Abstract
β-Defensins are cationic peptides with broad-spectrum antimicrobial activity that may play a role in mucosal defenses of several organs. They have been isolated in several species, and in humans, two β-defensins have been identified. Here, we report the identification of two genes encoding β-defensin homologues in the rat. Partial cDNAs were found by searching the expressed-sequence-tag database, and primers were designed to generate full-length mRNA coding sequences. One gene was highly similar to the human β-defensin-1 (HBD-1) gene and mouse β-defensin-1 gene at both the nucleic acid and amino acid levels and was termed rat β-defensin-1 (RBD-1). The other gene, named RBD-2, was homologous to the HBD-2 and bovine tracheal antimicrobial peptide (TAP) genes. The predicted prepropeptides were strongly cationic, were 69 and 63 residues in length for RBD-1 and RBD-2, respectively, and contained the six-cysteine motif characteristic of β-defensins. The β-defensin genes mapped closely on rat chromosome 16 and were closely linked to the α-defensins genes, suggesting that they are part of a gene cluster, similar to the organization reported for humans. Northern blot analysis showed that both RBD-1 and RBD-2 mRNA transcripts were ∼0.5 kb in length; RBD-1 mRNA was abundantly transcribed in the rat kidney, while RBD-2 was prevalent in the lung. Reverse transcription-PCR indicated that RBD-1 and RBD-2 mRNAs were distributed in a variety of other tissues. In the lung, RBD-1 mRNA expression localized to the tracheal epithelium while RBD-2 was expressed in alveolar type II cells. In conclusion, we characterized two novel β-defensin homologues in the rat. The rat may be a useful model to investigate the function and contribution of β-defensins to host defense in the lung, kidney, and other tissues.
β-Defensins are cationic antimicrobial peptides that comprise part of the innate immune system at mucosal surfaces (6, 14). Epithelial β-defensins were first described for cows (8) and subsequently identified in several species, including sheep (22), pigs (43), mice (1, 21, 28), and humans (3, 17). Two β-defensins of epithelial origin were recently identified in human cells: human β-defensin-1 (HBD-1) and HBD-2 (3, 17). HBD-1 expression was most abundant in the urogenital tract (38, 44), while HBD-2 expression predominated in skin and in the lung (2, 17, 32).
The pulmonary epithelium acts as a barrier to keep the normal lung free from infection from inhaled or aspirated microorganisms. The innate mucosal defenses of the lung include the production of several factors with antimicrobial properties, such as proteins and peptides (30). Cystic fibrosis, a disease characterized by chronic airway infection in the absence of any demonstrable systemic immune defect (40), may represent a human disease caused by defective pulmonary mucosal defenses. One proposed mechanism for the susceptibility of cystic fibrosis patients to chronic respiratory infection and inflammation is the inactivation of salt-sensitive antimicrobial peptides, such as β-defensins, by an elevated airway surface liquid salt content (16, 33). However, the relative contribution that β-defensins make to host defenses in the lung is unknown.
To investigate the role the β-defensins play in pulmonary defenses, it would be advantageous to identify genes for homologous peptides in laboratory animal models. Here, we report the discovery of rat genes encoding two new peptides homologous to the human β-defensins. Both rat gene products are expressed in the lung. The rat may be a useful model to study the activity and function of β-defensins in the lung and other epithelium-lined organs.
MATERIALS AND METHODS
Cloning of RBD-1 and RBD-2.
The HBD-1 mature peptide sequence (DHYNCVSSGGQCLYSACPIFTKIQGTCYRGKAKCCKS) was used to screen the GenBank dbest database of expressed sequence tags (ESTs) by using the National Center for Biotechnology Information BLAST program. The search identified three identical rat ESTs from a normalized rat kidney cDNA library with high sequence similarity to HBD-1. Three clones contained part of the open reading frame (ORF) for putative rat β-defensin-1 (RBD-1). Primer sequences designed for the partial RBD-1 sequence were used to clone the full-length cDNA from a rat kidney cDNA library by PCR. Similar methods were used to search for a homologue of HBD-2. The full HBD-2 amino acid sequence ( M RVLYLLFSFLFI FLMPLPGVFGGIGDPVTCLKSGAICHPVFC PRRYKQIGTCGLPGTKC) was used to screen the database, and two identical ESTs from a rat lung cDNA library were found. One clone contained the entire 192-bp ORF for putative RBD-2. A set of primers designed from the RBD-2 sequence was used to clone the cDNA from rat lung by reverse transcription (RT)-PCR.
β-Defensin mapping.
The rat radiation hybrid panel was purchased commercially (Research Genetics, Huntsville, Ala.). The following primers were selected for PCR: RBD-1-forward (5′-GGACGCAGAACAGATCAATACCGA-3′), RBD-1-reverse (5′-TCTTCAAACCACTGTCAACTCCTG-3′), RBD-2-forward (5′-TTAATTTGGTTTGTTTTGTGCAT-3′), RBD-2-reverse (5′-CATGCCTGACCAAAGGAGGCGTA-3′), rat α-defensin-3 (RAD-3)-forward (5′-TACCGACTCTGTTGCTGAGCA-3′), RAD-3-reverse (5′-GTCTAGGACACAACATACTAC-3′), RAD-4-forward (5′-TGGCCGCATCTACCGTCTCTG-3′), and RAD-4-reverse (5′-AACATGGGACAAGTAACAGTC-3′).
