ABSTRACT
Many negative-sense RNA viruses, including measles virus (MeV), are thought to carry out much of their viral replication in cytoplasmic membraneless foci known as inclusion bodies (IBs). The mechanisms by which IBs facilitate efficient viral replication remain largely unknown but may involve an intricate network of regulation at the host-virus interface. Viruses are able to modulate such interactions by a variety of strategies including adaptation of their genomes and “hijacking” of host proteins. The latter possibility broadens the molecular reservoir available for a virus to enhance its replication and/or antagonize host antiviral responses. Here, we show that the cellular 5′–3′ exoribonuclease, XRN1, is a host protein hijacked by MeV. We found that upon MeV infection, XRN1 is translocated to cytoplasmic IBs where it acts in a proviral manner by preventing the accumulation of double-stranded RNA (dsRNA) within the IBs. This leads to the suppression of the dsRNA-induced innate immune responses mediated via the protein kinase R (PKR)-integrated stress response (ISR) pathway.
IMPORTANCE Measles virus remains a major global health threat due to its high transmissibility and significant morbidity in children and immunocompromised individuals. Although there is an effective vaccine against MeV, a large population in the world remains without access to the vaccine, contributing to more than 7,000,000 measles cases and 60,000 measles deaths in 2020 (CDC). For negative-sense RNA viruses including MeV, one active research area is the exploration of virus-host interactions occurring at cytoplasmic IBs where viral replication takes place. In this study we present evidence suggesting a model in which MeV IBs antagonize host innate immunity by recruiting XRN1 to reduce dsRNA accumulation and subsequent PKR kinase activation/ISR induction. In the absence of XRN1, the increased dsRNA level acts as a potent activator of the antiviral PKR/ISR pathway leading to suppression of global cap-dependent mRNA translation and inhibition of viral replication.
KEYWORDS: 5′-3′ exoribonuclease XRN1, inclusion body, integrated stress response, measles virus, protein kinase PKR
INTRODUCTION
Measles virus (MeV) belongs to the Paramyxoviridae family that includes multiple important pathogens including parainfluenza viruses, respiratory syncytial virus, Hendra virus, Nipah virus, rinderpest virus, canine distemper virus, Newcastle disease virus, and Sendai virus (1). The MeV genome is single-stranded, non-segmented RNA of negative polarity (about 15.9 kb) and encodes 6 viral structural proteins, a feature shared by most viruses of the Paramyxoviridae family (1). The genome is encapsidated by the viral nucleocapsid protein (N). Together with the viral RNA-dependent RNA polymerase complex consisting of the catalytic large protein (L) and the essential cofactor phosphoprotein (P), these ribonucleoprotein (RNP) complexes constitute the functional unit for viral mRNA transcription and genome replication (1), both of which occur in the cytoplasm of infected cells. Formation and budding of mature virions are initiated by the matrix (M) protein, which directly interacts with MeV N and 2 membrane embedded viral glycoproteins, the hemagglutinin (H) and the fusion (F) proteins, to recruit viral RNPs to the plasma membrane where virion release takes place by budding. Unlike other viral proteins that are translated in the cytoplasm, both H and F are synthesized and form a complex in the endoplasmic reticulum (ER), then are transported via the Golgi apparatus to reach the plasma membrane. MeV F is initially present in a native inactive form known as F0 in the ER, which is cleaved by the host furin endoprotease in the Golgi to produce the active F1 and F2 fragments that are linked by disulfide bonds (2). During the initiation of infection, binding of MeV H to the host surface receptor activates the F1 fragment that leads to the fusion of the viral envelope with the host plasma membrane. The viral RNPs are released into the cytoplasm (3). In addition to the above structural proteins, MeV also encodes 2 additional proteins, C and V (4, 5), that facilitate viral transcription/replication through functions ranging from increasing the fidelity of the viral polymerase to antagonizing host innate immune responses (6–10).
A hallmark of infection by MeV, as well as by many other cytoplasmic replicating non-segmented negative-sense RNA viruses, is the biogenesis of viral inclusion bodies (IBs) in the cytoplasm of infected cells (11). IBs are membraneless structures where viral replication components, such as N, L, and P proteins, the single-stranded RNA viral genome and its replication intermediates, are concentrated (7, 12–19). Several studies (20–22), including ours with MeV (23), established that the formation and progression of IBs is mediated via liquid-liquid phase separation (LLPS), a physiochemical process to organize intracellular membraneless structures (24–28). A liquid-like organelle also has been described for Influenza A virus (a segmented negative-sense RNA virus) and is suggested to facilitate the viral genome assembly (29). Recent reports have further identified LLPS compartments formed by a broad range of additional viruses from different families including SARS-CoV-2 (a positive-sense RNA virus) (30, 31), HIV-1 (a retrovirus) (32), and HSV-1 (a DNA virus) (33). Considering that these membraneless structures play a pivotal role during the viral life cycle, one speculation is that they represent potential “hot spots” of virus-host macromolecular interactions.
We present evidence herein supporting a model in which MeV IBs utilize the host 5′–3′ exoribonuclease, XRN1, to downregulate the host innate immune response by impairing the accumulation of dsRNA. XRN1 is known for its canonical role in mRNA degradation within processing bodies (PBs) (34, 35). IBs and PBs are both cytoplasmic membrane-less organelles that are assembled via a liquid-liquid phase separation process, but they differ from each other by composition and function (11, 34, 36). IBs are supramolecular viral replication centers whose formation is triggered by viral proteins and RNA (11) whereas PBs are storage sites of translationally repressed mRNAs and the RNA degradation machinery that are constitutively formed and upregulated during stress responses (34, 37).
XRN1’s ability to degrade RNA raises an intriguing question as to what role XRN1 may play in viral infections where large quantities of exogenous RNA are produced during viral transcription and replication processes. Indeed, XRN1 has been shown to affect the replication of cytoplasmic replicating positive-sense RNA viruses. Prior studies indicated that XRN1 mediates the subgenomic RNA production of flaviviruses that is essential for pathogenesis (38, 39) and that XRN1 is hijacked by SARS-CoV to degrade cellular mRNAs and induce host shutoff (40). On the other hand, subsequent investigation revealed that XRN1 negatively impacts the replication of hepatitis C virus (41) and encephalomyocarditis virus (42). Compared to cytoplasmic positive-sense RNA viruses, knowledge about the role of XRN1 in cytoplasmic negative-sense RNA virus replication is more limited. One study using Newcastle disease virus (NDV) suggested that the antiviral activity of XRN1 only becomes detectable in the absence of an interferon response (42). Moreover, depletion of XRN1 modestly increases reporter gene expression when inserted into the genome of vesicular stomatitis virus (VSV) (43). Whether XRN1 broadly affects the replication of cytoplasmic negative-sense RNA viruses remains an important and, as of yet, unresolved question. This prompted our examination of whether XRN1 affected the replication of MeV. We found that XRN1 localized to the viral IBs and downregulated the level of dsRNA during MeV infection, and furthermore functioned as a potent proviral host factor by prevention of activation of the protein kinase R (PKR)-mediated integrated stress response (ISR) innate pathway.
RESULTS
XRN1 is a proviral host factor for MeV replication.
