Abstract
Familial dilated cardiomyopathy (DCM) is among the most prevalent forms of inherited heart disease. Here, two human-induced pluripotent stem cell (iPSC) lines were generated from peripheral blood mononuclear cells (PBMCs) from DCM patients carrying different mutations in the phospholamban encoding-gene (PLN). Both iPSC lines exhibited normal morphology, karyotype, pluripotency marker expression, and differentiation into the three germ layers. These patient-specific iPSC lines serve as valuable in vitro models for DCM pathology caused by PLN mutations.
Keywords: iPSC, Stem cell, Dilated cardiomyopathy, Phospholamban
1. Resource table
| Unique stem cell lines identifier | 1. SCVIi049-A |
| 2. SCVIi050-A | |
| Alternative name(s) of stem cell lines | 1. SCVIi049-A / SCVI104 |
| 2. SCVIi050-A / SCVI2486 | |
| Institution | Stanford Cardiovascular Institute, Stanford, CA, US |
| Contact information of distributor | Joseph C. Wu, joewu@stanford.edu |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC | Age: 44 (SCVIi049-A) and 30 (SCVIi050-A) |
| Sex: male | |
| Ethnicity if known: Not Hispanic or Latino | |
| Cell Source | Fibroblast (SCVIi049-A), PBMC (SCVIi050-A) |
| Clonality | Clonal |
| Method of reprogramming | Nonintegrating Sendai virus expression of human OCT4, SOX2, KLF4, and c-MYC |
| Genetic Modification | Yes |
| Type of Genetic Modification | Spontaneous mutation |
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | RT-qPCR |
| Associated disease | Dilated cardiomyopathy (DCM) |
| Gene/locus | PLN (6q22.31) |
| SCVIi049-A: heterozygous PLN (c.25C > T) | |
| SCVIi050-A: heterozygous PLN (c.40_42delAGA) | |
| Date archived/stock date | SCVIi049-A: 09/10/2019 |
| SCVIi050-A: 12/03/2021 | |
| Cell line repository/bank | https://hpscreg.eu/cell-line/SCVIi049-A |
| https://hpscreg.eu/cell-line/SCVIi050-A | |
| Ethical approval | The generation of the lines was approved by the Administrative Panel of Human Subjects Research (IRB) under IRB #29904 “Derivation of Human Induced Pluripotent Stem Cells” |
2. Resource utility
Patients carrying pathogenic (c.40_42delAGA) and likely pathogenic (c.25 C>T) mutations in the PLN gene developed dilated cardiomyopathy (DCM). Generation of iPSC lines carrying these mutations provides an unlimited source for differentiation into cardiac cell types (e.g., cardiomyocytes, endothelial cells, fibroblasts), thus providing an excellent tool for in vitro modeling of DCM pathogenesis, testing of candidate therapies, and advancement of personalized medicine (see Table 1).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Photography bright field | Normal | Fig. 1A |
| Phenotype | Qualitative analysis | Positive expression of pluripotency markers by immunocytochemistry: NANOG, SOX2, and OCT3/4 | Fig. 1B |
