ABSTRACT
Human norovirus (HNoV) accounts for one-fifth of all acute viral gastroenteritis worldwide and an economic burden of ~$60 billion globally. The lack of treatment options against HNoV is in part due to the lack of cultivation systems. Recently, a model of infection in biopsy-derived human intestinal enteroids (HIE) has been described: 3D-HIE are first dispersed in 2D-monolayers and differentiated prior to infection, resulting in a labor-intensive, time-consuming procedure. Here, we present an alternative protocol for HNoV infection of 3D-HIE. We found that 3D-HIE differentiated as efficiently as 2D-monolayers. In addition, immunofluorescence-based quantification of UEA-1, a lectin that stains the villus brush border, revealed that ~80% of differentiated 3D-HIE spontaneously undergo polarity inversion, allowing for viral infection without the need for microinjection. Infection with HNoV GII.4-positive stool samples attained a fold-increase over inoculum of ~2 Log10 at 2 days postinfection or up to 3.5 Log10 when ruxolitinib, a JAK1/2-inhibitor, was added. Treatment of GII.4-infected 3D-HIE with the polymerase inhibitor 2′-C-Methylcytidine (2CMC) and other antivirals showed a reduction in viral infection, suggesting that 3D-HIE are an excellent platform to test anti-infectives. The transcriptional host response to HNoV was then investigated by RNA sequencing in infected versus uninfected 3D-HIE in the presence of ruxolitinib to focus on virus-associated signatures while limiting interferon-stimulated gene signatures. The analysis revealed upregulated hormone and neurotransmitter signal transduction pathways and downregulated glycolysis and hypoxia-response pathways upon HNoV infection. Overall, 3D-HIE have proven to be a highly robust model to study HNoV infection, screen antivirals, and to investigate the host response to HNoV infection.
IMPORTANCE The human norovirus (HNoV) clinical and socio-economic impact calls for immediate action in the development of anti-infectives. Physiologically relevant in vitro models are hence needed to study HNoV biology, tropism, and mechanisms of viral-associated disease, and also as a platform to identify antiviral agents. Biopsy-derived human intestinal enteroids are a biomimetic of the intestinal epithelium and were recently described as a model that supports HNoV infection. However, the established protocol is time-consuming and labor-intensive. Therefore, we sought to develop a simplified and robust alternative model of infection in 3D enteroids that undergoes differentiation and spontaneous polarity inversion. Advantages of this model are the shorter experimental time, better infection yield, and spatial integrity of the intestinal epithelium. This model is potentially suitable for the study of other pathogens that infect intestinal cells from the apical surface but also for unraveling the interactions between intestinal epithelium and indigenous bacteria of the human microbiome.
KEYWORDS: human norovirus, 3D-human intestinal enteroids, polarity inversion, RNA sequencing
INTRODUCTION
Diarrheal diseases are the fourth cause of death worldwide and the second cause of morbidity in children less than 5 years old (1). In particular, human norovirus (HNoV) is the main causative agent of viral gastroenteritis worldwide, with a clinical burden of nearly 200,000 hospitalizations in Europe (2) and an economic burden of $65 billion (U.S.) per year, worldwide (1). The virus has been poorly characterized in terms of its viral life cycle, tropism, and pathophysiology due to the lack of easy-to-use cell culture systems that support robust viral replication and enable the production of cell culture-derived viral stocks. Consequently, the development of prophylactic and therapeutic agents has been challenging. The human intestinal enteroid (HIE) culture model, a biopsy-derived model of the intestinal epithelium, is an emerging tool to study enteric viruses that have been refractory to transformed cell culture systems. It proved efficacious to support the cultivation of a wide range of enteric pathogen, including HNoV (3). However, drawbacks of this systems are the time-consuming nature of the infection protocol, the elevated costs for the maintenance of the culture, the lack of long-term passage of HNoV, and the inability to generate a high-titer viral stock. The published protocol of infection includes the dispersion of HIE from 3D spheres into 2D monolayer on collagen-coated plates (3). For one well of a 96-well plate, it is recommended to use 100,000 cells, 10× more than transformed cell lines. Two to 6 days later, when HIE 2D-monolayers reach confluence, cells are differentiated by withdrawal of Wnt3a for 5 to 6 days before infection is initiated. Thus, the experimental time from HIE splitting to infection is 15 days, on average. Another disadvantage to the use of HIE 2D monolayers is the loss of 3D spatial organization that is crucial for intestinal physiology (4). For all these reasons, we sought to investigate whether differentiated 3D-HIE are amenable to HNoV infection. However, 3D-HIE are organized with the apical surface of the cells toward the lumen of the sphere; therefore, the exposure to a pathogen might be limited. Recently, a protocol to invert HIE polarity was published (Apical-Out HIE) (5), whereby the removal of basal membrane extract (BME) stimulates polarity reversal. The mechanism is dependent on extracellular matrix protein (ECM) concentration because addition of ECM to HIE in the absence of BME blocks polarity reversion (5). Apical-Out HIE thus provide a potential solution to ensure greater accessibility of 3D-HIE to pathogens.
The induction of the host innate response plays an essential role in the suppression of pathogen infection. In the case of enteric pathogens, interferon responses are highly upregulated in vivo and in vitro (6, 7). We previously reported that human astrovirus VA1, another enteric virus and causative agent of viral gastroenteritis, induces interferon and interferon-stimulated genes (ISG) in the HIE system but not in an immortalized cell line, the Caco2 cells (8). This also suggests that, intrinsically, the HIE culture restricts enteric virus infection more efficiently than immortalized cell lines. For this reason, a JAK inhibitor, ruxolitinib, has been successfully added to the infection protocol to prevent stimulating the innate immune response and thereby increasing the yield of HNoV in HIE (9).
Here, we describe the establishment of a HNoV infection protocol in 3D-HIE. We characterized the differentiation state of 3D-HIE compared with 2D-HIE and the degree of polarity inversion in 3D-HIE upon differentiation. We demonstrate that 3D-HIE are amenable to infection with HNoV with a reduction of experimental time of (on average) 3 days, increased yield (fold increase over inoculum), and reproducibility. We also characterized the host response of 3D-HIE treated with ruxolitinib to HNoV infection in culture by RNA sequencing. Altogether, we describe an adapted protocol for HNoV infection in 3D-HIE that is amenable to antiviral discovery and virus-host interaction studies. In addition, this model may facilitate future studies of other enteric pathogens/microbes that infect the apical surface of the intestinal epithelium.
RESULTS
3D-HIE undergo terminal differentiation similar to 2D-HIE, but infection with HNoV results in better yields.
