Abstract
We report the complete 119,443-bp sequence of the pgm locus from Yersinia pestis and its flanking regions. Sequence analysis confirms that the 102-kb unstable pgm locus is composed of two distinct parts: the pigmentation segment and a high-pathogenicity island (HPI) which carries virulence genes involved in iron acquisition (yersiniabactin biosynthetic gene cluster). Within the HPI, three genes coding for proteins related to phage proteins were uncovered. They are located at both extremities indicating that the entire HPI was acquired en bloc by phage-mediated horizontal transfer. We identified, within the pigmentation segment, two novel loci that may be involved in virulence: a fimbriae gene cluster and a locus probably encoding a two component regulatory system similar to the BvgAS regulatory system of Bordetella pertussis. Three genes containing frameshift mutations and two genes interrupted by insertion element insertion were found within this region. To investigate diversity among different Y. pestis and Yersinia pseudotuberculosis strains, the sequence of selected regions of the pgm locus and flanking regions were compared from 20 different Y. pestis and 10 Y. pseudotuberculosis strains. The results showed that the genes interrupted in Y. pestis are intact in Y. pseudotuberculosis. However, one of these mutations, in the bvgS homologue, is only present in Y. pestis strains of biovar Orientalis and not in those of the biovars Antiqua and Medievalis. The results obtained by analysis of variable positions in the sequence are in accordance with historical records, confirming that biovar Orientalis is the most recent lineage. Furthermore, sequence comparisons among 29 Yersinia strains suggest that Y. pestis is a recently emerged pathogen that is probably entering the initial phase of reductive evolution.
The genus Yersinia contains 11 species, three of which are pathogenic for humans. Yersinia pestis, the causative agent of plague, is transmitted primarily by fleas and has been responsible for devastating epidemics throughout history. Yersinia pseudotuberculosis and Y. enterocolitica are food- and waterborne pathogens that cause a much more benign enteric disease in humans. Despite these profoundly different pathogenesis strategies, Y. pestis and Y. pseudotuberculosis are very closely related phylogenetically and are nearly identical genetically, with >90% identity at the DNA level (6, 50). They are indistinguishable by standard DNA hybridization methods and share 99.7% nucleotide sequence identity in 16S ribosomal DNA (6, 37). Because of this and also physiological and antigenic similarities, it has been proposed that the two species be reclassified as subspecies of a single species (6). The striking difference in pathogenicity of these two bacteria, the presence of two additional plasmids in Y. pestis, and security reasons led to the decision not to reclassify them into one species. Y. pestis is a phenotypically homogeneous species with only one serotype, one lysotype, and three biovars (56). Strains of these three biovars exhibit no difference in their virulence or pathology in animals or humans (58).
One of the distinct features of Yersinia pathogenesis is the ability to scavenge iron from the host via a siderophore called yersiniabactin (Ybt). Eleven genes important for Ybt biosynthesis have been identified (4, 25, 26, 55). Distinct functions for the products of these genes were proposed and in most cases demonstrated. Genetic analyses demonstrated that these genes are chromosomally encoded and clustered on a high-pathogenicity island (HPI) in all three highly pathogenic species of Yersinia. The presence of this HPI distinguishes Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biotype 1B from Y. enterocolitica biotypes 2 to 5. The latter are pathogenic for humans but not able to disseminate in the mammalian host (i.e., they are not lethal for mice at low doses) (15).
In Y. pestis it was recently demonstrated that this HPI, whose size was estimated at ∼35 kb, is contained within a 102-kb unstable chromosomal region (12, 33, 46), termed the pgm (pigmentation) locus first described by Fetherston et al. (24). The remaining ∼68-kb region within the pgm locus was named the “pigmentation segment.” It includes the hms (hemin storage) locus (41), which confers a pigmented phenotype on colonies grown on Congo red-agar plates. It was shown that this locus is important for transmission of Y. pestis by the flea vector (35, 40). Both parts of this region carry genes important either for virulence (Ybt system) or for disease transmission (hms locus). The pgm locus deletes spontaneously en bloc at a frequency of 10−5 (10), probably by homologous recombination between its two flanking isertion element (IS) IS100 copies (24). Contradictory results have been obtained when the virulence of mutants deleted of the 102-kb unstable region was tested in the mouse model. Une and Brubaker (70) observed a loss of virulence only via a subcutaneous route of infection, while Iteman et al. (38) showed a strong decrease (106-fold) when strains were also injected intravenously. These contradictory results may be due to differences between the strains tested or to the fact that within the pgm locus different spontaneous deletions may occur, leading to a nonpigmented phenotype (12). Our work sought to identify new genes on the pigmentation segment possibly involved in the marked loss of virulence of Y. pestis upon deletion of the 102-kb pgm locus and to improve knowledge about the organization and the origin of the HPI of Y. pestis. We were also interested in studying genetic variations within the region sequenced among different Y. pestis and Y. pseudotuberculosis strains.
This study describes the first complete DNA sequence analysis of the 102-kb pgm locus in Y. pestis that has long been associated with plague pathogenesis. We present further evidence for the phage origin of the HPI and demonstrate an almost 100% identity of the yersiniabactin system among different Y. pestis strains. Analyses of the base composition of the 102-kb pgm locus and flanking regions confirm that the HPI of Y. pestis is distinct from the pigmentation segment and the flanking regions. Within the pigmentation segment we identified structural genes possibly involved in the synthesis of fimbriae that may be important for virulence and also regulatory genes encoding a two-component system. Investigation of genetic variations among different strains of Y. pestis and Y. pseudotuberculosis brings new insight into the phylogenetic relationship of these two closely related species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The characteristics of the Y. pestis and the Y. pseudotuberculosis strains used in this study are listed (see Table 2). Y. pestis 6/69M was isolated in Madagascar from a patient suffering from bubonic plague. The Escherichia coli strains used were electrocompetent DH10B (64) for the construction of the BAC library and E. coli XL2-Blue ultracompetent cells (Stratagene) for the construction of the shotgun libraries. Yersinia strains were grown for 24 h (peptone broth) or 48 h (Trypticase soy agar plates) at 28°C. E. coli DH10B transformants were grown at 37°C for 24 h on Luria-Bertani (LB) agar plates containing 12.5 μg of chloramphenicol per ml; E. coli XL2-Blue transformants were grown on LB medium containing 100 μg of ampicillin per ml.
TABLE 2.