| Genotype | Karyotype (G-banding) and resolution | Karyostat™ Assay, resolution 1–2 Mb: Normal karyotype 46, XY for both lines. | Fig. 1H |
| Identity | Microsatellite PCR (mPCR) or STR analysis | Not performed | N/A |
| 22 loci tested, 100% identical | Submitted in archive with journal | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | SCVIi049-A: heterozygous c.25C > T | Fig. 1F |
| SCVIi050-A: heterozygous c.40_42delAGA | |||
| Southern blot OR WGS | Not performed | Not performed | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence: Negative (p10 and above) | Supplemental Fig. 1A |
| Differentiation potential | Embryoid body formation or Teratoma formation or Scorecard or directed differentiation | Directed differentiation, positive expression of germ layer markers | Fig. 1G |
| List of recommended germ layer markers | Expression of these markers has to be demonstrated at mRNA (RT PCR) or protein (IF) levels, at least 2 markers need to be shown per germ layer | Positive expression of germ layer markers: | Fig. 1G |
| Ectoderm: Pax6, Otx2 | |||
| Mesoderm: Brachyury, Tbx6 | |||
| Endoderm: Sox17, Foxa2 | |||
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | Not performed |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | Not performed |
| HLA tissue typing | Not performed | Not performed | |
3. Resource details
Dilated cardiomyopathy (DCM), with a prevalence of nearly 1:2,500 people, is the most common cause of heart failure after coronary artery disease and the leading indication for heart transplantation (Maron et al., 2006). Clinical hallmarks of DCM include contractile dysfunction and thinning of the myocardium. Intracellular Ca2+ handling is the central coordinator of cardiac contraction and relaxation. Phospholamban, encoded by the PLN gene, is an abundant, 52 amino acid transmembrane SR phosphoprotein that regulates cardiomyocyte calcium handling as the primary inhibitor of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) (Schmitt et al., 2003). Several disease variants in the PLN gene have been described in heart failure, but no specific therapies exist beyond standard heart failure treatments or heart transplantation (Eijgenraam et al., 2020). The underlying mechanisms of PLN mutations in DCM remain incompletely understood. Using small animal modeling to study mutation-specific studies is historically a laborious, expensive, time-consuming strategy, taking years before the results of a single treatment may be evaluated (Eijgenraam et al., 2020). The advent of iPSC technology makes in vitro modeling of cardiac diseases possible. Here, cardiovascular cell types derived from patient-specific iPSCs with mutations in PLN present a valuable research opportunity to model DCM disease mechanisms.
