Abstract
Several bat species act as asymptomatic reservoirs for many viruses that are highly pathogenic in other mammals. Here, we have characterized the functional diversification of the protein kinase R (PKR), a major antiviral innate defense system. Our data indicate that PKR has evolved under positive selection and has undergone repeated genomic duplications in bats in contrast to all studied mammals that have a single copy of the gene. Functional testing of the relationship between PKR and poxvirus antagonists revealed how an evolutionary conflict with ancient pathogenic poxviruses has shaped a specific bat host-virus interface. We determined that duplicated PKRs of the Myotis species have undergone genetic diversification, allowing them to collectively escape from and enhance the control of DNA and RNA viruses. These findings suggest that viral-driven adaptations in PKR contribute to modern virus-bat interactions and may account for bat-specific immunity.
Exceptional adaptations in bat PKR drive host-specific interactions with modern viruses and potent antiviral response in bats.
INTRODUCTION
The present architecture of host innate immunity is the result of long-standing conflicts with ancient pathogenic viruses that continually adapted and counteradapted to defeat or evade the antiviral defense of their host (1, 2). Hallmarks of these virus-host conflicts are the disproportionate accumulation of nonsynonymous mutations and genetic novelties over evolutionary times at the interface of host antiviral effectors and viruses. While being the result of past viral pressure, these adaptations may explain why hosts are susceptible—or resistant—to modern-day viruses and may also enlighten the functional diversity of host antiviral defenses (1). Therefore, comparative functional genomics of hosts and viruses are of utmost importance to better understand what drives the specificity of virus-host interactions, particularly in wild host reservoirs of zoonotic viral pathogens.
As the second most diverse and geographically widespread mammalian order, bats are outstanding among mammals because of their unique capability of powered flight and their propensity to host a substantial viral richness (3). Several bat species are natural reservoirs for viruses that are highly virulent in other mammals, such as Marburg virus, Nipah virus, and severe acute respiratory syndrome (SARS) coronaviruses, without themselves showing symptoms (3). These differences between bats and other mammals, particularly humans and nonhuman primates, have recently gathered considerable efforts to characterize the antiviral mechanisms of these flying mammals (3). Bats may have evolved unique adaptations in their inflammasome components and signaling factors [e.g., NOD-like receptor family, pyrin domain containing 3 (NLRP3) (4), Stimulator of interferon genes (STING) (5), interferon (IFN) regulatory factor 3 (IRF3) (6), and receptor-interacting protein kinase 1 (7)] that mitigate flight’s detrimental effects and dampen excessive inflammation, thereby presumably increasing viral tolerance. Furthermore, with more than 1200 species and approximately 60 million years of divergence (8), bats have coevolved with a large diversity of viral pathogens. As a result, specific adaptive changes may also enable bats to efficiently control viral infections (9). For example, a handful of bat antiviral factors bear signatures of strong positive selection and gene duplications (9), including key restriction factors, such as Apolipoprotéins B mRNA editing enzyme, catalytic polypeptide-like proteins (APOBECs) (9), MX family guanosine triphosphatases (10), interferon-induced transmembrane protein 3 (IFITM3) (11), and Tripartite Motif Containing 5 and 22 (TRIM5, TRIM22) (12). Nevertheless, efforts to broadly and comprehensively characterize the functional diversification of bat restriction factors, compared to other mammals, remain very limited. In particular, conclusions from most functional studies on bat immunity are primarily drawn from a specific bat species and a virus system. In-depth comparative and functional studies of bat antiviral effectors based on representative species are thus needed to decipher the diversity and specificities of chiropteran antiviral immune mechanisms.
Among the innate antiviral mechanisms, activation of the protein kinase R (PKR) constitutes one of the first line of mammalian antiviral defense. PKR is a keystone immune sensor and a broad restriction factor of a multitude of DNA and RNA viral families, such as Poxviridae, Herpesviridae, and Orthomyxoviridae. Upon sensing of viral double-stranded RNA (dsRNA), PKR phosphorylates the α subunit of eukaryotic initiation factor 2α (eIF2α), leading to a potent cap-dependent translational shut down and viral inhibition. The importance of PKR in immunity is further highlighted by the fact that viruses have, in turn, developed various and strong antagonism mechanisms to circumvent PKR function (13). One remarkable example is the mimicry of eIF2α by the poxvirus antagonist protein K3, which directly interacts with PKR to block eIF2α phosphorylation (14). Over evolutionary times, PKR has continually been under pathogen’s pressure, as exemplified by its rapid adaptive evolution in primates and rabbits (15–17).
In bats, how PKR has genetically and functionally evolved and how its past diversification contributes to modern bat-virus interplays remain unknown. Here, we report deep functional adaptive changes and exceptional gene duplications in bat PKR that broaden escape mechanisms to viral antagonism, including from poxviruses, orthomyxoviruses, and herpesviruses, and enhance viral control in Myotis bats. Using an evolutionary-guided functional approach, we show that long-standing genetic conflicts with viral pathogens have driven the rapid evolution and duplication of bat PKRs, and the resulting adaptive changes account for modern host-virus antagonism specificity.
RESULTS
PKR has been the target of strong diversifying selection and unusual gene duplication events in bats
The scarcity of bat genome sequences limits the study of their immunity, their virus-host interface, and evolutionary history. To increase the robustness of our evolutionary analyses of bat PKRs, we sequenced and cloned additional coding sequences from 15 additional bat species (see Materials and Methods). Overall, 33 bat orthologous sequences of PKR have been included in our positive selection analyses, spanning 62 million years of evolution (8). We compared models that disallow positive selection (models M1 and M7) to those allowing for positive selection (M2 and M8) using the Phylogenetic Analysis by Maximum Likelihood (PAML) Codeml package (18). We found that PKR has evolved under strong and recurrent positive selection during bat evolution, leading to significant adaptive signatures at both the gene and the codon levels (PAML codeml M1 versus M2 and M7 versus M8, P = 4.4 × 10−83 and 7.7 × 10−86, respectively; table S1).
To determine whether this adaptive evolution is common to all mammals, we extended our analysis to four other major mammalian orders: Primata, Rodentia, Artiodactyla, and Carnivora. We showed that rapid and recurrent evolution of PKR is common, with significant evidence of positive selection in all tested mammals (PAML codeml M1 versus M2 and M7 versus M8, P < 4.2 × 10−4 and 2.5 × 10−5, respectively; table S1). However, comparative analyses suggest more frequent signatures of adaptive changes in bat PKR and marked differences in the location of the evolutionary footprints compared to other orders. While most of the rapidly evolving sites are concentrated in the kinase domain of primate, artiodactyl, and rodent PKRs (Fig. 1A), the fast-evolving codons are scattered across bat PKR, with three remarkable hotspots in the second dsRNA binding domain, the linker region, and the kinase domain (Fig. 1A). Because bat lineages may have evolved under different selective pressures, we used a branch-specific model [adaptive Branch-Site Random Effects Likelihood (aBSREL)] (19) to test for episodic positive selection in PKR during bat evolution. We found that several bat lineages have been the targets of intensive episodic positive selection, particularly in the Yangochiroptera infraorder (Fig. 1B), indicating differential pressure during bat evolution. Extending the branch analysis to other mammals showed that bat PKRs were among the most important targets of episodic positive selection across mammals (fig. S1).
Fig. 1. PKR has been the target of strong diversifying positive selection and original duplication in bats.
(A) Sites under positive selection in mammalian protein kinase R (PKR). Graphic panels represent the posterior probabilities of positive selection [Bayes empirical Bayes (BEB)] (y axis) in the M2 model (ω > 1) for each codon (x axis). Red bars indicate sites with BEB of >0.95. Numbers in brackets are the total species analyzed. Viral antagonists: Herpes virus US11, influenza A (IAV) virus NS1A, poxvirus E3 and K3 (green striped boxes), hepatitis C virus (HCV) NS5A, and human immunodeficiency virus Tat, reported to directly interact with PKR. (B) Maximum likelihood phylogeny of bat PKR indicating the branches under positive selection (P < 0.05, in red). Brackets, estimated values of ω and the proportion of sites under positive selection. Scale bar, number of substitutions per site. (C) Maximum likelihood phylogeny of Myotis PKR paralogous transcripts, with Murina aurata, E. fuscus, Lasiurus borealis, and Pipistrellus kuhlii as outgroups (collapsed for visualization). PKR1L and PKR2L may be paralogs or splicing variants of PKR1 and PKR2, respectively. Colors indicate the duplicated PKRs isolated from one individual. Bootstrap values of ≥0.7 are shown. Scale bar, number of substitutions per site. (D) Canonical locus of EIF2AK2/PKR in mammals. The EIF2AK2 genes (black and grey arrows), the EIF2AK2 pseudogene (striped arrow), and the adjacent genes (white arrows) are shown. The genomic coordinates are indicated. (E) Expression pattern of PKR paralogs upon basal and IFNα treatment of M. velifer fibroblasts. Boxplots represent the number of reads in log10 scale for each condition and PKR copy (for exons found in both genes).
