Abstract
Genetic admixture is central to primate evolution. Here, we combine 50 years of field observations of immigration and group demography with genomic data from ~9 generations of hybrid baboons to investigate the consequences of admixture in the wild. Despite no obvious fitness costs to hybrids, we find signatures of selection against admixture similar to those described for archaic hominins. These patterns are concentrated near genes where ancestry is strongly associated with gene expression. Our analyses also show that introgression is partially predictable across the genome. They demonstrate the value of integrating genomic and field data for revealing how “genomic signatures of selection” (e.g., reduced introgression in low recombination regions) manifest in nature, and underscore the importance of other primates as living models for human evolution.
One Sentence Summary:
Coupled genomic and field data indicate selection against baboon hybrids, despite no overt fitness costs in the wild.
The ancestors of modern humans intermixed with Neanderthals and other close, now-extinct lineages, leaving a genetic legacy that continues to shape human trait variation today (1-3). Even as these findings reshape our conception of human origins, they also bring us more closely in line with our primate relatives, where hybridization is observed in many species (4, 5). Studies of other living primates therefore provide context for understanding admixture dynamics in our own lineage. Field studies in hybrid zones, for instance, offer the opportunity to integrate demographic (e.g., reproductive success, immigration/emigration), phenotypic, and genomic data on early generation hybrids, which studies in humans suggest experienced the greatest fitness costs (6, 7).
Thus far, studies suggest that ancestry frequently predicts trait variation in primate hybrid zones, but admixture often does not result in overt fitness costs (8-11). However, field observations have not been combined with population and functional genomic analyses to investigate both the organismal and molecular consequences of admixture in primates. Here, we take such an approach to investigate whether selection against introgression (i.e., alleles introduced by gene flow from one distinct lineage to another) is compatible with apparently healthy hybrids, investigate the functional consequences of introgressed alleles, and follow the course of hybridization and natural selection across generations.
We focus on admixture between yellow baboons (Papio cynocephalus) and anubis baboons (P. anubis): large-bodied, terrestrial primates long used as a model for human biology and evolution (12). Although baboon taxonomy has undergone many revisions over time, six extant baboon species are currently recognized based on distinct phenotypic differences and a pattern of phylogenetic divergence supported by recent whole-genome sequencing data (12-14). This phylogeny establishes two major baboon lineages (the “northern” and “southern” clades) that separated ~1.4 million years ago, although the complex evolutionary history of baboons means that they may have experienced episodes of gene flow since that time (14-16). Anubis and yellow baboons belong to the northern and southern clades, respectively, and both phylogenetic and population genomic analyses confirm their divergence into distinct taxa (13, 14). Nevertheless, they interbreed to produce viable and fertile offspring where their ranges meet (Fig. 1A).
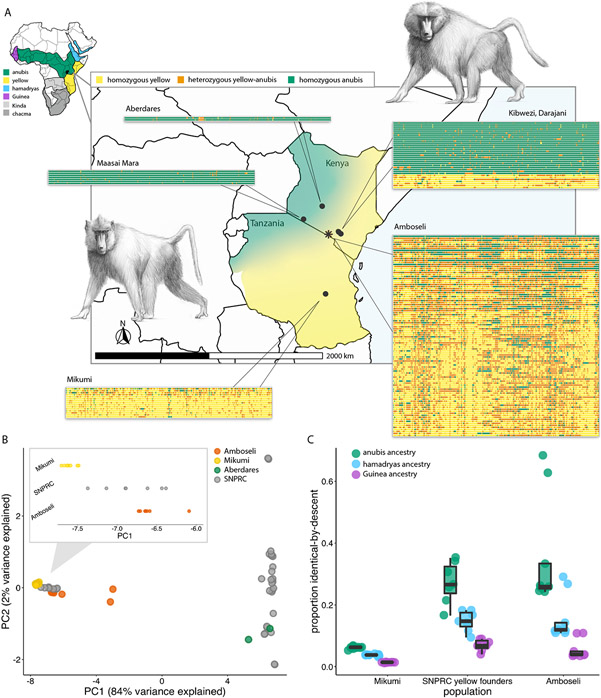
Figure 1: The structure of the baboon hybrid zone in Amboseli and the surrounding region.
(A) Geographic locations and local ancestry estimates for baboons in this study (black asterisk=Amboseli). For each population, each row corresponds to the first 20 Mb of chromosome 1 for one individual, organized vertically by global ancestry. For Amboseli, a subsample of 100 individuals is shown. Central map: ranges of yellow baboons and anubis baboons in Kenya and Tanzania; small map: ranges of all six African baboons (47), modified from a map by Kenneth Chiou (CC BY 3.0 license). Baboon drawings by Christopher Smith. (B) PCA of genotype data for high coverage genomes. Inset: distribution of “yellow-like” individuals along PC1. SNPRC yellow baboon founders resemble admixed Amboseli baboons. (C) IBDMix (26) results for three sets of yellow or majority yellow baboons. SNPRC yellow baboon founders and Amboseli baboons exhibit substantial identity-by-descent (IBD) with anubis baboons, while IBD estimates for Mikumi yellow and anubis baboons are low. The excess IBD in the SNPRC and Amboseli samples points to the contribution of gene flow beyond residual incomplete lineage sorting.
We concentrate on the region in and around the Amboseli basin of Kenya, where data from fifty years of continuous observation on a population near the center of the hybrid zone are available (17). Members of this majority-yellow baboon population include descendants of historical admixture prior to the start of monitoring in 1971, as well as descendants of a directly observed, recent wave of admixture beginning in 1982 (15, 18). In Amboseli, hybrids do not experience obvious fitness costs, and anubis ancestry may in fact confer benefits, including accelerated maturation, increased mating success, and higher rates of male-female affiliation (19-21). However, field and microsatellite data indicate that the hybrid zone is narrow (22), suggesting that natural selection may act to limit gene flow.
