Abstract
Salmonella enterica subspecies enterica serovar Infantis (S. Infantis) is a non-typhoid, zoonotic and foodborne serovar with worldwide distribution, and often associated with increasing antimicrobial resistance. Efflux pumps are antimicrobial resistance mechanisms able to promote and increase resistance levels to multiple distinct drug classes. Heavy metal tolerance genes have been demonstrated to promote resistance against these compounds and act in the co-selection of antimicrobial resistant strains. Despite the relevance of S. Infantis in clinical and non-clinical fields, few studies worldwide have investigated the occurrence of such genes in strains from diverse sources. Therefore, the present study aimed at determining the prevalence of antimicrobial efflux pump and heavy metal tolerance genes and their genomic relatedness through core-genome multi-locus sequence typing (cgMLST) of 80 S. Infantis strains isolated from food, environmental, human and animal sources from 2013 to 2018 in Brazil. Twenty efflux pump encoding genes were detected, with 17 of these (acrA, acrB, baeR, crp, emrB, emrR, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR and soxS) detected in all strains studied, golS in 98.75%, mdfA in 58.75% and tet(A) in 37.5%. Tolerance genes to arsenic (arsR) were detected in 100% of the strains, gold (golS and golT) in 98.75%, silver (silABCDEFPRS) in 36.25% and mercury (merR and merT) in 1.25%. cgMLST demonstrated a closer genetic relationship among strains harboring similar profiles of heavy metal and efflux pump encoding genes, despite their origin. In conclusion, the high prevalence of some efflux pump and heavy metal tolerance encoding genes alert us about the importance of strong surveillance measures to monitor resistance and the transmission of S. Infantis among diverse sources in Brazil.
1. Introduction
Non-typhoid Salmonella enterica (NTS) serovars are among the four leading bacterial pathogens associated with human foodborne diseases in the world [1]. In association with its zoonotic nature, increasing antimicrobial resistance rates have been constantly reported about these pathogens, leading the U.S. Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) to classify drug-resistant NTS as a serious threat and high priority pathogens in the control and prevention of antimicrobial resistance [2, 3].
Salmonella enterica subspecies enterica serovar Infantis (S. Infantis) is a non-typhoid serovar of global distribution and a ubiquitous nature, associated with human infections and present in foods, food-producing animals, farm and industry environments [4–6]. Increasing resistance rates have been reported for this serovar to drugs-of-choice employed in the treatment of human salmonellosis, such as fluoroquinolones and third- and fourth-generation cephalosporins, and drugs employed mainly in the veterinary field for animal therapy or as illegal growth promoters, such as aminoglycosides, phenicols and tetracycline [4–6].
In Gram-negative bacteria, such as NTS, antimicrobial resistance can be achieved by several types of mechanisms encoded by chromosomal mutations or horizontal transfer via plasmid-borne genes, which are usually responsible in conferring resistance to unique drug classes instead of multiple drug classes [7]. However, antibiotic-specific resistance genes are not the only ones capable of promoting drug resistance. Several genes encode the formation of efflux pumps in the cell membrane of Gram-negative bacteria, which are structures known for their capacity to promote and increase antimicrobial resistance levels to one or multiple distinct classes of antimicrobial molecules through pumping of the latter from the interior of bacterial cells to their exterior environment [8, 9].
Non-antibiotic compounds have also been demonstrated to play an important role in bacterial resistance. Heavy metals (such as arsenic, copper, mercury, silver and zinc) commonly occurr in nature, and some also are considered as environmental contaminants as a result of human pollution [10, 11]. Derivative products of these metals also have been employed in the treatment and prevention of bacterial infections in clinical fields, such as antiseptics or included in medical devices, and in non-clinical areas, such as disinfectants, food preservatives and feed additives for food-producing animals [10, 11]. Several genes have been reported to act in the bacterial tolerance to several types of heavy metals and also play an important role in the co-selection of antibiotic resistance strains [12].
In recent years, advancements in whole-genome sequencing (WGS) have significantly contributed to the study of zoonotic bacteria such as in the epidemiological monitoring of outbreaks and the tracking of antimicrobial resistance burden through genomics methods [13, 14]. Brazil is currently one of the world’s largest global meat exporters [15]. Despite this, and the common reports of S. Infantis in food items, environmental sources, humans and food-producing animals, few studies have employed genomics to investigate the presence of antimicrobial resistance determinants and the genomic relatedness among strains isolated in the country [6, 16, 17]. Also, few studies have conducted worldwide investigations on the occurrence of antimicrobial efflux pump and heavy metal tolerance encoding genes in S. Infantis [18–22].
Therefore, the aims of this study were: (I) to assess the prevalence of antimicrobial efflux pump and heavy metal tolerance encoding genes, and (II) to evaluate the genomic relatedness of sequenced S. Infantis strains isolated in Brazil from various sources (food, the environment, humans and animals) from 2013 to 2018.
2. Material and methods
2.1. Bacterial genomes
A total of 80 whole-genome sequenced S. Infantis strains isolated from food (n = 27), farm and industry environments (n = 24), humans (n = 19), animals (n = 7) and animal feed (n = 3) sources were analyzed in this study. These strains were provided by the Salmonella reference laboratory collection of the Oswaldo Cruz Foundation of Rio de Janeiro (FIOCRUZ-RJ) and were isolated from 2013 to 2018, in states of the South (Santa Catarina, Rio Grande do Sul and Paraná), Southwest (São Paulo and Minas Gerais), Midwest (Mato Grosso do Sul and Goiás) and Northwest (Alagoas and Maranhão) regions of Brazil (Table 1).
