Abstract
Hypertension is a leading risk factor of cardiovascular disease and mortality in the population worldwide. Recently, hundreds of genomic loci were reported for hypertension by GWAS, however, the most SNPs are located in intergenic regions of genome, where a functional cause is difficult to determine. In the current study, a TWAS of hypertension was conducted using 452,264 individuals including 84,640 patients. KEGG and GO enrichment analyses were performed for the hypertension-related genes identified via TWAS. PPI network analysis based on the STRING database was also performed to detect TWAS-identified genes in hypertension. We have identified 18,420 genes from the GWAS summary data, and of those 1010 non-overlapping genes expression were significantly associated with hypertension after FDR correction (PFDR <0.05) in four tissues (left heart ventricle, aorta, whole blood, and peripheral blood). The KEGG and GO terms were mostly related to autoimmune mechanisms, and the autoimmune-related pathways have also been enriched using GO analysis for PPI genes. We further performed Mendelian randomization analysis, and the results supported a significant association between autoimmunity and hypertension. Moreover, 15 novel hypertension-susceptible genes were identified in all tissues, and five of the genes (RBM6, HLA-DRB5, UHRF1BP1, LYZ, and TMEM116) were associated with autoimmune system, which provide further evidence supporting an autoimmune mechanism in hypertension. In summary, our study supports that an autoimmune mechanism plays an important role in the development of hypertension, and these findings will provide new biological insights that will assist in deciphering the molecular etiology of hypertension.
Keywords: TWAS, Hypertension, GWAS, Autoimmunity, BP
Highlights
-
•
This is the first systematic TWAS for essential hypertension.
-
•
15 novel genes were identified in four tissues, and five of the genes were associated with the autoimmune system.
-
•
Our study supports that an autoimmune mechanism plays an important role in the development of hypertension.
1. Introduction
Hypertension refers to a clinical syndrome characterized by increased systemic arterial blood pressure (BP), which may be accompanied by functional or organic damage to organs such as the heart, brain, and kidneys, and is the most common chronic disease [1,2]. It is a leading risk factor of cardiovascular disease and mortality, affecting approximately 27% of the population worldwide [3,4]. Genetic, estimated to have a 30–60% heritability, and environmental factors and their interaction determine the hypertension risk for an individual [5].
In 2009, two large-scale meta-analyses of genome-wide association studies (GWAS) have identified 13 genomic loci significantly associated with blood pressure variation [6,7]. Subsequently, the International Consortium for Blood Pressure Genome-Wide Association Studies and Kato et al. identified 16 novel loci and four new loci, respectively, in different populations [2,8]. The UK Biobank Cardio-metabolic Traits Consortium Blood Pressure Working Group reported 107 independent loci and related biological pathways for BP in 2017 [9]. To date, hundreds of genomic loci have been reported for hypertension by GWAS [4,[10], [11], [12]]. Unfortunately, the most associated single nucleotide polymorphism (SNP) markers of GWAS are not located in or near genes but rather in intergenic regions of genome, where the functional role of these variants is difficult to be elucidate. Integrating gene expression or other omics may be a better way to investigate underling biological mechanisms while overcoming the challenges in GWAS [13]. Recently, transcriptome-wide association studies (TWAS) integrating GWAS and expression quantitative trait loci (eQTLs) data have been proposed to identify gene-trait associations [14,15]. Gusev et al. performed a TWAS integrating the schizophrenia GWAS with expression data from brain, blood and adipose tissues, and identified 35 novel genes which were involved in 157 TWAS significant genes [16]. TWAS has been utilized to interrogate the genetics of many diseases including prostate cancer, ankylosing spondylitis, low-density-lipoprotein cholesterol, Crohn's, and may be more useful in prioritizing candidate causal genes than GWAS [15,[17], [18], [19]].
In the current study, a large-scale hypertension GWAS data set was utilized to identify genetic loci that may be associated with hypertension by TWAS. Integrating of bioinformatic analysis including enrichment analysis, protein-protein interaction (PPI) network analysis, and Mendelian randomization (MR) analysis, were performed on candidate genes to identify hypertension-related genes and biological pathways. This study has discovered new genetic loci related to hypertension and provided new clues for understanding the molecular mechanism of hypertension.
2. Materials and methods
This study was approved by the Medical Ethics Committee of Xi'an Jiaotong University (Xi'an, China).
2.1. GWAS data for hypertension
GWAS summary data for essential hypertension was obtained from UK Biobank samples (UK Biobank field: 20002). Briefly, information on hypertension phenotypes was collected from each participant in the UK Biobank cohort, which included a total of 452,264 white British individuals, including 84,640 patients with essential hypertension. All study subjects had blood samples taken at the subject's visit to the UK Biobank Assessment Centre, and DNA extraction and genotyping were done at the Affymetrix Research Services Laboratory. This dataset contains 9,113,133 filtered imputation variants and the IMPUTE4 program was used to perform the imputation (http://jmarchini.org/software/). Details on subjects, genotyping, attribution and quality control can be found in previous publication [20].
2.2. TWAS of hypertension
We used the FUnctional Summary-based ImputatiON (FUSION) method (http://gusevlab.org/projects/fusion/) for TWAS analysis of hypertension [14]. To measure significant SNP-trait associations, all genome-wide testing burdens have been corrected to ensure that the TWAS false positive rate is well-controlled. The software program FUSION (default settings) was used for the TWAS and joint analyses of regions containing multiple significant associations [21]. The most popular TWAS methods, such as PrediXcan, TWAS-Fusion, and SMR, test causal relationships between gene-expression levels and complex traits [22], among which, the TWAS-Fusion method is used more often. Briefly, Bayesian sparse linear-mixed models [23] were used to calculate SNP expression weights for specific genes at the 1-Mb cis position and estimate the association of predicted expression levels with hypertension using the following formula: Ztwas = w + Z/(w[Lw]1/2) [14], where w denotes the weight, Z denotes the Z-score, and L denotes the SNP correlation matrix (definition, LD). Each feature expanded in 100,000 bp was defined contiguous. The Minium p-value to include feature in the joint model was 0.05. Features with r2 greater than 0.9 would be considered identical, and Features with r2 less than 0.008 would be considered independent. The diagnosis of hypertension relies on blood pressure tests, which depends on the ability of left ventricle to eject, aortic pressure, and blood volume [24]. Thus, we used the gene expression weights for the left heart ventricle, aorta, whole blood, and peripheral blood as references, and they can be downloaded from the FUSION website (http://gusevlab.org/projects/fusion/). All P values are then subjected to multiple testing correction using the Benjamini-Hochberg procedure to gather Q values, which represent the minimum False Discovery Rate (FDR) threshold at which the contact is deemed significant.
