Abstract
Background
Advanced gastrointestinal stromal tumour (GIST) is characterised by genomic perturbations of key cell cycle regulators. Oncogenic activation of CDK4/6 results in RB1 inactivation and cell cycle progression. Given that single-agent CDK4/6 inhibitor therapy failed to show clinical activity in advanced GIST, we evaluated strategies for maximising response to therapeutic CDK4/6 inhibition.
Methods
Targeted next-generation sequencing and multiplexed protein imaging were used to detect cell cycle regulator aberrations in GIST clinical samples. The impact of inhibitors of CDK2, CDK4 and CDK2/4/6 was determined through cell proliferation and protein detection assays. CDK-inhibitor resistance mechanisms were characterised in GIST cell lines after long-term exposure.
Results
We identify recurrent genomic aberrations in cell cycle regulators causing co-activation of the CDK2 and CDK4/6 pathways in clinical GIST samples. Therapeutic co-targeting of CDK2 and CDK4/6 is synergistic in GIST cell lines with intact RB1, through inhibition of RB1 hyperphosphorylation and cell proliferation. Moreover, RB1 inactivation and a novel oncogenic cyclin D1 resulting from an intragenic rearrangement (CCND1::chr11.g:70025223) are mechanisms of acquired CDK-inhibitor resistance in GIST.
Conclusions
These studies establish the biological rationale for CDK2 and CDK4/6 co-inhibition as a therapeutic strategy in patients with advanced GIST, including metastatic GIST progressing on tyrosine kinase inhibitors.

Subject terms: Sarcoma, Targeted therapies, Cancer genomics, Molecular medicine, Cancer therapeutic resistance
Background
Gastrointestinal stromal tumour (GIST) is the most common mesenchymal neoplasm of the GI tract. Approximately 85% of GISTs are initiated by gain-of-function mutations in the KIT [1] or PDGFRA [2] genes resulting in constitutive tyrosine kinase (TK) activity. Tyrosine kinase inhibitors (TKI), such as imatinib, result in dramatic clinical responses in patients with advanced GIST [3, 4], but the majority of patients eventually develop resistant disease caused by the emergence of numerous subclonal TK mutations [5, 6]. The development of genomic heterogeneity with polyclonal inter- and intra-tumoral TK resistance mutations leads to fatal outcomes [5, 6] and highlights the need to target additional therapeutic vulnerabilities conserved across the metastatic burden in a given patient.
Malignant transformation in GIST is driven by a sequential series of genomic events that build upon the initiating KIT or PDGFRA mutation: early on, genomic inactivation of the 14q-tumour suppressor MAX is a highly recurrent mechanism causing transcriptional downregulation of p16 and thereby early cell cycle dysregulation in GIST [7], followed by subsequent inactivation of the 22q-tumour suppressor DEPDC5 [8], 1p deletion [9], genomic inactivation of cell cycle regulators such as CDKN2A, RB1 or TP53 [10], 15q deletion [9], inactivation of dystrophin [11], and finally, the emergence of KIT or PDGFRA resistance mutations during TKI therapy [5, 6]. Importantly, three of these eight steps incrementally target the CDK4/6 pathway (Supplementary Fig. 1). While the p16INK4A and p14ARF tumour suppressors are both encoded by CDKN2A, they use alternate reading frames, and their protein sequences have no similarity. p16 inhibits the activity of cyclin D-CDK4/6 complexes, whereas p14 prevents MDM2-mediated degradation of p53, enabling cell cycle arrest or p53-mediated apoptosis. Both pathways converge on RB1 which is inactivated through mono-phosphorylation by cyclin D-CDK4/6 complexes and hyperphosphorylation by cyclin E-CDK2 complexes to enable G1/S phase progression through E2F-mediated expression of target genes [12].
In non-metastatic GIST with high propensity to metastasise, genomic perturbations of key cell cycle regulators frequently target the CDK2 and CDK4/6 pathways [10, 13, 14] and result in RB1 inactivation and cell cycle progression. In contrast to the inter-tumoral heterogeneity of polyclonal TKI resistance mutations in individual patients, cell cycle regulator aberrations are conserved across the metastatic burden in many GIST patients, independent of TKI response [10], which highlights the restoration of cell cycle control as a potentially compelling therapeutic strategy for advanced GIST. The CDK4/6 inhibitors palbociclib [15], abemaciclib [16] and ribociclib [17] have been FDA-approved for the treatment of breast cancer, in combination with aromatase inhibitors or oestrogen receptor antagonists, with impressive clinical activity, although the primary impact appears to be the induction of cell cycle arrest in G1. A clinical trial failed to demonstrate any clinical activity of palbociclib monotherapy in advanced TKI-resistant CDKN2A/p16-deleted GIST [18], indicating that CDK4/6 inhibition alone is inadequate for disease control in this population.
Combined inhibition of CDK2 and CDK4/6 has recently been reported to synergistically inhibit cell proliferation in preclinical models of breast cancer, suggesting CDK2 inhibition as a potential therapeutic strategy to overcome palbociclib resistance [19].
Herein, we use genomic sequencing and multiplexed protein imaging to detect co-dysregulation of the CDK2 and CDK4/6 pathways in GIST clinical specimens. We evaluate strategies of therapeutic cell cycle restoration through combined inhibition of CDK2 and CDK4/6 in vitro, and explore mechanisms of acquired CDK-inhibitor resistance in GIST.
Methods
Tumour and tissue samples
Frozen discarded, deidentified tumour specimens were obtained at Brigham and Women’s Hospital (BWH). Paraffin-embedded and formalin-fixed (FFPE) tissue samples were retrieved from the BWH surgical pathology files. In total, 18 GISTs from 18 patients were included on a tissue microarray block with two 1.5-mm cores per tumour. Cases were selected to include GISTs of various risk categories and genomic subtypes with material available for genomic and protein evaluations.
Cell lines
GIST430, GIST430/654 and sub-lines, GIST48, GIST882 and LMS05 were established in the Fletcher laboratory. GIST-T1 was obtained from Kochi Medical School (Kochi, Japan). All cell lines were validated against the initial biopsy material by molecular cytogenetics and sequencing verification of known unique gene mutations. All cultures were shown to be mycoplasma-free. BT474 was obtained from ATCC (cat. no. HTB-20).
Multiplexed and amplified in situ protein imaging (Immuno-SABER)
Primary antibodies were conjugated to bridge oligos as detailed in Supplementary Table 2 using the general protocol previously described [20]. Primary concatemers were extended from 1 µM primers prepared in vitro by Primer Exchange Reaction (PER) [21] to an average length of 500–750 nucleotides as detailed before [20] following the modular SABER design [22]. Staining was performed as described before [20] with minor modifications (Supplementary Table 2). Multi-channel images were registered based on the DAPI channel and stitched (only for tissues) using the ASHLAR algorithm [23]. The resulting pyramid images in ome-tiff form were manually thresholded and prepared into figures using Omero [24].
Immunohistochemistry
IHC was performed on 4-µm-thick formalin-fixed paraffin-embedded TMA sections following pressure cooker antigen retrieval (Target Retrieval Solution; pH 6.1; Dako, Carpinteria, CA, USA) using antibodies listed in Supplementary Table 3, controls and staining conditions as described previously [25].
Protein blotting and immunoprecipitation
Whole-cell lysates were prepared as described previously [26]. Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA, USA, cat. no. 5000006). Electrophoresis, immunoblotting and chemiluminescence detection were performed as described previously [26] using antibodies listed in Supplementary Table 3. Protein expression was calibrated using the Multi Gauge V2.3 software (Fujifilm, Tokyo, Japan).
Targeted sequencing
Targeted sequencing was performed using the OncoPanel platform as described previously [27, 28], interrogating the exonic sequences of 447 cancer-associated genes for mutations and copy number variations, and 191 introns across 60 genes for gene rearrangements. DNA was extracted from FFPE or frozen tissue using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Construction of hybrid-capture libraries, sequencing using the Illumina HiSeq 2500 (Illumina, San Diego, CA), and sequence data analysis was performed as previously described [27]. All detected alterations were reviewed manually and annotated.
3’ Rapid amplification of cDNA ends (3’RACE) and polymerase chain reaction (PCR)
3’ RACE was performed using a 3’ RACE system (Thermo Fisher Scientific, cat. no. 18373019) using gene-specific forward primers against CCND1 exon 3 and 4 shown in Supplementary Table 4 and an abridged universal amplification primer. Nested PCR was performed on the PCR product using primers against CCND1 exon 5 and the 3’ flanking sequence of AP003555.2 shown in Supplementary Table 4, followed by Sanger sequencing (GeneWiz, South Plainfield, NJ, USA).
PCR and RT-PCR analysis
Genomic PCR and Sanger sequencing of RB1 exons 1 and 24 (NM_000321.3) was performed using primers shown in Supplementary Table 4. RT-PCR of the CCND1 fusion was performed using total RNA extracted using Qiagen RNeasy Plus Universal mini kit following the manufacturer’s instructions (Qiagen) using a SuperScript IV One-Step RT-PCR kit (Thermo Fisher Scientific, cat. no. 12594025) with primers listed in Supplementary Table 4.
Cell culture and reagents
Cell lines were maintained in RPMI 1640 medium plus 3% foetal bovine serum (FBS) containing penicillin/streptomycin and L-glutamine. GIST430/654 resistance cultures were created through subjection to palbociclib for at least 10 months. Palbociclib was obtained from LC Laboratories (Woburn, MA, USA, cat. no. P7788), abemaciclib from Selleck Chemicals (Houston, TX, USA, cat. no. S5716), PF-06873600 from MedChemExpress (Monmouth Junction, NJ, USA, cat. no. HY-114177), the CDK2 inhibitor II from Santa Cruz (CAS 222035-13-4, cat. no. sc-221409), alisertib from Selleck Chemicals (cat. no. S1133), the Skp2 inhibitor C1 from Selleck Chemicals (cat. no. S8652), and volasertib from Selleck Chemicals (cat. no. S2235).