Characteristics of the Yersinia strains used in this study
| Strain | Geographical region | Biovar or serotypea | Yr of isolation | Origin | Presence of mutation:
|
|||
|---|---|---|---|---|---|---|---|---|
| yp3 | yp48 | yp35 | hmsF | |||||
| Y. pestis | ||||||||
| 6/69 | Madagascar | Orientalis | 1969 | Human | + | + | + | + |
| 501 | Hamburg | Orientalis | 1964 | + | + | + | + | |
| 506 | Vietnam | Orientalis | 1955 | + | + | + | + | |
| 509 | Vietnam | Orientalis | 1963 | Human | + | + | + | + |
| 510 | Vietnam | Orientalis | 1963 | + | + | + | + | |
| 513 | Vietnam | Orientalis | 1964 | + | + | + | + | |
| 518 | Kurdistan | Medievalis | 1947 | Human | + | + | − | + |
| 522 | Senegal | Orientalis | 1944 | Human | + | + | + | + |
| 523 | Senegal | Orientalis | 1944 | Human | + | + | + | + |
| 530 | Madagascar | Orientalis | 1939 | Human | + | + | + | + |
| 531 | Vietnam | Orientalis | 1955 | + | + | + | + | |
| 532 | Vietnam | Orientalis | 1955 | + | + | + | + | |
| 537 | Kenya | Antiqua | 1952 | + | + | − | NDb | |
| 538 | Kenya | Antiqua | 1952 | + | + | − | + | |
| 539 | Kenya | Antiqua | 1952 | + | + | − | + | |
| 540 | Kenya | Antiqua | 1952 | + | + | − | + | |
| 543 | Rep. Congo | Antiqua | 1953 | Human | + | + | − | ND |
| 544 | Kenya | Antiqua | Human | + | + | − | + | |
| 554 | Kenya | Antiqua | Human | + | + | − | + | |
| 562 | Kurdistan | Medievalis | 1947 | Rat | + | + | − | + |
| 564 | Kurdistan | Medievalis | 1948 | Rat | + | + | − | + |
| Y. pseudotuberculosis | ||||||||
| IP32637 | France | I | 1983 | Human | − | − | − | + |
| IP32953 | France | I | 1990 | Human | − | − | − | + |
| IP32941 | France | I | 1990 | Human | − | − | − | + |
| IP32949 | France | I | 1990 | Human | − | − | − | + |
| IP32934 | France | II | 1989 | Monkey | − | − | − | + |
| IP32951 | France | II | 1990 | Human | − | − | − | + |
| IP32945 | Argentina | III | 1990 | Cattle | − | − | − | + |
| IP32937 | Argentina | III | 1989 | Cattle | − | − | − | + |
| IP31830 | Great Britan | IV | 1969 | Human | − | − | − | + |
| IP32817 | Japan | V | 1986 | − | − | − | + | |
Biovar for Y. pestis strains, serotype for Y. pseudotuberculosis strains.
ND, not determined.
Bacterial artificial chromosome (BAC) library construction.
Preparation of Y. pestis 6/69M DNA in agarose plugs was conducted as previously described (13). Plugs were stored in 0.2 M EDTA at 4°C and washed three times in 0.1% Triton X-100 buffer prior to use. pBeloBAC11 was kindly provided by H. Shizuya, Departement of Biology, California Institute of Technology (Pasadena, Calif.). Preparation of pBeloBAC11 was carried out as described by Une and Brubaker (71). Partial digestion of the Y. pestis DNA was carried out on plugs, each containing approximately 10 μg of high-molecular-weight DNA, after three 1-h equilibration steps in 50 μl of 1× HindIII digestion buffer (Boehringer Mannheim) plus 0.1% Triton X-100. The buffer was then removed and replaced by ice-cold HindIII enzyme buffer (1 ml/plug) containing 20 U of HindIII (Boehringer Mannheim). After 2 h of incubation on ice, the plugs were transferred to a 37°C water bath for 30 min. Digestions were stopped by adding 100 μl of 250 mM EDTA (pH 8.0). For size selection, the partially digested DNA was subjected to contour-clamped homogeneous electric field electrophoresis on a 1% agarose gel by using a DR III apparatus (Bio-Rad, Hercules, Calif.) in 1× Tris-acetate-EDTA buffer at 13°C, with a ramp from 3 to 15 s at 6 V/cm for 16 h. Agarose slices from 50 to 100 kb and from 100 to 150 kb were excised from the gel and stored in Tris-EDTA at 4°C. For ligation and transformation, the agarose slices containing the excised DNA were melted at 65°C for 10 min and digested with Gelase (Epicentre Technologies, Madison, Wis.) by using 1 U per 100 μl of gel slice. Then, 25 to 100 ng of the size-selected DNA was ligated to 10 ng of HindIII-digested, dephosphorylated pBeloBAC11 by using 10 U of T4 DNA ligase (New England Biolabs, Beverly, Mass.) at 16°C for 20 h. Ligation mixtures were heated at 65°C for 15 min and then drop dialyzed against Tris-EDTA with VS 0.025-mM-pore-size membranes (Millipore, Bedford, Mass.).
Fresh electrocompetent E. coli DH10B cells were harvested from 200 ml of a mid-log-phase culture grown in SOB medium. Cells were washed three times in ice-cold water and finally resuspended in ice-cold water to a cell density of 1011 cells/ml. One microliter of the ligation mix was used for electroporation of 30 μl of electrocompetent E. coli DH10B in an Easyject Plus electroporator (Eurogentec, Seraing, Belgium), with settings of 2.5 kV, 25 μF, and 99 Å in 2-mm-wide electroporation cuvettes. After electroporation, cells were resuspended in 600 μl of SOC medium, allowed to recover for 45 min at 37°C with gentle shaking, and then plated on LB agar containing chloramphenicol (12.5 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid; 50 μg/ml) and IPTG (isopropyl-β-d-thiogalactopyranoside; 25 μg/ml). The plates were incubated overnight and recombinants were picked manually in duplicate to 96-well plates containing 100 μl of 2YT-chloramphenicol at 12.5 μg/ml per well and incubated overnight. Then, 100 μl of 80% glycerol were added and the plates were stored at −80°C.
Shotgun cloning and DNA sequencing.
To generate a shotgun library in pcDNA2.1, 5 μg of DNA purified by a modified Birnboim and Doly protocol (9) from the BAC clones was sheared by nebulization, end repaired by using T4 DNA polymerase (Boehringer Mannheim), and then ligated to BstXI adaptors (Invitrogen). Ligated DNA was then fractionated by agarose gel electrophoresis prior to a second ligation of 1- to 3-kb fragments into BstXI-digested pcDNA2.1 (Invitrogen). Recombinant pcDNA2.1 templates were used for cycle sequencing reactions. For each sequencing reaction, 3 μl of DNA (200 to 300 ng), 2 μl primer (3.2 pmol), 4 μl of reaction mix of the Taq BigDye Terminator cycle Sequencing Kit (Applied Biosystems), and 3 μl of distilled water were used. After 26 cycles (96°C for 30 s, 56°C for 15 s, and 60°C for 4 min) in a thermocycler (Perkin-Elmer 9700), DNA was precipitated by using 70 μl of 70% ethanol–0.5 mM MgCl2, centrifuged, rinsed with 70% ethanol, dried, and dissolved in 2 μl of formamide-EDTA buffer. Samples were loaded onto a 96-lane, 4% polyacrylamide gels, and electrophoresis was performed on a model ABI PRISM 377 automatic DNA sequencer (Perkin-Elmer) for 10 h. In the closure phase, sequencing reactions were performed by using as templates clones from the shotgun library, BAC DNA, or PCR fragments and then by using custom-made oligonucleotides as primers.
Primers and PCR amplifications.
The sequences for the sense (SP) and antisense primers (ASP) used in this study for PCR analysis of the different frameshift mutations and variable positions were as follows: (i) yp3, SP, 5′-CTGCATATGGAAATAAGAAATCGTC-3′ and ASP, 5′-CCCTACCGTGCACGGCGG-3′; (ii) yp35, SP, 5′-TCATTGCCCGTCGTACCGG-3′, and ASP, 5′-AAGACCAAGAGCAGCCCCG-3′; (iii) hmsF, SP, 5′-ATTGCTAACCCACAGGGGAAT-3′; and ASP, 5′-CGCCAATACCGGCATCCAG-3′; and (iv) yp48, SP, 5′-GCCCTACGTCATCACCCAAC-3′, and ASP, 5′-GTTTGTTGCTGGTTTTATTGGGT-3′. PCR reactions were performed with Taq polymerase (Boehringer Mannheim). Reactions contained 5 μl of 10× PCR buffer (100 mM β-mercaptoethanol, 600 mM Tris HCl [pH 8.8]), 20 mM MgCl2, 170 mM (NH4)2SO4, 5 μl of 20 mM nucleotide mix, 0.2 μM concentrations of each primer, 10 to 50 ng of template DNA, 10% dimethyl sulfoxide, 0.5 U of Taq polymerase, and sterile distilled water to 50 μl. Thermal cycling was performed on a TouchDown amplifier (Hybaid, Ltd.) with an initial denaturation step of 90 s at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 55°C, and 2 min at 72°C.
