Abstract
One of the main obstacles in prevention and treatment of COVID-19 is the rapid evolution of the SARS-CoV-2 Spike protein. Given that Spike is the main target of common treatments of COVID-19, mutations occurring at this virulent factor can affect the effectiveness of treatments. The B.1.617.2 lineage of SARS-CoV-2, being characterized by many Spike mutations inside and outside of its receptor-binding domain (RBD), shows high infectivity and relative resistance to existing cures. Here, utilizing a wide range of computational biology approaches, such as immunoinformatics, molecular dynamics (MD), analysis of intrinsically disordered regions (IDRs), protein-protein interaction analyses, residue scanning, and free energy calculations, we examine the structural and biological attributes of the B.1.617.2 Spike protein. Furthermore, the antibody design protocol of Rosetta was implemented for evaluation the stability and affinity improvement of the Bamlanivimab (LY-CoV55) antibody, which is not capable of interactions with the B.1.617.2 Spike. We observed that the detected mutations in the Spike of the B1.617.2 variant of concern can cause extensive structural changes compatible with the described variation in immunogenicity, secondary and tertiary structure, oligomerization potency, Furin cleavability, and drug targetability. Compared to the Spike of Wuhan lineage, the B.1.617.2 Spike is more stable and binds to the Angiotensin-converting enzyme 2 (ACE2) with higher affinity.
Keywords: SARS-CoV-2, COVID-19, Spike, Mutation, B.1.617.2, Computational biology
Graphical abstract
1. Introduction
The SARS-CoV-2 caused an outbreak of severe acute respiratory syndrome in Wuhan, Hubei Province, China, in December 2019. The virus quickly spread around the world, resulting in an unexpected pandemic and leading governments to establish a global quarantine [1]. As of 12 November 2022, >641,715,848 SARS-CoV-2 infections and associated 6,620,827 deaths were recorded [2].
The members of the coronavirus family (CoVs) have relatively large single-stranded positive-sense RNA (+ssRNA) genomes (27–32 kb), which are longer than genomes of any other RNA viruses. SARS-CoV-2, a recently emerged coronavirus family member, has a genome size of about 29.9 kb [3]. The SARS-CoV-2 particles are enveloped and pleomorphic, and this pleomorphism causes SARS-CoV-2 to range in size from 60 to 140 nm [4]. This virus has four structural proteins and sixteen nonstructural proteins (NSPs). Structural proteins include nucleocapsid protein (N), membrane protein (M), Spike protein (S), and envelope protein (E). Outside the genome, the nucleocapsid protein forms the capsid, and the genome is further packed by an envelope containing three structural proteins (M, S, and E) [5]. Host infection, membrane fusion, viral assembly, morphogenesis and virus particle release are controlled by structural proteins [6]. Nonstructural proteins promote viral replication and transcription besides other activities [7], [8].
CoVs express a 3′-to-5′ exoribonuclease (NSP14-ExoN) that possesses a proofreading function and reduces their error rate during RNA copying by 100–1000 times compared to other RNA viruses [9]. However, errors still occur faster in the CoV genome than in eukaryotic cells, allowing for the accumulation of mutations in the viral genome when combined with high replication rates [10]. SARS-CoV-2 genome studies have shown mutations that may modify the virulence and transmissibility of viruses [11], [12]. Genome mutations occur due to errors during copying RNA to a new cell, and it can change over time, resulting in many SARS-CoV-2 variants circulating around the world [13]. However, many mutations do not affect the virus's capacity to transmit or cause disease because they do not affect the secondary and tertiary structure of a protein and do not change viral protein functionality. In contrast, mutations that modify the structure of functional proteins are more likely to change the virus pathobiology [14], [15]. The B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), P.1 (Gamma), and B.1.1.529 (Omicron) variants of SARS-CoV-2 that are currently circulating in the world are classified as variants of concern [16].
Lineage B.1.1.7 (also known as the UK variant of concern) was discovered in the UK in September 2020. This lineage carries a wide range of mutations, some of which, such as N501Y and P681H, have been shown to greatly affect the function of the Spike protein [17], [18]. These mutations can affect Spike properties and its IDRs pattern. IDRs are polypeptide elements that are lacking acceptable hydrophobic residues to intercede in cooperative folding. In return, these protein fragments commonly possess a higher ratio of polar or charged residues [19]. The N501Y substitution on the Spike protein has been demonstrated to improve ACE2 binding and cell infectivity in animal models [20], whereas the P681H substitution on the Spike protein affects the Furin-cleavage site [21]. Lineage B.1.351 (South Africa lineage) appeared in December 2020 in South Africa (SA) [22]. In this variant, the most significant amino acid substitutions are N501Y, K417N, and E484K, located in the receptor-binding motif (RBM) of the RBD region. They may induce conformational changes in the Spike protein [23]. Lineage P.1 (also known as B.1.1.28 or Brazilian variant) was discovered in December 2020, with the N501Y, E484K, and K417T (known to be of biological importance in the S protein) substitutions as the most important amino acid mutations [24], [25].
The South African National Institute for Communicable Diseases (NICD) verified the finding of a novel SARS-CoV-2 strain in November 2021, named B.1.1.529. The Technical Advisory Group (TAG) on SARS-CoV-2 Virus Evolution (TAG-VE) classified B.1.1.529 as variants of concern (VoC), naming it Omicron (https://www.ecdc.europa.eu/en/covid-19/variants-concern), on November 26, 2021 [26]. There are >50 mutations in the Omicron variant, of which approximately 30 mutations are found in the Spike protein. What's more concerning is the 15 mutated sites in the RBD that interact with human cells prior to entry, potentially increasing transmissibility (how easy they spread) [18], [27], [28].
B.1.617 has three sublineages, including B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.617.3. The variant that has caused the most concern is B.1.617.2, initially discovered in India in December 2020 [29]. B.1.617.1 is sometimes known as the “Indian double mutant,” although this name is misleading, because compared to previous versions it contains around 15 mutations. The term “double mutant” refers to two mutations (L452R and E484Q) in the Spike protein of this variant that are of particular significance in virus infectivity. These two mutations may reduce the effectiveness of antibodies to previous SARS-CoV-2 variants or existing vaccinations [30], [31].
