Abstract
DNA origami technology has rapidly developed into an ideal means to programmably crystallize nanoparticles. However, most existing DNA origami three-dimensional platforms normally used a single type of DNA origami unit, which greatly limits the types of nanoparticle superlattices that can be synthesized. Here, we report a universal strategy to vastly enrich the library of nanoparticle superlattices, based on multiple-unit (≥4 units) DNA origami platforms, which were constructed by programmably cocrystallizing three different DNA origami octahedral “homologs.” Through selectively inserting nanoparticles into DNA origami monomers, numerous nanoparticle superlattices can be synthesized on the basis of the same platform. In this work, we obtained 85 types of DOF/AuNP (DNA origami frame/gold nanoparticle) superlattices using three different DNA origami platforms as examples. We believe that our strategy can provide possible access to fabricate virtually endless types of nanoparticle superlattices and promote the construction of functional materials with special properties.
A general way is provided to enrich the nanoparticle lattices by cocrystallizing three different types of DNA origami frames.
INTRODUCTION
In nanomaterial science, because of the collective effect induced by nanoparticles within three-dimensional (3D) architectures, a diverse range of attractive properties can be generated, which can be applied to a series of regions involving optics, catalysis, electricity, and so on (1–7). However, because of the forces provided by thermodynamics and entropy, extant methods adopted for crystallizing nanoparticles are mainly creating closely packed structures with high symmetry (8–10). Therefore, nanoparticle superlattices currently available are mostly restricted to simple cubic, body-centered cubic, face-centered cubic, hexagonal close-packed, and some other specific crystal structures, regardless of how to tune the size, shape, component, ligand type of the engaged nanoparticles, and corresponding solution environments (11–19). DNA origami frames (DOFs) (20–26), which have an excellent capacity to accurately manipulate nanoparticles in a 3D manner, offer other possibilities to assemble or crystallize nanoparticles indirectly (27–30). Using this approach, nanoparticles, whether organic or inorganic, do not need to have the ability to be crystallized by themselves in solutions (31–36). By inserting nanoparticles into DOFs, the driving force to realize nanoparticle crystallization is transferred from the encaged nano-objects to the external DOFs. In these cases, DOFs serve as the skeleton of the crystals, while the nanoparticles are always “encapsulated” inside the DOFs. Hence, the binding modes induced by the external DNA origami monomers will mainly determine the packing behavior of nanoparticles caged. Therefore, this unique assembly concept can maximize the designability, programmability, and addressability of DOFs, providing a universal method to crystallize nanoparticles into diversified superlattices by varying the shapes, binding modes, or the number of monomer types of the DOF building blocks and also offering a general way to enrich the nanoparticle superlattice library. Nevertheless, in recent reports, researchers have still been too cautious to rationally assemble the nanoparticles when using DOFs, which has enormously restricted the diversity of nanoparticle superlattices that can be produced.
To this end, a general strategy has been developed to simultaneously cocrystallize multitype DOFs in a programmable manner to further synthesize more types of nanoparticle superlattices based on selectively binding nanoparticles to DOFs. In this work, a series of DOFs with similar octahedral shapes were synthesized. Each type of DOF can be encoded with specified sticky ends (SEs) at vertices for further assembly via vertex-to-vertex hybridization. In addition, a series of DOFs functionalized with SEs are termed “polychrome DNA origami homologs.” Similar to playing with toy bricks, diverse 3D DNA platforms could be programmably built by selectively assembling the DNA origami octahedral “homologs” in designed ways. Meanwhile, on the basis of the platforms constructed, we can artificially anchor nano-objects inside the homologs to fabricate polychrome superlattices. In this work, we deliberately built three unique 3D DNA platforms as examples by assembling DNA origami octahedral homologs as designed. By selectively inserting gold nanoparticles (AuNPs) into the DNA origami octahedral homologs, 85 types of DOF/AuNP superlattices with highly ordered arrangements have been obtained. Therefore, this universal approach can remarkably enrich the library of nanoparticle superlattices guided by DNA and effectively fill the gaps of current crystal structures by engaging more DNA origami octahedral homologs.
RESULTS
Polychrome DNA origami octahedral homologs
In this work, three different octahedral DOFs were used to programmably construct 3D DNA platforms, which consisted of regular octahedron (R_oct), elongated octahedron (E_oct), and partially elongated octahedron (P_oct) (Fig. 1A). R_oct DOF comprises 12 identical bundles with a length of ~28.56 nm. For E_oct DOF, the eight edges outside the middle square plane are altered to be longer (~35.70 nm). In the case of P_oct DOF, only the four bundles below the middle plane are elongated to ~35.70 nm, while the other eight bundles are invariant (~28.56 nm; see design details in fig. S1).
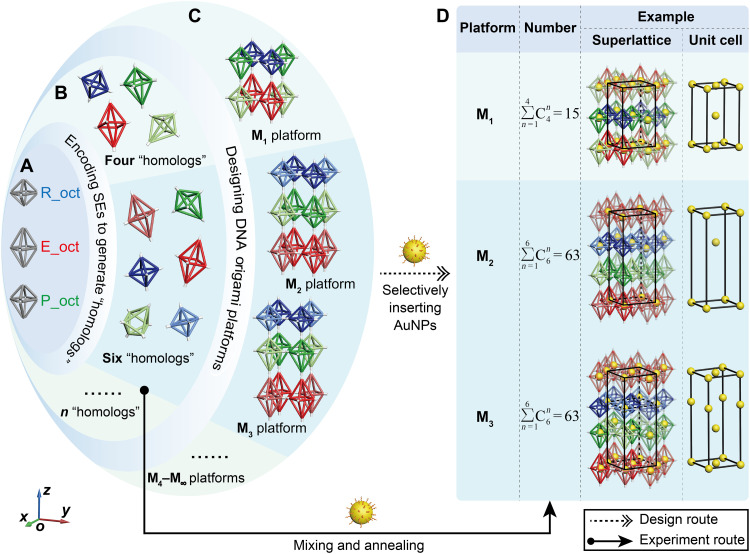
Fig. 1. Schematic illustration showing the process to assemble the DOF platforms and nanoparticle superlattices, including the design route and the experiment route.