Mass spectrometry
GeLC-MS/MS (gel-based liquid chromatography-based tandem mass spectrometry) samples were prepared as described previously [29, 30].
Cell-proliferation assays
Cells were plated in 96-well plates at 5000 (GIST430, GIST430/654 and sub-lines, GIST48) or 10,000 (GIST882) cells/well in growth medium and incubated overnight. Drug treatment was performed for 48 h. BrdU was added to the cells for 24 h. BrdU incorporation, fixation and detection were performed using a BrdU Cell Proliferation ELISA as per the manufacturer’s protocol (Roche Diagnostics, Indianapolis, IN, USA). BrdU incorporation was presented as fold change of DMSO control. All cell response assays were performed in triplicate wells.
Cell viability and apoptosis assays
GIST was plated at 15,000 cells/well in a 96-well flat-bottomed plate (Falcon, Lincoln, NJ) and cultured for 24 h before treatment with palbociclib or PF-06783600. Cell viability studies were performed after 6 days of treatment using a CellTiter-Glo luminescence assay (Promega, Madison, WI). Apoptosis studies were performed after 48 h of treatment using a Caspase-Glo 3/7 luminescence assay (Promega).
BrdU incorporation assay
Cells were pulsed with BrdU (10 µM; BD Biosciences) for 2 h and subsequently trypsinized, washed with phosphate-buffered saline (PBS), and fixed with 80% ethanol (EtOH) overnight at 4 °C. Next, DNA was hydrolysed by incubation with 2 N HCl/0.5% Triton X-100 for 30 min at room temperature (RT) and neutralised by adding 0.1 M Na2B4O7 (pH 8.5). Subsequently, cells were incubated in 100 µL of PBS/1% bovine serum albumin (BSA) containing fluorescein isothiocyanate-conjugated anti-BrdU antibody (BD Biosciences, cat. no. 556028) 1:5 for 30 min at RT. Next, cells were washed twice with PBS, stained with 20 µL of 7-AAD (7-Amino-Actinomycin D) (BD Biosciences), resuspended in PBS, and analysed by flow cytometry. Data acquisition was performed on a BD LSRFortessa Flow Cytometry Analyzer, and data analysis was done with FlowJo (v10.8.1).
Growth over time analysis
Cells were plated in regular growth media at 1.5 Mio cells/well in six-well plates, and treatment with palbociclib (31.25 nM) versus DMSO control was started one day after plating (day 0). Cells were trypsinized and counted on days 0, 1, 3, 7 and 10 using an automated cell counter (TC20, Bio-Rad TC20, Hercules, CA).
IC50 calculations, dose–response curves and drug synergy analysis
IC50 values were calculated using a Quest Graph ED50 Calculator (AAT Bioquest, Inc. 2022, https://www.aatbio.com/tools/ed50-calculator). Drug synergy testing was performed using SynergyFinder [31], which generated combination sensitivity scores [32].
Whole-exome sequencing
DNA was isolated using a QIAamp DNA Mini Kit (Qiagen) and submitted for paired-end 150-bp whole-exome sequencing performed on an Illumina HiSeq platform (GeneWiz) with ~350 M, a mean quality score of 39.10 and 94.68% of targets covered at a depth of ≥30× and aligned to the Homo Sapiens GRCh37 (hg19) reference genome. Somatic variants were called using the Illumina Dragen Bio-IT Platform in somatic mode. Variants were further filtered and annotated with Ensembl Variant Effect Predictor (VEP) v95. BAM files containing CCND1 and AP003555.2 coding and flanking sequence data were reviewed manually in IGV.
RNA sequencing
RNA library preparation, sequencing reaction and bioinformatic analysis were conducted at GeneWiz, LLC. (South Plainfield, NJ, USA) after 24 h treatment with palbociclib (31.25 nM) vs. DMSO. Total RNA was extracted using Qiagen RNeasy Plus Universal mini kit following the manufacturer’s instructions (Qiagen), and a sequencing library was prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina using the manufacturer’s instructions (NEB, Ipswich, MA, USA). The samples were sequenced using a 2 × 150-bp Paired-End (PE) configuration and overall read amount per HiSeq lane of ~350 M, and trimmed reads were mapped to the Homo Sapiens GRCh38 (hg38) reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Gene expression between the groups was compared using DESeq2. The Wald test was used to generate P values and log2 fold changes. Genes with adjusted P values <0.05 and absolute log2 fold changes >1 were called as differentially expressed genes for each comparison. Gene ontology analysis was performed on the statistically significant genes set by implementing the software GeneSCF. The goa_human GO list was used to cluster the set of genes based on their biological process and determine their statistical significance.
Constructs and virus infection
A custom CCND1::chr11.g:70025223 fusion constructs inserted into empty vector pLKO.1puro was obtained from GenScript (Piscataway, NJ, USA). Transfections and lentiviral harvesting were carried out as described before [33]. Following transduction, cells were selected with 1.5 μg/mL (GIST430/654, BT474) or 2 μg/mL (LMS05) puromycin. Culture images were obtained using Spot RT Slider Camera and Spot Software (Version 4.6 for Windows) and a Nikon Eclipse TE2000-S inverted microscope.
Statistics
Statistical analyses were performed using GraphPad Prism Software and Stata IC15 software, and a one-way ANOVA to compare two data sets. A P value of ≤0.05 was considered statistically significant.
Results
Frequent dysregulation of the CDK2 and CDK4/6 pathways in clinical samples from metastatic TKI-resistant GIST patients
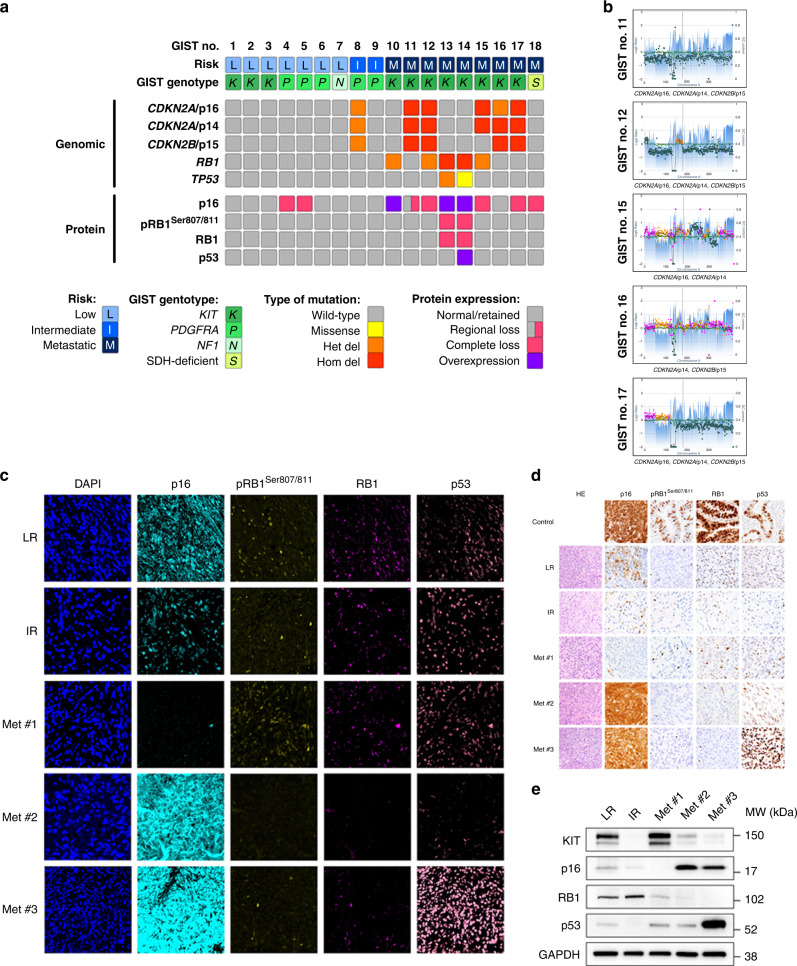
To determine the frequency of cell cycle regulator aberrations, particularly those co-dysregulating the CDK2 and CDK4/6 pathways in GIST and thereby establish a biological rationale for therapeutic co-targeting, we evaluated genomic and protein expression in GIST clinical specimens across risk categories and genomic subtypes (N = 18). Patients with metastatic KIT-mutant GISTs had progressing disease at the time of surgery after 2–4 lines of TKI therapy. While previous studies describe low levels of CDKN2A/p16 and CDKN2A/p14 mRNA expression in 24–55% of samples [13, 14], presumably due to homozygous co-deletions affecting CDKN2A/p16 and CDKN2A/p14 [14], the CDKN2A and CDKN2B genes have not been evaluated for inactivating genomic aberrations by targeted NGS, particularly with the concomitant evaluation of RB1 predictive of response to CDK4/6 inhibitor therapies. Our targeted NGS results demonstrated homozygous deletions of CDKN2A/p16 in 4 (22%), CDKN2A/p14 in 5 (28%), CDKN2B/p15 in 4 (22%), and RB1 in 2 (11%) of 18 GISTs, and a homozygous TP53 missense mutation in one (6%) of 18 GISTs (Fig. 1a). A homozygous deletion inactivating CDKN2A/p16, CDKN2A/p14 and CDKN2B/p15 coding regions was identified in three cases, and a homozygous deletion inactivating CDKN2A/p16 and CDKN2A/p14 or CDKN2A/p14 and CDKN2B/p15 was found in one case each, overall resulting in co-dysregulation of the CDK2 and CDK4/6 pathways in five of seven RB1-intact metastatic TKI-resistant GISTs (Fig. 1b). Multiplexed protein imaging with in situ signal amplification (Immuno-SABER) [20] (Fig. 1c) and validation by conventional immunohistochemistry (Fig. 1d) and immunoblotting (Fig. 1e) confirmed the results on the protein level and demonstrated abnormal expression of p16, RB1, and p53 in 7 (39%), 2 (11%) and 1 (6%) of 18 GISTs, respectively. The results demonstrate high concordance between genomic and protein expression evaluations: of seven p16-negative GISTs, p16 loss resulted from CDKN2A/p16 homozygous deletion in four metastatic GISTs, from 14q deletion and MAX homozygous deletion, respectively, in two low-risk GISTs, and from presumed epigenetic inactivation in one primary SDH-deficient GIST. Both GISTs with RB1 loss had RB1 homozygous deletion, and the GIST with p53 overexpression had a TP53 missense mutation. Abnormal protein expression of at least one cell cycle regulator was identified in eight of nine metastatic GISTs, in none of two intermediate-risk GISTs, and in two of seven low-risk GISTs. In addition, we observed compensatory p16 overexpression in two cases with RB1 homozygous deletion and in one case with RB1 heterozygous deletion. Identical aberrations of p16, RB1 and p53 were present in three of three metastatic lesions from a given patient (N = 3) (Supplementary Fig. 2).