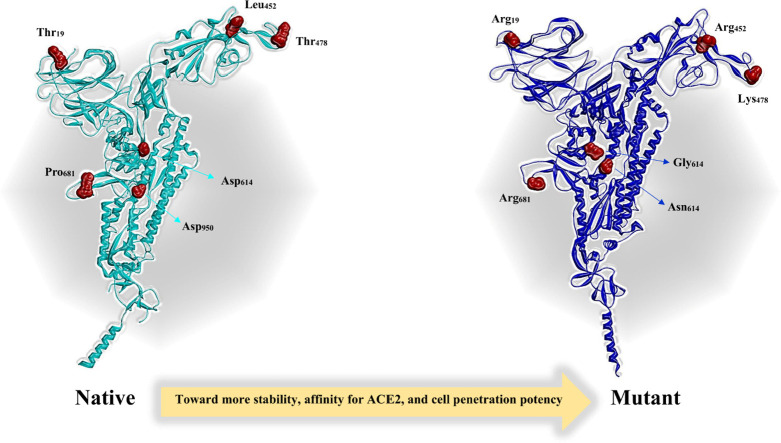
SARS-CoV-2 variant B.1.617.2 first appeared in India and is now widely spread globally. Infection with this variant in the UK and around the globe also confirms its higher transmission potential than other variants [32]. T19R, 157-158Del, L452R, T478K, D614G, P681R, and D950N are the Spike protein mutations observed in the Delta variant [33]. The T478K mutation in the RBD is specific to Delta and is located inside the epitope region of powerful neutralizing mAbs classified as Class 1. This mutation is similar to the E484K mutation, which promotes escape from neutralizing antibodies [34], [35], [36]. The L452R and P681R mutations are found in all three B.1.617 sublineages (B.1.617.1, B.1.617.2, and B.1.617.3). The P681R mutation in the S1–S2 cleavage site of the Delta variant causes enhanced transmissibility. However, Delta lacks mutations at amino acid positions 501 and 484 of its ACE2 receptor-binding region, which are associated with VoC (https://www.ecdc.europa.eu/en/covid-19/variants-concern) or resistance to neutralizing antibodies [37], [38]. More mutations within the Delta sublineage created a variety of variants, known as ‘Delta AY.1 (Also known as B.1.617.2.1 or Delta plus) and Delta AY.2,’ that have been discovered in India and other countries and that pose a danger to existing vaccines and treatments [39]. The Spike protein in AY.1 and AY.2 variants contains another mutation, K417N, which might lead to immune escape [40], [41], [42], [43], [44].
At the early stages of the COVID-19 outbreak, efforts were mainly focused on protecting health services from collapse by adopting non-pharmacological interventions, such as social distance, using protective masks, avoiding touching and handling objects, and sterilizing materials to reduce the transmission of SARS-CoV-2 [45]. In addition to preventive efforts to limit virus spread, efforts are currently being made to anticipate subsequent mutations and design a vaccine to counteract these mutations. Therefore, it is vital to establish a reliable paradigm for predicting and mitigating the impact of SARS-CoV-2 mutations on vaccinations and antibody treatments. We still do not know how mutations and following structural changes result in reduced vaccine effectiveness against SARS-CoV-2 variant B.1.617.2. Due to their great mutability compared to other lineages, B.1.617.2 and B.1.1.529 variants pose an additional hazard to human health. Hence, the study of structural changes in these strains and consequently virus neutralization is required. In the present study, our group by employing computational biology capabilities tracked the structural and pathobiology function changes of the B.1.617.2 Spike protein.
2. Materials and methods
2.1. Protein modeling and immunogenicity evaluation
Recent advances in immunoinformatics algorithms have made it possible to predict the immunogenicity of biological molecules with greater accuracy [46], [47]. As mentioned, the Delta variant of SARS-CoV-2 contains as many mutations. Since these mutations and evolutionary behavior can affect immune responses, virulence (the harm caused by pathogen infection) [48], and viral loads, we examined the immunogenicity potency of native and mutated Spike protein. For this purpose, the sequence of native Spike (accession no. P0DTC2) was retrieved in FASTA format from UniProt [3]. In addition, the B.1.617.2 Spike sequence carrying all desired mutations was constructed using SnapGene viewer (v5.2.4) [51]. Ultimately, sequences of native and mutant Spikes were conducted to linear epitope examination workflow.
To identify structural epitopes, the 3D structure of the native Spike (PDB ID: 6VYB) was downloaded from the RCSB database [52], while for the analysis of the B.1.617.2 Spike protein, the structure containing the desired mutations (PDB ID: 7V7N) was employed. All missing residues in the native and mutant structures were modeled using Modeller software (v10.0) [53]. The modeling process was implemented by the Pymod (v3) plugin of the PyMOL (v2.5) [54]. Since the Spike is a large homotrimer structure, we focused on one of its monomers to facilitate the study process. During the modeling, the top 10 structures that were most similar to the target structures were used as templates, and the structures that have the lowest DOPE score were chosen as the best models [55], [56], [57]. For resolving structure clashes and quality assessment of modeled structures, Chiron and Gaia servers were utilized, respectively. Chiron uses discrete molecular dynamics to minimize the energy of protein molecules. Also, Gaia assesses the folding and covalent geometry of an input structure by comparison of the input structure to high-resolution crystal structures. For epitope prediction, Immune Epitope Database (IEDB) server was utilized [9]. MHC-I and MHC-II epitopes were investigated by using T-cell epitope mapping tools of IEDB [58]. Moreover, continuous (consisting of consecutive residues) B-cell epitopes were examined by applying the antigen sequence properties (ASP) tool (Bepipred Linear Epitope Prediction method). Besides, for discontinuous epitopes (short fragments that are spread in the protein structure), ElliPro and DiscoTope were employed [59], [60], [61], [62].
2.2. MD simulation and IDRs prediction
Molecular dynamics is an all-embracing approach for studying molecular structure, behavior, and function at the atomic scale [63], [64], [65]. In our investigation, GROMACS (v2021.2) which is a versatile package for studying the dynamics of biomolecules was utilized for tracking Spike characteristics in native and mutant forms [66]. We performed molecular dynamics for two purposes and in two steps. At the first step of MD, we investigated the effects of observed mutations in the B.1.617.2 lineage on the structural properties of the Spike (monomeric form). Besides, at the second step of MD, the interaction ability of the native and mutant Spikes (RBD domains) with ACE2 was examined [56]. All structures were parametrized using CHARMM36 all-atom force field [67]. The structures (PDB ID: 6YVB and 7V7N) retrieved from the RCSB “with their missing residues modeled using Modeller (v10.0)” were conducted to the first step of MD. For the second step of MD, the complexes of Spike RBD domain with ACE2 that received the highest score during the docking procedure were utilized.
For solvation, the TIP3P water model was used, and Na+ and Cl− ions at 0.15 M concentrations were added to neutralize the systems. The number of atoms (protein, ions, and water) in the first step of MD for native and mutant simulated systems were 284,396 and 307,778, respectively. As well, for complexes of native and mutant Spikes RBD domains with ACE2 which were simulated in the second step of the MD, the number of atoms for native and mutant structures were 108,541 and 108,567, respectively. Before MD production to equilibration of systems, a canonical ensemble step (NVT) for temperature coupling at 310 K by V-rescale method for 200 ps was carried out. Furthermore, an isothermal–isobaric ensemble (NPT) by Parrinello-Rahman algorithm for 1 ns was accomplished to pressure coupling at 1 bar [68], [69]. In MD production, equilibrated systems were simulated for 200 ns and 30 ns at the first and second steps of MD, respectively. MD trajectory files were examined for analysis of structural attributes of B.1.617.2 Spike in terms of root-mean-square deviation (RMSD) [70], Ramachandran plot [71], root-mean-square fluctuation (RMSF) [72], the radius of gyration (Rg) [73], solvent accessible surface area (SASA) [74], free energy landscape (FEL) [75], principal component analysis (PCA) [76], secondary structure [77], probability density function [78], hydrogen bonds [79], minimum distance and number of contacts [80]. The md.gro file which was generated in the last step of the simulation and structures extracted from the MD trajectories were conducted to the other analysis steps. For prediction of IDRs which have significant effects on protein folding and function, the ODiNPred server (https://st-protein.chem.au.dk/odinpred) was utilized [81].