2.3. Functional exploration analysis
A Venn diagram was used to identify the common and tissue-specific genes that were expressed among the left heart ventricle, aorta, whole blood, and peripheral blood. The Kyoto Encyclopedia of Genes and Genomes (KEGG) [25] and Gene Ontology (GO) [26] enrichment analyses were performed to identify and confirm related biological processes. The Venn diagram, KEGG and GO enrichment were produced using the R packages “ggplot2”, “org.Hs.eg.db”, and “clusterProfiler” (R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/). PPI network was analyzed by using the STRING v11.5 database (STRING, https://string-db.org), which required a confidence score of 0.15, and “active interaction sources” according to a previous study [27]. Cytoscape was used to visualize all the interaction networks [28].
2.4. MR analysis
MR analysis refers to the use of genetic variants in observational epidemiology to infer the variable risk factors for the disease and health-related outcomes [29]. In this study, MR analysis was used to evaluate the causal relationship between autoimmune diseases (exposure, ID: finn-b-AUTOIMMUNE) and hypertension (outcome). We carried out inverse variance weighted (IVW) methods. The significant dietary patterns identified by LDSC analysis were checked and included in the subsequent analysis. The SNPs were included as instrumental variables after filtering out SNPs whose distance was within 10,000 kb and r2 > 0.001. The number of SNPs included and the effect values (confidence intervals) and P values were reported. MR analysis, heterogeneity and multiple validity tests were conducted by R packages, including “TwoSampleMR”, “RVAideMemoire”, and “MRPRESSO”. P values < 0.05 were considered significant.
2.5. Further analysis for TWAS-identified genes overlapped in four tissues
To identify articles that study the relationship between 15 overlapped TWAS-identified genes and autoimmune, PUBMED (http://www.ncbi.nlm.nih.gov), and SCOPUS (http://www.scopus.com) (up to July 2022) were surveyed with “gene name”, “autoimmunity” as keywords. The articles were read entirely to assess their appropriateness for cited in this study. We also collected the expression information of these 15 identified genes in different tissues in Genecard database (https://www.genecards.org/).
3. Results
3.1. TWAS analysis of hypertension
TWAS analysis identified 18,420 genes from the GWAS summary data, and of those 1387 genes expression were significantly associated with hypertension after FDR correction (PFDR < 0.05), including 422 genes from the left heart ventricle (Fig. 1A), 509 genes in aorta (Figs. 1B), 283 genes in whole blood (Figs. 1C), and 173 genes in peripheral blood (Fig. 1D). Finally, a total of 1010 non-overlapping differentially expressed genes were selected for subsequent analysis after eliminating overcounted genes.
Fig. 1.
Manhattan plots of the association results from the hypertension transcriptome-wide association study. The dashed horizontal lines represent P = 5.00 × 10−2. The solid horizontal lines represent P = 1.00 × 10−3. Each dot represents the genetically predicted expression of one specific gene in the left heart ventricle, aorta, whole blood, and peripheral blood. The X axis represents the chromosome (Chr) encoding the corresponding gene, and the Y axis represents the negative logarithm of the association PTWAS value. A: Gene-expression weights for the left heart ventricle. B: Gene-expression weights for the aorta. C: Gene-expression weights for the whole blood. D: Gene-expression weights for the peripheral blood.
3.2. Functional exploration of TWAS-identified genes associated with hypertension
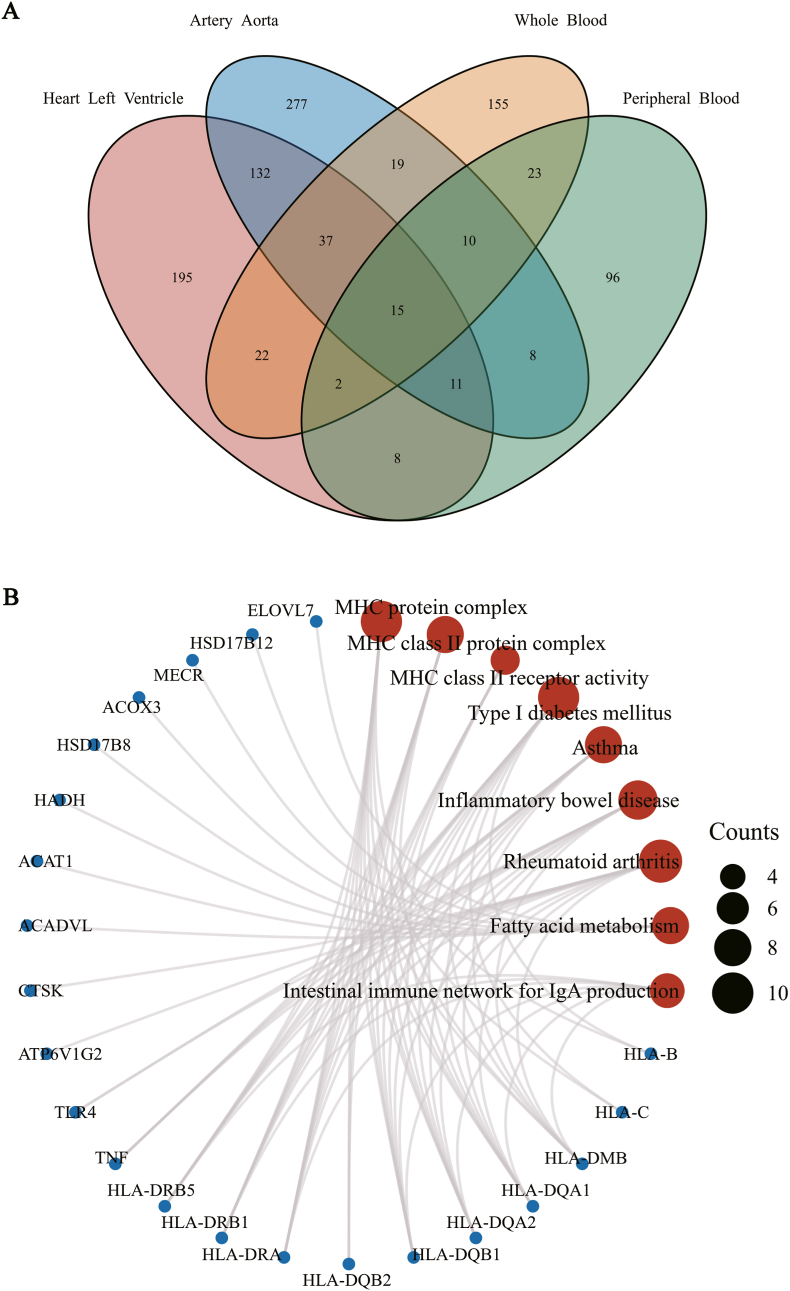
We performed an overlap analysis of the significantly associated genes in different tissues to identify the most representative genes, as every tissue have unique gene-expression profile. Fig. 2A shows the result of the Venn diagram, which indicates the number of genes expressed in one or more tissues. Finally, 15 novel hypertension-susceptible genes were identified by TWAS in all tissues, and they were Glutamine-Fructose-6-Phosphate Transaminase 1 (GFPT1), RNA-binding motif protein 6 (RBM6), Zinc Finger Protein 589 (ZNF589), Major Histocompatibility Complex, Class II, DR Beta 5 (HLA-DRB5), UHRF1 Binding Protein 1 (UHRF1BP1), Nei Like DNA Glycosylase 2 (NEIL2), NOP2/Sun RNA Methyltransferase 6 (NSUN6), Sideroflexin 4 (SFXN4), Fast Skeletal Type, Troponin T3 (TNNT3), Lysozyme (LYZ), Transmembrane Protein 116 (TMEM116), Cell Division Cycle 16 (CDC16), Exosome Component 6 (EXOSC6), Cytokine Receptor Like Factor 3 (CRLF3), Zinc Finger Protein 100 (ZNF100) (Table 1). The expression information of these 15 identified genes in different tissues were shown in Supplementary File 1.