Structure of the baboon hybrid zone
To assess selection against introgression in hybrid anubis-yellow baboons, we used whole-genome resequencing data to evaluate ancestry patterns for animals sampled in and near the Amboseli hybrid zone (Fig. 1 and table S1). We generated resequencing data from 430 wild baboons from Amboseli and Mikumi National Park in Tanzania (17 high coverage [mean=22.51x]; 413 low coverage [mean=1.04x]), which we combined with published baboon genomes from Amboseli (n=22), Mikumi (n=5), the Maasai Mara National Reserve (n=7), and the Aberdares region of central Kenya (n=2) (14, 15, 23). In Amboseli, our sample included 442 baboons born between 1969 and 2016. Finally, we also included the genomes of 39 captive baboons from the Southwest National Primate Research Center (SNPRC; n=33, n=31 of whom were colony founders) and the Washington National Primate Research Center (WNPRC; n=6) (14, 15, 24).
We estimated global and local ancestry for each individual using a composite likelihood method suitable for low coverage data, LCLAE, which uses genotype likelihoods across genomic windows rather than requiring genotypes at specific variants (13, 15). These results confirm that admixture is minimal or absent in the anubis baboon founders of the SNPRC colony, anubis baboons from Maasai Mara, and yellow baboons from Mikumi (Fig. 1 and fig. S5, although we cannot exclude ancient bouts of admixture that affect all living baboons). In contrast, all baboons from Amboseli are admixed (Fig. 1A; mean=30-37% genome-wide anubis ancestry ± 10% s.d.), including many whose ancestry can be traced to anubis-like immigrants within the last seven generations. These results closely match F4-ratio estimates (25) (<2% difference for nine high coverage Amboseli genomes), indicating that putative anubis ancestry in Amboseli reflects admixture, not incomplete lineage sorting (13).
We also detected a signal of ~17% mean anubis ancestry in the putatively yellow baboon founders of the SNPRC colony, who were previously thought to be unadmixed (Fig. 1A-C) (24). Identity-by-descent (IBD) analysis using IBDMix (26) confirms this pattern (Fig. 1C). As IBD between Mikumi yellow baboons and anubis baboons is <5%, these findings also implicate admixture rather than incomplete lineage sorting (Fig. 1C; (13)). Combined with evidence for yellow ancestry in a central Kenyan anubis baboon (13, 14), our results indicate that gene flow has been a common feature of baboon evolution in east Africa.
Selection against introgression in Amboseli
To investigate whether selection restricts gene flow between anubis and yellow baboons, we focused on the multigenerational data set from Amboseli. We replicated three analyses used to infer selection against Neanderthal or Denisovan introgression in humans (27-29). First, we tested for a relationship between anubis ancestry in Amboseli and yellow-anubis genetic divergence (based on unadmixed populations: (13)). Because the Amboseli population is largely of yellow baboon origin, if hybridization is deleterious, selection is expected to be less permissive of anubis alleles that are more diverged from their yellow counterparts. Indeed, anubis ancestry is systematically lower in regions of the genome with more fixed differences (Fig. 2A; Spearman’s rho=−0.119, p=8.05 x 10−34). In Amboseli, anubis alleles are 6.7% less common in the most diverged percentile relative to the least diverged percentile of the baboon genome. These results are similar to the negative correlation between the density of fixed human-Neanderthal differences and introgressed Neanderthal ancestry in modern humans (27) (Fig. 2B and table S2).
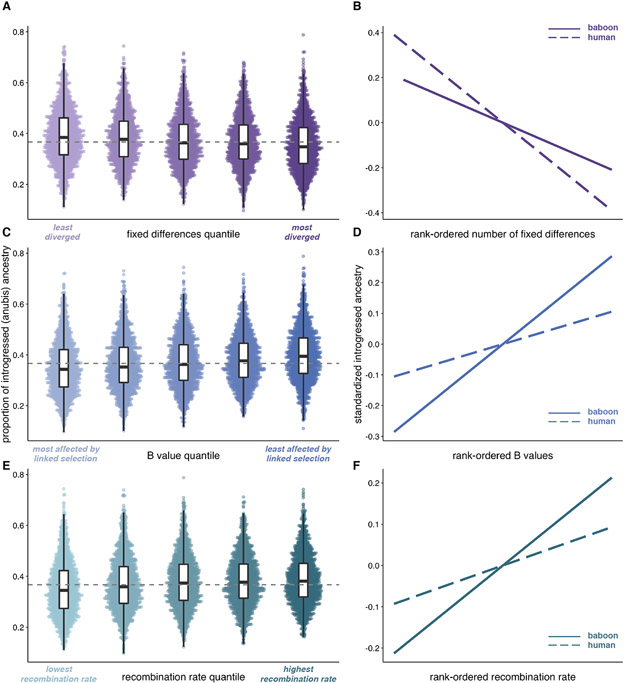
Figure 2: Selection against introgression in the Amboseli baboons mirrors patterns described for archaic hominin admixture.
(A, C, E) Proportion of introgressed (anubis) ancestry in Amboseli in 250 kb windows (n=10,324 total windows) as a function of (A) fixed differences between yellow and anubis baboons (Spearman’s rho=−0.119, p=8.05 x 10−34), (C) mean B statistic (rho=0.168, p=1.73 x 10−66), and (E) mean recombination rate (rho=0.127, p=2.49 x 10−38), divided into quintiles for visualization purposes only. Dashed grey lines show median anubis ancestry across all windows. (B, D, F) Predicted relationships between introgressed ancestry and all three measures are observed for both anubis ancestry in the Amboseli baboons (solid lines) and Neanderthal ancestry in modern human genomes (dashed lines) (27-29), consistent with selection against introgression. Panels show the relationship between introgressed ancestry and the rank-ordered (B) number of fixed differences, (D) mean B statistic, and (F) mean local recombination rate. Mean introgressed ancestry is centered on 0 and divided by the standard deviation for each species to facilitate comparison.
