Abstract
To further understand the epidemiology of enterotoxigenic Bacteroides fragilis (ETBF), 89 extraintestinal B. fragilis strains from Seoul, Korea, were examined for secretion of B. fragilis toxin (BFT) by the HT29/C1 biologic assay and for the B. fragilis toxin gene (bft) by colony blot hybridization and PCR. Complete agreement between the three techniques was found. Overall, 34 B. fragilis strains (38%) were identified as ETBF. Eleven of the 34 ETBF strains (32%) expressed a new isoform of BFT (Korea-BFT). This new isoform is more related to BFT-2 than to BFT-1. Like BFT-1 and BFT-2, Korea-BFT cleaves E-cadherin, the zonula adherens protein.
Strains of enterotoxigenic Bacteroides fragilis (ETBF) have been associated with diarrheal diseases of animals, young children, and adults (15, 16, 18–20, 23, 24, 30). The secretory response to ETBF is attributed to the expression of a ca. 20-kDa protein (termed B. fragilis toxin or BFT), which stimulates fluid accumulation in lamb ligated ileal loops and alters the morphology of human intestinal cells in vitro, especially HT29/C1 cells (2, 4, 11, 14, 17, 21, 22, 24, 27).
Sequencing of the bft gene and substrate analysis in vitro indicates that BFT is a zinc-dependent metalloprotease and that there are two isoforms of bft, bft-1 and bft-2. The two bft isoforms have 92% amino acid sequence identity (6, 10, 12). ETBF strains produce one, but not both, of these BFTs (6). Studies performed to date indicate that (i) purified BFT-2 has modest but consistently greater biological activity than purified BFT-1 when tested on HT29/C1 cells and (ii) BFT-1 and BFT-2 elute with different concentrations of NaCl from a high-resolution anion-exchange column (MonoQ) and exhibit different electrophoretic mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (28). Both BFTs act in a reversible manner to alter the morphology and physiology of polarized epithelial cells (HT29/C1, MDCK, and T84) (2, 17). Recently, it was determined that both BFTs cleave the zonula adherens protein, E-cadherin (29).
The bft gene is contained in a pathogenicity island of 6 kb, which contains another metalloprotease gene, termed mpII (5, 13). The protein encoded by mpII exhibits a zinc-binding metalloprotease motif similar to BFT-1 and BFT-2; however, recombinant MPII neither has biologic activity on HT29/C1 cells nor cleaves the E-cadherin protein (4a). It has been proposed that putative B. fragilis toxins fall into two categories (25). The first category (class I toxins) includes BFT-1 and BFT-2, which act by cleavage of the E-cadherin protein. The second category (class II toxins) at present contains only the MPII protein. As yet, no biological activity has been identified for MPII.
In this study, we identified a third isoform of bft class I (Korea-bft) in ETBF strains isolated from extraintestinal samples in Seoul, Korea. Eleven of 34 ETBF strains (32%) isolated in Korea contained Korea-bft. However, this third isoform of bft was not identified in a collection of ETBF strains isolated in the United States.
Detection of ETBF strains.
