Abstract
Background and Aims
Paediatric inflammatory bowel disease [IBD] is characterized by altered immunological and metabolic pathways. Metabolomics may therefore increase pathophysiological understanding and could develop into characterization of biomarkers for diagnosis and IBD treatment response. However, no uniform metabolomic profiles have been identified to date. This systematic review aimed to identify faecal metabolomic signatures in paediatric IBD vs controls, and to describe metabolites associated with disease activity and treatment response.
Methods
A literature search was performed in Embase, Medline, Web of Science and Cochrane Library. Studies assessing faecal metabolomics in paediatric patients < 18 years with IBD [de novo, active, inactive] with comparative groups [IBD vs non-IBD; responders vs non-responders] were included. The quality of included studies was assessed according to the Newcastle–Ottawa Scale.
Results
Nineteen studies were included [540 patients with IBD, 386 controls], assessing faecal short-chain fatty acids [SCFA] [five studies], amino acids [AA] [ten studies], bile acids [BA] [eight studies] and other metabolites [nine studies] using various methodologies. Significantly increased levels of AA [particularly phenylalanine], primary BA and lower levels of secondary BA were described in paediatric IBD compared to controls. Faecal SCFA results varied across studies. Additionally, responders and non-responders to exclusive enteral nutrition and infliximab showed differences in baseline faecal metabolites [based on BA, AA].
Conclusions
This systematic review provides evidence for distinct faecal metabolomic profiles in paediatric IBD. However, results varied across studies, possibly due to differences in study design and applied analytical techniques. Faecal metabolomics could provide more insight into host–microbial interactions in IBD, but further studies with standardized methodologies and reporting are needed.
Keywords: Inflammatory bowel disease, paediatrics, metabolomics
1. Introduction
Inflammatory bowel disease [IBD], consisting of the two main entities Crohn’s disease [CD] and ulcerative colitis [UC], is a chronic relapsing–remitting disorder of the gastrointestinal [GI] tract.1 Over the past few decades, a steady increase in incidence of paediatric IBD has been observed2,3 and a further rise is expected.4 Although the pathogenesis of IBD is not completely understood, the most widely accepted hypothesis is that IBD results from an inappropriate immune response following the interplay between host and the gut microbiota, in a genetically susceptible host.5 Changes in the gut microbiota composition have been reported extensively in IBD,6 commonly characterized by depletion of Firmicutes such as Faecalibacterium prausnitzii and Roseburia hominis and an increase in Proteobacteria.7 These shifts in gut microbiota composition may provoke changes in metabolic pathways.8
Metabolomics is a relatively novel omics technique that can shed light on the complex interplay between the host, gut microbiota and environmental factors such as diet. Metabolomic analysis allows for quantification of small molecules in biological samples such as urine, serum or faeces.9 Discriminatory metabolites in different conditions such as diabetes, liver disease and different cancers have been identified using metabolomics.10 Differences in several metabolite classes have also been found in patients with IBD compared to healthy individuals.5,11–13 The main metabolite classes that have been described are short-chain fatty acids [SCFA], amino acids [AA] and bile acids [BA]. These findings have improved our understanding of the metabolic pathways involved and host–microbial interactions in IBD, and indicate a potential role for metabolomic analysis to serve as a [adjuvant] diagnostic biomarker.
The faecal metabolome in particular is considered to provide a functional readout of gut microbial activity.14 Several longitudinal studies have observed alterations in faecal metabolites linked to clinical remission and response to therapy.15–17 This illustrates that faecal metabolomic analysis also has the potential to serve as a [adjuvant] biomarker to monitor disease activity in IBD, and potentially even to predict treatment response. Consequently, more personalized, tailored disease management strategies could be developed with identification of clear metabolomic signatures.
Although the growing interest in host–microbial interactions has resulted in an expansion of metabolomics studies, previous studies on the faecal metabolome in children with IBD are characterized by mostly small cohorts. So far, no disease-specific metabolomic signature has been found. Therefore, the aim of this systematic review was to summarize the available evidence on the differences in faecal metabolomics between children with IBD vs controls, with the aim of finding an IBD signature or identifying general characteristics of the metabolome. Secondly, we reviewed the role of metabolomic analysis to predict treatment response and monitor disease activity in paediatric IBD.
2. Materials and Methods
2.1 Search strategy
A literature search was performed based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses [PRISMA] statement.18 To identify all relevant publications, systematic searches in the bibliographic databases Medline [PubMed], Embase [Ovid], Web of Science [Core Collection] and Wiley/Cochrane Library were conducted from inception to November 24, 2021, by a medical information specialist. The following terms were used [including synonyms and closely related words] as index terms or free-text words: ‘Inflammatory Bowel Diseases’, ‘Metabolomics’, ‘Multiomics’, ‘Children’, ‘Adolescent’. The references of the identified articles were searched for relevant publications. Duplicate articles were excluded. The full search strategies for all databases can be found in the Supplementary material.
2.2 Study selection and patient population
Studies were included if they met the following criteria: [1] studies consisting of patients with [newly diagnosed, active and inactive] IBD (including the phenotypes CD, UC and IBD-unclassified [IBD-U]); [2] paediatric patients < 18 years old; [3] studies on faecal metabolomic analysis; and [4] inclusion of a comparative group [i.e. IBD vs non-IBD; responders vs non-responders].
Studies were excluded if they were systematic reviews, guidelines, editorials, or case series and case reports; studies comprising adults with IBD and studies with no comparative group; transcriptomic and proteomics studies; and in languages other than Dutch, English, Spanish or French. Animal studies and conference proceeding abstracts were also excluded.
Two reviewers [J.Z.J. and C.M.V.] independently screened all potentially relevant titles and abstracts for eligibility. If necessary, the full-text article was checked for the eligibility criteria. Differences in judgement were resolved through a consensus procedure.
2.3 Outcome assessment
The primary outcome was the difference in faecal metabolites [including metabolite classes SCFA, AA and BA] between children with IBD as compared to controls. Secondary outcomes were faecal metabolites associated with disease activity, and faecal metabolites associated with response or non-response to therapy in paediatric patients with IBD. We accepted all measurement methods for metabolomic analysis including liquid chromatography [LC], high-pressure LC [HPLC], gas chromatography [GC] coupled with mass spectrometry [MS] or nuclear magnetic resonance [NMR] spectroscopy.
2.4 Eligibility assessment and data extraction
Two reviewers [J.Z.J. and C.M.V.] independently screened abstracts for eligibility of each article. Resulting full-text articles were independently reviewed for complete analysis. Relevant data from papers as well as any available supplements were extracted independently by the two reviewers [J.Z.J. and C.M.V.] using predefined data extraction forms. Disagreements were resolved by discussion between the reviewers and remaining discordance was resolved by consulting a third author.
The following data were extracted from the selected studies: author names, the year of study, country, number of participants, IBD subtype [CD, UC or IBD-U] and control type, disease activity, disease localization, use of concomitant medication, metabolomics approach and analytical technique, and details regarding treatment response [definition and type of treatment]. Data on faecal metabolomics including all metabolites assessed and concentrations of unique metabolites were collected.
2.5 Methodological quality appraisal
The Newcastle–Ottawa Scale [NOS] was used to assess the quality of included studies. This scale consists of three domains with a maximum of nine stars and is applied to case-control and cohort studies. For case-control studies the domains include selection of cases and controls, comparability of cases and controls, and ascertainment of exposure, and for cohort studies they include selection of cohorts, comparability of cohorts and assessment of outcome. Two reviewers [J.Z.J. and C.M.V.] applied the NOS independently and differences were resolved by consensus.