Computer analysis.
The sequence was assembled by using PHRED (19, 20) and PHRAP (P. Green, unpublished); for the editing, CONSED (27) was used. Sequence analysis and annotation was managed by DIANA (Display and Analysis; Sanger Center) and the IMAGENE package (48). For identification of repeated sequences XSIP from the Staden sequence software analysis package was used. Determination of similarities with known proteins involved interrogation of BLAST2N, BLAST2X, and BLAST2P (1) and FASTA (54). tRNA genes were located and identified by using the tRNAscan (45). The sequence data have been submitted to the DDJ, EMBL, and GenBank databases under accession number AL031866.
RESULTS
Construction of a BAC library of Y. pestis 6/69M.
The BAC cloning system is capable of stably propagating large, complex DNA inserts in E. coli (65). A BAC library containing Y. pestis chromosomal DNA was constructed by using the pBeloBAC11 vector (39) as described above. The library contains 1,500 clones, which represents a 30-fold coverage of the Y. pestis chromosome, estimated to be 4.4 Mb in size. Size determination of randomly selected, NotI cleaved BACs via pulsed-field gel electrophoresis showed that the insert size for the majority of the clones ranged between 100 and 130 kb. Since the pgm locus spans 102 kb (12, 23), it was expected that some BAC clones would contain this entire chromosomal fragment. In order to identify such clones, 800 BAC clones of the library were hybridized successively with two probes (B0.9 and a PCR product specific to the psn gene (11), known to be present as single copies in the Y. pestis chromosome and to be located at opposite ends of the 102-kb region. DNA from six clones positive for both probes was prepared and subjected to restriction enzyme analysis with EcoRI. Southern hybridization with different clones encompassing the pgm locus indicated that these BAC clones contained the complete 102-kb region, differing only in the size of the fragment cloned outside the two IS100 insertion sequences. To detect eventual minor rearrangements due to cloning artefacts, the sequence was obtained from two BAC clones: 1F12 and 3B11. No differences were detected between the sequences.
DNA sequence analysis of the 102-kb unstable region.
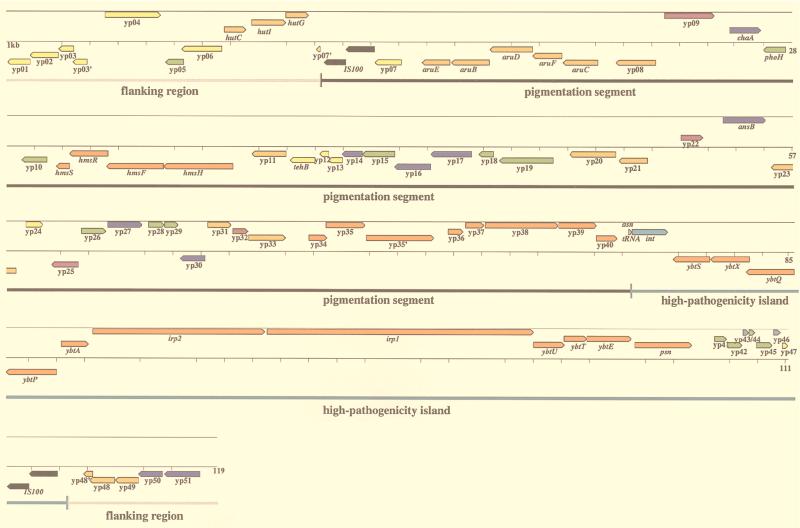
We determined a 119,443-bp long sequence of the Y. pestis chromosome of strain 6/69M encompassing the 102-kb pgm locus. Computer analysis of this sequence revealed the presence of 83 putative coding sequences (CDSs). In three cases (yp3, yp35, and yp48), two successive open reading frames (ORFs) were predicted to encode proteins similar to the N- and C-terminal parts, respectively, of the same protein (Fig. 1 and Table 1). This observation led us to predict possible frameshift mutations for these genes. Two CDSs (yp7 and yp47) are interrupted by IS100 insertion (Fig. 1). Among the 83 ORFs which may correspond to coding sequences, only 8 encode proteins lacking similarity to any known protein in the EMBL, GenBank, or Swissprot database, and two are similar to protein sequences of unknown function (yp5 and yp10). A putative function could therefore be attributed to 73 of 83 CDSs (Table 1). In a previous study (12) based on hybridization analyses, two repeated sequences (RS), RS4 and RS5, were identified in the pgm locus. A search for these RS in the sequence revealed that RS5 corresponds to a sequence of 167 bp (located between yp19 and yp20) which is repeated more than 60 times within the chromosome of Y. pestis (identified by a search in the Sanger Centre Y. pestis database). In contrast, we could not identify a sequence that would explain the hybridization results obtained previously that indicated the presence of RS4, suggesting that it does not correspond to a true repeated sequence.
FIG. 1.
Map of the pgm locus and flanking regions showing the position and orientation of known genes and CDSs. We used the following functional categories: related to virulence (red), phage-related functions (light blue), transport proteins (dark blue), regulatory proteins (purple), nitrogen and carbon metabolism (brown), miscellaneous (yellow), insertion elements (black), and proteins of unknown function (green). The thick lines below the gene map indicate flanking regions (gray), pigmentation segment (black), and HPI (blue). Numbers at the right end of the map indicate the scale in kilobases.
TABLE 1.