Design route: A-B-C-D. (A) Models of the octahedral DOFs including the R_oct, E_oct, and P_oct. (B) Encoding SEs to vertices of DOFs to generate various DNA origami octahedral homologs. (C) Examples of 3D DNA platforms that can be constructed, including M1 (composed of four homologs) and M2 and M3 (both consisting of six homologs) designed from DNA origami octahedral homologs as shown in (B). We emphasize here that more platforms can be achieved using more DNA origami octahedral homologs. (D) By selectively binding AuNPs to DNA origami octahedral homologs, various nanoparticle superlattices can be fabricated. The total numbers of superlattices that could be created by M1, M2, and M3 platforms are 15, 63, and 63, respectively. The right part shows three representative nanoparticle superlattices designed based on the M1, M2, and M3 platforms, respectively, with the corresponding unit cells of AuNPs shown beside it. Experiment route: B-D. The actual process to construct the superlattices in the experiments are shown, where the superlattices can be constructed by simply mixing corresponding DNA origami octahedral homologs with AuNPs directly and then causing them to undergo a specific annealing procedure.
Each vertex of the octahedral DOFs is artificially encoded with four 30–nucleotide (nt) SEs terminating in a devisable region for further assembly (Fig. 1B). For clarity, the SE model at each vertex is simplified as a colored cone or cylinder in the illustrations, where the cone and cylinder of the same color can connect with each other (fig. S2). Borrowing the concept of homologs from organic chemistry, which generally refers to the organic compounds with similar structure yet different molecular composition differing by several “CH2” atomic groups, DNA origami units with SEs designed in this work are termed polychrome DNA origami homologs, as these units are all octahedral shapes with different symmetries by altering the length of DNA bundles. It is worth emphasizing that DOFs with identical shapes but encoded with different SEs are also treated as homologs in our systems, which should be estimated from both the shapes and SEs installed. This step encodes the DOF monomers with functions to assemble with each other forming 3D DNA platforms composed of different DNA origami octahedral homologs (Fig. 1B). Each type of DOF functionalized with SEs was correctly synthesized with a high yield, as demonstrated by the transmission electron microscopy (TEM) (fig. S3). Theoretically speaking, a series of unique DOF platforms can be designed by flexibly taking advantage of the structural complementarity that will occur between the three types of DOF monomers and the specificity of the SEs installed at the vertices (fig. S4). Here, platform M1 consisting of four DNA origami octahedral homologs and platforms M2 and M3 both composed of six DNA origami octahedral homologs were selected as examples to prove this general approach (Fig. 1C). Platform M1 is composed of one type of R_oct and E_oct homologs and two types of P_oct homologs (same shape but with different SEs), while both platforms M2 and M3 require one more type of R_oct and E_oct homologs. Hence, diverse nanoparticle superlattices can be achieved by selectively inserting nanoparticles into DNA origami octahedral homologs. In this work, we adopted the manner of positioning AuNPs in the middle square plane of the DNA origami octahedral homologs to artificially create the connections in between. The number of nanoparticle superlattices that can be constructed is found to be equal to the number of ways to insert the AuNPs. According to the principle of combination number, and , more than 100 nanoparticle superlattices can be obtained from platforms M1 (15), M2 (63), and M3 (63) (Fig. 1D). Representative nanoparticle superlattices based on each DNA platform are shown from the top to bottom with the corresponding unit cells of AuNPs shown in the right part of Fig. 1D. Notably, experimentally, DNA origami octahedral homologs with the different preexperimental designs were directly mixed with the AuNPs, and then the mixture underwent a careful annealing process to fabricate the desired 3D superlattices (solid arrow in Fig. 1), which was much simpler than the design path described above (dotted arrow in Fig. 1). This proactive approach to design experiments underscores the advantages of DNA nanotechnology.
Crystallizing AuNPs via distinct 3D DNA platforms
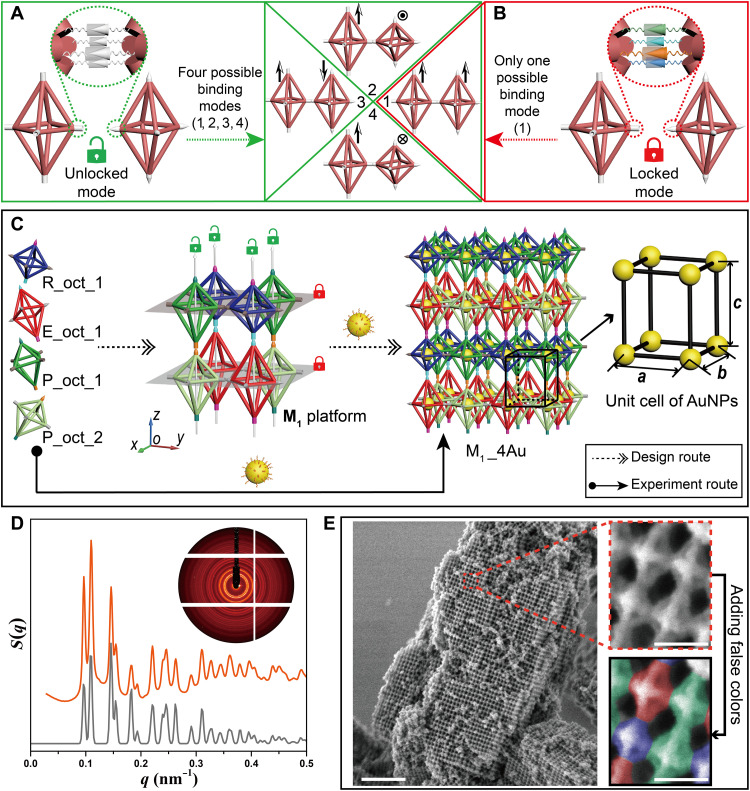
Compared with the conventional crystallization strategy guided by DNA, more precise spatial orientation control over monomers is highly in demand to construct platforms composed of three different anisotropic DOFs. Therefore, the SEs projected from the vertices should be designed with caution, which can both provide the original driving force and impose restrictions on relative positions between the DNA origami octahedral homologs during the assembly process. As shown in figures, the SEs at the vertices are simplified as models of cones and cylinders for clarity and can bind with each other when having the same color. According to the structures of the DOF platforms designed in our case, we adopt two different modes in SE design, “unlocked mode” (green opened lock) and “locked mode” (red closed lock). The binding modes between the two E_oct DOFs are taken as examples. In unlocked mode, the SEs installed at the same vertex are designed as identical, and four possible binding modes, numbered from 1 to 4, can occur when the E_oct DOFs bind with each other (Fig. 2A, drawn with the green boxes); while in locked mode, the SEs in the vertices are different from each other, in which case, only one possible binding mode can be generated (number 1; as shown in Fig. 2B, with red boxes). In the figure, the arrows are shown beside the binding modes for better differentiation of the relative positions between the E_oct DOFs. Therefore, if the four binding modes produced can be considered equivalent during the assembly, then unlocked mode would be preferred, which can simplify the types of SEs. However, if the binding modes between DOFs are strictly restricted, then the locked mode would be required.
