Summary
CD47 is an ubiquitously expressed surface molecule with significant impact on immune responses. However, its role for antiviral immunity is not fully understood. Here, we revealed that the expression of CD47 on immune cells seemed to disturb the antiviral immune response as CD47-deficient mice (CD47−/−) showed an augmented clearance of influenza A virus (IAV). Specifically, we have shown that enhanced viral clearance is mediated by alveolar macrophages (aMФ). Although aMФ displayed upregulation of CD47 expression during IAV infection in wildtype mice, depletion of aMФ in CD47−/− mice during IAV infection reversed the augmented viral clearance. We have also demonstrated that CD47 restricts hemoglobin (HB) expression in aMФ after IAV and severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) infection, with HB showing antiviral properties by enhancing the IFN-β response. Our study showed a negative role for CD47 during antiviral immune responses in the lung by confining HB expression in aMФ.
Subject areas: Immunology, Virology
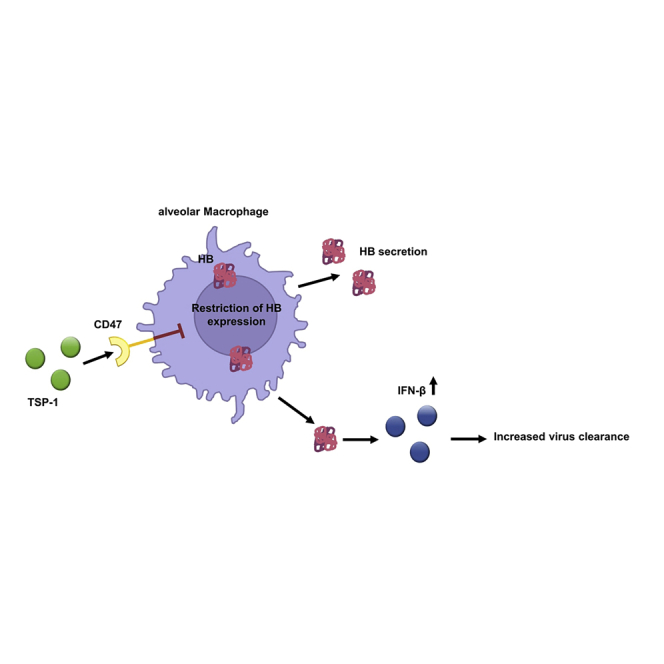
Graphical abstract
Highlights
-
•
CD47 restricts influenza A virus (IAV) clearance by alveolar macrophages (aMФ)
-
•
Hemoglobin (HB) expression is restricted by CD47 in aMФ after IAV infection
-
•
HB shows antiviral properties against IAV and SARS-CoV2
Immunology; Virology
Introduction
Influenza A viruses (IAV) are a common cause of lower respiratory infections. During seasonal epidemics up to 5 million people suffer from severe IAV infections with up to 650.000 deaths annually.1 Although vaccines as well as antiviral drugs against IAV are available to date, their efficiency is highly limited because of the antigenic variability of the virus and the occurrence of drug-resistant strains.2,3 Nevertheless, relevant immune, viral, and host factors defining either the clearance or the severity of the disease are still unclear. Same applies to the precise mechanisms defining why some patients only experience mild symptoms while others struggle with life-threatening viral pneumonia.4 Thus, novel treatment strategies consider the manipulation of IAV-related immune responses to ameliorate the clearance of the virus.
Different lung resident as well as recruited immune cells are involved in the clearance of influenza viruses. For example, alveolar macrophages (aMФ) reside in the alveolar lumen and recognize invading pathogens such as IAV.5,6 Yet, pro-inflammatory immune responses were demonstrated to contribute to the severity of IAV infection-associated pathology as well.7,8 Thus, pulmonary immunity needs to be highly regulated. Type I interferons (IFN I) partially take over this task. IFN I are released after recognition of pathogen associated molecular patterns (PAMPs) via innate receptors like toll-like receptor (TLR) 7, retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated protein 5 (MDA5).9 During influenza virus infection, interferon-β (IFN-β) modulates a type I immunity.10
CD47 is an ubiquitously expressed cell surface protein with immunoregulatory function. It is best noted for its function as a “don’t eat me signal” exploited by tumor cells to avoid proper antitumor immunity via the interaction with signal regulatory protein α (SIRPα),11 an inhibitory receptor expressed on phagocytes. Another known ligand of CD47 is thrombospondin 1 (TSP-1), an extracellular homotrimeric matrix protein, which links CD47 to cellular processes such as adhesion and apoptosis.12,13 Although previous studies implicated CD47 in antimicrobial immune responses,14 its role for antimicrobial immunity remains controversial and our current understanding of the effect of CD47 on antiviral immunity is even more fragmentary. Along this line, the oxygen carrying protein hemoglobin (HB) exerts multiple functions, including the regulation of innate immunity and resistance to the invasion of pathogens.15,16 Furthermore, it was recently described that HB could exert antiviral properties by enhancing recognition of viral RNA by RIG-I and thus the IFN I response during viral infection.17 However, its involvement in the antiviral immune response against IAV infection remains undefined.
In the present study, we addressed the impact of CD47 on immunity on acute IAV infection. Of interest, CD47-deficient mice (CD47−/−) showed an augmented clearance of IAV which was absent when aMФ were depleted. aMФ from infected CD47−/− mice surprisingly exhibited an elevated expression of HB compared to wildtype (WT) aMФ. We observed that HB has actually antiviral capacity against influenza virus infection, probably by inducing an enhanced IFN-β response. Of note, we were also able to demonstrate an antiviral effect of HB for severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), suggesting that the mode of action of the HB activity is virus-non-specific. Thus, our study is the first to claim CD47 as a negative regulator of antiviral aMФ immunity and to describe an antiviral role for HB during acute IAV infection.
Results
CD47 limits the clearance of influenza A virus during acute infection
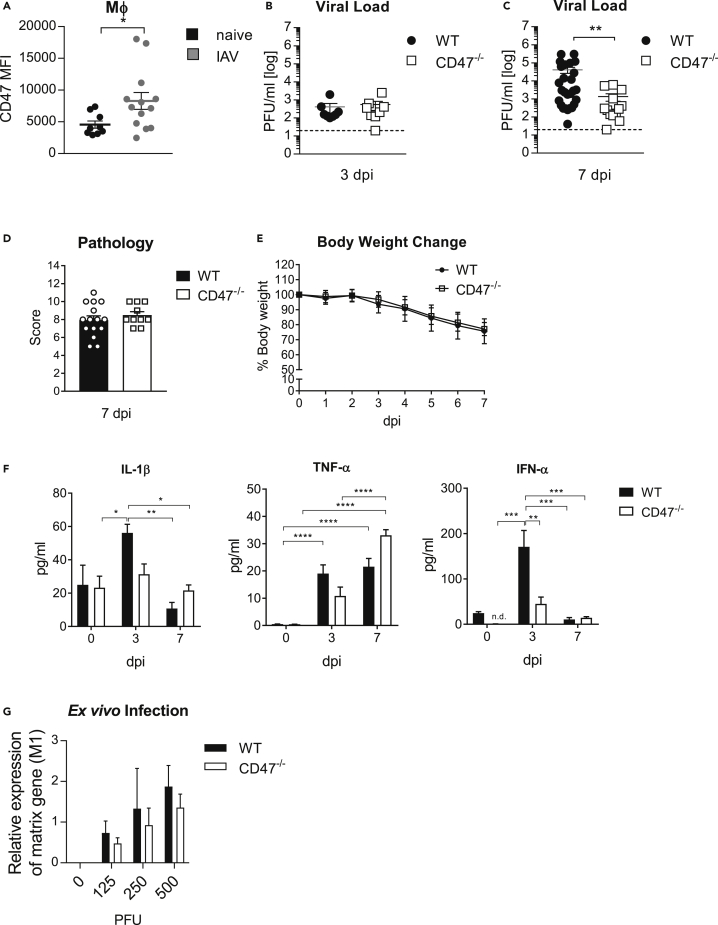
An augmented expression of CD47 was suggested to limit antibacterial or antiviral immunity during infection.18,19 To define a possible immunoregulatory function of CD47 during IAV infection, we infected C57BL/6 WT mice. Of interest, we found CD47 highly expressed on macrophages (MФ) in the lung of mice infected with a high dose of IAV (75 PFU) by flow cytometry analysis compared to MФ from uninfected mice (Figure 1A). However, no differences in the viral loads were observed at an early stage 3 days post infection (dpi) betweenCD47−/− mice and WT mice (Figure 1B). Besides MФ, significantly elevated levels of CD47 were also found on the surface of neutrophils (PMN) and CD8+T cells (Figure S1A). Moreover, CD47−/− mice showed significantly reduced viral loads in the lung 7 dpi compared to WT mice (Figure 1C). Yet, the genetic ablation of CD47 seems to rather not enhance the frequency or effector function of CD8+T cells in IAV infected CD47−/− mice compared to WT 7 dpi (Figures S1B–S1D). In addition, IAV-specific antibody titers in the bronchoalveolar lavage fluid (BALF) were the same in both CD47−/− and WT mice 7 dpi (Figure S1E). Furthermore, we did not see any differences in the pathology of the lung (Figures 1D and S1F) or body weight loss between infected WT and CD47−/− mice (Figure 1E). Of note, infection with a sublethal dose (8 PFU) resulted in only slight differences in viral loads and no difference in body weight change between infected WT and CD47−/− mice (Figures S2Aand S2B). Of interest, pro-inflammatory cytokine levels such as IL-1β, tumor necrosis factor-α (TNF-α), and interferon-α (IFN-α) were significantly decreased in the BALF of CD47−/− compared to WT mice at 3 days after high dose infection, whereas cytokine levels continued to decrease by 7 dpi, with the exception of TNF-α (Figure 1F). To exclude an effect of CD47 on IAV infectivity and life cycle, we exposed alveolar type II epithelial cells (AEC II), isolated from WT and CD47−/− mice to different doses of IAV. Importantly, a decrease in viral RNA was not found on ex vivo infection of CD47−/− AEC II compared to WT cells implying a regulatory role of CD47 for viral clearance rather than viral replication (Figure 1G). Thus, the results suggest an immune regulatory role of CD47 which interferes the clearance of IAV independent of adaptive immunity.
Figure 1.
CD47−/− mice have an enhanced clearance of influenza virus
Wildtype (WT) and CD47−/− mice were i.n. infected with a single dose of 75 PFU of influenza virus A/PR8/34.
(A–C) (A) F4/80+ macrophages (MΦ) in the lung were analyzed 3 days post infection (dpi) via flow cytometry regarding the mean fluorescence intensity (MFI) of CD47. Viral loads in the lung of mice infected with 75 PFU IAV was determined at day 3 (B) and 7 (C) after infection via plaque assay.
(D) Immune induced histopathology in the lung was determined at 7 dpi.
(E) Body weight change after infection.
(F) Pro-inflammatory cytokines including TNF-α, IL-1β and IFN-α were determined in the BALF at indicated timepoints via Luminex and ELISA.
(G) Alveolar type II epithelial cells were isolated from the lung and ex vivo infected with indicated doses. Virus titers were assessed by qRT-PCR 2 dpi (n = 4). Results are pooled from two or more independent experiments.
