Abstract
The nucleoprotein (NP) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is abundantly expressed during infection, making it a diagnostic target protein. We analyzed the structure of the NP in solution using a recombinant protein produced in E. coli. A codon-optimized Profinity eXact™-tagged NP cDNA was cloned into pET-3d vector and transformed into E. coli T7 Express. The recombinant protein was first purified via chromatographic step using an affinity tag-based system that was followed by tag cleavage with sodium fluoride, resulting in proteolytic removal of the N-terminal tag sequence. The digested sample was then loaded directly onto a size exclusion chromatography run in the presence of L-Arg-HCl, resulting in removal of host nucleic acids and endotoxin. The molecular mass of the main NP fraction was determined by mass photometry as a dimeric form of NP, consistent with the blue native PAGE results. Interestingly, analysis of the purified NP by our newly developed agarose native gel electrophoresis revealed that it behaved like an acidic protein at low concentration despite its alkaline isoelectric point (theoretical pI = 10) and displayed a unique character of concentration-dependent charge and shape changes. This study should shed light into the behavior of NP in the viral life cycle.
Keywords: SARS-CoV-2, Nucleoprotein, Agarose native, Arginine, High level soluble expression
Graphical abstract
1. Introduction
The nucleoprotein (NP) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a dimeric protein and assembles into nucleocapsid that protects the viral nucleic acids [[1], [2], [3]] and is a potential antigenic site of viral detection [4]. It consists of five domains, three of which are intrinsically disordered regions (IDR) [5]. The molecular weight, structure and assembly in solution have been studied by various techniques, including X-ray crystallography [6,7] and NMR [[8], [9], [10]], while the purity and molecular weight of the polypeptide constituting the complex SARS-CoV-2 structure have been studied by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [11]. Different from SDS-PAGE, native gel electrophoresis is a simple high-throughput technology, although not quantitative, for studying the heterogeneity of protein structure and changes in size in solution [12]. It does not require a sophisticated instrument and analytical software. While a native polyacrylamide gel (Native-PAGE) based on Tris-Glycine buffer system is commercially available, it is essentially designed to analyze acidic proteins with isoelectric points below the pH of Tris-Glycine running buffer of 8.3. The basic proteins with a pI above 8.3 may be analyzed by reversing the polarity, which however does not always give reliable data [[13], [14], [15]].
To overcome the shortcomings associated with the Tris-Glycine Native-PAGE, we have developed a native gel electrophoresis using agarose gel and histidine/MES buffer system at pH 6.1 [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]], in which samples are applied in the center position of the horizontal (flat) gel so that the acidic proteins migrate toward the anode and the basic proteins migrate toward the cathode. We then explored its applications for the analysis of protein complexes using various analytical techniques, including blotting, zymography and Ferguson plot [[21], [22], [23], [24]]. We have applied the agarose native gel electrophoresis to study solution properties of highly purified NP expressed in E. coli and analyzed the electrophoresis results along with the solution molecular weight of the purified NP by mass photometry [25], as reported in this paper.
2. Materials and methods
Bovine Serum Albumin (BSA) with molecular weight (MW) of 66,500 and isoelectric point (pI) of 4.7 was given from Proliant Biologicals (Ankeny, IA, USA).
UltraPure agarose was purchased from Thermo-Fisher Scientific (Waltham, MA, USA). Research grade 2-(N-morpholino) ethanesulfonic acid (MES) was purchased from Chemical DOJIN (Kumamoto, Japan). TCEP, GelGreen fluorescence dye, and Quick CBB (Coomassie brilliant blue) PLUS were purchased from FUJIFILM Wako Pure Chemical (Osaka, Japan). l-Histidine (His) was purchased from Nacalai Tesque (Kyoto, Japan).
2.1. Construction of the expression vector
The synthetic gene coding for N-terminal Profinity eXact tagged mature NP was designed on the basis of codon preference in E. coli [26,27]. A codon optimized Profinity eXact tag was added to the 5’ end of the gene coding for the mature NP (Accession number MN908947) (Fig. S1). This synthetic gene was digested with NcoI and BamHI restriction enzymes (TaKaRa Bio, Kusatsu, Japan) and then subcloned into NcoI/BamHI digested pET-3d expression vector (Merck, Rahway, NJ, USA) to produce the pET-3d-eXact-NP that encodes the eXact tag-SARS-CoV-2 NP fusion protein (Fig. S1A).
2.2. Expression and purification of recombinant SARS-CoV-2 nucleoprotein
The construct of pET-3d-eXact-NP was transformed into E. coli T7 Express lysY/Iq (NEB). Positive clones were selected on a Luria-Bertani (LB) agar plate with antibiotics (100 μg/mL ampicillin and 34 μg/mL chloramphenicol). For recombinant fusion protein expression, the selected clones were grown overnight in 10 mL of 2 × YT broth containing 2% glucose and antibiotics (2 × YTG) at 37 °C. The overnight cultures were then diluted 1:50 in fresh 200 mL of 2 × YTG and grown at 37 °C until the OD 600 nm reached approximately 0.6–0.8. Recombinant fusion protein expression was then induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the cells were incubated for 3 h at 30 °C with 125 rpm shaking. To determine the expression level of target protein, the total amounts of cell protein were analyzed on 10–20% SDS-polyacrylamide gel (e-PAGEL, ATTO, Tokyo, Japan) followed by protein staining with commassie brilliant blue (Aproscience, Tokushima, Japan).
The induced cells were harvested by centrifugation at 8,000 g for 3 min at 4 °C. The cell pellets were washed with 20 mM HEPES-NaOH pH 7.1, 0.3 M sodium acetate (binding buffer) and frozen at −80 °C until protein purification. Prior to purification, frozen samples were suspended in 20 mM HEPES-NaOH pH 7.0, 0.3 M sodium acetate, 0.5 M L-Arg, 1 × Bugbuster (Merck), protease inhibitor cocktail (Nacalai Tesque) and sonicated on ice for 10 min by a sonicator, UD-100 (TOMY SEIKO, Tokyo, Japan), until it was completely fragmented. The supernatant was recovered by centrifugation at 15,000 g for 20 min at 4 °C and used for the purification. The expressed fusion proteins were purified by chromatography, the Profinity eXact affinity column (Superflow crosslinked agarose coupled to a unique substilisin mutant, BioRad Laboratories) according to the manufacturer's instructions. Briefly, supernatant was loaded to the column equilibrated with binding buffer and incubated with rotating for 20 min at room temperature (RT). Then the column was extensively washed with the 20 mM HEPES-NaOH pH 7.1, 0.3 M sodium acetate, 0.5 M L-Arg-acetate pH 7.0 and incubated with 20 mM HEPES-NaOH pH 7.1, 0.3 M sodium acetate, 0.5 M L-Arg-acetate pH 7.0, 0.1 M sodium fluoride as elution buffer for 60 min at RT to cleave NP from the prodomain/Profinity eXact fusion tag by immobilized S189 subtilisin BPN. The tag-free recombinant protein was released at RT and analyzed on SDS-PAGE. NP was concentrated by ultrafiltration using Vivaspin Turbo 4 10,000 MWCO (SARTORIUS, Goettingen, Germany). Profinity eXact elution fraction was applied to the Vivaspin Turbo and centrifuged at 7,500 g at RT. Then, concentrated fraction was collected.
2.3. Size exclusion chromatography
NP (2–3 mg/mL, 500 μL) was separated by Superdex 200 increase 10/300 GL column (Cytiva, Marlborough, MA, USA) using AKTA pure 25 system. The column was eluted using the buffer (50 mM Tris-HCl, 150 mM NaCl, 400 mM L-Arg-HCl, pH 7.6) at a flow rate of 0.5 mL/min and 0.5 mL of fractions were collected. Highly purified recombinant protein was analyzed on SDS-PAGE and stained with commassie brilliant blue (CBB) or SilverQuest Silver Staining (Thermo Fisher Scientific).
