Abstract
The recently discovered SARS-CoV-2 variant Omicron (B.1.1.529) has rapidly become a global public health issue. The substantial mutations in the spike protein in this new variant have raised concerns about its ability to escape from pre-existing immunity established by natural infection or vaccination. In this review, we give a summary of current knowledge concerning the antibody evasion properties of Omicron and its subvariants (BA.2, BA.2.12.1, BA.4/5, and BA.2.75) from therapeutic monoclonal antibodies and the sera of SARS-CoV-2 vaccine recipients or convalescent patients. We also summarize whether vaccine-induced cellular immunity (memory B cell and T cell response) can recognize Omicron specifically. In brief, the Omicron variants demonstrated remarkable antibody evasion, with even more striking antibody escape seen in the Omicron BA.4 and BA.5 sub-lineages. Luckily, the third booster vaccine dose significantly increased the neutralizing antibodies titers, and the vaccine-induced cellular response remains conserved and provides second-line defense against the Omicron.
Keywords: SARS-CoV-2, Omicron, Vaccine, Antibody evasion, Cellular immunity
Highlights
-
•
Omicron and its sub-lineages significantly enhanced their transmissibility and immune evasion.
-
•
Omicron BA.2.12.1, BA.4/5, and BA.2.75 sub-lineages show more significant antibody escape than BA.1 and BA.2.
-
•
Third booster dose vaccination is important, but the need for the fourth dose remains to be determined.
-
•
The SARS-CoV-2 specific B cell and T cell response was conserved and able to cross-recognize the Omicron.
1. Introduction
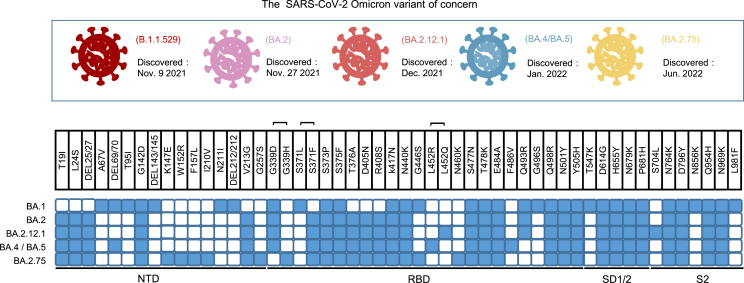
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been a pandemic worldwide since 2019 (Zhou et al., 2020). After that, a variety of SARS-CoV-2 variants such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) emerged with varying transmissibility and disease severity (Harvey et al., 2021; Tao et al., 2021; Tegally et al., 2021). On November 24, 2021, World Health Organization (WHO) announced the sequence of a novel SARS-CoV-2 variant Omicron (B.1.1.529) (Callaway, 2021) and rapidly classified it as the fifth variant of concerns (VOCs) (Viana et al., 2022). The Omicron variant hosts 34 mutations in the spike protein and showed unprecedented transmissibility and striking immune evasion. Within a few months, the Omicron variant (mainly BA.1 subvariants) rapidly replaced Delta VOC and became the dominant strain (Lambrou et al., 2022). Subsequently, a series of Omicron sub-lineages continuously emerged: mainly including BA.2, BA.2.12.1, BA.4, and BA.5, which exhibit different characteristics of transmission and immune evasion capacity (Cao et al., 2022c; Desingu et al., 2022; Iketani et al., 2022). More recently, a new prevalent Omicron subvariant, BA.2.75, was discovered in India and Japan, and reported in multiple other countries (Tan et al., 2022). This new BA.2.75 variants harbored nine additional mutations in the spike protein compared to BA.2, including G446S, R493Q, and N460K (Shaheen et al., 2022) (Fig. 1). These mutations significantly increased BA.2.75's affinity to ACE2 and enhanced cell-cell fusion, which may set off a new wave of epidemic peak (Zappa et al., 2022; Qu et al., 2022).
Fig. 1.
The spike mutations within the Omicron sub-lineages. The mutated residues in the spike protein among Omicron sub-lineages are colored blue.
In this review, we first describe the epidemic properties of SARS-CoV-2 Omicron and its subvariants by analyzing its transmissibility and pathogenicity. We then focus on evaluating the antibody evasion ability of Omicron sub-lineages by convalescent sera, vaccine recipients’ sera, and therapeutic monoclonal antibodies, and exploring the underlying mechanism behind immune evasion. Lastly, we describe Omicron-reactive B cell and T cell responses in COVID-19 vaccine recipients and propose potential strategies for the SARS-CoV-2 Omicron variants' threat.
2. The SARS-CoV-2 Omicron
2.1. The SARS-CoV-2 Omicron transmission
Epidemiological evidence from different countries suggests that the Omicron variant is substantially more transmissible than prior VOCs (Kumar et al., 2022; Ren et al., 2022). According to previous studies, the original Omicron variant has an average basic reproduction number and effective reproduction number of 8.2 and 3.6, respectively (Liu et al., 2022), and triggered 3.8 and 2.5 times higher transmissibility than the Delta variant (Nishiura et al., 2022; Liu and Rocklöv, 2022). The doubling time is also very short among populations that have achieved herd immunity, ranging from 3.2 days to 3.6 days (Grabowski et al., 2022). These data validated Omicron's high transmission rate.
Although the original Omicron variant has shown unprecedented transmissibility, the subsequent Omicron sublineages seem to be much more transmissible. It was reported that the BA.2 strain is spreading about 1.5 times faster than the Omicron BA.1 strain (Rahimi et al., 2022). The BA.2.12.1 lineage, branched off from BA.2, was reported to have 25% higher transmissibility than BA.2 (Beheshti et al., 2022). The BA.4 and BA.5 (BA.4/5), which are sister variants, harboring L452R, F486V, and R493Q mutations in the spike RBD, show even greater growth potential (Cao et al., 2022c). After a large wave of infections dominated by BA.1 and BA.2 (Rahimi et al., 2022), BA.4/5 has swept the globe and surpassed BA.1 and BA.2 as the most prevalent subtype in the world. Currently, data are limited for the transmissibility of the Omicron BA.2.75 variant. The preliminary study showed that the effective reproduction number of BA.2.75 is greater than that of BA.5 (Saito et al., 2022), which was expected to be more transmissible over other Omicron sub-lineages (Sugano et al., 2022).
A study revealed that the largely increased infectivity of original Omicron could be attributed to the accumulated mutations that created new salt bridges and hydrogen between Omicron and ACE2 receptors, enhancing the binding of Omicron spikes protein with human ACE2 (Mannar et al., 2022). Besides, the lower thermal stability of Omicron RBD and the altered lower dependence on transmembrane serine protease 2 (TMPRSS2) utilization also accounted for Omicron's increased transmission (Yin et al., 2022).
2.2. The SARS-CoV-2 Omicron pathogenicity
In contrast to the high transmissibility, the SARS-CoV-2 Omicron appears to have attenuated disease severity and mortality compared with previous VOCs. Most cases diagnosed with original Omicron manifested asymptomatic or mild disease, with more pronounced upper respiratory symptoms but rarely pneumonia (Meo et al., 2021). One significant real-world study reveals that the hospitalization and death rates of Omicron patients were reduced by 41% and 79% respectively when compared to Delta patients (Lewnard et al., 2022). In a large national cohort containing 1.5 million cases, the risk of severe outcomes is also substantially lower for Omicron infection than for Delta infection, suggesting that Omicron variants had a significantly reduced clinical severity (Nyberg et al., 2022). This observed milder clinical severity may be related to the immune barrier formed by vaccination. However, even in patients who had not been vaccinated, the Omicron infection group had substantially lower rates of emergency department visits, hospitalizations, ICU admissions, and the need for mechanical ventilation than the Delta group, confirming Omicron's attenuated pathogenicity (Wang L et al., 2022).
A series of studies shed light on the reason why the original Omicron infection showed attenuated pathogenicity. One of the most important reasons was that the Omicron variant exhibited much less damage to the lungs compared with other VOCs (Halfmann et al., 2022). Firstly, the replication ability of the Omicron variant in human lung and intestinal epithelial cells is significantly reduced (Shuai et al., 2022). Secondly, the lung pathological lesions and mortality rate of Omicron-infected mice models were lower than those of other VOCs infections (Uraki et al., 2022). Furthermore, although the TMPRSS2 protein is extensively expressed on the surface of lung cells, the Omicron reduced efficiency in utilizing the host TMPRSS2 to help the virus enter into cells (Shuai et al., 2022). These findings may explain why the Omicron infection was associated with reduced hospitalizations and deaths.
Several studies evaluated the pathogenicity between different Omicron sub-lineages. Despite BA.2 having higher transmissibility and fitness than BA.1, the risk of severe outcomes was comparable to BA.1/BA.1.1 (Wolter et al., 2022; Obermeyer et al., 2022). Similarly, there is no evidence suggesting that BA.2.12.1 causes more severe disease than the Omicron BA.1 strain. Although the BA.4/5 variants exhibit more pathogenic in hamster models infected with Omicron sublineages (Kimura et al., 2022), they did not increase the risk of critical illness in the real-world investigation (Davies et al., 2022). To date, it remains unclear whether novel BA.2.75 causes more severe clinical disease. In the lungs of infected hamsters, BA.2.75 showed a stronger replication capacity than BA.5 and caused the most severe focal viral pneumonia (Uraki et al., 2022). However, further large clinical studies are required to corroborate the disease severity of BA2.75 infection.