Other forms of genetic changes may be adaptive during evolutionary virus-host arms races. Notably, gene duplication and recombination are among the most important mechanisms underlying the diversification of the mammalian antiviral repertoire. We thus investigated how the gene encoding bat PKR, EIF2AK2, has evolved at the genomic level. Analyzing the publicly available genomes, we found distinct PKR-like sequences in the Myotis bats that suggested gene duplication of EIF2AK2 specifically in this chiropteran genus. However, because most of the publicly available PKR sequences from Myotis species are of low quality (i.e., highly fragmented and low coverage) and the PKR locus in the Myotis myotis genome (9) is incomplete, we sampled seven new Myotis species (M. bechsteinii, M. emarginatus, M. nigricans, M. riparius, M. myotis, M. mystacinus, and M. velifer; see Materials and Methods for details) and sequenced their complete PKR mRNA transcripts, as well as two genomic DNA (gDNA) fragments of the EIF2AK2 locus. This allows identification of potential differences in intronic regions between the putative PKR duplicates, which would be evidence of authentic genomic duplication and not splicing variants. Combining our results from mRNA and gDNA data, we found that EIF2AK2 has experienced repeated duplication events in a species-specific manner, leading to gene copy number variation across Myotis species (Fig. 1C and fig. S2). In particular, we detected evidences of PKR duplicates in four Myotis species (M. nigricans, M. riparius, M. myotis, and M. velifer), including a pseudogenized retrocopy of PKR that is specifically present in the New World clade of Myotis (Fig. 1C and figs. S2 to S6). M. myotis encodes two paralogous copies with intact open reading frames (referred to as PKR1 and PKR2) and one transcript variant (PKR1L) that may be a paralog or a splicing variant of PKR1 (Fig. 1C and figs. S3 and S4). In M. velifer, we isolated three distinct PKR sequences, two of which are paralogs (PKR1 and PKR2), while the third one is an isoform of PKR2 (figs. S3 and S5). The same pattern was found in M. riparius, although the complete coding sequence of PKR was solely obtained for one copy, while the others were partial sequences. The other Myotis species (M. bechsteinii, M. emarginatus, and M. mystacinus) had a single copy of PKR, although technical limits could have impaired the detection of PKR duplicates. Together, the phylogenetic and genomic analyses indicate a complex evolutionary history in the Myotis EIF2AK2/PKR locus, involving an ancient duplication of PKR before the diversification of Myotis genus around 30 Ma ago. This duplication event was then presumably followed by independent lineage-specific duplications (Fig. 1, C to D).
To characterize the genomic localization of EIF2AK2 duplicates, we analyzed the genomic locus of EIF2AK2 in M. velifer from an ongoing genomic sequencing project of the species. We mapped two EIF2AK2 copies in tandem in the M. velifer draft genome (Fig. 1D), while the pseudogene was located outside the canonical locus in the same chromosome. However, whereas one copy had an integral structure spanning from the 5′ untranslated region (5′UTR) to the 3′UTR, the second gene lacked the 5′UTR and the first four exons, probably resulting from technical issues at the assembly step (Fig. 1D and fig. S5). In addition, mRNA expression from RNA sequencing (RNA-seq) analyses of PKRs in M. velifer cells further showed that the two PKR copies are expressed in basal conditions and their expression is stimulated upon type I IFNα treatment (Fig. 1E).
Genetic arms races have shaped a specific bat PKR–poxviral K3 interface
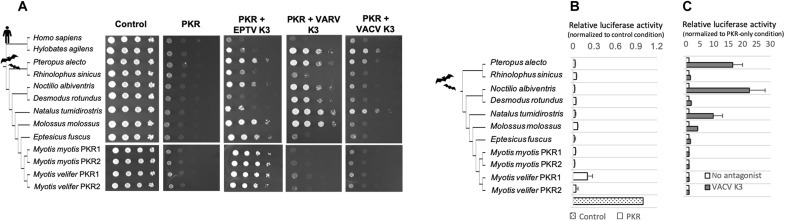
Such extensive molecular and genomic changes in bat PKR could be the result of pathogenic virus-driven selective pressure. Specifically, because (i) we identified, in bat PKR, a hotspot of positive selection at residues known to interface with poxvirus antagonist K3 in primate PKR (15, 16) and (ii) poxviruses are now circulating in bats (20–23), we investigated the specificity of bat PKR–poxvirus K3 interface in heterologous virus-host assays (24). On the virus side, we used a panel of K3 antagonists isolated from (i) Eptesipox virus (EPTV) (22), which naturally infects the bat Eptesicus fuscus, (ii) the archetypal poxvirus vaccinia virus (VACV), and (iii) the well-known human pathogen variola virus (VARV). On the host side, we tested the bat PKR-duplicated copies and seven orthologs from representative species, spanning 60 million years of chiropteran divergence, to capture the functional diversification of bat PKR-virus interface. We first used a surrogate yeast system in which the ability of PKR to drive translational shutoff in presence or absence of active antagonists can be directly assessed by measuring yeast growth rates (15, 16, 24). First, we found that the PKR paralogs and orthologs were all able to shut down protein synthesis in yeast, indicating that bat PKRs, including the duplicated PKR genes in Myotis, encode for functional proteins and retain their primary protein synthesis shutdown function (Fig. 2, A and B, and fig. S7). Second, while the PKR paralogs had the same phenotype to K3 antagonism, the orthologous PKRs differed in their ability to escape poxviral K3s in a host species-specific manner (Fig. 2, A and C). Last, we identified marked differences for PKR antagonism between the tested K3s, revealing virus-specific determinants of poxvirus K3 antagonism (Figs. 2A and 3A). To test whether this was also the case in a mammalian cellular system, we used HeLa PKR–knockout (KO) cells in which we transiently coexpressed PKR ± K3 and a luciferase expression plasmid as a reporter system for cell translation. We obtained similar results (Fig. 2, B and C), thereby validating the reliability of our yeast system. In this assay, all PKRs showed comparably strong repression of luciferase expression, with the exception M. velifer PKR2, which showed somewhat weaker activity (Fig. 2B).
Fig. 2. Species specificity in bat PKR resistance and poxvirus K3 antagonism.
(A) Species-specific sensitivity of PKR to distinct poxvirus K3 proteins in yeast assays. Plasmids expressing PKR paralogous copies (PKR1 and PKR2) and orthologous variants under a galactose-inducible promoter were introduced into a wild-type yeast strain or yeast strains expressing VACV, VARV, or EPTV K3. PKR variants from human and gibbon were used as positive control. Tenfold serial dilutions of transformants were spotted on plates containing either 2% glucose (control) or galactose. The Western blots for the expression of PKR and K3 proteins in yeasts are shown in fig. S6. (B) Luciferase reporter assay showing that the PKR orthologs and paralogs inhibit the protein expression at comparable levels, except M. velifer PKR1 showing slight differences. Luciferase activity was normalized to the no-PKR condition, in which cells were transfected with luciferase and empty vector. (C) Luciferase reporter assay confirming the differential sensitivity of PKR variants to VACV K3. Three independent experiments were conducted for bat PKR variants. Luciferase activity was normalized to the control condition in which cells were transfected with luciferase, PKR, and empty vector (x axis). Error bars indicate SEM.
Fig. 3. Evolutionary-guided functional approach reveals adaptive within-protein epistasis in bat PKR and a unique C-terminal extension in the bat poxvirus K3.
(A) Yeast spot assays of D. rotundus PKR mutants assaying PKR susceptibility to EPTV and VARV K3. Mutants D1 to D4 are chimeras between D. rotundus PKR and M. myotis PKR2. (B) Western blot of PKR and K3 in yeast. (C) Alignment of the residues underlying PKR-EPTV K3 interface. Left: Species cladogram of corresponding PKRs. Right: PKR protein alignment. Colors indicate site variations compared to the consensus (25% threshold). Codon numbers based on D. rotundus PKR sequence. Triangles: Residues under positive selection. (D) 3D protein structure of D. rotundus and M. myotis PKR2 kinase domain, obtained by I-TASSER. Red, residues identified by our assays in Helix αG; magenta, those in the kinase insert. (E) Multiple alignment and comparison of poxvirus K3s. Colors indicate site variations compared to the consensus (25% threshold). Sequence numbering based on EPTV K3. Right: 3D protein structure of VACV (Protein Data Bank 1LUZ) and EPTV K3 (I-TASSER). Dark blue, C-terminal insertion of EPTV K3. (F) Yeast assays of bat PKRs challenged with EPTV K3 and K3ΔCter (C-terminal Δ85 to 120 amino acids). Right: EPTV K3 and K3ΔCter. Rsin, R. sinicus; Mmol, M. molossus; Drot, D. rotundus; Mmyo, M. myotis. (G) Protein-protein complex structure between M. myotis PKR2 kinase domain and EPTV K3, inferred by Hdock. Gray, PKR kinase domain; red and pink, residues at the interface according to (D). Green, EPTV K3; blue, residues at the interface (dark blue, predicted contact residues; light blue, vaccinia K3-PKR binding residues) (63).
