Abstract
In Tibetans, noncoding alleles in EPAS1—whose protein product hypoxia-inducible factor 2α (HIF-2α) drives the response to hypoxia—carry strong signatures of positive selection; however, their functional mechanism has not been systematically examined. Here, we report that high-altitude alleles disrupt the activity of four EPAS1 enhancers in one or more cell types. We further characterize one enhancer (ENH5) whose activity is both allele specific and hypoxia dependent. Deletion of ENH5 results in down-regulation of EPAS1 and HIF-2α targets in acute hypoxia and in a blunting of the transcriptional response to sustained hypoxia. Deletion of ENH5 in mice results in dysregulation of gene expression across multiple tissues. We propose that pleiotropic adaptive effects of the Tibetan alleles in EPAS1 underlie the strong selective signal at this gene.
Disruption of a hypoxia-sensitive enhancer by adaptive mutations has highly pleiotropic effects in vitro and in vivo.
INTRODUCTION
During their dispersal across the globe, humans have adapted to a variety of environmental stressors. Efforts to identify locally adaptive alleles have largely relied on population genetics approaches designed to detect strong selection driving an allele to high frequency. Despite the identification of compelling examples [reviewed in (1)], growing evidence (2–4) indicates that this mode of adaptation has not been common in human populations. Moreover, even in the few cases that are well established, the underlying genetic mechanism remains largely unexplored; therefore, what distinguishes those exceptions is of great interest for understanding phenotypic evolution (5).
Here, we focus on one of the strongest signals of local adaptation in humans: the alleles found in Tibetans at the EPAS1 gene that codes for the hypoxia-inducible factor 2α (HIF-2α) transcription factor (6–8). Indigenous populations of the Tibetan plateau have experienced high-altitude hypoxic stress since the late Pleistocene and exhibit a suite of unique phenotypes in addition to better health and reproductive outcomes relative to acclimatized lowlanders (9–14). For example, Tibetans experience lower rates of hypoxia-induced pulmonary hypertension (PH) compared to lowlanders at the same altitude (15). Moreover, unlike lowlanders, Tibetans experience only a mild increase in hemoglobin (Hb) concentration at high altitude (16). The alleles in EPAS1 with strong and highly replicated selection signals are also associated with reduced Hb and oxygenated Hb (oxyHb) concentration in Tibetans (6, 8, 17). This observation suggested that this phenotype provides an advantage, possibly because it reduces the detrimental effects of high blood viscosity resulting from long-term polycythemia. However, as a consequence of relatively low Hb, yet normal oxygen saturation, Tibetans are severely hypoxemic at high altitude (12, 18). This raises the question of whether the benefit provided by unelevated Hb is sufficient to explain the signals of strong selection observed at the EPAS1 locus or whether this haplotype influences additional beneficial traits at high altitude.
RESULTS
To shed light on this question, we examined the results of a genome-wide association study (GWAS) focusing on oxyHb conducted in Tibetans and of a scan for adaptive allele frequency divergence between Tibetans and Han Chinese, summarized by the population branch statistic (PBS) (17). Combining these two sets of results, we honed in on a 31-kb region (chr2: 46570550 to 46601850; hg19) where association and selection signals colocalize for further analysis. We also included an upstream segment (ENH1; chr2: 46552014 to 46552396; hg19) spanning a single-nucleotide polymorphism (SNP) that was previously proposed to have allelic effects in a luciferase reporter promoter assay (19). Because all SNPs in these regions are noncoding, we investigated their effects on the regulation of EPAS1 and downstream consequences in multiple cell types. We chose to start our analyses in the endothelium because it is the primary site of EPAS1 expression. As the first point of contact for oxygenated blood, the endothelium is a crucial tissue in the physiological and pathological response to hypoxia (20–22). Moreover, HIF-2α activity in the endothelium helps regulate a wide range of functions from vasoconstriction/dilation to angiogenesis, endothelial barrier integrity, inflammation, and metabolism (23–25). Several Tibetan phenotypes also implicate the endothelium as a target of adaptation to hypoxia, particularly the low rates of hypoxia-induced PH observed in Tibetans (15). HIF-2α activity in the endothelium is essential for the pathogenesis of hypoxia-induced PH, and reduction or deletion of HIF-2α in the endothelium results in protection from PH (21).
The regulatory landscape of EPAS1 in endothelial cells
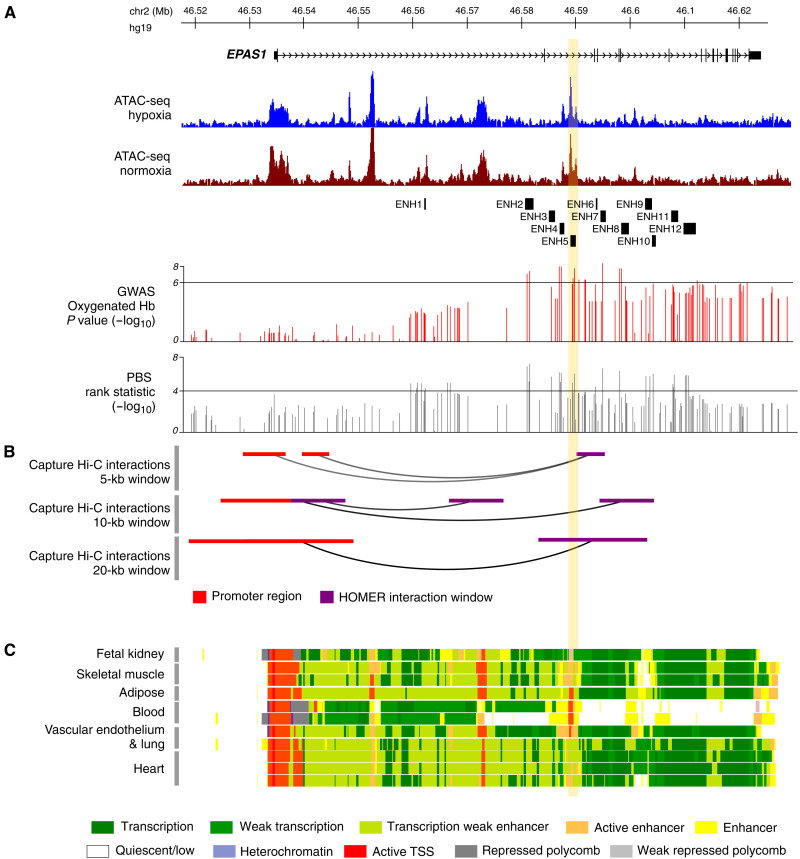
To characterize the regulatory landscape of EPAS1 in endothelial cells, we first performed assay for transposase-accesible chromatin seqeuncing (ATAC-seq) and RNA sequencing (RNA-seq) in primary human aortic endothelial cells (HAECs) cultured in normoxia and hypoxia (20 and 1% O2, respectively, for 24 hours; fig. S1 and table S1). Because hypoxia is a strong cellular stress, which causes broad changes in gene expression and chromatin accessibility (26, 27), we asked whether genomic regions within the EPAS1 locus containing signatures of selection changed chromatin accessibility in response to hypoxia. We did not identify differentially accessible regions within the EPAS1 gene, but we detected several regions of accessible chromatin (peaks shown in Fig. 1A, coordinates listed in table S1); multiple SNPs with strong selection and association signals are located near these regions (Fig. 1A). Next, we characterized the local topology of enhancer-promoter interactions by performing promoter capture Hi-C in HAECs to examine how chromatin loops to the promoter of EPAS1 and other nearby genes (Fig. 1B). In agreement with published data (28), we found evidence of extensive looping between the EPAS1 promoter and upstream genomic regions, including some within the Protein Kinase C Epsilon (PRKCE) gene (fig. S2A). Consistent with the role of HIF-2α in inducing erythropoietin (EPO) expression, dozens of GWAS signals for hematological traits observed in European cohorts and eQTLs for EPAS1 are found in these looping regions (29, 30). Focusing on the introns of EPAS1—including the 31-kb region of colocalized association and selection signals—we identified five segments, three of which are overlapping (ascertained in different window sizes), that form long-range interactions with the EPAS1 promoter [denoted as Hypergeometric Optimization of Motif EnRichment (HOMER) interaction windows, shown in purple] (Fig. 1B). Crucially, the large area of interaction, defined by the union of the three distal overlapping interaction windows (chr2: 46573123 to 46594333, hg19), encompasses nearly all the strongest oxyHb association signals, indicating that this region may have effects on both blood and endothelial traits (Fig. 1A and figs. S2 and S3A). This union region also loops to the EPAS1 promoter in other publicly available datasets, indicating the strength of this interaction across multiple cell types, including cardiomyocytes, preadipocytes, and CD34+ hematopoietic progenitor cells (31–34). Moreover, it contains chromatin Hidden Markov Model evidence of multiple regulatory regions across a range of tissue types (Fig. 1C).
Fig. 1. Regulatory landscape of EPAS1.
The EPAS1 promoter interacts with a region of open chromatin in HAECs, which spans the segment of colocalized association and selection signals and the enhancer elements active in multiple tissues. (A) The genomic structure of the EPAS1 gene and ATAC-seq peaks in hypoxia (blue) and normoxia (brown). Also shown is the position of all enhancer (ENH) regions tested in luciferase reporter assays. Below, two tracks show, respectively, the −log10 P value for the GWAS of oxyHb and the −log10 PBS rank statistic of all SNPs tested in EPAS1 by Jeong et al. (17). The horizontal bar in each indicates the cutoff used for defining candidate SNPs (table S2). (B) Results of the promoter capture Hi-C HOMER analysis showing all regions within the gene that interact with the EPAS1 promoter ascertained in three window sizes. (C) Chromatin state tracks from the NIH Roadmap Epigenomics Mapping Consortium. The vertical yellow bar indicates the position of ENH5.