3. Qualitative and quantitative analysis of NP product
The purified NP was further analyzed on agarose native gel electrophoresis (see below) for DNA/RNA detection using CBB and GelGreen fluorescent dye staining. The fluorescent signals were monitored using ImageQuant LAS500 analyzer (Cytiva). The analytical assay for dsDNA and RNA quantification was done using Qubit dsDNA or RNA HS (High sensitivity) and on the Qubit Fluorometer according to the kit protocols (Thermo Fisher Scientific). UV absorbance spectroscopy of the NP samples was performed using DS-11+ Spectrophotometer (DeNovix, Wilmington, DE).
3.1. Nuclease treatment
The purified NP was 10-diluted with TBS (50 mM Tris-HCl, pH7.6, 0.15 M NaCl) + 5 mM MgCl2 and treated with 0.28 units/μL of Benzonase (Merck, USA) at 25 °C for 1 h. The NP samples (250 ng/lane) before and after nuclease treatment were analyzed by 1% agarose-native gel electrophoresis (see below) for protein and DNA/RNA analysis.
3.2. Mass photometry
Mass photometry (MP) is based on interferometric scattering mass spectrometry (iSCAMS), as a means for detecting single protein particles and measuring their mass or the complexes they form in solution [25]. The MP experiments were carried out on an OneMP instrument (Refeyn, Oxford, UK) calibrated with calibrated with BSA (67 kDa), human IgG (150 kDa), catalase (236 kDa), and urease (552 kDa) at RT, i.e. approximately 20 °C. Microscope coverslips (24 × 50 mm, Fisher Scientific, and 24 × 24 mm, Globe Scientific) were prepared by washing the coverslips consecutively with ethanol and water and then drying under a stream of clean nitrogen. The high concentration protein stocks were diluted with PBS buffer and filtered through 0.22 μm filters. Purified NP (2.5 μg/mL = 55 nM) were incubated after dilution for approximately 10 min at RT before being loaded into flow chambers formed by stacked coverslips: note that such a low protein concentration was necessary to obtain scattering signals from each molecule or complex (or also termed here particle). Data from 3 μm × 10 μm instrument field of view were collected for 100 s at a 1 kHz frame rate. At least 5 × 103 particles (e.g., protein molecule or their complex) were detected in each acquisition. Images were processed using the LabView software provided by the instruments manufacture (Refeyn).
3.3. Agarose native gel electrophoresis
Agarose native gel electrophoresis in horizontal mode was performed using the Mupid-2 Mini-Gel System (Mupid, Tokyo, Japan) as previously reported [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]]. Agarose was dissolved at 1% in hot 0.1 M His/0.1 M MES buffer at pH 6.1 and cast onto a flat bed tray with a comb in the center position. After loading the sample, electrophoresis was run at RT or on ice for 30–45 min under a constant voltage of 100 V: care should be exercised for elevated gel temperature. The agarose native gel electrophoresis with running buffer (25 mM Tris, 192 mM Glycine, pH 8.3) was performed similarly.
3.4. Blue-Native-PAGE
Blue native polyacrylamide gel electrophoresis (BN-PAGE) on a Novex® NativePAGE Bis-Tris gels, 4–16%, 15 well gel (Thermo Fisher Scientific) was performed according to the vendor's manual [28]. The cathode side was filled with 200 mL of 20X NativePAGE Dark Blue cathode buffer diluted 20-fold with deionized water, and the anode side was filled with a total of 200 mL of 10 mL of 20X NativePAGE running buffer and 10 mL of 20X NativePAGE Cathode Additive after diluting them 20-fold with deionized water. Samples were prepared by mixing with Blue-NativePAGE Sample Buffer (4X) (Thermo Fisher Scientific) and run for 120 min under a constant voltage of 150 V.
3.5. ELISA
A potential ELISA assay was examined to detect the purified NP using a sandwich assay. To construct the sandwich assay, two antibodies that bind to different regions of the NP were generated. The two rabbit monoclonal antibodies were prepared by immunizing the rabbits with an NP-specific peptide antigen, whose epitope information is not available and picked for an optimal sandwich format: note that these rabbit antibodies are commercially available from Abwiz Bio Inc. (San Diego, CA, USA) as indicated below. Multi-well plate (MaxiSorp; Thermo Fisher Scientific) was coated with phosphate-buffered saline containing 1 μg/mL purified anti-NP rabbit monoclonal antibody #84C4a (#2486, Abwiz Bio Inc.) overnight at 4 °C, which was used to capture the NP to the plate. The plate was then washed three times with 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, and 0.05% Tween 20 (TBS-T; TaKaRa Bio) and blocked at RT for 1 h with StabilGuard (Surmodics, Eden Prairie, MN, USA) blocking solution diluted 4-fold with PBS. After washing three times with TBS-T, the recombinant NP antigen were seriously diluted with TBS-T and incubated with the antibody-coated plate at RT for 1 h. Next, the plate was washed, and 10 ng/mL of biotinylated rabbit monoclonal antibody #75G5a (#2482 Abwiz Bio Inc.) was added and incubated at RT for 1 h to detect the captured NP. After washing the plate, HRP-conjugated streptavidin (Thermo Fisher Scientific) (1:4,000) was added and incubated at RT for 1 h.
For fluorescence detection of NP at wide range concentrations, we used the QuantaBlu Fluorogenic Peroxidase Substrate Kit (Thermo Fisher Scientific). The substrate solution was added to the wells and incubated for 11.5 min at RT, followed by the addition of an equal volume of QuantaBlu stop solution. Then, the samples were transferred to a new well of a black plate (Black MaxiSorp; Thermo Fisher Scientific) and scanned using a 325/420 nm excitation/emission filter set by multimode plate reader EnSpire (PerkinElmer, Waltham, MA, USA). This assay was performed with n = 6.
For decision of Limit of detection (LOD), color development was performed with 3,3′,5,5′-tetramethylbenzidine chromogen substrate (Surmodics) for 11.5 min at RT and stopped by adding the Stop Solution (Surmodics). The optical density (OD) at 450 nm was measured by the above multimode plate reader. This assay was performed with n = 4.
3.6. Gel image analysis
Agarose gels stained with CBB were photographed by image scanner GT-X980 (EPSON, Suwa, Japan). The presented data are representative of at least two experiments.
3.7. In silico analysis
Prediction of the three dimensional structures of NP and tagged NP was obtained by AlphaFold2 [29].
Analysis of pH titration curve by each domain and the entire NP molecule was used by mailway software SEDNTERP3 (http://www.jphilo.mailway.com/sednterp.htm) kindly provided by Dr. John Philo [30].
4. Results and discussion
4.1. Expression and purification of SARS-CoV-2 nucleoprotein
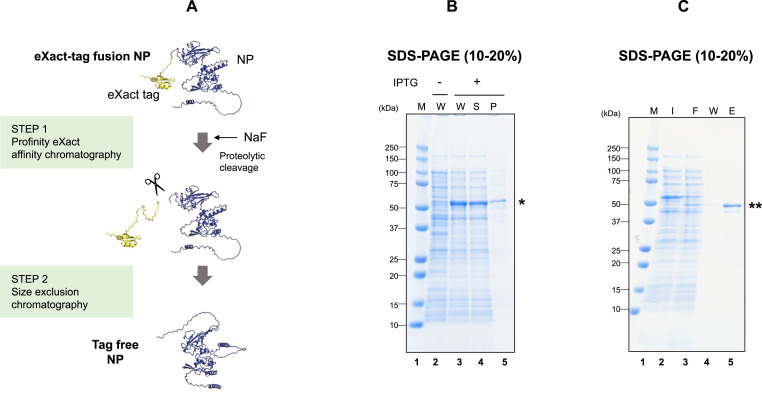
NP was expressed in E. coli as a fusion to eXact-tag. Fig. 1 A depicts a schematic flowchart of the purification of the NP, in which NP (blue) was expressed as a fusion to eXact-tag (orange), purified by affinity chromatography and processed in STEP 1 and polished in STEP 2. The structures of the NP and the tag were constructed by the AI software, AlphaFold2, consistent with the folded and disordered domains predicted from the sequence analysis. Fig. 1B shows expression of NP, mostly in the soluble fraction after IPTG induction (see asterisk). The expressed NP was bound to the affinity column through eXact-tag, which was subsequently cleaved by subtilisin on the column. Fig. 1C shows the affinity purification and proteolytic cleavage of NP. The NP eXact-tag fusion protein expressed in the whole lysate bound to the column, as is evident from no NP fusion protein in the flow-through (FT) fraction. A new band slightly below the fusion was observed after cleavage and elution (see double asterisks), expected from the loss of the eXact-tag sequence that caused the protein to elute and become smaller by the mass of the tag sequence.