Second, we tested whether introgressed anubis ancestry is depleted in genomic regions that are likely to be affected by linked selection, as summarized by B statistic values calculated for the baboon genome (13, 30) (i.e., due to high gene density per recombination distance). Again paralleling the case of Neanderthal ancestry in modern humans (28), anubis ancestry is most common in regions that are predicted to be least affected by linked selection (Fig. 2C-D; Spearman’s rho=0.168, p=1.73 x 10−66). Consequently, anubis ancestry per individual is reduced, on average, by 7.03% in protein-coding regions relative to random, size-matched regions of the genome (±4.20% s.d.; n=442 Amboseli baboons). Reductions in promoters and putative peripheral blood mononuclear cell enhancers were 5.56% (±4.10% s.d.) and 6.22% (±4.20%), respectively.
Third, we tested whether introgressed anubis ancestry is positively correlated with local recombination rate. This relationship is predicted if recombination influences the rate at which natural selection eliminates deleterious introgressed ancestry and uncouples deleterious from neutral introgressed variants. This prediction, documented across diverse taxa (29, 31, 32), is also observed in baboons (Fig. 2E; Spearman’s rho=0.127, p=1.48 x 10−38), with a magnitude similar to that reported for Neanderthal and Denisovan gene flow into modern humans (Fig. 2F; Spearman’s rho=0.17 and 0.14 for Neanderthal and Denisovan ancestry, respectively (29)).
To investigate these patterns further, we took advantage of the dynamic history of admixture within the Amboseli population. At the beginning of monitoring in 1971, all Amboseli animals were considered to be yellow baboons (33). Phenotypically anubis and admixed animals immigrated into the population starting in 1982, and the proportion of hybrid animals increased over the following decades (18, 34). Whole-genome data recapitulate these patterns, documenting an increase of 11.8% anubis ancestry from 1971 (23.1-29.6%) to 2020 (34.9-41.4%) (Fig. 3A). However, animals with no known anubis ancestors during the 50-year field study are also clearly admixed (Fig. 3B). Additionally, while immigrant males are more anubis-like than the study population as a whole (Fig. 3A), one immigrant male was among the most yellow-like in our sample (78.8% yellow ancestry), indicating ongoing gene flow involving both parental taxa.
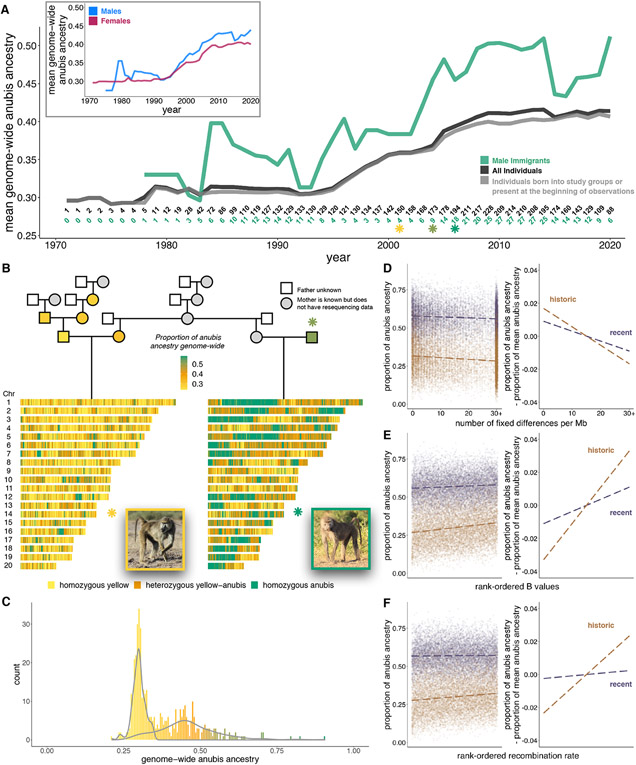
Figure 3: Recent and historic hybrid ancestry in Amboseli.
(A) Mean genome-wide anubis ancestry in the Amboseli population has increased since the 1970s. Numbers above the x-axis indicate the number of individuals used to calculate annual ancestry (black=all individuals, green=male immigrants). (B) Pedigree and ancestry estimates for example historical (left) and recent (right) hybrids. Pedigree individuals with resequencing data are colored based on ancestry. The two examples share a maternal grandmother and were born a few years apart (yellow and bright green asterisks in [A]). The father of the recent hybrid immigrated in 2004 (olive green asterisk in [A]). (C) Genome-wide anubis ancestry in Amboseli, with density plots overlaid for historical and recently admixed individuals. (D-F) The relationships between introgressed anubis ancestry and the rank-ordered (D) number of fixed anubis-yellow differences, (E) mean B statistic, and (F) mean local recombination rate. All relationships are stronger for historic hybrids than recent hybrids. Right-hand panels show anubis ancestry within each data set mean-centered to 0.
The Amboseli population today therefore contains individuals who descend from ancient, unobserved admixture events as well as those affected by recent hybridization, generating a bimodal distribution of genome-wide ancestry (Fig. 3C) (15). By integrating local ancestry calls, pedigree information, and field observations, we identified 188 “recently” admixed individuals whose ancestors include at least one anubis-like immigrant within the last 0-7 generations (mean=1.7 generations, although due to historical gene flow, these animals are not classical F1 or F2 hybrids). We also classified 214 baboons as “historically” admixed, as their genomes only contain anubis ancestry from before 1971. Forty baboons could not be assigned to either hybrid class (13). Based on a single-pulse model of admixture using DATES (35), historical admixture is dated to 283 ± 242 s.d. generations ago (n=7 high-coverage genomes), in contrast to 5 and 21 generations ago for two recent hybrids sequenced to high coverage.