Fig. 2. Using unlocked mode and locked mode in SE design to assemble the M1 platform and nanoparticle superlattice M1_4Au.
(A and B) Connections between the E_oct homologs are taken as an example to explain the two modes in SE design, in which SEs are indicated as colorful cones and cylinders that can bind with each other when having the same color. Unlocked mode (green opened lock): SEs (white) installed at the vertex are the same, resulting in four possible binding modes numbered from 1 to 4 (A). Locked mode (red closed lock): SEs (colorful) stretched out from the vertex are different from each other, which can cause only one possible binding mode (numbered 1) (B). Arrows are shown beside the binding modes for better differentiation of the relative positions between E_oct DOFs. (C) The process to construct the nanoparticle superlattice M1_4Au, including the design route (dotted arrows) and experiment route (solid arrow). R_oct_1, E_oct_1, P_oct_1, and P_oct_2 homologs encoded with specific SEs can assemble to form the M1 platform, in which the unlocked mode and locked mode are applied along the z axis and in the xoy plane, respectively. Nanoparticle superlattice M1_4Au can be fabricated by binding the AuNPs to all DNA origami octahedral homologs, and the corresponding unit cell of the AuNPs is shown beside it. (D) 2D small-angle x-ray scattering (SAXS) pattern (inset) and 1D experimental SAXS curve (orange) of the nanoparticle superlattice M1_4Au with corresponding fitting curve shown below (gray). (E) Left: Lower-magnification scanning electron microscopy (SEM) image of lattice M1_4Au after it was coated with a thin layer of silica. Scale bar, 0.5 μm. Right: Close-up view of the area framed in the red dotted box (top right), where the R_oct, E_oct, and P_oct homologs are falsely colored in blue, red, and green, respectively, for clear observation (bottom right). Scale bars, 50 nm.
The M1 platform is constructed by alternately stacking two types of composite DOF layers, which are composed of R_oct_1, P_oct_1, and E_oct_1, P_oct_2 (Fig. 2C, left). By applying the design principle as discussed above, the unlocked mode and locked mode of the SEs are, respectively, assigned to the vertical and horizontal directions in the M1 platform. Hence, four types of SE pairs (cyan, orange, magenta, and teal) with unlocked modes are needed to realize the correct assembly between P_oct_1 and P_oct_2 and between R_oct_1 and E_oct_1 along the z axis, which can guarantee that the correct connections occurred between the specific vertices. By contrast, the locked mode of the SEs is applied at each vertex in the xoy plane of the two types of composite DOF layers to prevent unnecessary binding modes. It is worth emphasizing that each vertex in the horizontal plane of the same DNA origami octahedral homologs is equivalent during assembly, which means that the vertices should be allowed to connect with the adjacent four vertices arbitrarily. Therefore, the SEs between the vertices in the xoy plane of the same DNA origami octahedral homologs should be identical, including the 8-nt complementary part and spatial placement. In general, 12 distinct SE pairs are needed in the M1 platform, including 4 pairs in the unlocked mode and the other 8 pairs used in the locked mode (see the detailed sequence in fig. S5). On the basis of the M1 platform, nanoparticle superlattice M1_4Au can be fabricated by binding AuNPs to all DNA origami octahedral homologs, and the corresponding unit cell of AuNPs is supposed to be a simple tetragonal structure (Fig. 2C, right). Here, the “M1” designation signifies that the nanoparticle superlattice is fabricated on the basis of the platform M1 that consists of four DNA origami octahedral homologs, and the “4Au” designation represents all of four DNA origami octahedral homologs that are populated by AuNPs. Please note that, because the platform M1 only consists of four different DNA origami octahedral homologs, M1_4Au refers to the system that all the four DNA origami octahedral homologs are containing particles. Moreover, because 10-nm AuNP is the only kind of the guest NPs used in this work, 4Au also refers to four same AuNPs. Experimentally, four types of DNA origami octahedral homologs encoded with different SEs were equally mixed with the AuNPs, which then underwent a slow annealing process to form the desired 3D superlattices (M1_4Au). Small-angle x-ray scattering (SAXS) and scanning electron microscopy (SEM) were used to characterize the structure details. As shown in Fig. 2D, the integrated experimental SAXS curve (orange) exhibited approximately 20 sharp scattering peaks, revealing the formation of a highly ordered crystalline material. The excellent agreement between the theoretical simulation (gray) and experimental curve further demonstrated that the fabricated nanoparticle superlattice was corresponding to the prescribed simple tetragonal structure with a = b = 57.50 nm and c = 65.00 nm. In addition, SEM was used to directly observe the arrangement of DNA origami octahedral homologs on the crystal surfaces after covering the DNA bundles of the 3D DNA platforms with a thin layer of silica shell, which can preserve the structures under the exposure of the electron beam (37–39). Under low magnification mode (Fig. 2E, left, and fig. S6), the superlattices can be easily found with an average size of ~1 to 2 μm. Further magnification of the region framed in the red dotted box showed that the DOF monomers were arranged as expected with the homologs of R_oct, P_oct, E_oct, and P_oct alternately packed (Fig. 2E, top right). Moreover, different types of DOFs were distinguished with distinct but obvious false colors for clear observation (Fig. 2E, bottom right). We should emphasize here that, the locked mode in the xoy plane is essential for the cocrystallization of the DNA origami octahedral homologs with desired arrangement. Unlocked or partial locked mode may result in incorrect connections in some regions (figs. S7 to S17).
