Abstract
In the direct competition for metabolic resources between cancer cells and tumor-infiltrating CD8+ T cells, the latter are bound to lose out. As such, these effector lymphocytes are rendered exhausted or dysfunctional. Emerging insights into the mechanisms of T cell unresponsiveness in the tumor microenvironment (TME) point towards epigenetic mechanisms as being crucial regulatory factors. In this review, we discuss the effects of characteristic components of the TME, i.e. glucose/amino acid dearth and high ROS concentration, on DNA methylation and histone modifications in CD8+ T cells. Subsequently, we take a closer look at the translational potential of epigenetic interventions that aim to improve current immunotherapeutic strategies, including the adoptive cell transfer of TCR- or CAR-engineered T cells.
The role of CD8+ T cells in tumor regression
CD8+ cytotoxic T lymphocytes (CTLs) are favored immune effector cells for targeting human cancers. Indeed, their infiltration in the tumor microenvironment (TME) of several tumor types (i.e. breast, colorectal and hepatocellular cancer) correlates with a positive prognosis [1]. Hence, two main immunotherapeutic strategies are founded on cancer cell killing by CTLs with unprecedented results in the treatment of certain malignancies: (i) immune checkpoint inhibitors, and (ii) adoptive cell transfer (ACT) [2,3]. Unfortunately, immune intervention does not yet offer a durable response in certain patients and in a variety of tumors. In the worst case, the tumor is completely refractory to treatment. The TME considerably influences therapeutic responses; this has been linked to the ability of the TME to compromise the performance and fate of CD8+ tumor-infiltrating lymphocytes (TILs) in favor of immunological tolerance [4]. While the major mechanisms leading to T cell exhaustion are still unclear, the harsh TME is now considered to be one of the important contributing factors. Therefore, knowledge on the hallmarks of truly protective CD8+ TILs (resistant to the immunosuppressive milieu), would represent a big leap forward in the field.
The metabolic alteration of the TME is a hallmark of cancer - a concept first introduced by Otto Warburg in the 1920’s [5]. Cancer cells redirect their metabolism to maintain an uninterrupted supply of energy and building blocks, safeguarding their own survival and growth. This occurs at the expense of the anti-tumor immune response, whereby CTLs need to survive with nutrient and oxygen dearth. These conditions profoundly shape immune cell fitness, localization, and phenotype [6]. Moreover, the combined occurrence of both hypoxia and glucose deprivation in the TME imposes non-compatible conditions on infiltrating T cells. Namely, whereas hypoxia reduces the use of oxidative phosphorylation (OXPHOS) and enhances glycolysis in CTLs, the reduced glucose availability in the TME cannot support this switch (Figure 1, Key Figure) [7]. Additionally, T cell dysfunction is endorsed by, among others, elevated concentrations of adenosine in the TME [8], macrophage-driven arginine depletion [9], glutamine scarceness [10], and tryptophan deprivation or kynurenine excess [11,12].
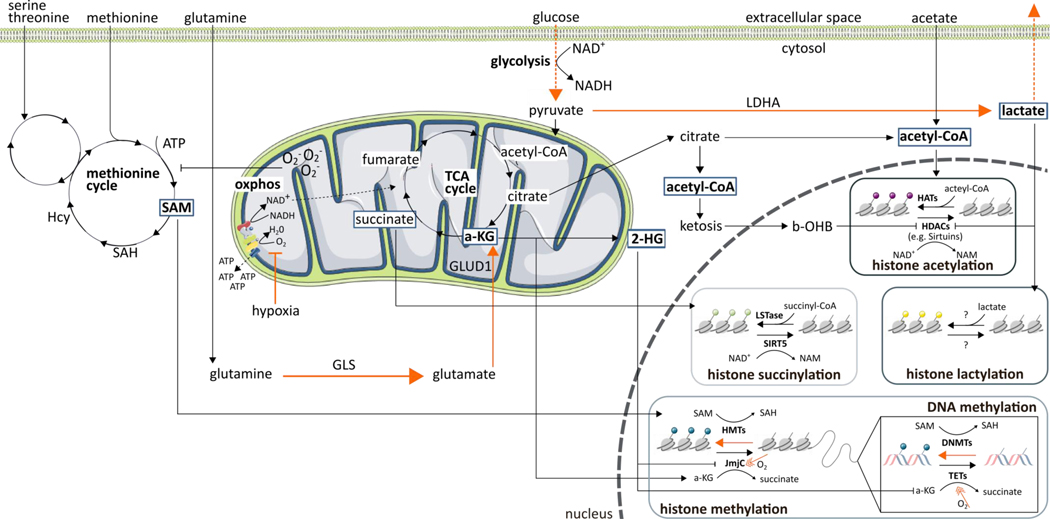
Figure 1. Relationship between immunometabolism, oxygen, and epigenetics.
Numerous metabolic intermediates as well as oxygen availability affect the cellular epigenome. The effects of hypoxia are indicated in orange.
Abbreviations: 2-HG, 2-hydroxyglutarate; a-KG, α-ketoglutarate; ATP, adenosine triphosphate;; b-OHB, β-hydroxybutyric acid; DNMT, DNA methyltransferases; GLS, glutaminase; GLUD1, glutamate dehydrogenase 1; HAT, histone acetyl transferase; Hcy, homocysteine; HDAC, histone deacetylases; HMT, histone methyltransferases; JmjC, Jumonji N/C terminal domains; LDHA, lactate dehydrogenase A; LSTase, lysine succyniltransferase; NAD+/NADH, nicotinamide adenine dinucleotide; NAM, nicotinamide; O2-, superoxide; oxphos, oxidative phosphorylation; SAH, S-adenosylhomocysteine; SAM, S-adenosyl-methionine; SIRT, sirtuin; TCA, tricarboxylic acid; TET, ten-eleven translocation methylcytosine dioxygenases
The field of immunometabolism is gaining momentum due to the recognition that metabolic remodeling triggers various aberrant immune responses. As such, the interaction between metabolic stress and immune dysfunction in cancer, and the potential to reprogram cell metabolism to bolster immune responses, has become one of the most exciting areas of translational research. In the last couple of years, accumulating evidence suggests that epigenetic remodeling is key in immunometabolic processes. Furthermore, the involvement of epigenetics (Box 1) in defining T cell functioning and fate has increasingly been recognized [13]. Hence, the question arises: can epigenetics be the enticing, missing link between the composition of the TME (embracing an array of metabolic features, cellular components, and cytokine surges), and CTL functioning, ultimately leading to overt clinical responses to immunotherapy? This review first highlights the strong link between the epigenetic machinery and the metabolic features of the TME. In a second part, the article focuses on past and future efforts to translate such findings into the rational reinforcement of current T cell-based immunotherapies.
Text Box 1 -. Basic epigenetic modifications at a glance.
As extensively outlined in multiple reviews [126–129], epigenetics refers to changes made “on top of” genetics; i.e. modifications made to DNA or chromatin that do not interfere with the DNA sequence itself, and include DNA methylation, the post-translational modification of histones (PTMs) and the effects of noncoding RNAs. This creates an opportunity for cells with identical underlying genomes to exhibit different phenotypes in response to environmental cues. It should, however, be noted that classifying noncoding RNAs under the heading of epigenetics is debated. Some researchers also advocate to define processes regulated by noncoding RNAs as non-genetic (rather than epigenetic) modifications.
DNA methylation
DNA methylation is a covalent modification catalyzed by a family of DNA methyltransferases (DNMTs); DNMT1, DNMT3a and DNMT3b. DNMTs add a methyl group to the 5-position of the cytosine ring, converting it to 5-methylcytosine (5mC). While the primary effect of DNA methylation is making the nucleosome relatively inaccessible (inhibiting transcription), the opposite effect can also be observed through binding of repressors, leading to gene activation. Active DNA demethylation is carried out by enzymes belonging to the ten-eleven translocation (TET) family; TET1, TET2 and TET3. They start with the conversion of 5mC to 5-hydroxymethylcytosine (5hmC).
Post-translational modification of histones
Acetylation, methylation, phosphorylation, and ubiquitination are well-known PTMs, but far from the only ones described. PTMs are catalyzed by histone-modifying enzymes, which add (writers), recognize (readers) or remove (erasers) PTMs. The resultant modification status of histones affects chromatin compaction and accessibility in two main ways: (i) neutralization of the charge of amino acids and (ii) recruitment of regulatory proteins. By way of illustration, histone acetylation defuses the positive charge of lysine residues, opening the chromatin structure. This is managed by histone acetyltransferases (HATs) and undone by histone deacetylases (HDACs).
Noncoding RNAs
The heterogeneous family of the noncoding RNAs consists of functional RNA molecules which are not further translated into proteins, including long noncoding RNA (lncRNA), piwi‐interacting RNAs (piRNAs), microRNA (miRNA), ribosomal RNA (rRNA), small interfering RNAs (siRNAs) and transfer RNA (tRNA). Since noncoding RNAs represent up to 60% of the transcriptional output in human cells, it may come as no surprise that they play an import role in regulating cellular processes, including chromatin remodeling, transcription and PTMs.
