Abstract
Microbial diversity can restrict the invasion and impact of alien microbes into soils via resource competition. However, this theory has not been tested on various microbial invaders with different ecological traits, particularly spore-forming bacteria. Here we investigated the survival capacity of two introduced spore-forming bacteria, Bacillus mycoides (BM) and B. pumillus (BP) and their impact on the soil microbiome niches with low and high diversity. We hypothesized that higher soil bacterial diversity would better restrict Bacillus survival via resource competition, and the invasion would alter the resident bacterial communities’ niches only if inoculants do not escape competition with the soil community (e.g. through sporulation). Our findings showed that BP could not survive as viable propagules and transiently impacted the bacterial communities’ niche structure. This may be linked to its poor resource usage and low growth rate. Having better resource use capacities, BM better survived in soil, though its survival was weakly related to the remaining resources left for them by the soil community. BM strongly affected the community niche structure, ultimately in less diverse communities. These findings show that the inverse diversity-invasibility relationship can be valid for some spore-forming bacteria, but only when they have sufficient resource use capacity.
Keywords: diversity-invasion relationship, invasion impact, microbial invasion, resource competition, soil functioning
This study evaluates the diversity-invasibility relationship of spore-forming Bacillus spp, and the impact of such invasion on the soil bacterial communities' niches.
Introduction
Microbial invasion is a common phenomenon, driven by natural dispersal and increased human activities such as shipping, trade, and farming (Chapman et al. 2017, Hallegraeff 1998, Mawarda et al. 2020), with significant consequences for the ecosystems (Thakur et al. 2019). In particular, microbial releases into the soil are an example of human-induced invasions, part of ecological approaches that increasingly happen in agroecosystems (de Souza et al. 2015, Florio et al. 2017). Research on microbial invasions has, in the recent years, increasingly addressed the traits of the invader and resident communities determining invasion success (De Roy et al. 2013, Eisenhauer et al. 2013, Hol et al. 2016, Mallon et al. 2015). This has highlighted key factors influencing the fate of microbial invasions, in particular the role of native community diversity and resource competition between the invader and the resident community (Mallon et al. 2015, Xing et al. 2020). The importance of these two factors is consistent with the basic principle of the diversity–invasibility relationship (DIR), whereby more diverse communities resist invasion better than less diverse ones. The theory suggests that as the species richness and evenness of the community increase, the community can better exploit and compete for available resources, hence leaving fewer resources available for the invader to establish (Mallon et al. 2015, Tilman 1977). The negative DIR has been proven in previous studies for Escherichia coli invasion (Mallon et al. 2015, van Elsas et al. 2012). In reality, the DIR is more nuanced, ranging from neutral to positive (Clark and Johnston 2011, Davies et al. 2007, Ferreira et al. 2021). Previous microbial invasion studies in soil have mainly used Gram-negative bacteria, such as Serratia liquefaciens (Jousset et al. 2011), Listeria monocytogenes (Vivant et al. 2013), Pseudomonas putida (Eisenhauer et al. 2013), and Escherichia coli (Mallon et al. 2015, 2018, van Elsas et al. 2012) as invader models. Never before has the DIR been examined in Gram-positive and spore-forming bacteria such as Bacillus spp., even though these are commonly introduced into soils as plant growth-promoting bacteria (Borriss 2015, McSpadden Gardener 2007). The ability of Bacillus to sporulate might allow it to escape the negative DIR, but this has never been tested.
While soil community diversity and resource use can control the survival and invasibility of a bacterial inoculant, bacterial invasion can, in turn, affect the composition and functioning of the soil native community (2013, Mallon et al. 2018, Mawarda et al. 2022, Trabelsi and Mhamdi 2013). A recent meta-analysis by Mawarda et al. (2020) showed that among 108 studies of microbial invasions, where the impact on the soil microbial community structure was measured, 71 revealed the inability of these communities to return to their initial state. However, it remains difficult to link changes in microbial community composition to functional consequences (Louca et al. 2018, Souza et al. 2016). Indeed, despite taxonomic variability, the functioning of microbial communities can remain stable and vice versa (Louca et al. 2017, Vanwonterghem et al. 2016, Wittebolle et al. 2008). Therefore, evaluating how a bacterial invader modifies the taxonomic diversity of the resident soil community is not sufficient to predict how it can change the soil microbiome's functioning. One possibility is to examine the impact of microbial invasion directly on the functionality of the resident soil community by targeting changes in their resource utilization profiles through high throughput phenotyping (metaphenomics) using the Biolog Gen III Plate. This approach has been widely used to measure microbial community functioning (Doolittle and Zhaxybayeva 2010). Specifically, Salles et al. (2009) showed that this method predicted about 70% of soil bacterial functioning, whereas the species richness of communities predicted only 14%. Moreover, this approach has been used to investigate whether resource competition drives invasion resistance (Mallon et al. 2015, Xing et al. 2020).
The purpose of this study was to examine (i) whether the survival of two spore-forming Bacillus spp. was determined by the diversity of soil bacterial community, particularly the community's resource utilization pattern compared to the inoculants’, and (ii) whether invasion by these Bacillus spp. affects the metabolic potential of the soil native community. We hypothesized that higher soil bacterial diversity would better restrict Bacillus survival via resource competition and that invasion would alter the resident bacterial communities' niche. However, an alternative hypothesis is that these effects would not hold for spore-forming inoculants if sporulation allows them to escape competition with the soil community. To test our hypotheses, we released soil-derived Bacillus mycoides M2E15 (referred to as BM) and Bacillus pumilus ECOB02 (referred to as BP), known to promote the growth of potato and grass, to native soil microcosms with either high or low diversity levels. Our results allow testing to what extent the fate of introduced spore-forming Bacillus in soil follows the negative DIR and whether this type of inoculant impacts native community niche as previously observed for non-spore-forming bacterial inoculants.
Materials and methods
Experimental set-up and design
The soil used for creating diversity gradients and gamma sterilized soil matrix is a sandy loam soil, sampled in Leeuwarden, Friesland, The Netherlands. To create a diversity gradient of soil microbiomes, sterile water and non-sterile soil were mixed and shaken vigorously in a ratio of 1 : 2 to detach and extract as many microbes as possible from the soil. This suspension was then 1 : 10 serially diluted in sterile water up to the 10−6 factor. Soil suspensions of 16 ml from 10−1, 10−3, 10−6 dilutions were aseptically transferred and homogenously stirred to inoculate microcosms containing 60 g of previously gamma sterilized soil (50kGy). This brought soil moisture to 65% of the water holding capacity (WHC). The microcosms were then incubated for 60 days at room temperature to allow microbial recolonization of soil. Previous studies also showed that 60-day incubation is enough to establish microbial network that avoids microbial invasions (Mallon et al. 2015, Mawarda et al. 2022, van Elsas et al. 2012). The soil moisture was checked and maintained regularly by measuring the weight of each microcosm and replenishing the water to maintain its level at 65% of the WHC until the first inoculation with a Bacillus strain.
