Abstract
Artificial intelligence (AI) and machine learning (ML) are increasingly used in radiology research to deal with large and complex imaging data sets. Nowadays, ML tools have become easily accessible to anyone. Such a low threshold to accessibility might lead to inappropriate usage and misinterpretation, without a clear intention. Therefore, ensuring methodological rigor is of paramount importance. Getting closer to the real-world clinical implementation of AI, a basic understanding of the main concepts should be a must for every radiology professional. In this respect, simplified explanations of the key concepts along with pitfalls and recommendations would be helpful for general radiology community to develop and improve their AI mindset. In this work, 22 key issues are reviewed within 3 categories: pre-modeling, modeling, and post-modeling. Firstly, the concept is shortly defined for each issue. Then, related common pitfalls and best practices are provided. Specifically, the issues included in this article are validity of the scientific question, unrepresentative samples, sample size, missing data, quality of reference standard, batch effect, reliability of features, feature scaling, multi-collinearity, class imbalance, data and target leakage, high-dimensional data, optimization, overfitting, generalization, performance metrics, clinical utility, comparison with conventional statistical and clinical methods, interpretability and explainability, randomness, transparent reporting, and sharing data.
Main points
Medical images are complex and include huge amounts of minable data, leading to the rise of a new field called radiomics.
High-dimensional data introduced by radiomics exceed not only human comprehension but also the capabilities of conventional statistical tools. In this respect, artificial intelligence (AI) seems to be an attractive alternative to handle such complex data sets.
Low threshold of accessibility to AI and machine learning (ML) tools might lead to inappropriate usage and misinterpretation, without a clear intention to do so.
Considering the recent advances of AI in the field of radiology, a basic understanding of the key issues of AI and ML should be a must for every radiology professional.
To develop and improve the AI mindset of the radiology community, simplified explanations of the key concepts along with pitfalls and recommendations would be helpful.
Medical images are complex and include huge amounts of minable data.1 This led to the rise of a new research field in medical imaging, namely, radiomics.2,3 Radiomics simply aims to extract high-dimensional data from clinical images, to find clinically meaningful correlations and models.2,3 However, complexity and high dimensionality introduced by radiomics exceed not only human comprehension but also the capabilities of traditional statistical tools. Artificial intelligence (AI) is now regarded as one of the attractive ways to analyze and make predictions on large and heterogeneous data sets as commonly seen with radiomic approaches.
As a subfield of AI, machine learning (ML) learns to automatically extract patterns from complex data to generate predictions on previously unseen instances, even without a theoretical model.4 AI has been increasingly used in radiology research over the years, as evidenced by the exponential increase of publications (Figure 1). Typical ML tasks in radiology can be like predicting whether a pathology or genomic feature is present or not,5-7 determining treatment response status,8,9 segmentation,10,11 image quality enhancement,12,13 contrast medium dose reduction,14 and radiation dose reduction.15,16
Figure 1.
Count of publications on artificial intelligence and machine learning in the field of radiology, between 2000 and 2021. Search syntax was (“artificial intelligence” OR “machine learning”) AND (“radiology” OR “computed tomography” OR “CT” OR “magnetic resonance imaging” OR “MRI” OR “ultrasound” OR “radiography” OR “X-ray”) on PubMed/MEDLINE. The last search date was the 12th of December 2021.
Nowadays, ML tools have become so easily accessible to anyone that ensuring methodological rigor is of paramount importance, especially in medical applications. Such a low threshold to accessibility might lead to inappropriate usage and misinterpretation, without a clear intention to do so. Getting closer and closer than ever to the clinical implementation of AI, a basic understanding of the key methodological concepts should be a must for every professional in the field of radiology.
This article aims to provide a simplified methodological perspective to radiologists about the key concepts, common related pitfalls, and best practices of AI and ML. To do so, 22 important issues are reviewed within 3 consecutive stages: pre-modeling, modeling, and post-modeling (Table 1).
Table 1.
Key concepts covered in this review
| Stages | Key concepts |
|---|---|
| Pre-modeling | Validity of scientific question |
| Unrepresentative samples | |
| Sample size | |
| Missing data | |
| Quality of reference standard | |
| Batch effect | |
| Reliability of features | |
| Feature scaling | |
| Multi-collinearity | |
| Class imbalance | |
| Data and target leakage | |
| High-dimensional data | |
| Modeling | Optimization |
| Overfitting | |
| Generalization | |
| Post-modeling | Performance metrics |
| Clinical utility | |
| Comparison with conventional statistical and clinical methods | |
| Interpretability and explainability | |
| Randomness | |
| Transparent reporting | |
| Sharing data |
Issues related to pre-modeling stage
Validity of scientific question
Concept
As with any scientific work, the true value of AI largely depends on whether a valid scientific clinical question has been formulated.
Pitfalls
Disregarding the real-world representation as a study goal is a very common pitfall, ultimately, resulting in the total failure in clinical utility.
Best practices
Before beginning an AI project, the study’s objective and how it would solve the clinical or scientific questions at hand must be defined very clearly.17 Having too many eligibility criteria generally signals unrealistic projects that are far away from clinical practice and production at scale. An AI project must be clinically meaningful and actionable, that is, its predictions should induce some action by the physicians or patients.18 For instance, a project tries to predict whether a solitary brain mass belongs to lung or breast metastasis. All too often, the model developed works with excellent predictive performance, even with the independent validation data from another institution. Despite being attractive at first glance, the model created seems problematic from a clinical perspective. In contrast, to use such a model in real life, one must question whether the lesion is primary in the first place. Furthermore, one should also question why other types of metastatic lesions are not included. Undoubtedly, the project might have been much more valid and useful if the primary purpose was to discriminate between primary and secondary neoplasms and then the multi-class prediction of metastatic subtypes like breast versus lung versus others.
Unrepresentative samples
Concept
The actual potential of an AI model is not only dependent on the valid scientific question but also on whether one has proper data to answer that question. Data used for AI modeling and future predictions must be representative of real-life scenarios in terms of distribution (Figure 2). The concept applies to both training and test data.
Figure 2.
Simplified illustration of representativeness of data. The unrepresentative sample includes different distributions compared to the target population.
Pitfalls
Focusing on the most common types of instances (e.g., lesions, tumors) and disregarding the uncommon ones that might constitute a considerable proportion is a widespread modeling pitfall. This can also be defined as spectrum bias.19,20 Such distribution differences or shifts are highly likely to cause significant performance problems in future predictions.
Best practices
Uncommon categories should not be ignored. Those can be grouped as a different category (e.g., others) to improve the representativeness of the data used. This issue is rather significant to detect particularly in the test dataset since main conclusions are usually based on the performance of the test data. Stratified sampling can be applied to split the data to preserve the distribution both in the training and test sets. In addition to simple summary statistics of clinical and demographic data, a comparison of the error patterns using the learning curve approach would simplify the identification of the unrepresentative sample problem.
Sample size
Concept
Data size problems affect medical imaging more than any other AI field. This is largely due to (i) the high-dimensional feature space of the medical images in that each pixel or voxel can represent a single feature and (ii) a very high number of hand-crafted radiomic features that can be extracted from original and derivative images.1 Such high dimensionality necessitates enormous data sets for proper training to achieve a particular level of stable performance and very low generalizability error.21-24
Pitfalls
Training a model with an extremely small data set is a common pitfall, leading to overfitting, noise, and outliers (Figure 3).