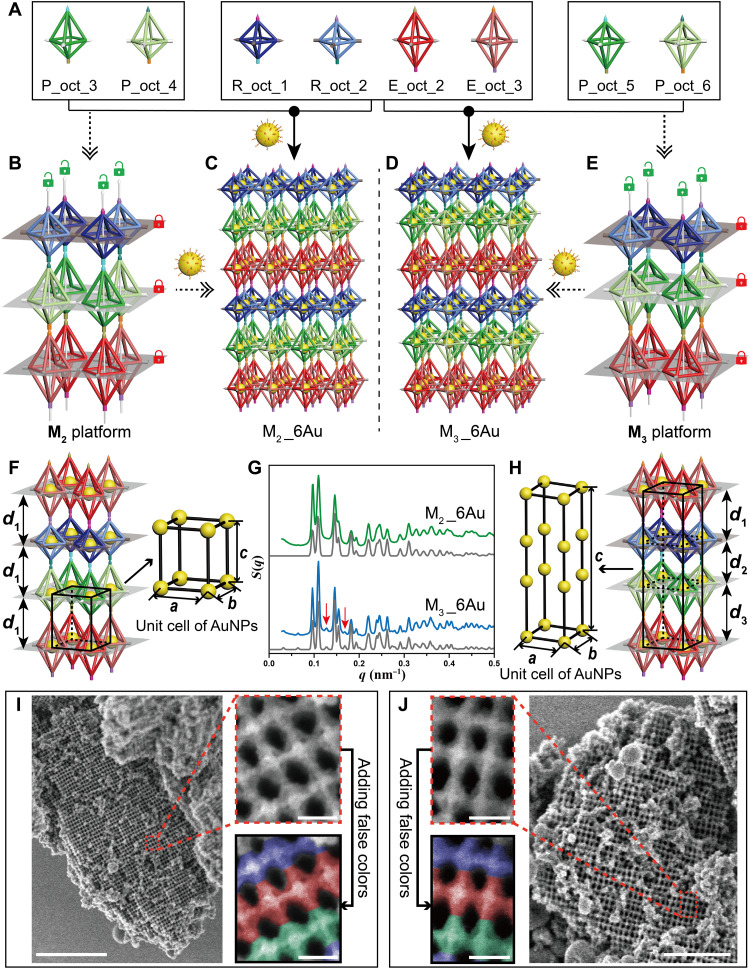
Other than the four-unit system, platforms composed of more DNA origami octahedral homologs can also be designed. As previously shown in Fig. 1, the M2 and M3 platforms are both designed to be constituted by six individual DNA origami octahedral homologs (R_oct_1, R_oct_2, E_oct_2, E_oct_3, P_oct_3, and P_oct_4 for the M2 platform; R_oct_1, R_oct_2, E_oct_2, E_oct_3, P_oct_5, and P_oct_6 for the M3 platform; see Fig. 3A for details). However, although the M2 and M3 platforms are both constructed by two R_oct, two E_oct, and two P_oct homologs and adopt the packing mode of the three different octahedral layers alternately arranged, we still obtain two completely different platforms by vertically rotating the P_oct layer 180°, as shown in Fig. 3 (B and E). The vertical flip of the P_oct layer is implemented by exchanging the SEs installed in the vertices along the z axis during the design process, which further changes the connecting logic between these octahedral layers. Similar to the SEs designed in the M1 platform, unlocked mode and locked mode are applied in the vertical and horizontal directions of the M2 and M3 platforms, respectively, and the pairs of complementary SE strands required to fabricate these structures are increased to 18 different types due to more assembly units engaged (see detailed sequences in fig. S18). Similar to the nanoparticle superlattice M1_4Au, M2_6Au and M3_6Au (with the same name definition as “M1_4Au” described above) can also be synthesized by binding the AuNPs to all DNA origami octahedral homologs; thus, constructing the M2 and M3 platforms (Fig. 3, C and D). Different from M1_4Au, because the M2 or M3 platform is composed of six different kinds of DNA origami octahedral homologs, M2_6Au refers to the system that all of the six DNA origami octahedral homologs are containing AuNPs. Because the opposite direction of the P_oct layers in these two platforms, the distances of the AuNPs between the adjacent octahedral layers of the M2 platform are equal to each other (d1; Fig. 3F), while for the M3 platform, the interlamellar distances of AuNPs are different (d1 ≠ d2 ≠ d3; Fig. 3H), which results in two different types of nanoparticle superlattices. For M2_6Au, the unit cell of the AuNPs is expected to be simple tetragonal (a = b ≠ c, c = d1; Fig. 3F), while for lattice M3_6Au, a more complex tetragonal unit cell of AuNPs could be obtained, as shown in Fig. 3H, where c = d1 + d2 + d3.
Fig. 3. Assembly of nanoparticle superlattices M2_6Au and M3_6Au on the basis of M2 and M3 platforms, respectively.
(A to E) The process to construct nanoparticle superlattices M2_6Au and M3_6Au, including the design route (dotted arrows) and the experiment route (solid arrows). DNA origami octahedral homologs R_oct_1, R_oct_2, E_oct_2, E_oct_3, P_oct_3, P_oct_4; and R_oct_1, R_oct_2, E_oct_2, E_oct_3, P_oct_5, P_oct_6 (A) can be assembled to form the M2 platform (B) and M3 platform (E), respectively, where unlocked mode (green opened lock) and locked mode (red closed lock) are applied along the z axis and in the xoy plane, respectively. Nanoparticle superlattices M2_6Au (C) and M3_6Au (D) can be synthesized on the basis of the M2 and M3 platforms, respectively, by inserting the AuNPs into all DNA origami octahedral homologs. (F) For M2_6Au, the interlayer spacing of the AuNPs is equal (d1); thus, the unit cell of the AuNPs is a simple tetragonal structure. (G) Experimental SAXS data of lattices M2_6Au (green) and M3_6Au (blue) with corresponding theoretical simulations shown below (gray). The obvious two differences in the peaks between the two nanoparticle superlattices are indicated by red arrows. (H) For M3_6Au, the interlayer spacing is different (d1, d2, and d3), and the unit cell of AuNPs is a more complex tetragonal structure. (I and J) Representative SEM images for M2_6Au (I) and M3_6Au (J) after coated with a thin layer of silica. Lower-magnification images (I, left and J, right). Higher-magnification images of the regions framed in the red dotted boxes (I, top right and J, top left). The R_oct, E_oct, and P_oct homologs are falsely colored in blue, red, and green, respectively, for easier identification (I, bottom right and J, bottom left). Scale bars, 1 μm (I, left), 0.5 μm (J, right) and 50 nm (I, right and J, left).