Fig. 1.
Expression of the purification of SARS-CoV-2 NP.
A. Flowchart of the expression and purification
The conformation predicted for AlphaFold2 is shown, where eXact tag is shown by orange and NP by blue.
B. Soluble expression of codon optimized eXact-NP fusion protein in E. coli.
Lane 1, molecular weight marker; Lane 2, total cell protein from E. coli T7Express pLysY/pET-3d-eXact-NP before IPTG induction (0 h); Lane 3, total cell protein 3 h after induction; Lane 4, soluble fraction after induction; Lane 5, insoluble fraction after induction. The single asterisk indicates the NP fusion protein.
C. Purificaion of NP by Profinity eXact affinity chromatography.
Lane 1, molecular weight marker; Lane 2, soluble fraction from supernatant of lysed and centrifuged cells; Lane 3, flow-through fraction of Profinity eXact chromatography; Lane 4, wash fraction; Lane 5, products after proteolytic cleavage: the double asterisk indicates tag free NP. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
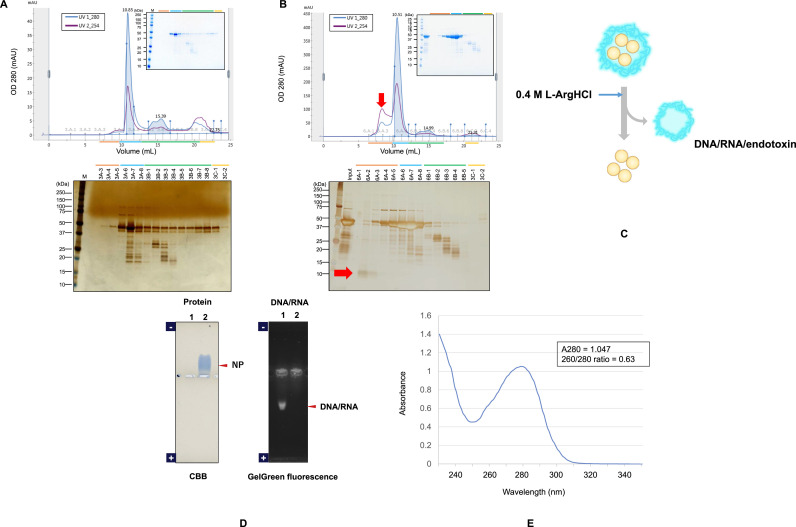
The above affinity-purified and tag-removed NP was further purified by size exclusion chromatography (SEC) in the presence of 0.4 M arginine hydrochloride (here termed simply arginine) (Fig. 2 ). It has been shown that arginine disrupts weak, non-specific interactions [[31], [32], [33]]. When TBS was used as a SEC running buffer, a major peak with a shoulder was observed followed by a few small peaks (Fig. 2A). The eluted fractions were analyzed by SDS-PAGE, showing a major peak containing the NP. However, this main peak also contained many bands co-eluting with the NP, suggesting that these contaminants were binding to the NP. It is evident that the NP was eluting not only in the main peak, but also throughout the total column volume or even exceeding it, indicating that NP interacted with the column non-specifically. Later fractions contained low molecular weight contaminants as expected. These results clearly show unique properties of NP that it can interact with other molecules or the surface of the SEC column. When 0.4 M arginine was included in the TBS buffer, a large peak before the major peak was observed, suggesting that arginine dissociated macromolecular complexes (Fig. 2B). Comparison with the above results with a simple TBS elution suggests that some of the contaminants that co-eluted with the NP as large complexes in the absence of arginine were dissociated by arginine. As shown in Fig. 2C by a cartoon, we speculated that NP (orange) was in complex with contaminants such as nucleic acids (blue), which can be disrupted by arginine, resulting in separation of contaminants and NP, consistent with the SEC results. The eluted fractions from the arginine elution were subjected to SDS-PAGE and stained by silver. The shoulder peak contained a major band most likely arising from LPS and nucleic acids that formed large complexes, upon dissociation from the NP, and hence eluted before the NP peak: note that nucleic acids can be detected by silver staining. The main peak contained primarily the NP, whose molecular weight was consistent with the sequence weight of 46 kDa upon SDS-PAGE. However, the molecular weight of the NP eluting in this peak actually is twice the value, or even larger than the dimeric size, as described later. Later eluting peaks contained low molecular weight contaminants, as has been observed in TBS-elution. Table 1 shows the purification summary. The expression of NP fusion was about 10 mg (6 mg without counting the fusion weight) from 150 mg cells, in which the soluble NP fusion accounted for 6 mg (4 mg without the fusion). After affinity chromatography, 2 mg of the cleaved NP was recovered. Size exclusion chromatography in 0.4 M arginine resulted in recovery of 1 mg NP. The purity of the final product was 95% with the overall yield of 25% from the soluble fraction and 17% from the total NP expressed.
Fig. 2.
Size exclusion chromatography and SDS-PAGE of NP.
A. Size exclusion chromatogram of the affinity-purified NP on Superdex 200 increase column in TBS (pH 7.6) running buffer. Fractions were analyzed by 10–20% SDS-PAGE and silver stain.
B. Size exclusion chromatogram of affinity-purified NP on Superdex 200 increase column in TBS (pH 7.6) running buffer containing 0.4 M L-Arg-HCl. Fractions were analyzed by 10–20% SDS-PAGE and silver stain. Red arrow shows contaminants in the shoulder.
C. Schematic illustration of NP complexed with contaminants that can be dissociated by arginine.
D. Analysis of DNA/RNA contained in purified NP by agarose gel electrophoresis.
One % flat agarose gels in 0.1 M His/0.1 M MES, pH 6.1 were run at 100 V for 30 min for protein and DNA/RNA analysis.
Lane 1: Fraction GA2 (see Fig. 2B) of size exclusion chromatography, 5 μL load.
Lane 2: Purified NP (1 mg/mL), 5 μg load.
Left: CBB-stained gel, NP indicated by arrow.
Right: Fluorescent GelGreen dye-stained gel, DNA/RNA indicated by arrow.
E. UV spectrum of the purified NP
The purified NP solution was measured in cuvette mode with Protein A280 appli using spectrophotometer. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Purification of SARS-CoV-2 NP fusion proteins overexpressed in E. coli T7 Express lysY/Iq.
| Purification step | Total proteina (mg) | NP fusion protein (tag-free protein)ab(mg) | dsDNAc (μg/mL) | Purityb (%) | Yieldd(%) |
|---|---|---|---|---|---|
| Cell lysate | 46 | 10 (6) | 41 | 22 | – |
| Soluble fraction | 26 | 6 (4) | 41 | 23 | 100 |
| Affnity chromatography | 4 | (2) | 0.37 | 60 | 50 |
| Size exclusion chromatography | 1 | (1) | N.D. | 95 | 25 |
Results are derived from 150 mg wet cell weight from 100 mL of culture Protein concentrations determined by BCA protein assay using BSA as a standard.