2.3. Protein-protein interaction analysis
One of the most widely used methods for studying protein-protein interactions is molecular docking. This approach has many aficionados due to its high speed and cost-effectiveness [82]. Due to the role of molecular docking in biological and pharmacological studies, so far various algorithms have been developed to investigate the pattern of protein interaction with other molecules, such as DNA, RNA, lipids, enzymes and small molecules [83]. Rosetta is one of the most well-known developed tools for molecular docking [84]. This tool is an open-source and accurate docking package that usually gets a competent score in the CAPRI ranking [85]. In the present study, Rosetta (v3.12) was utilized to examine the pattern and affinity of B.1.617.2 Spike protein in complex with ACE2. Docking was done using the docking_protocol.linuxgccrelease module of Rosetta in a site-specific manner for Spike (RBD domain)-ACE2 complexes. During docking number of outputs (−nstruct), rotation and translation are set at 2000, 8°, and 3 Å, respectively. For increased accuracy, the crystal structure of Spike-ACE2 (PDB ID: 7DF4) was used as the native structure in docking script.
2.4. Topology and residues contact maps analysis
Structural and topological features such as tunnels and cavities around the amino acids designate an essential function in proteins behaviors [86]. Any alteration in these structural attributes can influence the biochemical properties of proteins such as protein folding and enzyme degradation [87]. Residue displacement is one of the main factors in changing the topological properties of proteins [88]. To analyse the mutated positions, contact and distance map was generated for all structures using Pymod (v3) [54]. Furthermore, tunnel and cavities around mutated residues in B.1.617.2 lineage were examined by CAVER (v3.0) [89]. Throughout this step, the mutation position was selected as the starting point.
2.5. Evaluation of cleavability and oligomerization potency
Since the P681R mutation is adjacent to the cleavage site I (CS1) [15], its effect on Spike cleavage may play a substantial role in its infectivity and thus was examined. For this reason, topological attributes of the region around P681R position, and interaction pattern of Furin complex with native and mutant Spikes were studied. Spike in monomeric form was used to investigate its cleavage by Furin. Among the Delta lineage mutations, D950N due to its proximity to the oligomerization site of Spike, likely affects the ability of this protein to form its homotrimer state. For this purpose, using the SymDock protocol [90] of the Rosetta package, the effect of the D950N displacement in Spike oligomerization was tracked. The symmetric profile for the Spike in the homotrimeric state was provided using the make_symmdef.py script.
2.6. Free energy and residue scanning
Free energy is an important function in studying the stability of a structure and finding the most stable complex of biomolecules [91]. We implemented free energy in two phases, the purpose of the first phase was to evaluate the stability of the B.1.617.2 Spike compared to the native structure as a control in monomeric configuration. In the second phase of the free energy study, the affinity and stability of the ACE2 complex with the RBD domain of native and mutant Spikes were investigated. To perform the first and second phases of free energy calculations, the results obtained in the first and second steps of the MD (Section 2.2) were employed. We executed the G_mmpbsa module of GROMACS to achieve the objectives of the first and second steps of free energy investigation [92]. For G_mmpbsa calculations (in the second phase), the ACE2 complexes with native and mutant Spikes (RBD domains) were simulated for 30 ns using GROMACS (v2021.2).
G_mmpbsa using the MMBSA algorithm [93] analyzes free energy at two levels; molecular mechanics potential energy and free energy of solvation. In this tool, free energy of solvation is decomposed into polar and nonpolar solvation energy. To calculate the free energy of Spike in the single form at the first phase of calculating -diff set at no, but at the second phase of free energy investigation to calculate the binding free energy, ACE2 and Spike were indexed. The Score module of Rosetta [94] simultaneously was used to study the energy of the native and mutant Spikes in the single form (not in complex with ACE2). Additionally, the residue scanning module of Schrodinger (v20.21.2) was implemented for residue scanning of mutated positions [95].
2.7. Antibody stability-affinity improvement
Sequence and structural changes have a significant effect on the properties of proteins that play an essential role in the biological pathways of the cell. Aside from the crucial role that proteins play in the cell, these biomolecules form a major part of biotechnology products to detect and treatment of disease [96], [97]. Despite the development of the SARS-CoV-2 vaccines to prevent its spread, COVID-19 treatment remains a significant concern. In various diseases, monoclonal antibodies (mAbs) are recognized as versatile diagnostic and therapeutic tools due to their high specificity, great sensitivity, and high stability [98]. The high immunogenicity potency of neutralizing antibodies and their ability to scale up production make them a promising treatment for COVID-19 [99]. The focus of antibodies research is to identify monoclonal antibodies capable of targeting Spike proteins, especiall the RBD of the Spike protein, or specific antibodies able to bind ACE2 to prevent virus entry [100], [101].
The Food and Drug Administration (FDA) has given emergency use authorizations (EUAs) to 3 categories of mAbs for the treatment of COVID-19, including Bamlanivimab plus Etesevimab, Casirivimab plus Imdevimab, and Sotrovimab [102]. On the other hand, investigation shows that Delta variants tend to be more resistant to neutralization by Bamlanivimab than the Alpha variant. Therefore, we used a robust computational workflow to implementation of stability and affinity improvement for Bamlanivimab to improve its efficiency against B.1.617.2 lineage. For this purpose, we used a general design framework available in Rosetta. Bamlanivimab-Spike complex (PDB ID: 7KMG) was conducted to Rosetta antibody design protocol for affinity and stability improvement. Desired mutations in Spike B.1.617.2 lineage were implemented using mutagenesis plugin of PyMOL (v2.5). Backbone and sidechains of input structures before stability and affinity improvement were optimized against structural clashes using Relax and Repack [103], [104] module of Rosetta, respectively. In order to implement the design process, at first, a resfile containing hot spots (residues in the Spike-ACE2 interface with a distance of fewer than 7 Å) was created. Then using rosetta_scripts.default.linuxgccrelease module of Rosetta package improvement was performed. In the resfile, for the design of hot spots residue positions are set to ALLAA, which means allowing the placement of all natural residues in that position during the design.
2.8. B.1.617.2 Spike targetability
In addition to antibodies, small molecules represent another class of therapeutics which have attracted attention for SARS-CoV-2 targeting. The logistics of peptides/proteins distribution in remote areas are not as efficient as small molecules. As well as small molecules production costs are lower [105]. Given that residue displacement alter the function [106], [107] and affinity of proteins to interact with other molecules [108], at this step of study the impact of B.1.617.2 Spike mutations on its interaction ability with desired compounds has been investigated.
Based on the National Institutes of Health website (https://clinicaltrials.gov/), various small molecule inhibitors were proposed as possible treatments for SARS-CoV-2 [109], [110]. During the review of the available data for SARS-CoV-2 inhibitors, we found that in terms of clinical trial studies Arbidol (Phase IV) and Isotretinoin (Phase III) are in the later phase and more information is available about them, so we have chosen these compounds to compare the targetability of native and mutant Spikes [111], [112]. It revealed that Isotretinoin binds to Spike proteins' fatty acid pocket and modifying the conformational changes required to attach to ACE2 receptors [113]. Also, Arbidol (also known as Umifenovir) inhibits the adhesion and adsorption of SARS-CoV-2 to the host cells by targeting the Spike and blocking its trimerization [114].
To investigate the interaction pattern of the chosen compounds with native and mutant Spikes, AutoDock Vina and MedusaDock were employed [116], [117]. Input structures were prepared for molecular docking using MGLTools (v1.5.6). All polar hydrogens were added to ligands and receptors and charges of ingredients were estimated by the Gasteiger algorithm. A grid box with the desired dimensions was generated for the native and mutant Spikes. The dimensions of the grid box were 40 × 40 × 40 Å and spacing was set at 0.357 Å. In addition, the genetic algorithm was applied to search for conformation optimization, besides the number of population size and GA runs at 20 and 300, respectively. Ultimately, the best poses were selected based on docking and Ki score and receptor-ligand interactions for them were examined and visualized using discover studio visualizer (v21.1.0).