Fig. 1. Frequent dysregulation of the CDK2 and CDK4/6 pathways in clinical samples from metastatic TKI-resistant GIST patients.
a Summary of cell cycle regulator aberrations detected by targeted NGS as well as Immuno-SABER and IHC protein studies in 18 GISTs. Results are shown for low-risk, intermediate-risk, and metastatic stages of GIST progression (risk classifications), and for GISTs initiated by KIT, PDGFRA, NF1, and SDH mutations. Het del: heterozygous deletion; hom del: homozygous deletion. b Targeted NGS reveals co-dysregulation of the CDK2 and CDK4/6 pathways through homozygous co-deletions of CDKN2A/p16, CDKN2A/p14, and/or CDKN2B/p15 coding sequences in five of seven RB1-retained metastatic GISTs. c Immuno-SABER multiplexed and amplified in situ protein imaging demonstrates normal (i.e., retained) expression of key cell cycle markers p16, pRB1 Ser 807/811, RB1, and p53 in representative low-risk (LR; case #1) and intermediate-risk (IR; case #9) GISTs. In contrast, metastatic (Met) GIST #1 (case #12) has a loss of p16 resulting from CDKN2A/p16 homozygous deletion; metastatic GIST #2 (case #13) has p16 overexpression and loss of pRB1 Ser 807/811 and RB1 resulting from RB1 homozygous deletion; and metastatic GIST #3 (case #14) has p16 overexpression and loss of pRB1 Ser 807/811 and RB1 resulting from RB1 homozygous deletion as well as p53 overexpression caused by TP53 missense mutation (p.G245S); retained expression in admixed nonneoplastic cells serves as internal control. Corresponding immunohistochemistry (IHC) (d) and immunoblotting studies (e) validate cell cycle regulator genomic and protein aberrations in GISTs as described above. Positive IHC controls are HPV-associated squamous cell carcinoma (HPV16/18 positive by in situ hybridisation) for p16 and colorectal adenocarcinoma for pRB1 Ser 807/811, RB1 and p53 (d).
Together, these findings highlight genomic events co-dysregulating the CDK2 and CDK4/6 pathways as frequent events in metastatic GISTs as a strong rationale for therapeutic co-targeting.
Co-targeting CDK2 and CDK4/6 is synergistic in GIST and inhibits RB1 hyperphosphorylation and cell proliferation
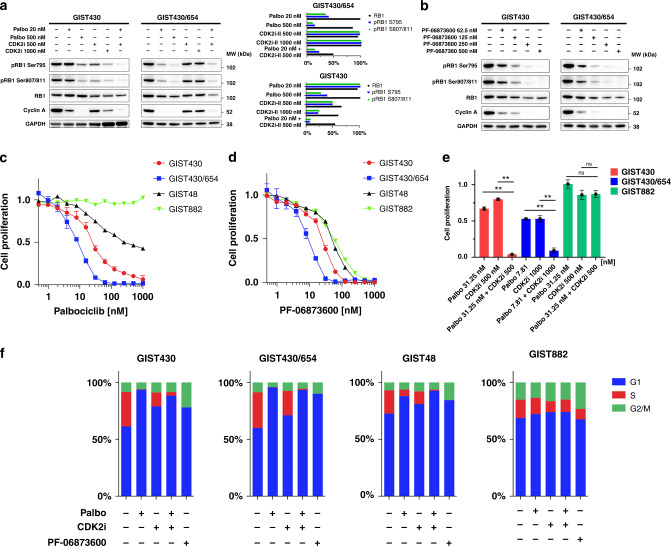
To determine the effects of therapeutic co-inhibition of CDK2 and CDK4/6 in vitro, we performed cell-proliferation assays and assessed RB1 hyperphosphorylation by immunoblotting in a range of KIT-mutant human GIST cell lines sensitive (GIST882, GIST430) or resistant (GIST430/654, GIST48) to imatinib, after treatment with inhibitors of CDK2 (CDK2 inhibitor II), CDK4/6 (palbociclib and abemaciclib), and CDK2/4/6 (PF-06783600) [34]. Immunoblotting studies demonstrated a dose response to single-agent palbociclib and CDK2 inhibitor II treatment in the imatinib-sensitive GIST430 and the imatinib-resistant GIST430/654 sub-line as measured by inhibition of RB1 hyperphosphorylation at Ser 795 and Ser 807/811 (pRB1) and inhibition of cyclin A levels indicative of cell proliferation (Fig. 2a). We then assessed whether combination therapies might add to the biological impact; the combination of palbociclib (20 nM) and CDK2 inhibitor II (500 nM) was synergistic with inhibition of RB1 hyperphosphorylation and extinction of cyclin A expression (Fig. 2a). No effect on cyclin A levels was found in GIST882 which has RB1 homozygous deletion (Supplementary Fig. 3). Likewise, 24-h treatment of GIST430 and GIST430/654 with the combined CDK2/4/6 inhibitor PF-06873600 inhibited RB1 hyperphosphorylation and cyclin A expression in a dose-dependent manner (Fig. 2b).
Fig. 2. Co-targeting CDK2 and CDK4/6 is synergistic in GIST and inhibits RB1 hyperphosphorylation and cell proliferation.
a Immunoblotting studies showing effects of low-dose palbociclib and low-dose CDK2 inhibitor II single-agent treatment (24 h) on RB1 hyperphosphorylation and cyclin A levels. High-dose palbociclib and CDK2 inhibitor II single-agent treatment (24 h) moderately inhibit pRB1 Ser 795, pRB1 Ser 807/8011 and cyclin A. Effects are maximised when combining low-dose palbociclib and CDK2 inhibitor II. GAPDH serves as loading control and the bar graph normalisations of GIST430 and GIST430/654 are with the higher value set to 100%. b Immunoblotting studies showing effects of treatment (24 h) with the combined CDK2/4/6 inhibitor PF-06873600 on pRB1 Ser 795, pRB1 Ser 807/8011, and cyclin A. Treatment results in inhibition of pRB1 Ser 795, pRB1 Ser 807/8011 and cyclin A along with a slight decrease in total RB1 expression. GAPDH serves as a loading control. Palbociclib (c) and PF-067836000 (d) reduce cell proliferation in GIST430, GIST430/654, and GIST48 as assessed by BrdU incorporation at 24 h (normalised to DMSO). GIST882 (RB1 homozygous deletion) is resistant to palbociclib (c) and shows limited sensitivity to PF-067836000 (d). e Combined inhibition of CDK2 and CDK4/6 using palbociclib and CDK2 inhibitor II was synergistic in GIST430 and GIST430/654, whereas GIST882 was resistant. ** indicates P < 0.01 (one-way ANOVA). Tests were performed in triplicate (mean +/− s.d). f BrdU incorporation assays highlight the fraction of cells in G1, S, and G2/M phase in GIST430, GIST430/654, GIST48 and GIST882 after treatment with DMSO control (first bar), palbociclib, CDK2 inhibitor II, palbociclib and CDK2 inhibitor II combination and PF-06873600 (1 μM each).
We next assessed the effects of CDK2 and CDK4/6 co-inhibition on cell proliferation. GIST430/654 was the cell line most sensitive to single-agent palbociclib (IC50 = 7.7 nM), followed by GIST430 and GIST48 (Fig. 2c and Table 1) with an in vitro IC50 of 40 nM in GIST430/654 corresponding to a clinical standard dose of 125 mg palbociclib QD (plasma mean Cmax for 125 mg QD: 116 ng/mL as per manufacturer; MW: 477.54 Daltons). GIST882 was resistant to palbociclib. Comparable IC50 values were achieved with abemaciclib (Supplementary Fig. 4A and Table 1). To determine the effects of single-agent CDK2 pathway inhibition, we evaluated GIST response to CDK2 inhibitor II and found IC50 values ranging from 2886.0-3842.9 nM in GIST430, GIST430/654 and GIST48 (Supplementary Fig. 4B and Table 1). GIST882 was resistant to single-agent CDK2 inhibition.
Table 1.
Proliferation IC50 values of CDK-inhibitor drugs in select GIST cell lines.
| Proliferation IC50 [nM] | ||||
|---|---|---|---|---|
| GIST cell line | Palbociclib | Abemaciclib | CDK2 inhibitor II | PF-06873600 |
| GIST430 | 24.2 | 16.78 | 3842.9 | 25.1 |
| GIST430/654 | 7.7 | 4.5 | 3320.7 | 8.6 |
| GIST48 | 42.1 | 17.6 | 2886.0 | 51.3 |
| GIST882 | N/A | N/A | 10738.5 | 60.1 |
We also evaluated the antiproliferative effects of combined CDK2 and CDK4/6 inhibition and found that GIST430/654 was most sensitive to PF-0687300 (IC50 = 8.6 nM), followed by GIST430, GIST48 and GIST882 (Fig. 2d and Table 1). Palbociclib and the CDK2 inhibitor II were synergistic and resulted in substantially increased inhibition of cell proliferation in GIST430 and GIST430/654, whereas GIST882 was resistant (Fig. 2e). Dose–response curves are shown in Supplementary Fig. 5. Combination sensitivity scores for palbociclib and the CDK2 inhibitor II were 92.97 in GIST430, in 80.21 GIST430/654, 69.19 in GIST48 and 15.93 in GIST882 (Supplementary Table 1 and Supplementary Fig. 6).
