Abstract
GGC repeat expansions within NOTCH2NLC have been identified as the genetic cause of neuronal intranuclear inclusion disease (NIID). To understand the molecular pathogenesis of NIID, here, we established both a transgenic mouse model and a human neural progenitor cells (hNPCs) model. Expression of the NOTCH2NLC with expanded GGC repeats produced widespread intranuclear and perinuclear polyglycine (polyG), polyalanine (polyA), and polyarginine (polyR) inclusions, leading to behavioral deficits and severe neurodegeneration, which faithfully mimicked the clinical and pathological features associated with NIID. Furthermore, conserved alternative splicing events were identified between the NIID mouse and hNPC models, among which was the enrichment of the binding motifs of hnRNPM, an RNA binding protein known as alternative splicing regulator. Expanded NOTCH2NLC-polyG and NOTCH2NLC-polyA could interact with and sequester hnRNPM, while overexpression of hnRNPM could ameliorate the cellular toxicity. These results together suggested that dysfunction of hnRNPM could play an important role in the molecular pathogenesis of NIID.
Expression of expanded GGC repeats in NOTCH2NLC gene produces multiple polypeptides and causes neurodegeneration.
INTRODUCTION
Polymorphic CGG repeats are more prevalent in the human genome than currently appreciated, and they can have a role in both neurodevelopmental and neurodegenerative disorders. In addition to paradigmatic CGG repeat expansions at the FMR1 locus associated with fragile X syndrome, several neuromuscular and neurodegenerative disorders with overlapping symptoms and similar pathological features have recently been shown to be caused by CGG repeat expansions in distinct loci (1, 2). Among them, neuronal intranuclear inclusion disease (NIID), a rare neurodegenerative disease characterized by widespread intranuclear inclusions in the nervous system as well as multiple visceral organs (3, 4), was found to be caused by expanded GGC repeats in the 5′ untranslated region (5′UTR) of the human-specific NOTCH2NLC gene by three independent research groups (1, 5, 6). Furthermore, the same mutation was then found in an increasing number of neurodegenerative and neuromuscular diseases, including essential tremor, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (ALS), frontotemporal dementia, leukoencephalopathy, multiple system atrophy, and oculopharyngodistal myopathy type 3 (5, 7–14).
NOTCH2NLC belongs to one of three NOTCH2 N-terminal–like (NOTCH2NL) paralogs (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC), which are only found in the human genome. Evolutionarily, the NOTCH2NL genes are derived from gene duplication and gene conversion of NOTCH2 and function in cortical neurogenesis exclusively in the human species (15, 16). Genetic analyses have shown that the extent of pathogenic GGC repeats in NOTCH2NLC ranges from about 60 to 500 (7). On the basis of the age of onset, NIID can be categorized into three subgroups: infantile, juvenile, and adult. To date, expanded GGC repeats in NOTCH2NLC have been reported in both juvenile and adult NIID (1, 5, 6, 17). Clinically, the manifestation of NIID is highly heterogeneous and may include dementia, muscle weakness, peripheral neuropathy, cerebellar ataxia, parkinsonism, seizure, and encephalitic episodes (5, 18–21). Immunohistochemically, the eosinophilic intranuclear inclusions in NIID are positive for p62 and ubiquitin. Although these findings in tissues of patients provide important clues for pathogenesis, the underlying molecular mechanism remains unclear due to the lack of genetic models. Current NIID mouse models were generated by overexpression of NOTCH2NLC with artificial pathogenic GGC repeats in the adult mouse brain via retro-orbital adeno-associated virus (AAV) injection (22) or by in utero electroporation in the mouse neocortex (23), making it difficult to define the specific effect of pathogenic GGC repeat expansions at different ages and in various types of tissues.
Here, we report the first transgenic mouse model in which the exon 1 of NOTCH2NLC gene is expressed, with either normal GGC repeats (17 GGC repeats) or expanded GGC repeats (98 GGC repeats) cloned from the genomic DNA of control and NIID patient, respectively. We show that GGC repeat expansions in NOTCH2NLC produced multiple polypeptides [polyglycine (polyG), polyalanine (polyA), or polyarginine (polyR) containing protein] through AUG-dependent translation, which together could cause neuronal toxicity. The NOTCH2NLC-(GGC)98 mice displayed severe neurodegeneration, motor dysfunction, and cognitive deficits, faithfully recapitulating the clinical and pathological features associated with NIID. Gene expression profiling revealed multiple pathways altered in different brain regions of the NOTCH2NLC mouse model, including the prefrontal cortex, cerebellum, and hippocampus. We further established human induced pluripotent stem cells (iPSCs) from patients with NIID and differentiated them into neural progenitor cells (NPCs). We also performed gene expression profiling of NPCs and compared it with that of our NOTCH2NLC mouse model. We identified overlapping patterns of alternative splicing (AS) events shared by the human NPCs (hNPCs) and mouse models. Among the AS sites, motif analyses revealed the enrichment of the RNA binding motifs of heterogeneous nuclear ribonucleoprotein M (hnRNPM). Mechanistically, we show that hnRNPM could bind to NOTCH2NLC-polyG and NOTCH2NLC-polyA and be sequestered into inclusions containing NOTCH2NLC-polyG and NOTCH2NLC-polyA. Furthermore, overexpression of hnRNPM could ameliorate cellular toxicity caused by expanded GGC repeats within NOTCH2NLC. These results together suggest that dysfunction of hnRNPM-mediated regulation could play a vital role in the molecular pathogenesis of NIID.
RESULTS
Expression of NOTCH2NLC-(GGC)98 produces polyG and polyA in an AUG-dependent manner and causes neuronal toxicity in vitro
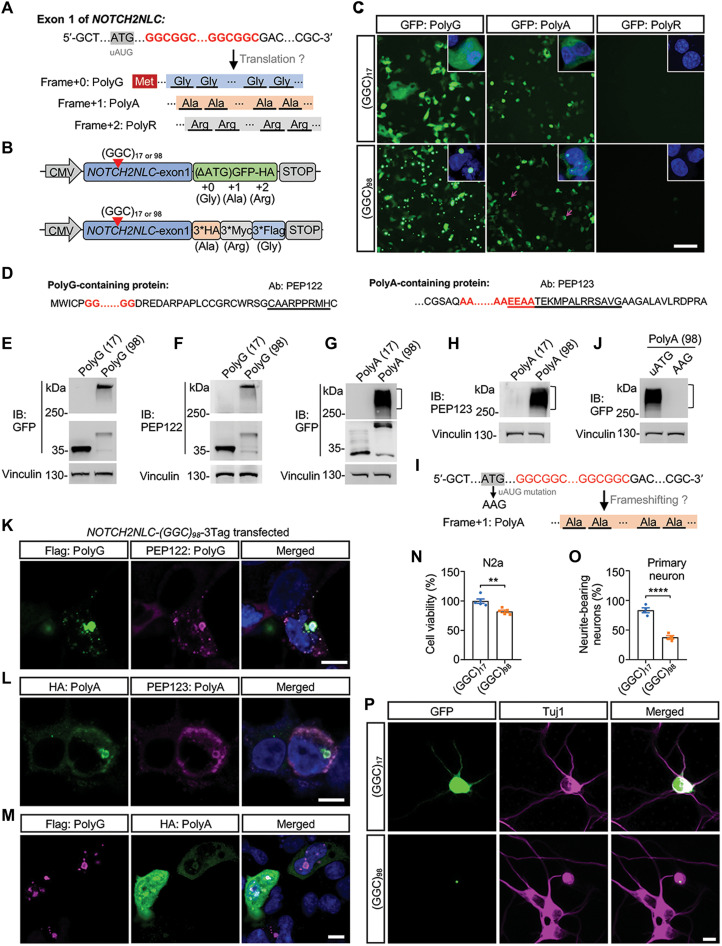
The human NOTCH2NLC gene has two different splicing isoforms, transcript variant 1 (TV1) and transcript variant 2 (TV2) (fig. S1A). We analyzed expression levels of the transcript variants using published human brain RNA sequencing (RNA-seq) datasets (24) and found that TV1 was the dominant transcript in different brain regions (fig. S1B). A similar pattern was also observed in human peripheral blood samples (fig. S1, C and D). Therefore, we focused on NOTCH2NLC TV1 and referred to it as NOTCH2NLC unless otherwise stated. The GGC repeat expansions reside in the 5′UTR of NOTCH2NLC. Notably, an unconventional AUG (uAUG) codon lies upstream and a TAA codon downstream, both in the same reading frame as the GGC repeat, which could produce a short polyG-containing protein named uN2CpolyG or N2NLCpolyG (Fig. 1A and fig. S1A) (22, 23). In addition, GGC repeat expansions could be translated into polyA or polyR via repeat-associated non-AUG (RAN) translation or ribosomal frameshifting (Fig. 1A), which had been reported in FMR1 with CGG repeat expansions (25, 26) or in C9ORF72 with G4C2 repeat expansions (27).
Fig. 1. Expression of NOTCH2NLC-(GGC)98 produces multiple polypeptides and causes cellular toxicity in vitro.
(A) Schematic diagram of the putative mutant NOTCH2NLC proteins. Glycine, Gly; alanine, Ala; arginine, Arg. (B) Schematic diagram of NOTCH2NLC constructs. NOTCH2NLC exon 1 with 17 GGC repeats [(GGC)17 for short] and 98 GGC repeats [(GGC)98 for short] were fused with a GFP tag (start codon removed) or 3*HA/3*Myc/3*Flag (3Tag) tag. (C) Direct GFP fluorescence of HEK293 cells transfected with NOTCH2NLC-GFP constructs. The magenta arrows indicate polyA aggregates. Green, GFP; blue, 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 100 μm. (D) Putative amino acid sequences of polyG-containing and polyA-containing proteins. The underlined sequences indicated the epitope of antibodies against polyG (PEP122) and polyA (PEP123). Ab, antibody. (E and F) Immunoblotting (IB) of HEK293 cells transfected with NOTCH2NLC-GFP (polyG frame) constructs using anti-GFP and PEP122 antibodies. (G and H) Immunoblotting of HEK293 cells transfected with NOTCH2NLC-GFP (polyA frame) constructs using anti-GFP and PEP123 antibodies. (I) Frameshifting hypothesis for polyA. The production of polyA may result from AUG-initiated ribosomal frameshifting translation. (J) Immunoblotting of HEK293 cells transfected with NOTCH2NLC-GFP (polyA frame with uAUG mutation) construct. (K to M) Coimmunostaining of HEK293 cells transfected with NOTCH2NLC-3Tag construct using anti-Flag and anti-PEP122 (K) or using anti-HA and anti-PEP123 (L) or using anti-Flag and anti-HA (M). Scale bars, 10 μm. (N) Cell viability of Neuro-2a cells transfected with NOTCH2NLC-GFP constructs was analyzed by the Cell Counting Kit-8 (CCK-8) method. Data are presented as means ± SEM. (N = 5 independent experiments, **P = 0.0036, two-tailed t test). (O and P) Cell viability of cultured mouse primary cortical neurons transfected with NOTCH2NLC-GFP constructs. Data are presented as means ± SEM (N = 4 independent experiments, at least 120 cells were counted per experiment, ****P < 0.0001, two-tailed t test). Green, GFP; magenta, Tuj1. Scale bar, 10 μm.
To test this possibility, we first cloned exon 1 of NOTCH2NLC harboring 17 GGC repeats or 98 GGC repeats from genomic DNA of control and NIID patient and fused the repeats with a green fluorescent protein–hemagglutinin (GFP-HA) tag (start codon removed) in each of three reading frames (Fig. 1B and fig. S2, A to C). As expected, when overexpressed in human embryonic kidney (HEK) 293 cells, homogeneous cytoplasmic GFP fluorescence was detected with the NOTCH2NLC-(GGC)17 construct in the polyG frame, while abundant intranuclear or perinuclear GFP fluorescent aggregates were found for NOTCH2NLC-(GGC)98, also in the polyG frame (Fig. 1C). Immunoblotting using anti-GFP antibody showed that NOTCH2NLC-(GGC)17 overexpression led to the appearance of bands around 35 kDa. Soluble NOTCH2NLC-(GGC)98–polyG appeared at about 40 kDa, while aggregated NOTCH2NLC-(GGC)98–polyG remained in the stacking gel (Fig. 1E). We developed a rabbit polyG antibody PEP122, whose epitope was identical to that of a 4D12 antibody (Fig. 1D). The 4D12 antibody was validated in both animal model and patients with NIID in a previous report (22). Immunoblotting using PEP122 antibody presented similar results with that using anti-GFP antibody (Fig. 1F).
