Abstract
Limited knowledge is available about the virulence mechanisms responsible for diarrheal disease caused by Salmonella typhimurium. To assess the contribution to diarrheal disease of virulence determinants identified in models of infection, we tested a collection of S. typhimurium mutants for their ability to cause enteritis in calves. S. typhimurium strains carrying mutations in the virulence plasmid (spvR), Salmonella pathogenicity island 2 (SPI-2) (spiB), or SPI-5 (sopB) caused mortality and acute diarrhea in calves. An S. typhimurium rfaJ mutant, which is defective for lipopolysaccharide outer core biosynthesis, was of intermediate virulence. Mutations in SPI-1 (hilA and prgH) or aroA markedly reduced virulence and the severity of diarrhea. Furthermore, histopathological examination of calves infected with SPI-1 or aroA mutants revealed a marked reduction or absence of intestinal lesions. These data suggest that virulence factors, such as SPI-1, which are required during intestinal colonization are more important for pathogenicity in calves than are genes required during the systemic phase of S. typhimurium infection, including SPI-2 or the spv operon. This is in contrast to the degree of attenuation caused by these mutations in the mouse.
Salmonella typhimurium has a murine reservoir (10) where it produces a typhoid fever-like disease (33). After oral infection of mice, bacteria invade the mucosa of the small intestine, displaying a tropism for M cells located in the follicle-associated epithelium of Peyer’s patches (21). Upon intestinal penetration, S. typhimurium causes a transient bacteremia and seeds systemic sites of infection, most importantly the liver and the spleen. S. typhimurium multiplies in splenic and hepatic tissues at a net growth rate of 0.5 to 1.5 log/day (19, 28). After bacterial numbers in liver and spleen reach between 108 and 109 CFU/organ, mice die from lipopolysaccharide (LPS)-induced damage of liver and spleen, which appears to be distinct from endotoxic shock (24).
In addition to causing illness in mice, S. typhimurium is frequently associated with disease in humans and livestock (42, 43, 45). The illness caused by S. typhimurium in these host species differs markedly, however, from murine typhoid. First, mice do not develop diarrhea, the prominent sign of disease during S. typhimurium infections in livestock and humans. Second, S. typhimurium infections in humans and livestock usually remain localized to the small intestine and mesenteric lymph node, while mice develop a systemic infection. The differences between murine typhoid and diarrheal disease raise the question whether S. typhimurium virulence mechanisms identified in the mouse are of equal importance during infections of humans or livestock.
S. typhimurium is a major cause of calf morbidity and mortality in the United States and in Europe (36, 40, 44). Furthermore, the infection in calves closely resembles illness caused by S. typhimurium in humans and can therefore serve as a model to study diarrheal disease. Calves infected experimentally with S. typhimurium develop diarrhea within 48 h (34). The bacteria invade the intestinal epithelium in the terminal ileum, resulting in exfoliation of epithelial cells and stunting of villi (11). Bovine enteritis caused by S. typhimurium is primarily an enteric infection with mortality resulting from dehydration and intestinal lesions. At necropsy of terminally infected calves, acute fibrinopurulent necrotizing enteritis is visible in both the villous ileum and Peyer’s patches (53). Histopathological examination reveals destruction of the mucosal epithelium, massive neutrophil infiltration, and depletion of lymphocytes in the germinal centers of intestinal lymphoid follicles (53).
At least 60 genes are required for full virulence of S. typhimurium in mice (15), but the roles of a only few of them, namely, aroA, dap, galE, and invH, during oral infection in cattle have been studied. Following oral inoculation of calves, S. typhimurium invH, dap, or aroA mutants show a marked reduction in the severity of enteritis (7, 46, 51). S. typhimurium aroA or aroA aroD mutants are furthermore attenuated sufficiently to be used as oral vaccines in calves (23, 46). Mutational inactivation of galE, on the other hand, results in attenuation of S. typhimurium in calves but does not render this pathogen unable to cause mortality when administered orally at a high dose (6). S. typhimurium strains carrying mutations in aroA, galE, dap, or invH elicit reduced fluid accumulation in bovine ligated ileal loops compared to that elicited by the wild type, suggesting that this assay is predictive of attenuation in calves (8, 51). Other mutations which reduce the ability of S. typhimurium to elicit fluid secretion in bovine intestinal loops include those in sirA and hilA (1). Furthermore, mutations in prgH, hilA, rfaJ, spvR, or spiB reduce the ability of S. typhimurium to colonize bovine tissues during competitive infection experiments (49). However, it is not clear from these experiments to what degree mutations in sirA, prgH, hilA, rfaJ, spvR, or spiB reduce S. typhimurium virulence or the severity of intestinal lesions during an oral infection of calves.