To investigate the genetic determinants underlying these phenotypic differences, we used an evolutionary-guided approach on both the host and the virus sides. On the host side, because Desmodus rotundus and M. myotis PKRs displayed opposite phenotypes to EPTV and VARV K3 antagonism, we generated a series of chimeras and mutants between these orthologs and tested their capacity to escape EPTV and VARV K3 antagonism (Fig. 3A and fig. S8). We showed that residues 475/476, located in the helix αG in D. rotundus, drive species specificity to variola K3 antagonism (Fig. 3A). This determinant is similar to the previously reported residue 496 in human PKR-VARV K3 interface (16). However, we further identified a yet undescribed within-protein epistatic (25) interaction between the residues 475 and 476 and 332 to 344 in the kinase insert of D. rotundus PKR (Fig. 3A) that represent specific determinants of susceptibility/resistance to EPTV K3. Swapping these sites significantly reduced K3-antagonist resistance of D. rotundus PKR and conversely in M. myotis PKR, without impeding their expression and their basal translation shutdown function (Fig. 3, A and B). These sites were among the fastest evolving codons in bat PKRs—with many substitutions and indels within the 332– to 344–amino acid stretch (Fig. 3C), substantially affecting the predicted three-dimensional (3D) structure of bat PKR (Fig. 3D). Therefore, accumulated mutations at these sites are adaptive in the context of bat-virus interactions and drive the host–poxvirus K3 specificity, supporting that ancient poxviruses that targeted these regions have been key drivers of PKR adaptation across bat species.
On the virus side, the protein alignment of orthopoxvirus K3 sequences revealed a unique structural C-terminal insertion in EPTV K3 (Fig. 3E). To investigate whether this insertion functionally contributes to bat PKR antagonism, we generated an EPTV K3 mutant, which lacks the C-terminal insertion, and tested its ability to antagonize PKRs compared to the wild-type K3. We found that the truncated K3 had a notable reduced anti-PKR activity, in a host-specific manner, without affecting K3 expression (Fig. 3F). This shows that the C-terminal insertion in EPTV K3 is essential for bat PKR antagonism and accounts for species specificity. Combining these functional assays with a protein-protein docking model, which was performed with the HDOCK software (26), we showed that the C-terminal insertion may be involved in PKR antagonism through direct contact with the residues 475-476, and 340, located in the helix αG and the kinase insert, respectively (Fig. 3G). In accordance with the functional assays, the protein complex between EPTV K3 and bat PKRs further depended on the bat PKR sequence and their 3D structure (fig. S9). Therefore, this C-terminal insertion may reflect a counteradaptation of EPTV K3 to maintain PKR antagonism.
The PKR paralogs functionally diverge in the ability to escape from poxvirus E3, cytomegalovirus TRS1, and influenza A virus NS1 antagonists
Because the PKR copies did not show phenotypic differences to poxvirus K3 antagonism, we tested whether they have evolved differences regarding their susceptibility to other viral antagonists encoded by poxviruses and other viral families that naturally infect bats or humans. In particular, we focused on the M. myotis paralogs and tested (i) E3 antagonist from EPTV (22) and VACV poxviruses, (ii) NS1 antagonist from human influenza A (IAV) H1N1 virus (27), (iii) NS5A proteins from human hepatitis C virus (HCV) and from bat hepaciviruses infecting Otomops martiensseni (Omar) (28) and Peropteryx macrotis (Pmac) (28), and (iv) TRS1 protein from human cytomegalovirus (29). Using the luciferase reporter assay, we showed that the Myotis PKR copies functionally and significantly differ in their ability to escape the viral antagonists NS1, TRS1, and E3 (Fig. 4, A and B, and fig. S10) under conditions where PKR1 and PKR2 block the luciferase activity to similar extent. Although the immunoblot analysis showed that PKR2 was more expressed than PKR1 in this assay, this latter could efficiently escape from the viral antagonists. In the case of TRS1 and VACV E3 antagonisms, the intraspecies differences between the PKR paralogs were of the order of magnitude of the interspecies PKR orthologs (fig. S11). Overall, this shows that adaptive duplication of PKR in Myotis bats has broadened escape mechanisms to antagonism from very diverse RNA and DNA viral families.
Fig. 4. Functional divergence of the PKR paralogs in their ability to escape from poxvirus E3, cytomegalovirus TRS1, and IAV NS1 antagonists.
(A and B) Relative luciferase activity in cells transfected with or without human PKR, M. myotis PKR1 or PKR2, in the presence or absence of putative viral antagonists: VACV E3 (A), and bat HCV NS5A (O. martiensseni and P. macrotis strains), human HCV NS5A (JFH1), human IAV NS1, human CMV TRS1, and EPTV E3 (B). The results shown are mean value of three and five independent experiments for (A) and (B), respectively. Luciferase activity was normalized to the control condition in which cells were transfected with luciferase, PKR, and empty vector. Error bars are SEM; asterisks indicate statistically significant differences between PKR1 and PKR2 (*P < 0.05). (C) Luciferase reporter assay of M. myotis PKR paralog chimeras, generated by swapping the kinase domain (chimera 1), the linker region (chimera 2), or the dsRNA domain (chimera 3) of M. myotis PKR1 (black) with that of PKR2 (gray). Luciferase activity was normalized to the luciferase-only condition. The graph represents the mean of three independent replicates. (D) Luciferase reporter assay of PKR chimeras challenged with human IAV NS1 (mean of three biological replicates). Error bars are SEM. Luciferase activity was normalized to the condition without antagonist. (E) Protein sequence alignment of M. myotis and M. velifer PKR duplicates and potential isoform. Black bars on top of alignment indicate positively selected residues that differ between the paralogs. Colors indicate site variations compared to the consensus within species (25% threshold). Sequence numbering based on M. myotis PKR1 sequence.
To decipher the underlying determinants of these functional differences, we engineered three chimeric PKR proteins, by swapping the dsRNA binding domain, the linker region, or the kinase domain of the duplicated copies, and we tested their susceptibility to human IAV NS1 antagonism. We showed that the linker region of the PKR paralogs drives the susceptibility or resistance to NS1, indicating that it is a key determinant for bat PKR antagonism by NS1 (Fig. 4, C and D). Most of the genetic intraspecies differences between the PKR copies are concentrated in the linker region (Fig. 4E). Mapping the positively selected sites (inferred from the interspecies analyses) on the PKR paralogs, we found that several of these sites have undergone amino acid replacement in the PKR duplicates (Fig. 4E), including in the linker region. Combined with our functional assays, these results indicate that (i) ancient viral pathogens from diverse RNA and DNA virus families may have contributed to the duplication and fixation of Myotis PKR paralogs and (ii) the resulting evolutionary patterns in PKR paralogs account for distinct interactions with modern viral proteins.
Bat PKR duplication leads to differential and potentially additive restriction of poxviral VACV and rhabdoviral VSV infections
The fact that the PKR duplicated copies (i) inhibit cellular translation, (ii) are up-regulated upon IFN stimulation, and (iii) are antagonized by diverse viral proteins suggests that both are potential antiviral restriction factors. To test this, we performed viral infection assays with two representative DNA and RNA viruses.
First, we created T-REx-293 PKR-KO cell lines expressing PKR1 or PKR2 from M. myotis or M. velifer, or E. fuscus PKR under doxycycline induction. For the infectivity assays with the DNA virus, we used a vaccinia poxvirus VC-R4 (30) lacking the viral K3 and E3 antagonists (VACV∆K3∆E3) and expressing a virus replication reporter enhanced green fluorescent protein (EGFP). We found that the PKR paralogs significantly differed in their capacity to restrict VC-R4 replication. Whereas M. myotis and M. velifer PKR2 effectively inhibited VC-R4, as also seen for E. fuscus PKR, M. myotis and M. velifer PKR1 had only weak and no effect on the EGFP signals, respectively (Fig. 5, A and B). This was the case despite comparable expression of the PKR paralogs (Fig. 5C). We further titrated VC-R4 replication in representative cell lines. T-REx-293 cells expressing E. fuscus PKR and M. velifer PKR2 were not included, as they showed the same level of EGFP suppression as M. myotis PKR2. The titration supported the differences in EGFP signals in the microscopy images, with M. myotis PKR2 expression conferring a 1000-fold reduction in VC-R4 titer, whereas only 3.6- and 1.2-fold titer reductions were observed for M. myotis PKR1 and M. velifer PKR1, respectively.
Fig. 5. Bat PKR duplication allows for differential and potential additive antiviral restriction of poxvirus and rhabdovirus infections.
(A) T-REx-293 PKR-KO cells stably expressing M. myotis and M. velifer PKR1 and PKR2 or E. fuscus PKR were infected with the VC-R4 (EGFP-VACVΔK3LΔE3L) at an MOI of 0.1, EGFP expression were imaged 24 hours postinfection (hpi). (B) Cells were infected as indicated above. Viruses in cell lysates were titrated in RK13+E3L+K3L cells. Error bars represent the SD from two independent infections. Fold differences in virus titers obtained with –doxycycline and +doxycycline are shown. (C) Expression of bat PKRs in the stable T-REx-293 PKR-KO cells. Cell lysates were separated on 10% SDS–polyacrylamide gel electrophoresis gels and analyzed by immunoblot analysis with anti-FLAG and anti–β-actin antibodies. (D) Hela PKR-KO cells were transfected with or without M. myotis or M. velifer PKR1 or PKR2 or with ISG20 (as a positive control of viral restriction). Infection was performed 24 hours after transfection with a VSV-GFP virus at an MOI of 3, and cells were fixed at 18 hpi for flow cytometry analyses. Infectivity is measured as the ratio of the mean of %EGFP+ cells in each condition relative to the vector control condition. Values represent the mean computed from five independent experiments and error bars are SD. Dox, doxycycline; WT, wild type; PFU, plaque-forming units.