Unexpectedly, a small quantity of intranuclear polyA inclusions were observed when the polyA frame NOTCH2NLC-(GGC)98 construct was expressed, while cytoplasmic GFP fluorescence was also detected with NOTCH2NLC-(GGC)17 in the polyA frame. The origin of this protein may be translation starting with an ATG initiation codon downstream of GGC repeats but upstream of the GFP sequence (Fig. 1C and fig. S2B). This was further confirmed by immunoblotting with anti-GFP antibody because both NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 constructs generated the same band size around 32 kDa. Same as polyG, NOTCH2NLC-(GGC)98–derived polyA had a soluble form around 48 kDa and also formed aggregates that remained in the stacking gel (Fig. 1G). To further confirm the existence of polyA-containing protein, we developed a rabbit antibody named PEP123, which was designed to recognize the C terminus of polyA (Fig. 1D). Immunoblotting showed that anti-PEP123 antibody could identify the same smeared polyA bands as anti-GFP (Fig. 1H). Moreover, liquid chromatography–tandem mass spectrometry analysis on HEK293 cells transfected with NOTCH2NLC-(GGC)98 identified partial peptide sequences of polyG and polyA proteins (fig. S2E). These results suggested that polyA-containing proteins were being produced. The production of polyA may result from at least two mechanisms: RAN translation or AUG-initiated ribosomal frameshifting translation. Because the production of NOTCH2NLC-polyG was uAUG dependent (22, 23), mutation of the uAUG codon should disturb the translation of polyA, if polyA was produced from ribosomal frameshifting. To this end, the uAUG codon of polyG frame in the NOTCH2NLC-(GGC)98–polyA–GFP construct was mutated to AAG (Fig. 1I). Immunoblotting showed that the polyA inclusion, which stayed in the stacking gel with the uAUG construct, disappeared when the uAUG was mutated (Fig. 1J). These results indicated that the translation of polyA was uAUG codon dependent.
In addition, immunostaining showed that both polyG and polyA inclusions colocalized with ubiquitin and P62 (fig. S3A), reminiscent of what has been observed in patients with NIID (5). The colocalization of polyG and polyA with ubiquitin and P62 led us to check whether polyG and polyA appeared in the same inclusions. To check this possibility, the GFP-HA tag of NOTCH2NLC-(GGC)98–GFP was replaced by a 3*HA/3*Myc/3*Flag (3Tag) tag, which was in reading frame with polyA, polyR, and polyG, respectively (Fig. 1B and fig. S2D). Seventy-two hours after transfection in HEK293 cells, coimmunostaining using anti-Flag and anti-PEP122 showed coimmunofluorescence of polyG (Fig. 1K); polyA also showed coimmunofluorescence when using anti-HA and anti-PEP123 antibodies (Fig. 1L). Coimmunostaining showed that polyG inclusions colocalized with polyA inclusions, and polyG-containing protein was aggregation prone while polyA-containing protein was more soluble (Fig. 1M). These results together indicated that GGC repeat expansions at the NOTCH2NLC locus could be translated into polyG- and polyA-containing proteins. Although two previous studies reported no polyA and polyR products coming from NOTCH2NLC in vitro, here, we identified the detection of polyA protein. The reasons for this discrepancy may be complicated. One possible reason is that our patient DNA-derived constructs harbor GGA interruptions within the GGC repeats, while the constructs Boivin et al. and Zhong et al. (22, 23) used contained homogeneous synthetic GGC repeats.
Next, we examined whether the products of NOTCH2NLC with GGC repeat expansions were toxic in vitro. Both NOTCH2NLC-(GGC)17– and NOTCH2NLC-(GGC)98–containing constructs were transiently transfected into Neuro-2a cells, followed by a cell viability assay. NOTCH2NLC-(GGC)98– transfected cells showed a significant decrease in viability when compared to NOTCH2NLC-(GGC)17–transfected cells (Fig. 1N). Transfected Neuro-2a cells were also treated with 25 mM retinoic acid for 72 hours to induce differentiation and promote neurite outgrowth. NOTCH2NLC-(GGC)98–expressing cells grew shorter neurites than NOTCH2NLC-(GGC)17–expressing cells (fig. S3, B and C). We further assessed cellular toxicity in cultured mouse primary cortical neurons by analyzing the proportion of GFP-positive neurons with neurites 48 hours after transfection. NOTCH2NLC-(GGC)98 caused more severe cell death when compared to NOTCH2NLC-(GGC)17 (Fig. 1, O and P). These results demonstrated neuronal toxicity associated with the expression of NOTCH2NLC-(GGC)98 in vitro.
Expression of NOTCH2NLC-(GGC)98 produces multiple polypeptides in NOTCH2NLC transgenic mice in an age-dependent manner
To systematically explore the pathogenic role of the GGC repeat expansions in the NOTCH2NLC gene underlying NIID, we generated two conditional transgenic NOTCH2NLC mouse models by inserting NOTCH2NLC exon 1 with 17 GGC repeats or 98 GGC repeats (with GGA interruption; fig. S2D) into the Rosa26 site. Contrasting with a previously reported AAV-injected mouse model in which only polyG was expressed (22), our transgenic mouse models may express both polyG and polyA, based on our in vitro observations (Fig. 1). Hence, the 3Tag tag was used for convenient detection of translated products of NOTCH2NLC in mice. Because intranuclear inclusions were found in the nervous system and peripheral tissues in patients with NIID, the conditional transgenic mice were crossed with EIIa-Cre mice to express NOTCH2NLC-(GGC)17 or NOTCH2NLC-(GGC)98 ubiquitously (figs. S4A and S5A). Genotyping was confirmed by polymerase chain reaction (PCR) and Sanger sequencing (fig. S4, B and C). The mRNA levels of NOTCH2NLC-(GGC)17 or NOTCH2NLC-(GGC)98 were comparable in two transgenic mice (fig. S4D). Because NOTCH2NLC is only present in the human genome, it was difficult to compare its expression level between the transgenic mice and human samples. By examining the fragments per kilobase of exon model per million mapped fragments (FPKM) of NOTCH2NLC, we found that the expression level of NOTCH2NLC in our mouse model was comparable to or lower than hNPCs, human differentiated neurons, and forebrain cortex from human postmortem brains (fig. S4E).
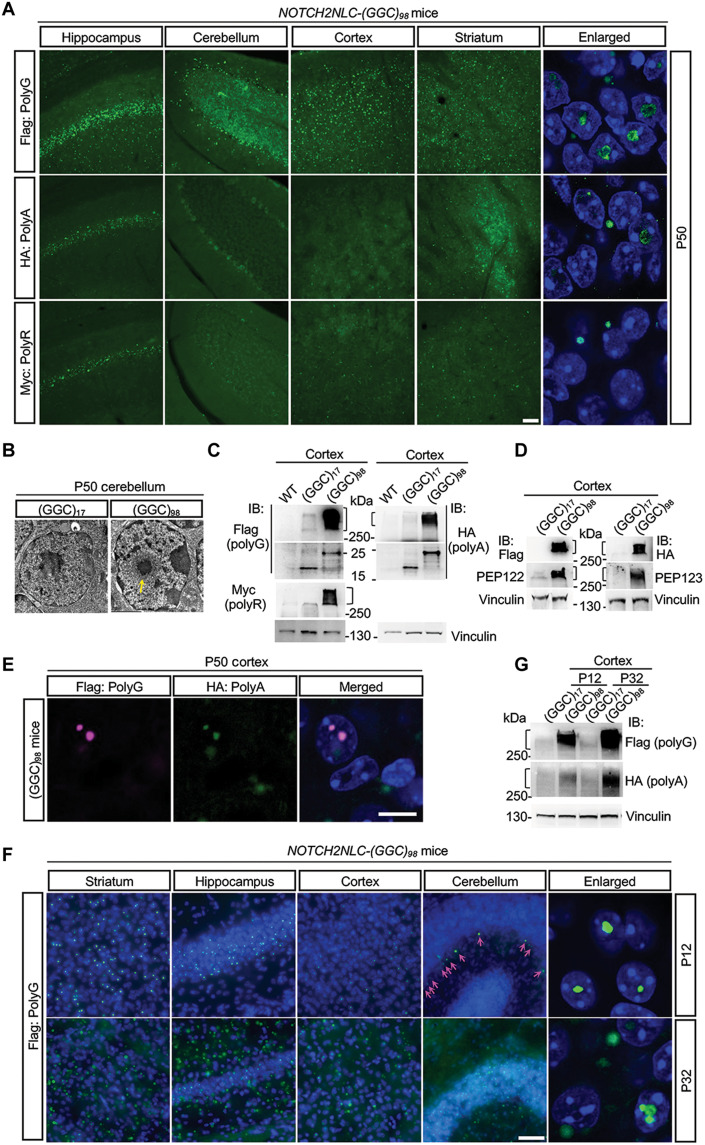
From birth to postnatal 40 days (P40), the wild-type (WT), NOTCH2NLC-(GGC)17, and NOTCH2NLC-(GGC)98 mice showed no overt anomalies in appearance. However, after 40 days of age, the NOTCH2NLC-(GGC)98 mice gradually developed a progressive phenotype characterized by inactivity, hunchback, aberrant posture, and ataxia (movie S1). NOTCH2NLC-(GGC)98 mice died at around 2 months, regardless of gender. Immunofluorescent staining of transgenic mouse brains at P50 showed that NOTCH2NLC-(GGC)98 formed a large amount of polyG inclusions throughout the brain, including the striatum, hippocampus, cerebellum, and cortex, while comparable inclusions were hard to detect in the NOTCH2NLC-(GGC)17 mice (Fig. 2A and fig. S5B). Furthermore, small amounts of polyA and polyR inclusions were also seen in the hippocampus and, to a lesser extent, in the striatum and cortex of the NOTCH2NLC-(GGC)98 mice (Fig. 2A); confocal images further indicated the presence of intranuclear or perinuclear polyG, polyA, and polyR inclusions in the mouse brains (Fig. 2A). PEP122 and PEP123 antibodies were also used to examine the inclusions. As expected, PEP122- and PEP123-positive inclusions colocalized with Flag and HA, respectively; most of them colocalized with P62 and ubiquitin as well (fig. S6). Electron microscopy images of NOTCH2NLC-(GGC)98 mice cerebellum showed obvious neuronal intranuclear inclusions exhibiting a pile of round dense filamentous materials without membrane structure (Fig. 2B), which were similar to those observed in patients with NIID (4, 5, 7). Meanwhile, Western blotting using anti-Flag, anti-HA, and anti-Myc antibodies showed abundant polyG, polyA, and polyR aggregates in the NOTCH2NLC-(GGC)98 mouse brains and soluble form of polyG and polyA with expected size (Fig. 2C). PolyG and polyA aggregates were further confirmed by anti-PEP122 and anti-PEP123 antibodies, respectively (Fig. 2D). Double immunostaining using anti-Flag and anti-HA antibodies showed the colocalization of the polyG inclusions with polyA inclusions (Fig. 2E).
Fig. 2. Expression of NOTCH2NLC-(GGC)98 produces multiple polypeptides in NOTCH2NLC transgenic mice in an age-dependent manner.
(A) Immunofluorescent staining against Flag (polyG), HA (polyA), and Myc (polyR) in the brain of NOTCH2NLC-(GGC)98 mice at P50. The intranuclear or perinuclear polyG, polyA, and polyR inclusions were observed under higher magnification in the hippocampus of NOTCH2NLC-(GGC)98 mouse. Green, Flag/HA/Myc; blue, DAPI. Scale bar, 50 μm. (B) Electron microscopy of cerebellar neurons from the NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50. Yellow arrow indicates the intranuclear inclusions. Scale bar, 2 μm. (C and D) Western blotting against Flag (polyG), HA (polyA), and Myc (polyR) tag (C) and PEP122 and PEP123 (D) in the cortex from control and NOTCH2NLC-(GGC)98 mice at P50. (E) Immunofluorescent staining against Flag (polyG) and HA (polyA) in cortex of NOTCH2NLC-(GGC)98 mice at P50. Green, Flag; magenta, HA. Scale bar, 10 μm. (F) Immunofluorescent staining against Flag (polyG) in the different brain regions of NOTCH2NLC-(GGC)98 mice at P12 and P32. Green, Flag; blue, DAPI. Magenta arrows indicate the intranuclear inclusions in the cerebellum. Scale bar, 50 μm. (G) Western blotting against Flag (polyG) and HA (polyA) in the cortex from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P12 and P32.
Because NOTCH2NLC mice died at around 2 months old, a point at which major brain development in mice is complete (28), we then examined expression patterns of polyG inclusions at earlier stages. Immunofluorescent staining of NOTCH2NLC-(GGC)98 mouse brain showed clearly distinct intranuclear polyG inclusions mainly in the striatum, hippocampus (CA1) and cortex, and less in the cerebellum at P12 (Fig. 2F and fig. S7). At P32, however, polyG inclusions had accumulated across the entire brain—not only in the nucleus but also outside the nucleus (Fig. 2F and fig. S7); Western blotting analysis of Flag (polyG) and HA (polyA) in the cortex at P12 and P32 of NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice demonstrated that polyG and polyA aggregates became more abundant over time (Fig. 2G). These results together suggested that NOTCH2NLC-(GGC)98–derived polypeptides accumulate in an age-dependent manner in vivo.
