ABSTRACT
Modification of the hepatitis C virus (HCV) positive-strand RNA genome by N6-methyladenosine (m6A) regulates the viral life cycle. This life cycle takes place solely in the cytoplasm, while m6A addition on cellular mRNA takes place in the nucleus. Thus, the mechanisms by which m6A is deposited on the viral RNA have been unclear. In this work, we find that m6A modification of HCV RNA by the m6A-methyltransferase proteins methyltransferase-like 3 and 14 (METTL3 and METTL14) is regulated by Wilms’ tumor 1-associating protein (WTAP). WTAP, a predominantly nuclear protein, is an essential member of the cellular mRNA m6A-methyltransferase complex and known to target METTL3 to mRNA. We found that HCV infection induces localization of WTAP to the cytoplasm. Importantly, we found that WTAP is required for both METTL3 interaction with HCV RNA and m6A modification across the viral RNA genome. Further, we found that WTAP, like METTL3 and METTL14, negatively regulates the production of infectious HCV virions, a process that we have previously shown is regulated by m6A. Excitingly, WTAP regulation of both HCV RNA m6A modification and virion production was independent of its ability to localize to the nucleus. Together, these results reveal that WTAP is critical for HCV RNA m6A modification by METTL3 and METTL14 in the cytoplasm.
IMPORTANCE Positive-strand RNA viruses such as HCV represent a significant global health burden. Previous work has described that HCV RNA contains the RNA modification m6A and how this modification regulates viral infection. Yet, how this modification is targeted to HCV RNA has remained unclear due to the incompatibility of the nuclear cellular processes that drive m6A modification with the cytoplasmic HCV life cycle. In this study, we present evidence for how m6A modification is targeted to HCV RNA in the cytoplasm by a mechanism in which WTAP recruits the m6A-methyltransferase METTL3 to HCV RNA. This targeting strategy for m6A modification of cytoplasmic RNA viruses is likely relevant for other m6A-modified positive-strand RNA viruses with cytoplasmic life cycles such as enterovirus 71 and SARS-CoV-2 and provides an exciting new target for potential antiviral therapies.
KEYWORDS: HCV, N6-methyladenosine, WTAP, METTL3, METTL14, RNA modification
INTRODUCTION
Hepatitis C virus (HCV) is a positive-sense single-stranded RNA (ssRNA) virus that represents a significant global health burden with over 1.5 million new infections and 400,000 estimated disease-related deaths annually (1). The ~9.6-kb RNA genome of HCV is translated in an internal ribosome entry site (IRES)-dependent manner as a single polyprotein, which is then cleaved by host and viral proteases into 10 individual viral proteins (2). These viral proteins include Core, the viral capsid protein that interacts with HCV RNA for virion production; NS5A, a key coordinator of viral RNA replication; and NS5B, the virally encoded RNA-dependent RNA-polymerase (RdRp) (2). As HCV is a positive-sense ssRNA virus, the viral RNA genome serves not only as the mRNA template for the translation of the viral proteins, but also as the template for RNA replication, and as the genetic material that is packaged into virions. Thus, spatial and temporal regulation of the viral genome is essential for successful viral replication (3). Indeed, the HCV RNA genome is regulated by several RNA elements such as miRNAs, secondary structures, and RNA-binding proteins (4). In addition, we have described previously how the RNA modification N6-methyladenosine (m6A) plays a crucial role in regulating the HCV life cycle (5).
The RNA modification m6A has now been shown to regulate infection by many viruses, through effects mediated by its presence on both viral RNA and cellular RNA (5–8). m6A is the most prevalent eukaryotic internal mRNA modification and regulates many aspects of RNA biology, such as mRNA stability, mRNA translation, and controlling interactions with RNA binding proteins (9–12). The addition of m6A to mRNA, which occurs within a consensus sequence motif, DRACH (D = G/A/U, R = G/A, and H = U/C/A), is catalyzed by an enzymatic protein complex made up of the enzyme methyltransferase-like 3 and its interacting cofactor methyltransferase-like 14 (METTL3 + 14), as well as accessory proteins, such as Wilms’ tumor 1-associating protein (WTAP), which colocalize at nuclear speckles (13–17). WTAP is essential for the function, localization, and RNA targeting of the m6A-methyltransferase complex and as such acts as a “central coordinator” of m6A (13, 16, 18). In this role, WTAP interacts with several proteins that influence the targeting of the m6A-methyltransferase complex to specific subcellular locations and mRNAs (13, 16, 18–20). RNA modification with m6A can be a reversible process with the removal of m6A from mRNA catalyzed by enzymes such as fat mass and obesity-associated protein (FTO) (21). Taken together, these m6A regulatory proteins have been shown to regulate diverse aspects of RNA virus infection, such as innate immune evasion, viral translation, and packaging of viral RNA into virions (5, 22–28). This regulation can also occur at the level of the host through m6A-mediated regulation of innate immunity or viral host factors (7, 8, 29, 30). During HCV infection, we previously found that the viral RNA genome is modified by m6A at multiple genomic sites and is bound by the known cellular m6A-binding YTHDF proteins (5, 31). Further, we found that m6A within the coding region of the HCV E1 gene negatively regulates viral particle production by preventing the interaction of the viral Core protein with the viral RNA (5). Others have since demonstrated how m6A modification of HCV RNA at other sites within the genome is important for viral RNA translation by enabling the recruitment of host translation factors or for promoting infection by shielding viral RNA from immune sensing by the RNA binding protein RIG-I (22, 23, 32).
The molecular mechanism of how the m6A-methyltransferase complex is targeted to the HCV RNA for m6A modification is still unclear (5). This is because the addition of m6A to cellular mRNA by METTL3 + 14 occurs in concert with RNA polymerase II-driven transcription in the nucleus, while HCV RNA replication is mediated by the RdRp NS5B and takes place in the cytoplasm (2, 3, 33, 34). Thus, m6A modification of HCV RNA by the m6A-methyltransferase complex must be occurring in a noncanonical manner in the cytoplasm. While WTAP and other members of the METTL3 + 14 m6A-methyltransferase complex are predominately localized to the nucleus, we and others have previously shown by biochemical fractionation that these proteins can be detected in the cytoplasm (5, 35). Further, METTL3 m6A-modification-independent functions in the cytoplasm have been described (36). In fact, when the m6A-methyltransferase complex member ZC3H13 is depleted, biochemical fractionation and immunofluorescence microscopy revealed that multiple members of the m6A-methyltransferase complex, including METTL3 and WTAP, relocalize away from the nucleus (35), suggesting that these proteins may have undescribed cytoplasmic roles. Interestingly, studies of RNA viruses modified with m6A have demonstrated that viral infection can alter WTAP, METTL3, and METTL14 localization to the cytoplasm (28, 37, 38). Here, we investigated the hypothesis that WTAP targets METTL3 + 14 to HCV RNA for m6A modification and m6A-mediated regulation of HCV infection. We found that WTAP increases in the cytoplasm following HCV infection and that it recruits METTL3 to HCV RNA for m6A modification. In addition, WTAP, like METTL3 + 14, negatively regulates the production of viral particles. Importantly, we also found the nuclear localization of WTAP was dispensable for m6A modification of HCV RNA and not required for regulation of infection. Overall, this work shows that WTAP actions in the cytoplasm control the METTL3 + 14-mediated m6A modification of HCV RNA.
RESULTS
HCV infection alters the subcellular localization of the m6A machinery accessory protein WTAP.