Second, to determine whether this pattern was virus dependent, we further tested the antiviral function of the paralogs against an RNA virus, the vesicular stomatitis virus (VSV) from the Rhabdoviridae family. We found that M. myotis and M. velifer PKR1s and PKR2s could all restrict VSV-GFP infection, although to varying extents (Fig. 5D), indicating that Myotis bats have two VSV restrictor PKR copies. Therefore, PKR genomic expansion and diversification in Myotis have led to at least two functional antiviral effectors, with potential specialization in the antipoxvirus activity and increased potency in the anti-VSV activity.
DISCUSSION
Combining in-depth phylogenetic and positive selection analyses with functional assays and experimental infections, we show how past genetic conflicts with pathogenic viruses have shaped chiropteran host antiviral immunity and susceptibility. In particular, we report extensive signatures of functional adaptation in PKR during bat evolution, with substantial molecular changes and genomic duplication, a novelty compared to other mammals. These adaptive changes now lead to species-specific interactions with contemporary viral pathogens and account for a specific, broad, and potent antiviral response in bats.
In primate PKR, single substitutions at specific residues in the helix αG are key determinants for vaccinia and variola K3 antagonism (15, 16). In contrast, the genetic basis and specificity of bat PKR sensitivity to EPTV K3 rely on a within-protein epistatic interaction (25) between two residues in the helix αG (475 and 476 in D. rotundus) and a stretch of amino acids in the kinase insert (332 to 344 in D. rotundus) of PKR. Although the role of this insert in PKR binding substrate was suggested in a previous study (31), its functional implication in bat PKR-K3 interaction indicates that it contributes to substrate discrimination in bats. Under virus-host conflicts, the flexible and disordered feature of the kinase insert may have been a source of evolutionary plasticity, allowing marked changes in PKR while maintaining eIF2α binding. In line with this, repeated deletions/insertions were found in the kinase insert of several bat species without negative cost on basal protein shutdown function. Such hotspots of variability in unstructured loops are also found in other antiviral proteins and are prime targets of viral antagonism while being essential for antiviral activity [e.g., (32)].
On the virus side, we showed that EPTV K3 evolved an adaptive C-terminal insertion that is essential for species-specific antagonism of bat PKR. This K3 C-terminal insertion was probably retained during EPTV evolution because of its increased PKR binding affinity through direct interaction with the kinase insert and the helix αG. Furthermore, because it increases PKR antagonism, the K3 insertion may not only drive the host range specificity but may also directly or indirectly contribute to EPTV virulence in bat host species. Comparing the EPTV K3 sequence to all other available mammalian poxvirus K3s showed that this C-terminal insertion was specific to EPTV. Its 86–amino acid length suggests that it could derive from gene transfer, as frequently observed in poxviruses (33). However, we failed to uncover the origin of this extension (i.e., no match in blat/blast searches), either from a parental host gene or from recombination of a viral sequence. One possible explanation is that the C-terminal sequence of EPTV K3 has substantially diverged from the parental one, such that their percentage of sequence similarity is negligible or nonexistent. Further studies will be important to determine whether other bat poxviruses have evolved similar adaptive changes and decipher the functional implication in poxvirus pathogenicity and epidemiology.
Apart from poxviruses, other viral pathogens have certainly contributed to the diversification of PKR in bats. These mammals are highly diverse and have evolved with many viral pathogens over million years. Therefore, the evolution of their PKR may reflect the selective pressure of different ancient epidemics. Notably, the fastest evolving codons in bat PKR are mapped with specific PKR-virus interfaces in primates, such as influenza virus NS1, cytomegalovirus TRS1, or hepacivirus NS5A (29, 34), in which homologs are encoded by bat-borne related viruses. Here, we found that ancient influenza- and cytomegalovirus-like viruses may have also been important drivers of PKR adaptation in bats, highlighting the diversity of viral selective pressures that have contributed to bat PKR evolution. In addition, the genetic differences between bat and other mammalian PKRs further suggest that specific bat-borne pathogens may be key actors and/or that related mammalian viruses may have evolved to antagonized different regions in bats.
Beyond substitutions or indels, we also found that gene duplication has diversified the bat antiviral repertoire in a lineage-specific manner. While all other studied mammals have one single gene encoding PKR, several bat species from the Myotis genus express at least two functional, genetically divergent copies of PKR. Expansions of genes encoding antiviral proteins were previously discovered in bat species, including the APOBEC3, IFITM3, and TRIM5 gene families, as well as the chimeric protein HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 5/6 (HERC5/6) (9, 11, 12, 35). However, these duplications involve known multigene immune families, which are prone to gene expansion in many mammals, in contrast to the EIF2AK2 (PKR) locus that is highly conserved in other mammals. Given the pleiotropic and central role of PKR in innate immunity, the duplication of PKR in Myotis bats reveals that major selective pressures have shaped bat evolution, leading to specific functional diversification in bat innate defense.
Before this study, independent PKR duplications were solely reported in amphibians and fishes (36). In the latter, a PKR-like protein, containing a Z-DNA binding domain, was described as a cooperator of fish PKR antiviral activity (37). In Myotis bats, the paralogs retained the typical structure of the mammalian PKR protein, with two dsRNA binding domains linked to a kinase domain, but they genetically differ through multiple amino acid changes and indels. Because the evolutionary fate of gene duplication depends on the benefits and costs associated with the duplicated copies, the fixation of PKR paralogs in Myotis genome suggests that they provide a functional selective advantage (38). Using two divergent RNA and DNA virus models (VSV and VACV, respectively) and various viral antagonists of PKR, we demonstrated that the PKR copies that could inhibit protein expression in a mammalian reporter assay differed in their capacity to restrict virus replication and escape viral antagonism, which may be an example of divergence with subfunctionalization (i.e., partitioning of the ancestral function between the copies). Therefore, the PKR paralogs have retained the basal function of the parental copy (i.e., translation shutdown) but have evolved specific roles in the host antiviral response, as reported in other cases of gene duplication [e.g., (39)]. One plausible explanation for this differential antiviral activity is that restriction and its potency is virus dependent. Alternatively, the PKR paralogs may have evolved to fill another functional niche. For example, PKR is positioned upstream of several important factors, such as the immune transcriptional regulator IRF-1 (40), or the inflammatory transcription factors nuclear factor κB (41) and signal transducer and activator of transcription 1 (42). Loosing one or some of these features could lower the overall antiviral response of one of the PKR paralogs, allowing specialization for another niche. Last, because PKR undergoes dimerization upon activation—which is essential for eIF2α phosphorylation, it is possible that PKR1 and PKR2 heterodimerize to confer a new function. Although we only tested the PKR paralogs independently, this remains possible, as we show that Myotis fibroblasts express both genes. Thus, one could postulate a synergistic functional interaction between the paralogs upon viral infection, which could modulate their function.
Overall, this study brings important clues on the functional diversification of bat antiviral repertoire. It was suggested that immune tolerance rather than increased viral control plays a key role in bat immunity (4, 5, 43). Here, the adaptive changes in bat PKRs increase the antiviral function and the viral evasion of PKR, which supports an adaptive enhancement for viral control in some species. This is in line with several studies reporting accelerated rate of evolution in bat restriction factors, indicating increased defense against virus infection (10–12, 44). Because each species has its own history of viral exposure, specific viral communities have certainly led to lineage-specific selection in bat’s antiviral immunity, highlighting the need to include multiple related species in comparative functional studies. Therefore, while dampening inflammatory response might be common to bats, strong episodic adaptations in antiviral factors, driven by ancient viral epidemics, may have shaped lineage-specific innate immune defenses in bats.
MATERIALS AND METHODS
Bat samples
Sampling was performed in France (Miniopterus schreibersii, M. emarginatus, M. myotis, and Rhinolophus ferrumequinum), French Guiana (D. rotundus, M. bechsteinii, M. riparius, M. nigricans, P. macrotis, Pteronotus rubiginosus, Tonatia saurophila, Natalus tumidirostris, Sturnira hondurensis, Molossus molossus, Noctilio albiventris, and Furipterus horrens), and Gabon (Hipposideros cf. ruber, Rousettus aegyptiacus). Authorizations were obtained from the Ministry of Ecology, Sustainable Development and Energy over the period 2015–2020 (approval no. C692660703 from the Departmental Direction of Population Protection, Rhône, France). Our methods for animal capture and management were approved by the Muséum national d’Histoire naturelle (MNHN), the Société française pour l’étude et la protection des mammifères (SFEPM), and the Direction de l’Environnement, de l’Aménagement et du Logement (DEAL) Guyane. African bat samples were approved by the Gabonese National Ethics Committee (Authorization N°PROT/0020/2013I/SG/CNE). Bat individuals were captured using harp traps at the entrance of caves or mist nests hoisted on the forest floor and in the tree canopy. The individuals were then released after sampling. All samples, including wing membrane and blood pellet, were conserved at −80°C until RNA extraction.