Expression of NOTCH2NLC-(GGC)98 causes neurodegeneration in mice
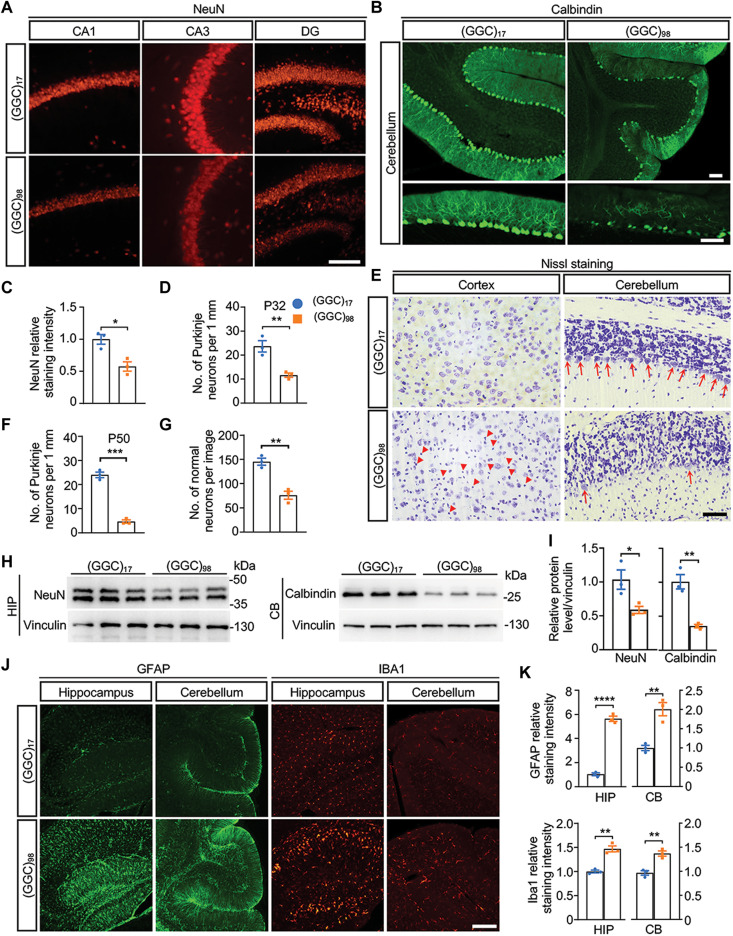
To determine the impact of widespread expression of NOTCH2NLC-(GGC)98, we first performed anatomical examinations on NOTCH2NLC-(GGC)98 mice and control mice. In general, we observed no remarkable abnormalities in the morphology of visceral organs from NOTCH2NLC-(GGC)98 mice at P50, regardless of gender. We then investigated whether there was neuronal loss in different brain regions of NOTCH2NLC-(GGC)98 mice. Immunofluorescent staining clearly showed that the numbers of NeuN-positive cells in the hippocampi of NOTCH2NLC-(GGC)98 mice were reduced significantly when compared to the control mice at P50 (Fig. 3, A and C, and fig. S8A). The numbers of calbindin-labeled Purkinje cells in the cerebellum were markedly reduced in NOTCH2NLC-(GGC)98 mice at P32 (Fig. 3, B and D). Nissl staining also confirmed that most of the Purkinje cells were lost at P50 (Fig. 3, E and F). In the cortex, a large number of dark shrunken neurons and a significantly reduced number of normal neurons indicated widespread neuronal death in NOTCH2NLC-(GGC)98 mice at P50 (Fig. 3, E and G). We also performed Western blotting of NeuN and calbindin in the hippocampus and cerebellum, respectively, which allowed for quantitative analysis of the relative expression level of NeuN and calbindin, and found a significant reduction in NOTCH2NLC-(GGC)98 mice (Fig. 3, H and I).
Fig. 3. Expression of NOTCH2NLC-(GGC)98 causes neurodegeneration in mice.
(A and B) Immunofluorescence against NeuN in the hippocampus (A) and calbindin in the cerebellum (B) from NOTCH2NLC mice. Red, NeuN; green, calbindin. Scale bars, 100 μm. (C and D) Quantification of the staining intensity of NeuN (C) and the number of Purkinje neurons (D). Data are presented as means ± SEM. [N = 3 mice per group, *P = 0.0152 (C), **P = 0.0086 (D), two-tailed t test]. (E) Nissl staining of the cortex and cerebellum from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50. Red arrowheads indicate dark shrunken neurons; red arrows indicate the Purkinje neurons. Scale bar, 50 μm. (F and G) Quantification of the number of Purkinje neurons in the cerebellum (F) and the number of normal neurons in the cortex (G). Data are presented as means ± SEM. [N = 3 mice per group, ***P = 0.0001 (F) and **P = 0.0033 (G), two-tailed t test]. (H and I) Western blotting against NeuN in the hippocampus (HIP) and against calbindin in the cerebellum (CB) from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50. Normalized relative expressions of NeuN and calbindin are present in (I). Data are presented as means ± SEM. (N = 3 mice per group, *P = 0.0434 and **P = 0.0038, two-tailed t test). (J and K) Immunofluorescence against GFAP and Iba1 in the HIP and CB from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50 (J). (K) Quantification of GFAP and Iba1 relative staining intensity. Data are presented as mean ± SEM. [N = 3 mice per group, ****P < 0.0001 (GFAP in HIP), **P = 0.0055 (GFAP in CB), **P = 0.0023 (Iba1 in HIP), and **P = 0.0048 (Iba1 in CB), two-tailed t test]. Green, GFAP; red, Iba1. Scale bar, 100 μm.
Next, we examined gliosis, another marker of neurodegeneration, which can be assessed by glial fibrillary acidic protein (GFAP) (astrocyte marker) and ionized calcium binding adapter molecule 1 (IBA1) (microglia marker) staining. As expected, activation of astrocytes and microglia was seen in the hippocampus, cerebellum, and striatum of NOTCH2NLC-(GGC)98 mice at P50 (Fig. 3, J and K, and fig. S8B). Together, our results suggested that the expression of NOTCH2NLC-(GGC)98 could cause neurodegeneration and inflammation in the nervous system.
Expression of NOTCH2NLC-(GGC)98 causes muscle degeneration in mice
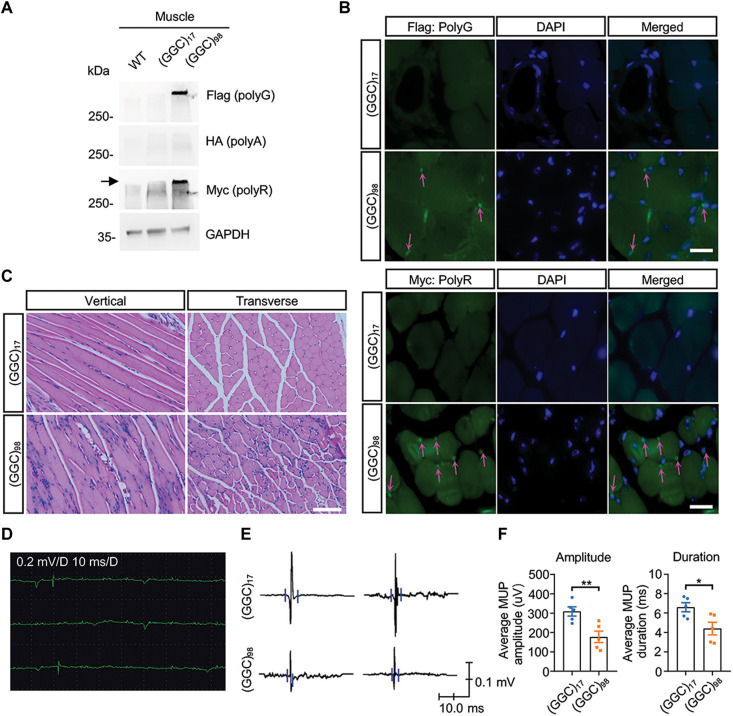
Because muscle weakness is one of the major clinical manifestations of NIID, we wanted to know whether the expression of NOTCH2NLC-(GGC)98 would influence muscle function in the mouse model. Immunoblotting of gastrocnemius muscle showed aggregated polyG and polyR in the NOTCH2NLC-(GGC)98 mouse (Fig. 4A). Then, we analyzed the distribution of NOTCH2NLC-derived polypeptides in the gastrocnemius muscle. Resembling the pattern observed in the brain, polyG and polyR also accumulated around the nuclei of muscle cells (Fig. 4B). Hematoxylin and eosin staining of gastrocnemius muscle revealed normal morphology in the NOTCH2NLC-(GGC)17 mice, presented as well-defined muscle cells in which the nuclei were localized peripherally (Fig. 4C). However, fragmented muscle morphology and multiple internalized or centralized nuclei, a feature of regenerated muscle cells after degeneration, were found in the NOTCH2NLC-(GGC)98 mice (Fig. 4C). The notable pathology we observed raised the possibility that the degeneration could also degrade muscle function. Electromyography (EMG) recordings of gastrocnemius muscle revealed fibrillation potentials and positive sharp waves, indicating denervated fibers (Fig. 4D). Moreover, a reduced average amplitude and duration of motor unit potential were observed in the NOTCH2NLC-(GGC)98 mice (Fig. 4, E and F), indicating muscle degeneration caused by expanded GGC repeats.
Fig. 4. Expression of NOTCH2NLC-(GGC)98 causes muscle degeneration in mice.
(A) Western blotting against polyG (Flag), polyA (HA), and polyR (Myc) in the gastrocnemius muscle from WT, NOTCH2NLC-(GGC)17, and NOTCH2NLC-(GGC)98 mice. (B) Immunofluorescent staining against Flag (polyG) and Myc (polyR) in the gastrocnemius muscle of NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50. Magenta arrows indicate the perinuclear inclusions. Scale bar, 20 μm. (C) Representative images of gastrocnemius muscle sections from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice at P50 using hematoxylin and eosin staining. Scale bar, 100 μm. (D to F) EMG recording for gastrocnemius muscle from NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 at P50. Spontaneous activity (fibrillation potentials and positive sharp waves) of the gastrocnemius muscle was observed in NOTCH2NLC-(GGC)98 mice (D). Representative images of motor unit potential (MUP) (E) and quantification of average amplitude and duration of MUP (F) in NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice. Data are presented as means ± SEM. N = 5 mice per group, **P = 0.0086 and *P = 0.0246.
NOTCH2NLC-(GGC)98 mice display behavioral phenotypes associated with NIID
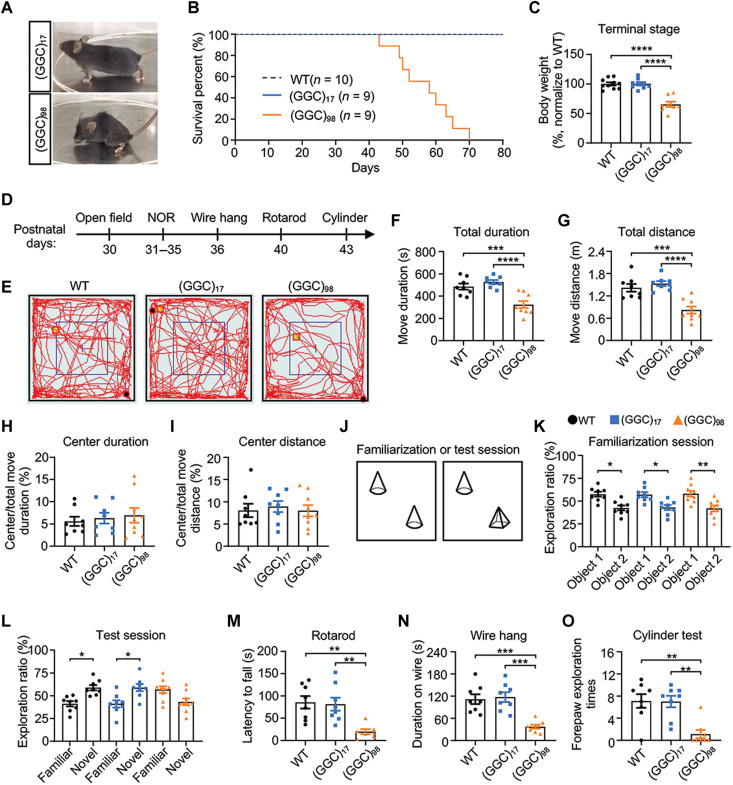
Because patients with NIID mainly display dementia and motor symptoms, we wanted to determine whether such deficits could be recapitulated in our mouse models. After 40 days of age, the NOTCH2NLC-(GGC)98 mice gradually developed progressive inactivity, hunchback, aberrant posture, ataxia (Fig. 5A and movie S1), and premature death (Fig. 5B), regardless of gender. Because the body weight of NOTCH2NLC-(GGC)98 mice decreased at the terminal stage (Fig. 5C), most behavioral tests were performed before P40 to avoid the effects of reduced body weight on locomotion (Fig. 5D).
