Abstract
Evidence from patients with Parkinson’s disease (PD) and our previously reported α-synuclein (SNCA) transgenic rat model support the idea that increased SNCA protein is a substantial risk factor of PD pathogenesis. However, little is known about the transcription control of the human SNCA gene in the brain in vivo. Here, we identified that the DYT6 gene product THAP1 (THAP domain-containing apoptosis-associated protein 1) and its interaction partner CTCF (CCCTC-binding factor) act as transcription regulators of SNCA. THAP1 controls SNCA intronic enhancers’ activities, while CTCF regulates its enhancer-promoter loop formation. The SNCA intronic enhancers present neurodevelopment-dependent activities and form enhancer clusters similar to “super-enhancers” in the brain, in which the PD-associated single-nucleotide polymorphisms are enriched. Deletion of the SNCA intronic enhancer clusters prevents the release of paused RNA polymerase II from its promoter and subsequently reduces its expression drastically in the brain, which may provide new therapeutic approaches to prevent its accumulation and thus related neurodegenerative diseases defined as synucleinopathies.
Transcription regulation of human SNCA gene by its intronic enhancers provides potential treatment target for Parkinson’s disease.
INTRODUCTION
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases in which α-synuclein (SNCA), encoded by the SNCA gene, is a major component of Lewy bodies (LBs) (1, 2). Although single point mutations in the SNCA gene can cause early-onset familial forms of autosomal dominant inherited PD (3, 4), several lines of evidence support the idea that increased dosage of wild-type human SNCA (hSNCA) protein level is critical to PD pathogenesis as well. Duplication, triplication, or rearrangements of the wild-type SNCA gene cause familial PD, and the severity of clinical phenotypes correlates with the number of SNCA copies (5–7). Furthermore, polymorphisms in SNCA gene regulatory regions that enhance SNCA expression are associated with sporadic PD (8, 9). Similarly, mouse and rat models overexpressing human wild-type SNCA protein showed α-synucleinopathies, nigrostriatal degeneration, and even inclusion body formation (10, 11). In addition, elevated levels of SNCA transcripts were detected in PD postmortem brains (12). To date, SNCA is one of the most validated and promising therapeutic targets for PD, and manipulations of SNCA levels have demonstrated a beneficial impact (13, 14). Thus, deciphering the transcription machinery of SNCA will help us to define new treatment targets to reduce its expression in patients with PD.
So far, only a few transcription factors of the SNCA gene have been reported. ZSCAN21 and ZNF219 can bind to the promoter or intron 1 of SNCA to activate or repress SNCA transcription (15). Other transcription factors such as GATA2, EMX2, NKX6-1, and CEBPD (CCAAT/enhancer-binding protein delta) bind either to the promoter or potential enhancers of the SNCA gene to regulate its expression (9, 16, 17). Poly(adenosine diphosphate–ribose) polymerase 1 is another transcription factor that regulates SNCA expression by binding to a polymorphic microsatellite region, called NACP-Rep1 (18). Other cis-regulatory elements (CREs) of the SNCA gene, such as enhancers, have been reported as well (19). However, the CREs of the SNCA gene and its transcription regulation machinery in the brain in vivo are not clear so far.
Our recent study showed dysregulation of SNCA in THAP1/DYT6 dystonia patients’ induced pluripotent stem cell (iPSC)–derived midbrain dopaminergic (mDA) neurons (20). In this study, we identified that the human DYT6 gene product THAP1 regulates SNCA expression in both human and rat brain through controlling its promoter and intronic enhancers’ activities, while THAP1’s interaction partner CTCF (CCCTC-binding factor) controls the enhancer-promoter loop formation of the SNCA gene. We further demonstrated that the SNCA intronic enhancers present cell lineage–dependent activities during neurodevelopment and form enhancer clusters similar to “super-enhancers” in the brain. Single-nucleotide polymorphisms (SNPs) highly associated with PD risk are enriched in these enhancer cluster regions. Deletion of the SNCA intronic enhancer cluster prevented its transcription elongation and subsequently reduced its expression drastically in the brain, suggesting that targeted editing of the SNCA intronic enhancers might be a new therapeutic approach to reduce SNCA expression and thus its accumulation in the brain, hopefully preventing SNCA-associated neurodegenerative diseases.
RESULTS
THAP1 mutations repress the expression of SNCA
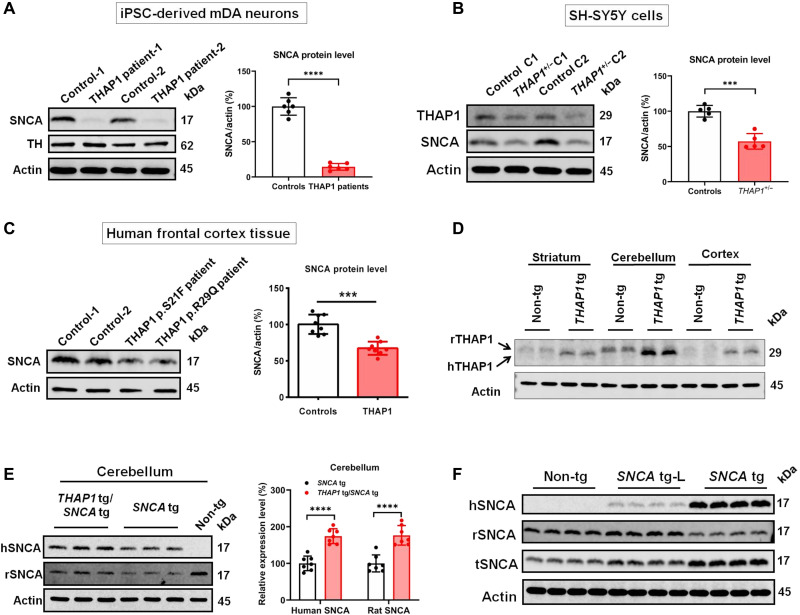
RNA sequencing (RNA-seq) analysis of iPSC-derived mDA neurons (iPSC-Neurons) from patients with THAP1/DYT6 dystonia and from THAP1 heterozygous knockout (THAP1+/−) dopaminergic SH-SY5Y neuronal cells showed significantly decreased expression of SNCA (20), indicating THAP1 as a potential transcriptional regulator of SNCA. Extended Western blot analysis confirmed decreased SNCA protein level in these cells (Fig. 1, A and B). In postmortem frontal cortex samples of patients with THAP1 mutation, we also observed significantly reduced SNCA protein level compared to the brain tissue of neurologically normal deceased controls (Fig. 1C). Conversely, overexpression of THAP1 in both SH-SY5Y and SK-N-AS neuroblastoma cells leads to a significant increase in SNCA expression (fig. S1A) (21). All these observations suggest that THAP1 may regulate the expression of SNCA.
Fig. 1. THAP1 regulates the expression of SNCA.
(A) Western blotting and statistical analysis show that the expression of SNCA in THAP1 patients’ iPSC-Neurons is significantly lower than in control iPSC-Neurons. (B) In THAP1 heterozygous knockout (THAP1+/−) dopaminergic SH-SY5Y cells, Western blotting and statistical analysis detect that the expression of THAP1 and SNCA is significantly lower than in control cells. (C) The expression of SNCA in THAP1 patients’ frontal cortex is lower than in control individuals by Western blotting and statistical analysis. (D) Western blot analysis shows the expression of hTHAP1 and rTHAP1 proteins in different brain regions of THAP1 transgenic rats (n = 3 rats in each group). (E) Western blotting and statistical analysis determine the protein level of human SNCA (hSNCA) and rSNCA in the cerebellum of human THAP1 and SNCA double-transgenic rats (THAP1 tg/SNCA tg), SNCA transgenic rats (SNCA tg), and nontransgenic wild-type rats (Non-tg) (n = 5 rats in each group). (F) Western blotting shows the protein level of hSNCA, rSNCA, and tSNCA in the cortex of Non-tg and two different SNCA-overexpressing transgenic rat lines (SNCA tg-L and SNCA tg) (n = 5 rats in each group). ***P < 0.001; ****P < 0.0001. Comparison by unpaired two-tailed t test.
Human THAP1 regulates human and rat SNCA expression in the brain of transgenic rats
To further prove the role of THAP1 in regulating SNCA expression in the brain, we generated THAP1-overexpressing transgenic rats (THAP1 tg) by pronuclear injection of the entire human THAP1 gene (12 kb) (fig. S1B). Both reverse transcription polymerase chain reaction (RT-PCR) and Western blot analysis confirmed the expression of wild-type human THAP1 (hTHAP1) in different brain regions of THAP1 tg rats (Fig. 1D and fig. S1C). The highest expression of hTHAP1 was seen in the cerebellum, similar to the expression patterns of endogenous rat THAP1 protein (rTHAP1) (Fig. 1D). We also observed that the molecular weight of hTHAP1 protein is slightly smaller than that of rTHAP1 protein (Fig. 1D), although the predicted molecular weight of hTHAP1 is 24.944 kDa, which is slightly larger than the predicted molecular weight of rTHAP1 (24.688 kDa) (www.uniprot.org). These data may indicate different posttranslational modifications of these two proteins, such as methylation (22).
We next crossbred the THAP1 tg rats with our previously generated human SNCA transgenic rats (SNCA tg). The SNCA tg rats harbor the entire human SNCA gene sequence (190 kb) (GenBank AF163864), including 30-kb upstream regulatory promoter sequences, all the intronic regions, and a 45-kb flanking downstream region (11). In the cerebellum of human SNCA and human THAP1 double-transgenic rats (SNCA tg/THAP1 tg), significantly increased expression of both rat SNCA (rSNCA) and hSNCA was detected when compared to SNCA tg rats (Fig. 1E). Increased expression of hSNCA and rSNCA was also seen in the striatum and cortex, but their changes were weaker than in the cerebellum (Fig. 1E and fig. S1, D and E). The expression change of hSNCA and rSNCA is correlated to the expression level of hTHAP1, as the cerebellum expresses more hTHAP1 protein than the striatum and cortex (Fig. 1D). These data further confirm the role of THAP1 in regulating the expression of SNCA in the brain in vivo.
When comparing the SNCA tg rats with nontransgenic rats (Non-tg), we observed decreased rSNCA protein level (Fig. 1E and fig. S1, D and E), which may suggest that overexpression of hSNCA protein represses the expression of endogenous rSNCA. We next analyzed the expression of rSNCA, hSNCA, and total SNCA (tSNCA) in two different SNCA transgenic rat lines, the SNCA tg line that expresses a very high level of hSNCA protein and the SNCA tg-L line that expresses a very low level of hSNCA protein, and observed that the rSNCA protein was decreased significantly only in the SNCA tg line but not in the SNCA tg-L line (Fig. 1F and fig. S1F). These data suggest the autoregulative function of SNCA, but it may happen only when the hSNCA protein reached a certain expression level.