We used previously published conditions for PCR (29) with the following modifications: the annealing temperature for RBD-1 was 65°C, and that for RBD-2 was 52°C. Mapping experiments were performed in duplicate for each marker, and each was scored independently. Discordant data were scored as unknown. The defensin radiation hybrid data were imported into the RHMAPPER (version 1.1) (35) program for analysis, along with data for genetic markers on rat chromosome 16. A retention frequency of 30% was used for the analysis, which was estimated from other unpublished data generated from this panel. The framework map was constructed with local support of greater than 1,000:1.
Tissue, cell, and RNA isolation.
Tissues were obtained from adult Sprague-Dawley rats for screening of β-defensin mRNA expression. In developmental studies, timed pregnant (sperm positive = day 0) Sprague-Dawley rats were obtained from Sasco (Omaha, Nebr.). Fetal lung tissues were obtained on days 16, 18, 19, and 21 of gestation and postnatal days 1, 3, 5, and 10. Isolated tissues were flash frozen in liquid nitrogen and stored at −80°C. Adult type II cells were isolated by using enzymatic digestion, differential adherence, and panning as described by Dobbs et al. (9). The majority of cells were tannic acid positive (>90%), and the viability was >98% as determined by trypan blue exclusion, as reported previously (25). The Animal Care and Use Committees of the University of Iowa and The Cleveland Clinic approved the protocols. Total RNA was isolated by standard methods as previously described (8, 26, 32).
Detection of β-defensin mRNA by RT-PCR.
RT-PCR was performed as follows. One microgram of total RNA from each sample was reverse transcribed with random hexamer primers by using the SuperScript transcription system (Gibco BRL) according to the manufacturer’s instructions. Specific primers were designed from the registered sequences of RBD-1 (GenBank accession no. AF068860) and RBD-2 (GenBank accession no. AF068861). The primer sequences used for cloning RBD-1 in rat tissue were as follows: forward, GACCCTGACTTCACCGACAT, and reverse, CCTGCAACAGTTGGGCTTAT. Additional RBD-1 primers used for detection were as follows: forward, CTTGGACGCAGAACAGATCA, and reverse, TCCACAAGTGCCAATCTGTC. The primer sequences for RBD-2 used in all experiments were as follows: forward, ATTTCTCCTGGTGCTGCTGT, and reverse, TCCACAAGTGCCAATCTGTC. The predicted product sizes were 225 bp for RBD-1 and 132 bp for RBD-2. Each RBD-1 reaction mixture contained an approximately 1.25 pM concentration of the primers, 3 mM Mg2+, and 5 μl of the RT reaction product for a total volume of 20 μl. Each RBD-2 reaction mixture contained an approximately 0.5 pM concentration of the primers, 3 mM Mg2+, and 10 μl of the RT reaction product for a total volume of 30 μl. After an initial denaturing step (95°C for 3 min), 30 cycles of denaturing (94°C for 1 min), annealing (60°C for 30 s for RBD-1 and 63°C for 30 s for RBD-2), and extending (72°C for 1 min), followed by 5 min at 72°C for elongation, were conducted. A 12-μl aliquot of the RBD-1 PCR product and an 18-μl aliquot of the RBD-2 product were electrophoresed on a 2% agarose gel and visualized with ethidium bromide. As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified in both RBD-1 and RBD-2 reactions with the following primers: forward, AGACAGCCGCATCTTCTTGT, and reverse, CTTGCCGTGGGTAGAGTCAT.
Northern blot analysis.
For Northern blot analysis, total RNA (10 μg) was denatured and separated on 1.2% agarose-formaldehyde gels and then blotted onto nucleic acid transfer membranes (Hybond-N+; Amersham). Northern blots were hybridized overnight with 32P-, end-labeled oligonucleotide probes in 35% (vol/vol) formamide–5× Denhardt’s solution–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% sodium dodecyl sulfate (SDS)–100 μg of yeast RNA per ml at 42°C. Membranes were washed with 2× SSC–0.1% SDS five times at room temperature for 50 min and then twice at 55°C for 15 min. The membranes were stripped of the oligonucleotide probes by incubation in 0.1× SSC–0.1% SDS at 70°C for 30 min. The sequences of the oligonucleotide probes were CCATTTTGGAGGCATCGGTATTGATCTGTTCTGCGTC (RBD-1), TGGAGGAGCAAATTCTGTTCATCCCATTGGTTCTTGG (RBD-2), CAGTTTTAGTCCCTTCATCTGTTTTTGGGGTAGGTTC (α-defensin; generously provided by Gill Diamond), and AGCCCCRGCCTTCTCCATGGTRGTGAAGACVCCR (GAPDH). Blots were exposed to X-Omat films (Eastman Kodak Co., Rochester, N.Y.) with intensifying screens at 80°C.
Southern blot analysis.