3. The significant antibody evasion of SARS-CoV-2 Omicron
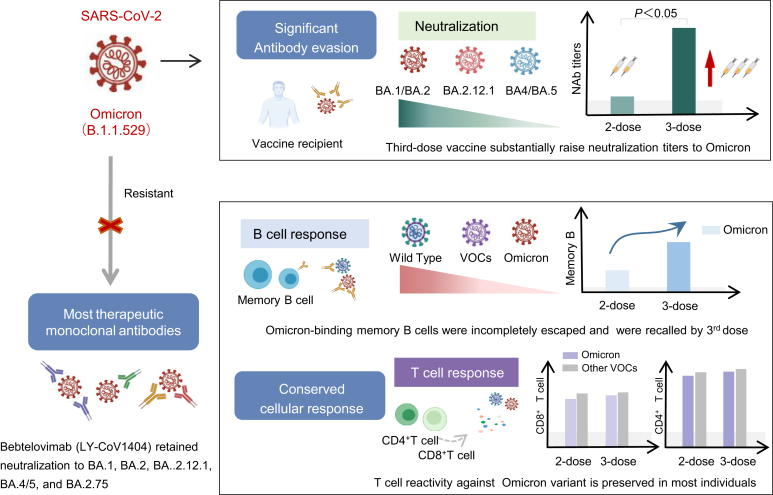
SARS-CoV-2 Omicron and its subvariants contain a large number of mutations in its spike protein and have occurred antigenic shift (McCallum et al., 2022). Since most SARS-CoV-2 vaccines and monoclonal antibodies are designed to target the wild-type (WT) spike protein, it's quite probable that the Omicron variant will drastically reduce the efficiency of pre-existing immunity induced by previously natural infection or current vaccines. Hence, it is necessary to investigate the antibody evasion potential of the original Omicron and its subvariants (BA.2, BA.2.12.1, BA.4/5, and BA.2.75) against convalescent COVID-19 individuals, vaccine recipients, and therapeutic monoclonal antibodies (Fig. 2).
Fig. 2.
The humoral and cellar immune evasion of Omicron. VOCs, variant of concerns.
3.1. Neutralization of Omicron by convalescent sera
Multiple studies suggested that there was a significant difference in neutralizing activity to Omicron between vaccinated and unvaccinated COVID-19 convalescent patients.
Serum obtained from most unvaccinated convalescent participants could not neutralize the original Omicron. In the early convalescent phase (less than three months) of WT SARS-CoV-2 infection, the majority of serum samples exhibited undetectable neutralizing activity against the original Omicron, with the seropositivity ranging from 20% to 26.7% (Carreño et al., 2022; Liu et al., 2022). The serum neutralizing titers against Omicron were significantly lower (8–80 fold) when compared with WT (Hoffmann et al., 2022; Lusvarghi et al., 2021; Zhang et al., 2022). Unsurprisingly, the neutralization titer exhibited a further substantial decline over time. Only 13%–39% of serum samples from COVID-19 convalescents had detectable neutralizing antibodies against the Omicron one year following recovery, whereas 71%–94% of serum still exhibited neutralizing antibody response against Delta (Planas et al., 2022; Ma et al., 2022). Of note, even convalescent individuals who have previously been infected with Alpha, Beta, or Delta variants can barely neutralize the Omicron (Dejnirattisai et al., 2022; Wratil et al., 2022; Zhang et al., 2021). In brief, both WT and VOCs-infected COVID-19 patients had very little neutralization capacity against Omicron, indicating considerable antibody escape.
However, SARS-CoV-2 vaccination dramatically improved neutralizing antibodies against the original Omicron in convalescent individuals. Several studies showed that almost all convalescent individuals (90%–100%) were able to neutralize the Omicron after a single dose vaccine (Cheng et al., 2022; Rössler et al., 2022; Sheward et al., 2022). The serum antibody titers against Omicron increased by 63–154 fold on average after vaccination (Schmidt et al., 2022; Wratil et al., 2022). Of note, compared to a single shot, vaccinated with a two-dose vaccine did not increase the neutralizing activity against Omicron in convalescent patients (Wratil et al., 2022). However, prolonging the interval between the second and third doses induced greater neutralization capacity against all SARS-CoV-2 VOCs, including Omicron (Wratil et al., 2022). The significant increase in neutralization capacity following convalescence-vaccination may be related to the consecutive antigen exposure rapidly induced broader neutralizing antibodies and affinity-matured memory B cell subsets (Wratil et al., 2022; Goel et al., 2022). This “hybrid immunity” achieved by infection-plus-vaccination or vaccination-plus-breakthrough infection resulted in repeated exposure to the original SARS-CoV-2 spike proteins, inducing high-quality and high-avidity neutralizing antibodies against various VOCs, including Omicron (Wratil et al., 2022). Supplementary Table S1 summarizes the neutralization of COVID-19 convalescent sera against Omicron variants before and after vaccination.
Several studies have described the antibody evasion capabilities among Omicron sub-lineages. Omicron BA.1 have a comparable antibody escape capacity to BA.2 (Ai et al., 2022; Yu et al., 2022), despite the BA.1 and BA.2 strain escaping neutralizing antibodies that target different epitopes (Cao et al., 2022b). However, those unvaccinated participants who had recovered from the Omicron BA.1 or BA.2 infection were unable to effectively cross-neutralize the Omicron BA.2.12.1, BA.4/5, and BA.2.75 sub-lineages (Khan et al., 2022; Cao et al., 2022a), suggesting the Omicron BA.2.12.1, BA.4/5, and BA.2.75 sub-lineages exhibit more striking antibody evasion over BA.1 or BA.2. The potential mechanism behind this phenomenon may be due to the humoral immune memory recalled by BA.1 breakthrough infections primarily target the SARS-CoV-2 wild type, and the neutralizing antibodies mainly enriched on spike epitopes that do not bind ACE2, thus were evaded by the BA.4/5 and BA.2.75 (Cao et al., 2022a). To date, BA.4/5 exhibited the greatest antibody resistance among known Omicron sub-lineages, followed by BA.2.75 (Wang Q et al., 2022b). In convalescents with BA.1 or BA.2 breakthrough infections, the neutralization titer against BA.2.75 was higher than that against BA.5 (Tan et al., 2022). Intriguingly, plasma from BA.5 breakthrough infection showed much lower neutralization against BA.2.75 than BA.5, this phenomenon may be accounted for BA.2.75's unique RBD and NTD-targeting antibody escape pattern (Cao et al., 2022a).
To sum up, those unvaccinated COVID-19 convalescent individuals showed an almost complete loss of neutralization to Omicron, requiring SARS-CoV-2 vaccination to boost their immunity against re-infection. The serum from Omicron BA.1 or BA.2 infection showed significant neutralization resistance to Omicron BA.4/5 sub-lineages, suggesting that Omicron natural infection will not be enough to protect us against emerging mutant strains.
3.2. Neutralization of Omicron by SARS-CoV-2 vaccinees
Most primary SARS-CoV-2 vaccine regimens consisted of two doses. The new emergence of Omicron variants poses a huge challenge to the effectiveness of two-dose vaccine-induced immunity. Increasing evidence demonstrates that Omicron and its sub-lineages greatly evade two-dose vaccine-induced antibodies in healthy individuals. For the mRNA vaccines, two-dose vaccination induced poor neutralization against Omicron (Dejnirattisai et al., 2022; Liu et al., 2022; Planas et al., 2022). The seropositivity of neutralizing antibodies ranges between 12.8% and 85% (Cheng et al., 2022; Lusvarghi et al., 2021; Muik et al., 2022) and the titer against Omicron dramatically decreased 43–122 fold when compared with WT (Garcia-Beltran et al., 2022). At 5–7 months after two-dose vaccination, neutralizing antibodies waned or were completely absent (Planas et al., 2022). Other primary vaccination regimens, such as an adenoviral-vectored vaccine or inactivated vaccines, displayed similar poor neutralization against Omicron (Cameroni et al., 2022; Lu et al., 2021; Meng et al., 2022). The above studies indicated that the immune barrier established by the primary vaccine regimen is too vulnerable to neutralize the Omicron.
Fortunately, the third booster dose significantly increased the neutralizing antibody against Omicron and its sub-lineages regardless of what vaccine platform was considered (Qu et al., 2022). Compared to the second dose, a third homologous mRNA-boosting vaccine substantially increased neutralization titers against the original Omicron by 23.4–34.2 fold, reaching approximately 100% seropositivity (Lusvarghi et al., 2021; Nemet et al., 2022; Planas et al., 2022). Compared to homologous boosting, a heterologous booster vaccination regimen can produce higher Omicron immunogenicity (Costa Clemens et al., 2022; GeurtsvanKessel et al., 2022). More importantly, the third-dose mRNA vaccine was efficient in neutralizing most Omicron sublineages, including BA.2, BA.3, BA.4, and BA.5 subtypes (Qu et al., 2022), despite some studies reporting that BA.4/5 show significant antibody escape to the sera from three-dose vaccine recipients, and was more prone to occur breakthrough infections (Tuekprakhon et al., 2022). In addition to substantially increasing the neutralizing antibodies, the third dose vaccine is effective in protecting individuals from serious COVID-19-related outcomes (Barda et al., 2021; Tseng et al., 2022). Some studies assessed the durability of neutralizing antibody responses against Omicron and its subvariants (BA.2.12.1 and BA.4/5) after the third booster dose (Qu et al., 2022; Lyke et al., 2022). It seems that the boosted peak titers decline rapidly within two months, but the effective antibody titer against Omicron is persistent for a long time (at least nine months) in the majority of the individuals (Qu et al., 2022; Lyke et al., 2022). The rate of decay of antibody titers was comparable between the Omicron sub-lineages (Qu et al., 2022). Supplementary Table S2 summarizes vaccine recipients' neutralizing capacity against Omicron variants before and after the third booster vaccination.
