraperitoneal injection of tamoxifen (Sigma-Aldrich, T5648) mixed and sonicated in 100% ethanol/sunflower seed oil (Sigma-Aldrich, S5007) at 1:9, v/v. Tamoxifen (40 mg/kg) was delivered daily over five days followed by a washout of at least 10 days before experimental measures.
Confocal microscopy
For frozen samples, fresh tissues were rapidly isolated and fixed overnight in 4% paraformaldehyde. Tissues were then cryoprotected over a second night with 30% sucrose followed by embedding in Optimal Cutting Temperature (OCT) compound (VWR, 102094-106). 5μm-7μm sections were cut with a cryostat (Leica Biosystems, CM 1950) and kept frozen until imaging. Slides were removed from −20°C and warmed to room temperature. OCT compound was removed by washing three times with 1x PBS. Auto-fluorescence was quenched with Vector® TrueVIEW® Autofluorescence Quenching Kit (Vector Laboratories, SP-8500-15) and the slides were cover-slipped with ProLong™ Gold Antifade Mountant (Thermo, P36930) containing DAPI stain and air-dried. Imaging was performed by a single person (OJS) who was blinded to the genotype for all images shown (save Fig. 3e and Extended Data Fig. 7) on either an Olympus IX81 or Zeiss LSM900 confocal microscope with 20x, 40x oil (z-stacked), and 60x oil (z-stacked) objectives and analyzed with Micro-manager 2.0 or ZEN 3.5 Blue software, respectively. Laser settings for confocal imaging were kept constant throughout. Final images were cropped to their published size in Adobe® Photoshop® with no digital enhancements, save the Testis panel corresponding to Itga8-CreERT2 in Extended Data Fig. 5a and the Itga8-CreERT2 data of Extended Data Fig. 6; the brightness was increased uniformly in each of these panels using Photoshop® to provide the reader with a better representation of GFP staining.
For paraffin sections, tissues were isolated and immersion-fixed in methanol-H2O-acetic acid (60/30/10, v/v) overnight, processed in a benchtop tissue processor (Leica Biosystems, TP1020), and embedded (Sakura, Tissue Tek TEC 5) in paraffin blocks. Sections were cut with a microtome (Microm, HM 355 S) at 5μm and baked overnight at 58°C. Slides were then deparaffinized in a Leica® Autostainer XL (Leica Biosystems, ST5010), stained with antibodies (Supplementary Table 5), quenched with Vector® TrueVIEW® Autofluorescence Quenching Kit (Vector Laboratories, SP-8500-15), and cover-slipped with ProLong™ Gold Antifade Mountant. Imaging was done on an Olympus IX81 or Zeiss LSM900 confocal microscope with 20x, 40x oil (z-stacked), and 60x oil (z-stacked) objectives and analyzed with Micro-manager 2.0 or ZEN 3.5 Blue software, respectively. Final paraffin sectioned images were processed in Photoshop® without any enhancements.
Assessment of Itga8-CreERT2/mTmG recombination in cerebral pericytes
Itga8-CreERT2/mTmG mice were induced with tamoxifen and anesthetized as described above. Once an anesthetic plane was reached, 200 μl of wheat germ agglutinin-AF647 (AlexaFluor-647; WGA-647 W32466 ®Fisher), suspended in sterile saline at 1mg/ml, was injected into the retroorbital sinus for five minutes for WGA-647 to distribute throughout the vasculature and bind to the endothelial glycocalyx. After five minutes, the mouse was euthanized by cervical dislocation and the brain removed from the skull. Sagittal slices of the brain were made with a razor blade and placed in Krebs buffer with 0.5% BSA. A piece of Sylgard 184® was used to weigh the brain slice against the coverslip bottom of a WPI FluoroDish Cell Culture Dish to prevent movement while imaging. We scanned the tissue for GFP+ cells that displayed the canonical pericyte morphology and acquired image stacks for tdTomato, GFP and WGA-AF647 with a Z-step of 0.33μm using a Leica 25x water objective (HC FLUOTAR L 25x/0,95 W VISIR) and an Andor Dragonfly scanning confocal head with a LEICA DMi8 inverted microscope. The color intensity and balance were uniformly adjusted to minimize brain autofluorescence and to highlight the Itga8-CreERT2 mediated GFP expression. Maximum projections were made using Fiji.62
Immunogold electron microscopy
Mice were perfusion fixed with 4% paraformaldehyde/2% glutaraldehyde in 0.1M cacodylate buffer (pH, 7.4), dissected and immersion fixed in the same fixative, dehydrated through a graded series of ethanol, and embedded in LR White Resin (Electron Microscopy Sciences [EMS], Ft. Washington, PA). Following facing and trimming of block, 75 nm sections were collected on Pioloform-coated nickel slot grids (Ted Pella, Redding, CA), floated on drops of etching solution (5% NaIO4 in PBS) for five minutes, washed in PBS, and then quenched for 30 minutes in 1M NH4Cl. Grids were blocked in Aurion Blocking Solution (EMS) for two to four hours at room temperature, floated on drops of rabbit anti-GFP (Novus Biologicals, Centennial, CO) at a dilution of 1:1000 overnight at 4°C and then washed in PBS. Grids were then floated on drops of anti-rabbit Nanogold (Nanoprobes, Yaphank, NY) reagent diluted 1:1000 in Aurion BSA-c buffer for three hours at room temperature. Nanogold particles were silver enhanced for 15 minutes using HQ Silver (Nanoprobes, Yaphank, NY), washed in ice-cold deionized water to halt enhancement, and stained with 2% uranyl acetate and lead citrate. Images were obtained with a JEOL 1400 Flash transmission electron microscope (JEOL USA Inc., Peabody, MA) at 120 kV, equipped with a One View CCD camera (Gatan Inc., Pleasanton, CA).