Epigenetic regulation of T cell exhaustion
As we move towards the point where we might instill T cells with desired, acquired characteristics that are suitable for cell-based therapies (i.e. capacity to infiltrate the tumor site, persistence), targeting their epigenetic programs offers a valuable perspective to improve their efficacy. In fact, the epigenetic signature of CD8+ T cells dictates their differentiation status [14,15]. Stemming from this, the question arises as to whether the exhausted state of T cells in the TME can be traced back to their epigenetic fingerprint (Box 2). The answer to this may open several new opportunities to therapeutically target exhaustion and functional impairment of T cells in the TME, and, conceivably, improve the response to immune checkpoint blockade. Even though immune checkpoint inhibitors are now able to temporarily restore T cell effector functions, the chromatin of treated cells may be left unchanged. However, experiments in murine models of lymphocytic choriomeningitis virus (LCMV) infection have shown that CD8+ T cells rapidly return to an exhausted state upon therapy withdrawal [16,17]. This suggests an irreversible installation of an exhaustion-specific genetic landscape, which can lead to defective tumor control. Moreover, it represents a likely mechanism of tumor resistance, which certainly merits further investigation.
Text Box 2 – Exhausted T cell differentiation states.
Exhausted T cells represent a range of subsets linked in a hierarchical developmental pathway [130]; (i) progenitor 1 exhausted T cells (Ly108+CD69+TCF1highTOXhigh) are quiescent and reside in lymphoid tissues, (ii) progenitor 2 exhausted T cells (Ly108+CD69-TCF1intTOXhigh) are highly proliferative and migrate into the circulation, (iii) intermediate exhausted T cells (Ly108-CD69-TCF1-TOXintT-bethigh) are mildly cytotoxic and are found in the circulation and blood-accessible organs, and (iv) terminally exhausted T cells (Ly108CD69+TCF1-TOXhighT-betlowEomeshigh), which are resident. These four phenotypes were found in exhausted CD8+ T cells of LCMV-clone-13-infected mice, in mouse B16 tumors and among TILs from human melanoma [130]. Hence, progenitor exhausted and terminally exhausted CD8+ TILs have distinct epigenetic states and functioning [131,132]. For example, only the former respond to PD-1 blockade and can be re-invigorated by the administration of checkpoint inhibitors. [132,133]. This knowledge has been a major advance in our understanding of T cell exhaustion; rather than being a homogenous CD8+ T cell phenotype, exhausted T cells are demarcated into different compartments in analogy with, for example, memory CD8+ T cell subsets. Therefore, an imminent question to be answered is how to enhance the rejuvenation of all these exhausted CD8+ T cells.
Although T cell receptor (TCR) signaling is a well-recognized contributor to CTL exhaustion, general insight into the mechanisms that reinforce exhausted T cells is lacking. Nevertheless, epigenetic remodeling is key. Various murine models of chronic infection or cancer have shown that TOX-driven epigenetic changes can turn memory precursor effector cells into progenitor exhausted CD8+ T cells [18,19]. The expression of TOX is in turn driven by chronic TCR stimulation and NFAT activation [20]. Furthermore, analysis of exhausted CD8+ T cell transcriptomes have revealed that the nuclear receptor transcription factor NR4a has an expression pattern similar to that of TOX [21]. Thus, NR4a has been identified as being crucial for the epigenetic and transcriptional program of CD8+ T cell exhaustion [21,22]. While this TCR-NFAT-TOX/NR4A axis contributes to guiding the CD8+ T cell exhaustion program and presents new avenues for the development of anticancer therapies [18,19], its relationship with the metabolic conditions of the TME remains unresolved. Since exhausted T cells exhibit metabolic insufficiency with suppressed mitochondrial respiration and glycolysis, poor metabolic fitness may well reinforce T cell exhaustion [23–26]. In line with this, two research groups recently reported that the soaring amounts of reactive oxygen species (ROS), generated under mitochondrial stress, can drive T cell terminal exhaustion [27,28]. As such, these data preliminarily suggest that the unique metabolic state of exhausted CD8+ T cells might not only be a consequence of its cellular differentiation status, but perhaps also be its cause.
Rewiring of the CD8+ T cell epigenome by the TME
Here, we discuss the major alterations in nutrient availability and utilization underlying certain differences in effector CD8+ T cells vs. exhausted CD8+ T cells, and address how these cells can rewire their epigenome and transcriptome under the metabolic cues of the TME. A solid understanding of such metabolic adaptation by CD8+ T cells may harbor important implications for achieving more effective tumor-targeting strategies.
Glucose
To fulfill the high bioenergetic and biosynthetic requirements of their effector functions, naïve CD8+ T cells can abruptly switch their metabolic program from oxidation of glucose, lipids, and amino acids, to robust consumption and metabolism of glucose and amino acids [29]. However, in the TME, the effector functions of CD8+ T cells are jeopardized due to glucose deprivation known as the “Warburg effect”[30]. Subsequently, a drop in the frequency of nucleo-cytosolic acetyl-CoA pools can be noticed. Indeed, intracellular acetyl-CoA concentrations are heavily dependent on the breakdown of glucose into pyruvate during glycolysis (Figure 1) [31]. This might contribute to explain the unique histone acetylation landscape that differentiates effector CD8+ T cells from exhausted T cells [32]. In line with this, acetate supplementation can promote histone acetylation and chromatin accessibility, thereby restoring IFN-γ production in T cells rendered hyporesponsive by glucose deprivation; this was observed in T cells isolated from both B16 melanoma tumors, and the blood of patients chronically infected with hepatitis C virus [31]. Of note, transitory glucose restriction was recently reported to enhance donor-derived CD8+ T cell tumor infiltration and function in a mouse B16 melanoma model [33]. This not only suggested sustained functional changes induced by nutrient availability, but also refined the general notion that glucose deprivation can harm T cell effector functions. Unfortunately, the authors did not explore the epigenetic aspects of their findings. In addition to histone acetylation, an elegant study shed some light on how lactate, as a byproduct of glycolysis, could be utilized in a new histone modification, histone lysine lactylation, and re-shape the epigenome of murine macrophages and human cancer cells [34]. The latter finding opened various questions on the broader roles of this modification. Indeed, elevated lactate concentrations in the TME can promote immunosuppression [35], and tumor-associated macrophages isolated from B16 melanoma and Lewis lung carcinoma tumors demonstrate a positive correlation between an anti-inflammatory phenotype and histone lactylation [34]. Thus, how are T cells affected by increased lactate amounts in the TME? Is there a connection between histone lysine lactylation in CD8+ TILs and an exhausted phenotype?
Apart from controlling the availability of substrates for epigenetic modifications, glucose metabolism strictly regulates the activity of epigenetic enzymes. For instance, in one study, under conditions of glucose restriction, the expression of methyltransferase EZH2 was restricted by microRNA (miRNA)26a and miRNA101 in mouse CTLs [36]. This led not only to reduced cytokine expression and diminished cytotoxicity [36], but also to general metabolic insufficiency and CTL exhaustion, relative to controls [37]. This influence of glucose availability on CD8+ T cell epigenetics and function is clinically relevant. High infiltration of EZH2+ CD8+ T cells in ovarian cancer patients correlates with a particularly long term survival [36]. Similarly, with nutrient deprivation, methyltransferase G9a can dissociate from the LC3B, WIPI1, and DOR promoters, whose products are required for the formation of autophagosomes [38,39] -- essential cellular structures for replenishing scarce nutrients. Collectively, these studies directly or indirectly highlight the importance of an epigenetic approach to overcome T cell dysfunction in a low-in-glucose TME (Box 3).
Box 3. Indirectly targeting epigenetic signatures by rewiring cellular metabolism.
An underexploited research avenue is the potential of metabolic interventions to reprogram the epigenome of exhausted T cells. As clearly emerges from the data discussed in this review, the T cell epigenome is heavily influenced by nutrient availability and the overall condition of the TME. This goes hand in hand with CD8+ T cell fitness. Also, the specific metabolic environment of the tumor may underlie the differentiation towards an exhausted phenotype in TILs. Hence, breaking the immune suppressive barrier of the TME by reprogramming the intrinsic metabolism of tumor-reactive T cells and preventing a pro-tumor epigenetic landscape to be established, is an emerging and promising area of cancer immunotherapy. Although still in its initial phase, in vivo epigenetic remodeling through a CRISPR-associated Cas9 system could be interesting in this context [134–136]. However, different questions will need to be answered first: which metabolic alterations in cancer cells/CD8+ T cells are able to sustain T cell fitness in the immunosuppressive TME? Which delivery method can be used in vivo? What can be said about tissue distribution and off-target effects? How do we prevent the unintended modification of other cells and tissues, potentially leading to unwanted side-effects?
Free fatty acids and cholesterol
Exogeneous free fatty acid uptake, lipid metabolism, and the concentration of lipids in the T cell plasma membrane can affect T cell functioning [40,41]. Even if glucose-derived acetyl-CoA is an important source of substrates for histone acetylation, lipid-derived acetyl-CoA obtained through β-oxidation has been suggested to be equally important [42]. However, this finding contradicts earlier claims [43], emphasizing the necessity to investigate the context-dependent role of free fatty acid oxidation in epigenetic regulation.