Stratifying individuals in the data set by admixture history reveals that signatures of selection against introgression are driven by historical admixture (i.e., genomes sampled dozens to hundreds of generations post-contact). Historically admixed individuals are more depleted of anubis ancestry in highly diverged and low B value regions of the genome than recently admixed animals (Fig. 3D-E). Further, the relationship between anubis ancestry and recombination rate is exclusive to the historically admixed data set, even based on recombination rates measured on chromosome-level scales (Fig. 3F and table S3, (13)). The weaker signature of selection in recent hybrids likely reflects intermittent gene flow in the last few generations and stochastic inheritance processes. In contrast, sufficient generations have passed since historical admixture to break apart large introgressed haplotypes, allowing us to observe non-random patterns of ancestry across the genome. This result emphasizes the importance of complementing field observations with genomic data, which provide insight into selective processes that operate over timescales longer than even the longest-running field studies.
Selection against regulatory divergence
Analyses of human-Neanderthal admixture suggest a consistent pattern of selection against regulatory variants (36). If so, the introgressed regions that persist in modern humans have likely been purged of many alleles with large regulatory effects (37, 38). However, direct comparisons between the effect sizes of retained versus lost archaic alleles are difficult, as only a fraction of archaic hominin alleles (e.g., Neanderthal, Denisovan, or other ghost lineages) segregate in modern human genomes today (28, 39). Extant primate populations, where hybridization and selection are ongoing, provide an opportunity to test this hypothesis.
To test for selection against gene regulatory divergence in baboons, we paired genetic ancestry data with blood-derived RNA-sequencing data from 145 individuals (n=157 samples (40-42); table S1). This data set includes whole blood and white blood cells, which were analyzed separately while controlling for age, sex, and kinship (13). Among 10,192 analyzed genes, we identified no significant associations between genome-wide ancestry and gene expression levels (10% FDR). In contrast, local ancestry predicted gene expression levels for 20.1% (2,046) of tested genes in one or both data sets (Fig. 4A), with concordant additive effects between data sets (Pearson’s R=0.43, p<10−200), and little evidence for non-additivity (13).
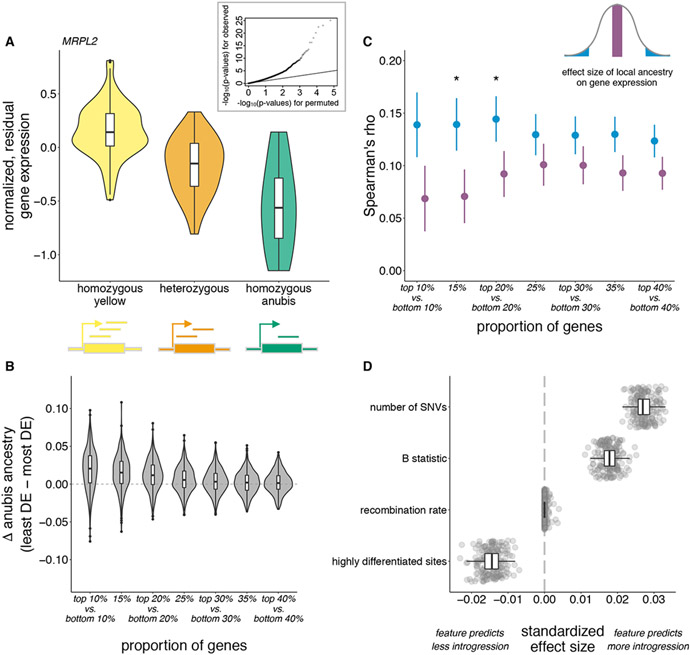
Figure 4: Selection against gene regulatory divergence and prediction of local introgression levels.
(A) Local ancestry predicts gene expression in the Amboseli population, as depicted for an example gene (MRPL2). Inset: quantile-quantile plot comparing p-value distributions for observed local ancestry effects (y-axis) to a permutation-based null (x-axis). (B) Difference in introgressed anubis ancestry between genes with the smallest versus largest local ancestry effects on gene expression. Violin plots show the distribution of differences across individuals; boxplots show the median and inter-quartile range (all p<0.05; table S4). (C) Correlation between anubis introgression and recombination rate calculated for sets of genes with the largest (blue) versus smallest (magenta) local ancestry absolute effect sizes. Asterisks denote bootstrapped p-value < 0.05 (table S4); error bars show standard deviations. (D) Distribution of effect sizes for features that consistently predict the extent of anubis introgression in Amboseli baboon genomes (table S5). The number of single nucleotide variants (SNVs) is derived from unadmixed yellow and anubis baboon genomes.
If introgressed alleles that perturb gene regulation are a primary target of selection, we reasoned that selection should purge anubis ancestry near genes where ancestry strongly affects gene expression. In support of this prediction, the top 15% of genes with the largest local ancestry effects on gene expression harbor 1.5% less anubis ancestry, on average, than the bottom 15% of genes with the smallest local ancestry effects (Fig. 4B; paired t-test p=1.10 x 10−36, n=442). This difference is exaggerated within historically admixed individuals (1.9% reduction, p=1.26 x 10−27, n=214; table S4). Further, the correlation between anubis ancestry and local recombination rate is larger for genes with the largest local ancestry effects than those with the smallest (Fig. 4C; rhodiff=0.07 for the top and bottom 15% of genes, bootstrapped p-value=0.027; table S4). Combined with the depletion of introgressed sequence in regulatory elements, these results support the hypothesis that introgressed alleles that affect gene regulation are nonrandomly purged post-hybridization. They are therefore consistent with the idea that natural selection removed archaic variants with large regulatory effects from the genomes of modern humans (37).