Recombination efficiency
Floxed mouse models carrying either Itga8-CreERT2 or Myh11-CreERT2 were treated with oil or tamoxifen as above. Following a standard 10-day washout period, tissues (aorta and intestine) were isolated for quantitative PCR of genomic DNA (mT/mG reporter) and Western blotting for the inducible Myocardin and floxed Srf mice, respectively. For popliteal vascular recombination efficiencies, mice were anesthetized with an injection of ketamine/xylazine (25/2.5 mg/ml, i.p.) and their legs shaved and cleaned. The popliteal artery and vein and the popliteal lymphatic vessel were excised from the tissue and kept in Krebs buffer with 0.5% BSA at 4°C until imaged. Each vessel was then cannulated and pressurized in an imaging chamber filled with Krebs buffer for further removal of adipose and connective tissue. Vessels were then equilibrated in calcium-free solution for 15 minutes to prevent movement during live imaging. At least two separate fields of view of each vessel were acquired with a Yokogawa CSU-X Spinning Disc Confocal Microscope on an inverted Olympus IX81 with a Hamamatsu Flash4 camera using both 20x and 40x objectives. Each live vessel acquisition was a Z-stack from the bottom edge of the vessel to the approximate midpoint for both the membrane GFP (488nm) and membrane tdTomato wavelengths (561nm). In Extended Data Fig. 7, maximum image projections from live popliteal artery and vein vessels were created from a confocal microscopy image stack using Fiji and the Z-Project Function. The resulting maximum projection was cropped to its final size, and intensity for each channel was adjusted across both image sets using Fiji to highlight GFP fluorescence due to ITGA8-CreERT2 activity. For the representative vessel stitching, the Fiji plugin for pairwise stitching was used to stitch three fields of view, acquired at 20x, together.63
Following the live image acquisition, the lymphatic popliteal vessels were fixed at room temperature with ice-cold 4% PFA for 15 minutes. The vessels were then washed with PBS and stored at 4°C for subsequent immunostaining. The fixed vessels were permeabilized with PBST (0.3% triton) for one hour and then blocked for three hours with BlockAid (Thermofisher). Vessels were stained overnight with anti-GFP at 1:200 (rabbit, ThermoFisher A-11122) and anti-smooth muscle actin (ACTA2) at 1:500 (mouse, Sigma A5228) in Blockaid buffer. The vessels were then washed with PBS and stained with Donkey anti-rabbit AF488 (A-21206, Thermofisher) and Donkey anti-mouse AF647 at 1:200 (A-31571, Thermofisher) for one hour at 4°C. Vessels were washed in PBS and then recannulated and imaged as described above.
To assess recombination efficiency in the smooth muscle layer of the popliteal vein and artery, the number of circumferential VSMCs expressing either GFP+ or tdTomato+ were tabulated from Z-stacks from 40x acquisitions. To assess recombination efficiency in the smooth muscle layer of popliteal lymphatic vessels the number of GFP+/ACTA2+ and GFP−/ACTA2+ lymphatic muscle cells were counted and tabulated from Z-stacks acquired at 40x. To create the differential images used as a visual aid, the RFP Z-stack and GFP Z-stack were converted to 32-bit images, the RFP Z-stack was divided by the GFP Z-stack in FIJI and a maximum projection was made of the resulting stack. The output image was merged with the GFP maximum projection and the threshold was adjusted to highlight the GFP negative but ACTA2 (SMA) positive cells in FIJI.