The samples were then synthesized as prescribed above, followed by a careful annealing protocol (see Materials and Methods for details). SAXS and SEM were used to measure the inner structure and surface details of the fabricated lattices. The characterization data were compared in parallel to make the differences between these two types of superlattices prominent. The 1D structure factor curves extracted from the SAXS data for M2_6Au (green curve) and M3_6Au (blue curve) both revealed the high quality of the crystals, with more than 15 sharp scattering peaks, and both show an excellent agreement with the corresponding theoretical fitting curves (gray curves), as shown in Fig. 3G. In addition, the essential distinction of the inner arrangement of the AuNPs between these two types of nanoparticle superlattices radically led to a difference in the number of scattering peaks. Compared with M2_6Au, M3_6Au had more scattering peaks, with some obvious ones indicated by the red arrows. Through calculation, the practical parameter c of the unit cell of the AuNPs was determined to be 65.00 nm for M2_6Au, while this parameter was 195.20 nm for M3_6Au. Parameters a and b were 57.60 nm for both lattices. To further observe the arrangement details of the lattices by SEM, the nanoparticle superlattices were also covered by a thin layer of silica on the DNA origami bundles, using a similar protocol as described above. Figure 3 (I and J) shows the highly ordered arrangement of the DNA origami octahedral homologs on the surfaces of the crystals for M2_6Au and M3_6Au, respectively, including both lower- and higher-magnification images (additional SEM and TEM images are shown in figs. S19 and S20). The configuration of octahedral homologs could be found as expected by magnifying the red dotted boxes (Fig. 3I, top right, and Fig. 3J, top left). For more convenient comparison, R_oct, E_oct, and P_oct were drawn in false colors with blue, red, and green, respectively, and the spatial orientation of P_oct in these two lattices could be easily found with contrary arrangements, which was consistent with the prescribed design.
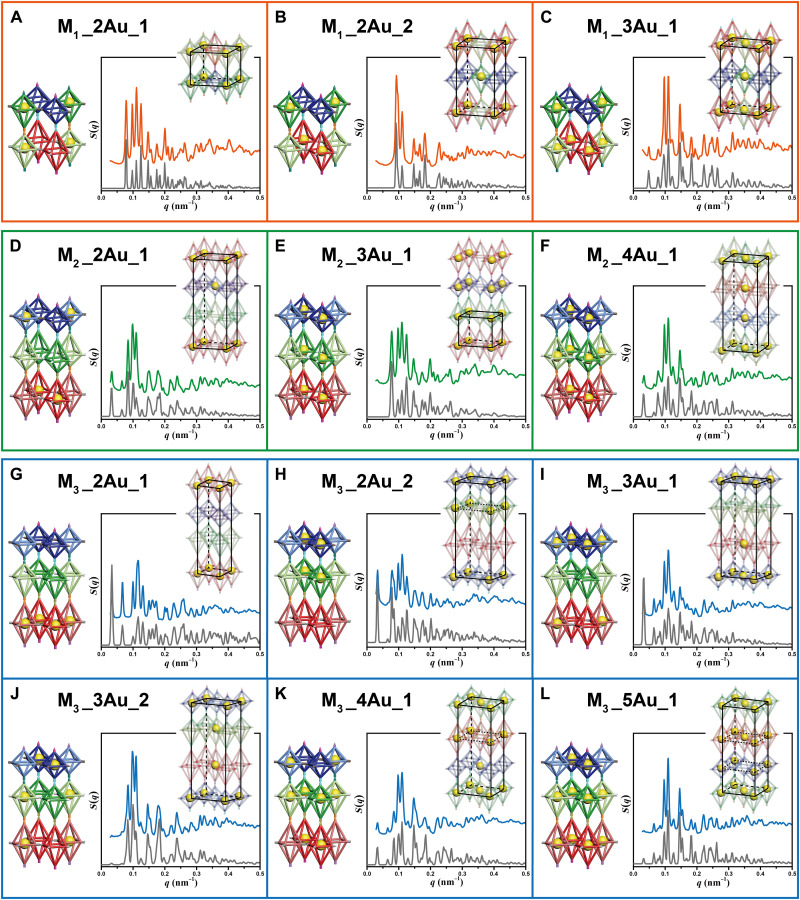
Compared with single-unit systems, the most significant advantage of the multiple-unit systems is that more building blocks within the platforms, coupled with the design principle of being able to selectively insert AuNPs into DNA origami octahedral homologs, could provide more packing possibilities for the nanoparticles, offering general access to fabricate diverse types of nanoparticle superlattices. In this work, AuNPs can be selectively inserted into every position of DNA origami octahedral homologs because of the strict binding rules we designed. We should stress here that every position means the positions of the different kinds of DNA origami octahedral homologs. For example, in the case of M1 platform, AuNPs can be arbitrarily inserted into any position of the four kinds of DNA octahedral monomers, while in the case of M2/M3 platform, AuNPs can be arbitrarily inserted into every position of the six kinds of DNA octahedral monomers. The superlattices fabricated in our cases could be viewed from two perspectives (fig. S21). Hence, two sets of criteria should be used to reasonably distinguish whether the nanoparticle superlattices fabricated by our approach belong to the same class. For criterion 1, we consider both the AuNP and the external DOF platforms, while for criterion 2, the arrangement of AuNPs is the only standard, as we could not gather the signal of DNA by SAXS measurements in the meanwhile. For ease of distinguishing, we call them “DOF/AuNP superlattices” when discussing the types of lattices under criterion 1. As previously mentioned in Fig. 1D, 15, 63, and 63 kinds of DOF/AuNP superlattices could be fabricated on the basis of platforms M1 (four homologs), M2 (six homologs), and M3 (six homologs), respectively. Among them, because of the equivalence of relationship between some homologs (generally with identical DNA shapes; as shown in fig. S22), 15, 35, and 35 typical types were well designed, prepared, and demonstrated by SAXS, respectively (for a total of 85 types), as shown in figs. S23 to S44. Tables S3 to S9 summarized the parameters of the unit cells and the coordination of AuNPs within these crystals. Among these, 12 representative superlattices were selected, as shown in Fig. 4, to demonstrate the diversity of the synthesized lattices with various arrangements of nanoparticles, including three types of M1 platform–based lattices (framed in orange), three types of M2 platform–based lattices (framed in green), and six types of M3 platform–based lattices (framed in blue). Through inserting AuNPs into different types and different numbers of DNA origami octahedral homologs, lattices with different types of AuNP unit cells could be fabricated. Specifically, different types of nanoparticle superlattices can be synthesized by inserting different numbers of AuNPs inside the same platform, such as superlattices M1_2Au_1 and M1_3Au_1 (Fig. 4, A and C) or M2_2Au_1, M2_3Au_1, and M2_4Au_1 (Fig. 4, D to F). For the definition of the name of the derived superlattices, such as M1_2Au_1, “M1_2Au” refers to the systems that only two of the four different types DNA origami octahedral homologs are containing AuNPs. M1_2Au may contain more than one system, because there would be different choices, so we differentiate them by adding a number in the end of the name, such as “M1_2Au_1.” Moreover, on the basis of the same platform, diverse lattices can also be fabricated by inserting the same number of AuNPs inside the lattices, such as superlattices M1_2Au_1 and M1_2Au_2 (Fig. 4, A and B) or M3_3Au_1 and M3_3Au_2 (Fig. 4, I and J). In addition, even if the same number of the same shape of DNA origami octahedral homologs is inserted AuNPs, we can obtain various superlattices on the basis of different platforms, such as M2_4Au_1 and M3_4Au_1 (Fig. 4, F and K). In general, merely from the standpoint of AuNPs (criterion 2), the types of nanoparticle superlattices fabricated in this work (regardless of DOFs) can be regarded as 38 types, because the arrangement of AuNPs among some of the 85 types of DOF/AuNP lattices is exactly the same, which also indicates that more than one path, based on our strategy, can be designed to fabricate identical nanoparticle superlattices.
