The Huntingtin protein, well known for its involvement in the neurodegenerative Huntington’s disease, is also expressed in skeletal muscle. The work presented here is dedicated to a better understanding of its function in skeletal muscle through the development and characterization of KO models.
Abstract
The expression of the Huntingtin protein, well known for its involvement in the neurodegenerative Huntington’s disease, has been confirmed in skeletal muscle. The impact of HTT deficiency was studied in human skeletal muscle cell lines and in a mouse model with inducible and muscle-specific HTT deletion. Characterization of calcium fluxes in the knock-out cell lines demonstrated a reduction in excitation–contraction (EC) coupling, related to an alteration in the coupling between the dihydropyridine receptor and the ryanodine receptor, and an increase in the amount of calcium stored within the sarcoplasmic reticulum, linked to the hyperactivity of store-operated calcium entry (SOCE). Immunoprecipitation experiments demonstrated an association of HTT with junctophilin 1 (JPH1) and stromal interaction molecule 1 (STIM1), both providing clues on the functional effects of HTT deletion on calcium fluxes. Characterization of muscle strength and muscle anatomy of the muscle-specific HTT-KO mice demonstrated that HTT deletion induced moderate muscle weakness and mild muscle atrophy associated with histological abnormalities, similar to the phenotype observed in tubular aggregate myopathy. Altogether, this study points toward the hypotheses of the involvement of HTT in EC coupling via its interaction with JPH1, and on SOCE via its interaction with JPH1 and/or STIM1.
Introduction
Huntingtin (HTT) is a large protein that acts as a scaffold in several cellular pathways, including endocytosis, microtubule-based transport, and the regulation of calcium homeostasis (Tang et al., 2003; Saudou and Humbert, 2016; Raymond, 2017). HTT modification has been linked with Huntington’s disease (HD), a dominant inherited adult neurodegenerative disorder caused by an abnormal expansion of a polyglutamine repeat (polyQ) in the HTT protein (Macdonald, 1993). The presence of this expansion on one allele of the gene has two consequences: a reduction in the amount of the normal HTT protein and the expression of a toxic mutant HTT protein containing an expanded polyQ repeat, which is prone to aggregation and cleavage (Ross, 2002). Except for its juvenile form, the symptoms of HD only emerge during mid-adulthood. Patients typically suffer from a triad of symptoms with movement, psychiatric, and cognitive impairments (Bates et al., 2015). HTT is ubiquitously expressed, and both wild-type (WT) and mutant HTT are expressed in the developing embryo; the mouse knockout is lethal at embryonic day 7.5 (Duyao et al., 1995; Nasir et al., 1995; Zeitlin et al., 1995). Although alterations of the central nervous system (CNS) are the most prominent clinical features of HD, patients also suffer from peripheral organ dysfunctions such as metabolic and immune disturbances, weight loss, cardiac failure, and skeletal muscle wasting (Zielonka et al., 2014). Significant body weight loss is measurable in HD patients at early as well as late stages, with a major contribution of skeletal muscle atrophy despite an adequate diet and feeding (Djousse et al., 2002; Trejo et al., 2004; Aziz et al., 2008). Moreover, both patients and animal models for the disease present muscle pathology with metabolic and mitochondrial defects and a significant reduction in lower limb muscle strength (Ribchester et al., 2004; Kosinski et al., 2007; Busse et al., 2008; Chaturvedi et al., 2009; Rodinova et al., 2019). Interestingly, it has been shown that skeletal muscle from R6/2 mice, one of the best-characterized transgenic models for HD (Mangiarini et al., 1996), presents altered calcium signaling and is hyperexcitable (Waters et al., 2013; Braubach et al., 2014; Khedraki et al., 2017). In addition, a form of HD not linked to alterations in the HTT but in the junctophilin 3 gene (JPH3) has been described (Seixas et al., 2012). Huntington’s disease-like-2 (HDL2) is a phenocopy of HD caused by CTG/CAG repeat expansion at JPH3 locus, and mice lacking JPH3 show a phenotype similar to HDL2 pathology (Nishi et al., 2002). JPH3 is the neuronal isoform of junctophilin family proteins that are involved in the formation of contacts between the plasma membrane and the endoplasmic reticulum to facilitate crosstalk between the cell surface and intracellular ion channels in excitable cells. The skeletal muscle isoform JPH1 is responsible for the formation of the triad, a specific structure on which calcium release relies to allow muscle contraction. In neurons, HTT was shown to be associated with microtubules and to participate in intracellular traffic and calcium homeostasis; HTT being ubiquitous, a similar role in muscle cells could be hypothesized. The understanding of the functions of HTT, and more specifically of its deletion, is of paramount importance as the phenotype of HD patients is the superposition of two pathophysiological mechanisms: on the one hand, the expression of a toxic mutant protein, and on the other hand a reduction in the expression level of the WT HTT. Besides substantially studied gain-of-function effects of the mutant HTT, a number of studies provide evidence that a loss-of-function (LOF) mechanism related to the reduction in the amount of WT HTT has major physiological consequences and contributes to the pathophysiology of HD (Cattaneo et al., 2005; Saudou and Humbert, 2016). Whereas the functions of the HTT have been extensively explored in neurons, little is known about the roles that HTT could play in muscle normal physiology.
Skeletal muscle is a striated tissue composed of multinucleated fibers that contract under the control of motoneurons. The efficient contraction of the muscle is based in part on the efficacy of the coupling between the electrical signal propagated on the plasma membrane and the intracellular calcium release, a mechanism known as excitation–contraction (EC) coupling (Marty and Fauré, 2016), which relies on the specific structure of the triad and the physical interaction between the two main proteins of the calcium release complex (CRC): the ryanodine receptor (RyR1) in the sarcoplasmic reticulum (SR) membrane and the dihydropyridine receptor (DHPR) in the T-tubule membrane (Marty et al., 1994). The tethering of the SR membrane to the T-tubule membrane is performed in part by the JPH1, which plays a major role in triad formation and function (Lehnart and Wehrens, 2022) and has been shown to interact with and regulate DHPR (Golini et al., 2011; Nakada et al., 2018).
The CRC is responsible for the release of the calcium stored within the SR upon electrical stimulation, but calcium homeostasis is also maintained by the store operated calcium entry (SOCE), responsible for the refilling of the SR calcium store upon Ca2+ depletion, and performed by the association of the two proteins ORAI1 and STIM1 (Dirksen, 2009).
This study has been performed to explore the involvement of HTT in skeletal muscle calcium release and contraction. We developed a human HTT-knock out (KO) muscle cell model and a mouse HTT partial LOF model with an inducible KO of Htt gene exclusively in adult skeletal muscle fibers. We characterized the calcium fluxes in HTT-KO muscle cells using calcium imaging and demonstrated the involvement of HTT in the regulation of EC coupling and SOCE. We performed a full characterization of skeletal muscles of the HTT partial LOF mice using histological staining, electron microscopy, and force measurements, and the in vivo data reinforce the hypothesis of the involvement of HTT in SOCE.
Materials and methods
Cell culture and HTT-KO cell line production
Immortalized human satellite cells (myoblasts) produced from a 25-yr control individual have been previously described and characterized (Mamchaoui et al., 2011; Cacheux et al., 2015; so called CTRL-HM for control human myoblasts in this study). The myoblasts were amplified in a proliferation medium composed of Ham’s F-10 (Life Technologies) supplemented with 20% FBS (Life Technologies), 2% Ultroser G (Pall-Sartorius), and 2% penicillin–streptomycin (Life Technologies). Differentiation into myotubes was induced by a shift to differentiation medium: DMEM (Life Technologies) supplemented with 2% heat-inactivated horse serum (Life Technologies) and 1% penicillin–streptomycin. Lentiviral-induced deletion of the HTT gene in these cells was performed using CRISPR/Cas9 according to the strategy described by Merienne et al. (2017). Two lentiviruses were produced, one encoding guide RNAs against the HTT and Cas9 genes (using plasmid #87919; Addgene) and one encoding Cas9 (using plasmid #87904; Addgene), by triple transfection of HEK 293T cells. The immortalized myoblasts were transduced with the two lentiviruses, and single-cell clones were isolated and further characterized by DNA sequencing. One CTRL clone (no modification in HTT gene, clone 1-CTRL) and two homozygote HTT-KO clones were further used (clone 5-KO and clone 8-KO) in addition to the initial cell line (CTRL-HM).
Animals
Animal care and experimental procedures were approved by the institutional ethics committee (CEEAGIN 04, N_134) and they followed the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. Mice were housed collectively (littermates of various genotypes and the same sex) in cages with environmental enrichment in a temperature-controlled room with a 12-h light/dark cycle and ad libitum access to water and food.
The HSA-Cre-ERT2 mouse line, in which the expression of the tamoxifen-dependent Cre-ERT2 recombinase is under the control of the human skeletal muscle α-actin (HSA) gene, has been described previously (Schuler et al., 2005). In this transgenic line, Cre-ERT2 is selectively expressed in skeletal muscle fibers and activated upon tamoxifen injection, thus Cre-ERT2-mediated recombination of the floxed target gene is skeletal muscle specific and strictly tamoxifen dependent. The HTT-flox mouse line, a gift from the Zeitlin lab (University of Virginia School of Medicine, Charlottesville, VA), has been created by the insertion of LoxP sequences 1.3 kb upstream of the Htt transcription initiation site and within intron 1. Upon Cre-mediated recombination, the promoter, exon 1, and a portion of the first intron are deleted, resulting in a null allele (Zeitlin et al., 1995). The two mouse lines were mated to create the HTTflox/flox::HSA-Cre-ERT2+/− mouse line. At birth, the HTTflox/flox::HSA-Cre-ERT2+/− mice were normal and at 6–8 wk of age, once young adults, recombination was induced by intraperitoneal injection of 1 mg tamoxifen/day for 5 d (from this time point, the recombined animals are called HTT-Rec). Control (CTRL) animals are littermates HTTflox/flox::HSA-Cre-ERT2−/− (without the HSA-Cre-ERT2 transgene) injected with tamoxifen. Males and females have been used in this study. The primers for genotyping are presented in Table 1.