Predicting the genomic landscape of introgression
Finally, we investigated our ability to predict the genomic locations most and least affected by introgression. We modeled mean anubis ancestry as a function of local recombination rate, SNP density in the reference yellow and anubis populations, yellow-anubis genetic divergence, gene and enhancer content, linked selection, and local ancestry-associated gene expression in blood. We iteratively trained an elastic net regression model on nonoverlapping 250 kb windows of the genome, representing 75% of the genome, and applied the model to a test set of windows in the remaining 25% (13). We found that our predicted values were consistently positively correlated with observed levels of anubis ancestry in the test sets (mean Pearson’s R=0.254 ± 0.016 s.d. vs. 0.014 ± 0.011 s.d. for models fit to permuted data), with frequent contributions from features capturing local recombination rate, linked selection, genetic variation, and sequence divergence (Fig. 4D and table S5). We consistently predicted anubis ancestry more accurately in historical hybrids than in recent hybrids (mean Pearson’s R=0.265 ± 0.017 s.d. vs. 0.177 ±0.018 s.d., bootstrapped p-value <10−3).
Our longitudinal data also indicate that increases in anubis ancestry across the fifty-year field study are non-randomly distributed throughout the genome. Controlling for the starting level of anubis ancestry in 1979, 100 kb windows characterized by lower FST and higher recombination rates experienced larger increases in anubis ancestry between 1979 and 2020, although both effect sizes are small (FST and recombination rate p-values <10−3, β=−2.965 x 10−4 and 1.020 x 10−4 respectively, n=25,797 windows; table S6). B statistic values did not predict temporal change in anubis ancestry independently of recombination rate.
Divergence and hybridization in primates
Our genomic analysis reveals evidence for selection against admixture that is remarkably consistent with results obtained for archaic introgression in humans. Our results also support a hypothesis that can only be indirectly tested in our own lineage: that natural selection has acted to eliminate introgressed alleles that strongly perturb gene regulation (37). These results contrast with the behavioral and life history evidence to date in Amboseli—one of the largest and longest-running primate field sites in the world—which indicates that hybrid baboons suffer no obvious fitness costs (19-21). Our results identify subtle selection against hybridization that may help explain the maintenance of primate taxonomic integrity in the face of frequent interspecific gene flow (4, 5). Ultimately, the outcome of this process will depend on the relative balance between this selection pressure, possible advantages to introgressed ancestry, migration rates, and demographic stochasticity—potentially explaining cases of nuclear swamping in baboons despite costs to hybridization (16).
The mode of selection against hybrids is unclear. Unlike in humans, hybridization load is unlikely to explain our results: yellow and anubis baboons harbor similar levels of genetic diversity compared to humans and Neanderthals (<50% difference in baboons compared to >3-fold between humans and Neanderthals (6, 14, 15, 27)). Both hybrid incompatibilities and ecological selection, however, could play a role. For example, some reports suggest that anubis and yellow baboons occupy distinct climatic niches (43). Previously described assortative mating by ancestry in the Amboseli baboons (20) may also limit introgression. Understanding the genetic and phenotypic mechanisms that influence interspecific gene flow, including the role of the X chromosome and adaptive introgression, remains an important goal for future work.
Combined, our findings illustrate the importance of contextualizing genomic data with phenotypic and demographic information to understand the evolutionary dynamics of admixture. Genomes harbor information about historical processes that stretch back many generations, and can capture subtle signatures of selection that may not be obvious in natural populations where demographic stochasticity is high, sample sizes are modest, and the specific phenotypes under selection may be unknown. Conversely, field data reveal the range of phenotypic and fitness outcomes that are compatible with genomic signatures of selection. Indeed, genomic evidence alone has led some researchers to posit that the costs of modern human-archaic hominin interbreeding must have been high, reflecting species at the brink of reproductive incompatibility (44, 45). Our results point to the limits of these inferences by indicating that qualitatively similar evidence for selection against introgression can be compatible with primate hybrids that thrive (19-21). This work therefore highlights the crucial role of other primates for understanding human evolution, especially for phenomena that are impossible to study in our lineage alone.
Supplementary Material
Acknowledgements:
We thank J. Altmann for her fundamental contributions to research on the Amboseli baboons. We thank the University of Nairobi, Institute of Primate Research, the National Museums of Kenya, the Amboseli-Longido pastoralist communities, the Enduimet Wildlife Management Area, Ker & Downey Safaris, Air Kenya, and Safarilink for assistance in Kenya; S. Sayialel, G. Marinka, B. Oyath, T. Wango, and V. Oudu as long-term members of the Amboseli Baboon Research Project (ABRP) team; and K. Pinc, N.H. Learn, and J.B. Gordon for contributions to the ABRP database. The Kenya-based research reported here was conducted with permission from the Kenya Wildlife Service (KWS), the National Environmental Management Authority (NEMA), and the National Commission for Science, Technology and Innovation (NACOSTI), which we currently renew on an annual basis. Samples were collected and exported under NEMA permits and CITES permits from KWS. We also hold a Memorandum of Agreement (MoA) with KWS, the University of Nairobi, the Institute of Primate Research, and the National Museums of Kenya that details the ABRP research mission and commitment to benefits sharing under the Nagoya Protocol. Finally, we thank G. McVicker, P. Moorjani, K. Samuk, S. He, and members of the Tung lab for discussion and assistance with code, and L. Barreiro, A. Goldberg, P. Moorjani, and G. Coop and members of the Coop lab for feedback on a previous version of this manuscript. The research in this study was approved by the Institutional Animal Care and Use Committees at Duke University, Princeton University, and the University of Notre Dame and adhered to the laws and guidelines of the Kenyan government.