High-altitude alleles reduce the activity of several tissue-shared enhancers
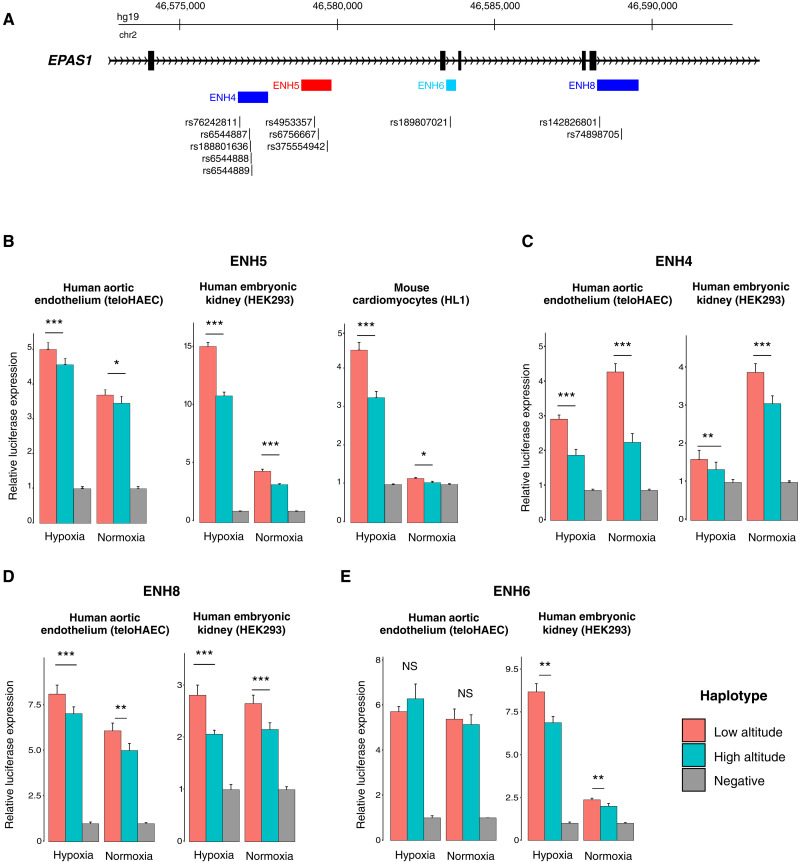
To systematically assess the regulatory effects of the candidate variants identified in the GWAS and selection scan, we generated 24 enhancer constructs spanning a total of 37 SNPs (fig. S2B and table S2) in 11 regions (ENH2 to ENH12), designed to tile across the segment of colocalized signals, and in an additional region (ENH1) implicated by a previous study (19) (fig. S2B). These constructs spanned candidate SNPs in the region, which we defined as having oxyHb P < 10−6, PBS values in the top 0.01% of the genome-wide distribution or sharing a derived allele with the Denisovan genome (reflecting a well-established adaptive introgression event) (35). For each of the 12 regions, we generated a high- and a low-altitude construct, each containing either high- or low-altitude allele(s) at all candidate SNPs within the construct. Each construct was tested in telomerase-immortalized HAECs (teloHAECs) cultured in parallel in normoxia and hypoxia (20% O2 and 1% O2, 48 hours). This analysis identified three enhancer regions—termed ENH4, ENH5, and ENH8—that show enhancer activity relative to the negative controls and significant differences between alleles under both normoxic and hypoxic conditions (6.77 × 10−06 < P < 4.33 × 10−02), with the high-altitude constructs consistently showing weaker activity than the corresponding low-altitude constructs (Fig. 2, A to D, and tables S3 and S4). All three enhancers were in, or near, regions of accessible chromatin identified by our ATAC-seq analysis and within the chromatin segment looping to the EPAS1 promoter identified by our capture Hi-C data, consistent with being active enhancers in this cell type (Fig. 1).
Fig. 2. EPAS1 enhancers with allele-specific activity.
Four genomic segments spanning candidate SNPs show significant differences in enhancer activity across high- and low-altitude alleles. (A) The locations of the four enhancer regions with allele-specific activity are shown for each enhancer along with the positions of the candidate SNPs within each construct. Enhancers are color-coded depending on the cell types in which significant allelic effects (P < 0.05) were observed: red for effects shared across kidney, endothelium, and cardiomyocytes; blue for kidney and endothelium; and cyan for kidney only. (B) Luciferase reporter assay results for ENH5 in endothelium, kidney, and cardiomyocytes. Results are for a minimum of two independent construct DNA preparations each with at least three independent transfection replicates in each condition and are depicted as mean relative expression + SEM. (C to E) Luciferase reporter assay results for ENH4 (C), ENH8 (D), and ENH6 (E). Relative expression data for each replicate can be found in table S3. Probability values for all paired t tests performed can be found in table S4. *P < 0.05, **P < 0.01, and ***P < 0.005. NS, not significant.
Because the alleles found within the endothelial enhancers are highly associated with Hb concentration in Tibetans (fig. S3A), we reasoned that these enhancers may be active also in the kidney, where HIF-2α activates EPO to stimulate the erythropoietic response (6, 36). Alternatively, other enhancers and alleles within the extended EPAS1 haplotype may be active in the kidney and may be responsible for the association with Hb concentrations. To distinguish between these two scenarios, we tested the same luciferase constructs in embryonic kidney cells [human embryonic kidney (HEK) 293T cells] and measured their activity in normoxia and hypoxia. We discovered that the same three enhancers characterized in endothelial cells also display consistent regulatory effects in embryonic kidney cells (Fig. 2, B to D, and tables S3 and S4). One additional region, ENH6, demonstrated enhancer activity and consistent allelic effects only in the HEK293T cells, again with the high-altitude allele showing lower enhancer activity compared to the low-altitude allele (Fig. 2E and tables S3 and S4). While all SNPs in ENH4 and ENH5 are shared with Denisovans, both SNPs in ENH6 are not, thus suggesting that mutations on the human lineage have functionally modified the introgressed haplotype. ENH8 contains two candidate SNPs of which one is shared with Denisovans; in this case, it is not possible to assign functional effects to archaic versus human mutation.
High-altitude alleles reduce the activity of ENH5, a pleiotropic and hypoxia-responsive enhancer
Of the three enhancers with shared activity in endothelium and kidney, we were most interested in further examining ENH5, which, in addition to consistent and robust allele-specific activity in both cell types, also exhibited hypoxia-responsive activity (Fig. 2B and table S4). Notably, both alleles displayed a significantly stronger enhancer activity under conditions of hypoxia than normoxia (kidney: low-altitude allele P = 6.02 × 10−14, high-altitude allele P = 1.41 × 10−12; endothelium: low-altitude allele P = 9.03 × 10−06, high-altitude allele P = 4.90 × 10−05; table S4), indicating that ENH5 is a hypoxia-responsive enhancer containing some of the strongest signals of selection in the Tibetan genome (table S2). HIF-2α protein abundance is known to be under tight posttranslational oxygen-dependent regulation (37); therefore, our finding of a hypoxia-sensitive transcriptional enhancer of EPAS1 points to a previously unknown layer of oxygen-dependent regulation of HIF-2α.
A closer examination of ENH5 in publicly available databases of inferred chromatin states (38–40) revealed that the region spanning ENH5 has regulatory activity in a multitude of cell types (Fig. 1C and fig. S3B). This finding indicates that this region may correspond to an enhancer with extensive tissue sharing and pleiotropic effects (41). We next investigated ENH5’s activity in cardiomyocytes because, like endothelial cells, they are important in both biological and pathological responses to hypoxia (42–45). In addition, the cardiovascular system is a target of adaptations to hypoxia, and polygenic tests detected an adaptation signal toward lower heart rate (17, 46). Published promoter capture Hi-C data from cardiomyocytes indicate that ENH5 loops to the EPAS1 promoter also in this cell type (32). We therefore tested the activity of ENH5 in cardiomyocytes by transfecting the same constructs into mouse cardiomyocytes (HL-1) followed by exposure to either normoxia or hypoxia, as with the kidney and endothelial cells. In HL-1 cells, ENH5 demonstrated the same patterns of expression as seen in kidney and endothelium (Fig. 2B and tables S3 and S4).
Among the three ENH5 SNPs that differ between the low- and high-altitude constructs (table S2), the high-altitude allele at rs375554942 was predicted to eliminate binding sites for the greatest number of transcription factors (table S5). To assess the contribution of this SNP in modulating ENH5 activity, we performed site-directed mutagenesis in the high-altitude construct, changing the high-altitude allele (derived, G) to the low-altitude allele (ancestral, A), and we performed reporter gene assays in HEK293T cells (fig. S3C and table S3). Results (shown in fig. S4) indicated that altering rs375554942 from the high- to low-altitude allele was sufficient to significantly increase enhancer activity (normoxia P = 0.0003, hypoxia P = 0.0058; table S6).
ENH5 regulates transcriptome-wide responses to chronic hypoxia
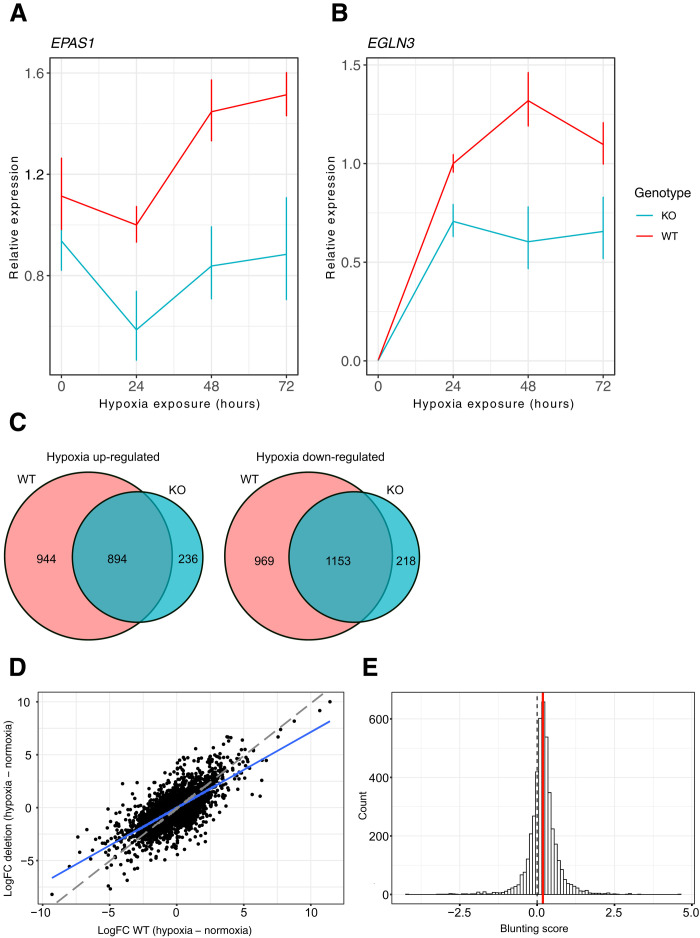
Because reporter gene assays do not assess regulatory activity in the endogenous chromatin context, we used CRISPR-mediated editing to delete an approximately 1-kb region (chr2: 46578867 to 46579857; hg19) spanning the entire ENH5 in teloHAECs and assessed the effects of the deletion on transcript levels in hypoxia over time (1% O2, 72 hours). Across hypoxia time points, EPAS1 transcript levels were significantly lower in the ENH5 knockout (KO) compared to the unedited [wild type (WT)] lines (P = 0.04465 at 24 hours, 0.0241 at 48 hours, and 0.0416 at 72 hours; Fig. 3A and table S7). In addition, consistent with ENH5 being a hypoxia-dependent enhancer, transcript levels markedly increase after 48 hours of hypoxia exposure in the unedited lines but do not in the deleted ones; EPAS1 transcript levels do not differ significantly across deleted and unedited lines in normoxia (P = 0.1968; table S7). These data indicate that ENH5, harboring variation strongly associated with adaptations in Tibetans, is necessary for the proper regulation of EPAS1 specifically in response to hypoxia.