Western blotting
Harvested cells or pulverized frozen tissues were washed with cold 1x PBS and lysed with RIPA Buffer (Sigma, R0278) containing complete protease inhibitor cocktail (Sigma-Aldrich, 4693132001). Protein concentration was determined (Bio-Rad, 5000202) and samples were boiled with NuPAGE LDS buffer (Thermo, NP0008). Samples were run in 4-20% SDS-PAGE gel (Bio-Rad, 4561094) in 1x Tris-Glycine running buffer (Bio-Rad, A0028). Proteins were transferred to PVDF membrane using Trans-blot Turbo transfer system and compatible transfer kit (Bio-Rad, 1704272). Membranes were blocked in blocking buffer (Bio-Rad, 12010020) and incubated with appropriate antibodies (Supplementary Table 5). Antibodies were validated by the respective providers for the applications described here; SRF and ITGA8 antibodies were further validated in the lab by loss-of-function and inclusion of positive/negative control tissues, respectively. Finally, chemiluminescent substrate (Bio-Rad, 1705061) and ChemiDoc imaging system (Bio-Rad, 120031) were used for digital imaging of Western blots. Molecular weight markers reflect the approximate size of proteins in kilodaltons. Urine from homozygous SrfItga8 and heterozygous SrfItga8 mice was collected and denatured in NuPAGE LDS buffer. Samples were loaded onto an SDS-PAGE gel and run in 1x Tris-Glycine running buffer. Gels were stained with Coomassie blue and images were documented as above. Purified bovine serum albumin was run alongside the urine samples and served as a positive control for the presence of protein.
RNA isolation and real-time quantitative RT-PCR
Total RNA was extracted using QIAzol® Lysis Reagent (Qiagen, 79306) followed by isopropanol precipitation and 70% ethanol washing. GlycoBlue™ Coprecipitant was applied for single aorta to facilitate the handling of small quantities of RNA. Air-dried RNA pellets were dissolved in RNase-free water. Reverse transcription was performed using Bio-Rad iScript cDNA synthesis kit (Bio-Rad, 1708891) after DNase I treatment (Thermo, AM2222). Real time quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, 1725121) with primers listed in Supplementary Table 4. Following delta delta Ct normalization to a housekeeping control, levels of normalized gene expression were plotted as fold-change from control samples.
Bulk RNA-seq
Homozygous SrfItga8 and heterozygous SrfItga8 mice (n=3 mice each) were administered tamoxifen at six weeks of age as above. Four weeks after the first dose of tamoxifen, aortas were collected and homogenized in RNeasy Plus Micro Kit with genomic DNA eliminator columns (Qiagen, 74034). Total RNA library preparation (TruSeq stranded) and high-throughput RNA-seq were performed on an Illumina HiSeq 4000 by Genomics Research Core of the University of Rochester. Gene Ontology analysis was done with DAVID (https://david.ncifcrf.gov/) and over-representation of transcription factor binding sites was determined with oPOSSUM.64 In a separate experiment, aortas from male wild type mice or age-matched mice with either Itga8-CreERT2 or Myh11-CreERT2 (n=3) were processed for total RNA isolation, library preparation, and sequencing on the Illumina NovaSeq 6000 at the University of Rochester. Standard data analysis, including principal component analysis and differential gene expression were done as described previously at the University of Rochester Genomics core.65
Nuclear-cytoplasmic RNA fractionation for LncRNA expression
Cultured mouse vascular smooth muscle cells (MOVAS) were fractionated using Protein and RNA isolation system kit according to the manufacturer’s protocol (Thermo Fisher, AM1921). A list of cell lines used is provided in Supplementary Table 6. Real-time qRT-PCR was done with primers to the LncRNA upstream of Itga8 as well as Actb primers, used as a cytoplasmic marker (Supplementary Table 4).
Flow Cytometry
Whole blood samples were collected from wild type, mTmG, Myh11-CreERT2/mTmG, Sm22-Cre/mTmG, and Itga8-CreERT2/mTmG mice and centrifuged for five minutes at 2000x g at room temperature. Buffy coats were carefully transferred to a fresh 1.5ml microfuge tube. 500ul of 1x ACK Lysing Buffer (Thermo, A1049201) was added to lyse red blood cells. Samples were then centrifuged for five minutes at 3000x g at room temperature. Supernatants were carefully aspirated and the pellets were suspended in 1x PBS. Cell suspensions were analyzed using an Accuri C6 flow cytometer (BD Biosciences) in the Flow Cytometry Core of the University of Rochester. Signals from the FITC channel were collected and gated for GFP-positive cells from each experimental condition. Please see Supplementary Fig. 5 and the Reporting Summary for more details.
Radio-telemetry recording of blood pressure
All physiological studies used male and female mice homozygous for floxed Srf and heterozygous for Itga8-CreERT2 (± tamoxifen). Following a 10-day washout period from the last tamoxifen (or oil) injection, male and female mice (13 weeks) were implanted with indwelling telemeters (carotid catheter) (DSI© Model #PA-C10, New Brighton, MN) under isoflurane anesthesia as previously described.66 After a seven day recovery period, baseline blood pressure (BP; mean arterial pressure, systolic and diastolic pressure) and heart rate measurements were consciously recorded for seven days. Mice were then implanted with subcutaneous mini-osmotic pumps (Alzet, model 1002, 14-day pump 0.25 ul/hour) containing angiotensin II (Phoenix Pharmaceuticals) diluted in isotonic saline at a dose of 490ng/kg/min as previously described.67 Blood pressure was recorded subsequently for 14 days.