3. Results and discussion
3.1. Models' quality and immunogenicity assessment
Available Spike structures in the RCSB server have some missing amino acids. All missing residues and mutations on target positions were modeled using Modeller (v10.0). Native and mutant modeled structures achieved DOPE scores −127,901.9 and −128,573.7, respectively. Following energy optimization utilizing Chiron, the quality of structures was checked by the Gaia server. The studies showed that the modeled structures have a good quality for structural analysis (Table 1 ).
Table 1.
Clash scores calculated for modeled structures by employing Gaia server.
| Query | Steric clashes (CE/C) | HBC in shell (%) | HBC in core (%) | Surface area (A2/aa0.8) | Voids (A3/aa) |
|---|---|---|---|---|---|
| Native | 0.018 | 11 | 0.1 | 165 | 0.619 |
| Mutant | 0.0176 | 11.5 | 0.9 | 168 | 0.61 |
| Benchmark distribution | 0.001–0.04 | 0–15 | 0–7 | 80–200 | 0–2 |
CE/C: Clash energy/Contact HBC: Hydrogen bond clashes aa: Amino acid length.
To investigate the influence of desired mutations on the immunogenicity potency of Spike protein, potential epitopes of native and mutant Spikes for MHC-I and MHC-II were examined. The length of the epitopes was set to 9 for MHC-I and 15 for MHC-II. Examination of the identified epitopes associated with MHC-I revealed that, except for position 19, mutation on the other positions reduced the affinity of Spike epitopes for binding to MHC-I. In the case of MHC-II, all mutations except the P681R diminished the affinity of Spike epitopes for interaction with the MHC-II epitope binding pocket. The identified epitopes and their characteristics are listed in Table 2 , higher score and lower percentile rank referred to better potency for interaction with MHC-I and MHC-II.
Table 2.
MHC-I and MHC-II identified epitopes for native and mutant Spikes.
| MHC type | Position | Sequence | Score |
|---|---|---|---|
| MHC-I (N) | 19 | TTRTQLPPA | 0.00121 |
| 452 | VGGNYNYLY | 0.094173 | |
| 478 | TPCNGVEGF | 0.002697 | |
| 614 | YQDVNCTEV | 0.037062 | |
| 681 | QTQTNSPRR | 0.007383 | |
| 950 | LQDVVNQNA | 0.005013 | |
| MHC-I (M) | 19 | RTRTQLPPA | 0.001245 |
| 452 | VGGNYNYRY | 0.060056 | |
| 478 | KPCNGVEGF | 0.001792 | |
| 614 | YQGVNCTEV | 0.000966 | |
| 681 | QTQTNSRRR | 0.003453 | |
| 950 | LQNVVNQNA | 0.000489 | |
| MHC type | Position | Sequence | Percentile rank |
|---|---|---|---|
| MHC-II (N) | 19 | QCVNLTTRTQLPPAY | 44 |
| 452 | YLYRLFRKSNLKPFE | 26 | |
| 478 | FERDISTEIYQAGST | 35 | |
| 614 | NQVAVLYQDVNCTEV | 28 | |
| 681 | GAGICASYQTQTNSP | 52 | |
| 950 | LGKLQDVVNQNAQAL | 30 | |
| MHC-II (M) | 19 | QCVNLRTRTQLPPAY | 51 |
| 452 | YRYRLFRKSNLKPFE | 26 | |
| 478 | FERDISTEIYQAGSK | 35 | |
| 614 | NQVAVLYQGVNCTEV | 65 | |
| 681 | GAGICASYQTQTNSR | 41 | |
| 950 | LQNVVNQNAQALNTL | 33 |
N: native; M: mutant.
Examination of linear and discontinuous B-cells epitopes was another step of our immunogenicity investigation. The mean score of linear epitopes for both native and mutant Spikes were 0.472. Mutations T19R, L452R and D950N increased the immunogenicity potency of Spike epitopes. However, displacement at positions 478, 614 and 681 discount immunogenicity potency of epitopes contain these mutations (Table 3 ).
Table 3.
Linear epitopes for native and mutant Spikes identified using ASP tool.
| Spike | Residue | Score | Method |
|---|---|---|---|
| Native | 19 | 0.536 | Bepipred Linear Epitope Prediction 2.0 |
| 452 | 0.482 | ||
| 478 | 0.572 | ||
| 614 | 0.427 | ||
| 681 | 0.691 | ||
| 950 | 0.420 | ||
| Mutant | 19 | 0.592 | |
| 452 | 0.536 | ||
| 478 | 0.552 | ||
| 614 | 0.402 | ||
| 681 | 0.688 | ||
| 950 | 0.422 |
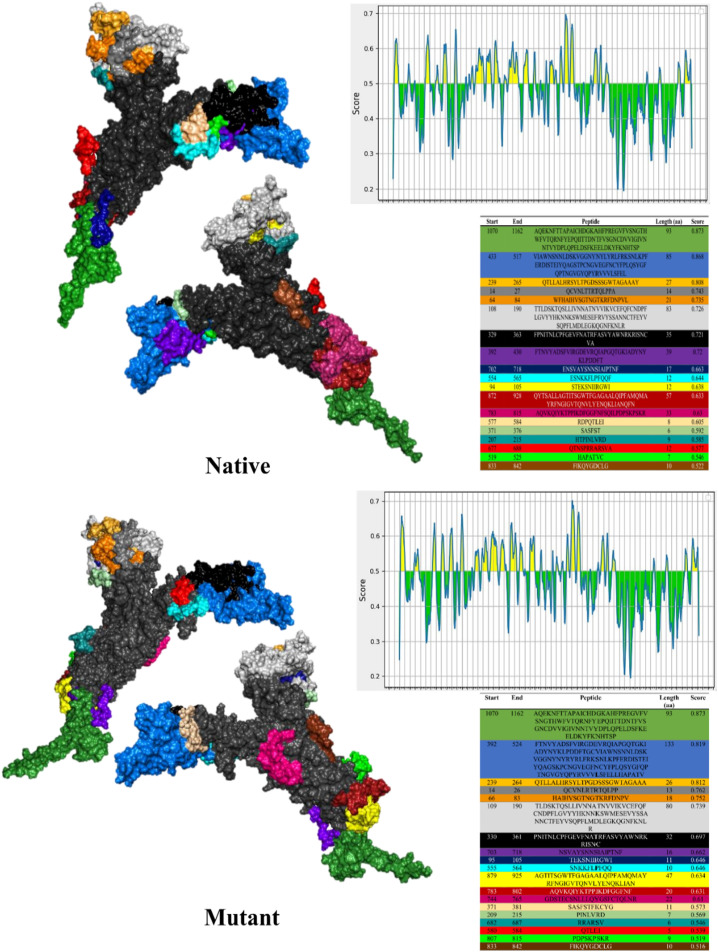
Unlike ASP, which identifies linear epitopes based on protein sequence, ElliPro examines linear epitopes using tertiary structure. ElliPro recognized 19 epitopes for both native and mutant Spikes. The mean scores of native and mutant structure epitopes were 0.675 and 0.660, respectively (Fig. 1 ).
Fig. 1.
Linear epitopes for native and mutant Spikes identified using ElliPro, tables display epitopes in the same color as structures.