Fig. 3. Deletion of ENH5 in endothelial cells results in dampening of transcriptional responses to acute and sustained hypoxia.
(A) EPAS1 and (B) EGLN3 transcript levels detected by quantitative polymerase chain reaction (qPCR) in the endothelial ENH5 KO and WT clones over 72 hours in hypoxia (1% O2). Mean relative expression of three homozygous KO and three homozygous WT clones is shown with red and blue lines, respectively. Error bars reflect the SEM. (C) Venn diagrams showing overlap of up- or -down-regulated genes in WT and KO clones after sustained hypoxia. Most differentially expressed genes in the KO are also differentially expressed in the WT. (D) Scatterplot showing the transcriptional response (logFC) to sustained hypoxia (14 days, 1% O2) in three ENH5 KO clones relative to that in three WT clones. The blue line shows the linear regression line of deletion log fold change (LFC) predicted by the WT LFC response (LFC_KO ~ LFC_WT + 0). The red dashed line represents a slope of 1. (E) Histogram of the distribution of blunting scores for differentially expressed genes in response to sustained hypoxia (46) showing an overall shift toward weaker response in ENH5 KO relative to WT clones (positive blunting score represents more extreme transcriptional changes in response to hypoxia in the WT relative to the KO). The dashed line marks a blunting score of 0 expected under the null hypothesis of no difference in transcriptional response across genotypes, while the green line depicts the observed mean score (0.185). Scores were tested in a two-sided, one-sample Student’s t test (HA: μ ≠ 0) with a P < 2.2 × 10−16.
To determine whether ENH5 deletion has functional consequences for downstream HIF-2α targets, we examined the expression of EGLN3, which is directly up-regulated by HIF-2α in the endothelium in response to hypoxia (37). We found that deletion of ENH5 leads to significant down-regulation of EGLN3 across multiple hypoxia time points (P = 0.0265 at 24 hours and 0.02365 at 48 hours; Fig. 3B and table S7), indicating that the changes observed in EPAS1 transcript levels have downstream consequences, blunting its response to acute hypoxia (Fig. 3, A and B). No significant changes in the expression of the PRKCE gene—upstream of EPAS1 but not regulated by HIF-2α—were detected in ENH5 KO lines relative to WT controls (fig. S5 and table S7).
To assess the impact of the ENH5 deletion under sustained—as opposed to acute—hypoxic conditions, we cultured the same deleted and unedited lines in 20% or 1% O2 for 14 days and then performed transcriptional profiling by RNA-seq. The KO lines display a markedly blunted response to sustained hypoxic stress compared to the WT lines, as demonstrated by the lower proportion of differentially expressed genes in response to hypoxia (30.7% versus 19.4% in WT and KO lines, respectively; table S8), and an overall pattern of blunting of the transcriptional response to hypoxia in the KO relative to the WT (Fig. 3, C and D). We quantified this pattern as the difference between the absolute values of the effect size in the transcriptional response to hypoxia across genotypes, which we refer to as “blunting score,” for differentially expressed genes (see Materials and Methods). We find a highly significant departure from the expectation that the mean blunting score is equal to zero (two-tailed one-sample t test P < 2.2 × 10−16), demonstrating that deletion of ENH5 results in a transcriptome-wide dampening of the response to sustained hypoxia (Fig. 3E). Collectively, these results point to a key role for transcriptional enhancer ENH5 in regulating the HIF-2α response to both acute and sustained hypoxic exposure.
ENH5 is a highly pleiotropic enhancer in vivo
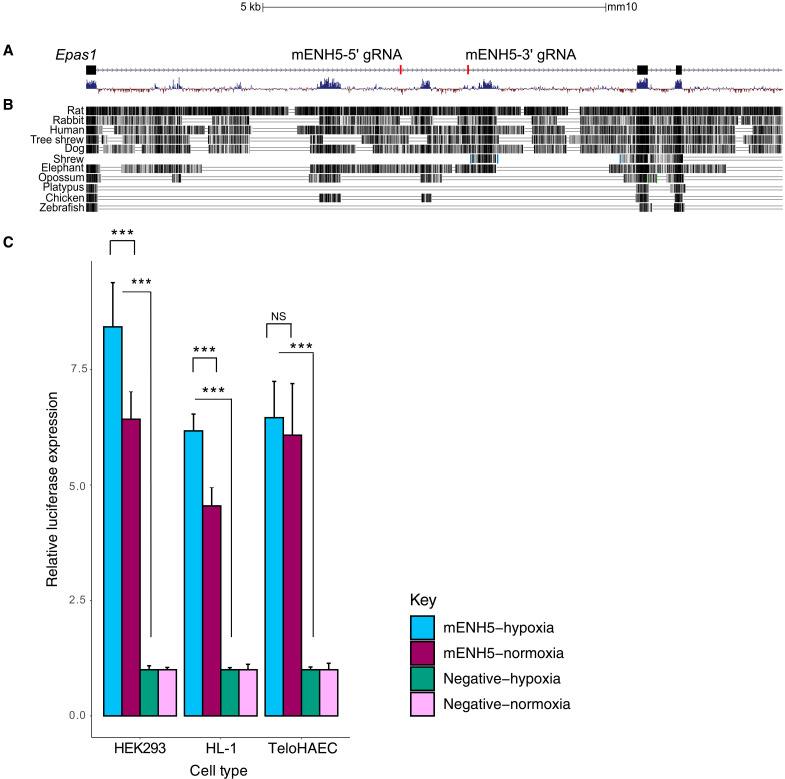
To better characterize the degree of pleiotropic activity of ENH5, we assessed the effect of an in vivo deletion of the orthologous ENH5 region in mice (mENH5, chr17: 86801737 to 86802738; mm10; Fig. 4A), conserved at the sequence level between mice and humans (Fig. 4B). Moreover, luciferase reporter assays of mENH5 demonstrated robust enhancer activity in teloHAEC, HEK293T, and HL-1 cells as well as evidence of hypoxia responsiveness, demonstrating that this region is also conserved at the functional level (Fig. 4C and tables S3 and S9). We deleted mENH5 using CRISPR editing and bred a stable deletion line from the resultant mice. We treated age- and cage-matched pairs of male mice overnight in hypobaric hypoxia (16 hours, 8% O2) and assessed the transcriptional profile by RNA-seq in seven tissues from each mouse: lung, kidney, left atrium, right atrium, left ventricle, right ventricle, and adrenal gland.
Fig. 4. mENH5 is a conserved, hypoxia-responsive enhancer.
(A) Genomic structure of the mouse EPAS1 region (chr17: 86,797,164 to 86,807,311, mm10) in a segment spanning the orthologous ENH5 enhancer. Red bars denote the location of CRISPR guide RNAs that flank mENH5. (B) ENH5 is conserved across multiple mammalian species as well as opossum and chicken at the sequence level. (C) ENH5 is functionally conserved between human and mouse. Luciferase results for mENH5 activity in kidney (HEK293), cardiomyocytes (HL-1), and endothelial (teloHAEC) cells are shown. The plots show means + SEM from three independent construct DNA preparations, each with three independent transfection replicates. * ***P < 0.005.
In independent single-tissue analyses, we identified a total 168 genes differentially expressed (adjusted P < 0.05) in at least one tissue, the majority of which were identified in the left atrium (table S10). Many of these, including Angpt1, Malat1, and Cox2, are up-regulated by HIF-2α and are all down-regulated in the left atrium of KOs relative to WT, as expected (47–49).
To exploit patterns of differential expression profile similarity across tissues, we used multivariate adaptive shrinkage (mashr) on the data from all seven tissues. Mashr leverages the combination of many effects (in our case gene expression differences between KO and WT) ascertained across many conditions (in our case many tissues) to model effect sharing across conditions and increase power (50). Significance in mashr is calculated in each tissue using the local false sign rate (lfsr). We identified 1329 genes that were differentially expressed in at least one tissue (lfsr < 0.05), with the largest numbers identified in lung, right atrium, and left ventricle [tables S11 and S12 and Supplementary Text (51)].
EPAS1 itself was not down-regulated at lfsr < 0.05 in any single tissue. Given the dynamic expression of EPAS1 observed in both our hypoxia time course (Fig. 3A) and the literature (22), detecting changes in the transcriptional targets of HIF-2α is a more reliable readout of the mENH5 deletion. Accordingly, differential expression analysis identified significant (lfsr < 0.05) differences across genotypes in multiple direct and indirect HIF-2α targets. Some of these genes (shown in table S13) are known to have clear functions in the modulation of heart rate, vasoconstriction/vasodilation, angiogenesis, microvascular maintenance, hypertension, inflammation, apoptosis, and/or to be targets of HIF-2α. Gene set enrichment analyses of all significantly (lfsr < 0.05) differentially expressed genes detected terms related to known HIF functions, including immune, inflammatory, angiogenic, metabolic, and apoptotic processes, as well as several relating to hypoxia response, stress response, hematopoiesis, and cell-cell adhesion (table S14). We also found enrichment for human phenotype ontology terms HP:0000822 “Hypertension” and HP:0002092 “Pulmonary arterial hypertension.” These sets include genes, such as COX2, COX3, Notch1, and Hspg2, that are up-regulated in PH models but are significantly down-regulated in multiple tissues in our KO mice (lfsr < 0.05) (52, 53). Conversely, genes whose up-regulation is protective against PH, including Id3, Socs3, and Dusp5, are significantly up-regulated in most tissues of our KO mice (lfsr < 0.05) (21, 54, 55). These findings support the hypothesis that down-regulation of EPAS1 through ENH5 may provide protection against PH. Consistent with this proposal, alleles in the high-altitude haplotype at EPAS1 have been associated with reduction of pulmonary arterial pressure in Tibetans, linking the high-altitude allele of ENH5 to PH protection (19). Overall, we find that deletion of mENH5 affects the expression of EPAS1 and its targets in multiple hypoxia-responsive tissues. Critically, these results implicate the disruption of ENH5 activity across multiple tissues in a wide range of possible phenotypic outcomes, including traits known to be common in Tibetans.