Table 1. Strain identifiers and isolation data of the 80 Salmonella Infantis strains studied isolated from food (n = 27), farm and industry environments (n = 24), humans (n = 19), animals (n = 7) and animal feed (n = 3) between 2013 and 2018 in Brazil.
| Strain identifiers and accession numbers | Isolation data | ||||
|---|---|---|---|---|---|
| Strain/Year | CFSAN no. | GenBank no. | Material | Source | State |
| SI 1348/13 | CFSAN107127 | AAWRHH000000000.1 | Human feces | Human | PR |
| SI 2385/13 | CFSAN107129 | AAWRGU000000000.1 | Soy | Food | PR |
| SI 2950/13 | CFSAN107130 | AAWRHS000000000.1 | Human feces | Human | AL |
| SI 2951/13 | CFSAN107131 | AAWRHN000000000.1 | Human feces | Human | AL |
| SI 3156/13 | CFSAN107132 | AAWRGH000000000.1 | Disposable shoe cover | Environment | SC |
| SI 5025/13 | CFSAN107133 | AAWRGA000000000.1 | Human feces | Human | SC |
| SI 124/14 | CFSAN107134 | AAWRDW000000000.1 | Swine feces | Animal | RS |
| SI 210/14 | CFSAN107136 | AAWREM000000000.1 | Dragging swab | Environment | SC |
| SI 212/14 | CFSAN107137 | AAWRDZ000000000.1 | Dragging swab | Environment | SC |
| SI 388/14 | CFSAN107138 | AAWRER000000000.1 | Soybean animal meal | Animal feed | SP |
| SI 583/14 | CFSAN107139 | AAWREP000000000.1 | Chicken carcass | Food | SC |
| SI 584/14 | CFSAN107140 | AAWREX000000000.1 | Pasta containing ham | Food | SC |
| SI 677/14 | CFSAN107141 | AAWRFG000000000.1 | Carcass cleaning wipe | Food | SC |
| SI 723/14 | CFSAN107142 | AAWRFD000000000.1 | Dragging swab | Environment | SC |
| SI 982/14 | CFSAN107143 | AAWRHV000000000.1 | Chicken feces | Animal | RS |
| SI 1143/14 | CFSAN107144 | AAWRHU000000000.1 | Chicken feces | Animal | RS |
| SI 1284/14 | CFSAN107145 | AAWRIM000000000.1 | Dragging swab | Environment | SC |
| SI 1380/14 | CFSAN107146 | AAWRIF000000000.1 | Chicken feces | Animal | RS |
| SI 1408/14 | CFSAN107148 | AAWRIL000000000.1 | Human feces | Human | RS |
| SI 1409/14 | CFSAN107149 | AAWRHF000000000.1 | Human feces | Human | RS |
| SI 1441/14 | CFSAN107150 | AAWRHL000000000.1 | Mayonnaise | Food | RS |
| SI 1711/14 | CFSAN107151 | AAYKFJ000000000.1 | Chicken feces | Animal | RS |
| SI 2378/14 | CFSAN107152 | AAWRHR000000000.1 | Truck swab | Environment | SC |
| SI 2430/14 | CFSAN107153 | AAWRHO000000000.1 | Mixed meat sausage | Food | SC |
| SI 2461/14 | CFSAN107154 | AAWRGI000000000.1 | Chicken carcass | Food | SC |
| SI 2463/14 | CFSAN107155 | AAYKFK000000000.1 | Chicken carcass | Food | SC |
| SI 2548/14 | CFSAN107156 | AAWRDS000000000.1 | Chicken feces | Animal | RS |
| SI 3836/14 | CFSAN107160 | AAXBHC000000000.1 | Dragging swab | Environment | RS |
| SI 4882/14 | CFSAN107164 | AAXBHW000000000.1 | Chicken carcass | Food | MG |
| SI 4892/14 | CFSAN107165 | AAXAKM000000000.1 | Chicken wings | Food | MG |
| SI 4895/14 | CFSAN107166 | AAXAKH000000000.1 | Chicken carcass | Food | MG |
| SI 4901/14 | CFSAN107167 | AAXAKN000000000.1 | Pig snout | Food | MG |
| SI 5247/14 | CFSAN107168 | AAXAKJ000000000.1 | Chicken upper leg and thigh | Food | MG |
| SI 342/15 | CFSAN107171 | AAXHSY000000000.1 | Swine heart | Food | SC |
| SI 444/15 | CFSAN107172 | AAXHRH000000000.1 | Pork filet | Food | SC |
| SI 447/15 | CFSAN107173 | AAXHRI000000000.1 | Smoked and salted pork meat | Food | SC |
| SI 1809/15 | CFSAN107179 | AAXHSE000000000.1 | Meat animal meal | Animal feed | SC |
| SI 1816/15 | CFSAN107180 | AAXHVG000000000.1 | Poultry viscera animal meal | Animal feed | SC |
| SI 2280/15 | CFSAN107182 | AAXHUK000000000.1 | Chicken carcass | Food | SC |
| SI 2302/15 | CFSAN107183 | AAXHUC000000000.1 | Cleaning wipe | Environment | SC |
| SI 2370/15 | CFSAN107185 | AAXHUH000000000.1 | Carcass cleaning wipe | Food | SC |
| SI 2869/15 | CFSAN107190 | AAXHUP000000000.1 | Chicken upper leg | Food | MG |
| SI 3056/15 | CFSAN107193 | AAXHUJ000000000.1 | Chicken carcass | Food | MG |
| SI 4764/15 | CFSAN107197 | AAXHVH000000000.1 | Cleaning wipe | Environment | SC |
| SI 5391/15 | CFSAN107200 | AAXHUD000000000.1 | Disposable shoe cover | Environment | SC |
| SI 5837/15 | CFSAN107201 | AAXHTN000000000.1 | Disposable shoe cover | Environment | SC |
| SI 5853/15 | CFSAN107202 | AAXJLL000000000.1 | Disposable shoe cover | Environment | SC |
| SI 5859/15 | CFSAN107203 | AAXHWB000000000.1 | Disposable shoe cover | Environment | SC |
| SI 5911/15 | CFSAN107204 | AAXHVK000000000.1 | Cleaning wipe | Environment | SC |
| SI 5912/15 | CFSAN107205 | AAYKGL000000000.1 | Cleaning wipe | Environment | SC |
| SI 5915/15 | CFSAN107206 | AAYKGJ000000000.1 | Cleaning wipe | Environment | SC |
| SI 5923/15 | CFSAN107207 | AAYKGQ000000000.1 | Cleaning wipe | Environment | SC |
| SI 220/16 | CFSAN107212 | AAYKGB000000000.1 | Cleaning wipe | Environment | SC |
| SI 3687/16 | CFSAN107222 | AAYKGA000000000.1 | Chicken carcass | Food | SC |
| SI 4447/16 | CFSAN107224 | AAYKGC000000000.1 | Pork sausage | Food | SC |
| SI 5946/16 | CFSAN107226 | AAYAAA000000000.1 | Pork rib | Food | SC |
| SI 6987/16 | CFSAN107229 | AAYAIC000000000.1 | Human feces | Human | MA |
| SI 7876/16 | CFSAN107233 | AAYAFO000000000.1 | Human feces | Human | RS |
| SI 11/17 | CFSAN107235 | AAYARD000000000.1 | Dragging swab | Environment | PR |
| SI 23/17 | CFSAN107237 | AAYAFK000000000.1 | Dragging swab | Environment | PR |