Before the Bacillus was inoculated, we checked that the culturable bacterial cells per gram of soil in microcosms had roughly reached the same abundance across the diversity levels after 60 days of incubation. This was done to ensure that the impact we observed was solely attributed to different diversity levels and avoid the bias resulting from different community sizes. The colonies were enumerated 4 days after dilution plating on Trypticase Soy Agar (TSA) and incubation at 28°C. After 60 days, total culturable bacteria reached the same level of abundance (Table S1; mean of 9.0 log CFU/g soil), the rifampicin-resistant strains of BM and BP (2.9 ml each) were introduced into the relevant soil microcosms at a level of 1 × 107 CFU per gram soil and stirred to ensure homogeneity. The fitness of the rifampicin-resistant strains is the same as the wild-type strains (Mawarda et al. 2022). The uninvaded controls were created by adding 2.9 ml of sterile water to soil microcosms in each diversity level. The inoculations raised soil moisture on each microcosm to 75% of WHC. The WHC value was kept constant by replenishing the water until the end of the experiments.
Destructive samplings were initially planned for days 0, 3, 10, 15, 30, and 90 for the microcosms invaded by BM or BP. However, the abundance of each Bacillus strain was below the detection limit (1 log CFU/g soil) three days after inoculation. We speculate that this was caused by a decrease in soil pH (to 4.9) upon inoculation since the pH of non-inoculated controls remained stable. For each experiment (i.e. either BP or BM), we, therefore, pooled invaded soils from all the remaining replicate jars together 7 days after the first inoculation (p.f.i) attempt. We then adjusted the soil pH to 7.0 by adding sterile 1 M Ca(OH)2, and transferred 39 g of soil at 65% WHC from each treatment to a new sterile jar. Meanwhile, the pH of uninvaded controls was not adjusted since the pH was stable at 7. The soil microcosms were incubated at room temperature (∼22°C) for 2 weeks. After verifying pH stability, either BP or BM were reinoculated into soil microcosms (at days 24 and 34 p.f.i, respectively) using the procedure described above.
Destructive soil sampling was done on days 0, 3, 34, 35, 37, 63, and 92 p.f.i for the BM experiment and on days 0, 3, 24, 25, 27, 56, and 84 p.f.i for the BP experiment. We conducted two separated experiments with different sampling timepoints based on the inoculant survival patterns observed from our previous study (Mawarda et al. 2022). The uninvaded control was sampled on day 0 and day 28. This comprised 144 soil microcosms (i.e. 3 dilution levels × 2 invaders × 7 sampling dates × 3 replicates and 3 dilutions for uninvaded control x 2 sampling dates x 3 replicates).
Determination of the initial soil community diversity
To determine the diversity of the resident bacterial community for each dilution on the day before the first inoculation, bacterial species richness was quantified by Illumina sequencing at the University of Minnesota Genomic Centre (UMGC) (Minneapolis, MN, USA). The sequencing was done on the Illumina Miseq (Illumina, San Diego, California) using 2 × 300 base paired-end reads and targeting the V4 region of the 16S rRNA gene (forward primer 16S-515F: 5′-GTGCCAGCMGCCGCGGTAA-3′; reverse primer 16S-806R: 5′-GGACTACHVGGGTWTCTAAT-3′) with a dual indexing method (Gohl et al. 2016). DNA was extracted from 0.5 g of soil using the DNeasy Powersoil Kit (Qiagen, Hilden, Germany) for each sampling date according to the manufacturer's instructions. DNA extract was quantified using the NanoDrop 2 000 spectrophotometer (Thermo Scientific, USA) and stored at −20°C before sequencing. The Quantitative Insights Into Microbial Ecology (QIIME 2, https://qiime2.org) software was used to process and analyze 16S rRNA gene sequences (see supplementary document A for detailed protocol).
Amplicon sequences data were then used to quantify bacterial diversity for each dilution. This was carried out in R v4.2.0 using the Phyloseq package. To minimize the effects of sequencing depth between samples when estimating species richness, 16S rRNA gene sequences of our samples were rarefied to a depth of 6002 sequences and 5775 sequences per sample for the BM and BP experiment, respectively. These rarefied data were used to calculate species richness and evaluate the differences between diversity treatments, using ANOVA and post-hoc Tukey Nemeyi tests.
Monitoring the survival of bacillus spp.
The total population of Bacillus was tracked via serial dilution plating on TSA medium supplemented with rifampicin (50 µg/mL) and cycloheximide (400 µg/mL). The abundance of the spores of each Bacillus was tracked by heating each diluted sample for 20 min at 80°C and plating on TSA containing the same antibiotics. Plates were incubated at 28 °C for 48 h. The survival of BP was tracked on days 0, 3, 24, 25, 27, 56, and 84. The survival of BM was monitored on days 0, 3, 34, 35, 36, 63, and 92. ANOVA was used to examine BP and BM survival differences stemming from time and diversity as factors.
Metabolic profile measurement and data analysis
To measure microbial communities' metabolic profile (i.e. resource use capacity), 3 destructive samplings were designed for BP at day 0 before the first inoculation, and 56 and 84 days p.f.i. For BM, this was done at day 0 (before the first inoculation) and 63 and 92 days p.f.i. The metabolic profiles of soil microbial communities and the Bacillus invaders were quantified by measuring their metabolic activity using the Biolog GEN III microtiter plate based on Biolog's high cell density phenotypic microarray protocol (Biolog, Hayward, California, USA). A detailed protocol can be found in supplementary document B. The experiments were conducted in triplicate. We did not measure the metabolic profile of non-invaded soil communities over time because previous studies have shown that incubation time had no impact on niche breadth and niche structure of resident bacterial communities (Mallon et al. 2018, Xing et al. 2020). Still, we measured the non-invaded community profile on day 28 and day 0 to verify this.