Rat genomic DNA (10 μg) was digested overnight with restriction endonucleases, size separated by agarose gel (1.0%) electrophoresis, and transferred to a nylon membrane (Hybond-N+). For reduced-stringency conditions, the membrane was hybridized with 32P-labeled RBD-2 cDNA overnight in 35% (vol/vol) formamide–5× Denhardt’s solution–5× SSC–1% SDS–100 mg of yeast RNA per ml at 42°C. The filter was washed with 2× SSC–0.1% SDS five times at room temperature for 10 min each and then twice at 58°C for 15 min. The membrane was exposed to film in the presence of an intensifying screen for 1 week. The filter was stripped of the probe by incubation in 0.5 M NaOH–1.5 M NaCl for 30 min at 37°C and then checked by autoradiography. For high-stringency conditions, the membrane was then hybridized with the probe overnight in 50% (vol/vol) formamide–5× Denhardt’s solution–5× SSC–1% SDS–100 mg of yeast RNA per ml at 42°C. The filter was washed with 2× SSC–0.1% SDS five times at room temperature for 10 min each and then twice with 0.1× SSC–0.1% SDS at 65°C for 15 min. The membrane was exposed to film in the presence of an intensifying screen for 10 days.
In situ hybridization.
We followed methods that were previously used to localize HBD-1 mRNA in urogenital tissues (38). Sense and antisense 35S-UTP, 35S-CTP (Amersham), and double-labeled RBD-1 and RBD-2 riboprobes were prepared from linearized cDNA plasmids and hybridized with frozen sections of rat kidney and lung tissue according to published methods (42). The sections were coated with NTB-2 autoradiography emulsion (Eastman Kodak Co.) diluted 1:1 in distilled H2O, air dried, and exposed for 4 to 12 weeks at 4°C before being developed. Sections were counterstained with toluidine blue, and the signals were localized by using bright-field and dark-field microscopy.
Assay of rat BAL fluid for antimicrobial activity.
Bronchoalveolar lavage (BAL) fluid was obtained from four rats by washing the lungs twice with 2-ml volumes of phosphate-buffered saline. The BAL fluid samples were pooled for analysis. A quantitative luminescence assay was used to screen rat BAL fluid for antimicrobial activity. The assay employs Escherichia coli DH5α containing the luminescence plasmid pCGLS1 (12, 32) and allows rapid quantitative assessment of bactericidal activity. We validated our assay by showing a correlation of luminescence with viable cell numbers. Luminescence is an energy-requiring activity that is directly related to bacterial viability as determined by plate counts (data not shown). Bacteria were grown at 30°C in Luria-Bertani medium, centrifuged, and resuspended at a concentration of 107 cells/ml in 10 mM potassium phosphate, pH 7.2, with 1% Luria-Bertani medium. Bacteria (106) were then incubated with mouse BAL fluid in 96-well plates (Optiplate; Packard Instruments) for 4 h, and luminescence was measured with a microtiter dish luminometer (Anthos). To assess the salt sensitivity of the BAL fluid activity, the assays were performed in the presence of 0 or 150 mM NaCl.
RESULTS
Cloning and structure of rat β-defensin cDNAs.
By searching the dbest database by using either the HBD-1 or HBD-2 peptide sequence, genes encoding two homologous rat proteins were found in normalized rat kidney or lung cDNA libraries. These sequences were termed RBD-1 and RBD-2, respectively. The RBD-1 clones identified in the database did not contain a full-length ORF. In order to obtain the entire coding sequence for RBD-1, anchored PCR was performed by using a kidney cDNA library template, part of the RBD-1 sequence as one primer, and a vector sequence as the other primer. The composite cDNA sequence contains an ORF 210 bp in length encoding a putative 69-amino-acid peptide which was ∼65% identical to HBD-1 and 92.5% identical to mouse β-defensin-1 (MBD-1) (Fig. 1). The RBD-2 cDNA contains a 192-bp ORF, and the putative RBD-2 peptide sequence is 53% identical to HBD-2 (Fig. 1).
FIG. 1.
(A) Nucleotide and putative amino acid sequences for the RBD-1 and RBD-2 cDNA. The predicted stop codon is indicated by an asterisk. (B) Amino acid alignment of putative prepropeptides for RBD-1 and RBD-2 with other known β-defensins.
Chromosomal localization and Southern blot analysis.