Vascular Reactivity
Thoracic aortas from mice utilized for telemetry blood pressure recording protocol (ANGII-infused) were excised and cleaned of excess adipose tissue. Aortas were cut in 2mm rings and mounted on pins of a DMT® wire myograph (Ann Arbor, MI) as previously described.66 Concentration response curves to phenylephrine (1nM −30μM concentrations) as well as maximum responses to KCl (80mM) were performed and recorded with LabChart® analysis software (AD Instruments®, Colorado Springs, CO).
Mapping and copy number determination of Myh11-CreERT2
Long-read libraries of (i) Ultra-Long DNA Sequencing (SQK-ULK001) and (ii) Cas9 Sequencing (SQK-CS9109) kits from ONT Nanopore were prepared following the manufacturer’s instructions (www.nanoporetech.com). For Ultra-Long DNA libraries, ultrahigh molecular weight genomic DNA was isolated with Circulomics kits (NB-900-601-01 and NB-900-701-01) following the manufacturer’s instructions (www.circulomics.com). For Cas9 libraries, high molecular weight genomic DNA was isolated with an NEB kit (T3060) following the manufacturer’s instructions (www.neb.com). Five males, including a male from Jackson Laboratory (www.jax.org/), were used as input for mapping as described.68 For Cas9 Sequencing (CRISPR-LRS),68 crRNAs were designed with CHOPCHOP using default parameters (https://chopchop.cbu.uib.no) (Supplementary Table 4). Long-read libraries were run on R9.4.1 flow cells on a minION Mk1B or a GridION Mk1 with fast5 to fastq read conversion using guppy (v4.2.2) on MinKNOW (v20.10.3) MinKNOW Core (v4.1.2) and fast base-calling option for the base-call model and minimum Q-score of 7 option for read filtering. Fastq files were mapped to reference sequences (improved Cre or segments of chromosome 16) using the Long-Read Support (beta) plugin in Qiagen CLC Genomics Workbench (www.qiagen.com) which uses open-source tool minimap2. Mapped, informative reads were manually queried against NCBI nr/nt, Refseq genome databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and UCSC genome browser with BLAT tool (https://genome.ucsc.edu), where a mini-tiled map of Myh11-CreERT2 integration locus was constructed.68 To determine the copy number of Myh11-CreERT2, qPCR was performed as above. Genomic DNA was diluted to 50 ng for input. Two primer sets targeting ERT2 of Myh11-CreERT2 were normalized to internal control primers (Supplementary Table 4). Real time quantitative PCR conditions were the following: step1 95°C for 3 min; step 2 95°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec for 40 cycles.
Statistical analysis
A Shapiro-Wilk test was performed to determine whether data were normally distributed. Paired or unpaired one- or two-tailed t-tests were used for comparisons between experimental and control conditions and one- or two-way ANOVA with Tukey’s posthoc testing for individual comparisons was done for multiple group comparisons. All data analyses were performed in GraphPad Prism 9 (GraphPad Software). Results are expressed as mean ± standard deviation. Probability values of p<0.05 were considered statistically significant.
Extended Data
Extended Data Fig. 1. Strategy and sequence validation of Itga8-CreERT2.
a, Partial Itga8 locus, overlapping long noncoding RNA, and general strategy for mouse ESC targeting. The 22 amino acid P2A self-cleaving cassette (GSGATNFSLLKQAGDVEENPGP) allows for concurrent detection of the CreERT2 fusion and the mCherry reporter; however, mCherry fluorescence was not easily detected and therefore not pursued in this study. Small horizontal arrows (p1-p4) denote primers for PCR genotyping of mice. b, Nucleotide sequence of targeted knock-in cassette following Flippase-mediated removal of the Neo gene. Colors correspond to the schematic in a. Bases highlighted in black boxes represent codon-optimized substitutions from original sequences. HR, homologous recombination.
Extended Data Fig. 2. ITGA8 protein expression in heterozygous Itga8-CreERT2 mice.
a, Western blots of ITGA8 protein in two independent experiments performed by two independent investigators with quantitative data at right. n=6 independent male aortas for each condition. b, qRT-PCR analysis of Itga8 mRNA and c, the antisense Ak086420 long noncoding RNA in wild type versus Itga8-CreERT2 heterozygous aorta. n=6 male aortas per genotype. d, qRT-PCR of cytosolic versus nuclear Ak086420 expression in wild type aorta (n=4 male aortas). Error bars represent the mean ± standard deviation; p-values determined by two-tailed, unpaired Student’s t-test. e, Western blot of ITGA8 and CreERT2 in aorta of seven week-old male and female Itga8-CreERT2 heterozygous mice (n=3 independent mice of each sex). Molecular weight markers at right here and below are in kilodaltons.