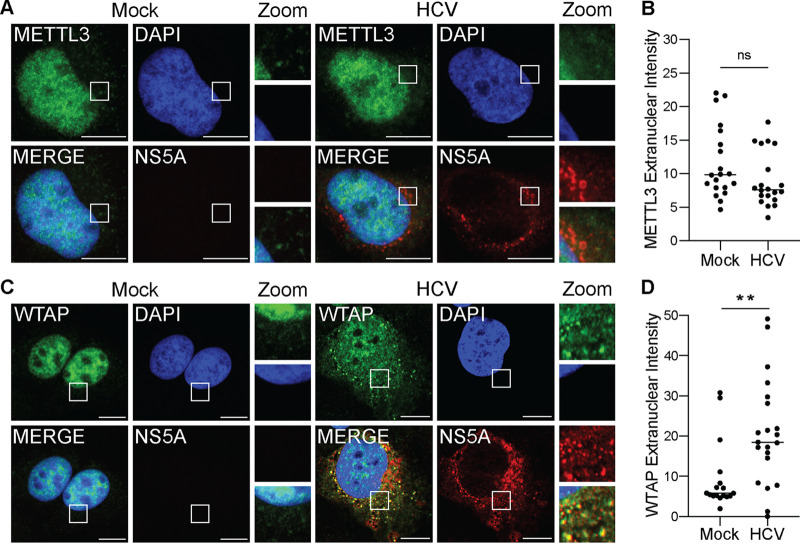
To determine if HCV infection alters the nuclear localization of proteins of the m6A-methyltransferase, we fixed and stained Huh7 liver hepatoma cells that were infected with HCV or mock infected for 48 h and analyzed METTL3 subcellular localization by confocal microscopy. We found that METTL3 was predominantly localized to the nucleus in both mock- and HCV-infected cells, with some distinct localization to the cytoplasm, and that this subcellular distribution of METTL3 did not change during infection (Fig. 1A and B). However, when we analyzed WTAP localization under the same conditions, we found that in response to HCV infection WTAP is present outside the nucleus and that it localizes in close proximity with the HCV NS5A protein, a marker of viral RNA replication compartments (39) (Fig. 1C). In contrast, mock-infected cells show only a limited level of WTAP in the cytoplasm (Fig. 1C). Indeed, quantification of the cytoplasmic and nuclear WTAP reveals increased WTAP in the cytoplasm in HCV-infected cells (Fig. 1D). Although, unlike WTAP, METTL3 localization is not changed with HCV infection, the fluorescence signal intensity of METTL3 in the cytoplasm is higher than those of WTAP in uninfected cells (Fig. 1B to D). These data reveal that HCV infection results in increased localization of WTAP to the cytoplasm near the sites of HCV replication and that METTL3 can be detected in the cytoplasm irrespective of HCV infection.
FIG 1.
HCV infection alters the subcellular localization of the m6A accessory protein WTAP. (A and C) Confocal micrographs of mock- or HCV (48 h, MOI of 0.3)-infected Huh7 cells stained with DAPI and antibodies against HCV NS5A and either METTL3 (A) or WTAP (C). Zoom is taken from area in the white box. (B and D) Quantification of the fluorescent signal intensity in the extranuclear region of Huh7 cells for METTL3 (B) or WTAP (D), as described in Materials and Methods for fields of mock (NS5A-negative)- or HCV-infected (NS5A-positive) cells. Scale bars = 10 μM. Graph shows mean ± SD; n = 21 fields. Data were analyzed by Welch’s unequal variances t test (**, P < 0.01; ns, not significant).
WTAP and METTL3 + 14 are essential for m6A modification of HCV RNA.
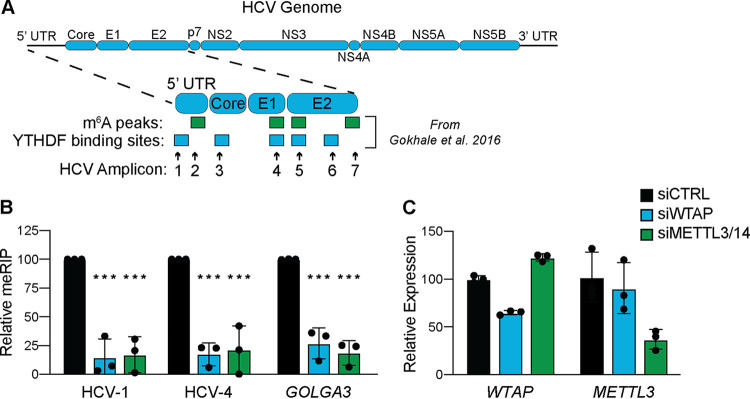
Previously, we found that abrogation of m6A at specific sites in the E1 coding region of the HCV RNA genome, as well as depletion of METTL3 + 14, increases the number of infectious viral particles by promoting viral RNA interaction with Core and packaging into virions (5). As METTL3 + 14 and WTAP are essential for m6A modification of cellular mRNA, we tested if they are similarly required for m6A modification of HCV RNA (13, 16). To accomplish this, we extracted RNA from Huh7 cells that were siRNA depleted of WTAP, METTL3 + 14, or nonspecific control and infected with HCV for 48 h. We then measured the m6A levels of previously identified m6A peaks or YTHDF protein binding sites on fragmented viral and host RNA by m6A-specific methylated RNA immunoprecipitation with quantitative PCR (meRIP-qPCR) (Fig. 2A) (5, 7, 15). We found that depletion of both WTAP and METTL3 + 14 led to reduced m6A levels at previously characterized HCV m6A sites on HCV RNA (Fig. 2A and B) (5, 23). Similarly, depletion of both WTAP and METTL3 + 14 led to a significant reduction in the m6A levels on GOLGA3, a transcript known to be m6A modified during HCV infection (Fig. 2B and C) (7). These data demonstrate that both WTAP and METTL3 + 14 are essential for the m6A modification of HCV RNA.
FIG 2.
WTAP and METTL3 + 14 are essential for m6A modification of HCV RNA. (A) Illustration of the HCV RNA genome with amplicons measured in this study and m6A peaks or YTHDF protein binding sites identified in reference 5. UTR, untranslated region. (B and C) Relative meRIP enrichment of the indicated viral or cellular amplicons from Huh7 cells treated with the indicated siRNAs and infected with HCV (48 h, MOI of 0. 3) (B), as determined as the percentage of input under each condition normalized to an m6A RNA spike-in with siCTRL samples normalized to 100%, and RT-qPCR analysis of indicated genes (C), relative to 18S rRNA. For panel B, graph shows mean ± SD (n = 3 biological replicates), while panel C is representative. Data were analyzed by two-way ANOVA with Šidák’s multiple-comparison test (***, P < 0.001).
METTL3 interacts with HCV RNA in a WTAP-dependent manner.
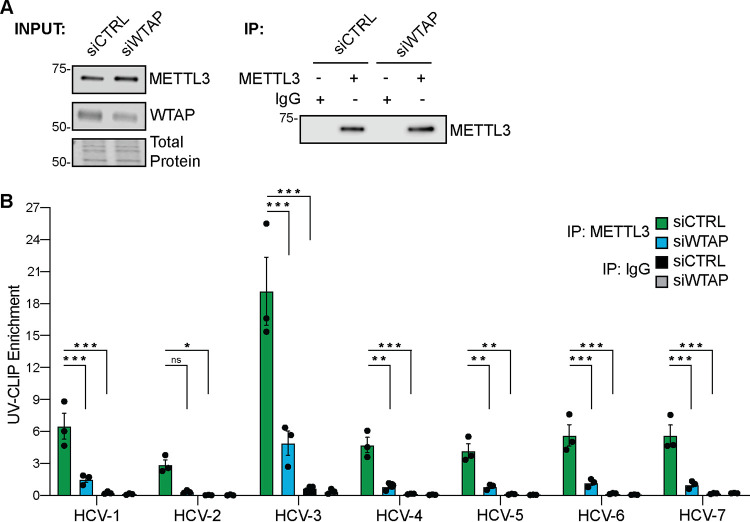
The interaction of METTL3 with its mRNA substrates requires WTAP (16, 40). Thus, we next tested whether METTL3 directly interacts with HCV RNA and if WTAP is required for this interaction. We used UV light to cross-link protein and RNA in Huh7 cells treated with siRNA against WTAP or control and infected with HCV for 72 h. RNA-protein complexes extracted from homogenized cells were immunoprecipitated with an antibody against METTL3, or IgG as a nonspecific control, followed by the capture of the bound complexes and stringent washing (Fig. 3A). Reverse transcriptase quantitative PCR (RT-qPCR) was then performed on extracted RNA with primers targeting previously described HCV RNA m6A sites or m6A-reader YTHDF protein binding sites (5). Immunoprecipitation of METTL3-RNA complexes enriched HCV RNA regions spanning the viral genome, while nonspecific IgG control did not (Fig. 3B). Importantly, depletion of WTAP abrogated METTL3 enrichment of many of these HCV RNA regions (Fig. 3B). Together, these data reveal that METTL3 interacts with HCV RNA and that WTAP is required for this interaction at several sites along the viral RNA genome.