We performed BrdU incorporation assays to determine the effect of CDK-inhibitor treatment on the cell cycle (Fig. 2f and Supplementary Fig. 7). The results demonstrate that palbociclib extinguished the fraction of cells in S phase in GIST430 and GIST430/654 and reduced the fraction of cells in S phase in GIST48. There was no effect of palbociclib in GIST882. CDK2 inhibitor II had some effect in all four cell lines. A combination of palbociclib and CDK2 inhibitor II extinguished the fraction of cells in S phase in GIST430 and GIST430/654 but also in GIST48. PF-06873600 extinguished the fraction of cells in S phase in GIST430 and GIST430/654 and GIST48 but not in GIST882. Together, these results demonstrate that inhibition of CDK2 and CDK4/6 arrests cells in G1 in GIST and that combined inhibition has synergistic effects.
To determine the effects of CDK-inhibitor treatment on apoptosis, we evaluated expression of apoptosis markers (i.e., cleaved caspase 3, cleaved caspase 9) and MCM7 by immunohistochemistry, performed Caspase-Glo 3/7 assays, and evaluated cell morphology after CDK-inhibitor treatment in the GIST430/654 and GIST882 cell lines. The results of these studies demonstrate no substantial effect on expression of apoptosis markers, no change in MCM7 expression, no substantial increase in caspase 3/7 activity, and no apoptosis after 24-h treatment by phase contrast in GIST430/654 and GIST882 (Supplementary Fig. 8). Together, these studies indicate that inhibition of CDK2 and CDK4/6 does not lead to increased apoptosis in GIST.
Assessment of cell growth over time demonstrated that palbociclib inhibited growth in RB1-intact cell lines (Supplementary Fig. 9). Evaluation of effects of CDK-inhibitor treatment on cell viability showed that palbociclib and PF-067836000 reduced cell viability in GIST430, GIST430/654 and GIST48 but not in GIST882 (Supplementary Fig. 10).
Our findings demonstrate that response to CDK inhibition in GIST is characterised by cell-proliferation arrest in G1.
Taken together, these findings highlight a dependency of GIST on CDK2 and CDK4/6 co-activation to induce oncogenic inactivation of RB1 via hyperphosphorylation, enabling cell cycle progression.
RB1 inactivation is a mechanism of acquired CDK-inhibitor resistance in GIST
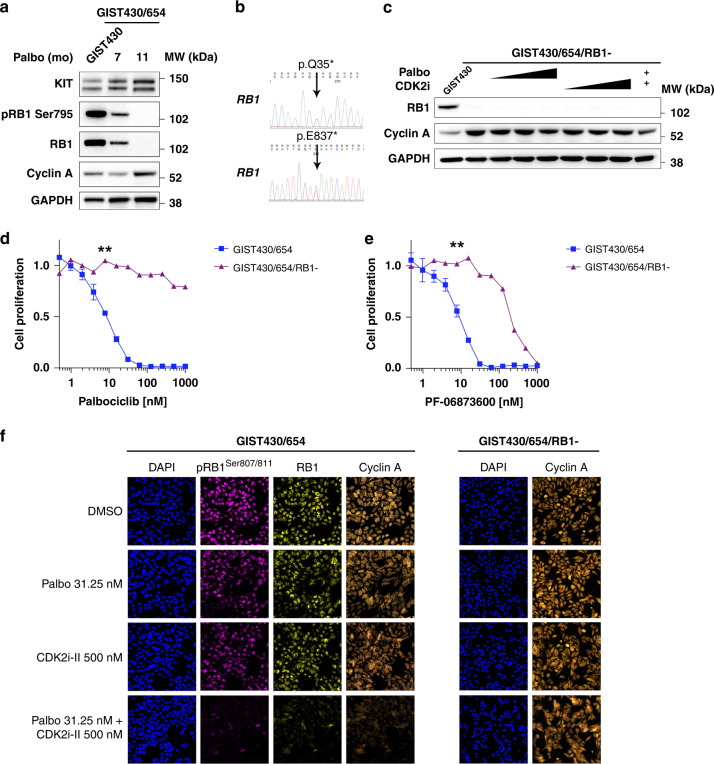
RB1 loss, oncogenic activation or amplification of CDK2, CDK6, cyclin E1 and E2 bypassing cyclin D1-CDK4/6 dependency have been described as mechanisms of acquired CDK4/6 inhibitor resistance in breast cancer and other cancer models [35–42]. To characterise mechanisms of acquired CDK4/6 inhibitor resistance in GIST in vitro, we exposed the palbociclib-sensitive GIST430/654 cell line to gradually increasing doses of palbociclib (20–100 nM). After 11 months of continued treatment, we observed accelerated cell growth in one sub-line and RB1 loss was identified (Fig. 3a), as well as two heterozygous RB1 mutations (p.Q35*, exon 1; p.E837*, exon 24) leading to complete RB1 inactivation (Fig. 3b). Treatment with palbociclib, CDK2 inhibitor II, and combination had no substantial effect on cyclin A levels in immunoblot studies (Fig. 3c). There was a significant shift in palbociclib (Fig. 3d) and PF-06873600 (Fig. 3e) proliferation IC50 values (P < 0.01 and P < 0.01, respectively) supporting the emergence of acquired CDK-inhibitor resistance. Immuno-SABER multiplexed imaging demonstrated that treatment of the RB1-intact GIST430 cell line with palbociclib, CDK2 inhibitor II, and the combination resulted in a dose-dependent inhibition of RB1 Ser 807/811 and cyclin A but had no effect on cyclin A expression in the resistant GIST430/654/RB1- sub-line (Fig. 3f). Together, these findings confirmed the emergence of acquired CDK2 and CDK4/6 resistance resulting from RB1 genomic inactivation.
Fig. 3. RB1 inactivation is a mechanism of acquired CDK-inhibitor resistance in GIST.
a Long-term exposure of GIST430/654 to palbociclib causes acquired resistance through RB1 loss as detected by weak RB1 expression at 7 months and complete loss of expression after 11 months by immunoblotting along with loss of pRB1 Ser 795 and a slight increase in cyclin A expression. KIT expression remained stable. GAPDH serves a loading control. b Sanger sequencing at 11 months identifies two new heterozygous RB1 inactivating mutations (c.103C > T; p.Q35*, exon 1; c.2509G > T; p.E837*, exon 24). c Treatment of the resistant GIST430/654/RB1- sub-line with palbociclib (at 31.25, 62.5 and 125 nM), CDK2 inhibitor II (at 500, 1000 and 2000 nM), and combination (palbociclib 31.25 nM + CDK2 inhibitor II 500 nM) has no substantial effect on cyclin A levels. GIST430 serves as RB1-intact control. GAPDH serves as a loading control. A significant shift in palbociclib (d) and PF-06873600 (e) proliferation IC50 values in the palbociclib-resistant GIST430/654/RB1- sub-line compared to parental GIST430/654 cells as assessed by BrdU incorporation at 24 h (normalised to DMSO). ** (7.81 nM) indicates P < 0.01 (one-way ANOVA). Tests were performed in triplicate. The error bars show s.d. f Multiplexed immunofluorescence by Immuno-SABER demonstrates that treatment with palbociclib, the CDK2 inhibitor II, and combination in the RB1-intact GIST430 cell line results in inhibition of RB1 Ser 807/811 and cyclin A expression but has no effect on cyclin A in the resistant GIST430/654/RB1- sub-line with RB1 inactivation.
While therapeutic inhibition of aurora kinase A (AURKA) [43, 44] has been reported to be synthetic lethal in other RB1-deficient cancer models, we did not identify growth inhibitory effects of the AURKA inhibitor alisertib in GIST430/654, the RB1-deficient GIST430/654 sub-line, and GIST882 as assessed by BrdU proliferation assays (Supplementary Fig. 4C). In addition, synthetic lethality of Skp2 inhibition has been described in RB1-deficient tumours [45], but we did not observe growth inhibitory effects of the Skp2 inhibitor C1 in GIST430/654, the RB1-deficient GIST430/654 sub-line, and GIST882 as assessed by BrdU proliferation assays (Supplementary Fig. 4D).