Figure 3.
Simplified illustration of the influence of data size on the performance of machine learning models.
Best practices
The optimal sample size for a particular AI algorithm applied to medical imaging data is often unknown. Nearly all methods for determining the optimal data size are empirical. A systematic literature search revealed the scarcity of research on data size calculation in ML applied to medical imaging (not only limited to radiologic and nuclear medicine images).25 In this recent analysis, such determination processes relied on (i) model-based considerations26,27 or (ii) generating predictive functions of model strength based on empirical testing at selected sample sizes, in other words, the learning curve-fitting approach.28,29 For tabular data, another general recommendation is to have a data size that is more than 10 times the number of features1,30 or that is with at least 50 samples.25 Unfortunately, gathering large quantities of high-quality medical image data is difficult.31,32 However, there are other ways to mitigate this problem: data augmentation (i.e., transforming original data like rotation, scaling, or generating images with generative adversarial networks, etc.),33,34 transfer learning (i.e., fine-tuning or training previously pre-trained models),35 and federated learning (i.e., developing models across institutions while protecting data privacy).36-38 No matter which technique is used, complex methods like deep learning still require a significant amount of data. To solve this data size problem, some data-efficient neural network techniques are on the horizon such as capsule systems adding viewpoint variance39 and vision transformers having self-attention mechanisms focusing on the most valuable components of the layers.40-42
Missing data
Concept
Most data sets in the real world contain missing data. Missing data can be anything from images with artifacts, missing sequence or phase in multi-parametric evaluations, incomplete features, and lacking clinical information. Missing values are rarely encountered in radiomics because features are computed from images and patients without imaging studies are usually excluded.
Pitfalls
Excluding all samples with the missing data may cause bias in the results.43
Best practices
Many ML algorithms expect complete data and fail to work when the data are incomplete. Having missing values is not necessarily a drawback, needing careful transformations. In addition to deletion methods, the missing data may require statistical imputation that can be simply defined as the replacement of missing data with new values. To impute, various methods have been developed such as mean/mode/median imputation, regression imputation, expectation–maximization, multiple imputation, and so on.43,44 Another alternative would be to use ML algorithms that accept missing values or even better algorithms that automatically detect and deal with missing data. Some algorithms can use the missing value as a unique and different value when building predictive models, such as classification and regression trees. For instance, a well-known and very powerful algorithm called XGBoost takes into consideration of any missing data.45
Quality of reference standard
Concept
In radiology, the reference standard (i.e., label) is generally an accepted test or a group of accepted criteria used in clinical practice. The reference standard may require subjective decisions, resulting in intra-rater and inter-rater inconsistencies. Quality and stability of reference standard should be ensured to achieve a precise evaluation of the proposed ML models and comparison with the other traditional tools.46
Pitfalls
Mislabeling and selecting an unstable reference standard are major problems that need to be avoided in developing AI models.
Best practices
The reference standard that is more insensitive to varying conditions (e.g., the dependency of experience) should be selected if options exist. In this context, intra-rater and inter-rater stability can be studied. Nonetheless, it is still very common to encounter reliability concerns. To alleviate this issue, consensus evaluations (i.e., combining interpretations by more than one evaluator) and voting (i.e., selection based on majority of decisions) should be considered.
Batch effect
Concept
The batch effect can be defined as technical variations that confound the determination of valuable features from data (Figure 4).47 In radiomics, this problem may be caused by the use of different scanners, varying acquisition parameters, and image processing factors.48 Thus, some subgroups might be under-represented, while others are unnecessarily over-represented. This is highly likely to be a problem for multi-center projects. Nonetheless, it can be seen in single-center studies.
Figure 4.
Simplified illustration of the batch effect. Tumor X and tumor Y from 2 different centers are classified according to 2 features as principal components. Please note the change of feature values following batch effect removal. PC, principal component
Pitfalls
Disregarding possible batch effects might inflate cross-validation accuracy which is the standard protocol for classifier evaluation in the absence of independent validation data sets.
Best practices
In practice, the batch effect is inevitable and needs to be dealt with in certain data preparation steps. The voxel size, slice thickness, and convolution kernel are relevant sources of radiomics variability. Data wrangling may include strategies that aim to reduce batch effects. Various techniques have been investigated in radiomics,48-52 of which, the ComBat strategy seems promising.48,49 In simple terms, this method creates a batch-specific transformation to represent the whole radiomic data in a common space lacking center or batch effects caused by scanner models, acquisition protocols, and/or image processing settings.
Reliability of features
Concept
The use of reliable features is important for the reproducibility of models. In radiomics, such concerns can be encountered in image acquisition and reconstruction level (e.g., vendor-based or technical differences),53,54 segmentation level (e.g., intra- and inter-rater segmentation differences, slice selection),55 and computation level (e.g., different mathematical formulas of feature extraction tools, different techniques for normalization, discretization, and resampling) (Figure 5).56
Figure 5.
Segmentation and labeling differences. (a) A low-grade glioma segmented by 2 different radiologists. (b) Image cropping or labeling for the same tumor with different sizes of boxes. Even slight differences in segmentation areas can result in significant changes in feature values or representations.
Pitfalls
Common pitfalls are lack of feature reliability assessment, use of whole data in reliability assessment, building AI models on non-robust features, and not being clear on the computational parameters of the features.
Best practices
Considering the variations in image acquisition and processing in radiomic features, it appears to be a difficult task to obtain constant and stable results. Therefore, a great effort should be exerted to minimize variations. The reliability of features can be assessed using several approaches such as intra- and inter-rater agreement analysis for the detection of segmentation differences,57 repeatability analysis for automatic methods,58 and reproducibility analysis with different image acquisition settings or even using phantom or simulation measurements.54,59 Automated techniques, particularly deep learning-based methods such as U-Net,60 might be a better option for segmentation purposes because it almost completely avoids rater’s variability. However, the generalizability of trained algorithms is currently a major limitation, and applying those algorithms to a different data set might result in complete failure. Regardless of the methodology, such reliability assessments should be performed on the training set without data leakage. For computational reliability, preprocessing steps (e.g., discretization, resampling, standardization, registration, filtering) must be transparently presented.
Feature scaling
Concept
Radiomic features come in widely different value ranges, influencing the performance of some AI algorithms. Feature scaling is the transformation of feature values to a standard range.
Pitfalls
Ignoring feature scaling may lead to overrepresentation or underrepresentation of some features. Another important pitfall is scaling features on the whole data set.
Best practices
Feature scaling matters for many of the algorithms such as support vector machines, k-nearest neighbors, and artificial neural networks. Most of these algorithms are based on the calculation of distance between data points. Feature scaling is also a key part of deep learning-based architectures.61 On the other hand, some other algorithms do not require feature scaling. Most of these are tree-based algorithms such as random forest and XGBoost. The procedure can be done in a couple of ways, for instance, standardization (i.e., z-score normalization), normalization (i.e., min-max normalization), or logarithmic transformation (Figure 6).62 AI practitioners should be cautious while performing feature scaling to avoid data leakage by separately performing the task for training and testing data.
Figure 6.
Simplified illustration of common feature scaling techniques: standardization and normalization.
Multi-collinearity
Concept
Multi-collinearity (i.e., collinearity) is a common statistical problem in which at least 2 variables are dependent upon each other such that one can be linearly predicted from the other with high accuracy.