Table 1.
Primers for genotyping
| Primers for genotyping | ||
|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |
| Cre-ERT2 | ATTTGCCTGCATTACCGGTC | ATCAACGTTTTCTTTTCGGA |
| HTT-Flox | CTAAAGCGCATGCTCCAGACTG | AGATCTCTGAGTTATAGGTCAGC |
| HTT-WT | CATTTGATTCTTACAGGTAGCCTG | AGATCTCTGAGTTATAGGTCAGC |
| Primers for qPCR | ||
| HTT | TGAGAGTCAGTGTGAAGG | CTACTGAGGATGGAGTAGAC |
| β-Actin | GACAGGATGCAGAAGGAGATTACTG | CTCAGGAGGAGCAATGATCTTGAT |
Antibodies and reagents
Rabbit polyclonal antibody against RyR1 has been previously described (Marty et al., 1994), as are antibodies against SERCA (kindly provided by Dr. M.J. Moutin; Moutin et al., 1994). Antibody against the alpha subunit of DHPR was from Abcam (ref ab2862), antibodies against HTT were from Abcam (ref ab109115, used for Western blot) and Euromedex/IGBMC (4C8, used for immunoprecipitation), antibodies against STIM1 from Merck Millipore (ref AB9870) and Ozyme (clone D88E10; ref 5668S; Cell Signaling Technology), antibodies against JPH1 from Sigma-Aldrich (ref. WH0056704M4) and Invitrogen (ref 40-5100), antibody against Calnexin from Enzo (ref ADI-SPA-860), antibody against tubulin from Sigma-Aldrich (ref Tub2.1), the antibody against cMyc from Santa Cruz (sc40), and the antibody against V5 from Thermo Fisher Scientific (ref R960-25). Wheat germ agglutinin (WGA) Alexa Fluor 594 conjugate was from Thermo Fisher Scientific. Secondary antibodies used for Western blot were labeled with HRP (Jackson Immuno Research), and the antibodies used for immunofluorescent staining were labeled with Alexa Fluor 488, Cy-3, or Cy-5 (Jackson Immuno Research). Tamoxifen was from Sigma-Aldrich.
Histological analysis
Tibialis anterior (TA) muscles were collected immediately after euthanasia, flash-frozen in liquid nitrogen, and embedded in an optimal cutting temperature compound (Tissue Tek, Sakura). Transversal cryosections (10 μm) were processed for hematoxylin and eosin, modified Gomori trichrome, and nicotinamide adenine dinucleotide (NADH) staining. Images were acquired using Axio Scan.Z1 (Zeiss), and the cross-sectional area (CSA) measurement and defects analysis were performed using ImageJ software. The CSA was measured in glycolytic and oxidative fibers from three different animals for each genotype at 3 and 10 mo of age. Briefly, the cross-sections of fibers between 25 and 75 μm diameter were segmented from WGA-labeled images with the help of a homemade ImageJ macro (whose code can be found at https://github.com/jbrocardplatim/Muscle-Fibers) until the satisfaction of a user blinded to the genotype of the animal. The contours of the fibers were then used by the macro to measure the mean area per muscle and detect aggregates of FITC-labeled SERCA. When the maximum FITC signal exceeded five times the median signal, the fiber was considered defective and automatically counted as such.
Biochemical analysis
Muscle (quadriceps or TA) and brain (cortex or cerebellum) homogenates were prepared from frozen tissues. Briefly, the tissue was homogenized at 4°C in 200 mM sucrose, 20 mM HEPES (pH 7.4), 0.4 mM CaCl2, 200 mM phenylmethylsulfonyl fluoride, and 1 mM diisopropyl fluorophosphate using a Minilys homogenizer (Bertin). Protein concentration was measured using the Folin–Lowry assay method. The presence and the amount of different proteins in tissue homogenates were assayed by Western blot analysis. After electrophoretic separation on a 5–15% gradient or 6% acrylamide gel and electrotransfer to Immobilon P (Bio-Rad), the membrane was incubated with primary antibodies and then HRP-labeled secondary antibodies (Jackson ImmunoResearch Laboratories). Signal quantification was performed using a ChemiDoc Touch apparatus (Bio-Rad) and the Image Lab software (Bio-Rad). The amount of the chosen protein in each sample was corrected for differences in loading using either the amount of tubulin or calnexin, or the total amount of protein using the stain-free trichloroethanol (TCE) technology (Bio-Rad).
Immunoprecipitations of HTT or JPH1 were performed using Protein G Dynabeads (Thermo Fisher Scientific) and 4C8 antibody (antipeptide 443–457, IGBMC; Cong et al., 2005) or JPH1 antibody (WH0056704M4; Sigma-Aldrich), respectively, in parallel with their respective isotypic controls anti-cMyc IgG1κ (sc40; Santa Cruz) and anti-V5 IgG2a (R960-25; Thermo Fisher Scientific) antibodies, from 500 μg mouse quadriceps homogenates lysed with ice-cold lysis buffer (20 mM Tris pH 7.5, 50 mM NaCl, 2 mM EGTA, 0.5% Triton, and protease inhibitor cocktail [cOmplete, EDTA-free, Roche]). Following an overnight incubation on a rotating wheel at 4°C, beads were washed in the same lysis buffer three times. Beads were resuspended in 2x SDS-PAGE gel loading buffer and boiled (same procedure for the controls and HTT or JP1 immunoprecipitations, with the same amount of beads, antibodies, and loading buffer). Western blot analysis of immunoprecipitated proteins was further performed with anti-HTT (ab109115; Abcam), anti-JPH1 (40-5100; Invitrogen), or anti-STIM1 (AB9870; Merck Millipore) antibody. All the Western blots for HTT and JPH1 have been performed with the antibodies from these providers. For STIM1, only the WB presented in Fig. 3 was performed with anti-STIM1 from Cell Signaling and all the other experiments were performed with anti-STIM1 from Merck Millipore.
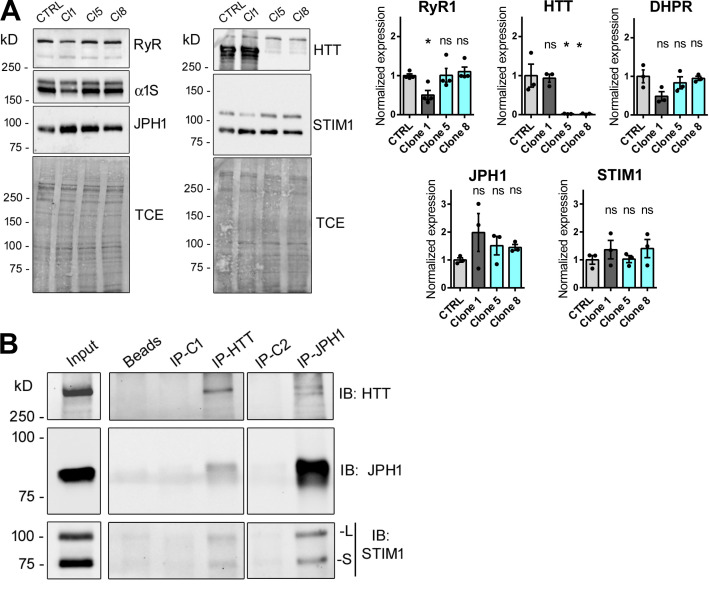
Figure 3.
Biochemical characterization of myotubes and muscle. (A) HTT-KO myotubes produced from CTRL-HM, Clone 1 (CTRL), Clone 5 (HTT-KO), and Clone 8 (HTT-KO) cell lines were analyzed by Western blot to quantify the amount of RyR1, HTT, α1 subunit of DHPR (the full length 210 kD and the truncated 170 kD are both observed), JPH1 (Invitrogen antibody), and STIM1 (STIM1L and STIM1S isoforms are both observed: cell signaling antibody), with the total amount of protein (TCE staining) as the loading control. The amount of each protein in Cl1, Cl5, and Cl8 was normalized to the amount in the CTRL-HM. Besides HTT, no modification has been observed in the amount of each protein between the control cells (CTRL-HM) and the HTT-KO groups, as illustrated in the quantification presented on the right panel, performed from three to four independent experiments. Statistical analysis: each clone versus CTRL was performed using one-way ANOVA with Dunnett’s correction for multiple comparison. Adjusted P value: RyR1 Clone 1 P = 0.0370, Clone 5 P = 0.9999, Clone 8 P = 0.8824; HTT Clone 1 P = 0.9808, Clone 5 P = 0.0057, Clone 8 P = 0.0056; DHPR Clone 1 P = 0.0587, Clone 5 P = 0.7092, Clone 8 P = 0.9844; JPH1 Clone 1 P = 0.2399, Clone 5 P = 0.6811, Clone 8 P = 0.7520; STIM1 Clone 1 P = 0.6329, Clone 5 P = 0.9999, Clone 8 P = 0.5676. (B) Immunoprecipitations were performed on WT mouse skeletal muscle homogenates (input) with antibodies against HTT (IP-HTT) or against JPH1 (IP-JPH1), or with their respective isotypic control antibody (IP-C1 as a control for IP-HTT and IP-C2 for IP-JPH1), or in absence of any antibody (beads). The Western blot of the immunoprecipitated proteins was performed with the antibodies mentioned on the right to detect HTT, JPH1, and STIM1 (both the long [L] and the short [S] isoform). The different panels presented were acquired in the same experiment with different exposition times to allow a good visualization of the different proteins. The images are representatives of three independent IP experiments. Source data are available for this figure: SourceData F3.
RT-qPCR analysis
Target gene transcript expression was measured in muscle homogenates by quantitative real-time polymerase chain reaction (RT-qPCR). Total RNA was extracted with TRIzol (Thermo Fisher Scientific) and RNA was reverse-transcribed using the iScript Reverse Transcription Supermix (Bio-Rad) following the manufacturer’s instructions. Gene expression was measured by RT-qPCR using the SsoAdvanced Universal Sybr green supermix (Bio-Rad) and the C1000 Touch Thermal Cycler–CFX96 Real-Time System (Bio-Rad). The list of specific primers (Eurofins) is provided in Table 1. Expression was normalized to the expression of three endogenous controls (β-actin, HPRT, and GAPDH) using the ∆∆Ct method as described previously (Pelletier et al., 2020).