Extended Data Fig. 3. Recombination activity of different SMC Cre mice in myeloid cells.
a, Representative flow cytometry of GFP labeled cells from wild type C57BL/6J mice (i); mT/mG reporter mice (ii); and mT/mG in Sm22-Cre (iii); tamoxifen-treated male Myh11-CreERT2 (iv); tamoxifen-treated male Itga8-CreERT2 (v); tamoxifen-treated female Itga8-CreERT2 (vi) mice. Quantitative data for GFP+ circulating cells in upper left quadrant Q1 (vii) and upper right quadrant Q2 (viii) are shown for each Cre driver line (n=3 male mice per Cre line, save the Itga8CreERT2 line which represent male, n=2, and female, n=2, samples pooled for the graphs shown). The horizontal axis was set to the same threshold for all panels according to the apparent two populations of cells in the positive control (Sm22-Cre-mT/mG). The vertical axis divides two apparent populations of different sized cells in the Sm22-Cre-mT/mG strain. Note, as we did not gate for any surface markers in this study, we cannot label any quadrant as to cell type. Source of cells was from circulation cleared of red blood cells. Error bars represent the mean ± standard deviation. One-way ANOVA and posthoc testing revealed indicated p-values. See Supplementary Figure 5 for more details. b, Bone marrow aspirates of 10-week-old male Itga8-CreERT2 (i, ii) and Myh11-CreERT2 (iii, iv) mice (n=2 mice/genotype) carrying mT/mG reporter and treated with tamoxifen. Arrows indicate multi-nucleated megakaryocytes. Scale bars are 20μm.
Extended Data Fig. 4. Comparative Cre activity in adult male and female blood vessels.
a, The GFP signal demonstrates recombination of the mT/mG reporter in medial SMCs of both the aorta and vena cava, whereas the red stained endothelium indicates the absence of recombination. Scale bar is 33μm for aorta and 100μm for vena cava. The aorta and vena cava of Itga8-CreERT2 was from a female mouse. b, Activity of each CreERT2 driver in indicated segments of male mouse aorta. Scale bars are 20μm for all panels. Lu, lumen of vessel wall. c, Tamoxifen-treated 54-week-old female (i-iii) and male (iv-vi) mouse thoracic aorta (i, iv), brain (ii, v), and heart (iii, vi). Each panel in a and b is representative of at least two independent male or female mice. Scale bars are all 20μm. d, Western blot of ITGA8 protein in 10-week-old versus 54-week-old female mouse aorta (n=3 independent mice per time point).
Extended Data Fig. 5. Comparative Cre activity in adult mouse tissues.
a, GFP signal restricted to VSMCs in blood vessels (arrows) of each indicated tissue type. b, GFP signal in VSMCs and non-VSMCs of indicated tissue types. White arrowheads, blood vessels of kidney and thymus; white arrows, glomeruli of kidney; yellow arrows, sinusoids of liver. Itga8-CreERT2-mediated GFP signal was also present in myoepithelial cells of mammary gland and thecal cells around mature follicle (white asterisk) of ovary. All images were processed the same except for the Testis panel under Itga8-CreERT2, which was uniformly enhanced to bring out more of the signal that otherwise would be too dark to visualize. Scale bar in a is 100μm for all images; scale bars in b are 50μm for kidney and thymus and 20μm for liver, mammary gland, and ovary. Data are representative of at least two independent male or female mice analyzed over the course of five years in two independent labs.
Extended Data Fig. 6. Leaky CreERT2 activity in aged mice.
Sections of aorta from 24-week-old or 54-week-old male Itga8-CreERT2 and 54-week-old Myh11-CreERT2 mice. The brightness in the 54-week Itga8-CreERT2 image was uniformly enhanced to better appreciate the GFP signal. Scale bars are 20μm for all panels. Images are representative of two independent mice.
Extended Data Fig. 7. Quantitative activity of Myh11-CreERT2 versus Itga8-CreERT2 in popliteal blood vessels.
Representative 2D maximum projections from confocal imaging of live GFP and tdTomato fluorescence popliteal artery (a, b) and vein (c, d) from Myh11-CreERT2 versus Itga8-CreERT2 with quantitation of each in panels e and f, respectively. n=4 male mice per genotype. Scale bars represent 50μm (a, b) and 100μm (c, d). Error bars in panels e-f represented by mean ± standard deviation; p-values determined by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 8. Intestinal phenotype in Myh11-CreERT2 versus Itga8-CreERT2 mediated knockout of serum response factor (Srf).
a, Western blot showing lack of effect of tamoxifen on SRF in wild type aorta (n=3 independent male mice per treatment). b, Quantitation of panel a represented by mean ± standard deviation; p-value determined by two-tailed, unpaired Student’s t-test. c, Anatomy of abdominal cavity in the indicated CreERT2 driver mice used for Srf inactivation. The two SrfMyh11 images were from mice 14 days following tamoxifen administration, whereas the SrfItga8 image was from a mouse 8 weeks after tamoxifen administration. d, Oil (panels i-iii) and Tamoxifen (panels iv-vi) treated SrfMyh11 mice with dissected gross intestine (i, iv), H&E stained intestine (ii, v), and immunostaining for SRF (green) and ACTA2 (red) in intestine (iii, vi). Scale bar is 100μm for ii, iii, v, and vi or 1mm for i and iv. Studies of panel d are representative of at least two independent male or female mice analyzed over the course of five years in two independent labs.