A second Salmonella serotype frequently associated with disease in cattle is Salmonella dublin (14, 44). Several loci, including aroA, invH, and the spv operon, contribute to virulence of S. dublin during oral infection of calves (26, 30, 37, 50, 51). Furthermore, mutations in the S. dublin invH, sopB (also known as sigD), sopD, sipB, pipA, pipB, or pipD gene result in reduced fluid accumulation in ligated ileal loops (13, 22, 51, 52). However, there are several differences between S. dublin and S. typhimurium during infection of cattle. While cattle inoculated orally with S. dublin develop a systemic infection (41), S. typhimurium causes an infection which is primarily enteric, with only occasional bacteremia (53). Furthermore, 75% of S. typhimurium infections occur in calves less than 2 months of age (42). In contrast, S. dublin is associated at similar frequencies with morbidity in both young and adult cattle (35, 42). Since differences between S. dublin and S. typhimurium infections are apparent, it is not clear whether mutations in orthologous genes result in identical virulence defects in both serotypes.
Although mutational inactivation of four genes, namely, aroA, dap, galE, and invH, has been shown to result in attenuation of S. typhimurium after oral infection of calves, evidence implicating other genes in S. typhimurium calf virulence is either indirect (based on data from bovine ligated ileal loops or competitive infection experiments) or has been inferred from studies performed with S. dublin. To obtain a more complete picture of the role in enteropathogenicity of virulence factors identified in various model systems, we have evaluated a collection of isogenic S. typhimurium mutants for virulence during oral infection of calves. This comparative analysis provided an opportunity to assess the relative importance of virulence mechanisms during diarrheal disease caused by S. typhimurium. To facilitate comparison of our results with published data, the collection contained the well-characterized S. typhimurium aroA vaccine, which has been characterized previously for calf virulence. In addition, the collection contained strains with mutations in major S. typhimurium virulence determinants, including Salmonella pathogenicity island 1 (SPI-1), SPI-2, SPI-5, the LPS outer core region, and the Salmonella plasmid virulence (spv) operon.
The invasion-associated type III secretion system encoded by SPI-1 is required for entry of S. typhimurium into intestinal epithelial cells and for colonization of murine Peyer’s patches (12). The contribution of SPI-1 to calf virulence was assessed by using two S. typhimurium mutants, STN61 (hilA) and STN162 (prgH) (49). STN61 carries a mutation in hilA, a gene required for efficient entry of S. typhimurium into HEp-2 cells (2). The hilA gene is located on SPI-1 at centisome 63 on the S. typhimurium chromosome and encodes a transcriptional activator required for expression of SPI-1 genes, including the prgHJIK operon (2, 20). PrgH is a component of the needle complex formed by the invasion-associated type III secretion apparatus, which is required for the export of effector proteins and for the entry of S. typhimurium into epithelial cells in vitro (3, 25). Strain STN162 carries a mutation in prgH which renders it defective for invasion of HEp-2 cells and for colonization of murine Peyer’s patches (49). Following secretion by the type III export machinery, effector proteins are translocated into the host cell cytosol, where they alter signaling pathways or interact directly with the cytoskeleton of the eukaryotic cell (9, 13, 17, 54). One of these effector proteins, SopB (also known as SigD), is encoded by a gene located on SPI-5 at centisome 20 of the S. typhimurium chromosome (13, 52). The contribution of SopB to S. typhimurium calf virulence was assessed by using a mutant (BA1567) carrying a transposon insertion in sopB (1) and a strain (SVM255) carrying a nonpolar frameshift mutation in sopB (18).
S. typhimurium possesses a second type III secretion system which is encoded by SPI-2 at 30.7 centisomes on the chromosome (32, 39). Mutations in genes encoding the centisome 30.7 type III secretion system reduce the ability of S. typhimurium to survive in macrophages in vitro and render bacteria unable to replicate in livers and spleens of mice (32, 38). A strain (STN119) carrying a mutation in spiB, a gene encoding a component of the type III secretion apparatus, was used to assess the contribution of SPI-2 to S. typhimurium calf virulence. Insertional inactivation of spiB renders S. typhimurium avirulent (more than 10,000-fold attenuated) for mice (32). A second virulence gene cluster required for bacterial growth in livers and spleens of mice, the spv operon, is encoded by the virulence plasmid of S. typhimurium (16). The first gene, spvR, encodes a transcriptional activator for the following four genes, spvABCD, whose functions are currently unknown (4). A Tn5 insertion in spvR results in a 1,000-fold-decreased colonization of murine splenic tissue by S. typhimurium (4). To assess the contribution of the spv operon to diarrheal disease, an S. typhimurium strain (STN272) carrying a transposon insertion in spvR was tested for virulence during oral infection of calves.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Derivatives of ATCC 14028, a bovine S. typhimurium isolate, are listed in Table 1. Strain IR715 is a spontaneous nalidixic acid-resistant derivative of 14028 (47), which is fully calf virulent (49). Bacteria were cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar (16 g/liter) plates. If appropriate, antibiotics were added at the indicated concentrations: kanamycin, 100 mg/liter; nalidixic acid, 50 mg/liter; tetracycline, 20 mg/liter.
TABLE 1.