Pitfalls
Creating models with highly correlated and clustered variables is a common problem in modeling. It is much more problematic in linear modeling.
Best practices
Since many radiomic features can be repetitively extracted using filtered and transformed images, leading to high correlation and clusters, radiomics-based modeling is likely to suffer from multi-collinearity.63 It inflates the variance of the coefficients, particularly in linear models. Nevertheless, non-linear algorithms can also be affected.64 Multi-collinearity might or might not influence the predictive performance of the ML models, rather it may affect the individual dependence on the features and interpretation of the final model. The most common method for detecting collinearity is bivariate correlation. However, bivariate correlation method disregards the collinearity of multiple variables together. In such circumstances, variance inflation factor and tolerance are well-known methods for removing multi-collinear features before modeling.64
Class imbalance
Concept
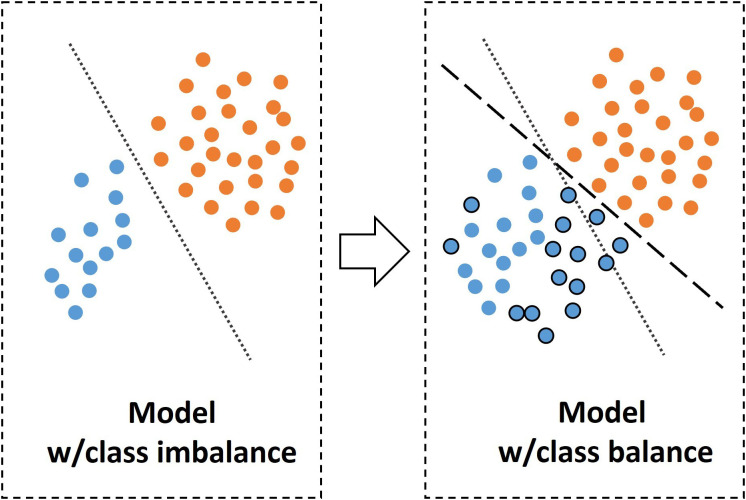
When the number of instances in one target class is higher than the other class or classes, there exists a class imbalance (Figure 7).65 Some ML algorithms are tended to vote for the class that constitutes the majority if a severe class imbalance is present.
Figure 7.
Simplified illustration of modeling with class balance and imbalance. Please note balancing the classes results in a new hyperplane and a different model. w, with.
Pitfalls
Class imbalance can lead to false impression of predictive performance due to bias toward one class. In medicine, another important pitfall is the balancing test set.
Best practices
In ideal conditions, ML algorithms perform better when the classes are equal in the training set.66 However, this is not common in the medical context. To fix the class imbalance problem, data sets can be rebalanced by resampling strategies such as under-sampling or over-sampling.67 Such sampling strategies are recommended only for the training data sets. An alternative approach might be optimization for detecting true-positive or false-negative instances depending on the clinical need, for instance, screening rather than diagnosis.68
Data and target leakage
Concept
Data leakage corresponds to the transfer of data among training, validation, and testing data sets, due to incorrect split of the data (Figure 8). This concept can also be referred to as data snooping bias. Target leakage is a special type of data leakage, and it happens if a model is trained with a feature that will not be available in real unseen data (Figure 9).
Figure 8.
Simplified machine learning pipeline highlighting the recommended and biased ways of dimension reduction along with possible data leakage scenarios. It is usually better to make the data split at the beginning. When needing calculations to determine the optimal parameters for a preprocessing step (e.g., optimal bin-width for radiomic feature extraction), such decisions should be made on training data only and transferred to the other splits. The recommended way for dimension reduction is to perform it within cross-validation folds.
Figure 9.
Simplified illustration of target leakage. Using data unavailable at the prediction time results in inflated and in turn misleading results. w, with; ROC, receiver operating characteristic; TPR, true-positive rate; FPR, false-positive rate.
Pitfalls
Leakage is a simple but major pitfall that must be avoided.69 Otherwise, it might lead to abnormally high performance of the model and significant generalizability issues.
Best practices
Data leakage can occur as early as the first steps of the pipeline. Therefore, the data split must be done at the beginning, before any transformation or processing of the images or data. Data snooping bias occurs when the test set directly or indirectly influences the training process. It should be double-checked whether there are no data from the same patient placed in training, validation, and test sets at the same time. If a model is extremely accurate, one should be suspicious of the results and further evaluate the pipeline for possible data and target leakage. For target leakage, features having a high correlation with the target can be determined with traditional statistical analysis.
High-dimensional data
Concept
High-dimensional data refers to having numerous features or very wide feature space. It forces the algorithms to learn several small feature details, causing overfitting and generalizability problems, particularly in situations in which the number of features is far greater than the number of instances,70-73 which is a common scenario in radiomics.
Pitfalls
Reducing the dimension of whole data is a common pitfall, which needs to be done only in the training set (Figure 8). Also, reducing the dimension before the cross-validation is an incorrect application, leading to bias.74
Best practices
Reducing the dimension of the data called feature selection or dimension reduction is a key step for producing valid and generalizable results. Several rules exist for defining optimal feature numbers for given sample size, but no true evidence exists in the literature. Several methods can be used for dimension reduction.75 Algorithm-based feature selection methods, intra-reader and inter-reader reliability analysis, multi-collinearity analysis, clustering, principal component analysis, and independent component analysis are the most common dimension reduction techniques. Another very good option would be using certain ML algorithms that have inherent feature selection capabilities, for instance, XGBoost. All dimension reduction techniques should be performed in the training phase, without data leakage. It is also best to select features within cross-validation folds, not before cross-validation.74
Issues related to modeling stage
Optimization
Concept
ML algorithms have hyperparameters that can be externally configured to optimize the model to the given data. The process of such configuration is called hyperparameter tuning or optimization. Optimization is a key step in modeling and has a significant impact on the model’s performance.
Pitfalls
The most common pitfall is not performing the optimization at all. Other common problems are the selection of wrong performance metrics to optimize and overfitting.
Best practices
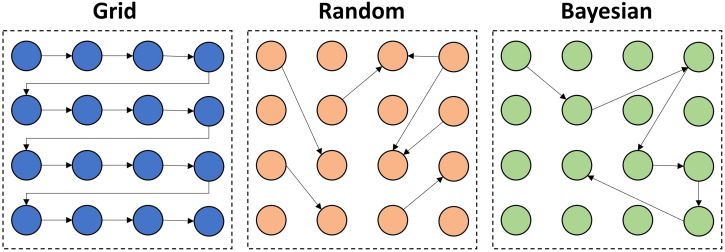
Optimization of the hyperparameters is a key part of any modern ML pipeline. Avoiding this step might result in significant performance problems on predictions of unseen data. Relying on the default configuration of the hyperparameters might be inappropriate for the classification or regression problem at hand. Optimization can be done to increase the performance of certain performance metrics such as sensitivity, specificity, accuracy, or area under the receiver operating characteristic curve depending on the purpose of the study, for instance, the screening of disease. Overfitting in optimization can be avoided by cross-validation or regularization techniques. Random search, grid search, and Bayesian optimization are popular methods that might result in different results (Figure 10).76,77
Figure 10.