Cholesterol is of vital importance as it specifically affects TCR clustering and the formation of an immunological synapse; pharmacologically inhibiting cholesterol esterification has resulted in an increased plasma membrane cholesterol concentration, boost in CTL function, and tumor control in mouse models of melanoma [44]. One of the main players preserving cholesterol metabolism equilibrium in human primary CD8+ T cells is the epigenetic regulator RORα: indeed, the RORα/ histone deacetylase (HDAC) complex acts as a transcriptional repressor of ACAT1/2 and ABCA1 [45]. Moreover, since the two-carbon acetate group of acetyl-CoA is used for cholesterol synthesis, alterations in cholesterol metabolism influence cellular acetyl-CoA pools in vitro [42,46], and thus, may affect the epigenetic histone acetylation landscape of CD8+ TILs. Nevertheless, the general scarcity of reports on cholesterol metabolism and epigenetics in cancer does not allow to make general statements to date. Hence, unraveling the epigenetic machinery of TILs might offer an answer to how cholesterol metabolism might support cancer cells while impairing immune cell functions.
Amino acids
Glutamine is a major fuel required for maintaining the tricarboxylic acid (TCA) cycle and a key source for lipid synthesis in both cancer cells and CTLs [47]. Consequently, the disrupted glutamine metabolism in CD8+ TILs will result in an imbalance of metabolic intermediates from the TCA cycle, such as α-ketoglutarate (α-KG). This in turn, influences the epigenome of CD8+ T cells (Figure 1) [47]. For instance, the presence of α-KG is crucial for the activity of histone and DNA demethylation enzymes such as Jumonji N/C terminal domains (JmjCs) and ten-eleven translocation (TETs) enzymes. Additionally, α-KG is oxidized within the TCA cycle and converted into succinyl-CoA. Then, succinyl-CoA can provide the necessary substrate for the histone modification known as histone succinylation [48,49], while also inhibiting TET- and JmJC-mediated demethylations [50]. This whole process has been deemed necessary for the maintenance of anti-tumor effector T cell functions[50]. Of note, JHU083 (developed in [10])-- a prodrug of the glutamine antagonist 6-diazo-5-oxo-L-norleucine-- reduced tumor growth and improved survival when injected subcutaneously in murine MC38 and CT26 colon cancer, EL-4 lymphoma, and B16 melanoma mouse models. It also demonstrated efficacy in boosting CD8+ T cells by enhancing their acetate metabolism to fuel the dysregulated TCA cycle [10], which might be interesting for future testing.
CD8+ T cells predominantly uptake exogenous methionine to maintain their cellular S-adenosyl-methionine (SAM) pools [51], the universal methyl donor for DNA, RNA and protein methyltransferases (Figure 1). As such, T cell activation seems to go hand in hand with methionine uptake and the generation of SAM, sustaining histone and RNA methylation [52,53]. However, due to the upregulation of methionine transporter SLC43A2 on both human and murine cancer cells, CD8+ TILs are known to undergo methionine deprivation, resulting in a decrease of histone methylation and cytokine production, relative to controls [52]. This illustrates again how activated T cells may share many metabolic similarities with cancer cells, and how the competition for metabolites deriving thereof can impair the anti-tumor effector functions of CD8+ TILs through epigenetic mechanisms. We should, however, be careful with ‘wearing CD8+ T cell blinders’. Although supplementation seems like a plausible solution, mice placed on a methionine-restricted diet have demonstrated slow tumor growth in two patient-derived xenograft mouse models of colorectal cancer with RAS mutation [54].
Potassium
Potassium is released from necrotic cancer cells into the TME, causing profound suppression of CD8+ TIL functioning [55]. Indeed, the concentrations of K+ in the interstitial fluid of mouse and human tumor tissues can be 5–10 times higher than in (normal) serum [55]. This results in an upsurge of intracellular K+ in CD8+ T cells, eventually blocking TCR-mediated activation of the Akt/mTOR signaling pathway, in a PP2A phosphatase-dependent manner (based on genetic disruption of PP2A via overexpression of a dominant-negative isoform (PP2A_DN) or via short-hairpin-mediated RNA interference of the PP2A subunit Ppp2r2d) [55]. Furthermore, altered transmembrane potassium concentration and membrane potential has led to functional caloric restriction in mouse CD8+ T cells, characterized by, among others, enhanced mitochondrial metabolism [56]. Consequently, nucleo-cytosolic acetyl-CoA concentrations were reduced, favoring its use in mitochondria for oxidative phosphorylation. The latter impacted histone acetylation at effector- and exhaustion- relevant loci, i.e. reduced H3K9 acetylation of Ifng, Pdcd1, Cd244, Havrc2, and Klrg1, (suppressing effector CTL programs). At the same time, the potassium-mediated starvation response also restricted the availability of methionine intermediates, curtailing methylation of histone marks that typically quell stemness-associated programs [56]. Of note, apart from the availability of metabolites, other components of the TME, including ion concentrations, are able to alter the metabolism of CD8+ TILs. Hence, new research projects should consider all the so-far unexplored components of the tumor interstitial fluid.
Hypoxia
The TME is characterized by areas of oxygen scarcity, directly impacting the activity of enzymes and substrates involved in epigenetic regulation. Hence, as hypoxia reduces the use of OXPHOS and enhances glycolysis in CTLs, a broad impact on the epigenetic level can be expected (Figure 1) [7]. For instance, low oxygen concentrations promote glutamine import, glutaminolysis, and the synthesis of α-KG [57]. However, tumor hypoxia subsequently induces the conversion of α-KG into 2-hydroxyglutarate (2-HG)-- a competitive inhibitor of α-KG-dependent enzymes, resulting in increased methylation of DNA and histone repressive marks, relative to controls [58]. In CD8+ T cells specifically, 2-HG has been shown to accumulate through HIF-1α-mediated LDHA expression inside the cells; and, the excess supply of 2-HG has led to altered DNA methylation patterns, particularly due to the inhibition of H3K27me2/3 demethylase KDM6A and TET enzymes [59]. Moreover, hypoxia can induce histone and DNA methylation in a HIF- and 2-HG-independent manner; indeed, certain histone demethylases, such as KDM5A and KDM6A, are oxygen sensitive [60,61]. Similarly, under pathophysiological oxygen concentrations found in tumors, TET enzyme activity has been reported to be reduced, relative to controls [62]. However, given that most work on the influence of hypoxia on immune cells has so far been conducted using in vitro systems, more work is needed to determine how these data can be translated to the in vivo setting in the context of the TME.
Oxidative stress
On the one hand, the TME is characterized by high concentrations of ROS that cause direct cellular damage, but also serve as signaling molecules [63]. The effect of ROS on epigenetic T cell regulation has recently been studied ex vivo and in LCMV infection in vivo mouse models [64]. Phosphatase of activated cells 1 (PAC1) was selectively upregulated in exhausted CD8+ TILs upon ROS exposure [64]. It accumulated on chromatin and recruited a HDAC1–HDAC2 complex, eliciting chromatin-closing enzymatic activity [64]. This is of translational importance, considering that multiple studies using PAC1 knockout mice have demonstrated attenuated cancer progression [65,66]; indeed, PAC1 expression is aligned with the expression of various inhibitory receptors [64], and its expression has also been associated with poor prognosis for certain cancer patients (e.g. colon cancer and ovarian carcinoma) [64,67]. Nitric oxide (NO) can also be present in the TME, produced, among others, by cancer cells themselves, as well as by NO synthase-positive tumor-infiltrating myeloid cells [68]. In a EG7 murine lymphoma model, it was reported that this NO might be essential for the anti-tumor activity of CTLs [69]. Possibly, this might be the result of an epigenetic modification, given that NO is capable of directly inhibiting the catalytic activity of the histone demethylase KDM3A [70]; however, this remains to be directly tested.
On the other hand, ROS are also generated as a byproduct of numerous enzymatic reactions in T cells themselves, playing an important role in cellular physiology [71]. This homeostasis can be disrupted in different malignancies. For example, glucose, glutamine, or pyruvate starvation, induce superoxide production. Since pyruvate functions as an antioxidant [72], a low concentration of the latter might contribute to enhanced ROS amounts in CTLs, warranting further investigation. In line with this, CD8+ TILs in renal cell carcinoma display reduced glucose uptake concomitant with hyperpolarized, fragmented mitochondria producing large amounts of ROS [25]. Enhanced concentrations of mitochondrial superoxide result in a decrease in total DNA methylation [71]. This might be caused by a disrupted methionine cycle, in view of its tight regulation with SAM’s metabolism; however, this remains hypothetical (Figure 1) [71]. Consequently, due to disrupted metabolic homeostasis, CD8+ T cell activation is flawed and CTLs are unable to appropriately perform their anti-tumor functions [25]. Of note, recently, one study demonstrated that antioxidants could reactivate gene expression of loci known to be inaccessible in exhausted T cells [27].
Finally, the reduction of nicotinamide adenine dinucleotide (NAD)+ to NADH is a limiting reaction for the glycolysis pathway-- a factor highly demanded in many other redox reactions, i.e. TCA cycle and fatty acid oxidation. Sirtuins (SIRT1-7) are NAD+-dependent class-III HDACs. In line with this, cytotoxic CD8+ T cells with enhanced glycolytic capacity exhibit decreased expression of SIRT1, promoting both glycolysis and secretion of granzyme B [73].
Oncometabolites
An important oncometabolite is 2-Hydroxyglutarate (2-HG). In cancer cells, mutations in the isocitrate dehydrogenase (IDH) enzyme result in the loss of its ability to convert isocitrate to α-KG. Instead, IDH acquires the ability to catalyze the NADPH-dependent reduction of α-KG to 2-HG [74,75]. Tumor cell-derived 2-HG is taken up by activated CD4+ and CD8+ T cells, where it interferes with α-KG-dependent demethylases (Figure 1) [76]. The latter can impair T cell tumor infiltration, suppress early TCR signaling events, and hamper T cell anti-tumor immunity (e.g. mouse and human gliomas) [77,78]. These data add to our understanding of how tumors can progress despite the infiltration of T cells that harbor the ability to destroy it.