FIG 3.
METTL3 directly binds HCV RNA in a WTAP-dependent manner. Huh7 cells were treated with the indicated siRNA and infected with HCV (72 h, MOI 1), followed by UV-CLIP with anti-METTL3 or nonspecific IgG. (A) Immunoblot analysis of input and immunoprecipitated UV-CLIP lysates. (B) Enrichment of indicated amplicons by METTL3 or IgG immunoprecipitation, relative to input, as measured by RT-qPCR. Graph shows mean ± SD; n = 3 biological replicates. Blot is representative of 3 independent experiments. Data were analyzed by two-way ANOVA with Šidák’s multiple-comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
WTAP negatively regulates HCV virion production.
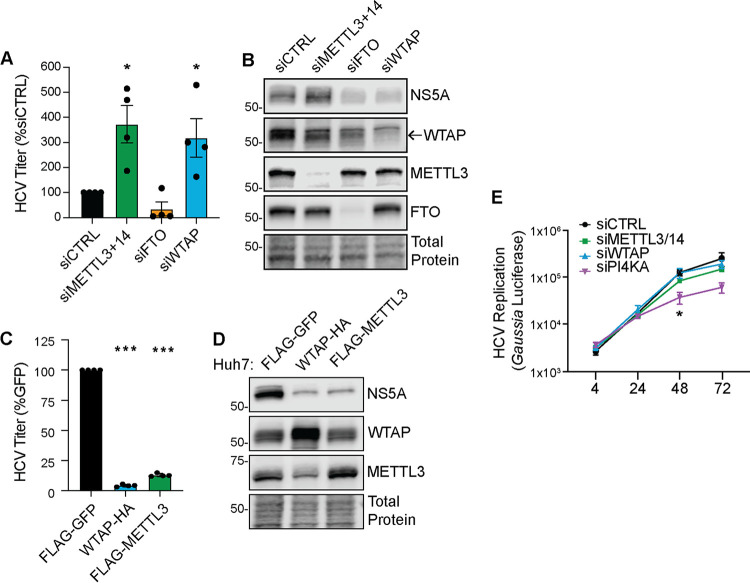
We previously showed that METTL3 + 14 negatively regulate HCV infection by decreasing the production of infectious viral particles (5). To determine whether WTAP also regulates HCV infection, we depleted WTAP by siRNA in Huh7 cells or generated Huh7 cells stably overexpressing WTAP and then used a focus-forming assay (FFA) to measure the production of infectious viral particles in the cellular supernatant harvested 48 h after HCV infection. Depletion of WTAP resulted in an increase in the production of infectious viral particles in comparison to cells treated with a nontargeting control siRNA (Fig. 4A). As we have shown before, depletion of the m6A-methyltransferase proteins METTL3 + 14 or the m6A demethylase FTO resulted in a similar increase, or decrease, in the production of viral particles, respectively (Fig. 4A) (5). Immunoblot analysis of cellular extracts revealed that WTAP depletion, unlike METTL3 + 14, resulted in decreased abundance of the HCV NS5A replicase protein compared to siRNA control (Fig. 4B) (5). Overexpression of both WTAP and METTL3 + 14 reduced infectious HCV particle production relative to cells overexpressing GFP (Fig. 4C). Immunoblot analysis of lysates from infected Huh7 cells overexpressing either WTAP or METTL3 + 14 revealed that the abundance of the HCV NS5A protein was reduced by overexpression of these proteins, in comparison to cells overexpressing GFP (Fig. 4D).
FIG 4.
WTAP negatively regulates HCV virion production. (A and B) Focus-forming assay of supernatant harvested from HCV-infected (48 h, MOI of 0.3) Huh7 cells depleted of indicated proteins (A), as well as immunoblot analysis of these lysates (B). (C and D) Focus-forming assay of supernatant harvested from HCV-infected (48 h, MOI of 0.3) Huh7 cells overexpressing the indicated proteins (C), as well as immunoblot analysis of these lysates (D). For WTAP and METTL3, protein-specific antibodies detect both endogenous and overexpressed proteins. (E) Gaussia luciferase values from the supernatant of Huh7.5-CD81 KO cells treated with the indicated siRNA and transfected with a full-length HCV RNA containing a Gaussia luciferase reporter cassette, measured at indicated hour posttransfection. Graphs show mean ± SD; n = 3 (C and E) or 4 (A) biological replicates. Blots are representative of 3 independent experiments. Data were analyzed by one-way ANOVA with Šidák’s multiple-comparison test (*, P < 0.05; ***, P < 0.001).
As we previously found that METTL3 + 14 negatively regulate the production of infectious HCV particles but not viral RNA replication (5), we next investigated if WTAP affected HCV RNA replication. For these experiments, we transfected HCV RNA encoding an internal Gaussia luciferase cassette as a reporter of viral replication (JFH1-QL/GLuc2A) into Huh7.5 cells in which the essential HCV entry factor CD81 had been deleted by CRISPR/Cas9 (Huh7.5-CD81 KO) (5, 41, 42). This allows for HCV RNA replication to be measured independently of virion production and spread. Depletion of WTAP did not alter the levels of HCV RNA replication relative to the control nontargeting siRNA over a time course (Fig. 4E). Similarly, as we have shown previously, METTL3 + 14 depletion did not significantly alter HCV RNA replication (5), whereas depletion of phosphatidylinositol 4-kinase alpha (PI4KA), a known host factor required for HCV RNA replication, decreased HCV RNA replication (Fig. 4E) (43). Together these data reveal that WTAP regulates the production of infectious HCV particles but does not impact viral RNA replication.
WTAP regulation of HCV RNA m6A modification is independent of its nuclear localization.