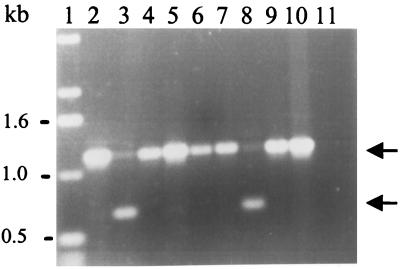
A total of 89 extraintestinal B. fragilis strains isolated between 1995 and 1997 from Severance Hospital, Seoul, Korea, were tested for BFT production by a tissue culture assay and for the presence of the bft gene by colony blot hybridization and PCR. For the tissue culture assay, HT29/C1 cells were used as previously described (14, 27). For colony blot hybridization, a 32P-labeled 1.2-kb SmaI-DraIII bft fragment containing bft-2 was used as a probe. For PCR, primers were designed from the published bft-2 sequence from ETBF 86-5443-2-2 (6). The sequence of the forward primer (primer 1) corresponded to bp 54 to 72 of bft with the addition of a sequence for an EcoRI restriction site at the 5′ end (5′-CGCGGAATTCATGTTCTAATGAAGCTGAT-3′), and the sequence of the reverse primer (primer 2) corresponded to the inverse complement of bp 10 to 38, downstream of the bft stop codon (5′-TTCCATTAATCGAACTTCGATTCTCACTC-3′). Complete agreement in detection of either BFT activity or bft sequence was found between the three assays. Overall, 34 B. fragilis strains (38%) identified by cell culture as producing BFT were also bft positive by colony blot hybridization and PCR. None of the 55 nontoxigenic B. fragilis (NTBF) strains, as determined by cell culture, were positive by colony blot or PCR. However, when primers 1 and 2 were used for PCR, 11 of the 34 ETBF strains identified by cell culture and colony blot produced a weak predicted product (ca. 1.2 kb) and also yielded a nonspecific 0.6-kb fragment (Fig. 1), suggesting that some differences might exist between the bft gene of these strains and the bft-2 sequence. The other 23 ETBF strains produced only the predicted 1.2-kb fragment. When primer 2 was replaced by primer 5, whose sequence was derived from the inverse complement of the last 19 nucleotides of bft (5′-TGGTCTCGAGATCGCCATCTGCTATTTCC-3′), all 34 ETBF strains yielded only the predicted 1.2-kb fragment (data not shown). ETBF strains were identified more often from blood (12 of 22 strains [54%]) than from the other extraintestinal sources (22 of 67 strains [33%]; P < 0.07 [by chi-square analysis]) (Table 1).
FIG. 1.
Representative results of PCR with primers 1 and 2 (sequences in text) for detection of ETBF from extraintestinal samples. All B. fragilis strains tested in this gel were identified as ETBF by cell culture assay and colony blot hybridization. Lanes: 1, DNA molecular weight marker (1-kb DNA ladder [Gibco BRL]); 2, ETBF 86-5443-2-2 (bft-2 positive control); 3, B. fragilis 419 (blood isolate); 4, B. fragilis 536 (pus isolate); 5, B. fragilis 723 (pleural fluid isolate); 6, B. fragilis 713 (right foot isolate); 7, B. fragilis 529 (blood isolate); 8, B. fragilis 586 (blood isolate); 9, B. fragilis 502 (blood isolate); 10, ETBF VPI 13784 (bft-1 positive control); 11, NTBF 077225-2 (negative control). The arrows show the predicted 1.2-kb product and the nonspecific 0.6-kb product found in strains 419 and 586.
TABLE 1.
Sources from which ETBF strains were isolated
| Source | No. of ETBF strains/ total no. of B. fragilis strainsa |
|---|---|
| Blood | 12/22 |
| Wound | 6/24 |
| Pus | 4/15 |
| Pleural fluid | 1/1 |
| Peritoneal fluid | 3/7 |
| Bile | 2/3 |
| Miscellaneous body sites | 6/17 |
| Total | 34/89 |
Strains were designated ETBF if the HT29/C1 cell assay and probe analysis for bft were positive.
Identification of the Korea-BFT subtype.
To determine the bft gene sequence of the ETBF strains that produced a weak predicted PCR product and an additional 0.6-kb band when primers 1 and 2 were used, the bft gene of one strain with this pattern (strain 419 [blood isolate]) was cloned and sequenced from a Lambda ZAP library by using the ZAP Express Vector Kit (Stratagene, La Jolla, Calif.). The B. fragilis 419 Lambda ZAP library was screened with a 519-bp PCR fragment yielded by primers BG1 (5′-ACGGTGTATGTGATTTGTCTGAG-3′) and BG2 (5′-CAACCGAGATTTTTAGCGATTA-3′). The sequences of the BG1 and BG2 primers were derived from bp 10 to 32 and from bp 506 to 527 of the published partial bft-1 sequence (12). Positive clones were isolated and excised as described by the manufacturer (Stratagene). The bft gene of one excised plasmid (pBFT-6) containing bft in a 4-kb BglII fragment (similar to bft-1 and bft-2 in strains VPI 13784 and 86-5443-2-2 [46]) was sequenced by the fluorescent dideoxy terminator method. The DNA sequence was analyzed with programs developed by the Genetics Computer Group of the University of Wisconsin (3) and the NCBI BLAST server (1). Like bft-1 and bft-2, the bft gene from strain 419 was predicted to encode a 397-residue holotoxin with a calculated molecular mass of ca. 44.5 kDa. However, alignment of the predicted amino acid sequence of the 419 bft gene with those of strains VPI 13784 (containing bft-1) and 86-5443-2-2 (containing bft-2) revealed that 419 BFT (termed Korea-BFT) has 93 and 96% identity with BFT-1 and BFT-2, respectively (Fig. 2). Alignment of Korea-BFT with the BFT-1 and BFT-2 proteins suggested that Korea-BFT is also synthesized as a preproprotein, where the initial 18 amino acids comprise a signal peptide followed by a 193-residue “pro” region and an active mature region of 186 residues containing the zinc-binding signature motif. Like BFT-1 and BFT-2 (6), the preproprotein domain of Korea-BFT was more conserved than the mature domain. Only 5 and 6 amino acid differences in the 211-amino-acid preproprotein region were detected between Korea-BFT and BFT-1 or BFT-2, respectively. In contrast, 21 and 11 amino acid changes were detected when the 186-amino-acid mature protein region of Korea-BFT was compared with those of BFT-1 and BFT-2, respectively. Sequence analysis indicated that Korea-BFT is more related to BFT-2 than to BFT-1; however, the amphipathic domain identified in the carboxy-terminal region of BFT-2 is not present in Korea-BFT (Fig. 2).