Funding:
This work was supported by NSF IOS 1456832 (S.C.A.), NIH R01AG053308 (S.C.A.), NIH R01AG053330 (E.A.A.), R01HD088558 (J.T.), P01AG031719 (S.C.A.), NSF BCS-1751783 (J.T., T.P.V.), NSF BCS-2018897 (J.T., A.S.F.), a Leakey Foundation Research Grant (T.P.V), and the North Carolina Biotechnology Center (2016-IDG-1013). A.S.F. was supported by NSF GRFP (DGE #1644868) and NIH T32GM007754. Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect the views of our funding bodies.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All sequencing data have been deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA755322 (reviewer link:https://dataview.ncbi.nlm.nih.gov/object/PRJNA755322?reviewer=vl5mnd4jvbuabjef068n3 rn2f6). Previously generated resequencing data are available under PRJNA308870, PRJNA433868, PRJNA54005, PRJNA20425, PRJNA162517, PRJNA54003, PRJNA54009, PRJNA54007, PRJNA251424, and PRJNA295782, and previously generated RNA-seq data are deposited under PRJNA269070, PRJNA480672, and PRJNA731520. Processed data and scripts are available on Zenodo (46) (reviewer link: https://zenodo.org/record/5912279?token=eyJhbGciOiJIUzUxMiIsImV4cCI6MTY3MjUyNzU5OS[…]reJQ5oi5ETgA_KoaoLOnmHoQYk6URQkieb-XwwqH3zs8AY-g9SOM9hjbrj6Rokg). All data and code required to reproduce or re-analyze these data are public. The biological samples are CITES-regulated and can only be shared with third parties with prior written authorization from KWS.
References and Notes
- 1.Huerta-Sánchez E et al. , Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonti CN et al. , The phenotypic legacy of admixture between modern humans and Neandertals. Science 351, 737–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeberg H, Pääbo S, The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587, 610–612 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Zinner D, Arnold ML, Roos C, The strange blood: natural hybridization in primates. Evol Anthropol 20, 96–103 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Tung J, Barreiro LB, The contribution of admixture to primate evolution. Curr Opin Genet Dev 47, 61–68 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Harris K, Nielsen R, The Genetic Cost of Neanderthal Introgression. Genetics 203, 881–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petr M, Pääbo S, Kelso J, Vernot B, Limits of long-term selection against Neandertal introgression. Proc Natl Acad Sci U S A 116, 1639–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolly CJ, Burrell AS, Phillips-Conroy JE, Bergey C, Rogers J, Kinda baboons (Papio kindae) and grayfoot chacma baboons (P. ursinus griseipes) hybridize in the Kafue river valley, Zambia. Am J Primatol 73, 291–303 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Bergman TJ, Phillips-Conroy JE, Jolly CJ, Behavioral variation and reproductive success of male baboons (Papio anubis x Papio hamadryas) in a hybrid social group. Am J Primatol 70, 136–147 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Detwiler KM, Mitochondrial DNA Analyses of Cercopithecus Monkeys Reveal a Localized Hybrid Origin for C. mitis doggetti in Gombe National Park, Tanzania. International Journal of Primatology 40, 28–52 (2018). [Google Scholar]
- 11.Kitchen DM et al. , Temporal but Not Acoustic Plasticity in Hybrid Howler Monkey (Alouatta palliata × A. pigra) Loud Calls. International Journal of Primatology 40, 132–152 (2017). [Google Scholar]
- 12.Fischer J et al. , Insights into the evolution of social systems and species from baboon studies. eLife 8, e50989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supplementary materials.
- 14.Rogers J et al. , The comparative genomics and complex population history of Papio baboons. Sci Adv 5, eaau6947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall JD et al. , Genomewide ancestry and divergence patterns from low-coverage sequencing data reveal a complex history of admixture in wild baboons. Mol Ecol 25, 3469–3483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinner D, Groeneveld LF, Keller C, Roos C, Mitochondrial phylogeography of baboons (Papio spp.): indication for introgressive hybridization? BMC Evol Biol 9, 83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberts SC, Altmann J, “The Amboseli Baboon Research Project: 40 Years of Continuity and Change” in Long-Term Field Studies of Primates, Kappeler PM, Watts DP, Eds. (Springer-Verlag, 2012), pp. 261–287. [Google Scholar]
- 18.Samuels A, Altmann J, Immigration of a Papio anubis male into a group of Papio cynocephalus baboons and evidence for an anubis-cynocephalus hybrid zone in Amboseli, Kenya. International Journal of Primatology 7, 131–138 (1986). [Google Scholar]
- 19.Charpentier MJ, Tung J, Altmann J, Alberts SC, Age at maturity in wild baboons: genetic, environmental and demographic influences. Mol Ecol 17, 2026–2040 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Tung J, Charpentier MJ, Mukherjee S, Altmann J, Alberts SC, Genetic effects on mating success and partner choice in a social mammal. Am Nat 180, 113–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogel AS et al. , Genetic ancestry predicts male–female affiliation in a natural baboon hybrid zone. Animal Behaviour 180, 249–268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charpentier MJ et al. , Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Mol Ecol 21, 715–731 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Snyder-Mackler N et al. , Efficient genome-wide sequencing and low-coverage pedigree analysis from noninvasively collected samples. Genetics 203, 699–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JA et al. , Analysis of 100 high-coverage genomes from a pedigreed captive baboon colony. Genome Res 29, 848–856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson N et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Wolf AB, Fu W, Li L, Akey JM, Identifying and interpreting apparent Neanderthal ancestry in African individuals. Cell 180, 677–687 e616 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Vernot B, Akey JM, Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Sankararaman S et al. , The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumer M et al. , Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360, 656–660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McVicker G, Gordon D, Davis C, Green P, Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5, e1000471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edelman NB et al. , Genomic architecture and introgression shape a butterfly radiation. Science 366, 594–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calfee E et al. , Selective sorting of ancestral introgression in maize and teosinte along an elevational cline. PLoS Genet 17, e1009810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altmann SA, Altmann J, Baboon ecology: African field research. (University of Chicago Press, 1970). [Google Scholar]