The absorbance was measured using a spectrophotometer at 590 nm and 750 nm at timepoints: 0, 12, 15, 18, 21, 24, 36, 39, 42, 45, and 48 h after adding the sample to the Biolog GEN III plate. The OD from each well was calculated using the following formula:
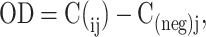 |
(1) |
where C represents the difference OD590–OD750 of well i, at time j and C(neg) represents the difference OD590–OD750 of the negative control well at time j. For wells with C- C(neg) < 0.1, the value was recorded as 0. The OD from each well was plotted against time to calculate the maximum slope, indicating the maximum resource utilization speed. This was carried out in R 1.2.5019 using the pipeline package. The area under the curve (AUC) from each well was calculated using the trapezoidal rule. Each AUC was normalized to 0–1 by dividing the value by the maximum value from the measurements across the 71 carbon sources in the plate. The data with AUC values were used to quantify the total amount of resource utilization (i.e. the sum of AUC), niche breadth (number of resources being used), and niche structure (matrix of the AUC for each resource). This was carried out in R v4.2.0 using the Vegan package. The remaining niche for the invader (i.e. the resources unused by the resident community and suitable substrates for the invader) was also quantified by subtracting the resource utilization of resident communities from that of the invader. When the values were negative for a given resource, the value was set to zero, indicating that the resident community could utilize the resource better than the invader (no remaining niche for the invader). Positive values indicated the resident community was less competitive than the invader on the resource, hence providing a vacant niche for the invader. The community exclusive niche (i.e. carbon resources exclusively used by the resident community) was also quantified by eliminating the data for all the carbon resources used by the invader. The community's exclusive niche breadth and structure were quantified in the same way as above.
T-test, Kruskal Wallis, pairwise Wilcoxon test, ANOVA and post-hoc Tukey Nemeyi tests were carried out to evaluate differences in metrics between treatments and their temporal changes. Variation in niche structure was assessed by calculating Bray-Curtis dissimilarity and visualized via a Principle Coordinates Analysis (PCoA). Temporal patterns of beta diversity on each diversity treatment were evaluated with PERMANOVA.
Results
Diversity of the soil bacterial community at the start of the experiment
The ASV richness of the soil bacterial community in the BM and BP experiments was significantly different between treatments (ANOVA, P<0.0001) (Fig. S1A&B). In both experiments, this value was significantly lower for the 10−6 than for the other treatments (Tukey's test, P<0.05), while ASV richness values were similar in the 10−3 and 10−1 treatments (Tukey, P>0.05). The soil bacterial community structure (16S rRNA gene sequencing), determined by Bray-Curtis dissimilarity, was also similar in the 10−1 and 10−3 treatments (pairwise-Adonis, P > 0.05), while the community structure of the 10−6 treatment was significantly different from that in the other treatments (PERMANOVA, P<0.0001; pairwise-Adonis, P < 0.05). Therefore, for all the analyses below, we grouped the 10−1 and 10−3 dilutions as the High-diversity treatment, while the 10−6 dilution was considered to represent the low-diversity treatment.
Survival of the introduced bacillus species
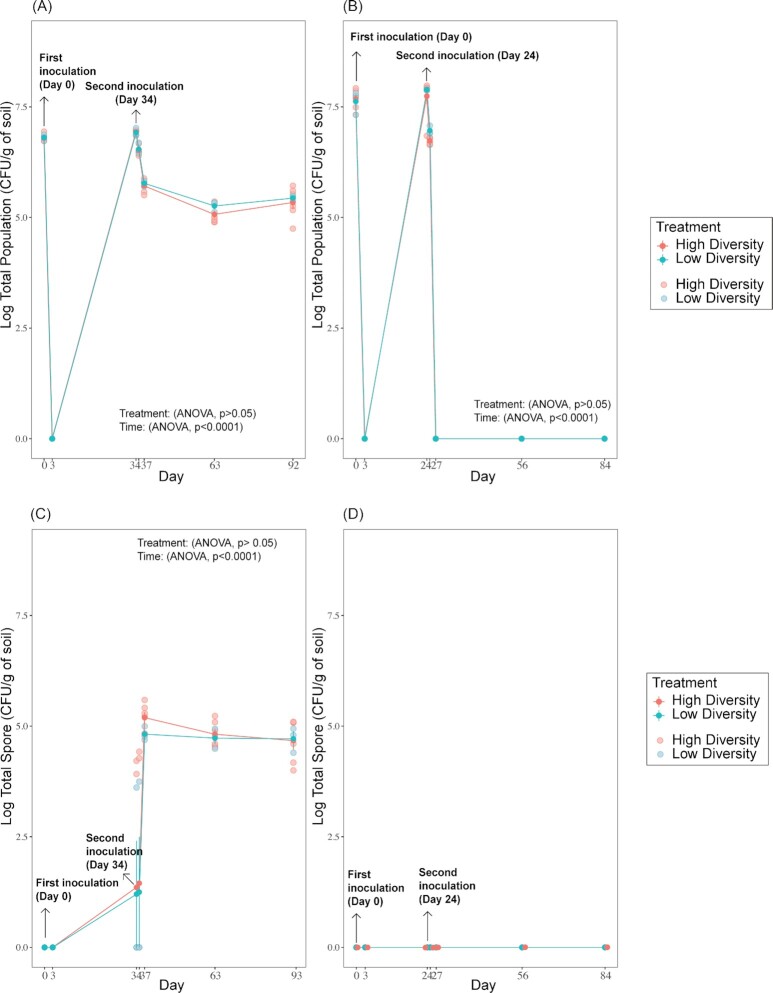
Concerning the inoculant survival dynamics, the total population of these invaders fell below the detection limit, meaning they were less than 1 log CFU/g soil 3 days post first inoculation (p.f.i; Fig. 1A). In the BM experiment, the second inoculation (at day 34 p.f.i) led to a successful establishment of the invader, detected at an abundance of 5–6 log CFU/g soil from day 35 to day 92 in both diversity treatments (Fig. 1A). Regarding the BM spore population, its abundance increased to around 1–2 log CFU/g soil after the first inoculation and stabilized to around 4–5 log CFU/g soil after the second inoculation in both high and low-diversity treatments (Fig. 1C: ANOVA, p(time)<0.0001).
Figure 1.
Survival of total population of B. mycoides M2E15 (A) and B. pumilus ECO-B-02 (B) and total spore number of B. mycoides M2E15 (C) and B. pumilus ECO-B-02 (D) for high and low community diversity treatments over time. Values represent the log CFU of the total population per gram of soil. Bars represent the standard error of the mean. Full symbols represent the average, whereas the light symbols indicate the values per replicate.
The survival of BP was poorer than BM as its total abundance was always below the detection limit of 1 log CFU/g soil after the first and second inoculations except at day 25 p.f.i (one day after the second inoculation) (Fig. 1B). The spores of BP were not found after the first and second inoculation events (Fig. 1D).