Derivatives of S. typhimurium ATCC 14028 used in this study
| Strain | Genotype | Reference |
|---|---|---|
| IR715 | 14028 nalidixic acid resistant, wild type | 47 |
| STN61 | IR715 hilA::mini-Tn5Km2 | 49 |
| STN119 | IR715 spiB::mini-Tn5Km2 | 49 |
| STN162 | IR715 prgH::mini-Tn5Km2 | 49 |
| STN166 | IR715 rfaJ::mini-Tn5Km2 | 49 |
| STN272 | IR715 spvR::mini-Tn5Km2 | 49 |
| SVM255 | 14028 sopB | 18 |
| BA1567 | 14028 sopB::MudJ | 1 |
| CL1509 | 14028 aroA::Tn10 | 48 |
Experimental animals.
Milk-fed male Friesian-Holstein calves (n = 42), aged 3 to 4 weeks, were obtained from a commercial dairy calf rearing operation. The weight of the calves ranged between 45 and 52 kg. Animals were cared for according to American Association for Accreditation of Laboratory Animal Care guidelines. Calves were fed milk replacer twice daily and were given water ad libitum. Prior to their use for experiments, calves were evaluated for the absence of elevated leukocyte counts, fever, and fecal excretion of Salmonella serotypes. Salmonella serotypes were detected in fecal swabs by enrichment in tetrathionate broth (Difco) and plating on brilliant green agar (BBL).
Challenge.
Oral infection of the calves was performed as described previously (26). In brief, the optical density of overnight cultures at 600 nm was determined, and volumes containing approximately equal numbers of each strain were added to 50 ml of a suspension of 5% magnesium trisilicate, 5% sodium bicarbonate, and 5% magnesium carbonate buffer. The inoculum was added to 950 ml of milk replacer and fed orally to calves. Serial 10-fold dilutions of the inoculum were spread on LB plates to determine the exact challenge dose per calf.
Examination of calves after challenge.
A previous report indicates that in calves which survive an S. typhimurium infection diarrhea does not persist beyond the 10th day postinfection (34). Calves were hence monitored for 10 days postinfection and then humanely euthanized. Weights and fecal scores of calves were recorded daily. Fecal score was used to assess the severity of diarrhea: 1 = normal feces; 2 = soft feces with loss of distinct conformation; 3 = diarrhea, loose feces with reduced solid matter; and 4 = diarrhea, aqueous feces with markedly reduced or little solid matter, blood, and shreds of fibrin. Shedding of S. typhimurium was monitored by taking daily fecal swabs, subsequently enriching them in tetrathionate broth (Difco), and plating them on brilliant green agar (BBL). When calves developed anorexia or were unable to stand, they were euthanized for humane reasons as described previously (26).
Histopathology.
Euthanasia and preparation of tissue samples for histopathology were performed as described previously (26). At necropsy, tissue samples from Peyer’s patches, ileum, and mesenteric lymph node were collected; a portion was homogenized in phosphate-buffered saline and plated in the presence of the appropriate antibiotics for enumeration of bacteria. The remaining tissue samples were coded for blind examination. Tissue samples were fixed in 10% formalin, embedded in paraffin, sectioned at 0.008 mm, and stained with hematoxylin and eosin for histopathological examination.
RESULTS
Mutations in aroA and SPI-1 reduce the severity of diarrhea in calves.
Mutations in sopB, prgH, hilA, rfaJ, spvR, or spiB have previously been implicated in enteropathogenicity (1, 22, 26, 49, 52), but their effect on S. typhimurium virulence during oral infection of calves is not known. In this study, the virulence of a collection of S. typhimurium strains, each carrying a mutation in one of these genes, was compared to that of the isogenic wild type (IR715) during oral infection of calves. Two S. typhimurium sopB mutants were used in this study (Table 1). Strain SVM255 carries a 4-bp deletion within the sopB coding sequence, and this mutation is nonpolar on the expression of the downstream gene pipC (also known as sigE) (18). Strain BA1567 carries the transposon MudJ inserted 100 bp downstream of the beginning of the sopB open reading frame (1). Strains STN61 and STN162 carry transposon insertions in the SPI-1 genes hilA (at bp 920 of the open reading frame) and prgH (at bp 129 of the open reading frame), respectively. The invasion-associated type III secretion system is nonfunctional in strains STN61 (hilA) and STN162 (prgH), as suggested by the reduced ability of these mutants to invade HEp-2 cells and to colonize murine Peyer’s patches (49). Strain STN166 is rough due to a mini-Tn5Km2 insertion at bp 564 of the rfaJ open reading frame (49). The rfaJ gene product is required for LPS outer core biosynthesis and mouse virulence (5, 27, 31). Strain STN272 carries a mini-Tn5Km2 insertion at bp 381 of the spvR open reading frame which renders this mutant defective for colonization of internal organs of mice (49). SpvR is a positive regulator of spvABCD expression and is required for full mouse virulence of S. typhimurium (4). Strain STN119 carries a transposon which is inserted at bp 909 of the spiB open reading frame (49). The spiB gene, which is part of the type III secretion apparatus encoded by SPI-2, is essential for S. typhimurium mouse virulence and growth within macrophages in vitro (32). Inactivation of spiB in strain STN119 results in a marked defect for colonization of internal organs of mice (49). Finally, an S. typhimurium aroA mutant (CL1509) was tested for calf virulence, since this mutation has previously been reported to result in avirulence and a reduced severity of diarrhea (46).