Electron microscopy
TA and extensor digitorum longus (EDL) muscles from 10-mo-old mice were fixed with glutaraldehyde (2.5%, pH 7.4), postfixed with osmium tetroxide (2%), dehydrated, and embedded in resin. Longitudinally oriented or transversally oriented ultrathin sections were obtained and stained with uranyl acetate and lead citrate. Observations were made using a transmission electron microscope (JEOL JEM 1200 EX), equipped with a digital camera (Veleta, SIS), at 80 kV.
Immunofluorescence
Cryosections of TA muscles were incubated with blocking solution (PBS, 0.1% Triton, 2% goat serum, 0.5% BSA) for 1 h at room temperature and then briefly washed three times in PBS. Cryosections were incubated with primary antibodies (diluted in blocking solution) overnight at 4°C. After four washes in PBS, cryosections were incubated for 2 h at room temperature with precleaned secondary antibodies and WGA–Alexa Fluor 594 conjugate diluted in a blocking solution. Sections were then washed five times in PBS, mounted with Fluorsave Reagent (Calbiochem, Millipore), and examined with the slide scanner microscope Axio Scan.Z1 (Zeiss), while confocal images were acquired with the inverted confocal microscopy LSM710 (Zeiss). Using Axioscan acquisition and a homemade plug-in for ImageJ, fibers containing a strong signal of SERCA1 were quantified out of the total number of fibers visualized with the WGA–Alexa Fluor 594 conjugate membrane staining; n = 3–4 mice per genotype.
Hanging test
The muscle strength was assessed using the hanging grid test. Mice were randomized and the operator was blinded to genotype. Briefly, the mouse was placed on a grid. The grid was shaken slightly three times to cause the mouse to grip the wires and then the grid was turned upside down. The latency to fall was recorded for a maximum time of 180 s. The test was performed once a week together with the body weight measurement on 9 mice/group.
Noninvasive investigation of gastrocnemius muscle function
Gastrocnemius muscle anatomy and mechanical performance were investigated noninvasively and longitudinally using magnetic resonance (MR) measurements in seven controls and five HTT-Rec littermates at 3, 6, and 10 mo after birth as described previously (Pelletier et al., 2020). Anesthetized mice were placed into a home-built cradle allowing the MR investigation of the left gastrocnemius muscle function and bioenergetics inside the 7-Tesla horizontal magnet of a preclinical 70/16 PharmaScan MR scanner (Bruker). This cradle is similar to that described previously (Giannesini et al., 2010). 10 consecutive axial anatomic images (1-mm thick, 0.25-mm spaced, and 0.078 × 0.078 mm2 spatial resolution) covering the region from the knee to the ankle were acquired at rest. Muscle function was evaluated with a dedicated ergometer during a fatiguing bout of exercise electrically induced by square-wave pulses (1-ms duration) using transcutaneous surface electrodes and consisting of 6 min of maximal isometric contractions repeated at a frequency of 2 Hz. Intracellular concentrations of high-energy phosphorylated compounds and pH were continuously measured using dynamic 31P-MR spectroscopy during 6 min of rest, exercise, and 16 min of postexercise recovery period. Gastrocnemius contractile force was recorded and processed using the Powerlab 35 series system driven by the LabChart v8.1 software (AD Instruments). To calculate muscle volume, MR images were processed using custom software code written in Python. For each MR image, the region of interest was manually outlined so that the corresponding CSA of the gastrocnemius muscle was measured.
Calcium imaging
Human myotubes (CTRL-HM, clone 1-CTRL, clone 5-KO, and clone 8-KO) were cultured for 7 to 8 d before intracellular calcium measurements. Changes in intracellular calcium were measured on the cultured myotubes using the calcium-dependent fluorescent dye Fluo 4-Direct (Molecular Probes) diluted in a differentiation medium, as described previously (Oddoux et al, 2009). Calcium imaging was performed in Krebs buffer (136 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES, pH 7.4). KCl stimulation (110 mM final concentration) was performed by application of 3.6 vol of Krebs in which NaCl was replaced by KCl (140 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES, pH 7.4). 4-Chloro-meta-cresol (4-CmC) stimulation (500 µm final concentration) was performed by the addition of 0.25 vol 4-CmC at 2.5 mM. To obtain a calcium-free Krebs solution, CaCl2 was left out, while 1 mM EGTA was added. Thapsigargin (Molecular Probes) was applied in calcium-free Krebs solution at 10 µM final concentration as described previously (Oddoux et al, 2009). Fluorescence was measured by video microscopy using a Leica-DMI 6000B operating system at 1 frame/s. Data are given as mean ± SEM, n represents the number of myotubes in each condition, from at least three different experiments.
Statistical analysis
The statistical analysis has been done with GraphPad Prism 6.0 software. The normal distribution of samples has been checked and the number of samples with the name of the parametric or nonparametric test applied is indicated in each figure legend. Results are considered significant when P < 0.05, the exact value for P is indicated in the text or figure legends, and significant results are labeled * on the graphs whatever the exact P value is. All data are shown as mean ± SEM.
Online supplemental material
Fig. S1 presents data on the mechanical performance and bioenergetics in gastrocnemius muscle using dynamic 31P-MR spectroscopy.
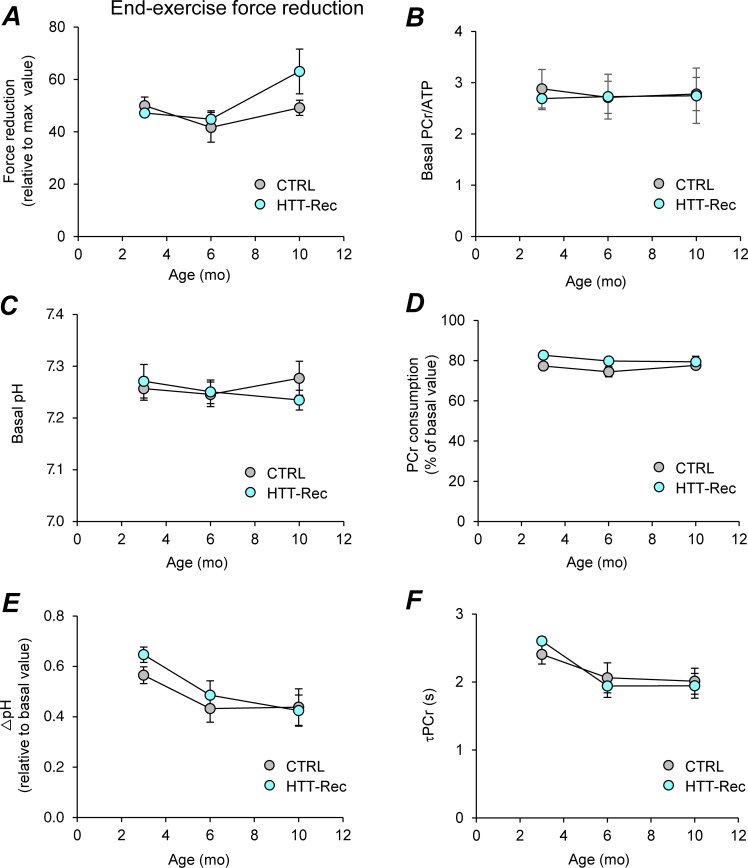
Figure S1.
Noninvasive investigation of mechanical performance and bioenergetics in gastrocnemius muscle using dynamic 31P-MR spectroscopy. (A) Force reduction at the end of the 6-min fatiguing bout of exercise and expressed as percent of the maximal force value reached during the exercise. (B and C) PCr/ATP (B) and pH (C) in resting muscle. (D and E) Extent of PCr consumption (D) and drop of intracellular pH (E) at the end of the 6-min exercise. (F) Time constant of PCr resynthesis (τPCr) during the post-exercise recovery period. Given that PCr synthesis during the post-exercise recovery period relies exclusively on oxidative ATP synthesis, τPCr is considered as an in vivo index of mitochondrial capacity. It is calculated by fitting the PCr resynthesis time-course to a monoexponential function with a least mean-squared algorithm: τPCr = −t/ln(PCrt/ΔPCr), where ΔPCr is the extent of PCr depletion measured at the start of the recovery period. Data are means ± SEM. Statistical analysis was performed using unpaired two-tailed Mann–Whitney tests. For each parameter, there is no difference between both groups whatever the age.
Results
Expression of HTT in skeletal muscle
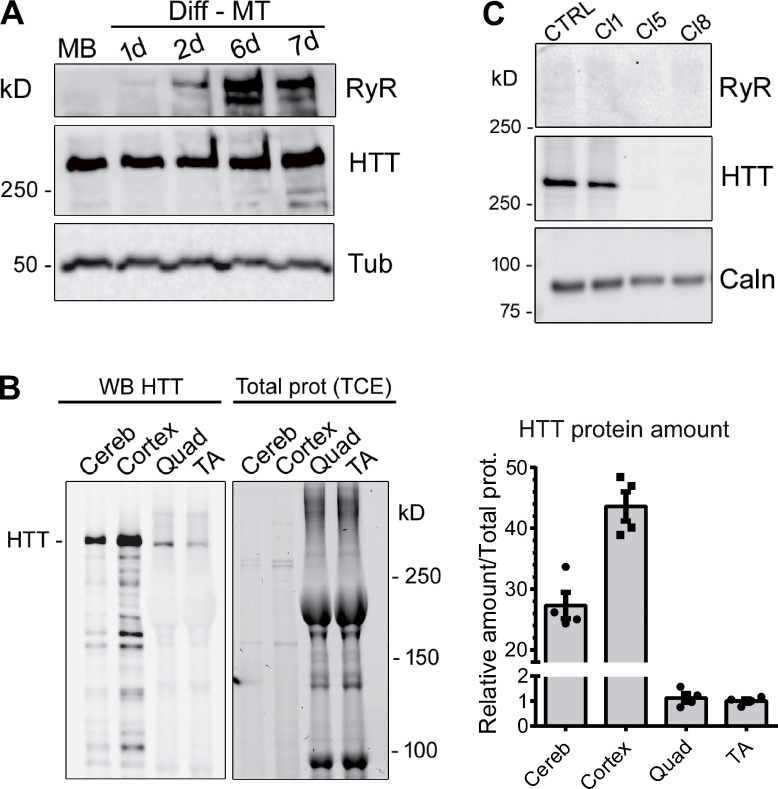
HTT expression is not restricted to the brain, but its abundance varies across tissues (Marques Sousa and Humbert, 2013). We measured the expression level of endogenous HTT protein during myoblasts differentiation and in skeletal muscle and compared it to its expression in CNS. Human immortalized myoblasts were induced in differentiation and collected at different time points. Whereas the calcium channel RyR1 is progressively expressed during differentiation (Fig. 1 A), HTT is expressed both in myoblasts and myotubes. Western blot analysis was also performed to compare the expression level of HTT in the cerebellum, cortex, quadriceps, and TA from adult WT mice. HTT signal is lower in skeletal muscles (quadriceps and TA) compared with CNS (Fig. 1 B), with the relative amount of HTT compared with the total amount of protein in skeletal muscle reaching about 3% of this amount in the cerebellum.
