Abstract
Background and Aims
We aimed to determine whether a targeted gene expression panel could predict clinical outcomes in paediatric ulcerative colitis [UC] and investigated putative pathogenic roles of predictive genes.
Methods
In total, 313 rectal RNA samples from a cohort of newly diagnosed paediatric UC patients (PROTECT) were analysed by a real-time PCR microfluidic array for expression of type 1, 2 and 17 inflammation genes. Associations between expression and clinical outcomes were assessed by logistic regression. Identified prognostic markers were further analysed using existing RNA sequencing (RNA-seq) data sets and tissue immunostaining.
Results
IL13RA2 was associated with a lower likelihood of corticosteroid-free remission (CSFR) on mesalamine at week 52 (p = .002). A model including IL13RA2 and only baseline clinical parameters was as accurate as an established clinical model, which requires week 4 remission status. RORC was associated with a lower likelihood of colectomy by week 52. A model including RORC and PUCAI predicted colectomy by 52 weeks (area under the receiver operating characteristic curve 0.71). Bulk RNA-seq identified IL13RA2 and RORC as hub genes within UC outcome-associated expression networks related to extracellular matrix and innate immune response, and lipid metabolism and microvillus assembly, respectively. Adult UC single-cell RNA-seq data revealed IL13RA2 and RORC co-expressed genes were localized to inflammatory fibroblasts and undifferentiated epithelial cells, respectively, which was supported by protein immunostaining.
Conclusion
Targeted assessment of rectal mucosal immune gene expression predicts 52-week CSFR in treatment-naïve paediatric UC patients. Further exploration of IL-13Rɑ2 as a therapeutic target in UC and future studies of the epithelial-specific role of RORC in UC pathogenesis are warranted.
Keywords: Paediatric inflammatory bowel disease, gene expression array, weighted gene co-expression network analysis
1. Introduction
The prevalence of ulcerative colitis [UC] in children is rising worldwide.1–4 Children with UC are initially treated with mesalamine or corticosteroids [CS] based on disease severity and therapy is escalated when corticosteroid-free remission [CSFR] is not achieved. Unsuccessful therapeutic trials expose patients to adverse drug effects and delay sustained remission. There is a need for predictors of treatment response that inform individualized treatment plans and target the safest and most effective medication to each unique patient.
Predicting Response to Standardized Pediatric Colitis Therapy [PROTECT] was a prospective clinical trial of standardized treatment with mesalamine with or without corticosteroids [based on disease severity] in paediatric patients newly diagnosed with UC. PROTECT demonstrated that 38% achieve CSFR at 1 year on mesalamine alone, but the remainder require a progression to thiopurines [19%], anti-tumour necrosis factor [TNF] biological agents [31%] or colectomy [6%].5 PROTECT produced a clinical predictive model for CSFR on mesalamine at 52 weeks that included baseline Pediatric Ulcerative Colitis Activity Index [PUCAI] score, haemoglobin and clinical remission at week 4. Notably, this predictive model relies on week 4 clinical status, and an opportunity exists to develop models from baseline clinical and molecular data alone.
Type 2 inflammation is defined by the production of interleukin [IL]-4, IL-5 and IL-13 cytokines by T helper 2 cells and group 2 innate lymphoid cells, which are involved in helminth eradication and allergic disease.6 Early studies of the mucosal immune response in human inflammatory bowel disease [IBD] identified an atypical type 2 immune response in adult UC, which was proposed to be pathogenic.7 However, collective evidence supports a mixed immune response in UC that also includes type 1 and type 17 inflammation, and the precise role of type 2 inflammation remains unclear.6 We previously showed that mucosal expression of type 2 and type 17 immune response genes, as assessed by a real-time reverse-transcription quantitative polymerase chain reaction [RT-qPCR] microfluidic array, distinguishes children with treatment-naïve UC from colonic Crohn’s disease, and that a type 2 immune gene expression profile characterized by detectable IL13 expression was associated with favourable treatment response in paediatric UC.8 However, this study was limited by the small cohort of UC patients studied and treatment heterogeneity.
The aim of the present study was to evaluate the prognostic ability of our RT-qPCR microfluidic array, focusing on type 1, 2 and 17 immune response genes as previously described,8 for predicting clinical outcomes in a larger paediatric UC cohort on standardized therapy. We opted to use a RT-qPCR array because of our prior smaller study indicating prognostic value,8 the high sensitivity of RT-qPCR for detecting gene expression, the targeted approach does not require as severe a multiple comparison statistical penalty compared to genome-wide expression approaches, and multiple RT-qPCR gene expression assays have been advanced to clinical use for informing cancer treatment.9 Identified prognostic gene expression markers were further studied using existing bulk and single-cell colon tissue RNA sequencing [RNA-seq] data and tissue immunostaining to identify their cellular sources and propose putative roles in disease pathogenesis.
2. Materials and Methods
2.1. PROTECT cohort rectal RNA samples
Rectal mucosal RNA, clinical and histopathologic data were obtained from the PROTECT study, a 29-centre cohort of 428 patients aged 4–17 years with a new diagnosis of UC initially treated with mesalamine with or without CS induction therapy and followed for 52 weeks.5 Eligibility criteria, key dates, study size and blinding approaches were previously published.5,10 Baseline rectal biopsies were obtained in 367 [85%] of PROTECT cohort patients and were processed for RNA and DNA isolation. A follow-up research endoscopy with biopsies at 52 weeks for assessment of endoscopic healing was obtained in 98 [23%] patients. Centralized Mayo Endoscopic scoring and histopathological assessment were performed at baseline and 52 weeks. We assayed high-integrity RNA from 313 baseline and 58 follow-up PROTECT samples. Biopsies from 20 patients without IBD and with normal rectal histopathology enrolled under a separate institutional review board [IRB]-approved protocol at Cincinnati Children’s Hospital Medical Center were included for analysis as controls.11
2.2. Ethical approval
The PROTECT study [ClinicalTrials.gov NCT 01536535] was approved by each centre’s IRB, and each participant provided informed consent or assent as applicable. Rectal mucosal biopsy tissue samples from paediatric UC and non-IBD patients used for immunostaining were collected under an IRB-approved protocol at Cincinnati Children’s Hospital Medical Center.
2.3. Real-time RT-qPCR
Rectal RNA was analysed by custom microfluidic RT-qPCR gene expression array [GEA] for expression of 24 genes related to type 1, type 2, type 17 and regulatory immune responses as previously described.8 In total, 200 ng of rectal RNA from diagnostic and week 52 follow-up endoscopies of PROTECT participants was analysed. RNA quality control was performed on 100 ng RNA from each PROTECT sample using an Agilent 2100 Bioanalyzer in the CCHMC Digestive Health Center. Only samples with an RNA integrity number [RIN] ≥ 7 were used. cDNA was synthesized from the rectal RNA samples, then RT-qPCRs were performed on custom TaqMan array 384-well microfluidic cards [ThermoFisher] and run on a 7900HT Fast Real-Time PCR System in the CCHMC Division of Gastroenterology. Twenty-four distinct gene expression assays related to type 1, type 2, type 17 and regulatory immune responses [Supplementary Table 1] were performed in duplicate. GAPDH was previously validated as the reference gene with the least variability between IBD and control patients, and expression relative to non-IBD controls was calculated using a modification of the 2–ΔΔCq method previously described.8
2.4. Outcomes
The primary outcome was the association of baseline gene expression with CSFR on mesalamine at 52 weeks, which was the PROTECT primary endpoint.5 CSFR was defined as PUCAI score < 10, no corticosteroids within 28 days of assessment, and no additional medical therapy or colectomy. Secondary outcomes included corticosteroid-naïve [never required CS] remission on mesalamine therapy [CSNR] at 12 and 52 weeks, escalation to infliximab [IFX] in patients with baseline moderate-to-severe disease [defined by the PROTECT study as initial treatment with oral CS and baseline PUCAI ≥ 45 or IV CS] by 52 weeks, colectomy in the baseline moderate-to-severe disease group by 52 weeks, CSFR on IFX in those who escalated to IFX therapy by 12 weeks and endoscopic healing [endoscopic Mayo score = 0] at 52 weeks.
2.5. Statistical analysis
SAS version 9.4 was used to conduct statistical analyses. Baseline characteristics of subgroup populations were compared using the chi-square test for categorical variables and Wilcoxon’s rank-sum non-parametric test for continuous variables. Differences in gene expression in UC patients compared to controls was assessed using a Wilcoxon rank-sum non-parametric test with false discovery rate correction for multiple comparisons. Unsupervised hierarchical clustering was used to identify patient clusters with unique gene expression patterns and Fisher’s exact test was used to assess associations between analyte clusters and outcomes. Correlations between mucosal gene expression and baseline mucosal eosinophil count and endoscopic scores were assessed by Spearman’s correlation coefficient. To mirror the PROTECT study analysis,5 univariable and multivariable logistic regression analyses were performed excluding a small subset of patients with protocol violations, with a final n = 284 for our GEA analysis. Univariable logistic regression was performed to assess the association of analytes and clinical variables against each outcome. Associations with a p-value < 0.1 were entered into multivariable analysis. When expression values of multiple genes were highly correlated, only the gene with the strongest significance was included in multivariable analysis. Results from multivariable analyses were integrated into existing PROTECT models or incorporated with clinical and histopathological data to develop new predictive models. Samples with missing data were excluded only from analyses that involved the variables for which they had missing information. Results were assessed with receiver operating characteristic [ROC] curves and corresponding area under the curve [AUC], sensitivity, specificity, positive predictive value [PPV] and negative predictive value [NPV]. We used a probability of 0.5 to set cut-off points for predictive performance characteristics for all outcomes except colectomy. A probability of 0.2 was used for colectomy, given the low incidence of colectomy at 52 weeks in this cohort. Differences in model performance were assessed using the likelihood ratio test. Complete univariable logistic regression associations are listed in Supplementary Table 4.