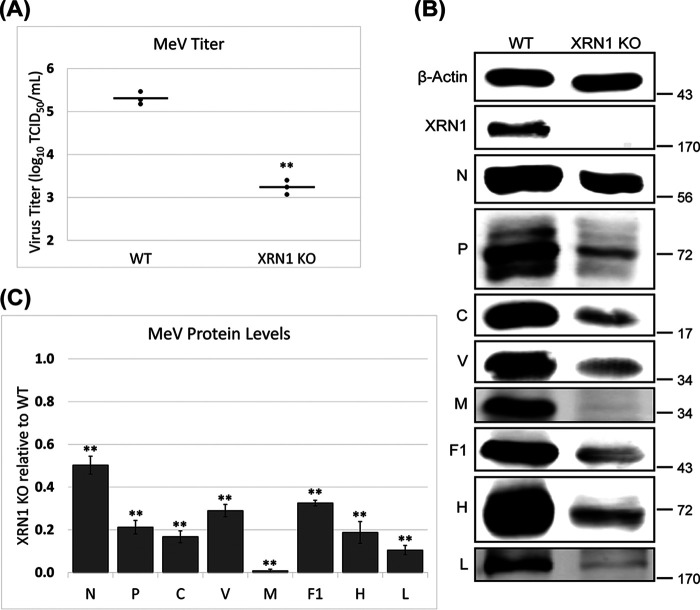
To test whether XRN1 affects MeV replication, we compared virus yields as well as viral protein production between a CRISPR engineered A549 cell line with XRN1 knocked out (KO) and parental wild-type (WT) cells (44). We found that, relative to WT control cells, the XRN1 KO mutant cells exhibited lower yields (reduced by up to ~120-fold) of infectious progeny virus (Fig. 1A). Consistent with the impaired virus yield, immunoblot analyses indicated that the XRN1 KO cells also produced reduced levels of MeV proteins compared to the WT control cells (Fig. 1B). Notably, the decrease in level of M was the greatest among the MeV proteins, with a reduction of over 95% (Fig. 1C). These data suggest that XRN1 acts as a proviral factor for MeV. As a control, the proliferation/viability of infected WT and XRN1 KO cells was assessed using an MTT colorimetric assay. The results showed that compared to the uninfected control cells, both infected WT and XRN1 KO cells exhibited a comparable level of reduction in their total metabolic activity, 23% +/− 0.91% for WT versus 20% +/− 2.3% for XRN1 KO (P > 0.14). Therefore, the difference in cell proliferation/viability did not contribute significantly to the reduced viral yield (Fig. 1A) observed with the KO cells under our experimental condition.
FIG 1.
Impaired MeV replication in XRN1 knockout cells. Parental wild-type (WT) and XRN1 knockout (KO) A549 cells were infected with MeV using an MOI of 1.0. At 24 hpi one set of cells were lysed for viral protein analysis and a parallel set were used for virus yield analysis. (A) Infectious viral yields were determined and are plotted on a scatterplot. (B) Representative blots of MeV proteins are shown. (C) Quantification of individual MeV protein levels relative to the WT cells are presented as a bar chart. Both the scatterplot and bar chart represent the averages from 3 independent experiments. P values were calculated using Microsoft Excel Student's t test: *, P < 0.05; **, P < 0.001; ns, not significant.
XRN1 suppresses the PKR-mediated integrated stress response and additional dsRNA sensing pathways during MeV infection.
Given the reduction of viral protein production seen across the MeV genome together with the cap-dependent translation of MeV mRNA (45–47), we investigated whether there was an involvement of ISR, an antiviral host response that causes the global repression of cap-dependent translation of infected cells (48). To assess the activity of ISR, we infected both WT and XRN1 KO cells as described in Fig. 1, and examined the phosphorylation of translation initiation factor eIF2α at serine 51 (p-eIF2α), a hallmark of ISR activation (49, 50). We found that under previously utilized infection conditions (Fig. 1), the KO of XRN1 resulted in an increased level of p-eIF2α without changing the amount of total eIF2α protein (Fig. 2A). This activation of ISR in XRN1 KO cells is further supported by the induction of GADD34 (Fig. 2A), a phosphatase cofactor for p-eIF2α that is upregulated during ISR activation as a negative feedback mechanism (51). Following the confirmation of ISR activation, we next interrogated the responsible kinase for the production of p-eIF2α. We found that the relative level of phosphorylated PKR at threonine 446 (p-PKR [T446]), a modification correlated with PKR activation (52, 53), was much higher in XRN1 KO cells compared to control cells, while the total PKR protein levels remained similar (Fig. 2A). There also appeared to be a modest increase of p-PKR (T446) level in uninfected XRN1 KO cells as well, although not nearly to the extent in infected cells (Fig. 2A).
FIG 2.

Enhanced activation of the PKR kinase and ISR pathway in XRN1 knockout cells following MeV infection. WT and XRN1 KO A549 cells were left uninfected (as a control) or were infected, and processed as described in Fig. 1. (A) Lysates were analyzed by quantitative western immunoblotting to probe for PKR and ISR activation with representative blots shown. (B) The bar charts represent the average from 3 independent infected sample sets. P values were calculated using Microsoft Excel Student's t test: *, P < 0.05; **, P < 0.001; ns, not significant. (C) Positive control lysates were analyzed by quantitative western immunoblotting to verify the decreased gel mobility (indicated by arrows) of activated PERK after treatment with thapsigargin (TG) at 400 nM for 4 h, and activated GCN2 after serum starvation for 24 h (top). WT and XRN1 KO cell lysates from (A) were then probed for activation of PERK and GCN2 (bottom).
No evidence was obtained for enhanced activation of 2 other stress-responsive eIF2α kinase pathways, PERK, and GCN2, following MeV infection of the XRN1 KO cells (Fig. 2C). Activation of PERK (54) and GCN2 (55, 56) kinases was assessed by testing for a shift in their gel mobility following phosphorylation. As controls, treatment of uninfected WT cells with thapsigargin (TG) or serum starvation caused selective activation of PERK or GCN2, respectively (Fig. 2C, upper). However, neither WT nor XRN1 KO cells displayed a shift in the mobility of either PERK or GCN2 protein following MeV infection relative to uninfected cells (Fig. 2C, lower).
The observed increase in PKR activity in MeV-infected cells lacking XRN1 (Fig. 2) led us to examine the impact of XRN1 KO on additional host dsRNA sensing pathways such as interferon (IFN) production and RNase L activation. We first tested whether XRN1 KO impacted the induction of beta interferon (IFN-β) or the action of IFN measured by Interferon-stimulated genes (ISG) expression. RNA was isolated from XRN1 KO and WT A549 cells, both uninfected and MeV-infected. Reverse transcription (RT)/quantitative (q) real-time PCR was used to determine the relative transcript levels. As shown in Fig. 3A, XRN1 KO cells expressed a higher basal level of IFN-β and ISG transcripts relative to the uninfected WT control, but both WT and XRN1 KO cells when infected displayed a robust increase in transcript amount for IFN-β and 5 ISGs, MxA, MxB, ISG56, ISG54, and ISG60. These data suggest that, whereas XRN1 can suppress the basal expression of IFN-β and ISGs, it does not prevent the induction of these transcripts upon MeV infection. While quantitative western analysis showed that XRN1 KO had no detectable effect on the protein level of ISG15, another gene that is highly regulated by interferon (Fig. 3B), it remains possible that the IFN pathway contributes to the proviral action of XRN1 given the small, but statistically significant, increase (2-4 fold) of IFN-β and ISG transcript levels observed in the infected XRN1 KO cells compared to the infected WT control (Fig. 3A). We also examined the effect of XRN1 KO on the host antiviral RNase L pathway that is activated in response to the IFN-induced 2′-5′ oligoadenylate synthetase (OAS)-dependent recognition of dsRNA and 2-5A synthesis. Once activated, RNase L catalyzes the degradation of cellular and viral RNAs (57–59). By using cleavage of 28S/18S rRNA as an indicator of RNase L activity (60, 61), we observed a small but detectable amount of rRNA cleavage in total RNA samples isolated from infected XRN1 KO cells; a poly(I·C) treated positive control was included for RNase L-generated cleavage products (62) (Fig. 3C).
FIG 3.
Increase of IFN-β induction and detectable RNase L activation in XRN1 knockout cells relative to parental wild-type cells following MeV infection. WT and XRN1 KO A549 cells were infected as described in Fig. 1. (A) and (C) At 24 hpi total RNA was extracted from uninfected and infected cells. (A) Total RNA from WT and XRN1 KO A549 cells were reverse transcribed into cDNA, and the relative IFN-β mRNA transcript levels determined using qPCR (top), as well as 5 different interferon-stimulated genes: MxA, MxB, ISG56, ISG54, and ISG60 (bottom). The bar charts represent the averages from 3 independent experiments. P values were calculated using Microsoft Excel Student's t test (top) or GraphPad Prism (version 9.2.0) one-way ANOVA and Šidák’s multiple-comparison test for selected pairs (bottom): *, P < 0.05; **, P < 0.001; ns, not significant. (B) At 24 hpi a parallel set of cells were lysed and the level of ISG15 was quantified using western immunoblotting. (C) Total RNA from WT and XRN1 KO A549 cells was analyzed using a Bioanalyzer on an RNA pico chip, including WT and RNase L KO A549 cells transfected with 1 μg/mL poly(I·C) for 6 h as a positive control for 28s/18s rRNA cleavage (indicated by arrows).