Over-simplified illustration of different hyperparameter tuning or optimization techniques. In grid search, all the hyperparameters are searched within a designated range. In random search, randomly selected hyperparameter values are experimented with, which can be limited to a maximum number of experiments. The random search takes less time than the grid search. Both grid search and random search methods are completely uninformed by the previous tuning experiments, which means that previous tuning experiments are not considered in the current or future tuning experiments. Bayesian optimization is a completely different and model-based approach, taking into account the previous experiments to find the best set of hyperparameters.
Overfitting
Concept
Overfitting occurs when an algorithm learns the patterns of limited data too well and is unable to make accurate predictions on unseen and new examples (Figure 11a).78 In other words, it memorizes certain individual patterns and noise related to each instance of the data set upon which the model is developed, making it inflexible to make predictions on real data. Most often, the algorithm is overfitted to the training data set, but overfitting can also occur on validation or test set in case one performs so many experiments on these data sets to find high-performing models. To better understand the concept of overfitting, one should be familiar with the concepts of bias and variance trade-off (Figure 11b).79-80
Figure 11.
Illustration of the concepts related to overfitting and bias-variance trade-off. (a) Learning and memorizing nearly every detail of the data results in overfitting, leading to failure to fit future data points. (b) Bias and variance are 2 sources of error in modeling. Bias is the error that is evident when a complex problem is represented by a simple model due to erroneous assumptions. Variance is the error that appears when a model is sensitive to very small alterations or noise in the data, producing unrealistic patterns. There should be a trade-off between bias and variance to achieve a better generalization beyond the training data. When the variance is high and bias is low, overfitting becomes inevitable.
Pitfalls
Overfitting itself is a common pitfall and needs to be dealt with appropriately. The main reason for high overfitting in medical models is their failure to represent the real-world situations in the data sets due to high degree of heterogeneity of medical data, varying demographic and biologic features even in the same disease state, differences in disease prevalence, vendor-based variations, and so on.81-84
Best practices
Overfitting is a multi-factorial issue and can be amended by following actions including using more training data to eliminate statistical bias, preventing information and target leakage, using resampling techniques, and regularization.85 Among these, reducing the complexity by regularization is probably the most important step in data science. Before introducing the sophisticated techniques for dealing with model complexity, it is noteworthy to mention that starting with simple models such as linear regression, least absolute shrinkage and selection operator (LASSO), and decision trees is generally regarded as the best practice in data science.86 Complex algorithms should only be spared only if the additional performance gain is required or relevant. Model complexity can be changed by adjusting the loss function (L1-L2 regularization, entropy regularization), sampling methods (data augmentation and cross-validation), and adjusting the training phase (dimension reduction or drop-out, adding noise, hyperparameter optimization, etc.). Of the regularization techniques, the most powerful strategies are loss function modifications and dimension reduction (feature selection and drop-out, etc.).
Generalization
Concept
Generalization in ML refers to the ability of the model to adapt to previously unseen and new data. Generalizability can be evaluated using internal or external test sets (Figure 12).
Figure 12.
Simplified illustration of optimal data split and generalizability assessment. Hyperparameter tuning needs a different split, namely, validation set (i.e., tuning set). Please note that it is much more valuable to perform testing both internally and externally in terms of generalizability assessment.
Pitfalls
Using only internal validation is highly likely to result in over-estimated generalization performance. Unrepresentativeness of the test data in terms of prevalence and distribution is a common pitfall.19,87 Inappropriate sampling strategies and data leakage are other common problems. Tuning the hyperparameters on the training data without using a tuning or validation set is another source of generalizability issue.
Best practices
In proper AI-based modeling, the data should include 3 partitions: training set, validation set, and test set. These data partitions are partly or wholly obtained and used with different sampling and validation strategies such as k-fold cross-validation, leave-one-out, hold-out, nested cross-validation, and so on. For the simplified schematic explanations of these methods, the readers are kindly referred to the following references.3,88,89 It is crucial to clearly understand the terms “validation” and “test.” A validation set is used to tune the hyperparameters of the algorithm. A test set is used to measure the generalization performance. The test set can be internal or external. AI models’ performance in the clinical environment might be different than that of the experiments done during development. Therefore, it is important and best to conduct a generalizability assessment with external data obtained directly in the target clinical environment.19,78,81,90,91 To further improve the generalizability of an AI model, additional training rounds and continuous learning capabilities might be incorporated into the model development stage using data from target hospitals and specific clinical environments where the end product will be used. For a true generalizability assessment, an independent test set must correctly represent the actual population of interest, for instance, in terms of disease prevalence and demographics.
Issues related to post-modeling stage
Performance metrics
Concept
Regardless of being a classification or a regression problem, the metrics are essential parts of performance evaluation in every ML pipeline. Appropriate selection of these metrics is important for proper performance evaluation. Due to several reasons such as randomness, data size, data perturbations, and class imbalance, the value of performance metrics is subject to considerable variability.
Pitfalls
Inappropriate selection of metrics and disregarding variability of metrics (i.e., single-point evaluation) are common pitfalls.
Best practices
Performance metrics should be selected based on the characteristics of the data in hand (e.g., class imbalance).92-94 Particularly, in classification tasks, it is important to evaluate confusion matrices that might provide broader insights about the capabilities of the models both in terms of overall performance and class-wise performance. In case of class imbalance, metrics like accuracy might be misleading. On the other hand, there are better metrics that can be used if class imbalance exists such as balanced accuracy, no information rate, the Matthews correlation coefficient, F1 measure, area under the receiver operating characteristic curve, and area under the precision-recall curve. Particularly, if the positive class constitutes the minority class, precision and recall can be used. Otherwise, if the negative class is the majority or there is balance, then, area under the receiver operating characteristic curve might be a much more appropriate metric due to lack of bias to the classes. Performance assessments should represent the variability limits of the study. The confidence interval, standard deviation, and standard error are common indicators of performance variability and should be included in the evaluation process.
Clinical utility
Concept
High predictive performance does not necessarily indicate that an AI model can improve clinical outcomes or does have high clinical utility. It is critical to directly assess the efficacy and utility of AI on clinical outcomes, in addition to its performance.95
Pitfalls
Assessment for clinical utility is often disregarded in ML-based classification tasks.
Best practices
The 2 most common tools to assess clinical utility are calibration statistics96,97 and decision curve analysis.98 Calibration statistics is the process of determining whether the predicted probability scores match the actual probability scores. Well-calibrated models are often much more useful than high-performing binary classification models, giving more insights to clinicians and patients about the probabilities (Figure 13). Decision curve analysis provides evidence about the net benefits of the models, considering both discriminatory predictive performance and calibration.99
Figure 13.
Clinical utility assessment with calibration statistics. (a) Calibration curves: poorly calibrated (i) and well-calibrated (ii) models. (b) Example use case. From the clinical perspective, it is much more useful to get well-calibrated probability scores (e.g., predicted probability of having a renal mass is 91.2%) rather than binary or multi-class predictions (e.g., the patient probably has a renal mass).
Comparison with conventional statistical and clinical methods
Concept
Due to media hype, AI might lead to unrealistic expectations. It should be known that traditional methods can outperform ML.100
Pitfalls
Evaluation of ML modeling without considering the traditional methods would not reflect the actual impact on real-world clinical practice. Disregarding the negative results is another problem while comparing the methods.