Fusion protein (54.0 kDa) and tag-free protein (45.6 kDa) as estimated by PAGE densitometry with Image Lab ver 6.0.1 (BIO-RAD).
dsDNA concentrations determined by Qubit assay (Thermo Fisher Scientific).
The yield is the amount of target protein at that step divided by the amount of target protein in the first step (defined as 100%) and the yield of tag-free protein was recalculated as tagged fusion protein.
The shoulder peak of SEC done in the presence of arginine (Fig. 2B) was further analyzed for nucleic acids on agarose native gel electrophoresis by GelGreen fluorescent dye staining. As shown in Fig. 2D (lane 1), CBB staining showed no NP band, but a fluorescent band due to nucleic acids was observed. On the contrary, the SEC purified NP sample (lane 2) showed a band upon CBB staining, but no GelGreen fluorescent dye staining. Also, the dsDNA and RNA content of the purified NP was below the detection limit as analyzed by the Qubit assay (Table 1). UV absorbance spectrum of the SEC purified NP used here showed a typical pure protein with 280 nm peak and without 260 nm shoulder, as in Fig. 2E. The 260/280 ratio of the purified NP was 0.63, which is close to 0.6 for common proteins. From these data, the SEC purified NP was shown to be free of nucleic acid contamination.
A similar approach has been reported for the purification of tag-free and highly pure recombinant NP in E. coli. For example, Vos et al. reported that soluble CASPON fusion tag NP expressed by their own E. coli enGene Xpress strain was treated with nuclease to remove nucleic acids, purified by native hydrophobic interaction chromatography hyphenated to multi-angle light scattering detection, and further processed to cleave and remove the tag with caspase-2 variant [34]. Carlson et al. reported that insoluble His-SUMO tag fusion NP was freed of contaminating nucleic acids in the presence of urea, purified by Ni-NTA purification and refolded, which was followed by cleavage of the tag by the recombinant Ulp1 catalytic domain [35]. Compared to such reports, the present method is considered to be a superior method that enables more effective purification with a simpler manufacturing process.
4.2. Analysis of NP structure in solution
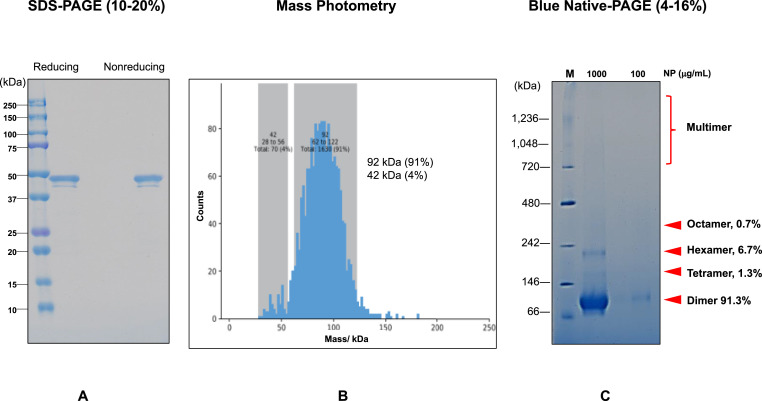
The purified NP was characterized by SDS-PAGE, Mass Photometry (MP) and native-PAGE. SDS-PAGE under reducing and non-reducing showed a major band with a faint band as shown in Fig. 3 A. The mobility was identical between both conditions, consistent with the absence of both intra- and inter-chain disulfide bonds: cleavage of the intra-chain disulfide bonds would expand the hydrodynamic size of SDS-protein complex, leading to a slower mobility on SDS-PAGE. A mass distribution by MP was determined at a low protein concentration of 2.5 μg/mL, which has been diluted from the solution containing 0.4 M arginine (L-Arg) into PBS. Fig. 3B shows the mass of the NP in PBS, indicating a major peak of 92 kDa, corresponding to a dimer and a shoulder of 42 kDa, corresponding to a monomer. The results demonstrate that the tertiary structure of NP in PBS is primarily a dimer at such a low protein concentration. On the other hand, the result of Blue-Native PAGE in Fig. 3C showed a main band both at 100 and 1,000 μg/mL, perhaps corresponding to the dimer, provided that the molecular weight standards used to calibrate the gel have the same charge density and hydrodynamic shape, upon binding the blue dye, with the NP. At 1,000 μg/mL, several low mobility bands, most likely corresponding to non-covalent oligomerization of NP, were observed. Combining both mass data and Blue-Native-PAGE results suggest a dimeric structure of NP at low protein concentration (below 100 μg/mL) and a possible larger oligomers at higher concentrations.
Fig. 3.
Analysis of molecular weight of NP in solution.
A. Reducing or non-reducing (10–20% SDS-PAGE).
B. Mass distributions histogram by mass photometry for 2.5 μg/mL NP in PBS. The major distribution peak represents the NP dimer.
C. Blue-Native PAGE (4–16%) of NP at 100 and 1,000 μg/mL.
Molecular weight standard of native proteins was also included. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
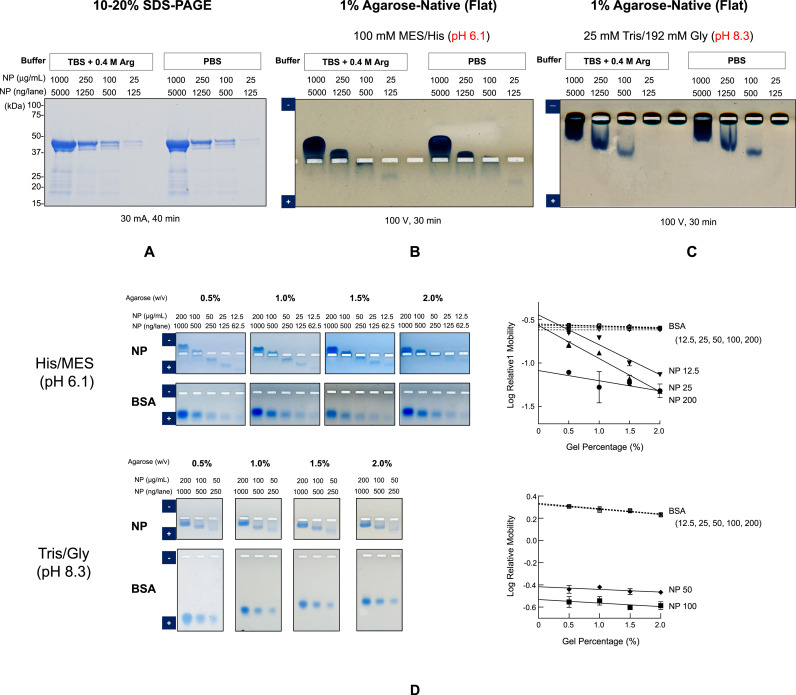
Such oligomer formation can be concentration-dependent and thus adequately characterized by native gel electrophoresis as a function of protein concentration. A stock solution of 1 mg/mL was diluted with either TBS containing 0.4 M arginine or PBS to 250, 100 and 25 μg/mL. Fig. 4 A shows the non-reducing SDS-PAGE with descending CBB staining intensity with decreasing protein concentration and hence loading amounts. The results were identical whether diluted with arginine-containing TBS or PBS. The mobility was independent of the concentration, as expected from monomeric structure of NP in the presence of SDS.
Fig. 4.
Analysis of NP by agarose native gel electrophoresis.
Different NP concentrations from 25 to 1,000 μg/mL were analyzed by three different electrophoresis methods.