The data presented above showed that the initial two-dose vaccination regimen resulted in a large reduction or complete loss of Omicron neutralization titers; however, the third booster vaccination substantially raised neutralization to Omicron and provided better protection. The third booster vaccine, regardless of homologous or heterologous regimen, should be extensively recommended for the general population, particularly high-risk groups.
Although the third booster dose has shown a satisfactory effect in increasing antibody titers and preventing hospitalization and death, the necessity for a fourth dose vaccine remains to be determined. The preliminary study in Israeli holds the opinion that the three-dose mRNA vaccines hit a “ceiling of immunity” (Regev-Yochay et al., 2022). Even though the fourth dose restores the maximal antibody titers that were achieved after three doses, it provides little additional protection against Omicron infection. The three-dose mRNA vaccines were efficient in preventing Omicron infection by 71.6% (Tseng et al., 2022), whereas the fourth dose was substantially less effective (11%–30%) (Regev-Yochay et al., 2022). Individuals who are young and healthy are unlikely to benefit significantly from a fourth dose, which mostly protects elderly high-risk groups (aged over 60 years) from COVID-19 hospitalization and death (Nordstrom et al., 2022; Arbel et al., 2022). Consistent with this study, Wang et al. reported that the fourth dose of inactivated COVID-19 vaccination was capable of recalling waned antibody titers, however, the peak value was lower than the third dose (Wang J et al., 2022). Furthermore, the fourth dose facilitates the shift of immune reactions from RBD to nucleocapsid protein (NP) and N-terminal domain (NTD) (Wang J et al., 2022); the shifted response was unable to neutralize Omicron due to significantly altered conformation. These studies revealed that immune responses could not be elevated indefinitely. More large studies are required in the future to determine the necessity for a fourth dose.
In summary, the above data highlighted the importance of the third vaccine dose. Facing the ongoing challenge of Omicron, the third booster vaccination is the best strategy in the context of the existing vaccine.
3.3. Neutralization of Omicron by therapeutic monoclonal antibodies
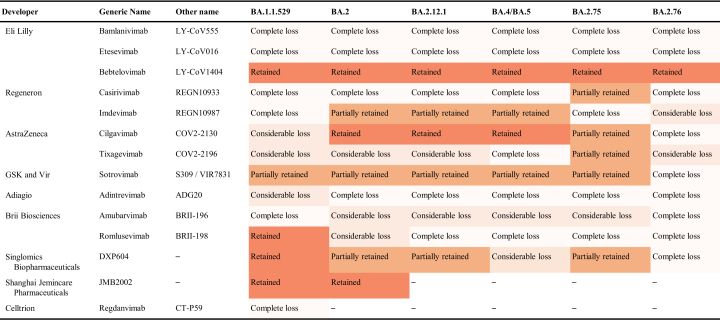
In addition to having a major impact on vaccines, Omicron created a great challenge to the therapy. Many studies estimated the neutralization efficacy of approved or developing antibodies against divergent Omicron sub-lineages, and the results were presented in Table 1. Overall, Omicron variants exhibit significant resistance to most of these monoclonal antibodies, including bamlanivimab (LY-CoV555), etesevimab (LY-CoV016), casirivimab (REGN10933), tixagevimab (COV2-2196/AZD8895), adintrevimab (ADG20), and amubarvimab (BRII-196). Among them, bamlanivimab (LY-CoV555) and etesevimab (LY-CoV016) completely lost their neutralizing capacity to all major Omicron sub-lineages (BA.1, BA.2, BA.2.12.1, BA.4/5, BA.2.75, and BA.2.76) (Cameroni et al., 2022; Dejnirattisai et al., 2022; Planas et al., 2022; Schulz et al., 2022). Fortunately, a part of the antibodies retained their neutralizing activity. For the BA.1 sub-lineages, sotrovimab (S309), romlusevimab (BRII-198), DXP604, and JMB2002 remained superior neutralizing activity (Cao et al., 2022b; Schulz et al., 2022; Yin et al., 2022). For the BA.2, BA.2.12.1, and BA.4/5 sub-lineages, bebtelovimab (LY-CoV1404), cilgavimab (COV2-2130/AZD1061), imdevimab (REGN10987), and sotrovimab (S309) retained or partially retained their neutralization (Wang Q et al., 2022a; Cao et al., 2022c). Despite the casirivimab (REGN10933) and tixagevimab (COV2-2196/AZD8895) showed a considerable or complete loss of neutralization to BA.1, BA.2, BA.2.12.1, BA.4/5, their neutralizing activities against BA.2.75 is partially restored due to R493Q reversion (Cao et al., 2022a; Qu et al., 2022). Of note, bebtelovimab (LY-CoV1404) was reported to potently neutralize major Omicron sub-lineages (BA.1, BA.2, BA.2.12.1, BA.4 and BA.5) (Ai et al., 2022; Iketani et al., 2022; Westendorf et al., 2022), including recently emerged BA.2.75 and BA.2.76 (Cao et al., 2022a).
Table 1.
Neutralization of Omicron by monoclonal antibodies.
Retained, IC50 < 100 (ng/mL); Partially retained, 100 < IC50 < 1000 (ng/mL); Considerable loss, 1000 < IC50 < 5000 (ng/mL); Complete loss,> 5000 (ng/mL).
Omicron's considerable antibody escape can be attributed to the numerous mutations accumulating on the spike epitopes targeted by these antibodies (Cao et al., 2022b). Compared to BA.2 variants, BA.4/5 and BA.2.75 bear four and nine additional mutations in their spike proteins respectively (Tan et al., 2022). The genetic and antigenic distance of Omicron sublineages is far away from the previous SARS-CoV-2 VOCs, especially WT strains, facilitating the Omicron's remarkable antibody evasion (Wang J et al., 2022). Due to crucial mutation sites like L452R and F486V being far from the bebtelovimab-RBD interface, LY-CoV1404 maintains potent neutralizing activity against all known Omicron sub-lineages (Zhou et al., 2022). Only K444Q, V445A, or P499 R/S mutation in spike protein will significantly decrease the activity of bebtelovimab (Westendorf et al., 2022). Apart from that, the alterations in local conformation and hydrophobic micro-environments drive epitope modification, leading to Omicron is not recognized by most RBD- and NTD-antibodies (Cui et al., 2022). Together, the antigenic shift in Omicron mediated considerable evasion to the majority of therapeutic monoclonal antibodies, emphasizing the importance of developing broadly neutralizing antibodies to combat the ongoing COVID-19 pandemic.
4. The conserved cellular immunity to Omicron
Given Omicron variants' striking antibody evasion, whether infection or vaccine-induced memory B cells or T cells may recognize Omicron is a major concern.
4.1. The Omicron RBD specific memory B cell response
Memory B cells constitute an important part of long-term immunity because memory B cells can rapidly differentiate and produce numerous new antibodies with higher affinity when exposed to a similar antigen. Several recent studies investigated whether memory B cells induced by two-dose mRNA vaccination can recognize Omicron. They found that only 40%–50% of memory B cells retained recognition of the original Omicron, which was substantially reduced compared with other variants (71% on average) (Goel et al., 2022; Sokal et al., 2022; Tarke et al., 2022). This is consistent with the findings that the neutralizing activity elicited by two-dose vaccination against Omicron is low. Nevertheless, the third booster dose significantly increased the Omicron neutralizing antibody titers, which is possible because the frequency of Omicron RBD-binding memory B cells significantly increased after the third dose (Goel et al., 2022; Muecksch et al., 2022). This increase was related to the continued expansion of existing memory B cell clones that were present after the second dose, as well as the appearance of new clones induced by the third dose. The existing resting RBD-binding IgG + memory B cells are steadily increasing the B cell receptor (BCR) breadth over time, which may enable the production of more broadly neutralizing antibodies (Kotaki et al., 2022). The newly produced memory B cell clones after the third dose targeted more conserved epitopes on Omicron spike protein, promoting the neutralizing antibodies encoded by these cells exhibited significantly increased breadth and potency (Garcia-Beltran et al., 2022; Muecksch et al., 2022). In addition to memory B cells, the long-lived plasma cells in the bone marrow also accumulated a high amount of somatic hypermutations, contributing to enhanced antibody affinity against Omicron (Kim et al., 2022).
Therefore, the immune evasion of Omicron to memory B cells is incomplete. Despite the Omicron variant exhibiting remarkable antibody escape, the memory B cell repertoire continues to evolve and can provide some protection in vaccinated individuals.
4.2. The preserved Omicron spike-specific T cell response
T cell response, particularly CD8+ T cell responses, is critical in preventing severe COVID-19 outcomes (Gao et al., 2021; Chandrashekar et al., 2022). Studies have shown that the case fatality ratio of Omicron infected patients with two-dose full vaccination was far lower than those with partial vaccination (Tan et al., 2020; He et al., 2022). Considering the neutralizing antibodies induced by two-dose vaccination having poor activity against Omicron, this protection against severe disease may be provided by T cells.
Many studies have shown that CD4+ and CD8+ T cell immunity induced by current vaccines is highly conserved to the original Omicron variants (Naranbhai et al., 2022; Liu et al., 2022; Mazzoni et al., 2022). No significant differences were found between the frequency of Omicron- and WT-specific polyfunctional CD4+/CD8+ T cells in two-dose vaccinated subjects (GeurtsvanKessel et al., 2022; Keeton et al., 2022). Even in six months post-vaccination, 84% (CD4+) and 85% (CD8+) of memory T cell responses preserved recognition of Omicron, demonstrating the long-term persistence of cross-reactive T cells (Tarke et al., 2022). Not only that, T cell repertoire analysis revealed that CD4+ and CD8+ T cells can recognize the majority of Omicron spike epitopes, with an average epitope preservation rate of 80%–85% (Tarke et al., 2022). After the third booster dose, the frequency of Omicron-specific CD4+ and CD8+ T cell responses remained stable or further increased in both healthy vaccinees and convalescents (Keeton et al., 2022; GeurtsvanKessel et al., 2022).