Data shown are mean ± SEM. Statistics were done by Student’s t test in (A). Two-way ANOVA with Sidak’s multiple-comparisons post-test was used in all other figures. one-way ANOVA with Tukey’s multiple-comparisons post-test was performed in (F). ∗ = p < 0.05, ∗∗ = p < 0.01.
CD47 influences macrophage responses in the lung during influenza virus infection
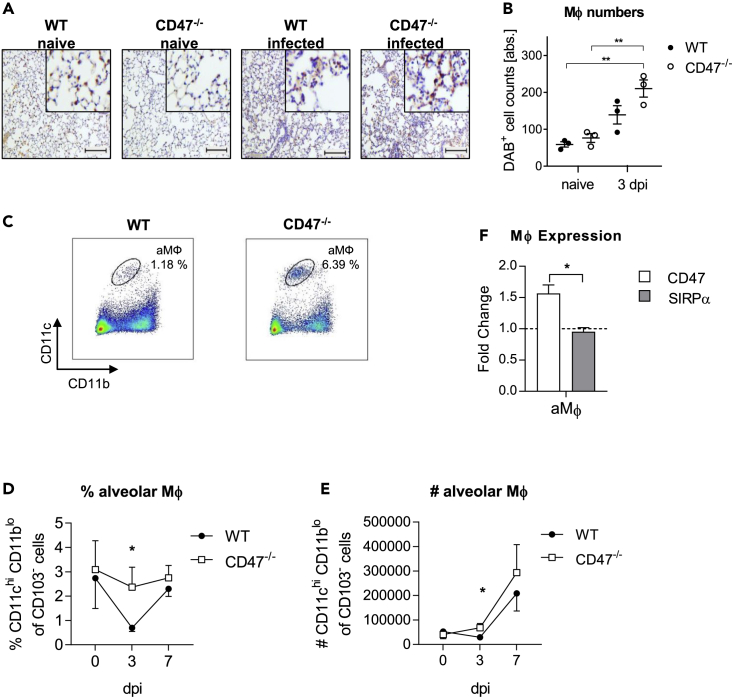
We next focused on MФ immunity to delineate the role of CD47 during IAV infection. By histological analysis of the lung, we noticed increased number of MФ in the alveolar lumen of CD47−/− compared to WT mice 3 dpi (Figures 2A and 2B). aMФ are among the first immune cells to encounter influenza virus infected cells because of their localization and are critical for modulating the disease severity. Therefore, we next analyzed the proportion of the aMФ population by flow cytometry. Of interest, the frequencies of aMФ in WT mice decreased significantly compared to CD47−/− animals (Figures S3, 2C, and 2D). At the same time, numbers of aMФ in CD47−/− were increased 3 dpi compared to WT mice (Figure 2E). Next, we also analyzed the expression of CD47 and SIRPα, the myeloid cell-expressed ligand of CD47 on aMФ of infected WT mice. aMФ displayed an increased expression of CD47 (Figure 2F). Likewise, expression level of SIRPα was constant on IAV infection. These results indicate that CD47 could influence aMФ during IAV infection.
Figure 2.
Alveolar macrophage frequencies are increased in CD47−/− mice after influenza virus infection
3 and 7 dpi MФ were analyzed in the lung of WT and CD47−/− mice.
(A) Mac3+ MФ were analyzed by immunohistology 3 dpi (scale bar = 100 μm). Representative slides are shown.
(B) Quantification of Diaminobenzidine+ (DAB) cell counts on slides stained for MAC-3 was performed by semi-quantitative analyses. Shown in the diagram are the absolute numbers of DAB+ cell counts. Each point represents the mean of ten random, non-overlapping fields of lung tissue from one specimen (n = 3).
(C) Representative dot plots of flow cytometry analysis of CD11chi CD11blo alveolar macrophages (aMФ) in the lung 3 dpi.
(D) Percentages of aMФ were analyzed by flow cytometry (n = 9).
(E) Total numbers of aMФ were calculated based on flow cytometry analysis (n = 9).
(F) aMФ from WT mice were analyzed regarding their mean fluorescence intensity (MFI) of CD47 and SIRPα 3 dpi (n = 22).
Results are pooled from three or more independent experiments. Data shown are mean ± SEM One-way ANOVA with Tukey’s multiple-comparisons post-test (B), or Student’s t test (D, E, and F) were performed. ∗ = p < 0.05, ∗∗ = p < 0.01.
CD47 deficiency enhances viral clearance by alveolar macrophages
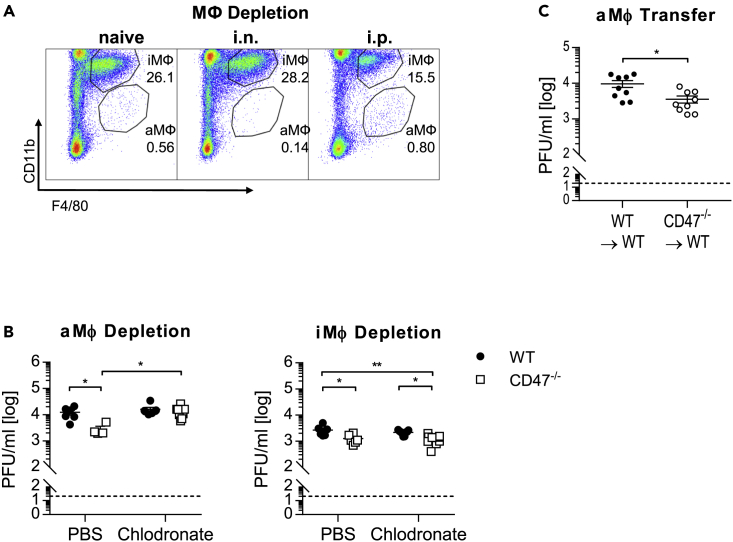
To assess whether elevated frequencies of aMФ are linked to reduced viral loads in CD47−/− mice,6,7 aMФ were specifically depleted by intranasal (i.n.) application of clodronate containing liposomes resulting in a decrease of ∼75% of aMФ from the lung (Figure 3A). Importantly, depletion of aMФ before infection reversed the beneficial effect on viral clearance in CD47−/− mice, whereas the viral load in WT mice was still unaffected (Figure 3B). Moreover, specific depletion of interstitial macrophages (iMФ) by intraperitoneal (i.p.) application of clodronate containing liposomes (efficiency ∼50%) did neither affect the viral load of WT nor CD47−/− mice. Hence, aMФ seemed to be responsible for the improved viral clearance in CD47−/− mice. Accordingly, adoptive transfer of aMФ isolated from CD47−/− but not WT donor mice into aMФ-depleted WT recipient mice before infection reduced the viral load of IAV (Figure 3C). Thus, genetic ablation of CD47 resulted in an enhanced viral clearance of IAV by aMФ.
Figure 3.
Alveolar macrophages are associated with enhanced influenza virus clearance from the lung
MФ populations were depleted by i.n. (aMФ) or i.p. (iMФ) application of clodronate containing liposomes 3 days prior to influenza virus infection.
(A) Representative flow cytometry plots depict the depletion of MФ populations in the lung (3 dpi).
(B) Viral loads were determined by plaque assay 5 dpi. C) aMФ were depleted by i.n application of clodronate liposomes in WT mice and recolonized with aMФ from naive WT or CD47−/− mice by adoptive transfer. Subsequently, mice were infected with influenza virus and viral titers were assessed by plaque assay 5 dpi.
Data shown are mean ± SEM. Two-way ANOVA with Sidak’s multiple-comparisons post-test (A and B) or Student’s t test were performed. ∗ = p < 0.05, ∗∗ = p < 0.01.
CD47 restricts the antiviral activity of alveolar macrophages independent of SIRPα
Although aMФ seem to be responsible for the enhanced viral clearance in CD47−/− mice, aMФ from IAV infected CD47−/− mice showed significantly less expression of costimulatory molecules compared to WT mice (Figure S4A). Also, similar values for the proportion of iNOS or arginase expressing aMФ between CD47−/− and WT mice were detected (Figure S4B).
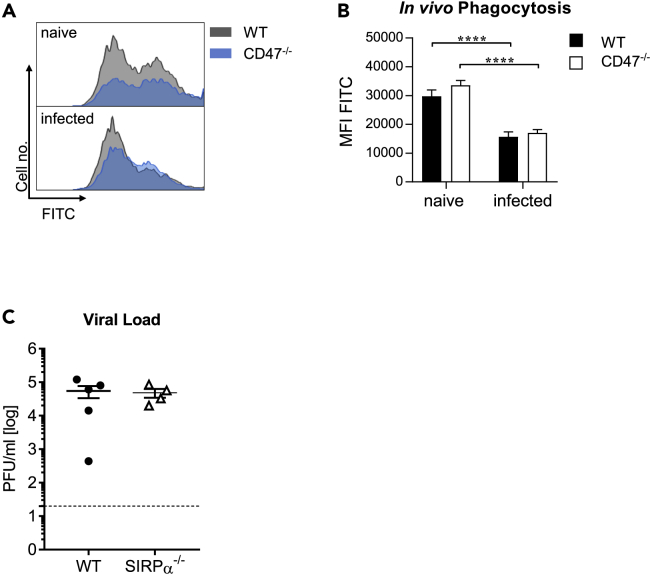
To address the impact of CD47 deficiency on IAV phagocytosis during infection, particles labeled with a pH-sensitive dye were intranasally applied into naive and infected (3 dpi) WT and CD47−/− mice (Figure 4A). The uptake of fluorescent particles by aMФ was determined by flow cytometry. Of interest, the phagocytic activity of aMФ was reduced on IAV infection, however the genetic deletion of CD47 expression did not influence the engulfment of foreign particles (Figure 4B). Thus, the observed protective function of CD47−/− aMФ on adoptive transfer into WT recipient mice is rather not because of enhanced phagocytosis. To further examine the role of CD47-SIRPα pathways, SIRPα−/− mice were infected with IAV. Importantly, the prevented interaction of CD47-SIRPα did not affect the clearance of IAV from infected tissue as similar virus titers were detected in WT and SIRPα−/− mice (Figure 4C). Thus, the antiviral effect of aMФ cannot be explained by an increase in phagocytosis and the disrupted CD47-SIRPα signaling does not seem to be responsible for the enhanced viral clearance in CD47−/− mice.
Figure 4.
CD47 restricts the antiviral activity of alveolar macrophages independent of SIRPα
At day 3 after influenza virus infection mice were i.n. treated with pH sensitive fluorescent particles and the phagocytosis of particles by aMФ was determined via flow cytometry after 1 h.
(A) The uptake of fluorescent particles is shown in representative histograms.
(B) The mean fluorescence intensity (MFI) for FITC of aMФ was determined (n = 10).
(C) The viral load in the lung of WT and SIRPα−/− mice was assessed by plaque assay 7 dpi. Representative results from one of two independent experiments are shown.
Data shown are mean ± SEM. Mann-Whitney test was performed. ∗∗∗∗ = p < 0.0001.