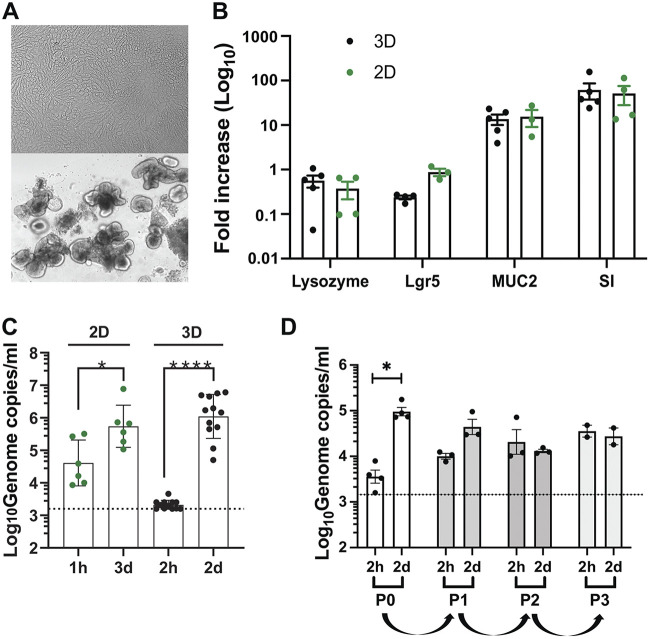
Differentiation of HIE is achieved by withdrawal of Wnt3a from the maintenance media that triggers the development of a heterogeneous, terminally differentiated epithelium by day 6, which is susceptible to HNoV (3, 8). Therefore, we first sought to determine if HIE derived from fetal ileum HT124 undergo terminal differentiation when maintained in 3D in BME. Towards that end, we monitored the mRNA transcript levels of selected differentiation markers by RT-qPCR. As a control, 2D-monolayers of HT124 HIE were also prepared and differentiated in collagen-coated plates using the previously published protocol (3). Representative images of differentiated 2D- and 3D-HIE are shown in Fig. 1A. Comparison of mRNA levels of Lgr5, lysozyme, mucin 2 (MUC2), and sucrase isomaltase (SI) as markers of stem-cells, Paneth cells, goblet cells, and mature enterocytes, respectively, revealed similar transcript levels in differentiated 2D versus 3D-HIE at 6 days postdifferentiation (Fig. 1B). This suggests that the withdrawal of Wnt3a triggered terminal differentiation of 3D-HIE in BME to a similar extent and time frame as 2D-HIE.
FIG 1.
3D-HIE undergo terminal differentiation and support HNoV replication. (A) Representative images of differentiated HT124 2D- and 3D-HIE at 5 and 6 days postdifferentiation, respectively. (B) Differentiation of 3D-HIE (3D), compared with 2D-HIE monolayers (2D). HIE from a fetal donor HT124 have been differentiated by Wnt3a removal in 3D-spheres or after dispersion in a collagen-coated plate. mRNA levels of selected host genes were quantified by qPCR. Lysoyzme, marker of Paneth cells; Lgr5, marker for crypt (stem)-cells; mucin 2, MUC2, marker of goblet cells and sucrase isomaltase; SI, marker for mature enterocytes. Each dot represents an independent experiment of the fold increase of mRNA of the selected transcript versus an undifferentiated control. (C) 2D and 3D-HIE were infected with HuNoV GII.4-positive stool sample (#233). 2D-HIE were infected (MOI ~5) according to the published protocol in the presence of ruxolitinib (2 μM) and harvested at 3 dpi. 3D-HIE were removed from BME and infected for 2 h at 37°C at MOI ~5, after which 3D HIE were washed twice prior to harvesting one set in TRI Reagent (2 h) while another set was seeded in BME and maintained in differentiation media with GCDCA (500 μM) and ruxolitinib (2 μM). Two dpi, 3D HIE were harvested in TRI Reagent (2 dpi), RNA was extracted from all the samples and RT-qPCR was used to establish viral titers. Each dot represents an independent biological replicate (independent infection). (D) HNoV-infected 3D-HIE were harvested at 2 dpi by freezing. After thawing, cells were centrifuged at 10,000 × g for 15 min at 4°C and half of the supernatant (200 μL) was used to infect differentiated 3D-HIE (passage 1). At 2 dpi the same procedure was repeated twice for passages 2 and 3. Viral genome copies were quantified by RT-qPCR. The dashed line represents the limit of detection. *, P value <0.05; ****, P value <0.0001; Student's t test calculated in GraphPad Prism.
To address whether differentiated 3D-HIE could support HNoV infection, GII.4-positive stool samples were used to infect 3D-HIE after removal from BME. After 2 h of incubation at 37°C in the presence of the bile acid GCDCA (500 μM), the inoculum was washed off; one set of 3D-HIE was harvested in TRI Reagent (2 h), while another set was re-embedded in BME and kept at 37°C for 2 days in differentiation media supplemented with 500 μM GCDCA and 2 μM the JAK inhibitor ruxolitinib. Differentiated 2D-HIE were infected in parallel according to the published protocol (3) in the presence of 500 μM GCDCA and 2 μM ruxolitinib. Quantification of HNoV yields by RT-qPCR revealed a 2.5 Log10 versus 1 Log10 increase in 3D-HIE versus 2D-HIE by 2 or 3 days postinfection (dpi), respectively (Fig. 1C). No cytopathic effect was observed in either culture condition. Because 3D-HIE health declined by 3 dpi and HNoV replication kinetics reach a plateau by 2 dpi (10), all subsequent infections were performed for 2 days. These data suggest that 3D-HIE differentiation was sufficient to support HNoV infection, and that in our system, infection of 3D-HIE resulted in higher yields (i.e., fold increase over inoculum) compared with the established 2D-HIE protocol. Lastly, to determine whether HNoV infection in 3D-HIE generates infectious viral progeny over multiple passages, we attempted to passage GII.4 HNoV in 3D-HIE. Significant amounts of viral genome were detected in passage 0 after 2 dpi (Fig. 1D). Infection of differentiated HIE with HNoV from passage 0 resulted in less than 1 Log10-fold increase, suggesting that infectious virus progeny was generated. However, no infection was detected at passage 2 or 3, potentially because virus titers were too low to establish a successful infection. Therefore, further optimization of the protocol may be necessary. One possibility is to adopt the passaging protocol by Ghosh et al. (11) in which successful passaging of HNoV in a human salivary gland cell line over multiple passages required isolation of HuNoV-containing extracellular vesicles prior to each passage.