Relationship between inoculant survival and species richness of the initial soil community
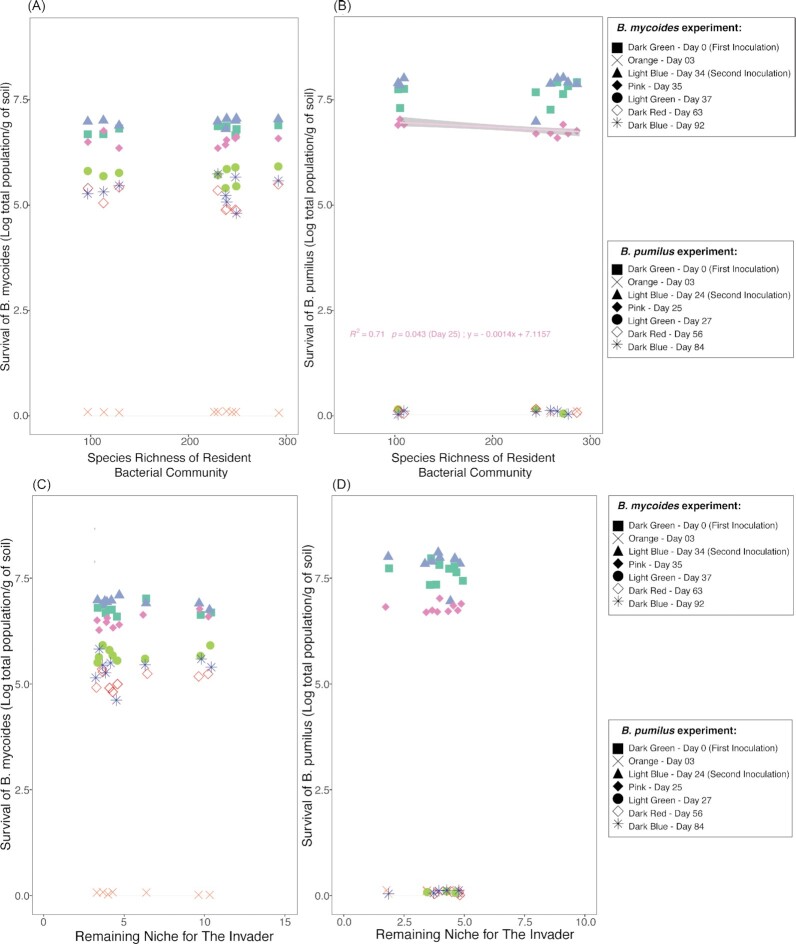
No relationship was observed between the survival of both Bacillus invaders and the species richness of the resident bacterial community at each sampling date (Fig. 2A-B). Even though there was a significant negative correlation on Day 25 (1 day after the second inoculation) in the BP experiment (Fig. 2B, p = 0.043), the slope was very low (less than 0.001). This indicates that the effect of species richness was (mathematically) too small to affect the survival of BP. No relationship was observed between the spore population for both Bacillus invaders and the resident bacterial community's species richness at each sampling day (data not shown).
Figure 2.
Correlations between the survival of B. mycoides M2E15 (A) and B. pumilus ECOB02 (B) and the species richness of the resident bacterial community in soil were observed on different sampling days (i.e. different symbols/colors). Correlation between the survival of B. mycoides M2E15 (C)and B. pumilus ECOB02 (D) and the remaining niche available for these inoculants, as observed at different sampling days (i.e. different symbols/colors). Values represent the log CFU of the total population of the inoculated bacteria per gram of soil.
Relationship between bacillus spp survival and initial soil community niche
To examine whether resource competition could govern the invasion of soil by the Bacillus spp., we calculated the remaining niche left by the soil microbiomes for the invader and the amount and speed of resource utilization by the invader compared to the community. For both Bacillus strains, invader abundance was never correlated with the remaining available niche (Fig. 2C-D). However, for BM, a relationship was observed at day 37 when focusing only on the 10−3 and 10−6 treatments.
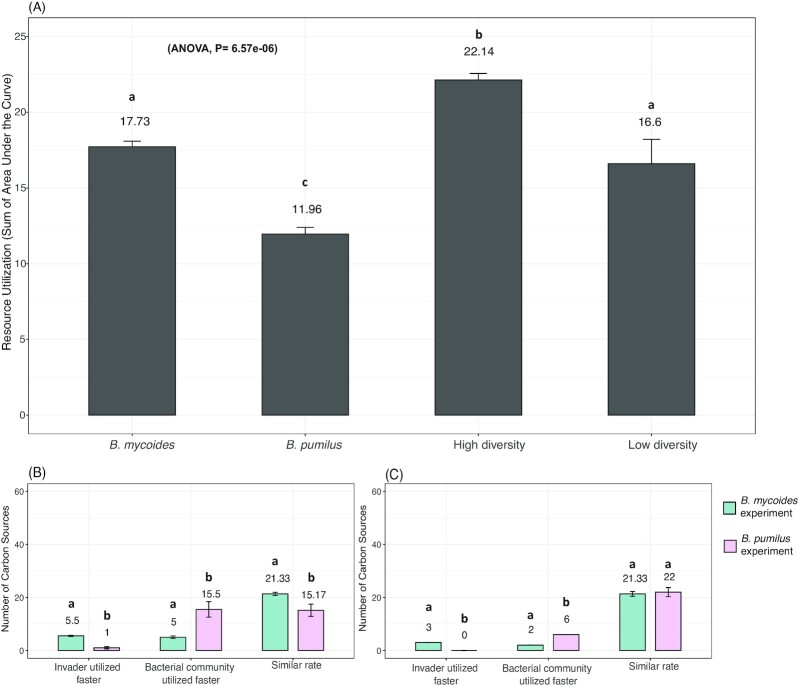
In addition, as depicted in Fig. 3A, the total resource utilization (i.e. the sum of AUC) was much lower for the BP than for the BM or soil community with low or high diversity (Tukey, P<0.01). Meanwhile, the high-diversity treatment community utilized more resources than the others (Tukey, P<0.01). The total resource utilization in the low-diversity treatment was similar to BM (Tukey, P>0.05; Fig. 3A). More specifically, the speed of resource utilization in the overlapping niches between invaders and communities (i.e. carbon sources used by each invader and the resident soil bacterial communities) was compared between BM and BP experiments. In both the high (Fig. 3B) and low-diversity treatments (Fig. 3C), there were nearly no carbon sources on which the BP invader could utilized faster C sources than the soil communities. In contrast, the number of resources used more quickly by the invader than the soil communities was higher for BM (T-test in high and low-diversity treatment, P<0.001). In addition, there were more carbon sources utilized faster by the bacterial community than the invader in the BP experiment than in the BM experiment (T-test for either high or low-diversity treatment, P<0.001). Finally, we also observed that BM was able to grow faster (at 0.068 h−1) in LB broth than BP (at 0.038 h−1) (Fig. S2, T-test, P<0.0001).
Figure 3.
Total resource use of each bacterial invader and the soil resident community before the invasion (A). The number of carbon sources used faster by the bacterial inoculants, faster by the soil community, or similarly by the inoculant and the resident community, distinguishing the B. mycoides experiment (blue) and the B. pumilus experiment (pink), in high (B) and low diversity (C)treatments. Bars represent the standard error of the mean. Different letters above the bars indicate significant differences within each panel.