FIG. 2.
Alignment of BFT-1, Korea-BFT (BFT-K), and BFT-2 sequences from ETBF VPI 13784, 419, and 86-5443-2, respectively. The signal peptide sequences are underlined, the arrowhead shows the start of the mature protein, residues forming the zinc-binding signature motif are in boldface type and marked with asterisks, and the carboxy-terminal 20 residues which are predicted to form an amphipathic region in BFT-2 are enclosed in a box. Modeling of this region in Korea-BFT did not yield an amphipathic domain.
Frequency of Korea-BFT.
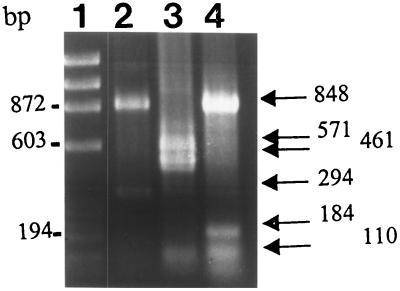
Our previous study showed that ETBF strains containing each subtype of BFT could be distinguished by hybridization with oligonucleotide probes specific for bft-1 or bft-2 (6). However, we found that the specific oligonucleotide probe used to detect ETBF strains containing bft-2 could not discriminate between this bft subtype and Korea-bft. To test the possibility that the restriction patterns of bft-1, bft-2, and Korea-bft might permit discrimination between the three subtypes, the nucleotide sequences of the three alleles were restriction mapped with a program developed by the Genetics Computer Group of the University of Wisconsin (3). We found that enzymatic digestion of the three bft subtypes with Sau3AI predicted different patterns. When primers 1 and 5 were used to generate the bft gene followed by Sau3AI, the fragments predicted by the program were 848 and 294 bp for bft-1; 571, 461, and 110 bp for bft-2; and 848, 184, and 110 bp for Korea-bft. These predicted fragments were obtained when the PCR-generated (primers 1 and 5) VPI 13784, 86-5443-2-2, and 419 bft genes were digested with the restriction enzyme Sau3AI (Fig. 3).
FIG. 3.
Agarose gel electrophoresis of fragments produced by Sau3AI digestion of the bft gene amplified by PCR with primers 1 and 5 (sequences in text). Lanes: 1, DNA molecular weight marker (φX174 RF DNA/HaeIII fragments [Gibco BRL]); 2, bft-1 from ETBF VPI 13784; 3, bft-2 from ETBF 86-5443-2-2; 4, Korea-bft from ETBF 419.
To determine the bft subtype of the 34 Korean ETBF strains identified by the HT29/C1 cell assay, colony blot, and PCR, the bft gene amplified by PCR with primers 1 and 5 was digested with the restriction enzyme Sau3AI. These data revealed that the 11 ETBF strains (32%) that yielded a weak PCR product when primers 1 and 2 were initially used exhibited the Korea-bft pattern after Sau3AI digestion of the PCR product of primers 1 and 5. Of the remaining 23 ETBF strains, 14 (41%), and 9 (22%) exhibited bft-1 and bft-2 patterns, respectively. These results were confirmed by colony blot hybridization with oligonucleotide probes specific for bft-1 (probe MTVPI, 5′-GGCGCTGAGCATACGGATAATT-3′) and bft-2/Korea-bft (probe MT086, 5′-GGTGCTAGGCATGCGGATGATC-3′). Probe MTVPI hybridized only with the 14 ETBF strains with the bft-1 pattern, and probe MT086 hybridized with the 20 ETBF strains with the bft-2 or Korea-bft pattern.
