Abstract
The coronavirus disease 2019 (COVID-19) pandemic underlines the urgent need for effective mRNA vaccines. However, current understanding of the immunological outcomes of mRNA vaccines formulated under different nanoplatforms is insufficient. Here, severe acute respiratory syndrome coronavirus 2 receptor binding domain mRNA delivered via lipid nanoparticle (LNP), cationic nanoemulsion (CNE), and cationic liposome (Lipo) was constructed. Results demonstrated that the structural and biochemical characteristics of nanoparticles shaped their tissue dissemination, cellular uptake, and intracellular trafficking, which eventually determined the activation of antiviral humoral and cellular immunity. Specifically, LNP was mainly internalized by myocyte and subsequently circumvented lysosome degradation, giving rise to humoral-biased immune responses. Meanwhile, CNE and Lipo induced cellular-preferred immunity, which was respectively attributed to the better lysosomal escape in dendritic cells and the superior biodistribution in secondary lymphoid organs. Overall, this study may guide the design and clinical use of mRNA vaccines against COVID-19.
mRNA vaccines based on different nanoplatforms elicit humoral immunity and cellular immunity with different preferences.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), is a highly transmissible, pathogenic, and mutagenic virus that challenges public health at an unprecedented scale (1–5). The outbreak of COVID-19 has prompted the development of prophylactic vaccines for the establishment of specific immune protection against virus infection. Of all vaccine modalities, mRNA-based ones have emerged as a promising candidate with advantages including fast and scalable manufacturing, potent immunogenicity, and good biosafety (6–8). However, the anionic, hydrophilic, and fragile nature of mRNA molecule greatly limits its bioavailability. Therefore, a delivery system is needed to promote the biochemical stability and site-specific accumulation of mRNA for effective mobilization of the antiviral immunity (9–12).
As the two main arms of adaptive immune system, humoral immunity and cellular immunity are intrinsically correlated, whose collaboration determines the effectiveness and sustainability of antiviral competence (13). Specifically, humoral immunity primarily depends on the antibody-mediated neutralization, which neutralizes virus infectivity and inhibits virus-mediated cell membrane fusion (14–17). On the other hand, cellular immunity is largely mediated by CD8+ T cells, which eliminates infected cells by T cell receptor recognition (18). Multifaceted immunological memory can be established during humoral and cellular immunoresponse to provide specific protection against virus invasion (19–21).
Antiviral humoral immunity and cellular immunity differ in the onset time, mode of action, magnitude, and persistence. Latest studies manifest that upon natural infection or vaccine inoculation, CD8+ T cells mediate an earlier (precedes the maturation of other effector arms), more vigorous, and more durable protection (22, 23). In contrast, antibody immunity has a delayed mobilization and quicker decline (a narrower protection window) (24, 25). It is reported that in the context of SARS-CoV-2 spike mRNA vaccine (BNT162b2, Pfizer-BioNTech), CD8+ T cells are quickly mobilized 1 week after prime vaccination, which undergo extensive clonal expansion and effector differentiation after boost vaccination. On the contrary, neutralizing antibodies are only readily detectable after boost (26). Characteristics from the ongoing pandemic indicate that CD8+ T cells are stably maintained to convey a long-term protection against SARS-CoV-2, while antibody-mediated response seems to attenuate over time (24, 27).
Lipid nanoparticle (LNP), cationic nanoemulsion (CNE), and cationic liposome (Lipo) are frequently used as mRNA delivery systems with unique characteristics (6, 10, 28–33). However, there is still a lack of systemic understanding on the mode of action of these nanocarriers in inducing antiviral humoral and cellular immunity. Here, SARS-CoV-2 receptor binding domain (RBD) mRNA–encapsulated/adsorbed LNP [which has been in clinical trials or granted a conditional marketing authorization (7, 22)], CNE [formulated on the basis of our previous studies (34)], and Lipo [prepared according to our previous works (35, 36)] were constructed, with their tissue dissemination, cellular uptake, intracellular trafficking, and immunological responses investigated. Results suggested that nanoparticles with different structural and biochemical characteristics elicited humoral immunity and cellular immunity with different preferences, which may instruct the design and clinical use of lipid nanoplatform-based mRNA vaccines against SARS-CoV-2.
RESULTS
Verification of the pharmacological activity of mRNA
Lipid-based nanoparticles are versatile carrier platforms facilitating the delivery of precise genetic information to target cells. At present, LNP-, nanoemulsion-, and liposome-formulated RNA vaccines against infectious diseases or malignancies have been shown in clinical trials to be safe and well tolerated (6, 10, 33, 37). Here, on the basis of the disclosed LNP prescription (by Pfizer/BioNTech) (30) and our previous works on nucleic acid nanoparticles (34, 36), LNP, CNE, and Lipo were prepared (Table 1) for mRNA delivery.
Table 1. Formulations of LNP, CNE, and Lipo.
DMG-PEG2000, 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DOTAP, dioleylhydroxypropyl-3-N,N,N-trimethylammonium; DSPC, 1,2-dioctadecanoyl-sn-glycero-3-phophocholine; DSPE-PEG2000, distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide (polyethylene glycol)-2000].
| Nanocarrier | Lipids | Composition (mass ratio) | Lipid concentration (mg/ml) | N/P ratio with mRNA |
| LNP | DLin-MC3-DMA:DMG-PEG2000:cholesterol:DSPC | 13:1.6:6.2:3.3 | 1.205 | 6 |
| CNE | DOTAP:lipoid E-80:vitamin E | 6:7:7 | 33.33 | 4.5 |
| Lipo | DOTAP:DOPE:DSPE-PEG2000:DSPE-PEG2000-Pardaxin | 1552:388:15:45 | 4 | 3.4 |
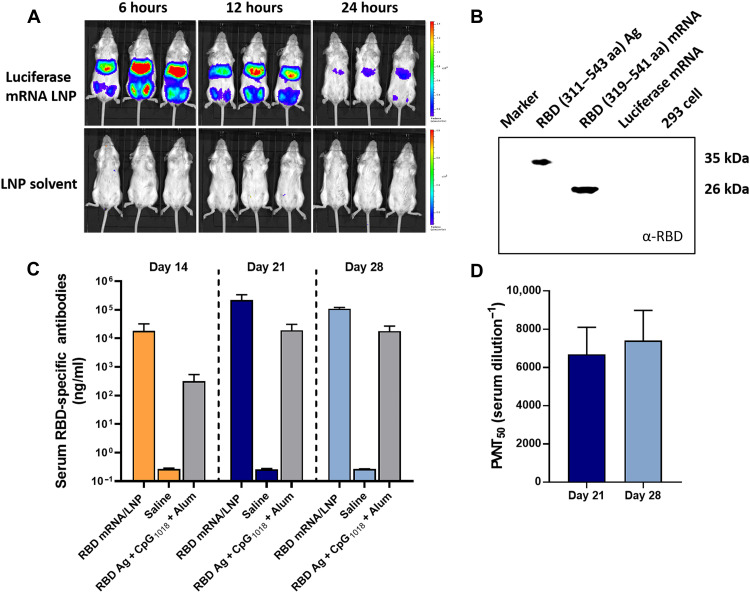
We first verified the pharmacological activity of mRNA using LNP-based delivery vehicle. Bioluminescence analysis suggested that when luciferase mRNA was used as the model nucleic acid and formulated with LNP, a robust luciferase signal was observed both at the injection site (i.e., on both hind legs) and the liver at 6 hours after intramuscular administration, which gradually declined within 24 hours (Fig. 1A). It should be mentioned that compared to the permanently charged cationic lipid, the ionizable lipid [(6Z,9Z,28Z,31Z)-heptatriacont-6,9,28,31-tetraene-19-yl 4-(dimethylamino) butanoate (DLin-MC3-DMA)] in LNP features a neutral to mildly cationic charge under physiological pH that contributes to a reduced toxicity and prolonged systemic circulation, which might explain for the dynamic turnover of luciferase in the liver. Then, the in vitro and in vivo transfection of SARS-CoV-2 RBD mRNA was evaluated. Western blot analysis showed that human embryonic kidney (HEK) 293 cells transfected with RBD (amino acids 319 to 541) mRNA for 36 hours successfully produced RBD protein in the whole-cell lysate (Fig. 1B). Meanwhile, BALB/c mice intramuscularly vaccinated with RBD mRNA–encapsulated LNP resulted in a higher serum RBD-specific immunoglobulin G (IgG) antibody concentration than that of RBD antigen (Ag) plus adjuvants (CpG1018 and Alum) on days 14, 21, and 28 (Fig. 1C), with serum pseudotyped virus neutralization titer 50% reaching some 1:6000 and 1:7000 on days 21 and 28, respectively (Fig. 1D). These results indicated that the SARS-CoV-2 RBD mRNA used in this work was pharmacologically valid to support our further exploration.