CDSs identified on the 119-kb chromosomal sequence of Y. pestis 6/69M
| CDSa designation | Size (aab residues) | Homologue as determined by FASTA | % Identity (aab overlap) |
|---|---|---|---|
| yp1 | 268 | GumP (X. campestris) | 33.7 (252) |
| yp2 | 338 | Oxidoreductase (E. coli) | 44.4 (331) |
| yp3c | 175 | GumO (X. campestris), C end | 39.5 (172) |
| yp3′c | 169 | GumO (X. campestris), N end | 38.9 (149) |
| yp4 | 658 | Anthranilate synthase (M. tuberculosis) | 34.3 (450) |
| yp5 | 190 | Hypothetical protein (E. coli) | 38.5 (179) |
| yp6 | 456 | Atrazine chlorohydrolase h. (M. xanthus) | 36.6 (421) |
| hutC | 255 | HutC (P. putida) | 63.2 (242) |
| hutI | 406 | HutI (P. putida) | 54.2 (408) |
| hutG | 268 | HutG (P. putida) | 59.6 (260) |
| ′yp7cd | 50 | OmpF (S. marcescens), C end | 66.0 (50) |
| IS100 | 259 | IS100 transposase (Y. pestis) | 100 (259) |
| IS100 | 340 | IS100 transposase (Y. pestis) | 100 (340) |
| yp7′c | 315 | OmpF (S. marcescens), N end | 58.0 (299) |
| aruE | 330 | AruE (P. aeruginosa) | 38.1 (322) |
| aruB | 447 | AruB (P. aeruginosa) | 60.8 (446) |
| aruD | 505 | AruD (P. aeruginosa) | 63.5 (491) |
| aruF | 350 | AruF (P. aeruginosa) | 52.9 (344) |
| aruC | 414 | AruC (P. aeruginosa) | 58.9 (416) |
| yp8 | 473 | Tyrosine aminotransferase (E. coli) | 56.8 (465) |
| yp9 | 593 | NarX (E. coli) | 47.9 (595) |
| chaA | 366 | Calcium/proton antiporter ChaA (E. coli) | 78.4 (356) |
| phoH | 262 | Phosphate starvation-inducible PhoH (E. coli) | 91.2 (262) |
| yp10 | 299 | Hypothetical protein (S. cerevisiae) | 42.6 (230) |
| hmsS | 155 | HmsS (Y. pestis KIM6+) | 100 (155) |
| hmsR | 457 | HmsR (Y. pestis KIM6+) | 99.8 (457) |
| hmsF | 673 | HmsF (Y. pestis KIM6+) | 96.9 (673) |
| hmsH | 822 | HmsH (Y. pestis KIM6+) | 100 (822) |
| yp11 | 405 | Proline peptidase (B. subtilis) | 25.7 (390) |
| tehB | 295 | Tellurite resistance protein (E. coli) | 61.3 (191) |
| yp12 | 104 | Cytochrome c553 precursor (D. vulgaris) | 29.8 (104) |
| yp13 | 165 | ResA (B. subtilis) | 28.8 (132) |
| yp14 | 237 | ABC transporter (E. coli) | 44.0 (225) |
| yp15 | 387 | Integral membrane protein (B. subtilis) | 22.9 (385) |
| yp16 | 430 | ABC transporter (E. coli) | 22.6 (442) |
| yp17 | 469 | H+-transporting ATPase (A. castellanii) | 26.7 (225) |
| yp18 | 175 | Membrane antigen precursor (T. pallidum) | 40.0 (170) |
| yp19 | 639 | Unknown | |
| yp20 | 546 | YfiU (B. subtilis) | 25.3 (538) |
| yp21 | 340 | Putative glutaminase (E. coli) | 42.5 (306) |
| yp22 | 256 | DeoR family, transcription regulator (B. subtilis) | 61.9 (252) |
| ansP | 508 | l-Asparagine permease (E. coli) | 79.6 (489) |
| yp23 | 386 | Aminotransferase (S. solfataricus) | 33.1 (393) |
| yp24 | 199 | NADH-ubiquinone oxidoreductase (H. influenzae) | 69.0 (197) |
| yp25 | 320 | LysR family, transcription regulator (E. coli) | 27.0 (293) |
| yp26 | 289 | Unknown | |
| yp27 | 409 | Transport protein (R. eutropha) | 44.2 (407) |
| yp28 | 186 | Unknown | |
| yp29 | 157 | Unknown | |
| yp30 | 294 | LysR family, transcription regulator (E. coli) | 40.8 (289) |
| yp31 | 280 | Citrate lyase beta-chain (E. coli) | 32.0 (291) |
| yp32 | 180 | Regulatory protein (M. tuberculosis) | 45.1 (173) |
| yp33 | 440 | 4-Hydroxybutyrate coenzyme A transferase (C. kluyveri) | 40.0 (440) |
| yp34 | 210 | BvgA (B. pertussis), FimZ (E. coli) | 48.3 (207) |
| yp35′c | 464 | BvgS, sensor protein (B. pertussis), N end | 26.3 (437) |
| ′yp35c | 803 | BvgS, sensor protein (B. pertussis, C end | 30.9 (783) |
| yp36 | 176 | Pilin precursor (E. coli) | 37.9 (177) |
| yp37 | 214 | HifB, periplasmic chaperone (H. influenzae) | 30.7 (228) |
| yp38 | 863 | FocD, outer membrane usher protein (E. coli) | 29.8 (857) |
| yp39 | 449 | HifA (H. influenzae) | 27.8 (187) |
| yp40 | 246 | F17d-D, chaperone (E. coli) | 33.1 (248) |
| asn tRNA | 88 | Asparagine tRNA synthetase (Y. pseudotuberculosis) | 100 (88) |
| int | 420 | Int (Y. pseudotuberculosis) | 100 (420) |
| IntB, p rophage P4 integrase (E. coli) | 54.1 (390) | ||
| Int, bacteriophage P4 | 49.5 (420) | ||
| ybtS | 434 | YbtS (Y. pestis KIM6+) | 100 (434) |
| ybtX | 462 | YbtX (Y. pestis KIM6+) | 100 (462) |
| ybtQ | 600 | YbtQ (Y. pestis KIM6+) | 100 (600) |
| ABC transporter, (M. tuberculosis) | 32.5 (587) | ||
| ybtP | 600 | YbtP (Y. pestis KIM6+) | 99.8 (600) |
| ABC transporter (M. tuberculosis) | 35.7 (577) | ||
| ybtA | 319 | YbtA AraC type regulator (Y. pestis KIM6+) | 100 (319) |
| irp2 | 2,041 | HMWP2 (Y. pestis KIM6+) | 100 (2041) |
| irp1 | 3,163 | HMWP1 (Y. pestis KIM6+) | 100 (3163) |
| ybtU | 365 | YbtU (Y. pestis KIM6+) | 100 (365) |
| Irp3 (Y. enterocolitica) | 97.8 (365) | ||
| ybtT | 267 | YbtT (Y. pestis KIM6+) | 100 (267) |
| ybtE | 525 | YbtE (Y. pestis KIM6+) | 100 (525) |
| psn | 673 | Psn, pesticin receptor (Y. pestis KIM6+) | 100 (673) |
| yp41 | 140 | Unknown | |
| yp42 | 168 | Unknown | |
| yp43 | 61 | ORF 88 (bacteriophage P4) | 36.1 (61) |
| yp44 | 54 | Unknown | |
| yp45 | 193 | Unknown | |
| yp46 | 69 | ORF 82 (bacteriophage P2), DNA-binding protein | 31.9 (69) |
| TraR (E. coli) | 33.8 (68) | ||
| yp47 | 59 | Hha, hemolysin expression modulator (E. coli) | 33.8 (57) |
| YmoA, modulating virulence expression | 35.8 (53) | ||
| IS100 | 259 | IS100 transposase (Y. pestis) | 100 (259) |
| IS100 | 355 | IS100 transposase (Y. pestis) | 100 (355) |
| ′yp48c | 113 | MalK C-terminal (E. coli) | 33.3 (87) |
| yp48′c | 296 | MalK N-terminal (E. coli) | 59.6 (265) |
| yp49 | 275 | MalG (E. coli) | 35.3 (266) |
| yp50 | 286 | ABC transporter (Rhizobium sp.) | 37.6 (269) |
| yp51 | 422 | ABC transporter (Sinorhizobium fredii) | 26.7 (419) |
Coding sequence.
aa, amino acid.
Designates an interrupted coding sequence.
yp7 is also highly similar to the E. coli PhoE protein.
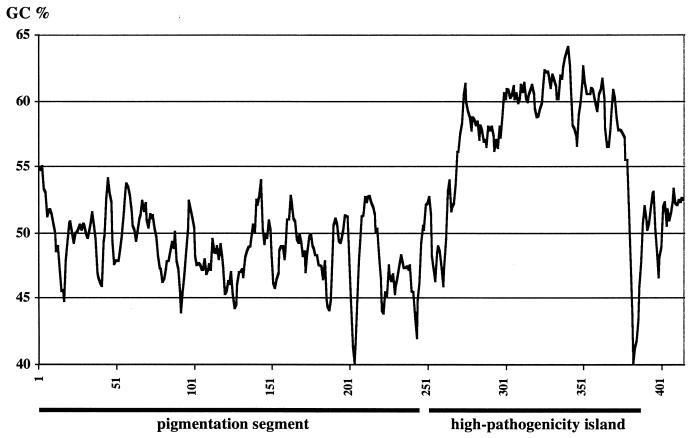
The base composition of the genes in the entire sequenced 119-kb region was found to be heterogeneous. An average GC content of 49%, which corresponds to the average GC content of the Y. pestis chromosome, was calculated for the CDSs located on the pigmentation segment and the flanking regions outside the IS100. In contrast, a significant increase in the GC content was observed for the CDSs on the 36.4-kb region defined as HPI. A similar observation was reported by Hare et al. (33) on partial sequences of the HPI. We found the average GC content of this region to be 59% (Fig. 2). Differences in GC content have been considered as indicators for the heterologous origin of PAIs. These results further substantiate that the 102-kb unstable region can be divided into two functional, distinct regions: the HPI and the pigmentation segment.