Changes in soil community niche following invasion by bacillus
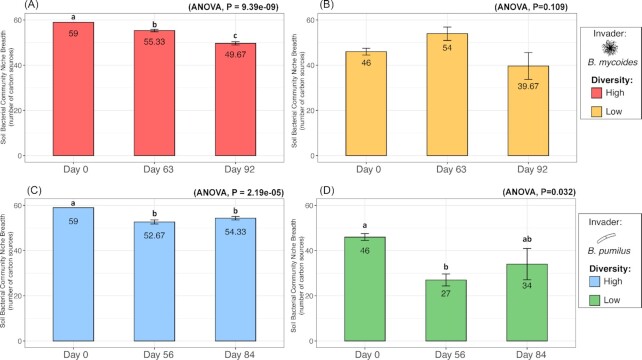
The full array of 71 carbon sources present in Biolog plates was examined to analyze the shift in the community niche breadth and structure induced by Bacillus invasion. The invasion by BM led to a significant decrease in the soil community niche breadth in the high-diversity treatment, the effect increasing with time (Fig. 4A: ANOVA, P<0.001), whereas niche breadth in the low-diversity treatment remained unchanged (Fig. 4B: ANOVA, P>0.05). Meanwhile, in the BP experiment, the invader decreased the niche breadth in both the high and low-diversity treatments (Fig. 4C for high-diversity treatment: ANOVA, P<0.001; Fig. 4D for Low-diversity treatment: ANOVA P<0.05). However, the impact was transient in the low-diversity treatment, and the initially reduced niche breadth returned to its pre-invasion state at Day 84 p.f.i (Tukey, P<0.001). The niche breadth of non-invaded control remained the same over time (Fig. S4a, T-test, P = 0.999). The results and patterns observed in total community niche breadth for the BM and BP experiments were similar to those observed in community exclusive niche breadth (not shown).
Figure 4.
Temporal changes in the total soil community niche breadth during the B. mycoides M2E15 (A, B) and B. pumilus ECO-B-02(C, D) invasion experiments for the high (A, C) and low diversity (B, D) treatments. Bars represent the standard error of the mean, and different letters above the bars indicate significant differences within each panel.
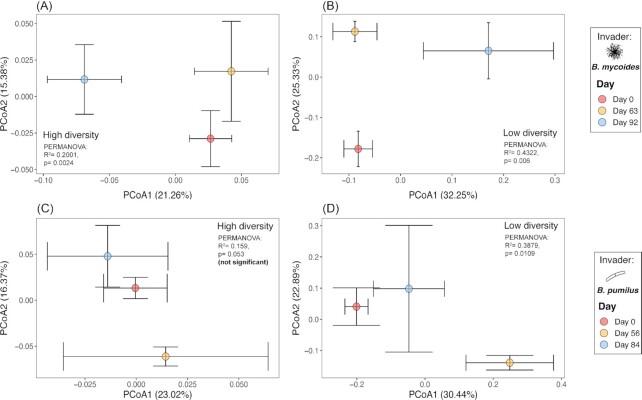
To further analyze the niche structure of the soil bacterial community in response to Bacillus invasion, we examined the Bray-Curtis dissimilarity of metabolic activity at each diversity level. In the BM experiment, the invader shifted the niche structure for both diversity levels (Fig. 5A, High-diversity treatment: ADONIS time, R2 = 0.20, P = 0.02; Fig. 5B, Low-diversity treatment: ADONIS time, R2 = 0.43, P = 0.006), and the niche structure shift was maintained until the end of the experiment. Meanwhile, the invasion by BP significantly altered the niche structure only in the low-diversity treatment (Fig. 5D ADONIS time, R2 = 0.36, P = 0.011); the niche structure of more diverse bacterial communities showing resistance to BP invasion (Fig. 5C, ADONIS time, P = 0.053). However, the impact of the low-diversity treatment was transient. The niche structure at day 56 p.f.i mainly shifted along the first axis (30.4% of variation) from the pre-invasion state and then shifted back to the initial state at Day 84 (Fig. 5D). The niche structure of non-invaded control remains the same over time (Fig. S4b, ADONIS time, R2 = 0.0004, P = 0.999).
Figure 5.
Temporal variation of the soil community niche structure, depicted by PCoA plot of Bray–Curtis dissimilarity, for the B. mycoides(A and B) and B. pumilus(C and D)invasion experiments, for the high (A, C) and low diversity (B, D) treatments. For each panel, the effect of invasion on the community niche structure, determined by PERMANOVA, indicated with the R2 and p values when significant. Each point corresponds to the centroid computed from the replicates and is presented with the standard errors (error bars) for the first two PCoA axes.
The results and patterns observed for the total community niche structure for BM and BP experiments were similar to those observed in the community exclusive niche structure (Fig. S3).
Discussion
Spore-forming bacterial invaders do not comply with the negative diversity-invasion relationship
Studies in microbial invasion highlight the importance of the resident community's diversity as the key factor that explains possible resistance to the invasion (van Elsas et al. 2007, 2012). The DIR suggests that highly diverse communities are less prone to invasion than less diverse communities (Elton 2020). In the framework of microbial invasion, this notion is tightly linked to biotic resistance imposed by the availability of resources and the ability of more diverse communities to exploit those resources better and hence compete with the invaders (Catford et al. 2009, Fargione and Tilman 2005, van Elsas et al. 2012). Our results show that these concepts do not apply well to the invasion of soil communities by spore-forming Gram-positive bacteria (i.e. two soil-derived Bacillus species, BM and BP) since the survival of these invaders was not related to the soil community species richness, nor the remaining niche left by the soil community to the invader. This contrasts with our previous work with E. coli, which showed that competition between the soil community and invader increases as community species richness increases, reducing the remaining niche available to the invader and ultimately decreasing the inoculant survival in soil (Mallon et al. 2015). The sporulation feature of Bacillus may explain the absence or poor applicability of the negative DIR concept in our study. A recent study by Blath and Tóbiás (2020) showed that dormancy is an effective survival strategy for spore-forming microbial invaders to withstand unfavorable conditions, increasing their survivability when facing resource scarcity, competitive pressure and extreme environmental fluctuations. Under oligotrophic conditions, the fitness of these microbial invaders may be mainly driven by their ability to stay alive rather than reproduce and grow. In our lab, for the BM experiment, resources were limited enough to trigger sporulation regardless of the differences in the remaining niche available across diversity treatments. Therefore, the survival of this type of spore-forming bacteria in soil may be largely independent of the diversity of recipient communities. This does not jeopardize the rationale of the DIR being related to competition for resources. Spore-forming bacteria such as BM may largely avoid competition with the resident community, thanks to their capacity to produce spores.