We derived two human iPSC lines (SCVIi049-A and SCVIi050-A) from peripheral blood mononuclear cells (PBMCs) and fibroblasts of two patients carrying variants in the PLN gene, including a 44-year-old East Asian male (SCVIi049-A, c.25 C>T encoding p.Arg9Cys, likely pathogenic), and a 30-year-old Caucasian male (SCVIi050-A, c.40_42delAGA encoding p.Arg14del, pathogenic) (Resource Table). Reprogramming of somatic donor cells to iPSCs was conducted using a non-integrating Sendai virus containing the four Yamanaka factors described previously (Liu et al., 2021). Both iPSC lines showed typical morphology (Fig. 1A, Table 1). SCVI049-A and SCVIi050-A demonstrated high expression of pluripotency markers, OCT3/4, NANOG, and SOX2 detected by immunofluorescence (Fig. 1B). The expression of pluripotency markers was confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Both SCVIi049-A and SCVIi050-A had comparable SOX2 and NANOG expression levels to the widely used positive control line, SCVI15 (Sun et al., 2012), but expressed much higher than iPSC-derived cardiomyocytes (iPSC-CMs) derived from SCVI15 (Fig. 1C–D). Furthermore, expression of the non-integrating Sendai virus, present at low passage numbers (SCVI15, p4), was absent in SCVIi049-A (p17) and SCVIi050-A (p20) measured by RT-qPCR (Fig. 1E).
Fig. 1.
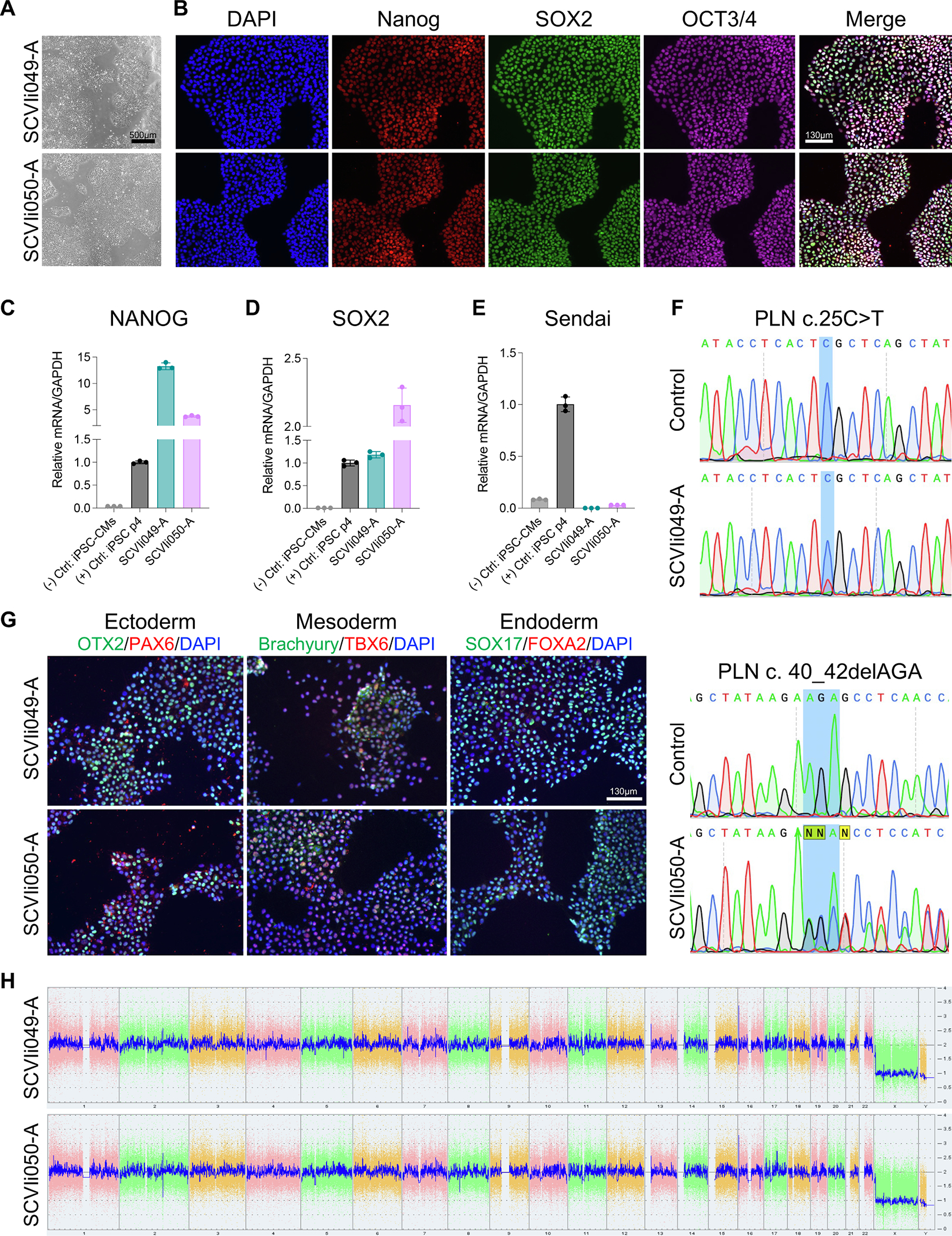
Characterization of patient-derived iPSC lines with c.25C>T and c.40_42delAGA mutations in PLN.
The heterozygous mutations of both iPSC lines were confirmed by Sanger sequencing (Fig. 1F). Short tandem repeat (STR) analysis confirmed that both SCVIi049-A and SCVIi050-A demonstrated overlapping profiles with their respective donor somatic cells (Submitted in archive with journal). Additionally, both iPSC lines could differentiate into all three – ectoderm, mesoderm, and ectoderm – germ layers visualized by immunocytochemistry (Fig. 1G). SCVIi049-A and SCVI050-A had normal karyotype results assessed by the KaryoStat™ assay (Fig. 1H). Both iPSC lines were mycoplasma-negative (Supplemental Fig. 1A).
4. Materials and methods
4.1. Reprogramming
Peripheral blood mononuclear cells (PBMCs) were isolated from patients’ blood by PercollR gradient separation. PBMCs were purified and replated as previously described (Liu et al., 2021). Briefly, PBMCs were cultured in 1 ml of Stem-Pro™-34 medium (100 ng/ml FLT3, 20 ng/ml IL-6, 20 ng/ml EPO, 20 ng/ml IL-3, and 100 ng/ml SCF). PBMCs were resuspended in 300ul of Stem-Pro™-34 medium and transduced with Sendai virus reprogramming cocktail (CytoTune®-iPSC Sendai Reprogramming Kit). After 24 h, cells were replated, and the medium was replaced every two days. On Day 7, 1 ml of supplemented StemMACS™ iPSC-Brew XF medium (Miltenyi Biotec) was added on top of Stem-Pro™-34 medium. On Day 8, the medium was replaced completely with StemMACS™ iPSC-Brew XF medium. Fresh StemMACS™ iPSC-Brew XF medium was replaced on Days 10–15 when colonies appeared.