Best practices
Comparisons can be made with traditional statistical models such as logistic regression or widely accepted clinical tools like expert readings. Potential gains should be meticulously evaluated in terms of discriminatory performance and clinical utility. It is noteworthy to mention that negative results are also as valuable as the positive ones.101
Interpretability and explainability
Concept
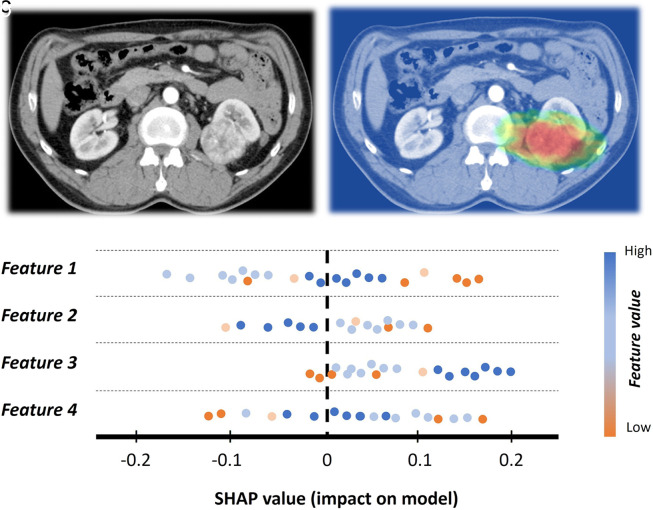
ML algorithms are usually perceived as black boxes, that is, lacking interpretability and explainability for the process that takes place from input to output.102 Interpretability is the ability to understand the workings of the ML method. Explainability is a kind of transparency in the processes when the ML model decides. Adding interpretability and explainability to the ML might provide trust and enhanced control (Figure 14).
Figure 14.
Example of interpretability in a patient with renal cell cancer. (a), Input image. (b) The input image is improved with activation mapping, adding some degree of interpretability to the model. It might also be useful in getting the attention of human readers. (c) Shapley additive explanations (SHAP) plot example for explainability. It simply summarizes how low or high feature values impact the model performance.
Pitfalls
Particularly in healthcare, failure in achieving improved interpretability and explainability will limit the potential impact of the model created.
Best practices
Some ML algorithms have the inherent characteristics of interpretability and explainability, for instance, linear regression, logistic regression, decision trees, and generalized linear models. However, the use of these algorithms may not be the best solution to capture the actual complexity of real-world problems. As ML models grow in complexity, generating interpretable and explainable models becomes increasingly difficult. Neural networks or ensemble models are complex models and need further tools to achieve interpretability and explainability. These goals in ML can be approached in 2 broad ways: local (i.e., explaining based on individual features such as local interpretable model-agnostic explanations (LIME)) and global (i.e., explaining based on a certain collection of features such as partial dependence plots (PDP)). Common methods for interpretability and explainability are permutation feature importance,103 PDP,104 LIME,105 Shapley additive explanations (SHAP),106 and activation atlases.107 Still, significant effort goes into creating new approaches to deal with this problem.
Randomness
Concept
In data science, the results are largely dependent on some random components such as the order of the instances, selection of samples or subsamples, initial weights that initialize the algorithms, and so on.
Pitfalls
If the randomness is not considered very well, it is highly likely to get different results even with the same data and settings, leading to uncertainty of the results (Figure 15).108 Not considering the possible effects of the uncertainty brought by randomness in pre-modeling and modeling is a major problem in terms of the reproducibility of ML models.
Figure 15.
Over-simplified illustration for the influence of randomness in machine learning modeling. Unstable (a) and stable (b) performances using different random states. Exp, experiment; w, with.
Best practices
Fixing the random number generator’s seed before model development is a key step to achieving reproducible results. It should be a default part of each experiment from sampling to modeling. Because of the dependency of randomness, there will be a range of several possible models, not a single model. Moreover, simply changing a seed value to a different one can drastically change the performance of the models created.109 Therefore, the performance should be evaluated within a range, not a single point value.
Transparent reporting
Concept
AI-based modeling is subject to considerable reproducibility and replicability issues.108 Considering the heavy parameter burden in AI models, slight changes in values can result in complete failure. The quality and replicability of the studies can be improved and maintained by systematic and transparent reporting strategies.
Pitfalls
Incomplete or non-systematic reporting of study parameters is the most common problem. Commonly missed parts are preprocessing details (e.g., image resizing, cropping, standardization, feature extraction parameters, etc.), model training details, and demographic and clinical characteristics.110
Best practices
Using checklists or frameworks is the most important best practice in transparent reporting. Some well-known frameworks are Radiomics Quality Score,2 checklist for artificial intelligence in medical imaging (CLAIM),17 transparent reporting of a multivariable prediction model for individual prognosis or diagnosis,111 and prediction model risk of bias assessment tool.112 Among them, CLAIM is specifically designed for medical imaging-related AI.17 There are also other detailed frameworks that can be used to improve the methodologic transparency of AI and ML-related works.20,88
Sharing data
Concept
AI research might be almost useless unless it is reusable, reproducible, and replicable.108 Therefore, sharing data in AI is important. Data sharing provides a basis for communication with a wider scientific community that can efficiently build upon the proposed project.
Pitfalls
No attempt to share data is a commonly occurring problem in AI studies.
Best practices
Data in AI can simply be grouped into pre-modeling (raw images, processed images, feature data, etc.) and post-modeling data (scripts, model files, etc.). There are several ways to share pre-modeling data, for instance, data pooling with anonymization and federated learning. AI teams should at least consider sharing post-modeling data such as scripts and resultant model files and using online repositories. These are important for better understanding, external validation, and improvement of the methodology.
Final Thoughts
AI and ML in medical imaging have several different methodological aspects, with their challenges and pitfalls. Even though it is not possible to cover all key concepts, common pitfalls, and best practices in a review article, the issues covered here will improve the AI mindset of the radiology community. As the community gains more awareness and experience on key methodological issues and related pitfalls, it will be easier to evaluate, compare, and select the most reasonable solutions for the radiologic problems in hand: ML versus traditional methods versus combination of techniques.
Footnotes
Acknowledgments The author would like to express his gratitude to the editor-in-chief, Mustafa Seçil, for his kind invitation to prepare this review. The author would also like to extend his gratitude to Aytül Hande Yardımcı and Ece Ateş Kuş for their internal peer-review and valuable recommendations.