- 34.Tung J, Charpentier MJ, Garfield DA, Altmann J, Alberts SC, Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Mol Ecol 17, 1998–2011 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Narasimhan VM et al. , The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Telis N, Aguilar R, Harris K, Selection against archaic hominin genetic variation in regulatory regions. Nat Ecol Evol 4, 1558–1566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy RC, Wakefield J, Akey JM, Impacts of Neanderthal-Introgressed Sequences on the Landscape of Human Gene Expression. Cell 168, 916–927 e912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colbran LL et al. , Inferred divergent gene regulation in archaic hominins reveals potential phenotypic differences. Nat Ecol Evol 3, 1598–1606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinrücken M, Spence JP, Kamm JA, Wieczorek E, Song YS, Model-based detection and analysis of introgressed Neanderthal ancestry in modern humans. Mol Ecol 27, 3873–3888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y, The genetic architecture of gene expression levels in wild baboons. eLife 4, e04729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lea AJ et al. , Dominance rank-associated gene expression is widespread, sex-specific, and a precursor to high social status in wild male baboons. Proc Natl Acad Sci U S A 115, E12163–E12171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson JA et al. , Distinct gene regulatory signatures of dominance rank and social bond strength in wild baboons. Philos Trans R Soc Lond B Biol Sci 377, 20200441 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chala D, Roos C, Svenning JC, Zinner D, Species-specific effects of climate change on the distribution of suitable baboon habitats - Ecological niche modeling of current and Last Glacial Maximum conditions. J Hum Evol 132, 215–226 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Currat M, Excoffier L, Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. Proc Natl Acad Sci U S A 108, 15129–15134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich D, Who we are and how we got here: Ancient DNA and the new science of the human past. (Oxford University Press, 2018). [Google Scholar]
- 46.Vilgalys TP et al. , Data and code for: Selection against admixture and gene regulatory divergence in a long-term primate field study. Zenodo; (2021): 10.5281/zenodo.5912279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C, Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol 150, 133–140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jordan VE et al. , A computational reconstruction of Papio phylogeny using Alu insertion polymorphisms. Mob DNA 9, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold ML, Meyer A, Natural hybridization in primates: one evolutionary mechanism. Zoology (Jena) 109, 261–276 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Cortés-Ortiz L, Roos C, Zinner D, Introduction to Special Issue on Primate Hybridization and Hybrid Zones. International Journal of Primatology 40, 1–8 (2019). [Google Scholar]
- 51.Alberts SC, Altmann J, Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. American Journal of Primatology 53, 139–154 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Phillips-Conroy JE, Jolly CJ, Changes in the structure of the baboon hybrid zone in the Awash National Park, Ethiopia. American Journal of Physical Anthropology 71, 337–350 (1986). [Google Scholar]
- 53.Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity (2011. https://www.cbd.int/abs/doc/protocol/nagoya-protocol-en.pdf).
- 54.Maples WR, McKern TW, “A preliminary report on classification of the Kenya baboon” in The Baboon in Medical Research, Vagtborg H, Ed. (University of Texas Press, 1967), vol. 2, pp. 13–22. [Google Scholar]
- 55.de Jong YA, Butynski TM, Photographic maps of the primates of Kenya and Tanzania: A tool for identification and conservation. Primate Conservation 25, 27–32 (2010). [Google Scholar]
- 56.Sapolsky RM, Altmann J, Incidence of hypercortisolism and dexamethasone resistance increases with age among wild baboons. Biological Psychiatry 30, 1008–1016 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Tung J et al. , Allele-specific gene expression in a wild nonhuman primate population. Mol Ecol 20, 725–739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A et al. , The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DePristo MA et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van der Auwera GA, O’Connor BD, Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. (O'Reilly Media, Inc., 2020). [Google Scholar]
- 61.Poplin R et al. , Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv, 10.1101/201178 (2018). [DOI] [Google Scholar]
- 62.Van der Auwera GA et al. , From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11.10.11–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krueger F, Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, (2012). [Google Scholar]
- 64.Broad Institute, Picard Toolkit. http://broadinstitute.github.io/picard/, (2019).
- 65.Batra SS et al. , Accurate assembly of the olive baboon (Papio anubis) genome using long-read and Hi-C data. Gigascience 9, giaa134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 1303.3997v1301 [q-bio.GN] (2013). [Google Scholar]
- 67.Morrissey MB, Wilson AJ, pedantics: an r package for pedigree-based genetic simulation and pedigree manipulation, characterization and viewing. Mol Ecol Resour 10, 711–719(2010). [DOI] [PubMed] [Google Scholar]
- 68.Lipatov M, Sanjeev K, Patro R, Veeramah KR, Maximum Likelihood Estimation of Biological Relatedness from Low Coverage Sequencing Data. bioRxiv, 10.1101/023374 (2015). [DOI] [Google Scholar]
- 69.Buchan JC, Alberts SC, Silk JB, Altmann J, True paternal care in a multi-male primate society. Nature 425, 179–181 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Alberts SC, Buchan JC, Altmann J, Sexual selection in wild baboons: from mating opportunities to paternity success. Animal Behaviour 72, 1177–1196 (2006). [Google Scholar]
- 71.Corbett-Detig R, Jones M, SELAM: simulation of epistasis and local adaptation during admixture with mate choice. Bioinformatics 32, 3035–3037 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Stephens ZD et al. , Simulating next-generation sequencing datasets from empirical mutation and sequencing models. PLoS One 11, e0167047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Corbett-Detig R, Nielsen R, A hidden markov model approach for simultaneously estimating local ancestry and admixture time using next generation sequence data in samples of arbitrary ploidy. PLoS Genet 13, e1006529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaefer NK, Shapiro B, Green RE, AD-LIBS: inferring ancestry across hybrid genomes using low-coverage sequence data. BMC Bioinformatics 18, 203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Danecek P et al. , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green RE et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reich D et al. , Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reich D, Thangaraj K, Patterson N, Price AL, Singh L, Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimin AV et al. , A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9, 20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfeifer SP, From next-generation resequencing reads to a high-quality variant data set. Heredity 118, 111–124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Danecek P et al. , Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang CC et al. , Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Purcell S et al. , PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Purcell S, PLINK v1.90b3.36. https://www.cog-genomics.org/plink/1.9/ [Google Scholar]
- 87.Petr M, Vernot B, Kelso J, admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 35, 3194–3195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cox LA, Mahaney MC, Vandeberg JL, Rogers J, A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics 88, 274–281 (2006). [DOI] [PubMed] [Google Scholar]
- 89.Cahill JA, Soares AE, Green RE, Shapiro B, Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc Lond B Biol Sci 371, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.UCSC Paleogenomics Lab, Chrom-Compare. https://github.com/Paleogenomics/Chrom-Compare (2014).