3. Results
3.1 Study selection
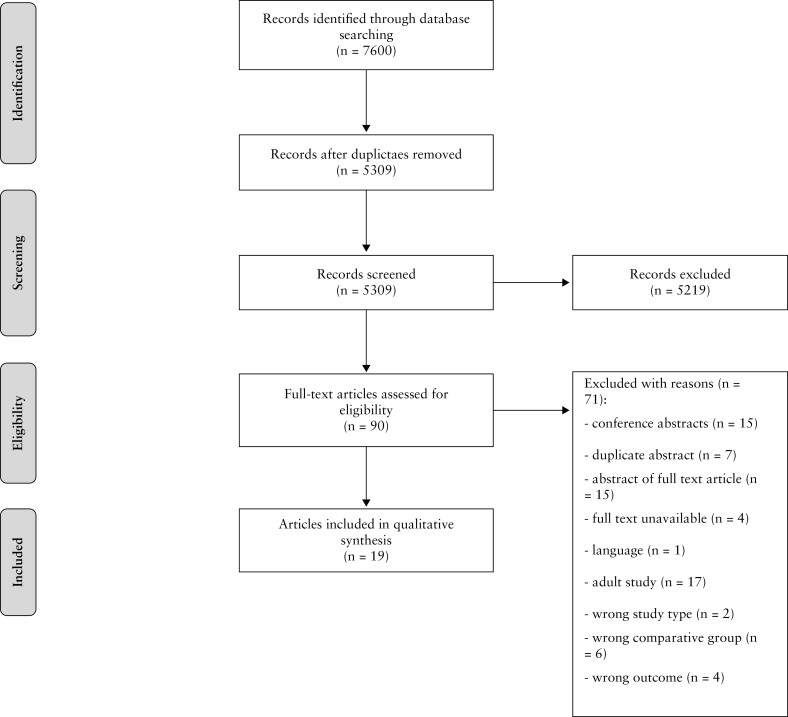
The literature search generated a total of 7600 citations: 2946 in Medline [PubMed], 2751 in Embase [Ovid], 1638 in Web of Science and 265 in Cochrane Library. After removing duplicates of citations that were selected from more than one database, 5309 references remained. In total, 5219 citations were excluded based on title and abstract relevance. The remaining 90 articles were reviewed, of which 19 were included for final analysis. The flow chart of the search and selection process is depicted in Figure 1. Reference lists of the included articles were searched for relevant publications. However, no additional relevant publications were identified. Fifteen relevant conference abstracts were identified for which the authors were contacted for additional data or possible full-text publication. This did not lead to additional inclusion of studies [no published full manuscript n = 8, abstract not found n = 4, contact details not found n = 1, no reply from author n = 2].
Figure 1.
Flowchart of the search and selection procedure of studies.
3.2 Study characteristics
This review included 19 studies comprising 540 unique children with IBD and 386 controls. Amongst these studies, nine studies assessed the faecal metabolome in both CD and UC, nine studies in children with CD and one study in children with UC only. Thirteen studies recruited healthy children as a control group, two studies recruited healthy unaffected siblings, and two studies recruited intention-to-diagnose controls who underwent endoscopy to exclude IBD. Regarding disease activity, 14 studies included patients with active IBD, two with patients in remission, and three with both active and inactive disease. Seven studies conducted metabolomic analysis in a cohort of newly diagnosed, treatment-naïve patients, while in the other 12 studies the IBD patients used medication. The characteristics of each individual study are depicted in Table 1. Only three studies19–21 explicitly reported on sample collection before bowel preparation.
Table 1.
The characteristics of the included studies [n = 19]
| First author [publication year] | Country | Disease [n] | Type | Disease localization | Medication | Approach | Analytical technique | Multiple comparison correction |
|---|---|---|---|---|---|---|---|---|
| Alghamdi [2018] | UK | CD [11] | Active [PCDAI > 10]Newly diagnosed, treatment-naïve [n= 7] | NR | EEN, AZA, 5-ASA | Untargeted | LC-MS | Yes |
| Controls [11] | Healthy | — | — | |||||
| Ashton [2017] | UK | CD [4] | Newly diagnosed, treatment-naïve | L3 [n= 4], L4 [n= 1] | None | Targeted [SCFA] | GC | No |
| UC [1] | Newly diagnosed, treatment-naïve | E4 [n= 1] | None | |||||
| IBD-U [1] | Newly diagnosed, treatment-naïve | NR | None | |||||
| Controls [3] | Healthy siblings | — | — | |||||
| Bosch [2018] | Netherlands | CD [15] | Newly diagnosed, treatment-naïve | L2 [n= 7], L3 [n= 8], L4 [n= 2], P [n= 3] | None | Targeted [AA] | HPLC | Yes |
| UC [15] | Newly diagnosed, treatment-naïve | E1 [n= 3], E2 [n= 2], E3 [n= 10] | None | |||||
| Controls [15] | Healthy | — | — | |||||
| Bosch [2020] | Netherlands | CD [5] | Newly diagnosed, treatment-naïve | L1 [n= 2], L3 [n= 3], L4 [n= 1], P [n= 2] | None | Targeted [AA] | HPLC | Yes |
| UC [6] | Newly diagnosed, treatment-naïve | NR | None | |||||
| Controls [8] | Intention-to-diagnose | — | — | |||||
| Bushman [2020] | USA | CD [20] | Mean PCDAI 18.3 [12.1 SD] | L1 [n= 2], L2 [n= 3], L3 [n= 14], L4 [n= 12] | NR | Untargeted | UPLC-MS | Yes |
| UC [3] IBD-U [4] | Mean PUCAI 19.3 [18.1 SD] | E1 [n= 1], E2 [n= 4], E4 [n= 2] | NR | |||||
| Controls [38] | Healthy | — | — | |||||
| Connors [2019] | Canada | CD [17] | Active | L1 [n= 3], L3 [n= 14] | AZA, MTX | Targeted [BA] | LC-MS-MS | No |
| Diederen [2020] | Netherlands | CD [43] | Newly diagnosed treatment-naïve | L1 [n= 5], L2 [n= 9], L3 [n= 29] | None | Untargeted and targeted [AA, BA] | 1H NMR spectroscopy and HPLC | Yes |
| Controls [18] | Healthy | — | — | |||||
| Ejderhamn [1991] | Sweden | CD [2] | Clinical remission [not specified] | Colonic [n= 1], Ileocolonic [n= 1] | NR | Targeted [BA] | GLC-MS | No |
| UC [16] | Clinical remission [not specified] | Distal colon only [n= 4], Pancolitis [n= 12] | Sulphasalazine | |||||
| Controls [5] | Healthy | — | — | |||||
| Gerasimidis [2014] | UK | CD [15] | Active, newly diagnosed, treatment-naïve [n= 11] | L1 [n= 1], L2 [n= 5], L3 [n= 9], L4 [n= 12] | Background medication [not specified], none | Targeted [SCFA] | GC | No |
| Controls [21] | Healthy | — | — | |||||
| Jacobs [2016] | USA | CD [17] | Clinical remission [HBI < 5] | Small intestine [n= 15], Colon involvement [n= 10], upper GI involvement [n= 6], perianal [n= 7], colon only [n= 2] | Anti-TNF, MTX, thiopurines, 5-ASA | Untargeted | UPLC/ToFMS | Yes |
| UC [4] | Clinical remission [Mayo score < 2] | NR | ||||||
| Controls [21] | Healthy siblings | — | — | |||||
| Jagt [2021] | Netherlands | CD [40] | Newly diagnosed, treatment-naïve | L1 [n= 4], L2 [n= 11], L3 [n= 25], L4 [n= 15], P [n= 6] | None | Targeted [AA] | HPLC | Yes |
| UC [38] | Newly diagnosed, treatment-naïve | E1 [n= 2], E2 [n= 6], E3 [n= 2], E4 [n= 28] | None | |||||
| Controls [105] | Healthy [n= 84] and GI symptoms [n= 21] | — | — | |||||
| Kolho [2017] | Finland | CD [13] | Newly diagnosed, treatment-naïve | NR | None | Untargeted | UPLC-MS | No |
| UC/IBD-U [10] | Newly diagnosed, treatment-naïve | NR | None | |||||
| Controls [14] | Intention-to-diagnose | — | — | |||||
| Ni [2017] | USA | CD [90]† | Active [PCDAI > 10] | Upper [n= 51], Ileum [n= 80], Colon [n= 84], Perianal [n= 15] | Antibiotics, 5-ASA, thiopurines, MTX, systemic steroids, rectal steroids | Untargeted and targeted [AA] | LC-MS and HPLC | Yes |
| Controls [26] | Healthy | — | — | |||||
| Rotondo-Trivette [2021] | USA | UC [23] | Active [mild–moderate] | NR | Biological, immunomodulator, 5-ASA, corticosteroids | Targeted [BA] | LC-MS | No |
| Controls [29] | Healthy | — | — | |||||
| Sundqvist [2019] | Sweden | CD [28] | Active [PCDAI > 15] | Small intestine [n= 5], small intestine/colon [n= 15], colon [n= 2], small intestine/colon/anal [n= 6] | EEN, 5-ASA, other [not specified] | Targeted [faecal tryptic activity, coprostanol, urobilinogen] | Spectrophotometry [faecal tryptic activity, urobilinogen], GLC [coprostanol] | Yes |
| Controls [28]‡ | Healthy | — | — | |||||
| Taylor [2020] | UK | CD [22] | Disease duration > 6 months | L1 [n= 1], L2 [n= 9]L3 [n= 10], L4 [n= 13], Perianal [n= 4] | Biological [infliximab/vedolizumab], thiopurines, MTX | Untargeted | 1H NMR spectroscopy | No |
| Tjellström [2012] | Sweden | CD [14] | Active [clinical index ≥ 10]§ | Small bowel/colon, perianal region | 5-ASA, AZA | Targeted [SCFA] | GLC | No |
| Controls [12] | Healthy | — | — | |||||
| Treem [1994] | USA | CD [22] | Active [mild–moderate, PCDAI ≥ 11] [n= 10], Inactive [PCDAI ≤ 10] [n= 12] | Patchy colonic, terminal ileum involvement | 5-ASA | Targeted [SCFA] | GLC | No |
| UC [17] | Inactive and mild [n= 12], moderate–severe [n= 5]¶ | Pancolitis | 5-ASA | |||||
| Controls [12] | Healthy | — | — | |||||
| Wang [2021] | China | CD [24] | Newly diagnosed, treatment-naïve | L2, L3, L4 | None | Untargeted | UPLC-MS | Yes |
| Controls [20] | Healthy | — | — |
Age < 20 years old.