Extended Data Fig. 9. Itga8-CreERT2 mediated inactivation of Srf in adult mouse tissues.
a, Immunofluorescence confocal microscopy of sections of carotid artery (i, iv), bladder (ii, v), and intestine (iii, vi) from tamoxifen-treated male mice carrying homofloxed Srf alleles in the absence (i-iii) or presence (iv-vi) of Itga8-CreERT2 (abbreviated Cre). Sections were stained with antibodies to ACTA2 (red), SRF (green), and DAPI. Arrows and arrowheads point to blood vessels and visceral SMCs, respectively. Scale bar is 20μm for all panels. Data from each panel are representative of at least two independent mice. b, SRF positive VSMCs were counted in sections of carotid arteries from Tam-administered homozygous floxed Srf mice without Cre (HoF+Tam, n=6 male mice per condition) or homozygous floxed Srf mice with Cre (HoF+Cre+Tam, n=5 male mice per condition). c, Same quantitative measures as in panel b only from intestine (n=3 mice per condition). Western blots of SRF in aorta (d) and bladder (e) of indicated genotypes, all treated with the same schedule of tamoxifen. Corresponding quantitative data are shown for mouse aorta (f) and bladder (g) (n=3 mice per genotype). HeF, heterozygous floxed Srf; HoF, homozygous floxed Srf. Error bars are mean ± standard deviation. Student unpaired, one-tailed t-test was applied in b and c, and one-way ANOVA with Tukey’s t-test for f and g to reveal indicated p-values in each graph.
Extended Data Fig. 10. Contractile activity of mesenteric artery in SrfItga8 conditional knockout mice.
a, Confocal immunofluorescence microscopy of SRF and DAPI alone (i, iii) and merged with ACTA2 (ii, iv) in mesenteric arteries of Oil control (i, ii) versus tamoxifen-treated (iii, iv) homozygous SrfItga8 mice; scale bars are 20μm. Results shown are of two independent mice for each condition. Western blots of SRF target, MYH11, in Oil versus Tam-induced SrfItga8 homozygous male (b) and female (c) mesenteric arteries. n=3 independent animals for each condition. Error bars represented by mean ± standard deviation; p-values determined by two-tailed, unpaired Student’s t-test.
Supplementary Material
Supplementary Data1 Source data for Supplementary Fig. 4
Supplementary Table 1. Floxed Alleles and associated SMC Cre Drivers
Supplementary Table 2. Differentially expressed genes in aorta of wild type versus Cre-containing mice
Supplementary Table 3. Mouse models
Supplementary Table 4. Oligonucleotide primers and sgRNAs for mapping Myh11-Cre transgene
Supplementary Table 5. Antibodies
Supplementary Table 6. Cell Lines
Acknowledgments
We thank members of the University of Rochester Medical Center Genomics Research Core for carrying out the RNA-seq experiments and generating Fig. 2e and the Electron Microscopy and Histology Core at Augusta University for performing the immunogold studies and generating Fig. 1b.
Sources of Funding
This work was supported by the American Heart Association post-doctoral grants 17POST3360938 to QL and CDA858380 to JLF; and National Institutes of Health grants R00HL146948 to JFL; HL155265 to EBC; HL122578 to MJD; HL122686 and HL139794 to XL; HL138987, HL136224, and HL147476 to JMM; EY026614 to LG; and R00-HL143198 to SDZ.
Footnotes
Competing Interests
A.A. is a co-founder of Tango Therapeutics, Azkarra Therapeutics, Ovibio Corporation and Kytarro; a member of the board of Cytomx and Cambridge Science Corporation; a member of the scientific advisory board of Genentech, GLAdiator, Circle, Bluestar, Earli, Ambagon, Phoenix Molecular Designs and Trial Library; a consultant for SPARC, ProLynx, and GSK; receives grant or research support from SPARC and AstraZeneca, and holds patents on the use of PARP inhibitors held jointly with AstraZeneca from which he has benefited financially (and may do so in the future). The patents are unrelated to any aspect of the current study. All other authors declare no competing interests.
Data availability
RNA-seq data are available through the Gene Expression Omnibus (GSE138824 and GSE199244). Long-read data (informative reads only) are available through the NCBI SRA database, which can be downloaded from www.ncbi.nlm.nih.gov/sra) under BioProject number PRJNA82551. Original source data are included in Supplementary materials. Updates to Supplementary Table 1 are available upon request.