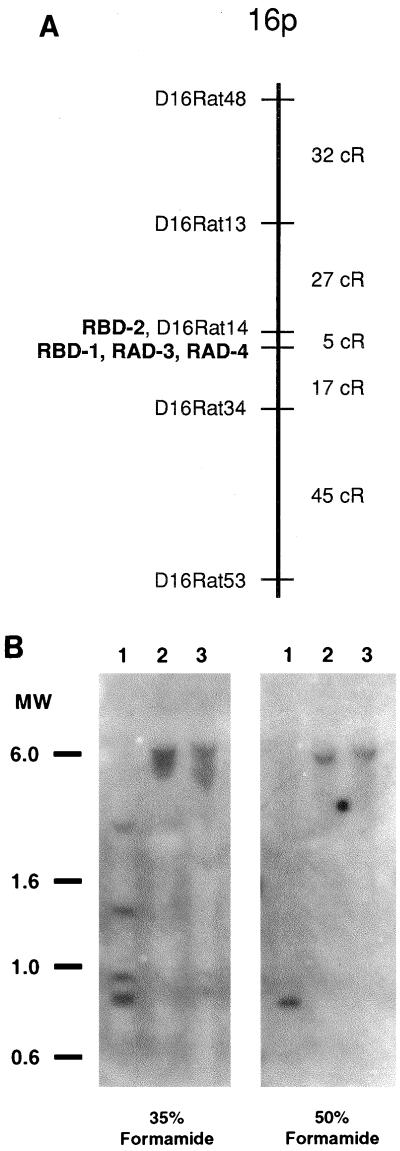
Radiation hybrid mapping demonstrated that the RBD-1 and RBD-2 genes were linked to each other at a distance of 3.8 centirads (cR) with a limits of detection (LOD) score of 24.32. A rough estimate of the physical distance between RBD-1 and RBD-2 may be obtained by determining the correlation between cR estimates and known physical distances. While such data do not exist for the rat panel we used, data from the human GeneBridge 4 panel, a radiation hybrid panel constructed with a similar radiation dose, suggest that 1 Mb of DNA is represented by roughly 3.7 cR. This metric suggests that the genes for RBD-1 and RBD-2 are separated by about 1 Mb of DNA. Based on syntenic groups of markers in the rat, human, and mouse, it was expected that these two rat β-defensin genes would be localized to rat chromosome 16. Pairwise analysis with both RBD-1 and RBD-2 demonstrated strong linkage to markers on chromosome 16. A framework map of five genetic markers was constructed for the distal 16p region by using multipoint analysis and is shown in Fig. 2A. RBD-2 and D16RAT14 were found to be completely linked and are shown occupying the same location in the framework map. Radiation hybrid mapping of rat neutrophil defensins RAD-3 and RAD-4 suggests that both genes are completely linked to RBD-1 (Fig. 2A).
FIG. 2.
(A) Radiation hybrid map of distal rat chromosome 16p. The RBD-2 gene and D16RAT14 are completely linked and thus are shown to occupy the same location within the framework map. The RBD-1, RAD-3, and RAD-4 genes are also completely linked and are also shown in the same map location. (B) Southern blot analysis of the RBD2 gene. Rat genomic DNA (10 μg) was digested with restriction endonucleases (lane 1, EcoRI and BamHI; lane 2, BglII and NotI; lane 3, EcoRI and HindIII), size separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with 32P-labeled RBD-2 cDNA. Probe hybridization buffer contained either 35 or 50% (vol/vol) formamide for reduced- or high-stringency conditions, respectively (see Materials and Methods). Numbers at the left are molecular sizes, in kilobases.
The RBD-2 cDNA was used as a probe in Southern blot analysis of rat genomic DNA. At a reduced level of stringency, four distinct bands with similar intensities were observed in DNA digested with the combination restriction enzymes EcoRI and BamHI (Fig. 2B, left panel, lane 1). Somewhat broader bands of probe hybridization were detected with two other pairs of restriction enzymes (Fig. 2B, left panel, lanes 2 and 3). In contrast, at high stringency, single sharp bands of hybridization with comparable intensities were detected in all three lanes. These data provide evidence that RBD-2 is present as a single copy gene in the rat and suggest that related genes or pseudogenes similar to RBD-2 may also be present in the rat genome.
Tissue distribution and ontogeny of rat β-defensin mRNA expression.
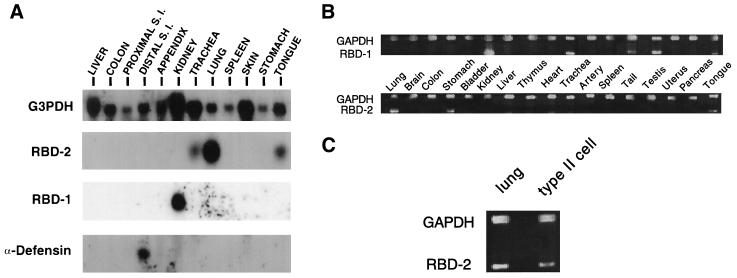
Total RNA from rat tissues was screened for rat β-defensin mRNA expression. Northern blot analysis (Fig. 3A) of the two rat β-defensin genes showed that both transcripts were ∼500 nucleotides in length. The predominant site of RBD-1 mRNA expression was the kidney, and that for RBD-2 was the lung. Lower levels of RBD-2 mRNA were also detected in the trachea and tongue. The expression of RBD-2 in the trachea was variable, as we failed to detect the message in samples by in situ hybridization or RT-PCR (see below). By contrast, a member of another subclass of defensin, an α-defensin expressed in small intestinal Paneth cells, showed a distinct, nonoverlapping pattern of mRNA expression.
FIG. 3.
Tissue distribution of RBD-1 and RBD-2. (A) Northern blot hybridization of selected rat tissue total RNA with subcloned RBD-1, RBD-2, or rat α-defensin oligonucleotide probes. Both RBD-1 and RBD-2 mRNA transcripts are about 0.5 kb in length. S. I., small intestine. (B) RT-PCR demonstration of rat β-defensin expression by using adult rat tissues as a template. Upper bands represent GAPDH transcript as an internal positive control; lower bands show the PCR products of RBD-1 and RBD-2. (C) RT-PCR demonstration of RBD-2 mRNA expression in rat lung and type II cells.