In addition to the wild field samples, immortalized fibroblast cells from wing tissue of E. fuscus and embryonic fibroblast cell lines from M. velifer were provided by the Feschotte Lab (Cornell University) (45). Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum, 1% penicillin/streptomycin, and 1% sodium pyruvate.
De novo sequencing of PKR (EIF2AK2) gene
Total genomic RNA was extracted from bat punches, fibroblast cells, and blood samples using Macherey-Nagel NucleoSpin RNA and RNA blood kits, respectively, following the manufacturer’s protocol. Total RNA was reverse transcribed into complementary DNA (cDNA) with random primers and oligo(dT), using the SuperScript III One-Step reverse transcription polymerase chain reaction (PCR) kit (Thermo Fisher Scientific, Poland). Species identification was first confirmed through PCR amplification and sequencing of cytochrome B gene (CytB), using the primers CytB-F and CytB-L/R (46). PKR mRNA was then amplified from each species using 30 ng of cDNA and different sets of primers (table S3) that were specifically designed using an alignment of publicly available PKR sequences. The PCR reactions were performed using the New England Biolabs (NEB) Q5 High-Fidelity DNA Polymerase, following the manufacturer’s protocol, including a final volume of 50 μl, a 0.5 μM primer concentration, and an annealing temperature of 58° to 60°C. PCR products with multiple bands were excised and purified from gel using the NucleoSpin Gel and PCR Clean-up Kit from Macherey-Nagel or cloned using the NEB PCR cloning kit (NEB) to obtain haplotype resolution. Sanger sequencing of PKR was performed by a commercial company (Genewiz, Azenta Life Sciences, Germany).
Collection of PKR orthologous sequences
To complete our dataset, orthologous coding sequences of bat PKR were retrieved from GenBank by BLASTn searches of the “Nucleotide,” “RefSeq Genome,” “Transcriptome Shotgun Assembly,” and “Whole-Genome Shotgun Contigs” databases, using the little brown bat (Myotis lucifugus) RefSeq coding sequence as query. In the case of unassembled bat genomes, PKR coding sequence was predicted from the genome contigs using Augustus (47) and GeneWise (48) with the little brown bat RefSeq protein as reference. In total, 19 bat PKR sequences were retrieved from public databases.
PKR coding sequences from primates (n = 29), rodents (n = 25), artiodactyls (n = 23), and carnivores (n = 19) were obtained by tBLASTn searches of the Nucleotide database from GenBank using human (Homo sapiens), mouse (Mus musculus), cow (Bos taurus), and dog (Canis lupus familiaris) PKR protein sequence as queries, respectively.
Phylogenetic and positive selection analysis of PKR orthologous sequences
PKR orthologous codon sequences were aligned for each mammalian group separately using the program PRANK (49), and the alignments were manually curated. We then built a phylogenetic tree, using the maximum likelihood method implemented in PhyML program (50). Selection of the best substitution model was performed with the Smart Model Selection (51) program in PhyML and was always: GTR + G + I. Node statistical support was computed through 1000 bootstrap replicates. The detection of recombination events was assessed with the Genetic Algorithm for Recombination Detection (GARD) implemented in the Hypothesis Testing using Phylogenies (HyPhy) package (52).
For positive selection analyses, models that disallow positive selection (models M1 and M7) were compared to those allowing for positive selection (M2 and M8) using the PAML Codeml package (18), with the following parameters: codon frequencies F61 and F3x4 and starting omega ω (dN/dS ratio) of 0.4. Comparison of each pair of models (M1 versus M2 and M7 versus M8) was then achieved with likelihood ratio tests. Bayes empirical Bayes of the dN/dS of >1 class in M2 or M8 models was used to assess positive selection at the codon level, with a posterior probability of ≥0.95 as significance threshold. The fast unbiased Bayesian approximation, the mixed effects model of evolution, and the fixed-effects likelihood, implemented in the HyPhy package (52), were also run to identify codons under significant positive selection. To ensure higher specificity, we considered that codons were under significant positive selection if they were identified by at least two methods. Moreover, to test whether the PKR domains (i.e., the dsRBD, the linker region and the kinase domain) have similarly been targets of positive selection, each domain was separately analyzed using the models M1, M2, M7, and M8 from the PAML package.
Last, we determined whether and how PKR experienced episodic selection during bat and mammalian evolution, using the branch-specific analysis aBSREL (19), implemented in the HyPhy package. This program allows testing the significance of positive selection and quantifying the dN/dS ratio for each branch independently. Sequences from perissodactyls (n = 3) and proboscidean (n = 1) were also analyzed. Tree visualization and annotation were performed with Interactive Tree Of Life webserver (https://itol.embl.de/).
Genomic and (phylo)genetic characterization of PKR paralogs in Myotis
Molecular identification of EIF2AK2 duplication was carried out in tissues from Myotis species, including M. myotis, M. velifer, M. riparius, M. nigricans, M. mystacinus, M. emarginatus, and M. bechsteinii. Total RNA and gDNA were extracted using the Macherey-Nagel NucleoSpin RNA tissue and gDNA kits, respectively, following the manufacturer’s instructions. Two complementary strategies were then used. First, PKR coding sequence was amplified from cDNA using the PKR “universal” primers designed in this study (table S3). Second, from gDNA, we PCR-amplified the genomic regions containing exons 1 to 3 (E1 to E3) and exons 4 to 6 (E4 to E6) of EIF2AK2 to identify potential differences in intronic regions between the putative PKR duplicates. Following PCR amplification, all PCR products from cDNA and gDNA were cloned into the pMiniT 2.0 Vector using the NEB Cloning Kit (NEB) and sequenced to ensure sequencing of a single DNA molecule. Phylogenetic reconstruction of the PKR paralogs followed the previously described phylogenetic analysis method.
We combined different methods to map and predict the EIF2AK2 locus in the M. velifer genome. First, we performed a BLASTN search (cutoff of 10−05) with PKR cDNA sequences from related Myotis species to identify the canonical locus and localization of EIF2AK2 gene copies. Second, we aligned the protein and RNA transcript sequences of PKR from related bat species on the M. velifer genome using the Fast Statistical Alignment software (53). Third, we integrated RNA-seq data (see methods below), by mapping the RNA-seq reads using HISAT2 (v.2.0.0) (54). Last, we de novo predicted the gene structure of each PKR copy using Augustus (47) in a single-genome mode with the human gene model. The final figure was generated with the R library, Gviz.
IFN cell treatment and transcriptomic analyses
M. velifer cells were seeded in six-well plates. Forty-eight hours later, they were treated or not with type I universal IFN (1000 U/ml; PBL Assay Science). Six hours after treatment, cells were collected, and the total RNA was extracted using the Macherey-Nagel NucleoSpin RNA. Six sample replicates (three without IFN treatment and three with) were then sent for library preparation and sequencing with Illumina NextSeq500, 150 paired-end, to the Institut de Génomique Fonctionnelle de Lyon sequencing platform.
We processed the RNA-seq data with the nonannotated draft M. velifer genome. The quality of the raw data was checked with FastQC and a Q20 threshold, and adapters were removed using Cutadapt 4.0 (55). The quality-controlled reads were then aligned to the M. velifer genome using HISAT2 (54). As a complement, we de novo predicted the gene from M. velifer genome using Augustus, with the human model as reference. We counted the number of reads that mapped to each gene (predicted by Augustus), in both basal and IFN conditions, with FeatureCounts (from the R package, Rsubread), and performed a differential analysis using DESeq2 (56). From this analysis, we obtained a list of genes with a significantly different number of reads between the two conditions. Because the 5′UTR and first four exons were not found in the present genome of M. velifer, we retained the number of reads from exons that were duplicated and specific to each paralog to assess the expression pattern of the PKR copies. The final figure representing the number of reads per paralog per condition was drawn via the R package, ggplot.
Protein structure prediction and docking models
The 3D protein structures of D. rotundus and M. myotis PKR kinase domain, as well as the EPTV K3 protein, were predicted using the Iterative Threading ASSEmbly Refinement (I-TASSER) server (57). The best model was carefully chosen on the basis of the C score, which assesses the quality of the models. The inferred protein structure of PKR was visualized and designed with Swiss-PdbViewer software (58).
Computational docking of bat PKR and EPTV K3 was performed to predict the complex structure between both proteins, using HDOCK webserver (26). This software uses a fast Fourier transform–based search strategy to model different potential binding means between the proteins, and then, each binding mode is evaluated using the scoring function ITScorePP. The 3D structure models of M. myotis, D. rotundus, and M. molossus PKR kinase domains, as well as EPTV K3, were obtained with I-TASSER. We kept the default parameters for computation, including a grid spacing set to 1.2 Å and the angle interval set to 15°A. We retained the first top three models and combined the docking results with our functional assays for final model selection.