References
- 1.Sauer B Inducible gene targeting in mice using the Cre/lox system. Methods 14, 381–392 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Gu H, Marth JD, Orban PC, Mossmann H & Rajewsky K Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265, 103–106 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Feil R et al. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A 93, 10887–10890 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majesky MW Developmental basis of vascular smooth muscle diversity. Arteriosclerosis, Thrombosis and Vascular Biology 27, 1248–1258 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Tasian G, Cunha G & Baskin L Smooth muscle differentiation and patterning in the urinary bladder. Differentiation 80, 106–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somlyo AP & Somlyo AV Signal transduction and regulation in smooth muscle. Nature 372, 231–236 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Kuriyama H, Kitamura K, Itoh T & Inoue R Physiological features of visceral smooth muscle cells, with special reference to receptors and ion channels. Physiol Rev 78, 811–920 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Owens GK, Kumar MS & Wamhoff BR Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological Reviews 84, 767–801 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty R et al. Promoters to study vascular smooth muscle: mistaken identity? Arteriosclerosis, Thrombosis and Vascular Biology 39, 603–612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angstenberger M et al. Severe intestinal obstruction on induced smooth muscle-specific ablation of the transcription factor SRF in adult mice. Gastroenterology 133, 1948–1959 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Mericskay M et al. Inducible mouse model of chronic intestinal pseudo-obstruction by smooth muscle-specific inactivation of the SRF gene. Gastroenterology 133, 1960–1970 (2007). [DOI] [PubMed] [Google Scholar]
- 12.He WQ et al. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology 135, 610–620 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albinsson S et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One 6, e18869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J et al. Myocardin is required for maintenance of vascular and visceral smooth muscle homeostasis during postnatal development. Proceedings of the National Academy of Sciences USA 112, 4447–4452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morano I et al. Smooth-muscle contraction without smooth-muscle myosin. Nature Cell Biology 2, 371–375 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Niessen P et al. Smoothelin-a is essential for functional intestinal smooth muscle contractility in mice. Gastroenterology 129, 1592–1601 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Halim D et al. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. Proceedings of the National Academy of Sciences USA 114, E2739–E2747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Wang DZ, Richardson JA & Olson EN The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proceedings of the National Academy of Sciences of the United States of America 100, 9366–9370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzzardo PM et al. A small cassette enables conditional gene inactivation by CRISPR/Cas9. Sci Rep 7, 16770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horii T et al. Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci Rep 7, 7891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurumurthy CB et al. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol 20, 171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossy B, Bossy-Wetzel E & Reichardt LF Characterization of the integrin a8 subunit: a new integrin b1-associated subunit, which is prominently expressed on axons and on cells in contact with basal laminae in chick embryos. EMBO Journal 10, 2375–2385 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnapp LM, Breuss JM, Ramos DM, Sheppard D & Pytela R Sequence and tissue distribution of the human integrin a8 subunit: a b1-associated a subunit expressed in smooth muscle cells. Journal of Cell Science 108, 537–544 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Hartner A, Schocklmann H, Prols F, Muller U & Sterzel RB Alpha8 integrin in glomerular mesangial cells and in experimental glomerulonephritis. Kidney International 56, 1468–1480 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Maltby KM & Miano JM A novel retinoid-response gene set in vascular smooth muscle cells. Biochem Biophys Res Commun 281, 475–482 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Zargham R & Thibault G a8b1 integrin expression in the rat carotid artery: involvement in smooth muscle cell migration and neointima formation. Cardiovascular Research 65, 813–822 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Zargham R, Touyz RM & Thibault G a8 integrin overexpression in de-differentiated vascular smooth muscle cells attenuates migratory activity and restores the characteristics of the differentiated phenotype. Atherosclerosis 195, 303–312 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kitchen CM, Cowan SL, Long X & Miano JM Expression and promoter analysis of a highly restricted integrin alpha gene in vascular smooth muscle. Gene 513, 82–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller U et al. Integrin a8b1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell 88, 603–613 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Z et al. Smooth muscle protein 22 alpha-Cre is expressed in myeloid cells in mice. Biochemical Biophysical Research Communications 422, 639–642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirth A et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nature Medicine 14, 64–68 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Miano JM et al. Restricted inactivation of serum response factor to the cardiovascular system. Proceedings of the National Academy of Sciences USA 101, 17132–17137 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Aznar A et al. Tamoxifen-independent recombination of reporter genes limits lineage tracing and mosaic analysis using CreER(T2) lines. Transgenic Res 29, 53–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanlandewijck M et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature (2018). [DOI] [PubMed] [Google Scholar]