Healthy controls aged 5–22 years.
Clinical index: consists of the three history items [abdominal pain, number of liquid stools and general well-being], three physical examination items [weight loss, abdominal examination and perirectal disease] of the PCDAI score, and no laboratory tests.
Based on Truelove–Witts criteria.
5-ASA, 5-aminosalicylic acid; AA, amino acids; AZA, azathioprine; BA, bile acids; CD, Crohn’s disease; E1, proctitis; E2, left-sided; E3, extensive; E4, pancolitis; EEN, enteral exclusive nutrition; GI, gastrointestinal; GLC, gas–liquid chromatography; HBI, Harvey–Bradshaw index; 1H NMR, proton nuclear magnetic resonance; HPLC, high-performance liquid chromatography; IBD-U, inflammatory bowel disease unclassified; L1, ileal; L2, colonic; L3, ileocolonic; L4, upper GI; LC-MS, liquid chromatography–mass spectrometry; MTX, methotrexate; NR, not reported; P, perianal; PCDAI, paediatric Crohn’s disease activity index; PUCAI, paediatric ulcerative colitis activity index; SCFA, short-chain fatty acids; ToFMS, time-of-flight mass spectrometry; UC, ulcerative colitis; UPLC-MS, ultra-performance liquid chromatography–mass spectrometry.
3.2.1 Metabolomic analysis and techniques
Of the 19 included studies, ten reported on AA, five on SCFA and eight on BA. Nine studies reported on metabolites outside of these categories. Included studies varied in their analytical approach, with 11 studies using a targeted approach,19,21–30 six using an untargeted approach16,20,31–34 and two that performed both.35,36 Analytical techniques varied across the different studies. Methods that were used were chromatography ([ultra] high-performance, gas or liquid) optionally coupled with MS, or proton NMR [H-NMR].37 An overview can be found in Table 2.
Table 2.
Analytical techniques used in selected metabolomics studies and their targets
| Method | Target | References |
|---|---|---|
| GC | SCFA | 22,25 |
| GLC | SCFA | 27,28 |
| GLC-MS | BA | 24 |
| 1H NMR | Untargeted | 16,35 |
| HPLC | Untargeted, AA, BA | 19,21,35,36 |
| LC-MS | Untargeted | 31,36 |
| LC-MS-MS | BA | 23 |
| Spectrophotometry | Other | 26 |
| UPLC-MS | Untargeted | 20,32–34 |
AA, amino acids; BA, bile acids; GC, gas chromatography; GLC, gas–liquid chromatography; 1H NMR, proton nuclear magnetic resonance; HPLC, high-performance liquid chromatography; LC-MS, liquid chromatography–mass spectrometry; SCFA, short-chain fatty acids; UPLC-MS, ultra-performance liquid chromatography–mass spectrometry.
The metabolomics workflow after sample acquisition consists of different analytical steps.38 Untargeted analysis allows broad assessment of metabolic changes across different metabolite classes, resulting in very high data output but allowing for discovery of unexpected molecules. In contrast, targeted analysis focuses on specific predefined metabolic classes leading to more accurate findings within that limited class.
Two main detection methods are commonly used in the metabolomics workflow: MS and NMR. MS is used to identify and quantify metabolites after separation with GC or LC. In MS, ions are accelerated by attraction and deflected by a magnetic field, after which they are detected and provide a spectrum.39 Samples detected by MS require separation prior to detection. The separation method used depends on the objected metabolite to be measured. GC is typically used for volatile chemicals such as SCFA. LC separates molecules in the liquid mobile phase, giving the advantage that a much wider range of analytes can be measured.38 NMR is a detection method that does not require prior separation of metabolites, and can measure different kinds of metabolites simultaneously. In NMR, an external magnetic field is applied, causing an energy transfer derived from nuclear spins of certain metabolites. The signal matching the energy transfer can be measured and processed, leading to a spectrum.40 However, NMR has a very low sensitivity, which leads to low-abundance metabolites being missed. Therefore, MS is nowadays more frequently used.
After this step, different analytical platforms are used for data interpretation due to the high-throughput data.41 This leads to varying annotation, interpretation and reporting across studies investigating the faecal metabolome.
3.3 Quality assessment
The NOS was applied to 16 case-control and three cohort studies. The quality assessment of included studies according to this scale is depicted in Supplementary Tables 1 and 2. The majority of the case-control studies gave a clear description of the selection [13/16 studies] and definition of paediatric IBD [11/16 studies] but this was less often the case for controls [selection 7/16 studies; definition 10/16 studies]. In eight out of the 19 included studies [42%], cases and controls were age-matched, while sex-matching was performed in 5/19 studies [26%]. Only three out of 16 case-control studies [19%] described if the metabolomics assessment was blinded.
3.4 Faecal metabolomics in IBD vs controls
3.4.1 Short-chain fatty acids
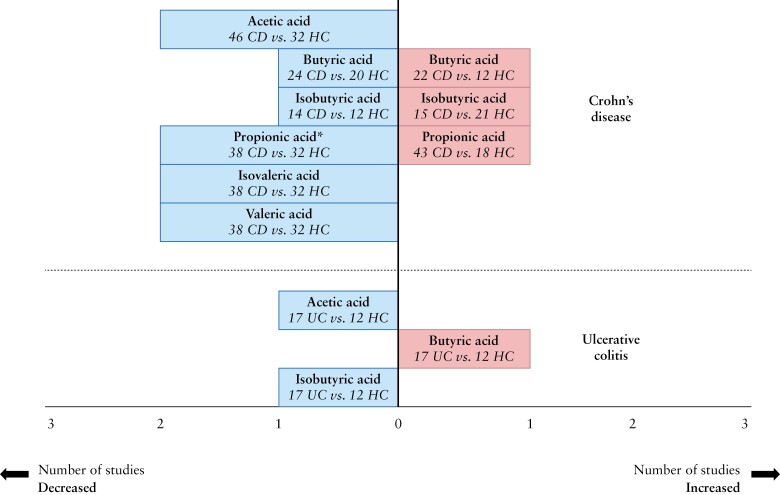
Five studies reported on faecal SCFA in 120 children with IBD [101 CD, 18 UC, one IBD-U] and 68 controls [65 healthy, three unaffected siblings] using different methodologies [GC, GLC, UPLC-MS].22,25,27,28,34 Overall, studies reporting on faecal SCFA levels in children with IBD [with concomitant medication] showed no statistically significant differences in cumulative [absolute] levels compared to healthy controls.25,28 Acetic, butyric and propionic acid levels were most prevalent in samples of children with IBD, similar to controls.22,25,27,28
Absolute acetic, isovaleric and valeric acid levels were generally significantly lower or showed no difference in children with CD compared to healthy controls.22,25,27,28,34 Gerasimidis et al.25 found that absolute values for isovaleric acid were similar, but the percentage contribution of isovaleric acid to the total SCFA pool was significantly higher in newly diagnosed children compared to healthy controls. Results for propionic, butyric and isobutyric acid were inconsistent across studies, reporting on significantly higher, lower or no difference in abundance, as shown in Figure 2.22,25,27,28,34 Tjellström et al.27 found both decreased and similar levels of propionic acid compared to controls depending on the primary disease location, while Diederen et al.35 found significantly higher concentrations in newly diagnosed CD irrespective of disease location. Concentrations of SCFA from individual studies are depicted in Supplementary Table 3.