Fig. 1. Transfection of mRNA in vitro and in vivo.
(A) Biodistribution and transfection of mRNA in vivo. BALB/c mice were intramuscularly (i.m.) administrated with luciferase mRNA–encapsulated LNP or LNP solvent (20 μg of mRNA per mouse) on both hind legs (n = 3). Bioluminescence was measured at 6, 12, and 24 hours using an in vivo imaging system. (B) Western blot analysis of the expression of RBD protein in the whole-cell lysate of HEK 293 cells at 36 hours after treatment. Here, 293 cells were respectively stimulated/transfected with 19.2 ng of RBD [amino acids (aa) 311 to 543] (Ag, positive control), 100 ng of RBD319–541 mRNA, 100 ng of luciferase mRNA (control mRNA), and culture medium (negative control). (C and D) BALB/c mice (n = 3) were intramuscularly vaccinated with RBD mRNA–encapsulated LNP (3 μg of mRNA), saline, or RBD Ag (3 μg) + Dynavax CpG1018 (10 μg) + Alum (50 μg). Serum concentration of RBD-specific IgG antibodies was determined using an ELISA kit on days 14, 21, and 28 (C). Meanwhile, serum SARS-CoV-2 pseudotyped virus neutralization titer 50% (PVNT50) of RBD mRNA LNP–treated mice was determined using COVID-19-Spike Protein Pseudovirus (catalog no. 11906ES50, Yeasen Biotechnology Co. Ltd., Shanghai, China) on days 21 and 28 (D). Data are presented as means ± SD.
Preparation and characterization of mRNA nanoparticles
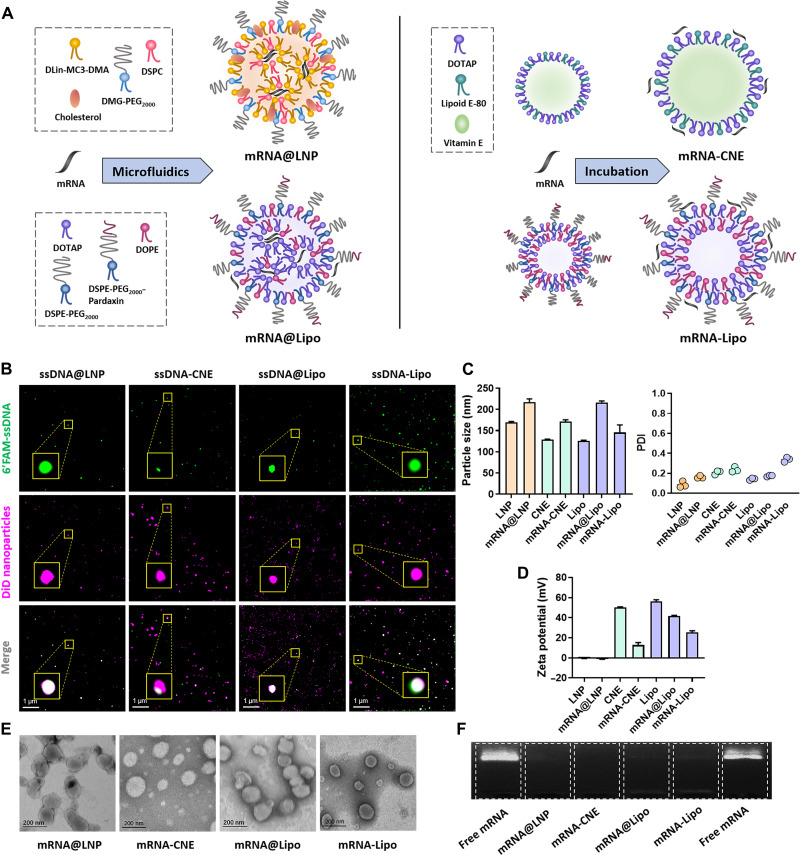
We next thought to compare the vaccine effects of mRNA prepared under different nanoplatforms and first characterized the physicochemical properties of each preparation. Specifically, mRNA was encapsulated inside the nanoparticles (mRNA@LNP and mRNA@Lipo) using microfluidic chip technique or adsorbed on the surface of cationic nanoparticles (mRNA-CNE and mRNA-Lipo) via electrostatic interactions (Fig. 2A). When fluorescein amidite (FAM)–labeled single-strand DNA (FAM-ssDNA) was used as the model nucleic acid and nanoparticles were labeled with the fluorescent dye 1,1’-dioctadecyl-3,3,3’,3’ tetramethylindodicarbocyanine,4-chlorobenzenesulfonate salt (DiD), structure illumination microscopy (SIM) images confirmed that FAM-ssDNA was successfully loaded by these nanoparticles via the two drug loading strategies, i.e., encapsulation and adsorption (Fig. 2B).
Fig. 2. Formulation and characterization of different mRNA nanoparticles.
(A) Schematic view of the structures of mRNA nanoparticles fabricated by microfluidics (mRNA@LNP and mRNA@Lipo) and postsynthetic surface adsorption (mRNA-CNE and mRNA-Lipo). (B) SIM observations of different ssDNA-loaded nanoparticles. Top, 6′FAM-ssDNA; middle, DiD-labeled nanoparticles; bottom, merged views. Scale bars, 1 μm. (C and D) Particle size, polydispersity index (PDI), and surface charge of LNP, CNE, and Lipo with/without enhanced green fluorescent protein (eGFP) mRNA (n = 3). All error bars are expressed as ±SD. (E) TEM images of mRNA@LNP, mRNA-CNE, mRNA@Lipo, and mRNA-Lipo. Scale bars, 200 nm. (F) mRNA encapsulation/complexation by agarose gel electrophoresis assay. Naked mRNA was used as a control. eGFP mRNA was used as the model nucleic acid in (C) to (F).
The association with enhanced green fluorescent protein (eGFP) mRNA led to an increased diameter and decreased surface charge of all nanoparticles (Fig. 2, C and D). Notably, RNA encapsulation resulted in a larger particle size, while RNA surface complexation caused a sharper decrease in the zeta potential. In addition, transmission electron microscopy (TEM) observations and agarose gel electrophoresis assay suggested that both loading approaches were able to generate well-defined nanosized particles with spherical morphology (Fig. 2E) and high mRNA loading efficiency (Fig. 2F).
Anti–SARS-CoV-2 humoral and cellular responses induced by mRNA nanoparticles
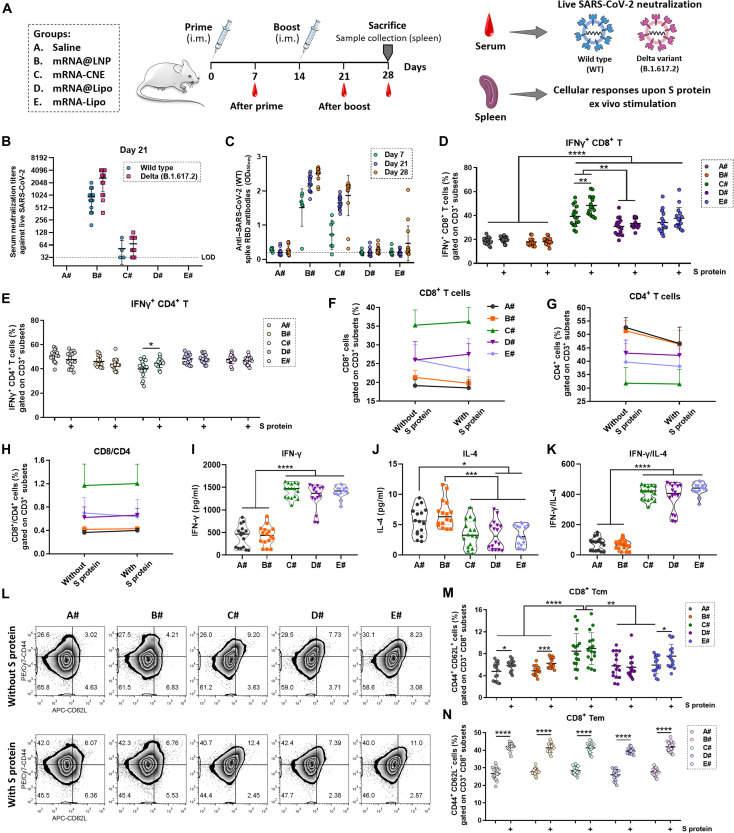
The spike (S) protein is an important structural protein of SARS-CoV-2 that binds with its receptor angiotensin-converting enzyme 2 (ACE2) to mediate viral entry into the host cells. As the smallest domain of S protein that directly binds to ACE2, RBD is frequently used as the immunogen to induce specific antibodies (block virus invasion) and effector T cells (eliminate infected cells). To date, RBD-based mRNA vaccines have been verified to produce effective immune protection in both animal models and human clinical trials (12, 38, 39). Here, nucleoside-modified mRNA encoding the RBD of SARS-CoV-2 S protein was used and formulated with LNP, CNE, and Lipo. BALB/c mice were intramuscularly administered with saline (A#), mRNA@LNP (B#), mRNA-CNE (C#), mRNA@Lipo (D#), or mRNA-Lipo (E#) (10 μg of RBD mRNA per mouse in B# to E#) on both hind legs on day 0 (prime) and day 14 (boost) (Fig. 3A).