Therapeutically tackling epigenetics in onco-immunology
During the last couple of years, epigenetic therapy has emerged as a promising strategy to combat malignancy, either on its own, or in combination with other treatments [81]. Hence, epigenetic modifiers could be used to harness the adaptive (antigen)-specific immune response against certain cancers. Two main strategies can be envisioned (i) targeting (tumor-infiltrating) CD8+ T cells in vivo (Table 1) and (ii) endorsing the (ex vivo) generation of superior anti-tumor T cell grafts for ACT.
Table 1.
The use of epigenetic drugs in the clinic (current clinical trials)a,b
| Class | Drug | Tumor type | Clinical phase | Rationale (CD8 T cells) | Setting | Clinical trial identifier |
|---|---|---|---|---|---|---|
| DNMT inhibitor | Guadecitabine | Non-small-cell lung cancer | II | DNMT inhibitors induce the expression of MHC class I molecules, increase the presentation level of tumor-associated antigens and the expression of chemokines that recruit CD8 T cells, promote Th1 polarization and CTL cytolytic activity [88,135]. | + carboplatin | NCT03913455 |
| refractory germ cell tumor | I | + cisplatin | NCT02429466 | |||
| Acute myeloid leukemia | II | + cladribine + idarubicin | NCT02096055 | |||
| Leukemia | II | After DLI | NCT02684162 | |||
| Leukemia | II | After HSCT | NCT03454984 | |||
| Leukemia | III | - | NCT02907359 | |||
| Leukemia rollover study | II | - | NCT03603964 | |||
| Central chondrosarcoma | II | + belinostat | NCT04340843 | |||
| Metastatic colorectal cancer | I | Aim of the combination: recruitment of CD45RO+ T cells into the tumor | + allogeneic colon cancer cell vaccine (GVAX) + yclophosphamide |
NCT01966289 | ||
| Leukemia | II | DNMT inhibitor + anti-CTLA4 [88], DNMT inhibitor+ anti-PD(L)1 [89–91] | + atezolizumab | NCT02935361 | ||
| Urothelial carcinoma | II | + atezolizumab | NCT03179943 | |||
| Kidney cancer | II | + durvalumab | NCT03308396 | |||
| Advanced Liver, pancreatic, bile duct or gallbladder cancer | I | + durvalumab | NCT03257761 | |||
| Lung cancer | I | + pembrolizumab + mocetinostat | NCT03220477 | |||
| Non-small-cell lung cancer Prostate cancer |
I | + pembrolizumab | NCT02998567 | |||
| Fallopian tube, ovarian or primary peritoneal carcinoma | II | + pembrolizumab | NCT02901899 | |||
| Lung cancer, melanoma | II | + ipilimumab + nivolumab | NCT04250246 | |||
| Metastatic colorectal cancer | II | + nivolumab | NCT03576963 | |||
| EZH2 inhibitor | CPI-1205 | B-cell lymphoma | I | EZH2 HMT inhibitors may reverse the suppression of CXCL9 and CXCL10 expression and subsequently improve effector T cell infiltration in the TME [136,137]. EZH2 is activated or overexpressed by several tumors, resulting in the silencing of cancer suppressor genes and diminished antigen presentation [138–140]. |
- | NCT02395601 |
| Metastatic castration resistant prostate Cancer | I | + enzalutamide or abiraterone/prednisone | NCT03480646 | |||
| Advanced solid tumors | II | + ipilimumab | NCT03525795 | |||
| Tazemetostat | B-cell non-hodgkin’s lymphoma | II | - | NCT03456726 | ||
| follicular lymphoma | III | - | NCT04224493 | |||
| Mesothelioma | II | - | NCT02860286 | |||
| Metastatic prostate cancer | I | + enzalutamide or biraterone/prednisone | NCT04179864 | |||
| Advanced sarcoma | III | + doxorubicin HCl | NCT04204941 | |||
| INI1-negative tumors or relapsed / refractory synovial sarcoma | II | NCT02601950 NCT02601937 | ||||
| Advanced solid tumors | II | - | NCT03213665 | |||
| Cancer rollover study | II | - | NCT02875548 | |||
| Lymphoma | I | EZH2 serves as a molecular switch controlling tumor immune escape. The combination with checkpoint blockade may result in synergistic effects [95,141] |
+ atezolimumab | NCT02220842 | ||
| Non-small-cell lung cancer | II | + atezolimumab | NCT03337698 | |||
| Urothelial carcinoma, metastatic bladder urothelial carcinoma | II | + pembrolizumab | NCT03854474 | |||
| SHR2554 | lymphoid neoplasms | I | - | NCT03603951 | ||
| luminal advanced breast cancer | II | + other treatments | NCT04355858 | |||
| solid tumors or B-cell lymphomas | II | The combination of EZH2 inhibitors and PD-L1/TGF-β blockade may enhance the efficiency of immunotherapy | + anti-PD-L1/TGF-β antibody SHR1701 | NCT04407741 | ||
| BET inhibitor | BMS-986158 | Pediatric cancer | I | BET inhibitors increase the presence of active cytotoxic CD8+ T cells in the TME [142,143]. | - | NCT03936465 |
| Advanced tumors | II | + nivolumab | NCT02419417 | |||
| CC-95775 | Non-Hodgkin lymphoma | I | - | NCT04089527 | ||
| CPI-061 | Leukemia | II | Ruxolitinib | NCT02158858 | ||
| Molibresib | Advanced and refractory solid tumors and lymphomas | I | + Entinostat | NCT03925428 | ||
| MK-8628 | Leukemia | I | - | NCT02698189 | ||
| ZEN-3694 | Metastatic castrationresistant prostate cancer | II | + enzalutamide, embrolizumab |
NCT04471974
NCT02711956 |
||
| HDACI inhibitor | Entinostat | hormone receptor-positive breast cancer | III | HDAC inhibitors have been shown to induce cancer cell apoptosis and to inhibit angiogenesis. HDAC3: negative regulator of CD8+ T cell cytotoxicity program. HDAC3 inhibition leads to the enhanced expression of granzyme B and the improved eradication of B16 melanoma target cells [144]. Note. HDAC inhibitors can synergize with checkpoint inhibitors [92–94] |
+ other treatments |
NCT02115282
NCT03538171 |
| HDACIII inhibitor | Nicotinamide | Non-small-cell lung cancer | III | + gefitinib or erlotinib | NCT02416739 | |
| HDACi | Abexinostat | Renal cell carcinoma | III | + pazopanib | NCT03592472 | |
| Chidamide | Leukemia | III | + other treatments |
NCT03564704
NCT03553238
NCT04231448 NCT04038411 NCT04040491 |
||
| Panobinostat | leukemia | III | + donor lymphocyte infusions | NCT04326764 | ||
| Myelofibrosis rollover study | III | + ruxolitinib | NCT02386800 | |||
| Vorinostat | Cutaneous T-cell lymphoma | III | or mogamulizumab | NCT01728805 | ||
| High grade glioma | III | Bevacizumab, temozolomide | NCT01236560 | |||
| Valproic acid | High grade gliomas | III | + temozolomide (or temozolomide + chloroquine) | NCT03243461 |
Note 1. The DNMT inhibitors azacytidine and decitabine are the most successful epigenetic drugs to date. Due to the overabundance of ongoing clinical trials with these drugs, only the novel hypomethylating agent guadecitabine is mentioned in this table.
Note 2. Considering the plethora of clinical trials ongoing with HDAD inhibitors, only the phase III clinical trials are mentioned
Improving in vivo CD8+ T cell responses
An important goal of cancer immunotherapy is to avert the exhausted phenotype of CD8+ TILs while promoting an effector state. Since a de novo DNA-methylation program controls the formation of fully exhausted T cells, the use of DNA methyltransferases (DNMT)-inhibiting cytosine nucleoside analogs, such as decitabine, could block the establishment of an exhaustion-associated epigenetic signature [16]. In a phase I/II clinical trial enrolling patients (n=100) with solid tumors or B cell lymphoma, treatment with decitabine promoted Th1 polarization and CTL cytolytic activity (Figure 2) (NCT01799083)II [82]. This study supports the feasibility of using epigenetic modulators as candidate anti-tumor therapeutics targeting CD8+ T cells in the TME.