A. 10–20% SDS-PAGE (reducing condition)
B. One% Agarose-native gel electrophoresis with 0.1 M His/MES running buffer (pH 6.1).
C. One% Agarose-native gel electrophoresis with Tris/Gly running buffer (pH 8.3).
D. Ferguson plot analysis of NP by agarose native gel electrophoresis in two different running buffers.
NP or BSA at different protein concentrations was run with 0.5–2% UltraPure agarose gel. The relative mobility of protein was calculated to the mobility of xylene cyanol or Orange G dye front.
(Linear regression line was drawn through the plots, as shown by the reasonably high correlation coefficients.)
E. Nuclease treatment of the purified NP.
Lane 1: Purified NP was 10-fold diluted with TBS-0.4 M L-ArgHCl.
Lane2: Purified NP was 10-fold diluted with TBS-5 mM MgCl2 and incubated at 25 °C for 1hr.
Lane 3: Purified NP was 10-fold diluted with TBS-5 mM MgCl2 –Benzonase and incubated at 25 °C for 1hr.
NP (250 ng/lane) before and after nuclease treatment were analyzed by 1% agarose-native gel electrophoresis for 45 min and CBB staining.
F. Model of NP structure with changes in protein concentration and pH.
G. Hypothesis of concentration dependent-conformational change of NP.
Concentration dependence of Ferguson plot and self-association and dimeric molecular weight by mass photometry at low protein concentration were taken into account to construct the model structures proposed here. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
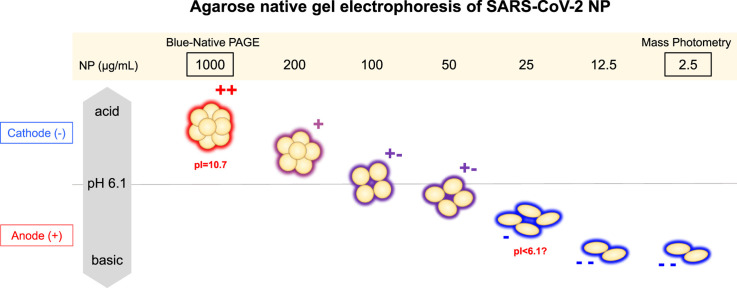
A surprising and unexpected result was observed with native gel electrophoresis.
Fig. 4B shows the results of agarose native gel electrophoresis with His/MES pH 6.1 buffer system. NP at 1,000 μg/mL showed mobility toward the cathode in both dilution methods (i.e., in the absence and presence of 0.4 M arginine), indicating that it was positively charged at the running pH of 6.1, consistent with its net charge as described below (see titration curve). The mobility decreased at 250 μg/mL and was near zero at 100 μg/mL. The NP at 25 μg/mL migrated toward the anode, indicating that it was negatively charged and hence totally unexpected from its titration curve. The mobility at 25 μg/mL appeared to be slightly faster in the absence of arginine. The observed results, regardless of the arginine concentration, clearly indicate that the charged states of NP undergo reversal around 100 μg/mL. Namely, the NP became negatively charged around this concentration range, which requires explanation. The results at pH 8.3 were qualitatively different from the results at pH 6.1. The NP was always negatively charged at the concentrations tested, i.e., between 100 and 1,000 μg/mL, inconsistent with the titration curve: no band was detected at 25 μg/mL under the conditions. The mobility increased with decreasing concentration, meaning that either the hydrodynamic size decreased with lowering protein concentration or the negative charge increased at lower concentration. The results were independent of the absence or presence of arginine. The observed increase in negative charges at pH 8.3 was at least in part consistent with the observed reversal of the charged state (from positive to negative) observed at pH 6.1.
Comparison of the native gel results at pH 6.1 and 8.3 gave some insight into the charged state of the NP. The NP at 250–1,000 μg/mL migrated toward the anode at pH 8.3, indicating that it was negatively charged, while it migrated toward the cathode at the same concentration range, indicating that the NP was positively charged within the above NP concentration range. Thus, it may be concluded that the net charge at 250–1,000 μg/mL was positive at pH 6.1 and negative at pH 8.3, meaning that the actual pI was between these two pH values. At low NP concentrations, the NP was always negatively charged.
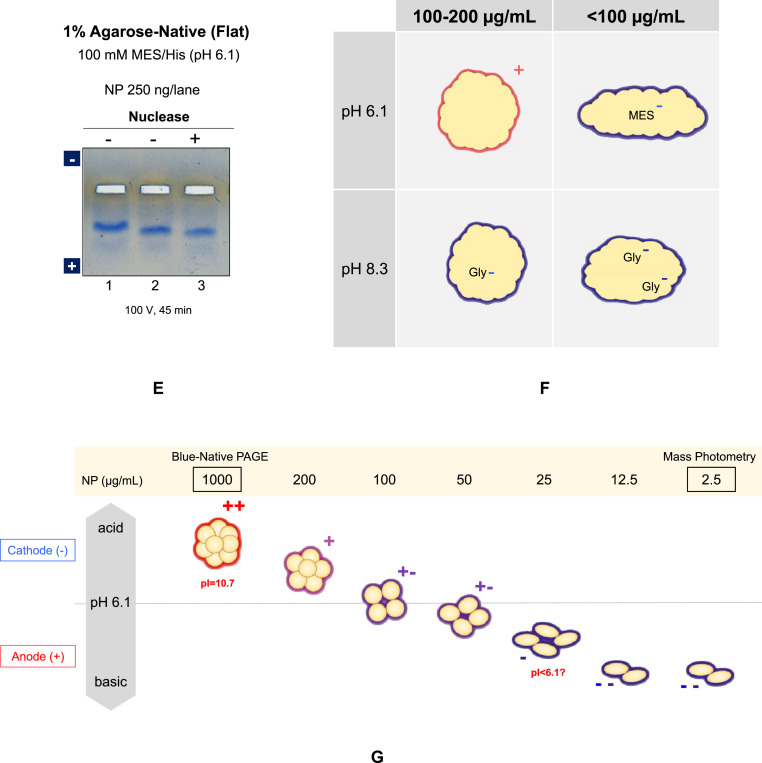
Can nucleic acid contamination, which was found unlikely as described above, explain the observed charge reversal at pH 6.1 and faster mobility at pH 8.3? It does appear so at least at low protein concentration. However, it cannot explain positively charged NP complex at pH 6.1 and its slow mobility at pH 8.3 when the protein concentration was increased. The behavior at high concentration may be explained by nucleic acids packed with counter cations within NP complex so that the effective charges of nucleic acids are at least partially neutralized. Thus, we examined this possibility. The UV absorbance spectrum of the NP preparation used indicated above negligible amount of nucleic acids. The SEC purified NP showed no fluorescent band staining due to nucleic acids. Mass photometry showed a molecular weight of NP as a dimer without significant contribution of nucleic acids to the mass. SDS-PAGE analysis in Fig. 2 showed removal of low molecular weight contaminants, which comprise at least in part nucleic acids, in the shoulder of the SEC done in 0.4 M arginine. No detectable amounts of dsDNA and RNA were present in the NP sample (Table 1). As a final proof, nuclease treatment was performed in the presence of arginine. As shown in Fig. 2C, inclusion of 0.4 M arginine after SEC purification was critical to maintain the NP solubility, but is likely to hamper nuclease activity. In order to make the reaction work, the sample was 10-fold diluted with TBS and treated with Benzonase nuclease, which can digest both DNA and RNA, for 1 h, followed by agarose gel electrophoresis. Nuclease treatment resulted in no change in charged state, i.e., the negatively charged state, as in Fig. 4E. Namely, the NP was still negatively charged after nuclease treatment.