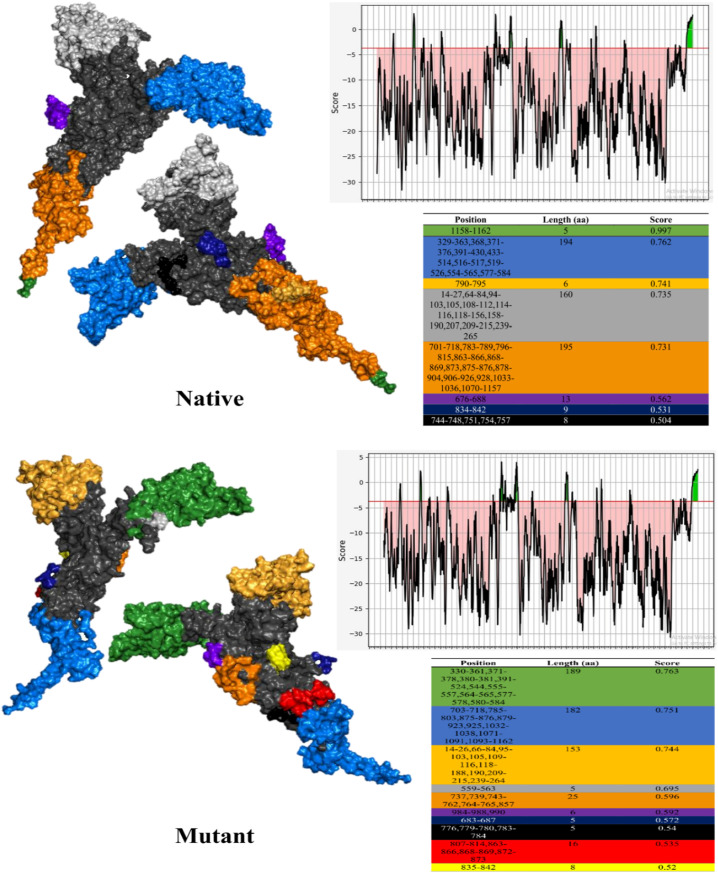
During the analysis of discontinuous epitopes using ElliPro 8 (mean score = 0.695) and 10 epitopes (mean score = 0.630) were recognized for the native and mutant Spikes, respectively (Fig. 2 ).
Fig. 2.
Discontinuous epitopes for native and mutant Spikes identified using ElliPro, tables display epitopes in the same color as structures.
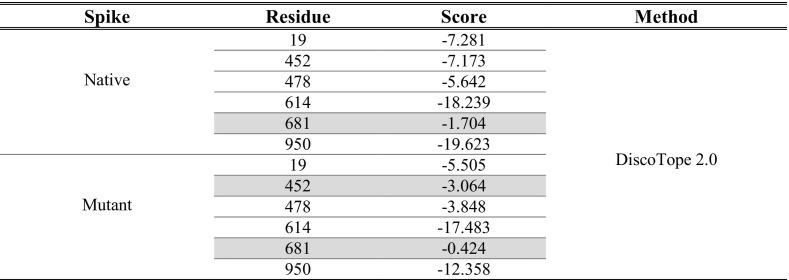
Besides ElliPro, DiscoTope was utilized for identifying discontinuous epitopes. Similar to ElliPro, DiscoTope recognizes epitopes by analyzing the tertiary structure of the protein. Examination of DiscoTope outputs showed that the immunogenicity score of B.1.617.2 Spike is higher than the native Spike (Table 4 ). Although the mutated positions have higher epitopic scores, since their scores were lower than the DiscoTope threshold, were considered as non-immunogenic regions.
Table 4.
Discontinuous epitopes identified using DiscoTope, immunogenic positions were highlighted.
Studies have shown that viral load in patients infected by B.1.617.2 is not different compared with other variants, while, the B.1.617.2 neutralization rate in treated patients with available antibodies and vaccines was lower than other variants [118], [119], [120]. Here, we found that the epitope pattern and immunogenicity potency of B.1.617.2 Spike protein are very different from native Spike (Wuhan). The identified epitopes of B.1.617.2 Spike protein have a lower tendency to interact with MHC-I and MHC-II binding pockets. In addition, examination of linear and discontinuous B-cells epitopes of the mutant showed that some mutations observed in B.1.617.2 reduce its immunogenicity. Indeed, change in the epitope pattern and binding affinity to interact with cells involved in immunogenic responses, can be considered as an efficient factor in shifting the pathogenicity and infectivity of the B.1.617.2 variant. The results of our immunogenicity investigation are consistent with Motozono et al. [121], which shows the behavior change of the B.1.617.2 in the face of the immune system.
3.2. MD simulation and IDRs evaluation
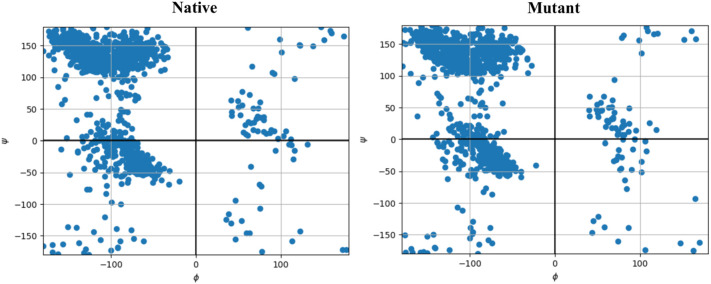
The first step of MD was performed to track the effects of the desired mutations on the structural characteristics and folding of the Spike. Analysis of the RMSD diagram revealed that following implementation of desired mutations on Spike protein, an extensive structural shift occurs in the structure. RMSD standard deviation of the last 50 ns was used to evaluate the convergence of MD simulation. RMSD deviations of native and mutant Spikes were 0.2 and 0.68 Å, respectively. The small values of RMSD standard deviation are indicative of system convergence [122], [123]. Consistent with RMSD, the distribution map of Phi and Psi torsion angles also confirmed the change in residues interaction characteristics that could justify the observed structural shift on the Spike tertiary structure (Fig. 3 ).
Fig. 3.
Ramachandranot plot of target structures extracted from MD trajectory files.
Structural fluctuation of Spike domains was another examined parameter. For this purpose, we extracted the RMSF diagram of residues throughout MD trajectory files. The average of RMSF for the native Spike was 0.514 nm, while for the mutant structure this value was 0.94 nm (Table 5 ). Increasing residues fluctuations can improve the flexibility of the structure and impact structural conformations. Also, the increase in RMSF can be related to the amount of Intrinsically disordered regions (IDRs), which was proven during the calculations of the ODiNPred server. We observed that following the mutations, the amount of the IDRs in the mutant Spike (9.9 %) increased compared to the wild structure (8.2 %). Increasing the size of the IDRs has an essential effect on Spike folding and SARS-CoV-2 pathogenesis.
Table 5.