DISCUSSION
Much of the previous research into EPAS1 variation in Tibetans has focused on either its connection to Hb concentration or its effects in a single-cell type (6, 19, 56). Here, we observe a much broader range of transcriptional effects, involving key hypoxia-sensitive tissues like heart, endothelium, and lungs. These findings raise the possibility that the strong signal of selection at EPAS1 reflects pleiotropic effects on multiple adaptive traits, summing up to a large overall net fitness; we refer to this property as adaptive pleiotropy. The observed pleiotropic effects on transcript levels derive from the disruption of a tissue-shared enhancer, ENH5, which we demonstrated in vitro and in vivo. Additional effects may be conferred by the SNPs, on the same haplotype, in ENH4, ENH6, and ENH8, based on our in vitro results. In particular, we show that adaptive alleles reduce ENH5 activity in kidney cells, leading to lower EPAS1 transcriptional response to hypoxia, and hence may act to reduce EPO expression and Hb concentrations. At the same time, in endothelial and vascular smooth muscle cells, these alleles may result in the protection against PH and infantile and gestational hypertension by altering the expression of genes involved in vasoconstriction/dilation and inflammation (21, 54, 55, 57). Like PH, gestational hypertension and early infantile hypertension occur at lower rates in Tibetan and Sherpa mothers at high altitude and are linked to the observed higher birth weight of their infants and lower infant mortality (10, 14). Other phenotypes implicated by the differential expression analyses in mice are also strong candidate adaptive traits under hypoxic conditions. For instance, multiple mitochondrial cytochrome c oxidase genes (58)—such as Cox6a1 and Cox8a—are dysregulated across multiple tissues, and the oxidative phosphorylation gene set is significantly enriched among the differentially expressed genes (adjusted P = 0.0027). These changes could be linked to metabolic alterations previously identified in Sherpa populations (9), which may, in turn, result in the greater vital capacity compared to Han high-altitude natives (11). While these phenotypes may, individually, contribute only moderate increases in fitness, they may collectively amount to a much larger fitness effect resulting in the observed strong signals of selection. Similar to EPAS1, an adaptive haplotype spanning the Eda gene in stickleback fish was shown to carry linked adaptive mutations and mutation(s) in a single 1.4-kb noncoding region with pleiotropic effects on multiple beneficial traits (59). In humans, a nonsynonymous allele in the EDAR gene carries a strong signature of selective sweep in East Asians. Modeling this variant in mice revealed multiple organismal phenotypes, many of which were proposed to confer increased fitness (5). These results suggest that pleiotropy is an important mechanism underlying a subset of well-established signals of strong positive selection. Additional studies, especially in vivo, of selective sweeps are needed to determine how common adaptive pleiotropy is among these signals.
This insight is important because several lines of evidence suggest that selective sweeps are not the main mode of adaptation in recent human evolution (3, 60). What, then, explains the existence of the few strong selective sweeps we do see? We propose that, where a strong selection signal is observed, adaptive pleiotropy provides a possible explanation. However, the a priori probability of adaptive pleiotropy depends on assumptions about the distribution of fitness effects conferred by the variant (the effect that the variant has on different traits and the effect that those traits have on fitness). For example, a pleiotropic variant that has small fitness effects in approximately balanced directions across traits is not expected to result in a strong selection signal. However, a pleiotropic variant with small but positive effects on several traits and limited deleterious consequences could confer a strong increase in overall fitness. Under this model, selective sweeps occur at the select few variants that happen to have correlated fitness effects across several traits.
While pleiotropy in general has been shown to be pervasive in humans (61), it is generally thought to reduce the rate of adaptation because a pleiotropic variant may have deleterious effects on a subset of traits. This notion has led to the expectation that adaptations are more likely to result from mutations with narrow phenotypic effects, such as tissue-specific cis-regulatory variants (62). We demonstrate that ENH5 is a key cis-regulatory element of EPAS1, making it a good candidate as a site of adaptive variation. However, because of its activity in diverse tissues combined with its regulatory effects on a key transcription factor with a large number of target genes, ENH5 variation is expected to be highly pleiotropic. Growing evidence shows that regulatory variation in enhancers is often shared across tissues (63) and that it can contribute to common disease risk (33, 41). Here, we showed that these pleiotropic enhancers may contribute also to adaptative traits. Future work will determine to what extent tissue-shared regulatory elements contribute to different modes of adaptation.
HIF-2α regulates the transcriptional cascade underlying the acclimatization response to hypoxia (37)—an array of multiorgan physiological changes aimed at mitigating transient drops in oxygen pressure. This capacity to respond to changes in environmental oxygen availability through changes in patterns of gene expression is an example of phenotypic plasticity. The impact of directional selection on phenotypic plasticity remains contentious (64–67). However, it has been proposed that phenotypic plasticity is maladaptive when organisms experience chronic rather than transient hypoxia and that selection would favor variants leading to a dampening of the response to hypoxia (64). By reducing the amount of a single transcription factor, in this case, HIF-2α, it is possible to induce multiple phenotypic effects that will tend to be associated with positive effects on fitness. For instance, in vitro studies of an Epas1 adaptive allele in high-altitude deer mice showed that it causes a defect in the interaction of HIF-2α with a transcriptional coactivator, with consequent decrease in the transcriptional response to hypoxia in the two tissues examined, i.e., left ventricle and adrenal gland (68). More recently, this Epas1 adaptive allele in high-altitude deer mice was shown to have organismal phenotypes such as reduction of ventilatory sensitivity and carotid body growth in chronic hypoxia (69). Our finding of a global dampening of the transcriptional response to sustained hypoxia in endothelial cells in the ENH5 KO relative to WT is consistent with an adaptive dampening of plasticity in high-altitude environments. Likewise, traits in Tibetans, such as unelevated Hb concentrations and lower pulmonary artery vasoconstriction, have been proposed to result from natural selection favoring a dampening of the response to chronic hypoxia (65, 70).
Limitations of this study include the use of HAECs and teloHAECs as an experimental system, as these are not fully representative of conduit blood vessels that may behave differently in terms of response to hypoxia (71). Similarly, mouse tissues were analyzed as bulk samples that could not disentangle the contribution of individual cell types. Last, because of the inherent challenges to studying the response to chronic hypobaric hypoxia in an animal model without the confounding due to repeated oxygen reperfusions for animal maintenance, we were unable to examine the long-term effects of ENH5 in hypoxia in vivo at the transcriptional or organismal level. Despite these limitations, this study adds significantly to our limited understanding of the adaptive mechanisms and organismal consequences of strong selection signals. More detailed studies will be needed to accurately parse the effects of ENH5 in specific cell types and more fully explore its phenotypic impacts in vivo.
MATERIALS AND METHODS
Cell culture
TeloHAEC cells were purchased from American Type Culture Collection (ATCC; CRL-4052), and HAEC cells were purchased form Lonza (CC-2535). Both cell types were cultured in endothelial cell growth medium -2 (EGM-2) (Lonza, CC-3162) supplemented with antibiotic-antimycotic (Gibco, 15240062). HEK293T cells (ATCC, CRL-3216) were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 2 mM l-glutamine and 10% fetal bovine serum (FBS). HL-1 cells were a gift from I. Moskowitz (University of Chicago, USA) and were cultured in Claycomb medium (Millipore Sigma, 51800C) supplemented with 10% FBS, penicillin-streptomycin, 0.1 mM norepinephrine, and 2 mM l-glutamine. For all experiments, cells were either cultured in a normoxic incubator (20% O2 and 5% CO2) or in an incubator inside of a hypoxia glove box (Coy Lab Products, O2 Control InVitro Glove Box; 1% O2 and 5% CO2).
ATAC-seq and RNA-seq in HAECs
Paired ATAC-seq and RNA-seq were performed in primary HAECs as described in (72). Triplicate samples of cells cultured in parallel for 24 hours in either normoxia (21% O2) or hypoxia (1% O2) were harvested and processed into ATAC-seq libraries using the Nextera DNA library prep kit (FC-121-1030) or RNA-seq libraries using the TruSeq RNA Library Preparation Kit v2, Set B (Illumina, RS-122-2002). Indexed samples were pooled and sequenced on an Illumina Hi-Seq 4000 to obtain single-end 50–base pair (bp) reads (averaging 37,133,304 reads per sample) at the University of Chicago Genomics Core.
Sequence alignment for RNA-seq libraries was performed using a standard pipeline. Briefly, reads were aligned using STAR, and read counts were performed with RSEM using default settings (73, 74). Differential expression analyses were performed using DEseq2 with default settings. Sequence alignment for ATAC-seq was performed with bwa (bwa 0.7.17-r1188). Reads were mapped to both the 1000 genomes Phase2 Reference Genome Sequence (hs37d5) and the Homo_sapiens_assembly.fasta. Mitochondrial reads were removed. All ATAC-seq results are given for the build hg19 assembly. Read filtering was performed with SAMtools (v1.10) (75) retaining only uniquely mapped reads with a mapping quality of ≥10. Polymerase chain reaction (PCR) duplicates were removed with the SAMtools markdup function. ATAC-seq peak calling was performed with MACS2 (2.1.0) (76). Differential peak calling was performed with the R package DiffBind (v 3.4.0) using RiP normalization and EdgeR differential expression testing with the ENCODE hg19-blacklist.v2 applied (77). Annotation was performed with HOMER v4.10.0. The genome-wide results of ATAC-seq and RNA-seq can be found in table S1.
Promoter capture Hi-C
In situ Hi-C was performed as described previously (78). Briefly, 5 million HAECs were grown in normoxia for 24 hours and then harvested and counted. Cells were resuspended in 1× Dulbecco's phosphate buffered saline (DPBS), and 37% formaldehyde was added to a final concertation of 1%. Formaldehyde treatment was carried out for 10 min to cross-link interacting DNA loci. Cross-linked chromatin was digested with Mbo I endonuclease (New England Biolabs, R0147). Subsequently, the restriction fragment overhangs were filled in, and the DNA ends were marked with biotin-14-dATP (Life Technologies, 19524-016). The biotin-labeled DNA was sheared and pulled down using Dynabeads MyOne Stretavidin T1 beads (Life Technologies, 65602). The in situ Hi-C library was amplified directly off of the T1 beads with 6 cycles of PCR, using Illumina primers and protocol (72°C for 3 min, 98°C for 30 s, 5 cycles of 98°C for 10 s, 63°C for 30 s, 72°C for 3-min hold at 10°C). Promoter capture was performed as described (32). The in situ Hi-C library was hybridized to 81,735 biotinylated 120-bp custom RNA oligomers (CustomArray Inc.; www.customarrayinc.com) targeting promoter regions (four probes/RefSeq transcription start sites). After hybridization, postcapture PCR (eight amplification cycles) was performed on the DNA bound to the beads via biotinylated RNA. Each library was sequenced on a full lane of an Illumina HiSeq 4000 machine. Standard promoter capture Hi-C analyses were performed as described in (32). Analyses were performed using HOMER (79). The quality and biological significance of identified interactions were assessed by integrating publicly available RNA-seq and H3K27Ac Chip-seq (40) data from the same cell type and examining the correlation between interacting genes, gene activity, and bound transcription factors. Summary statistics and validation information on promoter capture Hi-C are included in the HiCUP Processing report (data file S1).