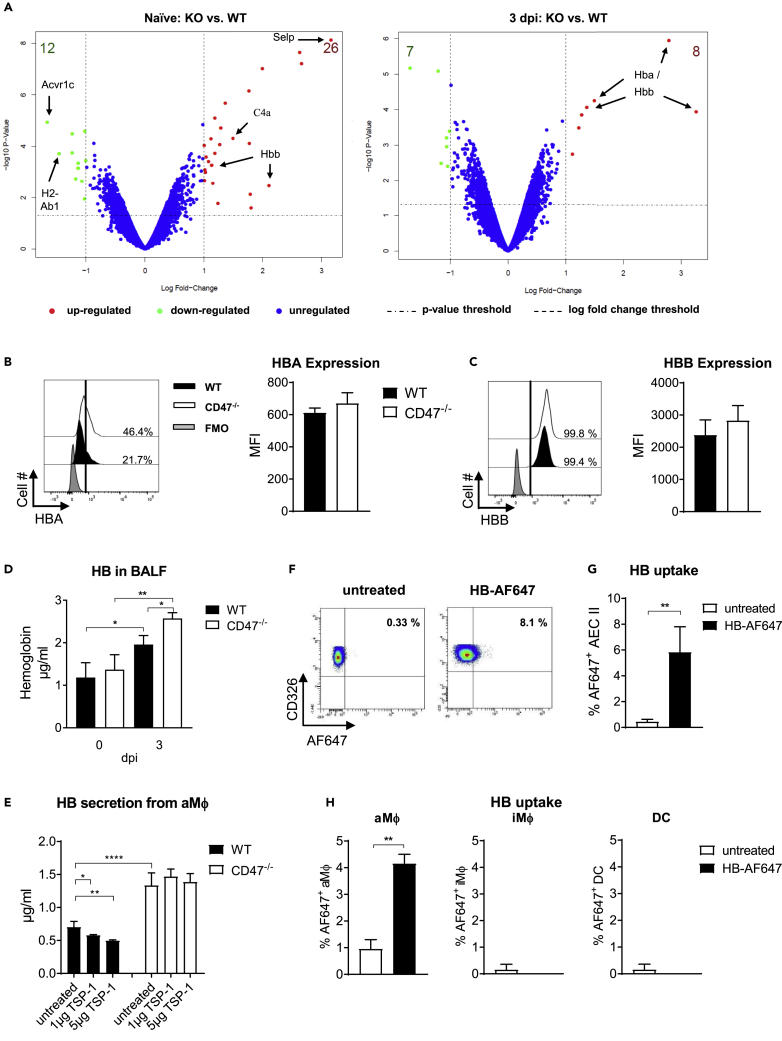
CD47 controls in the expression of hemoglobin in alveolar macrophages
Our previous experiments demonstrate an inhibitory role of CD47 on antiviral function, especially on aMФ during IAV infection. To define the mechanism of how CD47 restrains aMФ during IAV infection in more detail, we performed transcriptome analyses from aMФ that were isolated from either naive or IAV infected WT and CD47−/− mice (3 dpi). Already at steady state, CD47−/− aMФ displayed an altered expression profile showing an increased expression of genes involved in cell adhesion (e.g., SELP = selectin platelet) and inflammation (e.g., C4a = complement component 4a). In contrast, genes involved in adaptive immune responses (e.g., H2-Ab1 = histocompatibility 2, class II antigen A, beta 1) and apoptosis (e.g., ACVR1C = activin A receptor, type IC) were downregulated in absence of CD47 (Figure 5A). Surprisingly, an increased expression of hemoglobin-β (HBB) was detected in naive CD47−/− aMФ compared to WT aMФ. This augmented expression of HB was even more prominent during IAV infection as both hemoglobin-α (HBA) and HBB chains were found to be increased 3 dpi (Figure 5A). Importantly, HBA and HBB were also detectable in aMФ by flow cytometry (Figures 5B and 5C). Although aMФ generally expressed HBA and HBB, the expression was slightly increased in CD47−/− aMФ compared to WT aMФ (Figures 5B and 5C). Other immune cell populations such as iMΦ, DCs, PMN, CD4+T cells or B cells also seemed to express HBA and HBB, albeit at a significantly lower level compared to aMΦ (Figures S5A and S5B). In accordance to this, we also detected a significantly higher concentration of HB in the BALF of CD47−/− mice compared to WT mice after IAV infection (Figure 5D). This HB could be derived from aMФ, because we were able to detect HB in the supernatants of ex vivo cultured aMФ from infected mice, whereas the supernatants of CD47−/− aMФ showed significantly higher HB concentrations compared to WT aMФ. To investigate whether HB expression is regulated by CD47 signaling, we additionally stimulated these aMФ with the CD47 ligand TSP-1. In fact, we found that TSP-1 leads to significantly decreased secretion of HB by WT aMФ in a dose-dependent manner (Figure 5E). Importantly, this was not the case when CD47−/− aMФ were stimulated with TSP-1. Hence, aMФ in CD47−/− mice could produce more HB and secrete it into their environment, where it could act on surrounding cells. To test this hypothesis, we isolated AEC II as well as pulmonary immune cells from naive WT mice and incubated them ex vivo with fluorescently labeled HB. Of interest, HB seems to be taken up by AEC II within 30 min (Figures 5F and 5G). aMФ also appeared to be able to take up HB efficiently (Figure 5H). Of interest, this was not the case for iMФ and DCs. Thus, although the precise mechanism is still elusive, our data suggests that CD47 is involved in the negative regulation of HB expression in aMФ.
Figure 5.
CD47 is involved in the regulation of hemoglobin expression in alveolar macrophages
Mice were i.n. infected with IAV. aMФ were isolated from WT and CD47−/− mice 3 dpi.
(A) Transcriptome analysis of isolated WT and CD47−/− aMФ were assessed by an Affymetrix MicroArray. Hemoglobin alpha (HBA) (B) and beta (HBB) (C) chains were detected by flow cytometry analysis in aMФ isolated from WT and CD47−/− mice. Representative histograms for HBA and HBB are shown. Mean fluorescence intensity (MFI) for HBA and HBB from aMФ are shown (n = 4).
(D) Whole hemoglobin (HB) concentration was assessed in BALF at indicated time points by ELISA (n = 7–17).
(E) aMФ were isolated from infected WT and CD47−/− mice 3 dpi and stimulated ex vivo with thrombospondin-1 (TSP-1). After 24 h, HB concentration in the supernatants was measured by ELISA (n = 4).
(F) Type II alveolar epithelial cells (AEC II) were isolated from the lung of naive WT mice and incubated with 1 μg/mL Alexa Fluor 647 (AF647)-labeled HB for 30 min. Representative flow cytometry plots are shown. F) Percentages of HB+ AEC II cells are shown (n = 4).
(G) Isolated cells from lungs of WT mice were incubated with 1 μg/mL AF647-labelled HB for 30 min.
Percentages of HB+ aMФ, iMФ, and DCs are shown. Results depicted were obtained from one (A) or two independent experiments (C, D, E, G, and H). Data shown are mean ± SEM. One-way ANOVA with Tukey’s multiple-comparisons post-test (D and E) or Student’s t test (F) were performed. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗∗ = p < 0.0001.
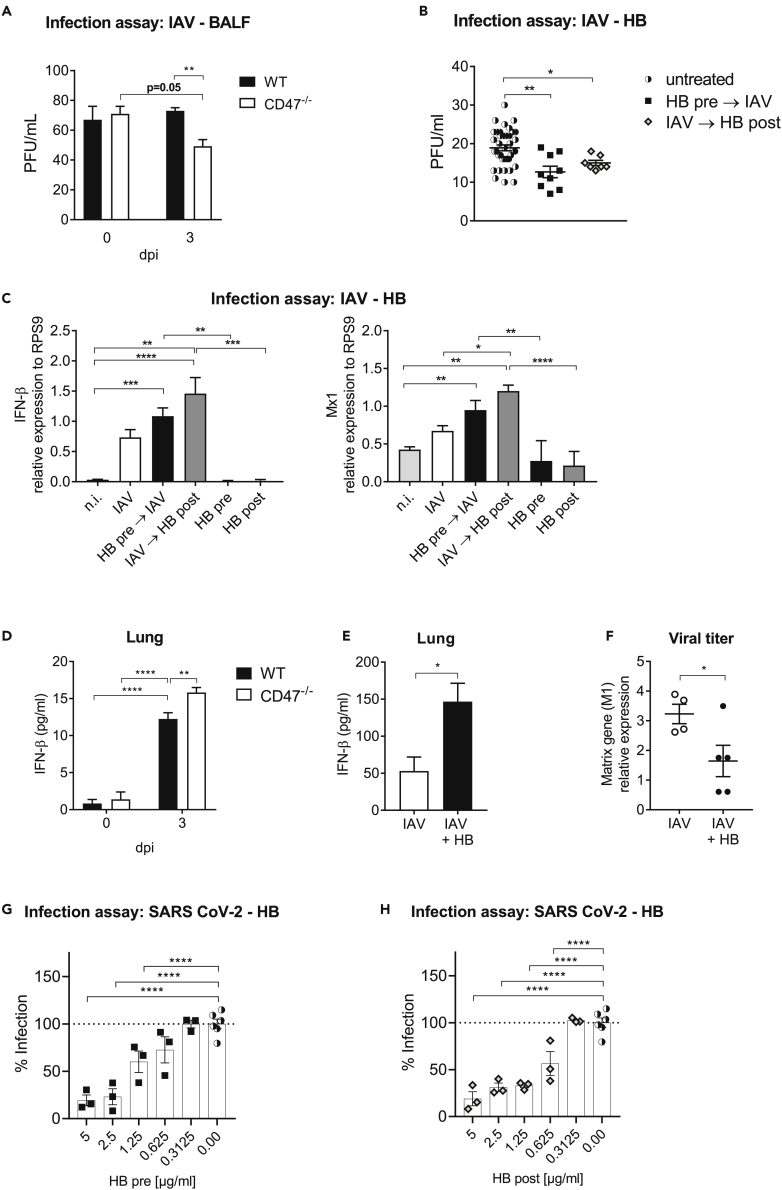
Hemoglobin restrains influenza virus infection
Although HB is mainly known for its role in oxygen transport, it was recently associated with antiviral responses against several RNA viruses.17 It is not yet known whether HB also has an antiviral function against IAV. We thus hypothesized, that HB might play a role in the defense against IAV infection and the corresponding inflammation. Therefore, we next examined whether BALF of infected CD47−/− mice could restrain IAV infection. Of interest, in vitro treatment of Madin-Darby canine kidney (MDCK) cells before IAV infection with BALF obtained from infected CD47−/− mice, but not WT mice suppressed the infectivity (Figure 6A). Because the antiviral activity of BALF from infected CD47−/− mice could be because of the increased HB concentrations, we next investigated the possible role of HB in the defense against IAV. To this end, we infected MDCK cells in presence or absence of HB prior or post infection. Pretreatment with HB was already sufficient to significantly decrease viral replication whereas the addition of HB post infection had a similar effect confirming an antiviral role for HB (Figure 6B). To analyze whether this effect could be related to an enhanced IFN I response induced by HB, we analyzed gene expression of IFN-β and the interferon inducible gene (ISG) Mx1 by MDCK cells 48 h after infection. Indeed, IFN-β and Mx1 expression were significantly increased after incubation of cells with HB prior or post infection in contrast to infection or HB treatment only (Figure 6C). Of interest, we were also able to determine an increased concentration of IFN-β in the lungs of CD47−/− compared to WT mice on day 3 after infection (Figure 6D). To investigate whether IFN-β can be specifically induced by administration of HB, we treated WT mice with HB during IAV infection. We observed, that HB treatment increases the secretion of IFN-β in the lungs of infected mice compared to untreated infected animals (Figure 6E). In addition, intranasal administration of HB reduced the viral loads in the lung compared to only infected mice (Figure 6F).