THAP1 regulates the activities of the SNCA promoter and intronic enhancers
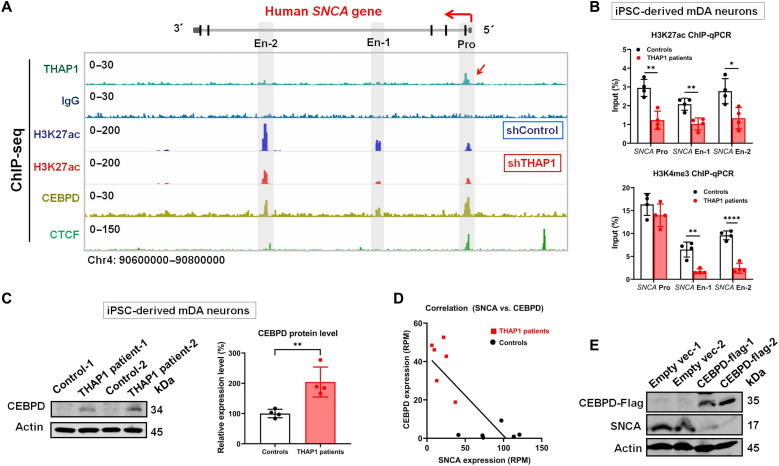
To understand how THAP1 regulates SNCA expression, we examined our THAP1 chromatin immunoprecipitation sequencing (ChIP-seq) data and observed weak binding signals to the SNCA promoter (Fig. 2A) (20). Further luciferase reporter assay analysis confirmed that wild-type THAP1 can up-regulate the activity of the entire SNCA promoter [10.7 kb upstream of translation start site (23)] (fig. S2A).
Fig. 2. Characterization of the mechanisms by which THAP1 regulates SNCA expression.
(A) THAP1 ChIP-seq detects its binding signal on the SNCA promoter (red arrow), which is not seen in IgG control (lanes 1 and 2). H3K27ac ChIP-seq defines the SNCA promoter and potential enhancers in SH-SY5Y cells and also shows its modification on the SNCA gene in control (shControl) and THAP1 knockdown (shTHAP1) SH-SY5Y cells (En-1: Chr4: 90720577–90722136; En-2: Chr4: 90673800–90675200) (lanes 3 and 4). CEBPD and CTCF ChIP-seq shows their binding sites on both the SNCA promoter and enhancers (lanes 5 and 6). (B) ChIP-qPCR analyzes their H3K27ac and H3K4me3 modifications on the SNCA promoter and enhancers in THAP1 patients’ iPSC-Neurons and in control iPSC-Neurons. (C) Western blot analysis shows the endogenous CEBPD protein level in THAP1 patients’ iPSC-Neurons and in control iPSC-Neurons. (D) Correlation analysis reveals that the CEPBD mRNA level is negatively correlated to the SNCA mRNA level in iPSC-Neurons [reads per million (RPM)] (equation: Y = −0.4196 * X + 43.28; P = 0.0014). (E) Western blotting determines the expression of CEBPD and SNCA in CEBPD-overexpressing SH-SY5Y cells. *P < 0.05; **P < 0.01; ****P < 0.0001. Comparison by unpaired two-tailed t test.
The acetylation at the 27th lysine residue of the histone H3 protein (H3K27ac) and trimethylation at the 4th lysine residue of the histone H3 protein (H3K4me3) are highly correlated with gene expression levels (24). H3K27ac is also an effective marker to determine active enhancers (25). We thus performed H3K27ac and H3K4me3 ChIP analyses to detect the activities of all SNCA regulatory elements. As the THAP1+/− SH-Y5Y cell lines still express close to normal levels of wild-type THAP1 protein due to its autoregulative function (20), we generated THAP1 knockdown SH-SY5Y cell lines (shTHAP1) in which the THAP1 protein was reduced to 30% (fig. S2B). Similarly decreased expression of SNCA was also observed in the shTHAP1 cell lines (fig. S2C). Significantly decreased H3K27ac modification on both SNCA promoter and potential enhancers, including enhancer 1 (En-1) and enhancer 2 (En-2), was seen in shTHAP1 cell lines compared to control cell lines (shControl) (Fig. 2A). The changes of SNCA enhancers’ activities are stronger than the changes of its promoter (fig. S2D). Similarly decreased H3K27ac and H3K4me3 modifications on SNCA promoters and enhancers were also seen in THAP1 patients’ iPSC-Neurons compared to control iPSC-Neurons, respectively (Fig. 2B).
As THAP1 has no direct binding site on SNCA enhancers (Fig. 2A), its regulation on the SNCA enhancers’ activities might be through a transcriptional repressor of SNCA, the CEBPD (CCAAT/enhancer-binding protein delta) (17), as both RNA-seq and Western blot analyses showed highly increased expression of CEBPD in THAP1 patients’ iPSC-Neurons compared to control neurons (Fig. 2C). The following data further support this hypothesis: (i) The expression level of SNCA is negatively correlated with the expression level of CEBPD in iPSC-Neurons (Fig. 2D); (ii) THAP1 binds to the CEBPD promoter (fig. S2E), and CEBPD binds to the SNCA promoter and enhancers (Fig. 2A); (iii) knockdown of THAP1 induced the expression of CEBPD and strengthened the binding activities of CEBPD on the SNCA promoter (fig. S2, F and G); and (iv) overexpression of CEBPD indeed repressed the expression of endogenous SNCA in SH-SY5Y cells (Fig. 2E).
THAP1’s interaction partner CTCF regulates SNCA expression
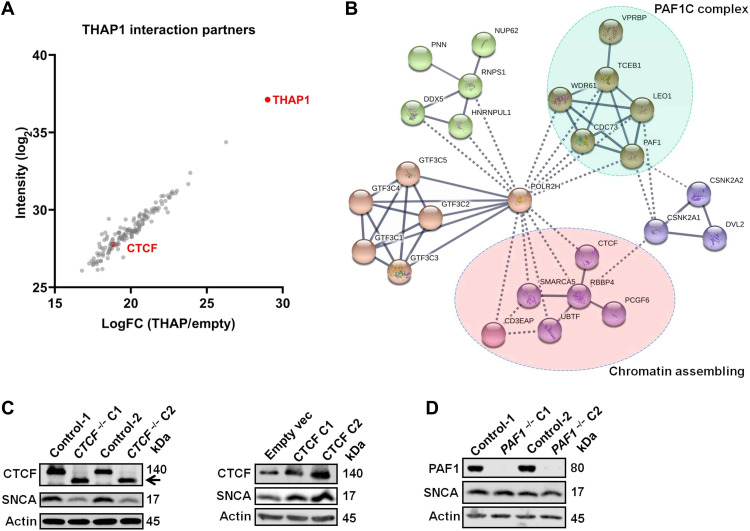
Next, we want to know whether THAP1’s interaction partners are also the transcription regulators of SNCA. We thus performed tandem affinity purification and mass spectrometry (TAP-MS) analysis to isolate the interaction partners of THAP1. In TAP-MS, we overexpressed flag-tagged THAP1 protein in human embryonic kidney (HEK) cells and pulled down THAP1 protein and its interaction partners using anti-flag agarose gel. Last, we performed MS analysis to identify all these proteins. Using TAP-MS analysis, we isolated a total of 149 proteins that strongly interact with THAP1 (with log2 > 15) (Fig. 3A). STRING protein-protein interaction networks analysis grouped these proteins into five functional complexes (Fig. 3B), such as the RNA polymerase II (Pol II)–associated factor 1 (PAF1) complex (PAF1C) and the chromatin assembling complex, including CTCF (Fig. 3B), which have a potential function in regulating mRNA expression. Western blot analysis confirmed the interactions between THAP1 and its interaction partners, such as CTCF (fig. S3A). ChIP-seq showed that CTCF binds to the SNCA promoter and a region near the SNCA En-2 (Fig. 2A). Knocking out CTCF down-regulates the expression of SNCA, while overexpression of CTCF leads to up-regulation of SNCA (Fig. 3C and fig. S3B). We also knocked out PAF1 in SH-SY5Y, a key component of PAF1C, but did not observe any expression change of SNCA (Fig. 3D and fig. S3C). All these data suggest that THAP1’s interaction partner CTCF, but not PAF1, is a transcription regulator of the SNCA gene. Further ChIP-qPCR analysis revealed that knocking out CTCF did not affect the activities of the SNCA promoter and enhancer (fig. S3D), which indicates that CTCF regulates the SNCA expression possibly through altering the enhancer-promoter contact, as it has a function in regulating enhancer-promoter interactions (26).
Fig. 3. THAP1’s interaction partner CTCF regulates the expression of SNCA.
(A) Dot plots present the interaction partners of THAP1 identified by TAP-MS analysis, and CTCF and THAP1 are marked in red color. LogFC, log fold change. (B) String-db network analysis annotates the functional complexes of THAP1 interaction partners. (C) Western blotting analyzes the protein level of CTCF and SNCA in CTCF knockout (left) and CTCF-overexpressing (right) SH-SY5Y cells. The black arrow indicates a small isoform of CTCF. (D) Western blotting detects the protein level of PAF1 and SNCA in PAF1 knockout SH-SY5Y cells.
SNCA enhancers in intron 4 make contact with its promoter and regulate its expression
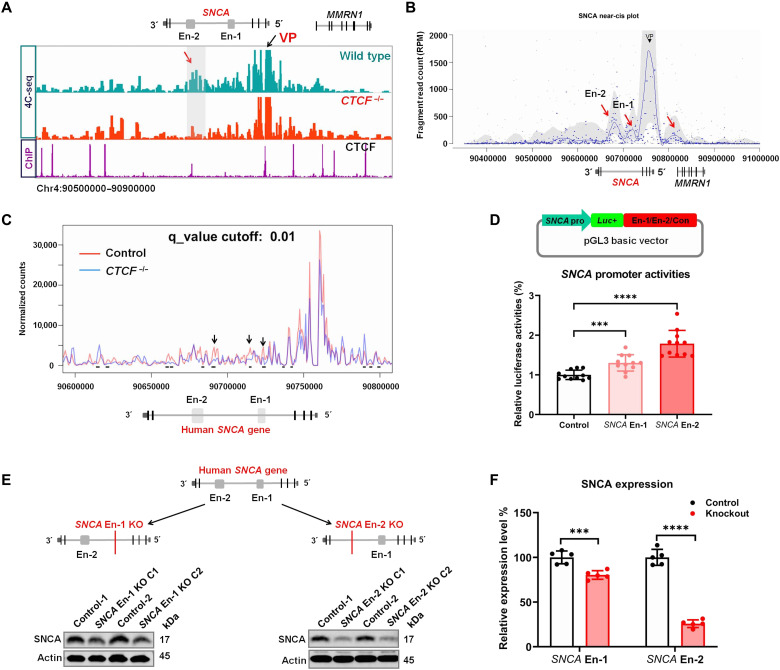
As THAP1 regulates SNCA expression through controlling its enhancers and promoter activities, we next wanted to know whether these enhancers indeed make physical contact with the SNCA promoter to regulate its expression and also to see whether CTCF regulates SNCA enhancer-promoter contacts. We thus performed circular chromosome conformation capture followed by sequencing (4C-seq) analysis in which the SNCA promoter was set as a view point (VP). We observed strong physical contact between the SNCA En-2 region and its promoter (Fig. 4, A and B). Other regions, such as the SNCA En-1 and MMRN1 promoter, also showed contact with the SNCA promoter when analyzing the 4C-seq data with the Basic4C package (Fig. 4B) (27). As CTCF binds to both SNCA promoter and SNCA enhancer En-2 (Fig. 4A), we investigated whether it also mediated the SNCA enhancer-promoter interaction. Knocking out CTCF reduced the contact frequencies between the VP (SNCA promoter) and several loci located on intron 4 of the SNCA gene (Fig. 4, A and C). Thus, CTCF regulates the structure of SNCA enhancer-promoter loops.
Fig. 4. SNCA intronic enhancers regulate its expression.