Calves infected with these S. typhimurium strains fell into two groups. Calves infected with strain IR715 (wild type), STN119 (spiB), STN166 (rfaJ), or STN272 (spvR) or either of two sopB mutants (SVM255 and BA1567) had aqueous diarrhea with feces containing various combinations of blood, fibrin, and mucus. These S. typhimurium strains could be detected in fecal swabs until day 9 postinfection unless lethal signs of disease developed earlier. The weight loss of animals infected at a dose of 1010 CFU/animal ranged between 1.5 and 5 kg, and terminal signs of illness, including anorexia and central nervous system depression, occurred between 1 and 3 days postinoculation.
In contrast, calves infected with strain CL1509 (aroA), STN61 (hilA), or STN162 (prgH) either had only soft feces for 3 to 4 days or had normal feces. Fecal shedding of these mutants ceased by day 6 postinfection in all but one calf. The weight of calves infected at a dose of 1010 CFU/animal remained constant or increased. After 10 days, all calves were healthy.
In conclusion, mutations in hilA, prgH, or aroA reduced the severity of diarrhea in calves, whereas animals infected with the wild type or strains carrying mutations in spiB, rfaJ, spvR, or sopB developed clinically severe diarrhea.
Mutations in aroA and SPI-1 result in a markedly reduced mortality in calves.
The 50% lethal morbidity dose of S. typhimurium IR715 for calves is approximately 6 × 108 CFU/animal (49). Calves were infected orally at a dose of 1010 CFU/animal (Table 2). At this dose, all calves infected with IR715 (wild type), STN119 (spiB), STN272 (spvR), BA1567 (sopB), or SVM255 (sopB) developed signs of terminal illness and were euthanized. In contrast, all calves infected with STN61 (hilA), STN162 (prgH), or CL1509 (aroA) survived the infection and remained healthy. No mortality was observed when groups of two calves were infected with strain STN119 (spiB), STN272 (spvR), STN61 (hilA), STN162 (prgH), or STN166 (rfaJ) at a dose of 109 CFU/animal, although fecal scores were similar at both doses. The wild type (IR715) caused lethal morbidity in six of eight calves infected orally with 109 CFU/animal. In conclusion, mutations in aroA, hilA, and prgH attenuated S. typhimurium to a much greater degree than did mutations in spiB, sopB, or spvR.
TABLE 2.
Virulence of S. typhimurium strains for 3- to 4-week-old Friesian-Holstein calves
| Strain | Genotype | Result for infection with 1010 CFU/animal
|
||
|---|---|---|---|---|
| No. dead/no. total | Avg time to death (days) | Avg fecal scorea | ||
| IR715 | Wild type | 8/8 | 3.1 | 3.1 |
| STN61 | hilA | 0/4 | NAb | 1.9 |
| STN119 | spiB | 2/2 | 2.3 | 3.8 |
| STN162 | prgH | 0/2 | NA | 2.1 |
| STN166 | rfaJ | 1/2 | 3 | 3.4 |
| STN272 | spvR | 2/2 | 1.2 | 4 |
| SVM255 | sopB | 2/2 | 4 | 3.8 |
| BA1567 | sopB | 2/2 | 4 | 3.1 |
| CL1509 | aroA | 0/2 | NA | 2.1 |
Fecal scores were used to assess the severity of diarrhea, where 1 = normal feces; 2 = soft feces with loss of distinct conformation; 3 = diarrhea, loose feces with reduced solid matter; and 4 = diarrhea, aqueous feces with markedly reduced or little solid matter, blood, or fibrin.
NA, not applicable.
Mutations in SPI-1, SPI-2, rfaJ, and aroA reduce the severity of intestinal lesions in calves.