Fig. 5. NOTCH2NLC-(GGC)98 mice display behavioral phenotypes associated with NIID.
(A) Representative image of NOTCH2NLC mice at P50. (B) Kaplan-Meier survival curve of WT and NOTCH2NLC mice. (C) Comparison of body weight of WT and NOTCH2NLC mice at terminal stage. (D) Schematic diagram of the timeline for behavioral tests. NOR, novel object recognition test. All mice used in behavioral tests were male. (E to I) Open field test in WT and NOTCH2NLC mice. (E) Representative image of moving trajectory in the open field test. Total duration (F) and total distance (G), central duration (H), and central distance (I) were measured. (J to L) Novel object recognition test in WT and NOTCH2NLC mice. Schematic diagram of novel object recognition test (J); exploration ratio, which was calculated as the ratio of time spent exploring one object over time spent exploring both objects, in familiarization session (K) and test session (L). Data are presented as means ± SEM. (M to O) Motor-related behavioral tests, including rotarod (M), wire hang (N), and cylinder test (O) in WT, NOTCH2NLC-(GGC)17, and NOTCH2NLC-(GGC)98 mice. Data are presented as means ± SEM. [WT, N = 8; NOTCH2NLC-(GGC)17, N = 8; NOTCH2NLC-(GGC)98, N = 9, *P < 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001; one-way analysis of variance (ANOVA) test with multiple comparisons].
We first performed the open-field test to measure physical/motor activity. The NOTCH2NLC-(GGC)98 mice showed markedly reduced total duration of activity and total distance traveled when compared to the control mice (Fig. 5, E to G), suggesting decreased motivation or motor ability. Meanwhile, the percentage of time spent and distance traveled in the central area displayed no significant differences, indicating that the NOTCH2NLC-(GGC)98 mice did not exhibit a heightened level of anxiety-like behaviors (Fig. 5, H and I).
The novel object recognition test is commonly used to evaluate cognitive function (29, 30). Because decreased activity of NOTCH2NLC-(GGC)98 mice was observed in the open-field test, the exploration ratio (time spent exploring one object over time spent exploring both objects) was analyzed. In familiarization sessions, two identical objects were placed in the box (Fig. 5J), and both NOTCH2NLC mice and control mice displayed a comparable preference for one object (Fig. 5K). However, when the less-preferred object was replaced by a novel object, both NOTCH2NLC-(GGC)17 mice and WT mice spent significantly more time exploring the novel object, while the NOTCH2NLC-(GGC)98 mice spent more time on the familiar object (Fig. 5L), which indicated impaired object recognition.
To evaluate motor functions, we conducted the rotarod test, wire-hang test, and cylinder test. In the rotarod test, the NOTCH2NLC-(GGC)98 mice exhibited notably poorer performance on the rod compared to the control mice (Fig. 5M). We also found that the length of time of NOTCH2NLC-(GGC)98 mice hanging on the inverted cage lid was shorter than that of control mice (Fig. 5N). In the cylinder test, the NOTCH2NLC-(GGC)98 mice could barely stand up to touch the cylinder with their forepaws (Fig. 5O). These results indicated locomotor deficits in the NOTCH2NLC-(GGC)98 mice.
Molecular signature associated with the expression of NOTCH2NLC-(GGC)98 in NOTCH2NLC transgenic mice
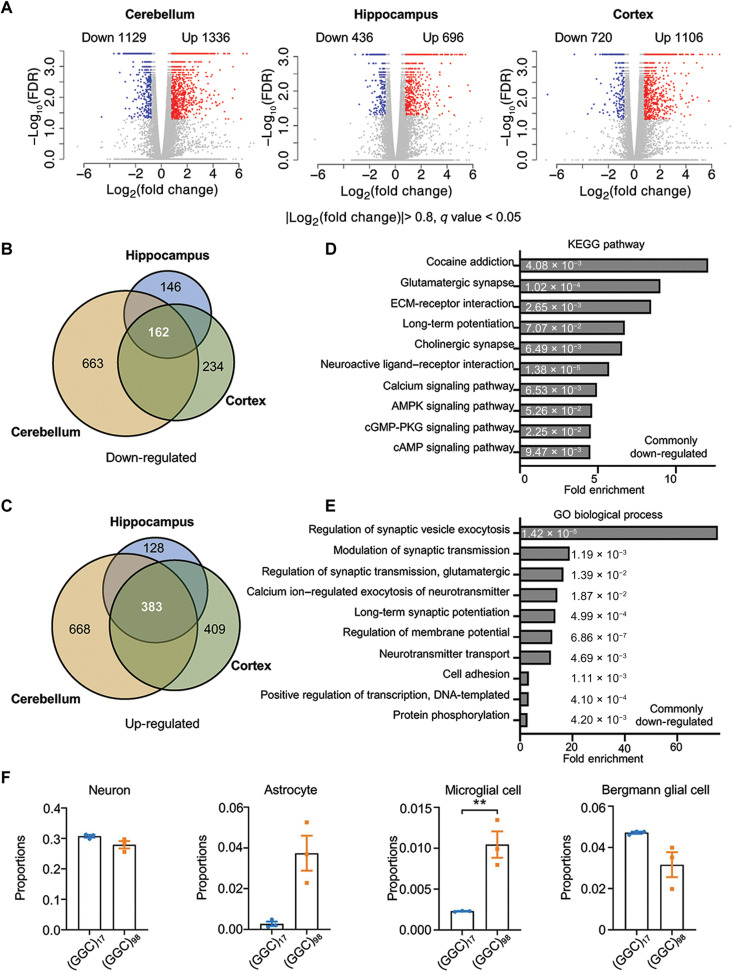
Both brain-preferential (22) and EIIa-Cre–induced ubiquitous expression of NOTCH2NLC-(GGC)98 led to NIID-like motor symptoms, suggesting that the brain is the main tissue involved in the pathogenesis of NIID. Therefore, we wanted to determine whether a molecular signature is associated with the expression of NOTCH2NLC-(GGC)98 in the brain. RNA-seq was performed on P50 tissue samples from major brain regions related to cognitive and motor functions: prefrontal cortex, cerebellum, and hippocampus. In total, 2465, 1132, and 1826 significantly differentially expressed genes (DEGs) were identified in the cerebellum, hippocampus, and cerebral cortex, respectively, when comparing NOTCH2NLC-(GGC)98 mice to the NOTCH2NLC-(GGC)17 mice (Fig. 6A and tables S1 to S3). These three major brain regions shared a large number of DEGs (Fig. 6, B and C). The commonly down-regulated genes were mainly associated with neuronal activities, such as modulation of synaptic transmission, long-term potentiation, and signal transduction, and especially involved in calcium signaling and adenosine 3′,5′-monophosphate (cAMP) signaling (Fig. 6, D and E, and fig. S9). The commonly up-regulated genes were particularly enriched in immune and cellular defense–related pathways (fig. S9A), which was consistent with increased gliosis seen in the NOTCH2NLC-(GGC)98 mouse brain (Fig. 3, J and K).
Fig. 6. Differential gene expression profiles in NOTCH2NLC-(GGC)98 mice.
(A) Shown are volcano plots of gene expression changes in the cerebellum, hippocampus, and prefrontal cortex of NOTCH2NLC-(GGC)98 mice compared to NOTCH2NLC-(GGC)17 mice. Colored dots indicate statistically significant DEGs [false discovery rate (FDR) < 0.05]. Blue dots are down-regulated genes [log2(fold change) < −0.8] and red dots are up-regulated genes [log2(foldChange) > 0.8]. Cuffdiff. N = 3 biological replicates per genotype per condition. (B and C) The numbers of down-regulated (B) and up-regulated (C) overlapping genes between the cerebellum, hippocampus, and cortex DEGs are shown in the Venn diagrams. (D and E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (D) and Gene Ontology (GO) biological process (E) enrichment analyses for the overlapping down-regulated DEGs identified from RNA-seq of NOTCH2NLC-(GGC)98 mice. The numbers on the bars are two-sided P values using Fisher’s exact. ECM, extracellular matrix; cGMP-PKG, cyclic guanosine 3’, 5’-monophosphate-dependent protein kinase G. (F) The cellular proportions of neurons, Bergmann glial cells, astrocytes, and microglial cells were estimated using MuSiC in the cerebellum samples from NOTCH2NLC-(GGC)98 mice and NOTCH2NLC-(GGC)17 mice. N = 3 mice per group, two-tailed t test, P = 0.1360 (neuron), P = 0.0540 (astrocyte), P = 0.0368 (microglia), and P = 0.1232 (Bergmann glia).
A limitation of bulk RNA-seq is that it typically measures average gene expression across many molecularly diverse cell types. Recent studies have shown that the proportions of different cell types in the brain might show correlations with various phenotypes, based on certain pathophysiological conditions (31). Hence, we investigated the proportion of each cell types in the prefrontal cortex, cerebellum, and hippocampus of NOTCH2NLC mice, using a cellular deconvolution approach that we developed previously (32–35). In all investigated brain regions, there were several prominent cell type alterations in NOTCH2NLC-(GGC)98 mice, affecting the proportion of cell types such as neurons, astrocytes, and microglia (Fig. 6F and fig. S10). A large proportional increase in astrocytes and microglial cells was consistent with the pathophysiological phenotypes of gliosis and excessive neuronal cell death (Fig. 3 and fig. S8A). Intriguingly, in the cerebellum, Bergmann glial cells appeared to decrease in proportion in NOTCH2NLC-(GGC)98 mice (Fig. 6F). Bergmann glial cells are proposed to contribute to plasticity and information processing in the cerebellum by actively interacting with Purkinje cells, through modulation of calcium signaling (36, 37). A decreased proportion of Bergmann glial cells was consistent with significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment in calcium signaling pathway genes in NOTCH2NLC-(GGC)98 mice (Fig. 6, D and E, and fig. S9, A and B).
Dysregulated AS is a common molecular alteration in both NOTCH2NLC-(GGC)98 mice and hNPCs
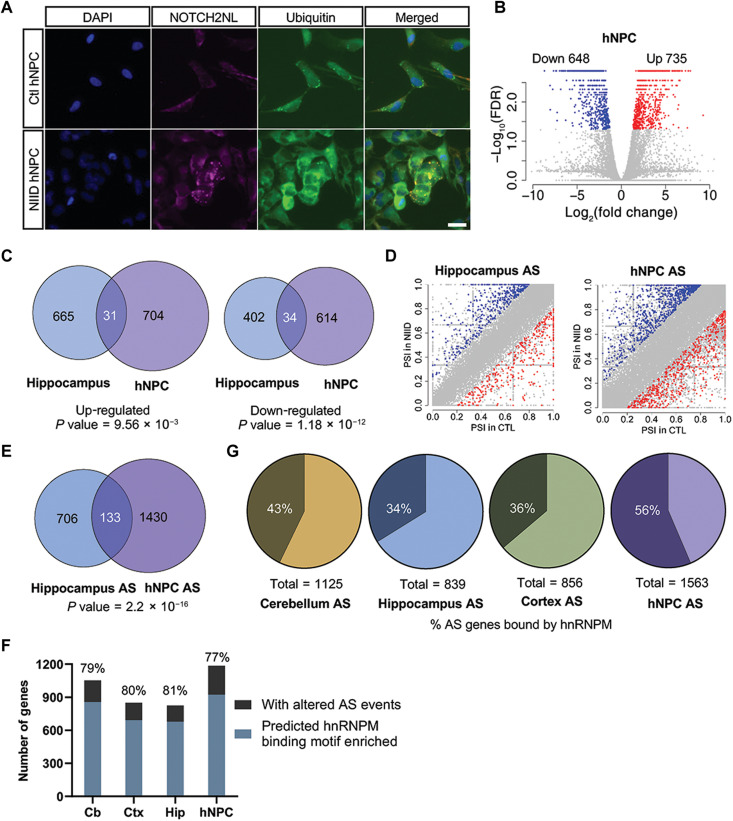
Because the NOTCH2NLC gene is only present in the human genome and not in mouse, we established iPSCs from patients with NIID and control individuals. These iPSC lines showed no obvious differences in proliferation and pluripotency gene expression (fig. S11, A and B; details about iPSCs lines were provided in table S4). The iPSC lines were then differentiated into hNPCs, which displayed ubiquitin and NOTCH2NL double-positive aggregates (Fig. 7A). In NIID hNPCs, expression of NOTCH2NLC TV1 was dominant, which was similar to what was observed in human brain tissue (fig. S11C). We performed gene expression profiling and compared these results with the findings from the NOTCH2NLC mice. Although hundreds of genes were differentially expressed in the hNPCs of NIID (Fig. 7B and table S5), we only observed a small number of overlapping DEGs between hNPCs and the NOTCH2NLC mouse model (4.2 to 8.6%; Fig. 7C and fig. S11D).