FIG. 2.
Distribution of the base composition of the CDS within the 119-kb region sequenced. The DNA sequence was analyzed for the base composition by using GMP Tool Box (unpublished) with a window size of 1,000 bases and was shifted each time by 250 bp. Because of the difference in average size of the genes present on the HPI and the pigmentation segment, the G+C content was calculated only from the CDSs. Intergenic regions were not considered. The percentage of the G+C is indicated. The thick line below indicates the region defined as HPI and as pigmentation segment.
The total sequence of the HPI of Y. pestis 6/69M reveals features most probably of phage origin.
We have identified 19 CDSs in the HPI of Y. pestis 6/69M. Of these, 11 that correspond to the Ybt biosynthetic gene cluster have been previously described in Y. pestis KIM6+ (21, 22, 25, 26). Four newly identified CDSs (yp41, yp42, yp44, and yp45) encode proteins with no significant similarities to proteins in the public databases. yp47 shows homology with Hha (Table 1), described as a small polypeptide in E. coli which modulates hemolysin expression (52), and with Y. enterocolitica YmoA, which is implicated in modulating the expression of several temperature-regulated virulence functions by changes in DNA supercoiling (61). However, yp47 does not seem to be functional since it contains an in-frame stop codon (codon 16) and appears to be disrupted at its 3′ end by the IS100 insertion sequence bordering the HPI. Disruption of this CDS by IS100 insertion was also reported for Y. pestis KIM6+ (56).
Three of the newly identified CDSs (int, yp43, and yp46) encode proteins related to phage proteins. Identification of an integrase gene in the HPI of Y. pseudotuberculosis and putative att sites bordering the HPI led us (11) and others (33) to assume phage-mediated integration of the HPI. The int gene sequenced on the HPI of Y. pestis 6/69M was identified, as in Y. pseudotuberculosis, as a putative integrase gene able to code for a protein of 420 amino acids (48 kDa). The sequence had one nucleotide difference compared to the integrase gene identified in the HPI of Y. pseudotuberculosis IP32637 (11). To obtain further evidence for a phage origin of the HPI, we searched the sequence for additional phage features. We identified two ORFs, yp43 and yp46, located 1,369 and 400 bp upstream of the IS100 insertion element respectively, that are of probable phage origin. yp43 is similar to ORF 88 from bacteriophage P4 (Table 1), which encodes a putative DNA-binding protein (32). The yp46 gene encodes a protein similar to ORF 82 of bacteriophage P2 (Table 1). The protein encoded by ORF 82 of bacteriophage P2 is also thought to have possible DNA-binding activity, since it contains two Cys-X-X-Cys motifs (43). The two Cys-X-X-Cys motifs found in ORF 82 of bacteriophage P2 are conserved at an equivalent position in yp46, which indicates that this protein might have DNA-binding activity.
Sequence of the pigmentation segment and the flanking regions of Y. pestis 6/69M.
With the exception of the hmsSFRH operon, nothing was known about the functions of genes located on the pigmentation segment and whether it is involved in the marked loss of virulence upon deletion of the 102-kb unstable region. In the pigmentation segment, 48 ORFs were predicted as coding. Outside the two IS100 insertion elements, a 11- and a 4-kb stretch of DNA was analyzed (Fig. 1). These flanking regions contain 16 CDSs. By using various database comparisons, we attributed probable functions to the predicted proteins of 58 of these CDSs. Two CDSs (yp5 and yp10) show similarities to hypothetical proteins in E. coli and Saccharomyces cerevisiae, respectively. The remaining four CDSs (yp19, yp26, yp28, and yp29) resemble no known proteins and may account for specific functions of Y. pestis (Table 1 and Fig. 1). Analysis of this region discloses several interesting features. A high density of genes involved in amino acid utilization is observed. Three genes (hutC, hutI, and hutG) are possibly involved in histidine utilization, and five (aruEBDFC) may be involved in arginine utilization. The yp8, yp11, yp21, and ansB genes code for proteins similar to tyrosine aminotransferases, proline peptidases, glutaminases, and l-asparagine permeases, respectively (Table 1 and Fig. 1). A number of genes involved in regulatory and transport functions were uncovered. Four CDSs (yp22, yp25, yp30, and yp32) code for proteins similar to transcriptional regulators of the DeoR (yp22) or LysR family (yp25 and yp30) or to a regulatory protein of Mycobacterium tuberculosis (yp32). Seven CDSs putatively code for different transport functions. The yp14 and yp16 genes code for proteins similar to ABC transporters, and chaA, yp17, yp27, yp50, and yp51 show similarities to transport proteins from various bacteria (Table 1).
Interestingly, the pigmentation segment contains two operons separated by 516 bp that are possibly involved in virulence: a pilin-like locus (yp36-yp40) and another locus (yp34-yp35) homologous to bvgA/bvgS of B. pertussis (3), which regulates virulence gene expression in B. pertussis (68). The pilin-like locus codes for proteins which show high similarities to the product of the F17-like fimbrial gene cluster from enterotoxigenic (accession number L77091) and invasive E. coli strains (18) and for the major pilus gene cluster hifABCDE of Haemophilus influenzae (49). It contains five CDSs (yp36 to yp40) organized in an operon (Fig. 1 and Table 1). The putative functions of these five CDSs, deduced from similarity searches with known proteins, are that yp36 appears to be the major fimbrial subunit protein, yp37 and yp40 are chaperone proteins, and yp38 is an outer membrane usher protein. The yp39 gene putatively codes for a protein that is weakly similar to major fimbrial subunit proteins. yp34 and yp35 are highly similar to the bvgAS locus of B. pertussis, which coordinately regulates the expression of virulence functions (3), and the evgAS locus of E. coli (65) (Fig. 1 and Table 1). The conclusion that yp34 and yp35 are also implicated in the regulation of expression of virulence genes and/or more specifically of the downstream operon is tempting. However, sequence analysis reveals a frameshift mutation due to a deletion of a thymidine residue at position 1163 in yp35, the homologue of bvgS, splitting it into two ORFs of 464 and 803 codons (see also below). Thus, one may conclude that this protein is not functional in Y. pestis 6/69M.
Comparison of the yersiniabactin biosynthetic gene cluster and the hms operon among Yersinia strains.
The Ybt region has already been sequenced in different Y. pestis strains. The availability of these sequences allowed us to investigate the degree of diversity of this region within three different strains isolated in different regions and belonging to different biovars. The strain we have sequenced and analyzed (Y. pestis 6/69M) is of biovar Orientalis and was isolated in Madagascar. KIM6+, the first strain for which the Ybt region was entirely sequenced (GenBank accession number AF091251) is of biovar Medievalis and was isolated in Kurdistan (4, 25). Strain CO92, whose genome is presently sequenced (62a) is of biovar Orientalis and was isolated in Colorado. Comparison of the three sequences revealed a striking conservation between them over a total of 35,806 nucleotides. The sequence of the Ybt region of Y. pestis 6/69M seems to be identical to that of Y. pestis CO92. However, the CO92 sequence still contains several ambiguities. Compared to Y. pestis KIM6+, 6/69M has only one nucleotide difference over a range of 35,806 residues. This one nucleotide difference, located in the ybtP gene, results in the change of alanine to threonine at position 445.