In contrast to antibody response, Omicron can hardly escape from T cell responses. One possible explanation is that T cells in each individual recognize a different fragment of the virus, moreover, T cell epitopes are distributed among structural and nonstructural proteins (Choi et al., 2022). These findings suggested that SARS-CoV-2 spike-specific T cell responses against Omicron were largely preserved, which could provide robust second-level protection against severe disease under the circumstances of substantially reduced neutralizing antibodies. The development of second-generation SARS-CoV-2 vaccines can fully utilize the role of T cells, including adding other immunogens such as SARS-CoV-2 nucleocapsid proteins, membrane proteins, or antigens in conserved regions to help develop pan-beta coronavirus vaccines.
5. Conclusions
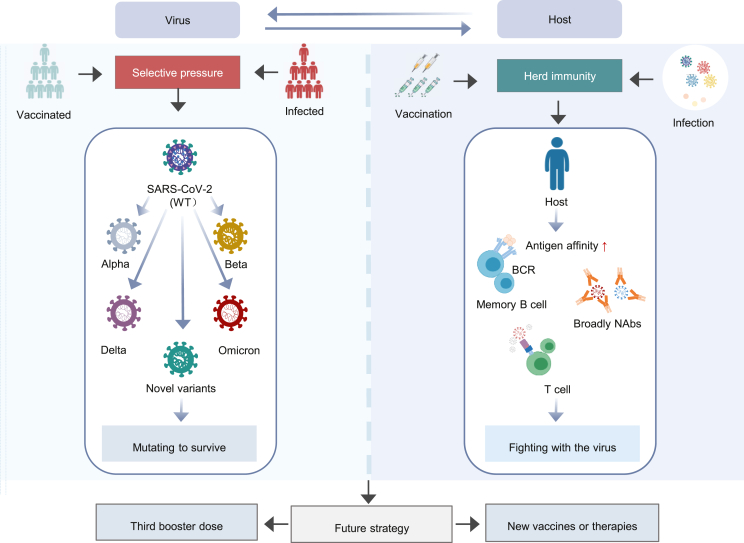
Over the past few months, the original Omicron variants have further evolved into multiple sub-lineages. Compared with previous VOCs, SARS-CoV-2 Omicron and its sub-lineages significantly enhanced their transmissibility and immune evasion. This may be a consequence of selective pressures arising from herd immunity due to mass vaccination and Omicron infection. The virus constantly evolves and mutates to survive, creating strains with higher affinity and stronger evasion capacity. Under the circumstances, being vaccinated with a third booster dose is extremely important. In addition to increasing the neutralizing antibody titers against various Omicron sub-lineages, the booster vaccine dose also improved the function of B cells and T cells to reduce the clinical severity of Omicron infection. Hence, active vaccination is essential regardless of a history of SARS-CoV-2 infection. With Omicron dominating COVID-19, strategies for developing Omicron-specific vaccines and novel therapeutics are still worth exploring (Fig. 3).
Fig. 3.
The interrelation between SARS-CoV-2 virus and host immunity. WT, wild-type; NAbs, neutralizing antibodies.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Science and Technology Major Project of China (92169121); the Applied Basic and Frontier Technology Research Project of Wuhan (2020020601012233), and the Key Biosafety Science and Technology Program of Hubei Jiangxia Laboratory (JXBS001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.11.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ai J., Wang X., He X., Zhao X., Zhang Y., Jiang Y., Li M., Cui Y., Chen Y., Qiao R., Li L., Yang L., Li Y., Hu Z., Zhang W., Wang P. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe. 2022;30:1077–1083. doi: 10.1016/j.chom.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel R., Sergienko R., Friger M., Peretz A., Beckenstein T., Yaron S., Netzer D., Hammerman A. Effectiveness of a second BNT162b2 booster vaccine against hospitalization and death from COVID-19 in adults aged over 60 years. Nat. Med. 2022;28:1486–1490. doi: 10.1038/s41591-022-01832-0. [DOI] [PubMed] [Google Scholar]
- Barda N., Dagan N., Cohen C., Hernán M.A., Lipsitch M., Kohane I.S., Reis B.Y., Balicer R.D. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398:2093–2100. doi: 10.1016/S0140-6736(21)02249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti N.A., Keikha M. BA.2.12.1 is a new omicron offshoot that is a highly contagious but not severe disease. Ann Med Surg (Lond). 2022;79 doi: 10.1016/j.amsu.2022.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P.E., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Snell G., Veesler D., Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., Zhu Q., Deng J., Fu W., Yu L., Zhang N., Wang J., Xiao T., An R., Wang J., Liu L., Yang S., Niu X., Gu Q., Shao F., Hao X., Jin R., Wang Y., Xie X.S., Wang X. Characterizations of enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30:1527–1539. doi: 10.1016/j.chom.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., Yu Y., Wang P., Zhang Z., Liu P., An R., Hao X., Wang Y., Wang J., Feng R., Sun H., Zhao L., Zhang W., Zhao D., Zheng J., Yu L., Li C., Zhang N., Wang R., Niu X., Yang S., Song X., Chai Y., Hu Y., Shi Y., Zheng L., Li Z., Gu Q., Shao F., Huang W., Jin R., Shen Z., Wang Y., Wang X., Xiao J., Xie X.S. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., Gonzalez-Reiche A.S., Dambrauskas N., Vigdorovich V., Alburquerque B., Amoako A.A., Banu R., Beach K.F., Bermúdez-González M.C., Cai G.Y., Ceglia I., Cognigni C., Farrugia K., Gleason C.R., van de Guchte A., Kleiner G., Khalil Z., Lyttle N., Mendez W.A., Mulder L.C.F., Oostenink A., Rooker A., Salimbangon A.T., Saksena M., Paniz-Mondolfi A.E., Polanco J., Srivastava K., Sather D.N., Sordillo E.M., Bajic G., van Bakel H., Simon V., Krammer F. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Chandrashekar A., Yu J., McMahan K., Jacob-Dolan C., Liu J., He X., Hope D., Anioke T., Barrett J., Chung B., Hachmann N.P., Lifton M., Miller J., Powers O., Sciacca M., Sellers D., Siamatu M., Surve N., VanWyk H., Wan H., Wu C., Pessaint L., Valentin D., Van Ry A., Muench J., Boursiquot M., Cook A., Velasco J., Teow E., Boon A., Suthar M.S., Jain N., Martinot A.J., Lewis M.G., Andersen H., Barouch D.H. Vaccine protection against the SARS-CoV-2 Omicron variant in macaques. Cell. 2022;185:1549–1555. doi: 10.1016/j.cell.2022.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., Ng S.S., Chan K.C.K., Ko F.W., Chen C., Yiu K., Lam B.H.S., Lau E.H.Y., Chan K.K.P., Luk L.L.H., Li J.K.C., Tsang L.C.H., Poon L.L.M., Hui D.S.C., Peiris M. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022;28:486–489. doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.J., Kim D., Noh J.Y., Kim S., Park S., Jeong H.W., Shin E. T cell epitopes in SARS-CoV-2 proteins are substantially conserved in the Omicron variant. Cell. Mol. Immunol. 2022;19:447–448. doi: 10.1038/s41423-022-00838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Clemens S.A., Weckx L., Clemens R., Almeida Mendes A.V., Ramos Souza A., Silveira M.B.V., Da Guarda S.N.F., de Nobrega M.M., de Moraes Pinto M.I., Gonzalez I.G.S., Salvador N., Franco M.M., de Avila Mendonça R.N., Queiroz Oliveira I.S., de Freitas Souza B.S., Fraga M., Aley P., Bibi S., Cantrell L., Dejnirattisai W., Liu X., Mongkolsapaya J., Supasa P., Screaton G.R., Lambe T., Voysey M., Pollard A.J., Bittaye M., Woods D., Davies S., Smith H., Ulaszewska M., Sanders H., Mabette R., Vernon S., Valliji Z., Mead G., Tejpal C., Park J., Beveridge A., Eldawi A., Felle S., Fraga M., Muniz Martins T., Martins Medrado C.L., de Arruda Cordeiro Matos L.J. Heterologous versus homologous COVID-19 booster vaccination in previous recipients of two doses of CoronaVac COVID-19 vaccine in Brazil (RHH-001): a phase 4, non-inferiority, single blind, randomised study. Lancet. 2022;399:521–529. doi: 10.1016/S0140-6736(22)00094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., Yang M., Xu G., Zhu B., Fu W., Chu T., Feng L., Wang Y., Pei X., Yang P., Xie X.S., Cao L., Cao Y., Wang X. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M.A., Morden E., Rosseau P., Arendse J., Bam J.L., Boloko L., Cloete K., Cohen C., Chetty N., Dane P., Heekes A., Hsiao N.Y., Hunter M., Hussey H., Jacobs T., Jassat W., Kariem S., Kassanjee R., Laenen I., Le Roux S., Lessells R., Mahomed H., Maughan D., Meintjes G., Mendelson M., Mnguni A., Moodley M., Murie K., Naude J., Ntusi N., Paleker M., Parker A., Pienaar D., Preiser W., Prozesky H., Raubenheimer P., Rossouw L., Schrueder N., Smith B., Smith M., Solomon W., Symons G., Taljaard J., Wasserman S., Wilkinson R.J., Wolmarans M., Wolter N., Boulle A. Int J Infect Dis; 2022. Outcomes of laboratory-confirmed SARS-CoV-2 infection during resurgence driven by Omicron lineages BA.4 and BA.5 compared with previous waves in the Western Cape Province, South Africa. S1201-9712(22)00615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., Nutalai R., Wang B., Dijokaite A., Khan S., Avinoam O., Bahar M., Skelly D., Adele S., Johnson S.A., Amini A., Ritter T.G., Mason C., Dold C., Pan D., Assadi S., Bellass A., Omo-Dare N., Koeckerling D., Flaxman A., Jenkin D., Aley P.K., Voysey M., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes Da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Baillie V., Serafin N., Kwatra G., Da Silva K., Madhi S.A., Nunes M.C., Malik T., Openshaw P.J.M., Baillie J.K., Semple M.G., Townsend A.R., Huang K.A., Tan T.K., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Constantinides B., Webster H., Crook D., Pollard A.J., Lambe T., Paterson N.G., Williams M.A., Hall D.R., Fry E.E., Mongkolsapaya J., Ren J., Schreiber G., Stuart D.I., Screaton G.R., Conlon C., Deeks A.S., Frater J., Frending L., Gardiner S., Jämsén A., Jeffery K., Malone T., Phillips E., Rothwell L., Stafford L., Baillie J.K., Semple M.G., Openshaw P.J., Carson G., Alex B., Andrikopoulos P., Bach B., Barclay W.S., Bogaert D., Chand M., Chechi K., Cooke G.S., Da Silva Filipe A., de Silva T., Docherty A.B., Dos Santos Correia G., Dumas M., Dunning J., Fletcher T., Green C.A., Greenhalf W., Griffin J.L., Gupta R.K., Harrison E.M., Hiscox J.A., Wai Ho A.Y., Horby P.W., Ijaz S., Khoo S., Klenerman P., Law A., Lewis M.R., Liggi S., Lim W.S., Maslen L., Mentzer A.J., Merson L., Meynert A.M., Moore S.C., Noursadeghi M., Olanipekun M., Osagie A., Palmarini M., Palmieri C., Paxton W.A., Pollakis G., Price N., Rambaut A., Robertson D.L., Russell C.D., Sancho-Shimizu V., Sands C.J., Scott J.T., Sigfrid L., Solomon T., Sriskandan S., Stuart D., Summers C., Swann O.V., Takats Z., Takis P., Tedder R.S., Thompson A.R., Thomson E.C., Thwaites R.S., Turtle L.C., Zambon M., Hardwick H., Donohue C., Griffiths F., Oosthuyzen W., Donegan C., Spencer R.G., Norman L., Pius R., Drake T.M., Fairfield C.J., Knight S.R., Mclean K.A., Murphy D., Shaw C.A., Dalton J., Girvan M., Saviciute E., Roberts S., Harrison J., Marsh L., Connor M., Halpin S., Jackson C., Gamble C., Plotkin D., Lee J., Leeming G., Law A., Wham M., Clohisey S., Hendry R., Scott-Brown J., Shaw V., McDonald S.E., Keating S., Ahmed K.A., Armstrong J.A., Ashworth M., Asiimwe I.G., Bakshi S., Barlow S.L., Booth L., Brennan B., Bullock K., Catterall B.W., Clark J.J., Clarke E.A., Cole S., Cooper L., Cox H., Davis C., Dincarslan O., Dunn C., Dyer P., Elliott A., Evans A., Finch L., Fisher L.W., Foster T., Garcia-Dorival I., Gunning P., Hartley C., Jensen R.L., Jones C.B., Jones T.R., Khandaker S., King K., Kiy R.T., Koukorava C., Lake A., Lant S., Latawiec D., Lavelle-Langham L., Lefteri D., Lett L., Livoti L.A., Mancini M., McDonald S., McEvoy L., McLauchlan J., Metelmann S., Miah N.S., Middleton J., Mitchell J., Moore S.C., Murphy E.G., Penrice-Randal R., Pilgrim J., Prince T., Reynolds W., Ridley P.M., Sales D., Shaw V.E., Shears R.K., Small B., Subramaniam K.S., Szemiel A., Taggart A., Tanianis-Hughes J., Thomas J., Trochu E., van Tonder L., Wilcock E., Zhang J.E., Flaherty L., Maziere N., Cass E., Carracedo A.D., Carlucci N., Holmes A., Massey H., Murphy L., McCafferty S., Clark R., Fawkes A., Morrice K., Maclean A., Wrobel N., Donnelly L., Coutts A., Hafezi K., MacGillivray L., Gilchrist T., Adeniji K., Agranoff D., Agwuh K., Ail D., Aldera E.L., Alegria A., Allen S., Angus B., Ashish A., Atkinson D., Bari S., Barlow G., Barnass S., Barrett N., Bassford C., Basude S., Baxter D., Beadsworth M., Bernatoniene J., Berridge J., Berry C., Best N., Bothma P., Chadwick D., Brittain-Long R., Bulteel N., Burden T., Burtenshaw A., Caruth V., Chadwick D., Chambler D., Chee N., Child J., Chukkambotla S., Clark T., Collini P., Cosgrove C., Cupitt J., Cutino-Moguel M., Dark P., Dawson C., Dervisevic S., Donnison P., Douthwaite S., Drummond A., DuRand I., Dushianthan A., Dyer T., Evans C., Eziefula C., Fegan C., Finn A., Fullerton D., Garg S., Garg S., Garg A., Gkrania-Klotsas E., Godden J., Goldsmith A., Graham C., Hardy E., Hartshorn S., Harvey D., Havalda P., Hawcutt D.B., Hobrok M., Hodgson L., Hormis A., Jacobs M., Jain S., Jennings P., Kaliappan A., Kasipandian V., Kegg S., Kelsey M., Kendall J., Kerrison C., Kerslake I., Koch O., Koduri G., Koshy G., Laha S., Laird S., Larkin S., Leiner T., Lillie P., Limb J., Linnett V., Little J., Lyttle M., MacMahon M., MacNaughton E., Mankregod R., Masson H., Matovu E., McCullough K., McEwen R., Meda M., Mills G., Minton J., Mirfenderesky M., Mohandas K., Mok Q., Moon J., Moore E., Morgan P., Morris C., Mortimore K., Moses S., Mpenge M., Mulla R., Murphy M., Nagel M., Nagarajan T., Nelson M., Norris L., O Shea M.K., Otahal I., Ostermann M., Pais M., Palmieri C., Panchatsharam S., Papakonstantinou D., Paraiso H., Patel B., Pattison N., Pepperell J., Peters M., Phull M., Pintus S., Pooni J.S., Planche T., Post F., Price D., Prout R., Rae N., Reschreiter H., Reynolds T., Richardson N., Roberts M., Roberts D., Rose A., Rousseau G., Ruge B., Ryan B., Saluja T., Schmid M.L., Shah A., Shanmuga P., Sharma A., Shawcross A., Sizer J., Shankar-Hari M., Smith R., Snelson C., Spittle N., Staines N., Stambach T., Stewart R., Subudhi P., Szakmany T., Tatham K., Thomas J., Thompson C., Thompson R., Tridente A., Tupper-Carey D., Twagira M., Vallotton N., Vancheeswaran R., Vincent-Smith L., Visuvanathan S., Vuylsteke A., Waddy S., Wake R., Walden A., Welters I., Whitehouse T., Whittaker P., Whittington A., Papineni P., Wijesinghe M., Williams M., Wilson L., Cole S., Winchester S., Wiselka M., Wolverson A., Wootton D.G., Workman A., Yates B., Young P. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desingu P.A., Nagarajan K., Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J. Med. Virol. 2022;94:1808–1810. doi: 10.1002/jmv.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Zhou J., Yang S., Wang L., Chen X., Yang Y., Li R., Pan Z., Zhao J., Li Z., Huang Q., Tang J., Hu L., Liu P., Zhang G., Chen Y., Ye L. The dichotomous and incomplete adaptive immunity in COVID-19 patients with different disease severity. Signal Transduct. Targeted Ther. 2021;6:113. doi: 10.1038/s41392-021-00525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., Feldman J., Roederer A.L., Gregory D.J., Poznansky M.C., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., Scherbeijn S., Gommers L., Sablerolles R.S.G., Nieuwkoop N.N., Rijsbergen L.C., van Dijk L.L.A., de Wilde J., Alblas K., Breugem T.I., Rijnders B.J.A., de Jager H., Weiskopf D., van der Kuy P.H.M., Sette A., Koopmans M.P.G., Grifoni A., Haagmans B.L., de Vries R.D. Divergent SARS-CoV-2 Omicron–reactive T and B cell responses in COVID-19 vaccine recipients. Science Immunology. 2022;7 doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Lundgreen K.A., Apostolidis S.A., Baxter A.E., Giles J.R., Mathew D., Pattekar A., Reynaldi A., Khoury D.S., Gouma S., Hicks P., Dysinger S., Hicks A., Sharma H., Herring S., Korte S., Kc W., Oldridge D.A., Erickson R.I., Weirick M.E., McAllister C.M., Awofolaju M., Tanenbaum N., Dougherty J., Long S., D Andrea K., Hamilton J.T., McLaughlin M., Williams J.C., Adamski S., Kuthuru O., Drapeau E.M., Davenport M.P., Hensley S.E., Bates P., Greenplate A.R., Wherry E.J. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022;185:1875–1887. doi: 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski F., Kochańczyk M., Lipniacki T. The spread of SARS-CoV-2 variant omicron with a doubling time of 2.0–3.3 Days can Be explained by immune evasion. Viruses. 2022;14:294. doi: 10.3390/v14020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., Foster S.L., Ying B., Case J.B., Chong Z., Whitener B., Moliva J., Floyd K., Ujie M., Nakajima N., Ito M., Wright R., Uraki R., Warang P., Gagne M., Li R., Sakai-Tagawa Y., Liu Y., Larson D., Osorio J.E., Hernandez-Ortiz J.P., Henry A.R., Ciuoderis K., Florek K.R., Patel M., Odle A., Wong L.R., Bateman A.C., Wang Z., Edara V., Chong Z., Franks J., Jeevan T., Fabrizio T., DeBeauchamp J., Kercher L., Seiler P., Gonzalez-Reiche A.S., Sordillo E.M., Chang L.A., van Bakel H., Simon V., Alburquerque B., Alshammary H., Amoako A.A., Aslam S., Banu R., Cognigni C., Espinoza-Moraga M., Farrugia K., van de Guchte A., Khalil Z., Laporte M., Mena I., Paniz-Mondolfi A.E., Polanco J., Rooker A., Sominsky L.A., Douek D.C., Sullivan N.J., Thackray L.B., Ueki H., Yamayoshi S., Imai M., Perlman S., Webby R.J., Seder R.A., Suthar M.S., García-Sastre A., Schotsaert M., Suzuki T., Boon A.C.M., Diamond M.S., Kawaoka Y. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Zhu S., Fu D., Xiao J., Zhao J., Lin Z., Liu T., Liang X., Ma W. SingaporeAssociation between COVID-19 Vaccination Coverage and Case Fatality Ratio:a Comparative Study - Hong Kong SAR, China and Singapore, December 2021-March 2022. China CDC Wkly. 2022;4:649–654. doi: 10.46234/ccdcw2022.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A., Winkler M.S., Lier M., Dopfer-Jablonka A., Jäck H., Behrens G.M.N., Pöhlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., Chik K.K.H., Yuen T.T.T., Yin M.T., Sobieszczyk M.E., Huang Y., Yuen K., Wang H.H., Sheng Z., Ho D.D. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., Madzorera S.V., Moyo-Gwete T., Mennen M., Skelem S., Adriaanse M., Mutithu D., Aremu O., Stek C., du Bruyn E., Van Der Mescht M.A., de Beer Z., de Villiers T.R., Bodenstein A., van den Berg G., Mendes A., Strydom A., Venter M., Giandhari J., Naidoo Y., Pillay S., Tegally H., Grifoni A., Weiskopf D., Sette A., Wilkinson R.J., de Oliveira T., Bekker L., Gray G., Ueckermann V., Rossouw T., Boswell M.T., Bhiman J.N., Moore P.L., Sigal A., Ntusi N.A.B., Burgers W.A., Riou C. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., Samsunder N., Sivro A., San J.E., Giandhari J., Tegally H., Pillay S., Naidoo Y., Mazibuko M., Miya Y., Ngcobo N., Manickchund N., Magula N., Karim Q.A., von Gottberg A., Abdool K.S., Hanekom W., Gosnell B.I., Lessells R.J., de Oliveira T., Moosa M.S., Sigal A. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13:4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Zhou J.Q., Horvath S.C., Schmitz A.J., Sturtz A.J., Lei T., Liu Z., Kalaidina E., Thapa M., Alsoussi W.B., Haile A., Klebert M.K., Suessen T., Parra-Rodriguez L., Mudd P.A., Whelan S.P.J., Middleton W.D., Teefey S.A., Pusic I., O Halloran J.A., Presti R.M., Turner J.S., Ellebedy A.H. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Yamasoba D., Tamura T., Nao N., Oda Y., Mitoma S., Ito J., Nasser H., Zahradnik J., Uriu K., Fujita S., Kosugi Y., Wang L., Tsuda M., Kishimoto M., Ito H., Suzuki R., Shimizu R., Begum M.M., Yoshimatsu K., Sasaki J., Sasaki-Tabata K., Yamamoto Y., Nagamoto T., Kanamune J., Kobiyama K., Asakura H., Nagashima M., Sadamasu K., Yoshimura K., Kuramochi J., Schreiber G., Ishii K.J., Hashiguchi T., Ikeda T., Saito A., Fukuhara T., Tanaka S., Matsuno K., Sato K. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA. subvariants, including BA.4 and BA.5. Cell. 2022;185 doi: 10.1016/j.cell.2022.09.018. 3992–4007.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki R., Adachi Y., Moriyama S., Onodera T., Fukushi S., Nagakura T., Tonouchi K., Terahara K., Sun L., Takano T., Nishiyama A., Shinkai M., Oba K., Nakamura-Uchiyama F., Shimizu H., Suzuki T., Matsumura T., Isogawa M., Takahashi Y. SARS-CoV-2 Omicron-neutralizing memory B cells are elicited by two doses of BNT162b2 mRNA vaccine. Science Immunology. 2022;7 doi: 10.1126/sciimmunol.abn8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94:1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- Lambrou A.S., Shirk P., Steele M.K., Paul P., Paden C.R., Cadwell B., Reese H.E., Aoki Y., Hassell N., Zheng X., Talarico S., Chen J.C., Oberste M.S., Batra D., McMullan L.K., Halpin A.L., Galloway S.E., MacCannell D.R., Kondor R., Barnes J., MacNeil A., Silk B.J., Dugan V.G., Scobie H.M., Wentworth D.E. Genomic surveillance for SARS-CoV-2 variants: predominance of the delta (B.1.617.2) and omicron (B.1.1.529) variants — United States, june 2021–january 2022. MMWR. Morbidity and Mortality Weekly Report. 2022;71:206–211. doi: 10.15585/mmwr.mm7106a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat. Med. 2022;28:1933–1943. doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Rocklöv J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J. Trav. Med. 2022;29 doi: 10.1093/jtm/taac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., Yu J., Collier A.Y., Barouch D.H. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K.H., Yuen T.T.T., Yoon C., To K.K.W., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K., Ho D.D. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Lu L., Mok B.W., Chen L., Chan J.M., Tsang O.T., Lam B.H., Chuang V.W., Chu A.W., Chan W., Ip J.D., Chan B.P., Zhang R., Yip C.C., Cheng V.C., Chan K., Jin D., Hung I.F., Yuen K., Chen H., To K.K. Neutralization of severe acute respiratory syndrome coronavirus 2 omicron variant by sera from BNT162b2 or CoronaVac vaccine recipients. Clin. Infect. Dis. 2021;75:e822–e826. doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Pollett S.D., Neerukonda S.N., Wang W., Wang R., Vassell R., Epsi N.J., Fries A.C., Agan B.K., Lindholm D.A., Colombo C.J., Mody R., Ewers E.C., Lalani T., Ganesan A., Goguet E., Hollis-Perry M., Coggins S., Simons M.P., Katzelnick L.C., Wang G., Tribble D.R., Bentley L., Eakin A.E., Broder C.C., Erlandson K.J., Laing E.D., Burgess T.H., Mitre E., Weiss C.D. bioRxiv; 2021. SARS-CoV-2 Omicron Neutralization by Therapeutic Antibodies, Convalescent Sera, and Post-mRNA Vaccine Booster. [Google Scholar]
- Lyke K.E., Atmar R.L., Islas C.D., Posavad C.M., Szydlo D., Paul C.R., Deming M.E., Eaton A., Jackson L.A., Branche A.R., El S.H., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., Mulligan M.J., Brady R.C., Frenck R.J., Backer M., Kottkamp A.C., Babu T.M., Rajakumar K., Edupuganti S., Dobrzynski D., Coler R.N., Archer J.I., Crandon S., Zemanek J.A., Brown E.R., Neuzil K.M., Stephens D.S., Post D.J., Nayak S.U., Suthar M.S., Roberts P.C., Beigel J.H., Montefiori D.C. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep Med. 2022;3 doi: 10.1016/j.xcrm.2022.100679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Chen X., Mei F., Xiong Q., Liu Q., Dong L., Liu C., Zou W., Zhan F., Hu B., Liu Y., Liu F., Zhou L., Xu J., Jiang Y., Xu K., Cai K., Chen Y., Yan H., Lan K. Drastic decline in sera neutralization against SARS-CoV-2 Omicron variant in Wuhan COVID-19 convalescents. Emerg. Microb. Infect. 2022;11:567–572. doi: 10.1080/22221751.2022.2031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni A., Vanni A., Spinicci M., Capone M., Lamacchia G., Salvati L., Coppi M., Antonelli A., Carnasciali A., Farahvachi P., Giovacchini N., Aiezza N., Malentacchi F., Zammarchi L., Liotta F., Rossolini G.M., Bartoloni A., Cosmi L., Maggi L., Annunziato F. SARS-CoV-2 spike-specific CD4+ T cell response is conserved against variants of concern, including omicron. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.801431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Czudnochowski N., Rosen L.E., Zepeda S.K., Bowen J.E., Walls A.C., Hauser K., Joshi A., Stewart C., Dillen J.R., Powell A.E., Croll T.I., Nix J., Virgin H.W., Corti D., Snell G., Veesler D. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375:864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., Zepeda S.K., Papa G., Kemp S.A., Ikeda T., Toyoda M., Tan T.S., Kuramochi J., Mitsunaga S., Ueno T., Shirakawa K., Takaori-Kondo A., Brevini T., Mallery D.L., Charles O.J., Baker S., Dougan G., Hess C., Kingston N., Lehner P.J., Lyons P.A., Matheson N.J., Ouwehand W.H., Saunders C., Summers C., Thaventhiran J.E.D., Toshner M., Weekes M.P., Maxwell P., Shaw A., Bucke A., Calder J., Canna L., Domingo J., Elmer A., Fuller S., Harris J., Hewitt S., Kennet J., Jose S., Kourampa J., Meadows A., O Brien C., Price J., Publico C., Rastall R., Ribeiro C., Rowlands J., Ruffolo V., Tordesillas H., Bullman B., Dunmore B.J., Gräf S., Hodgson J., Huang C., Hunter K., Jones E., Legchenko E., Matara C., Martin J., Mescia F., O Donnell C., Pointon L., Shih J., Sutcliffe R., Tilly T., Treacy C., Tong Z., Wood J., Wylot M., Betancourt A., Bower G., Cossetti C., De Sa A., Epping M., Fawke S., Gleadall N., Grenfell R., Hinch A., Jackson S., Jarvis I., Krishna B., Nice F., Omarjee O., Perera M., Potts M., Richoz N., Romashova V., Stefanucci L., Strezlecki M., Turner L., De Bie E.M.D.D., Bunclark K., Josipovic M., Mackay M., Butcher H., Caputo D., Chandler M., Chinnery P., Clapham-Riley D., Dewhurst E., Fernandez C., Furlong A., Graves B., Gray J., Hein S., Ivers T., Le Gresley E., Linger R., Kasanicki M., King R., Kingston N., Meloy