2.6. Weighted gene co-expression network analysis
Weighted gene co-expression network analysis [WGCNA] was applied as previously described12,13 to available PROTECT study RNA-seq data, to generate co-expression modules for outcome correlation and downstream network and functional annotation analyses. An adjacency matrix was constructed using a power value of 5 for soft thresholding. Co-expressed gene modules were dynamically identified with module size set to 30 and deepSplit to 4 to control the sensitivity for cluster splitting. Gene expression profiles within each module were represented by the first principal component, or eigengene, and correlated with phenotypic traits to identify candidate modules.13 Modules with similar expression profiles were merged and the eigengenes were clustered using the ‘average’ method with threshold set to 0.1. Both connectivity- and phenotype-based significance scores were used to identify hub genes within candidate modules, as previously described, and the Pearson correlation coefficient was calculated.12 Functional enrichment analysis was performed using the ToppFun application of the ToppGene Suite,14 with the top 200 correlated genes to IL13RA2 and RORC from their respective black and brown modules as the input gene sets. Functional enrichment network visualizations for the IL13RA2 and RORC gene sets were generated utilizing the top ten enriched terms and the Cytoscape application.15
2.7. Single-cell RNA-seq analysis
We analysed a publicly available adult human colon UC single-cell data set to examine cell-specific expression of gene co-expression modules of interest.16 Genes of interest along with their 20 most highly correlated genes within modules identified by WGCNA were assessed for cell-specific expression amongst subsets of colon epithelial, immune and stromal cells to assess the probable cellular source of each co-expression module. To further determine what cells were driving disease-associated differential expression of genes of interest, normalized cell-specific expression in cells expressing those genes were compared between disease states using the Kruskal–Wallis test with Dunn’s post-test for multiple comparisons.
2.8. IL13-Rα2 and RORγ immunostaining
For immunofluorescence analysis, formalin-fixed paraffin-embedded inflamed rectum, sigmoid or descending colon tissue sections from ten PROTECT patients and histologically normal tissue sections from eight non-IBD controls were deparaffinized and rehydrated. Antigen retrieval was performed using Tris-based antigen unmasking solution [Vector Laboratories] for 10 min in a pressure cooker. Sections were blocked with 5% donkey serum for 2 h at room temperature and then incubated with primary antibody mouse anti-IL13-Rα2 [Abcam, ab55275] and rabbit anti-CD3 [Abcam, ab135372] or rabbit anti-vimentin [Abcam, ab92547] in 5% donkey serum overnight at 4°C. Sections were washed with 0.1% BSA and 0.05% Tween/PBS and incubated with donkey anti-mouse Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 647 [Invitrogen] for 2 h at room temperature. Slides were washed in PBS and counterstained with DAPI/Supermount G solution [Southern Biotechnology Associates]. Images were acquired using an Olympus BX51 microscope with a DP80 camera [Olympus America] and CellSens Dimension digital imaging software [Olympus Corporation, version 1.18]. Images were merged using ImageJ 1.52q [FIJI] software [NIH, https://imagej.nih.gov/ij/]. Cells co-expressing IL13-Rα2 and vimentin or CD3 were counted on at least five high-power fields [40× magnification] per patient.
For RORγ immunohistochemistry, paraffin-embedded rectal mucosal tissue sections from five UC patients with active endoscopic activity [endoscopic Mayo score 2–3] and histologically normal tissue sections from five non-IBD controls were deparaffinized and rehydrated. Antigen was retrieved at high pressure for 12 min in 10 mM sodium citrate buffer plus 0.05% Tween-20 [pH 6] using a pressure cooker [Bio SB]. The sections were blocked in 10% normal rabbit serum and 1% BSA and incubated with RORγ antibody [Thermo Fisher Scientific, 14-6988-82] for 75 min at room temperature. Endogenous peroxidase activity was quenched with Bloxall [Vector Labs] and incubated with a biotinylated rabbit anti-rat IgG secondary antibody for 60 min at room temperature [Vectastain, BA-4000]. Signals were amplified with the Avidin-Biotin Complex [ABC] Kit [Vector Labs] and RORγ was detected using 3,3ʹ-diaminobenzidine [DAB, Vector Labs]. The nuclei were counterstained with haematoxylin. Similarly stained serial sections from the same tissues lacking only the primary antibody served as the negative control for each sample. Images of five fields per sample were acquired [20× and 60× objectives] using a Keyence BZ-X800 inverted microscope for further analyses.
Quantitative analysis of RORγ in epithelial cells was carried out using ImageJ Fiji software as previously described with modifications.17 Briefly, the contribution of DAB and haematoxylin staining was calculated and digitally separated with the colour deconvolution plugin.18 Each deconvoluted image with DAB staining was adjusted to lower and upper thresholds set at 0 and 122, respectively. The freehand tool was used to select the epithelium as the region for analysis. The expression level of RORγ in the epithelium was estimated by measuring the percentage area fraction with DAB staining. In the lamina propria, RORγ+ cells were manually counted in five high-power field images per section.
3. Results
3.1. Altered rectal mucosal gene expression in UC
Baseline characteristics of the UC and non-IBD participants and the relevant treatment or disease severity subgroups are listed and compared with those of the entire PROTECT cohort in Table 1. Importantly, the PROTECT patients with rectal RNA in this study were similar to the overall PROTECT cohort.
Table 1.
Demographic and clinical characteristics of study population by subgroups.
|
n
428 |
Full PROTECT cohort | Gene expression array [GEA] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
n
313 |
Complete GEA cohort |
n
284 |
Analysed GEA cohort |
n
88 |
Mesalamine only |
n
169 |
Moderate-to-severe disease |
n
20 |
Controls | |||
| Demographic | ||||||||||||
| Age, years | 428 | 12.7 ± 3.3 | 313 | 12.9 ± 3.2 | 284 | 12.8 ± 3.1 | 88 | 12.9 ± 3.1 | 169 | 12.9 ± 3.1 | 20 | 13.8 ± 3.5 |
| Sex | 428 | 313 | 284 | 88 | 20 | |||||||
| Female | 212 [50] | 147 [47] | 132 [46] | 43 [49] | 169 | 76 [45] | 11 [55] | |||||
| Male | 216 [50] | 166 [53] | 152 [54] | 45 [51] | 93 [55] | 9 [45] | ||||||
| Race | 420 | 307 | 278 | 86 | 165 | 20 | ||||||
| White | 351 [84] | 259 [84] | 237 [85] | 69 [80] | 144 [87] | 17 [85] | ||||||
| Non-white | 69 [16] | 48 [16] | 41 [15] | 17 [20] | 21 [13] | 3 [15] | ||||||
| Ethnicity | 424 | 310 | 283 | 87 | 169 | 20 | ||||||
| Hispanic or Latino | 38 [9] | 27 [9] | 24 [8] | 11 [13] | 10 [6] | 0 | ||||||
| Not Hispanic or Latino | 386 [91] | 283 [91] | 259 [92] | 76 [87] | 159 [94] | 20 [100] | ||||||