| SI 238/17 | CFSAN107238 | AAYAFN000000000.1 | Dragging swab | Environment | PR |
| SI 872/17 | CFSAN107239 | AAYAFR000000000.1 | Chicken carcass | Food | MG |
| SI 1171/17 | CFSAN107242 | AAYAFL000000000.1 | Soil | Environment | SP |
| SI 1256/17 | CFSAN107243 | AAYAFP000000000.1 | Soil | Environment | SP |
| SI 2580/17 | CFSAN107259 | AAYKFO000000000.1 | Human feces | Human | SC |
| SI 2953/17 | CFSAN107261 | AAYKFZ000000000.1 | Human fecal swab | Human | GO |
| SI 2954/17 | CFSAN107262 | AAYKFE000000000.1 | Human fecal swab | Human | GO |
| SI 3380/17 | CFSAN107263 | AAYKFP000000000.1 | Human fecal swab | Human | GO |
| SI 3877/17 | CFSAN107264 | AAYKFX000000000.1 | Chicken wings | Food | MG |
| SI 3906/17 | CFSAN107265 | AAYKFS000000000.1 | Sieve residue | Environment | SP |
| SI 4065/17 | CFSAN107266 | AAYKFR000000000.1 | Human feces | Human | PR |
| SI 4067/17 | CFSAN107267 | AAYKGD000000000.1 | Human feces | Human | PR |
| SI 4069/17 | CFSAN107268 | AAYKFD000000000.1 | Human blood | Human | PR |
| SI 52/18 | CFSAN107270 | AAYKFI000000000.1 | Chicken carcass | Food | MG |
| SI 331/18 | CFSAN107273 | AAYKFT000000000.1 | Human fecal swab | Human | GO |
| SI 623/18 | CFSAN107279 | AAYKFY000000000.1 | Human feces | Human | SC |
| SI 661/18 | CFSAN107280 | AAYKFW000000000.1 | Human feces | Human | MS |
| SI 942/18 | CFSAN107281 | AAYKFM000000000.1 | Human fecal swab | Human | RS |
| SI 1634/18 | CFSAN107284 | AAYKFQ000000000.1 | Yellowtail amberjack fish meat | Food | SC |
| SI 2676/18 | CFSAN107285 | AAYKFF000000000.1 | Avian reproductive matrix | Animal | GO |
Cleaning wipe: similar to synthetic tissues for domestic cleaning sold commercially; employed in the isolation procedure of microorganisms from surfaces in Brazil.
AL, Alagoas; BA, Bahia; GO, Goiás; MA, Maranhão; MG, Minas Gerais; MS, Mato Grosso do Sul; PE, Pernambuco; PR, Paraná; RS, Rio Grande do Sul; SC, Santa Catarina; SP, São Paulo.
These data are available in details at Vilela et al. 2021 [23].
The extraction of the genomic DNA was performed by the phenol-chloroform-isoamyl alcohol method, as previously described [23], and 1ng of the extracted DNA was used for the preparation of libraries with the Nextera XT DNA kit (Illumina, San Diego, CA). Genomes were sequenced in the Illumina MiSeq platform using the 2 X 150-bp paired-end MiSeq Reagent Kit version 3 (Illumina, San Diego, CA). Genome drafts were assembled with SKESA 2.2. Quality control was performed in the MicroRunQC workflow.
The complete isolation data, accession numbers and metadata of the genomic sequences of the 80 S. Infantis strains analyzed have been published in Vilela and collaborators [23] and are also partially displayed in Table 1.
2.2. Search of efflux pump coding genes
The Resistance Gene Identifier (RGI) tool of the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca/analyze/rgi) [24] was used to search for resistance genes responsible for the coding of antimicrobial efflux pumps for each of the 80 sequenced S. Infantis strains studied (Table 1). Default parameters were applied in the analysis. Only genes related to antimicrobial efflux and showing ≥80% identity/length were included in the results.
2.3. Search of heavy metal tolerance genes
The “Stress genotypes” filter, one of the automatic features of the Isolate Browser of NCBI’s Pathogen Detection database utilizing AMRFinderPlus curated database (https://www.ncbi.nlm.nih.gov/pathogens/isolates/), was used to detect heavy metal tolerance genes in each of the 80 sequenced S. Infantis studied (Table 1). Only genes related to heavy metal tolerance were included in the results.
2.4. Phylogenetic analysis
The genomic relatedness of the 80 sequenced S. Infantis strains was accessed by core genome Multi-locus Sequence Typing (cgMLST) and was performed from a set of reads in the cgMLSTFinder 1.1 tool (available at https://cge.cbs.dtu.dk/services/cgMLSTFinder/) using the Salmonella (Enterobase) filter [25]. The complete genome of the chicken isolate SINFA (Genbank accession number LN649235.1), isolated in the United Kingdom in 1973, also was included for comparison purposes.
Two different subsets were phylogenetically analyzed. The first analysis included the 80 S. Brazilian strains to provide an overview of the genomes studied in relation to the efflux pump and heavy metal tolerance genes found. In the second analysis, the 80 strains were analyzed in combination with 40 additional S. Infantis genomes from eight countries to provide a more global view of the genomes studied. These 40 genomes were selected in NCBI’s Pathogen Detection and were selected from strains of diverse years and sources including isolates from Canada, Ecuador, Germany, Mexico, Peru, South Africa, United Kingdom and the United States. Detailed descriptions and accession numbers of the 40 additional S. Infantis genomes are displayed in S2 Table.
2.5. Statistical analysis
The results of the search for antimicrobial efflux pump and heavy metal tolerance encoding genes were expressed in percentages. The Chi-square test was employed using the software GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA) in order to verify possible associations between specific profiles of interest among the antimicrobial efflux pumps and heavy metal tolerance genes found among the strains studied.