S., Moulton A., Muldoon F., Ovington N., Papadia S., Penkett C.J., Phelan I., Ranganath V., Paraschiv R., Sage A., Sambrook J., Scholtes I., Schon K., Stark H., Stirrups K.E., Townsend P., Walker N., Webster J., Butlertanaka E.P., Tanaka Y.L., Ito J., Uriu K., Kosugi Y., Suganami M., Oide A., Yokoyama M., Chiba M., Motozono C., Nasser H., Shimizu R., Kitazato K., Hasebe H., Irie T., Nakagawa S., Wu J., Takahashi M., Fukuhara T., Shimizu K., Tsushima K., Kubo H., Kazuma Y., Nomura R., Horisawa Y., Nagata K., Kawai Y., Yanagida Y., Tashiro Y., Tokunaga K., Ozono S., Kawabata R., Morizako N., Sadamasu K., Asakura H., Nagashima M., Yoshimura K., Cárdenas P., Muñoz E., Barragan V., Márquez S., Prado-Vivar B., Becerra-Wong M., Caravajal M., Trueba G., Rojas-Silva P., Grunauer M., Gutierrez B., Guadalupe J.J., Fernández-Cadena J.C., Andrade-Molina D., Baldeon M., Pinos A., Bowen J.E., Joshi A., Walls A.C., Jackson L., Martin D., Smith K.G.C., Bradley J., Briggs J.A.G., Choi J., Madissoon E., Meyer K.B., Mlcochova P., Ceron-Gutierrez L., Doffinger R., Teichmann S.A., Fisher A.J., Pizzuto M.S., de Marco A., Corti D., Hosmillo M., Lee J.H., James L.C., Thukral L., Veesler D., Sigal A., Sampaziotis F., Goodfellow I.G., Matheson N.J., Sato K., Gupta R.K. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo S.A., Meo A.S., Al-Jassir F.F., Klonoff D.C. Omicron SARS-CoV-2 new variant: global prevalence and biological and clinical characteristics. Eur. Rev. Med. Pharmacol. Sci. 2021;25:8012–8018. doi: 10.26355/eurrev_202112_27652. [DOI] [PubMed] [Google Scholar]
- Muecksch F., Wang Z., Cho A., Gaebler C., Ben Tanfous T., DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., Raspe R., Schaefer-Babajew D., Shimeliovich I., Daga M., Yao K., Schmidt F., Millard K.G., Turroja M., Jankovic M., Oliveira T.Y., Gazumyan A., Caskey M., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Lui B.G., Wallisch A., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., Güimil Garcia R.D.L.C., Poran A., Shpyro S., Finlayson A., Cai H., Yang Q., Swanson K.A., Türeci Ö., Şahin U. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine–elicited human sera. Science. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranbhai V., Nathan A., Kaseke C., Berrios C., Khatri A., Choi S., Getz M.A., Tano-Menka R., Ofoman O., Gayton A., Senjobe F., Zhao Z., St Denis K.J., Lam E.C., Carrington M., Garcia-Beltran W.F., Balazs A.B., Walker B.D., Iafrate A.J., Gaiha G.D. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041–1051. doi: 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Kliker L., Lustig Y., Zuckerman N., Erster O., Cohen C., Kreiss Y., Alroy-Preis S., Regev-Yochay G., Mendelson E., Mandelboim M. Third BNT162b2 vaccination neutralization of SARS-CoV-2 omicron infection. N. Engl. J. Med. 2022;386:492–494. doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Ito K., Anzai A., Kobayashi T., Piantham C., Rodríguez-Morales A.J. Relative reproduction number of SARS-CoV-2 omicron (B.1.1.529) compared with delta variant in South Africa. J. Clin. Med. 2022;11:30. doi: 10.3390/jcm11010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom P., Ballin M., Nordstrom A. Effectiveness of a Fourth Dose of mRNA COVID-19 Vaccine against All-Cause Mortality in Long-Term Care Facility Residents and in the Oldest Old: A Nationwide, Retrospective Cohort Study in Sweden. Lancet Reg Health Eur. 2022;21:100466. doi: 10.1016/j.lanepe.2022.100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., Hinsley W., Bernal J.L., Kall M., Bhatt S., Blomquist P., Zaidi A., Volz E., Aziz N.A., Harman K., Funk S., Abbott S., Hope R., Charlett A., Chand M., Ghani A.C., Seaman S.R., Dabrera G., De Angelis D., Presanis A.M., Thelwall S., Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., Hinsley W., Lopez Bernal J., Kall M., Bhatt S., Blomquist P., Zaidi A., Volz E., Abdul Aziz N., Harman K., Funk S., Abbott S., Hope R., Charlett A., Chand M., Ghani A.C., Seaman S.R., Dabrera G., De Angelis D., Presanis A.M., Thelwall S. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399:1303–1312. doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeyer F., Jankowiak M., Barkas N., Schaffner S.F., Pyle J.D., Yurkovetskiy L., Bosso M., Park D.J., Babadi M., MacInnis B.L., Luban J., Sabeti P.C., Lemieux J.E. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science. 2022;376:1327–1332. doi: 10.1126/science.abm1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W., Porrot F., Staropoli I., Lemoine F., Péré H., Veyer D., Puech J., Rodary J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina-Bokalanga T., Martí-Carreras J., Cuypers L., Sève A., Hocqueloux L., Prazuck T., Rey F.A., Simon-Loriere E., Bruel T., Mouquet H., André E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Qu P., Evans J.P., Zheng Y.M., Carlin C., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.L. Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.09.015. 1518–1526.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P., Faraone J.N., Evans J.P., Zheng Y.M., Carlin C., Lozanski G., Saif L.J., Oltz E.M., Gumina R.J., Liu S.L. 2022. Durability of the Neutralizing Antibody Response to mRNA Booster Vaccination against SARS-CoV-2 BA.2.12.1 and BA.4/5 Variants. bioRxiv, 2022.07.21.501010. [Google Scholar]
- Qu P., Faraone J.N., Evans J.P., Zou X., Zheng Y., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., Oltz E.M., Mohler P.J., Gumina R.J., Liu S. Differential Evasion of Delta and Omicron Immunity and Enhanced Fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. bioRxiv. 2022 [Google Scholar]
- Rahimi F., Talebi Bezmin Abadi A. The Omicron subvariant BA.2: birth of a new challenge during the COVID-19 pandemic. Int. J. Surg. 2022;99 doi: 10.1016/j.ijsu.2022.106261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Yochay G., Gonen T., Gilboa M., Mandelboim M., Indenbaum V., Amit S., Meltzer L., Asraf K., Cohen C., Fluss R., Biber A., Nemet I., Kliker L., Joseph G., Doolman R., Mendelson E., Freedman L.S., Harats D., Kreiss Y., Lustig Y. Efficacy of a fourth dose of covid-19 mRNA vaccine against omicron. N. Engl. J. Med. 2022;386:1377–1380. doi: 10.1056/NEJMc2202542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Wang W., Gao R., Zhou A. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases. 2022;10:1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent persons. N. Engl. J. Med. 2022;386:698–700. doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Kimura I., Ito J., Nasser H., Toyoda M., Nagata K., Uriu K., Kosugi Y., Fujita S., Yamasoba D., Shofa M., Begum M.M., Oda Y., Suzuki R., Ito H., Nao N., Wang L., Tsuda M., Yoshimatsu K., Yamamoto Y., Nagamoto T., Asakura H., Nagashima M., Sadamasu K., Yoshimura K., Ueno T., Schreiber G., Takaori-Kondo A., Shirakawa K., Sawa H., Irie T., Takayama K., Matsuno K., Tanaka S., Ikeda T., Fukuhara T., Sato K. Virological characteristics of the SARS-CoV-2 omicron BA.2.75. bioRxiv. 2022:2022–2028. doi: 10.1016/j.chom.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S.R., Hoffmann M., Roth E., Pracht K., Burnett D.L., Mazigi O., Schuh W., Manger B., Mielenz D., Goodnow C.C., Christ D., Pöhlmann S., Jäck H.M. Augmented neutralization of SARS-CoV-2 Omicron variant by boost vaccination and monoclonal antibodies. Eur. J. Immunol. 2022;52:970–977. doi: 10.1002/eji.202249841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen N., Mohamed A., Soliman Y., Abdelwahab O.A., Diab R.A., Desouki M.T., Rababah A.A., Khaity A., Hefnawy M.T., Swed S., Shaheen A., Elfakharany B., Shoib S. Could the new BA.2.75 sub-variant lead to another COVID-19 wave in the world? - Correspondence. Int. J. Surg. 2022;105 doi: 10.1016/j.ijsu.2022.106861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Ehling R.A., Pankow A., Castro Dopico X., Dyrdak R., Martin D.P., Reddy S.T., Dillner J., Karlsson Hedestam G.B., Albert J., Murrell B. Neutralisation sensitivity of the SARS-CoV-2 omicron (B.1.1.529) variant: a cross-sectional study. Lancet Infect. Dis. 2022;22:813–820. doi: 10.1016/S1473-3099(22)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., Huang X., Yoon C., Hu J., Liu H., Shi J., Liu Y., Zhu T., Zhang J., Hou Y., Wang Y., Lu L., Cai J., Zhang A.J., Zhou J., Yuan S., Brindley M.A., Zhang B., Huang J., To K.K., Yuen K., Chu H. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- Sokal A., Broketa M., Barba-Spaeth G., Meola A., Fernández I., Fourati S., Azzaoui I., de La Selle A., Vandenberghe A., Roeser A., Bouvier-Alias M., Crickx E., Languille L., Michel M., Godeau B., Gallien S., Melica G., Nguyen Y., Zarrouk V., Canoui-Poitrine F., Noizat-Pirenne F., Megret J., Pawlotsky J., Fillatreau S., Simon-Lorière E., Weill J., Reynaud C., Rey F.A., Bruhns P., Chappert P., Mahévas M. Analysis of mRNA vaccination-elicited RBD-specific memory B cells reveals strong but incomplete immune escape of the SARS-CoV-2 Omicron variant. Immunity. 2022;55:1096–1104. doi: 10.1016/j.immuni.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano A., Takaoka Y., Kataguchi H., Ohta M., Kimura S., Araki M., Morinaga Y., Yamamoto Y. SARS-CoV-2 Omicron BA.2.75 Variant May Be Much More Infective than Preexisting Variants Based on In Silico Model. Microorganisms. 2022;10:2090. doi: 10.3390/microorganisms10102090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F., Feng Z., Gao G.F. China CDC Weekly: A Trusted Resource on Public Health. China CDC Wkly. 2020;2:933–934. doi: 10.46234/ccdcw2020.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Lim B.L., Young B.E., Yeoh A.Y., Yung C.F., Yap W.C., Althaus T., Chia W.N., Zhu F., Lye D.C., Wang L.F. Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet Microbe. 2022;22:S2666–5247. doi: 10.1016/S2666-5247(22)00220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., Sidney J., Filaci G., Weiskopf D., Da Silva Antunes R., Crotty S., Grifoni A., Sette A. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.Y., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenco J., Alcantara L., Kosakovsky P.S., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., Lee G.S., Choi S.K., Takhar H.S., Aragones M., Qian L. Effectiveness of mRNA-1273 against SARS-CoV-2 omicron and delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., Das R., Skelly D., Ritter T.G., Amini A., Bibi S., Adele S., Johnson S.A., Constantinides B., Webster H., Temperton N., Klenerman P., Barnes E., Dunachie S.J., Crook D., Pollard A.J., Lambe T., Goulder P., Paterson N.G., Williams M.A., Hall D.R., Fry E.E., Huo J., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R., Conlon C., Deeks A., Frater J., Frending L., Gardiner S., Jämsén A., Jeffery K., Malone T., Phillips E., Rothwell L., Stafford L. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R., Kiso M., Iida S., Imai M., Takashita E., Kuroda M., Halfmann P.J., Loeber S., Maemura T., Yamayoshi S., Fujisaki S., Wang Z., Ito M., Ujie M., Iwatsuki-Horimoto K., Furusawa Y., Wright R., Chong Z., Ozono S., Yasuhara A., Ueki H., Sakai-Tagawa Y., Li R., Liu Y., Larson D., Koga M., Tsutsumi T., Adachi E., Saito M., Yamamoto S., Hagihara M., Mitamura K., Sato T., Hojo M., Hattori S., Maeda K., Valdez R., Bennett-Baker P., Chu Z., Davis D., Kowalski-Dobson T., Eckard A., Gherasim C., Gremel W., Lindsey K., Manthei D., Meyers A., Moya J.Z., Rico A., Stoneman E., Blanc V., Sneeringer S., Warsinske L., Okuda M., Murakami J., Duong C., Godbole S., Douek D.C., Maeda K., Watanabe S., Gordon A., Ohmagari N., Yotsuyanagi H., Diamond M.S., Hasegawa H., Mitsuya H., Suzuki T., Kawaoka Y. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022;607:119–127. doi: 10.1038/s41586-022-04856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., Choga W.T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S.L., Kraemer M.U.G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R.J., Lockman S., Lucaci A.G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M.B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M.S., Mphoyakgosi T., Msomi N., Mwangi P.N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J.E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., van Wyk S., Weaver S., Wibmer C.K., Wilkinson E., Wolter N., Zarebski A.E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O.G., Bhiman J., Glass A., Martin D.P., Rambaut A., Gaseitsiwe S., von Gottberg A., de Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Deng C., Liu M., Liu Y., Li L., Huang Z., Shang L., Jiang J., Li Y., Mo R., Zhang H., Liu M., Peng S., Xiao H. Four doses of the inactivated SARS-CoV-2 vaccine redistribute humoral immune responses away from the Receptor Binding Domain. medRxiv. 2022 [Google Scholar]
- Wang L., Berger N.A., Kaelber D.C., Davis P.B., Volkow N.D., Xu R. Incidence rates and clinical outcomes of SARS-CoV-2 infection with the omicron and delta variants in children younger than 5 Years in the US. JAMA Pediatr. 2022;176:811. doi: 10.1001/jamapediatrics.2022.0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., Shah J.G., Nguyen N., Chen Z., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D.D. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization e4. of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022;30:1512–1517. doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., Sauder J.M., Kraft L., Hwang Y., Siegel R.W., Chen J., Heinz B.A., Higgs R.E., Kallewaard N.L., Jepson K., Goya R., Smith M.A., Collins D.W., Pellacani D., Xiang P., de Puyraimond V., Ricicova M., Devorkin L., Pritchard C., O Neill A., Dalal K., Panwar P., Dhupar H., Garces F.A., Cohen C.A., Dye J.M., Huie K.E., Badger C.V., Kobasa D., Audet J., Freitas J.J., Hassanali S., Hughes I., Munoz L., Palma H.C., Ramamurthy B., Cross R.W., Geisbert T.W., Menachery V., Lokugamage K., Borisevich V., Lanz I., Anderson L., Sipahimalani P., Corbett K.S., Yang E.S., Zhang Y., Shi W., Zhou T., Choe M., Misasi J., Kwong P.D., Sullivan N.J., Graham B.S., Fernandez T.L., Hansen C.L., Falconer E., Mascola J.R., Jones B.E., Barnhart B.C. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter N., Jassat W., von Gottberg A., Cohen C. Clinical severity of omicron lineage BA.2 infection compared with BA.1 infection in South Africa. Lancet. 2022;400:93–96. doi: 10.1016/S0140-6736(22)00981-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratil P.R., Stern M., Priller A., Willmann A., Almanzar G., Vogel E., Feuerherd M., Cheng C., Yazici S., Christa C., Jeske S., Lupoli G., Vogt T., Albanese M., Mejías-Pérez E., Bauernfried S., Graf N., Mijocevic H., Vu M., Tinnefeld K., Wettengel J., Hoffmann D., Muenchhoff M., Daechert C., Mairhofer H., Krebs S., Fingerle V., Graf A., Steininger P., Blum H., Hornung V., Liebl B., Überla K., Prelog M., Knolle P., Keppler O.T., Protzer U. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat. Med. 2022;28:496–503. doi: 10.1038/s41591-022-01715-4. [DOI] [PubMed] [Google Scholar]
- Yin W., Xu Y., Xu P., Cao X., Wu C., Gu C., He X., Wang X., Huang S., Yuan Q., Wu K., Hu W., Huang Z., Liu J., Wang Z., Jia F., Xia K., Liu P., Wang X., Song B., Zheng J., Jiang H., Cheng X., Jiang Y., Deng S., Xu H.E. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science. 2022;375:1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., Hachmann N.P., Surve N., Nampanya F., Chandrashekar A., Barouch D.H. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappa M., Verdecchia P., Angeli F. Knowing the new Omicron BA.2.75 variant (’Centaurus’): a simulation study. Eur. J. Intern. Med. 2022;105:107–108. doi: 10.1016/j.ejim.2022.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu S., Wu B., Yang Q., Chen A., Li Y., Zhang Y., Pan T., Zhang H., He X. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct. Targeted Ther. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li Q., Liang Z., Li T., Liu S., Cui Q., Nie J., Wu Q., Qu X., Huang W., Wang Y. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microb. Infect. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang L., Misasi J., Pegu A., Zhang Y., Harris D.R., Olia A.S., Talana C.A., Yang E.S., Chen M., Choe M., Shi W., Teng I.T., Creanga A., Jenkins C., Leung K., Liu T., Stancofski E.D., Stephens T., Zhang B., Tsybovsky Y., Graham B.S., Mascola J.R., Sullivan N.J., Kwong P.D. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science. 2022;376:n8897. doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.