To further analyze the frequency of Korea-bft, we determined if this subtype of bft was present in bft-2-positive ETBF strains isolated in the United States. Seventeen ETBF strains containing bft-2 isolated from extraintestinal and intestinal samples in the United States (4b, 6) were characterized by enzymatic digestion with Sau3AI. We found that all 17 ETBF strains yielded the typical restriction pattern of bft-2 (data not shown).
Korea-BFT purification and biologic activity.
The Korea-BFT protein from strain 419 was purified as previously described to determine its biological activity and properties (26, 29). During purification steps with Q-Sepharose and phenyl-agarose chromatography, Korea-BFT did not differ significantly from BFT-1 and BFT-2 purified from strains VPI 13784 and 86-5443-2-2, respectively (28). In the final purification step with a high-resolution anion-exchange column (MonoQ), the Korea-BFT protein eluted at the same concentration of NaCl (ca. 0.18 M) as did BFT-2 purified from strain 86-5443-2-2 (28).
In Western blot analysis using anti-BFT-2 serum as the primary antibody, we found that purified Korea-BFT had the same electrophoretic mobility on SDS-PAGE as did BFT-2 purified from ETBF 86-5443-2-2 (data not shown); in contrast, BFT-1 purified from VPI 13784 had a different electrophoretic mobility (Fig. 4A).
FIG. 4.
(A) Electrophoretic mobility of purified BFT-1 from ETBF VPI 13784 (lane 1) and purified Korea-BFT from ETBF 419 (lane 2) on SDS-PAGE, analyzed by a Western blot probed with polyclonal anti-BFT-2 serum. (B) Cleavage of E-cadherin by BFT. HT29/C1 cells were treated with BFT (100 ng/ml) for 3 h, lysed in 1× SDS gel loading buffer, and examined by Western blotting with E2 antibodies against the cytoplasmic domain of E-cadherin. Lane 1, untreated HT29/C1 cells; lane 2, HT29/C1 cells treated with BFT-2 purified from ETBF 86-5443-2; lane 3, HT29/C1 cells treated with Korea-BFT purified from ETBF 419. Loss of staining intensity of intact E-cadherin (120 kDa) is observed in HT29/C1 cells treated with BFT-2 and Korea-BFT.
Purified Korea-BFT was active on HT29/C1 cells assay at a concentration similar to that observed with purified BFT-1 or BFT-2 (reference 28 and data not shown). Cleavage of E-cadherin on HT29/C1 cells by purified Korea-BFT (100 to 200 ng/ml) was determined by Western blotting with antibodies to the cytoplasmic domain of E-cadherin, as described previously (29). Like purified BFT-1 and BFT-2, Korea-BFT cleaved HT29/C1 E-cadherin after 3 h of incubation (Fig. 4B and data not shown).
This report extends our understanding of the molecular epidemiology of ETBF strains and indicates that the toxins secreted by these strains are more complex than previously identified. Intriguingly, Kato (9) recently reported data identifying a unique isoform of bft in 14% of intestinal B. fragilis strains isolated from children and adults in Japan. Subsequent data indicate that this bft isoform identified in Japan and the Korea-bft isoform reported herein are identical (9a). The absence of this newly identified bft isoform in ETBF strains isolated in the United States raises interesting questions regarding the molecular evolution of ETBF strains and indicates that further studies to clarify the global distribution of ETBF strains and their toxin subtypes are warranted. In addition, our data confirm prior reports of Kato et al. (7, 8) in which a high percentage of extraintestinal B. fragilis strains, particularly bloodstream isolates (ca. 30%), in Japan were identified as ETBF strains. In contrast, ETBF strains isolated from extraintestinal sites were rare (6%) in a collection of strains isolated at the Johns Hopkins Hospital (14). One hypothesis to potentially explain this difference is that the Asian ETBF strains are more virulent, by as-yet-unidentified mechanisms, than are ETBF strains examined to date in the United States. Alternatively, the prevalence of intestinal ETBF strains may be higher in Japan and Korea due to either gut ecologic factors (e.g., dietary influence on intestinal flora) or earlier evolution of ETBF strains in these countries. These data, combined with the association of ETBF with diarrheal illnesses in children greater than 1 year old and, more recently, in adults (30), indicate that further studies of the epidemiology and clinical impact of ETBF are necessary.