Emergence of a novel CCND1 fusion through palbociclib-selective pressure
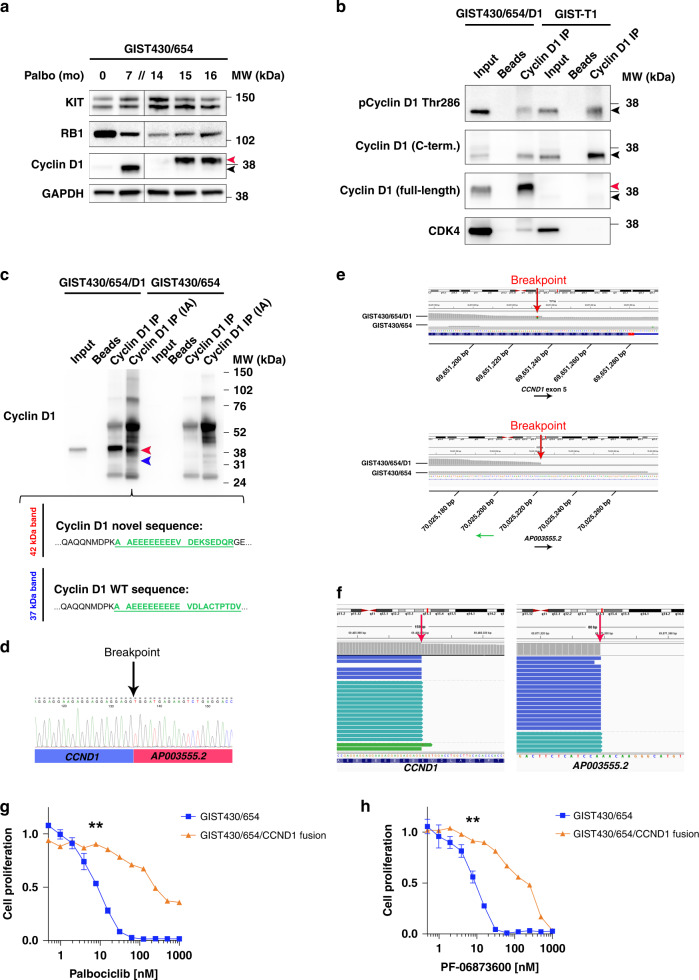
A second GIST430/654 resistance culture emerged after 14 months of palbociclib exposure, which was characterised by an abnormal form of cyclin D1 (Fig. 4a). The fact that the cells continued to grow in palbociclib 100 nM in the presence of preserved RB1 led us to hypothesise the emergence of a novel CDK4/6 inhibitor resistance mechanism caused by an oncogenic form of cyclin D1. To investigate the protein sequence of the abnormal 42-kDa cyclin D1, we performed cyclin D1 immunoprecipitation (IP) and used immunoblot antibodies directed against full-length cyclin D1, the C-terminus and against phospho-cyclin D1 (pCyclin D1) threonine (Thr) 286 located within the C-terminus. The results demonstrated that the 42-kDa cyclin D1 band was detected only by the full-length cyclin D1 antibody, but not by those directed against the C-terminus or pCyclin D1 Thr 286 (Fig. 4b). Co-IP studies revealed pull down of CDK4 in the cyclin D1 IP lane, indicating protein interactions between the 42-kDa cyclin D1 and CDK4, suggesting a preserved CDK4 binding domain (Fig. 4b). Taken together, these studies suggested that specifically the C-terminus was altered in the 42-kDa form of cyclin D1. Mass spectrometry on cyclin D1 IP bands isolated from a Coomassie-stained gel identified a novel peptide sequence (VDEKSEDQR) after amino acid (AA) 279 suggestive of an underlying gene fusion (Fig. 4c).
Fig. 4. Emergence of a novel CCND1 fusion through palbociclib-selective pressure.
a After 14 months of palbociclib treatment, an abnormal form of cyclin D1 (red arrowhead) (termed GIST430/654/D1) emerged in GIST430/654 evidenced by a faint band at 42 kDa, 5 kDa higher than regular cyclin D1 (37 kDa) (black arrowhead); KIT and RB1 expression were preserved; half the amount of protein was loaded for GIST430/654 control because of strong RB1 staining. GAPDH serves as a loading control. b Immunoprecipitation studies demonstrate detection of the abnormal 42-kDa cyclin D1 band (red arrowhead) in input and cyclin D1 IP lanes in the palbociclib-resistant sub-line termed GIST430/654/D1 by an antibody directed against full-length cyclin D1 (sc-20044), but not by those directed against the C-terminus (sc-718) or pCyclin D1 Thr 286. Background levels of wild-type cyclin D1 (black arrowheads) are detected by the full-length, C-terminal and pCyclin D1 Thr 286 antibodies in this cell line and in input and cyclin D1 IP lanes in GIST-T1 which has weak levels of cyclin D1 and serves as control. Co-IP detects interaction of the 42-kDa cyclin D1 protein with CDK4 in GIST430/654/D1. Beads-only lanes are negative control. c Mass spectrometry on cyclin D1 IP bands isolated from a Coomassie-stained gel with and without iodoacetamide (IA) treatment. Alkylation improved separation on SDS-PAGE and facilitated mass spectrometry analysis. Analysis of the isolated 42-kDa band (red) identifies a novel cyclin D1 peptide sequence after amino acid (AA) 279 (green). Analysis of the isolated 37-kDa cyclin D1 band (blue) reveals wild-type peptides (green). Parental GIST430/654 with weak expression of wild-type cyclin D1 serves as a reference. Beads-only lanes are negative controls. d 3’ rapid amplification of cDNA ends (RACE) followed by nested polymerase chain reaction (PCR) and Sanger sequencing identifies the presence of an intragenic rearrangement between CCND1 exon 5 and the 3’ flanking sequence of AP003555.2, a long non-coding RNA located ~300 kb 3’ of CCND1. e RNA sequencing confirms the breakpoints in CCND1 and the 3’ flanking sequence of AP003555.2 (red arrows). f Whole-exome sequencing detects the CCND1::chr11.g:70025223 rearrangement (breakpoints indicated by red arrows). A significant shift in palbociclib (g) and PF-06873600 (h) proliferation IC50 values in the palbociclib-resistant GIST430/654/D1 sub-line compared to parental GIST430/654 cells as assessed by BrdU incorporation at 24 h (normalised to DMSO). ** (7.81 nM) indicates P < 0.01 (one-way ANOVA). Tests were performed in triplicate. The error bars show s.d.
We next performed 3’ rapid amplification of cDNA ends (RACE) followed by nested polymerase chain reaction (PCR) and Sanger sequencing (Fig. 4d) and thereby confirmed the presence of an intragenic rearrangement between CCND1 exon 5 (breakpoint: 69,651,232, GRCh38/hg38) and the 3’ flanking sequence of AP003555.2 (breakpoint: 70,025,223, GRCh38/hg38), encoding a long non-coding RNA located ~300 kb downstream of CCND1. RNA sequencing (Fig. 4e) identified a breakpoint locating to CCND1 nucleotide 839 (encoding AA 280) and a second breakpoint in the AP003555.2 3’ flanking sequence. Whole-exome sequencing confirmed the presence of the CCND1::chr11.g:70025223 rearrangement (Fig. 4f). BrdU cell-proliferation assays after treatment with palbociclib (Fig. 4g) and PF-06873600 (Fig. 4h) and identified a significant shift in proliferation IC50 values (P < 0.01 and <0.01, respectively), thereby confirming acquired CDK-inhibitor resistance.
The novel oncogenic form of cyclin D1 decreases CDK4/6 inhibitor sensitivity when overexpressed in cell lines
Our results credential the first known example of an oncogenic cyclin D1 resulting from an intragenic rearrangement (CCND1::chr11.g:70025223) (Fig. 5a), leading to loss of cyclin D1 critical C-terminal phosphorylation sites, namely Thr 286 and Thr 288, which govern cyclin D1 proteasomal degradation (Fig. 5a). Phosphorylation of Thr 286 through GSK3β, p38SAPK2 and ERK2 regulates cyclin D1 nuclear export and stability [46], and phosphorylation of Thr 288 also regulates cyclin D1 stability and is mediated by the MIRK/DYRK1B kinase [46].
Fig. 5. The novel oncogenic form of cyclin D1 decreases CDK4/6 inhibitor sensitivity when overexpressed in cell lines.
a Schematic of the fusion between CCND1 exon 5 and the 3’ flanking sequence of AP003555.2, which results in a novel oncogenic form of cyclin D1. The 3’ flanking sequence of AP003555.2 is inserted in the opposite (i.e., 3’–5’ instead of 5’–3’) read direction (top). Replacement of the cyclin D1 C-terminus by the novel AA sequence results in loss of Thr 286 and Thr 288, two residues essential for cyclin D1 phosphorylation and proteasomal degradation (bottom). b Treatment with palbociclib (100 nM) from day 6–13 post infection results in growth inhibition of GIST430/654 cells infected with pLKO.1 empty vector but has no effect on the GIST430/654 cells infected with the CCND1 fusion construct. c Expression of the CCND1 fusion construct in the RB1-intact GIST430/654, LMS05 and BT474 cell lines at 4, 10 and 10 days post infection, respectively. No expression of cyclin D1 is detected in the untreated and empty vector controls. GAPDH serves as a loading control. d Treatment of GIST430/654 with palbociclib for 26 days further increases expression of the CCND1::chr11.g:70025223 construct and mildly decreases expression of pRB1 Ser 795. Total RB1 expression is unchanged. Shown are regular (1 min) and short (15 sec) exposures for cyclin D1. GAPDH serves as a loading control. e Decreased sensitivity to palbociclib in GIST430/654, LMS05 and BT474 infected with the CCND1::chr11.g:70025223 construct compared to empty vector-infected cells as assessed by BrdU incorporation at 24 h (normalised to DMSO). * indicates P < 0.05; ** indicates P < 0.01 (one-way ANOVA). Tests were performed in triplicate. The error bars show s.d.
We next performed lentiviral transduction of the CCND1::chr11.g:70025223 construct in GIST430/654, the leiomyosarcoma (LMS05), and ductal breast carcinoma (BT474) cell lines. Treatment with palbociclib (100 nM) resulted in growth inhibition of GIST430/654 cells infected with pLKO.1 empty vector but had no effect on the GIST430/654 cells infected with the fusion construct (Fig. 5b), which was expressed in all three cell lines (Fig. 5c). Treatment of GIST430/654 cells infected with the fusion construct with palbociclib for 26 days had no significant effect on RB1 hyperphosphorylation and further increased expression of the fusion protein over time (Fig. 5d). There was a significant shift in the palbociclib IC50 values in the cells infected with the fusion construct compared to p.LKO1 empty vector (P < 0.01 each) (Fig. 5e), confirming that the novel oncogenic form of cyclin D1 was associated with acquired resistance to CDK4/6 inhibition.
We next compared the response to CDK2 and CDK4/6 inhibition in early passages of GIST430 characterised by physiologically low levels of cyclin D1 expression and a later passage with expression 96 times as high, and demonstrated that the cyclin D1-low cell line was more sensitive to palbociclib, CDK2 inhibitor II, RAD001, and select combinations than the cyclin D1-high cell line, indicating roles of wild-type cyclin D1 expression for response to CDK inhibition (Supplementary Fig. 11A).