Luciferase reporter enhancer assays
Luciferase reporter enhancer assays were performed using the pGL4.23[luc2/minP] (Promega, cat: E8411) vector backbone. Twelve ~1000-bp windows were selected spanning all SNPs meeting one or more of the following criteria: strongly associated with oxyHb levels in a GWAS (with oxyHb P < 10−6), high PBS values (top 0.01% of the genome-wide distribution), sharing a derived allele with the Denisovan genome (figs. S2B and S3 and table S2) (17). One additional construct (ENH1) spanned a SNP implicated in a previous study (19). One SNP with strong association and selection signals but not shared with the Denisovan genome was not tested for technical reasons (table S2). For each region, portions of the Tibetan genome containing the high-altitude or low-altitude alleles were amplified from Tibetan DNA samples stored in the A.D.R. laboratory and described in (17). The low- and high-altitude alleles were amplified and added to the PGL4.23 vector (Addgene plasmid no. 60323) using pENTR/D-TOPO (Invitrogen, K240020) and Gateway cloning or Gibson Assembly (New England Biolabs, E5510S). The vectors were transfected together with the SV40-pRL Renilla Luciferase control vector (GenBank accession number AF025844) into teloHAECs using the Neon Electroporation system or into HEK cells (HEK293T) using jetPRIME (PolyPlus, 114-07). Follow-up transfections were performed in mouse immortalized atrial-derived cardiomyocytes (HL-1) using jetPRIME. HL-1s were chosen as a model for heart cells because human cardiomyocytes are difficult to transfect and maintain in culture over multiple passages (80). Following transfection, cells were placed in either normoxia or hypoxia for 48 hours to recover and then lysed and assessed for firefly luciferase expression, which was normalized to the Renilla luciferase expression using the Dual Luciferase Reporter Assay system from Promega (catalog no. E1910). The 48-hour time point was chosen on the basis of tests with green fluorescent protein transfections to optimize expression of transfected vectors in this cell type. Assays were run on the GloMax-Multi Detection System with Dual Injector (E7031). In addition to the EPAS1 constructs, additional cells were transfected with a negative control: the PGL4 vector containing a gene desert segment—chr9: 6,161,550 to 6,162,024—with no evidence of active chromatin marks (33). Normalized expression from the EPAS1 constructs was analyzed relative to the negative control in the corresponding condition (normoxia or hypoxia), and paired t tests were used to test for differences between alleles. To account for experimental variability, triplicate construct DNA preparations were made for each allele, and DNA from each preparation was transfected in triplicate; each genomic segment was tested in a minimum of two independent batches of cells (for a total of minimum six transfections per vector; tables S3 and S4). This level of replication was necessary to detect subtle but robust differences in allelic enhancer effect with statistical confidence. Statistical significance of the difference between alleles was determined using a one-tailed Student’s paired t test among independent transfection replicates (table S4). Statistical significance between normoxia and hypoxia was assessed using a two-tailed Student’s t test.
Site-directed mutagenesis
Transcription factor binding site positional weight matrix (PWM) scores were calculated across ENH5 with either the reference sequence (ref) or any combination of the high-altitude alleles at the three candidate SNPs (mut 00: rs375554942; mut01: rs4953357; mut02: rs6756667; mut03: rs375554942 and rs4953357; mut04: rs375554942 and rs6756667; mut05: rs4953357 and rs6756667; and mut06: rs375554942, rs4953357, and rs6756667). Table S5 shows all transcription factors for which the reference sequence is predicted to have a binding site (P < 1 × 10−04) that is disrupted by one or more of the high-altitude alleles. This analysis was performed using the HOCOMOCO database of PWMs (81). P values were generated by FIMO MEME suite 5.4.1. On the basis of these results, we performed site-directed mutagenesis on rs375554942.
Site-directed mutagenesis was performed in the high-altitude haplotype of the ENH5 luciferase vector using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, catalog no. E0554S) according to the manufacturer’s instructions. Briefly, using nonoverlapping, custom, mutagenic primers, plasmid DNA from the high-altitude ENH5 luciferase vector was edited to contain a low-altitude allele for SNP rs375554942. Results were confirmed with Sanger sequencing. This vector was transfected in triplicate, in parallel with the original high-altitude vector and low-altitude vector into HEK293T cells cultured in both normoxia and hypoxia (20 and 1% O2, respectively). These results are shown in fig. S4. Statistical significance was assessed using a one-tailed Student’s t test.
Generation and characterization of CRISPR-Cas9 deletion endothelial lines
ENH5 deletion lines were generated in teloHAECs (ATCC) using a modified IDT ALT-R Ribonucleoprotein (RNP) Protocol. Four CRISPR RNAs (crRNA) were designed to flank the region containing ENH5 using IDT’s Custom ALT-R CRISPR-Cas9 guide RNA design tool (table S15). Guides were selected on the basis of optimizing location and predicted efficacy while minimizing the possibility of off-target effects. The four crRNAs were annealed separately to tracrRNA to create four complete guide RNAs. RNPs were complexed by incubating the guides with purified Alt-R S.p. Cas9 Nuclease V3 (IDT, 1081058). Guides and RNPs were generated as described in the IDT ALT-R RNP Protocol. The pool of four RNPs was transfected into teloHAECs using jetCRISPR (PolyPlus, 502-07). Because of the difficulty of transfecting endothelial cells, this transfection was serially performed two to four times as described in (82) as cells were expanded and periodically checked by PCR for the presence of a deletion band (table S4). Once a strong deletion band was evident, the pool of edited cells was single-cell sorted into a 96-well plate on the FACSARIAIII at the University of Chicago Flow Core. Clones were genotyped using primers designed to flank the deletion site (table S16). Of all clones isolated, three KO clones and three WT controls from the same pool of cells were randomly selected for further analysis.
Real-time quantitative PCR
The three independent clonal isolates of ENH5 KO and WT control endothelial lines were expanded and split into four six-well cell culture plates and allowed to adhere overnight in normoxic incubators (37°C, 5% CO2, 21%O2). The next morning, the first plate of cells was lysed in plate using RLT Plus buffer from Qiagen under normoxic conditions. The remaining three plates were put into a normobaric hypoxia chamber in which neutral nitrogen gas is used to displace oxygen to obtain the desired oxygen concentration (Coy Lab Products, O2 Control InVitro Glove Box). Cells were placed in a humidified incubator contained within the box, and the entire box was maintained at 37°C, 5% CO2, 1%O2. At 24, 48, and 72 hours, one plate of cells was lysed inside the chamber under hypoxic conditions, and the lysate immediately frozen at −80°C. Once all samples had been harvested, lysates were thawed together, and RNA was extracted in parallel using an RNeasy Plus Mini kit (Qiagen, 74134) in a Qiagen Qiacube. cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad, 1708891), and reverse transcription quantitative PCR (qPCR) was performed on a QuantStudio 6 Flex machine using qPCRBIO SyGreen Blue Mix Lo-ROX (PCR Biosystems, PB20.11-05). Primers used are listed in table S16. Because hypoxia is an extreme stress on cells and causes a global change in gene expression, special considerations were needed to be taken for qPCR analysis. Many typically used qPCR control genes, such as GAPDH, are known to be down-regulated by hypoxic stress (83). Therefore, GAPDH would be a poor internal control for detecting gene expression differences at different hypoxic time points. Instead, we chose RPLP0 as an internal control as this gene has been shown to be stably expressed in hypoxia in the endothelium (83). qPCR cycling conditions were 1 cycle 95°C for 2 min, followed by 40 cycles of 95° for 5 s and 63° for 25 s. Statistical significance was determined using a one-tailed Student’s t test comparing the mean values of three technical replicates per clone. Results are displayed as means ± SEM.
Sustained hypoxia experiments
For sustained hypoxia experiments, three independent clonal isolates of ENH5 KO and WT control teloHAECs were plated at very low confluence (12,500 cells per well) in two sets of six-well plates and allowed to adhere overnight. The following day, one set of plates were placed in a humidified incubator contained within the box, and the entire box was maintained at 37°C, 5% CO2, 1% O2. The cell culture medium was changed every 2 days for 14 days, and then the cells were harvested as described above. RNA-seq library prep and sequencing were performed at the University of Chicago Genomics core and run together on the Illumina NovaSEQ (100 bp, paired end) with an average of 33.6M paired-end reads per sample. Analysis was performed using a standard RNA-seq pipeline. Briefly, reads were aligned using STAR, and read counts were performed with RSEM using default settings (73, 74). Differential expression analyses were performed using limma-voom with default settings (84, 85). All data were processed together and transcript levels modeled as ~βgenotypeGenotypei + βConditionConditioni + βinteractionGenotypei : Conditioni. To statistically assess the observed pattern of blunted response to hypoxia, we developed a blunting score. We had two criteria for including genes in our blunting score analysis: an adjusted P value < 0.05 for differential expression in one of the two genotypes and a consistent direction of effect for log fold change (LFC) in response to hypoxia in both genotypes. For up-regulated genes (those with LFC > 0 for both genotypes), we defined the blunting score to be equal to the difference in LFC in WT relative to KO. For down-regulated genes (those with LFC < 0 for both genotypes), we defined the blunting score to be equal to the difference in the absolute values of LFC in WT relative to KO. The blunting score is thus equivalent to the interaction term (βinteractionGenotypei : Conditioni) for up-regulated genes and negative of the interaction term (−βinteractionGenotypei : Conditioni) for down-regulated genes. We performed a two-tailed, one-sample Student’s t test to assess whether the mean blunting score differed significantly from 0. A positive mean blunting score would indicate a larger absolute effect size in the WT in response to hypoxia.
Generation and characterization of WT and KO mENH5 mice
All mouse work was approved by the Institutional Animal Care and Use Committee of the University of Chicago. All mice used were the C57BL/6J strain originally obtained from Charles River Laboratories Inc. (Wilmington, Massachusetts, USA). To test its functional conservation, the mouse orthologous ENH5 region (mENH5) was cloned from C57BL/6 mouse DNA into the same reporter gene assay vector and transfected into the same three cell types used to test the human ortholog. Similar to the approach we used to generate CRISPR-modified teloHAECs, mouse crRNA was designed to flank the orthologous ENH5 region in the mouse genome (~800 bp). RNPs (table S15) were generated as described above but were resuspended in nuclease-free water rather than resuspension buffer before use. The mixture of two RNPs was given to the University of Chicago Mouse Core for microinjection of C57BL/6 fertilized oocytes and implantation into a pseudopregnant female. Founder mice were genotyped using both internal and external primers relative to the deletion site (table S16). The internal primers were necessary as external primers would preferentially amplify the deletion band, and heterozygous mice were challenging to identify. A total of seven heterozygous mice were identified to form the founder population and were bred for at least two generations before downstream analyses. Sanger sequencing around the deletion site was performed in the homozygous founders to ensure proper deletion without small insertion events at the cut site.