3. Results
3.1. Antimicrobial efflux pump encoding genes
A total of 20 genes have been detected, with 17 of these genes harbored by 100% of the 80 S. Infantis strains studied (Table 2; S1 Table) including genes acrA, acrB, baeR, crp, emrB, emrR, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR and soxS. Other genes were variably observed including golS in 79 strains (98.75%), mdfA in 47 strains (58.75%), and tet(A) in 30 strains (37.5%). According to CARD, these genes are able to promote resistance to one up to sixteen classes of antibiotics (aminocoumarins, aminoglycosides, carbapenems, cephalosporins, cephamycins, diaminopyrimidine compounds, glycylcyclines, monobactams, penams, penems, phenicols, quinolones, rifamycins, tetracyclines, macrolides and nitroimidazoles), biocides (benzalkonium chloride and triclosan), dyes (rhodamine) and antibiotic peptides (S3 Table). The combination of multiple genes detected among the strains studied resulted in four different efflux pump encoding gene profiles, that are displayed in Table 2.
Table 2. Antimicrobial efflux pump encoding gene (ARG) profiles detected among the 80 Salmonella Infantis strains studied isolated from food (n = 27), farm and industry environments (n = 24), humans (n = 19), animals (n = 7) and animal feed (n = 3) in Brazil between 2013 and 2018.
| Profile no. | Gene profiles | Isolation sources | Total | ||||
|---|---|---|---|---|---|---|---|
| FO | EN | HU | AN | AF | |||
| 1 | acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS | 14 | 11 | 14 | 6 | 1 | 46 |
| 2 | acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS, tet(A) | 12 | 11 | 4 | 1 | 2 | 30 |
| 3 | acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS | 1 | 2 | - | - | - | 3 |
| 4 | acrA, acrB, baeR, crp, emrB, emrR, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS | - | - | 1 | - | - | 1 |
FO, food; EN, environment; HU, human; AN, animal; AF; animal feed.
The complete distribution of the genes in all 80 strains is demonstrated in S2 Table and the specific spectrum of drug resistance promoted by each of the 20 genes detected is shown in S3 Table.
3.2. Heavy metal tolerance encoding genes
All the 80 S. Infantis strains studied (100%) harbored the arsR gene, related to arsenic tolerance. A total of 79 strains (98.75%) harbored golS and golT, related to gold tolerance. A complete sil operon (silABCDEFPRS), related to silver tolerance, was detected in 29 strains (36.25%), while a unique strain (1.25%) harbored only silE gene. A single strain (1.25%) also harbored genes merR and merT, related to mercury tolerance. The distribution of the genes related to arsenic, gold, silver and mercury tolerance are displayed for each of the 80 S. Infantis strains analyzed in S2 Table.
3.3. Phylogenetic analyses
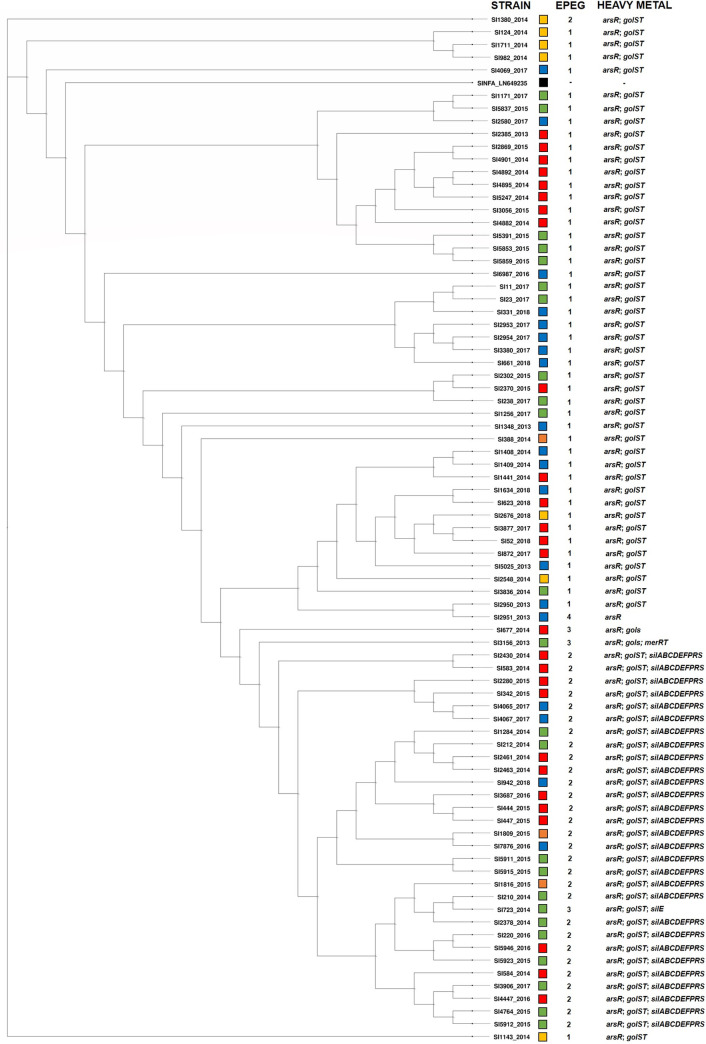
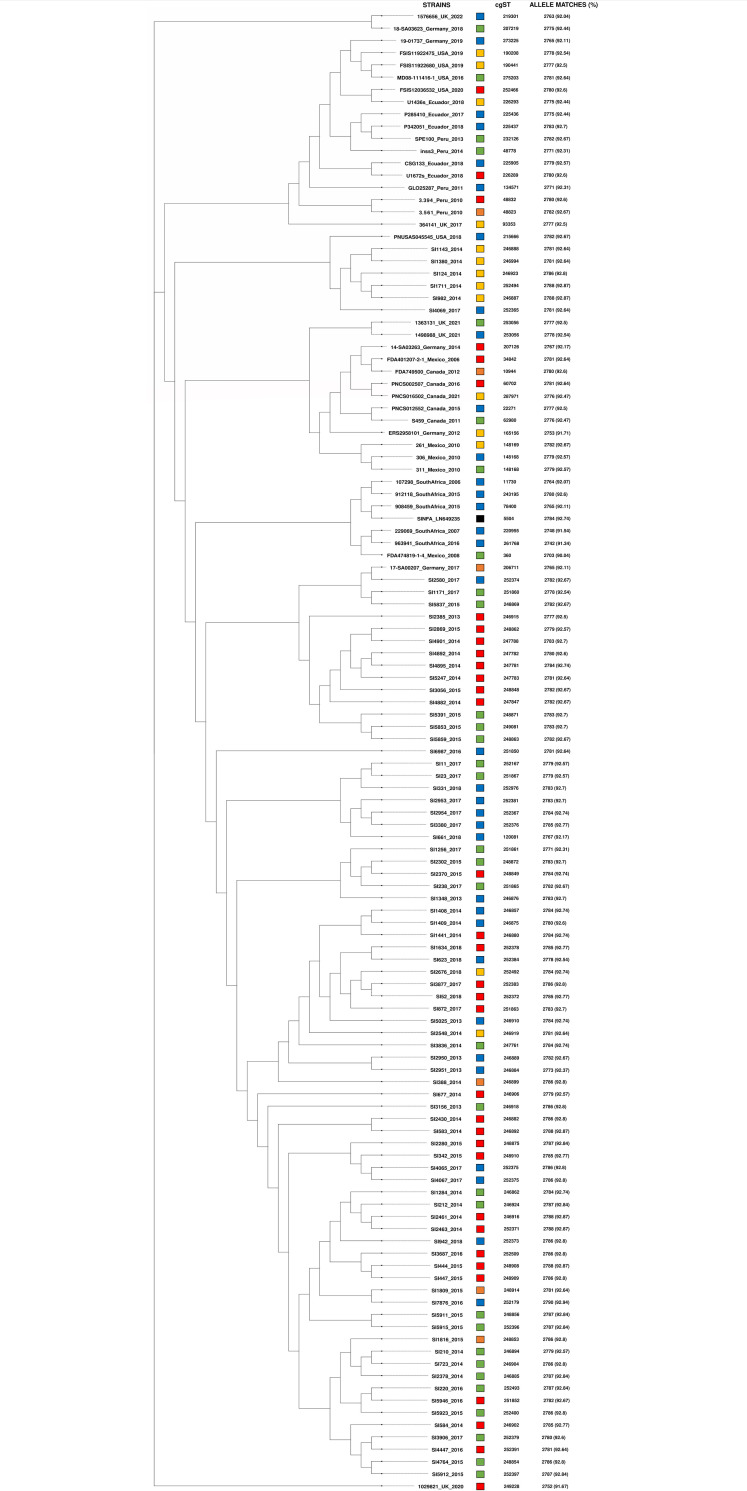
The phylogenetic trees based on the cgMLST analyses of 3,002 genes common to the Salmonella genus are presented in Figs 1 and 2. In Fig 1, the 80 S. Infantis strains are displayed alongside the profiles of efflux pump (Table 2) and heavy metal tolerance encoding genes. In Fig 2, the 120 S. Infantis strains are displayed along with the core genome sequence types (cgSTs) identified and numbers and percentages of the matched alleles found.
Fig 1. Phylogenetic tree based on the core-genome multi-locus sequence typing (cgMLST) analysis of the 80 whole-genome sequenced S. Infantis strains studied, isolated from food (red squares; n = 27), farm and industry environments (green squares; n = 24), humans (blue squares; n = 19), animals (yellow squares; n = 7) and animal feed (orange squares; n = 3) between 2013 and 2018 in Brazil.