PLK1 inhibition has been reported to have anti-tumoral activity in CCND1-driven breast cancer metastases with acquired palbociclib resistance [47]. However, we did not observe substantial antiproliferative activity of the PLK1 inhibitor volasertib in GIST430/654 parental cells and in the resistant sub-lines with RB1 inactivation or CCND1 fusion (Supplementary Fig. 11B).
RNA sequencing confirms that RB1 inactivation and a novel oncogenic form of cyclin D1 confer palbociclib resistance
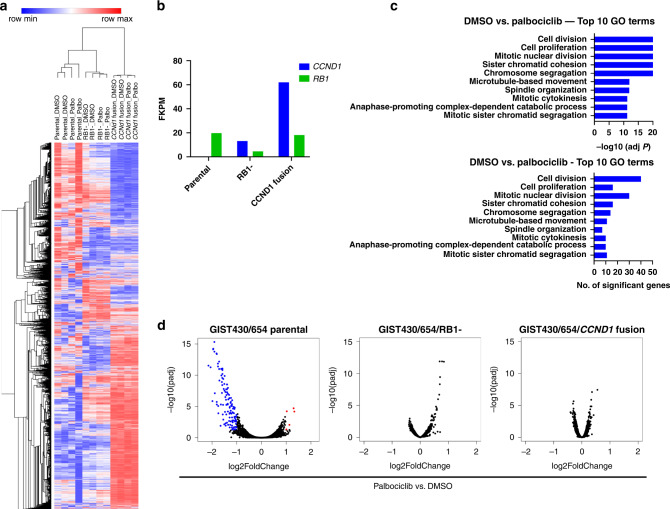
Next, we performed RNA sequencing analysis to determine the effects of CDK4/6 inhibition on the transcriptomic landscape in GIST430/654 parental cells, and the palbociclib-resistant GIST430/654/RB1- and GIST430/654/CCND1 fusion sub-lines. Unsupervised hierarchical clustering demonstrated that DMSO-control and palbociclib-treated conditions clustered together within each cell line (Fig. 6a), indicating that RB1 inactivation and the CCND1 fusion induced distinct pan-transcriptomic changes that superseded effects of palbociclib. Gene expression results by FKPM confirmed the near-absence of cyclin D1 expression in GIST430/654 parental cells (0.3 FKPM), low levels in those with RB1 inactivation (13.2 FKPM), and very high levels in those with the CCND1 fusion (62.0 FKPM) (Fig. 6b). RB1 expression was detected at moderate levels in GIST430/654 parental cells (19.8 FKPM) and those with CCND1 fusion (18.1 FKPM) and was very low in those with RB1 inactivation (4.6 FKPM) (Fig. 6b). AMBRA1 has been identified as a critical regulator of the stability of D-type cyclins and AMBRA1 loss reduced sensitivity to CDK4/6 inhibitors by promoting the formation of complexes of D-type cyclins with CDK2 [48]. However, our RNA sequencing studies did not identify differences in AMBRA1 expression levels in GIST430/654 parental cells (FKPM 7.2) versus GIST430/654 with CCND1 fusion (FKPM 6.0).
Fig. 6. RNA sequencing confirms that RB1 inactivation and a novel oncogenic form of cyclin D1 confer palbociclib resistance.
a Unsupervised hierarchical clustering demonstrates that DMSO-control and palbociclib-treated conditions cluster together within GIST430/654 parental (parental), the sub-line with RB1 inactivation (RB1-), and the sub-line with CCND1 fusion. b Gene expression results by FKPM show near-absence of cyclin D1 expression in GIST430/654 parental (parental), low levels in the sub-line with RB1 inactivation (RB1-), and very high levels in the sub-line with CCND1 fusion. RB1 expression is detected at moderate levels in GIST430/654 parental and the sub-line with CCND1 fusion and is very low in the sub-line with RB1 inactivation. c Gene ontology (GO) analysis in GIST430/654 parental after treatment with palbociclib detects enrichment of cell cycle/growth-associated GO terms, specifically, cell division, cell proliferation, mitotic nuclear division, sister chromatid segregation, and chromosome segregation GO terms (top). The number of significant genes per category are shown at the bottom. No enrichment of GO terms is detected in the sub-lines with RB1 inactivation and the CCND1 fusion. d Volcano plots demonstrate a number of significantly differentially expressed genes GIST430/654 parental and none in those with RB1 inactivation and the CCND1 fusion confirming the presence of palbociclib resistance on the RNA transcriptomic level.
Gene ontology (GO) analysis after palbociclib treatment demonstrated enrichment of cell cycle and growth-associated GO terms in GIST430/654 parental cells, specifically, cell division, cell proliferation, mitotic nuclear division, sister chromatid segregation, and chromosome segregation GO terms (Fig. 6c). In contrast, no enrichment of GO terms was detected in the sub-lines with RB1 inactivation and CCND1 fusion (Fig. 6c). Together with volcano plots demonstrating several significantly differentially expressed genes in GIST430/654 parental cells upon palbociclib treatment and none in those with RB1 inactivation and CCND1 fusion (Fig. 6d), these findings confirmed the presence of palbociclib resistance on the RNA transcriptomic level conferred by RB1 inactivation or CCND1 fusion.
Discussion
Frequent co-dysregulation of the CDK2 and CDK4/6 pathways in advanced GIST creates oncogenic co-dependency and offers a strong biological rationale for targeted combination therapies aimed at restoring cell cycle control. Such aberrations are conserved across the metastatic burden in many patients and therefore provide a compelling therapeutic target in patients with metastatic GIST progressing on TKI therapies due to polyclonal TK resistance mutations. The patients with metastatic GIST included in our study were progressing after therapy with at least two TKIs at the time of surgery, illustrating the challenges of effectively targeting polyclonal TKI resistance mutations.
A recent clinical trial failed to demonstrate clinical activity of palbociclib monotherapy in advanced TKI-resistant CDKN2A/p16-deleted GIST [18]. Of note, the authors excluded patients with RB1 gene deletion detected by array-comparative genomic hybridisation but did not assess RB1 by NGS or RB1 protein expression [18]. It is therefore likely that a subset of GISTs included in this study were RB1-deficient and therefore resistant to CDK4/6 inhibition at baseline.
Extending insights from earlier studies [10, 13, 14], our NGS results demonstrate homozygous co-deletions of CDKN2A/p16 and CDKN2A/p14 or CDKN2B/p15 and CDKN2A/p14 that dysregulate the CDK2 and CDK4/6 axis in five of seven RB1-intact metastatic TKI-resistant GISTs. We here demonstrate that co-targeting CDK2 and CDK4/6—either through the combination of inhibitors of CDK2 and CDK4/6 or the combined CDK2/4/6 inhibitor—is synergistic in GIST and inhibits RB1 hyperphosphorylation and cell proliferation in RB1-intact GIST models in vitro. Of note, the palbociclib single-agent in vitro IC50 can realistically be achieved in the clinical setting and would allow the reduction of palbociclib doses to increase tolerability. In addition, we showed that PF-06873600 effectively inhibited RB1 hyperphosphorylation and proliferation in RB1-intact GIST models. Of note, the RB1-deficient GIST882 cell line showed some response to PF-06873600 (IC50 = 60.1 nM) (Fig. 2d), whereas the GIST430/654 sub-lines with RB1 inactivation and CCDN1 rearrangement were substantially less sensitive (IC50 > 200 nM) (Figs. 3e and 4h). Resistance to palbociclib was more pronounced in all RB1-deficient cell lines (Figs. 2d, 3e and 4h). Freeman-Cook et al. describe that PF-06873600 retained antiproliferative activity in palbociclib-resistant breast cancer cell lines and suggest that RB1-independent CDK2 functions within the cell cycle, i.e., a non-G1 cell cycle checkpoint, may be inhibited by this compound [49]. It remains to be determined in subsequent studies which mechanisms are responsible for residual response to CDK2/4/6 inhibition in RB1-inactivated cell lines. PF-06873600 is currently being evaluated in a clinical trial in combination with endocrine therapy for patients with a range of metastatic solid cancers (NCT03519178). The results of our study suggest that PF-06873600 might be worth evaluating in patients with RB1-intact advanced GIST. In contrast to secondary TKI resistance mutations in GIST, which are often polyclonal in nature and challenging to target effectively with a single TKI, cell cycle regulator mutations are conserved across the metastatic burden in most patients, providing a compelling target for CDK-inhibitor therapies in metastatic, TKI-resistant GIST.
Our findings are in line with previous reports highlighting the CDK2 pathway as key mediator of CDK4/6 inhibitor resistance. Kumarasamy et al. previously reported that increased CDK2 activity due to amplification of cyclin E1 can promote CDK4/6 inhibitor resistance in breast cancer [50]. The authors showed that RB1 can act as a central player in coupling CDK4/6 kinase activity with CDK2 to mediate cellular response to palbociclib, suggesting CDK2-targeted therapy as an approach to rescue CDK4/6 inhibitor resistance [50]. They demonstrated that PF-06873600 ceased cell proliferation in vitro an in vivo [50]. Similarly, Pandey et al. reported that the combined targeting of CDK2 and CDK4/6 synergistically inhibited cell proliferation in breast cancer [19].
The results of our studies demonstrate that CDK2 and CDK4/6 inhibition arrests cells in G1 phase (Fig. 2f). These findings are in line with observations made in RB1-proficient breast cancer where CDK4/6 inhibitors induce cytostasis (G1 cell cycle arrest) [51]. While CDK4/6 inhibition has been shown to induce a phenotype resembling senescence in luminal breast cancer cells [52] characterised by cellular enlargement and flattening, and increased β-galactosidase activity [53], it remains to be determined whether CDK inhibition causes a senescent phenotype in GIST. The results of our studies demonstrate that cell proliferation is inhibited through G1 arrest. CDK2 and CDK4/6 inhibitors are expected to have therapeutic benefit in patients with TKI-resistant GIST for whom no effective targeted therapies exist. Similar to insights from breast cancer, where CDK inhibitors are combined with anti-hormonal therapies to achieve durable responses, these drugs might be combined with a second biologically relevant drug such as a tyrosine kinase inhibitor in GIST. Therefore, achieving convincing cytostatic effects with CDK-inhibitor therapies would represent a major therapeutic advancement in the GIST field.