Collectively, these data show a similar differentiation profile upon Wnt3a withdrawal and greater replication of HNoV in 3D- versus 2D-HIE.
Differentiation of 3D-HIE induces spontaneous inversion of polarity that enables HNoV accessibility.
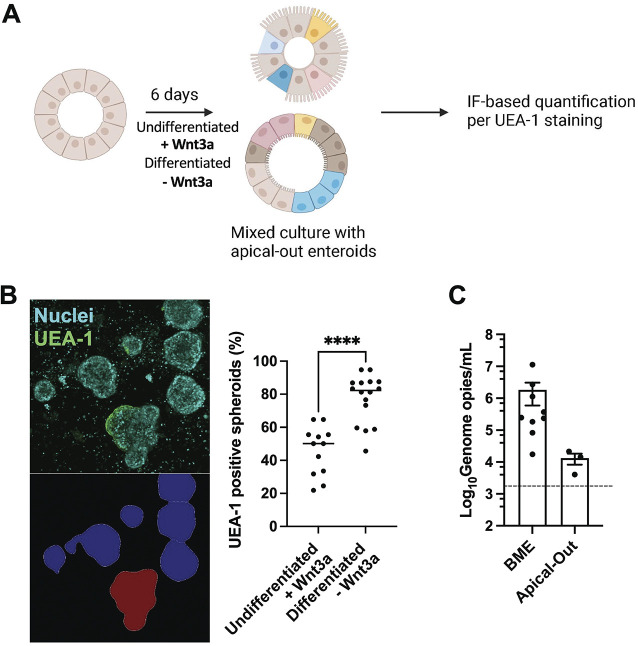
Efficient infection of 3D-HIE by HNoV suggested that the virus has access to the apical surface of enterocytes. Interestingly, 3D-HIE have been described to revert polarity spontaneously after removal of BME. The inversion is dependent on the percentage of extracellular matrix (ECM) proteins in the media (5). Because our differentiation protocol starts 3 to 4 days after passaging, resulting in 3D-HIE being embedded in BME for ~9 to 10 days, we hypothesized that the consequent reduction in % ECM proteins (caused by medium changes) could trigger spontaneous inversion. We also sought to determine whether the process of terminal differentiation initiated by Wnt3a withdrawal may further enhance polarity inversion. To this end, we differentiated 3D-HIE for 6 days and as a control, we kept 3D-HIE in proliferation media in parallel (Fig. 2A). Next, differentiated (–Wnt3A) and undifferentiated (+Wnt3a) 3D-HIE were removed from BME, fixed with paraformaldehyde (4% in PBS–/–) and stained with DAPI for nuclear detection and Ulex Europeus Agglutinin-1 (UEA-1), a lectin that binds the α-L-fucosyl residues of glycoproteins and glycolipids on the apical surface of the intestinal epithelium (8). To ensure that HIE stuck to the well during imaging, 10 to 20 3D-HIE were transferred into a poly-lysine-coated well of a black 96-well plate and subjected to confocal microscopy imaging with the CellVoyager CQ1 high-content microscope (Yokogawa). Image segmentation by CellProfiler (see representative image in Fig. 2B, bottom left) and analysis of the UEA-1 positive 3D-HIE (see representative image Fig. 2B, top left) revealed that after 10 days of culturing in proliferation media, about 50% of 3D-HIE spontaneously inverted polarity. In differentiation media, the proportion of UEA-1-positive spheroids rose to ~80% (Fig. 2B). Because a protocol of polarity inversion was recently published (5), we next wanted to compare the efficacy of replication in 3D-HIE generated with the published protocol (i.e., 3D-HIE without BME, Apical-Out) or with our protocol (i.e., 3D-HIE embedded in BME). Interestingly, the efficacy of replication at 2 dpi was >2 Log10 higher in BME 3D-HIE than in HIE generated with the Apical-Out protocol (Fig. 2C). These data suggested that the apical surface in a large proportion of differentiated 3D-HIE was exposed, and explained why these cultures were amenable to infection with HNoV without the need for microinjection. In addition, BME embedded 3D-HIE supported greater HuNoV infection compared to Apical-Out HIE.
FIG 2.
3D-HIE spontaneously revert polarity upon differentiation. (A) Schematic of 3D-HIE differentiation and pipeline for quantification of 3D-HIE with inverted polarity by immune fluorescence after staining with the lectin Ulex Europaeus Agglutinin-1 (UEA-1). (B) 3D-HIE were cultured for 6 days with maintenance or differentiation media. The 3D spheres were then collected and seeded in a black 96-well poly-lysine-coated plate. 3D-HIE were fixed for 20 min with 4% PFA and stained with UEA-1 and DAPI. Plates were imaged with the CellVoyager CQ1 scanning disk, automated, high content imaging microscope (Yokogawa). From each well, images from 16 fields were collected, covering ~80% of the surface area with a 10× objective. Representative images of UEA-1 stained HIE (top left) and automated image segmentation of 3D-HIE performed by CellProfiler (bottom left) are shown. The graph (right) represents quantification of 3D-HIE spheres, positive for UEA-1 after image analysis with CellProfiler software. Each dot represents a technical replicate from three independent experiments. ****, P value <0.0001. Student's t test calculated in GraphPad Prism. (C) 3D-HIE were first subjected to the apical-out protocol (Apical-Out), then differentiated and infected with GII.4-positive stool sample, #20942 (MOI ~5). In parallel, 3D-HIE were differentiated in BME and infected with #20942 (MOI ~5). Cells were harvested at 2 dpi and viral genome copies were quantified by RT-qPCR. Each dot represents an independent biological replicate. The dashed line represents the limit of detection.
HNoV infection of 3D-HIE is robust, dose-dependent, and sensitive to antiviral treatment.