(A) Distribution of SNCA 4C-seq raw reads in wild-type and CTCF−/− SH-SY5Y cells (lanes 1 and 2). CTCF ChIP-seq detects its binding sites around the SNCA gene (lane 3). The SNCA promoter region was set as VP. The red arrow indicates the SNCA En-2 region that is strongly in contact with the VP in wild-type cells, while this interaction disappeared in CTCF−/− SH-SY5Y cells (En-1 and En-2 indicate the position of the two SNCA intronic enhancers, respectively). (B) The Basic4C package was used to analyze the 4C-seq data. Red arrows indicate the regions that are in contact with the VP (SNCA promoter) (from left to right: SNCA En-2, SNCA En-1, and MMRN1 promoter). (C) The 4C-ker package analyzes the change of interactions between the SNCA promoter and its intron 4 region (including its two enhancers) in wild-type and CTCF knockout (CTCF−/−) SH-SY5Y cells (at least two biological replicates of each sample were performed). Black arrows and short black lines both show the regions with statistically changed interactions with the SNCA promoter (4C-ker analysis default setting: K = 10, q_value = 0.01). (D) Schematic diagram shows the strategies of cloning vectors to characterize the role of SNCA enhancers (En-1 and En-2) in regulating its promoter (SNCA pro) activities. A genomic unrelated region was used as a negative control (Con) (top). Luciferase reporter assay analysis determines the role of SNCA enhancers in regulating its promoter activities (bottom). (E) Schematic graphs present the strategies using the CRISPR-Cas9 technique to knock out SCNA En-1 or En-2 in SH-SY5Y cells (top). Western blotting (bottom) shows the SNCA protein level in En-1 or En-2 knockout SH-SY5Y cells. (F) Statistical analysis shows the SNCA protein level in SNCA En-1 or En-2 knockout SH-SY5Y cells. ***P < 0.001; ****P < 0.0001. Comparison by unpaired two-tailed t test.
To further validate the function of SNCA intronic enhancers, we subcloned the SNCA En-1 and En-2 fragments into the C terminus of the luc+ gene in pGL3 basic vector, respectively, in which the expression of luc+ gene was driven by the SNCA core promoter (Fig. 4D). Compared to genomic unrelated fragment (control), both SNCA En-1 and En-2 fragments can up-regulate the activity of the SNCA promoter, while the fragment En-2 has a stronger function than the En-1 fragment (Fig. 4D). Next, we knocked out SNCA En-1 and En-2 in SH-SY5Y cells to see how much these enhancers control the expression of SNCA (fig. S4, A and B). Knocking out SNCA En-1 significantly reduced its expression by about 20%, while knocking out En-2 drastically repressed 75% of its expression (Fig. 4, E and F). We also knocked out the SNCA En-1 in SK-N-AS neuroblastoma cell line, in which the SNCA En-1 is inactivated (fig. S4C). Unexpectedly, we did not observe any expression changes of SNCA after knocking out its En-1 region in this cell line (fig. S4C). These data suggest that both SNCA enhancers can regulate its expression but only when they are activated. The SNCA En-2 plays a predominant function in controlling the expression of SNCA in SH-SY5Y cells.
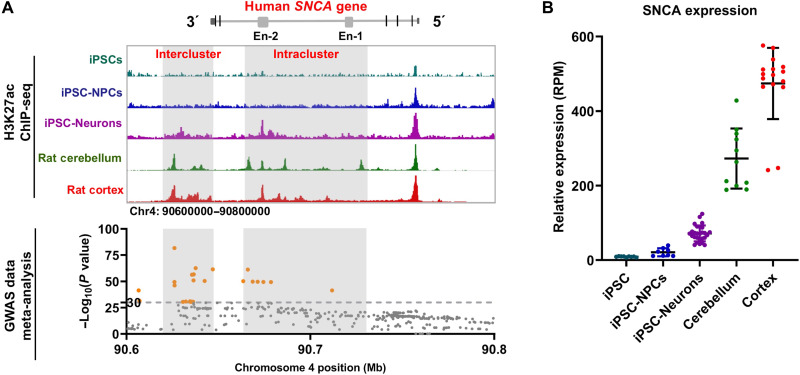
Lineage-specific SNCA enhancers control its expression during neurodevelopment
Next, we sought to know whether the activation of the human SNCA enhancer is associated with its expression in neurons and in the brain. We first analyzed the activation of these enhancers during neuronal differentiation of iPSCs by using H3K27ac ChIP-seq analysis. We observed that the activities of SNCA En-2 present cell lineage–dependent changes (Fig. 5A and fig. S5A). For example, it is inactivated in iPSCs and iPSC-derived neural precursor cells (iPSC-NPCs) but moderately activated in iPSC-Neurons. However, in the cortex and cerebellum of SNCA tg rats, this intronic enhancer region is highly activated and expanded over 30 kb to form enhancer clusters (defined as intracluster), similar to super-enhancers (Fig. 5A and fig. S5B) (28, 29). Further ChIP analysis revealed the enrichment of transcription factors P300 in this region (fig. S5C), and total RNA-seq analysis of the human frontal cortex detected the expression of enhancer RNA (eRNA) in this region (fig. S5B), which further support the characteristics of these intronic enhancer clusters as super-enhancers (30). Similar enhancer clusters on the human SNCA gene and the rat Snca gene were observed in the human and rat brain, respectively (fig. S5, D and E) (31). Furthermore, the region downstream of the SNCA gene also showed cell lineage–dependent changes and formed enhancer clusters in the brain (defined as intercluster in Fig. 5A).
Fig. 5. Lineage-dependent activation of SNCA enhancers is associated with its expression and PD risk.
(A) The status of H3K27ac modification of the human SNCA gene in iPSCs, iPSC-derived neural precursor cells (iPSC-NPC), iPSC-Neurons, frontal cortex of SNCA tg rats (rat cortex), and cerebellum of SNCA tg rats (rat cerebellum) (top). Dot plots show the distribution of PD risk–associated SNPs on the SNCA gene by meta-analysis of GWAS data. Orange dots indicate SNPs with an extremely higher association with PD risk [−log10(P values) > 30] (bottom). Intercluster and intracluster indicate the enhancer clusters downstream of the SNCA gene and the intronic enhancer clusters, respectively. (B) Relative expression level of SNCA in different cells and tissues determined by RNA-seq analysis.
In parallel with the activation of SNCA enhancers, its expression level increased correspondingly (Fig. 5B). Very low levels of SNCA were observed in iPSCs and iPSC-NPCs, while moderate SNCA expression was seen in iPSC-Neurons. Because the expression levels of hSNCA in different SNCA transgenic rat lines are varied (Fig. 1F), we thus analyzed the expression of SNCA mRNA in the human brain. Extremely high expression levels of SNCA were observed in the human cerebellum and frontal cortex (Fig. 5B). These data further suggest that the activities of SNCA enhancers may be responsible for the elevated expression of SNCA in neurons and in the brain. Genome-wide association study (GWAS) data meta-analysis revealed that the SNPs with the highest association with PD risk [−log10 (P value) > 30] are enriched in the enhancer cluster regions of SNCA (intracluster and intercluster in Fig. 5A, bottom) (32). As these enhancer regions can regulate SNCA expression, the PD risk–associated SNPs within these regions may affect the enhancer activities and SNCA expression, which thus contributes to sporadic PD risk.
SNCA intronic enhancer cluster predominantly controls the expression of SNCA in brain
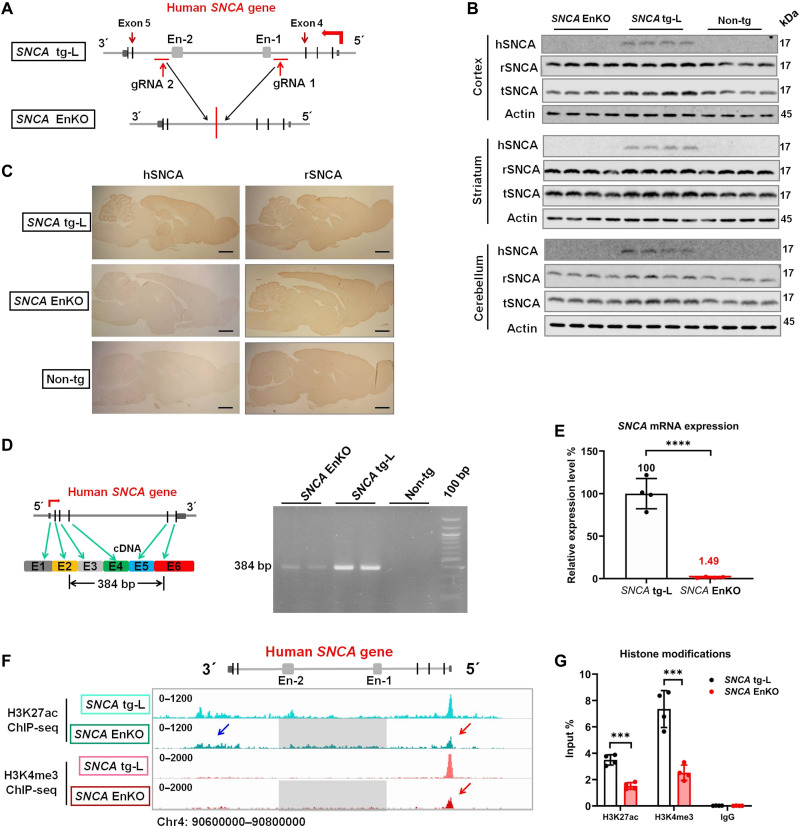
Next, we want to know whether the intronic enhancer clusters also predominantly regulate the expression of SNCA in the brain, as the SNCA En-2 region controls SNCA expression in dopaminergic SH-SY5Y cells (Fig. 4, E and F). For this purpose, we want to knock out these intronic enhancer clusters (intracluster: a region that includes SNCA En-1 to En-2) of the human SNCA gene in our SNCA transgenic rat model (Fig. 6A). To exclude the copy number changes/rearrangement during enhancer knockout, we analyzed the copy number of the human SNCA gene in our two different human SNCA transgenic rat lines (Fig. 1F) using whole-genome sequencing data (with the same sequencing depth). We observed that the copy number of the human SNCA gene in SNCA tg rats is over 10 times more than the copy number in SNCA tg-L rats (fig. S6A). We also observed that the SNCA tg-L rats harbor only one copy of the human SNCA gene, as the read intensity of the rat Snca gene (two copies) is two times more than the intensity of the human SNCA gene in the same rat (fig. S6B). Using the CRISPR-Cas9 technique, we generated an SNCA EnKO rat model by knocking out the intronic enhancer clusters in the SNCA tg-L rats (Fig. 6A and fig. S6C). Both the whole-genome sequencing and Sanger sequencing confirmed the DNA break point at the guide RNA (gRNA) target sites and religation of two residual ends in the SNCA EnKO rats, respectively (fig. S6C). These data proved that the SNCA EnKO rat model was successfully generated.
Fig. 6. The human SNCA intronic enhancer clusters predominantly control its expression in brain.