| BMI Z score | 428 | −0.2 ± 1.3 | 313 | −0.2 ± 1.3 | 284 | −0.2 ± 1.3 | 88 | −0.1 ± 1.2 | 169 | −0.3 ± 1.4 | 20 | 0.2 ± 1.6 |
| Initial treatment | 428 | 313 | 284 | 88 | 169 | |||||||
| Mesalamine | 136 [32] | 101 [32] | 88 [31] | 88 [100] | 0 | |||||||
| Oral CS | 144 [34] | 106 [34] | 97 [34] | 0 | 70 [41] | |||||||
| IV CS | 148 [35] | 106 [34] | 99 [35] | 0 | 99 [59] | |||||||
| Clinical | ||||||||||||
| Disease extent | 428 | 313 | 284 | 88 | 169 | |||||||
| Proctosigmoiditis | 29 [7] | 21 [7] | 18 [6] | 16 [18] | 2 [1] | |||||||
| Left-sided colitis | 44 [10] | 35 [11] | 30 [11] | 20 [23] | 5 [3] | |||||||
| Extensive/pancolitis | 355 [83] | 257 [82] | 236 [83] | 52 [59] | 162 [96] | |||||||
| PUCAI score [0–89] | 428 | 50 ± 20 | 313 | 50 ± 20 | 284 | 51 ± 20 | 88 | 31 ± 12 | 169 | 64 ± 13 | ||
| <45 | 156 [36] | 116 [37] | 102 [36] | 70 [80] | 5 [3] | |||||||
| ≥45 | 272 [64] | 197 [63] | 182 [64] | 18 [20] | 164 [97] | |||||||
| Mayo endoscopy sub-score [range 0–] | 428 | 2.2 ± 0.7 | 313 | 2.2 ± 0.7 | 284 | 2.2 ± 0.7 | 88 | 1.8 ± 0.6 | 169 | 2.4 ± 0.6 | ||
| 1 = Mild | 59 [14] | 45 [14] | 39 [14] | 28 [32] | 6 [3] | |||||||
| 2 = Moderate | 224 [52] | 163 [52] | 145 [51] | 48 [54] | 81 [48] | |||||||
| 3 = Severe | 145 [34] | 105 [34] | 100 [35] | 12 [14] | 82 [49] | |||||||
| Hgb, g/dL | 402 | 294 | 268 | 80 | 162 | |||||||
| Mean | 11.4 ± 2.2 | 11.5 ± 2.2 | 11.5 ± 2.2 | 12.4 ± 1.8 | 11 ± 2.2 | |||||||
| <10 | 98 [24] | 64 [22] | 62 [23] | 6 [8] | 52 [32] | |||||||
| CRP | 315 | 237 | 59 | 140 | ||||||||
| >ULN | 144 [46] | 112 [47] | 103 [47] | 10 [17] | 82 [59] | |||||||
| >2× ULN | 97 [31] | 79 [33] | 73 [33] | 6 [10] | 60 [43] | |||||||
| ESR, mm/h | 385 | 285 | 258 | 77 | 158 | |||||||
| Median | 25 [12-42] | 24 [12-39] | 24 [12-39] | 14 [7-23] | 30 [17-46] | |||||||
| Albumin, g/dL | 422 | 311 | 282 | 87 | 168 | |||||||
| Mean | 3.7 ± 0.7 | 3.7 ± 0.7 | 3.7 ± 0.7 | 4.0 ± 0.7 | 3.5 ± 0.7 | |||||||
| <3.5 | 138 [33] | 100 [32] | 92 [33] | 13 [15] | 69 [41] | |||||||
| Vit D, ng/mL | 393 | 296 | 268 | 85 | 158 | |||||||
| Median | 29 [24-35] | 28 [24-34] | 28 [24-34] | 28 [24-34] | 28 [23-34] | |||||||
| <20 | 42 [11] | 34 [11] | 32 [12] | 10 [12] | 21 [13] | |||||||
| 20–30 | 183 [46] | 140 [47] | 127 [47] | 44 [52] | 71 [45] | |||||||
| ≥30 | 168 [43] | 122 [41] | 109 [41] | 31 [36] | 66 [42] | |||||||
| Baseline faecal calprotectin, –g/g | 239 | 181 | 165 | 49 | 101 | |||||||
| Median | 2352 [1202–3928] | 2634 [1212–4022] | 2480 [1212–4022] | 1629 [772–3632] | 3333 [1421–4384] | |||||||
| >250 | 226 [95] | 171 [94] | 155 [94] | 40 [82] | 100 [99] | |||||||
| Rectal eosinophils, >32/hpf | 367 | 210 [57] | 300 | 161 [54] | 272 | 144 [53] | 84 | 45 [54] | 162 | 80 [49] | ||
Results are expressed as mean ± SD, n [%] or median [interquartile range]. Analysed GEA cohort excludes patients with protocol violations per PROTECT. IFX, infliximab; PUCAI, Pediatric Ulcerative Colitis Activity Index; BMI, body mass index; CS, corticosteroids; IV, intravenous; Hgb, haemoglobin; CRP, C-reactive protein; ULN, upper limit of normal; ESR, erythrocyte sedimentation rate; vit D, vitamin D; hpf, high-power field.
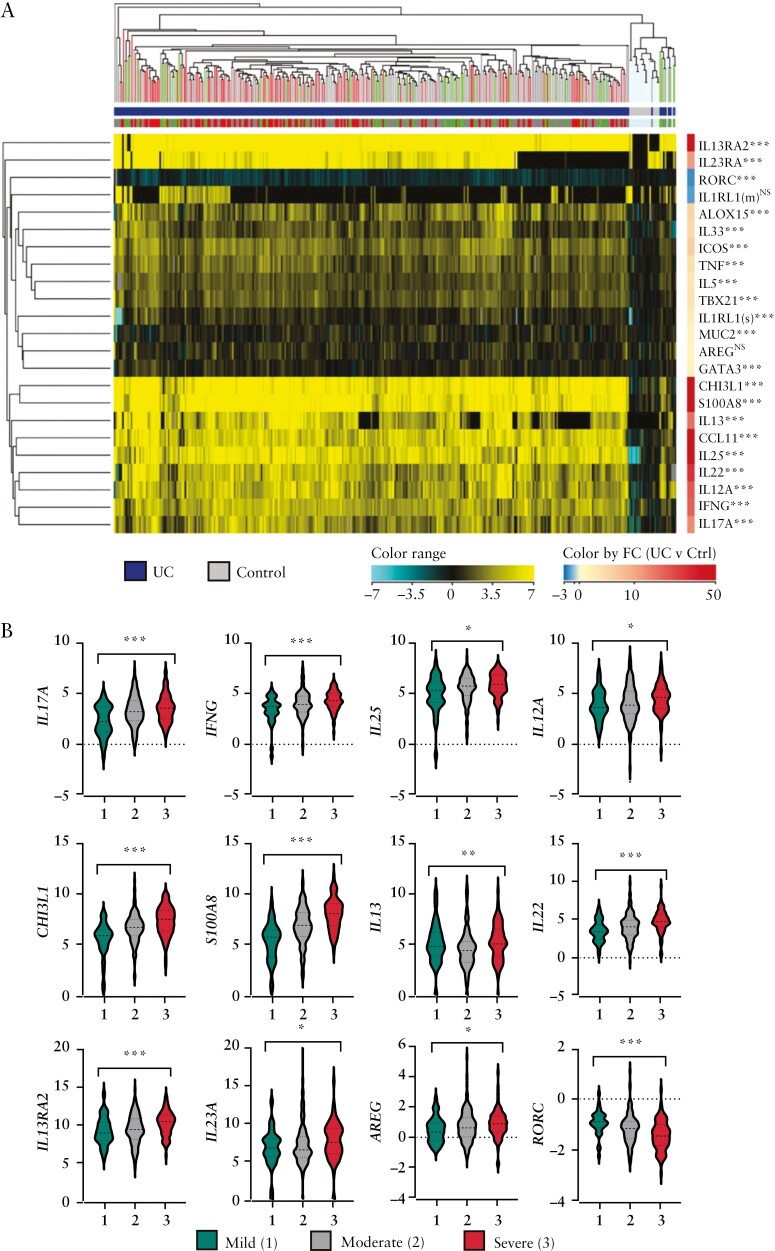
Compared to non-IBD controls, UC patients showed significantly altered rectal expression of all but two studied transcripts: IL1RL1[m], which encodes the membrane-bound IL-33 receptor, and AREG, which encodes the type 2 immune response growth factor amphiregulin19 [Figure 1A]. Most genes were upregulated in the inflamed UC rectum except for RORC [which encodes the nuclear receptor RORγ and its isoform RORγt, the canonical Th17 transcription factor], which was downregulated compared to controls.
Figure 1.
Targeted rectal mucosal gene expression of treatment-naïve paediatric UC patients at diagnosis compared to non-IBD patients. [A] Dendrogram and heatmap of unsupervised hierarchical clustering based on normalized expression of 23 genes as assessed by microfluidic RT-qPCR gene expression array. All p-values are false discovery rate-corrected. NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 vs non-IBD controls. [B] Violin plots showing analytes significantly associated with endoscopic Mayo score. The y-axis units are log2 gene expression relative to controls. *p < 0.05; **p < 0.01; ***p < 0.001 for Spearman’s correlation coefficient across endoscopic Mayo scores 1–3 .
We previously observed that hierarchical clustering of patients based on expression of 15 genes differentially expressed between UC, non-IBD and Crohn’s colitis identified a cluster of patients with higher likelihood of clinical remission. This cluster was primarily defined by detectable IL13 expression.8 Application of the same analysis using these 15 genes did not reveal clearly defined subclusters, and outcomes were not different between clusters [Supplementary Table 3, Supplementary Figure 1]. Therefore, we assessed expression of individual genes for association with treatment-specific outcomes.
3.2. IL13RA2 expression predicts corticosteroid-free remission on mesalamine at week 52 [CSFR]
In total, 36% of patients with high-quality RNA available achieved CSFR at week 52 on mesalamine alone, which is comparable to the proportion in the overall PROTECT cohort [38%]. Univariable logistic regression revealed that increased expression of IL13RA2 and S100A8 were associated with alower likelihood of achieving CSFR [Table 2 and Supplementary Table 4]. There was a trend toward an association of reduced RORC expression with lower likelihood of CSFR. IL13RA2 encodes a decoy receptor for the type 2 cytokine IL-13 that reduces its bioactivity,20 and S100A8 encodes a subunit of calprotectin. Since IL13RA2 and S100A8 expression were highly correlated, we assessed whether IL13RA2 expression alone improved the PROTECT published model for CSFR. Incorporating IL13RA2 modestly increased the area under the curve [AUC, from 0.72 to 0.74, p = .052] for the published PROTECT model [Table 2]. The existing model includes week 4 remission as a variable, which only allows use of the model after 4 weeks of observation. We found that a baseline data-only model including rectal IL13RA2 expression, haemoglobin, vitamin D and PUCAI exhibited similar performance [AUC = 0.70] to the published PROTECT model, which includes week 4 remission status [Table 2]. Furthermore, inclusion of IL13RA2 expression in this baseline model exhibited superior predictive value over a model of haemoglobin, vitamin D and PUCAI alone [p = .029]. Of note, baseline faecal calprotectin, a standard measure of mucosal disease severity used in clinical practice, did not predict CSFR by univariable analysis. Furthermore, although histological severity as measured by the Nancy histology index was associated with CSFR by univariable analysis [Supplementary Table 4], it did not add predictive value to the multivariable model. Therefore, IL13RA2 expression provides more prognostic information that these measures of mucosal disease severity.