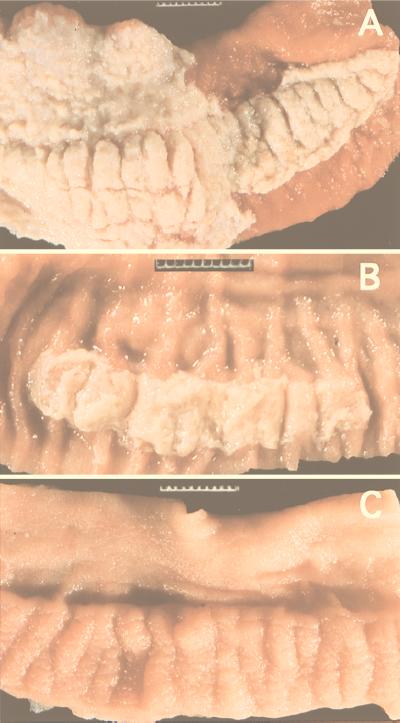
To more fully understand the disease processes and outcomes, it is necessary to study the development and spectrum of lesions in the target tissues. At necropsy, gross pathologic examination was performed with calves infected at a dose of 1010 CFU/animal and tissue samples from Peyer’s patches, ileum, and mesenteric lymph node were collected for histopathologic examination. The wild type, strain STN272 (spvR), and to a lesser extent strain SVM255 (sopB) produced macroscopically severe acute fibrinopurulent necrotizing enteritis with segmental or continuous pseudomembrane deposition (Fig. 1A) of the terminal 5 m of the ileum, Peyer’s patches of the middle to terminal ileum, and the cranial 1 to 2 m of the colon. Strains STN119 (spiB) and STN166 (rfaJ) produced lesions of moderate to marked subacute fibrinopurulent enteritis often confined to Peyer’s patches (Fig. 1B) of the terminal one-third of the ileum. No gross pathological lesions were detected during necropsy of calves infected with STN61 (hilA), STN162 (prgH), or CL1509 (aroA) (Fig. 1C).
FIG. 1.
Representative examples of the gross pathology of Peyer’s patch and mucosa of the terminal ileum of calves inoculated orally with 1010 CFU of different S. typhimurium strains. (A) Severe acute fibrinopurulent necrotizing enteritis with segmental or continuous pseudomembrane formation of calves inoculated with wild type (IR715), strain STN272 (spvR), or strain SVM255 (sigD). (B) Moderate to marked subacute fibrinopurulent enteritis often confined to Peyer’s patches of calves inoculated with STN119 (spiB) or STN166 (rfaJ). (C) Normal Peyer’s patch and ileal mucosa of calves inoculated with STN61 (hilA), STN162 (prgH), or CL1509 (aroA) or of uninoculated controls. Bar = 1 cm.
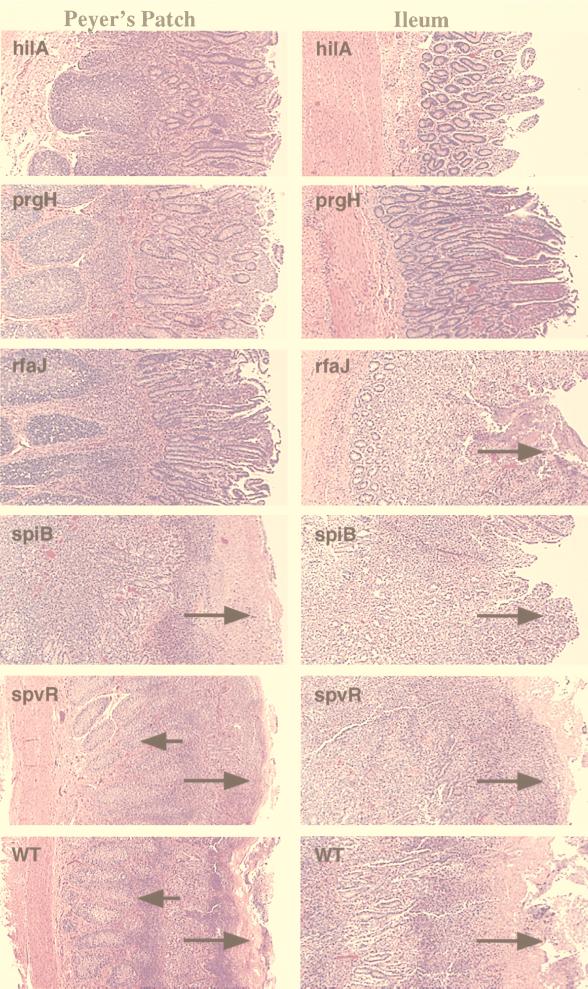
Histopathologic analysis revealed severe lymphoid depletion and confirmed the severe acute fibrinopurulent necrotizing enteritis extending from the muscularis mucosa to the luminal surface with pseudomembrane formation at the luminal surface of Peyer’s patches and the terminal ileum of calves infected with the wild type (IR715), strain STN272 (spvR), and strain SVM255 (sopB) (Fig. 2). Extensive lymphoid depletion also occurred in the germinal centers of the mesenteric lymph nodes of cattle infected with these mutants. The spiB mutant (STN119) and the rfaJ mutant (STN166) caused intestinal lesions of reduced severity with only mild lymphoid depletion (Fig. 2). Microscopic examination revealed that intestinal lesions were either negligible or absent in calves infected with STN61 (hilA), STN162 (prgH), or CL1509 (aroA) (data not shown). Occasional crypt abscesses were detected, and in addition, STN61 (hilA) caused slight lymphoid depletion of germinal centers in the mesenteric node.
FIG. 2.