(A) Schematic graph shows the position of gRNAs and the human SNCA gene structure in SNCA tg-L and SNCA EnKO rats. (B) Western blotting confirms the expression of hSNCA, rSNCA, and tSNCA in different brain regions of SNCA EnKO, SNCA tg-L, and Non-tg rats (n = 5 rats in each group). (C) Immunohistochemical staining of hSNCA and rSNCA in the brain of SNCA tg-L, SNCA EnKO, and Non-tg rats (n = 3 rats in each group). Scale bars, 2 mm. (D) Schematic representation of the primers used to amplify full-length human SNCA mRNA (384 bp in length) (left). RT-PCR determines the expression of full-length human SNCA mRNA in SNCA EnKO, SNCA tg-L, and Non-tg rats (right) (n = 3 rats in each group). (E) RT-qPCR determines the expression level of human SNCA mRNA in SNCA EnKO rats. SNCA tg-L rats were used as a control (n = 4 rats in each group). (F) H3K27ac and H3K4me3 ChIP-seq reveals the status of histone modifications of the human SNCA gene in SNCA tg-L and SNCA EnKO rats (blue arrow: H3K27ac modification downstream of the human SNCA gene in SNCA EnKO rat; red arrow: H3K27ac and H3K4me3 modification on the promoter of the human SNCA gene in SNCA EnKO rat; gray squares indicate the region that was deleted from the human SNCA gene in SNCA EnKO rat). (G) ChIP-qPCR quantitatively determines the histone modifications on the SNCA promoter in SNCA tg-L and SNCA EnKO rats (n = 4 rats in each group). ***P < 0.001; ****P < 0.0001. Comparison by unpaired two-tailed t test.
We next tested both rSNCA and hSNCA expression in the SNCA EnKO rat. Unexpectedly, both Western blot analysis and immunohistochemical staining did not detect hSNCA protein in the entire brain of SNCA EnKO rats (Fig. 6, B and C). As both methods are not sensitive to detect extremely low levels of target protein, we thus analyzed the mRNA level of human SNCA in the SNCA EnKO rat model using RT-PCR and RT-qPCR. Both methods detected an extremely low level of human SNCA mRNA in SNCA EnKO rat compared to SNCA tg-L rats (1.49%) (Fig. 6, D and E). Confirming Sanger sequencing excluded mutations, aberrant splicing, or exon skipping of the human SNCA mRNA in SNCA EnKO rat (fig. S6D). These data suggest that knocking out the intronic enhancer clusters drastically reduces the expression of hSNCA in the brain.
SNCA intronic enhancer cluster regulates its expression via the release of paused RNA Pol II
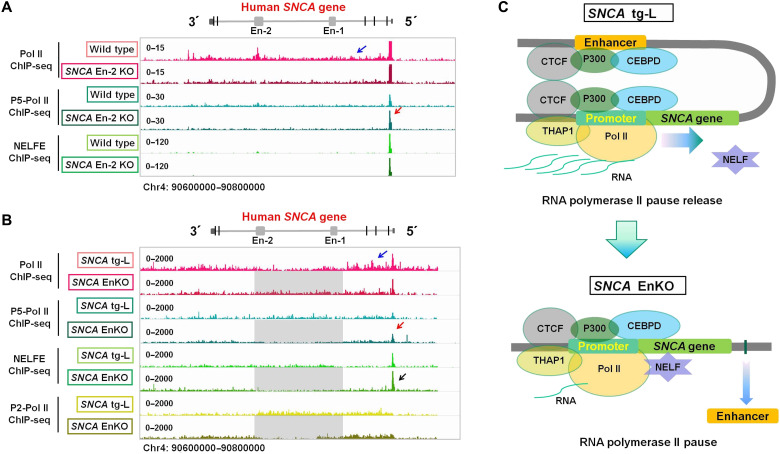
Enhancers could induce gene expression through promoting transcription initiation, the release of paused Pol II, or productive elongation. To see how the SNCA intronic enhancer clusters control its expression in the brain, we next performed H3K27ac and H3K4me3 ChIP-seq and ChIP-qPCR to check the human SNCA promoter activities in SNCA EnKO rats. Deletion of the intronic enhancer clusters leads to around 50% reduction of the H3K27ac and H3K4me3 modifications on the human SNCA promoter (Fig. 6, F and G). However, the change of promoter activity does not match the change of mRNA expression, because the human SNCA mRNA decreased to 1.49% after knocking out the intronic enhancer clusters (Fig. 6E). We thus want to know how the intronic enhancers regulate the expression of hSNCA. We first performed Pol II and S5 phosphorylated Pol II (P5-Pol II) ChIP-seq to examine the transcription initiation and elongation of the human SNCA gene in wild-type and SNCA En-2 KO SH-SY5Y cells (Fig. 7A). We observed that Pol II is highly enriched in the entire SNCA gene in wild-type SH-SY5Y cells compared to SNCA En-2 KO SH-SY5Y cells (Fig. 7A), which indicates decreased SNCA transcription elongation in SNCA En-2 KO SH-SY5Y cells. Increased enrichment of P5-Pol II on the SNCA promoter in SNCA En-2 KO SH-SY5Y cells was detected when compared to wild-type SH-SY5Y cells (Fig. 7A); these data suggest that the transcription initiation of the SNCA gene in SNCA En-2 KO SH-SY5Y cells is slightly higher than in wild-type SH-SY5Y cells. Similar changes were also observed in the frontal cortex of SNCA tg-L and SNCA EnKO rats. Higher enrichment of Pol II in the SNCA gene body was detected in the SNCA tg-L cortex compared to the SNCA EnKO cortex, while the SNCA EnKO rat cortex presents higher enrichment of P5-Pol II and negative elongation factor E (NELFE), an essential component of the NELF complex (33), compared to the SNCA tg rat cortex (Fig. 7B and fig. S7, A to C). Further ChIP-qPCR showed stronger binding activities of Pol II on exons 2 and 4 of the human SNCA gene in the SNCA tg-L rat cortex compared to that in the SNCA EnKO rat cortex (fig. S7A). All these data strongly support the notion that knocking out the SNCA intronic enhancer (SNCA En-2 alone or of the whole SNCA intraclusters) leads to RNA Pol II pausing on the SNCA promoter, which causes suppressed transcription elongation and reduced SNCA expression (Fig. 7C and fig. S7D). Thus, the SNCA intronic enhancer clusters predominantly regulate the expression of SNCA mainly through promoting the release of paused RNA Pol II and transcription elongation.
Fig. 7. The human SNCA intronic enhancer clusters promote release of paused RNA Pol II.
(A) ChIP-seqs show the binding profiles of RNA Pol II, serine-5 phosphorylated Pol II (P5-Pol II), and NELFE in wild-type and SNCA En-2 KO SH-SY5Y cells. (B) ChIP-seqs show the binding profiles of Pol II, P5-Pol II, NELFE, and serine-2 phosphorylated Pol II (P2-Pol II) in the SNCA tg-L rat cortex and SNCA EnKO rat cortex. (A and B) The blue arrow indicates Pol II binding signal on the human SNCA gene body. The red arrow shows the increased binding signal of P5-Pol II on the SNCA promoter in SNCA En-2 KO SH-SY5Y cells and SNCA EnKO rat cortex compared to wild-type SH-SY5Y and SNCA tg-L rat cortex, respectively. The black arrow shows that the NELFE binding signal is stronger in the SNCA En KO rat cortex than in the SNCA tg-L cortex. (C) Schematic graphs show the structures and transcription regulators of the human SNCA gene in wild-type (SNCA tg-L) and in enhancer cluster knockout (SNCA EnKO) conditions. The human SNCA promoter was bound by many transcription factors and makes contact with its intronic enhancer (Enhancer). The contact between promoter and enhancer promotes the release of Pol II pause and transcription elongation (top). Knocking out the intronic enhancer clusters (enhancer) of the human SNCA gene loses the contact between enhancer and promoter and increases the enrichment of NELF on its promoter, which causes RNA Pol II pausing (bottom).
DISCUSSION
We found two new transcription regulators of the human SNCA gene, the DYT6 dystonia gene product THAP1 and its interaction partner CTCF. THAP1 regulates the activities of the SNCA promoter and enhancers through both direct and indirect pathways, while CTCF mediates the enhancer-promoter interactions. During the neuronal development, the SNCA intronic enhancer En-2 presents cell lineage–dependent activities. In the brain, this enhancer expanded and formed enhancer clusters that predominantly up-regulate the expression of SNCA in the brain through promoting the release of RNA Pol II pause, while knocking out these intronic enhancer clusters drastically reduces its expression in the brain in vivo.
Decreased expression of SNCA may somehow be related to the phenotype of DYT6 dystonia, but it is most likely not the key node that is responsible for the DYT6 dystonia phenotype. Complete knockout of Snca in mice leads to reduction of striatal dopamine and an attenuation of DA-dependent locomotor response to amphetamine (34, 35). Similarly, deficiency of locomotor activities and abnormalities of amphetamine-induced firing frequency of striatal cholinergic interneurons were observed in our DYT6 rats (20). However, SNCA is not the only dysregulated gene found in DYT6 dystonia, and THAP1 mutations cause gene expression changes mainly through its target SP1 (20). Furthermore, the expression change of SNCA might not be common in other types of primary dystonia except DYT6 dystonia. In TAF1/DYT3 dystonia, patients’ fibroblasts and iPSC that differentiated into neurons did not show dysregulation of SNCA (36). Also, brain tissues from TorsinA knock-in mice also did not show expression changes of either CEBPD or SNCA (37).
Several studies confirmed that a high level of wild-type SNCA protein is a causative factor contributing to PD pathogenesis. Characterizing its transcription regulation might help us to find out new treatment targets for this disease. In this study, we found two new transcription regulators of the SNCA gene, THAP1 and CTCF. Both in vitro and in vivo data support the role of THAP1 in regulating the expression of SNCA in neuronal cells and in the brain, respectively. THAP1 can directly regulate the SNCA promoter activities or indirectly regulate its enhancer activities through CEBPD. CTCF is the first transcription factor identified so far that can regulate the SNCA gene structure, such as the enhancer-promoter loops.
In this study, we detected two enhancer clusters of the human SNCA gene in the brain and proved its function in vivo, the intraclusters (located in the intron 4 region, including the SNCA En-1 and SNCA En-2) and the interclusters (located in the 3′ downstream of the SNCA gene) (Fig. 5A). The PD risk–associated SNPs identified by GWAS meta-analyses are enriched in these two enhancer cluster regions. Although the exact roles of these SNPs are not well characterized so far, they may up-regulate the expression of endogenous SNCA either by modifying enhancer activity (38) or altering the enhancer-promoter contact (39) and thus contribute to PD pathogenesis. For example, the PD risk top hit rs356182 located in the intercluster region causes increased expression of SNCA in the human frontal cortex (40). Higher levels of SNCA mRNA were observed in the substantia nigra (SN) of patients with PD carrying risk SNPs located in the interclusters, such as rs356165, rs356219, and rs11931074, compared to control individuals (41, 42). Human iPSC-derived neurons harboring PD risk–associated SNPs located in intraclusters (rs356168) express a higher level of SNCA compared to control iPSC-derived neurons (9). PD risk–associated SNPs within the SNCA promoter (SNCA-Rep1) may also up-regulate the expression of SNCA in dopaminergic SH-SY5Y cells and in transgenic mouse brain (8, 43). However, this alteration could not be replicated in some other studies using different tissues. Decreased or unchanged mRNA levels were observed in blood from patients with PD carrying the SNCA-Rep1 variant or blood from its transgenic mice (41, 43). In the temporal cortex from patients with PD carrying the rs356168 variant, the expression level of SNCA-mRNA was decreased instead of increased (44). All these observations might indicate tissue-dependent changes of SNCA expression in patients with PD carrying risk SNPs, and the increased expression of SNCA may occur predominantly in the affected brain regions of patients with PD, such as in the midbrain or SN. In general, increased SNCA-mRNA was observed predominantly in the midbrain or SN of patients with PD (45, 46), while in other tissues/regions, such as in the temporal/frontal cortex or cerebrospinal fluid (CSF), even decreased protein levels of SNCA were detected, which was consistently reported by many different studies (44, 45, 47–49). Similarly, decreased CSF SNCA protein levels were also observed in patients with dementia with LBs or with multiple system atrophy (47). Decreased CSF SNCA level might be due to a possible increase in the rate of SNCA uptake from the CSF into neurons and oligodendroglia (47). Nevertheless, all these observations support a tissue-dependent expression change of SNCA in patients with PD, while the role of PD risk–associated SNPs in regulating SNCA expression in different tissues, as well as in the pathogenesis of PD, needs further systematic characterization.