Inhibition of PKR-dependent integrated stress response provides a mechanism to explain the proviral activity of XRN1.
Following our observation that XRN1 impaired PKR autophosphorylation/eIF2α phosphorylation and RNase L activation upon MeV infection, we next asked whether this inhibition contributed to the proviral role displayed by XRN1. In order to test this possibility, we utilized an A549 cell line in which both PKR and RNase L were knocked out in the background of the XRN1 knockout (designated as TKO for triple KO). An A549 cell line with only PKR and RNase L knocked out (referred to as DKO for double KO) was included as a control (44). As shown in in Fig. 4A, there was efficient rescue of viral protein expression across the MeV genome in the TKO cells (to an extent comparable to that observed in WT and DKO cells), with the exception of the mature MeV F1 protein. F1 expression surprisingly was not rescued in the TKO when compared to XRN1 KO (Fig. 4B). Accompanying the lack of rescue of MeV F1 expression in TKO cells, we observed an ~1.6-fold accumulation of the pro-protein form F0, the uncleaved version of F1. Consistent with the increased viral protein expression seen in the TKO cells, infectious virus yields produced by TKO were also significantly increased in comparison to XRN1 KO cells (Fig. 4C). The incomplete rescue of virus growth seen with the TKO may be due to the inefficient processing of MeV F protein, and future study would be needed to elucidate the mechanism involved. Taken together, our data indicate that the deficiency of both PKR and RNase L largely restored the MeV growth in the XRN1 KO background. To further delineate the roles of PKR and RNase L in the observed MeV rescue with TKO cells, we used RNAi to individually knockdown (KD) each protein in both WT and XRN1 KO cells and then assessed how their KDs affected MeV growth. Although both siRNAs effectively depleted their targets (>90% knockdown), PKR KD significantly enhanced MeV protein synthesis in the XRN1 KO cells, whereas RNase L KD showed little to no rescue (Fig. 4D).
FIG 4.
Significant restoration of MeV replication in XRN1/PKR/RNase L triple knockout cells compared to XRN1 knockout cells. (A) to (C) WT, XRN1 KO, PKR/RNase L double KO (DKO), and XRN1/PKR/RNase L triple KO (TKO) A549 cells were infected and analyzed as described in Fig. 1. (A) Representative blots of MeV proteins. (B) Bar charts showing the relative average levels of F0 and F1 proteins produced in the different cell lines as determined from 3 independent experiments. (C) Infectious viral yields were determined and are plotted as a scatterplot. Each graph represents the average from 3 independent experiments. P values were calculated using Microsoft Excel Student's t test: *, P < 0.05; **, P < 0.001; ns, not significant. (D) WT and XRN1 KO A549 cells were transfected with a non-targeting control or siRNA targeting either RNase L or PKR on days 1, 3, and 5. Transfected cells were infected on day 7, lysed on day 8, and analyzed as described in Fig. 1. Representative blots are shown.
We also utilized an alternative pharmacological inhibitor approach to examine the role of PKR/ISR in inhibiting MeV in the XRN1 KO cells. Consistent with the lack of PERK and GCN2 activation (Fig. 2C), PKR KD essentially abolished the XRN1 KO-induced eIF2α phosphorylation after MeV infection (Fig. 5A), suggesting that PKR is the major ISR kinase activated in MeV-infected XRN1 KO cells. To directly determine how ISR affected MeV replication in the XRN1 KO cells, we next examined the effect of ISRIB, an ISR inhibitor (54), on the levels of multiple MeV proteins in infected XRN1 KO cells. We found that this drug effectively enhanced viral protein expression (Fig. 5B and C). However, the incomplete restoration of MeV protein expression by ISRIB was not unexpected. ISRIB is capable of inhibiting ISR only during the early infection stage in the case of encephalomyocarditis virus when the eIF2α phosphorylation remains below a critical threshold (63). Collectively, these results suggest that the PKR-mediated ISR is an important contributor to the inhibition of MeV growth seen in the XRN1 KO cells.
FIG 5.

Enhanced MeV protein synthesis in XRN1 knockout cells after treatment with an ISRIB inhibitor. (A) Lysates from Fig. 4D were probed for eIF2α phosphorylation using western immunoblotting. Representative blots are shown. (B) and (C) WT and XRN1 KO A549 cells were seeded at comparable confluence and pretreated for 12 h with either 1 μM ISRIB inhibitor or an equal volume of DMSO solvent as a control. Cells were then infected and processed as described in Fig. 1. The ISRIB treatment continued throughout the 24 h of infection. (B) Representative blots of selected MeV protein levels are shown. (C) Quantification of selected MeV protein levels relative to the DMSO-treated WT cells is presented as a bar chart, representing the average from 3 independent experiments. P values were calculated using Microsoft Excel Student's t test: *, P < 0.05; **, P < 0.001; ns, not significant.
The effect of XRN1 during MeV infection is independent of its role in processing bodies.
To assess whether XRN1’s effect on ISR during MeV infection is dependent upon its role in processing bodies (PBs), we utilized the approach of disrupting PBs without effecting total XRN1 expression and examined whether this resulted in a similar phenotype as the XRN1 KO alone. For this purpose, we disrupted PBs through siRNA knockdown of a core PB structural protein, the eukaryotic translation initiation factor 4E-transporter protein (eIF4E-T) that has been shown to be necessary for PB formation/stability (64). The knockdown efficiency of two independent eIF4E-T siRNAs was verified by both immunofluorescence and western analyses (Fig. 6A and B). We furthermore confirmed that knockdown of eIF4E-T efficiently depleted the PBs in A549 cells (Fig. 6A) without affecting the level of XRN1 (Fig. 6B). However, unlike what was observed in XRN1 KO cells, neither eIF4E-T siRNA caused ISR activation nor had any detectable impact on MeV N or P protein expression (Fig. 6B). These results suggest that the effect XRN1 exerts on MeV is not dependent upon its function in PBs.
FIG 6.
Disruption of processing bodies does not affect MeV replication. WT A549 cells were transfected with a non-targeting control or each of the 2 independent eIF4E-T siRNAs on days 1, 3, and 5. (A) Transfected cell were fixed and stained using antibodies specific to eIF4E-T (to verify the knockdown efficiency) and DCP1A (to label the processing bodies). (B) Transfected cells were infected on day 7, lysed on day 8, and analyzed as described in Fig. 1. Representative blots for ISR activation and selected MeV protein levels are shown.
XRN1 is recruited to viral inclusion bodies in MeV-infected cells.
Because the effect of XRN1 on MeV replication was not dependent upon its PB localization, we examined whether there was any alternative localization of XRN1 within the cytoplasm of MeV-infected cells. For this analysis, we used live-cell imaging because comparison of the staining patterns between WT and XRN1 KO A549 cells revealed that the XRN1 antibody available for the western analyses exhibited a nonspecific IB staining (data not shown). Moreover, live-cell imaging avoids potential artifacts associated with cell fixation. We first validated the ability of transfected EGFP fusion of XRN1 (65) to localize to the PBs in uninfected A549 cells (Fig. 7A). We next conducted live-cell imaging utilizing A549 cells, stably expressing either EGFP (as a negative control) or EGFP-XRN1 fusion, infected by a recombinant MeV expressing an mCherry reporter carrying a nuclear localization signal. We found that EGFP-XRN1, but not EGFP alone, was enriched within large cytoplasmic puncta that are similar to viral IBs but distinguishable from PBs that formed in infected mCherry-positive cells (Fig. 7B). To determine whether these cytoplasmic puncta are viral IBs, we transfected constructs expressing either EGFP or EGFP-XRN1 fusion into A549 cells, and then infected the transfected cells with MeV before examining them by live-cell microscopy. This was done using an A549 cell line stably expressing a WDR5-mCherry fusion protein in order to have a live reference for MeV-induced IBs. It was previously shown that the host protein, WDR5, translocates to viral IBs during MeV infection (12). Interestingly, we found that when compared to EGFP alone, EGFP-XRN1 was more enriched within WDR5-mCherry puncta (Fig. 7C) demonstrating that XRN1 is recruited to MeV IBs.