Histopathology of Peyer’s patches and ileum from perinatal calves inoculated per os with 1010 CFU of the indicated strains of S. typhimurium per animal, photographed at a 40× magnification. Shown are hematoxylin-and-eosin-stained sections of Peyer’s patch (left column) and terminal ileum (right column). Short arrows indicate areas of marked lymphoid depletion; long arrows indicate zones of variable degrees of fibrinopurulent necrotizing ileitis at the mucosal surface.
The severity of intestinal lesions correlated in most cases with the ability of a mutant to colonize bovine intestinal tissues. S. typhimurium hilA or prgH mutants, which caused only minor lesions (Fig. 1C and 2), have a more than 350-fold-reduced ability to colonize bovine Peyer’s patches (49). A mutation in rfaJ reduces the ability to colonize bovine Peyer’s patches 40-fold (49) and leads to lesions of intermediate severity (Fig. 1B and 2). An spvR mutant does not display a significant colonization defect for bovine Peyer’s patches (49) and causes severe lesions (Fig. 1A and 2). Similarly, an S. dublin sopB mutant invades the intestinal mucosa in bovine loops at wild-type level (13), and an S. typhimurium sopB mutant produced severe lesions. However, an exception to this trend was seen with the spiB mutant (STN119), which has no significant colonization defect for bovine Peyer’s patches (49) and yet caused lesions of reduced severity (Fig. 1B).
DISCUSSION
Virulence is a complex phenotype, which is only fully expressed during host-pathogen interaction in vivo. Therefore, the elucidation of mechanisms by which S. typhimurium produces diarrhea requires the use of an animal model, which resembles the natural course of infection and the typical signs of disease. The animal models commonly utilized to study virulence mechanisms of S. typhimurium are inbred mouse lineages, most frequently BALB/c. The disease produced by S. typhimurium in mice is similar to typhoid fever caused by Salmonella typhi in humans (33). However, mice do not develop diarrhea, and therefore, murine typhoid does not accurately model the enteritis caused by S. typhimurium in humans. Calves infected with S. typhimurium, on the other hand, develop acute enteritis and hence are better suited for the study of diarrheal disease (14). The high cost of the calf model precludes its use for screening a large number of putative virulence determinants. As a consequence, to date mutations in only four genes (aroA, invH, dap, and galE) have been shown to attenuate S. typhimurium during oral infection of cattle (6, 7, 46, 51). The goal of the present study was to extend this analysis to include other S. typhimurium genes, which have been suggested to be important for virulence in calves.
Recently, the invasion-associated type III secretion system located on SPI-1 has been implicated in enteropathogenicity of S. typhimurium and S. dublin. For instance, a mutation in invH, a gene located on SPI-1, causes attenuation during oral infection of calves (51). Inactivation of sipB, a SPI-1 gene encoding a type III secreted effector protein, reduces S. dublin-induced fluid secretion in bovine intestinal loops (13). Furthermore, a mutation in hilA, a positive regulator of genes located on SPI-1, reduces the ability of S. typhimurium to cause fluid accumulation in bovine ligated ileal loops (1) and to colonize intestinal tissue of calves during a competitive infection with the wild type (49). Similarly, an S. typhimurium prgH mutant is recovered in lower numbers than is the wild type from bovine intestinal tissue (49). Consistent with these reports, we found that mutations in hilA and prgH markedly reduced the severity of diarrhea during an S. typhimurium infection in calves (Table 2). Calves infected with STN61 (hilA) and STN162 (prgH) passed loose stools for 3 to 5 days and did not lose weight. While SPI-1-mediated colonization of intestinal tissues appears to be essential for bovine enteritis, it is less important during infection of mice. For instance, a mutation in hilA does not attenuate S. typhimurium ATCC 14028 during intragastric infection of mice (1) but caused marked attenuation (≥47-fold) after oral infection of calves (Table 2). Since bovine enteritis closely resembles S. typhimurium infections in humans, it is likely that virulence determinants important for enteropathogenicity in calves (e.g., SPI-1) are of greater importance for human disease than predicted by the mouse data.
In addition to SPI-1, SPI-5 has recently been implicated in calf enteropathogenicity of S. dublin. For example, S. dublin-mediated fluid secretion in bovine ligated ileal loops requires sopB (sigD), a gene located on SPI-5 which encodes a protein whose injection into epithelial cells is mediated by an SPI-1-dependent mechanism (13, 52). Surprisingly, calves inoculated with either S. typhimurium sopB mutant (SVM255 or BA1567) developed severe diarrhea, severe intestinal lesions, and lethal morbidity at wild-type levels (Table 2). A mutation in the S. dublin sopB gene causes a relatively weak reduction in fluid accumulation in bovine ligated ileal loops compared to that elicited by a sipB mutant (13) or a sopB sopD mutant (22). Hence, it is possible that the observation that an S. dublin sopB mutant elicits reduced fluid secretion in bovine intestinal segments while an S. typhimurium sopB mutant causes severe diarrhea during an oral infection may be explained by the differences in sensitivity of the loop assay and oral infection. Alternatively, the relative importance of sopB for S. dublin and S. typhimurium calf virulence may differ. Nevertheless, this result illustrates the importance of performing oral infection experiments to conclude that a mutation markedly reduces the virulence of S. typhimurium for calves.