Figure 1.
Western blot analysis of HTT protein. (A) Western blot analysis of immortalized human control myoblasts (MB) and after 1, 2, 6, and 7 d in differentiation medium (Diff-MT). The HTT protein is stably expressed in myoblasts or myotubes. The RyR1 protein expression level increases during differentiation. Tubulin is used as the loading control. (B) 5 µg mouse brain (cerebellum or cortex) was analyzed by Western blot, together with 50 µg mouse skeletal muscle (quadriceps, Quad; tibialis anterior, TA). HTT protein amounts (right) were normalized to the total protein amounts determined with TCE. Data are presented as mean ± SEM of four mice. (C) Western blot analysis of HTT amount in human immortalized myoblasts CTRL and clone 1-CRTL (Cl1), and in HTT-KO clone 5 (Cl5) and 8 (Cl8). Calnexin is used as the loading control. Source data are available for this figure: SourceData F1.
Analysis of calcium fluxes in HTT-KO human muscle cell
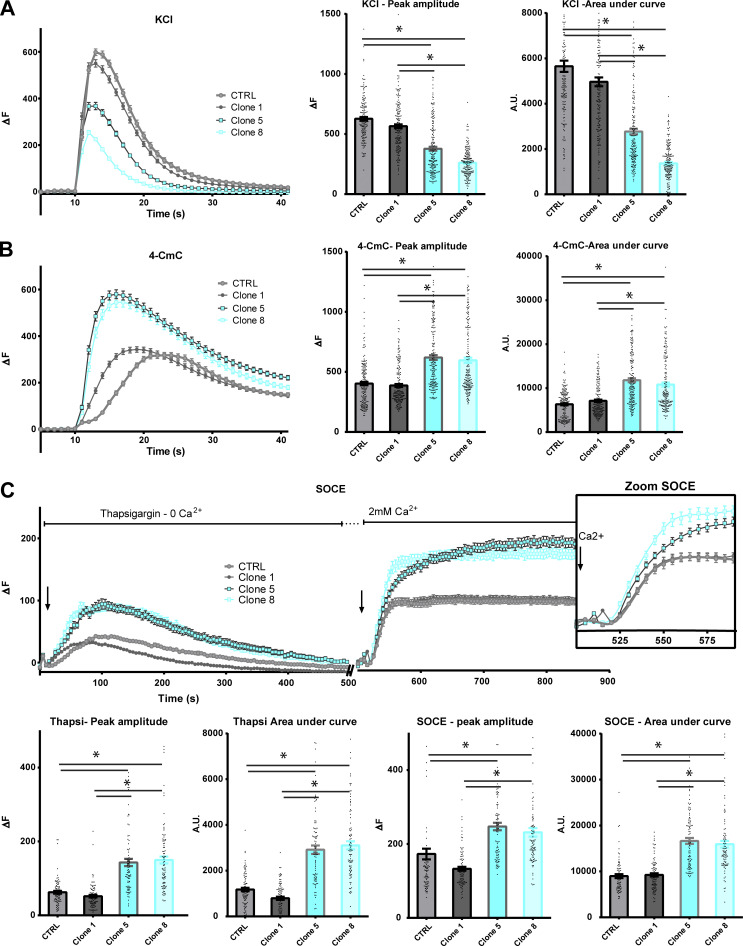
Human immortalized skeletal muscle cells (myoblasts) were used to produce HTT-KO muscle cells using lentiviral-mediated CRISPR/Cas9 expression according to the procedure previously described (Merienne et al., 2017; Beaufils et al., 2022). Three clones were further analyzed compared with the initial immortalized human myoblasts (CTRL-HM): one nonmodified clone (clone 1-CTRL) and two HTT-KO clones, clone 5-KO and clone 8-KO (Fig. 1 C). The modification in calcium fluxes using different stimuli was studied in myotubes from these cell lines, as well as the amount of the main proteins involved in these calcium fluxes. Stimulation of the whole CRC by membrane depolarization (exposure to 110 mM KCl) induced a large intracellular calcium release of similar maximal amplitude in CTRL-HM cells and clone 1-CTRL, whereas the calcium release was decreased in the two HTT-KO clones (1.5–2-fold reduction; Fig. 2 A), both in the maximal amplitude and in the amount of calcium released (area under the curve), pointing to either (1) an alteration in the EC coupling, (2) a reduction in the amount of calcium in the intracellular stores, or (3) a dysfunction in RyR1 or a reduced amount of RyR1. Direct stimulation of RyR1 was further induced by 4-CmC stimulation, and an approximately twofold increase in the amplitude of the peak of calcium was observed in the two HTT-KO clones (clones 5-KO and 8-KO; Fig. 2 B) compared with the controls (CTRL-HM and clone 1-CTRL), ruling out the hypothesis of a defective RyR1 and pointing to (1) a possible increase in the amount of calcium stored in the SR or (2) a possible increase in the amount of RyR1 protein. To discriminate between these different hypotheses, the amount of calcium stored in the SR was estimated by inhibition of calcium reuptake by the Ca2+-ATPase SERCA with thapsigargin. SERCA inhibition resulted in the leak of the calcium stored in the SR, which was about twofold larger in HTT-KO clones 5 and 8 compared with controls (Fig. 2 C). To determine if this increase in the SR calcium content was related to the hyperactivity of the SOCE mechanism, the application of thapsigargin in the absence of extracellular calcium was followed by incubation in the presence of 2 mM extracellular calcium. The resulting extracellular calcium influx via SOCE was also two times larger in HTT-KO clones as compared with controls (Fig. 2 C). In addition, the amount of RyR1, DHPR, JPH1, and STIM1, the central SR protein triggering SOCE, was estimated using quantitative Western blot (Fig. 3 A). No modification was observed in the amount of any of these proteins in the HTT-KO clones 5 and 8 compared with the control cells (CTRL-HM). Taken together, these results suggested that HTT deletion in skeletal muscle cells resulted in alterations in calcium fluxes with a reduction in the EC coupling, an increase in the amount of Ca2+ in the SR, and the hyperactivity of SOCE.
Figure 2.
Calcium imaging in CTRL or HTT-KO cultured human myotubes. All values are means ± SEM. In each condition, n = 90–180 myotubes have been analyzed, from at least three different experiments. Statistical significance was determined using ordinary one-way ANOVA with Tukey correction for multiple comparison. (A) KCl depolarization (110 mM) was induced in the presence of 2 mM external calcium. The left curves represent the fluorescence variation in control (gray curves) or HTT-KO myotubes (blue curves), produced from CTRL-HM, Clone 1 (CTRL), Clone 5 (HTT-KO), and Clone 8 (HTT-KO) cell lines. The histogram on the right presents the mean ± SEM of the peak of each curve (each dot represents one myotube) and the mean ± SEM area under the curve. For peak amplitude, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. For the area under the curve Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. (B) 4-CmC (500 µM) was used to directly stimulate RyR in the presence of 2 mM external calcium. The left curves represent the fluorescence variation in control (gray curves) or HTT-KO myotubes (blue curves), produced from CTRL-HM, Clone 1 (CTRL), Clone 5 (HTT-KO), and Clone 8 (HTT-KO) cell lines. The histogram on the right presents the mean ± SEM of the peak of each curve (each dot represents one myotube) and the mean ± SEM area under the curve. For peak amplitude, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. For the area under the curve, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001, Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. (C) The amplitude of the SOCE was determined using a two-step stimulation. The cells were first exposed to thapsigargin (10 µM) in absence of extracellular calcium to induce slow extrusion of the SR calcium and estimate the amplitude of the calcium store, and the cells were subsequently exposed to 2 mM extracellular calcium to induce an influx of extracellular calcium by the STIM1–ORAI1 complex. The inset on the right is a zoom on the curves between 500 and 600 s, to better visualize the kinetics of calcium influx by STIM1–ORAI1 upon the addition of external calcium. The amplitude of the peak of thapsigargin-induced calcium release or the extracellular calcium influx, as well as the area under each curve, are presented below each curve. For the thapsigargin peak amplitude, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. For the thapsigargin area under the curve, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. For the SOCE peak amplitude, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001. For the SOCE area under the curve, Clone 5 versus CTRL P < 0.0001; Clone 8 versus CTRL P < 0.0001; Clone 5 versus Clone 1 P < 0.0001; Clone 8 versus Clone 1 P < 0.0001.
To study the relationship between HTT and the SOCE complex (STIM1–ORAI1) on one hand and the CRC (RyR1–DHPR) on the other hand, immunoprecipitations were performed on WT mouse muscle homogenates. Immunoprecipitation of HTT led to the coimmunoprecipitation of JPH1, a structural protein of the triad, and STIM1 (both long and short isoforms; Fig. 3 B). Conversely, immunoprecipitation of JPH1 led to the coimmunoprecipitation of HTT and of STIM1 (both isoforms; Fig. 3 B). Therefore our results demonstrate an association of HTT with essential components of both CRC (JPH1) and SOCE (STIM1).