Fig. 7. Dysregulated AS is a common molecular alteration in both NOTCH2NLC-(GGC)98 mice and hNPCs.
(A) Immunostaining of NOTCH2NLC aggregates merged with ubiquitin in NIID and control hNPCs. Blue, DAPI; magenta, NOTCH2NL; green, ubiquitin. Scale bar, 10 μm. Each experiment was repeated at least three times independently. (B) Shown is a volcano plot of gene expression changes in NIID hNPCs compared to control hNPCs. Colored dots indicate statistically significant genes. Blue dots are down-regulated genes [log2(fold change) < −0.8] and red dots are up-regulated genes [log2(fold change) > 0.8] in NIID hNPCs compared to control hNPCs. Cuffdiff. N = 4 for NIID group; N = 3 for control group. (C) The overlaps of up-regulated and down-regulated DEGs between hNPCs and the hippocampus of NOTCH2NLC-(GGC)98 mouse are shown in the Venn diagrams (P value = 9.56 × 10−03 for up-regulated DEG overlaps and P value = 1.18 × 10−12 for down-regulated DEG overlaps. The P value was obtained by chi-square test). (D) Shown are plots of differential AS events in hNPCs and the hippocampus of NOTCH2NLC-(GGC)98 mice compared to the corresponding controls. N = 4 for NIID group; N = 3 for control group. P value < 0.05, inclusion level difference > 0.2, rMATS. (E) The overlaps of differential AS events between hNPCs and the hippocampus of NOTCH2NLC-(GGC)98 mice are shown in the Venn diagram. (P value = 2.2 × 10−16. The P value was obtained by chi-square test). (F) The prediction of hnRNPM binding on SE events was performed using RBPmap with high stringency. (G) The proportions of differential AS events of each sample [the cerebellum, hippocampus, or cortex of NOTCH2NLC-(GGC)98 mice and hNPCs] bound by hnRNPM are shown in the pie charts.
We next investigated alterations of AS in mouse cerebellum, cortex, hippocampus, and hNPCs upon expression of expanded GGC within NOTCH2NLC (Fig. 7D and fig. S12A). AS events were compared between mice and hNPCs (Fig. 7E, fig. S12B, and tables S6 to S9). Unlike the low overlapping rate of DEGs between them, about 15.9 to 17.3% of differential AS events in the hippocampus, cerebellum, and cortex of NOTCH2NLC-(GGC)98 mice overlapped with those in the hNPCs. To further evaluate AS changes, five types of AS events, including skipped exons (SEs), retained introns, alternative 5′ splice sites (A5SS), alternative 3′ splice sites (A3SS), and mutually exclusive exon (MXE), were analyzed by replicate multivariate analysis of transcript splicing (rMATS) on the RNA-seq data of the NOTCH2NLC mice. In total, 1125 genes in the cerebellum, 839 genes in the hippocampus, and 856 genes in the cortex showed 1532, 1089, and 1147 differential AS events, respectively (fig. S12C). In our deconvolution analysis, most of the cell population changes occurred in astrocytes and microglial cells (Fig. 6F and fig. S10, A and B). Approximately 30% of 856 genes with significantly altered AS were differentially expressed in NOTCH2NLC(GGC)98 mouse cortex.
Because the interaction partners of expanded repeat-related polypeptides have been reported to contribute to pathogenesis across several repeat expansion disorders (38), we then looked for proteins that interact with NOTCH2NLC-derived polypeptides. We overexpressed NOTCH2NLC-(GGC)17 or NOTCH2NLC-(GGC)98 in SH-SY5Y cells using lentivirus infection and then performed immunoprecipitation and mass spectrometry to identify associated proteins. A total of 41 proteins were identified exclusively in NOTCH2NLC-(GGC)98 group (fig. S13A). Intriguingly, GO enrichment analysis found that these proteins were exclusively associated with translation and RNA processing (fig. S13B), especially mRNA splicing. In this, group of 41 is the hnRNPM.
HnRNPM has been reported to be a component of the spliceosome complex and a splicing regulator for multiple genes (39–41). To gain insight into its possible role in altered patterns of AS, we examined the previously published large-scale cross-linking immunoprecipitation sequence (CLIP-seq) dataset (42). HnRNPM binding sites were enriched in UGUUG and ACAAC motifs, based on CLIP-seq data from human K562 and HepG2 cells (43). Because the exons in human and mouse are highly conserved, we used published hnRNPM CLIP-seq data from human K562 and HepG2 to look for hnRNPM binding sites flanking AS sites in mouse. The hnRNPM CLIP-seq peak files were downloaded from ENCODE (42). The hnRNPM binding sites reported in all replicates were selected by “bedtools intersect” and converted to mm9 by LiftOver. In cerebellum, about 9% of splice-out events labeled as red dots with higher percent spliced-in (PSI) in control than NIID were associated with hnRNPM, while only 5% of the splice-in events, which were labeled as blue dots in fig. S12A, were associated with hnRNPM, suggesting that hnRNPM promotes slightly more splice-out than splice-in (chi-square test P value = 0.024). Similar trends were also observed in cortex and hippocampus with no statistical significance (Fig. 7D and fig. S12A).
The predicted binding motifs of hnRNPM were highly enriched in genes with altered AS events at average 79% in all samples including the hippocampus, cerebellum, cortex of NOTCH2NLC-(GGC)98 mice, and NIID hNPCs by the analysis using RBPmap with high stringency and three consensus motifs, GGUUGGUU, UGUUGU, and UGUGU (Fig. 7F) (44). Moreover, we observed that about 34 to 43% of genes with altered AS events identified in NIID mice and as many as 56% of genes with altered AS events in hNPCs overlapped with the previously reported mRNAs bound by hnRNPM (Fig. 7G) (42). The high enrichment of hnRNPM binding sites/motifs regardless of species and consistent overlapping percentage of AS events across the samples suggested that dysregulated AS could be a common molecular alteration in both NIID mice and hNPCs.
Expanded NOTCH2NLC polypeptides interact with and sequester hnRNPM, while hnRNPM overexpression alleviates cellular toxicity
To verify the interaction of hnRNPM and NOTCH2NLC polypeptides, we performed coimmunoprecipitation and found that hnRNPM was associated with both NOTCH2NLC-polyG and NOTCH2NLC-polyA (Fig. 8A and fig. S14, A and B). Immunostaining also showed that hnRNPM was colocalized with NOTCH2NLC-polyG– and NOTCH2NLC-polyA–positive inclusions in the NOTCH2NLC-(GGC)98 mice (Fig. 8, B and C). In addition, hnRNPM was also colocalized with ubiquitin-positive inclusions in both mice and the hNPCs (Fig. 8D and fig. S14C). These results suggested that the inclusions of NOTCH2NLC-polyG and NOTCH2NLC-polyA might sequester hnRNPM from its normal function.
Fig. 8. Expanded NOTCH2NLC polypeptides interact with and sequester hnRNPM while hnRNPM overexpression alleviates cellular toxicity.
(A) Interaction of NOTCH2NLC-polyG and endogenous hnRNPM in HEK293 cells transfected with NOTCH2NLC-(GGC)17-GFP-HA or NOTCH2NLC-(GGC)98-GFP-HA was determined by coimmunoprecipitation. (B and C) Shown are the images of immunofluorescence staining against hnRNPM and NOTCH2NLC-polyG (B) or NOTCH2NLC-polyA (C) in the hippocampus of NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice. Green, NOTCH2NLC-polyG/polyA; magenta, hnRNPM; blue, DAPI. Scale bars, 20 μm. (D) Shown are representative images of immunofluorescence staining against hnRNPM in hNPCs. Green, hnRNPM; magenta, ubiquitin; blue, DAPI. Scale bar, 20 μm. (E) Cell viability of Neuro-2a cells transfected with NOTCH2NLC-GFP plus control or hnRNPM constructs was analyzed by the CCK-8 method. Data are presented as means ± SEM. (N = 4 independent experiments, one-way ANOVA followed with Tukey’s multiple comparisons test was performed, *P = 0.0327). n.s., not significant. (F and G) Cell viability of cultured mouse primary cortical neurons transfected with NOTCH2NLC-GFP plus control or hnRNPM constructs. Data are presented as means ± SEM (N = 4 independent experiments, at least 120 cells were counted per experiment, one-way ANOVA followed with Tukey’s multiple comparisons test was performed, ***P < 0.0005, ****P < 0.0001). (F) Representative immunofluorescence image. Green, GFP; magenta, Tuj1; gray, His. Scale bar, 10 μm.
If the sequestration of hnRNPM could affect its physiological functions and cause neuronal toxicity, then overexpression of hnRNPM might mitigate toxicity associated with the expanded GGC repeats. We tested this possibility using our established neuronal models. HnRNPM was transiently cotransfected with NOTCH2NLC-(GGC)17 or NOTCH2NLC-(GGC)98 constructs into cultured primary mouse neurons and Neuro-2a cells using the same evaluation criteria (Fig, 1, N to O, and fig. S3, B and C). The expression of hnRNPM with NOTCH2NLC-(GGC)98 could increase cellular viability and significantly improve neurite outgrowth as compared to expressing the NOTCH2NLC-(GGC)98 alone (Fig. 8, E to G). These results together confirmed the involvement of hnRNPM in neurodegeneration associated with the expression of expanded GGC repeats in NOTCH2NLC.
DISCUSSION
NIID, a progressive neurodegenerative disorder characterized by widespread intranuclear inclusions in the nervous system and multiple visceral organs, is caused by the expansion of GGC repeats in the 5′UTR of the human-specific NOTCH2NLC gene. The underlying molecular mechanism of NIID remains unclear. With the first transgenic mouse model of NIID, we have demonstrated that the expression of expanded GGC repeats leads to widespread intranuclear inclusions, severe neurodegeneration, motor dysfunction, and cognitive deficit. Furthermore, we established an hNPC cell model of NIID and identified a pattern of overlapping AS events between the NIID mouse and hNPC cell models by transcriptome analysis. Motif analyses revealed the enrichment for the binding motifs of hnRNPM among the AS sites. Mechanistically, we showed that NOTCH2NLC polypeptides might sequester hnRNPM and disturb its function, while overexpression of hnRNPM could ameliorate cellular toxicity caused by expanded GGC repeats within NOTCH2NLC. These results together suggested that dysfunction of hnRNPM could play a vital role in the molecular pathogenesis of NIID.
In this study, we developed an exon 1 but not the full gene model based on the following factors. Although NOTCH2NLC has two different splicing isoforms, our results suggested that TV1 was the dominant isoform in human brain, hNPCs, and peripheral blood, while the 5′UTR (exon 1 plus partial exon 2) and coding sequences (CDS) of TV1 may produce two separate proteins (figs. S1 and S11C). Furthermore, evidence from our study and others indicated that the 5′UTR sequence of TV1 restrained the expression of CDS of NOTCH2NLC (23). Therefore, we hypothesized that the 5′UTR of TV1 may produce the dominant product of the NOTCH2NLC gene. Although partial exon 2 sequences (36 bp) were not included in the model, our models faithfully mimicked the clinical manifestation and pathological features associated with NIID patients.
Compared to the previously reported AAV-injected mouse model (22), our transgenic mouse model displayed more severe pathological and behavioral phenotypes and premature death (around 2 months old), which may occur for several reasons. First, the expression of NOTCH2NLC with pathogenic GGC repeats began at 2 months of age in the AAV-injected mouse model, whereas expression started during embryonic development in the genetic mouse model, which was more consistent with the expression timing of NOTCH2NL genes in humans (15, 16). Second, in our genetic mouse model, NOTCH2NLC with pathogenic GGC repeats produced not only polyG- but also polyA- and polyR-containing proteins, which might contribute to the phenotypes of the mice. Third, NOTCH2NLC is expressed more broadly in the genetic mouse model. Consequently, NOTCH2NLC polypeptide inclusions could cause lesions in other tissues besides the brain such as the muscle, a tissue linked to NOTCH2NLC-related GGC repeat disorders (45, 46). Last, the constructs we used were directly cloned from a patient with NIID and thus harbor GGA interruptions, while the constructs Boivin et al. (22) used were homogeneous synthetic GGC repeats. It has been reported that different expansion compositions may cause different phenotypes in patients with NIID (6). Thus, the matched expansion composition and spatiotemporal expression pattern of NOTCH2NLC in the transgenic NIID mouse made it a suitable genetic model to investigate the pathogenesis of NIID. The issue that disease-related phenotypes progressed rapidly in our EIIa; NOTCH2NLC mouse model should be treated with caution, as we could not rule out toxic effects in other tissues. In the future, spatiotemporally directed expression of NOTCH2NLC may help to investigate the role of NOTCH2NLC with expanded GGC repeats in specific cell types or tissues or at different ages.