Table 2.
Significant variables associated with outcomes and predictive models.
| Gene expression univariable analysis | Multivariable predictive models | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | p value | OR | p value | AUC | Sen | Spec | LR+ | LR− | LR test* | ||
| CSFR [week 52] | PROTECT Model | ||||||||||
| IL13RA2 | 0.83 [0.74–0.94] | 0.002 | PUCAI < 45 | 1.87 [1.08–3.24] | 0.024 | 0.72 [0.66–0.79] | 35% [25–45] | 86% [81–91] | 2.56 [1.61–4.06] | 0.76 [0.65–0.88] | NA |
| S100A8 | 0.84 [0.74-0.96] | 0.010 | Hgb ≥ 10 | 3.55 [1.00–12.63] | 0.082 | ||||||
| RORC | 1.49 [0.98-2.28] | 0.065 | Wk4 Rem | 12.01 [3.51–41.14] | <0.001 | ||||||
| PROTECT Model + IL13RA2 | |||||||||||
| PUCAI < 45 | 1.81 [1.04–3.14] | 0.030 | 0.74 [0.68–0.80] | 45% [35–55] | 82% [76–87] | 2.48 [1.69–3.64] | 0.67 [0.56–0.81] | 0.052 | |||
| Hgb ≥ 10 | 3.48 [0.98–12.45] | 0.090 | |||||||||
| Wk4 Rem | 11.19 [3.25–38.50] | <0.001 | |||||||||
| IL13RA2 | 0.89 [0.78–1.00] | 0.046 | |||||||||
| Model without Wk4 data | |||||||||||
| PUCAI < 45 | 2.20 [1.25–3.87] | 0.006 | 0.70 [0.63–0.77] | 33% [23–43] | 89% [83–93] | 2.95 [1.74–5.00] | 0.76 [0.65–0.88] | 0.029 | |||
| Hgb ≥ 10 | 2.28 [1.10–4.74] | 0.026 | |||||||||
| Vit D level | 1.05 [1.01–1.09] | 0.010 | |||||||||
| IL13RA2 | 0.87 [0.76–0.99] | 0.032 | |||||||||
| CSNR [week 12] | |||||||||||
| IL13 | 1.28 [1.06–1.54] | 0.009 | IL13 | 1.28 [1.06–1.54] | 0.009 | 0.68 [0.57–0.79] | 67% [51–80] | 65% [50–79] | 1.92 [1.22–3.01] | 0.51 [0.32–0.82] | NA |
| ALOX15 | 1.38 [0.99–1.92] | 0.059 | |||||||||
| Escalation to IFX [week 52] | |||||||||||
| IL25 | 1.39 [1.06–1.82] | 0.016 | IL33 | 0.66 [0.47–0.93] | 0.017 | 0.66 [0.58–0.74] | 41% [29–53] | 74% [64–83] | 1.58 [1.02–2.47] | 0.80 [0.64–1.00] | NA |
| IL13RA2 | 1.25 [1.04–1.50] | 0.016 | IL13RA2 | 1.28 [1.06-1.54] | 0.010 | ||||||
| IL33 | 0.68 [0.49–0.94] | 0.020 | |||||||||
| ALOX15 | 0.77 [0.61–0.99] | 0.040 | |||||||||
| Colectomy [week 52] | |||||||||||
| RORC | 0.28 [0.10–0.78] | 0.015 | RORC | 0.36 [0.14–0.95] | 0.040 | 0.71 [0.57–0.85] | 22% [6–46] | 92% [86–96] | 2.63 [0.94–7.34] | 0.86 [0.68–1.09] | 0.033 |
| ALOX15 | 0.70 [0.48–1.01] | 0.056 | PUCAI < 45 | 0.14 [0.02–0.97] | 0.050 | ||||||
95% confidence intervals of predictive statistics are presented in parentheses. Probability cutoffs: 0.5 [CSFR, CSNR, Escalation to IFX]; 0.2 [Colectomy].
CSFR, corticosteroid-free remission; CSNR, corticosteroid-naïve remission; IFX, infliximab; OR, odds ratio; PUCAI, Pediatric Ulcerative Colitis Activity Index; Hgb, haemoglobin; Wk4 Rem, week 4 remission; OR, odds ratio; AUC, area under the curve; Sen, sensitivity; Spec, specificity; LR, likelihood-ratio; NA, not applicable.
LR test compares the indicated model to the same model including only clinical variables without rectal mucosal gene expression data.
3.3. IL13 expression predicts corticosteroid-naïve remission at week 12 [CSNR]
In total, 48% and 41% of patients who started on mesalamine alone achieved CSNR at week 12 and week 52, respectively. Univariable logistic regression revealed that increased expression of IL13, which encodes a type 2 effector cytokine, was significantly associated with the likelihood of achieving CSNR at week 12 [Table 2]. We did not detect any association between expression of genes on the panel and CSNR at week 52.
3.4. IL13RA2 and IL33 expression predict escalation to IFX by week 52
In total, 73 [43%] of the 169 patients with baseline moderate-to-severe disease were escalated to IFX by week 52. Univariable logistic regression revealed that increased expression of IL13RA2 and IL25, and decreased expression of ALOX15 and IL33, were associated with the likelihood of escalating to IFX [Table 2]. Multivariable logistic regression revealed that expression of IL13RA2 and IL33 [type 2 instructive cytokine] were independently associated with escalation to IFX and predicted this outcome with an AUC of 0.66. The published PROTECT model for escalation to IFX was validated in this subset of patients, but the addition of IL13RA2 or IL33 gene expression did not improve model performance.
3.5. No detected gene expression predictors of response to early IFX therapy
Forty-one patients received early treatment with IFX within 12 weeks of diagnosis, and 12 [29%] achieved CSFR on IFX at 52 weeks. We did not detect any significant associations between gene expression and this outcome, potentially due to the low patient numbers.
3.6. RORC expression predicts colectomy by week 52
Nineteen [11%] of the 169 patients with moderate-to-severe disease activity at baseline underwent colectomy by week 52. Univariable logistic regression revealed that increased expression of RORC was associated with a lower likelihood of colectomy. There was a trend toward an association of higher ALOX15 expression with reduced risk for colectomy. A multivariable model including RORC and PUCAI predicted risk for colectomy with an AUC of 0.71 [Table 2]. A model including PUCAI alone was inferior to a model including both PUCAI and RORC [p = 0.033].
3.7. Changes in rectal mucosal gene expression are associated with endoscopic healing
Twelve of the 24 genes correlated with baseline endoscopic Mayo score. Most of the analytes were positively correlated, except RORC, which was inversely correlated [Figure 1B].
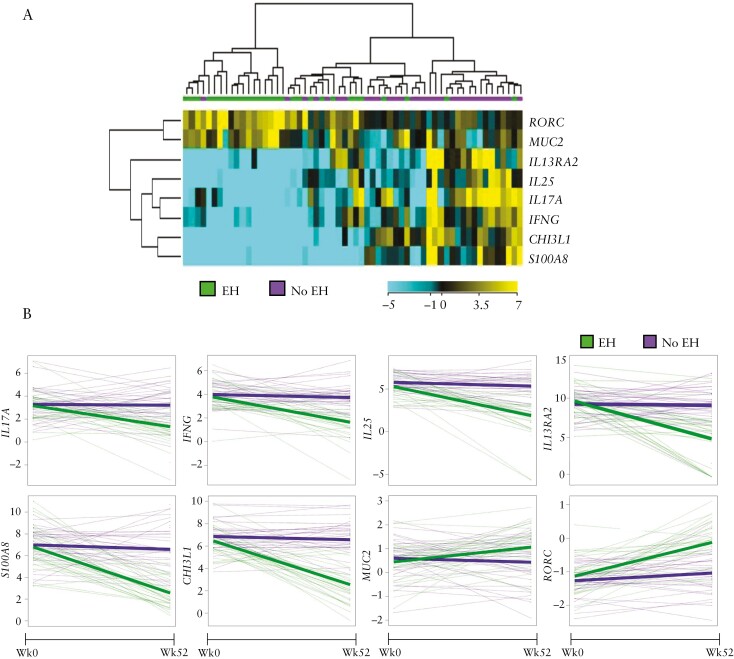
Twenty-eight [48%] of the 58 patients with 52-week follow-up rectal biopsies achieved endoscopic healing. Baseline characteristics of the patients who underwent follow-up endoscopy were similar to those of the entire cohort [Supplementary Table 2]. No analytes predicted endoscopic healing at 52 weeks for this smaller subset of patients. To determine if endoscopic healing is associated with dynamic changes in gene expression, we compared changes in gene expression from baseline to 52 weeks between patients with and without endoscopic healing. Increasing expression of RORC and MUC2, and decreasing expression of IL13RA2, IL25, IL17A, IFNG, CHI3L1 and S100A8 from week 0 to week 52 were associated with endoscopic healing [Figure 2].
Figure 2.