RNA expression profiling in hypoxia-treated mice
All WT and KO mice were age-, cage-, and sex-matched. The average weight of these animals was 22.67 ± 0.06 g for the KOs and 22.17 ± 0.06 g for the WTs. At age 9 weeks, the mice were placed in pairs, one KO and one WT, into a custom hypobaric hypoxia chamber as described in (86). The pressure was set to 280 to 290 mmHg, which is equivalent to ~8% O2, and the mice were left overnight (~16.5 hours). The following morning, both mice were removed together and euthanized one at a time in a randomly selected order via urethane injection as described in (87). Mice were rapidly dissected, and left kidney, right adrenal gland, left lung, left and right atria, and left and right ventricles were all independently flash-frozen in either RLT Plus buffer (kidney, adrenal gland, and lung) or TRIzol (all heart tissues) and stored at −80°C. Once all tissues had been collected from all three pairs of mice, tissues were removed from −80°C and homogenized. RNA was extracted one tissue at a time using the RNeasy Plus Mini kit (Qiagen, 74134) in a Qiagen Qiacube for all samples stored in RLT buffer and by hand using the RNeasy plus mini kit with Qiagen deoxyribonuclease (DNase) treatment (RNase-Free DNase Set, catalog no. 79254). RNA was extracted and quality was assessed on an Agilent BioAnalyzer. The RNA integrity number (RIN) was recorded for each sample. RNA-seq library preparation was performed using the TruSeq RNA Library Preparation Kit v2, Set B (Illumina, RS-122-2002). RNA-seq libraries were submitted to the University of Chicago Genomics Core, where they were sequenced across seven lanes of the Illumina HiSeq4000 (one organ per lane, balanced by genotype) with an average of 69.6M 50-bp single end reads per sample.
Analysis of mRNA expression data was performed independently in each tissue using a standard RNA-seq pipeline. Briefly, reads were aligned using STAR, and read counts were assessed with RSEM. Data were filtered to remove unannotated and lowly expressed genes using the filter_by_expression() function in EdgeR, and differential expression analyses were performed using limma-voom with default settings (84, 85). Transcript amounts were modeled as ~0 + βgenotypeGenotypei + βBatchBatch + βRINRIN (or Time) where Batch is the pairs of mice that were treated together, and Time is the approximate time that the mice spent alive in normoxia before euthanasia. P values determined by limma from each tissue were converted into z scores and merged into a single matrix containing genes that appeared in all seven datasets. This matrix of z scores was used as an input for mashr V0.2-11, which uses data from all seven tissues to infer patterns of differential gene expression (50). Correlation due to sample overlap among tissues was accounted for by using estimate_null_correlation_simple(). Both canonical and data-driven covariance matrices were tested to generate a final dataset of differentially expressed (DE) genes. We analyzed the results using data-driven covariance matrices to demonstrate the best fit as measured by log-likelihood comparisons among models. Gene Ontology analyses on the significantly DE gene list from mashr was performed using the g:Profiler online platform, which uses a cumulative hypergeometric test for functional enrichment (1, 88). The platform offers multiple data sources including gene ontology (89), Kyoto Encyclopedia of Genes and Genomes pathway and WikiPathways (90, 91), and Human Phenotype Ontology (92), all of which were considered. All annotated mouse genes were used as the background universe.
Acknowledgments
We thank members of M. Stephens laboratory and particularly P. Carbonetto for advice on applying mashr to our data, M. Krause for technical support on protocols for endothelial culture and editing, F. Lee for advice and insights on modeling Tibetan mutations in mice, and L. Barreiro for advice on bioinformatics tools. We are grateful to J. Berg, J. Pritchard, and M. Przeworski for helpful comments on the manuscript and to three anonymous reviewers for comments that allowed us to improve our manuscript.
Funding: This work was supported by NIH grant to A.D.R. (HL119577). O.A.G. was supported by NIH Training Grants (GM007197 and HL07605) and NIH F31 (HL142146).
Author contributions: A.D.R., O.A.G., and M.A.N. conceived and supervised the study. O.A.G. performed ATAC, and D.W. performed the analysis. D.R.S. performed Capture Hi-C, and N.J.S. performed the analysis. O.A.G., J.J., and Y.F. designed, performed, and analyzed the luciferase. O.A.G. and D.W. performed CRISPR/qPCR/RNA-seq. O.A.G., J.Y., N.R.P., and Y.-J.P. designed, performed, and analyzed mouse/RNA-seq/analysis. O.A.G. and A.D. wrote the manuscript with comments from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Data described in this paper can be found in the Supplementary Materials or in publicly available databases including the UCSC genome browser and Epigenome Roadmap. All sequencing data are deposited in the GEO accession database (www.ncbi.nlm.nih.gov/geo/); accession no. GSE197527 (HAEC ATAC-seq/RNA-seq: GSE197523; teloHAEC RNA-seq: GSE197525; Capture Hi-C: GSE197526, mouse RNA-seq: GSE197524).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S6
Tables S6, S9, and S12 to S16
References
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S5, S7, S8, S10, and S11
Data file S1
REFERENCES AND NOTES
- 1.S. Fan, M. E. B. Hansen, Y. Lo, S. A. Tishkoff,Going global by adapting local: A review of recent human adaptation. Science 354,54–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.G. Coop, J. K. Pickrell, J. Novembre, S. Kudaravalli, J. Li, D. Absher, R. M. Myers, L. L. Cavalli-Sforza, M. W. Feldman, J. K. Pritchard,The role of geography in human adaptation. PLOS Genet. 5,e1000500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.R. D. Hernandez, J. L. Kelley, E. Elyashiv, S. C. Melton, A. Auton, G. McVean; 1000 Genomes Project, G. Sella, M. Przeworski,Classic selective sweeps were rare in recent human evolution. Science 331,920–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Murphy, E. Elyashiv, G. Amster, G. Sella, “Broad-scale variation in human genetic diversity levels is predicted by purifying selection on coding and non-coding elements”. bioRxiv 10.1101/2021.07.02.450762 (2021). [DOI] [PMC free article] [PubMed]
- 5.Y. G. Kamberov, S. Wang, J. Tan, P. Gerbault, A. Wark, L. Tan, Y. Yang, S. Li, K. Tang, H. Chen, A. Powell, Y. Itan, D. Fuller, J. Lohmueller, J. Mao, A. Schachar, M. Paymer, E. Hostetter, E. Byrne, M. Burnett, A. P. McMahon, M. G. Thomas, D. E. Lieberman, L. Jin, C. J. Tabin, B. A. Morgan, P. C. Sabeti,Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 152,691–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. M. Beall,Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107,11459–11464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. S. Simonson, Y. Yang, C. D. Huff, H. Yun, G. Qin, D. J. Witherspoon, Z. Bai, F. R. Lorenzo, J. Xing, L. B. Jorde, J. T. Prchal, R. Ge,Genetic evidence for high-altitude adaptation in Tibet. Science 329,72–75 (2010). [DOI] [PubMed] [Google Scholar]
- 8.X. Yi, Y. Liang, E. Huerta-Sanchez, X. Jin, Z. X. P. Cuo, J. E. Pool, X. Xu, H. Jiang, N. Vinckenbosch, T. S. Korneliussen, H. Zheng, T. Liu, W. He, K. Li, R. Luo, X. Nie, H. Wu, M. Zhao, H. Cao, J. Zou, Y. Shan, S. Li, Q. Yang, P. N. Asan, G. Tian, J. Xu, X. Liu, T. Jiang, R. Wu, G. Zhou, M. Tang, J. Qin, T. Wang, S. Feng, G. Li, Huasang, J. Luosang, W. Wang, F. Chen, Y. Wang, X. Zheng, Z. Li, Z. Bianba, G. Yang, X. Wang, S. Tang, G. Gao, Y. Chen, Z. Luo, L. Gusang, Z. Cao, Q. Zhang, W. Ouyang, X. Ren, H. Liang, H. Zheng, Y. Huang, J. Li, L. Bolund, K. Kristiansen, Y. Li, Y. Zhang, X. Zhang, R. Li, S. Li, H. Yang, R. Nielsen, J. Wang, J. Wang,Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329,75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. A. Horscroft, A. O. Kotwica, V. Laner, J. A. West, P. J. Hennis, D. Z. H. Levett, D. J. Howard, B. O. Fernandez, S. L. Burgess, Z. Ament, E. T. Gilbert-Kawai, A. Vercueil, B. D. Landis, K. Mitchell, M. G. Mythen, C. Branco, R. S. Johnson, M. Feelisch, H. E. Montgomery, J. L. Griffin, M. P. W. Grocott, E. Gnaiger, D. S. Martin, A. J. Murray,Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. U.S.A. 114,6382–6387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. Miller, C. Tudor, V. Thorsten, U. U. Nyima, U. Sonam, U. Droyoung, L. Wright, M. Varner,Comparison of maternal and newborn outcomes of Tibetan and Han Chinese delivering in Lhasa, Tibet. J. Obstet. Gynaecol. Res. 34,986–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. F. Sun, T. S. Droma, J. G. Zhang, J. X. Tao, S. Y. Huang, R. G. McCullough, R. E. McCullough, C. S. Reeves, J. T. Reeves, L. G. Moore,Greater maximal O2 uptakes and vital capacities in Tibetan than Han residents of Lhasa. Respir. Physiol. 79,151–161 (1990). [DOI] [PubMed] [Google Scholar]
- 12.C. M. Beall,Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104,8655–8660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. W. Bigham, F. S. Lee,Human high-altitude adaptation: Forward genetics meets the HIF pathway. Genes Dev. 28,2189–2204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. G. Moore, D. Young, R. E. McCullough, T. Droma, S. Zamudio,Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am. J. Hum. Biol. 13,635–644 (2001). [DOI] [PubMed] [Google Scholar]
- 15.B. M. Groves, T. Droma, J. R. Sutton, R. G. McCullough, R. E. McCullough, J. Zhuang, G. Rapmund, S. Sun, C. Janes, L. G. Moore,Minimal hypoxic pulmonary hypertension in normal Tibetans at 3,658 m. J. Appl. Physiol. (1985) 74,312–318 (1993). [DOI] [PubMed] [Google Scholar]
- 16.N. Petousi, Q. P. P. Croft, G. L. Cavalleri, H.-Y. Cheng, F. Formenti, K. Ishida, D. Lunn, M. McCormack, K. V. Shianna, N. P. Talbot, P. J. Ratcliffe, P. A. Robbins,Tibetans living at sea level have a hyporesponsive hypoxia-inducible factor system and blunted physiological responses to hypoxia. J. Appl. Physiol. (1985) 116,893–904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. Jeong, D. B. Witonsky, B. Basnyat, M. Neupane, C. M. Beall, G. Childs, S. R. Craig, J. Novembre, A. D. Rienzo,Detecting past and ongoing natural selection among ethnically Tibetan women at high altitude in Nepal. PLOS Genet. 14,e1007650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. M. Beall,Quantitative genetic analysis of arterial oxygen saturation in Tibetan highlanders. Hum. Biol. 69,597–604 (1997). [PubMed] [Google Scholar]