Fig. 3. Anti–SARS-CoV-2 humoral and cellular immunity induced by different mRNA nanoparticles.
(A) Schematic outline of the experimental protocol. (B) On day 7 (after prime) and day 21 (after boost), sera from vaccinated mice were collected, serially diluted, and assayed for neutralization activities against infectious live SARS-CoV-2 [both wild type (WT) and mutant Delta, n = 15]. LOD, below the limit of detection. (C) On days 7 (n = 7), 21 (n = 15), and 28 (n = 15), serum anti–SARS-CoV-2 (2019-nCoV) spike RBD IgG antibodies were assayed using an ELISA kit. OD450nm, optical density at 450 nm. (D and E) Flow cytometric analysis of the frequency of IFN-γ+ CD8+ T cells (D) and IFN-γ+ CD4+ T cells (E) in splenic lymphocytes ex vivo stimulated with or without S protein (n = 16). (F to H) Changes in the frequency of CD8+ T cells (F), CD4+ T cells (G), and CD8/CD4 T cells (H) in splenic lymphocytes upon S protein stimulation (n = 16). (I to K) Determination of IFN-γ (I) and IL-4 (J) in the culture supernatant of S protein–activated splenic lymphocytes using cytokine kits (n = 16). The secretion of IFN-γ was divided by that of IL-4 (K) to investigate the T helper type 1 (TH1)–biased immunity. (L to N) Representative flow cytometric pictures (L) and summary plot (M and N) of data showing the frequency of CD8+ Tcm (CD44+ CD62L+ CD8+ T cells) and CD8+ Tem (CD44+ CD62L− CD8+ cells) in splenic lymphocytes stimulated with or without S protein in vitro (n = 16). All error bars are expressed as ±SD. Significant at *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Serum virus neutralization assay represents the gold standard for evaluating humoral immunity–mediated protection in vaccinated individuals. Results suggested that antiviral antibodies were significantly elicited in mRNA@LNP-vaccinated mice, as the immune sera from B# broadly neutralized the authentic infection of SARS-CoV-2 [both the wild-type strain and the Delta (B.1.617.2) variant] to Vero E6 cells on day 21 (after boost) (Fig. 3B), which also displayed the highest anti–SARS-CoV-2 (2019-nCoV) spike RBD IgG levels on days 7, 21, and 28 (Fig. 3C). Meanwhile, antiviral antibody responses were also induced in mice activated with mRNA-CNE, although insufficiently (Fig. 3, B and C). By contrast, CNE- and Lipo-based mRNA nanoparticles induced a cellular-biased immunity, with interferon-γ–positive (IFN-γ+) CD8+ T cells in splenic lymphocytes significantly produced (Fig. 3D and fig. S1). Besides, IFN-γ+ CD8+ and IFN-γ+ CD4+ T cells were actively generated in these groups after in vitro S protein restimulation (Fig. 3, D and E, and fig. S1). Notably, these mRNA nanoparticle–vaccinated mice exhibited a CD8+ T–favored and CD4+ T helper type 1 (TH1)–skewed immune response upon antigen reexposure, in which antiviral-specific CD8+ T cells, instead of CD4+ T cells, were extensively mobilized (Fig. 3, F to H), and typical TH1/TH2 cytokine secretion profiles [IFN-γ+/interleukin-4–positive (IL-4+)] by lymphocytes were observed (Fig. 3, I to K). Immunologic memory T cell response toward SARS-CoV-2 spike reencounter demonstrated that CD8+ T cells underwent phenotypic change from central memory (Tcm) to effector memory (Tem) in all groups (Fig. 3, L to N). Specifically, CD8+ Tcm cells were efficiently induced in CNE- and Lipo-based vaccines, especially in the mRNA-CNE group, which might contribute to a rapid and robust elimination of infected cells (40).
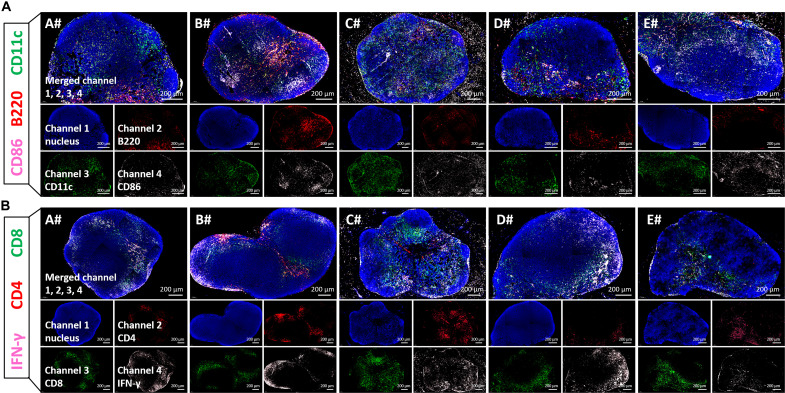
On the other hand, the inguinal lymph nodes (LNs) near the injection site were collected after boost for analyzing the activation (CD86+) of B cells (B220+) and dendritic cells (DCs) (CD11c+) (Fig. 4A), as well as the effector elicitation (IFN-γ+) of CD4+ T cells and CD8+ T cells (Fig. 4B). In line with the above results, immunofluorescence staining pictures revealed that LNP-based mRNA vaccine effectively activated B cells and CD4+ effector T cells in the draining LNs, while CNE- and Lipo-based vaccines (especially mRNA-CNE) induced more activated DCs and CD8+ effector T cells (Fig. 4).
Fig. 4. Immunofluorescence staining of inguinal LNs from different groups.
BALB/c mice were intramuscularly administered with saline (A#), mRNA@LNP (B#), mRNA-CNE (C#), mRNA@Lipo (D#), or mRNA-Lipo (E#) (5 μg of RBD mRNA per mouse) on both hind legs at day 0 (prime) and day 21 (boost). The inguinal LNs were collected on day 24 for analyzing the activation (CD86+, pink) of B cells (B220+, red) and DCs (CD11c+, green) (A), as well as the effector (IFN-γ+, pink), CD4+ (red), and CD8+ (green) T cells (B) by immunofluorescence staining of histological sections. Scale bars, 200 μm.
Together, these data indicated that different nanoplatforms elicited humoral immunity and cellular immunity with different preferences. Specifically, LNP-based mRNA vaccine induced a humoral-biased immune response, while CNE- and Lipo-based ones gave rise to cellular-preferred immunity. Here, compared to mRNA@Lipo, mRNA-Lipo was more representative in eliciting cellular immunity, which was thereby selected for the follow-up studies.