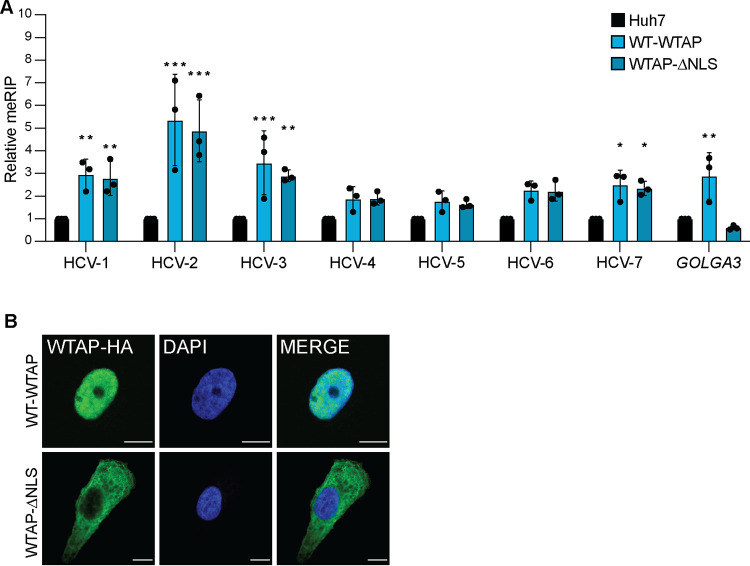
Cellular mRNA is m6A modified by the m6A-methyltransferase complex in the nucleus (13, 16, 17). As WTAP positively regulates HCV RNA m6A modification and relocalizes to the cytoplasm during HCV infection, we hypothesized that WTAP regulation of HCV RNA m6A modification is independent of its nuclear localization. To test this, we generated an Huh7 cell line overexpressing WTAP lacking its described nuclear localization signal (NLS; WTAP-ΔNLS) and measured HCV m6A modification on fragmented viral and host RNA by meRIP-qPCR (Fig. 5A) (17). The m6A levels of the cellular mRNA GOLGA3, which we have previously shown to have increased m6A during HCV infection, are increased by overexpression of wild-type WTAP but not by WTAP-ΔNLS (Fig. 5A) (7). Excitingly, the m6A levels of multiple HCV sites across the genome are similarly increased by overexpression of either wild-type WTAP or WTAP-ΔNLS, which does not localize to the nucleus (Fig. 5A and B). Taken together, these data reveal that WTAP regulation of HCV RNA m6A methylation, in contrast to cellular mRNA, can occur independently of its nuclear localization.
FIG 5.
WTAP regulation of HCV RNA m6A modification is independent of its nuclear localization. (A) Relative meRIP enrichment of indicated amplicons in HCV-infected (72 h, MOI of 1) parental Huh7-, wild-type (WT)-WTAP-HA-, or WTAPΔNLS-HA-overexpressing cells. (B) Confocal micrographs of Huh7 cells overexpressing WT-WTAP-HA or WTAPΔNLS-HA stained as indicated. Graph show mean ± SD; n = 3 biological replicates for panel A with micrographs of localization shown in panel B. Scale bars = 10 μM. Data were analyzed by two-way ANOVA with Šidák’s multiple-comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
WTAP regulation of HCV virion production requires METTL3 interaction but not nuclear localization.
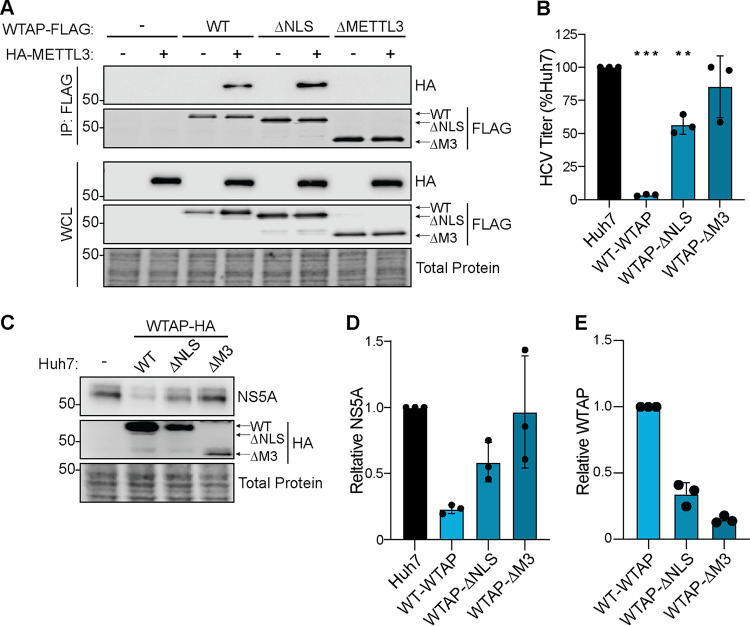
As our data reveal that WTAP regulates HCV RNA m6A modification and that this regulation is independent of WTAP nuclear localization, we next sought to determine the features of WTAP required for the regulation of HCV virion production. To accomplish this, we used Huh7 cells overexpressing wild-type WTAP, WTAP-ΔNLS, or a newly generated cell line expressing a mutant WTAP that does not interact with METTL3 (WTAP-ΔMETTL3), as seen by coimmunoprecipitation (17, 44) (Fig. 6A). We then measured the production of infectious viral particles in the cellular supernatant 48 h after HCV infection by FFA. We found that WTAP requires its METTL3 interaction domain to negatively regulate HCV particle production (Fig. 6B). However, WTAP lacking its nuclear localization signal still reduced HCV particle production, although not as much as wild-type WTAP (Fig. 6B). Immunoblot analysis of these lysates revealed that the levels of HCV NS5A protein are decreased by both wild-type WTAP and WTAP-ΔNLS but not by WTAP-ΔMETTL3, which corroborates the results of the FFA (Fig. 6C and D). Although WTAP is not expressed equally between the mutants, the difference in expression is not equal to the magnitude of the reduction in infectious viral particles (Fig. 6B and E). Taken together, these data reveal that WTAP features essential for its regulation of HCV RNA m6A modification, but not those needed for cellular mRNA modification (Fig. 5), are required for negative regulation of HCV virion production.
FIG 6.
WTAP regulation of HCV virion production requires METTL3 interaction but not nuclear localization. (A) Immunoblot analysis of anti-FLAG-immunoprecipitated lysates from Huh7 cells cotransfected with HA-METTL3 and indicated WTAP-FLAG constructs, either wild-type, ΔNLS, or ΔMETTL3 interaction. (B to E) Focus-forming assay of supernatant harvested from HCV-infected (48 h, MOI of 0.3) Huh7 cells overexpressing the indicated protein (B), as well as immunoblot analysis of lysates (C) and quantification of NS5A and WTAP levels and relative total protein (D and E). Graphs show mean ± SD; n = 3 biological replicates. Blots are representative of 3 independent experiments. Data were analyzed by one-way ANOVA with Šidák’s multiple-comparison test (**, P < 0.01; ***, P < 0.001).
DISCUSSION
Previously, we found that the HCV RNA genome is modified by m6A and that both abrogation of specific m6A sites on the HCV RNA genome and depletion of METTL3 + 14 regulate viral particle production (5). Others have shown that m6A sites on HCV RNA also can promote viral translation or evasion of RIG-I sensing (23, 32). While these studies reveal that m6A on HCV RNA regulates the viral life cycle, the mechanisms by which the HCV RNA in the cytoplasm is targeted by METTL3 + 14 for m6A modification have remained unclear. This is in part because METTL3 + 14 are described as functioning in the nucleus, and the mechanisms by which they are known to target mRNA for m6A modification would not apply to targeting HCV RNA in the cytoplasm (33, 34, 45, 46). For example, METTL3 + 14 localize to chromatin and interact with RNA polymerase II to m6A methylate mRNA in a cotranscriptional process, but HCV RNA replication does not involve RNA polymerase II or occur in the nucleus near chromatin (33). While our previous biochemical fractionation experiments did reveal that METTL3 + 14 could be localized in the cytoplasm, and others have shown the METTL3 has cytoplasmic functions, neither total cellular nor cytoplasmic levels of METTL3 + 14 change with HCV infection, and so how these proteins could be repurposed for cytoplasmic addition of m6A was unclear (5, 36). Here, we set out to determine how METTL3 + 14 are targeted to HCV RNA for m6A modification in the cytoplasm. We found that WTAP, the METTL3 + 14 interacting protein that coordinates RNA targeting by the broader m6A-methyltransferase complex, has increased localization to the cytoplasm during HCV infection. Importantly, we found that WTAP is essential for METTL3 interaction with HCV RNA and its m6A modification and that WTAP negatively regulates the production of infectious HCV particles, like METTL3 + 14 (5). Curiously, we found that depletion of WTAP, but not METTL3 + 14, resulted in decreased levels of the viral NS5A protein (Fig. 4B). While others have found that m6A on the HCV IRES promotes viral translation, they also found that depletion of METTL3 + 14 resulted in increased levels of NS5A, which is inconsistent with its predicted impact on IRES m6A modification (23). It remains to be determined why WTAP and METTL3 + 14 have seemingly different roles in regulating HCV protein production and how m6A deposition on HCV RNA, including at the IRES m6A site, affects the levels of HCV proteins during infection. Nonetheless, our work does reveal that WTAP is important to repurpose the m6A-methyltransferase complex to regulate HCV RNA m6A modification and control HCV infection.