5. Ferguson plot
Ferguson empirically showed that the size sieving effects of gel electrophoresis increase with gel concentration and the relative mobility decreases linearly with gel %, when the relative mobility is defined as a ratio of the mobility of the sample macromolecules to a reference standard, normally a tracking dye [[36], [37], [38]]. The relative mobility decreases to a greater degree for larger macromolecules than the smaller ones, namely the slope of the Ferguson plot is steeper for the larger macromolecules. This was done for the same NP preparations in a narrower protein concentration range, as the band at 1,000 μg/mL was too broad to define the band position. The mobility of NP was determined as a function of agarose concentration at 0.5, 1, 1.5 and 2.0% as shown in Fig. 4D. The gel electrophoresis was done with agarose in both His/MES (pH 6.1) and Tris/Gly (pH 8.3) systems. BSA was also analyzed as a control. The trend of electrophoretic profiles were similar for each buffer systems between different agarose concentrations and also similar to the results at 1% gel in Fig. 4B and C. For His/MES system, the charged state was reversed around 50–100 μg/mL at any agarose concentration. The mobility significantly decreased with gel %. Interestingly, the BSA mobility also showed slight concentration dependence, being slightly slower at higher BSA concentration, at each agarose concentration. This may be due to increasing friction at higher BSA concentration, causing retardation of its mobility. Nevertheless, it should be pointed out that such increased friction at higher protein concentration was not within the magnitude of the observed concentration dependence of the mobility for NP. Similarly, the protein concentration dependence of the mobility in Tris/Gly system showed the same trend at each agarose concentration. Since the band intensity was too weak to clearly define the mobility of NP at 12.5 and 25 μg/mL, they were omitted from the Ferguson analysis. Higher NP concentration resulted in less mobility at any agarose % with retarded mobility with increasing agarose concentration. In this buffer system, BSA showed little concentration dependence at each agarose concentration, implying that increased friction at higher BSA concentration appeared to be insignificant with this buffer system for unknown reason. BSA mobility showed significant decrease with increasing gel %.
Ferguson plot was made for those sample that showed significant mobilities, i.e., 12.5, 25 and 200 μg/mL for His/MES system and 50 and 100 μg/mL for Tris/Gly system. The mobility of XC (xylene cyanol) or Orange G dye was used as a reference standard to determine the relative mobility. The relative mobility of NP at each concentration was plotted against the agarose concentration, showing a linear relationship, although a large scattering was observed at 200 μg/mL for His/MES system due to low mobility. The slope of the pH 6.1 data showed that the slope was steeper in the order of 12.5 = 25 > 200 μg/mL. This suggests that the size of NP was greater at lower concentrations (12.5–25 μg/mL), unanticipated from the concentration-dependent self-association. Namely, the larger sizes at lower protein concentration is due to a factor other than a simple concentration-mediated self-association, for example, more expanded structure at lower concentration. For a control, the relative mobility of BSA was also included, showing much weaker dependence on gel %, nearly independent of the BSA concentration. The concentration-independent molecular size of BSA is expected from its high solubility and stable monomeric structure. The slope of BSA was much shallower than NP at 12.5–25 μg/mL and even at 200 μg/mL, indicating that the size of NP is greater than BSA.
Ferguson plot of NP in Tris/Gly (pH 8.3) system is also shown in Fig. 4D, in which the samples were diluted with the running buffer before loading on the gel. Interestingly, the slope was almost identical for NP at 50 and 100 μg/mL and for BSA, indicating that NP at pH 8.3 has a similar size to the BSA. This in turn suggests that NP at pH 8.3 has a similar molecular weight to BSA, provided that their hydrodynamic shapes are similar.
Another important parameter of Ferguson plot is the intercept (Y-intercept) of the plot at zero agarose concentration, which corresponds to the mobility of NP in free solution and hence closely follows the charged state in solution. The Y-intercept of NP at 12.5 and 25 μg/mL was similar to that of BSA, suggesting that the NP at pH 6.1 was similarly negatively charged to BSA in terms of charge density. The free mobility of NP was much slower at 200 μg/mL than at lower concentrations, suggesting that it was less charged. It should be emphasized here that the NP at 200 μg/mL was positively charged at pH 6.1, as it migrated toward the cathode, different from the NP at lower concentrations. Namely, the positive charge density of NP at 200 μg/mL was less than the negative charge density of NP at 12.5 and 25 μg/mL (Fig. 4F). With regard to the Y-intercept at pH 8.3, the value was much smaller for NP at 50 and 100 μg/mL than for BSA, indicating that the NP at pH 8.3 was far less negatively charged than BSA.
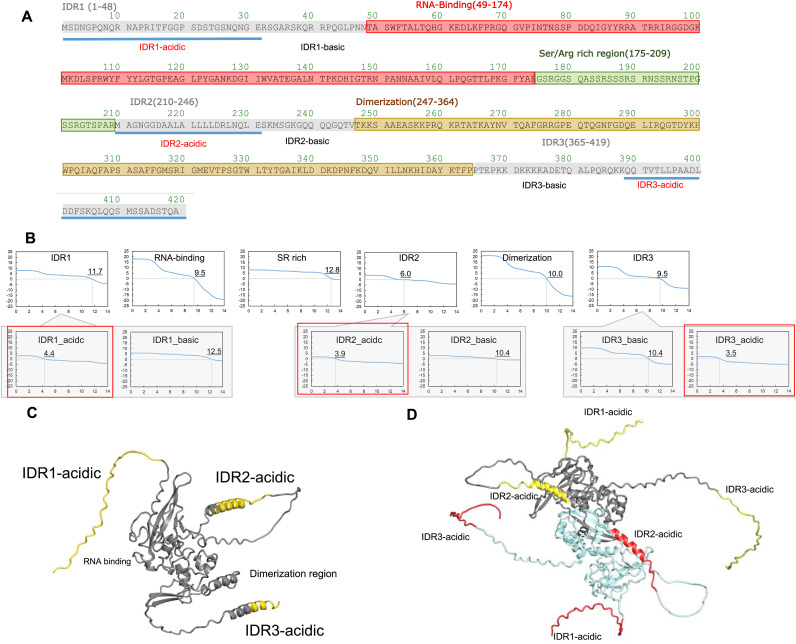
6. Discussion on different mobilities based on amino acid sequence analysis
NP is a highly basic protein with the isoelectric point of ∼10 as shown in Fig. 5 A and B by theoretical titration curve based on its amino acid sequence or composition. It was thus expected that NP would migrate toward the cathode both at pH 6.1 and 8.3. The observed mobility of NP on the pH 6.1 agarose native gel was consistent only above ∼200 μg/mL. The results on the pH 8.3 agarose native gel electrophoresis was totally inconsistent at any NP concentration examined, as it all migrated toward the anode. Combining the electrophoretic profiles at pH 6.1 and 8.3 suggests that the NP has a pI between 6.1 and 8.3 above ∼200 μg/mL and below 6.1 at lower concentrations. A question is how the charged state of this highly basic protein was made neutral and then acidic by simply varying the protein concentration. We suggested that the structure of NP may be concentration dependent, although at this stage there is no experimental support for this conclusion. Assuming a conformational change, it may be related to the unique amino acid sequence of NP, which is composed of many different domains, including basic and acidic clusters and intrinsically disordered and structured domains (Figs. 1 and 5C and D). It seems impossible to speculate that only structure changes can drastically alter the charged state from positively charged to negatively charge by simply decreasing the protein concentration. We believe that binding of negative ions, most likely stemming from the running buffer, i.e., MES (His/MES buffer) or glycine (Tris/Gly buffer), occurs to basic NP dependent on the NP concentration and alters the structure and charged state of NP, in particular at pH 6.1. Thus, we propose a scheme shown in Fig. 4F, which combined both changes in the charged state and hydrodynamic size based on the Ferguson plot. Namely, it shows possible ion binding and structure expansion as a function of protein concentration.
Fig. 5.
Structural analysis of NP sequence.
A. Domain architecture of NP, which is composed of many different domains, including basic and acidic clusters and three intrinsically disordered region (IDR) and structured domains.