Fig. 2.
Functional exploration of the TWAS-identified genes associated with hypertension. A: Venn diagram revealed overlapping TWAS-significant genes in different tissues. Red, left heart ventricle; blue, aorta; orange, whole blood; green, peripheral blood. B: Network plot of enriched KEGG and GO terms for the TWAS-significant genes. Counts: the number of genes involved in the corresponding signaling pathway. (Red represents pathways, and blue represents genes.)
Table 1.
Causal analysis results between Autoimmune Diseases and Hypertension.
| Exposure | Outcome | Number of SNP | Method | OR (95% CI) | P value |
|---|---|---|---|---|---|
| Autoimmune Diseases | Hypertension | 44 | IVW | 1.1134 (1.059–1.171) | 2.48E-05 |
| WM | 4.30E-04 | ||||
| MR Egger | 5.83E-03 |
We subjected the 1010 TWAS-identified associated genes to KEGG and GO analysis (Fig. 2B). The significantly enriched KEGG and GO terms including MHC protein complex, MHC class II protein complex, MHC class II receptor activity, type I diabetes mellitus, asthma, inflammatory bowel disease, rheumatoid arthritis, fatty acid metabolism, intestinal immune network for IgA production. Furthermore, the P values for GO and KEGG analyses were shown in Supplementary Table 1.
3.3. PPI network of the TWAS-identified genes
1010 hypertension-associated genes were used for PPI analysis, and 736 protein-coding genes were successfully revealed. The PPI genes were used for enrichment analysis (Fig. 3). Several autoimmune-related pathways have been enriched, such as adaptive immune system, regulation of cellular response to stress, cellular responses to stimuli, regulation of hormone levels. Furthermore, PPI network of the 736 TWAS-identified genes was shown in Supplementary Fig. 1.
Fig. 3.
PPI network and functional exploration. A: PPI enrichment analysis, colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. B: PPI enrichment analysis, colored by P-value, where terms containing more genes tend to have a more significant P-value.
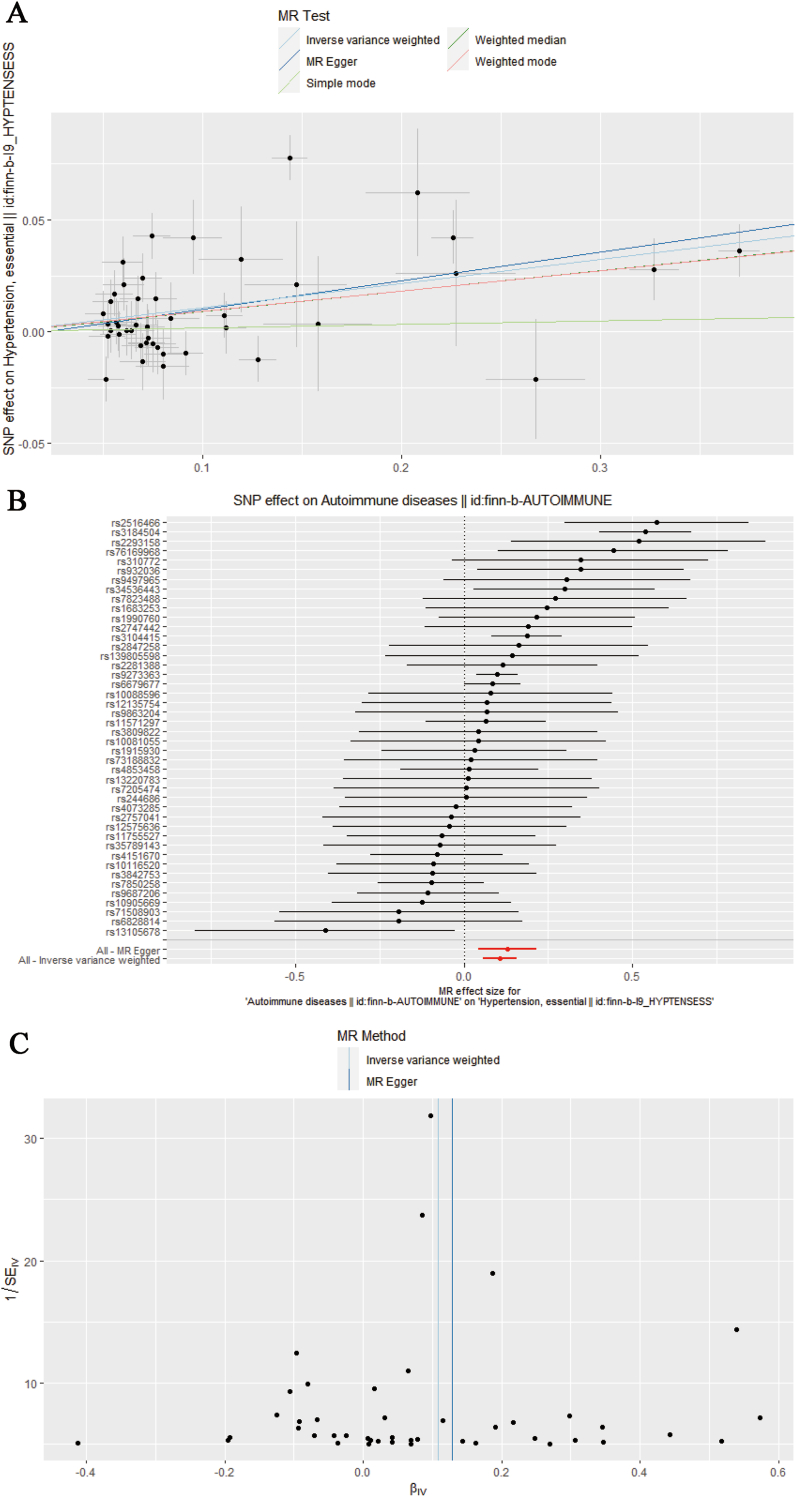
3.4. Causal relationships between autoimmune diseases and hypertension
We also identified causal relationships between autoimmune diseases and hypertension using MR (odds ratio (OR) = 1.1134, 95% confidence interval (CI) = (1.059, 1.171), P value = 2.48E-05) (Table 1). Our study suggested that autoimmune diseases play varied roles in the progression of hypertension (Fig. 4). The result did not satisfy heterogeneity and pleiotropy after testing.