Figure 2.
Findings on faecal SCFA in children with IBD vs controls [six studies in Crohn’s disease22,25,27,28,34,35 and one study in ulcerative colitis28. All described SCFA were included in this figure. The x-axis represents the number of studies in which the SCFA were increased or decreased, in the IBD phenotype compared to controls. The y-axis represents the unique SCFA.
*The study by Tjellström et al. is included, in which propionic acid was increased in Crohn’s disease patients with perianal disease but not in children with only small bowel involvement.
Only one study reported on specific SCFA in relation to disease localization in children with active CD with ongoing concomitant treatment [azathioprine, 5-aminosalicylate]. In primary small bowel and colonic localization, decreased levels of isobutyric acid, isovaleric acid and valeric acid were observed, while decreased amounts of propionic acid, isobutyric acid, isovaleric acid and valeric acid were found in a small subgroup of patients with primary perianal disease. No differences in acetic and butyric acid levels were observed between disease locations.27
Treem et al. 28 reported on the only study that specifically assessed SCFA levels in children with UC with pancolitis compared to controls. This study showed increased butyric acid levels while isobutyric acid and acetic acid were decreased compared to healthy controls [Figure 2]. This study also compared patients with CD to UC in terms of SCFA: patients with CD had significantly higher isobutyric and isovaleric acid than patients with UC, while all other SCFA were not significantly different between the two groups.28
A summary of the SCFA results is displayed in Figure 2.
3.4.2 Amino acids and their derivatives
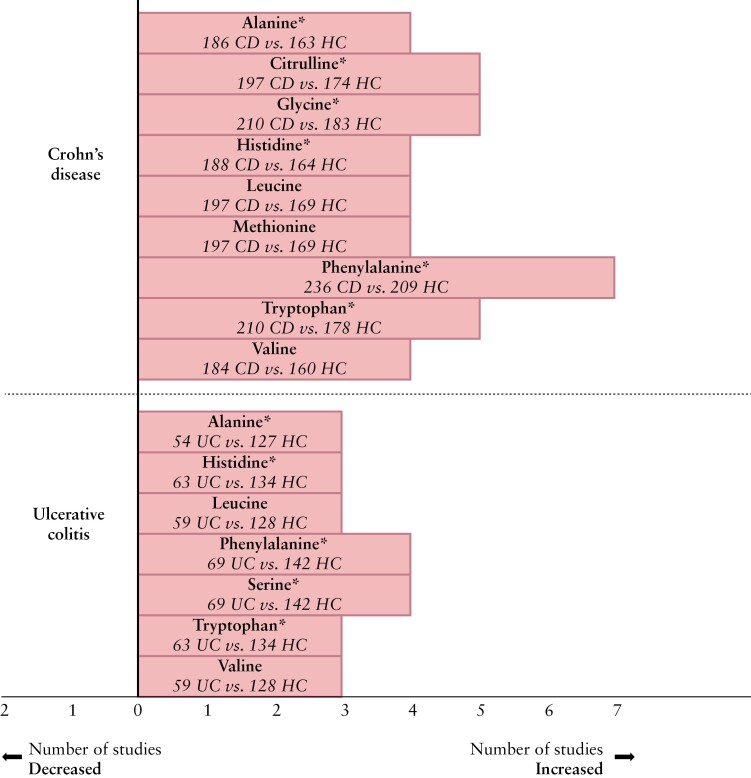
Ten studies assessed faecal AA comprising 354 children with IBD [278 CD, 72 UC, four IBD-U] and 276 controls [233 healthy, 43 intention-to-diagnose cases], by using different analytical approaches and techniques.19–21,29,31–36
Four studies reported the AA in IBD [n = 146] vs controls [n = 166].19,21,29,32 In IBD, most consistently increased AA concentrations included leucine,19,21,29,32 phenylalanine,19,21,29,32 valine,19,21,29,32 glutamine,19,29,32 tryptophan21,29,32 and tyrosine,19,21,29 measured by HPLC19,21,29 or UPLC-MS.32 Two studies19,21 reported on areas under the ROC curve [AUCs] of multiple AA for the discrimination between newly diagnosed, treatment-naïve IBD and controls [AUCs varying from 0.78 to 0.99] as shown in Supplementary Table 4.
In CD vs controls, phenylalanine levels were significantly increased in six studies,21,29,31,34–36 whereas another study demonstrated no difference19 [Figure 3]. Other faecal AA that were observed at higher levels in CD are shown in Figure 3. Conflicting findings were reported for tyrosine,29,31,35,36 threonine,20,29,35,36 taurine20,29,31 and aspartic acid.20,29,35,36 Differences in absolute AA concentrations between patients with CD and healthy controls are depicted in Supplementary Table 5.
Figure 3.
Findings on faecal AA in children with IBD vs controls [nine studies in Crohn’s disease19–21,29,31,33–36 and four studies in ulcerative colitis19–21,29]. The x-axis represents the number of studies in which the AA were increased, in the IBD phenotype compared to controls. The y-axis represents the unique AA. Only AA that were increased in three or more studies were included in this figure.
*The study by Kolho et al. did not test for significance.
Compared with controls, faecal samples of patients with UC contained increased AA levels. The most consistently increased AA are depicted in Figure 3. Jagt and colleagues29 showed that children with newly diagnosed UC could be correctly discriminated from controls based on 29 unique faecal AA, with an accuracy of 90%. Furthermore, levels of tryptophan, valine and histidine were significantly positively associated with more extended disease [pancolitis] in children with UC. Predictive values of faecal AA for diagnosis of UC are shown in Supplementary Table 4.
Regarding phenotype description, three studies19,21,29 found no significant differences in faecal AA concentrations between children with newly diagnosed UC and CD. Kolho et al.20 observed increased AA including asparagine, aspartate and 5-hydroxytryptophan [tryptophan derivative] in UC as compared to CD, based on a partial least squares discriminant analysis [PLS-DA].
3.4.3 Bile acids
Seven studies including 179 children with IBD [119 CD, 46 UC, four IBD-U, ten UC/IBD-U] and 145 controls [110 healthy, 21 siblings, 14 intention-to-diagnose cases] reported on faecal BA levels.20,24,30,32–35 Most studies used LC-MS for BA quantification but often did not report on BA levels separately.
For IBD regardless of phenotype, two studies reported on significantly higher concentrations of primary BA in children with IBD compared to healthy controls24,32 although it should be noted that one study comprised predominantly patients with CD32 [20 CD, three UC, four IBD-U] and one predominantly patients with UC24 [two CD, 16 UC] in their cohort. Bushman and colleagues32 did not report on other differences in BA composition [outside the increased primary BA] in children with IBD compared to controls.