FIG 7.
Induced translocation of EGFP-XRN1 to inclusion bodies following infection. (A) Validation of utilizing the transfected EGFP-XRN1 fusion for the live-cell imaging reporter as shown by its expected enrichment within the processing bodies (PBs) marked by DCP1A. (B) WT A549 cell lines stably expressing either EGFP or EGFP-XRN1 were infected with MeV expressing mCherry (containing a nuclear localization signal for reference of infected cells) using an MOI of 1.0. At 24 hpi live cells were imaged under a fluorescence microscope to capture representative images of EGFP-XRN1’s MeV-induced localization to large cytoplasmic puncta. (C) WT A549 cells stably expressing WDR5-mCherry fusion were transfected with either EGFP or EGFP-XRN1 plasmids, and then infected using an MOI of 1.0. At 24 hpi live transfected cells were imaged under a fluorescence microscope to capture representative images of EGFP-XRN1’s colocalization with viral IBs using WDR5-mCherry as a live marker for MeV IB.
MeV-induced inclusion bodies accumulate dsRNA more frequently in XRN1 KO than WT cells.
The IB localization of XRN1 prompted us to consider whether XRN1 suppresses PKR activation during infection via a mechanism by which XRN1 acts to impair accumulation of RNA species that have the propensity to form dsRNA structures within IBs. To test this possibility, we used immunofluorescent staining to visualize both dsRNA abundancy within IBs with the dsRNA specific monoclonal antibody J2. MeV P protein was utilized as a marker for viral IBs. The J2 antibody was previously used with MeV to demonstrate dsRNA accumulation in IBs during infection with MeV C KO, a mutant virus known to overproduce viral dsRNA during viral replication in the form of defective interfering RNA (DI-RNA) (7). After infection and staining of both WT and XRN1 KO A549 cells, we found that there was a significant increase in the percentage of infected cells with IBs enriched with dsRNA as measured by J2 staining, increasing from ~8% in WT A549 to ~50% in XRN1 KO cells (Fig. 8A). The time course analysis further indicated that this accumulation of dsRNA in the XRN1 KO cells took place predominantly in the later stages of the infection cycle, with most IB-associated dsRNA enrichment occurring between 24 hpi and 30 hpi (Fig. 8B). Given that viral IBs are thought to represent replication factories for cytoplasmic replicating negative-sense RNA viruses (11), the dsRNA that accumulated in the XRN1 KO cells most likely was of viral origin.
FIG 8.
Increased accumulation of dsRNA within MeV inclusion bodies in XRN1 knockout cells. (A) WT and XRN1 KO A549 cells were infected using an MOI of 0.1. At 36 hpi cells were fixed and stained using antibodies specific to dsRNA (J2) or MeV P (to label viral IBs), and then imaged under a fluorescence microscope to capture representative images (top). The bar chart shows the average percentage of infected cells with dsRNA enriched IBs from 3 independent experiments with 200 infected cells counted for each group per experiment (bottom). P values were calculated using Microsoft Excel Student's t test: *, P < 0.05; **, P < 0.001; ns, not significant. (B) WT and XRN1 KO A549 cells were infected using an MOI of 0.1. Cells were fixed and stained using the J2 and P antibodies and scored at different time intervals of infection (18, 20, 24, 30, and 36 hpi).
DISCUSSION
Viruses have evolved strategies to hijack host cell components to antagonize innate immune responses and subsequently enhance their replication. Herein we demonstrate that XRN1 is a potent proviral factor that enhances the replication of MeV. The conclusion that maximal replication of MeV is dependent upon the presence of XRN1 is based on the reduced viral protein production and reduced infectious virus yields observed in A549 XRN1 CRISPR KO cells infected by MeV (Fig. 1 and 4). We further show that, unlike what has been described for influenza A virus (a negative-sense RNA virus that replicates in the nucleus) (66), the proviral ability of XRN1 to enhance MeV replication was independent of its PB localization (Fig. 6). Instead, we found that XRN1 was recruited to MeV IBs (Fig. 7) where the accumulation of dsRNA is impaired and hence leads to the suppression of dsRNA-mediated innate immunity (Fig. 8). Moreover, our data suggest that the PKR/ISR response represents a key dsRNA-dependent pathway modulated by XRN1 (Fig. 4 and 5), although a role for additional pathways, such as IFN and RNase L (Fig. 3), cannot be ruled out. Moreover, some paramyxoviruses, exemplified by MeV, Sendai virus and simian virus, do not encode PKR antagonists, and are generally assumed to tightly regulate their replication process via virally-encoded C (7, 10, 67) or P/V (68) proteins to prevent PKR activation. Our study demonstrates that in addition to the viral components, MeV requires at least some host factors, including XRN1, to effectively evade the PKR-mediated innate response.
Interestingly, similar to our findings for MeV, XRN1 has been found to promote the replication of vaccinia virus (VV), a DNA virus that also replicates in the cytoplasm, by reducing virus-derived double-stranded RNA (dsRNA) and antagonizing innate immunity (44, 69). However, in contrast to our findings with MeV and earlier work with vaccinia virus, an antiviral role for XRN1 has been suggested in the case of Newcastle disease virus (NDV) (42) and vesicular stomatitis virus (VSV) (43). It is not yet understood why some cytoplasmic negative-sense RNA viruses, such as NDV and VSV, show an antiviral response whereas MeV, also a cytoplasmic negative-sense RNA virus, shows a proviral response to XRN1. Possible contributing parameters include the relative production of dsRNA by the virus that triggers sensors such as PKR, and the extent of the degradation of genomic/antigenomic RNA by XRN1, both determined in part by the particular RNA structure which may vary among the specific viral RNAs. For example, if there is more dsRNA produced which is degraded by XRN1, the effect would be proviral as ISGs such as PKR and OAS would not be activated as efficiently. On the other hand, if genomic/antigenomic RNA is sensitive to degradation the effect may be antiviral as the functional viral RNA would be reduced in concentration (42). The differential ability of viral N proteins to bind and shield viral genomic/antigenomic RNAs from degradation by XRN1 may also be important. Perhaps in the case of MeV, genomic/antigenomic RNA is more completely protected from XRN1 degradation due to more efficient co-replicational encapsidation by N than is the case of NDV or VSV RNAs. Moreover, whereas XRN1 is generally assumed to degrade the bulk mRNA, studies have shown that it may preferentially target specific transcripts (70–72), raising the possibility that XRN1 may display different specificity of various types of virus-derived RNAs among different viruses. Finally, one may speculate that the role of XRN1 in viral infection is modulated by the differential thresholds of innate responses present in different tissue/cell types, illustrated by ADAR1 (73–76).
Since viral IBs are thought to function as cytoplasmic replicating factories for negative-sense RNA viruses, it is tempting to speculate that the dsRNA which accumulates within IBs of MeV-infected XRN1 KO cells is derived from the virus. However, the specific identity of the dsRNA affected by XRN1 has not yet been elucidated. One candidate is defective interfering viral RNA (DI-RNA), a dsRNA error by-product of MeV replication which was previously shown to accumulate within viral IBs and that was found to be a strong activator of PKR especially in cells infected by the MeV C KO mutant (7, 77). Another factor consistent with the notion that DI-RNA is the likely substrate of XRN1 comes from the observation that the fraction of dsRNA enriched IBs in XRN1 KO infected cells showed the greatest increase at late times after infection, between 24 and 30 hpi (Fig. 8B). This coincides with the time of MeV infection when replication of the genome exceeds that of the antigenome and when DI-RNA accumulation within IBs begins to be detected (7, 78). Other possibilities for viral RNA include the leader RNA and mRNAs. Despite the short length, it has suggested that the leader RNA of MeV can activate the RIG I-mediated interferon response (79). Whereas none of the MeV mRNAs were found to induce IFN (79), their effect on PKR activation was not examined.