- 19.Y. Peng, C. Cui, Y. He, Ouzhuluobu, H. Zhang, D. Yang, Q. Zhang, Bianbazhuoma, L. Yang, Y. He, K. Xiang, X. Zhang, S. Bhandari, P. Shi, Yangla, Dejiquzong, Baimakangzhuo, Duojizhuoma, Y. Pan, Cirenyangji, Baimayangji, Gonggalanzi, C. Bai, Bianba, Basang, Ciwangsangbu, S. Xu, H. Chen, S. Liu, T. Wu, X. Qi, B. Su,Down-regulation of EPAS1 transcription and genetic adaptation of tibetans to high-altitude hypoxia. Mol. Biol. Evol. 34,818–830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. E. Green, A. M. Turner,The role of the endothelium in asthma and chronic obstructive pulmonary disease (COPD). Respir. Res. 18,20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C.-J. Hu, J. M. Poth, H. Zhang, A. Flockton, A. Laux, S. Kumar, B. McKeon, G. Mouradian, M. Li, S. Riddle, S. C. Pugliese, R. D. Brown, E. M. Wallace, B. B. Graham, M. G. Frid, K. R. Stenmark,Suppression of HIF2 signalling attenuates the initiation of hypoxia-induced pulmonary hypertension. Eur. Respir. J. 54,1900378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R. Bartoszewski, A. Moszyńska, M. Serocki, A. Cabaj, A. Polten, R. Ochocka, L. Dell’Italia, S. Bartoszewska, J. Króliczewski, M. Dąbrowski, J. F. Collawn,Primary endothelial cell–Specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. FASEB J. 33,7929–7941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. L. Krock, N. Skuli, M. C. Simon,Hypoxia-induced angiogenesis: Good and evil. Genes Cancer 2,1117–1133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.P. I. Aaronson, T. P. Robertson, G. A. Knock, S. Becker, T. H. Lewis, V. Snetkov, J. P. T. Ward,Hypoxic pulmonary vasoconstriction: Mechanisms and controversies. J. Physiol. 570,53–58 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. Claesson-Welsh, E. Dejana, D. M. McDonald,Permeability of the endothelial barrier: Identifying and reconciling controversies. Trends Mol. Med. 27,314–331 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Batie, L. del Peso, S. Rocha,Hypoxia and chromatin: A focus on transcriptional repression mechanisms. Biomedicine 6,47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. B. Johnson, M. C. Barton,Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat. Res. 618,149–162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.I. Jung, A. Schmitt, Y. Diao, A. J. Lee, T. Liu, D. Yang, C. Tan, J. Eom, M. Chan, S. Chee, Z. Chiang, C. Kim, E. Masliah, C. L. Barr, B. Li, S. Kuan, D. Kim, B. Ren,A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 51,1442–1449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Li, J. T. Glessner, H. Zhang, C. Hou, Z. Wei, J. P. Bradfield, F. D. Mentch, Y. Guo, C. Kim, Q. Xia, R. M. Chiavacci, K. A. Thomas, H. Qiu, S. F. A. Grant, S. L. Furth, H. Hakonarson, P. M. A. Sleiman,GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 22,1457–1464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. Buniello, J. A. L. MacArthur, M. Cerezo, L. W. Harris, J. Hayhurst, C. Malangone, A. McMahon, J. Morales, E. Mountjoy, E. Sollis, D. Suveges, O. Vrousgou, P. L. Whetzel, R. Amode, J. A. Guillen, H. S. Riat, S. J. Trevanion, P. Hall, H. Junkins, P. Flicek, T. Burdett, L. A. Hindorff, F. Cunningham, H. Parkinson,The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47,D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Y. Wang, F. Song, B. Zhang, L. Zhang, J. Xu, D. Kuang, D. Li, M. N. K. Choudhary, Y. Li, M. Hu, R. Hardison, T. Wang, F. Yue,The 3D genome browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19,151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. E. Montefiori, D. R. Sobreira, N. J. Sakabe, I. Aneas, A. C. Joslin, G. T. Hansen, G. Bozek, I. P. Moskowitz, E. M. McNally, M. A. Nóbrega,A promoter interaction map for cardiovascular disease genetics. eLife 7,e35788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. R. Sobreira, A. C. Joslin, Q. Zhang, I. Williamson, G. T. Hansen, K. M. Farris, N. J. Sakabe, N. Sinnott-Armstrong, G. Bozek, S. O. Jensen-Cody, K. H. Flippo, C. Ober, W. A. Bickmore, M. Potthoff, M. Chen, M. Claussnitzer, I. Aneas, M. A. Nóbrega,Extensive pleiotropism and allelic heterogeneity mediate metabolic effects of IRX3 and IRX5. Science 372,1085–1091 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.B. Mifsud, F. Tavares-Cadete, A. N. Young, R. Sugar, S. Schoenfelder, L. Ferreira, S. W. Wingett, S. Andrews, W. Grey, P. A. Ewels, B. Herman, S. Happe, A. Higgs, E. LeProust, G. A. Follows, P. Fraser, N. M. Luscombe, C. S. Osborne,Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47,598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 35.E. Huerta-Sánchez, X. Jin, Asan, Z. Bianba, B. M. Peter, N. Vinckenbosch, Y. Liang, X. Yi, M. He, M. Somel, P. Ni, B. Wang, X. Ou, Huasang, J. Luosang, Z. X. P. Cuo, K. Li, G. Gao, Y. Yin, W. Wang, X. Zhang, X. Xu, H. Yang, Y. Li, R. Nielsen,Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512,194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. J. Percy, P. W. Furlow, G. S. Lucas, X. Li, T. R. J. Lappin, M. F. McMullin, F. S. Lee,A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N. Engl. J. Med. 358,162–168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.N. L. Downes, N. Laham-Karam, M. U. Kaikkonen, S. Ylä-Herttuala,Differential but complementary HIF1α and HIF2α transcriptional regulation. Mol. Ther. 26,1735–1745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W. J. Kent, C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, D. Haussler,The human genome browser at UCSC. Genome Res. 12,996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Ernst, M. Kellis,ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9,215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.X. Zhou, D. Li, B. Zhang, R. F. Lowdon, N. B. Rockweiler, R. L. Sears, P. A. F. Madden, I. Smirnov, J. F. Costello, T. Wang,Epigenomic annotation of genetic variants using the roadmap epigenome browser. Nat. Biotechnol. 33,345–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. C. Joslin, D. R. Sobreira, G. T. Hansen, N. J. Sakabe, I. Aneas, L. E. Montefiori, K. M. Farris, J. Gu, D. M. Lehman, C. Ober, X. He, M. A. Nóbrega,A functional genomics pipeline identifies pleiotropy and cross-tissue effects within obesity-associated GWAS loci. Nat. Commun. 12,5253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. C. Simon, L. Liu, B. C. Barnhart, R. M. Young,Hypoxia-induced signaling in the cardiovascular system. Annu. Rev. Physiol. 70,51–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. Naeije,Physiological adaptation of the cardiovascular system to high altitude. Prog. Cardiovasc. Dis. 52,456–466 (2010). [DOI] [PubMed] [Google Scholar]
- 44.L. Bernardi, "Heart Rate and Cardiovascular Variability at High Altitude" in 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE, Lyon, France, 2007;http://ieeexplore.ieee.org/document/4353892/), pp. 6678–6680. [DOI] [PubMed] [Google Scholar]
- 45.F. León-Velarde, F. C. Villafuerte, J.-P. Richalet,Chronic mountain sickness and the heart. Prog. Cardiovasc. Dis. 52,540–549 (2010). [DOI] [PubMed] [Google Scholar]
- 46.J. E. Crawford, R. Amaru, J. Song, C. G. Julian, F. Racimo, J. Y. Cheng, X. Guo, J. Yao, B. Ambale-Venkatesh, J. A. Lima, J. I. Rotter, J. Stehlik, L. G. Moore, J. T. Prchal, R. Nielsen,Natural selection on genes related to cardiovascular health in high-altitude adapted andeans. Am. J. Hum. Genet. 101,752–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.X. Li, Z.-N. Han, Y. Liu, L. Hong, B.-R. Cui, X. Cui,Endogenous ET-1 promotes ANP secretion through activation of COX2-L-PGDS-PPARγ signaling in hypoxic beating rat atria. Peptides 122,170150 (2019). [DOI] [PubMed] [Google Scholar]
- 48.V. L. Dengler, M. Galbraith, J. M. Espinosa,Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 49,1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.X. Xue, Y. M. Shah,Hypoxia-inducible factor-2α is essential in activating the COX2/mPGES-1/PGE2 signaling axis in colon cancer. Carcinogenesis 34,163–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. M. Urbut, G. Wang, P. Carbonetto, M. Stephens,Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 51,187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O. A. Gray, J. Yoo, D. R. Sobreira, J. Jousma, D. Witonsky, Y.-J. Peng, N. R. Prabhakar, Y. Fang, M. A. Nóbrega, “Supplementary Materials for: A pleiotropic hypoxia-sensitive EPAS1 enhancer is disrupted by adaptive alleles in Tibetans.” [DOI] [PMC free article] [PubMed]
- 52.X. Yang, K. K. K. Sheares, N. Davie, P. D. Upton, G. W. Taylor, J. Horsley, J. Wharton, N. W. Morrell,Hypoxic induction of Cox-2 regulates proliferation of human pulmonary artery smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 27,688–696 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Y.-T. Chang, C.-N. Tseng, P. Tannenberg, L. Eriksson, K. Yuan, V. A. de Jesus Perez, J. Lundberg, M. Lengquist, I. R. Botusan, S.-B. Catrina, P.-K. Tran, U. Hedin, K. Tran-Lundmark,Perlecan heparan sulfate deficiency impairs pulmonary vascular development and attenuates hypoxic pulmonary hypertension. Cardiovasc. Res. 107,20–31 (2015). [DOI] [PubMed] [Google Scholar]
- 54.L. Bai, Z. Yu, G. Qian, P. Qian, J. Jiang, G. Wang, C. Bai,SOCS3 was induced by hypoxia and suppressed STAT3 phosphorylation in pulmonary arterial smooth muscle cells. Respir. Physiol. Neurobiol. 152,83–91 (2006). [DOI] [PubMed] [Google Scholar]