Plasmids
Expression in yeast cells
Bat EPTV (Washington strain) (22) K3L and E3L sequences were synthetized (Genewiz) with an integrated C-terminal hemagglutinin (HA) epitope tag and cloned into the yeast LEU2 integrating plasmid pSB305, which contains a galactose promoter, using the Xho I and Not I restriction sites. Bat PKR cDNAs from divergent chiropteran families (Pteropus alecto, Rhinolophus sinicus, D. rotundus, N. tumidirostris, M. molossus, N. albiventris, M. myotis, and E. fuscus) were cloned into the yeast pGAL expression plasmid, pSB819 (URA), using the Xho I and Not I restriction sites. The human and gibbon PKR expression vectors were previously described (16). D. rotundus × M. myotis PKR chimeras were synthetized and subcloned into pSB819. The chimeras include the swap of amino acids 268 to 344, 345 to 380, 381 to 420, and 421 to 530 in D. rotundus, referred as D1 to D4, respectively. PKR site–specific mutants and epteK3∆227-508 were generated by PCR mutagenesis using the QuickChange Lightning Mutagenesis Kit (Agilent) and primers holding the desired mutations/deletions, following the manufacturer’s protocol. PKR mutants and chimeras were N-terminal HA-tagged by PCR, using integrated HA-tag PKR primers (table S3).
Expression in human cells
Bat PKR orthologs and paralogs were subcloned from pSB819 into the expression vector pSG5, by means of Kpn I and Xho I sites introduced into PKR primers. Cloning of human PKR was previously described (15). Full-length NS5A proteins from human HCV (JFH1), bat hepacivirus from O. martiensseni (NC_031947.1) (28), and bat hepacivirus L from P. macrotis (NC_031916.1) (28) were synthetized and cloned into the expression plasmid pCDNA3.1+ N-terminal HA-tag using Bamh I and Not I restriction sites. The EPTV E3 and vaccinia E3 genes were cloned into the expression vector pSG5. The human IAV virus A/England/195/2009(H1N1) NS1 expressing-plasmid (pCAGGS V5-tag) (27) and the human cytomegalovirus TRS1 plasmid (pCDNA3.1 V5-tag) (29) were provided by W. Barclay and A. Geballe, respectively.
Yeast strains and growth assays
To determine whether bat PKR variants differed in their ability to escape poxviral antagonism, we used a heterologous yeast growth assay (24). This method relies on the recognition and phosphorylation of yeast eIF2α by PKR, which leads to yeast growth arrest. However, coexpression with poxvirus K3 or E3 that are able to antagonize PKR leads to growth rescue. Yeast growth assays were performed in two steps.
First, yeast strain H2557 was modified for stable expression of bat poxvirus K3 and E3 proteins following the standard yeast transformation protocol (59). EPTV K3 (eptK3) or EPTV E3 (eptE3) was integrated into H2257 at the LEU2 locus under the gal promoter, using subcloned pSB305 plasmids linearized with EcoRV. The resulted strains H2557-eptK3, H2557-eptE3, and H2557-pteE3 were confirmed through PCR amplification and sequencing of K3 and E3, using the universal primers M13 F and M13R. Yeast strains expressing vaccinia and variola HA-K3 as well as the wild-type control (HM3, with integrated empty vector) were previously described (16).
Second, the modified yeast strains were transformed with 100 ng of PKR expression plasmids pSB19. For each transformation, four colonies were selected and streaked on S-leu-ura medium (yeast minimal complete medium with amino acids minus uracil and leucine) containing 2% glucose (SD) or galactose (SGal) and grown at 30°C for 3 days. Representative transformants colonies were then grown to saturation in SD-leu-ura medium and plated in dilution series (D600 of 3.0, 0.3, 0.03, and 0.003) on SD and SGal-leu-ura medium for 3 days. All yeast assays were conducted in biological triplicate experiments.
Cell lines
HeLa PKR-KO cells (provided by A. Geballe) (29) were maintained in DMEM supplemented with 5% fetal bovine serum and puromycin (1 μg/ml; Sigma-Aldrich). RK13+E3L+K3L cells (rabbit) (60) were maintained in DMEM supplemented with 5% fetal bovine serum, penicillin/streptomycin (100 IU/ml), geneticin (500 μg/ml), and zeocin (300 μg/ml; Gibco). Wild-type (Invitrogen) and PKR-KO T-REx-293 cells were grown in DMEM supplemented with 10% fetal bovine serum, penicillin/streptomycin (100 IU/ml), zeocin (100 μg/ml), and blasticidin (15 μg/ml; Gibco). The T-REx-293 cells stably expressing the bat PKRs were under constant selection with blasticidin (15 μg/ml) and hygromycin (50 μg/ml; Invitrogen).
Luciferase reporter assays
Luciferase assays were carried out following the protocol described in (61). Briefly, 50,000 HeLa PKR-KO cells were seeded per well in 24-well plates and transfected 16 hours after seeding with 350 ng of PKR expression vector or empty control, 350 ng of viral antagonist expression plasmid (NS1, NS5A, EPTV E3, or TRS1) or empty control, and 5 ng of FFLuc firefly luciferase reporter plasmid, using Trans-IT-LT1 (Muris Bio) following the manufacturer’s protocol. Cells were lysed 48 hours after transfection by means of the reporter lysis 5× buffer (Promega), and then, the luciferase substrate (Promega) was added following the manufacturer’s recommendations. Luciferase reporter quantitation was carried out with a LUMIstar Omega microplate reader optima (BMG Labtech). All luciferase assays were conducted in triplicate in at least five independent experiments. For the luciferase assays with VACV K3 and E3 antagonists, 50,000 HeLa PKR-KO cells per well were transfected 24 hours after seeding with 200 ng of PKR expression vector, 200 ng of VACV E3 expression plasmids (VACV K3 and E3), and 50 ng of pGL3 firefly luciferase expression vector (Promega) using GenJet (SignaGen) at a DNA to GenJet ratio of 1:2 following the manufacturer’s protocol. Cells were lysed 48 hours after transfection with mammalian lysis buffer (GE Healthcare), and then, the luciferase substrate (Promega) was added following the manufacturer’s recommendations. Luciferase reporter quantitation was carried out with a GloMax luminometer (Promega). All luciferase assays were conducted in triplicate in at least three independent experiments.
Generation of doxycycline-inducible bat PKR-expressing 293 cells
Bat PKRs (E. fuscus, the two M. myotis paralogs, and the two M. velifer paralogs) were cloned into the pcDNA5/FRT/TO expression vector with two C-terminal FLAG tag sequences. T-REx-293 PKR-KO cells were stably transfected with each bat PKR plasmid by GenJet (SignaGen) according to the manufacturer’s instructions, and polyclonal pools of the transfected cells were selected by their resistance to hygromycin.
Poxvirus infection
Generation of VC-R4, a derivative of VACV Copenhagen strain, was described (30). A total of 500,000 of T-REx bat PKR expressing cells were seeded per well in 12-well plates and induced with doxycycline (1 μg/ml) for 24 hours. Forty-eight hours after seeding, each well was infected by VC-R4 at a multiplicity of infection (MOI) of 0.1. Fluorescent pictures were taken with an inverted fluorescent microscope (Leica) at the indicated time after infection. For the virus replication assay, cells and supernatants were collected 24 hours after infection and subjected to three rounds of freezing at −80°C and thawing at 37°C. Lysates were sonicated for 15 s, 50% amplitude (Qsonica Q500). Viruses were titrated by 10-fold serial dilutions on confluent RK13+E3L+K3L cells in 12-well plates. One hour after infecting RK13+E3L+K3L cells with the dilutions, the medium was replaced with DMEM containing 1% carboxymethylcellulose. After 48 hours, cells were stained with 0.1% crystal violet and counted for plaques. Infections and viral titer were performed in duplicate.
VSV infection
A total of 200,000 Hela PKR-KO cells were seeded per well in 12-well plates and transfected with either empty pSG5 plasmid, M. myotis PKRs or M. velifer PKRs, using Trans-IT-LT1. An interferon-stimulated exonuclease gene 20 (ISG20)–encoding plasmid (62) was used as a positive control because of its established antiviral activity against VSV. Infection was performed 24 hours after transfection, with a VSV-GFP virus at an MOI of 3 (62), and cells were fixed at 18 hours after infection with 4% paraformaldehyde. Single-cell analysis was performed using BD FACSCanto II Flow Cytometer to quantify VSV infectivity as the percentage of GFP+ cells. Fold change results are normalized to the empty (no PKR or ISG20) condition from five independent experiments.
Western blots
To examine the yeast expression of bat PKR and poxvirus K3 proteins, yeast transformants were grown overnight in 2% glucose S-leu-ura medium, followed by induction with 2% galactose for 15 hours. Cell lysates were treated with 0.1 M of sodium hydroxide (NaOH) for 5 min and then lysed in 2× SDS–polyacrylamide gel electrophoresis buffer supplemented with protease inhibitor cocktail (Roche) and 355 mM β-mercaptoethanol (Sigma-Aldrich) at 95°C for 5 min. PKR was then precipitated at 65°C for 45 min, frozen overnight, and reprecipitated. Proteins were resolved by 12% Mini-PROTEAN GTX polyacrylamide gel (Bio-Rad) and then transferred to polyvinylidene difluoride (PVDF) membranes. Proteins were probed with rabbit anti-HA (1:1000; Sigma-Aldrich, H3663) or anti–β-actin as loading control (1:10,000; Sigma-Aldrich, A5441) primary antibody and then with goat anti-rabbit secondary antibody. Blots were visualized using the Image Lab Touch Software (version 2.0.0.27, ChemiDoc Imaging System from Bio-Rad) or film.