RMSF value for mutated positions throughout MD simulation.
| Spike | Residue | RMSF (nm) | Total value (nm) |
|---|---|---|---|
| Native | 19 | 0.735 | 0.514 |
| 452 | 0.785 | ||
| 478 | 1.279 | ||
| 614 | 0.4 | ||
| 681 | 1.096 | ||
| 950 | 0.367 | ||
| Mutant | 19 | 0.737 | 0.94 |
| 452 | 1.186 | ||
| 478 | 1.715 | ||
| 614 | 0.48 | ||
| 681 | 0.539 | ||
| 950 | 1.238 |
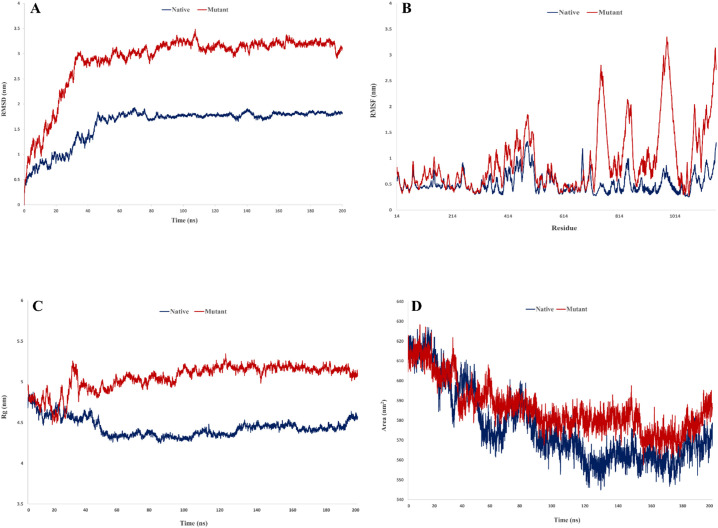
Two structural parameters include Rg and SASA were employed to track the effect of occurred mutations in B.1.617.2 on the compactness and surface area of the Spike protein. Investigation of Rg which refers to atoms' distance from the center of target molecule, showed that the amount of compression in mutant Spike (average of Rg = 5.05 nm) decreased compared to the native structure (average of Rg = 4.44 nm) (Fig. 4 ). The average of SASA values for native and mutant Spikes were 576.3 and 586.1 nm2, respectively. An increase in SASA value indicates more surface for interaction of protein with its receptor. As shown in Fig. 4, all utilized functions were equilibrated throughout simulation which refers to the sufficiency of the MD time.
Fig. 4.
Structural dynamic functions obtained from trajectories, A) RMSD, B) RMSF, C) Rg, D) SASA.
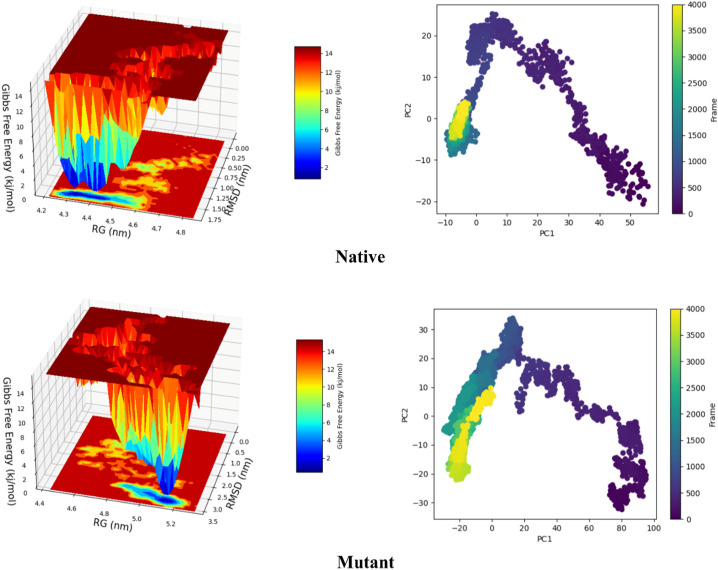
Thereafter, to further investigate mutations effect on structure flexibility and compactness, PCA and FEL functions were implemented. The results of PCA in accordance with RMSF showed that the rate of structural mobility increased in the mutant structure. As well, FEL analysis explained that the mutated protein has obtained more optimal energy following the reduction of compactness. These results confirmed Rg and SASA outputs and refer to a discount in compactness of B.1.617.2 Spike compared to the Wuhan lineage (Fig. 5 ).
Fig. 5.
The left panel shows the free energy landscape values for the conformations throughout MD, while the right panel represents principal component analysis for dihedral angels space.
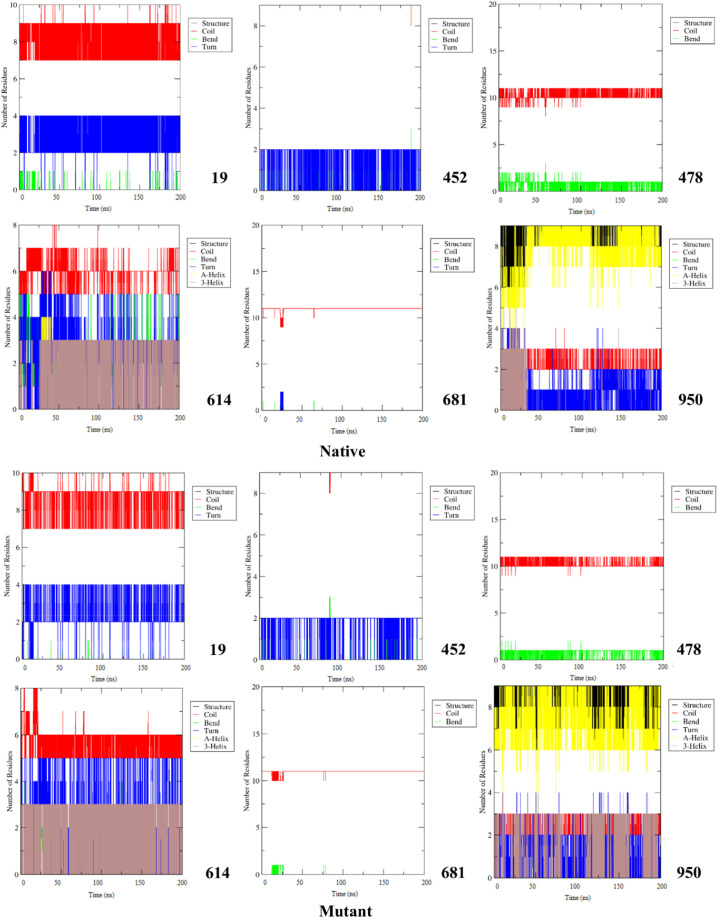
During the examination of secondary structure, we noticed that the shape of regions around the mutated residues was changed extensively. For example, following the T19R mutation, the tendency for Coil and Bend configuration was decreased. Also, in position 614, displacement of Asp with Gly limited the formation of A-Helix and Bend elements. In the case of P681R, the tendency to appear Turn in the mutant structure was limited, although the chance of Coil occurrence raised. Besides, investigation of areas around the D950N position revealed that the 3-Helix element had increased in the mutant structure (Fig. 6 ).
Fig. 6.
Secondary structure patterns of mutation positions along with their surrounded residues for native and mutant Spikes.