Biodistribution of different nanoparticles
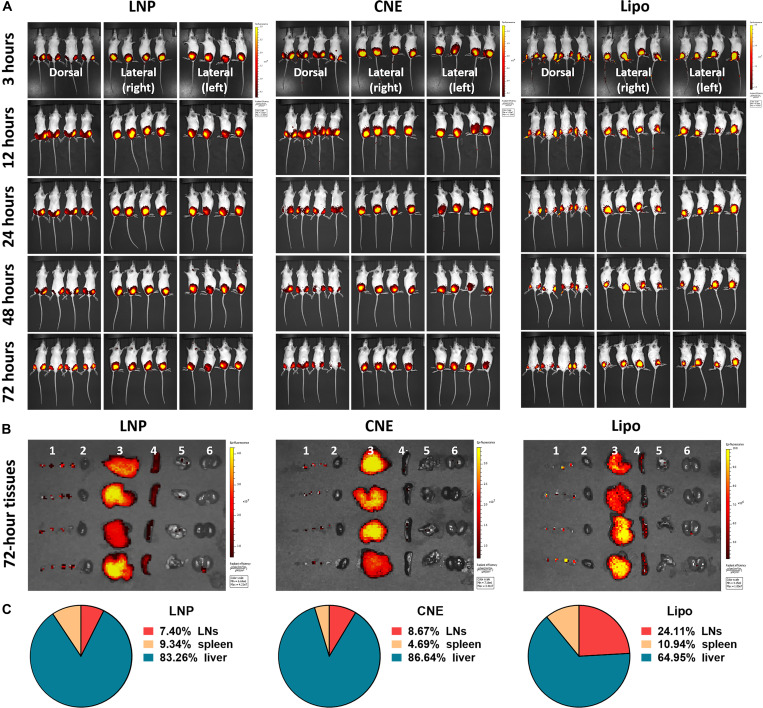
To illustrate the underlying mechanisms, biodistribution patterns of different LNPs (i.e., LNP, CNE, and Lipo) were investigated. Following a single dose of intramuscular administration into both hind legs, all the tested nanoparticles primarily resided at the injection site (i.e., muscle) within 72 hours (Fig. 5A). It should be mentioned that the fluorescence intensity of Lipo was remarkably lower than that of CNE and LNP, possibly due to the signal interference of polypeptide (i.e., Pardaxin) surface modification. Further ex vivo dissection (Fig. 5, B and C) showed that apart from the muscle tissues, there was a robust accumulation of LNPs in the livers (83.26% in LNP, 86.64% in CNE, and 64.95% in Lipo), spleens (9.34% in LNP, 4.69% in CNE, and 10.94% in Lipo), and bilateral axillary and inguinal LNs (7.40% in LNP, 8.67% in CNE, and 24.11% in Lipo). Compared to LNP (16.74%) and CNE (13.36%), Lipo (35.05%) had a better secondary lymphoid organ (SLO; i.e., LNs and spleen)–draining ability, which might contribute to its preferred elicitation of cellular immunity.
Fig. 5. Tissue dissemination of different nanoparticles in vivo.
Biodistribution (A) of DiR-labeled LNP, CNE, and Lipo (a total of 3 μg of DiR per mouse) at 3, 12, 24, 48, and 72 hours after intramuscular injection (on both hind legs) (n = 4). At 72 hours, mice were euthanized with major organs: (1) Bilateral axillary and inguinal LNs, (2) heart, (3) liver, (4) spleen, (5) lung, and (6) kidney were collected for evaluating the tissue dissemination of different nanoparticles (B). Fluorescence intensity at the region of interest representing the accumulative distribution of nanoparticles in LNs, spleen, and liver from each specimen is shown in (C) (n = 4).
Cellular uptake, lysosome escape, and transfection of mRNA nanoparticles
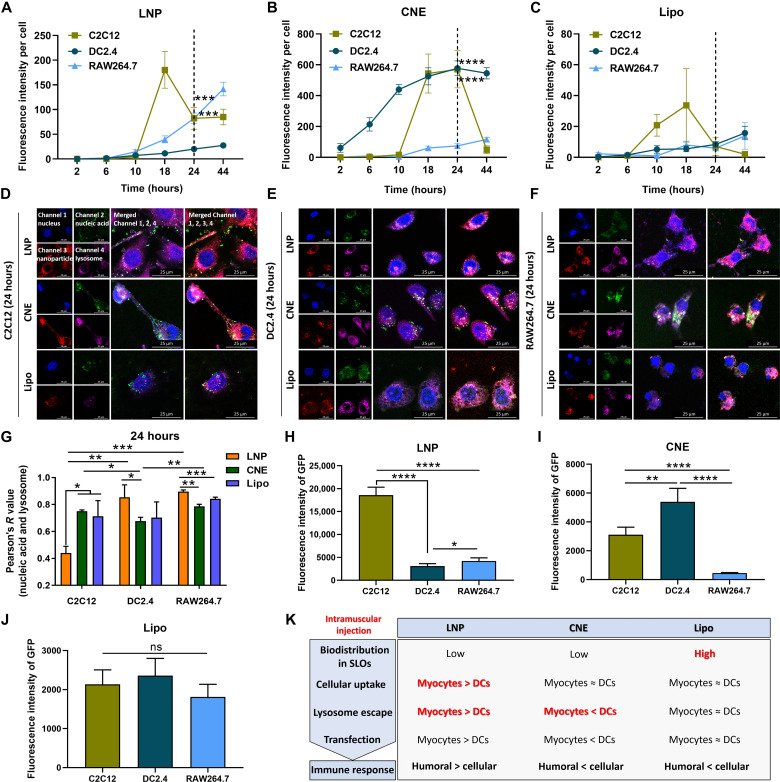
It was reported that somatic cells abundant at sites of administration including myocytes can be positively vaccinated by mRNA vaccines, which served as indirect immune elicitors by transferring antigen to the nearby antigen-presenting cells (APCs), magnifying the breadth and magnitude of adaptive immune response (34). Therefore, we further investigated the cellular uptake, intracellular trafficking, and mRNA transfection of different nanoparticles in myocytes (C2C12) and APCs (DC DC2.4 and macrophage RAW264.7).
The in vitro uptake behaviors showed that myocytes displayed a burst accumulation of all nanoparticles at 10 to 18 hours, which was readily excreted/eliminated at 18 to 44 hours. On the contrary, APCs had a sustained intracellular accumulation of all LNPs within 44 hours (Fig. 6, A to C, and fig. S2). The intracellular localization of cargos by different nanoplatforms was observed by confocal laser scanning microscopy with FAM-ssDNA used as the model nucleic acid and nanoparticles visualized by DiD fluorescent labeling. Results indicated that at 24 hours after treatment (no significant intergroup difference was observed at 6 hours; fig. S3), most ssDNA loaded by LNP circumvented lysosome and diffused into the cytoplasm in myocytes, while CNE and LNP displayed a better lysosomal escape in APCs (Fig. 6, D to G). Next, eGFP mRNA was used to evaluate the transfection efficiency of different carrier systems. As a result, compared to that in APCs (DCs in particular), the expression of mRNA at 24 hours in C2C12 was higher with LNP but lower with CNE. Meanwhile, the mRNA transfection efficiency in C2C12 was comparable to that in APCs with Lipo (Fig. 6, H to J, and fig. S4).
Fig. 6. In vitro cellular uptake, intracellular trafficking, and mRNA transfection of different nanoparticles by myocytes and APCs.
(A to C) Cellular uptake of LNP (A), CNE (B), and Lipo (C) by C2C12, DC2.4, and RAW264.7. Fluorescence images were taken at 2, 6, 10, 18, 24, or 44 hours after administration and further analyzed by ImageJ to semi-quantitate the average fluorescence intensity of DiD-labeled nanoparticles (n = 4). (D to G) Confocal microscopic observation of the lysosomal escape of nucleic acid–loaded nanoparticles in C2C12 (D), DC2.4 (E), and RAW264.7 (F) at 24 hours after treatment. Nucleus (channel 1, blue), 6′FAM-ssDNA (channel 2, green), DiD-labeled nanoparticle (channel 3, red), and lysosome (channel 4, pink). Scale bars, 25 μm. Images were analyzed to calculate the colocalization ratio of lysosomes and nanoparticles based on Pearson’s correlation coefficient (G) (n = 3). (H to J) Transfection of eGFP mRNA in C2C12, DC2.4, and RAW264.7 with LNP (H), CNE (I), and Lipo (J) at 24 hours. The transfection efficiency was determined by ImageJ (n = 4). All error bars are expressed as ±SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns, not significant. (K) Summary table depicting the characteristics of different LNPs. These experiments were performed independently twice.
Structural and biochemical characteristics of mRNA nanoparticles determined humoral and cellular immunity
It should be mentioned that LNP, CNE, and Lipo displayed different cellular internalization, lysosome escape, and mRNA expression in myocytes and APCs at 24 hours after treatment, which, together with the aforementioned biodistribution profiles, might partially explain their selective induction of humoral immunity and cellular immunity (Fig. 6K): For LNP, the accumulative endocytosis of nanoparticles in C2C12 was similar to that in RAW264.7 and significantly higher than that in DC2.4 (cellular uptake: myocytes ≈ macrophages > DCs; Fig. 6A). Intracellularly, the lysosome escape of LNP in C2C12 outplayed that in DC2.4 and RAW264.7 (lysosome escape: myocytes > macrophages ≈ DCs; Fig. 6G), which might contribute to its superior mRNA expression in C2C12 (transfection: myocytes > macrophages > DCs; Fig. 6H). Possibly, the translated protein of interest in myocytes was positively secreted or passively released (e.g., cells undergo apoptosis) into the extracellular space to be captured by locally recruited APCs (34) or entered the lymphatic drainage and/or blood circulation to activate APCs in SLOs. In this case, antigen was internalized by APCs in the form of exogenous substance, which favored the major histocompatibility complex (MHC) class II–restricted presentation that activated specific CD4+ T cells and B cells to elicit humoral-biased immune responses (41).