Regardless of the identity of the IB-associated dsRNA, given that XRN1 only recognizes 5′-monophosphorylated RNA (35), we anticipate that XRN1-mediated reduction in amount of IB-associated dsRNA requires the accessory factors. Candidates include factors that remove the 5′-cap (of MeV mRNAs [45–47]) or 5′ pyrophosphate (of MeV leader RNA [79], or full-length viral RNAs produced during MeV replication and their abortive products such as DI RNAs [79–81]) to generate the 5′-monophosphorylated RNA. In this regard, the VV genome is known to encode 2 of its own decapping protein homologues (D9/D10) that would produce suitable substrates for XRN1 degradation of VV viral mRNA thereby preventing dsRNA accumulation (69). Since MeV does not encode either a viral decapping factor or pyrophosphatase, we presume that the modification requires a host accessory protein that functions together with XRN1. In human cells, the DXO exonuclease and 8 (DCP2, NUDT2, NUDT3, NUDT12, NUDT15, NUDT16, NUDT17, and NUDT19) members of the NUDIX hydrolases have been suggested to be capable of carrying out the decapping and/or pyrophosphatase activities (43, 82, 83). We are currently assessing the ability of these candidates to affect PKR activity and MeV growth.
Our study raises the intriguing question of the relationship between the formation of viral IBs and the triggering of innate immune responses. Recent reports have suggested that in addition to functioning as a viral replication factory, viral IBs also provide a platform to permit viruses to escape the detection of host innate immunity. Disruption of SARS-CoV-2 IBs through inhibition of LLPS was shown to activate the innate immune response through the mitochondrial antiviral-signaling protein (MAVS) pathway (84). Respiratory syncytial virus sequesters the host p65 subunit of NF-κB within its IBs to inhibit an integral step of the antiviral NF-κB activation (85). We provide evidence herein that MeV hijacks XRN1 by localizing the protein to viral IBs as a strategy to suppress innate immunity. Future work is needed to determine whether XRN1 is recruited to viral IBs through a protein-protein or protein-RNA interaction. Another important question concerns the mechanism by which the IB-associated dsRNA results in the activation of PKR, either through the PKR protein gaining access to the dsRNA within IBs or the dsRNA moving out of the IBs and activating the PKR in the cytoplasm. The answer to this question is anticipated to shed insight into the role that viral IBs play in the recognition of innate signals (e.g., dsRNA) by their sensors (e.g., PKR, OAS, MDA5, RIG1, and ADAR1) (86).
Lastly, compared to other MeV proteins, the nearly complete loss of MeV M protein production after XRN1 KO (Fig. 1) was unexpected. M protein is crucial for the formation and budding of progeny MeV virions (87). This drastic decrease in protein production provides an explanation for the large reduction in infectious viral yield seen with XRN1 KO compared to WT A549 cells. The observation that the level of MeV M protein was fully restored in the TKO cells further suggests that a hypersensitivity of MeV M mRNA translation to the PKR/ISR pathway may contribute significantly to the XRN1 KO-induced ablation of MeV M, a model consistent with the efficient rescue of MeV M protein production after siRNA KD of PKR in XRN1 KO cells as well as treatment of XRN1 KO cells with ISRIB (Fig. 4 and 5). Further research is necessary to elucidate the underlying mechanism. Isolation of MeV proteins from the brains of patients with subacute sclerosing panencephalitis (SSPE), a progressive and deadly neurodegenerative disease characterized by persistent MeV infection of the central nervous system, shows a significant depletion of MeV M in parallel with observations of increased fusion between neighboring cells (88). Although evidence suggesting that the deficiency of MeV M protein seen in SSPE specimens is the result of hyperediting of M gene mRNA potentially by the A-to-I ADAR adenosine deaminase (4, 89), our study also raises the possibility of a translational regulation of MeV mRNA in the SSPE patients.
MATERIALS AND MEHTODS
Cells and virus.
A549 cells were cultured with advanced Dulbecco’s modified Eagles medium (DMEM; HyClone DMEM-RS) supplemented with 4% fetal bovine serum (Neuromics), 2 mM GlutaMAX (Gibco), and penicillin (100 μg/mL)-streptomycin (100U/mL) (HyClone). The A549 cell lines stably expressing EGFP, EGFP-XRN1, and WDR5-mCherry were grown in the same medium with an additional 3 μg/mL puromycin. Dissociation of cells was done using Trypsin-EDTA 0.25% (Gibco). XRN1 KO, RNase L KO, PKR/RNase L double KO, and XRN1/PKR/RNase L triple KO, A549 cell lines as well as its parental WT A549 control cells were generously provided by Bernard Moss (NIAID).
For MeV infection we used recombinant mCherry-expressing vac2 MeV with a nuclear localization signal (mCherry NLS) isogenic to the Moraten vaccine strain as previously described (77). Cells were infected with a multiplicity of infection of either 0.1 or 1.0 as indicated. For determination of infectious yields, 6.7 X 105 cells as indicated were infected with an MOI of 1.0, harvested at 24 hpi unless otherwise specified, and stored frozen at −80°C overnight. Cells were then subjected 3 cycles of freeze (liquid nitrogen)/thaw (37°C water bath) followed by centrifugation at 3000 g for 3 min. Vero cells were used to determine infectious virus titers of the supernatant solution using the Spearman-Kärber method (90, 91).
Cell viability assay.
The MTT colorimetric assay was done similarly as described by Mosman (92). Cells were infected with an MOI of 1.0 and after 24 hpi 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT reagent, Invitrogen) was added to the medium with cells from a prepared stock of 5 mg/mL in water to a final concentration of 0.5 mg/mL. Afterwards, cells were incubated at 37°C for 1 h, medium was removed, and MTT dye was released with DMSO at an equal volume to the original medium volume. Cells were incubated at room temperature for 30 min in the dark, the DMSO was then collected, and the cells had their absorbance read at 540 nm with an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences).
DNA and RNAi transfections.
The recombinant plasmid PT7-EGFP-XRN1 expressing the EGFP-XRN1 fusion protein was previously described (65). TurboFect transfection reagent (Thermo Fisher) was used for transfection of plasmids and poly(I·C) at a ratio of 3 μL TurboFect per 1 μg of DNA/poly(I·C).
Small interfering RNA knockdown of eIF4E-T, PKR, and RNase L was performed using Silencer Select Predesigned siRNA from Ambion; eIF4E-T siRNA11 (s32160; target sequence, 5′-CUA CCU CAG UGA UUC GUA ATT-3′), eIF4E-T siRNA12 (s32162; target sequence, 5′-GGA UAG UAG AUC CAC GAG ATT-3′), PKR siRNA (s11185; target sequence, 5′-UAA CGU AAG UCU UUC CGU CAA-3′), and RNase L siRNA (s12066; target sequence, 5′-UUG CAG ACA AGA GAC UUC CCG-3′). The siRNAs were transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen) at a working concentration of 5 nM. To obtain optimal knockdown, cells were transfected with siRNA a total of 3 times (on days 1, 3, and 5).
Antibodies.
Primary antibodies used for western analyses (WB) and immunofluorescence (IF) were as follows; mouse anti-β-actin (BD Transduction Lab. no. 612657; WB, 1:4000), rabbit anti-eIF4E-T (Bethyl no. A300-706A; WB 1:2500), mouse anti-eIF4E-T (Santa Cruz Biotechnology no. sc-393788; IF 1:100), mouse anti-DCP1A (Santa Cruz Biotechnology no. sc-100706; IF 1:250), mouse anti-XRN1 (Santa Cruz Biotechnology no. sc-165985; WB, 1:1000), rabbit anti-eIF2α (Cell Signaling Technology no. 9722; WB, 1:1000), rabbit anti-p-eIF2α(S51) (Abcam no. Ab32157; WB, 1:1500), rabbit anti-GADD34 (Proteintech no. 10449-1-AP; WB, 1:750), mouse anti-PKR (Santa Cruz Biotechnology no. sc-100378; WB, 1:1000), rabbit anti-p-PKR (T446) (Abcam no. Ab32036; WB, 1:1000), mouse anti-RNase L (Santa Cruz Biotechnology no. sc-74405; WB, 1:1000), mouse anti-ISG15 (Santa Cruz Biotechnology no. sc-166755; WB, 1:500), rabbit anti-PERK (Cell Signaling Technology no. 5683; WB, 1:1000), rabbit anti-GCN2 (Cell Signaling Technology no. 3302; WB, 1:750), mouse anti-J2 (English & Scientific Consulting no. J2-1102; IF, 1:250), rabbit anti-MeV N505 (WB, 1:5000), rabbit anti-MeV P254 (WB, 1:5000, IF, 1:5000), rabbit anti-MeV C2 (WB, 1:4000), rabbit anti-MeV VCT (WB, 1:3000), rabbit anti-MeV M81 (WB, 1:2000), rabbit anti-MeV F (WB, 1:4000), rabbit anti-MeV HCYT (WB, 1:4000), rabbit anti-MeV L2170 (WB, 1:2000).