Fig. 4.
MR analysis results. A: Scatter plot of MR. B: Forest plot of MR. C: Funnel plot of MR.
3.5. Five TWAS-identified genes were reported to associate with autoimmune
Five TWAS-identified genes (RBM6, HLA-DRB5, UHRF1BP1, LYZ, and TMEM116) were identified to associate with autoimmunity by the combined search, which is shown in Table 2.
Table 2.
15 TWAS-identified genes associated with hypertension in four tissues.
| Gene | Chromosome | BEST.GWAS.ID |
PFDR value |
Associated with autoimmunitya | |||
|---|---|---|---|---|---|---|---|
| Left Ventricle | Aorta | Whole Blood | Peripheral Blood | ||||
| GFPT1 | 2 | rs4852868 | 6.38E-03 | 1.88E-03 | 2.95E-02 | 1.04E-02 | NR |
| RBM6 | 3 | rs6785549 | 1.32E-03 | 2.23E-03 | 6.30E-03 | 3.00E-03 | Yes |
| ZNF589 | 3 | rs6800730 | 1.56E-02 | 1.07E-04 | 3.32E-04 | 9.65E-05 | NR |
| HLA-DRB5 | 6 | rs3130342 | 4.88E-03 | 1.25E-02 | 4.44E-04 | 6.52E-05 | Yes |
| UHRF1BP1 | 6 | rs205262 | 5.48E-05 | 5.54E-05 | 3.77E-05 | 5.88E-06 | Yes |
| NEIL2 | 8 | rs11998678 | 4.30E-04 | 1.06E-03 | 2.69E-02 | 6.24E-05 | NR |
| NSUN6 | 10 | rs11014171 | 1.19E-02 | 6.69E-03 | 3.54E-02 | 8.86E-03 | NR |
| SFXN4 | 10 | rs915272 | 2.53E-02 | 2.15E-02 | 1.73E-02 | 2.56E-02 | NR |
| TNNT3 | 11 | rs4980379 | 2.18E-09 | 2.95E-06 | 2.21E-12 | 2.40E-07 | NR |
| LYZ | 12 | rs618470 | 2.75E-03 | 5.13E-03 | 2.82E-02 | 1.55E-02 | Yes |
| TMEM116 | 12 | rs653178 | 1.00E-03 | 1.98E-02 | 2.60E-02 | 1.13E-02 | Yes |
| CDC16 | 13 | rs11617448 | 1.58E-02 | 1.72E-04 | 7.73E-04 | 2.36E-04 | NR |
| EXOSC6 | 16 | rs3790085 | 7.17E-03 | 1.18E-02 | 1.39E-02 | 1.20E-02 | NR |
| CRLF3 | 17 | rs3760318 | 4.25E-03 | 9.56E-03 | 2.00E-02 | 1.20E-02 | NR |
| ZNF100 | 19 | rs2968084 | 1.85E-04 | 2.89E-04 | 9.57E-05 | 1.74E-04 | NR |
NR: No Reported.
4. Discussion
The genetic architecture of BP includes more than 30 genes and 1400 common SNPs to date. The related signaling pathways involve the renin-angiotensin-aldosterone system and the adrenal glucocorticoid pathway. Most of the BP SNPs identified by GWAS show pleiotropic associations, and the effect of each SNP on BP is small. Further investigation of these loci may identify novel targets for the molecular etiology of hypertension and the prevention of cardiovascular disease [12,30].
In the current study, we have identified 1010 susceptibility genes of essential hypertension with four tissues (left heart ventricle, aorta, whole blood, and peripheral blood) from 18,420 candidate genes of GWAS utilizing a TWAS framework. To our knowledge, this is the first systematic TWAS for essential hypertension.
The enrichment analysis was carried out to identify and confirm related signaling pathways of TWAS-identified genes. The KEGG and GO terms were mostly related to autoimmune mechanisms, including MHC protein complex, MHC class II protein complex, MHC class II receptor activity, type I diabetes mellitus, asthma, inflammatory bowel disease, rheumatoid arthritis, fatty acid metabolism, intestinal immune network for IgA production, which is consistent with the report of Wolf et al. [31]. Evidence supporting a role for immunity and inflammation in the development of hypertension began as early as the late 1950s [32]. With the emergence and development of gene editing technology, the study of the immune system using knockout mice has developed rapidly [33]. Further understanding of immune factors, immune cells, and the role of the immune system in hypertension has also been gained [34]. Research findings show that long-term low-intensity inflammatory response and persistent activation of the immune system play an important role in the development of hypertension [35]. In addition, the accumulation of immune cells in blood vessels, kidneys, heart, and brain promotes chronic inflammatory responses that impair blood pressure regulation in these organs, and can also lead to hypertension [[36], [37], [38], [39]].