In this study, we observed that the human SNCA intronic enhancers also present lineage-dependent activations during neuronal differentiation of human iPSCs. During differentiation, many genes are switched on via interactions between tissue- and developmental stage–specific enhancers and their cognate promoters (50). However, how enhancers control the transcription cycle at promoters during differentiation is not well understood so far. Enhancers can regulate transcription initiation through which the expressions of genes are predominantly regulated during differentiation (50). In this study, we showed that the neuronal differentiation–associated enhancers can promote the release of paused Pol II from its promoter through which the expression of SNCA was predominantly regulated. Enhancers promoting the release of RNA Pol II pause have been reported for some other genes as well (51); one potential mechanism could be that the enhancer expresses eRNA that can facilitate NELF complex release and thus promote transcription elongation (52). The expression of the SNCA eRNA in the human brain also supports the role of its intronic enhancer clusters in promoting RNA Pol II release. As the human SNCA enhancers are activated specifically in some cells/tissues, such as in neurons or in the brain, the SNCA enhancer–mediated Pol II pause release might also be cell lineage dependent.
Because of the role of high levels or mutant SNCA in the pathogenesis of PD and other neurodegenerative disorders, many treatment studies in animals or humans have been postulated by applying small interfering RNA or short hairpin RNA (shRNA) with the aim to reduce the level of endogenous SNCA (53). Antisense oligonucleotides such as amido-bridged nucleic acid–modified antisense oligonucleotides were also implicated as potential treatment targeting the SNCA gene (54). Although reduction of the SNCA expression through different methods can protect neurodegeneration induced by hSNCA (55), rotenone (56), or 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (57), side effects such as toxicity or inflammation caused by vectors or injection itself and reduced dopamine content and TH neurons due to the extremely lower level of endogenous SNCA were also observed (55–57). Similarly, targeting general transcription factors or modifying histone modification, such as H3K27ac (58), might not be an ideal treatment strategy for PD, as both strategies will cause global gene expression changes and subsequently may cause unknown side effects. The SNCA promoter and CRE-like enhancers directly regulate its expression and could be used as treatment targets. Using a CRISPR-dCas9–based technique to modify the DNA methylation or histone modification on the SNCA promoter has been reported as a treatment through which the SNCA expression in culture neurons was down-regulated (14, 59). However, the in vivo and long-term treatment effects of these virus vector–based treatments are all unknown. Furthermore, the brain-specific SNCA expression was regulated by its intronic enhancer cluster; thus, targeting the promoter might not be sufficient enough to down-regulate the expression of SNCA in the brain. As the transcription machinery of the human SNCA gene and its expression level in the brain are different from that in cultured neurons, the use of cultured neurons or iPSC-derived neurons as a model system to study the transcription regulation of the human SNCA gene or the effects of PD risk–associated SNPs needs to be interpreted with caution.
Our data and previous report both support that the SNCA En-1 is specifically activated in dopaminergic neuronal cells (19). Our results proved that knocking out activated SNCA enhancers (such as En-1) in dopaminergic neuronal cells will moderately repress its expression, but in En-1–inactivated neuronal cells such as SK-N-AS cells, its deletion does not affect SNCA expression. On the basis of the role of intronic enhancer clusters in regulating the brain-specific expression of SNCA, targeted editing of a small region of the intronic enhancer clusters, such as SNCA En-1, using the in vivo CRISPR-Cas9 gene editing system to moderately reduce the expression of SNCA specifically in the brain or dopaminergic neurons could be used as a new therapeutic strategy to hopefully prevent SNCA accumulation in SNCA-associated neurodegenerative diseases.
MATERIALS AND METHODS
Cell culture
All cell lines were authenticated by using polymorphic short tandem repeat loci before they are used in experiments (20). Mycoplasma contamination was tested regularly every 3 months. HEK-293 [American Type Culture Collection (ATCC) CRL-1573] and SH-SY5Y (ATCC CRL-2266) cells were purchased from Sigma-Aldrich (85120602 and 94030304) (RRID: CVCL_0045 and CVCL_0019) and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 1% GlutaMAX at 37°C in a humidified atmosphere containing 5% CO2.
Generation of shTHAP1 SH-SY5Y cell lines
Stable knockdown of THAP1 in SH-SY5Y cells was performed by stable transfection of THAP1 shRNA expression vector (SureSilencing shRNA plasmids KH07515N, QIAGEN). Two shRNA expression vectors were used for stably knocking down THAP1 in SH-SY5Y cells (target 1: GACGTATCAGAAAGAGGTTAT; target 2: GCCTGTAAAGGAGTTTCATTT). The expression of THAP1 was determined by both real-time PCR and Western blot. Control cell lines were stably transfected with negative control shRNA (sequence: GGAATCTCATTCGATGCATAC).
Plasmid vectors
hTHAP1 complementary DNA (cDNA) open reading frame was amplified by PCR from the cDNA of HEK-293 cells and subcloned into pcDNA3.1 or pcDNA 3.1/Flag plasmid (Invitrogen). CEBPD expression vector was purchased from Genscript (OHu17203D; Genscript). pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from F. Zhang (Addgene plasmid #42230; http://n2t.net/addgene:42230). pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from F. Zhang (Addgene plasmid #62988). pBABE-puro was a gift from H. Land, J. Morgenstern, and B. Weinberg (Addgene plasmid #1764; http://n2t.net/addgene:1764). CTCF expression vector was generated by amplifying its cDNA from pDONR223_CTCF_WT (a gift from J. Boehm, W. Hahn, and D. Root; Addgene plasmid #81789) and subcloning into pcDNA3.1/V5tag vector. All subcloned vectors were verified by Sanger sequencing.
Generation of iPSCs and differentiation into mDA neurons
Generation of iPSCs and differentiation into mDA neurons have been described in our previous publication (20). The ethical approval for the development of and research pertaining to patient-derived cell lines has been given by the National Committee for Ethics in Research (Luxembourg). In brief, native fibroblasts were obtained from skin biopsies with the written informed consent of donors. Fibroblasts were maintained and cultured as previously described (60). The information of patients and control individuals was published previously (20). Fibroblasts were reprogrammed to pluripotency using the Simplicon RNA reprogramming kit (Merck). Molecular karyotyping and identity analysis were performed using HumanOmni 2.5 Exome-8 DNA Analysis BeadChip (Life&Brain GmbH). The iPSCs were cultured as previously published (60, 61).
mDA neurons were differentiated from iPSCs according to the previously reported protocols (61). iPSCs were first derived into NPCs using N2B27 media as described before (20). The NPCs were directly differentiated in N2B27 media containing 1 μM purmorphamine, 200 μM ascorbic acid, and FGF8b (100 ng/ml; PeproTech, 100-25). After 8 days of differentiation, the culture medium was changed to N2B27 supplemented with 0.5 μM purmorphamine and 200 μM ascorbic acid for another 2 days. From 10 days of directed differentiation until the designated experimental time point (day 45 onward), the following substances were added to N2B27 media: brain-derived neurotrophic factor (10 ng/ml; PeproTech, 450-02), glial cell line–derived neurotrophic factor (10 ng/ml; PeproTech, 450-10), transforming growth factor–β3 (1 ng/ml; PeproTech, 100-21), adenosine 3′,5′-monophosphate (500 μM; Applichem, A0455), and ascorbic acid (200 μM).
Human postmortem brain tissues
The use of human brain tissue for the experiment has been approved in accordance with guidelines established by the Institutional Animal Care and Use Committee at the University of Tuebingen, Germany (project number: 565/2019BO2). Frozen postmortem frontal cortex tissues were obtained from two patients with THAP1 mutation (with THAP1 p.S21F and p.R29Q mutation, respectively) and three neurologically normal control individuals as described previously (20).
CRISPR-Cas9 knockout of CTCF and PAF1
Knockout of CTCF and PAF1 was performed using the CRISPR-Cas9 technique (62). gRNA was selected by CRISpick (https://portals.broadinstitute.org/gppx/crispick/public). gRNA oligos synthesized by biomers (www.biomers.net) were annealed and subcloned into pX459 vector. Transfection was performed by cotransfection of pX459/gRNA and pBabe empty vector (ratio, 10:1) into SH-SY5Y cells. Twenty-four hours after transfection, cells were selected by puromycin (final concentration, 2 μg/ml) for another 48 hours. The surviving cells were subcultured in 10-cm dishes until single clone formation. Sanger sequencing and Western blot were used to select positive cell clones and verify target protein expression. All gRNA sequence and genotype primers are listed in table S1.
CRISPR-Cas9 knockout of SNCA En-1 and En-2
SNCA En-1 and En-2 knockout was performed using the CRISPR-Cas9 system. Two gRNAs target flanking SNCA En-1 (SNCA En-1 gRNA 1 and gRNA 2; table S1) or SNCA En-2 (SNCA En-2) (SNCA En-2 gRNA 1 and gRNA 2; table S1) were designed by CRISPick (https://portals.broadinstitute.org/gppx/crispick/public) and subcloned into pX459 vector. Two paired gRNAs and pBabe were cotransfected (ratio, 1:1:0.2) into SK-N-AS or SH-SY5Y cells to knock out SNCA En-1 or En-2. Twenty-four hours after transfection, cells were selected by puromycin (final concentration, 2 μg/ml) for another 48 hours. The surviving cells were subcultured on 10-cm dishes until single clone formation. Genotyping of single clone was done by PCR. Positive and negative genotype PCR primers are listed in table S2.
Animal studies
All animal (rat) experiments were approved by the local Ethics Review Committees (Regierungspräsidium Tübingen) (HG6/14, HG03/20G, and HG01/21G). Experimental procedures involving animals were conducted in accordance with guidelines established by the Institutional Animal Care and Use Committee at the University of Tuebingen. All the animal studies were performed according to the “3R” principle (replace, reduce, and refine). For protein analysis, RNA analysis, and immunohistochemistry staining, at least three to five animals (3 months old) of each group were used. All experiments, including protein analysis, RNA-seq, and ChIP-seq, were performed using 3-month-old male rats. The following animal models were generated for this study.
hTHAP1 transgenic rat model (hSNCA tg)
To generate the human THAP1 gene transgenic rat model, the whole THAP1 gene (12 kb) was amplified from human genomic DNA using the following PCR primers: forward primer: CCTCATGAATGGCTTTGAGT; reverse primer: AAGCCAAGTAGCAATTCAGG. The PCR product was subcloned into pUC19 vector using Kpn I and Sal I restriction enzymes. After validating the sequences of positive clones by Sanger sequencing, the THAP1 gene fragment was digested and purified. High-quality THAP1 DNA fragments were directly injected into the pronucleus of rat zygotes to generate the hTHAP1 transgenic rat model. The expression of hTHAP1 was analyzed at both mRNA and protein levels.
Human SNCA and hTHAP1 double-transgenic rat model (SNCA/THAP1 tg)
The hSNCA/hTHAP1 double-transgenic rat model was generated by crossbreeding our previously published human SNCA tg rats (11) with the newly generated human THAP1 tg rats.