4.2. Cell culture
Patient-derived iPSCs were cultured in StemMACS iPS-Brew XF medium. Rock inhibitor (10uM, Y27632 Selleck Chemicals) was added up to 24 h after passage. Medium was replaced every two days until confluency. Cells were maintained in a 37 °C incubator with 5% CO2 and 20% O2.
4.3. Karyotyping
Patient-derived iPSCs were analyzed using the KaryoStat™ assay (ThermoFisher Scientific) at p10 (SCVIi049-A) and p8 (SCVIi050-A).
4.4. Trilineage differentiation
STEMdiff™ Trilineage Differentiation Kit (STEMCELL Technologies #05230) was used to induce differentiation into endoderm and ectoderm. Mesoderm differentiation was induced with RPMI + glucose medium with B27 minus insulin. Differentiation was performed at p10 (SCVIi049-A) and p8 (SCVIi050-A).
4.5. Immunofluorescence staining
At room temperature, cells were fixed in 4% paraformaldehyde, then permeabilized with 50 ug/ml digitonin (Sigma Aldrich #D141) for 10 min each. Cells were incubated with a blocking solution (1% BSA) for 30 min. Cells were incubated with primary antibodies (Table 2) overnight at 4 °C. The following day, cells were washed 3 times. Cells were incubated in secondary antibodies (Table 2) for 30 min at room temperature, then washed 3 times. Nuclei were stained with Molecular Probes NucBlue (ThermoFisher Scientific #R37606) for 10 min at room temperature. Cells were washed 3 times, then imaged using a confocal light microscope. Immunostaining was carried out at p16 (SCVIi049-A) and p19 (SCVIi050-A).
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry |
||||
| Antibody | Dilution | Company Cat # | RRID | |
|
| ||||
| Pluripotency Marker | Mouse IgG2b kAnti-OCT-3/4 | 1:100 | Santa Cruz Biotechnology Cat# sc-5279 | RRID: AB_628051 |
| Pluripotency Marker | Rabbit Anti-NANOG | 1:100 | Protein Tech Cat# 142951-1-AP | RRID: AB_1607719 |
| Pluripotency Marker | Mouse IgG1 kAnti-SOX2 | 1:100 | RRID: AB_10842165 | |
| Santa Cruz Biotechnology Cat# sc-365823 | ||||
| Differentiation Marker (Ectoderm) | Goat Anti-OTX2 | 1:200 | R&D Systems Cat#963273 | RRID: AB_2157172 |
| Differentiation Marker (Ectoderm) | Rabbit Anti-PAX6 | 1:200 | Thermo Fisher Scientific Cat#42-6600 | RRID: AB_2533534 |
| Differentiation Marker (Endoderm) | Goat Anti-SOX17 | 1:200 | R&D Systems Cat#963121 | RRID: AB_355060 |
| Differentiation Marker (Endoderm) | Rabbit Anti-FOXA2 | 1:250 | Thermo Fisher Scientific Cat#701693 | RRID: AB_2576439 |
| Differentiation Marker (Mesoderm) | Goat Anti-Brachyury | 1:200 | R&D Systems Cat#963427 | RRID: AB_2200235 |
| Differentiation Marker (Mesoderm) | Rabbit Anti-TBX6 | 1:200 | Thermo Scientific Cat#PA5-35102 | RRID: AB2552412 |
| Secondary Antibody | Alexa Fluor 488 Goat Anti-Mouse IgG1 | 1:1000 | Thermo Fisher Scientific #A-21121 | RRID: AB_2535764 |
| Secondary Antibody | Alexa Fluor 647 Goat Anti-Mouse IgG2b | 1:250 | Thermo Fisher Scientific #A21242 | RRID: AB_2535811 |
| Secondary Antibody | Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L) | 1:500 | Thermo Fisher Scientific #A-21428 | RRID: AB_141784 |
| Secondary Antibody | Alexa Fluor 488 Donkey Anti-Goat IgG (H+L) | 1:1000 | Thermo Fisher Scientific #A-11055 | RRID: AB_2534102 |
| Primers Target | Size of band | Forward/Reverse primer (5′-3′) | ||
|
| ||||
| Genotyping | SCVIi049-A: c.25 C>T | 376 bp | F: TTTTACATTCCAGGCTACCTAAAAG | |
| R: TCTACTCAGGAAGTGGTCTGT | ||||
| Genotyping | SCVIi050-A: c.40_42delAGA | 376 bp | F: TTTTACATTCCAGGCTACCTAAAAG | |
| R: TCTACTCAGGAAGTGGTCTGT | ||||
| Sendai virus plasmid (RT-qPCR) | Sendai virus genome | 181 bp | Mr042698800_mr | |
| Pluripotency markers (RT-qPCR) | SOX2 | 258 bp | Hs04234836_s1 | |
| Pluripotency markers (RT-qPCR) | NANOG | 327 bp | Hs02387400_g1 | |
| Housekeeping genes (RT-qPCR) | GAPDH | 471 bp | Hs02786624_g1 | |
4.6. RT-qPCR
RNA was extracted using the Direct-zol™ RNA Miniprep Kit (ZYMO Research #3R2061). To generate cDNA, iScript™ cDNA Synthesis Kit (BioRad #1708891) was used as follows: 5 min at 25 °C, 20 min at 46 °C, and 1 min at 95 °C. Expression of SOX2, NANOG, and SEV was amplified using commercial primers (Table 2) and TaqMan™ Gene expression Assay (Applied Biosystems™ #4444556).