Molecular consequences of HTT deletion in muscle
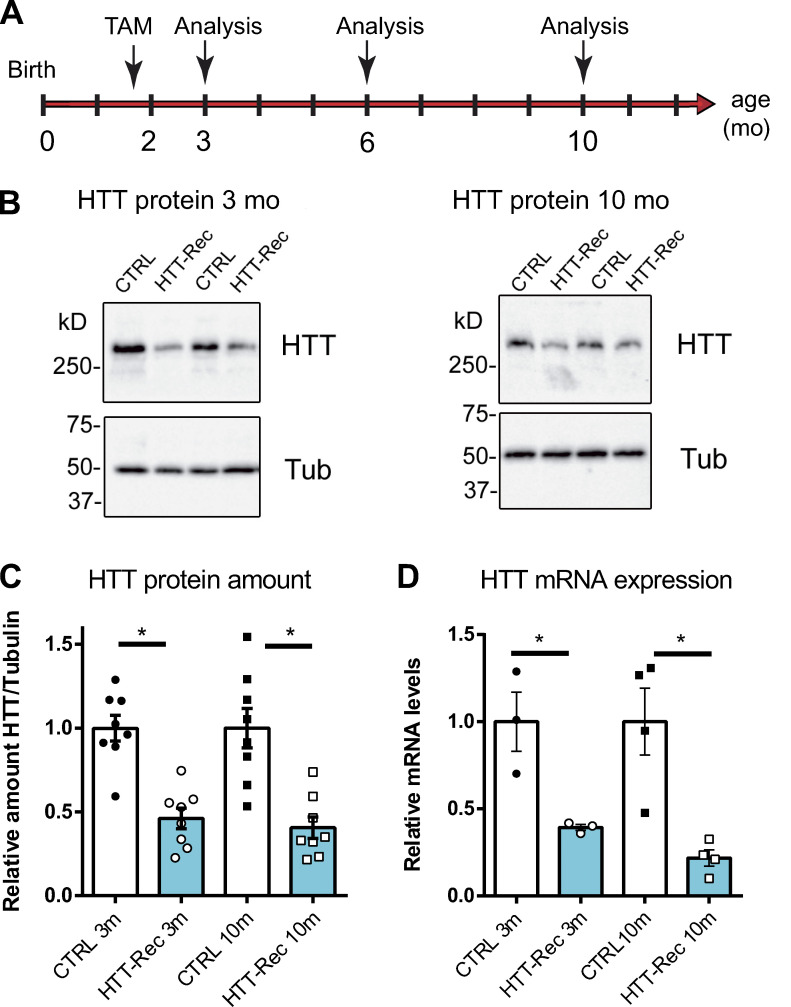
We further studied the consequences of HTT deletion in skeletal muscle using a mouse model. As the full KO of HTT is embryonically lethal, we developed an inducible and muscle-specific HTT-KO mouse line, the HTTflox/flox::HSA-Cre-ERT2 mouse line, in which HTT knockout is induced exclusively in adult skeletal muscle fibers upon tamoxifen injection. At 6–8 wk of age, mice were injected with tamoxifen to induce the recombination of the HTT gene (Fig. 4 A). Mice expressing the recombinase Cre were thereafter named HTT-Rec while their negative littermates for Cre expression were used as CTRL. The amount of HTT protein was further analyzed using quantitative Western blot in TA muscle homogenates (Fig. 4, B and C). At 3 mo of age, the amount of HTT protein in HTT-Rec muscle reached 46 ± 6% compared with CTRL (100 ± 8%). At 10 mo of age, a level of 40 ± 6% was quantified compared with CTRL (100 ± 12%). Together, the recombination induced by tamoxifen caused a drop in HTT protein expression of ∼50–60%. A similar reduction was also observed in other skeletal muscles, such as quadriceps and gastrocnemius (data not shown).
Figure 4.
The HTT-Rec mouse line. (A) Mice were injected with tamoxifen to induce the recombination at 6–8 wk of age and were analyzed at variable times thereafter (3, 6, and 10 mo of age). (B) Representative Western blots of HTT and tubulin in TA homogenates of two different CTRL and two different HTT-Rec mice at 3 and 10 mo. (C) Quantification of the relative amount of HTT protein compared with β-tubulin at 3 mo (P < 0.0001, HTT-Rec versus CTRL) and 10 mo (P = 0.0006, HTT-Rec versus CTRL) on eight mice in each group. (D) Real-time qPCR analysis of HTT transcript in quadriceps of CTRL and HTT-Rec mice at 3 mo (P = 0.0235, HTT-Rec versus CTRL) and 10 mo (P = 0.0074, HTT-Rec versus CTRL, n = 3–4 mice in each group), compared with β-actin, HPRT, and GAPDH as reference genes. The amount in CTRL littermate was set to 1. The quantification was performed using the ΔΔCt method. Graphs represent mean ± SEM. Statistical analysis: unpaired t test. Source data are available for this figure: SourceData F4.
The relative amount of HTT at the mRNA level was then analyzed in quadriceps muscle using RT-qPCR in CTRL and HTT-Rec mice (Fig. 4 D). A significant decrease in HTT mRNA level was observed in 3-mo-old HTT-Rec animals compared with CTRL (39 ± 2 versus 100 ± 17%) and in older animals (22 ± 5 versus 100 ± 19%). The amount of HTT mRNA was not completely abolished because of the ubiquitous HTT expression and the muscle fiber–specific HSA-induced recombination and the skeletal muscle containing not only muscle fibers but also satellite cells, adipocytes, fibroblasts, and vascular cells in which the recombination did not occur.
Physiological analysis of HTT-Rec mice
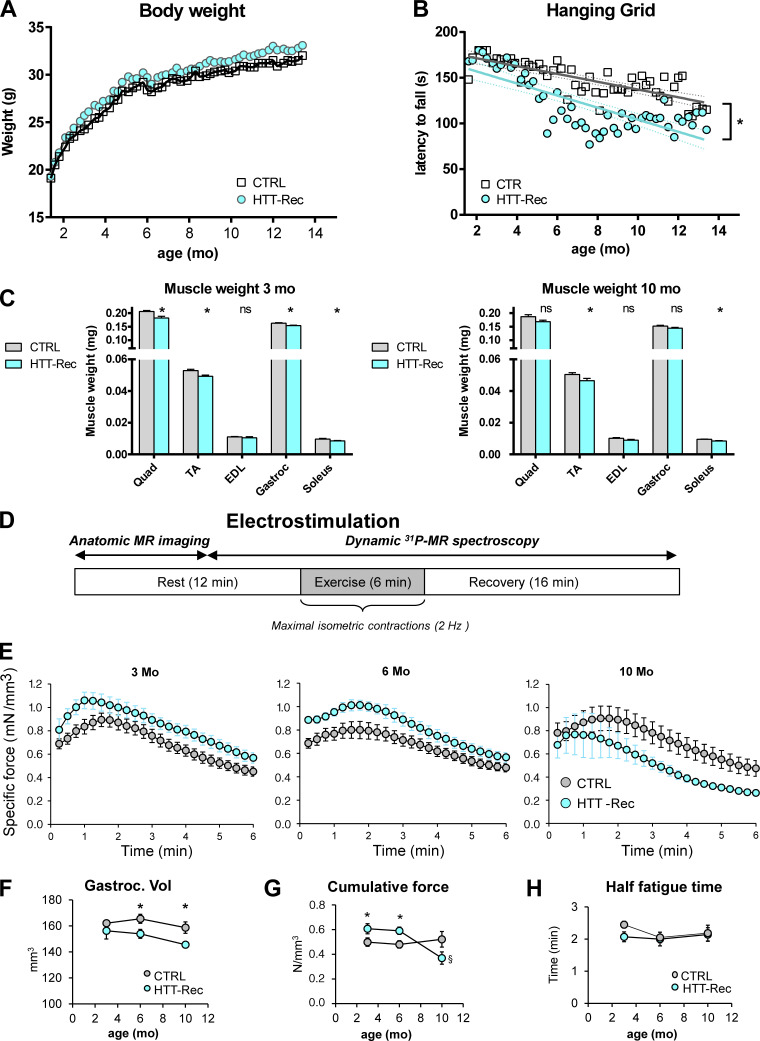
The body weight (BW) of the animals was recorded every week after recombination over a year (Fig. 5 A). There were no significant differences in the BW between the HTT-Rec animals and the CTRL. Muscle force was assessed by the hanging grid task (Fig. 5 B). While CTRL mice showed a slow and progressive deterioration of their performance over time as they got older, the HTT-Rec mice showed a more marked deterioration in their performance starting from week 15 after the tamoxifen injection. We wondered whether the decrease in muscle force observed in the HTT-Rec animals was associated with muscle fiber atrophy. Muscle weight was measured right after dissection at 3 and 10 mo of age (Fig. 5 C). A mild weight loss (6–13%) was observed in all muscles tested when comparing the HTT-Rec mice to the CTRL. To confirm this moderate loss of muscle force, the animals were submitted to a 6-min noninvasive electrostimulation protocol of the gastrocnemius muscle coupled to anatomic MR imaging and 31P-spectroscopy under general anesthesia (Fig. 5 D) at 3, 6, and 10 mo of age. A small reduction in the gastrocnemius volume was observed at 6 and 10 mo in HTT-Rec animals compared with CTRL (Fig. 5 F). Whereas the specific muscle strength is slightly larger at 3 and 6 mo in the HTT-Rec group than in the CTRL group, an inversion is observed at 10 mo, in the HTT-Rec animals presenting a smaller specific force than the CTRL (Fig. 5 E), and the cumulative force during the 6 min exercise being significantly reduced in the HTT-Rec group (Fig. 5 G). Force reduction at the end of the 6-min fatiguing bout of exercise did not significantly differ between CTRL and HTT-Rec animals (Fig. 5 H and Fig. S1 A), thereby indicating that HTT deletion did not affect muscle fatigability. Furthermore, there was no difference between both groups for bioenergetics parameters measured dynamically using 31P-MR spectroscopy throughout the resting and exercising periods (Fig. S1, B–E), which demonstrates that basal bioenergetics status and energy demand during exercise were not disturbed in mice lacking HTT. Interestingly, the fact that the time constant of PCr resynthesis (τPCr) was similar in both groups (Fig. S1 F) leads to the conclusion that HTT deletion does not affect skeletal muscle mitochondrial function.