Human SNCA intronic enhancer cluster knockout (SNCA EnKO) rat model
The SNCA intronic enhancer cluster knockout rat model was generated by using the CRISPR-Cas9 system. Two gRNAs were designed by targeting two ends of the large intronic cluster region (about 48.6 kb), which includes the SNCA En-1 and SNCA En-2 regions (gRNA1: CTGATACCACCTGCCTGCAC; gRNA2: GCATGGTCACAACTTACTAA). Briefly, two synthesized gRNAs together with single-stranded oligodeoxynucleotide template and Cas9 protein were injected into the pronucleus of rat zygotes. The genotyping was done by positive PCR and negative PCR analyses. Positive PCR was performed using primers that target two ends of residuals after knockout (forward primer: GGCCAAAATGAACTAAATGAACAGGG; reverse primer: GAACTTGGCTGAATCTGAGTCCATTC). The negative PCR was performed using primers targeting the sequencing upstream and downstream of gRNA2 site (forward primer: CGTATATGAAGGCTTTAAGGGTGTGC; reverse primer: GAACTTGGCTGAATCTGAGTCCATTC).
Western blot
The use of human brain tissue and iPSC-differentiated mDA neurons for protein analysis was approved by the ethics committee of the University Hospital Tuebingen, Tuebingen, Germany. Western blot was performed as previously described (20). Briefly, culture cells were lysed in radioimmunoprecipitation assay (RIPA) buffer [1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mmol NaCl, 50 mmol tris (pH 8.0), and 1× proteinase inhibitor]. After measuring protein concentration by a Bicinchoninic acid kit (Thermo Fisher Scientific), protein samples were separated by SDS gel and blotted onto a nitrocellular membrane (Millipore). After incubating with primary antibody (working concentrations of different antibodies are listed in table S2) and secondary antibody [1:15,000 in concentration; IRDye 800CW goat anti-rabbit immunoglobulin G (IgG) (H + L) or IRDye 680RD goat anti-mouse IgG (H + L)], blot membranes were visualized under the Odyssey Fc Imaging System (Li-Cor). Each Western blot was repeated at least three times.
Immunohistochemistry staining
Immunohistochemistry staining was performed as reported previously (63). Brains of male rats at 3 months of age were fixed by transcardiac perfusion with 4% paraformaldehyde in phosphate-buffered saline (PBS), followed by postfixation with the same fixatives overnight at 4°C. The half brain was embedded in paraffin and subjected to microtome sectioning. DAB (3,3′-diaminobenzidine) staining was used to stain the hSNCA and rSNCA in SNCA tg rats and SNCA EnKO rats. In detail, the whole procedure was carried out at room temperature. The endogenous peroxidase activity of brain sections was blocked by using 0.5% sodium borohydride, followed by section permeabilization in tris-buffered saline containing 0.4% Triton X-100. The incubation in primary antibody anti-SNCA (anti-hSNCA, 1:250; anti-rSNCA, 1:500) was performed overnight followed by secondary antibody staining using biotinylated goat anti-rabbit at a dilution of 1:1000 (Vector Laboratories). Then, sections were treated with an avidin-biotin complex (Vector Laboratories, BA-1000) and exposed to DAB-H2O2 (0.01% DAB and 0.001% hydrogen peroxide) until a suitable staining intensity had developed.
Luciferase reporter assay
Luciferase reporter assays were performed as previously described (64). The full SNCA promoter region (10.7 kb) (23) was amplified by PCR and subcloned into pGL3 empty vector. Activities of firefly and Renilla luciferase were measured at 48 hours after cotransfection of pGL3 vector with pcDNA3-THAP1 or pcDNA3-Empty vector by incubating with the Dual Luciferase Reporter Assay System (Promega) in a Mithras luminometer (Berthold Technologies). The TOR1A promoter was used as a positive control as THAP1 can repress the activity of the TOR1A promoter (64). All experiments were verified at least in five independent replicates.
To detect the interactions between SNCA enhancers and promoter, the core promoter region (1.34 kb) of the SNCA gene was amplified from the whole SNCA promoter using the primers AGCTAGCAAAGGCTTTCTGCTA (forward) and TCTCGAGGGTGGAAAGGCAGAA (reverse) and subcloned into pGL3 basic vector (Promega) using Nhe I and Xho I restriction enzymes. The SNCA En-1 region [1560 base pairs (bp); forward primer: CAAACTCACAGCGTAGACAA; reverse primer: AATTTCCTGGATGCTCAGTG], SNCA En-2 region (1496 bp; forward primer: TAAAGGGGACTGGGAAAGTT; reverse primer: AAAACCATCACCACAGTTCC), and negative control region (1504 bp; forward primer: AATGCACTGGAAAGGTTGTT; reverse primer: TCTTTTTCCCCATGAGGTCT) were amplified from human genomic DNA and subcloned into pGL3/SNCA core promoter vector, respectively. The subcloned pGL3 vector carrying both the SNCA promoter and enhancer was cotransfected with pRL vector (Promega) into SH-SY5Y cells to analyze the interaction between the SNCA promoter and enhancer.
Nuclear protein extraction
Nuclear proteins extracted from HEK cells, SH-SY5Y cells, or brain tissues were used for analyzing THAP1 protein level or for TAP-MS experiments. Briefly, the cytoplasmic fraction was collected using buffer A [10 mM Hepes (Sigma-Aldrich), 1 mM EDTA (Applichem), 0.1 mM EGTA (Sigma-Aldrich), 10 mM KCl (Carl-Roth), 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), phosphatase inhibitor cocktail 2 and 3 (1 μg/ml; Sigma-Aldrich), 1 mM dithiothreitol (DTT; Merck), and complete protease inhibitor (1 μg/ml; Roche)]. After 15 min of incubation, NP-40 (Roche) was added to the final concentration of 0.1% to break the cell membrane. Following centrifugation at 10,000g for 5 min at 4°C, the supernatant was removed (cytoplasmic fraction), and the nuclei were harshly resuspended in buffer C [20 mM Hepes, 0.2 mM EDTA, 0.1 mM EGTA, 25% glycerol (Carl-Roth), 420 mM NaCl (Merck), 1.5 mM MgCl2 (Sigma-Aldrich), 1 mM PMSF, phosphatase inhibitor cocktail 2 and 3 (1 μg/ml), 1 mM DTT, and complete protease inhibitor (1 μg/ml)] and rotated on an intellimixer for 20 min at 4°C. After centrifugation at 10,000g for 5 min at 4°C, the supernatant containing nuclear proteins was collected.
Tandem affinity purification and mass spectrometry
TAP-MS was performed as reported previously (65). TAP was performed in HEK-293 cells overexpressing Strep/FLAG-tagged THAP1 protein. Nuclear protein was extracted as described before and incubated with anti-Flag M2 agarose affinity gel (Sigma-Aldrich). During the incubation, 300 U of benzonase nuclease (Sigma-Aldrich) per milligram of nuclear extract was added to disturb the DNA- or RNA-mediated interactions. The eluted protein solution by flag peptide was treated by trypsin for quantitative tandem MS analysis (liquid chromatography–tandem MS). Raw data were quantified by building ratios of the peak area intensities from specific fragment ions of the corresponding samples. Each group (empty vector group and wild-type THAP1 group) was repeated five times.
ChIP and sequencing
ChIP sample preparation and high-throughput sequencing were done as previously described (20). For brain tissues, frozen brain tissues were thawed and cut into less than 1-mm3 pieces. PBS with 1% formaldehyde (Thermo Fisher Scientific) was added to cross-link tissues for 15 min. After adding glycine and incubating for another 5 min, tissues were collected by centrifugation. For cell samples, cells were cross-linked by directly adding formaldehyde into the culture medium to the final concentration of 1% and incubated for 10 min at room temperature. After adding glycine to the final concentration of 0.125 M and incubating for 5 min, cells were collected. Cell pellets/tissue pellets were sonicated in RIPA buffer [10 mM tris-HCl, 1 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, and 1× protease inhibitor cocktail (pH 8.0)] to 200 to 500 bp using Covaris S220. After centrifugation, chromatin was incubated with an antibody-bead complex, which was prepared 1 day before by incubating antibody with goat anti-rabbit IgG/mouse IgG magnetic beads (Thermo Fisher Scientific) overnight. After incubation of the chromatin and antibody-bead complexes overnight, the complexes were washed by low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer. The ChIP DNA–protein complex was eluted from beads and decross-linked overnight. Last, ChIP DNA was extracted using a PCR purification kit (QIAGEN). At least 5 ng of immunoprecipitated DNA was used for the library preparation using the NEB Ultra II DNA Library Preparation Kit for Illumina (NEB, E7103L). Next-generation sequencing was done on an Illumina NextSeq500/NovaSeq 6000 system using the 75/100-bp single-end high-output sequencing kit.
Chromatin immunoprecipitation quantitative polymerase chain reaction
Quantitative analysis of ChIP results was performed using ChIP-qPCR. The ChIP-qPCR was performed using QuantiTect PCR Kits (QIAGEN) as previously described (64). The ChIP input sample was used to calculate ChIP binding signal (as input %). IgG ChIP sample was used as a negative control. Each experiment was repeated at least three times. Primers used for ChIP-qPCR are listed in table S3. To analyze the RNA Pol II pause, SNCA promoter primers were used to detect the Pol II binding on the promoter region, and the SNCA Exon 2 and SNCA Exon 4 were used to detect the Pol II binding on the SNCA gene body.
mRNA-seq and total RNA-seq
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) or RNeasy Lipid Tissue Mini Kit (QIAGEN) following the manufacturer’s protocol. RNA samples used for library preparation presented a high integrity number (RNA integrity number > 9). For mRNA-seq, a total of 100 ng of total RNA was subjected to polyadenylate enrichment, and cDNA libraries were constructed using the resulting mRNA and the NEBNext Ultra II Directional RNA library Preparation Kit for Illumina (NEB, #E7760L). Libraries were sequenced as paired-end 100-bp reads on a NovaSeq6000 (Illumina) with a depth of approximately 20 million reads each. Library preparation and sequencing procedures were performed by the same individual, aiming to minimize technical batch effects.
Reverse transcription PCR
The expression level of SNCA mRNA in SNCA EnKO rats was analyzed by RT-PCR and RT-qPCR as previously described (66). In brief, brain tissue total RNA was extracted using the RNeasy Lipid Tissue Mini Kit (QIAGENE) and reverse-transcribed into cDNA using the Omniscript RT Kit. Total human SNCA cDNA transcript was amplified using primers targeting exon 2 and the 3′ untranslated region (3′UTR) of the SNCA gene (forward primer: AAGCAGCAGGAAAGACAAAA; reverse primer: ACATCTGTCAGCAGATCTCA). RT-qPCR of the human SNCA mRNA was performed using QuantiTect PCR Kits (QIAGEN) with the primers targeting exon 5 and 3′UTR of the SNCA gene (forward primer: CTGTGGATCCTGACAATGAG; reverse primer: CAAGAAACTGGGAGCAAAGA). Rat housekeeping gene actin was used as an internal control (forward primer: AGAAGGAGATTACTGCCCTG; reverse primer: CCACCAATCCACACAGAGTA).