In our study, we observed a characteristic expression pattern for NOTCH2NLC-derived proteins in NIID cellular and animal models. In the cellular model, NOTCH2NLC-(GGC)98 produced polyG and polyA but not polyR, while in the mouse model, protein production of NOTCH2NLC-(GGC)98 displayed a tissue or cell type–specific expression pattern. Three types of polypeptides (polyG/A/R) were produced in the brain, but only polyG and polyR were observed in the muscle. In addition, we observed age-dependent and brain region–specific expression of polypeptides. Specifically, polyG inclusions were first found in the nucleus and then appeared both inside and outside the nucleus with age. It is reasonable to postulate that large aggregates cannot pass through the nuclear pore and remain in the cytoplasm, eliciting complex and adverse effects on multiple cellular functions. In our NOTCH2NLC mouse model, polypeptide inclusions were not detected at P2 and began to appear in the striatum and hippocampus at P12, while obvious neuronal loss was observed at P32, indicating that the generation and accumulation of polypeptide inclusions precede neurological symptoms. These results might be ascribed to age-related cellular stress, because misfolded proteins are known to accumulate with age and this accumulation amplifies the level of cellular toxicity, as is seen with other proteins associated with neurodegenerative diseases, such as Huntingtin and C9ORF72 dipeptide repeat proteins (47, 48).
Using the NIID mouse model developed here, we performed detailed transcriptome analysis on the cerebellum, cerebral cortex, and hippocampus, the major brain regions involved in movement, coordination, learning, and memory. Intriguingly, these brain regions shared a large number of DEGs and enriched GO and KEGG pathway categories among dysregulated genes, converging on calcium signaling and cAMP signaling pathways. It is well documented that subtle changes in calcium homeostasis can cause aberrant alterations in intracellular Ca2+, contributing to cellular and especially neuronal dysfunction, and the onset of a range of human diseases like ALS and Huntington’s disease (49). These results suggested the potential involvement of the abnormal calcium signaling pathway in NIID pathogenesis.
Our cellular and mouse models—based on NPCs and p50 mouse brain—reflect early and mature stages of brain development, respectively. Comparative transcriptomic analyses of our NIID mouse model and hNPCs revealed a small number of overlapping DEGs, but they shared a higher proportion of aberrant AS events. The high percentage of overlapping AS events in two models representing different developmental stages and from different species implies that dysregulated AS may play a vital role in the pathogenesis of NIID.
The spliceosome machinery that regulates AS is composed of a large number of elements, such as small nuclear ribonucleoproteins and RNA binding proteins like serine and arginine-rich proteins and hnRNPs, including hnRNPM (50). Here, we found that hnRNPM could bind to the AS sites that we identified in NIID hNPCs and mice. Meanwhile, hnRNPM was colocalized with polyG and polyA inclusions in the NIID mice and interacted with both polyG and polyA in vivo. These data suggested that polypeptides produced by NOTCH2NLC with expanded GGC repeats could potentially sequester hnRNPM and affect hnRNPM-mediated RNA splicing. Consistent with this model, coexpression of hnRNPM could ameliorate cellular toxicity caused by expanded GGC repeats within NOTCH2NLC. These findings together indicated a key role for hnRNPM in NIID pathogenesis.
In summary, we have developed both mouse and hNPCs models for NIID, which faithfully mimicked the clinical manifestations and pathological features of patients with NIID. Our molecular analyses revealed that dysregulated AS was involved in the neurodegeneration and identified hnRNPM as the fulcrum molecule in the pathogenesis of NIID, providing a promising therapeutic target for the treatment of NIID.
MATERIALS AND METHODS
Human samples
This study was approved by the Ethics Committee of Xiangya Hospital of the Central South University in China. Human skin samples used for generation of fibroblast lines and human peripheral blood were collected in a previous study (5). Written informed consent was obtained from all individuals.
Animals
The conditional transgenic NOTCH2NLC-(GGC)17 and NOTCH2NLC-(GGC)98 mice were generated by a CRISPR-Cas9 system in a C57BL/6J mouse background. Briefly, Cas9 mRNA was in vitro–transcribed with the mMESSAGE mMACHINET7 Ultra Kit (Ambion, TX, USA) according to the manufacturer’s instructions. 5′-GGG GAC ACA CTA AGG GAG CT-3′ was chosen as the Cas9-targeted single guide RNA (sgRNA) and in vitro–transcribed using the MEGAshortscript Kit (Thermo Fisher Scientific, USA). The Rosa26 targeting vector, sgRNA, and Cas9 mRNA were microinjected into C57BL/6J fertilized eggs. The microinjection was performed by Shanghai Model Organisms Center Inc. F0 generation mice positive for homologous recombination were crossed with C57BL/6J mice to obtain germline conditional transgenic mice. To express NOTCH2NLC-(GGC)17 or NOTCH2NLC-(GGC)98 ubiquitously, conditional transgenic mice were crossed with EIIa-Cre mice. The EIIa-Cre mice was obtained from The Jackson Laboratory and backcrossed into the C57BL/6 background for 13 generations. Primers used for genotyping are listed in the Supplementary Materials. All mice were bred and maintained in the animal facility under specific pathogen–free conditions in accordance with the institutional guidelines of the Animal Care and Use Committee at Central South University.
Derivation of iPSC and differentiation to NPCs
The generated fibroblasts from patient skin biopsies were reprogrammed into iPSCs using a CytoTune-iPS 2.0 Sendai reprogramming kit (Thermo Fisher Scientific, A16517) to derive integration-free iPSCs. Briefly, three Sendai virus–based reprogramming vectors, including Yamanaka factors, klf4, oct4, sox2, hc-Myc, and hKlf4, were proportionally mixed and applied to the fibroblasts (51). The transduced cells were cultured in fibroblast medium [Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal bovine serum (FBS) and 1% minimum essential medium (MEM) nonessential amino acid solution] for 8 days, and transduced fibroblasts were passed to mouse embryo fibroblast (MEF) dishes with fibroblast medium on day 7. Medium was switched to iPSC medium [DMEM/F-12 with GlutaMAX supplemented with 20% KnockOut Serum Replacement, 1% MEM nonessential amino acid solution, 100 μM β-mercaptoethanol, 1% penicillin/streptomycin, and basic fibroblast growth factor (10 ng/ml)] from day 8. The emergence of iPSC colonies was monitored, and they were manually picked and passed to 24-well MEF plates. After that, iPSCs were expanded to Matrigel-coated six-well plates and maintained on Matrigel in mTeSR1 medium (STEMCELL Technologies, 85850) and passaged with ReLeSR (STEMCELL Technologies, 05873) to selectively pass the undifferentiated iPSCs. Then, iPSCs were differentiated to NPCs using the STEMdiff SMADi Neural Induction Kit (STEMCELL Technologies, 08581) via a monolayer protocol. The generated NPCs were maintained in neural progenitor medium (STEMCELL Technologies, 05833) on Martigel and used for the immunocytochemistry, RNA-seq, and Western blot experiments. HEK293 and Neuro-2a cells were cultured in DMEM medium supplemented with 10% FBS and penicillin/streptomycin (100 U/ml) at 37°C with 5% CO2.
Constructs
The human exon 1 of NOTCH2NLC with 17 GGC repeats or 98 GGC repeats were cloned into pCDNA3.1(+) vector using polyG frame–, polyA frame–, and polyR frame–specific primers, respectively. A GFP-HA tag was then fused to the aforementioned three frames to obtain NOTCH2NLC-(GGC)17-GFP and NOTCH2NLC-(GGC)98-GFP constructs (polyG, polyA, and polyR frames, respectively). Primers used for constructing NOTCH2NLC constructs are listed as follows: Exon 1_Common_F: ATA GGT ACC ACC GGT GCT GAG GCG GCG GCC GAG GAG CG, Exon 1_polyG frame_R: ATA GGA TCC GCG CGG GGG TCG CGC AGC AC, Exon 1_polyA frame_R: ATA GGA TCC TGC GCG GGG GTC GCG CAG CAC, Exon 1_polyR frame_R: ATA GGA TCC ATG CGC GGG GGT CGC GCA GCA, polyG frame_ATG to AAG_F: TCC CCA AGT GGA TCT GCC CA, and polyG frame_ATG to AAG_R: GGG GGT CCC GAT AGA GGA GC. To make the Rosa26 donor vector, the GFP-HA tag of NOTCH2NLC-GFP-HA (polyA frame) was replaced with a 3*HA/3*Myc/3*Flag (3Tag) tag. Then, NOTCH2NLC-(GGC)17-3Tag or NOTCH2NLC-(GGC)98-3Tag were digested by Age I and Ecor V and inserted into the Rosa26 targeting vector, which contains cytomegalovirus immediate enhancer/b-actin (CAG)–loxP-stop-loxP–Age I–Ecor V–woodchuck hepatitis virus posttranscriptional regulatory element–bovine growth hormone (bGH) poly(A). The hnRNPM construct was generated by cloning complementary DNA (cDNA) of human hnRNPM (NM_ 005968) from human first-strand cDNA into a pCDNA3.1-3*Flag vector with the following primers: hnRNPM_F: CG GGA TCC ATG GCG GCA GGG GTC GAA; and hnRNPM_R: GC TCT AGA TTA AGC GTT TCT ATC AA. All constructs were confirmed by sequencing.
Antibodies
Primary antibodies used in this study include the following: Flag (Sigma-Aldrich, F1804), HA (Cell Signaling Technology, 3724S), Myc (Cell Signaling Technology, 2276S), calbindin (Sigma-Aldrich, C9848), vinculin (Sigma-Aldrich, V9131), NeuN (Cell Signaling Technology, 24307s), GFAP (Cell Signaling Technology, 3670S), IBA1 (Wako, 019-19741), ubiquitin (Sigma-Aldrich, 05-1307), P62 (BD Biosciences, 610832), GFP (Invitrogen, A-11122), β-actin (Cell Signaling Technology, 3700S), Tuj1 (Abcam, ab18207), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Proteintech, 60004-1-Ig), NOTCH2NL (Gentex, GTX117659), and hnRNPM (Santa Cruz Biotechnology, sc-20002). Secondary antibodies were donkey anti-rabbit and donkey anti-mouse Alexa Fluor 488 or 594 from Jackson ImmunoResearch.
Generation of anti-NOTCH2NLC polypeptide antibodies
Rabbit polyclonal anti–NOTCH2NLC-polyG (PEP122) and anti–NOTCH2NLC-polyA (PEP123) antibodies were developed commercially. Briefly, rabbits were sequentially immunized with synthetic peptide CAARPPRMH (polyG) and EEAATEKMPALRRSAVG (polyA) for three times. One week after the third injection, the sera were tested by enzyme-linked immunosorbent assay. Rabbits with eligible titer were immunized again, and the sera were purified through affinity purification and further validated by Western blot. The sera from the rabbit (PEP122) and rabbit (PEP123) were collected and purified to generate anti–NOTCH2NLC-polyG (PEP122) and anti–NOTCH2NLC-polyA (PEP123) antibodies.
Cell culture, transfection, and cell viability assay
HEK293 cells, Neuro-2a cells, and SH-SY5Y cells were obtained from the National Collection of Authenticated Cell Culture of China (HEK293 accession number: GNHu43; Neuro-2a accession number: TCM29; SH-SY5Y accession number: SCSP-5014). HEK293 cells and Neuro-2a cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin (100 U/ml) at 37°C with 5% CO2. Plasmid transient transfection on HEK293 cells and Neuro-2a cells was performed using Lipofectamine 2000 reagent following the manufacturer’s instruction. Cell viability assay was performed using the Cell Counting Kit-8 (CCK-8) following the manufacturer’s instruction. Twenty-four hours after transfection, Neuro-2a cells were cultured in DMEM with 1% FBS and supplemented with retinoic acid at final concentration of 25 mM for another 72 hours to induce neurite outgrowth. The Neuro-2a cells were fixed with 4% paraformaldehyde and stained with Tuj1. The neurite length of GFP-positive cells was measured by ImageJ software using a previously reported method (52). SH-SY5Y cells were cultured in DMEM/F-12 supplemented with 10% FBS and penicillin/streptomycin (100 U/ml) at 37°C with 5% CO2. Twenty-four hours after seeding, lentivirus infection was performed at a multiplicity of infection (20) for 12 hours. For mouse primary cortical neuron culture, the cortex from embryonic day 16 (E16) and E17 mice was dissected and digested by 0.025% trypsin (Thermo Fisher Scientific) at 37°C for 25 min. After washing twice with Hanks’ balanced salt solution, the cortical tissue was triturated with Pasteur pipettes, and cells were plated on Lab-Tek chamber slides (Thermo Fisher Scientific) in Neurobasal medium (Thermo Fisher Scientific) supplemented with B27, GlutaMAX, and penicillin/streptomycin. For cell viability and morphology, primary cortical neurons were transfected at days in vitro 2 using Lipofectamine 2000 reagent following the manufacturer’s instruction. After 48 hours, neurons were fixed with 4% paraformaldehyde and stained with Tuj1.