For CNE, the cellular uptake in C2C12 and DC2.4 was remarkably better than that in RAW264.7 (cellular uptake: DCs ≈ myocytes > macrophages; Fig. 6B), while the lysosome escape was better in DC2.4 than in other cells (lysosome escape: DCs > myocytes ≈ macrophages; Fig. 6G). Consequently, the mRNA transfection efficacy of CNE in C2C12 and DC2.4 was higher than that in RAW264.7 (transfection: DCs > myocytes > macrophages; Fig. 6I). We believed that the escaped lysosome degradation of payloads in DCs promoted MHC class I–restricted presentation of the endogenously synthesized antigen, which activated CD8+ T cells and facilitated the development of cellular immunity.
In terms of Lipo, there was no significant difference among all three cell lines in cellular uptake, lysosome escape, and mRNA transfection (myocytes ≈ DCs ≈ macrophages; Fig. 6, C, G and J). However, Lipo outperformed its counterparts (i.e., LNP and CNE) in SLO trafficking (Fig. 5), which may contribute to the development of MHC class I–associated antigen presentation and cellular-preferred immune responses (42, 43).
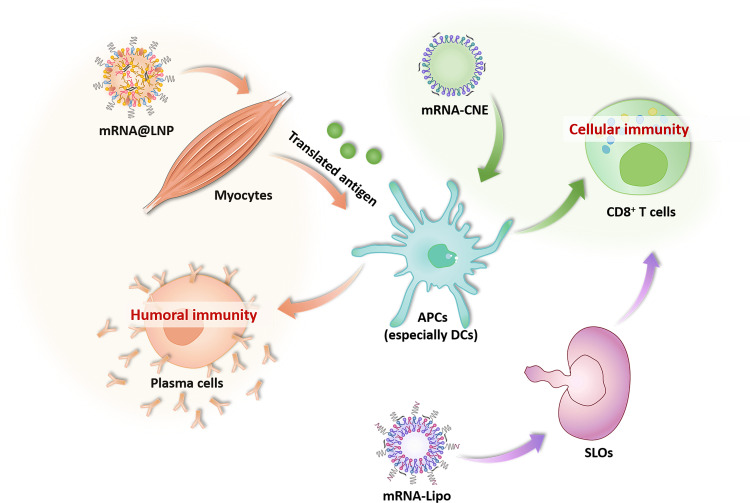
In summary, on the basis of the in vivo and in vitro results, we proposed the hypothesis that LNP-based mRNA vaccine might benefit from the superior endocytosis and lysosome escape in myocytes, which promoted the intracellular translation and intercellular transfer (from myocytes to nearby APCs) of antigen to elicit humoral-biased immunity. In contrast, CNE and Lipo induced cellular-preferred immune responses, which mainly depended on a better lysosome escape in DC and SLO draining, respectively (Fig. 7).
Fig. 7. Potential mechanisms regulating the different mobilization of humoral and cellular immunity by LNP, CNE, and Lipo.
DISCUSSION
A multidimensional mobilization of the humoral and cellular immunity is needed to establish effective antiviral protection against COVID-19. Compared to other RNA viruses, SARS-CoV-2 has a relatively high mutation rate that drives viral genetic variability, facilitating a rapid antigenic shift to escape host immunity (44). Most neutralizing antibodies target the RBD of SARS-CoV-2 S protein. Mutations in the RBD were shown to partially, even completely, ablate antibody affinity and evade neutralization (45–48). Moreover, such mutations could become prevalent over a longer period of evolution as virus gradually accumulates genetic variation (49), contributing to a magnified transmission dynamics and more overwhelming infection rate. Meanwhile, SARS-CoV-2 may also evade CD8+ T cell surveillance through point mutations in the viral MHC class I epitopes, leading to a reduced peptide–MHC-I binding and altered cellular immunity (2). Therefore, a broad-spectrum elicitation of the adaptive immunity is of vital importance to build up an all-round protection against virus immune evasion.
Notably, the safety of antiviral immune responses depends heavily on an appropriate engagement of the humoral and cellular immunity. Several studies have characterized that compared to healthy individuals, bulk T cells from patients with COVID-19 displayed impaired effector function (reduced cytokine production), hyperactivation [high expression of CD38, CD39, programmed cell death protein 1 (PD-1), and human leukocyte antigen-DR isotype (HLA-DR)], constrained reactivation, and limited tissue migration (50). To make it worse, such hyperactivation profiles of T cells may trigger the development of organ-specific autoimmunity, bone marrow infection, and even severe pneumonia with sepsis (51, 52). On the other hand, anti–SARS-CoV-2 antibody-based vaccines and therapeutics may exacerbate the severity of COVID-19 through antibody-dependent enhancement (ADE). Increasing evidence suggests that ADE promotes viral infections via immune complex formation, complement deposition, and local immune activation (53, 54). In these regards, measures to reduce the risks of T cell malfunction and ADE from immunotherapies are urgently needed to ensure vaccine safety in the large-scale medical interventions against COVID-19.
In summary, the efficacy and safety of antiviral therapeutics rely heavily on a well-balanced mobilization of the humoral and cellular arms of immunity. In this work, our data demonstrate that different mRNA nanoparticles elicit humoral immunity and cellular immunity with different preferences, suggesting that proper structural and biochemical optimization of lipid nanocarriers might integrate multiple superiorities and induce a broad-spectrum activation of the adaptive immune protection against SARS-CoV-2 with satisfactory efficacy and safety.
MATERIALS AND METHODS
Cell lines and animals
HEK 293 cells (American Type Culture Collection CRL-1573), mouse C2C12 (H-2k) myoblasts cells (BeNa Culture Collection, Beijing, China), immortalized DC DC2.4, and macrophage RAW264.7 (Shanghai Cell Bank, Chinese Academy of Sciences) were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; GNM12800, JiNuo Biotechnology Co. Ltd., Zhejiang, China) supplemented with 10% fetal bovine serum (FBS) (16140071, Gibco Life Technologies, New York, USA). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 (Heraeus, Germany).
Female BALB/c (H-2Kd) mice were purchased from Slaccas Experimental Animal Co. Ltd. (Shanghai, China) and bred under pathogen-free conditions. All animal experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of Zhejiang University and the Tab of Animal Experimental Ethical Inspection of the First Affiliated Hospital Zhejiang University School of Medicine (reference number: 2021-1374).
Preparation and characterization of LNPs
mRNA@LNP was synthesized by mixing one volume of lipid mixture of DLin-MC3-DMA (AVT Co. Ltd., Shanghai, China), 1,2-dioctadecanoyl-sn-glycero-3-phophocholine (AVT Co. Ltd., Shanghai, China), cholesterol, and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (AVT Co. Ltd., Shanghai, China) (50:10:38.5:1.5, molar ratio) in ethanol and three volumes of mRNA [1:25 (w/w) mRNA to lipid, nitrogen/phosphate (N/P) ratio = 6] in citrate buffer (pH 4.0; 10 mM). Lipids and mRNA were injected into a microfluidic mixing device (ZX-LS-32, FluidicLab, Shanghai, China) at a combined flow rate of 1.5 ml/min. The resultant mixture was dialyzed against phosphate-buffered saline (PBS; pH 7.4; 10 mM) for 16 hours to remove ethanol.
mRNA@Lipo was prepared by mixing one volume of lipid mixture of DOTAP (dioleylhydroxypropyl-3-N,N,N-trimethylammonium; Avanti Co. Ltd., Birmingham, UK), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; AVT Co. Ltd., Shanghai, China), DSPE-PEG2000 {distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide (polyethylene glycol)-2000]; AVT Co. Ltd., Shanghai, China}, and DSPE-PEG2000-Pardaxin [synthesized according to our previous works (35, 55)] (77.6:19.4:0.75:2.25, w/w) in ethanol and three volumes of mRNA [1:10 (w/w) mRNA to lipid, N/P ratio = 3.4] in diethyl pyrocarbonate (DEPC)–treated water. Lipids and mRNA were injected into a microfluidic mixing device at a total flow rate of 1.5 ml/min. The aqueous and ethanol phases were mixed at a flow rate ratio of 3 where the lipid solution experienced a rapid increase in polarity. At a critical polarity, the precipitates form as LNPs. The resultant mixture was evaporated under reduced pressure for 2 hours to remove ethanol.