In HeLa-KO cells, protein expression was assayed with 400,000 cell per well in six-well plates and transfected with 1.4 μg of the indicated PKR and viral antagonist–expressing plasmids. Cells were lysed after 48 hours with 1% SDS in phosphate-buffered saline (VWR), and then, proteins were separated on 4 to 12% Mini-PROTEAN GTX polyacrylamide gel (Bio-Rad) and transferred to nitrocellulose membranes. Proteins were probed with mouse anti-FLAG (1:5000; Sigma-Aldrich, F3165) or anti–β-actin as loading control (1:10,000; Sigma-Aldrich, A5441) and then with anti-mouse immunoglobulin G peroxidase–conjugated secondary antibody (1:10,000; Sigma-Aldrich, cat. A9044). Proteins were visualized as described above. To detect the expression of bat PKRs in the stably transfected T-REx-293 cells, 600,000 cells were seeded per well in six-well plates and induced by doxycycline (1 μg/ml) 24 hours after seeding. Cells were lysed 24 hours after induction with 1% SDS, and then, proteins were separated on 10% TGX FastCast Acrylamide gel (Bio-Rad) and transferred to PVDF membranes. Proteins were probed with mouse anti-FLAG (1:5000; Sigma-Aldrich, F3165) or anti–β-actin (1:5000; Sigma-Aldrich, A1978) primary antibody and then with donkey anti-mouse (1:10,000; Thermo Fisher Scientific, 715-035-150) secondary antibody. Images were taken using the iBright Imaging System (Invitrogen).
Statistical analyses
Expression data and differences between conditions were statistically analyzed using the nonparametric Mann-Whitney Wilcoxon test with R software. For each of these tests, the P value was considered significant when inferior to 0.05. Error bars in graphics are SEM or SD.
Acknowledgments
We are particularly grateful to the Poitou-Charentes association as well as the volunteers and field workers who have helped us during the field sessions: V. Alt, M. Bely, G. Chagneau, M. Dorfiac, S. Dufour, C. Gizardin, G. Leblanc, M. Leuchtmann, E. Loufti, A. Le Guen, and L. Trebucq. We thank B. Larsen, M. Bucci, and R. Ledgister for the help with M. velifer sample collection; Pima County, AZ, USA for sampling permission; and Dovetail Genomics for sequencing. For the advices and the RNA-seq analysis that we used for the PKR transcripts, we thank M. Sémon, M. Cariou, C. Rey, C. Dechaud, and R. Bulteau, the students from the Master UE NGS and the Master Biosciences, École Normale Supérieure de Lyon, Université Claude Bernard Lyon 1, Université de Lyon, 69342 Lyon Cedex 07, France; as well as B. Gillet and S. Hughes from the IGFL platform for the library preparation and the sequencing. We thank C. Feschotte and R. Cosby (Cornell University, NY) for sharing M. velifer and E. fuscus cell lines. We also thank A. Geballe (Fred Hutchinson Cancer Center, WA) and W. Barclay (Imperial College London, UK) for sharing the reagents (the references are given herein). We acknowledge the contribution of the SFR Biosciences (UAR3444/CNRS, US8/Inserm, ENS de Lyon, UCBL) AniRA-Cytometry platform, especially V. Barateau for the help. We also thank all the contributors of the LBBE bioinformatic server, of the publicly available bioinformatic programs, and of the publicly available genomic sequences. We thank all LP2L Ecoépidémiologie Evolutionniste team members for feedback on this work.
Funding: D.P. and L.E. are supported by the ANR LabEX ECOFECT [ANR-11-LABX-0048 of the Université de Lyon, within the program Investissements d’Avenir (ANR-11-IDEX-0007) operated by the French National Research Agency]. D.P., L.E., and S.J. are supported by the French Agence Nationale de la Recherche (ANR), under grant ANR-20-CE15-0020-01 (project “BATantiVIR”). L.E. and D.E. are supported by a grant from the joint program between the CNRS and the University of Arizona. L.E. is further supported by the CNRS and by grants from the French Research Agency on HIV and Emerging Infectious Diseases ANRS/MIE (nos. ECTZ19143 and ECTZ118944). D.P. is supported by the CNRS, the European Regional Development Fund (ERDF), and the ANR EBOFAC. S.J. is also supported by the Fondation L’Oréal-Unesco “For Women In Science.” M.E.L. is supported by NSF Rules of Life Postdoctoral Research Fellowship in Biology (NSF 2010884). S.R. was supported by grant R01 AI114851 (from the National Institute of Allergy and Infectious Diseases). N.C.E. and M.C. are supported by NIH grants R35 GM134936 and F30 GM146410 (from the National Institute of General Medical Sciences).
Author contributions: Study conceptualization: S.J., L.E., and D.P. Methodological design: S.J., S.R., N.C.E., D.P., and L.E. Field sampling: J.-B.P., O.F.-C., B.N., J.D., and D.P. Phylogenetic and genomic analyses: S.J. RNA-seq processing and analysis: S.J., A.E.F., C.D., and L.E. Technical support: C.D. Genome sampling, sequencing and analysis: M.E.L. and D.E. Yeast assays: S.J., M.C., C.V., and C.M.C. Luciferase assays: S.J., C.Z., and C.P. Infectivity assays: C.Z., C.D.L.M.M., and G.B. Study coordination and supervision: L.E. and D.P. Further supervision: S.J., S.R., and N.C.E. Funding: S.J., L.E., D.E., D.P., A.C., S.R., and N.C.E. Writing—original draft: S.J. Writing—first editing: L.E. and D.P. Writing—review and editing: All the authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All de novo PKR sequences were deposited in GenBank under accession numbers OP006534 to OP006561. Dataset for Fig. 1 with alignments and phylogenetic trees is publicly available at https://figshare.com/projects/Datasets_for_Jacquet_et_al_2022/142388. The Flp-In T-REx 293 PKR-KO cells were provided by R. Bruneau.