Figure 6.
Hemoglobin has antiviral function and reduces influenza virus replication
(A) MDCK cells were incubated with the BALF of WT and CD47−/− mice isolated at the indicated time points. After 30 min cells were infected with 1000 PFU of influenza virus and 3 dpi the antiviral activity of BALF samples was determined based on plaque count (n = 8).
(B) MDCK cells were incubated with 1 μg/mL hemoglobin (HB) for 30 min. Afterward, cells were infected and 3 dpi the antiviral activity of HB was determined based on plaque count. For post infection treatment, HB was added 2 h after infection.
(C) MDCK cells were incubated prior or after infection with 1 μg/mL HB as described. 48 h after infection cells were harvested and analyzed for IFN-β and Mx1 gene expression by qRT-PCR (n = 6). Non infected (n.i.) cells served as controls.
(D) Mice were i.n. infected with IAV. 3 dpi IFN-β concentrations were assessed in lung homogenates (n = 4). One representative experiment of two is shown.
(E) IAV-infected mice were intranasally treated with 50 μg HB protein on day 0 and 3 post infection. IFN-β expression by ELISA was analyzed 6 dpi from lung homogenates (n = 3). One representative experiment of two is shown.
(F) Virus titers were assessed by qRT-PCR from lung homogenates of HB treated and untreated mice 6 dpi. One representative experiment of two is shown.
(G) A549-AT cells were incubated with indicated HB concentrations for 30 min. Afterward, cells were infected and 24 h later the antiviral activity of HB was determined by in cell ELISA.
(H) For post infection treatment, HB was added after SARS CoV-2 infection of A549-AT cells. Two independent experiments are shown unless otherwise noted.
Data shown are mean ± SEM. One-way ANOVA with Tukey’s multiple-comparisons post-test was performed in (A–C, G, and H). Two-way ANOVA with Sidak’s multiple-comparisons post-test was performed in (D). Student’s t test was performed in (E and F). ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
The antiviral abilities of HB might not be IAV specific, but also applicable to other (RNA) viruses such as SARS-CoV-2. Therefore, we tested whether HB can exert an antiviral effect on SARS-CoV-2 infection and added HB to A549-AT cells before or after infecting target cells with SARS-CoV-2. Importantly, we found dose-dependent significant reductions in infection under both conditions (Figures 6G and 6H). This again confirms the potent antiviral effect of HB even across different virus types. Thus, enhanced expression of antiviral HB in aMФ of CD47−/− mice might augment the clearance of IAV during infection by enhanced IFN-β induction, which means that disruption of CD47 signaling could elevate its expression.
Discussion
A deeper understanding of the immunological mechanisms during an influenza virus infection remains an urgent need, as the efficiency of existing vaccines or antiviral drugs is limited. Here, the manipulation of antiviral immune responses is of special interest, as they contribute to disease severity.7,8 Among the known immunoregulatory proteins, CD47 is currently widely studied, as it appeared to be a promising target in cancer therapy,11 whereas its impact on antiviral immunity is a matter of debate. Within the present study, we defined the role of CD47 for innate and adaptive immune responses during an acute IAV infection. Upon infection with IAV, significantly lower viral loads were observed in CD47−/− compared to WT mice. Of interest, CD47 did not seem to limit the adaptive immune response but restricted antiviral aMФ immunity. Our data suggests that CD47 regulates the expression of HB by aMФ, which might exert antiviral activity in the lung during IAV infection.
We found increased cell surface levels of CD47 during acute IAV infection on different immune cells. Inflammatory stimuli such as pro-inflammatory cytokines (e.g., TNF-α, IFN) or activation of pattern recognition receptors (e.g., TLR7) have been described to induce the expression of CD47 during cancer as well as infection.18,20 Although the role of CD47 as an innate immune checkpoint is unambiguous during cancer,21 its effect on the course of infectious diseases is varying depending on the infectious agent and the site of infection. On the one hand, CD47 was shown to be involved in protective immunity against systemic bacterial (Escherichia coli) or fungal (Candida albicans) infection14,22 as well as the clearance of chronic LCMV infection.23 On the other hand, blockade or genetic ablation of CD47 were shown to ameliorate the clearance of Plasmodium yoelii and acute LCMV infection.19,24 Within the present study, we detected significantly reduced viral loads in CD47−/− compared to WT mice at day 7 after IAV infection implying a repressive role of CD47. This is interesting because CD47 was previously described as a negative regulator of type 1 immune responses.25,26 Commonly, bone marrow chimeric mice would be used to unravel the distinct role of CD47 expressed on tissue and immune cells. Yet, as CD47 functions as a marker of self this approach is difficult to implement because a stable transfer of CD47−/− cells into WT mice has not been achieved so far.27 Instead, we used ex vivo infection of CD47−/− and WT AEC II to confirm the regulatory role of CD47 on viral clearance rather than viral replication. In line with our hypothesis, similar accumulation of viral RNA was observed in CD47−/− and WT epithelial cells independent of the infective dose. However, we did not observe any differences in body weight loss or histopathology between IAV infected WT and CD47−/− mice indicating comparable inflammation. Of interest, we found significantly enhanced virus clearance especially after high-dose infection, whereas low-dose infection seemed to result in trend differences in virus clearance. It has previously been shown that the level of the infectious dose in IAV infection can determine the type of immune response initiated through TLR7 and RIG-I recognition.28 In this context, the initially high number of viral particles and their replication could indeed be important for the effects observed by us.
In contrast to previous studies showing a major effect of CD47 on CTL responses by the inhibition of antigen phagocytosis and subsequent presentation,18,19,21 CD8+T cell frequencies were not enhanced in the lung of CD47−/− mice during IAV infection. Instead, a reduced expression of cytotoxic effector molecules (e.g., GzmB) was observed in absence of CD47. Thus, CD47 might have a dual role for CTL immunity during infectious diseases depending on the location of and time point of infection. In line with this, CD47 was described to both contribute to and limit T cell activation.29,30 The underlying mechanisms appear to be individual and have yet to be elucidated. In accordance with a previous study, which investigated the role of CD47 during vaccination we did not find augmented levels of IgA, IgM and IgG antibodies in the BALF of CD47−/− compared to WT mice after IAV infection.31
Of interest, we determined a shift in the proportion of aMФ within the lung at day 3 after influenza infection. Specifically, frequencies and numbers of aMФ in the lung of CD47−/− increased compared to WT mice 3 dpi. Ultimately, this could also be because of increased retention of aMΦ in the event of CD47 loss. aMФ are a highly specialized subset of MФ, which derive during embryogenesis and form a self-renewing population.32 Because of their specific localization in the alveolar lumen they are the first immune cells to encounter invading influenza viruses and significantly shape subsequent immunity.5 The application of clodronate containing liposomes is a suitable method to deplete MФ in vivo.33 However, repeated intranasal treatment worsened condition of the mice, so that we had to measure the viral load here as early as day 5 after infection. Importantly, viral loads were already decreased 5 dpi in the lung of CD47−/− mice treated with PBS containing liposomes. However, depletion of aMФ by intranasal application of clodronate containing liposomes impaired this viral clearance in IAV infected CD47−/− mice. The fact that aMФ depletion in WT mice had no such effect on viral loads suggests a crucial role of CD47 for the antiviral function of aMФ. This is also in accordance with our finding that the transfer of CD47−/− aMФ into WT recipient mice was able to significantly augment the clearance of IAV suggesting a negative role for CD47 in the regulation of antiviral function of aMФ.
In this regard, aMФ have been shown to contribute to the clearance of IAV by the phagocytosis of aggregated viral particles as well as the engulfment of dying or dead infected epithelial cells.34,35 Although the binding of CD47 to SIRPα was previously described to restrain the phagocytic activity of MФ,36 no enhanced phagocytosis by CD47−/− aMФ were observed within this study. Furthermore, the viral clearance after IAV infection was not improved in SIRPα deficient mice compared to WT mice. Thus, other proteins such as surfactant proteins A and D might still interact with SIRPα in the alveolar lumen in absence of CD47.37,38 At the same time, it suggests that the negative regulation of the antiviral function of aMФ could rely on another ligand of CD47, such as TSP-1. TSP-1 is a multi-domain matrix glycoprotein that has been shown to be involved in inflammation.39 Different functions of TSP-1 for macrophages have been described so far controlling pro-inflammatory immunity.40 Well in line, in our experimental setting TSP-1 decreased aMФ mediated HB secretion from WT but not from CD47−/− mice. Thus, our data strongly suggests TSP-1 as a negative regulator of the HB expression and antiviral function of macrophages via CD47. To what extent TSP-1 is involved in the antiviral role of CD47 during IAV infection requires further investigation. However, transcriptome analysis revealed elevated transcript levels of HBA and HBB in CD47−/− aMФ compared to WT aMФ before and after IAV infection. This was surprising because previous studies did not indicate an induction of HB expression, i.e., by AEC II after IAV infection.41
HB is essential for oxygen transport by erythrocytes. However, the gene expression of non-erythroid HB in macrophages was initially described in 1999.42 Our current study confirms now that HB is expressed and exists at the protein level in aMФ. The fact that we could not see any significant differences in the HB protein level between WT and CD47−/− MФ here could possibly be because of ingested HB. In fact, the uptake of fluorescently labeled HB by aMФ and AEC II that we noticed in our study indicates that HB can both be secreted and taken up by these cells. Moreover, additional functions of HB in antimicrobial and antiviral defense have been demonstrated. For example, C-terminal fragments of HBB have been shown to inhibit entry of herpes simplex virus 2 (HSV-2) and to limit the growth of different bacteria.43,44 In addition, HBB has been demonstrated to restrict infectivity of different RNA viruses by regulating RIG-I mediated type I IFN response.17 Influenza viruses are also recognized via RIG-I.9 Importantly, to our best knowledge we show here for the first time that HB inhibited influenza virus replication in vitro and in vivo, which was accompanied by enhanced IFN-β expression. Beside acceleration of viral clearance by modulating type 1 immunity, IFN-β mediates inhibition of viral replication by induction of ISGs such as Mx1 that interestingly cannot be compensated for by IFN-α.10,45 Of note, although we used mouse-derived HB, sequence analyses revealed that there are high similarities between mouse, dog and human HB. This suggests that HB mediates an improved antiviral response via similar mechanisms as described by Yang et al.17 Hence, our data indicate, that the CD47−/− aMФ do not directly enhance the clearance of virus, e.g., via phagocytosis of infected cells, but alter the cytokine response (i.e., IFN-β) possibly in target cells such as AEC II via HB secretion. This indirect effect of CD47−/− aMФ could result in a delayed effect on the antiviral response and explain the discrepancy between the late decreased viral load in the CD47−/− mice at day 7 post-infection and the early altered aMФ response at day 3. However, we were able to detect significant reduction in viral load in lungs of CD47−/− compared to WT mice as early as day 5 after infection (Figure 3B), indicating that the enhanced antiviral response in CD47−/− mice must develop directly after the 3 dpi time point examined. Thus, our data suggests that aMФ initiate the enhanced virus clearance indirectly. A MФ-specific knockdown of HB could clarify the connection between CD47-deficient macrophages and the increased antiviral effect via HB. Unfortunately, such transgenic mice that would allow to specifically delete HB in aMФ are not available. Thus, future studies still need to further specify the role of MФ-derived HB. However, the direct treatment of IAV-infected WT mice with HB led to the effects we postulate: reduced viral load with simultaneously increased IFN-β concentration in the lungs.