In this study, 15 novel hypertension-susceptible genes (GFPT1, RBM6, ZNF589, HLA-DRB5, UHRF1BP1, NEIL2, NSUN6, SFXN4, TNNT3, LYZ, TMEM116, CDC16, EXOSC6, CRLF3, and ZNF100) were identified by TWAS in all tissues, and the genes (RBM6, HLA-DRB5, UHRF1BP1, LYZ, and TMEM116) were associated with the autoimmune, which provide further evidence supporting the involvement of an autoimmune mechanism in hypertension. While studying the role of somatic mutations in an autoimmune disease, multiple sclerosis (MS), Valori et al. found that the RBM6 plays an important role in autoimmunity, providing an interesting and unexplored role for subsequent research [40]. HLA-DRB5 was reported to regulate TH1, TH2, and TH17 cell differentiation signaling pathways in T cells in primary Sjögren's syndrome (pSS), which is one of the most common autoimmune diseases that mainly affect middle-aged and older women [41]. As a chronic autoimmune disease with heterogeneous presentation and complex etiology, systemic lupus erythematosus (SLE) was studied by GWASs, and UHRF1BP1 and TMEM116 were identified as new susceptibility loci in different studies, although the role of these genes in SLE was unknown [42,43]. Graves' disease (GD) is an autoimmune inflammatory disease of the eye, and Fairfax et al. found that the level of LYZ in the tears of patients was significantly increased, which provided a new marker for differentiating the degree of clinical symptoms in patients [44].
Further GO enrichment analysis of PPI genes also detected several GO terms with autoimmunity, such as the adaptive immune system, regulation of cellular response to stress, cellular responses to stimuli, regulation of hormone levels. This supports the important role of autoimmunity in hypertension. Moreover, we performed MR analysis, and the results also supported a significant association between autoimmunity and hypertension [29].
Finally, our results should be used with caution as the GWAS data in this study are predominantly from the UK Biobank cohort with European ancestry, therefore, limiting the generalisability of the findings. In addition, there are few studies on the five autoimmune-related hypertension susceptibility genes identified by TWAS, and the link between them can not be well elucidated.
In summary, we integrated the GWAS datasets of hypertension from the UK Biobank and mRNA expression profiling together to complete a TWAS in four tissues, and our results support that an autoimmune mechanism plays an important role in the development of hypertension. Our findings will provide new biological insights into BP control and help us to further untangle the molecular etiology of hypertension.
Data availability statement
The datasets analyzed during the current study are available from the Database of Genomic Variants (http://projects.tcag.ca/variation/); the URLs for Consortia and Groups (https://www.preventcd.com); the BioGPS (http://biogps.gnf.org); and the UK biobank (http://geneatlas.roslin.ed.ac.uk/) (fields: 20002).
Author contributions
LH: data collection and analysis, manuscript drafting; QZ: data collection, KEGG and Go analysis; RV: conceptualization, manuscript revision; JX: PPI analysis and interpretation; FY: bioinformatic analysis, and data representation; JM: conceptualization, manuscript drafting and revision.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We are indebted to all individuals who have participated in, or helped, with our work. This study was supported by Natural Science Basic Research Plan in Shaanxi Province of China (No.2022JM-440); National Natural Science Foundation of China (No. 31371298; 81301151); Xi'an Science and Technology Planning Project (No.21YXYJ0039).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101387.
Contributor Information
Fang Yan, Email: yanfang06292022@163.com.
Jie Ma, Email: majie_article@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kearney P.M., Whelton M., Reynolds K., Muntner P., Whelton P.K., He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.S. International Consortium for Blood Pressure Genome-Wide Association. Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., Pihur V., Vollenweider P., O'Reilly P.F., Amin N., Bragg-Gresham J.L., Teumer A., Glazer N.L., Launer L., Zhao J.H., Aulchenko Y., Heath S., Sober S., Parsa A., Luan J., Arora P., Dehghan A., Zhang F., Lucas G., Hicks A.A., Jackson A.U., Peden J.F., Tanaka T., Wild S.H., Rudan I., Igl W., Milaneschi Y., Parker A.N., Fava C., Chambers J.C., Fox E.R., Kumari M., Go M.J., van der Harst P., Kao W.H., Sjogren M., Vinay D.G., Alexander M., Tabara Y., Shaw-Hawkins S., Whincup P.H., Liu Y., Shi G., Kuusisto J., Tayo B., Seielstad M., Sim X., Nguyen K.D., Lehtimaki T., Matullo G., Wu Y., Gaunt T.R., Onland-Moret N.C., Cooper M.N., Platou C.G., Org E., Hardy R., Dahgam S., Palmen J., Vitart V., Braund P.S., Kuznetsova T., Uiterwaal C.S., Adeyemo A., Palmas W., Campbell H., Ludwig B., Tomaszewski M., Tzoulaki I., Palmer N.D., consortium C.A., Consortium C.K., KidneyGen C., EchoGen c., consortium C.