Blank CNE and Lipo were prepared as previously described (34, 36) and stored at 4°C before use:
1) CNE (high-energy emulsification method): 15 mg of DOTAP, 17.5 mg of lipoid E-80 (egg lecithin-80, Avanti Co. Ltd., Birmingham, UK), and 17.5 mg of vitamin E-acetate (DL-alpha-tocopheryl acetate, BASF) were dissolved in 25 μl of ethanol as the oil phase. Meanwhile, 1.5 ml of DEPC-treated water was used as the aqueous phase and added dropwise to the oil phase with vigorous stirring via vortex to produce a primary emulsion, which was further probe-sonicated (30%, work for 2 s, pause for 3 s, 5 min, three to four rounds) on an ice bath to generate uniform nanoemulsions.
2) Lipo (thin-film rehydration method): Lipid mixtures composed of 6.208 mg of DOTAP, 1.552 mg of DOPE, 0.06 mg of DSPE-PEG2000, and 0.18 mg of DSPE-PEG2000-Pardaxin were dissolved in 3 ml of ethanol-chloroform solution and evaporated under reduced pressure to make a thin film of lipids. Lipo was fabricated by probe ultrasound (10%, work for 2 s, pause for 3 s, 4 min, two rounds) following the hydration of lipid film with 2 ml of DEPC-treated water.
For mRNA complexation, blank Lipo and CNE were incubated with mRNA at a N/P ratio of 3.4 and 4.5 in DEPC-treated water, respectively, for 30 min at 4°C to generate mRNA electrostatically adsorbed Lipo and CNE. The morphology of nanoparticles was viewed by TEM (JEOL JEM-1400 microscopes, Tokyo, Japan) and SIM (N-SIM, Nikon, Tokyo, Japan), while their particle size and zeta potential in Milli-Q water were measured using dynamic light scattering (Malvern Zeta sizer Nano-ZS instrument, UK).
Nucleic acid complexation
The mRNA encapsulation/complexation ability of LNPs was assayed by an agarose retardation assay. A total of 300 ng of eGFP-encoding mRNA (5moU) (eGFP mRNA, 996 nucleotides, catalog no. L-7201, TriLink Biotechnologies, San Diego, California) was encapsulated or complexed with LNP, Lipo, and CNE, and electrophoresis was performed using 2% (w/v) agarose gels stained with Golden View for 20 min at 110 V. Images were then acquired using a Bio-Rad ChemiDoxXRS system.
In vivo vaccination
For RBD mRNA vaccination studies, groups of 6- to 8-week-old female BALB/c mice (n = 16) were primed intramuscularly (50 μl in both hind legs, 10 μg of RBD mRNA per mouse) with saline, mRNA@LNP, mRNA-CNE, mRNA@Lipo, or mRNA-Lipo on day 0 and boosted on day 14 with the same dose. RBD (319 to 541 amino acids) mRNA was synthesized by Ausper Biopharma Inc. (Hangzhou, China), and the sequence of RBD mRNA was mainly based on the research findings by Zhang et al. (32). At indicated time points, mice were bled, and serum was collected for live SARS-CoV-2 neutralization assay by observing the cytopathic effects (CPEs) to Vero E6 cells, the serum was also analyzed for specific IgG level using a SARS-CoV-2 (2019-nCoV) spike RBD antibody titer assay kit (catalog no. KIT006, Sino Biological, Beijing, China) according to the product specification (humoral immunity). On day 28, mice were all euthanized with spleen single-cell suspensions collected for the evaluation of immunologic effector and memory T cell responses toward SARS-CoV-2 spike protein and measurement of cytokine secretion (cellular immunity). Briefly, splenocytes (1 × 107/ml) from naive or immunized mice were incubated with/without SARS-CoV-2 (2019-nCoV) Spike RBD recombinant protein (5 μg/ml; catalog no. 40592-VNAH, Sino Biological, Beijing, China) in complete 1640 medium supplemented with mouse recombinant IL-2 (10 ng/ml; BioLegend, California, USA), 2 mM l-glutamine, 10 mM nonessential amino acids, 1 mM sodium pyruvate, and 50 mM β-mercaptoethanol at 37°C for 48 hours. Then, culture supernatants were harvested and analyzed for TH1/TH2 cytokine secretion profiles using commercially available enzyme-linked immunosorbent assay (ELISA) kits [mouse IFN-γ (EK280/3) and IL-4 (EK204/2) ELISA kits, MultiSciences Biotech Co. Ltd., Zhejiang, China]. Meanwhile, splenocytes were collected and incubated with mouse antibodies against fluorescein isothiocyanate (FITC) anti-CD3 (100204), phycoerythrin (PE) anti-CD4 (100408), allophycocyanin–anti-CD8a (100712), and PE/Cy7 anti–IFN-γ antibodies (505826), or FITC anti-CD3, PE anti-CD8a (100708), allophycocyanin-conjugated anti-mouse CD62L (104412), and PE/Cyanine7 anti-CD44 (103030) antibodies (antibodies were all from BioLegend), and the proportions of CD3+/CD8+/IFN-γ+, CD3+/CD4+/IFN-γ+, CD3+/CD8+/CD44+/CD62L+, and CD3+/CD8+/CD44+/CD62L− T cells were assayed by flow cytometric detection (BD Fortessa, New Jersey, USA). Data were further analyzed with FlowJo V10 software to investigate the memory commitment of T cells. The experiments were performed independently twice, with similar trends of results observed, and all displayed data were representative.
Serum neutralization of live SARS-CoV-2
All mouse sera were heat-inactivated at 56°C for 30 min before use. The working stocks of live SARS-CoV-2 [both the wild-type strain (EPI_ISL_12040150) and the Delta (B.1.617.2) variant] were obtained from sputum samples and propagated by infection of Vero cells as previously described (56). To analyze the serum neutralizing activity, 50 μl of serially diluted serum samples (serial twofold dilutions, starting at 1:32) was mixed with the same volume of 100 50% tissue culture infective dose (calculated by Spearman-Karber method) units of SARS-CoV-2, and the resultant serum-virus mixtures were added into a 96-well microtiter plate and incubated at 37°C in 5% CO2 for 2 hours. Then, 100 μl of Vero E6 cell suspension was seeded into the plate at a density of 1 × 104 cells per well and cultured at 35°C in 5% CO2 for 3 days. Here, virus mixture without serum and untreated Vero E6 cells were respectively used as positive and negative controls. Microplates were observed under the microscope for the presence of virus-induced CPE on the cell monolayer from day 2 to 3, and the serum neutralization titers were calculated on day 3 as the reciprocal of serum dilution resulting in a 100% reduction of the CPE.
Biodistribution
C57BL/6 mice were intramuscularly injected (50 μl in both hind legs) with DiR-labeled LNP, CNE, and Lipo [3 μg of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR) for each mouse]. The biodistribution of different preparations at 3, 12, 24, 48, and 72 hours was observed and analyzed with an in vivo imaging system (Maestro EX, CRI Inc., Woburn, MA).
Cellular uptake
C2C12, DC2.4, and RAW264.7 were seeded in 24-well dish with a confluence of 70 to 80% in 500 μl of complete medium per well and treated with 5 μl of DiD-labeled LNP, CNE, and Lipo. At 2, 6, 10, 18, 24, and 44 hours, cells were washed twice with ice-cold PBS, and nucleus was visualized using Hoechst 33342 (10 μg/ml; 20 to 30 min, at room temperature, Beyotime Co. Ltd., Shanghai, China). After extensive wash with PBS, cells were fixed with 4% formaldehyde (10 min at 4°C), and fluorescence images were taken with an inverted fluorescence microscope (AIR, Nikon, Tokyo, Japan) under constant laser intensity. Images were analyzed by graphic processing software ImageJ to semi-quantitate the average fluorescence intensity of DiD.
mRNA transfection
Cells were seeded in a 48-well dish with a confluence of 60 to 70% and transfected with eGFP mRNA (1 μg/ml) formulated under different nanoplatforms in serum-free DMEM for 4 hours at 37°C before FBS supplementation. Cells were cultured for another 20 hours and viewed with a fluorescence microscope under constant laser intensity. Images were further analyzed by ImageJ to semi-quantitate the fluorescence intensity of GFP.
Subcellular colocalization
Cells (1 × 105 cells per well) were seeded onto 12-mm glass coverslips in a 24-well plate overnight and treated with 6′FAM-ssDNA (1.5 μg/ml; sequence: 6-FAM-5′-CAGACCGACTGGATCT-3′; Shanghai Sangon Biotech Co. Ltd., Shanghai, China)–loaded and DiD-labeled nanoparticles for 24 hours before lysosome staining (50 nM LysoTracker Red; 30 min at 37°C, Beyotime Co. Ltd., Shanghai, China). Then, cells were fixed with 4% paraformaldehyde and poststained with Hoechst. Last, the slides were imaged using a confocal microscope (Leica TCS SP8, Leica, Germany). Different fluorescent pictures were analyzed by software LAS X.