Unlike S. dublin, which causes a systemic disease in calves, S. typhimurium produces primarily an enteric infection (41, 53). The virulence plasmid of S. dublin is not involved in the enteric phase of infection in calves but is required for full virulence and persistence at systemic sites (50). The finding that an S. typhimurium spvR mutant (STN272) produced severe diarrhea, marked intestinal lesions, and lethal morbidity in calves confirmed, therefore, that growth at systemic sites of infection is not critical to the pathogenesis of bovine enteritis caused by this pathogen (Table 2 and Fig. 2).
S. typhimurium SPI-2 mutants are avirulent (more than 10,000-fold attenuated) in mice (32, 39), because they are unable to grow in liver and spleen (38). Furthermore, SPI-2 mutants are immunogenic for mice and capable of effective delivery of foreign antigen (29). In contrast, an S. typhimurium SPI-2 (spiB) mutant caused lethal morbidity and diarrhea comparable to those caused by the wild type in calves infected with 1010 CFU/animal. This example illustrates that attenuation in mice is not always predictive of the degree to which a mutation will reduce pathogenicity for calves. In addition, these data suggest that the use of the mouse model for the development of live attenuated S. typhimurium vaccines biases the selection of new candidates toward those unable to grow in liver and spleen, since this defect leads to an optimal attenuation in mice. However, such candidates may not be sufficiently attenuated to serve as oral vaccines in cattle, since growth at systemic sites of infection is not as critical to the pathogenesis of bovine enteritis.
ACKNOWLEDGMENTS
We thank V. L. Miller for providing strain SVM255, B. M. Ahmer for providing strain BA1567, C. Tanksley and T. Parsons for care of animals, R. Barthel and J.-A. Gutiérrez-Pabello for assistance with necropsies, and R. A. Kingsley and M. Manson for critical comments on the manuscript.
This work was supported by USDA/NRICGP grant 9702568 to R.M.T., USDA Formula Animal Health Funding to T.A.F. and A.J.B., and USDA/NRICGP grant 9802610 and PHS grant AI40124 to A.J.B.
REFERENCES
- 1.Ahmer B M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. HilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell A L, Gulig P A. The Salmonella typhimurium virulence plasmid encodes a positive regulator of a plasmid-encoded virulence gene. J Bacteriol. 1991;173:7176–7185. doi: 10.1128/jb.173.22.7176-7185.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carstenius P, Flock J I, Lindberg A. Nucleotide sequence of rfaI and rfaJ genes encoding lipopolysaccharide glycosyl transferases from Salmonella typhimurium. Nucleic Acids Res. 1990;18:6128. doi: 10.1093/nar/18.20.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke R C, Gyles C L. Galactose epimerase mutants of Salmonella typhimurium as live vaccines for calves. Can J Vet Res. 1986;50:165–173. [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke R C, Gyles C L. Vaccination of calves with a diaminopimelic acid mutant of Salmonella typhimurium. Can J Vet Res. 1987;51:32–38. [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke R C, Gyles C L. Virulence of wild and mutant strains of Salmonella typhimurium in ligated intestinal segments of calves, pigs, and rabbits. Am J Vet Res. 1987;48:504–510. [PubMed] [Google Scholar]
- 9.Collazo C M, Galan J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 10.Edwards P R, Bruner D W. The occurrence and distribution of Salmonella types in the United States. J Infect Dis. 1943;72:58–67. doi: 10.1093/infdis/83.3.220. [DOI] [PubMed] [Google Scholar]
- 11.Frost A J, Bland A P, Wallis T S. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet Pathol. 1997;34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- 12.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 14.Gibson E A. Salmonellosis in calves. Vet Rec. 1961;73:1284–1296. [PubMed] [Google Scholar]
- 15.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 16.Gulig P A, Doyle T J. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardt W D, Chen L M, Schuebel K E, Bustelo X R, Galan J E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 18.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hormaeche C E. The in vivo division and death rates of Salmonella typhimurium in the spleens of naturally resistant and susceptible mice measured by the superinfecting phage technique of Meynell. Immunology. 1980;41:973–979. [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 21.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones P W, Dougan G, Hayward C, Mackensie N, Collins P, Chatfield S N. Oral vaccination of calves against experimental salmonellosis using a double aro mutant of Salmonella typhimurium. Vaccine. 1991;9:29–34. doi: 10.1016/0264-410x(91)90313-u. [DOI] [PubMed] [Google Scholar]