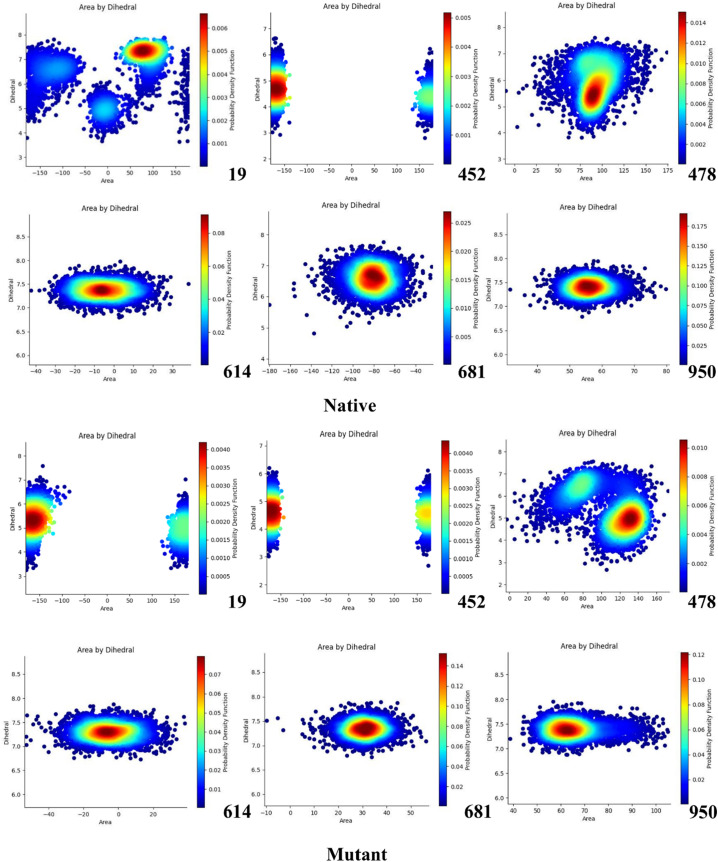
Examination of dihedral angles and triangles area distribution can provide comprehensive information about protein structural changes due to mutation happening. Here, the PDF function showed the extensive changes by examining the dihedral angles and triangles area for mutated positions (Fig. 7 ).
Fig. 7.
PDF plots for mutated positions and their surrounding residues.
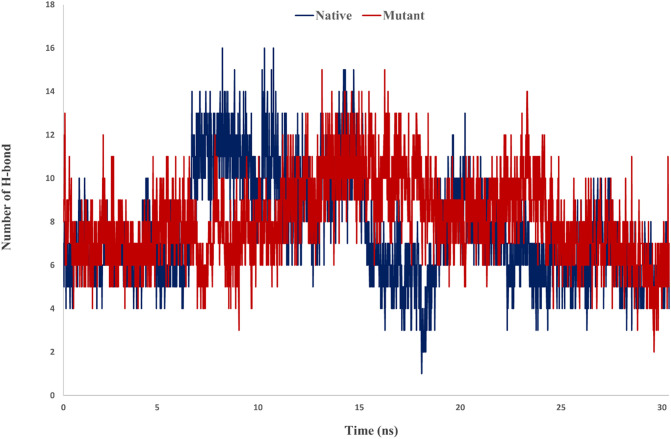
To investigate the interaction potency of Spike-ACE2 trajectory files of the second step of the MD were examined. In this step three functions including minimum distance, number of contacts and number of hydrogen bonds were studied. Examination number of hydrogen bonds for Spike-ACE2 complexes demonstrated in mutant Spike a greater number of hydrogen bonds were formed. The average of hydrogen bonds for native and mutant complexes were 7.59 and 8.09, respectively (Fig. 8 ).
Fig. 8.
Number of H-bonds for native and mutant Spikes-ACE2 complexes.
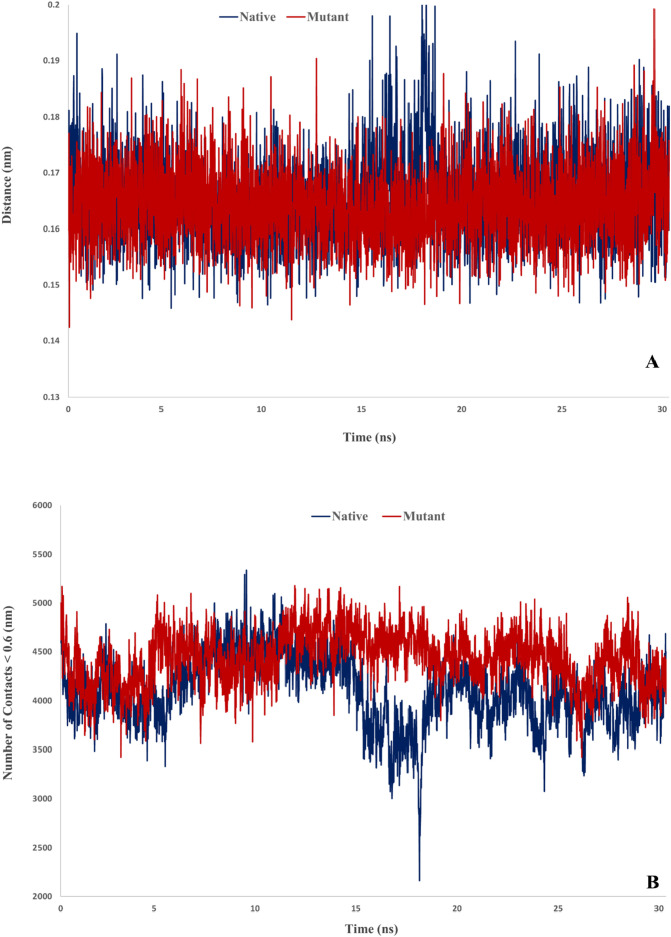
Furthermore, analysis of the minimum distance and number of contacts between native and mutant Spikes with ACE2 indicated that mutant Spike has great potency for interaction with ACE2. The average of minimum distance for native and mutant complex were 0.166 and 0.164 nm, respectively. Also, the average of number of contacts for native and mutant complex were 4105.5 and 4434.4, respectively (Fig. 9 ).
Fig. 9.
Minimum distance and number of contacts for desired complexes.
3.3. Pattern of Spike-ACE2 interaction
The binding pattern and affinity of Spike-ACE2 in native and mutant forms were investigated using the Rosetta protein docking protocol. For this purpose, MD output structures were optimized using relax and prepack modules of Rosetta. Thereafter, prepared structures were examined by Rosetta docking protocol. In Rosetta's output structures, the energy score was reported as REU, which is an arbitrary scale. The docking procedure revealed that following mutation in Spike protein, its affinity for ACE2 was improved. The mean binding energies for native and B.1.617.2 Spike-ACE2 complex were −1907.06REU and −1936.3REU, respectively. Some of the docking energies terms are listed in Table 6 .
Table 6.
Some docking energy terms for native and mutant Spikes-ACE2 complexes (all energy terms are in REU).