Statistical analysis
All data were evaluated and plotted using GraphPad Prism (version 8.0.1). Comparisons between two or several groups were analyzed using unpaired Student’s t tests or one-way analysis of variance (ANOVA; Tukey’s multiple comparisons test), respectively. In addition, a value of P < 0.05 was considered to be statistically significant.
Acknowledgments
We thank Y. Zhang from the P3 (biosafety protection level III) Laboratory of Zhejiang Provincial Center for Disease Control and Prevention and H. Yao from the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases for the support on live SARS-CoV-2 neutralization analysis.
Funding: This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LD22H190003), Zhejiang Provincial Science and Technology Department Key Technologies R&D Program (2022C03020), and National Health Commission Science Foundation of China-Zhejiang Provincial Major Health Technology Projects (WKJ-ZJ-2206).
Author contributions: Conceptualization: Y.S., Y.Li., J.Y., and Y.Lo. Methodology: Y.S., J.H., and Y.Li. Investigation: Y.S., J.H., and Y.Li. Visualization: Y.S. Funding acquisition: Y.Q. and Y.Lo. Project administration: Y.S., J.H., Y.Li., J. Liu, X.G., J. Li, L.G., X.Z., and G.C. Supervision: Y.Q., Y.Lo., and J.Y. Writing—original draft: Y.S. Writing—review and editing: Y.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Hu B., Guo H., Zhou P., Shi Z.-L., Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L. F., Smyth M., Popa A., Genger J.-W., Endler L., Florian D. M., Mühlgrabner V., Graninger M., Aberle S. W., Husa A.-M., Shaw L. E., Lercher A., Gattinger P., Torralba-Gombau R., Trapin D., Penz T., Barreca D., Fae I., Wenda S., Traugott M., Walder G., Pickl W. F., Thiel V., Allerberger F., Stockinger H., Puchhammer-Stöckl E., Weninger W., Fischer G., Hoepler W., Pawelka E., Zoufaly A., Valenta R., Bock C., Paster W., Geyeregger R., Farlik M., Halbritter F., Huppa J. B., Aberle J. H., Bergthaler A., SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 6, eabg6461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghedin E., Sengamalay N. A., Shumway M., Zaborsky J., Feldblyum T., Subbu V., Spiro D. J., Sitz J., Koo H., Bolotov P., Dernovoy D., Tatusova T., Bao Y., George K. S., Taylor J., Lipman D. J., Fraser C. M., Taubenberger J. K., Salzberg S. L., Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437, 1162–1166 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Liu J., Plante K. S., Plante J. A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Widen S. G., Menachery V. D., Shi P.-Y., Weaver S. C., The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602, 294–299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., Iwatsuki-Horimoto K., Ito M., Yamayoshi S., Loeber S., Tsuda M., Wang L., Ozono S., Butlertanaka E. P., Tanaka Y. L., Shimizu R., Shimizu K., Yoshimatsu K., Kawabata R., Sakaguchi T., Tokunaga K., Yoshida I., Asakura H., Nagashima M., Kazuma Y., Nomura R., Horisawa Y., Yoshimura K., Takaori-Kondo A., Imai M., Chiba M., Furihata H., Hasebe H., Kitazato K., Kubo H., Misawa N., Morizako N., Noda K., Oide A., Suganami M., Takahashi M., Tsushima K., Yokoyama M., Yuan Y., Tanaka S., Nakagawa S., Ikeda T., Fukuhara T., Kawaoka Y., Sato K., Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baden L. R., el Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T.; COVE Study Group , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F., A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 5, 237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., Zhang C., Yang S., Xiao W., Zheng Q., Song X., The investigation of mRNA vaccines formulated in liposomes administrated in multiple routes against SARS-CoV-2. J. Control Rel. 335, 449–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang R., Deng Y., Huang B., Huang L., Lin A., Li Y., Wang W., Liu J., Lu S., Zhan Z., Wang Y., A R., Wang W., Niu P., Zhao L., Li S., Ma X., Zhang L., Zhang Y., Yao W., Liang X., Zhao J., Liu Z., Peng X., Li H., Tan W., A core-shell structured COVID-19 mRNA vaccine with favorable biodistribution pattern and promising immunity. Signal Transduct. Target. Ther. 6, 213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun W., He L., Zhang H., Tian X., Bai Z., Sun L., Yang L., Jia X., Bi Y., Luo T., Cheng G., Fan W., Liu W., Li J., The self-assembled nanoparticle-based trimeric RBD mRNA vaccine elicits robust and durable protective immunity against SARS-CoV-2 in mice. Signal Transduct. Target. Ther. 6, 340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R., Johnson A. J., Liggitt D., Bevan M. J., Cellular and humoral immunity against vaccinia virus infection of mice. J. Immunol. 172, 6265–6271 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L. M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Hilker R., Becker D., Eller A.-K., Grützner J., Boesler C., Rosenbaum C., Kühnle M.-C., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Karikó K., Palanche T., Fischer B., Schultz A., Shi P.-Y., Fontes-Garfias C., Perez J. L., Swanson K. A., Loschko J., Scully I. L., Cutler M., Kalina W., Kyratsous C. A., Cooper D., Dormitzer P. R., Jansen K. U., Türeci Ö., COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Levi-Schaffer F., Marco A., Coronavirus disease 2019 and the revival of passive immunization: Antibody therapy for inhibiting severe acute respiratory syndrome coronavirus 2 and preventing host cell infection: IUPHAR review 31. Br. J. Pharmacol. 178, 3359–3372 (2021). [DOI] [PubMed] [Google Scholar]
- 16.VanBlargan L. A., Goo L., Pierson T. C., Deconstructing the antiviral neutralizing-antibody response: Implications for vaccine development and immunity. Microbiol. Mol. Biol. Rev. 80, 989–1010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lederer K., Castaño D., Gómez Atria D., Oguin T. H., Wang S., Manzoni T. B., Muramatsu H., Hogan M. J., Amanat F., Cherubin P., Lundgreen K. A., Tam Y. K., Fan S. H. Y., Eisenlohr L. C., Maillard I., Weissman D., Bates P., Krammer F., Sempowski G. D., Pardi N., Locci M., SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53, 1281–1295.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janice Oh H.-L., Ken-En Gan S., Bertoletti A., Tan Y.-J., Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 1, e23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widge A. T., Rouphael N. G., Jackson L. A., Anderson E. J., Roberts P. C., Makhene M., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A. B., Flach B., Lin B. C., Doria-Rose N. A., O’Dell S., Schmidt S. D., Neuzil K. M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.-V., Floyd K., Suthar M. S., Buchanan W., Luke C. J., Ledgerwood J. E., Mascola J. R., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 384, 80–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng O.-W., Chia A., Tan A. T., Jadi R. S., Leong H. N., Bertoletti A., Tan Y.-J., Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 34, 2008–2014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodda L. B., Netland J., Shehata L., Pruner K. B., Morawski P. A., Thouvenel C. D., Takehara K. K., Eggenberger J., Hemann E. A., Waterman H. R., Fahning M. L., Chen Y., Hale M., Rathe J., Stokes C., Wrenn S., Fiala B., Carter L., Hamerman J. A., King N. P., Gale M., Campbell D. J., Rawlings D. J., Pepper M., Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 184, 169–183.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T. W., Gentzke A. S., Neff L. J., Glidden E. V., Jamal A., Park-Lee E., Ren C., Cullen K. A., King B. A., Hacker K. A., Disposable E-cigarette use among U.S. youth—An emerging public health challenge. N. Engl. J. Med. 384, 1573–1576 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Kalimuddin S., Tham C. Y. L., Qui M., de Alwis R., Sim J. X. Y., Lim J. M. E., Tan H.-C., Syenina A., Zhang S. L., le Bert N., Tan A. T., Leong Y. S., Yee J. X., Ong E. Z., Ooi E. E., Bertoletti A., Low J. G., Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med. (N Y) 2, 682–688.