Y. pseudotuberculosis serotype I strains are also known to possess the Ybt system (11, 33, 60), but only the pesticin-yersiniabactin receptor psn and parts of the irp2 gene have been sequenced. We compared the sequence of the psn gene of Y. pestis 6/69M with that of Y. pseudotuberculosis, available in the public database (accession number Z35107). It shows only one nucleotide substitution over a range of 2,017 residues. When the partial sequence of irp2 (accession number Z35451) of Y. pseudotuberculosis was compared with that of Y. pestis 6/69M, also only one difference (over a range of 275 nucleotides) was noted.
Y. enterocolitica biotype 1B also harbor the Ybt region. Rakin et al. (59) reported the sequence of the pesticin-Ybt receptor gene fyuA (accession number Z29675), Pelludat et al. (55) reported the sequence of the irp1, irp3/ybtU, irp4/ybtT, and irp5/ybtE genes (accession number Y12527), and Guilvout et al. (29) reported the sequence of the irp2 gene (accession number L18881). We compared these known sequences with that of Y. pestis 6/69M. Gene similarity of the Ybt region of Y. pestis with these regions sequenced from Y. enterocolitica ranged between 94.5% (ybtT/irp4) and 98.9% (psn/fyuA). These results show that the Ybt system of the three pathogenic yersiniae is therefore nearly identical.
Comparison of the hms operon sequence between Y. pestis 6/69M and Y. pestis CO92 (62a), both biovar Orientalis strains, revealed 100% identity over a range of 6,320 residues. When the sequence of the hms operon of strain Y. pestis 6/69M was compared with that of Y. pestis KIM6+ (accession number U22837) (biovar Medievalis), we found that hmsS and hmsH were identical. In hmsR one nucleotide difference was noted accounting for an amino acid change from an alanine to a valine at position 50. Another observation was made on hmsF: an insertion of 19 codons in strains Y. pestis 6/69M and CO92 or a deletion of 19 codons in strain Y. pestis KIM6+. Sequencing of the 403 bp surrounding this region in 18 strains of Y. pestis of different biovars and 10 Y. pseudotuberculosis strains from different serotypes (Table 2) reveals a sequence identical to that determined here for Y. pestis 6/69M. Subsequent to this study it has been shown that the sequence of hmsF of strain KIM6+ is also identical to the sequence we found for strain 6/69M and all the others characterized here (55a).
Comparisons of Y. pestis and Y. pseudotuberculosis strains.
The comparison of our sequence with already-published sequences from other Y. pestis strains led to the discovery of only one variable position within 42,126 nucleotides. Surprisingly, the conservation within the hms operon was comparable to the conservation within the yersiniabactin region. In order to investigate whether this nearly 100% nucleotide sequence conservation is also found outside of the yersiniabactin region and the hms operon, which are known to be important for virulence or disease transmission, we attempted to identify possible variable positions within other regions of the 119-kb segment analyzed. The frameshifts revealed within yp3, yp35, and yp48 were attractive candidates for such variations.
Primarily, we clarified whether these mutations are specific to strain 6/69M or whether they are also present in other strains of Y. pestis. For this purpose, we determined the sequence of these three regions in 20 selected strains (Table 2). DNA was prepared from each strain, and fragments of 349, 665, and 270 bp for yp3, yp35, and yp48, respectively, covering the region where the mutation occurred, were amplified by PCR and sequenced. Since Y. pestis and Y. pseudotuberculosis are genetically highly similar, we could also amplify and determine the nucleotide sequence of the three regions in 10 selected strains of Y. pseudotuberculosis (Table 2) by using the same primers as in Y. pestis.
The sequences of the BAC clones of strain 6/69M and the PCR fragments obtained from genomic DNA were found to be identical, confirming that the three mutations are genomic and are not due to cloning artifacts. Comparison with the Y. pseudotuberculosis sequences reveals that these three genes are intact in Y. pseudotuberculosis (Table 2). This result allowed us to determine the nature of the genetic event leading to these frameshifts in Y. pestis. yp3 is a CDS of 1,032 bp in Y. pseudotuberculosis which is interrupted in Y. pestis at position 522 by the insertion of a thymidine leading to the introduction of a stop codon. In yp35, a CDS of 3,801 bp, the frameshift is due to the deletion of a thymidine at position 1163. For yp48 of Y. pestis, we found a deletion of 25 bp from position 887 to position 912.
When we compared the four regions studied in different Y. pestis strains (yp3, yp35, yp48, and hmsF), covering 1,686 bp, only one variable position was detected. All 20 strains investigated here harbor the frameshift mutation within yp3 and yp48, but only 11 had the frameshift mutation in yp35 (including strain 6/69M), whereas 9 strains had an intact yp35 gene (Table 2). Further exploration of the data reveals a correlation between the biovar and the presence of the frameshift mutation. All strains belonging to the biovar Antiqua and Medievalis had an intact yp35 gene, whereas all strains belonging to the biovar Orientalis had the same frameshift as in strain 6/69M.
DISCUSSION
The 102-kb chromosomal DNA fragment of Y. pestis 6/69M and its flanking regions have been sequenced and analyzed. This region is known to carry virulence genes (the Ybt biosynthetic gene cluster) and to delete spontaneously en bloc (23, 24). Although this arrangement of pathogenicity associated genes flanked by ISs superficially resembles a pathogenicity island (30), it was first suggested by Fetherston and Perry (23) that not the whole region has been transferred horizontally. Recently, it has been shown by physical mapping and hybridization studies that only the ∼35-kb region carrying the Ybt gene cluster should be considered an HPI per se (12, 33). Our sequence analyses confirm this result and reveal further evidence that the 102-kb region encompasses two distinct parts, with only one being a HPI according to the definition of Hacker et al. (30). One of the major characteristics of pathogenicity islands (PAI) is a GC content that is different from that of the genome average. A significant difference in the base composition between genes of the 36.4-kb region defined as HPI (59% GC) and those in the pigmentation segment and the flanking regions (49% GC) was observed (Fig. 2), substantiating the heterologous origin of the region carrying the Ybt biosynthetic gene cluster.
The presence of a prophage integrase next to a tRNA coding sequence containing a putative att site suggests acquisition of PAIs from a phage by horizontal transfer. Previous hybridization studies indicated the presence of a tRNA and an integrase gene within the 102-kb region of Y. pestis (11). In our sequence an asn tRNA gene and a putative integrase gene highly similar to that of bacteriophage P4 were identified at one extremity of the HPI and 17-bp sequences identical to a putative attB site are present at both extremities of the HPI. Furthermore, two CDSs related to phage proteins of bacteriophage P4 and bacteriophage P2 were identified at the other extremity of the HPI. To our knowledge, this is the first report of phage related CDSs at both extremities of a PAI. This finding strongly argues in favor of the hypothesis that the entire 36.4-kb region carrying the Ybt system was acquired en bloc via horizontal transfer from a bacteriophage. This is different to what is seen, for instance, for the Salmonella pathogenicity island 2 (SPI-2), which is reported to have mosaic structure and to be composed of at least two different genetic elements (34) or the SPI-3, for which a multistep process in the evolution of SPI-3 sequences was proposed (8). The fact that the entire region seems to be acquired en bloc may explain why the Yersinia PAI encodes only one virulence factor: the yersiniabactin iron transport system.
In Y. pestis, the HPI appears always to be associated with the pigmentation segment (12, 23). This is different from the situation in Y. pseudotuberculosis. It has been shown that the HPI of Y. pseudotuberculosis can precisely excise and insert in any of the three copies of asn tRNA present in the chromosome (11). Deletion of the HPI of Y. pseudotuberculosis probably occurs by homologous recombination between two 17-bp direct repeats located at each extremity of the HPI (11). In contrast, for the HPI of Y. pestis, no evidence of precise excision has been described (12, 23), although the organization with two 17-bp direct repeats located at both extremities is similar to the situation seen in Y. pseudotuberculosis. The fact that in Y. pestis the organization (HPI linked to the pigmentation segment) is well conserved might indicate an important difference between Y. pestis and Y. pseudotuberculosis. It suggests that one or some genes encoded on the pigmentation segment are important or necessary for gene expression or regulation of gene(s) located on the HPI.