More extensive tissue distribution studies were conducted by RT-PCR. As shown in Fig. 3B, the RBD-1 transcript was detected in the kidney, trachea, tail, testis, and tongue. RBD-2 mRNA was expressed in the lung, stomach, and tongue (Fig. 3B). In some animals, RBD-1 was detected in the brain (data not shown). Thus, RBD-1 was abundantly expressed in the kidney, while RBD-2 was readily detected in the lung. Further RT-PCR studies were performed on freshly isolated lung cells to determine the cell types expressing RBD-2. As shown in Fig. 3B, RBD-2 mRNA was detected in alveolar type II cells. Alveolar macrophages and lung fibroblasts were also screened and showed no evidence of RBD-2 expression (data not shown). The RBD-1 transcript was not detected in type II cells (data not shown).
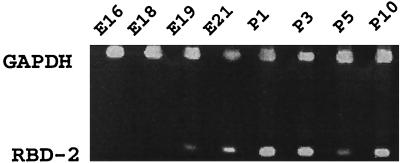
To determine the ontogeny of RBD-2 mRNA expression in the lung, fetal and neonatal tissues were screened by RT-PCR. As shown in Fig. 4, RBD-2 mRNA expression was not detected before 19 days of gestation and markedly increased between fetal day 19 and postnatal day 1.
FIG. 4.
Developmental expression of RBD-2 mRNA in fetal and postnatal rat lungs, identified by RT-PCR. Embryonic (E) rat lungs were collected at days 16, 18, 19, and 21 of gestation; postnatal (P) lungs were collected at days 1, 3, 5, and 10.
Localization of β-defensin mRNA by in situ hybridization.
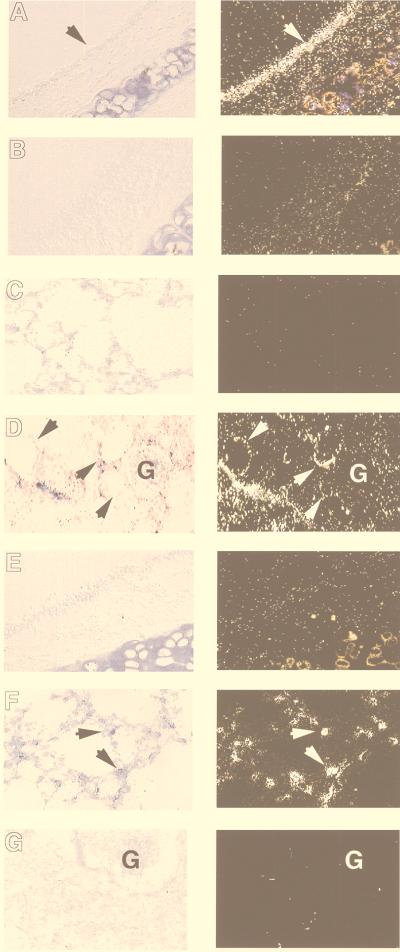
To localize the expression of RBD-1 and RBD-2 mRNAs to specific cell types, we performed in situ hybridization on tissue sections from the trachea, lung, and kidney. As shown in Fig. 5A, RBD-1 mRNA expression was detected in the tracheal epithelium and kidney cortex epithelia but not in the distal lung. In the trachea, expression was confined to the tracheal epithelium and was diffuse. Expression in the kidney cortex was restricted to epithelia of the distal tubules. In contrast, RBD-2 expression was noted in a population of cuboidal cells in the alveolus consistent with alveolar type II cells but not in the tracheal epithelium or kidney (Fig. 5B).
FIG. 5.
Localization of RBD-1 (A to D) and RBD-2 (E to G) mRNA expression in the trachea, lung, and kidney by in situ hybridization. Bright-field (left panels) and dark-field (right panels) images are presented for each section. (A) Antisense probe shows RBD-1 mRNA expression localized to the tracheal epithelium (arrow). (B) Control sense probe shows no specific RBD-1 mRNA signal over trachea. (C) Antisense probe on lung tissue shows no specific signal. (D) Antisense probe showing RBD-1 mRNA localization to the kidney cortex distal tubules (arrows). There was no expression in the glomerulus (indicated on the panel by the letter G). (E) Antisense RBD-2 probe demonstrates no expression in trachea. (F) RBD-2 mRNA expression in the lung localized to cells in the alveolus consistent with type II cells (arrows, sense probe). (G) Antisense probe recognized no specific signal in the kidney (G, glomerulus). Sense probe controls for all samples showed only nonspecific backgrounds similar to that shown in panel B.
Rat BAL fluid exhibits salt-sensitive antimicrobial activity.
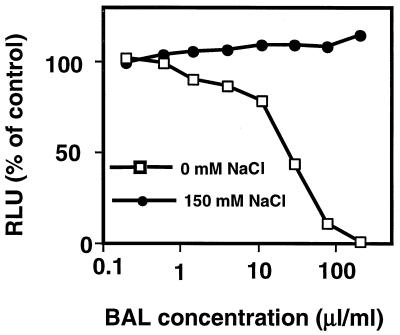
The antimicrobial activity of β-defensins is characteristically inhibited by salt (14). We hypothesized that if the RBD-1 or RBD-2 gene products were secreted into airway surface liquid, salt-sensitive antimicrobial activity would be present in rat BAL fluid. We isolated BAL fluid from normal rats and tested its ability to kill bacteria in the presence or absence of NaCl. As shown in Fig. 6, rat BAL fluid killed E. coli in a concentration-dependent manner. When the BAL fluid samples were tested in the presence of 150 mM NaCl, the antimicrobial activity was lost. Thus, rat BAL fluid, like human airway surface liquid (33), exhibited antimicrobial activity that was inhibited by salt.