Change in targeted rectal mucosal gene expression from paediatric UC patients between diagnosis and week 52. [A] Dendrogram and heatmap denoting significantly altered gene expression between week 0 and week 52 as assessed by microfluidic RT-qPCR gene expression array for study patients [n = 58] with endoscopic healing [EH, endoscopic Mayo score 0] compared to not healed [no EH, endoscopic Mayo score 1–3]. [B] Spaghetti plots with overlying growth curves showing differential change in gene expression from week 0 to week 52 between those with and without endoscopic healing at week 52. The y-axis units are log2 gene expression relative to non-IBD control patients.
3.8. Rectal mucosal expression of type 2 genes is associated with mucosal eosinophils
Eosinophils are recruited or activated by type 2 cytokines.6,7 Published analyses from the PROTECT study have demonstrated that a rectal biopsy eosinophil count of ≤32 eosinophils per high-power field [hpf] is associated with treatment escalation or colectomy by 12 weeks in those treated with intravenous corticosteroids and escalation to IFX by 52 weeks in patients with moderate-to-severe disease.5,10 We found ALOX15 [lipoxygenase expressed by eosinophils] and CCL11 [eosinophil-specific chemokine] expression were significantly associated with a higher rectal eosinophil count, and IL13 and IL33 trended toward significance [Supplementary Table 5].
3.9. IL13RA2 and RORC are hubs within UC-associated gene co-expression modules
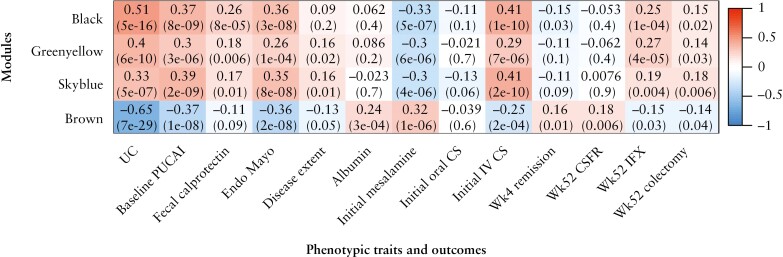
To further examine the putative roles of IL13RA2 and RORC in paediatric UC, we applied WGCNA to bulk RNA-seq data from the PROTECT study [n = 206 UC and n = 20 controls]11 to determine whether these outcome-associated genes were contained within disease-relevant gene networks. We identified 16 distinct gene co-expression modules [named with unique colours] significantly correlated with UC. A heatmap of selected modules along with their trait correlations is shown in Figure 3, and a complete heatmap including all module-trait correlations is shown in Supplementary Figure 2.
Figure 3.
Association of PROTECT RNA-seq gene co-expression modules with phenotypic traits and clinical outcomes. Heatmap of the correlation of selected modules derived from weighted gene co-expression network analysis [WGCNA] of PROTECT RNA-seq data with phenotypic traits and clinical outcomes. The black module contains IL13RA2, and the brown module contains RORC. p-values are shown in parentheses beneath the corresponding Pearson correlation coefficient in each box. UC, ulcerative colitis; PUCAI, Pediatric Ulcerative Colitis Activity Index; Endo, endoscopic; CS, corticosteroids; Wk, week; CSFR, corticosteroid-free remission; IFX, infliximab.
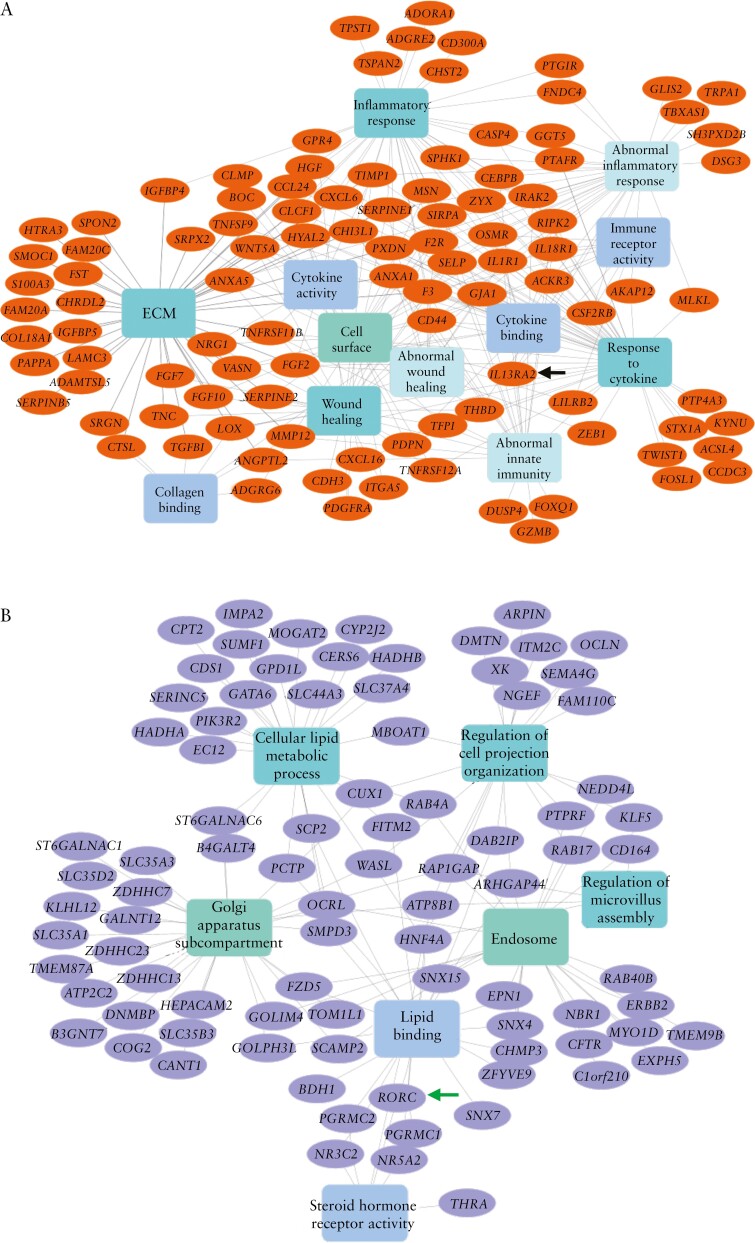
IL13RA2 was a hub gene, based on both connectivity and phenotype significance scores, within the black module, which positively correlated with measures of disease severity and poor outcomes [escalation to IFX, colectomy; Figure 3]. IL13RA2 co-expressed genes within the black module were associated with extracellular matrix, collagen binding, cytokine activity, inflammatory response and abnormal innate immunity [Figure 4A].
Figure 4.
Network representation of significantly enriched terms [rectangles] of IL13RA2 and RORC top-correlated genes [ovals] from the black and brown modules, respectively. [A] Black module containing IL13RA2, indicated by the black arrow. [B] Brown module containing RORC, indicated by the green arrow.
RORC was a hub gene within the brown module. Similar to RORC gene expression alone, the brown module negatively correlated with disease severity and poorer outcomes [escalation to IFX, colectomy] and positively correlated with superior outcomes [week 4 remission and CSFR at 52 weeks]. RORC co-expressed genes within the brown module were involved in lipid binding, lipid metabolic processes, steroid hormone receptor activity and microvillous assembly [Figure 4B].
Two additional modules, greenyellow and skyblue, showed strong positive correlations with disease severity and poorer outcomes. The greenyellow module was associated with host entry and immune response, and the skyblue module was associated with cytokine production and leukocyte chemotaxis [Supplementary Figure 3].
3.10. Single-cell RNA-seq reveals cell-specific dysregulation of RORC and IL13RA2 in adult UC
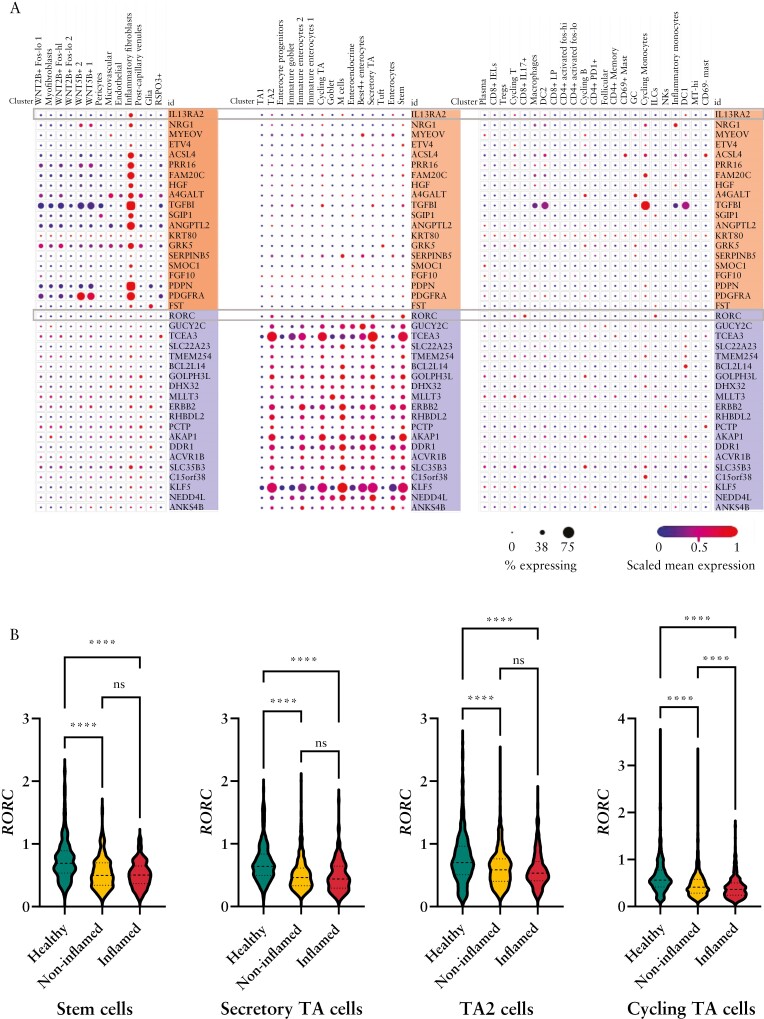
To gain an understanding of the cellular sources of RORC and IL13RA2 and their associated gene expression modules in UC, we analysed a publicly available adult colon single-cell RNA-seq data set including inflamed and non-inflamed UC tissues, and tissues from non-IBD controls.16 That study previously described that IL13RA2 expression marked a distinct subset of inflammatory fibroblasts associated with UC. Accordingly, we found that expression of IL13RA2 and its 20 most highly correlated genes within the black module were expressed in inflammatory fibroblasts [Figure 5A].