Fig. 4. SAXS characterization of 12 representative nanoparticle superlattices constructed by selectively inserting the AuNPs into specific kinds of DNA origami octahedral homologs within the M1, M2, and M3 platforms.
(A to C) Nanoparticle superlattices fabricated on the basis of the M1 platform, M1_2Au_1 (A), M1_2Au_2 (B), and M1_3Au_1 (C). (D to F) Nanoparticle superlattices fabricated on the basis of the M2 platform, M2_2Au_1 (D), M2_3Au_1 (E), and M2_4Au_1 (F). (G to L) Nanoparticle superlattices fabricated on the basis of the M3 platform, M3_2Au_1 (G), M3_2Au_2 (H), M3_3Au_1 (I), M3_3Au_2 (J), M3_4Au_1 (K), and M3_5Au_1 (L). From left to right, each panel contains corresponding DNA platforms in which the AuNPs are selectively inserted into different types of DNA origami octahedral homologs, the 1D experimental SAXS data (orange, green, and blue for superlattices based on the M1, M2, and M3 platforms, respectively), the simulation curve (gray), and the unit cell of the AuNPs outlined by the black lines.
DISCUSSION
In summary, we have demonstrated a universal strategy for programmably assembling three kinds of anisotropic DOFs to fabricate highly ordered superlattices. These lattices were designed to be composed of multiple types of octahedral homologs (≥4). On the basis of the M1, M2, and M3 platforms constructed in this work, by inserting the different numbers of AuNPs inside different types of DNA origami octahedral homologs, up to 85 distinct DOF/AuNP hybrid superlattices with 38 different types of nanoparticle superlattices have been realized, which greatly increased the databases of DNA-guided crystals. In addition, more relative positions between DNA origami octahedral homologs and AuNPs could be adopted to enrich the nanoparticle superlattice library, including sitting on the bundles or circled inside the faces of the octahedron. We believe that the crystals synthesized using this strategy could be more diversified by extending the versatility of guest objects to other nanomaterials with appropriate sizes and shapes, such as quantum dots, proteins, or other types of organic or inorganic nanoparticles, and even the mixture of them. Homologs of other types of DNA origami units, such as cube, tetrahedron, etc., can also be cocrystallized using a similar strategy as we proposed in this work to further increase the diversity of nanoparticle lattices, such as diamond family crystals, monoclinic crystals, triclinic crystals, and so on. In addition, a strict design of the locked mode along specific dimension needs to be carefully considered when different homologs are cocrystallized to create proposed patterns. More general design rules for crystallization of arbitrarily designed DNA origami shapes are potentially interesting to be summarized after more types of DNA origami crystals could be successfully fabricated. Some potential limitations when using the proposed strategy for fabricating more complicated DNA origami platforms are also needed to be considered. For example, most of the DNA origami crystals reported are linked vertices by vertices, which greatly limits the diversity and complexity of the final assemblies. More binding methods, such as edge-to-edge, face-to-face, and even the vertex-to-edge may be helpful for fabricating more complicated and function-guided DNA structures. Moreover, the progress made in this work provides an accessible way to assemble nanomaterials with a more complex way and lays a solid foundation for designing and fabricating functional materials with exciting plasmonic, catalytic, or photonic properties.
MATERIALS AND METHODS
Design and synthesis of DNA origami monomers
R_oct, E_oct, and P_cot DOFs were all designed by caDNAno software, which can be found at http://cadnano.org/. In our design, each octahedral DOF has 12 edges composed of a six-helix bundle (6HB). For R_oct, each edge is 84–base pair long (~28.56 nm). For E_oct, the four edges in the middle square plane are around 28.56 nm (84 base pairs) and others are about 35.70 nm (105 base pairs). For P_oct, the four edges below the middle square plane are 105 base pairs (~35.70 nm), while the other edges are 84 base pairs (~28.56 nm) (see fig. S1 for details). In this design, four SEs can be stretched out from each vertex of the DOF, with one sticking out from each 6HB (see fig. S2 for details). Meanwhile, each edge in the middle square plane was designed to extend inner strands to capture AuNPs.
All DNA origami octahedral homologs are folded by mixing the M13mp18 scaffold DNA (Bayou Biolabs, LLC) with corresponding staple strands and SEs in a ratio of 1:10:10 and then going through a slow annealing from 90° to 20°C for about 22 hours. The final solution contains 10 nM M13mp18, 100 nM staples, 100 nM SEs, 1 mM EDTA, 12.5 mM magnesium acetate, and 40 mM tris acetate. If necessary, then additional inner strands will be added in a ratio of 7.5:1 with M13mp18.