- 91.Li H, Durbin R, Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu FL et al. , A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS Biol 18, e3000838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL, Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet 10, e1004410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin SH, Davey JW, Salazar C, Jiggins CD, Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol 17, e2006288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wright S, The genetical structure of populations. Ann Eugen 15, 323–354 (1951). [DOI] [PubMed] [Google Scholar]
- 96.Chiou KL et al. , High-altitude adaptation and incipient speciation in geladas. bioRxiv, 10.1101/2021.09.01.458582 (2021). [DOI] [Google Scholar]
- 97.Moore CM et al. , Cytogenetic and fertility studies of a rheboon, rhesus macaque (Macaca mulatta) x baboon (Papio hamadryas) cross: further support for a single karyotype nomenclature. Am J Phys Anthropol 110, 119–127 (1999). [DOI] [PubMed] [Google Scholar]
- 98.Chan AH, Jenkins PA, Song YS, Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet 8, e1003090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Browning SR, Browning BL, Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boissinot S, Alvarez L, Giraldo-Ramirez J, Tollis M, Neutral nuclear variation in Baboons (genus Papio) provides insights into their evolutionary and demographic histories. Am J Phys Anthropol 155, 621–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stevison LS et al. , The time scale of recombination rate evolution in great apes. Mol Biol Evol 33, 928–945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O'Leary NA et al. , Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44, D733–745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hinrichs AS et al. , The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34, D590–598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Auton A et al. , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aken BL et al. , Ensembl 2017. Nucleic Acids Res 45, D635–D642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hinch AG et al. , The landscape of recombination in African Americans. Nature 476, 170–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prüfer K et al. , The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prüfer K et al. , A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mafessoni F et al. , A high-coverage Neandertal genome from Chagyrskaya Cave. Proc Natl Acad Sci U S A 117, 15132–15136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Max Planck Institute for Evolutionary Anthropology, Pääbo S, Genotypes of high coverage Neanderthal genomes. http://ftp.eva.mpg.de/neandertal/. [Google Scholar]
- 113.Dobin A, Gingeras TR, Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics 51, 11.14.11–11.14.19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Anders S, Pyl PT, Huber W, HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Law CW, Chen Y, Shi W, Smyth GK, voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ritchie ME et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akdemir D, Okeke U, EMMREML: Fitting mixed models with known covariance structures. R package 3.1, (2015). [Google Scholar]
- 120.Storey JD, Tibshirani R, Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Snyder-Mackler N et al. , Social status alters immune regulation and response to infection in macaques. Science 354, 1041–1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Landry CR et al. , Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171, 1813–1822 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coolon JD, Wittkopp PJ, “cis- and trans-regulation in Drosophila interspecific hybrids” in Polyploid and Hybrid Genomics, Chen ZJ, Birchler JA, Eds. (Wiley-Blackwell, 2013), pp. 37–57. [Google Scholar]
- 124.Mack KL, Campbell P, Nachman MW, Gene regulation and speciation in house mice. Genome Res 26, 451–461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dion-Côté AM, Renaut S, Normandeau E, Bernatchez L, RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol Biol Evol 31, 1188–1199 (2014). [DOI] [PubMed] [Google Scholar]
- 126.Renaut S, Bernatchez L, Transcriptome-wide signature of hybrid breakdown associated with intrinsic reproductive isolation in lake whitefish species pairs (Coregonus spp. Salmonidae). Heredity 106, 1003–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muggeo VMR, segmented: an R package to fit regression models with broken-line relationships. R News 8/1, 20–25 (2008). [Google Scholar]
- 128.Davidson JH, Balakrishnan CN, Gene regulatory evolution during speciation in a songbird. G3 Genes∣Genomes∣Genetics 6, 1357–1364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meiklejohn CD, Coolon JD, Hartl DL, Wittkopp PJ, The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res 24, 84–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dannemann M, Prüfer K, Kelso J, Functional implications of Neandertal introgression in modern humans. Genome Biol 18, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sankararaman S, Mallick S, Patterson N, Reich D, The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Urbut SM, Wang G, Carbonetto P, Stephens M, Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat Genet 51, 187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Friedman JH, Hastie T, Tibshirani R, Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software 33, 22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 134.Zou H, Hastie T, Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 67, 301–320 (2005). [Google Scholar]
- 135.Larsen F, Gundersen G, Lopez R, Prydz H, CpG islands as gene markers in the human genome. Genomics 13, 1095–1107 (1992). [DOI] [PubMed] [Google Scholar]
- 136.Rice P, Longden I, Bleasby A, EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.