Importantly, we could also demonstrate for the first time an antiviral effect of HB on SARS-CoV-2 infection, indicating that the observed effects are IAV non-specific processes induced by HB and thus could be of therapeutic interest to treat acute viral respiratory infections. The fact that we identified increased levels of HB after IAV infection in the BALF of CD47−/− mice compared to samples from WT mice could indicate a secretion of HB by aMФ and subsequent uptake by surrounding cells, such as AEC II and DCs. Our uptake studies demonstrate the general possibility of uptake of secreted HB. The uptake of HB by innate immune cells may then lead to increased recognition of influenza virus or SARS-CoV-2 RNA, which ultimately results in increased IFN-β expression. In line with this, we were able to detect higher IFN-β concentrations in the lung of infected CD47−/− compared to WT mice as well as after the treatment of infected WT mice with HB. However, subsequent studies have yet to identify the exact source of this IFN-β.
Taken together, our study is the first to demonstrate CD47 as a negative regulator of antiviral aMΦ immunity during acute IAV infection. Improved viral clearance of IAV in absence of CD47 seemed to be related to the expression of HB. Yet, further studies are of need to define the detailed mechanisms of the induction and function of antiviral HB. Importantly, due to the non-specific nature of CD47 signaling as well as the evolutionary conservation of HB, CD47 targeting therapies or application of HB may allow the treatment of various respiratory infections such as IAV, SARS-CoV-2 or even the respiratory syncytial virus (RSV).
Limitations of the study
HB is a tetramer protein. Although we were able to demonstrate an antiviral effect of the entire HB protein, it remains unclear whether this effect can also be achieved by the α- or β-subunit. The commercial availability of murine HB-α and HB-β is currently limited. Transgenic mice that would allow to specifically delete HB in aMФ are not available to investigate the direct relationship between MФ-derived HB and antiviral effects.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | Cat# |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD47 antibody, PeCy7 | Biolegend | 127524; RRID:AB_2629545 |
| Anti-mouse CD19 antibody PE PeCy7 |
BD Biosciences BD Biosciences |
557399; RRID:AB_396682 552854; RRID:AB_394495 |
| Anti-mouse CD31 antibody, PE | Biolegend | 102407; RRID:AB_312902 |
| Anti-mouse CD11b antibody PE PerCP-Cy5.5 APC |
BD Biosciences BD Biosciences ThermoFisher Scientific |
557397; RRID:AB_396680 550993; RRID:AB_394002 17-0112-82; RRID:AB_469343 |
| CD16/CD32 monoclonal antibody, PE | eBioscience | 12-0161-82; RRID:AB_465568 |
| Anti-mouse CD45 antibody, PE | BD Biosciences | 553081; RRID:AB_394611 |
| F4/80 monoclonal antibody APC Pacific Blue PE |
ThermoFisher Scientific | 17-4801-82; RRID:AB_2784648 MF48028; RRID:AB_10373419 12-4801-82; RRID:AB_465923 |
| Anti-mouse CD4 antibody PerCP PE |
BD Biosciences BD Biosciences |
561090; RRID:AB_10562560 553653; RRID:AB_394973 |
| Anti-mouse CD93 antibody, APC | Biolegend | 136509; RRID:AB_2275879 |
| Anti-mouse CD11c antibody APC V450 |
BD Biosciences BD Biosciences |
550261; RRID:AB_398460 560521; RRID:AB_1727423 |
| TruStain fcX™ (anti-mouse CD16/32) antibody | Biolegend | 101320; RRID:AB_1574975 |
| Anti-mouse CD172a (SIRPa) antibody APC BV421 |
Biolegend BD Bioscineces |
144014; RRID:AB_2564061 740071; RRID:AB_2739835 |
| Anti-mouse CD8 antibody, Pacific Blue | BD Biosciences | 558106; RRID:AB_397029 |
| Anti-mouse Ly6G antibody APC FITC |
Biolegend BD Biosciences |
127614; RRID:AB_2227348 561105; RRID:AB_10562567 |
| Granzyme B monoclonal antibody, APC | ThermoFisher Scientific | MHGB05; RRID:AB_10373420 |
| Anti-mouse CD107a antibody, PE | BD Biosciences | 558661; RRID:AB_1645247 |
| iNOS monoclonal antibody, PeCy7 | ThermoFisher Scientific | 25-5920-82; RRID:AB_2573499 |
| Human/mouse Arginase 1 antibody, APC | Bio-Techne | IC5868A; RRID:AB_2810265 |
| Anti-mouse CD80 antibody, V450 | BD Biosciences | 560523; RRID:AB_1727515 |
| Anti-mouse CD86 antibody, PeCy7 | BD Biosciences | 560582; RRID:AB_1727518 |
| Anti-mouse I-A/I-E antibody, BV510 | Biolegend | 107636; RRID:AB_2734168 |
| Anti-rabbit IgG DyLight 649 | Biolegend | 406406; RRID:AB_1575135 |
| Anti-hemoglobin alpha (HBA) antibody | Novus Biologicals | NBP2-67716; RRID:AB_2809609 |
| Anti-hemoglobin beta (HBB) antibody | Invitrogen | PA-576877; RRID:AB_2720604 |
| Anti-SARS-CoV-2-NP | Bioss | ABIN6952435; RRID:AB_2890255 |
| Peroxidase-AffiniPure Goat Anti-Mouse IgG (H+L) | Dianova | 115-035-003; RRID:AB_10015289 |
| Anti-mouse MAC-3 | Biolegend | 108502; RRID:AB_313383 |
| HRP-Goat Anti-Rat IgG (H+L) | ThermoFisher Scientific | A18865; RRID:AB_2535642 |
| Bacterial and virus strains | ||
| Influenza A/PR/8/34 | Prof. Dr. P. Stäheli (Department of Virology, University Freiburg) | N/A |
| SARS-CoV-2 B.1.1.10 | Patient material (Heilingloh et al.) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 2N H2SO4 | Roth | X873 |
| Avicel | Sigma Aldrich | 11365 |
| Bovine Serum Albumin (BSA) | Biomol | 01400.100 |
| Brefeldin A | Sigma Aldrich | B 7651 |
| Clodronate Liposomes & Control Liposomes (PBS) | Liposoma B.V. | N/A |
| Collagenase D | Roche Life Science | 11088866001 |
| Corning® Dispase | Corning | 354235 |
| Crystal violet | Roth | T 123.1 |
| Diaminobenzidine (DAB) | Dako Agilent | K3467 |
| Dispase | Corning | 354235 |
| DNase I | Roche Life Science | 10104159001 |
| DNase I from bovine prancreas | Merck KGaA | D4263 |
| Ethylenediaminetetraacetic acid (EDTA) | Roth | 8043.2 |
| Fixable viability dye, eFluor 780 | ThermoFisher Scientific | 65-0865-18 |
| Gentamycin | Sigma Aldrich | G 1272 |
| Goat Serum | Invitrogen | 31872 |
| GoTaq® DNA Polymerase | Promega | M3001 |
| Igepal | Sigma Aldrich | I8896 |
| Ionomycin | Sigma Aldrich | I 0634 |
| L-Glutamine | Sigma Aldrich | G 7513 |
| LE Agarose | Biozym | 840004 |
| M-MLV Reverse Transcriptase | Promega | M 3683 |
| Matrigel (Basement Membrane Matrix) Matrigel | Corning | 354230 |
| Murine Hemoglobin protein | Fitzgerald Industries | FGI-30-1135 |
| Paraformaldehyde (PFA) | Roth | 0335.2 |
| pHrodo Green E. coli BioParticles | ThermoFisher Scientific | P35366 |
| Recombinant Mouse Thrombospondin-1 (TSP-1) | R&D Systems | 7859-TH-050 |
| RNase-Free DNase Set | Qiagen | 79254 |
| ROTI Histofix | Roth | P087.3 |
| TMB Substrate Solution | Biolegend | 421101 |
| Triton X-100 | Roth | 3051.4 |
| Trypsin/EDTA | Sigma Aldrich | T4174 |
| Trypsin from bovine prancreas | Sigma Aldrich | T8802 |
| Tween 20 | Roth | 9127 |
| Critical commercial assays | ||
| AlexaFluor 647 Protein Labeling Kit | ThermoFisher Scientific | A20173 |
| FoxP3 Staining Buffer Set | eBioscience | 00-5523-00 |
| Luminex Assay | Bio-Techne | N/A |
| LumiKine Xpress mIFN-α/-β 2.0 ELISA Kit | Invivogene | Luex-mifnav2 / Luex-mifnbv2 |
| Maxima SYBR Green qPCR Kit | ThermoFisher Scientific | K0222 |
| MouseFree Hemoglobin ELISA Kit | Novus Biologicals | NBP2-59999 |
| Nucleo Spin RNA XS | Macherey-Nagel | 740902.250 |
| RNeasy Mini Kit | Qiagen | 74106 |
| Deposited data | ||
| MicroArray data | This paper | GEO: GSE201825 |
| Experimental models: Cell lines | ||
| A549-AT | Dr. Marek Widera (Institute of Virology, University Frankfurt) | N/A |
| Madin-Darby canine kidney (MDCK) | Prof. Dr. P. Stäheli (Department of Virology, University Freiburg) | N/A |
| Experimental models: Organisms/strains | ||
| CD47−/− mice | Jackson Laboratory | 003173 |
| C57BL/6 wildtype mice | Envigo | 057 |
| SIRPα−/− mice | Riken Bioresource Center | RBRC01544 |
| Oligonucleotides | ||
| Forward primer for matrix gene M1 5′- CTTCTAACCGAGGTCGAAACG-3′ |
Eurofins | N/A |
| Reverse primer for matrix gene M1 5′-AGGGCATTTTGGACAAAGTCGTCTA-3′ |
Eurofins | N/A |
| Forward primer for Rps9 5′-CTGGACGAGGGCAAGATGAAGC-3′ |
Eurofins | N/A |
| Reverse primer for Rps9 5‘-TGACGTTGGCGGATGAGCACA-3‘ |
Eurofins | N/A |
| Forward primer for canis Ifnb 5‘-CAGTTCCAGAAGGAGGACA-3’ |
Eurofins | N/A |
| Reverse primer for canis Ifnb 5′-TGTCCCAGGTGAAGTTTTCC-3′ |
Eurofins | N/A |
| Forward primer for canis Mx1 5′- GAATCCTGTACCCAATCATGTG-3′ |
Eurofins | N/A |
| Reverse primer for canis Mx1 5′-TACCTTCTCCTCATATTGGCT-3′ |
Eurofins | N/A |
| Forward primer for canis Rps9 5′-GGCCAAGTCCATCCACCAT-3′ |
Eurofins | N/A |
| Reverse primer for canis Rps9 5′-GGCGGCCACCCCCATAC-3′ |
Eurofins | N/A |
| Software and algorithms | ||
| BD FACSDivaTM software v 8.0.1 | BD Biosciences | N/A |
| Fiji | Schindelin et al., 201246 | N/A |
| Prism 7 | GraphPad Software | N/A |
| Other | ||
| Dulbecco’s Modified Eagle Medium | Invitrogen | 41966029 |
| Opti-MEM | Invitrogen | 31985047 |
| Fetal Bovine Serum | Sigma Aldrich | F7524-500ML |
| Rosswell Memorial Park Institute Medium | Invitrogen | 72400-021 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Torben Knuschke (torben.knuschke@uk-essen.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Microarray data were deposited in NCBI’s Gene Expression Omnibus and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental models and subjectdetails
Cell culture
Madin-Darby canine kidney (MDCK) cell line was a kind donation from Professor Peter Staeheli, Department of Virology, University of Freiburg. MDCK cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) medium complete containing 10% endotoxin-free fetal bovine serum (FBS) and 50 μg/mL penicillin/streptomycin. Cell lines were maintained in a humidified 5% CO2 atmosphere at 37°C.