Functionalizing AuNPs with single-stranded DNA
Thiol-modified single-stranded DNA (ssDNA) purchased from Sangon Biotech are first mixed with TCEP [tris(2-carboxyethyl) phosphine] and incubated in ice for 1.5 hours to reduce disulfide bonds, and the size exclusion column (G-25, GE Healthcare) is used to remove excess small molecules. Then, the 10-nm spherical AuNPs (Ted Pella Inc.) are mixed with purified DNA strands in a ratio of 1:300 and incubated at room temperature for 1.5 hours. Next, the mixture is buffered to get the 10 mM phosphate buffer solution and incubated for another 1.5 hours. After that, the NaCl solution is gradually added to the mixed solution until the concentration of NaCl in solution reaches 0.3 M. Last, the mixed solution is aged at room temperature for at least 18 hours. The excess DNA strands are removed by four times of centrifugation, and the solution is washed using 0.1 M NaCl and 10 mM phosphate buffer.
Fabrication of DNA DOF/nanoparticle complex crystals
Corresponding DNA origami building blocks are mixed in equal proportion, followed by adding excess AuNPs modified with ssDNA. Then, the DOF/nanoparticle crystals can be achieved by annealing the mixture twice using the specific procedures (0.2°C hour−1 from 50° to 40°C, 0.1°C hour−1 from 40° to 25°C, and 0.2°C hour−1 from 25° to 20°C).
Preparation of silica-coated DOF/nanoparticle complex crystals
The DOF/nanoparticle complex crystals cannot maintain stability after leaving the salt solution. Therefore, it is necessary to encapsulate the crystal with a thin layer of silica shell when characterized under electron microscopy. First, because the nanoparticle superlattices fabricated settles at the bottom of the tube, we wash the samples by removing the supernatant and filling the buffer solution containing 1 mM EDTA, 12.5 mM magnesium acetate, and 40 mM tris acetate for seven to eight times to remove impurities without losing much of the materials, and the volume is lastly made up to 15 to 25 μl. Next, the solution is mixed with 0.4 to 0.7 μl of TMAPS (N-trimethoxysilylpropyl-N,N,N-trimethylammonium chloride) and shaken at room temperature at 400 rpm for 20 min. Subsequently, 0.3 to 0.6 μl of TEOS (tetraethyl orthosilicate) is added, and the mixture is shaken at the same temperature at 500 rpm for 30 min. Last, the mixed solution is left steadily for 12 hours and washed by deionized water.
TEM sample preparation and characterization
For DNA origami octahedral homologs, the carbon-coated copper grids are first glow-discharged for 30 s using PELCO easiGlow (Ted Pella Inc.). Subsequently, 5 μl of sample is dropped onto the copper grid and deposited for 5 min. The excess sample is wicked away from the edge of the copper grid using filter paper. After washed by deionized water twice, the sample is stained using 5 μl of 2% (w/v) uranyl acetate solution for 10 s. The excess uranyl acetate is removed using filter paper. The prepared samples were characterized under JEOL JEM-2800 at 200 kV.
For silica-coated DOF/nanoparticle complex crystals, the carbon-coated copper grids are first glow-discharged for 30 s using PELCO easiGlow (Ted Pella Inc.). Then, 5 μl of sample is dropped onto the copper grid and deposited for 5 min. The excess sample is wicked away from the edge of copper grid using filter paper. After washed by deionized water one to two times, the sample is dried in the air directly. JEOL JEM-2800 TEM was used to observe the prepared samples at 200 kV.
SEM sample preparation and characterization
Three- to 5-μl silica-coated DOF/nanoparticle complex crystals are dropped onto silicon slice, which has been cleaned with ethanol in advance, and then the silicon slice with samples is transferred to infrared lamp for drying. The prepared samples are characterized through the HITACHI Regulus 8100 SEM.
Small-angle x-ray scattering
SAXS experiments were performed at BL16B1 and BL19U2 beamlines of the Shanghai Synchrotron Radiation Facility. The nanoparticle superlattices loaded into the capillary are fixed on the sample stage for testing. The 2D scattering data are collected on area detectors and integrated into 1D I(q) scattering curve as a function of the scattering vector q, where , λ is the wavelength of the incident x-ray, and θ is the scattering angle. The structure factor S(q) is obtained by dividing I(q) by the corresponding particle form factor P(q).
Acknowledgments
We thank the staff from BL16B1 beamline at Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank the staff from BL19U2 beamline of the National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for assistance during data collection.
Funding: This work is supported by the National Natural Science Foundation of China (grant no. 92056114, grant no. 21971109, and grant no. 21834004) and the Program for Innovative Talents and Entrepreneur in Jiangsu. W.X. acknowledges financial support from the National Key R&D Program of China (grant no. 2020YFA0406104) and the National Natural Science Foundation of China (grant no. 22173044).