Circular chromosome conformation capture followed by sequencing
4C-seq was performed as previously described (27) with the following modification: (i) Nla III (New England Biolabs) and Bfa I (Thermo Fisher Scientific) were selected as the first and second restriction enzymes, respectively; (ii) before generating 4C-seq libraries for sequencing, the 4C template of viewer point (VP: SNCA promoter) was amplified by PCR of six cycles using specific primers (forward primer: ACGAATGGTCGTGGGCACCG; reverse primer: CCTCTCTTGGGCCCCTTCTG). Final 4C-seq libraries were generated by PCR of 24 cycles using indexed primers (P5 and P7; forward: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNACGAATGGTCGTGGGCACCG; reverse: CAAGCAGAAGACGGCATACGAGATGCCGTCCTCCTCCTCCTAG; NNNNNN: barcode sequences used for isolating reads of different samples). Next-generation sequencing was done on an Illumina NextSeq500 system using the 150-bp single-end high-output sequencing kit.
Data analysis
ChIP-seq data analysis
Briefly, ChIP-seq raw reads were aligned to the hg19 genome or rn6 genome assembly using Bowtie2. Only reads that uniquely mapped to the genome were used for further analysis. ChIP-seq peak calling was analyzed using MACS2. Bam files were converted into coverage bigWig file using deeptools (version 3.3.2) and samtools (version 1.9) package and viewed using Integrative Genomics Viewer (IGV) browsers (version 2.12.2).
RNA-seq data analysis
The quality of RNA-seq data in fastq files was assessed using QoRTs (v1.2.37) to identify sequencing cycles with low average quality, adapter contamination, or repetitive sequences from PCR amplification. Reads were aligned using HISAT2 (version 2.2.1), allowing gaped alignments to account for splicing against a custom-built genome composed of the Ensembl Homo Sapiens GRCh37, and alignment quality was analyzed using samtools (v1.1) and visually inspected in the IGV (version 2.12.2). Normalized read counts for all genes were obtained using featureCount (v3.18.1). Bam files were converted into coverage bigWig file using the deeptools (version 3.3.2) and samtools (version 1.9) package and viewed using IGV browsers (version 2.12.2).
4C-seq data analysis
Raw reads were separated into individual sample FastQ files by barcode sequences. Raw reads of each sample were filtered by retaining VP-specific PR amplicons (GGAGGGGGTGGTGCTGC), trimmed to remove nucleotide sequences belonging to the VP restriction fragment, and subsequently mapped to the hg19 genome assembly using Bowtie2. Only uniquely mapped reads were used for further analyses with Basic4C (27) and 4C-ker packages (67). Mapped raw reads were saved as bedgraph files and viewed using IGV browsers (version 2.12.2).
Statistical analyses
All the data presented in this study were analyzed using two-tailed unpaired t test when two groups were compared. Statistical analyses were performed using the Prism software 10.0 (GraphPad Software, La Jolla, CA). Significant differences were considered when *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Acknowledgments
We would like to thank the patients with DYT6 dystonia for donating skin tissue for this research. We also want to thank NICHD Brain and Tissue Bank for Developmental Disorders, Baltimore, MD, USA, and Harvard Brain Tissue Resource Center (HBTRC), McLean Hospital, Belmont, MA, USA, for providing DYT6 brain tissues for this study. We acknowledge computing support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC, and the German Research Foundation (DFG) through grant no. INST 37/935-1 FUGG.
Funding: The research was partially supported by the fortüne junior grant, University of Tuebingen (FC: 2407-0-0), the Deutsche Forschungsgemeinschaft (DFG; OR: RI 682/19-1 AOBJ663994; MS: SH 599/6-1), the William N. and Bernice E. Bumpus Foundation (Innovation Award to P.A.B.), the Luxembourg Fond National de Recherche (FNR) within the PEARL (to R.K.) (FNR/P13/6682797), the INTER programme (to R.K. and P.A.B.) (INTER/LEIR/18/12719318), the MotaSYN (12719684), and the Deutsche Forschungsgemeinschaft (OR: NGS Competence Center Tübingen INST 37/1049-1).
Author contributions: F.C. and O.R. contributed to the conception of the project and experiment design. F.C. and W.Z. performed experiments and data analysis. C.L. performed 4C-seq data analysis.P.A.B. and R.K. contributed to iPSC generation and differentiation. L.Y.-T. performed the immunohistochemical staining. N.C. and J.A. conducted the next-generation sequencing and raw data processing. J.H.-S. performed the protein analysis and interpretation. H.H. contributed to the patient brain samples and demographic data collection. M.S. conducted the data extraction of PD-GWAS data. K.B., K.J., and M.U. performed MS method and data interpretation. T.O. and K.G.-H. conducted the animal model generation. F.C. wrote the manuscript.
Competing interests: O.R. and F.C. are inventors on a patent application related to this work filed by the University of Tuebingen (no. EP22196721, filed 20 September 2022). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The following datasets from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) were reused in this study (GSE136656, GSE158382, GSE126358, GSE140016, GSE95344, GSE122429, and GSE62193) to analyze the SNCA expression in iPSCs and iPSC-derived NPC or neurons. The RNA-seq, ChIP-seq, and 4C-seq datasets generated in this study are available through the GEO via accession GSE141278 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141278) and GSE184961 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184961). The animal models used in this study can be provided by the Institute of Medical Genetics and Applied Genomics, University of Tuebingen pending scientific review and a completed material transfer agreement. Requests for the animals should be submitted to the corresponding author.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.Spillantini M., Schmidt M., Lee V. Y., Trojanowski J. Q., Jakes R., Goedert M., α-Synuclein in lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., Trojanowski J. Q., Iwatsubo T., Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879–884 (1998). [PMC free article] [PubMed] [Google Scholar]
- 3.Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L., Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O., Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 18, 106–108 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K., α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Chartier-Harlin M. C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., Waucquier N., Defebvre L., Amouyel P., Farrer M., Destée A., Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364, 1167–1169 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ibáñez P., Lesage S., Janin S., Lohmann E., Durif F., Destée A., Bonnet A. M., Brefel-Courbon C., Heath S., Zelenika D., Agid Y., Dürr A., Brice A.; French Parkinson’s Disease Genetics Study Group , α-Synuclein gene rearrangements in dominantly inherited parkinsonism: Frequency, phenotype, and mechanisms. Arch. Neurol. 66, 102–108 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Chiba-Falek O., Nussbaum R. L., Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum. Mol. Genet. 10, 3101–3109 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Soldner F., Stelzer Y., Shivalila C. S., Abraham B. J., Latourelle J. C., Barrasa M. I., Goldmann J., Myers R. H., Young R. A., Jaenisch R., Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masliah E., Rockenstein E., Veinbergs I., Mallory M., Hashimoto M., Takeda A., Sagara Y., Sisk A., Mucke L., Dopaminergic loss and inclusion body formation in alpha-synuclein mice: Implications for neurodegenerative disorders. Science 287, 1265–1269 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Nuber S., Harmuth F., Kohl Z., Adame A., Trejo M., Schonig K., Zimmermann F., Bauer C., Casadei N., Giel C., Calaminus C., Pichler B. J., Jensen P. H., Muller C. P., Amato D., Kornhuber J., Teismann P., Yamakado H., Takahashi R., Winkler J., Masliah E., Riess O., A progressive dopaminergic phenotype associated with neurotoxic conversion of α-synuclein in BAC-transgenic rats. Brain 136, 412–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gründemann J., Schlaudraff F., Haeckel O., Liss B., Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson’s disease. Nucleic Acids Res. 36, e38 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi M., Suzuki M., Fukuoka M., Fujikake N., Watanabe S., Murata M., Wada K., Nagai Y., Hohjoh H., Normalization of overexpressed α-synuclein causing Parkinson’s disease by a moderate gene silencing with RNA interference. Mol. Ther. Nucleic Acids 4, e241 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Kantor B., Tagliafierro L., Gu J., Zamora M. E., Ilich E., Grenier C., Huang Z. Y., Murphy S., Chiba-Falek O., Downregulation of SNCA expression by targeted editing of DNA methylation: A potential strategy for precision therapy in PD. Mol. Ther. 26, 2638–2649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clough R. L., Dermentzaki G., Stefanis L., Functional dissection of the alpha-synuclein promoter: Transcriptional regulation by ZSCAN21 and ZNF219. J. Neurochem. 110, 1479–1490 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Scherzer C. R., Grass J. A., Liao Z., Pepivani I., Zheng B., Eklund A. C., Ney P. A., Ng J., McGoldrick M., Mollenhauer B., Bresnick E. H., Schlossmacher M. G., GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proc. Natl. Acad. Sci. U.S.A. 105, 10907–10912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valente T., Dentesano G., Ezquerra M., Fernandez-Santiago R., Martinez-Martin J., Gallastegui E., Domuro C., Compta Y., Martí M. J., Bachs O., Márquez-Kisinousky L., Straccia M., Solà C., Saura J., CCAAT/enhancer binding protein δ is a transcriptional repressor of α-synuclein. Cell Death Differ. 27, 509–524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiba-Falek O., Kowalak J. A., Smulson M. E., Nussbaum R. L., Regulation of alpha-synuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am. J. Hum. Genet. 76, 478–492 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClymont S. A., Hook P. W., Soto A. I., Reed X., Law W. D., Kerans S. J., Waite E. L., Briceno N. J., Thole J. F., Heckman M. G., Diehl N. N., Wszolek Z. K., Moore C. D., Zhu H., Akiyama J. A., Dickel D. E., Visel A., Pennacchio L. A., Ross O. A., Beer M. A., McCallion A. S., Parkinson-associated SNCA enhancer variants revealed by open chromatin in mouse dopamine neurons. Am. J. Hum. Genet. 103, 874–892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng F., Zheng W., Barbuti P. A., Bonsi P., Liu C., Casadei N., Ponterio G., Meringolo G., Admard J., Dording C. M., Yu-Taeger L., Nguyen H. P., Grundmann-Hauser K., Ott T., Houlden H., Pisani A., Krueger R., Riess O., DYT6 mutated THAP1 is a cell type dependent regulator of the SP1 family. Brain 2022, awac001 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Cheng F., Walter M., Wassouf Z., Hentrich T., Casadei N., Schulze-Hentrich J., Barbuti P., Krueger R., Riess O., Grundmann-Hauser K., Ott T., Unraveling molecular mechanisms of THAP1 missense mutations in DYT6 dystonia. J. Mol. Neurosci. 70, 999–1008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz-Virumbrales M., Ruiz M., Hone E., Dolios G., Wang R., Morant A., Kottwitz J., Ozelius L. J., Gandy S., Ehrlich M. E., Dystonia type 6 gene product Thap1: Identification of a 50 kDa DNA-binding species in neuronal nuclear fractions. Acta Neuropathol. Commun. 2, 139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touchman J. W., Dehejia A., Chiba-Falek O., Cabin D. E., Schwartz J. R., Orrison B. M., Polymeropoulos M. H., Nussbaum R. L., Human and mouse alpha-synuclein genes: Comparative genomic sequence analysis and identification of a novel gene regulatory element. Genome Res. 11, 78–86 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlić R., Chung H. R., Lasserre J., Vlahovicek K., Vingron M., Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U.S.A. 107, 2926–2931 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., Jaenisch R., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren G., Jin W., Cui K., Rodrigez J., Hu G., Zhang Z., Larson D. R., Zhao K., CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell 67, 1049–1058.