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K. J. A., Hemmings O., O’Byrne A., Kouphou N., Galao R. P., Betancor G., Wilson H. D., Signell A. W., Winstone H., Kerridge C., Huettner I., Jimenez-Guardeño J. M., Lista M. J., Temperton N., Snell L. B., Bisnauthsing K., Moore A., Green A., Martinez L., Stokes B., Honey J., Izquierdo-Barras A., Arbane G., Patel A., Tan M. K. I., O’Connell L., O’Hara G., MacMahon E., Douthwaite S., Nebbia G., Batra R., Martinez-Nunez R., Shankar-Hari M., Edgeworth J. D., Neil S. J. D., Malim M. H., Doores K. J., Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 5, 1598–1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaebler C., Wang Z., Lorenzi J. C. C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T. Y., Cipolla M., Viant C., Barnes C. O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K. G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D. F., Zhao Z., Gazumyan A., Schwartz R. E., Hatziioannou T., Bjorkman P. J., Mehandru S., Bieniasz P. D., Caskey M., Nussenzweig M. C., Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., Wild K., Basho K., Marinescu M. S., Fuchs J., Topfstedt F., Janda A., Sogukpinar O., Hilger H., Stete K., Emmerich F., Bengsch B., Waller C. F., Rieg S., Sagar, Boettler T., Zoldan K., Kochs G., Schwemmle M., Rizzi M., Thimme R., Neumann-Haefelin C., Hofmann M., Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature 597, 268–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonifacius A., Tischer-Zimmermann S., Dragon A. C., Gussarow D., Vogel A., Krettek U., Gödecke N., Yilmaz M., Kraft A. R. M., Hoeper M. M., Pink I., Schmidt J. J., Li Y., Welte T., Maecker-Kolhoff B., Martens J., Berger M. M., Lobenwein C., Stankov M. V., Cornberg M., David S., Behrens G. M. N., Witzke O., Blasczyk R., Eiz-Vesper B., COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity 54, 340–354.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brito L. A., Chan M., Shaw C. A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G. R., Beard C. W., Dey A. K., Lilja A., Valiante N. M., Mason P. W., Mandl C. W., Barnett S. W., Dormitzer P. R., Ulmer J. B., Singh M., O’Hagan D. T., Geall A. J., A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 22, 2118–2129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou G., Anderluzzi G., Schmidt S. T., Woods S., Gallorini S., Brazzoli M., Giusti F., Ferlenghi I., Johnson R. N., Roberts C. W., O’Hagan D. T., Baudner B. C., Perrie Y., Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: The impact of cationic lipid selection. J. Control Rel. 325, 370–379 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Tenchov R., Bird R., Curtze A. E., Zhou Q., Lipid nanoparticles─From liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano 15, 16982–17015 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Guan S., Rosenecker J., Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 24, 133–143 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., Guo Y., Sun S.-H., Fan H., Zu S.-L., Chen Q., He Q., Cao T.-S., Huang X.-Y., Qiu H.-Y., Nie J.-H., Jiang Y., Yan H.-Y., Ye Q., Zhong X., Xue X.-L., Zha Z.-Y., Zhou D., Yang X., Wang Y.-C., Ying B., Qin C.-F., A thermostable mRNA vaccine against COVID-19. Cell 182, 1271–1283.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samaridou E., Heyes J., Lutwyche P., Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 154–155, 37–63 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Shi Y., Lu Y., Qin B., Jiang M., Guo X., Li X., Liu Y., Huang J., Zhang J., Luo Z., Yin H., Liu H., Guan G., Du Y., Peng L., You J., Antigen transfer from non-APCs to APCs impacts the efficacy and safety of protein- and mRNA-based vaccines. Nano Today 41, 101326 (2021). [Google Scholar]
- 35.Qin B., Yuan X., Jiang M., Yin H., Luo Z., Zhang J., Zhu C., Li X., Shi Y., Luo L., Du Y., You J., Targeting DNA to the endoplasmic reticulum efficiently enhances gene delivery and therapy. Nanoscale 12, 18249–18262 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Yin H., Yuan X., Luo L., Lu Y., Qin B., Zhang J., Shi Y., Zhu C., Yang J., Li X., Jiang M., Luo Z., Shan X., Chen D., You J., Appropriate delivery of the CRISPR/Cas9 system through the nonlysosomal route: Application for therapeutic gene editing. Adv. Sci. (Weinh). 7, 1903381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahin U., Oehm P., Derhovanessian E., Jabulowsky R. A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A. N., Omokoko T., Kranz L. M., Diken M., Kreiter S., Haas H., Attig S., Rae R., Cuk K., Kemmer-Brück A., Breitkreuz A., Tolliver C., Caspar J., Quinkhardt J., Hebich L., Stein M., Hohberger A., Vogler I., Liebig I., Renken S., Sikorski J., Leierer M., Müller V., Mitzel-Rink H., Miederer M., Huber C., Grabbe S., Utikal J., Pinter A., Kaufmann R., Hassel J. C., Loquai C., Türeci Ö., An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585, 107–112 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Amanat F., Krammer F., SARS-CoV-2 vaccines: Status report. Immunity 52, 583–589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le T. T., Andreadakis Z., Kumar A., Román R. G., Tollefsen S., Saville M., Mayhew S., The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 19, 305–306 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Jameson S. C., Masopust D., Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fehres C. M., Unger W. W. J., Garcia-Vallejo J. J., van Kooyk Y., Understanding the biology of antigen cross-presentation for the design of vaccines against cancer. Front. Immunol. 5, 149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song T., Xia Y., Du Y., Chen M. W., Qing H., Ma G., Engineering the deformability of albumin-stabilized emulsions for lymph-node vaccine delivery. Adv. Mater. 33, e2100106 (2021). [DOI] [PubMed] [Google Scholar]
- 43.An M., Li M., Xi J., Liu H., Silica nanoparticle as a lymph node targeting platform for vaccine delivery. ACS Appl. Mater. Interfaces 9, 23466–23475 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Giovanetti M., Benedetti F., Campisi G., Ciccozzi A., Fabris S., Ceccarelli G., Tambone V., Caruso A., Angeletti S., Zella D., Ciccozzi M., Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biochem. Biophys. Res. Commun. 538, 88–91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greaney A. J., Starr T. N., Gilchuk P., Zost S. J., Binshtein E., Loes A. N., Hilton S. K., Huddleston J., Eguia R., Crawford K. H. D., Dingens A. S., Nargi R. S., Sutton R. E., Suryadevara N., Rothlauf P. W., Liu Z., Whelan S. P. J., Carnahan R. H., Crowe J. E., Bloom J. D., Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C. D., Caskey M., Robbiani D. F., Rice C. M., Nussenzweig M. C., Hatziioannou T., Bieniasz P. D., Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes C. O., Jette C. A., Abernathy M. E., Dam K.-M. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey-Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P., Bjorkman P. J., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W., Xu C., Wang P., Anashkina A. A., Jiang Q., Impact of mutations in SARS-COV-2 spike on viral infectivity and antigenicity. Brief. Bioinform. 23, bbab375 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong A. H. M., Tomlinson A. C. A., Zhou D., Satkunarajah M., Chen K., Sharon C., Desforges M., Talbot P. J., Rini J. M., Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat. Commun. 8, 1735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gangaev A., Ketelaars S. L. C., Isaeva O. I., Patiwael S., Dopler A., Hoefakker K., de Biasi S., Gibellini L., Mussini C., Guaraldi G., Girardis M., Ormeno C. M. P. T., Hekking P. J. M., Lardy N. M., Toebes M., Balderas R., Schumacher T. N., Ovaa H., Cossarizza A., Kvistborg P., Identification and characterization of a SARS-CoV-2 specific CD8+ T cell response with immunodominant features. Nat. Commun. 12, 2593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saini S. K., Hersby D. S., Tamhane T., Povlsen H. R., Hernandez S. P. A., Nielsen M., Gang A. O., Hadrup S. R., SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 6, eabf7550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. Gagiannis, J. Steinestel, C. Hackenbroch, M. Hannemann, V. Umathum, N. Gebauer, M. Stahl, H. Witte, K. Steinestel, COVID-19-induced acute respiratory failure—An exacerbation of organ-specific autoimmunity? medRxiv 10.1101/2020.04.27.20077180 (2020). [DOI]
- 53.Lee W. S., Wheatley A. K., Kent S. J., DeKosky B. J., Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 5, 1185–1191 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Arvin A. M., Fink K., Schmid M. A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H. W., A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584, 353–363 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Yuan X., Qin B., Yin H., Shi Y., Jiang M., Luo L., Luo Z., Zhang J., Li X., Zhu C., Du Y., You J., Virus-like nonvirus cationic liposome for efficient gene delivery via endoplasmic reticulum pathway. ACS Cent. Sci. 6, 174–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lou Y., Zhao W., Wei H., Chu M., Chao R., Yao H., Su J., Li Y., Li X., Cao Y., Feng Y., Wang P., Xia Y., Shang Y., Li F., Ge P., Zhang X., Gao W., Song G., Du B., Liang T., Qiu Y., Liu M., Cross-neutralization of RBD mutant strains of SARS-CoV-2 by convalescent patient derived antibodies. Biotechnol. J. 16, e2100207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4