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S11
REFERENCES AND NOTES
- 1.Duggal N. K., Emerman M., Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 12, 687–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daugherty M. D., Malik H. S., Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 46, 677–700 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Irving A. T., Ahn M., Goh G., Anderson D. E., Wang L.-F., Lessons from the host defences of bats, a unique viral reservoir. Nature 589, 363–370 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Ahn M., Anderson D. E., Zhang Q., Tan C. W., Lim B. L., Luko K., Wen M., Chia W. N., Mani S., Wang L. C., Ng J. H. J., Sobota R. M., Dutertre C.-A., Ginhoux F., Shi Z.-L., Irving A. T., Wang L.-F., Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 4, 789–799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie J., Li Y., Shen X., Goh G., Zhu Y., Cui J., Wang L. F., Shi Z. L., Zhou P., Dampened STING-dependent interferon activation in bats. Cell Host Microbe 23, 297–301.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A., Rapin N., Bollinger T., Misra V., Lack of inflammatory gene expression in bats: A unique role for a transcription repressor. Sci. Rep. 7, 2232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cariou M., Picard L., Guéguen L., Jacquet S., Cimarelli A., Fregoso O. I., Molaro A., Navratil V., Etienne L., Distinct evolutionary trajectories of SARS-CoV-2-interacting proteins in bats and primates identify important host determinants of COVID-19. Proc. Natl. Acad. Sci. U.S.A. 119, e2206610119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agnarsson I., Nadia C. M. Z., Laura P. F., A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLOS Curr. 3, RRN1212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jebb D., Huang Z., Pippel M., Hughes G. M., Lavrichenko K., Devanna P., Winkler S., Jermiin L. S., Skirmuntt E. C., Katzourakis A., Burkitt-Gray L., Ray D. A., Sullivan K. A. M., Roscito J. G., Kirilenko B. M., Dávalos L. M., Corthals A. P., Power M. L., Jones G., Ransome R. D., Dechmann D. K. N., Locatelli A. G., Puechmaille S. J., Fedrigo O., Jarvis E. D., Hiller M., Vernes S. C., Myers E. W., Teeling E. C., Six reference-quality genomes reveal evolution of bat adaptations. Nature 583, 578–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs J., Hölzer M., Schilling M., Patzina C., Schoen A., Hoenen T., Zimmer G., Marz M., Weber F., Müller M. A., Kochs G., Evolution and antiviral specificities of interferon-induced Mx proteins of bats against Ebola, influenza, and other RNA viruses. J. Virol. 91, e00361–e00317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benfield C. T., MacKenzie F., Ritzefeld M., Mazzon M., Weston S., Tate E. W., Teo B. H., Smith S. E., Kellam P., Holmes E. C., Marsh M., Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci. Alliance 3, e201900542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes A. P., Águeda-Pinto A., Pinheiro A., Rebelo H., Esteves P. J., Evolution of TRIM5 and TRIM22 in bats reveals a complex duplication process. Viruses 14, 345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesaro T., Michiels T., Inhibition of PKR by viruses. Front. Microbiol. 12, 757238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dar A. C., Sicheri F., X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol. Cell 10, 295–305 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Rothenburg S., Seo E. J., Gibbs J. S., Dever T. E., Dittmar K., Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 16, 63–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elde N. C., Child S. J., Geballe A. P., Malik H. S., Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457, 485–489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess H. M., Mohr I., Evolutionary clash between myxoma virus and rabbit PKR in Australia. Proc. Natl. Acad. Sci. U.S.A. 113, 3912–3914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Smith M. D., Wertheim J. O., Weaver S., Murrell B., Scheffler K., Kosakovsky Pond S. L., Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 32, 1342–1353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson G. L., Nordhausen R., Garner M. M., Huckabee J. R., Johnson S., Wohrle R. D., Davidson W. B., Wilkins K., Li Y., Doty J. B., Gallardo-Romero N. F., Metcalfe M. G., Karem K. L., Damon I. K., Carroll D. S., Novel poxvirus in big brown bats, northwestern United States. Emerg. Infect. Dis. 19, 1002–1004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lelli D., Lavazza A., Prosperi A., Sozzi E., Faccin F., Baioni L., Trogu T., Cavallari G. L., Mauri M., Gibellini A. M., Chiapponi C., Moreno A., Hypsugopoxvirus: A novel poxvirus isolated from hypsugo savii in Italy. Viruses 11, 568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu S. L., Nakazawa Y., Gao J., Wilkins K., Gallardo-Romero N., Li Y., Emerson G. L., Carroll D. S., Upton C., Characterization of Eptesipox virus, a novel poxvirus from a microchiropteran bat. Virus Genes 53, 856–867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker K. S., Leggett R. M., Bexfield N. H., Alston M., Daly G., Todd S., Tachedjian M., Holmes C. E. G., Crameri S., Wang L. F., Heeney J. L., Suu-Ire R., Kellam P., Cunningham A. A., Wood J. L. N., Caccamo M., Murcia P. R., Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441, 95–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dever T. E., Chen J.-J., Barber G. N., Cigan A. M., Feng L., Donahue T. F., London I. M., Katze M. G., Hinnebusch A. G., Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. U.S.A. 90, 4616–4620 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan Y., Tao H., He J., Huang S.-Y., The HDOCK server for integrated protein-protein docking. Nat. Protoc. 15, 1829–1852 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Elderfield R. A., Watson S. J., Godlee A., Adamson W. E., Thompson C. I., Dunning J., Fernandez-Alonso M., Blumenkrantz D., Hussell T.; The MOSAIC Investigators, Zambon M., Openshaw P., Kellam P., Barclay W. S., Lyles D. S., Accumulation of human-adapting mutations during circulation of A(H1N1)pdm09 influenza virus in humans in the United Kingdom. J. Virol. 88, 13269–13283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quan P. L., Firth C., Conte J. M., Williams S. H., Zambrana-Torrelio C. M., Anthony S. J., Ellison J. A., Gilbert A. T., Kuzmin I. V., Niezgoda M., Osinubi M. O. V., Recuenco S., Markotter W., Breiman R. F., Kalemba L., Malekani J., Lindblade K. A., Rostal M. K., Ojeda-Flores R., Suzan G., Davis L. B., Blau D. M., Ogunkoya A. B., Castillo D. A. A., Moran D., Ngam S., Akaibe D., Agwanda B., Briese T., Epstein J. H., Daszak P., Rupprecht C. E., Holmes E. C., Lipkin W. I., Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U.S.A. 110, 8194–8199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Child S. J., Früh K., Malouli D., Hickson S. E., Geballe A. P., Bayer A., Antagonism of the protein kinase R pathway in human cells by rhesus cytomegalovirus. J. Virol. 92, e01793-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vipat S., Brennan G., Park C., Haller S. L., Rothenburg S., Rapid, seamless generation of recombinant poxviruses using host range and visual selection. J. Vis. Exp. 159, e61049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M. S., Wang D., Morimoto H., Yim H. C., Irving A. T., Williams B. R., Sadler A. J., Molecular dynamics reveal a novel kinase-substrate interface that regulates protein translation. J. Mol. Cell Biol. 6, 473–485 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Mitchell P. S., Patzina C., Emerman M., Haller O., Malik H. S., Kochs G., Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12, 598–604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fixsen S. M., Cone K. R., Goldstein S. A., Sasani T. A., Quinlan A. R., Rothenburg S., Elde N. C., Poxviruses capture host genes by LINE-1 retrotransposition. eLife 11, e63332 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S., Min J. Y., Krug R. M., Sen G. C., Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349, 13–21 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Jacquet S., Pontier D., Etienne L., Rapid evolution of HERC6 and duplication of a chimeric HERC5/6 gene in rodents and bats suggest an overlooked role of HERCs in mammalian immunity. Front. Immunol. 11, 605270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenburg S., Deigendesch N., Dey M., Dever T. E., Tazi L., Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: Varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. 6, 12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T.-K., Zhang Y.-B., Liu Y., Sun F., Gui J.-F., Cooperative roles of fish protein kinase containing Z-DNA binding domains and double-stranded RNA-dependent protein kinase in interferon-mediated antiviral response. J. Virol. 85, 12769–12780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Evolution by gene duplication: An update. Trends Ecol. Evol. 18, 292–298 (2003). [Google Scholar]
- 39.Daugherty M. D., Schaller A. M., Geballe A. P., Malik H. S., Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 5, e14228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirchhoff S., Koromilas A. E., Schaper F., Grashoff M., Sonenberg N., Hauser H., IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene 11, 439–445 (1995). [PubMed] [Google Scholar]
- 41.Yu H., Peng C., Zhang C., Stoian A. M. M., Tazi L., Brennan G., Rothenburg S., Maladaptation after a virus host switch leads to increased activation of the pro-inflammatory NF-κB pathway. Proc. Natl. Acad. Sci. U.S.A. 119, e2115354119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong A. H.-T., Tam N. W. N., Yang Y.-L., Cuddihy A. R., Li S., Kirchhoff S., Hauser H., Decker T., Koromilas A. E., Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 16, 1291–1304 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavlovich S. S., Lovett S. P., Koroleva G., Guito J. C., Arnold C. E., Nagle E. R., Kulcsar K., Lee A., Thibaud-Nissen F., Hume A. J., Mühlberger E., Uebelhoer L. S., Towner J. S., Rabadan R., Sanchez-Lockhart M., Kepler T. B., Palacios G., The Egyptian Rousette genome reveals unexpected features of bat antiviral immunity. Cell 173, 1098–1110.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkins J. A., Kaczmarek M. E., Müller M. A., Drosten C., Press W. H., Sawyer S. L., A metaanalysis of bat phylogenetics and positive selection based on genomes and transcriptomes from 18 species. Proc. Natl. Acad. Sci. U.S.A. 116, 11351–11360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cosby R. L., Judd J., Zhang R., Zhong A., Garry N., Pritham E. J., Feschotte C., Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371, eabc6405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin D. M., Kocher T. D., Wilson A. C., Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32, 128–144 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Hoff K. J., Stanke M., Predicting genes in single genomes with AUGUSTUS. Curr. Protoc. Bioinformatics 65, e57 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Birney E., Clamp M., Durbin R., GeneWise and genomewise. Genome Res. 14, 988–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Löytynoja A., Goldman N., webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11, 579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Lefort V., Longueville J.-E., Gascuel O., SMS: Smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pond S. L. K., Frost S. D. W., Muse S. V., HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Bradley R. K., Roberts A., Smoot M., Juvekar S., Do J., Dewey C., Holmes I., Pachter L., Fast statistical alignment. PLOS Comput Biol. 5, e1000392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 56.Love M., Anders S., Huber W., Differential analysis of count data—the DESeq2 package. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., Zhang Y., Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics 52, 5–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson M. U., Zoete V., Michielin O., Guex N., Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC bioinformatics. 13, 173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R. D. Gietz, R. A. Woods, Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method, in Methods in Enzymology (Elsevier, 2002), vol. 350, pp. 87–96. [DOI] [PubMed] [Google Scholar]
- 60.Rahman M. M., Liu J., Chan W. M., Rothenburg S., McFadden G., Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLOS Pathog. 9, e1003465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng C., Haller S. L., Rahman M. M., McFadden G., Rothenburg S., Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc. Natl. Acad. Sci. U.S.A. 113, 3855–3860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu N., Nguyen X.-N., Wang L., Appourchaux R., Zhang C., Panthu B., Gruffat H., Journo C., Alais S., Qin J., Zhang N., Tartour K., Catez F., Mahieux R., Ohlmann T., Liu M., Du B., Cimarelli A., The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLOS Pathog. 15, e1008093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies M. V., Chang H. W., Jacobs B. L., Kaufman R. J., The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67, 1688–1692 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S11