Moreover, our results show that RB1 inactivation is predictive of primary CDK-inhibitor resistance in GIST as would be expected [37, 39, 41, 42]. Together, these studies suggest that combination therapies targeting CDK2 and CDK4/6, in a biomarker-selected population of advanced GIST with intact RB1—such as assessment of RB1 status by immunohistochemistry [25]—should be evaluated.
At the same time, we explored mechanisms of acquired resistance to CDK2 and CDK4/6 inhibition and identified RB1 genomic inactivation as a key driver of CDK-inhibitor resistance in GIST, in line with observations made in breast cancer [37, 39, 41, 42]. In addition, we discovered a novel form of cyclin D1 which represents the first known example of an oncogenic and functional cyclin D1 resulting from an intragenic rearrangement (CCND1::chr11.g:70025223). We show that the CCND1 fusion caused loss of critical C-terminal phosphorylation sites, namely Thr 286 and Thr 288, that govern proteasomal degradation, possibly leading to impaired degradation and overexpression of the cyclin D1 fusion protein. In addition, the proximity of the fusion gene to the ANO1 super-enhancer region (Supplementary Fig. 12), which has been implicated in orchestrating oncogenic programmes in SDH-deficient GISTs [54] and has been involved in genetic rearrangements in GIST [55], may have contributed to overexpression of the fusion protein. Cyclin D1 overexpression has been described as a resistance mechanism in CDK4/6 inhibitor-treated breast cancer [50, 56] and non-small cell lung cancer [56]. While cyclin D1 upregulation has been identified by our group in GISTs developing KIT independence during TKI therapy [33], the novel oncogenic cyclin D1 occurred in a different biological context and was not accompanied by KIT independence.
Lentiviral transfection of the CCND1 fusion gene into palbociclib-sensitive GIST430/654, leiomyosarcoma, and breast cancer cells reduced sensitivity to palbociclib; however, not as dramatically as in the original GIST430/654 sub-line with CCND1 fusion, possibly due to oncogenic stress and activation of compensatory mechanisms caused by the introduction of a strong cyclin D1 oncogenic signal. While this is the first description of a CCND1 rearrangement in GIST associated with CDK-inhibitor resistance, similar aberrations or other means of cyclin D1 oncogenic activation are expected to emerge when CDK inhibitors are evaluated in GIST patients. Our observation of reduced CDK-inhibitor sensitivity in GIST430/654 cells with physiologically higher levels of cyclin D1 expression suggests that cyclin D1 overexpression might contribute to CDK-inhibitor resistance. Hence, evaluation of cyclin D1 expression, along with RB1, is expected to yield informative insights for predicting treatment response. Further studies are needed to determine the functional impact of the novel CCND1 fusion to identify opportunities for therapeutic targeting. Whether the CCND1 rearrangement will evolve as mechanism of acquired resistance in patients treated with CDK inhibitors remains to be validated in clinical trials.
Inhibition of AURKA, Skp2 or PLK1 did not have antiproliferative activity in the palbociclib-resistant sub-lines and additional studies are needed to identify synthetic lethal targets in GIST with RB1 inactivation or oncogenic cyclin D1 activation.
In summary, these results demonstrate frequent genomic aberrations in cell cycle regulators causing co-activation of the CDK2 and CDK4/6 pathways in advanced GIST. We show that therapeutic co-targeting of CDK2 and CDK4/6 is synergistic in GIST cell lines with intact RB1. Moreover, we identify RB1 inactivation and a novel oncogenic cyclin D1 resulting from an intragenic rearrangement as mechanisms of acquired CDK-inhibitor resistance in GIST. These studies suggest CDK2 and CDK4/6 co-inhibition as a therapeutic strategy in patients with advanced GIST, including patients with metastatic TKI-resistant GIST. In addition, the findings expand the spectrum of potential CDK-inhibitor resistance mechanisms with translational potential for improving cell cycle targeted therapies in other cancer types.
Supplementary information
Permission from coauthors to add two new authors
Acknowledgements
The authors thank Dr. Peter Sicinski, Dana-Farber Cancer Institute, for facilitating the cell cycle phase analyses and for critical review of the manuscript and Dr. Geoffrey I. Shapiro, Dana-Farber Cancer Institute, for critical review of the manuscript.
Author contributions
IMS and JAF designed the study. IMS, MLH, MZL, MPS, IG, JAP, AME, WBO and SKS performed the experiments and data analysis with assistance from SPG, NL and JLH. IMS and JAF supervised the study. JAP, SPG, SG, JAM, MMB, ETA, AME, JLH, CPR, GDD, SKS and JAF provided resources. IMS drafted the manuscript. MLH, MZL, JAP, SG, MMB, AME, WBO, GDD, SKS and JAF edited the manuscript. All authors commented on the manuscript.
Funding
This work is supported by the NCI K08CA241085 grant (IMS), SARC (Sarcoma Alliance for Research through Collaboration) Young Investigator Award (IMS), the Wyss Institute Validation Project Program and the Office of Naval Research grant N00014-18-1-2549 (PY), the NIGMS R01GM132129 grant (JAP), the NIH UH3CA25513303 and 1DP1GM133052 grants (SKS, IG, MPS, and PY), Pfizer and Lilly through the Alliance for Clinical Trials in Oncology (MMB) and the European Molecular Biology Laboratory (EMBL) core funding (SKS).
Data availability
The data generated in this study are publicly available in GenBank (NCBI) at accession no. 2511709, Gene Expression Omnibus (GEO) at accession number GSE206257, and within the article and its Supplementary data files. As per genomic data sharing guidelines, sequence and assay data are deposited in a publicly available database sponsored by the American Association for Cancer Research (AACR) Project GENIE. Variant calls and a limited clinical dataset from patients are sent to the Synapse platform, developed by Sage Bionetworks, where the data are harmonised and protected health information (PHI) removed in a secure Health Insurance Portability and Accountability Act (HIPAA)-the compliant environment that provides data governance.
Competing interests
SKS and PY are inventors for a US patent application for the multiplexed imaging technology used in this work. PY is a co-founder of Ultivue, Inc. and NuProbe Global. SG serves as consultant/advisory board member to Deciphera Pharmaceuticals, Blueprint Medicines, Daiichi-Sankyo, Kayothera, Immunicum, Eli Lilly, Bayer, Ayala; reports research funding to the institution by Deciphera Pharmaceuticals, Blueprint Medicines, Daiichi-Sankyo, Theseus Pharmaceuticals, Merck, Eisai, Springworks, Pfizer and Bayer; holds equity at Abbott Laboratories; and receives royalties from Wolters Kluwer/UpToDate. MMB serves on the Board of Directors of Natera, Inc., and Leap Therapeutics. JLH serves as a consultant to Aadi Biosciences and TRACON Pharmaceuticals. GDD serves as a Board of Directors member with minor equity holding in Blueprint Medicines; serves as co-founder with minor equity holding in IDRX; serves as consultant/SAB member with minor equity holding in G1 Therapeutics, Caris Life Sciences, Erasca Pharmaceuticals, RELAY Therapeutics, Bessor Pharmaceuticals, CellCarta, IKENA Oncology, Kojin Therapeutics, Acrivon Therapeutics; serves as a scientific consultant with sponsored research to Dana-Farber to Bayer, Pfizer, Novartis, Roche/Genentech, Janssen, PharmaMar, Daiichi-Sankyo, AdaptImmune; serves as a scientific consultant to GlaxoSmithKline, EMD-Serono, MEDSCAPE, Mirati, WCG/Arsenal Capital, RAIN Therapeutics; and receives Novartis royalty to Dana-Farber for use patent of imatinib in GIST. The remaining authors declare no competing interests.
Ethics approval and consent to participate
Patient sample collection and analysis were conducted following protocols approved by the Dana-Farber/Brigham and Women’s Hospital Institutional Review Board. Written informed consent was obtained from patients for use of samples. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01990-5.