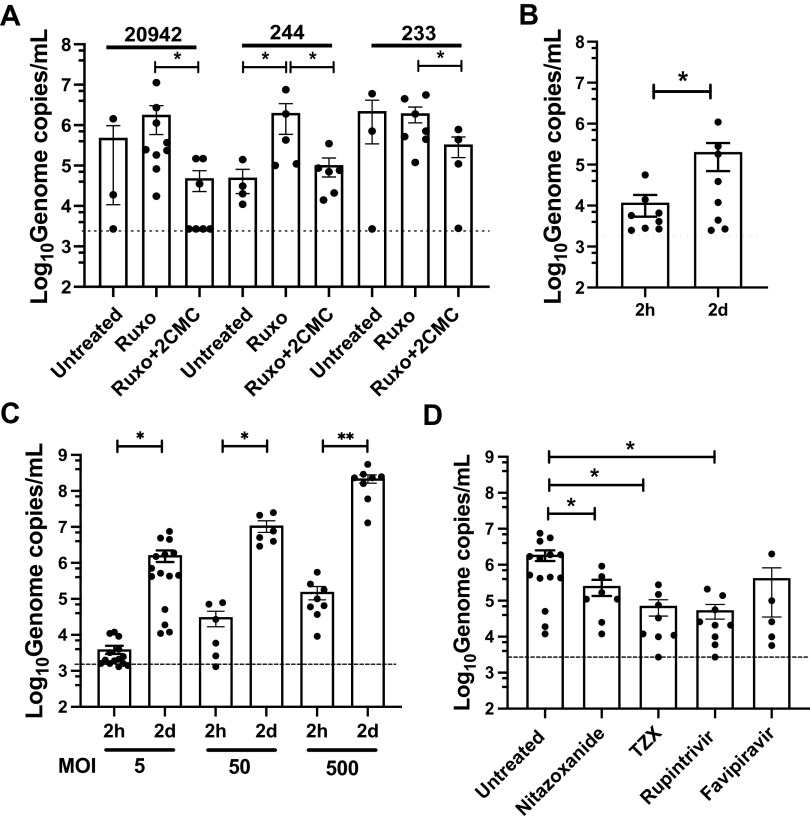
To characterize the infection of 3D-HIE in further detail, we evaluated the efficacy of infection of several HNoV-positive stool samples in the presence or absence of ruxolitinib, a JAK inhibitor that limits interferon responses and the nucleoside inhibitor 2′-C-Methylcytidine (2CMC). Replication of stool samples 20942 and 244, but not 233, was improved by adding ruxolitinib (Fig. 3A), further substantiating the critical importance of the interferon response in restricting HNoV infection in HIE (9). In addition, treatment with the nucleoside inhibitor 2′-C-Methylcytidine (2CMC) reduced replication for all three stool samples at 2 dpi (Fig. 3A). To confirm translatability of findings to other HIE, we also assessed HNoV replication in an HIE line derived from adult jejunal tissues (J2). We observed an ~1 Log10 increase at 2 dpi (Fig. 3B), suggesting that infection also occurred in this adult line in the 3D-format but is reduced compared with the fetal line HT124. We next infected 3D-HIE with increasing doses of HNoV-positive stool samples (Fig. 3C). A dose-dependent increase of replication was observed at 2 dpi. However, the fold increase over inoculum remained constant at ~2.5 Log10, suggesting that the system supports a defined amount of infection regardless of the initial infectious dose. To test whether the infection model could be used to test antivirals, we lastly infected 3D-HIE in the presence of compounds that were previously described to exhibit antiviral activity against norovirus in vitro or in vivo in HNoV-infected patients: Nitazoxanide, and its active metabolite TZX (12), the protease inhibitor Rupintrivir (13); and the polymerase inhibitor Favipiravir (Fig. 3D). The highest nontoxic concentration (in HIE) of selected drugs was used to ensure drug penetrance in 3D-HIE embedded in BME. We could detect a significant reduction in HNoV replication in the case of Nitazoxanide, TZX, and Rupintrivir. Altogether, these data clearly demonstrate that 3D-HIE are amenable to infection with HNoV and to medium-throughput screening of anti-infectives.
FIG 3.
3D-HIE constitute a robust platform for HNoV infection and screening of anti-infectives. (A) 3D-HIE were infected with filtered stool positive for GII.4 (#233, #244, and #20942) at MOI ~5 and after washing, 3D-HIE were seeded in BME and maintained in differentiation media with GCDCA (Untreated), with ruxolitinib (2 μM, Ruxo) or with the nucleoside inhibitor 2’-C-methylcytidine (50 μM, Ruxo + 2CMC). Cells were harvested 2 dpi and viral genome copies were calculated by RT-qPCR. (B) 3D-HIE derived from the J2 adult jejunum line were infected with stool positive for HNoV GII.4 at MOI ~5 (data from different isolates was combined) and harvested at 2 h and 2 dpi. (C) 3D-HIE were infected with increasing MOI of HNoV GII.4 (data from different isolates was combined) and were harvested at 2 h and 2 days for viral genome copies quantification by RT-qPCR. (D) 3D-HIE were infected with HNoV GII.4 at MOI ~5 (data from different isolates was combined) and treated with selected compounds, Nitazoxanide (50 μM) and its active metabolite tizoxanide (TZX, 50 μM), the enterovirus 3C-protease inhibitor rupintrivir (50 μM), the polymerase inhibitor Favipiravir (250 μM). The small molecule antivirals were added after infection in differentiation media. For A to D, each dot represents a technical replicate of at least three independent biological experiments. The dashed line represents the limit of detection. Statistical analysis was performed with GraphPad Prism. *, P value <0.05. Student's t test.
Host response signature upon infection suggests a functional activation of enteroendocrine cells.