SDS-PAGE and Western blotting.
Radioimmunoprecipitation (RIPA) buffer was used for lysis (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with 1X Halt Protease and Phosphatase Inhibitor Single-Use Cocktail (Thermo Fisher), and 5 mM EDTA. Protein concentrations of lysates were determined with the CB-X protein assay (G-Biosciences). Lysates were analyzed on SDS polyacrylamide gels at either equal volume of lysate (for analysis of viral proteins) or equal amount of total protein (for analysis of host proteins). Protein transfer was done with Immobilon-FL 0.45-μm polyvinylidene difluoride (PVDF) membranes (Millipore) and a mini PROTEAN 3 Cell machine (Bio-Rad). Membranes were incubated overnight in the primary antibody diluted in 1X Western Blotting blocking buffer (Rockland). Membranes were then washed 3 times for 5 min each with 1X PBST (1X PBS supplemented with 0.1% Tween 20), incubated in the dark for 45 min with the DyLight 680 conjugated secondary antibody (Thermo Fisher), 3 times for 5 min each with 1X PBST, washed one time for 5 min with 1X PBS, air dried in the dark, and analyzed on an Odyssey infrared imaging system (LI-COR Biosciences). In the case of a phosphor-specific primary antibody, 1X TBST (1X TBS supplemented with 0.1% Tween 20) and 1X TBS were used instead of 1X PBST and 1X PBS, respectively.
RNA extraction, reverse transcription and quantitative real-time PCR.
RNA extraction was performed on 6.7 X 105 cells at 24 hpi at an MOI of 1.0 using the RNeasy Plus minikit (Qiagen). A total of 1 μg RNA from each sample was converted to cDNA by RT-PCR using the Verso cDNA Synthesis Kit (Thermo Fisher) following the suggested thermal cycling program (42°C for 30 min and 95°C for 2 min). The primers used for RT-PCR of mRNA were a 3:1 (vol/vol) blend of random hexamers and anchored oligo-dT. The cDNA product was then diluted 10-fold with H2O and 2 μL were used for subsequent quantitative PCRs (qPCR). Quantitative PCR was performed using PowerUP SYBR green Master Mix (Thermo Fisher) and primers for IFN-β (forward 5′-AAA CTC ATG AGC AGT CTG CA-3′; reverse 5′-AGG AGA TCT TCA GTT TCG GAG G-3′), MxA (forward 5′-GGC TGT TTA CCA GAC TCC GAC A-3′; reverse 5′-CAC AAA GCC TGG CAG CTC TCT A-3′), MxB (forward 5′-AAA AGC AGC CCT GTG AGG CAT G-3′; reverse 5′-GTG ATC TCC AGG CTG ATG AGC T-3′), ISG56 (forward 5′-GCC TTG CTG AAG TGT GGA GGA A-3′; reverse 5′-ATC CAG GCG ATA GGC AGA GAT C-3′), ISG54 (forward 5′-GGA GCA GAT TCT GAG GCT TTG C-3′; reverse 5′-GGA TGA GGC TTC CAG ACT CCA A-3′), ISG60 (forward 5′-CCT GGA ATG CTT ACG GCA AGC T-3′; reverse 5′-GAG CAT CTG AGA GTC TGC CCA A-3′), GAPDH (forward 5′-AGC AAG AGC ACA AGA GGA AGA G-3′; reverse 5′-GAG CAC AGG GTA CTT TAT TGA TGG-3′) with a Bio-Rad CFX96 Real-Time System (one cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min, and plate read for real-time quantification). The relative values for mRNA levels were determined by normalizing the Ct values to that of GAPDH in each sample, and relative expression levels were calculated using the ΔΔCt method.
Bioanalyzer RNA analysis.
Total RNA samples were analyzed using an Agilent 2100 Bioanalyzer on an RNA 6000 pico chip according to the manufacturer’s instructions.
Immunofluorescent microscopy.
Immunofluorescent staining of dsRNA was performed on infected A549 cells (0.1 MOI; 18, 20, 24, 30 and 36 hpi) as described (7). Coverslips were fixed using 3% formaldehyde for 20 min, quenched with 1.0 M glycine solution for 10 min, permeabilized with 0.5% (vol/vol) Triton X-100 for 10 min, blocked in a buffer consisting of 2.5% (wt/vol) nonfat dry milk and 0.1% Triton X-100 for 30 min, incubated with primary antibody in PBS with 0.1% Triton X-100 for 2 h, washed 3 times with PBS, incubated with fluorescent-conjugated secondary antibodies (Jackson ImmunoResearch) in PBS consisting of 0.1% Triton X-100 for 1.5 h, washed 3 times with PBS and 3 times with nanopure H2O, and air dried in the dark. Coverslips were then mounted on slides with ProLong Gold Antifade Mount with DAPI (Invitrogen) and left in the dark at room temperature for 24 h before imaging. Immunofluorescent staining of DCP1A and eIF4E-T were conducted using the same protocol except the following modifications: 3% formaldehyde for 10 min, permeabilized with 0.1% (vol/vol) Triton X-100 for 7.5 min, blocked in 1X Blocker BSA (10%) in PBS (Thermo Fisher) supplemented with 5% normal goat serum (Jackson ImmunoResearch), and incubated with primary and secondary antibodies in the above blocking solution.
Slides were imaged using an Olympus IX-81 microscope with a Plan-Apochromat 60× oil fluorescence objective (numeric aperture, 1.45; Olympus), Semrock excitation and emission filters, and an X-Cite Exacte illumination source (Lumen Dynamics). Images were captured using a monochrome digital camera (Evolution QEi; Media Cybernetics) controlled with in vivo acquisition software, v.3.2.0 (Media Cybernetics). Image analysis was performed using ImageJ (NIH), and Illustrator (Adobe).
Live-cell fluorescence microscopy.
Glass-bottomed Delta T dishes (Bioptechs) were coated with rat tail collagen type I (BD Biosciences) in 20 mM acetic acid at a concentration of 55 μg/mL in preparation for cell attachment. Cells were imaged at 24 hpi using an Olympus IX-81 microscope while being kept at 37°C with 5% CO2 perfusion (Bioptechs).
Statistical analysis.
Statistical tests were performed by Student's t test using Microsoft Excel. Unless otherwise specified, data are presented as means with bars showing standard errors of the means (SEM). For the ISG qPCR analyses, a one-way ANOVA followed by Šidák’s multiple-comparison test for selected pairs was performed.
ACKNOWLEDGMENTS
We are grateful to Roberto Cattaneo (Mayo Clinic, Rochester, USA) for the MeV antibodies; Cátia Igreja (Max-Planck-Institut, Tübingen, Germany) for the EGFP-XRN1 recombinant plasmid; and Bernard Moss (NIAID, Bethesda, USA) for A549 cell lines (WT, XRN1 KO, PKR/RNase L KO, RNase L KO, and XRN1/PKR/RNase L KO). The work was supported by a University of California Santa Barbara Faculty Research Grant, and by Undergraduate Research and Creative Activities Grants.
We declare that there are no conflicts of interest.
E.B., C.K.P., C.E.S., and D.M. designed the experiments and wrote the manuscript, and E.B., Y.P., and D.M. performed the experiments.