References
- 1. Gillies RJ, Kinahan PE, Hricak HH. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563 577. 10.1148/radiol.2015151169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambin P, Leijenaar RTH, Deist TM.et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749 762. 10.1038/nrclinonc.2017.141) [DOI] [PubMed] [Google Scholar]
- 3. Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. 2019;25(6):485 495. 10.5152/dir.2019.19321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects, science. American Association for the Advancement of Science; 2015;349(6245):255–260. 10.1126/science.aaa8415) [DOI] [PubMed] [Google Scholar]
- 5. Pasquini L, Napolitano A, Lucignani M.et al. AI and high-grade glioma for diagnosis and outcome prediction: do all machine learning models perform equally well? Front Oncol. 2021;11:601425. 10.3389/fonc.2021.601425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xv Y, Lv F, Guo H.et al. Machine learning-based CT radiomics approach for predicting WHO/ISUP nuclear grade of clear cell renal cell carcinoma: an exploratory and comparative study. Insights Imaging. 2021;12(1):170. 10.1186/s13244-021-01107-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Izquierdo C, Casas G, Martin-Isla C.et al. Radiomics-based classification of left ventricular non-compaction, hypertrophic cardiomyopathy, and dilated cardiomyopathy in cardiovascular magnetic resonance. Front Cardiovasc Med. 2021;8:764312. 10.3389/fcvm.2021.764312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng J, Huang J, Huang G, Zhang J. Predicting the initial treatment response to transarterial chemoembolization in intermediate-stage hepatocellular carcinoma by the integration of radiomics and deep learning. Front Oncol. 2021;11:730282. 10.3389/fonc.2021.730282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng Y, Chen L, Liu M, Wu J, Yu R, Lv F. Prediction of clinical outcome for high-intensity focused ultrasound ablation of uterine leiomyomas using multiparametric MRI radiomics-based machine leaning model. Front Oncol. 2021;11:618604. 10.3389/fonc.2021.618604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosas-Gonzalez S, Birgui-Sekou T, Hidane M, Zemmoura I, Tauber C. Asymmetric ensemble of asymmetric U-net models for brain tumor segmentation with uncertainty estimation. Front Neurol. 2021;12:609646. 10.3389/fneur.2021.609646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baressi Šegota S, Lorencin I, Smolić K.et al. Semantic segmentation of urinary bladder cancer masses from CT images: a transfer learning approach. Biology. 2021;10(11):1134. 10.3390/biology10111134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bash S, Johnson B, Gibbs W, Zhang T, Shankaranarayanan A, Tanenbaum LN. Deep learning image processing enables 40% faster spinal MR scans which match or exceed quality of standard of care: a prospective multicenter multireader study. Clin Neuroradiol. 2022;32(1):197 203. 10.1007/s00062-021-01121-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen XV, Oztek MA, Nelakurti DD.et al. Applying artificial intelligence to mitigate effects of patient motion or other complicating factors on image quality. Top Magn Reson Imaging. 2020;29(4):175 180. 10.1097/RMR.0000000000000249) [DOI] [PubMed] [Google Scholar]
- 14. Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging. 2018;48(2):330 340. 10.1002/jmri.25970) [DOI] [PubMed] [Google Scholar]
- 15. Nagayama Y, Sakabe D, Goto M.et al. Deep learning–based reconstruction for lower-dose pediatric CT: Technical principles, image characteristics, and clinical implementations. radiographics. Radiol Soc North America. 2021;41(7):1936 1953. 10.1148/rg.2021210105) [DOI] [PubMed] [Google Scholar]
- 16. Immonen E, Wong J, Nieminen M.et al. The use of deep learning towards dose optimization in low-dose computed tomography: a scoping review. Radiography (Lond). 2022;28(1):208 214. 10.1016/j.radi.2021.07.010) [DOI] [PubMed] [Google Scholar]
- 17. Mongan J, Moy L, Kahn CE. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;2(2):e200029. 10.1148/ryai.2020200029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seneviratne MG, Shah NH, Chu L. Bridging the implementation gap of machine learning in healthcare. BMJ Innov. 2020;6(2):45 47. 10.1136/bmjinnov-2019-000359) [DOI] [Google Scholar]
- 19. Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286(3):800 809. 10.1148/radiol.2017171920) [DOI] [PubMed] [Google Scholar]
- 20. England JR, Cheng PM. Artificial intelligence for medical image analysis: a guide for authors and reviewers. AJR Am J Roentgenol. 2019;212(3):513 519. 10.2214/AJR.18.20490) [DOI] [PubMed] [Google Scholar]
- 21. Gorban AN, Tyukin IY. Blessing of dimensionality: mathematical foundations of the statistical physics of data. Trans R Soc Math Phys Eng Sci. Royal Society. 2018;376(2118):20170237. 10.1098/rsta.2017.0237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell Syst. 2009;24(2):8 12. 10.1109/MIS.2009.36) [DOI] [Google Scholar]
- 23. Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med. 2016;375(13):1216 1219. 10.1056/NEJMp1606181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raudys SJ, Jain AK. Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Trans Pattern Anal Mach Intell. 1991;13(3):252 264. 10.1109/34.75512) [DOI] [Google Scholar]
- 25. Balki I, Amirabadi A, Levman J.et al. Sample-size determination methodologies for machine learning in medical Imaging Research: a systematic review. Can Assoc Radiol J. 2019;70(4):344 353. 10.1016/j.carj.2019.06.002) [DOI] [PubMed] [Google Scholar]
- 26. Baum EB, Haussler D. What size net gives valid generalization? Neural Comput. 1989;1(1):151 160. 10.1162/neco.1989.1.1.151) [DOI] [Google Scholar]
- 27. Vapnik VN, Chervonenkis A. On the uniform convergence of relative frequencies of events to their probabilities. In: Vovk V, Papadopoulos H, Gammerman A.eds. Meas Complex Festschr Alexey Chervonenkis. Cham: Springer International Publishing; 2015:11 30. 10.1007/978-3-319-21852-6_3) [DOI] [Google Scholar]
- 28. Perlich C. Learning curves in machine learning. In: Sammut C, Webb GI.eds. Encycl Mach Learn. Boston, MA: Springer US; 2010:577 580. 10.1007/978-0-387-30164-8_452) [DOI] [Google Scholar]
- 29. Hoiem D, Gupta T, Li Z, Shlapentokh-Rothman MM. Learning curves for analysis of deep networks. Cs Stat. 2021. Available at: http://arxiv.org/abs/2010.11029. Accessed November 1, 2021. [Google Scholar]
- 30. Sollini M, Cozzi L, Antunovic L, Chiti A, Kirienko M. PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep. 2017;7(1):358. 10.1038/s41598-017-00426-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ithapul VK, Singh V, Okonkwo O, Johnson SC. Randomized denoising autoencoders for smaller and efficient imaging based AD clinical trials. Med Image Comput Comput Assist Interv. 2014;17(2):470 478. 10.1007/978-3-319-10470-6_59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moody A. Perspective: the big picture. Nature. 2013;502(7473):S95. 10.1038/502s95a) [DOI] [PubMed] [Google Scholar]
- 33. Shin H-C, Tenenholtz NA, Rogers JK.et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. Cs Stat. 2018. Available at: http://arxiv.org/abs/1807.10225. Accessed November 1, 2021. [Google Scholar]
- 34. Tucker A, Wang Z, Rotalinti Y, Myles P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. npj Digit Med. 2020;3(1):147. 10.1038/s41746-020-00353-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. Lecture Notes in Computer Science. 2018. 10.1007/978-3-030-01424-7_27) [DOI] [Google Scholar]