S. Infantis reference strain SINFA LN649235.1 (black square) was included for comparison purposes. Additional information regarding the isolation sources, efflux pump encoding genes (EPEG) and heavy metal tolerance genes are included. Profile 1 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS); Profile 2 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS, tet(A)); Profile 3 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS); Profile 4 (acrA, acrB, baeR, crp, emrB, emrR, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS).
Fig 2. Phylogenetic tree based on the core-genome multi-locus sequence typing (cgMLST) analysis of the 120 genomes of S. Infantis strains from Brazil, Canada, Ecuador Germany, Mexico, Peru, South Africa, United Kingdom (UK) and United States (US).
The 120 strains were isolated from food (red squares), environments (green squares), humans (blue squares), animals (yellow squares) and animal feed (orange squares). S. Infantis reference strain SINFA LN649235.1 (black square) was included for comparison purposes. Additional information regarding the isolation sources, core-genome sequence types (cgSTs) and the number and percentages of the identified alleles are included.
Regarding the analysis in Fig 1, it was possible to observe a strong genetic association also demonstrated statistically (p < .00001) among 46 strains harboring golS, golT, arsR and the profile 1 of efflux pump encoding genes, as well as 29 strains harboring golS, golT, arsR, the sil operon and the profile 2 of efflux pump encoding genes (Fig 1; Table 2; S1 Fig), regardless of their isolation sources, materials and locations.
As illustrated in Fig 2, an extensive variation of cgSTs was observed among both Brazilian and international S. Infantis genomes (Fig 2). Of the 120 genomes analyzed, only six shared the same cgSTs. Two Brazilian strains isolated in 2017 from humans (SI4065 and SI4067) belonged to cgST 252375. Two genomes of human and environmental sources isolated in 2010 in Mexico (306 and 311) were assigned to cgST 148168. Finally, two genomes of human and environmental sources isolated in 2021 from the United Kingdom belonged to cgST 253056 (Fig 2).
4. Discussion
The increasing rates of drug-resistant NTS have become a public health and food safety concern worldwide [2, 3]. S. Infantis is a major ubiquitous NTS serovar, present in food, the environment, humans and animal sources, and also associated with increasing resistant rates to antimicrobial compounds of human and veterinary use [4–6, 16]. Previously, the 80 S. Infantis strains studied were analyzed using ResFinder (Center for Genomic Epidemiology) and AMRFinder (Pathogen Detection—NCBI) for resistance gene detection [6], where acquired resistance genes conferring resistance to β-lactams, diaminopyrimidine compounds, amphenicols, aminoglycosides, tetracycline, sulfonamide, and chromosomal point mutations associated to quinolone and antimicrobial peptide resistance, have been detected (S2 Table).
Antimicrobial efflux pumps have been described as important mechanisms of antimicrobial resistance among Gram-negative bacteria [8, 9]. However, despite the clinical and veterinary importance and the increasing resistance rates, there is little information about the frequency and diversity of antimicrobial efflux pump encoding genes in S. Infantis. According to the current published literature, only one study has evaluated the genetic variability among the sequences of acr, mar and sox genes in S. Infantis mutants and its correlation to quinolone resistance [18].