B. pH titration for each domain of NP.
C. Position of acid cluster (yellow colour) in IDR1, 2, and 3 on 3D structure of NP monomer predicted by AlphaFold2.
D. Position of acid cluster in IDR1, 2, and 3 on 3D structure of NP dimer predicted by AlphaFold2. Three IDRs of one protein in the dimer are shown in yellow colour and those of another protein in red colour. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
It should be noted here that mass photometry indicated mostly dimeric structure at 2.5 μg/mL, 10-fold lower than 25 μg/mL for the lowest concentration of native gel analysis. Although the buffer (PBS) used for mass photometry was different from the native gel analysis (His/MES and Tris/Gly), it may be safe to state that the observed bands at 25 μg/mL could be a dimeric form. The results of Blue-Native-PAGE, along with known concentration-dependent self-association of NP, suggest formation of larger oligomers at higher concentration. This scenario is schematically depicted in Fig. 4G. We assumed that the shape of the subunit was more spherical for larger oligomers and expanded for the smaller oligomers to take the Ferguson analysis into consideration in Fig. 4F. It should be noted, however, that NP may undergo different post-translational modifications in infected host cells from those in E. coli and thus may have different physical properties from those shown here. Nevertheless, the observed results should give an insight into the physical behavior of NP in solution.
7. ELISA
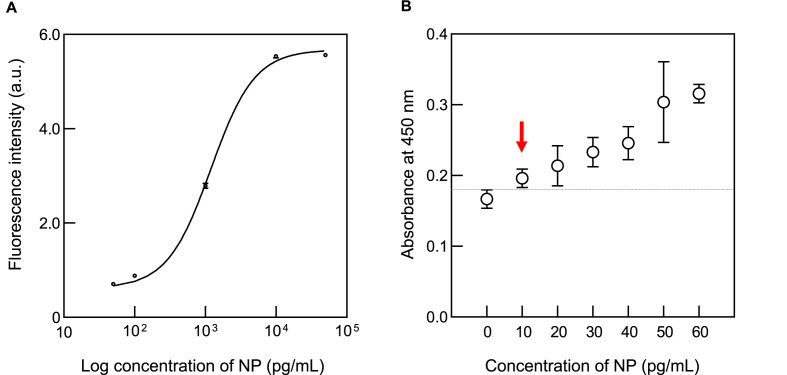
Finally, biological activity of NP was evaluated by sandwich ELISA. The assay was performed using purified NP antigen and specific rabbit monoclonal antibodies. The specific rabbit antibodies were generated by immunization of the rabbits with SARS-CoV-2 NP specific peptide antigens. Unfortunately, their epitope information is confidential. Fig. 6 A shows the dose response curve of ELISA signal at the NP concentration in the 50 pg/mL to 50 ng/mL range. This assay enabled us to distinguish the signal difference caused by a 200-fold increase in NP concentration from 50 pg/mL to 10 ng/mL. The sensitivity of assay was further evaluated in low pg/mL range as shown in Fig. 6B, in which the error bars of the ELISA signals were provided. Limit of detection (LOD) was defined as the minimum amount of antigen that gave a significant signal as compared to the signal from the mean blank plus 2.6-fold standard deviations, indicating that it detected 10 pg/ml of NP. Therefore, it was shown that the highly purified NP prepared here can be used as a standard antigen for ELISA and can also be applied to the evaluation of antibodies.
Fig. 6.
ELISA using purified NP antigen
A. Standard curve of the ELISA using recombinant NP
Stock NP solution of 1 mg/mL was diluted serially in Tris-buffered saline supplemented with Tween 20 (TBS-T) (blank) at the concentrations shown in the figure and applied to an in-house sandwich ELISA. Anti-NP rabbit monoclonal antibody #84C4a was used as a capture antibody, and biotinylated #75G5a was used as a detection antibody. Fluorescence Intensity was measured by detecting fluorogenic substrates reacted with streptavidin-HRP. The average emission at each point is indicated on the Y-axis, whereas the X-axis indicated the concentration of NP. The sigmoidal curve was fitted line to the stoichiometric binding model. The error bar indicates standard deviation (SD).
B. The limit of detection of ELISA.
NP was diluted serially with TBS-T. The diluted samples (10, 20, 30, 40, 50, and 60 pg/mL) and TBS-T (blank) were measured five times using in house sandwich ELISA. The average of the absorbance values for each measurement were plotted; error bars represent 2.6 SD. The dotted line represents the average absorbance value of the blank plus 2.6 SD (0.179). The LOD was 10 pg/mL shown by red arrow. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
8. Conclusion
Our newly developed agarose native gel electrophoresis method revealed that SARS-CoV-2 NP have unique properties, shown by concentration-dependent changes in charge and shape. The theoretical isoelectric point of NP indicates that it is a basic protein, but at high concentrations it behaves like an acidic protein. Experiments using purified NP that does not contain tag sequence, host cell proteins, host DNA, and endotoxin should enable us to understand not only the characteristics of NP in solution but also its possible role in the viral life cycle.
CRediT authorship contribution statement
Ryo Sato: Visualization. Yui Tomioka: Visualization. Chiaki Sakuma: Visualization. Masataka Nakagawa: Visualization, Validation. Yasunori Kurosawa: Validation. Kohei Shiba: Visualization, Validation, Writing. Tsutomu Arakawa: Writing – review & editing, Supervision. Teruo Akuta: Writing – review & editing, Supervision.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ab.2022.114995.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 2.Solomon M., Liang C. Human coronaviruses: the emergence of SARS-CoV-2 and management of COVID-19. Virus Res. 2022;319 doi: 10.1016/j.virusres.2022.198882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Coronavirus Dis. 2020;2019:23–31. COVID-19) [Google Scholar]
- 4.Lu R.M., Ko S.H., Chen W.Y., Chang Y.L., Lin H.T., Wu H.C. Monoclonal antibodies against nucleocapsid protein of SARS-cov-2 variants for detection of COVID-19. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222212412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., 3rd, Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39 doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinesh D.C., Chalupska D., Silhan J., Koutna E., Nencka R., Veverka V., Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redzic J.S., Lee E., Born A., Issaian A., Henen M.A., Nichols P.J., Blue A., Hansen K.C., D'Alessandro A., Vögeli B., Eisenmesser E.Z. The inherent dynamics and interaction sites of the SARS-CoV-2 nucleocapsid N-terminal region. J. Mol. Biol. 2021;433(15) doi: 10.1016/j.jmb.2021.167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessa L.M., Guseva S., Camacho-Zarco A.R., Salvi N., Maurin D., Perez L.M., Botova M., Malki A., Nanao M., Jensen M.R., Ruigrok R.W.H., Blackledge M. The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a. Sci. Adv. 2022;8(3) doi: 10.1126/sciadv.abm4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39(24) doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arndt C., Koristka S., Feldmann A., Bachmann M. Native polyacrylamide gels. Methods Mol. Biol. 2019;1855:87–91. doi: 10.1007/978-1-4939-8793-1_8. [DOI] [PubMed] [Google Scholar]
- 13.Arakawa T., Haniu M., Narhi L.O., Miller J.A., Talvenheimo J., Philo J.S., Chute H.T., Matheson C., Carnahan J., Louis J.C., Yan Q., Welcher A.A., Rosenfeld R. formation of heterodimers from three neurotrophins, nerve growth factor, neurotrophin-3, and brain-derived neurotrophic factor. J. Biol. Chem. 1994;269:27833–27839. [PubMed] [Google Scholar]