- 36. Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Brainlesion. 2019;11383:ArXiv181004304. 10.1007/978-3-030-11723-8_9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheller MJ, Edwards B, Reina GA.et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep. 2020;10(1):12598. 10.1038/s41598-020-69250-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang K, Balachandar N, Lam C.et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc. 2018;25(8):945 954. 10.1093/jamia/ocy017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sabour S, Frosst N, Hinton GE. Dynamic routing between capsules. Proc 31st Int Conf Neural Inf Process Syst. Red Hook, NY, USA: Curran Associates Inc.; 2017:3859 3869. [Google Scholar]
- 40. Matsoukas C, Haslum JF, Söderberg M, Smith K. Is it time to replace CNNs with transformers for medical images? CS. 2021. Available at: http://arxiv.org/abs/2108.09038. Accessed November 1, 2021. [Google Scholar]
- 41. Dosovitskiy A, Beyer L, Kolesnikov A.et al. An image is worth 16x16 words: transformers for image recognition at scale. CS. 2021. Available at: http://arxiv.org/abs/2010.11929. Accessed November 1, 2021. [Google Scholar]
- 42. Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M. Transformers in vision: A survey. ACM Comput Surv. 2021. 10.1145/3505244) [DOI] [Google Scholar]
- 43. Donders ART, van der Heijden GJMG, Stijnen T, Moons KGM. Review: a gentle introduction to imputation of missing values. J Clin Epidemiol. 2006;59(10):1087 1091. 10.1016/j.jclinepi.2006.01.014) [DOI] [PubMed] [Google Scholar]
- 44. Kang H. The prevention and handling of the missing data. Korean J Anesthesiol. 2013;64(5):402 406. 10.4097/kjae.2013.64.5.402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Proc 22nd ACM SIGKDD Int Conf Knowl Discov Data Min. New York, NY: ACM; 2016:785 794. 10.1145/2939672.2939785) [DOI] [Google Scholar]
- 46. Krause J, Gulshan V, Rahimy E.et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125(8):1264 1272. 10.1016/j.ophtha.2018.01.034) [DOI] [PubMed] [Google Scholar]
- 47. Lazar C, Meganck S, Taminau J.et al. Batch effect removal methods for microarray gene expression data integration: a survey. Brief Bioinform. 2013;14(4):469 490. 10.1093/bib/bbs037) [DOI] [PubMed] [Google Scholar]
- 48. Ligero M, Jordi-Ollero O, Bernatowicz K.et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur Radiol. 2021;31(3):1460 1470. 10.1007/s00330-020-07174-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology. 2019;291(1):53 59. 10.1148/radiol.2019182023) [DOI] [PubMed] [Google Scholar]
- 50. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat (Oxf Engl). 2007;8(1):118 127. 10.1093/biostatistics/kxj037) [DOI] [PubMed] [Google Scholar]
- 51. Lucia F, Visvikis D, Vallières M.et al. External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2019;46(4):864 877. 10.1007/s00259-018-4231-9) [DOI] [PubMed] [Google Scholar]
- 52. Chatterjee A, Vallières M, Dohan A.et al. Creating robust predictive radiomic models for data From independent institutions using normalization. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):210 215. 10.1109/TRPMS.2019.2893860) [DOI] [Google Scholar]
- 53. Berenguer R, Pastor-Juan MDR, Canales-Vázquez J.et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288(2):407 415. 10.1148/radiol.2018172361) [DOI] [PubMed] [Google Scholar]
- 54. Meyer M, Ronald J, Vernuccio F.et al. Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology. 2019;293(3):583 591. 10.1148/radiol.2019190928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kocak B, Ates E, Durmaz ES, Ulusan MB, Kilickesmez O. Influence of segmentation margin on machine learning-based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol. 2019;29(9):4765 4775. 10.1007/s00330-019-6003-8) [DOI] [PubMed] [Google Scholar]
- 56. Moradmand H, Aghamiri SMR, Ghaderi R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys. 2020;21(1):179 190. 10.1002/acm2.12795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kocak B, Durmaz ES, Kaya OK, Ates E, Kilickesmez O. Reliability of single-slice–based 2D CT texture analysis of renal masses: influence of intra- and interobserver manual segmentation variability on radiomic feature reproducibility. AJR Am J Roentgenol. 2019;213(2):377 383. 10.2214/AJR.19.21212) [DOI] [PubMed] [Google Scholar]
- 58. Estrada S, Lu R, Conjeti S.et al. FatSegNet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal Dixon MRI. Magn Reson Med. 2020;83(4):1471 1483. 10.1002/mrm.28022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jha AK, Mithun S, Jaiswar V.et al. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci Rep. 2021;11(1):2055. 10.1038/s41598-021-81526-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. CS. 2015. 10.1007/978-3-319-24574-4_28) [DOI] [Google Scholar]
- 61. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. CS. 2015. Available at: http://arxiv.org/abs/1502.03167. Accessed November 5, 2021. [Google Scholar]
- 62. KumarSingh B, Verma K, Thoke S. Investigations on impact of feature normalization techniques on classifier’s performance in breast tumor classification. Int J Comput Appl. 2015;116:11 15. 10.5120/20443-2793) [DOI] [Google Scholar]
- 63. van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11(1):91. 10.1186/s13244-020-00887-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Garg A, Tai K. Comparison of statistical and machine learning methods in modelling of data with multicollinearity. Int J Modell Identif Control. 2013;18(4). 10.1504/IJMIC.2013.053535) [DOI] [Google Scholar]
- 65. He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263 1284. 10.1109/TKDE.2008.239) [DOI] [Google Scholar]
- 66. Haixiang G, Yijing L, Shang J, Mingyun G, Yuanyue H, Bing G. Learning from class-imbalanced data: review of methods and applications. Expert Syst Appl. 2017;73:220 239. 10.1016/j.eswa.2016.12.035) [DOI] [Google Scholar]
- 67. Storkey A. When training and test sets are different: characterizing learning transfer. Dataset shift. Mach Learn. 2009:3 28. 10.7551/mitpress/9780262170055.003.0001) [DOI] [Google Scholar]
- 68. Bae SH, Yoon KJ. Polyp detection via imbalanced learning and discriminative feature learning. IEEE Trans Med Imaging. 2015;34(11):2379 2393. 10.1109/TMI.2015.2434398) [DOI] [PubMed] [Google Scholar]
- 69. Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46(13):2638 2655. 10.1007/s00259-019-04391-8) [DOI] [PubMed] [Google Scholar]
- 70. Kristensen VN, Lingjærde OC, Russnes HG, Vollan HKM, Frigessi A, Børresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014;14(5):299 313. 10.1038/nrc3721) [DOI] [PubMed] [Google Scholar]
- 71. Clarke R, Ressom HW, Wang A.et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8(1):37 49. 10.1038/nrc2294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jain AK, Duin RPW, Mao J. Statistical pattern recognition: a review. IEEE Trans Pattern Anal Mach Intell. 2000;22(1):4 37. 10.1109/34.824819) [DOI] [Google Scholar]
- 73. Ferté C, Trister AD, Huang E.et al. Impact of bioinformatic procedures in the development and translation of high-throughput molecular classifiers in oncology. Clin Cancer Res. 2013;19(16):4315 4325. 10.1158/1078-0432.CCR-12-3937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Demircioğlu A. Measuring the bias of incorrect application of feature selection when using cross-validation in radiomics. Insights Imaging. 2021;12(1):172. 10.1186/s13244-021-01115-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157 1182. [Google Scholar]