Animals
Wildtype C57BL/6 mice were purchased from Envigo Laboratories (Envigo CRS GmbH). CD47−/− mice were purchased from Jackson laboratories (JAX stock #003173; Jackson Laboratory). SIRPα−/− mice used in the present study were kindly provided by K. Lang (Institute for Immunology, University Hospital Essen). Male and female mice were used in the experiments. All mice used in the experiments were 7–9 weeksold at the time point of infection and housed under specific-pathogen-free conditions in the Laboratory Animal Facility of the University Hospital Essen. Animal experiments were performed in strict accordance with the German regulations of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). The experimental protocols were approved by the North Rhine-Westphalia State Agency for Nature, Environment and Consumer Protection (LANUV). All efforts were made to minimize suffering.
Viruses
Infection studies were performed by using the Influenza A/PR/8/34 (H1N1) strain (a kind donation by Professor Peter Stäheli, Department of Virology, University of Freiburg) or the SARS-CoV-2 strain B.1.1.10.
Method details
Influenza A virus infection
Wildtype mice, CD47−/− mice and SIRPα−/− were anesthetized by ketamine (100 mg/kg body weight)/xylazine (5 mg/kg body weight) and intranasally infected with sublethal (8 PFU) or lethal dose (75 PFU) influenza virus A/PR/8/34 (25 μL). Upon infection mice were monitored daily and weight loss was recorded.
Histopathological analysis
Lungs were rinsed in ice-cold PBS and fixed in 4% paraformaldehyde (PFA), followed by embedding in paraffin and sectioning at 4 μm thickness. After staining with hematoxylin and eosin (H&E) the severity of the histopathology was scored in a blinded manner according to the inflammation markers, including inflammatory cell infiltration, cell necrosis, granulocyte infiltration and histiocytosis. Each marker was scored from 0 (= no signs of inflammation) to 4 (= severe signs of inflammation).
Ex vivo AEC II infection assay
The specific isolation of alveolar epithelial cells was performed as described previously.47 Briefly, lungs were perfused with phosphate buffer saline (PBS) and filled with Dispase Solution (Corning) followed by 1% Agarose Solution (Biozym). After incubation for 2 min at 4°C, lungs were dissected and digested for 45minat RT in Dispase Solution. Next, the lobes were rigorously minced and DNase I (Merck KGaA) as well as TruStain fcX™ (Biolegend) were added. The samples were incubated for 10minat RT on a rocker and the obtained cell suspension was sequentially filtered through a 100, 70, and 30 μm strainer. Erythrocyte lysis was performed using Ammonium-Chloride-Potassium (ACK) buffer and cells were stained with following antibodies: CD19-PE, CD31-PE, CD11b-PE, CD16/32-PE, CD45 PE, F4/80-APC, CD93-APC and CD11c-APC. The AEC II cells were sorted with the BD FACSAria II and isolated AEC II cells were plated on a 96-well plate, which was coated with 3 mg/mL Matrigel (Corning). Before further use the AEC II were allowed to rest for 12hat 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS, 50 U/mL penicillin and 25 mM HEPES (Invitrogen). Next, AEC II were infected with different doses of IAV and incubated for 48hat 37°C. Thereafter the cells were lysed, and the RNA was isolated in accordance with the manufacturer’s instructions of NucleoSpin® RNA XS kit (Macherey-Nagel). The obtained RNA was transcribed into cDNA and the viral titer of IAV in infected AEC II was determined by qRT-PCR using the Maxima SYBR Green qPCR Kit (Thermo Scientific). A conserved sequence in the matrix gene (M1) of influenza A virus was detected using the specific primers (5′- CTTCTAACCGAGGTCGAAACG-3′; 5′-AGGGCATTTTGGACAAAGTCGTCTA-3′) described elsewhere.48
Cell recovery
To obtain single cell suspensions of pulmonary immune cells, mice were euthanized at indicated time points, the lung tissue was perfused with PBS and dissected. Digestion of the tissue was performed at 37°C in Iscove’s Modified Dulbecco’s Medium (IMDM) (Invitrogen) containing 0.5 mg/mL DNase I (Roche Life Science), 0.16 mg/mL Collagenase D (Roche Life Science) and 5% heat-inactivated FBS. After 45 min the tissue was placed on a 70 μm nylon cell strainer and erythrocytes were lysed using ACK buffer. Finally, the cells were resolved in PBS containing 2% heat-inactivated FBS and 2 mM Ethylenediaminetetraacetic acid (EDTA) for further use.
For generation of BALF, the trachea was surgically exposed and intubated with a syringe catheter. The lungs underwent lavage with 1 mL pre-warmed PBS containing 2 mM EDTA and 0.5% FBS for 5 times. In total 5 mL BALF was obtained from each mouse and cells in BALF were pelleted by centrifugation (1.200 rpm for 10 min at 4°C). Cells were then BAL cells were erythrocyte-depleted using ACK buffer and resuspended in PBS containing 2% heat-inactivated FBS and 2 mM EDTA or medium.
Flow cytometric analysis
According to Misharin et al. aMФ were classified by the expression of CD11chi and CD11blo49 (Figure S3) for investigation of the proportion of aMФ. The gating was adjusted by previously gating on CD103- cells to exclude DCs. For staining of CD107a lymphocytes were restimulated for 4 h with PMA and ionomycin in the presence of brefeldin A and CD107a antibody. Subsequent intracellular staining for granzyme B was performed according to the Foxp3 Staining Buffer Set (eBioscience). For intracellular HBA/HBB staining cells were fixed with 2% PFA (Roth) for 30minat 4°C. Afterward, cells were washed and treated for 10 min with 0.1% Igepal (Sigma Aldrich) at room temperature. Subsequently, intracellular staining for HB alpha and beta was performed for 30minat 4°C followed by staining with anti-rabbit IgG (Biolegend) for 30minat 4°C. For all stainings a Fixable Viability Dye (Thermo Scientific) was used to exclude dead cells from analysis. Data was acquired by using a LSR II or Canto™ instrument together with DIVA software (BD Biosciences Pharmingen).
Immunohistology
Immunohistochemical staining of MФ responses were performed as described previously.50 Briefly, the lungs were perfused and fixed, embedded in paraffin and sectioned with a thickness of 5 μm. Subsequently, the paraffin was removed from the sections and the samples were steam boiled in citrate buffer (pH 6). Endogenous peroxidase activity was blocked with 3% H2O2 and MФ were labeled based on the expression of MAC-3. Visualization of labeled cells was accomplished via horseradish peroxidase (HRP) coupled secondary antibodies and a subsequent diaminobenzidine (DAB) staining. Moreover, the sections were counterstained with hematoxylin.
Influenza virus plaque assay and suppression assay
Lungs were obtained from IAV infected mice at indicated time points and subsequently homogenized in PBS containing 0.3% bovine serum albumin (BSA). Viral loads were determined by plaque assay as described previously.51 Homogenized lungs were centrifuged for 8minat 8.000 RPM. Lung supernatants were serially diluted in Opti-MEM (Invitrogen) containing 0.3% BSA and added to MDCK cells on a 12 well plate for 1hat room temperature. Afterward, supernatants were discarded and DMEM complete containing 3% Avicel, trypsin (0.4 μg/mL) and gentamycin (100 μg/mL) was added for 72hat 37°C. To count the plaques MDCK cells were fixed for 30 min with 4% PFA. Afterward MDCK cell layer was stained with crystal violet (Roth).
In vivo macrophage depletion
Specific depletion of macrophages was achieved by administration of dichloromethylene bisphosphonate (clodronate) containing liposomes (Liposoma B.V. - Amsterdam, Netherlands). A single intranasal instillation of 75 μL of clodronate containing liposomes was performed to deplete aMФ from the alveolar lumen. Depletion of iMΦ was achieved by twice intraperitoneal administration of 200 μL of clodronate containing liposomes at an interval of 48 h.52
Adoptive transfer of murine alveolar macrophages
Adoptive transfer of aMФ was accomplished by an initial depletion of macrophages in the alveolar lumen of recipient mice as described before. After 48 h aMФ isolated from wildtype and CD47−/− donor mice were transferred intranasally into recipient mice. Isolation of murine aMФ was achieved by repeated BAL and a subsequent sorting of the cells based on their autofluorescence via the BD FACSAria II to obtain an untouched aMФ population. The purity of the population was checked by staining for F4/80+ and CD11chi cells as described previously.53
In vivo phagocytosis assay
The in vivo phagocytic activity of macrophages in the lung was determined by intranasal administration of 0.05 mg pHrodo Green E. coli BioParticles (Thermo Scientific) in 75 μL of PBS. Anesthesia was performed as mentioned above and after 1 h cells of the BAL were isolated for flow cytometric analyses. The pH-sensitive pHrodo Green dye enables a specific determination of phagocytosed BioParticles as it emits green light in the acidic environment of phagosomes.
In vivo treatment of mice with hemoglobin
Mice were infected with 75 PFU as described and intranasally treated on day 0 and 3 post infection with 50 μg murine hemoglobin protein (Fitzgerald Industries) diluted in PBS.
Transcriptome analysis
Global gene expression of WT and KO aMФ was assessed by Affymetrix MicroArray. Quality and integrity of total RNA was controlled on Agilent Technologies 2100 Bioanalyzer (Agilent Technologies; Waldbronn, Germany). RNA was extracted using Qiagen RNeasy according to manufacturer’s manual. 2–10 ng of total RNA were used for biotin labeling according GeneChip Pico Kit (Affymetrix). 5.5 μg of biotinylated cDNA were fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. Samples were hybridized to an identical lot of Affymetrix Clariom™ S (400 Format) for 17 h at 45°C. Hybridization was done for 16hat conditions recommended by the manufacturer. Clariom™ S chips were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned using the Affymetrix GCS 3000. Image analysis was done by Affymetrix® GeneChip® Command Console® Software (AGCC) and Affymetrix® Expression Console™ Software.