The comparison of the HPI of Y. pestis to those of Y. pseudotuberculosis and Y. enterocolitica 1B reveals a striking conservation within the sequences as well as in its organization, suggesting relatively recent horizontal transfer. Nevertheless, there are differences between the Yersinia HPIs. In Y. pseudotuberculosis a deletion of 268 bp between the IS100 and the putative att site was noted. In contrast, the HPI of Y. enterocolitica does not contain an IS100 element at its extremity but contains IS1400, RS3, and IS1328 elements and is also reported to be ∼10 kb longer than that of the two other Yersinia species (14). These results indicate that the HPI of the three highly pathogenic Yersinia spp. consists of a conserved core region that contains the Ybt biosynthetic, regulation, and uptake genes, a region important for integration and/or excision of the HPI (integrase gene and putative att sites) and a part with no defined functions which diverges minimally in Y. pseudotuberculosis serogroup I and to a greater degree in Y. pseudotuberculosis serogroup III and Y. enterocolitica 1B.
With the exception of the hms locus, no information exists with regard to the functions encoded by the pigmentation segment. Although the GC content indicates that it should not be considered an HPI, we identified at least two loci which might be important for virulence, a pilin-like locus and a locus putatively coding for a two-component regulatory system.
The interaction of Y. pestis with host macrophages has long been thought to be important in the pathogenesis of bubonic plague (16), although little is known about this interaction. Y. pseudotuberculosis and Y. enterocolitica express different adhesins termed YadA, invasin, and PsaA/Myf. Y. pestis produces neither Inv (66) nor YadA (67). Recently, it was shown that the plasminogen activator Pla, encoded on the 9.5-kb plasmid of Y. pestis, enhances bacterial adhesion to extracellular matrix but that Y. pestis also exhibits a low level of Pla-independent adhesion (44). Another putative adhesin expressed by Y. pestis is PsaA (pH6 antigen), which forms virulence-associated fimbriae on the bacterial surface (5, 42). However, lack of the pH6 antigen causes only a relatively small decrease in virulence of Y. pestis (42). We identified, within the pigmentation segment of Y. pestis, a locus which putatively codes for a major fimbrial subunit protein, two chaperone proteins, an outer membrane usher protein, and a protein related to fimbrial subunit proteins. A number of pili from other bacteria, like the major pilus gene cluster of H. influenzae (49) or the type 4 pili from Neisseria spp. (7, 51, 62) have tip-localized, pilus-associated proteins functioning as adhesins. We could not identify, in the fimbrial gene cluster identified in Y. pestis, a putative adhesin. However, it was reported for Pa type 4 pili of Pseudomonas aeruginosa that the pilin subunit itself carries the adherence function (31). Therefore, the identified pilus-like gene cluster of Y. pestis described here might be organized in a similar way to the Pa type 4 pili of P. aeruginosa. A challenging question for the future is to determine the possible impact of these fimbriae on virulence of Y. pestis.
A second locus which is putatively related to virulence has been identified in this study. It encodes two proteins highly homologous to two-component regulatory systems. Surprisingly, the coding sequence of the sensor protein contains a frameshift mutation. However, this mutation was present only in Y. pestis strains belonging to the biovar Orientalis and not in those belonging to the biovars Medievalis and Antiqua. Y. pseudotuberculosis strains also show intact CDSs for the sensor protein. However, this locus could also be involved in the control of genes unrelated to virulence. Therefore, functional studies should elucidate whether this locus regulates virulence gene expression of Y. pestis, as it was demonstrated for the B. pertussis BvgA/S two component regulatory system (63). Should this locus be shown to regulate the expression of virulence genes in Y. pestis biovar Antiqua and biovar Medievalis and in Y. pseudotuberculosis, it would be of great interest to determine the effect of the frameshift mutation on Y. pestis biovar Orientalis. It should indeed be noted that inactivation of a sensor protein does not exclude cross talk between a nonpartner sensor of similar sequence and the regulatory protein, as shown for instance in the case of NtrB of E. coli (53).
Analyses of the DNA sequence and comparison of selected regions between Y. pestis and Y. pseudotuberculosis strains led to several hypotheses concerning the evolutionary history of this pathogen. The fact that only one variable position was identified when comparing four loci within 20 Y. pestis strains indicates that Y. pestis is a recently emerged pathogen. This conclusion is further substantiated by the following observation: within the entire yersiniabactin biosynthetic gene cluster (35,806 nucleotides), as well as within the hms operon (6,320 nucleotides), only one nucleotide difference each was present when three Y. pestis strains belonging to two different biovars were compared.
The fact that three genes interrupted by a frameshift mutation in Y. pestis are functional in Y. pseudotuberculosis indicates that Y. pseudotuberculosis is the progenitor of Y. pestis. Furthermore, the variable position detected in the bvgS homologue reveals a striking feature: the polymorphism is strictly correlated with the biovar of the strains. All strains of biovar Antiqua and strains of biovar Medievalis had an intact CDS, while the strains of biovar Orientalis display the frameshift mutation in the bvgS homologue. These findings indicate that biovar Orientalis is the most recent biovar. In 1951, Devignat proposed an association between the three biovars of Y. pestis and the three plague pandemics (17). However, he acknowledged that these observations were based on historical data and remained speculative in the absence of other evidence. Remarkably, the results obtained in our analysis of genetic markers are in complete accordance with historical records.
Previous studies showed that some genes, particularly those thought to be involved in the virulence of Yersinia spp., are not functional in Y. pestis. This observation was reported for the yadA gene, which presents a 1-bp deletion in Y. pestis compared to the sequence of Y. pseudotuberculosis (67). The inv gene, intact in Y. pseudotuberculosis and Y. enterocolitica, was reported to be interrupted by the insertion of IS1541 (66), and the ail gene was shown to be interrupted by IS285 insertion in Y. pestis EV76-51F (69). Recently, it was reported that ylpA, a gene encoded on the low-Ca2+-response plasmid, is likely to be a pseudogene in Y. pestis (36, 57). By systematic sequencing of a 119,443-bp sequence, which is less than 3% of the chromosome, we discovered two genes disrupted by IS insertions and three genes disrupted by a frameshift mutation. All of the genes harboring the frameshift mutation are intact in Y. pseudotuberculosis. In Rickettsia prowazekii, mutations which inactivate an expendable gene initiate a sequence of events where subsequent mutations freely transform the inactive gene, by degrees, from a pseudogene to an unrecognizable sequence and then to small fragments and finally to extinction (2). Similar results were reported for M. leprae (28). It is possible that the inactivation or deletion of genes in Y. pseudotuberculosis provides an evolutionary pathway to enhance virulence as it evolved into Y. pestis similar to the observation reported by Maurelli et al. (47), who showed that deletion of the cadA gene in Shigella spp. may have enabled it to evolve into a more-pathogenic form. As such, the inactivated genes revealed in this study and also reported for other genes (57, 66, 67, 69) may indicate that Y. pestis is in an initial stade of a “reductive evolution.”
ACKNOWLEDGMENTS
We thank Steve Gordon for the critical reading of the manuscript and Cathrine Soravito-Urban for technical assistance.