- 36.Shimshek DR et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Nicholls PK, Bellott DW, Cho TJ, Pyntikova T & Page DC Locating and characterizing a transgene integration site by nanopore sequencing. G3 (Bethesda) 9, 1481–1486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue B, Pamidimukkala J & Hay M Sex differences in the development of angiotensin II-induced hypertension in conscious mice. Am J Physiol Heart Circ Physiol 288, H2177–2184 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Park C et al. Loss of serum response factor induces microRNA-mediated apoptosis in intestinal smooth muscle cells. Cell Death Dis 6, e2011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Retailleau K et al. Selective involvement of serum response factor in pressure-induced myogenic tone in resistance arteries. Arteriosclerosis, Thrombosis and Vascular Biology 33, 339–346 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Galmiche G et al. Inactivation of serum response factor contributes to decrease vascular muscular tone and arterial stiffness in mice. Circ Res 112, 1035–1045 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Herring BP, Hoggatt AM, Gupta A & Wo JM Gastroparesis is associated with decreased FOXF1 and FOXF2 in humans, and loss of FOXF1 and FOXF2 results in gastroparesis in mice. Neurogastroenterol Motil 31, e13528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rashbrook VS, Brash JT & Ruhrberg C Cre toxicity in mouse models of cardiovascular physiology and disease. Nature Cardiovascular Research (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthaei KI Genetically manipulated mice: a powerful tool with unsuspected caveats. J Physiol 582, 481–488 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry MN et al. Annotated expression and activity data for murine recombinase alleles and transgenes: the CrePortal resource. Mamm Genome 33, 55–65 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buelow B & Scharenberg AM Characterization of parameters required for effective use of tamoxifen-regulated recombination. PLoS One 3, e3264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandlesh P, Juang T, Safina A, Higgins MJ & Gurova KV Uncovering the fine print of the CreERT2-LoxP system while generating a conditional knockout mouse model of Ssrp1 gene. PLoS One 13, e0199785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meinke G, Bohm A, Hauber J, Pisabarro MT & Buchholz F Cre recombinase and other tyrosine recombinases. Chemical Reviews 116, 12785–12820 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Clayton JA Applying the new SABV (sex as a biological variable) policy to research and clinical care. Physiol Behav 187, 2–5 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Liao M et al. An X-linked Myh11-CreER(T2) mouse line resulting from Y to X chromosome-translocation of the Cre allele. Genesis 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan AP, Bell TA, Crowley JJ & Pardo-Manuel de Villena F Instability of the pseudoautosomal boundary in house mice. Genetics 212, 469–487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodwin LO et al. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res 29, 494–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zargham R, Pepin J & Thibault G a8b1 integrin is up-regulated in the neointima concomitant with late luminal loss after balloon injury. Cardiovascular Pathology 16 212–220 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Fuchs H et al. Mouse phenotyping. Methods 53, 120–135 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Nishimichi N et al. Induced hepatic stellate cell integrin, alpha8beta1, enhances cellular contractility and TGFbeta activity in liver fibrosis. J Pathol 253, 366–373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corliss BA et al. Myh11 lineage corneal endothelial cells and ASCs populate corneal endothelium. Invest Ophthalmol Vis Sci 60, 5095–5103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Armulik A, Abramsson A & Betsholtz C Endothelial/pericyte interactions. Circ Res 97, 512–523 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Hess DL et al. Perivascular cell-specific knockout of the stem cell pluripotency gene Oct4 inhibits angiogenesis. Nat Commun 10, 967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orlich MM et al. Mural cell SRF controls pericyte migration, vessel patterning and blood flow. Circ Res 131, 308–327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miano JM Serum response factor: toggling between disparate programs of gene expression. Journal of Molecular Cellular Cardiology 35, 577–593 (2003). [DOI] [PubMed] [Google Scholar]
- 61.George SH et al. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proceedings of the National Academy of Sciences USA 104, 4455–4460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Preibisch S, Saalfeld S & Tomancak P Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT & Wasserman WW oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Research 35, W245–W252 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao P et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol 22, 83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faulkner JL et al. Lack of suppression of aldosterone production leads to salt-sensitive hypertension in female but not male Balb/C mice. Hypertension 72, 1397–1406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J et al. Inflammation and mechanical stretch promote aortic stiffening in hypertension through activation of p38 mitogen-activated protein kinase. Circ Res 114, 616–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bryant WB et al. CRISPR-LRS for mapping transgenes in the mouse genome. bioRxiv, 2022.2001.2005.475144 (2022). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data1 Source data for Supplementary Fig. 4
Supplementary Table 1. Floxed Alleles and associated SMC Cre Drivers
Supplementary Table 2. Differentially expressed genes in aorta of wild type versus Cre-containing mice
Supplementary Table 3. Mouse models
Supplementary Table 4. Oligonucleotide primers and sgRNAs for mapping Myh11-Cre transgene
Supplementary Table 5. Antibodies
Supplementary Table 6. Cell Lines
Data Availability Statement
RNA-seq data are available through the Gene Expression Omnibus (GSE138824 and GSE199244). Long-read data (informative reads only) are available through the NCBI SRA database, which can be downloaded from www.ncbi.nlm.nih.gov/sra) under BioProject number PRJNA82551. Original source data are included in Supplementary materials. Updates to Supplementary Table 1 are available upon request.