In the present study, the S. Infantis strains analyzed harbored 20 different types of efflux pumps encoding genes using the CARD tool that are capable to promote resistance from one to sixteen classes of antibiotics, biocides, dyes and antibiotic peptides (S3 Table). Previously, ResFinder and AMRFinder were also employed to determine the genotypic resistance profile of the same set of strains, and were able to identify the efflux pump encoding genes msdA, mdsB, tet(A) and point mutations in acrB. Interestingly, both acrA and tet(A) were detected at the same frequencies (S2 Table) as found herein [6]. Therefore, the results here obtained in comparison to data previously generated by other analysis tools suggested that differences in gene content may occur from platform to platform, and it possibly may be due to differences in the gene content deposited in their databases by their curators.
The acrA and acrB genes partially encode the well-studied AcrAB-TolC tripartite efflux system, which has been demonstrated to confer significant resistance to several antibiotic classes in NTS serovars [26]. However, the necessity of an intact set of genes in the efflux system’s and the possible occurrence of mutations may influence the resistance levels promoted [29]. In addition, the genes marAR and soxRS have been demonstrated to act in AcrAB-TolC system’s regulation and to be essential to its multidrug resistance action [27]. The ermB and ermR genes act in the promotion of increased quinolone resistance, mainly related to nalidixic acid, but its conjunct action with chromosomal point mutations and plasmid-borne genes can also favor the development of high resistance levels to fluoroquinolone drugs, such as ciprofloxacin [28]. The mdfA and mdtK genes have been reported and characterized to encode expressive resistance to tetracycline, chloramphenicol, norfloxacin, doxorubicin, acriflavine and biocides in the NTS serovar S. Typhimurium [29]. The kpnF gene, initially described in Klebsiella pneumoniae, also promotes significant resistance to several classes of antibiotics, antiseptics and disinfectants [30]. Finally, according to the CARD database, it is also important to mention genes like baeR, crp, golS, hns, rsmA and sdiA, which do not encode specific efflux pumps, but act as important regulators of acr, erm and mdt efflux systems.
In this way, this diverse occurrence of efflux pump encoding genes with broad spectrum of resistance found in high frequencies in S. Infantis strains may be a concern. The presence of these genes, in association to other genetic resistance determinants, could influence the resistance to drugs-of-choice for the treatment of human salmonellosis (such as fluoroquinolones and cephalosporins) [31, 32] and also to drugs less employed in human therapy but broadly employed in the veterinary area (such as tetracycline, phenicols and aminoglycosides) [33]. Additionally, the presence of genes able to promote resistance to non-antibiotic compounds demonstrate that antiseptics, disinfectants and related products could also show some inefficacy against S. Infantis, facilitating a possible co-selection of antibiotic-resistant strains.
The wide presence of heavy metal tolerance genes in bacteria has been described as a result of the selective pressure due to the combination of the occurrence of these compounds in nature, environmental pollution, and its applications in medical devices, disinfectants, antiseptics and preservatives [10–12]. In the present study, arsenic (arsR), gold (golS and golT), silver (silABCDEFPRS) and mercury (merR and merT) tolerance genes were detected among the 80 S. Infantis strains analyzed.
The sil operon (silABCDEFPRS) encodes the formation of the SilE periplasmic silver binding-protein, SilP and SilABC silver efflux pumps, SilF and SilG chaperones and SilS and SilR two-component signal transduction pair [11, 12, 34]. The first detailed characterization of the sil operon was provided after a major hospital outbreak with high mortality rates among burned patients caused by a broadly resistant S. Typhimurium clone [35], and since then, these genes have been reported in a wide range of clinical and environmental bacteria [11, 34]. Gold tolerance genes golS and golT are described in NCBI’s Pathogen Detection Gene Catalog (https://www.ncbi.nlm.nih.gov/pathogens/refgene/) to encode a sensor transcriptional regulator and a gold translocating P-type ATPase, respectively. The arsenic tolerance gene arsR encodes a responsive trans-acting transcriptional repressor and is part of the minimum arsenic tolerance operon aRBC [11, 12], whose genes have been reported previously among S. Infantis strains [22]. Related to regulatory and transport functions, the mercury tolerance genes merR and merT belong to the mer operon [11, 12], and have been reported among S. Infantis strains from several countries in association to the pESI mega plasmid [19–21].
Silver, gold and mercury compounds have been explored over the years in diverse applications for human medicine due to their high toxicity for bacteria [12, 36]. For example, silver can be found in the coating of medical devices like catheters and endotracheal tubes, as part of dental amalgam, and used for the treatment of wounds and prophylaxis of gonococcal ophthalmia neonatorum [12]. Gold has been employed in medical imaging devices, in the treatment of human diseases, and more recently, in the development of nanoparticles, including those with antimicrobial potential [12, 36]. In the environmental and veterinary field, silver has been applied for water treatment, while organic arsenic compounds have been employed as pesticides and feed supplements for food-producing animals [11]. Finally, despite its toxicity, mercury has also been used in agriculture and in human medicine in the form of organic and inorganic compounds, respectively [11, 12].
It is important to mention that the acquisition and transference of resistance genes in bacteria have been largely associated to their presence in plasmids [21, 22]. However, as demonstrated previously [6], the 80 S. Infantis strains studied have a low frequency and diversity of plasmids. These data, when compared with the profiles of efflux pump and heavy metal tolerance encoding genes here detected (S2 Table) suggest that these genes may have a chromosomal location in the strains analyzed. For example, as mentioned above, mercury tolerance genes merR and merT are highly associated to S. Infantis pESI endemic plasmid [19–21]. However, this correlation was not observed in this study (S2 Table), since the strain showing this gene profile has been previously demonstrated not to carry this type of plasmid [6].
Therefore, the presence of genes conferring tolerance to heavy metals employed in the medical, veterinary and environmental field among S. Infantis strains may also warn us on the necessity of increased surveillance of these genetic traits, due to their potential role in the development of resistance against these non-antibiotic compounds and the co-selection of drug-resistance strains of this serovar.