References
- 1.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–10. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 3.Joensuu H, Roberts PJ, Sarlomo-Rikala M, Andersson LC, Tervahartiala P, Tuveson D, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344:1052–6. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich MC, Rankin C, Blanke CD, Demetri GD, Borden EC, Ryan CW, et al. Correlation of long-term results of imatinib in advanced gastrointestinal stromal tumors with next-generation sequencing results: analysis of phase 3 SWOG intergroup trial S0033. JAMA Oncol. 2017;3:944–52. doi: 10.1001/jamaoncol.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liegl B, Kepten I, Le C, Zhu M, Demetri GD, Heinrich MC, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216:64–74. doi: 10.1002/path.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wardelmann E, Merkelbach-Bruse S, Pauls K, Thomas N, Schildhaus HU, Heinicke T, et al. Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin Cancer Res. 2006;12:1743–9. doi: 10.1158/1078-0432.CCR-05-1211. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer IM, Wang Y, Liang CW, Bahri N, Quattrone A, Doyle L, et al. MAX inactivation is an early event in GIST development that regulates p16 and cell proliferation. Nat Commun. 2017;8:14674. doi: 10.1038/ncomms14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang Y, Xie F, Cao H, Wang C, Zhu M, Liu X, et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc Natl Acad Sci USA. 2019;116:22746–53. doi: 10.1073/pnas.1914542116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wozniak A, Sciot R, Guillou L, Pauwels P, Wasag B, Stul M, et al. Array CGH analysis in primary gastrointestinal stromal tumors: cytogenetic profile correlates with anatomic site and tumor aggressiveness, irrespective of mutational status. Genes Chromosomes Cancer. 2007;46:261–76. doi: 10.1002/gcc.20408. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich MC, Patterson J, Beadling C, Wang Y, Debiec-Rychter M, Dewaele B, et al. Genomic aberrations in cell cycle genes predict progression of KIT-mutant gastrointestinal stromal tumors (GISTs) Clin Sarcoma Res. 2019;9:3. doi: 10.1186/s13569-019-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Marino-Enriquez A, Bennett RR, Zhu M, Shen Y, Eilers G, et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2014;46:601–6. doi: 10.1038/ng.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife. 2014;3:e02872. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller F, Gunawan B, von Heydebreck A, Schwager S, Schulten HJ, Wolf-Salgo J, et al. Prognostic role of E2F1 and members of the CDKN2A network in gastrointestinal stromal tumors. Clin Cancer Res. 2005;11:6589–97. doi: 10.1158/1078-0432.CCR-05-0329. [DOI] [PubMed] [Google Scholar]
- 14.Perrone F, Tamborini E, Dagrada GP, Colombo F, Bonadiman L, Albertini V, et al. 9p21 locus analysis in high-risk gastrointestinal stromal tumors characterized for c-kit and platelet-derived growth factor receptor alpha gene alterations. Cancer. 2005;104:159–69. doi: 10.1002/cncr.21113. [DOI] [PubMed] [Google Scholar]
- 15.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 16.Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J Clin Oncol. 2017;35:3638–46. doi: 10.1200/JCO.2017.75.6155. [DOI] [PubMed] [Google Scholar]
- 17.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N Engl J Med. 2016;375:1738–48. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 18.Toulmonde M, Blay JY, Bouche O, Mir O, Penel N, Isambert N, et al. Activity and safety of palbociclib in patients with advanced gastrointestinal stromal tumors refractory to imatinib and sunitinib: a biomarker-driven phase II study. Clin Cancer Res. 2019;25:4611–5. doi: 10.1158/1078-0432.CCR-18-3127. [DOI] [PubMed] [Google Scholar]
- 19.Pandey K, Park N, Park KS, Hur J, Cho YB, Kang M, et al. Combined CDK2 and CDK4/6 inhibition overcomes palbociclib resistance in breast cancer by enhancing senescence. Cancers. 2020;12:3566. [DOI] [PMC free article] [PubMed]
- 20.Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol. 2019;37:1080–90. doi: 10.1038/s41587-019-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishi JY, Schaus TE, Gopalkrishnan N, Xuan F, Yin P. Programmable autonomous synthesis of single-stranded DNA. Nat Chem. 2018;10:155–64. doi: 10.1038/nchem.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishi JY, Lapan SW, Beliveau BJ, West ER, Zhu A, Sasaki HM, et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods. 2019;16:533–44. doi: 10.1038/s41592-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muhlich J, Chen Y-A, Russell D, Sorger PK. Stitching and registering highly multiplexed whole slide images of tissues and tumors using ASHLAR. Bioinformatics. 2022:btac544. 10.1093/bioinformatics/btac544. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 24.Burel JM, Besson S, Blackburn C, Carroll M, Ferguson RK, Flynn H, et al. Publishing and sharing multi-dimensional image data with OMERO. Mamm Genome. 2015;26:441–7. doi: 10.1007/s00335-015-9587-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer IM, Lundberg MZ, Demicco EG, Przybyl J, Matusiak M, Chibon F, et al. Relationships between highly recurrent tumor suppressor alterations in 489 leiomyosarcomas. Cancer. 2021;127:2666–73. doi: 10.1002/cncr.33542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, et al. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–21. [PubMed] [Google Scholar]
- 27.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–8. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 29.Paulo JA. Sample preparation for proteomic analysis using a GeLC-MS/MS strategy. J Biol Methods. 2016;3:e45. doi: 10.14440/jbm.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S, Wang W, Aldahdooh J, Malyutina A, Shadbahr T, Tanoli Z, et al. SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Genomics Proteomics Bioinformat. 2022. 10.1016/j.gpb.2022.01.004. [DOI] [PMC free article] [PubMed]
- 32.Malyutina A, Majumder MM, Wang W, Pessia A, Heckman CA, Tang J. Drug combination sensitivity scoring facilitates the discovery of synergistic and efficacious drug combinations in cancer. PLoS Comput Biol. 2019;15:e1006752. doi: 10.1371/journal.pcbi.1006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou WB, Ni N, Zuo R, Zhuang W, Zhu M, Kyriazoglou A, et al. Cyclin D1 is a mediator of gastrointestinal stromal tumor KIT-independence. Oncogene. 2019;38:6615–29. doi: 10.1038/s41388-019-0894-3. [DOI] [PubMed] [Google Scholar]
- 34.Freeman-Cook KD, Hoffman RL, Behenna DC, Boras B, Carelli J, Diehl W, et al. Discovery of PF-06873600, a CDK2/4/6 inhibitor for the treatment of cancer. J Med Chem. 2021;64:9056–77. doi: 10.1021/acs.jmedchem.1c00159. [DOI] [PubMed] [Google Scholar]
- 35.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D, et al. Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23:5561–72. doi: 10.1158/1078-0432.CCR-17-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandarlapaty S, Razavi P. Cyclin E mRNA: assessing cyclin-dependent kinase (CDK) activation state to elucidate breast cancer resistance to CDK4/6 inhibitors. J Clin Oncol. 2019;37:1148–50. doi: 10.1200/JCO.19.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res. 2016;76:2301–13. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min A, Kim JE, Kim YJ, Lim JM, Kim S, Kim JW, et al. Cyclin E overexpression confers resistance to the CDK4/6 specific inhibitor palbociclib in gastric cancer cells. Cancer Lett. 2018;430:123–32. doi: 10.1016/j.canlet.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Guarducci C, Bonechi M, Benelli M, Biagioni C, Boccalini G, Romagnoli D, et al. Cyclin E1 and Rb modulation as common events at time of resistance to palbociclib in hormone receptor-positive breast cancer. NPJ Breast Cancer. 2018;4:38. doi: 10.1038/s41523-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36:2255–64. doi: 10.1038/onc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Condorelli R, Spring L, O’Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29:640–5. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 42.O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8:1390–403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong X, Du J, Parsons SH, Merzoug FF, Webster Y, Iversen PW, et al. Aurora A kinase inhibition is synthetic lethal with loss of the RB1 tumor suppressor gene. Cancer Discov. 2019;9:248–63. doi: 10.1158/2159-8290.CD-18-0469. [DOI] [PubMed] [Google Scholar]
- 44.Lyu J, Yang EJ, Zhang B, Wu C, Pardeshi L, Shi C, et al. Synthetic lethality of RB1 and aurora A is driven by stathmin-mediated disruption of microtubule dynamics. Nat Commun. 2020;11:5105. doi: 10.1038/s41467-020-18872-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Z, Bauzon F, Fu H, Cui J, Zhao H, Nakayama K, et al. Skp2 suppresses apoptosis in Rb1-deficient tumours by limiting E2F1 activity. Nat Commun. 2014;5:3463. doi: 10.1038/ncomms4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montaudon E, Nikitorowicz-Buniak J, Sourd L, Morisset L, El Botty R, Huguet L, et al. PLK1 inhibition exhibits strong anti-tumoral activity in CCND1-driven breast cancer metastases with acquired palbociclib resistance. Nat Commun. 2020;11:4053. doi: 10.1038/s41467-020-17697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simoneschi D, Rona G, Zhou N, Jeong YT, Jiang S, Milletti G, et al. CRL4(AMBRA1) is a master regulator of D-type cyclins. Nature. 2021;592:789–93. doi: 10.1038/s41586-021-03445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freeman-Cook K, Hoffman RL, Miller N, Almaden J, Chionis J, Zhang Q, et al. Expanding control of the tumor cell cycle with a CDK2/4/6 inhibitor. Cancer Cell. 2021;39:1404–21.e1411. doi: 10.1016/j.ccell.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Kumarasamy V, Vail P, Nambiar R, Witkiewicz AK, Knudsen ES. Functional determinants of cell cycle plasticity and sensitivity to CDK4/6 inhibition. Cancer Res. 2021;81:1347–60. doi: 10.1158/0008-5472.CAN-20-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watt AC, Goel S. Cellular mechanisms underlying response and resistance to CDK4/6 inhibitors in the treatment of hormone receptor-positive breast cancer. Breast Cancer Res. 2022;24:17. doi: 10.1186/s13058-022-01510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–51. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575:229–33. doi: 10.1038/s41586-019-1668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbini M, Astolfi A, Indio V, Nannini M, Schipani A, Bacalini MG, et al. Gene duplication, rather than epigenetic changes, drives FGF4 overexpression in KIT/PDGFRA/SDH/RAS-P WT GIST. Sci Rep. 2020;10:19829. doi: 10.1038/s41598-020-76519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong T, Xue Y, Cencic R, Zhu X, Monast A, Fu Z, et al. eIF4A inhibitors suppress cell-cycle feedback response and acquired resistance to CDK4/6 inhibition in cancer. Mol Cancer Ther. 2019;18:2158–70. doi: 10.1158/1535-7163.MCT-19-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Permission from coauthors to add two new authors
Data Availability Statement
The data generated in this study are publicly available in GenBank (NCBI) at accession no. 2511709, Gene Expression Omnibus (GEO) at accession number GSE206257, and within the article and its Supplementary data files. As per genomic data sharing guidelines, sequence and assay data are deposited in a publicly available database sponsored by the American Association for Cancer Research (AACR) Project GENIE. Variant calls and a limited clinical dataset from patients are sent to the Synapse platform, developed by Sage Bionetworks, where the data are harmonised and protected health information (PHI) removed in a secure Health Insurance Portability and Accountability Act (HIPAA)-the compliant environment that provides data governance.