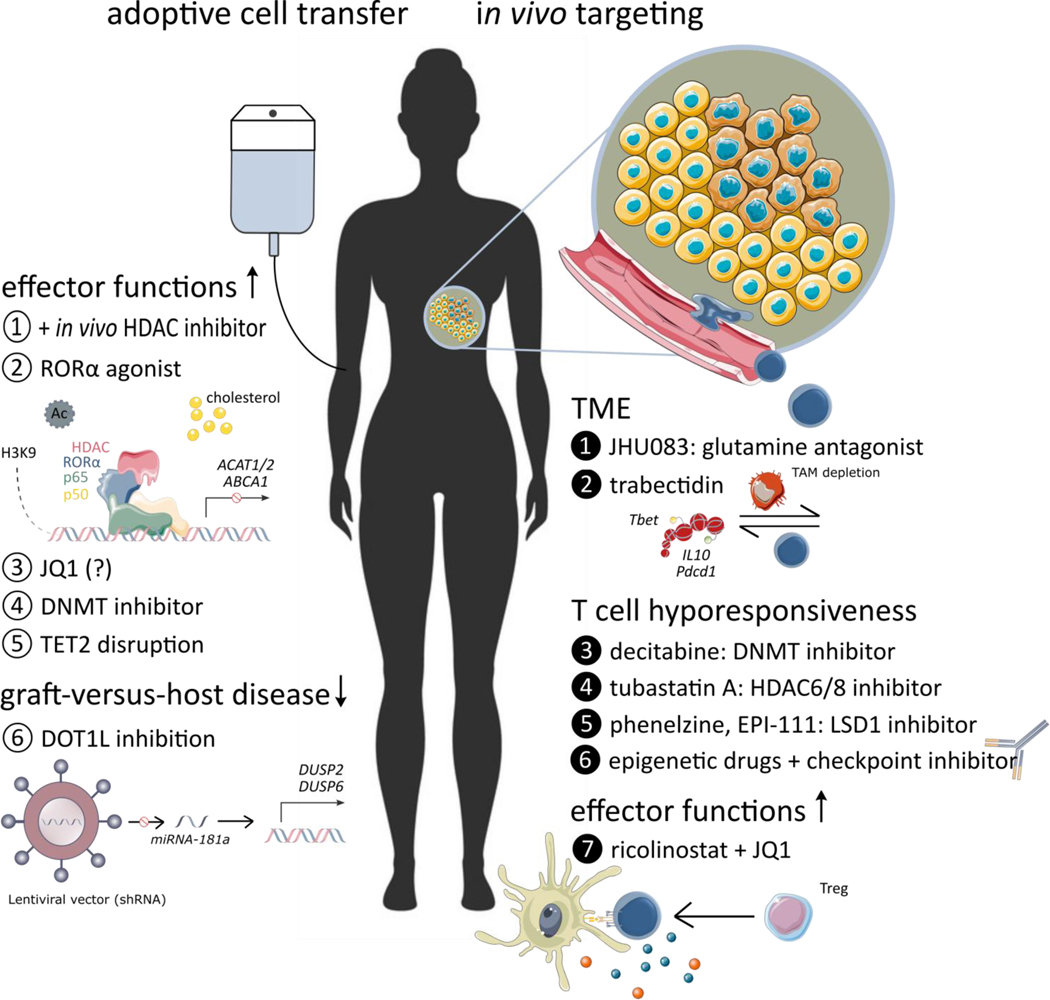
Figure 2. Explored therapeutic interventions based on epigenetic modifications.
The diagram depicts the generation of superior anti-tumor CD8+ T cell grafts for adoptive cell transfer (ACT) (left), and targeting (tumor-infiltrating) CD8+ T cells in vivo (right). The remaining challenges with ACT, both at the level of efficacy and at the level of toxicity, might potentially be solved with epigenetic drugs (epidrugs). In vivo, epidrugs might also be at the center of reinvigorating the adaptive immune response, overcoming tumor immune escape, and resistance to therapy.
Note. Data on the effects of JQ1 are not unequivocal, (designated by ‘?’)
Abbreviations: DNMT, DNA methyltransferases; HDAC, histone deacetylase; miRNA, microRNA; TET, ten-eleven translocation
Alternatively, the reversal of the exhaustion status of CD8+ TILs can be envisioned. By focusing on DNA and histone methylation, targeting the phosphorylated LSD1 pathway might be promising, since it is enriched in exhausted T cells. LSD1 forms nuclear complexes with Eomesodermin (EOMES) in dysfunctional CD8+ T cells of immunotherapy-resistant melanoma and breast cancer patients [83]. It demethylates and acetylates key lysine residues within the nuclear localization sequence motif or DNA-binding motif of EOMES to restrict nuclear translocation (Figure 2) [83]. This finding is of particular interest, considering it is, to our knowledge, the first report to tackle the so far considered “immunotherapy-resistant” terminally exhausted T cell population (Box 2), and harness this population back into the anti-tumor immune response [16,17].
Regarding histone acetylation, a first exciting target is the serine/arginine-rich splicing factor 2 (SRSF2)-- a regulator of the expression of multiple immune checkpoint molecules. SRSF2 controls their transcription by modifying the H3K27ac status of the relevant gene promoters in human renal cell carcinoma CD8+ T cells [84]. A second strategy includes reinvigorating Bhlhe40-deficient CD8+ T cells [85]. Bhlhe40 is a stress-responsive transcription factor, essential for maintaining effector gene acetylation via its support of TCA-cycle activity and OXPHOS. Bhlhe40 expression in CD8+ TILs was suppressed by local PD-1 signaling in a B16 mouse melanoma model, hampering CD8+ T cell fitness and polyfunctionality [85]. Upon administration of Tubastatin A, a HDAC6/8 inhibitor, or the supplementation of acetate, effector functions and persistence of CD8+ TILs could be enhanced (Figure 2) [85].
Finally, some noncoding RNAs have been implicated in modulating the exhaustion state of CD8+ T cells [86]. For instance, miRNA-155 can restrict CD8+ T cell functional exhaustion by promoting the expression of Phf19 via phosphorylated AKT. This leads, in turn, to enhanced PRC2 functionality, and ultimately restrains T cell senescence by inhibiting key transcription factors driving terminal differentiation and exhaustion [87]. This suggests that targeting these miRNAs might be considered as a putative therapeutic strategy for certain cancers, for example, by investigating miRNA mimetics or miRNA antagonists. Collectively, with a better understanding of the epigenetic states and the molecular pathways that drive CD8+ T cell exhaustion in tumors, epigenetic drugs could have important roles by synergizing with other anti-cancer therapies or in reversing acquired tumor resistance.
Combination therapy
Synergistic effects of epigenetic drugs and established immunotherapeutics are foreseeable. To further improve the success rate of immune checkpoint blockade, different clinical trials are currently ongoing combining epigenetic modifiers and checkpoint inhibitors. This has been the result of encouraging data observed in murine tumor models where several combinations have been tested: bromodomain and extraterminal domain (BET) inhibitor + anti-PD-1 monoclonal antibodies (mAbs) (Ab) [88,89], DNMT inhibitor + anti-CTLA-4 mAbs [90], DNMT inhibitor+ anti-PD-(L)1 mAbs [16,91–93], HDAC inhibitor + antiPD-1 mAbs [94–96] and EZH2 inhibitor + anti-CTLA-4 mAbs [97]. Agonistic mAbs targeting costimulatory molecules constitute an alternative to checkpoint blockade [98]. Urelumab, a fully human IgG4k mAb agonist of CD137/4–1BB, regulates DNA methylation, poising CD8+ T cells to respond more robustly upon antigen rechallenge [99].
Epigenetic drugs have also been studied in combination with adoptive T cell therapy, improving the activity and expansion of adoptively transferred CD8+ T cells. In a B16 melanoma model, the addition of HDAC inhibitor dacinostat to T cell vaccination therapy promoted its therapeutic action through (i) effects on target cancer cells, (ii) a decrease in competing endogenous lymphocytes and (iii) an upgrade of adoptively transferred lymphocyte functional activity [100]. Similar results were obtained with panobinostat (Figure 2) [101].
A more indirect approach is also possible. For example, the combination of two epigenetic modifiers; ricolinostat, a HDAC6 inhibitor, and JQ1, a specific inhibitor of BET proteins, shows potential for the treatment of non-small cell lung cancer. This combination promotes T cell-mediated anti-tumor immunity and, as such, immune-mediated tumor growth arrest. Ricolinostat improves the functioning of antigen-presenting cells, leading to an enhanced activation of CD8+ TILs and secretion of effector cytokine IFN-γ. At the same time, the suppressive functions of regulatory T cells are reduced by JQ1, increasing the CD8+ T cells / regulatory T cell ratio [102]. In line with this, the combination of HDAC6 and BET inhibition shows potential in a multiple myeloma model. It efficiently reduces c-MYC expression and counteracts the BET inhibition-mediated HDAC6 upregulation, associated with cancer progression and drug resistance [103].
Currently, although the rationale and data seem promising, it is still too soon to make a ruling on the translatability of these findings to humans and, as such, on their clinical significance. Considering the vast number of ongoing clinical trials with epigenetic drugs, in combination with other (immuno)therapeutics or alone, the upcoming years are expected to inform on the subject. Here, special attention will be required in selecting patients who might gain the greatest benefit. Might there be a role for epigenetic biomarkers? Further research should address this possibility.