However, this did not apply to BP, which did not sporulate and quickly ‘vanished’ below the detection limit. A disclaimer should be placed here in that we cannot fully address how the introduced populations behaved in terms of establishment, survival, and platability (the ability to form colonies in agar plate) when taken out of the soil onto a rifampicin-containing growth medium. In other words, the exact fate of the introduced cells, as to their physiological status (injured, dying, temporarily viable-but-nonculturable), remains unknown. This can be explained by examining the nature of dormancy itself (Hutchison et al 2014) since switching to a metabolically inactive state (sporulation) requires resources (Atrih and Foster 2001, Bressuire-Isoard et al. 2018) and hence leads the invader to compete with the resident bacterial communities, at least in a transient manner (Blath and Tóbiás 2020). Our data showed that BP has much less competitive capacity than BM, which is exemplified by BP's inability to outcompete with even less diverse communities on all resources and a lower growth rate on LB medium. It is thus very likely that BP experienced stronger competition in the soil system than BM following the invasion. This might explain why BP could not gain enough resources to sporulate and disappear quickly from the soil, regardless of the diversity level.
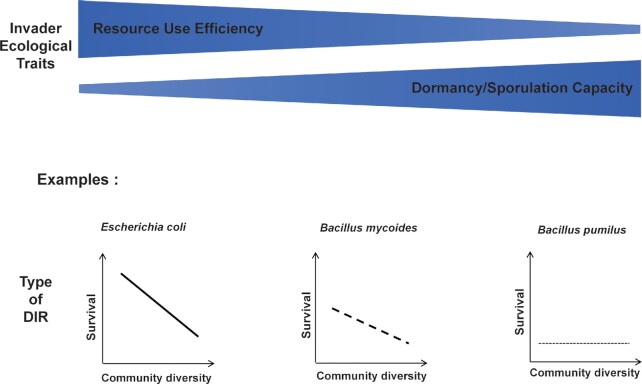
Overall, our findings align with previous studies reporting that the DIR varied from neutral to positive (Belote et al. 2008, Ferreira et al. 2021, von Holle 2013). Our results highlight that understanding DIR implies that one must distinguish various invaders’ lifestyles and evolutionary strategies. The summation of conceptual analysis about this finding is available in Fig. 6.
Figure 6.
Conceptual synthesis depicting the outcome of diversity-invasibility relationship which depends on the ecological strategies and functional traits of each invader (i.e resource use efficiency and dormancy/sporulation capacity).
The bacillus invader competitiveness influences their impact on the soil community functioning
The results above deduce that Bacillus invasions could alter the metabolic potential of a soil resident bacterial community, regardless of invader survival. This result corroborates previous findings that, despite invader death and defeat, unsuccessful invasions caused tangible changes in the diversity (Amor et al. 2020, Buchberger and Stockenreiter 2018) and functionality of resident soil communities (Derrien and van Hylckama Vlieg 2015, Mallon et al. 2018). Indeed, the microbiome modulation is one of the most important responses following the introduction of microbial inoculants (Berg et al. 2021, Mawarda et al. 2020). In particular, numerous in situ studies have shown that Bacillus inoculants could modify the indigenous soil and plant microbiome structure and functionality (Erlacher et al. 2014, Kröber et al. 2014). However, our result showed that this impact depends on the ecological ‘strength’ of the invader. In line with our previous studies (Mawarda et al. 2022), the invasion by BM led to a more significant impact, as they had a higher competitive ability which allowed them to establish and survive in the soil. In contrast, BP could hardly compete efficiently for resources with the resident soil community. Indeed, the BP invasion did not lead to major changes in the soil community niche structure, with no significant difference for high-diversity communities and only transitory change for low diversity communities.
This finding is similar to what was found in previous studies by Mallon et al. (2018) and Xing et al. (2020), who found niche shifts for the resident bacterial communities upon E. coli invasion. Since BM could better compete for some resources than the resident community, we speculate that resident taxa using the same carbon sources as the invader might have been outcompeted, leading to the decline of these taxa's abundance, thereby reducing these taxa's abundance in the niche breadth of resident bacterial communities. Such a decline in diversity following invasion has been observed in micro- (Mawarda et al. 2020) and macroorganisms (Carboni et al. 2021, Sapsford et al. 2022), which often leads to the changes in their functional diversity (Mallon et al. 2018, Matsuzaki et al. 2016, Souza et al. 2021, Xing et al. 2020). This type of footprint of microbial invaders on soil communities has to be accounted for to fully appreciate the impact of invasion on soil functioning and services (Liu et al. 2022).
In the BP experiment, the niche structure of the invaded communities in the low-diversity treatment was first altered following the invasion and then returned to their initial state. This was likely due to the recovery and regrowth of previously impacted taxa, reoccupying the niche that the invader just transiently occupied. Consistently, using successional analysis, it was shown that some decreasing taxa following invasion by Bacillus reappeared in later stages after the invaders’ abundance waned off (Mawarda et al. 2022). Similarly, Orlewska et al. (2018) found that the niche shift following Pseudomonas putida introduction restored to its initial balance on day 90 after the invasion. Meanwhile, the invasion by BP showed more prominent impact on the niche breadth than the niche structure, as they significantly reduced the niche breadth in both diversity treatments. Our previous study showed that even though BP did not survive, their arrival could still reduce the abundance of some copiotrophic bacteria that did not recover after invasion (Mawarda et al. 2022). These decreasing taxa may explain the reduction of communities’ niche breadth following the BP invasion.
However, it is important to mention that measuring microbial community functionality using the Biolog technology has its own limitations, especially when dealing with soil and other natural environments. A previous study by Smalla et al. (1998) showed that only copiotrophic bacteria growing under such inoculation regime and incubation conditions, contributed to the substrate utilization patterns. That is to say the responding organisms on the plate may not reflect the whole in-situ dynamics of the soil sample. However, despite this drawback, Biolog technology is still an effective way to gauge the potential resource use and metabolic structure of microbial communities (Borglin et al. 2012, Mackie et al. 2014). Numerous studies have proven that this approach is able to differentially characterize microbial communities in the artic soil (Kumar et al. 2016), rhizosphere (Velasco et al. 2009), wastewater treatment (Jałowiecki et al. 2017), even in the case of multi drug resistance bacteria (Blanco et al. 2018).
Taken together, our findings reinforce the notion that the response of resident microbial communities to microbial invaders depends on the type and magnitude of the invasion disturbance, which is related to the ecological strategies and functional traits of each invader (Fig. 6). All the previous studies on microbial invasion in the soil only used Gram-negative bacteria as invader models. Meanwhile, Bacillus is Gram-positive and capable of changing its physiology from an active to dormant state depending on the availability of nutrients. These features likely explain why we found different invasion patterns and biodiversity-stability relationships for Bacillus invaders compared to previously studied invaders. In terms of practical application, our results call for more systematic investigations of microbial traits (Krause et al. 2014) when selecting microbial inoculants, going beyond the traits directly linked to e.g. plant-inoculant interactions and including traits determining the outcome of competition with the resident community. Specifically, our results suggest that the ability of Bacillus inoculants to outcompete soil resident communities on a sufficient amount of resources is essential for making inoculation successful or not. This requires profiling inoculant candidates based on these capacities to avoid the use of poor competitors. Furthermore, since commercial products of Bacillus inoculants are practically applied in a spore-form, future experiment should introduce Bacillus strains as spores to examine whether the data would corroborate or reject the current findings. Finally, it could be tested to what extent resource pulses (i.e. punctual additions of resources usable by the inoculant) and the presence of plants could improve Bacillus survival in soil, and to what extent these could lead to the finding of a more classical, negative DIR.