FIG. 6.
Antimicrobial activity of rat BAL fluid determined by E. coli luminescence assay. BAL fluid from normal rats exhibited antimicrobial activity that was inhibited in the presence of 150 mM NaCl. Results represent pooled BAL fluid samples from four rats. RLU, relative light units.
DISCUSSION
In this study we have identified the first β-defensin genes in the rat. The gene products, RBD-1 and RBD-2, are homologous to β-defensins characterized in other species, including humans. The rat α- and β-defensins localized to chromosome 16 and were closely linked. Investigation of the tissue distribution of the rat β-defensin mRNAs showed an organ-specific distribution. RBD-1, like its human homologue, was abundantly expressed in the kidney; RBD-2 was most abundant in the lung. In the lung, RBD-1 was expressed in the tracheal epithelium while RBD-2 was expressed in tracheal epithelia and alveolar type II cells. We found that rat BAL fluid exhibited salt-sensitive microbicidal activity. Members of the β-defensin class of antimicrobial peptides are salt sensitive (14, 32); therefore, the rat β-defensins may contribute to the activity in BAL fluid. Since both RBD-1 and RBD-2 were expressed in the lung, the rat may be a useful small animal model for further study of the role β-defensins play in the pulmonary innate defense system.
The RBD-1 and RBD-2 sequences were highly similar to other published β-defensin sequences, including those of the human genes (3, 8, 17). Interestingly, the RBD-2 amino acid sequence differs from other known sequences in the spacing between the second and third cysteine residues (Fig. 1). Other β-defensins reported to date have had four intervening amino acids between these cysteine residues. In contrast, the putative RBD-2 amino acid sequence has only three residues between the cysteines and diverges from the consensus motif. The six cysteine residues are otherwise conserved in both RBD-1 and RBD-2.
Radiation hybrid panel results revealed that RBD-1 and RBD-2 are closely linked on rat chromosome 16 and near the α-defensins. This suggests that the rat α- and β-defensin genes lie as a cluster on chromosome 16, in a region syntenic to human chromosome 8. The defensin loci in humans (18, 23, 24), mice (21), cattle (13), and sheep (22) are also known to consist of gene clusters. The observation that defensin loci exist as gene clusters in several species has led to the speculation that these genes arose from the duplication of an ancestral gene (4, 24). Our data from the rat add further support to that hypothesis.
In some species, notably cattle and sheep, there are a collection of several HBD-2-like genes (7, 20, 31, 36). These genes are very similar in sequence and genomic organization but have distinct patterns of tissue expression. For example, bovine TAP, lingual antimicrobial peptide (LAP), and enteric β-defensin (EBD) have approximately 90% sequence identity, but the predominant sites of expression are tracheal mucosa, tongue epithelia, and distal enteric mucosa, respectively. Our Southern blot analysis suggests that the rat genome may also contain additional RBD-2-related genomic sequences (similar genes or pseudogenes), a possibility currently under investigation.
The rat β-defensin genes were expressed in a tissue-specific manner. RBD-1, like its human (3, 38, 44) and mouse (1, 21, 28) homologues, was abundantly expressed in the kidney. The RBD-1 mRNA localized to the distal tubules of the kidney cortex, as previously noted for MBD-1 (1) and HBD-1 (38). RBD-1 was also detected in the tracheal epithelium, similar to MBD-1 (1, 21, 28). However, in contrast to MBD-1, RBD-1 expression was absent from the lung. The predominant site of RBD-2 expression was the lung, specifically the alveolar type II cells. The type II cells are also a site of lysozyme production in the rat (15). The human homologue of RBD-2 is also expressed in the lung (2, 17, 32). The human (2, 32) and bovine (7) homologues of RBD-2 are also expressed in the respiratory tract but predominantly in airway epithelia. This feature may suggest a distinction of some details regarding innate host defense in rodents compared with other mammals.
RBD-2 mRNA expression was developmentally regulated in the lung. The transcript was undetectable until just prior to birth (day 19). This pattern of regulation is reminiscent of several pulmonary genes important in perinatal adaptation, including the genes for surfactant protein A (34), surfactant protein B (41), and the epithelial sodium channel subunits (37, 39). This suggests that RBD-2 contributes to host defenses in the perinatal period and later.
Rat BAL fluid exhibited salt-sensitive microbicidal activity. The activity of β-defensin peptides studied to date is characteristically salt sensitive (14). Recent studies suggest that disruption of mucosal innate immunity may contribute to the development of chronic pulmonary infection in cystic fibrosis (16, 32, 33). Alterations in airway surface liquid NaCl concentration may inhibit the activity of salt-sensitive antimicrobial factors, including the β-defensins (16, 32, 33). Several other antimicrobial factors in airway surface liquid, including lysozyme and secretory leukocyte protease inhibitor, behave in a salt-sensitive fashion (5, 10, 11, 19, 27, 33); thus, the relative contribution of the rat β-defensins to the killing activity in airway surface liquid is unknown. Further study is needed to determine the role of RBD-1 and RBD-2 in the microbicidal activity of rat BAL fluid.
The availability of a small animal model expressing homologues of HBD-1 and HBD-2 may facilitate further study of the role these peptides play in the mucosal defenses of the lung, kidney, and other organ systems.