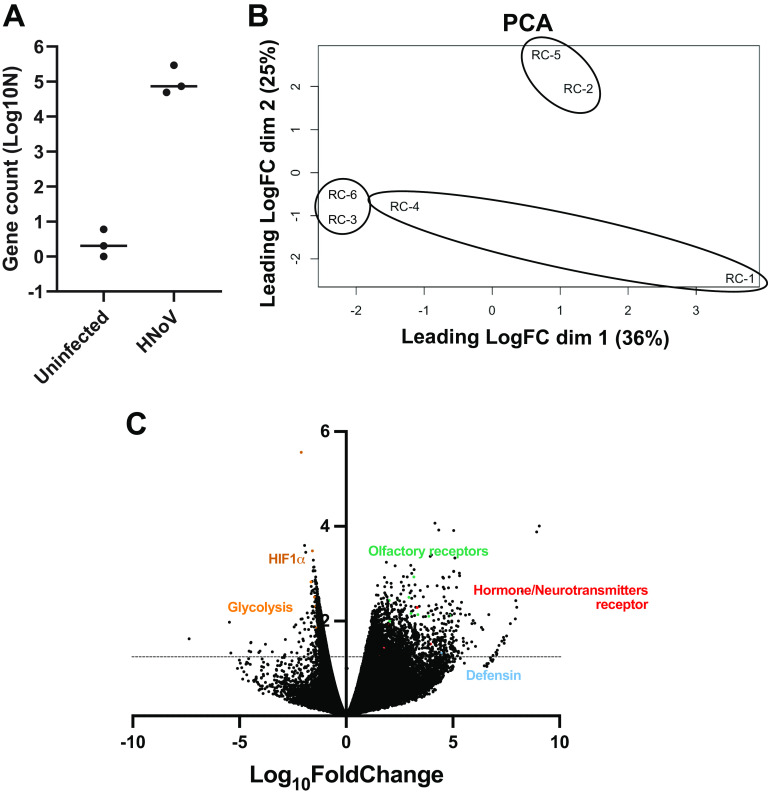
To evaluate the relevance of this system in addressing fundamental questions about the host response against HNoV infection, we performed a population-wide RNA sequencing experiment on HNoV-infected and uninfected 3D-HIE. The experiment was repeated independently three times and the efficacy of infection was evaluated by RT-qPCR before sample submission (data not shown). Because of the preponderant interferon (IFN) signature that was detected in previous RNAseq data sets (9, 14), we included ruxolitinib to enrich for less represented, virus-associated pathways. First, we confirmed infection by detecting virus sequences in the HNoV-infected samples. We found 72,984, 293,942, and 48,704 viral reads in the infected samples compared with 0, 2, and 6 reads in uninfected samples, respectively (Fig. 4A). In addition, we performed a multidimensional scaling analysis on the biological replicates that highlighted great variability between replicates and coclustering of infected and uninfected samples from each independent experiment (RC4-RC1, RC5-RC2, and RC6-RC3; Fig. 4B). For this reason, we pairwise analyzed the differentially expressed genes (DEGs) in infected versus uninfected 3D-HIE (Fig. 4C). Due to the ruxolitinib treatment, IFN-stimulated genes were not differentially expressed between HNoV-infected and uninfected samples, although some IFN genes were upregulated upon infection (IFNA14 with fold change [FC] = 5; IFNW1 with FC = 4; and IFNL3 and L4 with FC of 2.3 and 1.8, respectively). A pathway analysis was next performed on significatively (P value <0.05) down- and upregulated genes (FC <-1.5 and >1.5, respectively) after removing genes with unknown coding product (Table 1). Only 65 genes were significantly downregulated upon infection, and they strongly clustered in hypoxia-response and metabolic pathways. On the other hand, over 500 genes were significantly upregulated in response to infection, but the pathway analysis did not highlight statistically significant signatures. However, the top upregulated pathways were olfactory transduction and response to hormones and neurotransmitters (dopamine, relaxine). Intriguingly, hormone and neurotransmitter receptors are mostly expressed on enteroendocrine cells (15) that were recently described as a target of HNoV infection (16).
FIG 4.
RNA sequencing of HNoV-infected 3D-HIE. 3D-HIE were infected with filtered stool positive for GII.4 (#20942) at MOI ~5 for 2 h at 37°C or kept uninfected. 3D-HIE were then washed twice in PBS, seeded in BME, and cultured for 2 days in the presence of GCDCA (500 μM) and ruxolitinib (2 μM). RNA was extracted with Direct-Zol RNA minikit and after quality control, the RNA was sequenced with the Illumina Hi-Seq 2500 platform. (A) Viral reads were aligned to a consensus GII.4 Sydney (GenBank accession no. JX459908.1) and the counts were plotted in Graph Pad Prism. Each dot represents a biological replicate. (B) Principal-component analysis was performed to compare the biological replicates. Circles represent the independent repeats of the infected samples RC-4, RC-5, and RC-6 with the corresponding uninfected samples RC-1, RC-2, and RC-3. (C) Volcano plot of differentially expressed genes. The y axis represents the Log10 P value. Above the dotted line lay genes with significant P value (P value < 0.05). Some genes of interest are highlighted in color.
TABLE 1.
RNA sequencing pathway analysis
| Differentially expressed genes | Name | P value | Adjusted P value | Genes |
|---|---|---|---|---|
| Upregulated (575 genes) | Olfactory transduction | 0.0003459 | 0.09513 | OR2C1, OR4Q2, OR8U1, OR2S2, OR2AE1, OR2W3, OR10W1, OR2AG2, OR11H6, OR56B1, OR8B4, OR5AP2, OR5B2, OR52B6, OR4D2, OR2B2, OR2T27, OR6J1, OR6N1, OR5P3, OR5P2, OR10A7, OR4K15, OR9A4, OR51T1, OR52A5 |
| Hormone ligand-binding G-protein coupled receptors | 0.003283 | 0.4514 | CGA, FSHR, GNRH2 | |
| Signaling by GPCR | 0.006266 | 0.5355 | CXCL6, OR2C1, DHH, OR4Q2, RXFP2, PIK3R6, OR8U1, OR2S2, OR2AE1, OR2W3, OR10W1, NPY, OR2AG2, OR56B1, OR8B4, OR5AP2, CGA, HCRT, DRD3, OR5B2, WNT2, INSL5, OR52B6, DRD5, NTSR2, CCR2, OR4D2, OR2B2, OR2T27, WNT10A, OR6J1, TAAR5, OR6N1, OR5P3, OR5P2, OR10A7, OR4K15, OR9A4, FSHR, OR51T1, GNRH2, OR52A5 | |
| Dopamine receptors | 0.007789 | 0.5355 | DRD5, DRD3 | |
| Relaxin receptors | 0.02059 | 1 | RXFP2, INSL5 | |
| Defensins | 0.03982 | 1 | DEFB121, DEFB124, DEFA6 | |
| Downregulated (65 genes) | HIF-1 transcriptional activity in hypoxia | 2.377E-06 | 0.0004137 | BHLHE40, SLC2A1, ADM, CA9, HK2 |
| BDNF signaling pathway | 2.171E-05 | 0.001889 | FOXC1, P4HA1, GDF15, IGFBP3, ADM, HSPA1B, HSPA1A | |
| Apoptosis modulation by HSP70 | 0.001716 | 0.05855 | HSPA1B, HSPA1A | |
| Glycolysis and gluconeogenesis | 0.001872 | 0.05855 | SLC2A1, PCK1, HK2 | |
| Transcriptional regulation of white adipocyte differentiation | 0.002019 | 0.05855 | CEBPB, ANGPTL4, PCK1 | |
| p53 signaling pathway | 0.001089 | 0.05855 | BNIP3L, GDF15, IGFBP3, HSPA1B |
Altogether, our data show that HNoV infection of 3D-HIE is a simplified and robust alternative to the established 2D protocol that is suitable for screening of anti-infectives and amenable to virus-host interaction studies.