When focusing on CD phenotype in treatment-naïve children, overall total BA levels were comparable to those of healthy controls, according to two studies.34,35 Primary BA levels [not specified] were, however, reported to be increased in three studies focusing on CD,33–35 in agreement with data from studies focusing on IBD in general,24,32 reported in children with both newly diagnosed and longer existing disease. One study found lower levels of secondary BA in children with newly diagnosed CD, corresponding to the earlier reported higher primary BA, resulting in a lower secondary/primary BA ratio.34 This study also showed higher levels of conjugated and lower levels of unconjugated BA in newly diagnosed, treatment-naïve children with CD compared to healthy controls.34 Jacobs and colleagues33 additionally found higher levels of sulphated derivatives of chenodeoxycholic as well as deoxycholic acid in children with CD in remission compared to their healthy siblings. In a study by Kolho et al.20 using a PLS-DA variable importance plot for distinguishing disease vs healthy controls, BA levels were evaluated but were not distinctive for disease presence when compared to other metabolites.
Ejderhamn et al.24 reported on an IBD cohort with predominantly patients with UC [16/18 UC; with 12/16 pancolitis and 4/16 distal colon involvement only], and found significantly increased concentrations of total BA in faeces of children with disease in remission compared to healthy controls. They also found significantly higher concentrations of unconjugated as well as glycine/taurine-conjugated BA compared to healthy controls.24 Similar results were described by Rotondo et al.30 in paediatric patients with UC and active disease vs controls. They found similar total BA levels, with increased primary and conjugated BA compared to healthy controls, similar to the differences found between active CD and healthy controls.30
3.4.4 Other faecal metabolites
Nine articles reported on other metabolites in patients with IBD and controls,20,25,26,31–36 which will be summarized below. Two studies32,35 consisting of 70 children with IBD [63 CD, three UC, four IBD-U] and 56 healthy children demonstrated higher concentrations of cadaverine in faecal samples of patients with IBD. Cadaverine is a polyamine produced by decarboxylation of lysine. None of the other studies, however, reported on this individual metabolite. Diederen et al.35 also found significantly higher levels of putrescine in de novo, treatment-naïve CD, putrescine being another polyamine related to cadaverine.
Amongst lipids, sphingomyelin and related metabolites, and a breakdown product named ceramide were found in higher concentrations in children with IBD31,32 compared with healthy controls. In the study by Bushman et al.,32 increased ceramide levels had the greatest impact on the random forest classifier for discriminating IBD from controls. Furthermore, Jacobs et al.33 observed that stercobilin [a haem degradation product] and boldione [product of steroid catabolism] were decreased in children with IBD vs non-IBD controls. Kolho and colleagues20 observed higher faecal levels of glyceraldehyde in both de novo CD and UC compared to controls.
Regarding children with CD, l-lactate levels were significantly higher compared with controls in one study ,25 while the other study showed no difference.34 Moreover, Wang et al.34 observed that two organic acids—methylmalonic acid and succinic acid—were included in the ten most discriminative metabolites for prediction of CD from healthy children. The study by Sundqvist et al.,26 consisting of children with active CD and healthy controls, was the only study that investigated the metabolites coprostanol and urobilinogen. These metabolites were decreased in patients with CD.
Kolho et al.20 observed metabolites that could discriminate UC from CD, including increased concentrations of guanosine and γ-glutamylcysteine in patients with UC based on PLS-DA. By contrast, levels of pyridoxine, orotate and 4-pyridoxate were decreased in children with UC as compared to CD.
3.5 Faecal metabolomics related to disease activity
3.5.1 Short-chain fatty acids
Taylor et al.16 found the branched SCFA valeric acid to be one of the principal discriminators in a machine-learning model when comparing high to low faecal calprotectin [FC] samples of children with CD [regardless of clinical disease activity], and correlated to ‘mucosal healing’, i.e. low calprotectin levels. Another study reported increased propionic acid levels at baseline to normalize in responders to exclusive enteral nutrition [EEN] [>50% reduction FC], but this normalization was not seen in non-responders.35 Treem and colleagues28 evaluated the differences in active (paediatric Crohn’s disease activity index [PCDAI] > 10) and inactive CD but found no significant differences in SCFA levels between the two groups. In patients with moderate to severe UC [Truelove–Witts criteria], total SCFA, acetic, isobutyric and isovaleric acid were significantly reduced compared to healthy controls. In contrast, children with mild or inactive UC had comparable concentrations of total SCFA, acetate and propionic acid compared to healthy controls. By contrast, children with inactive–mild UC had higher faecal n-butyric acid concentrations than controls and patients with severe UC.28
3.5.2 Amino acids and their derivatives
Bosch et al.21 found no significant correlations between faecal AA levels and inflammatory markers including FC and C-reactive protein [CRP] in children with IBD. Another study by Bosch et al.,19 however, observed that citrulline, leucine, phenylalanine and valine were significant positively correlated with FC.
Regarding CD, Ni et al.36 observed positive correlations between faecal AA and their derivatives, and FC. These correlations were reduced in patients with CD who used antibiotic treatment at the time of sampling. Taylor et al.16 demonstrated that lysine was increased in children with CD [using biologic therapy] with high FC [≥ 100 µg/g] vs low FC [< 100 µg/g]. Three studies20,21,29 found no significant correlations between FC and AA in children with de novo, treatment-naïve CD. Wang et al.34 observed that several AA such as alanine, aspartic acid, glutamic acid, histidine, leucine, proline, phenylalanine and glycine were positively correlated with PCDAI and other serum indexes of inflammation.
In children with UC, one study20 showed significant positive correlations between aminoadipic acid, taurine and methionine, and FC whereas the other two studies21,29 showed no significant correlations between AA levels and disease activity.
3.5.3 Bile acids
One study34 found glycochenodeoxycholic acid [conjugated primary BA] to be positively correlated with PCDAI, erythrocyte sedimentation rate, white blood cells and CRP in children with newly diagnosed CD. Deoxycholic acid, a secondary BA, correlated negatively with these inflammatory markers [measured by Spearman correlations].34
3.5.4 Other metabolites
In patients with IBD, the metabolite carnosine [a hybrid peptide] was significantly positively correlated with FC.20 This metabolite, together with ribose-5-phosphate, choline and glyceraldehyde, was also positively correlated with FC in patients with UC only.20
Amongst CD, one study20 found that allantoin was significant positively correlated with FC. Wang et al.34 observed that two dicarboxylic acids [methylmalonic acid and succinic acid] and four organic acids [cis-aconitic acid, trans-aconitic acid, citric acid and isocitric acid] were positively correlated with PCDAI and serum inflammatory markers.
3.6 Faecal metabolomics for predicting treatment response
Three studies including 84 children with CD reported on faecal metabolites associated with response or non-response to therapy [EEN and biologic agents].
Faecal AA levels before initiation of EEN could not predict response to this nutritional therapy in children with de novo, treatment-naïve CD.35 However, at the end of EEN, responders [based on reduction in FC of > 50%] had lower concentrations of serine, glycine and alanine as compared to non-responders.
Connors et al.23 found that all children who did not sustain remission [weighted PCDAI < 12.5] from 12 to 24 weeks after EEN therapy had a primary BA dominant profile with a significantly higher percentage of primary BA contribution compared to patients with sustained remission. Interestingly, this primary BA dominance was not seen in patients who did not reach remission at week 12. Total BA concentration differences were not found between groups at each time point.23 Diederen and colleagues35 found no differences between BA concentration, BA hydrophobicity and the fraction of secondary BA at baseline between responders [>50% reduction in FC] and non-responders to EEN.
Wang et al.34 investigated the relationship between faecal metabolomic profiles and therapeutic outcomes of infliximab [IFX] therapy in children with CD. They found higher faecal levels of glycine and the metabolites linoleic acid and l-lactic acid at baseline [prior to IFX treatment] in patients who achieved sustained remission as compared to non-sustained remission patients. Enriched metabolites in non-sustained remission patients [at baseline] included N-acetylserotonin, methylglutaric acid, adipic acid, 4-aminohippuric acid, citramalic acid, isovaleric acid, nicotinic acid, pentadecanoic acid and N-acteylglutamine.