ACKNOWLEDGMENTS
We thank Bento Soares and Val Sheffield for helpful advice and assistance with this project. We acknowledge Anne Black (Medical College of Wisconsin) for providing unpublished rat radiation hybrid data and Ann McClain (University of Iowa) and Dennis Wilk and Yang Yu (The Cleveland Clinic Foundation) for technical assistance.
This work was supported in part by the following grants and organizations: NIH P50 HL-61234-01 SCOR (P.B.M. and B.F.T.), NIH HL61234 (C.L.B.), AI-32234 (C.L.B.), AI-32738 (C.L.B.), Cystic Fibrosis Foundation (97ZO; P.B.M.), and the Children’s Miracle Network Telethon (P.B.M.). P.B.M. is the recipient of a Career Investigator Award from the American Lung Association.
REFERENCES
- 1.Bals R, Goldman M J, Wilson J M. Mouse β-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect Immun. 1998;66:1225–1232. doi: 10.1128/iai.66.3.1225-1232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson J M. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Investig. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensch K W, Raida M, Magert H J, Schulz-Knappe P, Forssmann W G. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 4.Bevins C L, Jones D E, Dutra A, Schaffzin J, Muenke M. Human enteric defensin genes: chromosomal map position and a model for possible evolutionary relationships. Genomics. 1996;31:95–106. doi: 10.1006/geno.1996.0014. [DOI] [PubMed] [Google Scholar]
- 5.Davies R C, Neuberger A, Wilson B M. The dependence of lysozyme activity on pH and ionic strength. Biochim Biophys Acta. 1969;178:294–305. doi: 10.1016/0005-2744(69)90397-0. [DOI] [PubMed] [Google Scholar]
- 6.Diamond G, Bevins C L. β-Defensins: endogenous antibiotics of the innate host defense response. Clin Immunol Immunopathol. 1998;88:221–225. doi: 10.1006/clin.1998.4587. [DOI] [PubMed] [Google Scholar]
- 7.Diamond G, Jones D E, Bevins C L. Airway epithelial cells are the site of expression of a mammalian antimicrobial peptide gene. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbs L G, Gonzalez R, Williams M C. An improved method for isolating type II cells in high yield and purity. Am Rev Respir Dis. 1986;134:141–145. doi: 10.1164/arrd.1986.134.1.141. [DOI] [PubMed] [Google Scholar]
- 10.Ellison R T, Giehl T J. Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Investig. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming A. On a remarkable bacteriolytic element found in tissues and secretions. Proc R Soc (London) 1922;93:306–319. [Google Scholar]
- 12.Frackman S, Anhalt M, Nealson K H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172:5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher D S, Jr, Ryan A M, Diamond G, Bevins C L, Womack J E. Somatic cell mapping of β-defensin genes to cattle syntenic group U25 and fluorescence in situ localization to chromosome 27. Mamm Genome. 1995;6:554–556. doi: 10.1007/BF00356177. [DOI] [PubMed] [Google Scholar]
- 14.Ganz T, Lehrer R I. Defensins. Pharmacol Ther. 1995;66:191–205. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 15.Gibson K F, Phadke S. Intracellular distribution of lysozyme in rat alveolar type II epithelial cells. Exp Lung Res. 1994;20:595–611. doi: 10.3109/01902149409031739. [DOI] [PubMed] [Google Scholar]
- 16.Goldman M J, Anderson M G, Stolzenberg E D, Kari P U, Zasloff M, Wilson J M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:1–9. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 17.Harder J, Bartels J, Christophers E, Schroder J-M. A peptide antibiotic from human skin. Nature. 1997;387:861–862. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 18.Harder J, Siebert R, Zhang Y, Matthiesen P, Christophers E, Schlegelberger B, Schroder J-M. Mapping of the gene encoding human β-defensin-2 (DEFB2) to chromosome region 8p22–p23.1. Genomics. 1997;46:472–475. doi: 10.1006/geno.1997.5074. [DOI] [PubMed] [Google Scholar]
- 19.Hiemstra P S, Maassen R J, Stolk J, Heinzel-Weiland R, Steffens G J, Dijkman J H. Antibacterial activity of antileukiprotease. Infect Immun. 1996;64:4520–4524. doi: 10.1128/iai.64.11.4520-4524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttner K M, Brezinski-Caliguri D J, Mahoney M M, Diamond G. Antimicrobial peptide expression is developmentally regulated in the ovine gastrointestinal tract. J Nutr. 1998;128:297S–299S. doi: 10.1093/jn/128.2.297S. [DOI] [PubMed] [Google Scholar]
- 21.Huttner K M, Kozak C A, Bevins C L. The mouse genome encodes a single homolog of the antimicrobial peptide human β-defensin 1. FEBS Lett. 1997;413:45–49. doi: 10.1016/s0014-5793(97)00875-2. [DOI] [PubMed] [Google Scholar]