It should also be addressed that additional future research in the field, such as targeted gene expression and phenotypic analyses, could greatly contribute to the elucidation of the role and presumed connections of the efflux pumps and heavy metal tolerance encoding genes here detected with antimicrobial resistance in S. Infantis.
Over the recent years, phylogenetic analyses based on genomic data have demonstrated their importance to epidemiology and tracking of antimicrobial resistance in pathogens of public health relevance [13, 14]. Among the various methods currently available for phylogenetic analyses, cgMLST is based in the allelic variations between the set of 3,002 conserved genes of the Salmonella genus [25], and has been developed and employed as an evolution of the legacy MLST, based in the allelic differences of seven housekeeping genes. While cgMLST has still been less employed in the study of S. Infantis strains [21, 37, 38], MLST has been largely employed over the years and shows a high global predominance of the sequence type ST32 for strains of this serovar [6, 16, 21, 22]. Previously, the dominance of ST32 has also been determined by MLST in the 80 Brazilian S. Infantis strains. In addition, pulsed-field gel electrophoresis (PFGE) showed the presence of three distinct clusters with a high overall similarity above 78%, and the single-nucleotide polymorphism (SNP) analysis of NCBI’s Pathogen Detection assigned the strains to 13 distinct SNP clusters, where seven of these also comprised international isolates and six were composed exclusively of Brazilian isolates [6].
Our Fig 1 did not demonstrate any clear distinction among the strains analyzed regarding their diverse isolation years, sources, materials or Brazilian states. However, some strains harbored combinations of specific profiles of heavy metal tolerance and efflux pump encoding genes were closely grouped in the phylogenetic tree and possessed a strong statistically significant association (Table 2; Fig 1; S1 Fig). Moreover, the results from Fig 2 revealed that the majority of the Brazilian strains possessed a significant proximity among each other and, in contrast, were clearly distinct from international isolates. Only five strains isolated from animals in 2014 (SI124, SI982, SI1143, SI1380, SI1711) and one from human in 2017 (SI4069) were closely located among international genomes (Fig 2).
Furthermore, based on the findings of cgMLST, an extensive diversity of cgSTs was recorded among both Brazilian and international S. Infantis genomes. It is interesting to note that previous studies also performed cgMLST to subtype S. Infantis isolated in European countries, and similarly reported the same high occurrence of MLST’s ST32 in contrast to a high diversity of cgSTs found by using cgMLST in related strains [21, 37, 38].
The results of the present study demonstrate that although, the S. Infantis strains studied isolated in Brazil may present a high overall genomic relatedness and a possible common occurrence in diverse sources, it was also possible to identify distinct strain subtypes in comparison to strains from other countries, as well as the presence of subgroups of strains showing different profiles for heavy metal tolerance and efflux pump encoding genes.
In conclusion, the high prevalence of diverse efflux pump encoding genes, the high occurrence of some heavy metal tolerance genes and the high genomic relatedness of S. Infantis strains sharing equal resistant profiles, regardless of their origin, warn us of the importance of strong surveillance measures to monitor resistance and possible transmission of strains of this serovar among diverse sources in Brazil. Together, these results provided new insights on the genomic diversity of S. Infantis strains circulating in Brazil using WGS.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank to the Kentucky Division of Lab Services, Centralized Lab Facility (Frankfort KY, USA) for performing the whole-genome sequencing of the strains studied, Maria Balkey from FDA/CFSAN for the support during this study, and Ludmilla Tonani for the helpful discussions regarding the statistical analysis.
Data Availability
All genomic sequences analyzed in this study are publicly available for consult and can be accessed through the accession numbers provided in Table 1, S1 Table, in previous publications (Vilela et al. 2021) and/or in other fields in the body of the paper.
Funding Statement
This study was supported by research grants from the FDA/Center for Food Safety and Applied Nutrition (CFSAN) under the supervision of M.W.A. and from the São Paulo Research Foundation (FAPESP; Proc. 2019/19338-8) under the supervision of J.P.F. During the course of this work, F.P.V. was supported by a PhD student scholarship from the National Council for Scientific and Technological Development (CNPq; Proc. 141017/2021-0) and J.P.F. received a Productive fellowship from CNPq (Proc. 304399/2018-3 and 304803/2021-9). This study was financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) – Finance Code 001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization (WHO). Salmonella (non-typhoidal). WHO. 2018. Feb 20 [Cited in 2022 June 05]. Available from: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal). [Google Scholar]
- 2.World Health Organization (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. WHO. 2017. Feb 27 [Cited in 2022 June 05]. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. [Google Scholar]
- 3.U.S. Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, Atlanta, GA: U.S. Department of Health and Human Services, CDC. 2019. [Google Scholar]
- 4.Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, Tate H, et al. CTX-M-65 Extended-Spectrum β-Lactamase-Producing Salmonella enterica Serotype Infantis, United States. Emerg Infect Dis. 2018;24(12): 2284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acar S, Bulut E, Stasiewicz MJ, Soyer Y. Genome analysis of antimicrobial resistance, virulence, and plasmid presence in Turkish Salmonella serovar Infantis isolates. Int J Food Microbiol. 2019;307: 108275. [DOI] [PubMed] [Google Scholar]
- 6.Vilela FP, Rodrigues DDP, Allard MW, Falcão JP. Genomic characterization and antimicrobial resistance profiles of Salmonella enterica serovar Infantis isolated from food, humans and veterinary-related sources in Brazil. J Appl Microbiol. 2022;132(4): 3327–3342. [DOI] [PubMed] [Google Scholar]
- 7.Michael GB, Butaye P, Cloeckaert A, Schwarz S. Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes Infect. 2006;8(7): 1898–914. [DOI] [PubMed] [Google Scholar]
- 8.Blair JM, Richmond GE, Piddock LJ. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9(10): 1165–77. doi: 10.2217/fmb.14.66 [DOI] [PubMed] [Google Scholar]
- 9.Colclough AL, Alav I, Whittle EE, Pugh HL, Darby EM, Legood SW, et al. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020;15: 143–57. doi: 10.2217/fmb-2019-0235 [DOI] [PubMed] [Google Scholar]
- 10.Mourão J, Novais C, Machado J, Peixe L, Antunes P. Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- clones circulating in Europe. Int J Antimicrob Agents. 2015;45(6): 610–6. doi: 10.1016/j.ijantimicag.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 11.Argudín MA, Hoefer A, Butaye P. Heavy metal resistance in bacteria from animals. Res Vet Sci. 2019;122: 132–47. doi: 10.1016/j.rvsc.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 12.Hobman JL, Crossman LC. Bacterial antimicrobial metal ion resistance. J Med Microbiol. 2015;64(Pt 5): 471–97. doi: 10.1099/jmm.0.023036-0 [DOI] [PubMed] [Google Scholar]
- 13.Gilmour MW, Graham M, Reimer A, Van Domselaar G. Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genomics. 2013;16(1–2): 25–30. doi: 10.1159/000342709 [DOI] [PubMed] [Google Scholar]
- 14.Allard MW, Bell R, Ferreira CM, Gonzalez-Escalona N, Hoffmann M, Muruvanda T, et al. Genomics of foodborne pathogens for microbial food safety. Current Opinion in Biotechnology. 2018;49: 224–9. doi: 10.1016/j.copbio.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 15.Brazilian Association of Animal Protein (ABPA). Annual Report 2021. 2021 [Cited in 2022 June 05]. Available from: http://abpa-br.org/relatorios/.