Figure 5.
Evolution of muscle strength of the HTT-Rec mice. (A and B) Evolution with the age of body weight and hanging grid task performance in CTRL and HTT-Rec mice. Data are presented as the mean of n = 9 mice/group. Statistical analysis: linear regression, P = 0.01142. (C). Muscle weight normalized to body weight in 3-mo-old CTRL (n = 5) or HTT-Rec (n = 4) and 10-mo-old CTRL (n = 6) or HTT-Rec (n = 6) male mice. Statistical analysis: Unpaired t test, HTT-Rec versus CTRL, at 3 mo Quad P = 0.0026, TA P = 0.0044, EDL P = 0.3659, Gastroc P = 0.0076, Sol P = 0.0384; at 10 mo Quad P = 0.0553, TA P = 0.0465, EDL P = 0.1175, Gastroc P = 0.0627, Sol P = 0.0014. (D) Protocol for the investigation of gastrocnemius muscle function and bioenergetics, performed on CTRL mice (n = 7 at each age) and HTT-Rec mice (n = 5 at 3 and 6 mo and n = 4 at 10 mo). Data are mean ± SEM. (E) Time course of specific force measured in vivo throughout the 6-min fatiguing bout of exercise at 3, 6, and 10-mo of age. (F) Gastrocnemius muscle volume measured from MR images: statistical analysis unpaired two-tailed Mann–Whitney t tests, HTT versus CTRL P = 0.030 at 6 mo and P = 0.029 at 10 mo. (G) The cumulative specific force, defined as the total amount of specific force produced during the whole 6-min exercise, was determined for each group. Statistical analysis was performed using unpaired two-tailed Mann–Whitney t tests. *, significant difference between HTT-Rec and CTRL groups at the same age (P = 0.048 at 3 mo, P = 0.019 at 10 mo); §, significant difference within the HTT-Rec group between 10 and 3 mo (P = 0.003), and 10 and 6 mo (P = 0.002). (H) Half-fatigue time, defined as the time required for the force to decline by half from its maximal value, is not significantly different in each group and at each age.
Morphological analysis of HTT-Rec mice muscles
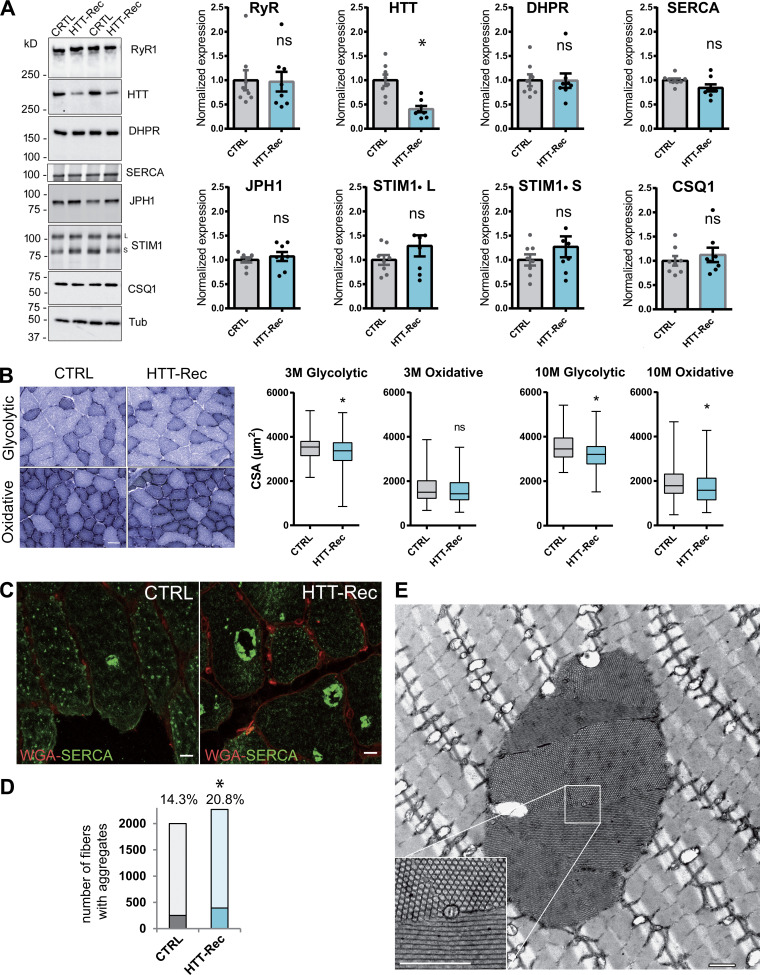
The muscles of the HTT-Rec mice were further analyzed using Western blot and histological staining. Western blot analysis of the different proteins involved in calcium homeostasis was performed, and no significant modification was observed in RyR1, DHPR, SERCA, JPH1, STIM1, and CSQ1 (Fig. 6 A). Analysis of the CSA of glycolytic and oxidative fibers showed that the mean CSA of glycolytic fibers decreased by 7 and 9% in 3- and 10-mo-old mice, respectively, whereas the mean CSA of oxidative fibers decreased by 4 and 13% in 3- and 10-mo-old mice, respectively (Fig. 6 B). Although mild, this reduction in fiber size is significant and in accordance with muscle weight loss and gastrocnemius volume reduction, with no major impact on BW. Muscle defects were further analyzed using different histology protocols. As STIM1 hyperactivity has been linked to tubular aggregate myopathy (Böhm et al., 2013), we looked for the presence of tubular aggregates. Tubular aggregates have been described in skeletal muscle as regular arrays of densely packed membrane tubules that contain SR proteins such as SERCA, RyR1, Calsequestrin, and STIM1 (Chevessier et al., 2004; Böhm et al., 2013). They occurred naturally in inbred old male mice but are enriched in tubular aggregate myopathy. The tubular aggregates in HTT-Rec mice were labeled with SERCA antibodies (Fig. 6, C and D), and their honeycomb structure was confirmed using electron microscopy (Fig. 6 E). Quantification of the number of fibers with tubular aggregates (Fig. 6 D) demonstrated a significant increase in the number of altered fibers from 14.5% in 10-mo-old CTRL to 21% in 10-mo-old HTT-Rec mice. No modification was observed in the overall triads’ structure, position, and number (data not shown).
Figure 6.
Morphological characterization of the muscles of HTT-Rec mice. (A) Western blot analysis of RyR1, HTT, DHPR-α1S, JPH1, STIM1, SERCA, and CSQ1 in TA muscle homogenates from CTRL and HTT-Rec 10 mo old mice. Tubulin is the loading control. Eight mice per group, each dot represents a mouse, the value for each mouse is the mean of three Western blots. Statistical analysis: unpaired t test, HTT-Rec versus CTRL, RyR1 P = 0.9208, HTT P = 0.0006, DHPR P = 0.9681, JPH1 P = 0.5164, STIM1L P = 0.2478, STIM1S P = 0.2856, SERCA P = 0.0547, CSQ1 P = 0.5036. (B) The CSA was measured in glycolytic and oxidative fibers from CTRL and HTT-Rec TA muscle stained with NADH at 3 mo of age. Representative images of TA sections in regions enriched in glycolytic fibers or oxidative fibers in CRTL or HTT-Rec at 3 mo, are presented. Scale bar, 50 µm. Quantification has been performed on n = 375 fibers from three CTRL mice and n = 495 fibers from three HTT-Rec mice. Student’s t test, P = 0.0038 for glycolytic fibers and P= 0.1667 for oxidative fibers. CSA was measured in glycolytic and oxidative fibers from CTRL and HTT-Rec TA muscle stained with NADH at 10 mo of age, on n = 399 fibers from three CTRL mice and n = 450 fibers from three HTT-Rec mice. Student’s t test, P < 0.0001 for glycolytic fibers and oxidative fibers. (C) Representative confocal images of tubular aggregates in TA of 10 mo old CTRL and HTT-Rec mice using WGA (red) and SERCA (green). Scale bar, 10 µm. (D) The number of fibers containing SERCA aggregates was determined out of the total number of fibers (WGA staining) on n = 3–4 mice per genotype. Of note: the labeling of the aggregates with the SERCA antibody is so strong that the labeling of SERCA in the whole SR is not visible. The χ2 test was used to determine whether there was a significant difference between the frequencies of fibers with aggregates: CTRL, 251 out of 1,749 (14.3%); HTT-Rec, 392 out of 1,878 (20.8%); χ2 = 26.414, P < 0.0001. (E) Representative EM picture of longitudinal TA section of HTT-Rec mouse. Scale bar, 1 µm. Source data are available for this figure: SourceData F6.