- 55.B. S. Ferguson, S. A. Wennersten, K. M. Demos-Davies, M. Rubino, E. L. Robinson, M. A. Cavasin, M. S. Stratton, A. M. Kidger, T. Hu, S. M. Keyse, R. A. McKnight, R. H. Lane, E. S. Nozik, M. C. M. Weiser-Evans, T. A. McKinsey,DUSP5-mediated inhibition of smooth muscle cell proliferation suppresses pulmonary hypertension and right ventricular hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 321,H382–H389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. Xin, H. Zhang, Y. He, Z. Duren, C. Bai, L. Chen, X. Luo, D.-S. Yan, C. Zhang, X. Zhu, Q. Yuan, Z. Feng, C. Cui, X. Qi, Ouzhuluobu, W. H. Wong, Y. Wang, B. Su,Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation. Nat. Commun. 11,4928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Quilley, Y.-J. Chen,Role of COX-2 in the enhanced vasoconstrictor effect of arachidonic acid in the diabetic rat kidney. Hypertension 42,837–843 (2003). [DOI] [PubMed] [Google Scholar]
- 58.S. A. Watson, G. P. McStay,Functions of cytochrome c oxidase assembly factors. Int. J. Mol. Sci. 21,E7254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. L. Archambeault, L. R. Bärtschi, A. D. Merminod, C. L. Peichel,Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus. Evol. Lett. 4,282–301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. K. Pritchard, A. Di Rienzo,Adaptation – Not by sweeps alone. Nat. Rev. Genet. 11,665–667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.K. Watanabe, S. Stringer, O. Frei, M. Umićević Mirkov, C. de Leeuw, T. J. C. Polderman, S. van der Sluis, O. A. Andreassen, B. M. Neale, D. Posthuma,A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51,1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 62.S. B. Carroll,Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134,25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 63.GTEx Consortium ,The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369,1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.T. D. Price, A. Qvarnström, D. E. Irwin,The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B Biol. Sci. 270,1433–1440 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.J. F. Storz, G. R. Scott, Z. A. Cheviron,Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 213,4125–4136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Merilä,Perplexing effects of phenotypic plasticity. Nature 525,326–327 (2015). [DOI] [PubMed] [Google Scholar]
- 67.C. K. Ghalambor, J. K. McKAY, S. P. Carroll, D. N. Reznick,Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21,394–407 (2007). [Google Scholar]
- 68.D. Song, A. W. Bigham, F. S. Lee,High-altitude deer mouse hypoxia-inducible factor-2α shows defective interaction with CREB-binding protein. J. Biol. Chem. 296,100461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.C. M. Ivy, J. P. Velotta, Z. A. Cheviron, G. R. Scott,Genetic variation in HIF-2α attenuates ventilatory sensitivity and carotid body growth in chronic hypoxia in high-altitude deer mice. J. Physiol. 600,4207–4225 (2022). [DOI] [PubMed] [Google Scholar]
- 70.C. L. Peichel, D. A. Marques,The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos. Trans. R. Soc. B Biol. Sci. 372,20150486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D. J. Green, J. H. Walsh, A. Maiorana, V. Burke, R. R. Taylor, J. G. O’Driscoll,Comparison of resistance and conduit vessel nitric oxide-mediated vascular function in vivo: Effects of exercise training. J. Appl. Physiol. (1985) 97,749–755 (2004). [DOI] [PubMed] [Google Scholar]
- 72.J. D. Buenrostro, B. Wu, H. Y. Chang, W. J. Greenleaf,ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109,21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras,STAR: Ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 29,15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.B. Li, C. N. Dewey,RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12,323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sequence Alignment/Map format and SAMtools | Bioinformatics | Oxford Academic, (https://academic.oup.com/bioinformatics/article/25/16/2078/204688).
- 76.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu,Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9,R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. S. Ross-Innes, R. Stark, A. E. Teschendorff, K. A. Holmes, H. R. Ali, M. J. Dunning, G. D. Brown, O. Gojis, I. O. Ellis, A. R. Green, S. Ali, S.-F. Chin, C. Palmieri, C. Caldas, J. S. Carroll,Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481,389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. S. P. Rao, M. H. Huntley, N. C. Durand, E. K. Stamenova, I. D. Bochkov, J. T. Robinson, A. L. Sanborn, I. Machol, A. D. Omer, E. S. Lander, E. L. Aiden,A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159,1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass,Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38,576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.W. C. Claycomb, N. A. Lanson Jr., B. S. Stallworth, D. B. Egeland, J. B. Delcarpio, A. Bahinski, N. J. Izzo,HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. U.S.A. 95,2979–2984 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.I. V. Kulakovskiy, I. E. Vorontsov, I. S. Yevshin, R. N. Sharipov, A. D. Fedorova, E. I. Rumynskiy, Y. A. Medvedeva, A. Magana-Mora, V. B. Bajic, D. A. Papatsenko, F. A. Kolpakov, V. J. Makeev,HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-seq analysis. Nucleic Acids Res. 46,D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.M. D. Krause, R.-T. Huang, D. Wu, T.-P. Shentu, D. L. Harrison, M. B. Whalen, L. K. Stolze, A. Di Rienzo, I. P. Moskowitz, M. Civelek, C. E. Romanoski, Y. Fang,Genetic variant at coronary artery disease and ischemic stroke locus 1p32.2 regulates endothelial responses to hemodynamics. Proc. Natl. Acad. Sci. U.S.A. 115,E11349–E11358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.S. Bakhashab, S. Lary, F. Ahmed, H.-J. Schulten, A. Bashir, F. W. Ahmed, A. L. Al-Malki, H. S. Jamal, M. A. Gari, J. U. Weaver,Reference genes for expression studies in hypoxia and hyperglycemia models in human umbilical vein endothelial cells. G3 (Bethesda) 4,2159–2165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.B. Phipson, S. Lee, I. J. Majewski, W. S. Alexander, G. K. Smyth,Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10,946–963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.C. W. Law, Y. Chen, W. Shi, G. K. Smyth,voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15,R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.B. Wang, Y.-J. Peng, X. Su, C. Zhang, J. S. Nagati, J. A. Garcia, N. R. Prabhakar,Olfactory receptor 78 regulates erythropoietin and cardiorespiratory responses to hypobaric hypoxia. J. Appl. Physiol. (1985) 130,1122–1132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.D. D. Kline, T. Yang, P. L. Huang, N. R. Prabhakar,Altered respiratory responses to hypoxia in mutant mice deficient in neuronal nitric oxide synthase. J. Physiol. 511,273–287 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.U. Raudvere, L. Kolberg, I. Kuzmin, T. Arak, P. Adler, H. Peterson, J. Vilo,g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47,W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.F. Cunningham, P. Achuthan, W. Akanni, J. Allen, M. R. Amode, I. M. Armean, R. Bennett, J. Bhai, K. Billis, S. Boddu, C. Cummins, C. Davidson, K. J. Dodiya, A. Gall, C. G. Girón, L. Gil, T. Grego, L. Haggerty, E. Haskell, T. Hourlier, O. G. Izuogu, S. H. Janacek, T. Juettemann, M. Kay, M. R. Laird, I. Lavidas, Z. Liu, J. E. Loveland, J. C. Marugán, T. Maurel, A. C. McMahon, B. Moore, J. Morales, J. M. Mudge, M. Nuhn, D. Ogeh, A. Parker, A. Parton, M. Patricio, A. I. Abdul Salam, B. M. Schmitt, H. Schuilenburg, D. Sheppard, H. Sparrow, E. Stapleton, M. Szuba, K. Taylor, G. Threadgold, A. Thormann, A. Vullo, B. Walts, A. Winterbottom, A. Zadissa, M. Chakiachvili, A. Frankish, S. E. Hunt, M. Kostadima, N. Langridge, F. J. Martin, M. Muffato, E. Perry, M. Ruffier, D. M. Staines, S. J. Trevanion, B. L. Aken, A. D. Yates, D. R. Zerbino, P. Flicek,Ensembl 2019. Nucleic Acids Res. 47,D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.M. Kanehisa, Y. Sato, M. Furumichi, K. Morishima, M. Tanabe,New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47,D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.D. N. Slenter, M. Kutmon, K. Hanspers, A. Riutta, J. Windsor, N. Nunes, J. Mélius, E. Cirillo, S. L. Coort, D. Digles, F. Ehrhart, P. Giesbertz, M. Kalafati, M. Martens, R. Miller, K. Nishida, L. Rieswijk, A. Waagmeester, L. M. T. Eijssen, C. T. Evelo, A. R. Pico, E. L. Willighagen,WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46,D661–D667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.P. N. Robinson, S. Köhler, S. Bauer, D. Seelow, D. Horn, S. Mundlos,The human phenotype ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83,610–615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.K. Yokogami, S. Yamashita, H. Takeshima,Hypoxia-induced decreases in SOCS3 increase STAT3 activation and upregulate VEGF gene expression. Brain Tumor Pathol. 30,135–143 (2013). [DOI] [PubMed] [Google Scholar]
- 94.A. Stahl, J.-S. Joyal, J. Chen, P. Sapieha, A. M. Juan, C. J. Hatton, D. T. Pei, C. G. Hurst, M. R. Seaward, N. M. Krah, R. J. Dennison, E. R. Greene, E. Boscolo, D. Panigrahy, L. E. H. Smith,SOCS3 is an endogenous inhibitor of pathologic angiogenesis. Blood 120,2925–2929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.I. Tirado-Hurtado, W. Fajardo, J. A. Pinto,DNA damage inducible transcript 4 Gene: The switch of the metabolism as potential target in cancer. Front. Oncol. 8,106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.J. L. Ebersole, M. J. Novak, L. Orraca, J. Martinez-Gonzalez, S. Kirakodu, K. C. Chen, A. Stromberg, O. A. Gonzalez,Hypoxia-inducible transcription factors, HIF1A and HIF2A, increase in aging mucosal tissues. Immunology 154,452–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.X. Jiang, W. Tian, A. B. Tu, S. Pasupneti, E. Shuffle, P. Dahms, P. Zhang, H. Cai, T. T. Dinh, B. Liu, C. Cain, A. J. Giaccia, E. C. Butcher, M. C. Simon, G. L. Semenza, M. R. Nicolls,Endothelial hypoxia-inducible factor-2α is required for the maintenance of airway microvasculature. Circulation 139,502–517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.S. Wingett, P. Ewels, M. Furlan-Magaril, T. Nagano, S. Schoenfelder, P. Fraser, S. Andrews,HiCUP: Pipeline for mapping and processing Hi-C data. F1000Research 4,1310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S6
Tables S6, S9, and S12 to S16
References
Tables S1 to S5, S7, S8, S10, and S11
Data file S1