Figure 5.
Cell-specific expression of IL13RA2 and RORC and their most closely correlated genes. [A] Expression of IL13RA2 and its 20 most closely correlated genes within the black module [shaded in orange] and RORC and its 20 most closely correlated genes from the brown module [shaded in purple] within stromal, epithelial and immune cell types from existing adult colon UC single-cell RNA-seq data.16 The size and colour of the dots are proportional to the percentage expressing the gene and the normalized expression level, respectively. [B] Further analysis of the same adult single-cell RNA-seq data set comparing RORC expression within non-zero expressing cells between healthy, non-inflamed UC, and inflamed UC from specific epithelial cell types. The y-axis units for RORC expression are log2[TP10K + 1]. NS, not significant; ****p < 0.0001. TA, transit amplifying.
RORC encodes the nuclear receptor RORγ, including its isoform RORγT, the canonical type 17 transcription factor. Given that expression of other type 17 genes [IL17, IL23] were increased in UC [Figure 1], but RORC and its brown module were associated with healthy colon and superior outcomes, we questioned whether disease-associated alterations in RORC were immune cell-intrinsic. Analysis of this single-cell RNA-seq data set showed that RORC and its brown module-correlated genes are expressed in colon epithelial cells, specifically stem cells and transit amplifying [TA] cells [Figure 5A].
We then examined disease-state differential expression of IL13RA2 and RORC in this data set. As expected, IL13RA2 was expressed in inflammatory fibroblasts from inflamed and non-inflamed UC tissues, but not non-IBD tissues [Supplementary Figure 4A]. RORC was detected in ~20% of epithelial stem cell and TA cell subsets [Figure 5A and Supplementary Figure 4B]. This may be related to stochastic detection of expression for low- to moderately expressed genes by single-cell RNA-seq pipelines.21 Therefore, we compared normalized RORC expression within epithelial stem cells, secretory TA cells, TA2 cells and cycling TA cells with detectable RORC expression between disease states. We observed reduced RORC expression in RORC-expressing stem cells, secretory TA cells, TA2 cells and cycling TA cells from inflamed and non-inflamed UC tissues compared to non-IBD tissues [Figure 5B]. These observations further support that the reduced tissue expression of RORC in UC is derived from reduced expression in epithelial stem and TA cells.
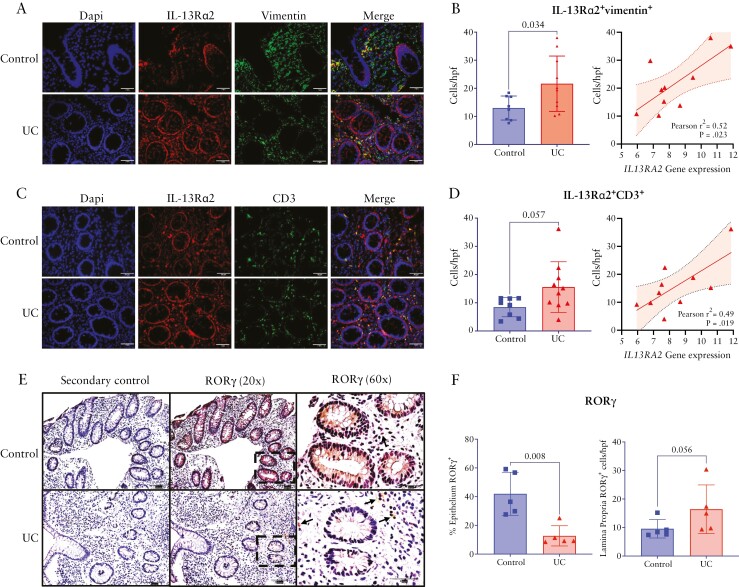
We performed immunofluorescence staining and immunohistochemistry to localize IL-13Rα2 and RORγ protein expression, respectively. Previous reports have localized IL-13Rα2 to fibroblasts or T cells.16,22,23 There were significantly more IL-13Rα2+vimentin+ fibroblasts populating the colon lamina propria in paediatric UC compared to non-IBD controls [p = 0.034, Figure 6A and 6B]. There was also a trend toward an increase in IL-13Rα2+CD3+ T cells in UC compared to non-IBD [p = 0.057, Figure 6C and 6D]. IL-13Rα2 was ubiquitously expressed in epithelial cells of both UC and non-IBD controls. Numbers of both IL-13Rα2+vimentin+ fibroblasts and IL-13Rα2+CD3+ T cells correlated with IL13RA2 gene expression in paediatric UC [Figure 6B and 6D]. Consistent with the bulk and single-cell RNA-seq analyses, the epithelium of UC tissues exhibited significantly less expression of RORγ compared to that of non-IBD control tissue [p = 0.008]. Also, consistent with our finding of increased IL17A expression in UC, there was a trend toward increased numbers of RORγ+ cells populating the lamina propria of UC tissues compared to that of non-IBD control tissues [p = 0.056; Figure 6E and 6F].
Figure 6.
IL-13Rα2 and RORγ protein localization in paediatric UC tissues. Representative immunofluorescence images showing IL-13Rα2 co-localization with vimentin [A] and CD3 [C]; 40× magnification [scale bars = 50 μm]. Comparison of the number of IL-13Rα2+vimentin+ [B] or IL-13Rα2+CD3+ cells [D] per high-power field between UC and non-IBD control patients and correlation with IL13RA2 gene expression within UC tissues. Representative images of immunohistochemical staining for RORγ protein expression [brown] in UC and non-IBD control tissues at 20× [scale bars = 100 μm] and 60× magnification [scale bars = 50 μm]. Images from staining with secondary antibody are displayed as a negative control. Location of the 60× high-power image is indicated by the dashed bordered box overlying the 20× magnification image. Arrows indicate RORγ+ lamina propria cells. Comparison of the percentage of the epithelium positively stained for RORγ and the number of RORγ+ lamina propria cells/hpf between non-IBD control and UC tissues [F]. hpf, high power field; UC, ulcerative colitis.
4. Discussion
In a well-established cohort of treatment-naive paediatric UC patients treated with standardized mesalamine and CS, we show that targeted assessment of baseline rectal mucosal immune gene expression predicts clinical outcomes. A model including IL13RA2 and only baseline clinical parameters was as accurate for predicting CSFR as an established clinical model that requires week 4 remission status. Furthermore, our findings validate the association of type 2 immune genes [IL13, IL33] with positive outcomes, and IL13RA2, which decreases the bioactivity of IL-13 [a key effector type 2 cytokine], with poorer outcomes, suggesting a protective role of type 2 cytokines in UC. We observed signals of IL13RA2 expression associated with poor outcomes and RORC with superior outcomes. Our analysis of publicly available bulk and single-cell data sets support that IL13RA2 is a hub gene within an inflammatory fibroblast gene co-expression module upregulated in UC associated with extracellular matrix and innate immune response. We found RORC to be a hub gene within an epithelial stem and TA cell co-expression module associated with lipid binding and metabolism, steroid hormone receptor activity, and microvillus assembly.
Studies of cytokine secretion from lamina propria immune cells of adult UC surgical tissues established the association of a type 2 immune response with UC.7,24 Our group has now demonstrated upregulation of IL13 expression in two treatment-naïve paediatric UC cohorts, and previously showed this was specific to paediatric UC over Crohn’s colitis.8 Previous studies have supported that IL-13 is produced by atypical natural killer T cells bearing IL-13Rα2 that exert epithelial cytotoxicity and are pathogenic in UC.22,25 However, there is an emerging understanding of the protective roles of type 2 immune responses in UC related to promotion of tissue repair and differentiation of mucin-secreting goblet cells.6,26 The negative results of clinical trials of two anti-IL-13 monoclonal antibodies for the treatment of UC contradicts the theory of a primary pathogenic role for IL-13.27,28 In fact, in a smaller treatment-naïve paediatric UC cohort treated with standard care, our group found that a tissue gene expression pattern marked by detectable expression of IL13 was associated with improved clinical outcomes.8 Here, in a larger independent treatment-naïve cohort in which we examined specific treatment pathways, we validate our previous findings with the observation that IL13 expression is associated with CSNR at 12 weeks in patients treated with mesalamine alone at diagnosis. Interestingly, amongst paediatric patients with baseline moderate-to-severe UC, expression of the type 2 instructive cytokine IL33 was also associated with a lower likelihood of escalation to IFX. Collectively, these findings support that a type 2 immune response in UC may be a protective response to inflammation that improves treatment outcomes.