- 16.Monte DF, Lincopan N, Berman H, Cerdeira L, Keelara S, Thakur S, et al. Genomic Features of High-Priority Salmonella enterica Serovars Circulating in the Food Production Chain, Brazil, 2000–2016. Sci Rep. 2019;9(1): 11058. doi: 10.1038/s41598-019-45838-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertani AMdJ, Cunha MPV, de Carvalho E, de Araújo LT, Dos Santos CA, Amarante AF, et al. Genomic characterization of a multi-drug resistant, CTX-M-65-producing clinical isolate of Salmonella Infantis isolated in Brazil. Microbes Infect. 2022: 104972. doi: 10.1016/j.micinf.2022.104972 [DOI] [PubMed] [Google Scholar]
- 18.Kehrenberg C, Cloeckaert A, Klein G, Schwarz S. Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J Antimicrob Chemother. 2009;64(6): 1175–80. [DOI] [PubMed] [Google Scholar]
- 19.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, et al. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol. 2014;16(4): 977–94. [DOI] [PubMed] [Google Scholar]
- 20.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, et al. Comparative Analysis of Extended-Spectrum-β-Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States. Antimicrob Agents Chemother. 2017;61(7): e00488–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alba P, Leekitcharoenphon P, Carfora V, Amoruso R, Cordaro G, Di Matteo P, et al. Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb Genom. 2020;6(5): e000365. doi: 10.1099/mgen.0.000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kürekci C, Sahin S, Iwan E, Kwit R, Bomba A, Wasyl D. Whole-genome sequence analysis of Salmonella Infantis isolated from raw chicken meat samples and insights into pESI-like megaplasmid. Int J Food Microbiol. 2021;337: 108956. [DOI] [PubMed] [Google Scholar]
- 23.Vilela FP, Pribul BR, Rodrigues DDP, Balkey M, Allard M, Falcão JP. Draft Genome Sequences of 80 Salmonella enterica Serovar Infantis Strains Isolated from Food, Environmental, Human, and Veterinary Sources in Brazil. Microbiol Resour Announc. 2021;10(24): e0031321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1): D517–D25. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14(4): e1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricci V, Tzakas P, Buckley A, Piddock LJ. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob Agents Chemother. 2006;50(1): 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari RG, Galiana A, Cremades R, Rodríguez JC, Magnani M, Tognim MC, et al. Expression of the marA, soxS, acrB and ramA genes related to the AcrAB/TolC efflux pump in Salmonella enterica strains with and without quinolone resistance-determining regions gyrA gene mutations. Braz J Infect Dis. 2013;17(2): 125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Y, Huang L, Wu C, Huang J, Hao H, Yuan Z, et al. The Evolution of Fluoroquinolone Resistance in Salmonella under Exposure to Sub-Inhibitory Concentration of Enrofloxacin. Int J Mol Sci. 2021;22(22): 12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59(1): 126–41. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan VB, Rajamohan G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother. 2013;57(9): 4449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miriagou V, Tassios PT, Legakis NJ, Tzouvelekis LS. Expanded-spectrum cephalosporin resistance in non-typhoid Salmonella. Int J Antimicrob Agents. 2004;23(6): 547–55. [DOI] [PubMed] [Google Scholar]
- 32.Cuypers WL, Jacobs J, Wong V, Klemm EJ, Deborggraeve S, Van Puyvelde S. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb Genom. 2018;4(7): e000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong W, Sun Y, Zeng Z. Antimicrobial use and antimicrobial resistance in food animals. Environ Sci Pollut Res Int. 2018;25(19): 18377–84. doi: 10.1007/s11356-018-1852-2 [DOI] [PubMed] [Google Scholar]
- 34.Mijnendonckx K, Leys N, Mahillon J, Silver S, Van Houdt R. Antimicrobial silver: uses, toxicity and potential for resistance. Biometals. 2013;26(4): 609–21. doi: 10.1007/s10534-013-9645-z [DOI] [PubMed] [Google Scholar]
- 35.McHugh GL, Moellering RC, Hopkins CC, Swartz MN. Salmonella Typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet. 1975;1(7901): 235–40. [DOI] [PubMed] [Google Scholar]
- 36.Faa G, Gerosa C, Fanni D, Lachowicz JI, Nurchi VM. Gold—Old Drug with New Potentials. Curr Med Chem. 2018;25(1): 75–84. doi: 10.2174/0929867324666170330091438 [DOI] [PubMed] [Google Scholar]
- 37.Mughini-Gras L, van Hoek AHAM, Cuperus T, Dam-Deisz C, van Overbeek W, van den Beld M, et al. Prevalence, risk factors and genetic traits of Salmonella Infantis in Dutch broiler flocks. Vet Microbiol. 2021;258:109120. [DOI] [PubMed] [Google Scholar]
- 38.Szmolka A, Wami H, Dobrindt U. Comparative Genomics of Emerging Lineages and Mobile Resistomes of Contemporary Broiler Strains of Salmonella Infantis and E. coli. Front Microbiol. 2021;12:642125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All genomic sequences analyzed in this study are publicly available for consult and can be accessed through the accession numbers provided in Table 1, S1 Table, in previous publications (Vilela et al. 2021) and/or in other fields in the body of the paper.