-H., Aspelund T., Garcia M., Chang Y.P., O'Connell J.R., Steinle N.I., Grobbee D.E., Arking D.E., Kardia S.L., Morrison A.C., Hernandez D., Najjar S., McArdle W.L., Hadley D., Brown M.J., Connell J.M., Hingorani A.D., Day I.N., Lawlor D.A., Beilby J.P., Lawrence R.W., Clarke R., Hopewell J.C., Ongen H., Dreisbach A.W., Li Y., Young J.H., Bis J.C., Kahonen M., Viikari J., Adair L.S., Lee N.R., Chen M.H., Olden M., Pattaro C., Bolton J.A., Kottgen A., Bergmann S., Mooser V., Chaturvedi N., Frayling T.M., Islam M., Jafar T.H., Erdmann J., Kulkarni S.R., Bornstein S.R., Grassler J., Groop L., Voight B.F., Kettunen J., Howard P., Taylor A., Guarrera S., Ricceri F., Emilsson V., Plump A., Barroso I., Khaw K.T., Weder A.B., Hunt S.C., Sun Y.V., Bergman R.N., Collins F.S., Bonnycastle L.L., Scott L.J., Stringham H.M., Peltonen L., Perola M., Vartiainen E., Brand S.M., Staessen J.A., Wang T.J., Burton P.R., Soler Artigas M., Dong Y., Snieder H., Wang X., Zhu H., Lohman K.K., Rudock M.E., Heckbert S.R., Smith N.L., Wiggins K.L., Doumatey A., Shriner D., Veldre G., Viigimaa M., Kinra S., Prabhakaran D., Tripathy V., Langefeld C.D., Rosengren A., Thelle D.S., Corsi A.M., Singleton A., Forrester T., Hilton G., McKenzie C.A., Salako T., Iwai N., Kita Y., Ogihara T., Ohkubo T., Okamura T., Ueshima H., Umemura S., Eyheramendy S., Meitinger T., Wichmann H.E., Cho Y.S., Kim H.L., Lee J.Y., Scott J., Sehmi J.S., Zhang W., Hedblad B., Nilsson P., Smith G.D., Wong A., Narisu N., Stancakova A., Raffel L.J., Yao J., Kathiresan S., O'Donnell C.J., Schwartz S.M., Ikram M.A., Longstreth W.T., Jr., Mosley T.H., Seshadri S., Shrine N.R., Wain L.V., Morken M.A., Swift A.J., Laitinen J., Prokopenko I., Zitting P., Cooper J.A., Humphries S.E., Danesh J., Rasheed A., Goel A., Hamsten A., Watkins H., Bakker S.J., van Gilst W.H., Janipalli C.S., Mani K.R., Yajnik C.S., Hofman A., Mattace-Raso F.U., Oostra B.A., Demirkan A., Isaacs A., Rivadeneira F., Lakatta E.G., Orru M., Scuteri A., Ala-Korpela M., Kangas A.J., Lyytikainen L.P., Soininen P., Tukiainen T., Wurtz P., Ong R.T., Dorr M., Kroemer H.K., Volker U., Volzke H., Galan P., Hercberg S., Lathrop M., Zelenika D., Deloukas P., Mangino M., Spector T.D., Zhai G., Meschia J.F., Nalls M.A., Sharma P., Terzic J., Kumar M.V., Denniff M., Zukowska-Szczechowska E., Wagenknecht L.E., Fowkes F.G., Charchar F.J., Schwarz P.E., Hayward C., Guo X., Rotimi C., Bots M.L., Brand E., Samani N.J., Polasek O., Talmud P.J., Nyberg F., Kuh D., Laan M., Hveem K., Palmer L.J., van der Schouw Y.T., Casas J.P., Mohlke K.L., Vineis P., Raitakari O., Ganesh S.K., Wong T.Y., Tai E.S., Cooper R.S., Laakso M., Rao D.C., Harris T.B., Morris R.W., Dominiczak A.F., Kivimaki M., Marmot M.G., Miki T., Saleheen D., Chandak G.R., Coresh J., Navis G., Salomaa V., Han B.G., Zhu X., Kooner J.S., Melander O., Ridker P.M., Bandinelli S., Gyllensten U.B., Wright A.F., Wilson J.F., Ferrucci L., Farrall M., Tuomilehto J., Pramstaller P.P., Elosua R., Soranzo N., Sijbrands E.J., Altshuler D., Loos R.J., Shuldiner A.R., Gieger C., Meneton P., Uitterlinden A.G., Wareham N.J., Gudnason V., Rotter J.I., Rettig R., Uda M., Strachan D.P., Witteman J.C., Hartikainen A.L., Beckmann J.S., Boerwinkle E., Vasan R.S., Boehnke M., Larson M.G., Jarvelin M.R., Psaty B.M., Abecasis G.R., Chakravarti A., Elliott P., van Duijn C.M., Newton-Cheh C., Levy D., Caulfield M.J., Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y., Tomlinson B., Chu T., Fang Y.J., Gui H., Tang C.S., Yip B.H., Cherny S.S., Hur Y.M., Sham P.C., Lam T.H., Thomas N.G. A genome-wide linkage and association scan reveals novel loci for hypertension and blood pressure traits. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botzer A., Grossman E., Moult J., Unger R. A system view and analysis of essential hypertension. J. Hypertens. 2018;36:1094–1103. doi: 10.1097/HJH.0000000000001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D., DeStefano A.L., Larson M.G., O'Donnell C.J., Lifton R.P., Gavras H., Cupples L.A., Myers R.H. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 6.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T., Aulchenko Y., Lumley T., Kottgen A., Vasan R.S., Rivadeneira F., Eiriksdottir G., Guo X., Arking D.E., Mitchell G.F., Mattace-Raso F.U., Smith A.V., Taylor K., Scharpf R.B., Hwang S.J., Sijbrands E.J., Bis J., Harris T.B., Ganesh S.K., O'Donnell C.J., Hofman A., Rotter J.I., Coresh J., Benjamin E.J., Uitterlinden A.G., Heiss G., Fox C.S., Witteman J.C., Boerwinkle E., Wang T.J., Gudnason V., Larson M.G., Chakravarti A., Psaty B.M., van Duijn C.M. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., Papadakis K., Voight B.F., Scott L.J., Zhang F., Farrall M., Tanaka T., Wallace C., Chambers J.C., Khaw K.T., Nilsson P., van der Harst P., Polidoro S., Grobbee D.E., Onland-Moret N.C., Bots M.L., Wain L.V., Elliott K.S., Teumer A., Luan J., Lucas G., Kuusisto J., Burton P.R., Hadley D., McArdle W.L., Wellcome Trust Case Control C., Brown M., Dominiczak A., Newhouse S.J., Samani N.J., Webster J., Zeggini E., Beckmann J.S., Bergmann S., Lim N., Song K., Vollenweider P., Waeber G., Waterworth D.M., Yuan X., Groop L., Orho-Melander M., Allione A., Di Gregorio A., Guarrera S., Panico S., Ricceri F., Romanazzi V., Sacerdote C., Vineis P., Barroso I., Sandhu M.S., Luben R.N., Crawford G.J., Jousilahti P., Perola M., Boehnke M., Bonnycastle L.L., Collins F.S., Jackson A.U., Mohlke K.L., Stringham H.M., Valle T.T., Willer C.J., Bergman R.N., Morken M.A., Doring A., Gieger C., Illig T., Meitinger T., Org E., Pfeufer A., Wichmann H.E., Kathiresan S., Marrugat J., O'Donnell C.J., Schwartz S.M., Siscovick D.S., Subirana I., Freimer N.B., Hartikainen A.L., McCarthy M.I., O'Reilly P.F., Peltonen L., Pouta A., de Jong P.E., Snieder H., van Gilst W.H., Clarke R., Goel A., Hamsten A., Peden J.F., Seedorf U., Syvanen A.C., Tognoni G., Lakatta E.G., Sanna S., Scheet P., Schlessinger D., Scuteri A., Dorr M., Ernst F., Felix S.B., Homuth G., Lorbeer R., Reffelmann T., Rettig R., Volker U., Galan P., Gut I.G., Hercberg S., Lathrop G.M., Zelenika D., Deloukas P., Soranzo N., Williams F.M., Zhai G., Salomaa V., Laakso M., Elosua R., Forouhi N.G., Volzke H., Uiterwaal C.S., van der Schouw Y.T., Numans M.E., Matullo G., Navis G., Berglund G., Bingham S.A., Kooner J.S., Connell J.M., Bandinelli S., Ferrucci L., Watkins H., Spector T.D., Tuomilehto J., Altshuler D., Strachan D.P., Laan M., Meneton P., Wareham N.J., Uda M., Jarvelin M.R., Mooser V., Melander O., Loos R.J., Elliott P., Abecasis G.R., Caulfield M., Munroe P.B. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato N., Takeuchi F., Tabara Y., Kelly T.N., Go M.J., Sim X., Tay W.T., Chen C.H., Zhang Y., Yamamoto K., Katsuya T., Yokota M., Kim Y.J., Ong R.T., Nabika T., Gu D., Chang L.C., Kokubo Y., Huang W., Ohnaka K., Yamori Y., Nakashima E., Jaquish C.E., Lee J.Y., Seielstad M., Isono M., Hixson J.E., Chen Y.T., Miki T., Zhou X., Sugiyama T., Jeon J.P., Liu J.J., Takayanagi R., Kim S.S., Aung T., Sung Y.J., Zhang X., Wong T.Y., Han B.G., Kobayashi S., Ogihara T., Zhu D., Iwai N., Wu J.Y., Teo Y.Y., Tai E.S., Cho Y.S., He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B., Ntalla I., Surendran P., Liu C., Cook J.P., Kraja A.T., Drenos F., Loh M., Verweij N., Marten J., Karaman I., Lepe M.P., O'Reilly P.F., Knight J., Snieder H., Kato N., He J., Tai E.S., Said M.A., Porteous D., Alver M., Poulter N., Farrall M., Gansevoort R.T., Padmanabhan S., Magi R., Stanton A., Connell J., Bakker S.J., Metspalu A., Shields D.C., Thom S., Brown M., Sever P., Esko T., Hayward C., van der Harst P., Saleheen D., Chowdhury R., Chambers J.C., Chasman D.I., Chakravarti A., Newton-Cheh C., Lindgren C.M., Levy D., Kooner J.S., Keavney B., Tomaszewski M., Samani N.J., Howson J.M., Tobin M.D., Munroe P.B., Ehret G.B., Wain L.V., International Consortium of Blood Pressure G.A., Consortium B., Lifelines Cohort S., Understanding Society Scientific g., Consortium C.H.D.E., Exome B.P.C., Consortium T.D.G., Go T.D.C., H. Cohorts for, B.P.E.C. Ageing Research in Genome Epidemiology, C. International Genomics of Blood Pressure, U.K.B.C.C.B.w. group, Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie G., Delles C. The future of "omics" in hypertension. Can. J. Cardiol. 2017;33:601–610. doi: 10.1016/j.cjca.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnett D.K., Claas S.A. Omics of blood pressure and hypertension. Circ. Res. 2018;122:1409–1419. doi: 10.1161/CIRCRESAHA.118.311342. [DOI] [PubMed] [Google Scholar]
- 12.Lip S., Padmanabhan S. Genomics of blood pressure and hypertension: extending the mosaic theory toward stratification. Can. J. Cardiol. 2020;36:694–705. doi: 10.1016/j.cjca.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharya A., Garcia-Closas M., Olshan A.F., Perou C.M., Troester M.A., Love M.I. A framework for transcriptome-wide association studies in breast cancer in diverse study populations. Genome Biol. 2020;21:42. doi: 10.1186/s13059-020-1942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A., Sullivan P.F., Nikkola E., Alvarez M., Civelek M., Lusis A.J., Lehtimaki T., Raitoharju E., Kahonen M., Seppala I., Raitakari O.T., Kuusisto J., Laakso M., Price A.L., Pajukanta P., Pasaniuc B. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., Bjorkegren J.L.M., Im H.K., Pasaniuc B., Rivas M.A., Kundaje A. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., Schizophrenia Working Group of the Psychiatric Genomics C., McCarroll S., Neale B.M., Ophoff R.A., O'Donovan M.C., Crawford G.E., Geschwind D.H., Katsanis N., Sullivan P.F., Pasaniuc B., Price A.L. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porcu E., Rueger S., Lepik K., e Q.C., Consortium B., Santoni F.A., Reymond A., Kutalik Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019;10:3300. doi: 10.1038/s41467-019-10936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L., Yang Y., Guo X., Shu X.O., Cai Q., Shu X., Li B., Tao R., Wu C., Nikas J.B., Sun Y., Zhu J., Roobol M.J., Giles G.G., Brenner H., John E.M., Clements J., Grindedal E.M., Park J.Y., Stanford J.L., Kote-Jarai Z., Haiman C.A., Eeles R.A., Zheng W., Long J., consortium P., Consortium C., Consortium B.P.C., Consortium C., Consortium P. An integrative multi-omics analysis to identify candidate DNA methylation biomarkers related to prostate cancer risk. Nat. Commun. 2020;11:3905. doi: 10.1038/s41467-020-17673-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng R., Lu M., Liu L., Xu K., Xu P. Transcriptome-wide association studies and integration analysis of mRNA expression profiles identify candidate genes and pathways associated with ankylosing spondylitis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.814303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canela-Xandri O., Rawlik K., Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pain O., Pocklington A.J., Holmans P.A., Bray N.J., O'Brien H.E., Hall L.S., Pardinas A.F., O'Donovan M.C., Owen M.J., Anney R. Novel insight into the etiology of autism spectrum disorder gained by integrating expression data with genome-wide association statistics. Biol. Psychiatr. 2019;86:265–273. doi: 10.1016/j.biopsych.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Quick C., Yu K., Barbeira A., Consortium G.T., Luca F., Pique-Regi R., Kyung Im H., Wen X. PTWAS: investigating tissue-relevant causal molecular mechanisms of complex traits using probabilistic TWAS analysis. Genome Biol. 2020;21:232. doi: 10.1186/s13059-020-02026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuo M.L., Huang Y., Chen J.Z., Sun L.H., Yang R.F., Chen H.Z., Lv X., Li H.L., Wei Y.S., Liu G., Zhang R., Ma T.M., Cai H., Hui R.T., Liu D.P., Liang C.C. Endothelium-specific overexpression of human IC53 downregulates endothelial nitric oxide synthase activity and elevates systolic blood pressure in mice. Cardiovasc. Res. 2009;84:292–299. doi: 10.1093/cvr/cvp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill D.P., Blake J.A., Richardson J.E., Ringwald M. Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies. Genome Res. 2002;12:1982–1991. doi: 10.1101/gr.580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H., Cheng S., Li C., Cheng B., Liu L., Yang X., Meng P., Yao Y., Pan C., Zhang J., Zhang H., Chen Y., Zhang Z., Wen Y., Jia Y., Zhang F. Dissecting the association between psychiatric disorders and neurological proteins: a genetic correlation and two-sample bidirectional Mendelian Randomization study. Acta Neuropsychiatr. 2022:1–24. doi: 10.1017/neu.2022.10. [DOI] [PubMed] [Google Scholar]