We observed that increased baseline rectal mucosal IL13RA2 expression was associated with a lower likelihood of CSFR, higher likelihood of escalating to IFX therapy by 1 year and higher baseline endoscopic severity. IL13RA2 expression was associated with increased disease severity as evidenced by correlations with endoscopic Mayo score and S100A8 expression. Nonetheless, it informed our predictive models better than baseline faecal calprotectin [which had no detectable association with clinical outcomes] and histological severity score. Several lines of evidence implicate IL13RA2 in the pathogenesis of IBD. IL13RA2 encodes an alternative receptor for IL-13 [classically described as a decoy receptor] with both membrane-bound and soluble forms that block the activity of IL-13.29 However, some have reported IL-13Rα2 signalling activity.30 Mucosal expression of IL13RA2 is associated with resistance to anti-TNF therapy in both adult UC and Crohn’s disease.31,32 Single-cell RNA-seq has revealed enrichment of inflammatory fibroblasts expressing IL13RA2 in adult UC that are similarly associated with anti-TNF resistance.16 Receptor–igand analysis proposed a strong interaction between inflammatory monocytes and IL13RA2+IL11+ inflammatory fibroblasts expressing the receptor for oncostatin M, another monocyte-derived cytokine implicated in treatment resistance.11,33,34 It is suspected that IL13RA2 negatively regulates IL-13 signalling and blocks its anti-inflammatory effects, supported by the observation that IL13RA2 knockout mice exhibit increased IL-13 activity and are protected against both acute and chronic models of colitis.20,35,36 Interestingly, IL13RA2 was not within the published PROTECT antimicrobial RNA-seq gene expression signature that strengthened the predictive models for CSFR and escalation to IFX therapy.5 Here, we may have been better poised to detect the association of IL13RA2 with poor outcomes due to our use of different technology [RT-qPCR microfluidic array] or the stringent multiple comparison correction required for higher order transcriptomic data analysed in PROTECT. Consistent with a role for IL13RA2 in inflammatory fibroblasts and monocyte interactions, we found that in UC patients, IL13RA2 was a hub gene with OSMR within a co-expression module associated with extracellular matrix, cytokine activity and innate immune response. As a whole, our findings corroborate adult IBD studies by demonstrating the utility of mucosal IL13RA2 for improving outcome prediction in paediatric UC over clinical models alone and provide further evidence that the role of IL13RA2 in UC pathogenesis involves expression by inflammatory fibroblasts.
RORC encodes the nuclear receptor RORγ and its isoform RORγT, the latter being the canonical Th17 transcription factor. Higher baseline RORC expression is associated with improved response to anti-TNF therapy and endoscopic healing in adult UC patients.37 One would expect RORC to be upregulated in UC compared to healthy controls given the upregulation of type 17 instructive and effector cytokines we observed. However, we found that RORC was net downregulated in UC and inversely associated with endoscopic severity. Furthermore, we observed that increased RORC expression was associated with improved outcomes, including a lower likelihood of colectomy at 1 year. In line with our findings, an independent analysis of PROTECT bulk RNA-seq data found RORC to be one of nine genes within a transcriptional risk score, derived from differential expression of UC risk genes that are also expression quantitative trait loci [eQTL], predictive of colectomy at 52 weeks.38 Our WGCNA of RNA-seq PROTECT data mirrored these findings, as the module containing RORC was associated with lower severity and positive outcomes. Interestingly, our functional enrichment analysis suggests that RORC is involved in epithelial cell lipid metabolic and structural processes rather than primarily immune cells. RORC and its co-expressed genes within this module mapped to epithelial stem cells and less-differentiated transit-amplifying cells in an adult UC single-cell RNA-seq data set. This observation is in line with a previous report of RORC expression in the colon epithelium at crypt bases in human colon and mouse models.39 Our analysis of published single-cell RNA-seq data and tissue immunostaining confirmed epithelial downregulation of RORγ protein in UC. Regarding other organ systems, there have been reports of RORγ expression localizing to epithelial cells in skin,40 and of RORγ mediating the epithelial–mesenchymal transition in breast cancer and hepatic fibrosis.41,42 One interesting hypothesis deserving of further study is that colon epithelial-intrinsic RORγ may have a role in epithelial wound repair.
A notable strength of our study was use of the large PROTECT inception cohort, free from confounding by previous therapy. Other strengths include the analysis of 52-week follow-up rectal biopsies, and confirmation of findings through RNA-seq WGCNA and single-cell analysis. We were able to validate our previously published association of IL13 expression with positive outcomes in this larger cohort and narrow its predictive capacity to mesalamine response. Limitations of our study include measuring mRNA gene expression rather than protein analysis, though previous studies have shown sufficient correlation between cytokine gene expression measured by real-time RT-qPCR and protein abundance.43 In addition, there is no comparable paediatric UC cohort for validation of new signals identified. This weakness is mitigated by our approach to examine signals through targeted GEA and RNA-seq, and by the observation that IL13RA2 has previously been associated with poor response to therapy in adults.31,44 Although, at the cut-off values chosen, our models exhibit low sensitivity, specificities were fairly high. Cut-off values were based on pre-selected probabilities and could be modified to favour sensitivity or specificity. For instance, specificity for the model for colectomy was quite high, and therefore a positive test may accurately identify patients at very high risk for colectomy who may benefit from early treatment with anti-TNF therapy.
In conclusion, in a large treatment-naïve paediatric UC inception cohort, targeted assessment of mucosal immune gene expression predicted clinical outcomes. A model including baseline gene expression and clinical factors was as accurate for predicting CSFR as an established clinical model, which requires week 4 remission status. Association of type 2 immune genes with improved outcomes, and the type 2 counteracting gene IL13RA2 with poorer outcomes, implies a beneficial role of type 2 cytokines in UC. Our findings support those of others associating IL13RA2 from inflammatory fibroblasts with the innate immune response and extracellular matrix functions and poor response to therapy. Increased expression of RORC, probably from crypt base stem and TA cells, is associated with lipid metabolism and cell structure and a lower likelihood of colectomy. These findings support further exploration of IL-13Rα2 as a therapeutic target in UC and future studies of the epithelial-specific role of RORC in colon mucosal homeostasis and UC pathogenesis.
Supplementary Material
Contributor Information
Kathryn Clarkston, Division of Gastroenterology, Hepatology and Nutrition; Division of Pediatric Gastroenterology, Children’s Mercy Hospital, University of Missouri-Kansas City School of Medicine, Kansas City, MO, USA.
Rebekah Karns, Division of Gastroenterology, Hepatology and Nutrition.
Anil G Jegga, Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Mihika Sharma, Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Sejal Fox, Division of Gastroenterology, Hepatology and Nutrition.
Babajide A Ojo, Division of Pediatric Gastroenterology, Department of Pediatrics, Stanford University School of Medicine, Stanford, CA, USA.
Phillip Minar, Division of Gastroenterology, Hepatology and Nutrition; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Thomas D Walters, Division of Pediatric Gastroenterology, Hospital for Sick Children, Toronto, ON, Canada.
Anne M Griffiths, Division of Pediatric Gastroenterology, Hospital for Sick Children, Toronto, ON, Canada.
David R Mack, Division of Gastroenterology, Hepatology and Nutrition, Children’s Hospital of Eastern Ontario and University of Ottawa, Ottawa, ON, Canada.
Brendan Boyle, Division of Gastroenterology, Hepatology, and Nutrition, Nationwide Children’s Hospital, Columbus, OH, USA.
Neal S LeLeiko, Department of Pediatrics, Columbia University Vagelos College of Physicians and Surgeons and NewYork-Presbyterian Morgan Stanley Children’s Hospital, New York, NY, USA.
James Markowitz, Division of Gastroenterology, Hepatology, and Nutrition, Cohen Children’s Medical Center of New York, New Hyde Park, NY, USA.
Joel R Rosh, Division of Gastroenterology, Hepatology, and Nutrition, Goryeb Children’s Hospital, Atlantic Health, Morristown, NJ, USA.
Ashish S Patel, Division of Gastroenterology, Phoenix Children’s Hospital, Phoenix, AZ, USA.
Sapana Shah, Division of Gastroenterology, Hepatology and Nutrition, UPMC Children’s Hospital of Pittsburgh, Pittsburgh, PA, USA.
Robert N Baldassano, Division of Gastroenterology, Hepatology and Nutrition, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Marian Pfefferkorn, Division of Gastroenterology, Hepatology, and Nutrition, Riley Children’s Hospital, Indianapolis, IN, USA.
Cary Sauer, Division of Gastroenterology, Hepatology and Nutrition, Department of Pediatrics, Emory University and Children’s Healthcare of Atlanta, Atlanta, GA, USA.
Subra Kugathasan, Division of Gastroenterology, Hepatology and Nutrition, Department of Pediatrics, Emory University and Children’s Healthcare of Atlanta, Atlanta, GA, USA.
Yael Haberman, Division of Gastroenterology, Hepatology and Nutrition; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA; Sheba Medical Center, Tel Hashomer, Israel.