4. Discussion
This systematic review provides a summary of the available evidence on faecal metabolomics in children with IBD vs healthy controls, with the aim of characterizing a metabolomic signature in paediatric IBD, both for disease activity and for treatment response. All included studies observed distinct metabolomic profiles in children with IBD compared to controls. However, based on the available data, no unique faecal metabolomic signature could be defined due to inconsistency of individual study results and lack of standardization of metabolomics assessment methods. The majority of studies reported on higher faecal AA levels in IBD as compared to controls. Additionally, most studies on BA observed higher levels of primary and lower levels of secondary BA in IBD, while contradictory results were found for SCFA. Although studies on metabolomics linked to therapy response were limited, significant differences in baseline faecal metabolites [BA and AA] between responders and non-responders to EEN and IFX have been described.
The most consistent findings were higher levels of faecal AA in children with IBD as compared to controls, yet this also was the most studied metabolite class. Phenylalanine—an essential aromatic amino acid—was significantly increased in CD, in UC and in both phenotypes combined in the majority of studies. Only one study demonstrated no significant difference in phenylalanine concentrations in CD vs controls, which could be explained by their small sample size [five CD, eight controls].19 These findings correspond with the results of a recent systematic review on metabolomic analysis in adults with IBD,42 which showed higher levels of AA, including branched-chain AA, phenylalanine, glycine, tyrosine, alanine and taurine in IBD compared with non-IBD controls.
The use of omics-techniques in IBD has been rising in the hope of reaching a more mechanistic and functional understanding of the disease. Lloyd-Price and colleagues7 reported on their multi-omics analysis of gut microbial ecosystems using stool, biopsies and blood samples in a cohort of 132 subjects with IBD comprising 61 children from different hospitals. They showed characteristic changes of the microbiome, and disruptions in microbial transcription, serum antibody levels and metabolite pools. Gallagher and colleagues42 conducted a systematic review describing metabolomic analysis in urine, stool and serum from adults with IBD. Reagrding stool metabolomics results, the authors described lower concentrations of SCFA [namely acetate, propionate and butyrate] compared to controls. This observation was similar to the results we found in children, but was not consistent across all studies.25,28,35 Interestingly, decreased levels of both primary and secondary BA have been described in adult IBD, although primary BA findings were not consistent across studies.7,43 We found increased levels of primary BA in children with IBD compared to controls, consistent with the pathophysiological between the dysbiotic microbiome and BA.44,45
The metabolome is closely related to the microbiome in that it mainly consists of end products of microbial metabolism. Hence, shifts in microbiome composition, i.e. dysbiosis, also affect faecal metabolite composition including AA, SCFA and BA.13 Aromatic AA such as phenylalanine, tryptophan and tyrosine are substrates for a pathway from the gut symbiont Clostridium sporogenes [phylum Firmicutes], generating aromatic amino acid metabolites. These metabolites affect intestinal permeability and systemic immunity.46 Additionally, tryptophan is partly metabolized by gut bacteria into indole metabolites which can serve as aryl hydrocarbon receptor [AhR] ligands.47 This receptor is considered to play a role in IBD pathogenesis, as shown by Lamas et al.48They observed that the gut microbiota of Card9−/− [an IBD susceptibility gene] mice were unable to metabolize tryptophan into AhR ligands. This inability resulted in a defective interleukin 22 [IL-22] activation and consequently led to increased sensitivity to colitis.48 It is important to note that higher faecal AA concentrations might hypothetically also result from intestinal malabsorption or protein loss through colonic leakage, rather than reflecting microbial changes.49
SCFA are another metabolite class known to interact with the gut microbiota. In this review, differences in SCFA concentrations were observed in children with IBD compared to controls. Specifically, decreased levels of acetic and valeric acid were found in CD, comparable to the results of a meta-analysis on SCFA characterization in adults with CD.50 SCFA are predominantly produced by bacterial anaerobic fermentation of dietary fibres in the intestine, making microbiome composition crucial in the production of SCFA.51 SCFA have been considered metabolites of interest due to their role as a primary energy source of colonocytes and immune homeostasis. Immunomodulation by SCFA can occur by regulation of regulatory T cells, activation of G protein-coupled receptors, inhibition of histone deacetylase and other described mechanisms.52–54 Butyrate, one of the predominant SCFA, has also been described to restore epithelial barrier function by activating genes coding for tight junction components.55 Butyrate oxidation accounts for more than 70% of the oxygen consumed by human colonic tissue.56 Depletion of SCFA therefore also leads to a decrease in available colonocyte energy and subsequent damage to intestinal epithelial cells. This has also been hypothesized to be the cause of diversion colitis, in which inflammation could be successfully treated with SCFA enemas.57
Butyrate production is often mediated by members of the phylum Firmicutes, while Bacteroides mediate propionate and acetate production.58,59 Dysbiosis in IBD has commonly been characterized by a lower abundance of certain SCFA producers, such as F. prausnitzii and R. hominis, and therefore lead to altered SCFA levels in IBD patients.60 However, IBD-specific SCFA signatures have not been defined so far, although conclusions seem to depend on clinical activity of subject samples.50 Faecal concentrations of SCFA are not only determined by microbiome composition but also by the type and amount of dietary intake. However, increased metabolic capacity of SCFA synthesis and increased butyrate levels at baseline have been shown to be important in maintaining anti-tumour necrosis factor [anti-TNF] and azathioprine remission in adults.15,61
Bile acids have been described to have different immunomodulatory functions, mainly by interaction with different host BA receptors.62,63 Bile acids are synthesized in the liver and further deconjugated and dehydroxylated by microbial species in the intestine.45 Changes in the gut bacterial community can thus alter BA composition, as has been described in IBD.44 In this review, we found significantly higher concentrations of primary BA in patients with IBD [specifically CD] as compared to controls in five studies.24,32–35 Bile acids, in particular secondary BA, have been shown to modulate intestinal immune responses.44,64 The increased concentrations of primary BA could at least partly be explained by an impaired BA absorption in the distal ileum due to inflammation in patients with CD.65 However, this observation could also be explained by the absence of microbial species with the ability for dehydroxylation [rather than deconjugation], which is a key step in primary to secondary BA conversion in the intestine.23 Duboc et al.44 observed significantly higher faecal-conjugated BA rates and significantly lower secondary BA rates in colonic IBD compared to controls. These authors also showed that dysbiosis in IBD results in BA dysmetabolism characterized by impaired deconjugation, transformation and desulphation.44
Amongst lipids, two of the included studies observed higher levels of sphingomyelin and related metabolites, and the breakdown product ceramide in paediatric IBD compared with controls. An enrichment in sphingolipids, particularly in ceramide and sphingomyelin, has been previously identified in stool of adults with IBD compared to controls.66 Sphingolipids are crucial components of intestinal membranes and are involved in a series of cellular actions such as cell differentiation, proliferation, migration and apoptosis, all contributing to a healthy gut.67 Additionally, sphingolipids are produced by commensal bacteria belonging to the phylum Bacteroidetes, and it has been observed that bacterial and host sphingolipid pathways are dysregulated during inflammation and IBD.68
Regarding treatment response, we found that a faecal primary BA-dominant profile at baseline has been associated with non-responders to EEN treatment. Furthermore, levels of several metabolites including multiple AA were found to be correlated with the response to IFX therapy in children with CD. In adult IBD studies, faecal BA, lipids and SCFA have been demonstrated to predict response to therapy with biologicals.69 Ding et al.17 observed that lower faecal levels of histidine [AA] and deoxycholic acid [secondary BA] are predictive of primary non-response to anti-TNF therapy in adults with CD. To date, only limited evidence based on small cohorts of paediatric patients with IBD is available and thus external validation in larger cohorts of patients is needed. If these potentially predictive metabolic features could be validated, this would contribute significantly to the development of personalized treatment strategies.