DISCUSSION
After 50 years from its discovery, there are still many knowledge gaps about HNoV tropism, biology, and pathophysiology. Similar to other strictly enteric viruses, the lack of in vitro cultivation systems has also hampered the development of successful therapeutics to combat these prevalent virus infections. The discovery of HNoV replication in human intestinal enteroids in 2016 (3) opened new avenues for norovirus research. However, limitations of this model exist in terms of labor, costs, and the lack of generation of a HNoV cell-derived stock. We sought to explore the possibility of simplifying the current protocol of HIE infection to overcome some of these limitations. In our hands, the dispersion of 3D-HIE into 2D-monolayer prior to differentiation has always been a variable step for a successful infection. The data presented herein demonstrate that HIE dispersion was not necessary for a successful HIE differentiation and that 3D-HIE spontaneously reverted polarity upon differentiation, even if they were kept in BME. This finding was puzzling as a recent report suggested that HIE polarity is dependent on % ECM proteins (5). We reasoned that in our protocol, the 3D-HIE are kept in BME for more than the canonical 6 days, and with every medium change, the concentration of ECM proteins would hence decrease. However, we also observed that in our model, terminal differentiation exacerbated the Apical-Out phenotype. In addition, β1 integrin was proposed to change HIE polarity in BME, suggesting that the presence of BME is not an a-priori obstacle for polarity reversion. Whether the observed spontaneous polarity and its kinetics is age and/or donor and/or intestinal segment dependent or whether additional factors also play a role in driving polarity inversion deserves further studies. However, successful infection (that can be used as proxy for successful differentiation and polarity inversion) was also observed in the adult jejunum line J2, albeit at lower efficiency, suggesting other factors also play a role in driving polarity inversion.
Importantly, we successfully established infection of differentiated 3D-HIE with different HNoV GII.4-positive stool samples and across three institutions (Wobus lab at the University of Michigan, Rocha-Pereira lab at the KU Leuven, and Taube lab at the University of Lübeck). As previously reported (9), ruxolitinib, a JAK inhibitor, increased HNoV infection while the nucleoside inhibitor 2CMC reduced infection, corroborating that HNoV actively replicates in this model. Interestingly, the inversion of polarity alone as with the Apical-Out HIE did not ensure a successful infection by HNoV, suggesting that the BME provides an environment that is beneficial for a sustained viral infection.
We further demonstrated that this model is amenable to antiviral studies as we successfully reported activity of compounds that are endowed with antiviral activity against HNoV, such as nitazoxanide and its prodrug TZX (12), and the protease inhibitor rupintrivir (13).
Lastly, we explored whether this model would also provide insights into viral-host interactions and to this end, we performed an RNA sequencing analysis. Previous studies on enteric virus infection of enteroids unequivocally showed a signature dominated by interferons and innate immune pathways (7). Typically, interferon responses are initiated by infected cells but mostly amplified by bystander cells (6). In order to focus our analysis on signatures and pathways in infected cells as a consequence of viral replication, we included ruxolitinib, an inhibitor of JAK 1/2, and the downstream STAT-1 signaling, that limits interferon signaling. As a consequence, genes under the control of STAT-1 that might be upregulated upon infection will be reduced in expression. To our surprise, among the downregulated pathways, we found HIF-1α response genes and glycolysis. The latter was previously described by us as an important host factor in the context of murine norovirus infection in macrophages (17). Hence, it is intriguing to wonder whether the previously observed “viral-induced” responses derived from actively replicating cells or whether they are a consequence of innate immune activation of uninfected bystander cells. Other possibilities are that macrophages and epithelial cells initiate metabolically different responses to infection because mature enterocytes exhibit low rates of glycolysis (18) or that inhibition of the inflammatory response by ruxolitinib suppresses the inflammation driven glycolytic reprogramming (19). Studies that dissect the role and the contribution of uninfected bystander cells in an infected sample by single cell RNA sequencing are hence urgently needed.
Our RNA sequencing analysis might also shed some light into mechanisms of HNoV pathogenesis. Unlike the recently reported upregulation of aquaporin (AQ) 1 upon HNoV infection of enteroids and potential involvement in HNoV-associated pathogenesis (20), we did not detect upregulation of AQ1 or other members of the AQ family in the RNAseq analysis. Instead, among the top upregulated pathways in response to infection, we found olfactory receptors and neurotransmitter signal transduction. Recently, HNoV antigen was detected in enteroendocrine (EEC, Chromogranin A positive) cells from infected intestinal biopsies (16). EEC are rare cells in the gut epithelia (~1%) and are chemosensors (15), hormone-producing cells in response to nutrients metabolites, bacterial metabolites, gut hormones, and neurotransmitters. A subtype of EEC, the enterochromaffin cells have been recently described to be electrically excitable, and able to modulate the serotonin-sensitive primary afferent nerve fibers via synaptic connections and directly affect gut motility (21). Our analysis suggests a mechanism of virally induced diarrhea by direct infection or by functional activation of enterochromaffin cells. It is also important to note that the pathways positively associated with infection were not strongly upregulated, suggesting that the signal from the rare EEC might be diluted in a bulk RNAseq analysis.
In summary, we describe here an adapted protocol for HIE infection by HNoV. This model is less time-consuming, less costly, more reproducible, and more scalable for medium-throughput studies of antiviral/vaccine efficacy. In addition, because of the spontaneous polarity inversion, this model is amenable to infection by enteric viruses and likely other microbes without the burden of microinjection. We demonstrated the utility of this model for antiviral testing and studies of virus-host interactions but it may also be useful for complex studies of HNoV-microbiome interactions.
MATERIALS AND METHODS
Cells, treatment, and virus.
Human intestinal enteroids derived from fetal ileum, HT124, and from adult jejunum, J2, were maintained at 37°C with 5% CO2 in BME (MatriGel, Corning, 354234 or Cultrex Ultimatrix, BME001-05) and maintenance media, CMGF+ (Advanced DMEM-F12, LWRN-conditioned medium, B27, N2, mrEGF, N-acetyl cysteine, Leu15-Gastrin, A-83-01, and SB202190), that was replaced every other day. Differentiation was triggered by Wnt3a removal with differentiation media (Advanced DMEM-F12, Noggin, B27, N2, mrEGF, N-acetyl cysteine, Leu15-Gastrin, and A-83-01) for 6 days with medium changes every other day. Stool samples positive for HNoV GII.4 were prepared as a 10% solution in Opti-MEM or PBS +/+ and then filtered through a 0.22-μm membrane. The filtrate was then titrated by RT-qPCR, according to the published protocol (22), and used for infection at selected multiplicity of infection (MOI).
Infection and viral quantification by RT-qPCR.