Supplementary Material
ACKNOWLEDGEMENTS
We thank EcoStyle for providing B. pumilus ECOB02, Jan Spoelder and HLB for providing B. mycoides M2E15 and Moniek Bloemsma for helping with the experiment. This work was supported by the Indonesia Endowment Fund for Education (LPDP, Departemen Keuangan, Republik Indonesia) scholarship to P.C.M. J.F.S was financed by the ERA-NET Cofund SusCrop project potatoMETAbiome (Grant No 771134), supported by EU Horizon 2020 research and innovation program and NWO, part of the Joint Programming Initiative on Agriculture, Food Security and Climate Change (FACCE-JPI). XLR was supported by the IMMINENT project funded by the French Agence Nationale pour la Recherche ANR (project ANR-20-CE02-0014-01).
Contributor Information
Panji Cahya Mawarda, Microbial Community Ecology Cluster, expertise group GREEN, Groningen Institute of Evolutionary Life Sciences (GELIFES), University of Groningen, Nijenborgh 7, 9747 AG, Groningen, The Netherlands; Research Center for Environment and Clean Technology, National Research and Innovation Agency Republic of Indonesia (BRIN), Komplek LIPI, Jalan Sangkuriang No 21, Bandung 40135, Indonesia.
Cyrus A Mallon, Microbial Community Ecology Cluster, expertise group GREEN, Groningen Institute of Evolutionary Life Sciences (GELIFES), University of Groningen, Nijenborgh 7, 9747 AG, Groningen, The Netherlands.
Xavier Le Roux, INRAE, CNRS, Université Lyon 1, Université de Lyon, VetAgroSup, Laboratoire d'Ecologie Microbienne LEM, UMR 1418 INRAE, UMR 5557 CNRS, 69622 Villeurbanne Cedex, France.
Jan Dirk van Elsas, Microbial Community Ecology Cluster, expertise group GREEN, Groningen Institute of Evolutionary Life Sciences (GELIFES), University of Groningen, Nijenborgh 7, 9747 AG, Groningen, The Netherlands.
Joana Falcão Salles, Microbial Community Ecology Cluster, expertise group GREEN, Groningen Institute of Evolutionary Life Sciences (GELIFES), University of Groningen, Nijenborgh 7, 9747 AG, Groningen, The Netherlands.
Conflicts of interests
The authors declare no conflict of interest.
References
- Amor DR, Ratzke C, Gore J. Transient invaders can induce shifts between alternative stable states of microbial communities. Sci Adv. 2020;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih A, Foster SJ. Analysis of the role of bacterial endospore cortex structure in resistance properties and demonstration of its conservation amongst species. J Appl Microbiol. 2001;91:364–72. [DOI] [PubMed] [Google Scholar]
- Belote RT, Jones RH, Hood SMet al. Diversity–invasibility across an experimental disturbance gradient in Appalachian forests. Ecology. 2008;89:183–92. [DOI] [PubMed] [Google Scholar]
- Berg G, Kusstatscher P, Abdelfattah Aet al. Microbiome modulation—toward a better understanding of plant microbiome response to microbial inoculants. Frontiers in Microbiology. 2021;12:803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco P, Corona F, Martínez JL. Biolog phenotype microarray is a tool for the identification of multidrug resistance efflux pump inducers. Antimicrob Agents Chemother. 2018;62:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blath J, Tóbiás A. Invasion and fixation of microbial dormancy traits under competitive pressure. Stochastic Proc Appli. 2020;130:7363–95. [Google Scholar]
- Borglin S, Joyner D, DeAngelis KMet al. Application of phenotypic microarrays to environmental microbiology. Curr Opin Biotechnol. 2012;23:41–8. [DOI] [PubMed] [Google Scholar]
- Borriss R. Bacillus, a plant-beneficial bacterium. Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture. 2015 : 379–91.
- Bressuire-Isoard C, Broussolle V, Carlin F. Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol Rev. 2018;42:614–26. [DOI] [PubMed] [Google Scholar]
- Buchberger F, Stockenreiter M. Unsuccessful invaders structure a natural freshwater phytoplankton community. Ecosphere. 2018;9:1–16. [Google Scholar]
- Carboni M, Livingstone SW, Isaac MEet al. Invasion drives plant diversity loss through competition and ecosystem modification. J Ecol. 2021;109:3587–601. [Google Scholar]
- Catford JA, Jansson R, Nilsson C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Diversity Distributions. 2009;15:22–40. [Google Scholar]
- Chapman D, Purse BV, Roy HEet al. Global trade networks determine the distribution of invasive non-native species. Global Ecol Biogeogr. 2017;26:907–17. [Google Scholar]
- Clark GF, Johnston EL. Temporal change in the diversity–invasibility relationship in the presence of a disturbance regime. Ecology Letters. 2011;14:52–7. [DOI] [PubMed] [Google Scholar]
- Davies KF, Harrison S, Safford HDet al. Productivity alters the scale dependence of the diversity–invasibility relationship. Ecology. 2007;88:1940–7. [DOI] [PubMed] [Google Scholar]
- De Roy K, Marzorati M, Negroni Aet al. Environmental conditions and community evenness determine the outcome of biological invasion. Nat Commun. 2013;4. 10.1038/ncomms2392 [DOI] [PubMed] [Google Scholar]
- de Souza R, Ambrosini A, Passaglia LMP. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol Biol. 2015;38:401–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Hylckama Vlieg JET. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;25:354–66. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Zhaxybayeva O. Metagenomics and the units of biological organization. Bioscience. 2010;60:102–12. [Google Scholar]
- Eisenhauer N, Schulz W, Scheu Set al. Niche dimensionality links biodiversity and invasibility of microbial communities. Funct Ecol. 2013;27:282–8. [Google Scholar]
- Elton CS. The Ecology of Invasions by Animals and Plants. Springer Nature, 2020. [Google Scholar]
- Erlacher A, Cardinale M, Grosch Ret al. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Front Microbiol. 2014;5:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargione JE, Tilman D. Diversity decreases invasion via both sampling and complementarity effects. Ecology Letters. 2005;8:604–11. [Google Scholar]
- Ferreira DA, da Silva TF, Pylro VSet al. Soil microbial diversity affects the plant-root colonization by arbuscular mycorrhizal fungi. Microb Ecol. 2021;82:100–3. [DOI] [PubMed] [Google Scholar]
- Florio A, Pommier T, Gervaix Jet al. Soil C and N statuses determine the effect of maize inoculation by plant growth-promoting rhizobacteria on nitrifying and denitrifying communities. Sci Rep. 2017;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Vangay P, Garbe Jet al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;9:942–9. [DOI] [PubMed] [Google Scholar]