To our knowledge, this is the first systematic review describing faecal metabolomics in paediatric IBD. However, there are several limitations to the identified studies and therefore to this systematic review. The included studies had small patient groups [12 out of 17 had fewer than 50 patients in total] and only a minority had age- and sex-matched controls. Studies differed in subject disease activity, treatment-naivety, stool consistency, sample handling, sample time points, diet and type of controls. Stool consistency [i.e. stool water content] reflects GI transit time and can severely impact the stool microbiome and metabolome.70,71 However, none of the studies took this into account when reporting their analyses. This is important to note when comparing children with IBD—often presenting with diarrhoea—to healthy controls with ‘normal’ stool consistency. Nutritional status also influences the stool metabolome, as we know that specific dietary products can influence the gut, target the microbiome, and subsequently alter metabolism and thus its metabolic readout. However, none of the included studies collected detailed information on dietary intake. Ideally, only newly diagnosed, treatment-naïve children would have been included to ensure the most unbiased metabolomic signatures, as different treatment strategies in IBD have been shown to influence metabolomic measurements of patients with IBD.72 All these factors inhibit identification of metabolomic signatures, especially for important subgroups such as active disease vs remission and response to therapy over time.
The lack of standardization in metabolomics analysis between studies should be considered. There was heterogeneity in the methods of metabolomics assessment, including the approach [targeted or untargeted] and techniques [NMR vs MS]. The use of NMR is often hampered by a relatively low sensitivity as compared to MS methods,73 which leads to differences in faecal metabolites that could be detected and quantification of the faecal metabolites across a number of studies. Even when similar methods were used, not all results were reported in the same manner [absolute vs relative, ratios, selective metabolites only]. Sample handling is particularly important when performing microbiome and metabolome analyses.74 Hence, different sample [handling and analysis] protocols can influence the results, making it impossible to correct for this variation especially when comparing between different studies.
Future studies should therefore comprise larger sample sizes with standardized time points and disease activity markers, using the same methodologies and analytical techniques for metabolomics interpretation in order to obtain consistent results and allow firm conclusions to be drawn. Additionally, further validation in independent cohorts is recommended to develop new metabolic biomarkers for diagnostics, and prediction of disease course and of treatment response in paediatric IBD. Available evidence has shown that faecal metabolomics provide valuable information on the metabolome and inherently the activity of the gut microbiota, which are influenced by host genetics and environment. Multi-omics studies, including microbiome analysis, should therefore be performed to improve knowledge on the host–microbial interactions in the disease pathogenesis of paediatric IBD.
In conclusion, faecal metabolomic profiles in paediatric IBD differ from healthy controls, particularly in AA. Moreover, distinct metabolic profiles for AA and BA were observed between responders and non-responders to EEN and IFX at baseline. However, the inconsistency of metabolic differences identified across studies and selective reporting hamper the determination of a clear metabolomic signature in paediatric IBD compared to controls, and response to treatment. This highlights the importance of further research with standardized methodologies in order to generate more consistent findings and gain more insight into metabolic alterations in IBD. Defining disease-associated signatures might contribute to the development of novel, non-invasive, personalized diagnostic and even therapeutic approaches in paediatric IBD.
Supplementary Material
Contributor Information
Jasmijn Z Jagt, Department of Paediatric Gastroenterology, Emma Children’s Hospital, Amsterdam UMC, Vrije Universiteit Amsterdam, HV Amsterdam, The Netherlands; Amsterdam UMC, Vrije Universiteit Amsterdam, Paediatric Gastroenterology, Amsterdam Gastroenterology Endocrinology Metabolism, De Boelelaan, Amsterdam, Netherlands.
Charlotte M Verburgt, Department of Paediatric Gastroenterology and Nutrition, Amsterdam University Medical Centres – location University of Amsterdam, Emma Children’s Hospital, AZ Amsterdam, The Netherlands; Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology Endocrinology Metabolism, University of Amsterdam, BK Amsterdam, The Netherlands; Amsterdam Reproduction & Development, AZ Amsterdam, The Netherlands.
Ralph de Vries, Medical Library, Vrije Universiteit Amsterdam, HV Amsterdam, The Netherlands.
Nanne K H de Boer, Department of Gastroenterology and Hepatology, Amsterdam Gastroenterology and Metabolism Research Institute (AGEM), Amsterdam University Medical Centre, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Marc A Benninga, Department of Paediatric Gastroenterology and Nutrition, Amsterdam University Medical Centres – location University of Amsterdam, Emma Children’s Hospital, AZ Amsterdam, The Netherlands.
Wouter J de Jonge, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology Endocrinology Metabolism, University of Amsterdam, BK Amsterdam, The Netherlands; Department of Surgery, University of Bonn, Bonn, Germany.
Johan E van Limbergen, Department of Paediatric Gastroenterology and Nutrition, Amsterdam University Medical Centres – location University of Amsterdam, Emma Children’s Hospital, AZ Amsterdam, The Netherlands; Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology Endocrinology Metabolism, University of Amsterdam, BK Amsterdam, The Netherlands; Department of Pediatrics, Dalhousie University, Halifax, NS, Canada.
Tim G J de Meij, Department of Paediatric Gastroenterology, Emma Children’s Hospital, Amsterdam UMC, Vrije Universiteit Amsterdam, HV Amsterdam, The Netherlands; Department of Paediatric Gastroenterology and Nutrition, Amsterdam University Medical Centres – location University of Amsterdam, Emma Children’s Hospital, AZ Amsterdam, The Netherlands.
Funding
This work was supported by the ‘Right on Time’ grant with number WO 19 – 25, from The Dutch Digestive Foundation [Maag Lever Darm Stichting; MLDS].
Conflict of Interest
N.dB. has served as a speaker for AbbVie and MSD and has served as consultant and/or principal investigator for TEVA Pharma BV and Takeda. He has received a [unrestricted] research grant from Dr. Falk, TEVA Pharma BV, MLDS and Takeda. All outside the submitted work. J.vL. reports consulting, travel and/or speaker fees and research support from AbbVie, Janssen, Nestlé Health Science and Novalac. J.J., C.V., R.dV., M.B., W.dJ. and T.dM. have nothing to declare.
Author Contribtions
Guarantor of the article: Jasmijn Z. Jagt. J.J. and C.V. conceptualized and designed the study, carried out the data collection and analysis, drafted the initial version of the paper, and reviewed and revised this paper. R.dV. carried out the data collection and analysis. N.dB., J.vL. and T.dM. conceptualized and designed this study, coordinated and supervised data collection, and critically reviewed this paper for important intellectual content. M.B. and W.dJ. critically reviewed this paper for important intellectual content. All authors approved the final version of the article, including the authorship list.
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.