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Werken H. J., de Vree P. J., Splinter E., Holwerda S. J., Klous P., de Wit E., de Laat W., 4C technology: Protocols and data analysis. Methods Enzymol. 513, 89–112 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Pott S., Lieb J. D., What are super-enhancers? Nat. Genet. 47, 8–12 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A., Young R. A., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold P. R., Wells A. D., Li X. C., Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front. Cell Dev. Biol. 7, 377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermunt M. W., Reinink P., Korving J., de Bruijn E., Creygthon P. M., Basak O., Geeven G., Toonen P. W., Lansu N., Meunier C., van Heesch S., Clevers H., de Laat W., Cuppen E., Creyghton M. P., Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 9, 767–779 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Nalls M. A., Pankratz N., Lill C. M., Do C. B., Hernandez D. G., Saad M., DeStefano A. L., Kara E., Bras J., Sharma M., Schulte C., Keller M. F., Arepalli S., Letson C., Edsall C., Stefansson H., Liu X., Pliner H., Lee J. H., Cheng R.; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s research: The organized GENetics initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group, Ikram M. A., Ioannidis J. P. A., Hadjigeorgiou G. M., Bis J. C., Martinez M., Perlmutter J. S., Goate A., Marder K., Fiske B., Sutherland M., Xiromerisiou G., Myers R. H., Clark L. N., Stefansson K., Hardy J. A., Heutink P., Chen H., Wood N. W., Houlden H., Payami H., Brice A., Scott W. K., Gasser T., Bertram L., Eriksson N., Foroud T., Singleton A. B., Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H., NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W. H., Castillo P. E., Shinsky N., Verdugo J. M., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A., Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25, 239–252 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Al-Wandi A., Ninkina N., Millership S., Williamson S. J., Jones P. A., Buchman V. L., Absence of α-synuclein affects dopamine metabolism and synaptic markers in the striatum of aging mice. Neurobiol. Aging 31, 796–804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aneichyk T., Hendriks W. T., Yadav R., Shin D., Gao D., Vaine C. A., Collins R. L., Domingo A., Currall B., Stortchevoi A., Multhaupt-Buell T., Penney E. B., Cruz L., Dhakal J., Brand H., Hanscom C., Antolik C., Dy M., Ragavendran A., Underwood J., Cantsilieris S., Munson K. M., Eichler E. E., Acuña P., Go C., Jamora R. D. G., Rosales R. L., Church D. M., Williams S. R., Garcia S., Klein C., Müller U., Wilhelmsen K. C., Timmers H. T. M., Sapir Y., Wainger B. J., Henderson D., Ito N., Weisenfeld N., Jaffe D., Sharma N., Breakefield X. O., Ozelius L. J., Bragg D. C., Talkowski M. E., Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 172, 897–909.e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beauvais G., Rodriguez-Losada N., Ying L., Zakirova Z., Watson J. L., Readhead B., Gadue P., French D. L., Ehrlich M. E., Gonzalez-Alegre P., Exploring the interaction between eIF2α dysregulation, acute endoplasmic reticulum stress and DYT1 dystonia in the mammalian brain. Neuroscience 371, 455–468 (2018). [DOI] [PubMed] [Google Scholar]
- 38.McVicker G., van de Geijn B., Degner J. F., Cain C. E., Banovich N. E., Raj A., Lewellen N., Myrthil M., Gilad Y., Pritchard J. K., Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corradin O., Cohen A. J., Luppino J. M., Bayles I. M., Schumacher F. R., Scacheri P. C., Modeling disease risk through analysis of physical interactions between genetic variants within chromatin regulatory circuitry. Nat. Genet. 48, 1313–1320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pihlstrøm L., Blauwendraat C., Cappelletti C., Berge-Seidl V., Langmyhr M., Henriksen S. P., van de Berg W. D. J., Gibbs J. R., Cookson M. R.; International Parkinson Disease Genomics Consortium; North American Brain Expression Consortium, Singleton A. B., Nalls M. A., Toft M., A comprehensive analysis of SNCA-related genetic risk in sporadic parkinson disease. Ann. Neurol. 84, 117–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchs J., Tichopad A., Golub Y., Munz M., Schweitzer K. J., Wolf B., Berg D., Mueller J. C., Gasser T., Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 22, 1327–1334 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Cardo L. F., Coto E., de Mena L., Ribacoba R., Mata I. F., Menéndez M., Moris G., Alvarez V., Alpha-synuclein transcript isoforms in three different brain regions from Parkinson’s disease and healthy subjects in relation to the SNCA rs356165/rs11931074 polymorphisms. Neurosci. Lett. 562, 45–49 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Cronin K. D., Ge D., Manninger P., Linnertz C., Rossoshek A., Orrison B. M., Bernard D. J., El-Agnaf O. M. A., Schlossmacher M. G., Nussbaum R. L., Chiba-Falek O., Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum. Mol. Genet. 18, 3274–3285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glenn O., Tagliafierro L., Beach T. G., Woltjer R. L., Chiba-Falek O., Interpreting gene expression effects of disease-associated variants: A lesson from SNCA rs356168. Front. Genet. 8, 133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiba-Falek O., Lopez G. J., Nussbaum R. L., Levels of alpha-synuclein mRNA in sporadic Parkinson disease patients. Mov. Disord. 21, 1703–1708 (2006). [DOI] [PubMed] [Google Scholar]
- 46.McLean J. R., Hallett P. J., Cooper O., Stanley M., Isacson O., Transcript expression levels of full-length alpha-synuclein and its three alternatively spliced variants in Parkinson’s disease brain regions and in a transgenic mouse model of alpha-synuclein overexpression. Mol. Cell Neurosci. 49, 230–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mollenhauer B., Locascio J. J., Schulz-Schaeffer W., Sixel-Döring F., Trenkwalder C., Schlossmacher M. G., α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: A cohort study. Lancet Neurol. 10, 230–240 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Mollenhauer B., Trautmann E., Taylor P., Manninger P., Sixel-Döring F., Ebentheuer J., Trenkwalder C., Schlossmacher M. G., Total CSF α-synuclein is lower in de novo Parkinson patients than in healthy subjects. Neurosci. Lett. 532, 44–48 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Kang J., Irwin D. J., Chen-Plotkin A. S., Siderowf A., Caspell C., Coffey C. S., Waligórska T., Taylor P., Pan S., Frasier M., Marek K., Kieburtz K., Jennings D., Simuni T., Tanner C. M., Singleton A., Toga A. W., Chowdhury S., Mollenhauer B., Trojanowski J. Q., Shaw L. M.; Parkinson’s progression markers initiative , Association of cerebrospinal fluid β-amyloid 1–42, T-tau, P-tau181, and α-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA Neurol. 70, 1277–1287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larke M. S. C., Schwessinger R., Nojima T., Telenius J., Beagrie R. A., Downes D. J., Oudelaar A. M., Truch J., Graham B., Bender M. A., Proudfoot N. J., Higgs D. R., Hughes J. R., Enhancers predominantly regulate gene expression during differentiation via transcription initiation. Mol. Cell 81, 983–997.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen F. X., Xie P., Collings C. K., Cao K., Aoi Y., Marshall S. A., Rendleman E. J., Ugarenko M., Ozark R. A., Zhang A., Shiekhattar R., Smith E. R., Zhang M. Q., Shilatifard A., PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357, 1294–1298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaukowitch K., Joo J. Y., Liu X., Watts J. K., Martinez C., Kim T. K., Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamori M., Junn E., Mochizuki H., Mouradian M. M., Nucleic acid-based therapeutics for Parkinson’s disease. Neurotherapeutics 16, 287–298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uehara T., Choong C. J., Nakamori M., Hayakawa H., Nishiyama K., Kasahara Y., Baba K., Nagata T., Yokota T., Tsuda H., Obika S., Mochizuki H., Amido-bridged nucleic acid (AmNA)-modified antisense oligonucleotides targeting α-synuclein as a novel therapy for Parkinson’s disease. Sci. Rep. 9, 7567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khodr C. E., Sapru M. K., Pedapati J., Han Y., West N. C., Kells A. P., Bankiewicz K. S., Bohn M. C., An α-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson’s disease, but displays toxicity in dopamine neurons. Brain Res. 1395, 94–107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zharikov A. D., Cannon J. R., Tapias V., Bai Q., Horowitz M. P., Shah V., El Ayadi A., Hastings T. G., Greenamyre J. T., Burton E. A., shRNA targeting α-synuclein prevents neurodegeneration in a Parkinson’s disease model. J. Clin. Invest. 125, 2721–2735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Javed H., Menon S. A., Al-Mansoori K. M., Al-Wandi A., Majbour N. K., Ardah M. T., Varghese S., Vaikath N. N., Haque M. E., Azzouz M., El-Agnaf O. M., Development of nonviral vectors targeting the brain as a therapeutic approach for Parkinson’s disease and other brain disorders. Mol. Ther. 24, 746–758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Mittal S., Bjørnevik K., Im D. S., Flierl A., Dong X., Locascio J. J., Abo K. M., Long E., Jin M., Xu B., Xiang Y. K., Rochet J. C., Engeland A., Rizzu P., Heutink P., Bartels T., Selkoe D. J., Caldarone B. J., Glicksman M. A., Khurana V., Schüle B., Park D. S., Riise T., Scherzer C. R., β2-Adrenoreceptor is a regulator of the α-synuclein gene driving risk of Parkinson’s disease. Science 357, 891–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guhathakurta S., Kim J., Adams L., Basu S., Song M. K., Adler E., Je G., Fiadeiro M. B., Kim Y. S., Targeted attenuation of elevated histone marks at SNCA alleviates α-synuclein in Parkinson’s disease. EMBO Mol. Med. 13, e12188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schöndorf D. C., Aureli M., McAllister F. E., Hindley C. J., Mayer F., Schmid B., Sardi S. P., Valsecchi M., Hoffmann S., Schwarz L. K., Hedrich U., Berg D., Shihabuddin L. S., Hu J., Pruszak J., Gygi S. P., Sonnino S., Gasser T., Deleidi M., iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat. Commun. 5, 4028 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Reinhardt P., Schmid B., Burbulla L. F., Schöndorf D. C., Wagner L., Glatza M., Höing S., Hargus G., Heck S. A., Dhingra A., Wu G., Müller S., Brockmann K., Kluba T., Maisel M., Krüger R., Berg D., Tsytsyura Y., Thiel C. S., Psathaki O. E., Klingauf J., Kuhlmann T., Klewin M., Müller H., Gasser T., Schöler H. R., Sterneckert J., Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12, 354–367 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grundmann K., Glöckle N., Martella G., Sciamanna G., Hauser T. K., Yu L., Castaneda S., Pichler B., Fehrenbacher B., Schaller M., Nuscher B., Haass C., Hettich J., Yue Z., Nguyen H. P., Pisani A., Riess O., Ott T., Generation of a novel rodent model for DYT1 dystonia. Neurobiol. Dis. 47, 61–74 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Cheng F. B., Feng J. C., Ma L. Y., Miao J., Ott T., Wan X. H., Grundmann K., Combined occurrence of a novel TOR1A and a THAP1 mutation in primary dystonia. Mov. Disord. 29, 1079–1083 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Gloeckner C. J., Boldt K., Schumacher A., Ueffing M., Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Methods Mol. Biol. 564, 359–372 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Cheng F. B., Ozelius L. J., Wan X. H., Feng J. C., Ma L. Y., Yang Y. M., Wang L., THAP1/DYT6 sequence variants in non-DYT1 early-onset primary dystonia in China and their effects on RNA expression. J. Neurol. 259, 342–347 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Raviram R., Rocha P. P., Müller C. L., Miraldi E. R., Badri S., Fu Y., Swanzey E., Proudhon C., Snetkova V., Bonneau R., Skok J. A., 4C-ker: A method to reproducibly identify genome-wide interactions captured by 4C-seq experiments. PLoS Comput. Biol. 12, e1004780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3