Adoptive cell transfer improvement
One of the current challenges of ACT is the improvement of persistence and durability response. The in vitro expansion of CD8+ T cells to attain the large numbers required for vaccination, inevitably goes hand in hand with T cell differentiation and loss of proliferative potential [104,105]. To obtain the essential proportion of stem cell memory T cells and central memory T cells in a graft, epigenetic modifiers can be employed. Bromodomain inhibitor JQ1 has been shown to maintain these T cell subsets with desired proliferative capacity. In an acute lymphoblastic leukemia (ALL) xenograft mouse model with Nalm-6 pre-BALL human cells, JQ1-treated T cells showed greater cell persistence and superior anti-tumor effects [105]. Data should, however, be treated carefully, since they contradict a more recent study, demonstrating reduced efficacy in a murine B16 melanoma model [106]. Treatment duration and inhibitor concentration could underlie this discrepancy. Timing, choice, and concentration of the epigenetic modifier may prove to be decisive. An alternate approach to reduce T cell exhaustion, improve effector functions and persistence, is the use of DNMT inhibitors during CD8+ T cell expansion [16,107]. The addition of decitabine during chimeric antigen receptor (CAR) T manufacturing has improved the tumor-homing ability and anti-tumor potential of the graft, in both Nalm-6 ALL and Raji non-Hodgkin’s lymphoma mouse models [108]. One case described a chronic lymphocytic leukemia (CLL) patient who achieved a complete response, with no evidence of CLL in his bone marrow, due to the expansion of one T cell clone with a CAR lentiviral integration site in the TET2 gene [109]. As such, the disruption of the TET2 gene and the resulting changes in the epigenetic landscape gave rise to a T cell clone with a significant long-life span, underpinning clinical efficacy of CAR T cell therapy in the concerning patient [109]. Further research on the effects of knocking out the TET2 gene and the effects on CAR T cell survival is therefore needed.
In view of the earlier discussed approaches involving DNMT and TET2 inhibition, we should briefly touch upon the current provocative nature of the topic. Loss-of-function mutations in DNMT and TET occur frequently in patients with, among others, ALL and acute myeloid leukemia, and are associated with poor prognosis [110,111]. Recent insights coming from clonal hematopoiesis studies suggest that null mutations in these genes allow for preservation of a stem-like state in hematopoietic cells [110,112–115]. Hence, although transformation to malignancy requires additional mutations, and as such, the DNMT and TET mutations do not drive malignancy per se, therapeutic approaches that center on modifying the enzymatic activity of these epigenetic modifiers should be carefully monitored for long-term effects.
The generation of a universal off-the-shelf CAR T cell product is another hurdle to be tackled to enhance the feasibility and diffusion of this treatment. To attenuate the development of graft-versus-host disease, epigenetic modification could yet again be a solution. DOT1L, a H3 lysine-79 specific methyltransferase, plays different roles in cancer as well as T cell differentiation in both mice and humans [116–118]. DOT1L has been shown to repress allogeneic T cell responses, while retaining potent anti-tumor activity [119]. Mechanistically, the inhibition of DOT1L reduces miRNA-181a expression, which is followed by an upregulation of the genes encoding phosphatases DUSP2 and DUSP6. In turn, DUSP6-mediated ERK dephosphorylation selectively ameliorates low-avidity T cell responses through the modulation of TCR sensitivity (Figure 2) [119]. Of note, HDAC11 associates with the Eomes and Tbet gene promoter regions in resting cells, inhibiting CD8+ T cell effector functions, and disassociates upon activation. Hence, T cells from HDAC11 knockout mice are hyperresponsive, mediating sturdy anti-tumor activity as well as more forceful graft-versus-host disease [120]. Including the data above, targeting T cell epigenetics holds great potential for cancer therapies. Still, caution is advised concerning acute or long-term toxicity. To cope with (un)known toxicities, in case of the ex vivo treatment of T cells with epigenetic drugs, a safety switch could be included in, for example, the CAR design; namely, CAR constructs that also express a suicide gene [121–123] or co-express a cell-surface elimination marker [124,125]. Both methods can result in the irreversible depletion of administered cells, and, as such, cope with their toxicity.
Concluding remarks
A compelling body of evidence endorses the potential of immunotherapy in oncology; however, ample, and robust cross-disciplinary work remains to be done to maximize its clinical efficacy for different malignancies. We find interesting that epigenetic mechanisms affect all aspects of the cancer-immunity cycle. As such, this review highlights the concept that epigenetic therapies might represent novel immunotherapies themselves, harnessing once again, the adaptive CD8+ T cell response in the fight against malignancy. Therefore, delving into CD8+ T cell epigenetics and its regulating role in T cell biology and function, can ideally result in the discovery of targeted pharmacological or genetic interventions, supporting current treatment strategies against cancer (Outstanding Questions Box). An important challenge will be to distinguish between the dysregulation of driver genes and those which result from changes in driver genes. Hence, further research should strive to elucidate the exact roles of epigenetic modifications in CD8+ T cell differentiation, effector functions, and exhaustion. In the context of cancer, we should try to address whether the epigenetic profiles of T cells influence T cell trafficking and homing into tumors, and once arrived in the TME, how the immunosuppressive imprinting might be reversed. Finally, since it has become clear that cancer treatments should progress from one-size-fits-all to personalized therapy, epigenetic biomarkers and epigenetic signatures could especially be of interest.
Outstanding Questions Box.
What are the epigenetic hallmarks of the truly protective CD8+ T cells in cancer patients? Namely, which epigenetic modifications render cytotoxic T cells resistant to the immunosuppressive milieu of the tumor micro-environment (TME)?
How do the intracellular concentrations and fluxes of different metabolites impact the epigenetic signature of CD8+ T cells? Are the types of post-translational modifications, a direct reflection of the metabolite content of the cell?
Can we reprogram the epigenome of exhausted CD8+ tumor-infiltrating lymphocytes (TILs) by altering the metabolic composition of the TME?
Is there a direct link between nutrient intake and the microbiome of a patient on the one hand, and the epigenetic modifications observed in his/her CD8+ TILs on the other?
Can we extrapolate any knowledge from the epigenetic profile of CD8+ TILs to treatment success in solid tumor patients? What might be inferred for assessing epigenetically-based biomarkers?
Highlights Box.
The tumor microenvironment forms a metabolic barrier against cytotoxic CD8+ T cells, hampering current immunotherapeutic strategies.
It is increasingly recognized that epigenetics bridge cellular metabolism with gene expression.
Tackling epigenetic modifications or cell metabolism to (i) target the epigenetic signature of (tumor-infiltrating) CD8+ T cells in vivo or (ii) potentiate the ex vivo generation of anti-tumor T cell grafts for adoptive cell transfer, has gained much interest as a promising strategy to combat malignancy.
Acknowledgements
Conceptualization, HHVA; Original draft preparation, HHVA and TS; review and editing, HHVA, TS, SM, SMK and MM; visualization, HHVA and TS; funding acquisition, MM. All authors have read and agreed to the published version of the manuscript. HHVA is supported by fundamental postdoctoral mandate of the Belgian Foundation against Cancer (Stichting tegen Kanker). The authors would like to thank prof. Sébastien Anguille for granting us the right to use his artwork in the current publication. The authors declare no conflict of interest.
Glossary
- ABCA1
This protein functions as a cholesterol efflux pump in the cellular lipid removal pathway
- ACAT1–2
Cholesterol esterification enzymes that convert free cholesterol to cholesteryl esters for storage
- Adoptive cell transfer (ACT)
CTL vaccination, with the goal of recognizing, targeting, and destroying tumor cells. With the ACT of T cell receptor (TCR)- or chimeric antigen receptor (CAR)-engineered T cells, expanded from readily available blood CD8+ T cells, it is now theoretically possible to target any tumor
- Autophagosomes
double-membrane sequestering vesicles contributing to the recycling of cytosolic components and organelles when nutrients are scarce
- BET proteins
epigenetic readers that control gene transcription by binding to acetylated lysine residues on histones
- Bhlhe40
also known as Bhlhb2, belongs to a family of basic helix-loop-helix transcriptional factors. Under cellular stress conditions (such as hypoxia), it translocates to the nucleus where it promotes gene transcription by binding to E-box elements
- Chimeric antigen receptor (CAR) T cells
T cells modified with artificial receptor proteins consisting of (i) an ectodomain binding directly a tumor-specific molecule on the cell surface, (ii) an extracellular hinge/spacer and a transmembrane domain spanning the membrane, and (iii) an endodomain providing T cell signaling
- Dacinostat
potent pan-HDAC inhibitor
- Decitabine
DNA methyltransferases-inhibiting cytosine nucleoside analog
- Eomesodermin (Eomes)
T-box transcription factor, implicated in CD8+ T cell exhaustion
- EZH2
Histone methyltransferase catalyzing the methylation of H3K27, resulting in chromatin compaction and repression of transcription
- G9a
Histone methyltransferase for repressive H3K9me2 marks
- Glycolysis
Cytoplasmatic ATP-yielding catabolism of glucose
- HIF-1α
The hypoxia signaling pathway is primarily governed by hypoxia inducible transcription factor-1α
- Immune checkpoint inhibitors
antibodies directed against checkpoint inhibitory receptors such as cytotoxic T lymphocytes antigen-4 (CTLA-4), programmed death-1 (PD-1) and programmed death-ligand 1 (PD-L1), with the goal of breaking immune tolerance and stimulate CD8+ T cell effector responses
- Immunological synapse
stable cell-cell junction between a leukocyte (e.g. T cell) and an antigen-presenting cell, allowing (contact-dependent) communication between two immune cells
- Jumonji N/C terminal domains (JmjCs) enzymes
important family of histone lysine demethylases (KDMs)
- JQ1
aspecific inhibitor of the BET protein family; notable for its high affinity for bromodomains
- LSD1
H3K4 and H3K9 demethylase, also targeting non-histone proteins (i.e. DNMT1, STAT3)
- Oncometabolites
Conventional products of metabolism aberrantly accumulating in cancer cells, possessing pro-oncogenic capabilities
- Phf19
PHD finger protein 19; a key component of PRC2
- PRC2
multisubunit protein complex; a methyltransferase regulating gene expression by catalyzing trimethylation of histone H3 on lysine 27 (H3K27me3), leading to transcription repression
- Oxidative phosphorylation (OXPHOS)
Mitochondrion-based oxidation of metabolites, generating ATP through complexes I–V
- Regulatory T cells
subset of CD4+ T cells, essential for maintaining peripheral tolerance via the secretion of negative regulatory (anti-inflammatory) cytokines, i.e. IL-10 and TGF-β
- Ricolinostat
selective inhibitor of HDAC class 6, also inhibiting HDAC class 1–3 at high concentrations