Contributor Information
Dzwokai Ma, Email: dzwokai.ma@lifesci.ucsb.edu.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Lamb RA, Parks GD. 2013. Paramyxoviridae:the viruses and their replication, p 957–995. In Fields BN, Knipe DM, Howley PM (eds), Fields Virology: Sixth Edition. Lippincott, Williams, and Wilkins. Philadelphia, PA, USA. [Google Scholar]
- 2.Brindley MA, Chaudhury S, Plemper RK. 2015. Measles virus glycoprotein complexes preassemble intracellularly and relax during transport to the cell surface in preparation for fusion. J Virol 89:1230–1241. 10.1128/JVI.02754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin LT, Richardson CD. 2016. The host cell receptors for measles virus and their interaction with the viral hemagglutinin (H) protein. Viruses 8:250. 10.3390/v8090250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattaneo R, Kaelin K, Baczko K, Billeter MA. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759–764. 10.1016/0092-8674(89)90679-X. [DOI] [PubMed] [Google Scholar]
- 5.Bellini WJ, Englund G, Rozenblatt S, Arnheiter H, Richardson CD. 1985. Measles virus P gene codes for two proteins. J Virol 53:908–919. 10.1128/JVI.53.3.908-919.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi K, Kadota SI, Takeda M, Miyajima N, Nagata K. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett 545:177–182. 10.1016/S0014-5793(03)00528-3. [DOI] [PubMed] [Google Scholar]
- 7.Pfaller CK, Radeke MJ, Cattaneo R, Samuel CE. 2014. Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J Virol 88:456–468. 10.1128/JVI.02572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parks GD, Alexander-Miller MA. 2013. Paramyxovirus activation and inhibition of innate immune responses. J Mol Biol 425:4872–4892. 10.1016/j.jmb.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson JB, Thomas D, Lewicki H, Billeter MA, Oldstone MB. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80–89. 10.1006/viro.1999.0118. [DOI] [PubMed] [Google Scholar]
- 10.Toth AM, Devaux P, Cattaneo R, Samuel CE. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J Virol 83:961–968. 10.1128/JVI.01669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su JM, Wilson MZ, Samuel CE, Ma D. 2021. Formation and function of liquid-like viral factories in negative-sense single-stranded RNA virus infections. Viruses 13:126. 10.3390/v13010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma D, George CX, Nomburg JL, Pfaller CK, Cattaneo R, Samuel CE. 2018. Upon infection, cellular WD repeat-containing protein 5 (WDR5) localizes to cytoplasmic inclusion bodies and enhances measles virus replication. J Virol 92:e01726-17. 10.1128/JVI.01726-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlos TS, Young DF, Schneider M, Simas JP, Randall RE. 2009. Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J Gen Virol 90:2147–2156. 10.1099/vir.0.012047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cifuentes-Muñoz N, Branttie J, Slaughter KB, Dutch RE. 2017. Human metapneumovirus induces formation of inclusion bodies for efficient genome replication and transcription. J Virol 91:e01282-17. 10.1128/JVI.01282-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, Blondel D. 2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J Virol 83:7948–7958. 10.1128/JVI.00554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog 6:e1000958. 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolnik O, Stevermann L, Kolesnikova L, Becker S. 2015. Marburg virus inclusions: A virus-induced microcompartment and interface to multivesicular bodies and the late endosomal compartment. Eur J Cell Biol 94:323–331. 10.1016/j.ejcb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Hoenen T, Shabman RS, Groseth A, Herwig A, Weber M, Schudt G, Dolnik O, Basler CF, Becker S, Feldmann H. 2012. Inclusion bodies are a site of ebolavirus replication. J Virol 86:11779–11788. 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rincheval V, Lelek M, Gault E, Bouillier C, Sitterlin D, Blouquit-Laye S, Galloux M, Zimmer C, Eleouet JF, Rameix-Welti MA. 2017. Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nat Commun 8:563. 10.1038/s41467-017-00655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudrière-Gesbert C, Gaudin Y, Blondel D. 2017. Negri bodies are viral factories with properties of liquid organelles. Nat Commun 8:58. 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinrich BS, Maliga Z, Stein DA, Hyman AA, Whelan SPJ. 2018. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio 9:e02290-17. 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guseva S, Milles S, Blackledge M, Ruigrok RWH. 2019. The nucleoprotein and phosphoprotein of measles virus. Front Microbiol 10:1832. 10.3389/fmicb.2019.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Su JM, Samuel CE, Ma D. 2019. Measles virus forms inclusion bodies with properties of liquid organelles. J Virol 93:e00948-19. 10.1128/JVI.00948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science (New York, NY) 357:eaaf4382. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 25.Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol 28:420–435. 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomes E, Shorter J. 2019. The molecular language of membraneless organelles. J Biol Chem 294:7115–7127. 10.1074/jbc.TM118.001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberti S, Dormann D. 2019. Liquid-liquid phase separation in disease. Annu Rev Genet 53:171–194. 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 29.Alenquer M, Vale-Costa S, Etibor TA, Ferreira F, Sousa AL, Amorim MJ. 2019. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun 10:1629. 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD, Zimmerman MI, Vithani N, Griffith D, Wagoner JA, Bowman GR, Hall KB, Soranno A, Holehouse AS. 2021. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun 12:1936. 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. 2020. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation stimulated by RNA and partitions into phases of human ribonucleoproteins. bioRxiv. 10.1101/2020.06.09.141101. [DOI]
- 32.Monette A, Niu M, Chen L, Rao S, Gorelick RJ, Mouland AJ. 2020. Pan-retroviral nucleocapsid-mediated phase separation regulates genomic RNA positioning and trafficking. Cell Rep 31:107520. 10.1016/j.celrep.2020.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metrick CM, Koenigsberg AL, Heldwein EE. 2020. Conserved outer tegument component UL11 from herpes simplex virus 1 is an intrinsically disordered, RNA-binding protein. mBio 11:e00810-20. 10.1128/mBio.00810-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Na Z, Slavoff SA. 2018. P-bodies: composition, properties, and functions. Biochemistry 57:2424–2431. 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagarajan VK, Jones CI, Newbury SF, Green PJ. 2013. XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829:590–603. 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 85:1581–1593. 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poblete-Durán N, Prades-Pérez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverría F. 2016. Who regulates whom? An overview of RNA granules and viral infections. Viruses 8:180. 10.3390/v8070180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bavia L, Mosimann AL, Aoki MN, Duarte Dos Santos CN. 2016. A glance at subgenomic flavivirus RNAs and microRNAs in flavivirus infections. Virol J 13:84. 10.1186/s12985-016-0541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazeaud C, Freppel W, Chatel-Chaix L. 2018. The multiples fates of the flavivirus RNA genome during pathogenesis. Front Genet 9:595. 10.3389/fgene.2018.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. 2012. A common strategy for host RNA degradation by divergent viruses. J Virol 86:9527–9530. 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Yamane D, Lemon SM. 2015. Dissecting the roles of the 5’ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J Virol 89:4857–4865. 10.1128/JVI.03692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng CS, Kasumba DM, Fujita T, Luo H. 2020. Spatio-temporal characterization of the antiviral activity of the XRN1-DCP1/2 aggregation against cytoplasmic RNA viruses to prevent cell death. Cell Death Differ 27:2363–2382. 10.1038/s41418-020-0509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laudenbach BT, Krey K, Emslander Q, Andersen LL, Reim A, Scaturro P, Mundigl S, Dächert C, Manske K, Moser M, Ludwig J, Wohlleber D, Kröger A, Binder M, Pichlmair A. 2021. NUDT2 initiates viral RNA degradation by removal of 5′-phosphates. Nat Commun 12:6918. 10.1038/s41467-021-27239-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, Moss B. 2016. Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR-Cas9 knockout cell lines and vaccinia virus mutants. J Virol 90:7864–7879. 10.1128/JVI.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshikawa Y, Mizumoto K, Yamanouchi K. 1986. Characterization of messenger RNAs of measles virus. The J General Virology 67 (Pt 12):2807–2812. 10.1099/0022-1317-67-12-2807. [DOI] [PubMed] [Google Scholar]
- 46.Hercyk N, Horikami SM, Moyer SA. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163:222–225. 10.1016/0042-6822(88)90253-X. [DOI] [PubMed] [Google Scholar]
- 47.Jacobson BA, Sadiq AA, Tang S, Jay-Dixon J, Patel MR, Drees J, Sorenson BS, Russell SJ, Kratzke RA. 2017. Cap-dependent translational control of oncolytic measles virus infection in malignant mesothelioma. Oncotarget 8:63096–63109. 10.18632/oncotarget.18656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y, Zhang Z, Li Y, Li Y. 2021. The regulation of integrated stress response signaling pathway on viral infection and viral antagonism. Front Microbiol 12:814635. 10.3389/fmicb.2021.814635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinnebusch AG. 2000. 5 Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. Cold Spring Harbor Monograph Archive 39:185–243. [Google Scholar]
- 50.Costa-Mattioli M, Walter P. 2020. The integrated stress response: from mechanism to disease. Science (New York, NY) 368:eaat5314. 10.1126/science.aat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kojima E, Takeuchi A, Haneda M, Yagi F, Hasegawa T, Yamaki K, Takeda K, Akira S, Shimokata K, Isobe K. 2003. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress - elucidation by GADD34-deficient mice. FASEB J 17:1573–1575. 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- 52.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol 18:2282–2297. 10.1128/MCB.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122:901–913. 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 54.Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, Okreglak V, Ashkenazi A, Hann B, Nader K, Arkin MR, Renslo AR, Sonenberg N, Walter P. 2013. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2:e00498. 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GCN2 Antibody. Cell signaling technology. https://www.cellsignal.com/products/primary-antibodies/gcn2-antibody/3302. Accessed 10 August 2022.