Our work shows that proteins beyond METTL3 are important to target the m6A-methyltransferase complex to HCV RNA for modification. This is supported by the fact that although METTL3 preferentially modifies DRACH motifs in vitro, not all consensus motifs are modified in cells (14, 15, 47–50). Indeed, METTL3 was originally identified as part of a complex of proteins with m6A-methyltransferase activity (51, 52). This protein complex is now known to include both METTL14 and WTAP, which have specific functions in regulating m6A deposition (13, 16). METTL14 interacts with METTL3 and targets METTL3 + 14 to sites of active transcription marked by histone H3 trimethylation at lysine 36 (53). METTL3 + 14 then interact with WTAP (13, 16, 17), and WTAP broadly controls RNA targeting of the m6A-methyltransferase complex, with targeting to specific mRNAs regulated by WTAP-interacting proteins such as RBM15, VIRMA, and ZC3H13 (18–20, 35, 54). This m6A-methyltransferase complex then interacts with RNA polymerase II to add m6A to nascent mRNA (33, 34). While this mechanism for m6A modification drives the bulk of m6A on mRNA, in some cases, METTL3 can be directly recruited to mRNA transcription start sites via the protein CEBPZ (46). HCV m6A modification must happen differently than cellular m6A modification because the HCV RNA is regulated differently than cellular RNA. First, HCV transcription is mediated by the viral RNA-dependent RNA-polymerase NS5B and not RNA polymerase II, and thus m6A modification of HCV RNA is not necessarily cotranscriptional (2). Second, HCV RNA is solely present in the cytoplasm, separate from nuclear chromatin and histones (3). As such, a unique mechanism must recruit METTL3 + 14 to HCV RNA. Our work shows that WTAP is important for METTL3 + 14 targeting to HCV RNA, that WTAP relocalizes out of the nucleus during infection, and that m6A modification of HCV RNA in the cytoplasm is driven by WTAP.
WTAP relocalization during HCV infection is likely a key factor that drives how viral RNA gets m6A modified in the cytoplasm; however, questions remain as to how this occurs. We do know that during infection, HCV remodels intracellular membranes to generate replication compartments (55). These compartments contain pores that are coated by nucleoporin proteins recruited from the nuclear envelope, and these nucleoporin proteins can mediate selective access for proteins involved in viral replication (56, 57). As such, we hypothesized that WTAP utilizes its NLS to access these replication compartments to facilitate m6A modification of HCV RNA. However, we found that the NLS of WTAP is dispensable for m6A modification of HCV RNA at the sites we tested (Fig. 5), while it seems to be partially required to regulate infection (Fig. 6). This suggests that WTAP regulation of HCV infection may also occur independent of its function in targeting HCV RNA for m6A modification. We do know that WTAP recruitment to HCV RNA occurs through a mechanism that does not require its NLS. While it could be that a viral protein recruits WTAP to the HCV RNA, this would not explain how HCV infection induces WTAP localization to the cytoplasm. Interestingly, the nuclear localization of WTAP can be regulated by the cellular protein ZC3H13 (35), and a prior screen for cellular-HCV protein interactions suggested that three HCV proteins may interact with WTAP (58). Thus, it is possible that a viral protein interacts with newly translated WTAP to prevent its interaction with ZC3H13 and keep WTAP in the cytoplasm. This viral protein could then bring WTAP and METTL3 + 14 to HCV RNA. In support of this model, HCV E1, NS3, and NS4B, the three proteins suggested to interact with WTAP, have all been shown to broadly interact with HCV RNA and thus are candidates for bringing a WTAP/METTL3 + 14 complex to viral RNA for m6A modification (59–61). In fact, for other RNA viruses known to be m6A modified, such as enterovirus 71, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), and human metapneumovirus, viral proteins do either interact or colocalize with METTL3, METTL14, or WTAP (37, 38, 62). This suggests that m6A targeting may be altered during infection; indeed, we have observed this previously (7). Thus, identifying which HCV and cellular proteins interact with the m6A-methyltransferase complex during infection will be critical to understanding how m6A modification is regulated during infection.
m6A modification of HCV RNA occurs at several positions across the genome, and several of these m6A sites regulate specific aspects of the HCV life cycle (5, 23, 32). Our initial study identified 16 high-confidence m6A sites across the HCV RNA genome, and 3 of these sites have unique functions during HCV infection (5, 23, 32). Our understanding of how m6A modification at each of these sites occurs in relation to each other is limited. It may be that there are unique mechanisms that recruit METTL3 + 14 to viral RNA to m6A modify specific sites, with differing effects on the HCV life cycle. This possibility may explain why METTL3 + 14 and WTAP depletion differentially affects NS5A expression, although it is unclear how m6A could both positively regulate IRES-mediated translation and negatively regulate the levels of HCV proteins (Fig. 4) (23). Importantly, current methods to identify m6A do not allow for specific mapping of the m6A profile for each copy of the viral RNA during infection. This m6A profile on individual viral RNAs may regulate distinct viral processes, such as translation, transcription, or virion production. As such, it may be that the HCV RNA molecules involved in active translation may have one set of m6A sites modified, whereas those involved in viral packaging have a different set of m6A sites modified. Each of these m6A profiles could arise from different viral RNA targeting factors. In fact, WTAP enhancement of m6A modification does not appear to be uniform for all tested regions of HCV RNA (Fig. 5). Thus, additional viral RNA targeting factors may be required for the modification of particular m6A sites.
Overall, this study reveals that WTAP is an important regulator of m6A modification of a cytoplasmically localized RNA. Specifically, WTAP regulates HCV RNA m6A modification and as such it regulates virion production. Importantly, this regulation by WTAP is independent of its ability to localize to the nucleus. Thus, this work supports a model by which HCV infection induces WTAP localization changes to mediate cytoplasmic m6A modification of viral RNA. Studies of how methylation of specific HCV m6A sites is controlled and how HCV RNA m6A modification is regulated throughout the viral life cycle will undoubtedly provide insight into the mechanisms involved. Our work reveals that we still have much to learn about the processes that govern m6A methylation, an RNA regulatory mechanism critical in cellular differentiation, numerous cancers, and infection by an ever-growing list of viruses, including those of global health concern such as SARS-CoV-2 and members of the Flaviviridae.
MATERIALS AND METHODS
Cell culture.
Huh7, Huh7.5 (gift of Michael Gale, Jr., University of Washington [63]), Huh7.5 CD81 KO (gift of Matthew Evans, Icahn School of Medicine at Mount Sinai [64]), and 293T cells were grown in Dulbecco’s modification of Eagle’s medium (DMEM; Mediatech), supplemented with 10% fetal bovine serum (HyClone), 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Thermo Fisher), and 1× nonessential amino acids (Thermo Fisher), referred to as complete DMEM (cDMEM). Cells were verified using the Promega GenePrint STR kit (DNA Analysis Facility, Duke University) and as mycoplasma free by the LookOut Mycoplasma PCR detection kit (Sigma-Aldrich).
Plasmids.