- 24.Khan S A, Everest P, Servos S, Foxwell N, Zahringer U, Brade H, Rietschel E T, Dougan G, Charles I G, Maskell D J. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 25.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 26.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes of the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyman M B, Steward J P, Roantree R J. Characterization of the virulence and antigenic structure of Salmonella typhimurium strains with lipopolysaccharide core defects. Infect Immun. 1976;13:1539–1542. doi: 10.1128/iai.13.6.1539-1542.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maw J, Meynell G G. The true division and death rates of Salmonella typhimurium in the mouse spleen determined with superinfecting phage P22. Br J Exp Pathol. 1968;49:597–613. [PMC free article] [PubMed] [Google Scholar]
- 29.Medina E, Paglia P, Nikolaus T, Müller A, Hensel M, Guzmán C A. Pathogenicity island 2 mutants of Salmonella typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect Immun. 1999;67:1093–1099. doi: 10.1128/iai.67.3.1093-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukkur T K, Walker K H, Jones D, Wronski E, Love D N. Immunizing efficacy of aromatic-dependent Salmonella dublin in mice and calves. Comp Immunol Microbiol Infect Dis. 1991;14:243–256. doi: 10.1016/0147-9571(91)90005-x. [DOI] [PubMed] [Google Scholar]
- 31.Nevola J J, Stocker B A, Laux D C, Cohen P S. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun. 1985;50:152–159. doi: 10.1128/iai.50.1.152-159.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ørskov J, Moltke O. Studien über den Infektionsmechanismus bei verschiedenen Paratyphus-Infektionen in weißen Mäusen. Z Immunitaetsforsch. 1929;59:357–405. [Google Scholar]
- 34.Rankin J D, Taylor R J. The estimation of the doses of Salmonella typhimurium for experimental production of disease in calves. Vet Rec. 1966;78:706–707. doi: 10.1136/vr.78.21.706. [DOI] [PubMed] [Google Scholar]
- 35.Rice D H, Besser T E, Hancock D D. Epidemiology and virulence assessment of Salmonella dublin. Vet Microbiol. 1997;56:111–124. doi: 10.1016/S0378-1135(96)01352-1. [DOI] [PubMed] [Google Scholar]
- 36.Rothenbacher H. Mortality and morbidity in calves with salmonellosis. J Am Vet Med Assoc. 1965;147:1211–1214. [PubMed] [Google Scholar]
- 37.Segall T, Lindberg A A. Salmonella dublin experimental infection in calves: protection after oral immunization with an auxotrophic aroA live vaccine. Zentbl Vetmed Reihe B. 1991;38:142–160. doi: 10.1111/j.1439-0450.1991.tb00857.x. [DOI] [PubMed] [Google Scholar]
- 38.Shea J E, Beuzon C R, Gleeson C, Mundy R, Holden D W. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith B P, Da Roden L, Thurmond M C, Dilling G W, Konrad H, Pelton J A, Picanso J P. Prevalence of salmonellae in cattle and in the environment on California dairies. J Am Vet Med Assoc. 1994;205:467–471. [PubMed] [Google Scholar]
- 41.Smith H W, Jones J E. Observations on experimental oral infection with Salmonella dublin in calves and Salmonella choleraesuis in pigs. J Pathol Bacteriol. 1967;93:141–156. doi: 10.1002/path.1700930114. [DOI] [PubMed] [Google Scholar]
- 42.Sojka W J, Field H I. Salmonellosis in England and Wales 1958–1967. Vet Bull. 1970;40:515–531. [Google Scholar]
- 43.Sojka W J, Wray C, Hudson E B, Benson J A. Incidence of salmonella infection in animals in England and Wales, 1968–73. Vet Rec. 1975;96:280–284. [Google Scholar]
- 44.Sojka W J, Wray C, Shreeve J, Benson A J. Incidence of salmonella infection in animals in England and Wales 1968–1974. J Hyg (London) 1977;78:43–56. doi: 10.1017/s0022172400055923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sojka W J, Wray C, Shreeve J E, Bell J C. The incidence of salmonella infection in sheep in England and Wales, 1975 to 1981. Br Vet J. 1983;139:386–392. doi: 10.1016/s0007-1935(17)30383-4. [DOI] [PubMed] [Google Scholar]
- 46.Stocker B A, Hoiseth S K, Smith B P. Aromatic-dependent “Salmonella sp.” as live vaccine in mice and calves. Dev Biol Stand. 1983;53:47–54. [PubMed] [Google Scholar]
- 47.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsolis R M, Bäumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsolis, R. M., S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Bäumler. Identification of a putative Salmonella typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 50.Wallis T S, Paulin S M, Plested J S, Watson P R, Jones P W. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun. 1995;63:2755–2761. doi: 10.1128/iai.63.7.2755-2761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Mutation of invH, but not stn, reduces salmonella-induced enteritis in cattle. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood M W, Jones M A, Watson P R, Hedges S, Wallis T S, Galyov E E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- 53.Wray C, Sojka W J. Experimental Salmonella typhimurium infection in calves. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]
- 54.Zhou D, Mooseker M S, Galan J E. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]