Nucleotide sequence accession number.
The Korea-bft gene sequence has been submitted to the GenBank database and assigned accession no. AF081785.
Acknowledgments
We thank Dwight Derr for tissue culture assistance, and we acknowledge the gifts of B. fragilis strains by Tracy Wilkins of the Virginia Polytechnic Institute (strain VPI 13784) and Lyle Myers (strain 86-5443-2-2) and the anti-E-cadherin E2 antibody by James Nelson, Stanford University.
This work was supported by National Research Award AI09863-01 (A.A.F.) and National Institutes of Health Award DK45496 (C.L.S.).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers F G, Koshy S S, Saidi R F, Clark D P, Moore R D, Sears C L. Bacteroides fragilis toxin exhibits polar activity on monolayers of human intestinal epithelial cells (T84 cells) in vitro. Infect Immun. 1997;65:3561–3570. doi: 10.1128/iai.65.9.3561-3570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donelli G, Fabbri A, Fiorentini C. Bacteroides fragilis enterotoxin induces cytoskeletal changes and surface blebbing in HT-29 cells. Infect Immun. 1996;64:113–119. doi: 10.1128/iai.64.1.113-119.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Franco, A. A., S. Wu, and C. L. Sears. Unpublished data.
- 4b.Franco, A. A., and C. L. Sears. Unpublished data.
- 5.Franco A A, Kaper J B, Sears C L. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Enterotoxigenic Bacteroides fragilis (ETBF) strains contain a potential pathogenicity island, abstr. B-426; p. 102. [Google Scholar]
- 6.Franco A A, Mundy L M, Trucksis M, Wu S, Kaper J B, Sears C L. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect Immun. 1997;65:1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato N, Karuniawati A, Jotwani R, Kato H, Watanabe K, Ueno K. Isolation of enterotoxigenic Bacteroides fragilis from extraintestinal sites by cell culture assay. Clin Infect Dis. 1995;20(Suppl. 2):S141. doi: 10.1093/clinids/20.supplement_2.s141. [DOI] [PubMed] [Google Scholar]
- 8.Kato N, Kato H, Watanabe K, Ueno K. Association of enterotoxigenic Bacteroides fragilis with bacteremia. Clin Infect Dis. 1996;23:S83–S86. doi: 10.1093/clinids/23.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 9.Kato N. Abstracts of the 2nd World Congress on Anaerobic Bacteria and Infections. 1998. Prevalence of enterotoxigenic Bacteroides fragilis in children, adults and elderly people in Japan, abstr. 5.002. Nice, France. [Google Scholar]
- 9a.Kato, N. Personal communication.