Raw data obtained after image analysis were further analyzed by R/BioConducter packages “oligo” and “Biobase”. Raw signal intensities of each probeset (gene feature) were summarized by median polish method. Summarized probeset data was log2 transformed followed by RMA normalization procedure. Finally, the obtained dataset was annotated by NetAffx (Affymetrix). Normalized datasets were filtered for informative genes (showing at least expression values > log2(10) in more than two samples). Datasets were tested across all groups (ANOVA) or pairwise using linear models to assess differential expression in context of the multifactorial designed experiment. For statistical analysis and assessing differential expression the R/BioConductor package “limma” was used that utilizes an empirical Bayes method to moderate the standard errors of the estimated log-fold changes. Functional analysis was performed by R package “clusterProfiler”.
Ex vivo TSP-1 stimulation of aMФ
WT and CD47−/− mice were infected with 75 PFU as described. 3 dpi aMФ were isolated by repeated BAL as described. 2×106 BAL cells were then incubated in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics for 90minat 37°C, 5% CO2 in cell culture plates.54 Non-adherent cells were then removed by extensively rinsing the surface with PBS. To verify effective purification, adherent cells were detached by treatment with 0.05% Trypsin/EDTA (Sigma Aldrich), stained with antibodies against F4/80 and CD11c and analyzed by flow cytometry. Cells were incubated with 1 μg/mL and 5 μg/mL murine TSP-1 (R&D) in 300 μL Medium on a 24 well plate for 24 h. Afterward, supernatants were collected and analyzed for HB concentration by ELISA.
Uptake of hemoglobin by cells
Murine hemoglobin protein was fluorescently labeled using the Alexa Fluor 647 (AF647) Protein Labeling Kit (Invitrogen) according to manufacturer’s instructions. To examine the uptake by AEC II the cells were isolated as described. 1 × 105 AEC II were then incubated with 1 μg/mL of AF647-labelled hemoglobin for 30minat 37°C. Afterward, cells were washed. CD45− CD326+ AF647+ cells were identified via flow cytometry as having taken up hemoglobin. For the investigation of the uptake of HB by pulmonary immune cells single cell suspensions were prepared as described. Then 2.5×105 pulmonary immune cells were incubated with 1 μg/mL of AF647-labelled HB for 30minat 37°C. Indicated AF647+ cells were identified as having taken up HB.
Suppression assay to determine the antiviral activity of hemoglobin
The suppressive activity of BALF samples and hemoglobin was determined based on plaque assay as described. Apart from this, MDCK cells were pretreated for 30minat room temperature with the BALF from WT and CD47−/− mice of indicated time points. BALF was diluted 1:3 in Opti-MEM Medium (Invitrogen). Subsequently BALF was removed and MDCK cells were infected with 1000 PFU of A/PR8/34 influenza virus for 1 h. 72 h later plaques were counted. The suppressive activity of the BALF samples was calculated relative to a medium-treated control.
To determine the antiviral activity of HB, MDCK cells were pretreated for 30minat room temperature with indicated concentrations of murine whole HB protein (Fitzgerald Industries). Afterward, HB was removed and MDCK cells were infected as described. 72 h later plaques were counted. For post infection treatment of MDCK cells 1 μg/mL HB was added to the cell culture 2 h after infection.
Generation of infectious SARS-CoV-2 stocks
The SARS-CoV-2 strain used in this study was isolated from a nasopharyngeal swab of a patient suffering from COVID-19 disease using a Virocult vial (Sigma, Germany). For propagation, the Virocult medium was then incubated on 2×106 VeroE6 cells that were seeded in a T75 flask and maintained in DMEM supplemented with 10% fetal bovine serum, L-glutamine, penicillin (100 IU/mL) and streptomycin (100 μg/mL). After 3 days of incubation, the supernatant was harvested and cell debris was removed by centrifugation. Aliquots of the supernatant were prepared and stored at −80°C. Viral titers were determined by endpoint dilution assay and the 50% tissue culture infective dose (TCID50) was calculated as previously reported.55 Virus stock was sequenced and assigned to B.1.1.10 according to Pangolin database56 accession number EPI_ISL_602518.
Stimulation of A549-AT cells with HB
A549 cells, which constitutively express ACE2 and TMPRSS2 (A549-AT) were kindly provided by Marek Widera (University Hospital Essen).57 A549-AT cells were seeded in 24 well plates. Next day, medium was removed and cells were either stimulated 30 min with HB prior to infection with 350 PFU/mL SARS-CoV-2 for 1 h (HB pre) or left untreated. Fort post treatment with HB, virus-containing medium was removed 1 h post infection and cells were stimulated with HB or left untreated. After 24 h of culture in 37°Cat 5% CO2 atmosphere cells were analyzed by in-cell ELISA.
In-cell ELISA
The in-cell (ic) ELISA was performed based on the previously published protocol.58 Briefly, infected and HB-treated VeroE6 cells were fixed with 4% ROTI Histofix (Roth) for 2hat room temperature to fix the cells and inactivate the virus 24 h post infection.
Afterward, the plate was washed thrice with PBS. The PBS was aspirated and 200 μL of freshly prepared permeabilization buffer (PBS, 1% Triton X-100 (Roth)) were added to the cells and the plate was incubated for 30minat room temperature under constant shaking. Subsequently, the permeabilization buffer was aspirated and 200 μL of blocking buffer (PBS, 3% FBS) were added for 1 h. Then, the blocking buffer was aspirated and 50 μL of primary antibody solution (anti-SARS-CoV-2-NP (RRID: AB_2890255) 1:5,000 diluted in PBS +1% FBS) was added to each well. The plate was incubated overnight at 4°C. The next day, the primary antibody solution was aspirated and the plate was washed thrice with wash buffer (PBS, 0.05% Tween 20 (Roth)). Thereafter, 50 μL of the secondary antibody solution (Peroxidase-AffiniPure Goat Anti-Mouse IgG (H + L) (RRID: AB_10015289) 1:2,000 in PBS, 1% FBS) was added to the wells and the plate was incubated for 2hat room temperature. After the incubation period, the wells were washed 4 times with 250 μL wash buffer. Afterward 100 μL of TMB substrate solution (Biolegend) were added and the plate was incubated about 20minat room temperature in the dark. The reaction was stopped by addition of 100 μL 2N H2SO4 (Roth). The absorbance was measured at 450 nm with a reference wavelength of 620 nm using Spark 10M multimode microplate reader (Tecan).
Quantification and statistical analysis
Quantitative data are expressed as the mean ± SEM. Statistical analyses were performed using Prism 7.0 software (GraphPad). One-way or two-way ANOVA with multiple-comparisons post-test, Student’s t test or Mann-Whitney test were performed as indicated in the figure legend. A p value of <0.05 was considered statistically significant. The details of the statistical analysis used in each experiment are found in the figure legends.
DAB quantification of slides stained for MAC-3 and counterstained with hematoxylin was performed by semi-quantitative analyses. Ten random, non-overlapping fields (magnification, x200) of lung tissue from each specimen were photographed and pictures were automated single cell counted for DAB using the "Fiji" version of ImageJ from http://fiji.sc.46 The following adjustments for automatic counting were used: threshold 140, particle size 200–8000, Circularity 0.14–1.00. From all fields, the mean counts were averaged to yield the final score for each specimen.
Acknowledgment
We thank Christina Liebig, Christian Fehring, Mechthild Hemmler-Rohloff, Witold Bartosik and Barbara Bleekmann for excellent technical assistance. We thank Marek Widera (University Hospital Frankfurt) and Sandra Ciesek (University Hospital Frankfurt) for providing us A549-AT cells. Funding: This work was supported by the Deutsche Forschungsgemeinschaft (RTG 1949 & BR2221/4-1).
Author contributions
T.K., A.M.W., and J.B. conceived of the study. T.K. and C.W. designed the experiments and wrote the paper. C.W., T.K., P.S., F.W., K.S., J.D.B., R.K., D.B. V.J., and R.G. substantially contributed to the acquisition and analysis of the data. C.W. and T.K. wrote the original manuscript and all authors edited and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105540.