- Hallegraeff GM. Transport of toxic dinoflagellates via ships ballast water: bioeconomic risk assessment and efficacy of possible ballast water management strategies. Marine Ecol Prog Ser. 1998;168:297–309. [Google Scholar]
- Hol FJH, Hubert B, Dekker Cet al. Density-dependent adaptive resistance allows swimming bacteria to colonize an antibiotic gradient. ISME J. 2016;10:30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison EA., Miller, DA.and Angert, ER.. Sporulation in Bacteria: Beyond the Standard Model. Microbiol Spectr. 2014;2:87–102.. 10.1128/microbiolspec.TBS-0013-2012 [DOI] [PubMed] [Google Scholar]
- Jałowiecki Ł, Chojniak J, Dorgeloh Eet al. Using phenotype microarrays in the assessment of the antibiotic susceptibility profile of bacteria isolated from wastewater in on-site treatment facilities. Folia Microbiol (Praha). 2017;62:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A, Schulz W, Scheu Set al. Intraspecific genotypic richness and relatedness predict the invasibility of microbial communities. ISME J. 2011;5:1108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, le Roux X, Niklaus PAet al. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol. 2014;5:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröber M, Wibberg D, Grosch Ret al. Effect of the strain bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol. 2014;5:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Männistö MK, van Elsas JDet al. Plants impact structure and function of bacterial communities in arctic soils. Plant Soil. 2016;399:319–32. [Google Scholar]
- Liu X, le Roux X, Salles JF. The legacy of microbial inoculants in agroecosystems and potential for tackling climate change challenges. Iscience. 2022;25:103821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Jacques SMS, Pires APFet al. High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol. 2017;1:1–12. [DOI] [PubMed] [Google Scholar]
- Louca S, Polz MF, Mazel Fet al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–43. [DOI] [PubMed] [Google Scholar]
- Mackie AM, Hassan KA, Paulsen ITet al. Biolog phenotype microarrays for phenotypic characterization of microbial cells. Methods Mol Biol. 2014;1096:123–30. [DOI] [PubMed] [Google Scholar]
- Mallon C, Poly F, le Roux Xet al. Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology. 2015;96:915–26. [DOI] [PubMed] [Google Scholar]
- Mallon CA, le Roux X, van Doorn GSet al. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader's niche. ISME J. 2018;12:728–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki SS, Sasaki T, Akasaka M. Invasion of exotic piscivores causes losses of functional diversity and functionally unique species in Japanese lakes. Freshwater Biol. 2016;61:1128–42. [Google Scholar]
- Mawarda PC, Lakke SL, Dirk van Elsas Jet al. Temporal dynamics of the soil bacterial community following bacillus invasion. Iscience. 2022;25:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawarda PC, le Roux X, van Elsas JDet al. Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol Biochem. 2020;148, 10.1016/j.soilbio.2020.107874 [DOI] [Google Scholar]
- McSpadden Gardener BB. Ecology of Bacillus and Paenibacillus spp. in Agricultural Systems. 2007;94:1252–8.. http://dx.doi.org/101094/PHYTO200494111252 [DOI] [PubMed] [Google Scholar]
- Orlewska K, Markowicz A, Piotrowska-Seget Zet al. Functional diversity of soil microbial communities in response to the application of cefuroxime and/or antibiotic-resistant pseudomonas putida strain MC1. Sustainability. 2018;10:3549. [Google Scholar]
- Salles JF, Poly F, Schmid Bet al. Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology. 2009;90. 10.1890/09-0188.1 [DOI] [PubMed] [Google Scholar]
- Sapsford SJ, Wakelin A, Peltzer DAet al. Pine invasion drives loss of soil fungal diversity. Biological Invasions. 2022;24:401–2014. [Google Scholar]
- Smalla K, Wachtendorf U, Heuer Het al. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza CP de, Rodrigues-Filho CA de S, Barbosa FARet al. Drastic reduction of the functional diversity of native ichthyofauna in a Neotropical lake following invasion by piscivorous fishes. Neotropical Ichthyol. 2021;19. [Google Scholar]
- Souza RC, Mendes IC, Reis-Junior FBet al. Shifts in taxonomic and functional microbial diversity with agriculture: how fragile is the Brazilian Cerrado?. BMC Microbiol. 2016;16. 10.1186/s12866-016-0657-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur MP, van der Putten WH, Cobben MMPet al. Microbial invasions in terrestrial ecosystems. Nat Rev Microbiol. 2019;17:621–31. [DOI] [PubMed] [Google Scholar]
- Tilman D. Resource competition between plankton algae: an experimental and theoretical approach. Ecology. 1977;58:338–48. [Google Scholar]
- Trabelsi D, Mhamdi R. Microbial inoculants and their impact on soil microbial communities: a review. Biomed Res Int. 2013;2013:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas J, Chiurazzi M, Mallon Cet al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA. 2012;109:1159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas JD, Hill P, Chroňková Aet al. Survival of genetically marked Escherichia coli O157:H7 in soil as affected by soil microbial community shifts. ISME J. 2007;1:204–14. [DOI] [PubMed] [Google Scholar]
- Vanwonterghem I, Jensen PD, Rabaey Ket al. Genome-centric resolution of microbial diversity, metabolism and interactions in anaerobic digestion. Environ Microbiol. 2016;18. 10.1111/1462-2920.13382 [DOI] [PubMed] [Google Scholar]
- Velasco A, Probanza A, Gutierrez Mañero FJet al. Functional diversity of rhizosphere microorganisms from different genotypes of Arabidopsis thaliana. Commun Ecol 2008 10 : 1. 2009;10:111–9. [Google Scholar]
- Vivant AL, Garmyn D, Maron PAet al. Microbial diversity and structure are drivers of the biological barrier effect against listeria monocytogenes in soil. PLoS One. 2013;8. 10.1371/journal.pone.0076991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Holle B. Environmental stress alters native-nonnative relationships at the community scale. Biological Invasions. 2013;15:417–27. [Google Scholar]
- Wittebolle L, Vervaeren H, Verstraete Wet al. Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl Environ Microbiol. 2008;74. 10.1128/AEM.01006-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Jia X, Wang Het al. The legacy of bacterial invasions on soil native communities. Environ Microbiol. 2020;23:1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.