Mass spectrometry analysis
Forty-eight hours after transfection, protein was extract from HEK293 cells (72 hours transfection for SH-SY5Y cells), treated with dithiothreitol, alkylated with iodoacetamide, reconstituted in Triethylammonium bicarbonate (TEAB), hydrolyzed with trypsin, and lastly desalted by C18 SPE column. The tryptic peptides were dissolved and directly loaded onto a reversed-phase analytical column. Peptides were separated with a gradient formic acid and acetonitrile on an EASY-nLC 1200 ultra performance liquid chromatography system (Thermo Fisher Scientific). The separated peptides were analyzed in Q Exactive HF-X (Thermo Fisher Scientific) with a nano-electrospray ion source. The resulting tandem mass spectrometry data were processed using MaxQuant search engine (v.1.6.15.0). Tandem mass spectrometry was searched against the human SwissProt database (20,422 entries) and homemade database of three potential translated peptides from exon 1 of NOTCH2NLC concatenated with reverse decoy database. The mass tolerance for precursor ions was set at 20 parts per million (ppm) in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set at 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification, and oxidation on Met was specified as variable modifications. False discovery rate was adjusted to <1%.
Mouse behavioral analysis
Mouse body weight was measured once every 2 days, and survival was monitored regularly. There was no gender difference in the survival time. At least eight male mice per genotype were included for the behavioral tests. Open field test was performed in a perspex box (70 cm by 70 cm by 31 cm) with the floor divided into 16 equal squares. The movement of mice during a 10-min test was filmed and analyzed by an automated behavior analysis system. Total moving duration and distance and the moving duration and distance in the central four squares were compared between different groups. Novel object recognition test was performed using a modified method (29, 30). Briefly, in habituation session, mice were habituated for 5 min in an empty perspex box (30 cm by 30 cm by 30 cm) for three consecutive days (habituation session). On the fourth day (familiarization session), two identical objects were placed at the opposite ends of the box. Mice were allowed free exploration for 10 min, and the amount of time exploring each object was recorded. Twenty-four hours later (test session), the less preferred object was replaced by a novel object, and again, the exploratory time to each object was recorded. The exploratory ratio was calculated as the ratio of time spent exploring one object over time spent exploring both objects. In the wire hang test, mice were placed on a wire lid of a mouse cage, which was then inverted. During a 3-min test, the latency to fall was recorded. Each mouse was tested three times, and the longest hang time was recorded. In the rotarod test, mice were trained for 3 min on the rotarod for three trials a day for three consecutive days (5 rpm for day 1, 10 rpm for day 2, and 15 rpm for day 3). On the testing day, a fixed-speed test at 20 rpm for 3 min was performed three times for each mouse (10-min intertrial interval), and latency to fall was recorded for each trial. In the cylinder test, mice were placed in a transparent cylinder and filmed with video from the side. The times of standing and touching the cylinder wall with forepaws for each mouse were recorded.
Immunofluorescent staining, Nissl staining, and electron microscopy
Mice were anesthetized and perfused intracardially with 0.9% saline solution, followed by 4% paraformaldehyde in 0.1 M phosphate buffer at pH 7.2. Isolated mouse brains were dehydrated in 30% sucrose at 4°C and sectioned at 30 μm for subsequent immunofluorescence study: Brain sections were blocked in 3% bovine serum albumin in 0.3% Triton X-100/phosphate-buffered saline (PBS) for 60 min, followed by incubation with primary antibodies overnight at 4°C. After incubation with the fluorescent secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI), the brain sections were mounted to coated glass slides and examined using a Leica DMi8 fluorescence microscope and a Zeiss LSM-880 confocal microscope. For hNPC immunostaining, cells grown on coverslips were fixed with 4% paraformaldehyde for 15 min at room temperature, and 5% normal goat serum and 0.5% Triton X-100 in PBS were used for whole staining processes. Incubation with primary antibodies was overnight at 4°C and followed by fluorescent secondary antibody incubation for 45 min at room temperature. Pictures were taken by a Zeiss AX10 fluorescence microscope. For Nissl staining, the paraffin sections were dewaxed with xylene and washed with graded ethanol, followed by incubation with 1% toluidine blue for 40 min and then washed with ultrapure water. After dehydration with gradient ethanol and vitrification by xylene for 5 min (twice), the slices were sealed with neutral gum. Images were taken by Leica DMi8. Electron microscopic study was performed as previously reported (5); prepared sections were examined and photographed on a Hitachi HT-7700 electron microscope.
Coimmunoprecipitation and Western blot
Methods for coimmunoprecipitation and Western blot were described previously (38). Cells were harvested in 1% NP-40 lysis buffer [150 mM NaCl, 1% NP-40, 2 mM EDTA, and 50 mM tris (pH 8.0)] with protease inhibitor and phosphatase inhibitor cocktails 48 hours after transfection. Approximately 500 μg of protein was incubated overnight at 4°C with the primary antibody. Protein A beads (20 μl) were used to capture the antibody-protein complex, which was then analyzed by Western blot.
Protein samples were separated by SDS–polyacrylamide gel electrophoresis and were then transferred to a polyvinylidene difluoride membrane (Millipore). After blocking, blots were probed with the appropriate antibodies overnight at 4°C. After PBS wash, protein bands were developed using an ECL Prime Chemiluminescence kit (GE Healthcare) on a Bio-Rad developing and imaging platform.
Electromyography
Five male mice per genotype were included for EMG. The concentric needle electrode was inserted without anesthesia into the quadriceps, and a ground electrode was placed on the tail. The EMG signal was amplified and recorded by DANTEC Keypoint 9033A07 machine (sweep speed = 10 ms/division; sensitivity = 0.2 mV/division), and 10 different motor unit potentials (MUPs) per mice were recorded. Average MUP amplitude and duration of motor unit action potential were calculated.
Reverse transcriptase PCR and quantitative real-time PCR
Total RNA was extracted from mouse brain tissue using a TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized from total RNA (1 μg) using the RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1622) and oligo dT primers. Quantitative real-time PCR was analyzed with the comparative cycle threshold method. Primers used for quantitative real-time PCR were listed as follows: for NOTCH2NLC, NOTCH2NLC_3Tag_F: GGG GAG TCG AGG CAT TTG and NOTCH2NLC_3Tag_R: CAC TCT CTC CTC CCC TCC TC; for GAPDH, GAPDH_F: CAT GGC CTC CAA GGA GTA AGA AAC and GAPDH_R: TGT GAG GGA GAT GCT CAG TG.
RNA-seq and analysis
Tissues from P50 male mice or hNPCs were used for RNA extraction. An amount of 1 μg of total RNA per sample was processed for library preparation using the NEBNext UltraTM RNA Library Prep Kit for Illumina (New England Biolabs, USA) following the manufacturer’s recommendations. Libraries were sequenced on a HiSeq with a read length configuration of 150 paired end (PE), targeting 100 M total reads per sample.
All total reads were mapped to hg38/mm9 by TopHat2 with default parameters, followed by gene expression quantification and differential expression analysis using Cuffdiff. Genes with an adjusted P value < 0.05 and |log2(fold change)| > 0.8 were assigned as significantly differentially expressed. KEGG pathways and GO enrichment analysis were performed by DAVID or PANTHER online. The cellular proportions of neurons, Bergmann glial cells, astrocytes, and microglial cells were estimated using MuSiC (33, 34).
The AS events, including SE, IR, MXE, A3SS, and A5SS, were reported by rMATS (53). Splicing events with a P value < 0.05 and inclusion level difference > 0.2 were considered as significant AS events. For the hnRNPM analysis, we used published hnRNPM CLIP-seq data from human K562 and HepG2 as hnRNPM binding sites in a flanking region of AS sites in a mouse. The hnRNPM CLIP-seq peak files were downloaded from ENCODE (42). The hnRNPM binding sites reported in all replicates were selected by bedtools intersect and converted to mm9 by LiftOver. The prediction of hnRNPM binding was performed using RBPmap with high stringency and three consensus motifs, GGUUGGUU, UGUUGU, and UGUGU (44).
Quantification and statistical analysis
To eliminate bias, images or animal analyses were either completely automated or blinded. All statistical analyses were performed with two-tailed Student’s t test or the one-way analysis of variance (ANOVA) analysis (Tukey’s multiple comparisons test). All quantification data were presented as means ± SEM. A P value of less than 0.05 was considered statistically significant.
Acknowledgments
We would like to thank K. Yuan and F. Chen from Xiangya Hospital, Central South University and J.-Q. Tan from School of Life Sciences, Central South University for providing advice and technical assistance to the study and Q. Eastman for critical reading of the manuscript and editing.
Funding: This work was supported, in part, by the National Natural Science Foundation of China (U20A20355 to B.T, 32071037 and 81701281 to Q.L., 82171843 to Y.P., and 81730036 to K.X.), the National Institutes of Health (HD104458 and HD104463 to P.J. and Z.W. and NS051630 and NS111602 to P.J.), and the FRAXA Research Foundation (to Y.K.).