HIE derived from fetal ileum, HT124, were maintained in BME (MatriGel, Corning, 354234 or Cultrex Ultimatrix, BME001-05) and maintenance media for 3 to 4 days after splitting, then differentiated by Wnt3a withdrawal (differentiation media) for 5 to 6 days (8 to 10 days postsplitting). The apical-out HIE were prepared according to the published protocol (5). On the day of infection, 3D-HIE were collected in a 15-mL falcon tube and BME was washed with CMGF– medium. After pelleting (100 × g for 3 min at 4°C), 3D-HIE were resuspended in differentiation media and transferred into a 1.5-mL Eppendorf tube and the HNoV clinical isolate was added at a defined MOI, according to the experiment. We estimated the MOI based on the number of spheroids per condition with an average of 300 cells/spheroid, as measured by immune fluorescence. Virus and cells were incubated in infection media (differentiation media with 500 μM GCDCA) for 2 h at 37°C with occasional mixing. Then, HIE were washed 3 times with CMGF–, the 2-h condition was harvested in TRI Reagent (Zymo Research, R2050-1) and the rest of infected HIE were resuspended in BME and plated in a prewarmed 24-well plate (3 × 10 μL drop/condition). Plates were left for 5 min at 37°C to ensure BME polymerization and infection media with 2 μM ruxolitinib and selected drugs were added. At 2 dpi, cells were harvested in TRI Reagent and RNA extraction (Zymo Research, R2051) was performed according to manufacturer’s instruction.
For the passaging experiment, HNoV-infected HIE were collected in a 1.5-mL tube at 2 dpi and subjected to one freeze-thaw cycle prior to centrifugation at 10,000 × g for 10 min at 4°C. Then, 200 μL of the supernatant (1/2 of the total volume) was used to infect newly differentiated 3D-HIE. HNoV-infected HIE (P1, passage 1) were harvested at 2 dpi and used to infect newly differentiated 3D-HIE. The procedure was repeated for three passages.
For infection of 2D-HIE, cells were kept in maintenance media for 4 to 5 days (postsplitting) prior to trypsin treatment to create a cell suspension. Cells were then seeded in collagen-coated wells of a 96-well plate at 105 cells/well. Once 2D-HIE formed a confluent monolayer, which could take between 2 and 5 days, 2D-HIE were differentiated by Wnt3a withdrawal for 5 to 6 days. Afterwards, 2D-HIE were infected with HNoV in the presence of 500 μM GCDCA for 1 h at 37°C. Once the viral inoculum was removed, cells were kept in differentiation media with GCDCA and 2 μM ruxolitinib for 3 days. At 3 dpi, cells were harvested in TRI Reagent and RNA extraction (Zymo Research, R2051) was performed according to manufacturer’s instruction. Viral quantification by RT-qPCR was performed as previously described (22).
Quantification of markers of differentiation by RT-qPCR.
After 6 days of differentiation following Wnt3a withdrawal from the culture media, 2D- and 3D-HIE were harvested in TRI Reagent (Zymo Research, R2050-1) and cellular mRNA was extracted with DirectZol RNA extraction kit (Zymo Research, R2051). Two-step RT-PCR was used to quantify the relative expression of intestinal cells markers (Sucrase Isomaltase for mature enterocytes, Mucin 2 for goblet cells, Lgr5 for stem-cells, and Lysozyme for Paneth cells). The iScript cDNA synthesis kit (Bio-Rad) was used for cDNA synthesis. Quantitative real-time PCR (qPCR) with specific primers (Sucrase Isomaltase [SI]: forward AATCCTTTTGGCATCCAGATT and reverse GCAGCCAAGAATCCCAAAT; Lgr5: forward CAGCGTCTTCACCTCCTACC and reverse TGGGAATGTATGTCAGAGCG; Mucin 2 [MUC2]: forward TGTAGGCATCGCTCTTCTCA and reverse GACACCATCTACCTCACCCG; Lysozyme: forward ACAAGCTACAGCATCAGCGA and reverse GTAATGATGGCAAAACCCCA; GAPDH: forward CTCTGCTCCTCCTGTTCGAC and reverse TTAAAAGCAGCCCTGGTGAC) was performed with thermal cycler (Bio-Rad) by using iTaq Universal SYBR green supermix (Bio-Rad) as previously described (8). GAPDH was used to normalize gene expression.
Immunofluorescence and image analysis.
3D-HIE in maintenance media and differentiation media (both 10 days after splitting) were collected and washed to remove BME. In suspension, 3D-HIE were fixed with 4% paraformaldehyde for 30 min and then stained with DAPI to identify nuclei, and Ulex Europeus Agglutinin-1 (UEA-1) to label the villus brush border at the apical surface of the enterocytes. After staining, for ease of experimentation, 10 to 20 3D-spheres were transferred in a poly-l-lysine coated black plate and subjected to automated high-content confocal imaging (CellVoyager, Yokogawa). Briefly, 16-maximum projection fields per well, covering more than 90% of the surface of the well, were acquired with a 10× objective. A Cell Profiler pipeline for image segmentation and a classifier for UEA-1 positive and negative 3D-HIE was then developed. The percentage of UEA-1 positive 3D-HIE was obtained for each field and averaged for each well (technical replicate).
RNA sequencing and statistical analysis.
HT124 were infected at MOI ~5 or kept uninfected for 2 days in the presence of GCDCA (500 μM) and ruxolitinib (2 μM). The infection experiment was performed three times. RNA was isolated using the DirectZol RNA extraction kit (Zymo Research, R2051). RNA library preparation and RNA-sequencing (single-end, 50-bp read length) were performed by the University of Michigan DNA Sequencing Core using the Illumina Hi-Seq 2500 platform. All RNA sequences were deposited in the NCBI GEO database and are cataloged under the accession number GSE205007. RNA sequences were generated for 151-bp paired-end reads according to the manufacturer’s protocol (Illumina NovaSeq) by the University of Michigan Advanced Genomics Core. Bcl2fastq2 Conversion Software was used to generate demultiplexed Fastq files. Raw sequences were mapped to a combined human genome GRCh38 (ENSEMBL) and the human norovirus GII.4 Sydney 2012 genome (GenBank accession no. JX459908.1) using STAR v2.5.2a (23). Aligned reads were then assigned count estimates to genes using featureCounts (24). Sample normalization and differential gene expression of the aligned sequences and all other analyses were calculated using the R package EdgeR (25). We performed unbiased clustering of the normalized read counts using multidimensional scaling. To test for differentially expressed genes between virally infected HIEs and mock-infected HIEs, by biological replicate, we applied the quasi-likelihood F-test using the glmQLFtest function, controlling for biological replicates as a covariate in analysis. The KEGG pathway enrichment analysis was performed using the kegga function. Plots were constructed using the package ggplot2 (26). Gene pathways were analyzed with the software Enrichr (27).