- RORα
member of the orphan nuclear receptor (ONR) family. It binds to hormone response elements upstream of several genes, boosting their expression
- S-adenosyl-methionine (SAM)
synthesized in the methionine pathway; universal methyl donor for DNA, RNA, and protein methyltransferases
- Stemness
capacity of a cell to perpetuate its lineage, having both the ability for self-renewal and differentiation
- Stem cell memory T cell
long-lived memory T cells gifted with stem-like properties (self-renewal and multipotency)
- TCA (tricarboxylic acid) cycle
Krebs cycle or citric acid cycle; stepwise oxidation of acetyl-CoA (fatty acid oxidation), glutamate (glutaminolyse) or pyruvate (glycolysis), producing NADH and FADH, which in turn fuel the electron transport chain
- T cell exhaustion
describes the functionally impaired differentiation state of T cells induced by persistent antigen stimulation
- Tumor microenvironment (TME)
composed of immune cells, extracellular matrix, fibroblasts, proteins, and blood vessels that surround and feed the tumor. The inherent inflammation within the TME and its nourishment by a failing vascular network enable tumor progression, metastasis, and resistance to therapies
- Ten-eleven translocation (TET) enzymes
Active DNA demethylation is carried out by enzymes belonging to the TET family; TET1, TET2 and TET3. They mediate the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC)
- Th1 polarization
Differentiation of CD4+ T cells into Th1 cells, characterized by the secretion of IFN-γ and TNF-β, promoting cell-mediated immune responses
- TOX
member of the HMG (high mobility group) transcription factors
- Urelumab
fully human IgG4k mAb agonist of CD137/4–1BB
- Warburg effect
aerobic glycolysis; describes the preference of tumor cells for aerobic glycolysis (followed by lactic acid fermentation) over OXPHOS, even in the presence of abundant oxygen
Footnotes
Resources
I This trial is listed in https://clinicaltrials.gov/ct2/show/NCT01799083
II The clinical trial identifiers listed in Table 1 are found in https://clinicaltrials.gov/ with their respective trial ID numbers (https://clinicaltrials.gov/ct2/show/NCT03913455, etc.).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fridman WH et al. (2017) The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 [DOI] [PubMed] [Google Scholar]
- 2.Havel JJ et al. (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timmers M et al. (2019) Chimeric antigen receptor-modified T cell therapy in multiple myeloma: beyond B cell maturation antigen. Front. Immunol. 10, 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y and Ertl HCJ (2016) Starved and asphyxiated: how can CD8+ T cells within a tumor microenvironment prevent tumor progression. Front. Immunol. 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ and Chandel NS (2020) We need to talk about the Warburg effect. Nat. Metab. 2, 127–129 [DOI] [PubMed] [Google Scholar]
- 6.Riera-Domingo C. et al. (2020) Immunity, hypoxia, and metabolism – the ménage à trois of cancer: implications for immunotherapy. Physiol. Rev. 100, 1–102 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y. et al. (2017) Enhancing CD8+ T cell fatty Acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 32, 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigano S. et al. (2019) Targeting adenosine in cancer immunotherapy to enhance T-cell function. Front. Immunol. 10, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiger R. et al. (2016) L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leone RD et al. (2019) Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prendergast GC et al. (2018) Indoleamine 2,3-dioxygenase and its therapeutic inhibition in cancer. Int. Rev. Cell Mol. Biol. 336, 175–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y. et al. (2018) Tumor-repopulating cells induce PD-1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell 33, 480–494 [DOI] [PubMed] [Google Scholar]
- 13.Su X. et al. (2016) Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 30, 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonseca R. et al. (2020) Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 21, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zebley CC et al. (2020) Rewriting history: epigenetic reprogramming of CD8+ T cell differentiation to enhance immunotherapy. Trends Immunol. 41, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghoneim HE et al. (2017) De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauken KE et al. (2016) Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan O. et al. (2019) TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao C. et al. (2019) Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott AC et al. (2019) TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo H. et al. (2019) TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U. S. A. 116, 12410–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J. et al. (2019) NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugiura A. and Rathmell JC (2018) Metabolic barriers to T cell function in tumors. J. Immunol. 200, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengsch B. et al. (2016) Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siska PJ et al. (2017) Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, e93411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menk AV et al. (2018) 4–1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immuno-therapeutic responses. J. Exp. Med. 215, 1091–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vardhana SA et al. (2020) Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21, 1022–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharping NE et al. (2021) Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma EH et al. (2019) Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870 [DOI] [PubMed] [Google Scholar]
- 30.Chang C-H et al. (2015) Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu J. et al. (2019) Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng M. et al. (2016) Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein Geltink RI et al. (2020) Metabolic conditioning of CD8+ effector T cells for adoptive cell therapy. Nat. Metab. 2, 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D. et al. (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ippolito L. et al. (2019) Lactate: a metabolic driver in the tumour landscape. Trends Biochem. Sci. 44, 153–166 [DOI] [PubMed] [Google Scholar]
- 36.Zhao E. et al. (2016) Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 17, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koss B. et al. (2020) Epigenetic control of Cdkn2a.Arf protects tumor-infiltrating lymphocytes from metabolic exhaustion. Cancer Res. 80, 4707–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowling SD and Macian F. (2018) Autophagy and T cell metabolism. Cancer Lett. 419, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artal-Martinez de Narvajas A. et al. (2013) Epigenetic regulation of autophagy by the methyltransferase G9a. Mol. Cell. Biol. 33, 3983–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y. et al. (2017) Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W. et al. (2016) Regulation of T cell signalling by membrane lipids. Nat. Rev. Immunol. 16, 690–701 [DOI] [PubMed] [Google Scholar]
- 42.McDonnell E. et al. (2016) Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 17, 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellen KE et al. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W. et al. (2016) Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee IK et al. (2020) RORα regulates cholesterol metabolism of CD8+ T cells for anticancer immunity. Cancers (Basel) 12, 1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salminen A. et al. (2016) AMPK/Snf1 signaling regulates histone acetylation: Impact on gene expression and epigenetic functions. Cell. Signal. 28, 887–895 [DOI] [PubMed] [Google Scholar]
- 47.Shyer JA et al. (2020) Metabolic signaling in T cells. Cell Res. 30, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurmi K. et al. (2018) Carnitine palmitoyltransferase 1A has a lysine succinyltransferase activity. Cell Rep. 22, 1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y. et al. (2017) KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang M. and Pollard PJ (2013) Succinate: a new epigenetic hacker. Cancer Cell 23, 709–711 [DOI] [PubMed] [Google Scholar]
- 51.Roy DG et al. (2020) Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 31, 250–266 [DOI] [PubMed] [Google Scholar]
- 52.Bian Y. et al. (2020) Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair LV et al. (2019) Antigen receptor control of methionine metabolism in T cells. Elife 8, e44210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao X. et al. (2019) Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eil R. et al. (2016) Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vodnala SK et al. (2019) T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, eaau0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y. et al. (2019) Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 10, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Intlekofer AM et al. (2015) Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 22, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyrakis PA et al. (2016) The immunometabolite S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Batie M. et al. (2019) Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226 [DOI] [PubMed] [Google Scholar]
- 61.Chakraborty AA et al. (2019) Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thienpont B. et al. (2016) Tumour hypoxia causes DNA hyper-methylation by reducing TET activity. Nature 537, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberg F. et al. (2019) Reactive oxygen species in the tumor microenvironment: an overview. Cancers (Basel) 11, 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dan Lu DL et al. (2020) The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity. Nat. Immunol. 21, 287–297 [DOI] [PubMed] [Google Scholar]
- 65.Kidger AM et al. (2017) Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proc. Natl. Acad. Sci. U. S. A. 114, E317–E326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Png CW et al. (2016) DUSP10 regulates intestinal epithelial cell growth and colorectal tumorigenesis. Oncogene 35, 206–217 [DOI] [PubMed] [Google Scholar]
- 67.Givant-Horwitz V. et al. (2004) The PAC-1 dual specificity phosphatase predicts poor outcome in serous ovarian carcinoma. Gynecol. Oncol. 93, 517–523 [DOI] [PubMed] [Google Scholar]
- 68.Marvel D. and Gabrilovich DI (2015) Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest. 125, 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marigo I. et al. (2016) T cell cancer therapy requires CD40–CD40L activation of tumor necrosis factor and inducible nitric-oxide-synthase-producing dendritic cells. Cancer Cell 30, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hickok JR et al. (2013) Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J. Biol. Chem. 288, 16004–16015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moshfegh CM et al. (2019) Mitochondrial superoxide disrupts the metabolic and epigenetic landscape of CD4+ and CD8+ T-lymphocytes. Redox Biol. 27, 101141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X. et al. (2007) Pyruvate protects mitochondria from oxidative stress in human neuroblastoma SK-N-SH cells. Brain Res. 1132, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeng MY et al. (2018) Metabolic reprogramming of human CD8+ memory T cells through loss of SIRT1. J. Exp. Med. 215, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berger RS et al. (2019) Degradation of D-2-hydroxyglutarate in the presence of isocitrate dehydrogenase mutations. Sci. Rep. 9, 7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dang L. et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu C. et al. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L. et al. (2018) D-2-hydroxyglutarate is an intercellular mediator in IDH-mutant gliomas inhibiting complement and T cells. Clin. Cancer Res. 24, 5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bunse L. et al. (2018) Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 [DOI] [PubMed] [Google Scholar]