Jeffrey S Hyams, Division of Digestive Diseases, Hepatology, and Nutrition, Connecticut Children’s Medical Center, Hartford, CT, USA.
Lee A Denson, Division of Gastroenterology, Hepatology and Nutrition; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Michael J Rosen, Division of Pediatric Gastroenterology, Department of Pediatrics, Stanford University School of Medicine, Stanford, CA, USA; Division of Gastroenterology, Hepatology and Nutrition; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health [R01DK117119 to M.J.R., U01DK095745 to J.S.H. and L.A.D., and T32DK007727 to K.C.]. This project was also supported in part by NIH P30 DK078392 Gene Analysis and Clinical Component Scores of the Digestive Diseases Research Core Center in Cincinnati, OH. Data management was further supported by the Center for Clinical and Translational Science and Training funded in part by the National Center for Advancing Translational Sciences [2UL1TR001425].
Conflict of Interest
MJR has served on an advisory board for Entasis Therapeutics. JSH has served on advisory boards for Janssen and AbbVie, and as a consultant for Pfizer, Lilly, Takeda, BMS and Thetis Pharmaceuticals. JRR has served on an advisory board for BMS and Lilly and has received grant/research support from AbbVie and Janssen. ASP has served as a consultant for AbbVie. NSL has served on a data monitoring board for AbbVie. TDW has served as a consultant for Janssen Canada, AbbVie Canada and Merck Canada. All other authors declare no competing interests.
Author Contributions
Conceptualization: MJR, LAD, RK, JSH, YH. Data curation: TDW, RK, KC, MJR. Formal analysis: KC, RK, AGJ, MS, YH. Funding acquisition: MJR, LAD, JSH. Investigation: KC, MJR, SF, BAO, PM, TDW, AMG, DRM, BB, NSL, JM, JRR, ASP, SS, RNB, MP, CS, SK, JSH, LAD. Methodology: RK, AGJ. Project administration: MJR, KC. Resources/provision of patient samples: PM, TDW, AMG, DRM, BB, NSL, JM, JRR, ASP, SS, RNB, MP, CS, SK, JSH, LAD, MJR. Supervision: MJR. Visualization: KC, RK, SF, BAO, AGJ, MS. Drafting of manuscript: KC, SF, BAO. Critical review and editing of manuscript: PM, TDM, AMG, DRM, BB, NSL, JM, JRR, ASP, SS, RNB, MP, CS, SK, YH, JSH, LAD, MJR.
Data Availability
Statement: Data from the PROTECT study will be available from the US National Institutes of Diabetes, and Digestive and Kidney Diseases data repository [https://repository.niddk.nih.gov/home/]. Requestors can apply for access via this website and must have approval from their institution’s review board.
References
- 1. Ye Y, Manne S, Treem WR, et al. Prevalence of inflammatory bowel disease in pediatric and adult populations: recent estimates from large national databases in the United States, 2007–2016. Inflamm Bowel Dis 2020;26:619–25. [DOI] [PubMed] [Google Scholar]
- 2. Roberts SE, Thorne K, Thapar N, et al. A systematic review and meta-analysis of paediatric inflammatory bowel disease incidence and prevalence across Europe. J Crohns Colitis 2020;14:1119–48. [DOI] [PubMed] [Google Scholar]
- 3. Kaplan GG, Ng SC.. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 2017;152:313–321.e2. [DOI] [PubMed] [Google Scholar]
- 4. Sykora J, Pomahacova R, Kreslova M, et al. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol 2018;24:2741–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hyams JS, Davis Thomas S, Gotman N, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet 2019;393:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol 2015;15:271–82. [DOI] [PubMed] [Google Scholar]
- 7. Fuss IJ, Neurath M, Boirivant M, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol 1996;157:1261–70. [PubMed] [Google Scholar]
- 8. Rosen MJ, Karns R, Vallance JE, et al. Mucosal expression of type 2 and type 17 immune response genes distinguishes ulcerative colitis from colon-only Crohn’s disease in treatment-naive pediatric patients. Gastroenterology 2017;152:1345–1357.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med 2018;379:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hyams JS, Davis S, Mack DR, et al. Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol Hepatol 2017;2:855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haberman Y, Karns R, Dexheimer PJ, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun 2019;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haberman Y, Iqbal NT, Ghandikota S, et al. Mucosal genomics implicate lymphocyte activation and lipid metabolism in refractory environmental enteric dysfunction. Gastroenterology 2021;160:2055–2071.e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang B, Horvath S.. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 14. Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–730.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crowe AR, Yue W.. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc 2019;9:e3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruifrok AC, Johnston DA.. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 2001;23:291–9. [PubMed] [Google Scholar]
- 19. Monticelli LA, Sonnenberg GF, Abt MC, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 2011;12:1045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karmele EP, Pasricha TS, Ramalingam TR, et al. Anti-IL-13Ralpha2 therapy promotes recovery in a murine model of inflammatory bowel disease. Mucosal Immunol 2019;12:1174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JK, Kolodziejczyk AA, Ilicic T, et al. Characterizing noise structure in single-cell RNA-seq distinguishes genuine from technical stochastic allelic expression. Nat Commun 2015;6:8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuss IJ, Joshi B, Yang Z, et al. IL-13Ralpha2-bearing, type II NKT cells reactive to sulfatide self-antigen populate the mucosa of ulcerative colitis. Gut 2014;63:1728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graham DB, Xavier RJ.. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuss IJ, Heller F, Boirivant M, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest 2004;113:1490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heller F, Florian P, Bojarski C, et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 2005;129:550–64. [DOI] [PubMed] [Google Scholar]
- 26. Monticelli LA, Osborne LC, Noti M, et al. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci USA 2015;112:10762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danese S, Rudzinski J, Brandt W, et al. Tralokinumab for moderate-to-severe UC: a randomised, double-blind, placebo-controlled, phase IIa study. Gut 2015;64:243–9. [DOI] [PubMed] [Google Scholar]
- 28. Reinisch W, Panes J, Khurana S, et al. Anrukinzumab, an anti-interleukin 13 monoclonal antibody, in active UC: efficacy and safety from a phase IIa randomised multicentre study. Gut 2015;64:894–900. [DOI] [PubMed] [Google Scholar]
- 29. Hershey GK. IL-13 receptors and signaling pathways: an evolving web.. J Allergy Clin Immunol 2003;111:677–90; quiz 691; quiz 691. [DOI] [PubMed] [Google Scholar]
- 30. Fichtner-Feigl S, Strober W, Kawakami K, et al. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006;12:99–106. [DOI] [PubMed] [Google Scholar]
- 31. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612–9. [DOI] [PubMed] [Google Scholar]
- 32. Verstockt B, Verstockt S, Creyns B, et al. Mucosal IL13RA2 expression predicts nonresponse to anti-TNF therapy in Crohn’s disease. Aliment Pharmacol Ther 2019;49:572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyams JS, Brimacombe M, Haberman Y, et al. Clinical and host biological factors predict colectomy risk in children newly diagnosed with ulcerative colitis. Inflamm Bowel Dis 2022;28:151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson MS, Ramalingam TR, Rivollier A, et al. Colitis and intestinal inflammation in IL10-/- mice results from IL-13Ralpha2-mediated attenuation of IL-13 activity. Gastroenterology 2011;140:254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verstockt B, Perrier C, De Hertogh G, et al. Effects of epithelial IL-13Ralpha2 expression in inflammatory bowel disease. Front Immunol 2018;9:2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Viazis N, Giakoumis M, Bamias G, et al. Predictors of tissue healing in ulcerative colitis patients treated with anti-TNF. Dig Liver Dis 2017;49:29–33. [DOI] [PubMed] [Google Scholar]
- 38. Mo A, Nagpal S, Gettler K, et al. Stratification of risk of progression to colectomy in ulcerative colitis via measured and predicted gene expression. Am J Hum Genet 2021;108:1765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Modica S, Gofflot F, Murzilli S, et al. The intestinal nuclear receptor signature with epithelial localization patterns and expression modulation in tumors. Gastroenterology 2010;138:636–48, 648.e1, 648 e1-12. [DOI] [PubMed] [Google Scholar]
- 40. Slominski AT, Kim TK, Takeda Y, et al. RORalpha and ROR gamma are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. FASEB J 2014;28:2775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oh TG, Wang SM, Acharya BR, et al. The nuclear receptor, RORgamma, regulates pathways necessary for breast cancer metastasis. EBioMedicine 2016;6:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim SM, Choi JE, Hur W, et al. RAR-related orphan receptor gamma [ROR-gamma] mediates epithelial–mesenchymal transition of hepatocytes during hepatic fibrosis. J Cell Biochem 2017;118:2026–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Flores MG, Zhang S, Ha A, et al. In vitro evaluation of the effects of candidate immunosuppressive drugs: flow cytometry and quantitative real-time PCR as two independent and correlated read-outs. J Immunol Methods 2004;289:123–35. [DOI] [PubMed] [Google Scholar]
- 44. Verstockt B, Perrier C, De Hertogh G, et al. Effects of epithelial IL-13Rα2 expression in inflammatory bowel disease. Front Immunol 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statement: Data from the PROTECT study will be available from the US National Institutes of Diabetes, and Digestive and Kidney Diseases data repository [https://repository.niddk.nih.gov/home/]. Requestors can apply for access via this website and must have approval from their institution’s review board.