Data availability.
Data from the RNA sequencing are available at the GEO database under accession number GSE205007 and in Data Set S1 in the supplemental material.
ACKNOWLEDGMENTS
The work was in part funded by the University of Michigan Biological Science Scholars Program to C.E.W. C.M. is supported by the Marie-Skłodowska Curie actions global fellowship (GA-841247). N.S.-F. is supported by a Horizon 2020-funded ITN program, OrganoVIR (Grant 812673). M.G.G. and J.A.C. are supported by the National Institutes of Health (P30ES017885 and P30DK034933). We thank Jana Van Dycke for her support and fruitful discussions.
Footnotes
Supplemental material is available online only.
Contributor Information
Carmen Mirabelli, Email: carmen.mirabelli@uni-luebeck.de.
Joana Rocha-Pereira, Email: joana.rochapereira@kuleuven.be.
Christiane E. Wobus, Email: cwobus@umich.edu.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. 2016. Global economic burden of norovirus gastroenteritis. PLoS One 11:e0151219. 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calduch EN, Cattaert T, Verstraeten T. 2021. Model estimates of hospitalization discharge rates for norovirus gastroenteritis in Europe, 2004–2015. BMC Infect Dis 21. 10.1186/s12879-021-06421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatehullah A, Tan SH, Barker N. 2016. Organoids as an in vitro model of human development and disease. Nat Cell Biol 18:246–254. 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 5.Co JY, Margalef-Català M, Li X, Mah AT, Kuo CJ, Monack DM, Amieva MR. 2019. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep 26. 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanifer ML, Guo C, Doldan P, Boulant S. 2020. Importance of type I and III interferons at respiratory and intestinal barrier surfaces. Front Immunol 10.3389/fimmu.2020.608645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cieza RJ, Golob JL, Colacino JA, Wobus CE. 2021. Comparative analysis of public RNA-sequencing data from human intestinal enteroid (Hies) infected with enteric RNA viruses identifies universal and virus-specific epithelial responses. Viruses 13:1059. 10.3390/v13061059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolawole AO, Mirabelli C, Hill DR, Svoboda SA, Janowski AB, Passalacqua KD, Rodriguez BN, Dame MK, Freiden P, Berger RP, Vu DL, Hosmillo M, O’Riordan MXD, Schultz-Cherry S, Guix S, Spence JR, Wang D, Wobus CE. 2019. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog 15:e1008057. 10.1371/journal.ppat.1008057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo BK, Merenda A, Lillestol R, Drumright L, Zilbauer M, Goodfellow I. 2020. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 11. 10.1128/mBio.00215-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin SC, Ayyar BV, Cortes-Penfield NW, Haga K, Neill FH, Opekun AR, Broughman JR, Zeng XL, Blutt SE, Crawford SE, Ramani S, Graham DY, Atmar RL. 2019. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses 11:638. 10.3390/v11070638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S, Kumar M, Santiana M, Mishra A, Zhang M, Labayo H, Chibly AM, Nakamura H, Tanaka T, Henderson W, Lewis E, Voss O, Su Y, Belkaid Y, Chiorini JA, Hoffman MP, Altan-Bonnet N. 2022. Enteric viruses replicate in salivary glands and infect through saliva. Nature 607:345–350. 10.1038/s41586-022-04895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang W, Xu L, Ma B, Chen S, Yin Y, Chang KO, Peppelenbosch MP, Pan Q. 2018. Nitazoxanide inhibits human norovirus replication and synergizes with ribavirin by activation of cellular antiviral response. Antimicrob Agents Chemother 62. 10.1128/AAC.00707-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocha-Pereira J, Nascimento MSJ, Ma Q, Hilgenfeld R, Neyts J, Jochmans D. 2014. The enterovirus protease inhibitor rupintrivir exerts cross-genotypic anti-norovirus activity and clears cells from the norovirus replicon. Antimicrob Agents Chemother 58:4675–4681. 10.1128/AAC.02546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SC, Qu L, Ettayebi K, Crawford SE, Blutt SE, Robertson MJ, Zeng XL, Tenge VR, Ayyar BV, Karandikar UC, Yu X, Coarfa C, Atmar RL, Ramani S, Estes MK. 2020. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci USA 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribble FM, Reimann F. 2016. Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu Rev Physiol 10.1146/annurev-physiol-021115-105439. [DOI] [PubMed] [Google Scholar]
- 16.Green KY, Kaufman SS, Nagata BM, Chaimongkol N, Kim DY, Levenson EA, Tin CM, Yardley AB, Johnson JA, Barletta ABF, Khan KM, Yazigi NA, Subramanian S, Moturi SR, Fishbein TM, Moore IN, Sosnovtsev S. 2020. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat Commun 11. 10.1038/s41467-020-16491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passalacqua KD, Lu J, Goodfellow I, Kolawole AO, Arche JR, Maddox RJ, Carnahan KE, O’Riordan MXD, Wobus CE. 2019. Glycolysis is an intrinsic factor for optimal replication of a norovirus. mBio 10. 10.1128/mBio.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. 2012. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci Rep 2:568. 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rath E, Haller D. 2022. Intestinal epithelial cell metabolism at the interface of microbial dysbiosis and tissue injury. Mucosal Immunol 15:595–604. 10.1038/s41385-022-00514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, Zhang B, Chen R, Li M, Zheng Z, Xu W, Zhang Y, Gong S, Hu Q. 2022. Human norovirus induces aquaporin 1 production by activating NF-κB signaling pathway. Viruses 14:842. 10.3390/v14040842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellono NW, Bayrer JR, Leitch DB, Castro J, Zhang C, O’Donnell TA, Brierley SM, Ingraham HA, Julius D. 2017. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 170:185–198.e16. 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taube S, Kolawole AO, Höhne M, Wilkinson JE, Handley SA, Perry JW, Thackray LB, Akkina R, Wobus CE. 2013. A mouse model for human norovirus. mBio 4. 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginestet C. 2011. ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A Stat Soc 174. 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
- 27.Xie Z, Bailey A, Kuleshov Mv, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A. 2021. Gene set knowledge discovery with Enrichr. Curr Protoc 1. 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download jvi.00855-22-s0001.xlsx, XLSX file, 4.1 MB (4.1MB, xlsx)
Data Availability Statement
Data from the RNA sequencing are available at the GEO database under accession number GSE205007 and in Data Set S1 in the supplemental material.