4.7. Short tandem repeat analysis
Genomic DNA (gDNA) from fibroblasts (SCVI049-A), PBMCs (SCVIi050-A), and iPSCs were purified using DNeasy Blood & Tissue Kit (Qiagen). STR analysis was performed using CLA Identifier™ Plus and Identifier™ Direct PCR Amplification Kits (Thermo Fisher) by the Stanford PAN Facility.
4.8. Sanger sequencing
PCR primers were designed to flank PLN mutations (Table 2) and used to amplify the genomic region using Q5® Hot Start High-Fidelity DNA Polymerase (New England BioLabs). The PCR reaction was performed as follows: 98 °C for 5 sec, 62 °C for 10 sec, 72 °C for 20 sec for 35 cycles. PCR products were purified using QIAquick Purification Kit (Qiagen) and sent to the Stanford PAN facility.
4.9. Mycoplasma detection
Mycoplasma contamination was evaluated using a MycoAlert Detection Kit (Lonza #LT07–318) at p17 (SCVIi049-A) and p20 (SCVIi050-A).
Supplementary Material
Acknowledgements
We thank Chelsea Lee, Celine Lai, McKay MS Mullen, James WS Jahng, Julio Vicente Guevara, and Yan Zhuge for the technical support of this manuscript. This work was supported by the National Institutes of Health (NIH) R01 HL130020, R01 HL163680, P01 HL141084 (to J.C. W), NIH Administrative Diversity Supplement 3R01HL130020-06S1 (to C.D.V), and American Heart Association (AHA) Postdoctoral Fellowship 908936 (to A.C) and AHA Research Supplement to Promote Diversity in Science 872244 (to G.M.P).
Footnotes
Declaration of Competing Interest
The authors declare the following financial interest/personal relationships which may be considered as a potential competing : JCW is a co-founder and board member of Greenstone Biosciences and Khloris Biosciences. The other authors declare no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2022.102855.
References
- Eijgenraam TR, Boukens BJ, Boogerd CJ, Schouten EM, van de Kolk CWA, Stege NM, te Rijdt WP, Hoorntje ET, van der Zwaag PA, van Rooij E, van Tintelen JP, van den Berg MP, van der Meer P, van der Velden J, Silljé HHW, de Boer RA, 2020. The phospholamban p.(Arg14del) pathogenic variant leads to cardiomyopathy with heart failure and is unresponsive to standard heart failure therapy. Scientific Rep. 10, 9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Shenoy SP, Jahng JWS, Liu Y, Knowles JW, Zhuge Y, Wu JC, 2021. Generation of two heterozygous MYBPC3 mutation-carrying human iPSC lines, SCVIi001-A and SCVIi002-A, for modeling hypertrophic cardiomyopathy. Stem Cell Res. 53, 102279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB, 2006. Contemporary definitions and classification of the cardiomyopathies. Circulation 113, 1807–1816. [DOI] [PubMed] [Google Scholar]
- Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, Kranias EG, MacLennan DH, Seidman JG, Seidman CE, 2003. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299, 1410–1413. [DOI] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC, 2012. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 4, 130ra147–130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