- 30.Padmanabhan S., Dominiczak A.F. Genomics of hypertension: the road to precision medicine. Nat. Rev. Cardiol. 2021;18:235–250. doi: 10.1038/s41569-020-00466-4. [DOI] [PubMed] [Google Scholar]
- 31.Wolf V.L., Ryan M.J. Autoimmune disease-associated hypertension. Curr. Hypertens. Rep. 2019;21:10. doi: 10.1007/s11906-019-0914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommers S.C., Relman A.S., Smithwick R.H. Histologic studies of kidney biopsy specimens from patients with hypertension. Am. J. Pathol. 1958;34:685–715. [PMC free article] [PubMed] [Google Scholar]
- 33.Trott D.W., Thabet S.R., Kirabo A., Saleh M.A., Itani H., Norlander A.E., Wu J., Goldstein A., Arendshorst W.J., Madhur M.S., Chen W., Li C.I., Shyr Y., Harrison D.G. Oligoclonal CD8+ T cells play a critical role in the development of hypertension. Hypertension. 2014;64:1108–1115. doi: 10.1161/HYPERTENSIONAHA.114.04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Iturbe B., Pons H., Johnson R.J. Role of the immune system in hypertension. Physiol. Rev. 2017;97:1127–1164. doi: 10.1152/physrev.00031.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy C.G., Goulopoulou S., Wenceslau C.F., Spitler K., Matsumoto T., Webb R.C. Toll-like receptors and damage-associated molecular patterns: novel links between inflammation and hypertension. Am. J. Physiol. Heart Circ. Physiol. 2014;306:H184–H196. doi: 10.1152/ajpheart.00328.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattson D.L. Immune mechanisms of salt-sensitive hypertension and renal end-organ damage, Nature reviews. Nephrology. 2019;15:290–300. doi: 10.1038/s41581-019-0121-z. [DOI] [PubMed] [Google Scholar]
- 37.Brooks W.W., Bing O.H., Conrad C.H., O'Neill L., Crow M.T., Lakatta E.G., Dostal D.E., Baker K.M., Boluyt M.O. Captopril modifies gene expression in hypertrophied and failing hearts of aged spontaneously hypertensive rats. Hypertension. 1997;30:1362–1368. doi: 10.1161/01.hyp.30.6.1362. [DOI] [PubMed] [Google Scholar]
- 38.Cha H.J., Kim H.Y., Kim H.S. Sulfatase 1 mediates the attenuation of Ang II-induced hypertensive effects by CCL5 in vascular smooth muscle cells from spontaneously hypertensive rats. Cytokine. 2018;110:1–8. doi: 10.1016/j.cyto.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 39.Santisteban M.M., Ahmari N., Carvajal J.M., Zingler M.B., Qi Y., Kim S., Joseph J., Garcia-Pereira F., Johnson R.D., Shenoy V., Raizada M.K., Zubcevic J. Involvement of bone marrow cells and neuroinflammation in hypertension. Circ. Res. 2015;117:178–191. doi: 10.1161/CIRCRESAHA.117.305853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valori M., Jansson L., Kiviharju A., Ellonen P., Rajala H., Awad S.A., Mustjoki S., Tienari P.J. A novel class of somatic mutations in blood detected preferentially in CD8+ cells. Clin. Immunol. 2017;175:75–81. doi: 10.1016/j.clim.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong X., Meng S., Tang D., Wang T., Ding L., Yu H., Li H., Liu D., Dai Y., Yang M. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4(+) T lymphocytes and a landscape of immune cells in primary sjogren's syndrome. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.594658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gateva V., Sandling J.K., Hom G., Taylor K.E., Chung S.A., Sun X., Ortmann W., Kosoy R., Ferreira R.C., Nordmark G., Gunnarsson I., Svenungsson E., Padyukov L., Sturfelt G., Jonsen A., Bengtsson A.A., Rantapaa-Dahlqvist S., Baechler E.C., Brown E.E., Alarcon G.S., Edberg J.C., Ramsey-Goldman R., McGwin G., Jr., Reveille J.D., Vila L.M., Kimberly R.P., Manzi S., Petri M.A., Lee A., Gregersen P.K., Seldin M.F., Ronnblom L., Criswell L.A., Syvanen A.C., Behrens T.W., Graham R.R. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y.F., Wei W., Tangtanatakul P., Zheng L., Lei Y., Lin Z., Qian C., Qin X., Hou F., Zhang X., Shao L., Satproedprai N., Mahasirimongkol S., Pisitkun P., Song Q., Lau Y.L., Zhang Y., Hirankarn N., Yang W. Identification of shared and Asian-specific loci for systemic lupus erythematosus and evidence for roles of type III interferon signaling and lysosomal function in the disease: a multi-Ancestral genome-wide association study. Arthritis Rheumatol. 2022;74:840–848. doi: 10.1002/art.42021. [DOI] [PubMed] [Google Scholar]
- 44.Aass C., Norheim I., Eriksen E.F., Bornick E.C., Thorsby P.M., Pepaj M. Establishment of a tear protein biomarker panel differentiating between Graves' disease with or without orbitopathy. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the Database of Genomic Variants (http://projects.tcag.ca/variation/); the URLs for Consortia and Groups (https://www.preventcd.com); the BioGPS (http://biogps.gnf.org); and the UK biobank (http://geneatlas.roslin.ed.ac.uk/) (fields: 20002).