Discussion
HTT expression is not restricted to the brain as shown in the early studies using Northern blot and in situ RNA hybridization performed throughout the brain, heart, liver, lung, kidney, pancreas, and muscle (Strong et al., 1993; Li et al., 1993), further confirmed by Western blot analysis on mouse tissues (Wood et al., 1996). With the goal to study HTT function exclusively in skeletal muscle, we first confirmed its expression in skeletal muscles and cultured myoblasts and myotubes. Although not as abundant as in the CNS, the expression of HTT was confirmed in human-cultured myoblasts and myotubes, as well as in adult mouse skeletal muscle. The characterization of calcium fluxes in cultured HTT-KO myotubes demonstrated a reduction in the efficiency of EC coupling and an increase in the amount of calcium stored within the SR, pointing to the involvement of HTT in the RyR1–DHPR coupling and the activity of the STIM1–ORAI1 complex. Thus, these results lead to the hypothesis of the involvement of HTT in calcium flux regulation via its role in cellular traffic and/or in the structuration of SR–plasma membrane contact points in skeletal muscle (triads). Indeed, both the CRC (RyR1–DHPR complex) and the SOCE complex (STIM1–ORAI1) are anchored to two different membranes (SR and T-tubule or plasma membrane), and JPH1 is involved in the tethering of these two membranes to form the triads. As we have evidenced an HTT–JPH1 interaction, the HTT-KO induced alterations in EC coupling could therefore be mediated by dysregulation of JPH1 function, in line with JPH1 involvement in muscle EC coupling (Perni and Beam, 2022). In addition, STIM1 has been shown to move from longitudinal SR, where it is localized in its inactive form when the SR calcium store is full, to the triad, where it interacts with ORAI1 in its oligomerized activated form when the calcium store is empty (Smyth et al., 2010; Wei-Lapierre et al., 2013). The HTT-KO induced modifications in SOCE could thus also rely on dysregulation of the HTT–JPH1 functional link, in line with the previously described regulation of SOCE by junctophilins (Hirata et al., 2006), or the alteration of STIM1 movement along the microtubules, which could directly involve HTT and its function in microtubule-based transport. Nevertheless, STIM1–ORAI1 has also been described as a preassembled complex at the triad, possibly because of the presence of the long isoform of STIM1-L (Darbellay et al., 2011), which does not require any SR remodeling or movement along the microtubules (Saüc et al., 2015) for its fast activation (Koenig et al., 2018). The dual opposite effects of HTT deletion (reduction in EC coupling and increase in SOCE) can therefore be explained with different hypotheses. It could be attributed to a direct effect of HTT deletion on the SOCE complex, and involved, for example, in the dissociation of STIM1 from ORAI1, leading to the deactivation of SOCE. A confirmation of this hypothesis would however require a precise analysis of the dynamics of STIM1 in HTT-KO cells. An alternative explanation would rely on the interaction of HTT with JPH1. Although no reduction in global JPH1 amount is observed in HTT-KO cells, the function or targeting of JPH1 could be altered, which would in turn lead to an alteration in the DHPR–RyR1 coupling (Nakada et al., 2018). The uncoupling of RyR1 from DHPR would result in a greater sensitivity of RyR1 to 4-CmC stimulation and an increased calcium leak upon thapsigargin stimulation because uncoupled RyR1 is more prone to uncontrolled opening, but as observed for JPH1 KO cells, without alteration in the junctional membrane (Nakada et al., 2018).
The in vivo deletion of HTT being lethal, the consequences of its exclusive reduction in adult skeletal muscle were studied in a mouse model with an inducible and muscle-specific HTT deletion. Muscle strength measurements and analyses of muscle morphology demonstrated mild alterations in the structure with muscle and muscle fiber atrophy, and mild reduction in muscle strength, resembling tubular aggregate myopathy. Altogether, our results lead to the hypothesis that HTT could play a role in skeletal muscle calcium homeostasis by regulating two major protein complexes, the RyR1–DHPR complex and the STIM1–ORAI1, both of which rely on the perfect association of the T-tubules membrane and the SR membrane, and therefore HTT could be involved in the precise structuration of the triads or the maintenance of this structure. It could also be involved in the reverse traffic of STIM1 from the triads to the longitudinal SR, a hypothesis that requires additional experiments. The absence of HTT would thus result in a mild myopathy with alteration in calcium fluxes and the development of tubular aggregates, which have up to now been linked to hyperactivity of the STIM1–ORAI1 complex. Such a myopathy could contribute to the muscle phenotype of HD patients. Interestingly, neuronal SOCE upregulation has also been observed in different HD models and proposed to contribute to the HD phenotype (Czeredys, 2020), thus this pathophysiological mechanism could affect both the CNS and the skeletal muscle. Altogether, our experiments demonstrate that HTT reduction in skeletal muscle resulted in a mild muscle weakness associated with a mild (10% reduction) muscle atrophy at 10 mo of age, corresponding to a moderate myopathy resembling tubular aggregate myopathy.
Supplementary Material
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 6.
Acknowledgments
Eduardo Ríos served as editor.
We thank the Myoline Plateform (Myology Institute, Paris, France) for the immortalized human control satellite cells, and M. Durut and B. Bin for their technical assistance.
The work was supported by grants from Association Française contre les Myopathies (AFM-Téléthon).
The authors declare no competing financial interests.
Author contributions: M. Chivet, A.S. Nicot, S. Humbert, F. Saudou, J. Fauré, and I. Marty designed the research studies; S. Humbert, F. Saudou, and I. Marty obtained the funding; M. Chivet, M. McCluskey, A.S. Nicot, J. Brocard, M. Beaufils, D. Giovannini, and B. Poreau conducted the experiments and acquired the data; M. Chivet, M. McCluskey, A.S. Nicot, J. Brocard, M. Beaufils, D. Giovannini, B. Poreau, J. Brocard, and I. Marty analyzed the data; M. Chivet and I. Marty wrote the manuscript; all the authors read, corrected, and approved the manuscript.
Footnotes
This work is part of a special issue on excitation–contraction coupling.
References
- Aziz, N.A., van der Burg J.M., Landwehrmeyer G.B., Brundin P., Stijnen T., and EHDI Study Group . 2008. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 71:1506–1513. 10.1212/01.wnl.0000334276.09729.0e [DOI] [PubMed] [Google Scholar]
- Bates, G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R., et al. 2015. Huntington disease. Nat. Rev. Dis. Prim. 1:15005. 10.1038/nrdp.2015.5 [DOI] [PubMed] [Google Scholar]
- Beaufils, M., Tourel A., Petiot A., Halmai N.B., Segal D.J., Rendu J., and Marty I.. 2022. Development of knock-out muscle cell lines using lentivirus-mediated CRISPR/Cas9 gene editing. J. Vis. Exp. 10.3791/64114 [DOI] [PubMed] [Google Scholar]
- Böhm, J., Chevessier F., Maues De Paula A., Koch C., Attarian S., Feger C., Hantaï D., Laforêt P., Ghorab K., Vallat J.M., et al. 2013. Constitutive activation of the calcium sensor STIM1 causes tubular-aggregate myopathy. Am. J. Hum. Genet. 92:271–278. 10.1016/j.ajhg.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braubach, P., Orynbayev M., Andronache Z., Hering T., Landwehrmeyer G.B., Lindenberg K.S., and Melzer W.. 2014. Altered Ca2+ signaling in skeletal muscle fibers of the R6/2 mouse, a model of Huntington’s disease. J. Gen. Physiol. 144:393–413. 10.1085/jgp.201411255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse, M.E., Hughes G., Wiles C.M., and Rosser A.E.. 2008. Use of hand-held dynamometry in the evaluation of lower limb muscle strength in people with Huntington’s disease. J. Neurol. 255:1534–1540. 10.1007/s00415-008-0964-x [DOI] [PubMed] [Google Scholar]
- Cacheux, M., Blum A., Sébastien M., Wozny A.S., Brocard J., Mamchaoui K., Mouly V., Roux-Buisson N., Rendu J., Monnier N., et al. 2015. Functional characterization of a central core disease RyR1 mutation (p.Y4864H) associated with quantitative defect in RyR1 protein. J. Neuromuscul. Dis. 2:421–432. 10.3233/JND-150073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo, E., Zuccato C., and Tartari M.. 2005. Normal huntingtin function: An alternative approach to Huntington’s disease. Nat. Rev. Neurosci. 6:919–930. 10.1038/nrn1806 [DOI] [PubMed] [Google Scholar]
- Chaturvedi, R.K., Adhihetty P., Shukla S., Hennessy T., Calingasan N., Yang L., Starkov A., Kiaei M., Cannella M., Sassone J., et al. 2009. Impaired PGC-1α function in muscle in Huntington’s disease. Hum. Mol. Genet. 18:3048–3065. 10.1093/hmg/ddp243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevessier, F., Marty I., Paturneau-Jouas M., Hantaï D., and Verdière-Sahuqué M.. 2004. Tubular aggregates are from whole sarcoplasmic reticulum origin: Alterations in calcium binding protein expression in mouse skeletal muscle during aging. Neuromuscul. Disord. 14:208–216. 10.1016/j.nmd.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Cong, S.Y., Pepers B.A., Roos R.A., Van Ommen G.J., and Dorsman J.C.. 2005. Epitope mapping of monoclonal antibody 4C8 recognizing the protein huntingtin. Hybridoma. 24:231–235. 10.1089/hyb.2005.24.231 [DOI] [PubMed] [Google Scholar]
- Czeredys, M. 2020. Dysregulation of neuronal calcium signaling via store-operated channels in Huntington’s disease. Front. Cell Dev. Biol. 8:611735. 10.3389/fcell.2020.611735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbellay, B., Arnaudeau S., Bader C.R., Konig S., and Bernheim L.. 2011. STIM1L is a new actin-binding splice variant involved in fast repetitive Ca2+ release. J. Cell Biol. 194:335–346. 10.1083/jcb.201012157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen, R.T. 2009. Checking your SOCCs and feet: The molecular mechanisms of Ca2+ entry in skeletal muscle. J. Physiol. 587:3139–3147. 10.1113/jphysiol.2009.172148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoussé, L., Knowlton B., Cupples L.A., Marder K., Shoulson I., and Myers R.H.. 2002. Weight loss in early stage of Huntington’s disease. Neurology. 59:1325–1330. 10.1212/01.WNL.0000031791.10922.CF [DOI] [PubMed] [Google Scholar]