Author contributions: Y.T. and M.J. conceived the project. M.J. and Z.Z. performed all the experiments. M.J., Z.Z., W.C., and N.M. contributed to characterizations and data analysis. M.J. contributed to the simulation. M.J., Z.Z., and Y.T. wrote and edited the manuscript. W.X. provided helpful suggestions. Y.T. supervised the project. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
DNA sequences
Figs. S1 to S48
Tables S1 to S9
REFERENCES AND NOTES
- 1.Sharma J., Chhabra R., Cheng A., Brownell J., Liu Y., Yan H., Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 323, 112–116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuzyk A., Schreiber R., Fan Z., Pardatscher G., Roller E.-M., Hoegele A., Simmel F. C., Govorov A. O., Liedl T., DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Liu Z., Yu G., Jiang W., Mao C., Self-assembly of molecule-like nanoparticle clusters directed by DNA nanocages. J. Am. Chem. Soc. 137, 4320–4323 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Jiang Q., Liu Q., Shi Y., Wang Z.-G., Zhan P., Liu J., Liu C., Wang H., Shi X., Zhang L., Sun J., Ding B., Liu M., Stimulus-responsive plasmonic chiral signals of gold nanorods organized on DNA origami. Nano Lett. 17, 7125–7130 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Man T., Ji W., Liu X., Zhang C., Li L., Pei H., Fan C., Chiral metamolecules with active plasmonic transition. ACS Nano 13, 4826–4833 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Fang W. N., Jia S. S., Chao J., Wang L. Q., Duan X. Y., Liu H. J., Li Q., Zuo X. L., Wang L. H., Wang L. H., Liu N., Fan C. H., Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Sci. Adv. 5, eaau4506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P., Huh J.-H., Park H., Yang D., Zhang Y., Zhang Y., Lee J., Lee S., Ke Y., DNA origami guided self-assembly of plasmonic polymers with robust long-range plasmonic resonance. Nano Lett. 20, 8926–8932 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Shevchenko E. V., Talapin D. V., Kotov N. A., O’Brien S., Murray C. B., Structural diversity in binary nanoparticle superlattices. Nature 439, 55–59 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Nykypanchuk D., Maye M. M., van der Lelie D., Gang O., DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Park S. Y., Lytton-Jean A. K. R., Lee B., Weigand S., Schatz G. C., Mirkin C. A., DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Jones M. R., Macfarlane R. J., Lee B., Zhang J., Young K. L., Senesi A. J., Mirkin C. A., DNA-nanoparticle superlattices formed from anisotropic building blocks. Nat. Mater. 9, 913–917 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Macfarlane R. J., Lee B., Jones M. R., Harris N., Schatz G. C., Mirkin C. A., Nanoparticle superlattice engineering with DNA. Science 334, 204–208 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., Macfarlane R. J., Young K. L., Choi C. H. J., Hao L., Auyeung E., Liu G., Zhou X., Mirkin C. A., A general approach to DNA-programmable atom equivalents. Nat. Mater. 12, 741–746 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Lu F., Yager K. G., Zhang Y., Xin H., Gang O., Superlattices assembled through shape-induced directional binding. Nat. Commun. 6, 6912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien M. N., Jones M. R., Lee B., Mirkin C. A., Anisotropic nanoparticle complementarity in DNA-mediated co-crystallization. Nat. Mater. 14, 833–839 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Paik T., Diroll B. T., Kagan C. R., Murray C. B., Binary and ternary superlattices self-assembled from colloidal nanodisks and nanorods. J. Am. Chem. Soc. 137, 6662–6669 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Tagawa M., Xin H. L., Wang T., Emamy H., Li H., Yager K. G., Starr F. W., Tkachenko A. V., Gang O., Diamond family of nanoparticle superlattices. Science 351, 582–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M. S., Alexander-Katz A., Macfarlane R. J., Nanoparticle assembly in high polymer concentration solutions increases superlattice stability. Small 17, 2102107 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Lee M. S., Yee D. W., Ye M., Macfarlane R. J., Nanoparticle assembly as a materials development tool. J. Am. Chem. Soc. 144, 3330–3346 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Douglas S. M., Dietz H., Liedl T., Hoegberg B., Graf F., Shih W. M., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D., Pal S., Nangreave J., Deng Z., Liu Y., Yan H., DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Ke Y., Voigt N. V., Gothelf K. V., Shih W. M., Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 134, 1770–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Y., Wang T., Liu W., Xin H. L., Li H., Ke Y., Shih W. M., Gang O., Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 10, 637–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerling T., Wagenbauer K. F., Neuner A. M., Dietz H., Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Jun H., Zhang F., Shepherd T., Ratanalert S., Qi X. D., Yan H., Bathe M., Autonomously designed free-form 2D DNA origami. Sci. Adv. 5, eaav0655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan X., Lu X., Shen C., Ke Y., Ni W., Wang Q., Au nanorod helical superstructures with designed chirality. J. Am. Chem. Soc. 137, 457–462 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Xin L., Zhou C., Duan X., Liu N., A rotary plasmonic nanoclock. Nat. Commun. 10, 5394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei H., Sha R. J., Wang X. W., Zheng M., Fan C. H., Canary J. W., Seeman N. C., Organizing end-site-specific SWCNTs in specific loci using DNA. J. Am. Chem. Soc. 141, 11923–11928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan P. F., Urban M. J., Both S., Duan X. Y., Kuzyk A., Weiss T., Liu N., DNA-assembled nanoarchitectures with multiple components in regulated and coordinated motion. Sci. Adv. 5, eaax6023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y., Zhang Y., Wang T., Xin H. L., Li H., Gang O., Lattice engineering through nanoparticle-DNA frameworks. Nat. Mater. 15, 654–661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., Gaitanaros S., Lee S., Bathe M., Shih W. M., Ke Y., Programming self-assembly of DNA origami honeycomb two-dimensional lattices and plasmonic metamaterials. J. Am. Chem. Soc. 138, 7733–7740 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Zhang T., Hartl C., Frank K., Heuer-Jungemann A., Fischer S., Nickels P. C., Nickel B., Liedl T., 3D DNA origami crystals. Adv. Mater. 30, e1800273 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ji M., Liu J., Dai L., Wang L., Tian Y., Programmable cocrystallization of DNA origami shapes. J. Am. Chem. Soc. 142, 21336–21343 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Tian Y., Lhermitte J. R., Bai L., Vo T., Xin H. L., Li H., Li R., Fukuto M., Yager K. G., Kahn J. S., Xiong Y., Minevich B., Kumar S. K., Gang O., Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 19, 789–796 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Ma N., Dai L., Chen Z., Ji M., Wang Y., Tian Y., Environment-resistant DNA origami crystals bridged by rigid DNA rods with adjustable unit cells. Nano Lett. 21, 3581–3587 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Auyeung E., Macfarlane R. J., Choi C. H. J., Cutler J. I., Mirkin C. A., Transitioning DNA-engineered nanoparticle superlattices from solution to the solid state. Adv. Mater. 24, 5181–5186 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Liu X., Zhang F., Jing X., Pan M., Liu P., Li W., Zhu B., Li J., Chen H., Wang L., Lin J., Liu Y., Zhao D., Yan H., Fan C., Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Dai L. Z., Ding Z. Y., Ji M., Liu J. L., Xing H., Liu X. G., Ke Y. G., Fan C. H., Wang P., Tian Y., DNA origami single crystals with Wulff shapes. Nat. Commun. 12, 3011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences
Figs. S1 to S48
Tables S1 to S9