- 22.Huttner K M, Lambeth M R, Burkin H R, Burkin D J, Broad T E. Localization and genomic organization of sheep antimicrobial peptide genes. Gene. 1998;206:85–91. doi: 10.1016/s0378-1119(97)00569-6. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Wang L, Jia H P, Zhao C, Heng H H Q, Schutte B C, McCray P B, Jr, Ganz T. Structure and mapping of the human β-defensin HBD-2 gene and its expression at sites of inflammation. Gene. 1998;222:237–244. doi: 10.1016/s0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Zhao C, Heng H H Q, Ganz T. The human β-defensin-1 and α-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics. 1997;43:316–320. doi: 10.1006/geno.1997.4801. [DOI] [PubMed] [Google Scholar]
- 25.Longo C, Tyler D, Mallampalli R K. Sphingomyelin metabolism is developmentally regulated in rat lung. Am J Respir Cell Mol Biol. 1997;16:605–612. doi: 10.1165/ajrcmb.16.5.9160843. [DOI] [PubMed] [Google Scholar]
- 26.McCray P B, Jr, Bentley L. Human airway epithelia express a β-defensin. Am J Respir Cell Mol Biol. 1997;16:343–349. doi: 10.1165/ajrcmb.16.3.9070620. [DOI] [PubMed] [Google Scholar]
- 27.Miller M. The susceptibility to lysozyme of beta-lactamase-producing and non-producing derivatives of Staphylococcus aureus strain 1030. J Med Microbiol. 1987;23:127–132. doi: 10.1099/00222615-23-2-127. [DOI] [PubMed] [Google Scholar]
- 28.Morrison G M, Davidson D J, Kilanowski F M, Borthwick D W, Crook K, Maxwell A I, Govan J R W, Dorin J R. Mouse beta defensin-1 is a functional homolog of human beta defensin-1. Mamm Genome. 1998;9:453–457. doi: 10.1007/s003359900795. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura D Y, Swiderski R E, Alward W L, Searby C C, Patil S R, Bennet S R, Kanis A B, Gastier J M, Stone E M, Sheffield V C. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds H Y. Integrated host defense against infections. In: Reynolds H Y, editor. The lung. New York, N.Y: Raven Press, Ltd.; 1997. pp. 2353–2365. [Google Scholar]
- 31.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 32.Singh P K, Jia H P, Wiles K, Hesselberth J, Liu L, Conway B D, Greenberg E P, Valore E V, Welsh M J, Ganz T, Tack B F, McCray P B., Jr Production of β-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 34.Snyder J M. The biology of the surfactant-associated proteins. In: Bourbon J, editor. Pulmonary surfactant: biochemical, functional, regulatory and clinical concepts. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 105–126. [Google Scholar]
- 35.Stein L, Kruglyak D, Slonim D, Lander E. “RHMAPPER” software. Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research. 1995. http://www.genome.wi.mit.edu/ftp/pub/software/rhmapper Available at http://www.genome.wi.mit.edu/ftp/pub/software/rhmapper and via anonymous ftp to ftp.genome.wi.mit.edu, directory /pub/software/rhmapper. and via anonymous ftp to , directory /pub/software/rhmapper. [Google Scholar]
- 36.Tarver A P, Clark D P, Diamond G, Russell J P, Erdjument-Bromage H, Tempst P, Cohen K S, Jones D E, Sweeney R W, Wines M, Hwang S, Bevins C L. Enteric β-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tchepichev S, Ueda J, Canessa C, Rossier B C, O’Brodovich H. Lung epithelial Na channel subunits are differentially regulated during development and by steroids. Am J Physiol. 1995;269:C805–C812. doi: 10.1152/ajpcell.1995.269.3.C805. [DOI] [PubMed] [Google Scholar]
- 38.Valore E V, Park C H, Quayle A J, Wiles K R, McCray P B, Jr, Ganz T. Human β-defensin-1, an antimicrobial peptide of urogenital tissues. J Clin Investig. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe S, Matsushita K, Stokes J B, McCray P B., Jr Developmental regulation of epithelial sodium channel subunit mRNA expression in rat colon and lung. Am J Physiol. 1998;275:G1227–G1235. doi: 10.1152/ajpgi.1998.275.6.G1227. [DOI] [PubMed] [Google Scholar]
- 40.Welsh M J, Boat T F, Tsui L-C, Beaudet A L. Cystic fibrosis. In: Scriver C R, Beaudet A L, Sly W S, Valle D, editors. The metabolic and molecular basis of inherited disease. New York, N.Y: McGraw-Hill, Inc.; 1995. pp. 3799–3876. [Google Scholar]
- 41.Whitsett J A, Nogee L M, Weaver T E, Horowitz A D. Human surfactant protein B: structure, function, regulation, and genetic disease. Physiol Rev. 1995;75:749–757. doi: 10.1152/physrev.1995.75.4.749. [DOI] [PubMed] [Google Scholar]
- 42.Wilcox J N. Fundamental principles of in situ hybridization. J Histochem Cytochem. 1993;41:1725–1733. doi: 10.1177/41.12.8245419. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Wu H, Shi J, Ganz T, Ross C R, Blecha F. Molecular cloning and tissue expression of porcine β-defensin-1. FEBS Lett. 1998;424:37–40. doi: 10.1016/s0014-5793(98)00134-3. [DOI] [PubMed] [Google Scholar]
- 44.Zucht H D, Grabowsky J, Schrader M, Liepke C, Jurgens M, Knappe P S, Forssmann W G. Human β-defensin-1: a urinary peptide present in variant molecular forms and its putative functional implication. Eur J Med Res. 1998;3:315–323. [PubMed] [Google Scholar]