C. Buchrieser received a grant from the Austrian Program for Advanced Research and Technology (APART). Sequence data for Y. pestis CO92 were obtained from the Sanger Centre website (62a).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C M, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Arico B, Miller J F, Roy C, Stibitz S, Monack D, Falkow S, Gross R, Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci USA. 1989;86:6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Efraim S, Aronson M, Bichowsky-Slomnicki L. New antigenic component of Pasteurella pestis formed under specific conditions of pH and temperature. J Bacteriol. 1961;81:704–714. doi: 10.1128/jb.81.5.704-714.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bercovier H, Mollaret H H, Alonso J M, Brault J, Fanning G R, Steigerwalt A G, Brenner D J. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Y. pseudotuberculosis. Curr Microbiol. 1980;4:225–229. [Google Scholar]
- 7.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 8.Blanc-Potard A B, Solomon F, Kayser J, Groisman E A. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosch R, Gordon S V, Billault A, Garnier T, Eiglmeier K, Soravito C, Barrell B G, Cole S. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brubaker R R. Mutation rate to nonpigmentation in Pasteurella pestis. J Bacteriol. 1969;98:1404–1406. doi: 10.1128/jb.98.3.1404-1406.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 12.Buchrieser C, Prentice M, Carniel E. The 102-kilobases unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchrieser C, Weagant S D, Kaspar C W. Molecular characterization of Yersinia enterocolitica by pulsed-field gel electrophoresis and hybridization of DNA fragments to ail and pYV probes. Appl Environ Microbiol. 1994;60:4371–4379. doi: 10.1128/aem.60.12.4371-4379.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter P B. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavanaugh D C, Randall R. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J Immunol. 1959;83:348–363. [PubMed] [Google Scholar]
- 17.Devignat R. Variétés de l’espèce Pasteurella pestis. Nouvelle hypothèse. Bull Org Mond Santé. 1951;4:247–263. [PMC free article] [PubMed] [Google Scholar]
- 18.el Mazouari K, Oswald E, Hernalsteens J P, Lintermans P, De Greve H. F17-like fimbriae from an invasive Escherichia coli strain producing cytotoxic necrotizing factor type 2 toxin. Infect Immun. 1994;62:2633–2638. doi: 10.1128/iai.62.6.2633-2638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 20.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 21.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 22.Fetherston J D, Lillard J W, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 24.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 25.Gehring A M, Demoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague—modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 26.Gehring A M, Mori I, Perry R D, Walsh C T. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 27.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 28.Gordon, S. V., K. Eigelmeier, R. Brosch, T. Garnier, N. Honore, B. G. Barrell, and S. T. Cole. Genomics of Mycobacterium tuberculosis and Mycobacterium leprae, in press. Blackwell Science, Oxford, England.
- 28a.Greene, P. Unpublished data.
- 29.Guilvout I, Mercereau-puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 31.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 32.Halling C, Calendar R, Christie G E, Dale E C, Deho G, Finkel S, Flensburg J, Ghisotti D, Kahn M L, Lane K B, Lin C S, Lindquist B H, Pierson L S, Six E W, Sunshine M G, Ziermann R. DNA sequence of satellite bacteriophage P4. Nucleic Acids Res. 1990;18:1649. doi: 10.1093/nar/18.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–303. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 34.Hensel M, Nikolaus T, Egelseer C. Molecular and functional analysis indicates mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 36.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahim A, Goebel B M, Liesack W, Griffiths M, Stackebrandt E. The phylogeny of the genus Yersinia based on 16S rDNA sequences. FEMS Microbiol Lett. 1994;114:173–177. doi: 10.1111/j.1574-6968.1993.tb06569.x. [DOI] [PubMed] [Google Scholar]
- 38.Iteman I, Guiyoule A, de Almeida A M P, Guilvout I, Baranton G, Carniel E. Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect Immun. 1993;61:2717–2722. doi: 10.1128/iai.61.6.2717-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim U J, Birren B W, Slepak T, Mancino V, Boysen C, Kang H L, Simon M I, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 40.Kutyrev V V, Filippov A A, Oparina O S, Protsenko O A. Analysis of Yersinia pestis chromosomal determinants Pgm(+) and Pst(s) associated with virulence. Microb Pathog. 1992;12:177–186. doi: 10.1016/0882-4010(92)90051-o. [DOI] [PubMed] [Google Scholar]
- 41.Lillard J W, Jr, Fetherston J D, Pedersen L, Pendrak M L, Perry R D. Sequence and genetic analysis of the hemin storage (Hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 42.Lindler L E, Klempner M S, Straley S C. Yersinia pestis pH-6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Saha S, Haggard-Ljungquist E. Studies of bacteriophage P2 DNA replication. The DNA sequence of the cis-acting gene A and ori region and construction of a P2 mini-chromosome. J Mol Biol. 1993;231:361–374. doi: 10.1006/jmbi.1993.1288. [DOI] [PubMed] [Google Scholar]
- 44.Lähteenmäki K, Virkola R, Emödy S A, L, Korhonen T K. Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect Immun. 1998;66:5755–5762. doi: 10.1128/iai.66.12.5755-5762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe T M, Eddy S R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic DNA. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucier T S, Brubaker R R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurelli A T, Fernandez R E, Bloch C A, Rode C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Médigue, C., F. Rechenmann, A. Danchin, and A. Viari. Imagene: an integrated computer environment for sequence annotation and analysis. Bioinformatics, in press. [DOI] [PubMed]
- 49.Mhlanga-Mutangadura T, Morlin G, Smith A L, Eisenstark A, Golomb M. Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore R L, Brubaker R R. Hybridization of deoxyribonucleotide sequences of Yersinia enterocolitica and other selected members of Enterobacteriaceae. Int J Syst Bacteriol. 1975;25:336–339. [Google Scholar]
- 51.Nassif X, Beretti J L, Lowy J, Stenberg P, O’Gaora P, Pfeifer J, Normark S, So M. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc Natl Acad Sci USA. 1994;91:3769–3773. doi: 10.1073/pnas.91.9.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieto J M, Carmona M, Bolland S, Jubete Y, de la Cruz F, Juarez A. The hha gene modulates haemolysin expression in Escherichia coli. Mol Microbiol. 1991;5:1285–1293. doi: 10.1111/j.1365-2958.1991.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 53.Ninfa A J, Ninfa E G, Lupas A N, Stock A, Magasanik B, Stock J. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc Natl Acad Sci USA. 1988;85:5492–5496. doi: 10.1073/pnas.85.15.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Perry, R. Personal communication.
- 56.Perry R D, Fetherston J D. Yersinia pestis-etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+ response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poland J, Barnes A. Plague in CRC Press 1979. In: Steele J H, editor. CRC handbook series in zoonoses. Vol. 1. Boca Raton, Fla: CRC Press, Inc.; 1979. pp. 515–550. [Google Scholar]
- 59.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 60.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 62.Rudel T, Scheurerpflug I, Meyer T F. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 62a.Sanger Centre. [Online.] http://www.sanger.ac.uk/Projects/Y_pestis. [18 March 1999, last date accessed.]
- 63.Scarlato V, Arico B, Prugnola A, Rappuoli R. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 1991;10:3971–3975. doi: 10.1002/j.1460-2075.1991.tb04967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheng Y, Mancino V, Birren B. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 1995;23:1990–1996. doi: 10.1093/nar/23.11.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simonet M, Riot B, Fortineau N, Berche P. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skurnik M, Wolf-Watz H. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol Microbiol. 1989;3:517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 68.Stibitz S, Aaronson W, Monack D, Falkow S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989;338:266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- 69.Torosian S T, Zsigray R M. Abstracts of the General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. The ail locus of Yersinia pestis EV76-51F is disrupted by IS285 insertion, abstr. B-213; p. 191. [Google Scholar]
- 70.Une T, Brubaker R R. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woo S S, Jiang J, Gill B S, Paterson A H, Wing R A. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]