- 56.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 57.Silverman RH, Weiss SR. 2014. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J Interferon Cytokine Res 34:455–463. 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennan-Laun SE, Ezelle HJ, Li XL, Hassel BA. 2014. RNase-L control of cellular mRNAs: roles in biologic functions and mechanisms of substrate targeting. J Interferon Cytokine Res 34:275–288. 10.1089/jir.2013.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman R. 1997. 2-5A-dependent RNase L: a regulated endoribonuclease in the interferon system, p 515–551. In D’Alessio G, Riordan JF (ed), Ribonucleases: Structure and Functions. Academic Press, Cambridge, MA USA. [Google Scholar]
- 61.Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. 1983. rRNA cleavage as an index of ppp(A2’p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol 46:1051–1055. 10.1128/JVI.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandey M, Bajaj GD, Rath PC. 2004. Induction of the interferon-inducible RNA-degrading enzyme, RNase L by stress-inducing agents in the human cervical carcinoma cells. RNA Biology 1:20–26. 10.4161/rna.1.1.896. [DOI] [PubMed] [Google Scholar]
- 63.Rabouw HH, Langereis MA, Anand AA, Visser LJ, de Groot RJ, Walter P, van Kuppeveld FJM. 2019. Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc Natl Acad Sci USA 116:2097–2102. 10.1073/pnas.1815767116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717–727. 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang CT, Muthukumar S, Weber R, Levdansky Y, Chen Y, Bhandari D, Igreja C, Wohlbold L, Valkov E, Izaurralde E. 2019. A low-complexity region in human XRN1 directly recruits deadenylation and decapping factors in 5′-3′ messenger RNA decay. Nucleic Acids Res 47:9282–9295. 10.1093/nar/gkz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu YC, Mok BW, Wang P, Kuo RL, Chen H, Shih SR. 2021. Cellular 5′-3′ mRNA exoribonuclease XRN1 inhibits interferon beta activation and facilitates influenza A virus replication. mBio 12:e0094521. 10.1128/mBio.00945-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J Virol 82:10102–10110. 10.1128/JVI.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gainey MD, Dillon PJ, Clark KM, Manuse MJ, Parks GD. 2008. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J Virol 82:828–839. 10.1128/JVI.02023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burgess HM, Mohr I. 2015. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17:332–344. 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souret FF, Kastenmayer JP, Green PJ. 2004. AtXRN4 Degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15:173–183. 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 71.Rymarquis LA, Souret FF, Green PJ. 2011. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17:501–511. 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takaoka S, Yanagiya A, Mohamed HMA, Higa R, Abe T, Inoue K, Takahashi A, Stoney P, Yamamoto T. 2021. Neuronal XRN1 is required for maintenance of whole-body metabolic homeostasis. iScience 24:103151. 10.1016/j.isci.2021.103151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.George CX, Ramaswami G, Li JB, Samuel CE. 2016. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem 291:6158–6168. 10.1074/jbc.M115.709014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, Gupte A, Keegan LP, George CX, Ramu A, Huang N, Pollina EA, Leeman DS, Rustighi A, Goh YPS, Chawla A, Del Sal G, Peltz G, Brunet A, Conrad DF, Samuel CE, O'Connell MA, Walkley CR, Nishikura K, Li JB, Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550:249–254. 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R. 2018. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol 16:e2006577. 10.1371/journal.pbio.2006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maurano M, Snyder JM, Connelly C, Henao-Mejia J, Sidrauski C, Stetson DB. 2021. Protein kinase R and the integrated stress response drive immunopathology caused by mutations in the RNA deaminase ADAR1. Immunity 54:1948–1960. 10.1016/j.immuni.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfaller CK, Mastorakos GM, Matchett WE, Ma X, Samuel CE, Cattaneo R. 2015. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol 89:7735–7747. 10.1128/JVI.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plumet S, Duprex WP, Gerlier D. 2005. Dynamics of viral RNA synthesis during measles virus infection. J Virol 79:6900–6908. 10.1128/JVI.79.11.6900-6908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One 2:e279. 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hefti E, Bishop DH. 1975. The 5’ nucleotide sequence of vesicular stomatitis viral RNA. J Virol 15:90–96. 10.1128/JVI.15.1.90-96.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leppert M, Kolakofsky D. 1978. 5’ Terminus of defective and nondefective Sendai viral genomes is ppp Ap. J Virol 25:427–432. 10.1128/JVI.25.1.427-432.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carreras-Puigvert J, Zitnik M, Jemth AS, Carter M, Unterlass JE, Hallström B, Loseva O, Karem Z, Calderón-Montaño JM, Lindskog C, Edqvist PH, Matuszewski DJ, Ait Blal H, Berntsson RPA, Häggblad M, Martens U, Studham M, Lundgren B, Wählby C, Sonnhammer ELL, Lundberg E, Stenmark P, Zupan B, Helleday T. 2017. A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat Commun 8:1541. 10.1038/s41467-017-01642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borbolis F, Syntichaki P. 2022. Biological implications of decapping: beyond bulk mRNA decay. FEBS J 289:1457–1475. 10.1111/febs.15798. [DOI] [PubMed] [Google Scholar]
- 84.Wang S, Dai T, Qin Z, Pan T, Chu F, Lou L, Zhang L, Yang B, Huang H, Lu H, Zhou F. 2021. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat Cell Biol 23:718–732. 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 85.Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. 2020. Respiratory syncytial virus sequesters NF-κB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J Virol 94:e01380-20. 10.1128/JVI.01380-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samuel CE. 2019. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. J Biol Chem 294:1710–1720. 10.1074/jbc.TM118.004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cox RM, Plemper RK. 2017. Structure and organization of paramyxovirus particles. Curr Opin Virol 24:105–114. 10.1016/j.coviro.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter MA, Cattaneo R. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J 17:3899–3908. 10.1093/emboj/17.14.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patterson JB, Cornu TI, Redwine J, Dales S, Lewicki H, Holz A, Thomas D, Billeter MA, Oldstone MB. 2001. Evidence that the hypermutated M protein of a subacute sclerosing panencephalitis measles virus actively contributes to the chronic progressive CNS disease. Virology 291:215–225. 10.1006/viro.2001.1182. [DOI] [PubMed] [Google Scholar]
- 90.Devaux P, Hodge G, McChesney MB, Cattaneo R. 2008. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J Virol 82:5359–5367. 10.1128/JVI.00169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kärber G. 1931. Beitrag zur kollektiven behandlung pharmakologischer reihenversuche. Archiv f Experiment Pathol u Pharmakol 162:480–483. 10.1007/BF01863914. [DOI] [Google Scholar]
- 92.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]