- 10.Kling J J, Wright R L, Moncrief J S, Wilkins T D. Cloning and characterization of the gene for the metalloprotease enterotoxin of Bacteroides fragilis. FEMS Microbiol Lett. 1997;146:279–284. doi: 10.1016/s0378-1097(96)00488-0. [DOI] [PubMed] [Google Scholar]
- 11.Koshy S S, Montrose M H, Sears C L. Human intestinal epithelial cells swell and demonstrate actin rearrangement in response to the metalloprotease toxin of Bacteroides fragilis. Infect Immun. 1996;64:5022–5028. doi: 10.1128/iai.64.12.5022-5028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moncrief J S, Obiso R, Jr, Barroso L A, Kling J J, Wright R L, Van Tassell R L, Lyerly D M, Wilkins T D. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect Immun. 1995;63:175–181. doi: 10.1128/iai.63.1.175-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moncrief J S, Duncan A J, Wright R L, Barroso L A, Wilkins T D. Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect Immun. 1998;66:1735–1739. doi: 10.1128/iai.66.4.1735-1739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundy L M, Sears C L. Detection of toxin production by Bacteroides fragilis: assay development and screening of extraintestinal clinical isolates. Clin Infect Dis. 1996;23:269–276. doi: 10.1093/clinids/23.2.269. [DOI] [PubMed] [Google Scholar]
- 15.Myers L L, Firehammer B D, Shoop D S, Border M M. Bacteroides fragilis: a possible cause of acute diarrheal disease in newborn lambs. Infect Immun. 1984;44:241–244. doi: 10.1128/iai.44.2.241-244.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers L L, Shoop D S, Stackhouse L L, Newman F S, Flaherty R J, Letson G W, Sack R B. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J Clin Microbiol. 1987;25:2330–2333. doi: 10.1128/jcm.25.12.2330-2333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obiso R J, Jr, Azghani A O, Wilkins T D. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect Immun. 1997;65:1431–1439. doi: 10.1128/iai.65.4.1431-1439.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pantosti A, Menozzi M G, Frate A, Sanfilippo L, D’Ambrosio F, Malpeli M. Detection of enterotoxigenic Bacteroides fragilis and its toxin in stool samples from adults and children in Italy. Clin Infect Dis. 1997;24:191–196. doi: 10.1093/clinids/24.1.12. [DOI] [PubMed] [Google Scholar]
- 19.Sack R B, Myers L L, Almeido-Hill J, Shoop D S, Bradbury W C, Reid R, Santosham Enterotoxigenic Bacteroides fragilis: epidemiologic studies of its role as a human diarrhoeal pathogen. J Diarrhoeal Dis Res. 1992;10:4–9. [PubMed] [Google Scholar]
- 20.Sack R B, Albert M J, Alam K, Neogi P K, Akbar M S. Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. J Clin Microbiol. 1994;32:960–963. doi: 10.1128/jcm.32.4.960-963.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saidi R F, Sears C L. Bacteroides fragilis toxin rapidly intoxicates human intestinal epithelial cells (HT29/C1) in vitro. Infect Immun. 1996;64:5029–5034. doi: 10.1128/iai.64.12.5029-5034.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saidi R F, Jaeger K, Montrose M H, Wu S, Sears C L. Bacteroides fragilis toxin rearranges the actin cytoskeleton of HT29/C1 cells without direct proteolysis of actin or decrease in F-actin content. Cell Motil Cytoskelet. 1997;37:159–165. doi: 10.1002/(SICI)1097-0169(1997)37:2<159::AID-CM8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.San Joaquin V H, Griffis J C, Lee C, Sears C L. Association of Bacteroides fragilis with childhood diarrhea. Scand J Infect Dis. 1995;27:211–215. doi: 10.3109/00365549509019011. [DOI] [PubMed] [Google Scholar]
- 24.Sears C L, Myers L L, Lazenby A, Van Tassell R L. Enterotoxigenic Bacteroides fragilis. Clin Infect Dis. 1995;20(Suppl. 2):S142–S148. doi: 10.1093/clinids/20.supplement_2.s142. [DOI] [PubMed] [Google Scholar]
- 25.Sears C L. Bacteroides fragilis toxins. In: Alouf J E, Freer J H, editors. Bacterial protein toxins: a comprehensive sourcebook, in press. London, England: Academic Press; 1999. [Google Scholar]
- 26.Van Tassell R L, Lyerly D M, Wilkins T D. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect Immun. 1992;60:1343–1350. doi: 10.1128/iai.60.4.1343-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weikel C S, Grieco F D, Reuben J, Myers L L, Sack R B. Human colonic epithelial cells, HT29/C1, treated with crude Bacteroides fragilis enterotoxin dramatically alter their morphology. Infect Immun. 1992;60:321–327. doi: 10.1128/iai.60.2.321-327.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Dreyfus L A, Tzianabos A O, Franco A A, Hayashi C, Sears C L. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Diversity of the metalloprotease toxin produced by enterotoxigenic Bacteroides fragilis, abstr. B-270; p. 101. [Google Scholar]
- 29.Wu S, Lim K-C, Huang J, Saidi R F, Sears C L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci USA. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G B, Weintraub A. Abstracts of the 2nd World Congress on Anaerobic Bacteria and Infections. 1998. Detection of enterotoxigenic Bacteroides fragilis (ETBF) in clinical samples from an adult population in Sweden, abstr. 3.018. Nice, France. [Google Scholar]