Supplemental information
References
- 1.Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D.M.E., Fouchier R.A.M. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 3.Burnet M., Fraser K.B., Lind P.E. Genetic interaction between influenza viruses. Nature. 1953;171:163–165. doi: 10.1038/171163a0. [DOI] [PubMed] [Google Scholar]
- 4.Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., García-Sastre A. Influenza. Nat. Rev. Dis. Primers. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 6.Tate M.D., Pickett D.L., van Rooijen N., Brooks A.G., Reading P.C. Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J. Virol. 2010;84:7569–7580. doi: 10.1128/JVI.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumpey T.M., García-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Pantin-Jackwood M.J., Schultz-Cherry S., Solórzano A., Van Rooijen N., Katz J.M., Basler C.F. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandes M., Klauschen F., Kuchen S., Germain R.N. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killip M.J., Fodor E., Randall R.E. Influenza virus activation of the interferon system. Virus Res. 2015;209:11–22. doi: 10.1016/j.virusres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo J.K., Baker D.P., Fish E.N. Interferon-beta modulates type 1 immunity during influenza virus infection. Antiviral Res. 2010;88:64–71. doi: 10.1016/j.antiviral.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Willingham S.B., Volkmer J.-P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., Wang J., Contreras-Trujillo H., Martin R., Cohen J.D., et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S.S., Liu Z., Uzunel M., Sundqvist K.G. Endogenous thrombospondin-1 is a cell-surface ligand for regulation of integrin-dependent T-lymphocyte adhesion. Blood. 2006;108:3112–3120. doi: 10.1182/blood-2006-04-016832. [DOI] [PubMed] [Google Scholar]
- 13.Lamy L., Foussat A., Brown E.J., Bornstein P., Ticchioni M., Bernard A. Interactions between CD47 and thrombospondin reduce inflammation. J. Immunol. 2007;178:5930–5939. doi: 10.4049/jimmunol.178.9.5930. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg F.P., Bullard D.C., Caver T.E., Gresham H.D., Beaudet A.L., Brown E.J. Decreased resistance to bacterial infection and granulocyte defects in IAP-deficient mice. Science. 1996;274:795–798. doi: 10.1126/science.274.5288.795. [DOI] [PubMed] [Google Scholar]
- 15.Gomes I., Dale C.S., Casten K., Geigner M.A., Gozzo F.C., Ferro E.S., Heimann A.S., Devi L.A. Hemoglobin-derived peptides as novel type of bioactive signaling molecules. AAPS J. 2010;12:658–669. doi: 10.1208/s12248-010-9217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coates C.J., Decker H. Immunological properties of oxygen-transport proteins: hemoglobin, hemocyanin and hemerythrin. Cell. Mol. Life Sci. 2017;74:293–317. doi: 10.1007/s00018-016-2326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q., Bai S.Y., Li L.F., Li S., Zhang Y., Munir M., Qiu H.J. Human hemoglobin subunit beta functions as a pleiotropic regulator of RIG-I/MDA5-Mediated antiviral innate immune responses. J. Virol. 2019;93 doi: 10.1128/JVI.00718-19. e00718-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tal M.C., Torrez Dulgeroff L.B., Myers L., Cham L.B., Mayer-Barber K.D., Bohrer A.C., Castro E., Yiu Y.Y., Lopez Angel C., Pham E., et al. Upregulation of CD47 is a host checkpoint response to pathogen recognition. mBio. 2020;11 doi: 10.1128/mBio.01293-20. e012933-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cham L.B., Torrez Dulgeroff L.B., Tal M.C., Adomati T., Li F., Bhat H., Huang A., Lang P.A., Moreno M.E., Rivera J.M., et al. Immunotherapeutic blockade of CD47 inhibitory signaling enhances innate and adaptive immune responses to viral infection. Cell Rep. 2020;31:107494. doi: 10.1016/j.celrep.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betancur P.A., Abraham B.J., Yiu Y.Y., Willingham S.B., Khameneh F., Zarnegar M., Kuo A.H., McKenna K., Kojima Y., Leeper N.J., et al. A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat. Commun. 2017;8:14802. doi: 10.1038/ncomms14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matlung H.L., Szilagyi K., Barclay N.A., van den Berg T.K. The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 2017;276:145–164. doi: 10.1111/imr.12527. [DOI] [PubMed] [Google Scholar]
- 22.Navarathna D.H.M.L.P., Stein E.V., Lessey-Morillon E.C., Nayak D., Martin-Manso G., Roberts D.D. CD47 promotes protective innate and adaptive immunity in a mouse model of disseminated candidiasis. PLoS One. 2015;10:e0128220. doi: 10.1371/journal.pone.0128220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nath P.R., Gangaplara A., Pal-Nath D., Mandal A., Maric D., Sipes J.M., Cam M., Shevach E.M., Roberts D.D. CD47 expression in natural killer cells regulates homeostasis and modulates immune response to lymphocytic choriomeningitis virus. Front. Immunol. 2018;9:2985. doi: 10.3389/fimmu.2018.02985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee R., Khandelwal S., Kozakai Y., Sahu B., Kumar S. CD47 regulates the phagocytic clearance and replication of the Plasmodium yoelii malaria parasite. Proc. Natl. Acad. Sci. USA. 2015;112:3062–3067. doi: 10.1073/pnas.1418144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouguermouh S., Van V.Q., Martel J., Gautier P., Rubio M., Sarfati M. CD47 expression on T cell is a self-control negative regulator of type 1 immune response. J. Immunol. 2008;180:8073–8082. doi: 10.4049/jimmunol.180.12.8073. [DOI] [PubMed] [Google Scholar]
- 26.Engelbertsen D., Autio A., Verwilligen R.A.F., Depuydt M.A.C., Newton G., Rattik S., Levinsohn E., Saggu G., Jarolim P., Wang H., et al. Increased lymphocyte activation and atherosclerosis in CD47-deficient mice. Sci. Rep. 2019;9:10608. doi: 10.1038/s41598-019-46942-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Madariaga M.L., Wang S., Van Rooijen N., Oldenborg P.-A., Yang Y.-G. Lack of CD47 on nonhematopoietic cells induces split macrophage tolerance to CD47null cells. Proc. Natl. Acad. Sci. USA. 2007;104:13744–13749. doi: 10.1073/pnas.0702881104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang I.K., Pillai P.S., Iwasaki A. Efficient influenza A virus replication in the respiratory tract requires signals from TLR7 and RIG-I. Proc. Natl. Acad. Sci. USA. 2013;110:13910–13915. doi: 10.1073/pnas.1303275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhold M.I., Lindberg F.P., Kersh G.J., Allen P.M., Brown E.J. Costimulation of T Cell activation by integrin-associated protein (CD47) is an adhesion-dependent, CD28-independent signaling pathway. J. Exp. Med. 1997;185:1–11. doi: 10.1084/jem.185.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latour S., Tanaka H., Demeure C., Mateo V., Rubio M., Brown E.J., Maliszewski C., Lindberg F.P., Oldenborg A., Ullrich A., et al. Bidirectional negative regulation of human T and dendritic cells by CD47 and its cognate receptor signal-regulator protein-α: down-regulation of IL-12 responsiveness and inhibition of dendritic cell activation. J. Immunol. 2001;167:2547–2554. doi: 10.4049/jimmunol.167.5.2547. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y.-T., Ko E.-J., Lee Y., Lee Y.-N., Bian Z., Liu Y., Kang S.-M. CD47 plays a role as a negative regulator in inducing protective immune responses to vaccination against influenza virus. J. Virol. 2016;90:6746–6758. doi: 10.1128/jvi.00605-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno S.G. Depleting macrophages in vivo with clodronate-liposomes. Methods Mol. Biol. 2018;1784:259–262. doi: 10.1007/978-1-4939-7837-3_23. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto Y., Moki T., Takizawa T., Shiratsuchi A., Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J. Immunol. 2007;178:2448–2457. doi: 10.4049/jimmunol.178.4.2448. [DOI] [PubMed] [Google Scholar]
- 35.Huber V.C., Lynch J.M., Bucher D.J., Le J., Metzger D.W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001;166:7381–7388. doi: 10.4049/jimmunol.166.12.7381. [DOI] [PubMed] [Google Scholar]
- 36.Okazawa H., Motegi S.i., Ohyama N., Ohnishi H., Tomizawa T., Kaneko Y., Oldenborg P.A., Ishikawa O., Matozaki T. Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. J. Immunol. 2005;174:2004–2011. doi: 10.4049/jimmunol.174.4.2004. [DOI] [PubMed] [Google Scholar]
- 37.Janssen W.J., McPhillips K.A., Dickinson M.G., Linderman D.J., Morimoto K., Xiao Y.Q., Oldham K.M., Vandivier R.W., Henson P.M., Gardai S.J. Surfactant proteins A and D suppress alveolar macrophage phagocytosis via interaction with SIRPα. Am. J. Respir. Crit. Care Med. 2008;178:158–167. doi: 10.1164/rccm.200711-1661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida M., Whitsett J.A. Alveolar macrophages and emphysema in surfactant protein-D-deficient mice. Respirology. 2006;11:S37–S40. doi: 10.1111/j.1440-1843.2006.00806.x. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Dee Z., Pidcock K., Gutierrez L.S. Thrombospondin-1: multiple paths to inflammation. Mediators Inflamm. 2011;2011:296069. doi: 10.1155/2011/296069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein E.V., Miller T.W., Ivins-O’Keefe K., Kaur S., Roberts D.D. Secreted thrombospondin-1 regulates macrophage interleukin-1β production and activation through CD47. Sci. Rep. 2016;6:19684. doi: 10.1038/srep19684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stegemann-Koniszewski S., Jeron A., Gereke M., Geffers R., Kröger A., Gunzer M., Bruder D. Alveolar type II epithelial cells contribute to the anti-influenza A virus response in the lung by integrating pathogen- and microenvironment-derived signals. mBio. 2016;7 doi: 10.1128/mBio.00276-16. e002766-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L., Zeng M., Stamler J.S. Hemoglobin induction in mouse macrophages. Proc. Natl. Acad. Sci. USA. 1999;96:6643–6647. doi: 10.1073/pnas.96.12.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodson D., Hirsch J.G. The antibacterial activity of hemoglobin. J. Exp. Med. 1958;107:167–183. doi: 10.1084/jem.107.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mak P., Wójcik K., Silberring J., Dubin A. Antimicrobial peptides derived from heme-containing proteins: hemocidins. Antonie Leeuwenhoek. 2000;77:197–207. doi: 10.1023/a:1002081605784. [DOI] [PubMed] [Google Scholar]
- 45.Koerner I., Kochs G., Kalinke U., Weiss S., Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J. Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gereke M., Autengruber A., Gröbe L., Jeron A., Bruder D., Stegemann-Koniszewski S. Flow cytometric isolation of primary murine type II alveolar epithelial cells for functional and molecular studies. J. Vis. Exp. 2012;10:4322. doi: 10.3791/4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fouchier R.A., Bestebroer T.M., Herfst S., Van Der Kemp L., Rimmelzwaan G.F., Osterhaus A.D. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 2000;38:4096–4101. doi: 10.1128/JCM.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misharin A.V., Morales-Nebreda L., Mutlu G.M., Budinger G.R.S., Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Leve S., Wirsdörfer F., Cappuccini F., Schütze A., Meyer A.V., Röck K., Thompson L.F., Fischer J.W., Stuschke M., Jendrossek V. Loss of CD73 prevents accumulation of alternatively activated macrophages and the formation of prefibrotic macrophage clusters in irradiated lungs. FASEB J. 2017;31:2869–2880. doi: 10.1096/fj.201601228R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knuschke T., Sokolova V., Rotan O., Wadwa M., Tenbusch M., Hansen W., Staeheli P., Epple M., Buer J., Westendorf A.M. Immunization with biodegradable nanoparticles efficiently induces cellular immunity and protects against influenza virus infection. J. Immunol. 2013;190:6221–6229. doi: 10.4049/jimmunol.1202654. [DOI] [PubMed] [Google Scholar]
- 52.Thepen T., Van Rooijen N., Kraal G. Alveolar macrophage elimination in vivo is associated with an increase in pulmonary immune response in mice. J. Exp. Med. 1989;170:499–509. doi: 10.1084/jem.170.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Désirée Boehme J., Pietkiewicz S., Lavrik I., Jeron A., Bruder D. Morphological and functional alterations of alveolar macrophages in a murine model of chronic inflammatory lung disease. Lung. 2015;193:947–953. doi: 10.1007/s00408-015-9797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boehme J.D., Pietkiewicz S., Lavrik I., Jeron A., Bruder D. Erratum to: morphological and functional alterations of alveolar macrophages in a murine model of chronic inflammatory lung disease. Lung. 2015;193:955. doi: 10.1007/s00408-015-9812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuhenn J., Meister T.L., Todt D., Bracht T., Schork K., Billaud J.N., Elsner C., Heinen N., Karakoese Z., Haid S., et al. Differential interferon-alpha subtype induced immune signatures are associated with suppression of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2111600119. e2111600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Widera M., Wilhelm A., Toptan T., Raffel J.M., Kowarz E., Roesmann F., Grözinger F., Siemund A.L., Luciano V., Külp M., et al. Generation of a sleeping beauty transposon-based cellular system for rapid and sensitive screening for compounds and cellular factors limiting SARS-CoV-2 replication. Front. Microbiol. 2021;12:701198. doi: 10.3389/fmicb.2021.701198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schöler L., Le-Trilling V.T.K., Eilbrecht M., Mennerich D., Anastasiou O.E., Krawczyk A., Herrmann A., Dittmer U., Trilling M. A novel in-cell ELISA assay allows rapid and automated quantification of SARS-CoV-2 to analyze neutralizing antibodies and antiviral compounds. Front. Immunol. 2020;11:573526. doi: 10.3389/fimmu.2020.573526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Microarray data were deposited in NCBI’s Gene Expression Omnibus and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.