- 76. Feurer M, Hutter F. Hyperparameter optimization. In: Hutter F, Kotthoff L, Vanschoren J.eds. Autom Mach Learn Methods Syst Chall. Cham: Springer International Publishing; 2019:3 33. 10.1007/978-3-030-05318-5_1) [DOI] [Google Scholar]
- 77. Yu T, Zhu H. Hyper-parameter optimization: a review of algorithms and applications. Cs Stat; 2020. Available at: http://arxiv.org/abs/2003.05689. Accessed November 17, 2021. [Google Scholar]
- 78. Mutasa S, Sun S, Ha R. Understanding artificial intelligence based radiology studies: what is overfitting? Clin Imaging. 2020;65:96 99. 10.1016/j.clinimag.2020.04.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Domingos P. A few useful things to know about machine learning. Commun ACM. 2012;55(10):78 87. 10.1145/2347736.2347755) [DOI] [Google Scholar]
- 80. James G, Hastie T. Generalizations of the Bias/Variance Decomposition for Prediction Error; 1997. [Google Scholar]
- 81. Park SH, Choi J, Byeon JS. Key principles of clinical validation, device approval, and insurance coverage decisions of artificial intelligence. Korean J Radiol. 2021;22(3):442 453. 10.3348/kjr.2021.0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17(1):195. 10.1186/s12916-019-1426-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ghassemi M, Naumann T, Schulam P, Beam AL, Chen IY, Ranganath R. Practical guidance on artificial intelligence for health-care data. Lancet Digit Health. 2019;1(4):e157 e159. 10.1016/S2589-7500(19)30084-6) [DOI] [PubMed] [Google Scholar]
- 84. Willemink MJ, Koszek WA, Hardell C.et al. Preparing medical imaging data for machine learning. Radiology. 2020;295(1):4 15. 10.1148/radiol.2020192224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kernbach JM, Staartjes VE. Machine learning-based clinical prediction modeling – A practical guide for clinicians. Cs Stat; 2020. Available at: http://arxiv.org/abs/2006.15069. Accessed November 28, 2021. [Google Scholar]
- 86. Claeskens G, Hjort NL. Model Selection and Model Averaging. Cambridge: Cambridge University Press; 2008. 10.1017/CBO9780511790485) [DOI] [Google Scholar]
- 87. Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20(3):405 410. 10.3348/kjr.2019.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kocak B, Kus EA, Kilickesmez O. How to read and review papers on machine learning and artificial intelligence in radiology: a survival guide to key methodological concepts. Eur Radiol. 2021;31(4):1819 1830. 10.1007/s00330-020-07324-4) [DOI] [PubMed] [Google Scholar]
- 89. Kocak B, Durmaz ES, Erdim C, Ates E, Kaya OK, Kilickesmez O. Radiomics of renal masses: systematic review of reproducibility and validation strategies. AJR Am J Roentgenol. 2020;214(1):129 136. 10.2214/AJR.19.21709) [DOI] [PubMed] [Google Scholar]
- 90. Bluemke DA, Moy L, Bredella MA.et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the radiology editorial board. Radiology. Radiol Soc North America; 2020;294(3):487 489. 10.1148/radiol.2019192515) [DOI] [PubMed] [Google Scholar]
- 91. Nsoesie EO. Evaluating artificial intelligence applications in clinical settings. JAMA Netw Open. 2018;1(5):e182658. 10.1001/jamanetworkopen.2018.2658) [DOI] [PubMed] [Google Scholar]
- 92. Daskalaki S, Kopanas I, Avouris N. Evaluation of classifiers for an uneven class distribution problem. Appl Artif Intell. 2006;20(5):381 417. 10.1080/08839510500313653) [DOI] [Google Scholar]
- 93. Luque A, Carrasco A, Martín A, de las Heras A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019;91:216 231. 10.1016/j.patcog.2019.02.023) [DOI] [Google Scholar]
- 94. Jiao Y, Du P. Performance measures in evaluating machine learning based bioinformatics predictors for classifications. Quant Biol. 2016;4(4):320 330. 10.1007/s40484-016-0081-2) [DOI] [Google Scholar]
- 95. Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK. SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26(9):1364 1374. 10.1038/s41591-020-1034-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dankers FJWM, Traverso A, Wee L, van Kuijk SMJ. Prediction modeling methodology. In: Kubben P, Dumontier M, Dekker A.eds. Fundam Clin Data Sci. Cham (CH): Springer; 2019. Available at: http://www.ncbi.nlm.nih.gov/books/NBK543534/ Accessed November 30, 2021. [PubMed] [Google Scholar]
- 97. Bella A, Ferri C, Hernández-orallo J, Ramírez-quintana MJ. Calibration of Machine Learning Models. [Google Scholar]
- 98. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565 574. 10.1177/0272989X06295361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Vickers AJ, van Calster B, Steyerberg EW. A simple, step-by-step guide to interpreting decision curve analysis. Diagn Progn Res. 2019;3:18. 10.1186/s41512-019-0064-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Austin PC, Tu JV, Lee DS. Logistic regression had superior performance compared with regression trees for predicting in-hospital mortality in patients hospitalized with heart failure. J Clin Epidemiol. 2010;63(10):1145 1155. 10.1016/j.jclinepi.2009.12.004) [DOI] [PubMed] [Google Scholar]
- 101. Taghavi M, Staal FC, Simões R.et al. CT radiomics models are unable to predict new liver metastasis after successful thermal ablation of colorectal liver metastases. Acta Radiol. 2021:2841851211060437. 10.1177/02841851211060437) [DOI] [PubMed] [Google Scholar]
- 102. Castelvecchi D. Can we open the black box of AI? Nature. 2016;538(7623):20 23. 10.1038/538020a) [DOI] [PubMed] [Google Scholar]
- 103. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340 1347. 10.1093/bioinformatics/btq134) [DOI] [PubMed] [Google Scholar]
- 104. Greenwell BM. Pdp: an R package for constructing partial dependence plots. R J. 2017;9(1):421 436. 10.32614/RJ-2017-016) [DOI] [Google Scholar]
- 105. Zafar MR, Khan NM. DLIME: A deterministic local interpretable model-agnostic explanations approach for computer-aided diagnosis systems. Cs Stat; 2019. Available at: http://arxiv.org/abs/1906.10263. Accessed December 28, 2021. [Google Scholar]
- 106. Nohara Y, Matsumoto K, Soejima H, Nakashima N. Explanation of machine learning models using improved Shapley additive explanation. Proc 10th ACM Int Conf Bioinforma Comput Biol Health Inform. New York, NY, USA. Association for Computing Machinery; 2019:546. 10.1145/3307339.3343255) [DOI] [Google Scholar]
- 107. Sivaramakrishnan R, Antani S, Xue Z, Candemir S, Jaeger S, Thoma GR. Visualizing abnormalities in chest radiographs through salient network activations in Deep Learning. IEEE Life Sci Conf LSC; 2017;2017:71 74. 10.1109/LSC.2017.8268146) [DOI] [Google Scholar]
- 108. Beam AL, Manrai AK, Ghassemi M. Challenges to the reproducibility of machine learning models in health care. JAMA. 2020;323(4):305 306. 10.1001/jama.2019.20866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Henderson P, Islam R, Bachman P, Pineau J, Precup D, Meger D. Deep reinforcement learning that matters. AAAI. Proc AAAI Conf Artif Intell. 2018;32(1). 10.1609/aaai.v32i1.11694) [DOI] [Google Scholar]
- 110. Roberts M, Driggs D, Thorpe M.et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3(3):199 217. 10.1038/s42256-021-00307-0) [DOI] [Google Scholar]
- 111. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350(jan07 4):g7594. 10.1136/bmj.g7594) [DOI] [PubMed] [Google Scholar]
- 112. Wolff RF, Moons KGM, Riley RD.et al. PROBAST: A Tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170(1):51 58. 10.7326/M18-1376) [DOI] [PubMed] [Google Scholar]



 Content of this journal is licensed under a
Content of this journal is licensed under a