- Duyao, M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., and MacDonald M.E.. 1995. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 269:407–410. 10.1126/science.7618107 [DOI] [PubMed] [Google Scholar]
- Giannesini, B., Vilmen C., Le Fur Y., Dalmasso C., Cozzone P.J., and Bendahan D.. 2010. A strictly noninvasive MR setup dedicated to longitudinal studies of mechanical performance, bioenergetics, anatomy, and muscle recruitment in contracting mouse skeletal muscle. Magn. Reson. Med. 64:262–270. 10.1002/mrm.22386 [DOI] [PubMed] [Google Scholar]
- Golini, L., Chouabe C., Berthier C., Cusimano V., Fornaro M., Bonvallet R., Formoso L., Giacomello E., Jacquemond V., and Sorrentino V.. 2011. Junctophilin 1 and 2 proteins interact with the L-type Ca2+ channel dihydropyridine receptors (DHPRs) in skeletal muscle. J. Biol. Chem. 286:43717–43725. 10.1074/jbc.M111.292755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, Y., Brotto M., Weisleder N., Chu Y., Lin P., Zhao X., Thornton A., Komazaki S., Takeshima H., Ma J., and Pan Z.. 2006. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys. J. 90:4418–4427. 10.1529/biophysj.105.076570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khedraki, A., Reed E.J., Romer S.H., Wang Q., Romine W., Rich M.M., Talmadge R.J., and Voss A.A.. 2017. Depressed synaptic transmission and reduced vesicle release sites in Huntington’s disease neuromuscular junctions. J. Neurosci. 37:8077–8091. 10.1523/JNEUROSCI.0313-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig, X., Choi R.H., and Launikonis B.S.. 2018. Store-operated Ca2+ entry is activated by every action potential in skeletal muscle. Commun. Biol. 1:31. 10.1038/s42003-018-0033-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski, C.M., Schlangen C., Gellerich F.N., Gizatullina Z., Deschauer M., Schiefer J., Young A.B., Landwehrmeyer G.B., Toyka K.V., Sellhaus B., and Lindenberg K.S.. 2007. Myopathy as a first symptom of Huntington’s disease in a Marathon runner. Mov. Disord. 22:1637–1640. 10.1002/mds.21550 [DOI] [PubMed] [Google Scholar]
- Lehnart, S.E., and Wehrens X.H.T.. 2022. The role of junctophilin proteins in cellular function. Physiol. Rev. 102:1211–1261. 10.1152/physrev.00024.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.H., Schilling G., Young W.S. III, Li X.J., Margolis R.L., Stine O.C., Wagster M.V., Abbott M.H., Franz M.L., Ranen N.G., et al. 1993. Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron. 11:985–993. 10.1016/0896-6273(93)90127-D [DOI] [PubMed] [Google Scholar]
- Macdonald, M., and The Huntington’s Disease Collaborative Research Group . 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 72:971–983. 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- Mamchaoui, K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L., et al. 2011. Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skeletal Muscle. 1:34. 10.1186/2044-5040-1-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini, L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., and Bates G.P.. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87:493–506. 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- Marques Sousa, C., and Humbert S.. 2013. Huntingtin: Here, there, everywhere!. J. Huntingtons Dis. 2:395–403. 10.3233/JHD-130082 [DOI] [PubMed] [Google Scholar]
- Marty, I., and Fauré J.. 2016. Excitation-contraction coupling alterations in myopathies. J. Neuromuscul. Dis. 3:443–453. 10.3233/JND-160172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty, I., Robert M., Villaz M., De Jongh K., Lai Y., Catterall W.A., and Ronjat M.. 1994. Biochemical evidence for a complex involving dihydropyridine receptor and ryanodine receptor in triad junctions of skeletal muscle. Proc. Natl. Acad. Sci. USA. 91:2270–2274. 10.1073/pnas.91.6.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne, N., Vachey G., de Longprez L., Meunier C., Zimmer V., Perriard G., Canales M., Mathias A., Herrgott L., Beltraminelli T., et al. 2017. The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Rep. 20:2980–2991. 10.1016/j.celrep.2017.08.075 [DOI] [PubMed] [Google Scholar]
- Moutin, M.J., Cuillel M., Rapin C., Miras R., Anger M., Lompré A.M., and Dupont Y.. 1994. Measurements of ATP binding on the large cytoplasmic loop of the sarcoplasmic reticulum Ca2+-ATPase overexpressed in Escherichia coli. J. Biol. Chem. 269:11147–11154. 10.1016/S0021-9258(19)78103-4 [DOI] [PubMed] [Google Scholar]
- Nakada, T., Kashihara T., Komatsu M., Kojima K., Takeshita T., and Yamada M.. 2018. Physical interaction of junctophilin and the CaV1.1 C terminus is crucial for skeletal muscle contraction. Proc. Natl. Acad. Sci. USA. 115:4507–4512. 10.1073/pnas.1716649115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir, J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., and Hayden M.R.. 1995. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 81:811–823. 10.1016/0092-8674(95)90542-1 [DOI] [PubMed] [Google Scholar]
- Nishi, M., Hashimoto K., Kuriyama K., Komazaki S., Kano M., Shibata S., and Takeshima H.. 2002. Motor discoordination in mutant mice lacking junctophilin type 3. Biochem. Biophys. Res. Commun. 292:318–324. 10.1006/bbrc.2002.6649 [DOI] [PubMed] [Google Scholar]
- Oddoux, S, Brocard Ju., Schweitzer A., Szentesi P., Giannesini B., Brocard Ja., Fauré J., Pernet-Gallay K., Bendahan D., Lunardi J., et al. 2009. Triadin deletion induces impaired skeletal muscle function. J. Biol. Chem. 284:34918–34929. 10.1074/jbc.M109.022442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, L., Petiot A., Brocard J., Giannesini B., Giovannini D., Sanchez C., Travard L., Chivet M., Beaufils M., Kutchukian C., et al. 2020. In vivo RyR1 reduction in muscle triggers a core-like myopathy. Acta Neuropathol. Commun. 8:192. 10.1186/s40478-020-01068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni, S., and Beam K.. 2022. Junctophilins 1, 2, and 3 all support voltage-induced Ca2+ release despite considerable divergence. J. Gen. Physiol. 154:e202113024. 10.1085/jgp.202113024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, L.A. 2017. Striatal synaptic dysfunction and altered calcium regulation in Huntington disease. Biochem. Biophys. Res. Commun. 483:1051–1062. 10.1016/j.bbrc.2016.07.058 [DOI] [PubMed] [Google Scholar]
- Ribchester, R.R., Thomson D., Wood N.I., Hinks T., Gillingwater T.H., Wishart T.M., Court F.A., and Morton A.J.. 2004. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington’s disease mutation. Eur. J. Neurosci. 20:3092–3114. 10.1111/j.1460-9568.2004.03783.x [DOI] [PubMed] [Google Scholar]
- Rodinova, M., Krizova J., Stufkova H., Bohuslavova B., Askeland G., Dosoudilova Z., Juhas S., Juhasova J., Ellederova Z., Zeman J., et al. 2019. Deterioration of mitochondrial bioenergetics and ultrastructure impairment in skeletal muscle of a transgenic minipig model in the early stages of Huntington’s disease. Dis. Model. Mech. 12:dmm038737. 10.1242/dmm.038737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, C.A. 2002. Polyglutamine pathogenesis: Emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 35:819–822. 10.1016/S0896-6273(02)00872-3 [DOI] [PubMed] [Google Scholar]
- Saüc, S., Bulla M., Nunes P., Orci L., Marchetti A., Antigny F., Bernheim L., Cosson P., Frieden M., and Demaurex N.. 2015. STIM1L traps and gates Orai1 channels without remodeling the cortical ER. J. Cell Sci. 128:1568–1579. 10.1242/jcs.164228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou, F., and Humbert S.. 2016. The biology of Huntingtin. Neuron. 89:910–926. 10.1016/j.neuron.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Schuler, M., Ali F., Metzger E., Chambon P., and Metzger D.. 2005. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 41:165–170. 10.1002/gene.20107 [DOI] [PubMed] [Google Scholar]
- Seixas, A.I., Holmes S.E., Takeshima H., Pavlovich A., Sachs N., Pruitt J.L., Silveira I., Ross C.A., Margolis R.L., and Rudnicki D.D.. 2012. Loss of junctophilin-3 contributes to Huntington disease-like 2 pathogenesis. Ann. Neurol. 71:245–257. 10.1002/ana.22598 [DOI] [PubMed] [Google Scholar]
- Smyth, J.T., Hwang S.Y., Tomita T., DeHaven W.I., Mercer J.C., and Putney J.W.. 2010. Activation and regulation of store-operated calcium entry. J. Cell Mol. Med. 14:2337–2349. 10.1111/j.1582-4934.2010.01168.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong, T.V., Tagle D.A., Valdes J.M., Elmer L.W., Boehm K., Swaroop M., Kaatz K.W., Collins F.S., and Albin R.L.. 1993. Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat. Gen. 5:259–265. 10.1038/ng1193-259 [DOI] [PubMed] [Google Scholar]
- Tang, T.S., Tu H., Chan E.Y., Maximov A., Wang Z., Wellington C.L., Hayden M.R., and Bezprozvanny I.. 2003. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 39:227–239. 10.1016/S0896-6273(03)00366-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo, A., Tarrats R.M., Alonso M.E., Boll M.C., Ochoa A., and Velásquez L.. 2004. Assessment of the nutrition status of patients with Huntington’s disease. Nutrition. 20:192–196. 10.1016/j.nut.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Wei-Lapierre, L., Carrell E.M., Boncompagni S., Protasi F., and Dirksen R.T.. 2013. Orai1-dependent calcium entry promotes skeletal muscle growth and limits fatigue. Nat. Commun. 4:2805. 10.1038/ncomms3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, C.W., Varuzhanyan G., Talmadge R.J., and Voss A.A.. 2013. Huntington disease skeletal muscle is hyperexcitable owing to chloride and potassium channel dysfunction. Proc. Natl. Acad. Sci. USA. 110:9160–9165. 10.1073/pnas.1220068110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, J.D., MacMillan J.C., Harper P.S., Lowenstein P.R., and Jones A.L.. 1996. Partial characterisation of murine huntingtin and apparent variations in the subcellular localisation of huntingtin in human, mouse and rat brain. Hum. Mol. Gen. 5:481–487. 10.1093/hmg/5.4.481 [DOI] [PubMed] [Google Scholar]
- Zeitlin, S., Liu J.-P., Chapman D.L., Papaioannou V.E., and Efstratiadis A.. 1995. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Gen. 11:155–163. 10.1038/ng1095-155 [DOI] [PubMed] [Google Scholar]
- Zielonka, D., Piotrowska I., Marcinkowski J.T., and Mielcarek M.. 2014. Skeletal muscle pathology in Huntington’s disease. Front. Physiol. 5:380. 10.3389/fphys.2014.00380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 6.