Author contributions: B.T. and P.J. designed the study, supervised the study, and revised the manuscript. Y.P. and Q.L. designed the study, performed cell and animal experiments, and wrote the manuscript. Y.K. and K.Z. performed iPSC induction, differentiation, and experiments on NPCs, led the molecular aspects of the project, and wrote the manuscript. P.D., Yujing Li., and Y.Z. contributed to plasmid construction. Yangping Li and Z.L. contributed to RNA-seq analysis. Yun. T., Q.S., and Yu. T. contributed to providing skin tissue and establishing fibroblasts. Q.M., J.-L.W., J.G., K.X., J.-D.L., H.J., L.S., R.D., E.G.A., Z.W., and B.Y. provided advice and technical assistance to the study.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: We have deposited the RNA-seq data into the Gene Expression Omnibus (GEO) at https://ncbi.nlm.nih.gov/geo/. The accession number is GSE182878. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S9
Movie S1
REFERENCES AND NOTES
- 1.Ishiura H., Shibata S., Yoshimura J., Suzuki Y., Qu W., Doi K., Almansour M. A., Kikuchi J. K., Taira M., Mitsui J., Takahashi Y., Ichikawa Y., Mano T., Iwata A., Harigaya Y., Matsukawa M. K., Matsukawa T., Tanaka M., Shirota Y., Ohtomo R., Kowa H., Date H., Mitsue A., Hatsuta H., Morimoto S., Murayama S., Shiio Y., Saito Y., Mitsutake A., Kawai M., Sasaki T., Sugiyama Y., Hamada M., Ohtomo G., Terao Y., Nakazato Y., Takeda A., Sakiyama Y., Umeda-Kameyama Y., Shinmi J., Ogata K., Kohno Y., Lim S.-Y., Tan A. H., Shimizu J., Goto J., Nishino I., Toda T., Morishita S., Tsuji S., Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 51, 1222–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Deng J., Yu J., Li P., Luan X., Cao L., Zhao J., Yu M., Zhang W., Lv H., Xie Z., Meng L., Zheng Y., Zhao Y., Gang Q., Wang Q., Liu J., Zhu M., Guo X., Su Y., Liang Y., Liang F., Hayashi T., Maeda M. H., Sato T., Ura S., Oya Y., Ogasawara M., Iida A., Nishino I., Zhou C., Yan C., Yuan Y., Hong D., Wang Z., Expansion of GGC repeat in GIPC1 is associated with oculopharyngodistal myopathy. Am. J. Hum. Genet. 106, 793–804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi-Fujigasaki J., Neuronal intranuclear hyaline inclusion disease. Neuropathology 23, 351–359 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Sone J., Mori K., Inagaki T., Katsumata R., Takagi S., Yokoi S., Araki K., Kato T., Nakamura T., Koike H., Takashima H., Hashiguchi A., Kohno Y., Kurashige T., Kuriyama M., Takiyama Y., Tsuchiya M., Kitagawa N., Kawamoto M., Yoshimura H., Suto Y., Nakayasu H., Uehara N., Sugiyama H., Takahashi M., Kokubun N., Konno T., Katsuno M., Tanaka F., Iwasaki Y., Yoshida M., Sobue G., Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain 139, 3170–3186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y., Wang J.-L., Huang W., Zeng S., Jiao B., Liu Z., Chen Z., Li Y., Wang Y., Min H.-X., Wang X.-J., You Y., Zhang R.-X., Chen X.-Y., Yi F., Zhou Y.-F., Long H.-Y., Zhou C.-J., Hou X., Wang J.-P., Xie B., Liang F., Yang Z.-Y., Sun Q.-Y., Allen E. G., Shafik A. M., Kong H. E., Guo J.-F., Yan X.-X., Hu Z.-M., Xia K., Jiang H., Xu H.-W., Duan R.-H., Jin P., Tang B.-S., Shen L., Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am. J. Hum. Genet. 105, 166–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sone J., Mitsuhashi S., Fujita A., Mizuguchi T., Hamanaka K., Mori K., Koike H., Hashiguchi A., Takashima H., Sugiyama H., Kohno Y., Takiyama Y., Maeda K., Doi H., Koyano S., Takeuchi H., Kawamoto M., Kohara N., Ando T., Ieda T., Kita Y., Kokubun N., Tsuboi Y., Katoh K., Kino Y., Katsuno M., Iwasaki Y., Yoshida M., Tanaka F., Suzuki I. K., Frith M. C., Matsumoto N., Sobue G., Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 51, 1215–1221 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sun Q.-Y., Xu Q., Tian Y., Hu Z.-M., Qin L.-X., Yang J.-X., Huang W., Xue J., Li J.-C., Zeng S., Wang Y., Min H.-X., Chen X.-Y., Wang J.-P., Xie B., Liang F., Zhang H.-N., Wang C.-Y., Lei L.-F., Yan X.-X., Xu H.-W., Duan R.-H., Xia K., Liu J.-Y., Jiang H., Shen L., Guo J.-F., Tang B.-S., Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. Brain 143, 222–233 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Yuan Y., Liu Z., Hou X., Li W., Ni J., Huang L., Hu Y., Liu P., Hou X., Xue J., Sun Q., Tian Y., Jiao B., Duan R., Jiang H., Shen L., Tang B., Wang J., Identification of GGC repeat expansion in the NOTCH2NLC gene in amyotrophic lateral sclerosis. Neurology 95, e3394–e3405 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Shi C. H., Fan Y., Yang J., Yuan Y. P., Shen S., Liu F., Mao C. Y., Liu H., Zhang S., Hu Z. W., Fan L. Y., Li M. J., Fan S. H., Liu X. J., Xu Y. M., NOTCH2NLC intermediate-length repeat expansions are associated with Parkinson disease. Ann. Neurol. 89, 182–187 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Yan Y., Cao L., Gu L., Zhang B., Xu C., Pu J., Tian J., Yin X., Zhang B., Zhao G., Assessing the NOTCH2NLC GGC expansion in essential tremor patients from eastern China. Brain 144, e1 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Okubo M., Doi H., Fukai R., Fujita A., Mitsuhashi S., Hashiguchi S., Kishida H., Ueda N., Morihara K., Ogasawara A., Kawamoto Y., Takahashi T., Takahashi K., Nakamura H., Kunii M., Tada M., Katsumoto A., Fukuda H., Mizuguchi T., Miyatake S., Miyake N., Suzuki J., Ito Y., Sone J., Sobue G., Takeuchi H., Matsumoto N., Tanaka F., GGC repeat expansion of NOTCH2NLC in adult patients with leukoencephalopathy. Ann. Neurol. 86, 962–968 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Fang P., Yu Y., Yao S., Chen S., Zhu M., Chen Y., Zou K., Wang L., Wang H., Xin L., Hong T., Hong D., Repeat expansion scanning of the NOTCH2NLC gene in patients with multiple system atrophy. Ann. Clin. Transl. Neurol. 7, 517–526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao B., Zhou L., Zhou Y., Weng L., Liao X., Tian Y., Guo L., Liu X., Yuan Z., Xiao X., Jiang Y., Wang X., Yang Q., Li C., Zhu Y., Zhou L., Zhang W., Wang J., Li Y., Gu W., Yang J., Xia J., Huang Q., Yin J., Xue J., Duan R., Tang B., Shen L., Identification of expanded repeats in NOTCH2NLC in neurodegenerative dementias. Neurobiol. Aging 89, 142.e1–142.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Yu J., Deng J., Guo X., Shan J., Luan X., Cao L., Zhao J., Yu M., Zhang W., Lv H., Xie Z., Meng L., Zheng Y., Zhao Y., Gang Q., Wang Q., Liu J., Zhu M., Zhou B., Li P., Liu Y., Wang Y., Yan C., Hong D., Yuan Y., Wang Z., The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain 144, 1819–1832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiddes I. T., Lodewijk G. A., Mooring M., Bosworth C. M., Ewing A. D., Mantalas G. L., Novak A. M., van den Bout A., Bishara A., Rosenkrantz J. L., Lorig-Roach R., Field A. R., Haeussler M., Russo L., Bhaduri A., Nowakowski T. J., Pollen A. A., Dougherty M. L., Nuttle X., Addor M.-C., Zwolinski S., Katzman S., Kriegstein A., Eichler E. E., Salama S. R., Jacobs F. M. J., Haussler D., Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki I. K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F., Detours V., Vanderhaeghen P., Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng J., Gu M., Miao Y., Yao S., Zhu M., Fang P., Yu X., Li P., Su Y., Huang J., Zhang J., Yu J., Li F., Bai J., Sun W., Huang Y., Yuan Y., Hong D., Wang Z., Long-read sequencing identified repeat expansions in the 5′UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J. Med. Genet. 56, 758–764 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Araki K., Sone J., Fujioka Y., Masuda M., Ohdake R., Tanaka Y., Nakamura T., Watanabe H., Sobue G., Memory loss and frontal cognitive dysfunction in a patient with adult-onset neuronal intranuclear inclusion disease. Intern. Med. 55, 2281–2284 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Sone J., Hishikawa N., Koike H., Hattori N., Hirayama M., Nagamatsu M., Yamamoto M., Tanaka F., Yoshida M., Hashizume Y., Imamura H., Yamada E., Sobue G., Neuronal intranuclear hyaline inclusion disease showing motor-sensory and autonomic neuropathy. Neurology 65, 1538–1543 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Nakamura M., Ueki S., Kubo M., Yagi H., Sasaki R., Okada Y., Akiguchi I., Kusaka H., Kondo T., Two cases of sporadic adult-onset neuronal intranuclear inclusion disease preceded by urinary disturbance for many years. J. Neurol. Sci. 392, 89–93 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Motoki M., Nakajima H., Sato T., Tada M., Kakita A., Arawaka S., Neuronal intranuclear inclusion disease showing intranuclear inclusions in renal biopsy 12 years earlier. Neurology 91, 884–886 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Boivin M., Deng J., Pfister V., Grandgirard E., Oulad-Abdelghani M., Morlet B., Ruffenach F., Negroni L., Koebel P., Jacob H., Riet F., Dijkstra A. A., McFadden K., Clayton W. A., Hong D., Miyahara H., Iwasaki Y., Sone J., Wang Z., Charlet-Berguerand N., Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: The polyG diseases. Neuron 109, 1825–1835.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong S., Lian Y., Luo W., Luo R., Wu X., Ji J., Ji Y., Ding J., Wang X., Upstream open reading frame with NOTCH2NLC GGC expansion generates polyglycine aggregates and disrupts nucleocytoplasmic transport: Implications for polyglycine diseases. Acta Neuropathol. 142, 1003–1023 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Wang T., Pan Q., Lin L., Szulwach K. E., Song C.-X., He C., Wu H., Warren S. T., Jin P., Duan R., Li X., Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 21, 5500–5510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Todd P. K., Oh S. Y., Krans A., He F., Sellier C., Frazer M., Renoux A. J., Chen K. C., Scaglione K. M., Basrur V., Elenitoba-Johnson K., Vonsattel J. P., Louis E. D., Sutton M. A., Taylor J. P., Mills R. E., Charlet-Berguerand N., Paulson H. L., CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krans A., Skariah G., Zhang Y., Bayly B., Todd P. K., Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome. Acta Neuropathol. Commun. 7, 152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabet R., Schaeffer L., Freyermuth F., Jambeau M., Workman M., Lee C. Z., Lin C. C., Jiang J., Jansen-West K., Abou-Hamdan H., Desaubry L., Gendron T., Petrucelli L., Martin F., Lagier-Tourenne C., CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun. 9, 152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semple B. D., Blomgren K., Gimlin K., Ferriero D. M., Noble-Haeusslein L. J., Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 106-107, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leger M., Quiedeville A., Bouet V., Haelewyn B., Boulouard M., Schumann-Bard P., Freret T., Object recognition test in mice. Nat. Protoc. 8, 2531–2537 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Gibson E. M., Nagaraja S., Ocampo A., Tam L. T., Wood L. S., Pallegar P. N., Greene J. J., Geraghty A. C., Goldstein A. K., Ni L., Woo P. J., Barres B. A., Liddelow S., Vogel H., Monje M., Methotrexate chemotherapy induces persistent tri-glial dysregulation that underlies chemotherapy-related cognitive impairment. Cell 176, 43–55.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avila Cobos F., Alquicira-Hernandez J., Powell J. E., Mestdagh P., De Preter K., Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 11, 5650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick E., Taga M., Ergun A., Ng B., Casazza W., Cimpean M., Yung C., Schneider J. A., Bennett D. A., Gaiteri C., De Jager P. L., Bradshaw E. M., Mostafavi S., Deconvolving the contributions of cell-type heterogeneity on cortical gene expression. PLoS Comput. Biol. 16, e1008120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Guo Z., Cheng Y., Jin P., Wu H., Robust partial reference-free cell composition estimation from tissue expression. Bioinformatics 36, 3431–3438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Park J., Susztak K., Zhang N. R., Li M., Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F., Xu Q., Wang W., Takano T., Nedergaard M., Bergmann glia modulate cerebellar Purkinje cell bistability via Ca2+−dependent K+ uptake. Proc. Natl. Acad. Sci. U.S.A. 109, 7911–7916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piet R., Jahr C. E., Glutamatergic and purinergic receptor-mediated calcium transients in Bergmann glial cells. J. Neurosci. 27, 4027–4035 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q., Huang S., Yin P., Yang S., Zhang J., Jing L., Cheng S., Tang B., Li X. J., Pan Y., Li S., Cerebellum-enriched protein INPP5A contributes to selective neuropathology in mouse model of spinocerebellar ataxias type 17. Nat. Commun. 11, 1101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hovhannisyan R. H., Carstens R. P., Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J. Biol. Chem. 282, 36265–36274 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Geuens T., Bouhy D., Timmerman V., The hnRNP family: Insights into their role in health and disease. Hum. Genet. 135, 851–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lleres D., Denegri M., Biggiogera M., Ajuh P., Lamond A. I., Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 11, 445–451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Nostrand E. L., Freese P., Pratt G. A., Wang X., Wei X., Xiao R., Blue S. M., Chen J.-Y., Cody N. A. L., Dominguez D., Olson S., Sundararaman B., Zhan L., Bazile C., Bouvrette L. P. B., Bergalet J., Duff M. O., Garcia K. E., Gelboin-Burkhart C., Hochman M., Lambert N. J., Li H., McGurk M. P., Nguyen T. B., Palden T., Rabano I., Sathe S., Stanton R., Su A., Wang R., Yee B. A., Zhou B., Louie A. L., Aigner S., Fu X.-D., Lécuyer E., Burge C. B., Graveley B. R., Yeo G. W., A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramesh N., Kour S., Anderson E. N., Rajasundaram D., Pandey U. B., RNA-recognition motif in Matrin-3 mediates neurodegeneration through interaction with hnRNPM. Acta Neuropathol. Commun. 8, 138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paz I., Kosti I., Ares M. Jr., Cline M., Mandel-Gutfreund Y., RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–W367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogasawara M., Iida A., Kumutpongpanich T., Ozaki A., Oya Y., Konishi H., Nakamura A., Abe R., Takai H., Hanajima R., Doi H., Tanaka F., Nakamura H., Nonaka I., Wang Z., Hayashi S., Noguchi S., Nishino I., CGG expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy with neurological manifestations. Acta Neuropathol. Commun. 8, 204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X. R., Tang B. S., Jin P., Guo J. F., The phenotypes and mechanisms of NOTCH2NLC-related GGC repeat expansion disorders: A comprehensive review. Mol. Neurobiol. 59, 523–534 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Li X. J., Li S., Proteasomal dysfunction in aging and Huntington disease. Neurobiol. Dis. 43, 4–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Root J., Merino P., Nuckols A., Johnson M., Kukar T., Lysosome dysfunction as a cause of neurodegenerative diseases: Lessons from frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol. Dis. 154, 105360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berridge M. J., The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol. Rev. 96, 1261–1296 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Nik S., Bowman T. V., Splicing and neurodegeneration: Insights and mechanisms. Wiley Interdiscip. Rev. RNA 10, e1532 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Ma Q., Hu Q. S., Xu R. J., Zhen X. C., Wang G. H., Protease Omi facilitates neurite outgrowth in mouse neuroblastoma N2a cells by cleaving transcription factor E2F1. Acta Pharmacol. Sin. 36, 966–975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen S., Park J. W., Lu Z. X., Lin L., Henry M. D., Wu Y. N., Zhou Q., Xing Y., rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S9
Movie S1