References
- 1. Neurath MF, Travis SP.. Mucosal healing in inflammatory bowel diseases: a systematic review. Gut 2012;61:1619–35. [DOI] [PubMed] [Google Scholar]
- 2. Roberts SE, Thorne K, Thapar N, et al. A systematic review and meta-analysis of paediatric inflammatory bowel disease incidence and prevalence across Europe. J Crohns Colitis 2020;14:1119–48. [DOI] [PubMed] [Google Scholar]
- 3. Sýkora J, Pomahačová R, Kreslová M, et al. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol 2018;24:2741–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coward S, Clement F, Benchimol EI, et al. Past and future burden of inflammatory bowel diseases based on modeling of population-based data. Gastroenterology 2019;156:1345–1353.e4. [DOI] [PubMed] [Google Scholar]
- 5. Imhann F, Vich Vila A, Bonder MJ, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pittayanon R, Lau JT, Leontiadis GI, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. [DOI] [PubMed] [Google Scholar]
- 7. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Visconti A, Le Roy CI, Rosa F, et al. Interplay between the human gut microbiome and host metabolism. Nat Commun 2019;10:4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicholson JK, Lindon JC.. Systems biology: Metabonomics. Nature 2008;455:1054–6. [DOI] [PubMed] [Google Scholar]
- 10. Gowda GA, Zhang S, Gu H, et al. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn 2008;8:617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bjerrum JT, Wang Y, Hao F, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics 2015;11:122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schirmer M, Garner A, Vlamakis H, et al. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lavelle A, Sokol H.. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223–37. [DOI] [PubMed] [Google Scholar]
- 14. Zierer J, Jackson MA, Kastenmüller G, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 2018;50:790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aden K, Rehman A, Waschina S, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology 2019;157:1279–1292.e11. [DOI] [PubMed] [Google Scholar]
- 16. Taylor H, Serrano-Contreras JI, McDonald JAK, et al. Multiomic features associated with mucosal healing and inflammation in paediatric Crohn’s disease. Aliment Pharmacol Ther 2020;52:1491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding NS, McDonald JAK, Perdones-Montero A, et al. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn’s disease. J Crohns Colitis 2020. [DOI] [PubMed] [Google Scholar]
- 18. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bosch S, El Manouni El Hassani S, Brizzio Brentar M, et al. Fecal amino acid profiles exceed accuracy of serum amino acids in diagnosing pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2020;71:371–5. [DOI] [PubMed] [Google Scholar]
- 20. Kolho KL, Pessia A, Jaakkola T, et al. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J Crohns Colitis 2017;11:321–34. [DOI] [PubMed] [Google Scholar]
- 21. Bosch S, Struys EA, van Gaal N, et al. Fecal amino acid analysis can discriminate de novo treatment-naïve pediatric inflammatory bowel disease from controls. J Pediatr Gastroenterol Nutr 2018;66:773–8. [DOI] [PubMed] [Google Scholar]
- 22. Ashton JJ, Colquhoun CM, Cleary DW, et al. 16S sequencing and functional analysis of the fecal microbiome during treatment of newly diagnosed pediatric inflammatory bowel disease. Medicine [Baltim] 2017;96:e7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Connors J, Dunn KA, Allott J, et al. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn’s disease. ISME J 2020;14:702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ejderhamn J, Rafter JJ, Strandvik B.. Faecal bile acid excretion in children with inflammatory bowel disease. Gut 1991;32:1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerasimidis K, Bertz M, Hanske L, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis 2014;20:861–71. [DOI] [PubMed] [Google Scholar]
- 26. Sundqvist T, Stenhammar L, Tjellström B, et al. Evidence of disturbed gut microbial metabolic activity in pediatric Crohn’s disease. Crohn’s & Colitis 360 2019;1:otz010. [Google Scholar]
- 27. Tjellström B, Högberg L, Stenhammar L, et al. Effect of exclusive enteral nutrition on gut microflora function in children with Crohn’s disease. Scand J Gastroenterol 2012;47:1454–9. [DOI] [PubMed] [Google Scholar]
- 28. Treem WR, Ahsan N, Shoup M, et al. Fecal short-chain fatty acids in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 1994;18:159–64. [DOI] [PubMed] [Google Scholar]
- 29. Jagt JZ, Struys EA, Ayada I, et al. Fecal amino acid analysis in newly diagnosed pediatric inflammatory bowel disease: a multicenter case-control study. Inflamm Bowel Dis 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rotondo-Trivette S, Wang B, Gayer C, et al. Decreased secondary faecal bile acids in children with ulcerative colitis and Clostridioides difficile infection. Aliment Pharmacol Ther 2021;54:792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alghamdi A, Gerasimidis K, Blackburn G, et al. Untargeted metabolomics of extracts from faecal samples demonstrates distinct differences between paediatric Crohn’s disease patients and healthy controls but no significant changes resulting from exclusive enteral nutrition treatment. Metabolites 2018;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bushman FD, Conrad M, Ren Y, et al. Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe 2020;28:422–433.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs JP, Goudarzi M, Singh N, et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol 2016;2:750–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Gao X, Zhang X, et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes 2021;13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diederen K, Li JV, Donachie GE, et al. Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn’s disease. Sci Rep 2020;10:18879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ni J, Shen TD, Chen EZ, et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 2017;9:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaddurah-Daouk R, Kristal BS, Weinshilboum RM.. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol 2008;48:653–83. [DOI] [PubMed] [Google Scholar]
- 38. Wang JH, Byun J, Pennathur S.. Analytical approaches to metabolomics and applications to systems biology. Semin Nephrol 2010;30:500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Di Girolamo F, Lante I, Muraca M, et al. The role of mass spectrometry in the ‘omics’ era. Curr Org Chem 2013;17:2891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emwas AH, Roy R, McKay RT, et al. NMR spectroscopy for metabolomics research. Metabolites 2019;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Worley B, Powers R.. Multivariate analysis in metabolomics. Curr Metabolomics 2013;1:92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gallagher K, Catesson A, Griffin JL, et al. Metabolomic analysis in inflammatory bowel disease: a systematic review. J Crohns Colitis 2020. [DOI] [PubMed] [Google Scholar]
- 43. Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One 2009;4:e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013;62:531–9. [DOI] [PubMed] [Google Scholar]
- 45. Wahlström A, Sayin SI, Marschall HU, et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016;24:41–50. [DOI] [PubMed] [Google Scholar]
- 46. Dodd D, Spitzer MH, Van Treuren W, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017;551:648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X, Zhang ZH, Zabed HM, et al. An insight into the roles of dietary tryptophan and its metabolites in intestinal inflammation and inflammatory bowel disease. Mol Nutr Food Res 0461;2020:e200. [DOI] [PubMed] [Google Scholar]
- 48. Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bosch S, de Meij TGJ, de Boer NK.. Altered tryptophan levels in patients with inflammatory bowel disease owing to colonic leakage, metabolism, or malabsorption? Gastroenterology 2018;154:1855–6. [DOI] [PubMed] [Google Scholar]
- 50. Zhuang X, Li T, Li M, et al. Systematic review and meta-analysis: short-chain fatty acid characterization in patients with inflammatory bowel disease. Inflamm Bowel Dis 2019;25:1751–63. [DOI] [PubMed] [Google Scholar]
- 51. Pascale A, Marchesi N, Marelli C, et al. Microbiota and metabolic diseases. Endocrine 2018;61:357–71. [DOI] [PubMed] [Google Scholar]
- 52. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vinolo MA, Rodrigues HG, Nachbar RT, et al. Regulation of inflammation by short chain fatty acids. Nutrients 2011;3:858–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martin-Gallausiaux C, Béguet-Crespel F, Marinelli L, et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep 2018;8:9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zheng L, Kelly CJ, Battista KD, et al. Microbial-derived butyrate promotes epithelial barrier function through il-10 receptor-dependent repression of claudin-2. J Immunol 2017;199:2976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 1980;21:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harig JM, Soergel KH, Komorowski RA, et al. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med 1989;320:23–8. [DOI] [PubMed] [Google Scholar]
- 58. Louis P, Flint HJ.. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009;294:1–8. [DOI] [PubMed] [Google Scholar]
- 59. Louis P, Flint HJ.. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017;19:29–41. [DOI] [PubMed] [Google Scholar]
- 60. Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- 61. Effenberger M, Reider S, Waschina S, et al. Microbial butyrate synthesis indicates therapeutic efficacy of azathioprine in IBD patients. J Crohns Colitis 2021;15:88–98. [DOI] [PubMed] [Google Scholar]
- 62. Chen ML, Takeda K, Sundrud MS.. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol 2019;12:851–61. [DOI] [PubMed] [Google Scholar]
- 63. Fiorucci S, Biagioli M, Zampella A, et al. Bile acids activated receptors regulate innate immunity. Front Immunol 2018;9:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sinha SR, Haileselassie Y, Nguyen LP, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020;27:659–670.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen F, Ma L, Sartor RB, et al. Inflammatory-mediated repression of the rat ileal sodium-dependent bile acid transporter by c-fos nuclear translocation. Gastroenterology 2002;123:2005–16. [DOI] [PubMed] [Google Scholar]
- 66. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abdel Hadi L, Di Vito C, Riboni L.. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Mediators Inflamm 2016;2016:3827684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown EM, Ke X, Hitchcock D, et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 2019;25:668–680.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bjerrum JT, Wang YL, Seidelin JB, et al. IBD metabonomics predicts phenotype, disease course, and treatment response. EBioMedicine 2021;71:103551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Degen LP, Phillips SF.. How well does stool form reflect colonic transit? Gut 1996;39:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crouwel F, Buiter HJC, de Boer NK.. Gut microbiota-driven drug metabolism in inflammatory bowel disease. J Crohns Colitis 2020;15:307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang A, Sun H, Wang P, et al. Modern analytical techniques in metabolomics analysis. Analyst 2012;137:293–300. [DOI] [PubMed] [Google Scholar]
- 74. De Spiegeleer M, De Graeve M, Huysman S, et al. Impact of storage conditions on the human stool metabolome and lipidome: Preserving the most accurate fingerprint. Anal Chim Acta 2020;1108:79–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.