- 79.Mohammad HP et al. (2019) Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 25, 403–418 [DOI] [PubMed] [Google Scholar]
- 80.Li X. et al. (2017) Increased IFNγ(+) T cells are responsible for the clinical responses of low-dose DNA-eemethylating agent decitabine antitumor therapy. Clin. Cancer Res. an Off. J. Am. Assoc. Cancer Res. 23, 6031–6043 [DOI] [PubMed] [Google Scholar]
- 81.Tu WJ et al. (2020) Targeting nuclear LSD1 to reprogram cancer cells and reinvigorate exhausted T cells via a novel LSD1–EOMES switch. Front. Immunol. 11, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Z. et al. (2020) Modulation of SRSF2 expression reverses the exhaustion of TILs via the epigenetic regulation of immune checkpoint molecules. Cell. Mol. Life Sci. 77, 3441–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li C. et al. (2019) The transcription factor Bhlhe40 programs mitochondrial regulation of resident CD8+ T cell fitness and functionality. Immunity 51, 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji Y. et al. (2015) miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic γc cytokines. Proc. Natl. Acad. Sci. U. S. A. 112, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji Y. et al. (2019) miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8+ T cell fate. Nat. Commun. 10, 2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hogg SJ et al. (2017) BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 18, 2162–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adeegbe DO et al. (2018) BET bromodomain inhibition cooperates with PD-1 blockade to facilitate antitumor response in Krasmutant non–small cell lung cancer. Cancer Immunol. Res. 6, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L. et al. (2015) Decitabine enhances lymphocyte migration and function and synergizes with CTLA-4 blockade in a murine ovarian cancer model. Cancer Immunol. Res. 3, 1030–1041 [DOI] [PubMed] [Google Scholar]
- 89.Taylor K. et al. (2020) An open-label, phase II multicohort study of an oral hypomethylating agent CC-486 and durvalumab in advanced solid tumors. J. Immunother. Cancer 8, e000883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu G. et al. (2019) Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell. Mol. Immunol. 16, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang KC-Y. et al. (2020) Decitabine augments chemotherapy-induced PD-L1 upregulation for PD-L1 blockade in colorectal cancer. Cancers (Basel) 12, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knox T. et al. (2019) Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and downregulation of immunosuppressive proteins in tumor cells. Sci. Rep. 9, 6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Woods DM et al. (2015) HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol. Res. 3, 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng H. et al. (2016) HDAC inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 22, 4119–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goswami S. et al. (2018) Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 128, 3813–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi X. et al. (2019) Optimization of 4–1BB antibody for cancer immunotherapy by balancing agonistic strength with FcγR affinity. Nat. Commun. 10, 2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aznar MA et al. (2018) CD137 (4–1BB) costimulation modifies DNA methylation in CD8+ T cell-relevant genes. Cancer Immunol. Res. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 98.Vo DD et al. (2009) Enhanced antitumor activity induced by adoptive T-cell transfer and adjunctive use of the histone deacetylase inhibitor LAQ824. Cancer Res. 69, 8693–8699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lisiero DN et al. (2014) The histone deacetylase inhibitor, LBH589, promotes the systemic cytokine and effector responses of adoptively transferred CD8+ T cells. J. Immunother. Cancer 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adeegbe DO et al. (2017) Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov. 7, 852–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carew JS et al. (2019) Rational cotargeting of HDAC6 and BET proteins yields synergistic antimyeloma activity. Blood Adv. 3, 1318–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alizadeh D. et al. (2019) IL15 enhances CAR-T cell antitumor activity by reducing mTORC1 activity and preserving their stem cell memory phenotype. Cancer Immunol. Res. 7, 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kagoya Y. et al. (2016) BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J. Clin. Invest. 126, 3479–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chee J. et al. (2020) Impaired T cell proliferation by ex vivo BET-inhibition impedes adoptive immunotherapy in a murine melanoma model. Epigenetics 15, 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crompton JG et al. (2016) Lineage relationship of CD8+ T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell. Mol. Immunol. 13, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y. et al. (2021) Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nat. Commun. 12, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fraietta JA et al. (2018) Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cimmino L. et al. (2017) Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoang N-M and Rui L. (2020) DNA methyltransferases in hematological malignancies. J. Genet. Genomics 47, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ito K. et al. (2019) Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Rep. 28, 2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loberg MA et al. (2019) Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia 33, 1635–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeong M. et al. (2018) Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 23, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ostrander EL et al. (2020) Divergent effects of Dnmt3a and Tet2 mutations on hematopoietic progenitor cell fitness. Stem Cell Rep. 14, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kwesi-Maliepaard EM et al. (2020) The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 117, 20706–20716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salgado C. et al. (2019) A novel germline variant in the DOT1L gene co-segregating in a Dutch family with a history of melanoma. Melanoma Res. 29, 582–589 [DOI] [PubMed] [Google Scholar]
- 116.Wong M. et al. (2017) The histone methyltransferase DOT1L promotes neuroblastoma by regulating gene transcription. Cancer Res. 77, 2522–2533 [DOI] [PubMed] [Google Scholar]
- 117.Kagoya Y. et al. (2018) DOT1L Inhibition attenuates graft-versus-host disease by allogeneic T cells in adoptive immunotherapy models. Nat. Commun. 9, 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Woods DM et al. (2017) T cells lacking HDAC11 have increased effector functions and mediate enhanced alloreactivity in a murine model. Blood 130, 146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Straathof KC et al. (2005) An inducible caspase 9 safety switch for T-cell therapy. Blood 105, 4247–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tiberghien P. et al. (2001) Administration of herpes simplexthymidine kinase-expressing donor T cells with a T-cell-depleted allogeneic marrow graft. Blood 97, 63–72 [DOI] [PubMed] [Google Scholar]
- 121.Sato T. et al. (2007) Engineered human tmpk/AZT as a novel enzyme/prodrug axis for suicide gene therapy. Mol. Ther. 15, 962–970 [DOI] [PubMed] [Google Scholar]
- 122.Ruella M. et al. (2020) A cellular antidote to specifically deplete anti-CD19 chimeric antigen receptor-positive cells. Blood 135, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koristka S. et al. (2019) Anti-CAR-engineered T cells for epitope-based elimination of autologous CAR T cells. Cancer Immunol. Immunother. 68, 1401–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Topper MJ et al. (2020) The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 17, 75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cavalli G. and Heard E. (2019) Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 [DOI] [PubMed] [Google Scholar]
- 126.Feinberg AP (2018) The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378, 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jones PA and Baylin SB (2007) The epigenomics of cancer. Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beltra J-C et al. (2020) Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang R. et al. (2019) Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol. 21, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller BC et al. (2019) Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buggert M. et al. (2014) T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 10, e1004251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weber J. et al. (2015) CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl. Acad. Sci. U. S. A. 112, 13982–13987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen M. et al. (2018) RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270 [DOI] [PubMed] [Google Scholar]
- 134.Liu Q. et al. (2019) Multistage delivery nanoparticle facilitates efficient CRISPR/dCas9 activation and tumor growth suppression in vivo. Adv. Sci. (Weinh.) 6, 1801423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li A. et al. (2017) Increased IFNγ+ T cells are responsible for the clinical responses of low-dose DNA-demethylating agent decitabine antitumor therapy. Clin. Cancer Res. 23, 6031–6043 [DOI] [PubMed] [Google Scholar]
- 136.Zou W. and Chen L. (2008) Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 8, 467–477 [DOI] [PubMed] [Google Scholar]
- 137.Curiel TJ et al. (2003) Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat. Med. 9, 562–567 [DOI] [PubMed] [Google Scholar]
- 138.Zhou L. et al. (2020) Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti-PD-1 resistance in head and neck cancer. Clin. Cancer Res. an Off. J. Am. Assoc. Cancer Res. 26, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burr ML et al. (2019) An evolutionarily conserved function of polycomb silences the mhc class i antigen presentation pathway and enables immune evasion in cancer. Cancer Cell 36, 385–401.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ennishi D. et al. (2019) Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 9, 546–563 [DOI] [PubMed] [Google Scholar]
- 141.Zingg D. et al. (2017) The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 20, 854–867 [DOI] [PubMed] [Google Scholar]
- 142.Riganti C. et al. (2018) Bromodomain inhibition exerts its therapeutic potential in malignant pleural mesothelioma by promoting immunogenic cell death and changing the tumor immune-environment. Oncoimmunology 7, e1398874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Erkes DA et al. (2019) The next-generation BET inhibitor, PLX51107, delays melanoma growth in a CD8-mediated manner. Pigment Cell Melanoma Res. 32, 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tay RE et al. (2020) Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J. Exp. Med. 217, e20191453 [DOI] [PMC free article] [PubMed] [Google Scholar]