The following plasmids were generated by subcloning PCR-generated amplicons from the indicated oligonucleotides from Table 1 into pEFtak or pLEX vector using In-fusion recombinase (TaKaRa) according to manufacturer’s instructions: pEFtak-WTAP-FLAG, pEFtak-WTAPΔNLS-FLAG, pEFtak-WTAPΔMETTL3-FLAG, pEFtak-HA-METTL3, pLEX-WTAP-HA, pLEX-FLAG-METTL3, pLEX-WTAPΔNLS-HA, pLEX-FLAG-GFP, and pLEX-WTAPΔMETTL3-HA. pJFH1-QL/GLuc2A was a gift from Stanley Lemon (University of North Carolina at Chapel Hill [42]).
TABLE 1.
Primer sequences for RT-qPCR
| Amplicon | Sense oligonucleotide (5′–3′) | Antisense oligonucleotide (5′–3′) |
|---|---|---|
| HCV-1 qPCR | AGAGCCATAGTGGTCTGCGG | CTTTCGCAACCCAACGCTAC |
| HCV-2 qPCR | GATAGGGTGCTTGCGAGTGCCC | GTCTTCTGGGCGACGGTTGGTG |
| HCV-3 qPCR | CATCCCCAAAGATCGGCGCTCC | CCAGCCGAGTCCCTCATTCCCA |
| HCV-4 qPCR | ATCTACCCTGGCACCATCAC | AGTAGGCCAAGCCGAACAT |
| HCV-5 qPCR | GGCTTTCTCGCGGCCTTGTTCT | CCCCACCCTATCCGGAAAGCCT |
| HCV-6 qPCR | TGACTTCAACGCCAGCACGGAC | CACTTTGGTGTGAGCCAGGGCC |
| HCV-7 qPCR | TGCTCATCTTGTTGGGCCAGGC | CTGATGTGCCAAGCTGCCACGA |
| GOLGA3 qPCR | CGTGAGACCCGAAGAACAAAAC | CACCAAAGAGGCTGTACAGTGA |
| pLEX FLAG-GFP | GCGGCCGCTATGGTGAGCAAGGGCGAGG | ACTCGAGTTAACCTTGTACAGCTCGTCCATGCC |
| pLEX WTAP-HA | GAGGATCCATGACCAACGAAGAACCTCTTCCC | GACTCGAGTTAGTAGTCTGGGACGTCGTATGGG |
| pLEX FLAG-METTL3 | CACAGATCTACCATGGATTATAAGGATGATGATG | CACGTCGACTTAAACCTATAAATTCTTAGGTTTAGAG |
| pLEX WTAPΔNLS-HA | CCGACTCTACTAGAGGATCCGCCACCATGAGTGAAACAGACTTCAAAGTTATGGC | GACTCGAGTTAGTAGTCTGGGACGTCGTATGGG |
| pEFtak-WTAP-FLAG | GGTACCATGACCAACGAAGAACCTCTT | TTATAATCAGCGGCCGCCAAAACTGAACCCTGTACATTTACAC |
| pEFtak-HA-METTL3 | CGTCCCAGACTACGCGGCCGCTTCGGACACGTGGAGCTCTATCC | CTGATCAGCGGGTTTAAACCTATAAATTCTTAGGTTTAGAGATGATACCATCTGGG |
| pEFtak-WTAPΔNLS-FLAG | CTTGGTACCATGAGTGAAACAGACTTCAAAGTTATGGCAAG | TTATAATCAGCGGCCGCCAAAACTGAACCCTGTACATTTACAC |
| pEFtak-WTAPΔMETTL3-FLAG | AGGTAAGCTTGGTACCATGTTCCTAAAAATGAAAGGTGAACTGGAACAG | AGGTAAGCTTGGTACCATGTTCCTAAAAATGAAAGGTGAACTGGAACAG |
| pLEX-WTAPΔMETTL3-HA | CCGACTCTACTAGAGGATCCGCCACCATGTTCCTAAAAATGAAAGGTGAACTGGAAC | CCGACTCTACTAGAGGATCCGCCACTGTTCCTAAAAATGAAAGGTGAACTGGAAC |
In vitro transcription.
Generation of HCV luciferase reporter RNA was accomplished with the MEGAscript T7 transcription kit (Invitrogen) using XbaI-linearized and Mung Bean Nuclease-treated (enzymes from New England Biolabs) JFH1-QL/GLuc2A plasmid (42), following the manufacturer’s instructions.
Viruses.
Infectious stocks of a cell culture-adapted strain of genotype 2A JFH-1 HCV (JFH-1 M9 [65]) were generated in Huh7.5 cells. Measurement of viral titers and virion production from infected supernatants was performed in Huh7.5 cells by FFA, as previously described (65). For viral infections, cells were incubated in a low volume of serum-free DMEM containing virus at the indicated multiplicity of infection (MOI) for 3 h, following which cDMEM was replenished.
Cell line generation.
293T cells were transfected with pLEX-WTAP-HA, pLEX-FLAG-METTL3, pLEX-WTAPΔNLS-HA, or pLEX-WTAPΔMETTL3-HA and the viral packaging plasmids psPAX2 and pMD2.G (Addgene numbers 12260 and 12259; gift of Duke Functional Genomics Facility), and the supernatant was harvested and filtered with a 0.22-μm filter 72 h after transfection. This filtered supernatant was then used to transduce Huh7 cells for 24 h. Transduced cells were then placed in 2 μg/μL puromycin for 72 h, and overexpression validated by immunoblotting, as described below. After selection and validation, cell lines were maintained in 1 μg/μL puromycin cDMEM until the time of experimentation.
siRNA treatment.
Cells were transfected with siRNA against indicated targets using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s protocol for 24 h before experimental infection or treatment. Depletion of siRNA targets was confirmed by immunoblot analysis. siRNAs (Qiagen) used included siWTAP (SI00069853), siMETTL3 (SI04317096), siMETTL14 (SI00459942), siFTO (SI04177530), siPI4KA (SI02777390), and siCTRL (1027281).
Luciferase assay.
Huh7.5 CD81 KO cells seeded into 12-well plates were transfected with siRNAs as described above. Cells were then transfected with 1 μg of JFH1-QL/GLuc2A in vitro transcribed RNA (1 μg) using polyethylenimine (Polysciences, Inc.) (Fig. 4E). For electroporation, 5 μg of in vitro transcribed viral RNA was electroporated into 4 × 106 cells at 250 V and 950 μF with a Gene Pulser Xcell system in Ingenio Electroporation Solution (Mirus). Supernatants were harvested and mixed with Renilla luciferase buffer and substrate, and luciferase values were measured according to the manufacturer’s instructions (Renilla Luciferase Assay System; Promega) using a BioTek Synergy 2 microplate reader.
Immunoblotting.
Cells were lysed in a modified radio immunoprecipitation assay (RIPA) buffer (10 mM Tris pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor cocktail (Millipore), and supernatants were collected after centrifugal clarification. Quantified protein, as determined by Bradford assay (Bio-Rad), was resolved by 10% (Fig. 3, 4D, and 6) or 4 to 20% (Fig. 4B) SDS-PAGE gels, transferred to nitrocellulose or polyvinylidene difluoride membranes using the Turbo-transfer system (Bio-Rad), stained with REVERT total protein stain (Licor Biosciences), and blocked with 3% bovine serum albumin (BSA) (Sigma-Aldrich) in phosphate-buffered saline (PBS) with 0.1% Tween (PBS-T). Membranes were probed with specific antibodies, washed with PBS-T, incubated with species-specific horseradish peroxidase (HRP)-conjugated antibodies (Jackson ImmunoResearch), washed again with PBS-T, and treated with Pico PLUS enhanced chemiluminescent reagent (Thermo Fisher). The signal was then captured by using a LICOR Odyssey FC.
Protein immunoprecipitation.