- 14.Li C., Arakawa T. Agarose native gel electrophoresis of proteins. Int. J. Biol. Macromol. 2019;140:668–671. doi: 10.1016/j.ijbiomac.2019.08.066. [DOI] [PubMed] [Google Scholar]
- 15.Tomioka Y., Nakagawa M., Sakuma C., Kurosawa Y., Nagatoishi S., Tsumoto K., Arakawa T., Akuta T. Analysis of bovine serum albumin unfolding in the absence and presence of ATP by SYPRO Orange staining of agarose native gel electrophoresis. Anal Biochem. 2022;654 doi: 10.1016/j.ab.2022.114817. [DOI] [PubMed] [Google Scholar]
- 16.Li C., Akuta T., Nakagawa M., Sato T., Shibata T., Maruyama T., Okumura C.J., Kurosawa Y., Arakawa T. Agarose native gel electrophoresis for characterization of antibodies. Int. J. Biol. Macromol. 2020;151:885–890. doi: 10.1016/j.ijbiomac.2020.02.185. [DOI] [PubMed] [Google Scholar]
- 17.Sakuma C., Sato T., Shibata T., Nakagawa M., Kurosawa Y., Okumura C.J., Maruyama T., Arakawa T., Akuta T. Western blotting analysis of proteins separated by agarose native gel electrophoresis. Int. J. Biol. Macromol. 2021;166:1106–1110. doi: 10.1016/j.ijbiomac.2020.10.265. [DOI] [PubMed] [Google Scholar]
- 18.Sakuma C., Tomioka Y., Li C., Shibata T., Nakagawa M., Kurosawa Y., Arakawa T., Akuta T. Analysis of protein denaturation, aggregation and post-translational modification by agarose native gel electrophoresis. Int. J. Biol. Macromol. 2021;172:589–596. doi: 10.1016/j.ijbiomac.2021.01.075. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa M., Tomioka Y., Sakuma C., Sato R., Shibata T., Kurosawa Y., Sato Y., Ono Y., Arakawa T., Akuta T. Optimization and application of silver staining of non-glycosylated and glycosylated proteins and nucleic acids for agarose native gel electrophoresis. Int. J. Biol. Macromol. 2021;189:869–878. doi: 10.1016/j.ijbiomac.2021.08.142. [DOI] [PubMed] [Google Scholar]
- 20.Tomioka Y., Arakawa T., Akuta T., Nakagawa M., Ishibashi M. Analysis of proteins by agarose native gel electrophoresis in the presence of solvent additives. Int. J. Biol. Macromol. 2022;198:26–36. doi: 10.1016/j.ijbiomac.2021.12.084. [DOI] [PubMed] [Google Scholar]
- 21.Arakawa T., Nakagawa M., Tomioka Y., Sakuma C., Li C., Sato T., Sato R., Shibata T., Kurosawa Y., Akuta T. Gel-electrophoresis based method for biomolecular interaction. Methods Cell Biol. 2022;169:67–69. doi: 10.1016/bs.mcb.2021.12.030. [DOI] [PubMed] [Google Scholar]
- 22.Sakuma C., Nakagawa M., Tomioka Y., Maruyama T., Entzminger K., Fleming J.K., Shibata T., Kurosawa Y., Okumura C.J., Arakawa T., Akuta T. Western blotting of native proteins from agarose gels. BioTechnique. 2022;72:207–218. doi: 10.2144/btn-2022-0012. [DOI] [PubMed] [Google Scholar]
- 23.Akuta T., Maruyama T., Sakuma C., Nakagawa M., Tomioka Y., Entzminger K., Fleming J.K., Sato R., Shibata T., Kurosawa Y., Okumura C.J., Arakawa T. A new method to characterize conformation-specific antibody by a combination of agarose native gel electrophoresis and contact blotting. Antibodies. 2022;11:36. doi: 10.3390/antib11020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomioka Y., Nakagawa M., Sakuma C., Nagatoishi S., Tsumoto K., Arakawa T., Akuta T. Ladder observation of bovine serum albumin by high resolution agarose native gel electrophoresis. Int. J. Biol. Macromol. 2022;215:512–520. doi: 10.1016/j.ijbiomac.2022.06.118. [DOI] [PubMed] [Google Scholar]
- 25.Young G., Hundt N., Cole D., Fineberg A., Andrecka J., Tyler A., Olerinyova A., Ansari A., Marklund E.G., Collier M.P., Chandler S.A., Tkachenko O., Allen J., Crispin M., Billington N., Takagi Y., Sellers J.R., Eichmann C., Selenko P., Frey L., Riek R., Galpin M.R., Struwe W.B., Benesch J.L.P., Kukura P. Quantitative mass imaging of single biological macromolecules. Science. 2018;360:423–427. doi: 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan B., Fisher K.E., Alexander P.A., Doroshko V., Bryan P.N. Engineering subtilisin into a fluoride-triggered processing protease useful for one-step protein purification. Biochemistry. 2004;43:14539–14546. doi: 10.1021/bi048177j. [DOI] [PubMed] [Google Scholar]
- 27.Akuta T., Kikuchi-Ueda T., Imaizumi K., Oshikane H., Nakaki T., Okada Y., Sultana S., Kobayashi K., Kiyokawa N., Ono Y. Expression of bioactive soluble human stem cell factor (SCF) from recombinant Escherichia coli by coproduction of thioredoxin and efficient purification using arginine in affinity chromatography. Protein Expr. Purif. 2015;105:1–7. doi: 10.1016/j.pep.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 29.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G.J., Bateman A., Evans R., Pritzel A., Figurnov M., Ronneberger O., Bates R., Kohl S.A.A., Potapenko A., Ballard A.J., Romera-Paredes B., Nikolov S., Jain R., Clancy E., Reiman D., Petersen S., Senior A.W., Kavukcuoglu K., Birney E., Kohli P., Jumper J., Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laue T.M., Shah B.D., Ridgeway T.M., Pelletier S.L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S.E., Rowe A.J., Horton J.C., editors. Royal Society of Chemistry; Cambridge: 1992. Computer-aided interpretation of analytical sedimentation data for proteins; pp. 90–125. [Google Scholar]
- 31.Tsumoto K., Ejima D., Kita Y., Arakawa T. Review: why is arginine effective in suppressing aggregation? Protein Pept. Lett. 2005;12:613–619. doi: 10.2174/0929866054696109. [DOI] [PubMed] [Google Scholar]
- 32.Ejima D., Yumioka R., Arakawa T., Tsumoto K. Arginine as an effective additive in gel permeation chromatography. J. Chromatogr. A. 2005;1094:49–55. doi: 10.1016/j.chroma.2005.07.086. [DOI] [PubMed] [Google Scholar]
- 33.Arakawa T., Ejima D., Tsumoto K., Obeyama N., Tanaka Y., Kita Y., Timasheff S.N. Suppression of protein interactions by arginine: a proposed mechanism of the arginine effects. Biophys. Chem. 2007;127:1–8. doi: 10.1016/j.bpc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 34.de Vos J., Aguilar P.P., Köppl C., Fischer A., Grünwald-Gruber C., Dürkop M., Klausberger M., Mairhofer J., Striedner G., Cserjan-Puschmann M., Jungbauer A., Lingg N. Production of full-length SARS-CoV-2 nucleocapsid protein from Escherichia coli optimized by native hydrophobic interaction chromatography hyphenated to multi-angle light scattering detection. Talanta. 2021;235 doi: 10.1016/j.talanta.2021.122691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80(6):1092–1103. doi: 10.1016/j.molcel.2020.11.025. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferguson K.A. Starch-gel electrophoresis—application to the classification of pituitary proteins and polypeptides. Matabolism. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- 37.Chrambach A. How far have we progressed toward automated electrophoresis in sieving media of the twenty-first century? Electrophoresis. 1993;14:1250–1254. doi: 10.1002/elps.11501401189. [DOI] [PubMed] [Google Scholar]
- 38.Ferguson K.A. The origin of the Ferguson plot. Electrophoresis. 2007;28:499–500. doi: 10.1002/elps.200600666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.