Fifty to 100 μg of protein extracted as above was incubated with 25 μL anti-FLAG M2 magnetic beads (Sigma-Aldrich) in modified 1× RIPA in a total volume of 300 μL at 4°C overnight with rotation. Beads were washed three times in PBS and eluted in 40 μL 2× Laemmli buffer with 1:20 β-mercapto-ethanol (Bio-Rad) at 95°C for 5 min. Eluates were resolved by SDS-PAGE and immunoblotting, as described above.
RT-qPCR.
Total cellular RNA was extracted using the Qiagen RNeasy kit (Life Technologies) or TRIzol extraction (Thermo Fisher). RNA was then reverse transcribed using the iSCRIPT cDNA synthesis kit (Bio-Rad) as per the manufacturer’s instructions. The resulting cDNA was diluted 1:5 in nuclease-free distilled H2O. RT-qPCR was performed in triplicate using the Power SYBR green PCR master mix (Thermo Fisher) and the Applied Biosystems QuantStudio 6 Flex RT-PCR system. Primer sequences for RT-qPCR are listed in Table 1.
meRIP.
For meRIP, total RNA was extracted from cells using TRIzol (Thermo Fisher) according to the manufacturer’s protocol and diluted to equivalent concentrations. Then, meRIP was performed as previously described (8). Following meRIP, cDNA from the input and immunoprecipitated RNA fractions was generated and analyzed by RT-qPCR as described above. The relative m6A level for each transcript was calculated as the percentage of input under each condition normalized to that of the respective positive-control m6A RNA spike-in, as described previously (8). Percent change of enrichment was calculated with siCTRL samples normalized to 100%.
UV-CLIP.
UV-CLIP was adapted as a modified version of formaldehyde CLIP (40). Briefly, Huh7 cells were plated in 10 cm dishes, treated with siRNA, and HCV infected as described above. For UV cross-linking, the supernatant was removed and replaced with 2.5 mL of 4°C PBS. Plates were then irradiated with 150 mJ/cm2 254 nm UV, and then cross-linked cells were harvested in 500 μL CLIP-RIPA buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 1% CA-630, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with 1 mM dithiothreitol (DTT), protease inhibitor, and RNAseIN+ (Promega). Next, the cross-linked cells were passed through a Qiashredder column (Qiagen) twice to generate homogenized lysates. These lysates were incubated with 2 μL Turbo DNase I and 1 μL 1:2,000 diluted RNase I for fragmentation for 25 min at 37°C with constant agitation and then clarified by centrifugation (enzymes from New England Biolabs). Equivalent amounts of lysates were then precleared for 4 h using protein A beads (Thermo Fisher) that were preblocked (1 μg of yeast tRNA and 1% BSA per 100 μL of beads in a total volume of 750 μL CLIP-RIPA buffer). RNA and protein inputs were reserved from these lysates and prepared as follows: for the RNA input, cross-links were removed by incubating equal amounts (50 μg) of precleared lysate in a total volume of 250 μL of CLIP-elution buffer (50 mM Tris-HCl pH 7.4, 5 mM EDTA, 10 mM DTT, 1% SDS, 1% RNAseIN+, 1:100 proteinase K) and incubated at 50°C for 1 h with constant agitation, with RNA extracted using TRIzol-LS (Thermo Fisher) and reserved for RT-qPCR; for the protein input, ~10 μg was reserved for immunoblotting. The remaining precleared lysates were divided and incubated at 4°C for >12 h with either METTL3-bound or IgG-bound preblocked protein A beads in 1 mL of CLIP-RIPA + 2 μL RNAseIN+. Then, these samples were washed 5× (CLIP-RIPA + 1 M NaCL and 1 M urea) and 1× (CLIP-RIPA) followed by resuspension in 100 μL CLIP-elution buffer without proteinase K. A portion of this eluate (10 μL) was reserved for immunoblotting, while CLIP-elution buffer (160 μL of CLIP-elution buffer) + proteinase K (2 μL) were added to the remaining 90 μL of beads, which were incubated for 1 h at 50°C with constant agitation, and RNA was extracted using TRIzol-LS and this and the input were analyzed by RT-qPCR.
Immunofluorescent microscopy.
Cells were fixed in 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 10% FBS in PBS. Slides were stained with indicated antibodies, incubated with conjugated Alexa Fluor secondary antibodies (Life Technologies), and mounted with ProLong Diamond + 4′,6-diamidino-2-phenylindole (Invitrogen). Imaging was performed on a Zeiss 880 laser scanning confocal microscope, using a 63×/1.25 oil objective using 405, 488, and 561 and laser lines at a 4× optical zoom with pinholes set to 1 airy unit for 561 (Light Microscopy Core Facility, Duke University) or a Leica DM4B widefield fluorescence microscope. Gain and offset settings were optimized, and final images were taken with a line averaging of 4. All images were processed with NIH Fiji/ImageJ (66). To quantify the cytoplasmic:nuclear ratios of proteins, 7 fields from each biological replicate with at least 5 cells each (21 fields total, >100 cells per condition) were analyzed in NIH Fiji/ImageJ (66) using the Intensity Ratio Nuclei Cytoplasm Tool RRID:SCR_018573 with protein (METTL3 or WTAP) signal intensity demarcated by the tool and then calculated as an average of all cells in each field. Importantly for the HCV-infected conditions, only fields containing NS5A-positive cells were quantified.
Antibodies.
Antibodies used in this study and their applications include HCV NS5A 9e10 (gift of Charles Rice; immunoblot, FFA, and immunofluorescence), FLAG-HRP (Sigma-Aldrich; A8592; immunoblot), HA (Sigma-Aldrich H6908; immunoblot), METTL3 (Novus Biologicals; AB_2687437; immunoblot), METTL3 (Abcam; ab195352; immunofluorescence and immunoprecipitation), FTO (Abcam; ab92821; immunoblot), WTAP (Proteintech; AB_10859484 or 10200-1-AP; immunoblot [Fig. 4]), WTAP (Abcam; ab195380; immunofluorescence [Fig. 1] and immunoblot [Fig. 3A and 6]), and nonspecific rabbit IgG (Cell Signaling Technologies; 2729S; immunoprecipitation).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 9. Data-appropriate statistical tests were performed, including one- and two-way ANOVA with post hoc testing (Fig. 2 and 3, 4, 5, 6) or Welch’s t test (Fig. 1). Values are presented as mean ± standard deviation of the mean for biological replicates (n = 3, or as indicated; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
This work was supported by Burroughs Wellcome Fund (to S.M.H.) and National Institutes of Health grants R01AI125416 (to S.M.H.) and T32-CA00911 (to M.T.S. and K.M.B.).
We thank those colleagues who generously provided reagents, including Matthew Evans, Michael Gale, Jr., Stanley Lemon, and Charlie Rice; the Duke Functional Genomics Core, the Duke Light Microscopy Core Facility, and members of the Horner Lab for valuable feedback and discussion, particularly Matthew Thompson.
Contributor Information
Stacy M. Horner, Email: stacy.horner@duke.edu.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Manns MP, Buti M, Gane E, Pawlotsky JM, Razavi H, Terrault N, Younossi Z. 2017. Hepatitis C virus infection. Nat Rev Dis Primers 3:17006. 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.Shulla A, Randall G. 2015. Spatiotemporal analysis of hepatitis C virus infection. PLoS Pathog 11:e1004758. 10.1371/journal.ppat.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niepmann M, Shalamova LA, Gerresheim GK, Rossbach O. 2018. Signals involved in regulation of hepatitis C virus RNA genome translation and replication. Front Microbiol 9:395. 10.3389/fmicb.2018.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20:654–665. 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams GD, Gokhale NS, Horner SM. 2019. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu Rev Virol 6:235–253. 10.1146/annurev-virology-092818-015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gokhale NS, McIntyre ABR, Mattocks MD, Holley CL, Lazear HM, Mason CE, Horner SM. 2020. Altered m(6)A modification of specific cellular transcripts affects flaviviridae infection. Mol Cell 77:542–555.e8. 10.1016/j.molcel.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, Xhemalce B, Mason CE, Horner SM. 2021. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep 34:108798. 10.1016/j.celrep.2021.108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399. 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arguello AE, DeLiberto AN, Kleiner RE. 2017. RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J Am Chem Soc 139:17249–17252. 10.1021/jacs.7b09213. [DOI] [PubMed] [Google Scholar]
- 12.Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen P, Rossa M, Muller M, Stunnenberg HG, He C, Carell T, Vermeulen M. 2017. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24:870–878. 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 15.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149:1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Rendtlew Danielsen JM, Liu F, Yang YG. 2014. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24:177–189. 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, Feederle R, Bruckmann A, Meister G. 2018. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24:499–512. 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. 2013. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 288:33292–33302. 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, Andrade-Navarro MA, Biggiogera M, Helm M, Soller M, Buhler M, Roignant JY. 2018. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev 32:415–429. 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep 12:562–572. 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim GW, Siddiqui A. 2021. The role of N6-methyladenosine modification in the life cycle and disease pathogenesis of hepatitis B and C viruses. Exp Mol Med 53:339–345. 10.1038/s12276-021-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim GW, Siddiqui A. 2021. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc Natl Acad Sci USA 118:e2022024118. 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira-Montecinos C, Toro-Ascuy D, Ananias-Saez C, Gaete-Argel A, Rojas-Fuentes C, Riquelme-Barrios S, Rojas-Araya B, Garcia-de-Gracia F, Aguilera-Cortes P, Chnaiderman J, Acevedo ML, Valiente-Echeverria F, Soto-Rifo R. 2022. Epitranscriptomic regulation of HIV-1 full-length RNA packaging. Nucleic Acids Res 50:2302–2318. 10.1093/nar/gkac062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li N, Rana TM. 2022. Regulation of antiviral innate immunity by chemical modification of viral RNA. Wiley Interdiscip Rev RNA e1720. 10.1002/wrna.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, Knight R, Smith D, Wang Y, Carlin AF, Rana TM. 2021. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep 35:109091. 10.1016/j.celrep.2021.109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim GW, Siddiqui A. 2021. Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc Natl Acad Sci USA 118:e2019455118. 10.1073/pnas.2019455118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao M, Dong Y, Wang Y, Liu H, Ma H, Zhang H, Zhang L, Cheng L, Lv X, Xu Z, Zhang F, Lei Y, Ye W. 2020. N(6)-methyladenosine modifications enhance enterovirus 71 ORF translation through METTL3 cytoplasmic distribution. Biochem Biophys Res Commun 527:297–304. 10.1016/j.bbrc.2020.04.088. [DOI] [PubMed] [Google Scholar]
- 29.Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N, Le-Trilling VTK, Trilling M, Mandelboim M, Hanna JH, Schwartz S, Stern-Ginossar N. 2019. Publisher Correction: m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 20:243. 10.1038/s41590-019-0314-4. [DOI] [PubMed] [Google Scholar]
- 30.Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. 2018. RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev 32:1472–1484. 10.1101/gad.319475.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y. 2014. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 24:1493–1496. 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim GW, Imam H, Khan M, Siddiqui A. 2020. N (6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem 295:13123–13133. 10.1074/jbc.RA120.014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slobodin B, Han R, Calderone V, Vrielink J, Loayza-Puch F, Elkon R, Agami R. 2017. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169:326–337.e12. 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE, Jr, Darnell RB. 2017. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31:990–1006. 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, Lan F, Shi YG, He C, Shi Y, Diao J. 2018. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell 69:1028–1038.e6. 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, Santisteban P, George RE, Richards WG, Wong KK, Locker N, Slack FJ, Gregory RI. 2018. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561:556–560. 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, Wang H, Zhang B, Qiu J, Deng F, Guan W. 2019. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res 47:362–374. 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J, Feng Z, Niewiesk S, Peeples ME, He C, Li J. 2020. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol 5:584–598. 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyre NS, Fiches GN, Aloia AL, Helbig KJ, McCartney EM, McErlean CS, Li K, Aggarwal A, Turville SG, Beard MR. 2014. Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. J Virol 88:3636–3652. 10.1128/JVI.02490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537:369–373. 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol 78:1448–1455. 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AH, Jr, Reid LM, Lemon SM. 2014. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 20:927–935. 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi M, Ohsugi M, Sasako T, Awazawa M, Umehara T, Iwane A, Kobayashi N, Okazaki Y, Kubota N, Suzuki R, Waki H, Horiuchi K, Hamakubo T, Kodama T, Aoe S, Tobe K, Kadowaki T, Ueki K. 2018. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis. Mol Cell Biol 38:e00116-18. [ 10.1128/MCB.00116-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C, Chen W, He J, Jin S, Liu Y, Yi Y, Gao Z, Yang J, Yang J, Cui J, Zhao W. 2020. Interplay of m(6)A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci Adv 6:eaba0647. 10.1126/sciadv.aba0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, Vakoc CR, Vassiliou GS, Kouzarides T. 2017. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552:126–131. 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harper JE, Miceli SM, Roberts RJ, Manley JL. 1990. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res 18:5735–5741. 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kane SE, Beemon K. 1985. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol 5:2298–2306. 10.1128/mcb.5.9.2298-2306.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. 1984. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci USA 81:5667–5671. 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narayan P, Rottman FM. 1988. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 242:1159–1162. 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 51.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. 1994. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 269:17697–17704. 10.1016/S0021-9258(17)32497-3. [DOI] [PubMed] [Google Scholar]
- 52.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 53.Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, Klemm L, Wu H, Zuo Z, Qin X, Dong Y, Zhou Y, Qin H, Tao S, Du J, Liu J, Lu Z, Yin H, Mesquita A, Yuan CL, Hu YC, Sun W, Su R, Dong L, Shen C, Li C, Qing Y, Jiang X, Wu X, Sun M, Guan JL, Qu L, Wei M, Muschen M, Huang G, He C, Yang J, Chen J. 2019. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567:414–419. 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, Wang F, Wang X, Shen B, Wang Y, Feng X, He C, Liu J. 2018. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4:10. 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul D, Madan V, Bartenschlager R. 2014. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579. 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M, Jr, Tyrrell DL, Wozniak RW. 2016. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog 12:e1005428. 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neufeldt CJ, Joyce MA, Levin A, Steenbergen RH, Pang D, Shields J, Tyrrell DL, Wozniak RW. 2013. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog 9:e1003744. 10.1371/journal.ppat.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M. 2015. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57:329–340. 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haddad JG, Rouille Y, Hanoulle X, Descamps V, Hamze M, Dabboussi F, Baumert TF, Duverlie G, Lavie M, Dubuisson J. 2017. Identification of novel functions for hepatitis C virus envelope glycoprotein E1 in virus entry and assembly. J Virol 91:e00048-17. [ 10.1128/JVI.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Einav S, Elazar M, Danieli T, Glenn JS. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol 78:11288–11295. 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439:105–108. 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, Hao H, Ma L, Zhang Y, Hu X, Chen Z, Liu D, Yuan J, Hu Z, Guan W. 2021. Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication. mBio 12:e0106721. 10.1128/mBio.01067-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hopcraft SE, Evans MJ. 2015. Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins. Hepatology 62:1059–1069. 10.1002/hep.27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aligeti M, Roder A, Horner SM. 2015. Cooperation between the hepatitis C Virus p7 and NS5B proteins enhances virion infectivity. J Virol 89:11523–11533. 10.1128/JVI.01185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]