| Complex | fa_atr | fa_rep | fa_sol | fa_intra_rep | lk_ball_wtd | fa_elec | omega | fa_dun | Total |
|---|---|---|---|---|---|---|---|---|---|
| Native | −4736.7 | 577.8 | 2899.03 | 8.8 | −140.15 | −1249.3 | 93.2 | 994 | −1907.06 |
| Mutant | −4712.8 | 548.2 | 2888.91 | 8.7 | −141.09 | −1235.9 | 92.4 | 996 | −1936.3 |
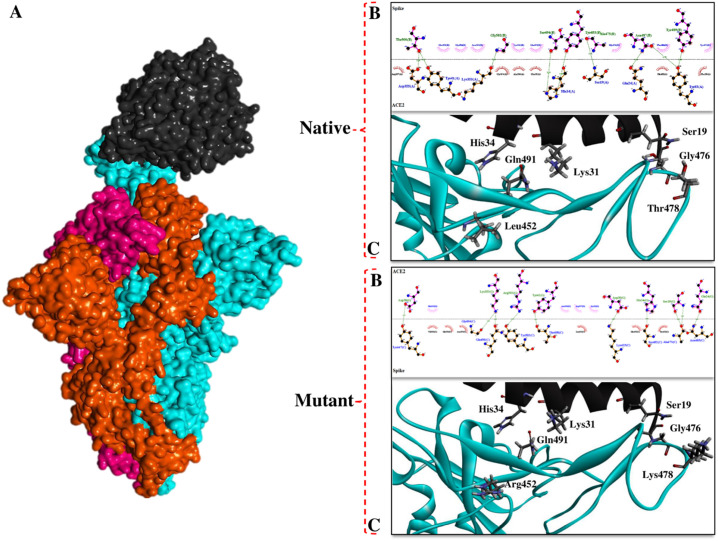
We found that longer side chains of Arg452 and Lys478 along with their positive effects on the conformation of Spike interface residues, improve the opportunity of interactions between Spike and ACE2 (Fig. 10 ). Furthermore, L452R and T478K mutations reduce distances in the Spike-ACE2 complex and allow the interface residues around the mutant positions to gain a better spatial arrangement to interact with the ACE2. Similar studies have confirmed the positive effect of B.1.617.2 Spike mutations on its affinity for the ACE2 [125], [126].
Fig. 10.
The interaction pattern of ACE2 with native and mutant Spikes. A) 3D structures of Spike-ACE2 (PDB ID: 7DF4), B) Interface residues of Spike-ACE2 complexes, C) Arrangement of mutated positions and surrendered residues.
3.4. Topology and contact maps analysis
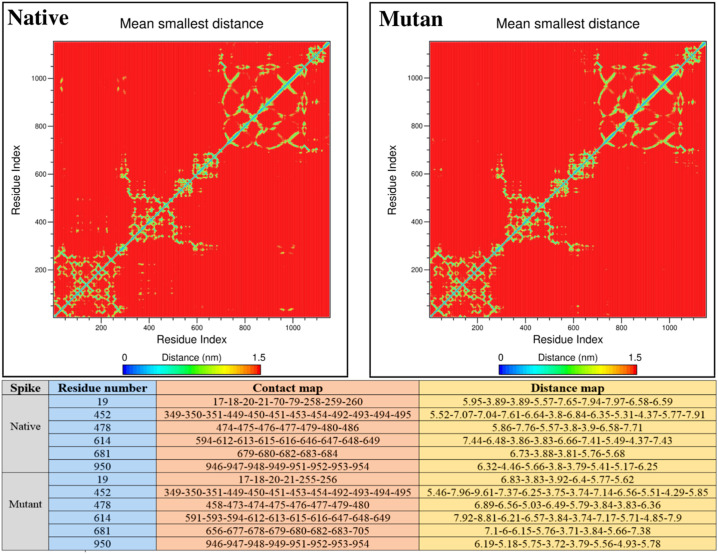
Mapping of residues contacts and their distances from adjacent positions provides a great insight into tertiary structure characteristics. Given that two types of mutation include deletion and displacement occurs in B.1.617.2 Spike, contacts and distances map for extracted structures from MD trajectories were examined to track the topological attributes of native and mutated proteins. In addition to the contact map, we found that the distance map pattern of mutated residues underwent extensive changes (Fig. 11 ). These differences can be a justification for the structural changes observed in the MD step.
Fig. 11.
Contact and distance map for native and mutant Spikes.
In addition to the amino acids sequence that plays a key role in the formation of protein primary structure, the shape of amino acids contacts and distance map dramatically affects the secondary and tertiary folding of the proteins [127]. The change in the contact and distance map of the residues disclosed at this stage can be a confirmation of structural changes (such as shifts in RMSD, RMSF, SASA, and Rg) observed in the MD step. Also, because residues contact and distance affect the formation of pockets around residues [128], it is expected that examined changes will affect Spike pharmacological behavior and its interaction with drugs designed to inhibit COVID-19.
Examination of topological attributes like cavities and tunnels around mutated residues also confirmed extensive changes in Spike structure. CAVER outputs revealed that tunnels and cavities around the mutant positions have increased compared to the native protein (Table 7 ). The results of this step were consistent with the Rg step which demonstrated that the protein compression was reduced in the B.1.617.2 Spike.
Table 7.
Tunnels and cavities around studied positions.
| Spike | Position | Avg_BR | Avg_L | Avg_C | Avg_t |
|---|---|---|---|---|---|
| Native | 19 | 1.026 | 1.999 | 1.102 | 0.84088 |
| 452 | 0.91 | 1.01 | 0.89 | 0.79 | |
| 478 | 0.83 | 1.05 | 0.83 | 0.81 | |
| 614 | 0.913 | 2.391 | 1.159 | 0.81687 | |
| 681 | 1.240 | 1.945 | 1.187 | 0.86768 | |
| 950 | 1.052 | 3.313 | 1.161 | 0.88291 | |
| Mutant | 19 | 1.328 | 1.898 | 1.012 | 0.90322 |
| 452 | 1.215 | 1.459 | 1.033 | 0.86510 | |
| 478 | 1.074 | 1.436 | 1.120 | 0.88172 | |
| 614 | 1.165 | 4.434 | 1.231 | 0.85288 | |
| 681 | 0.921 | 2.569 | 1.315 | 0.87482 | |
| 950 | 1.137 | 3.951 | 1.013 | 0.80552 |
Avg_BR: Average radius, Avg_L: Average length, Avg_C: Average curvature, Avg_t: Average throughput.
3.5. Evaluation of oligomerization and cleavability potency
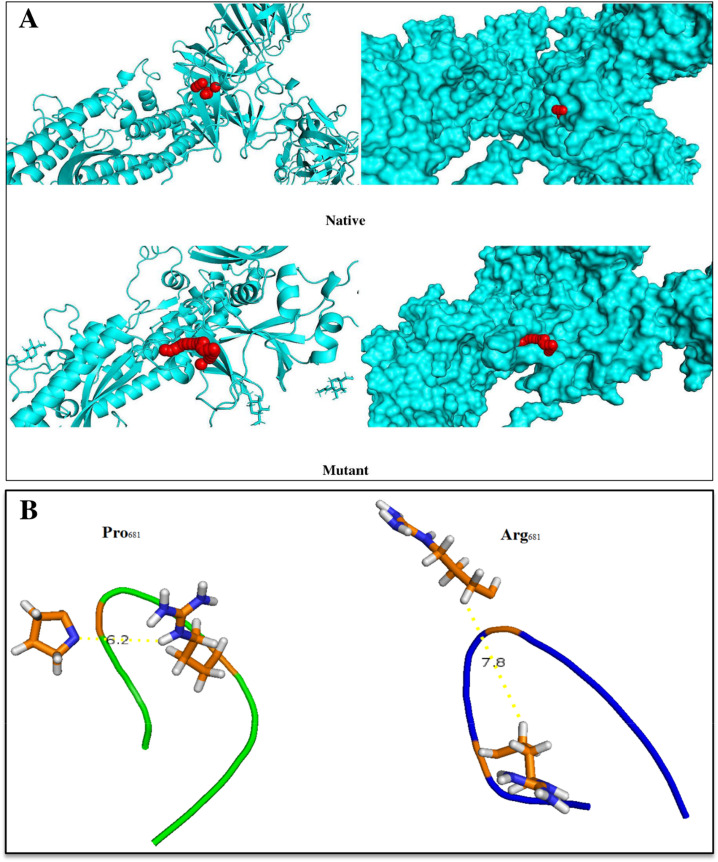
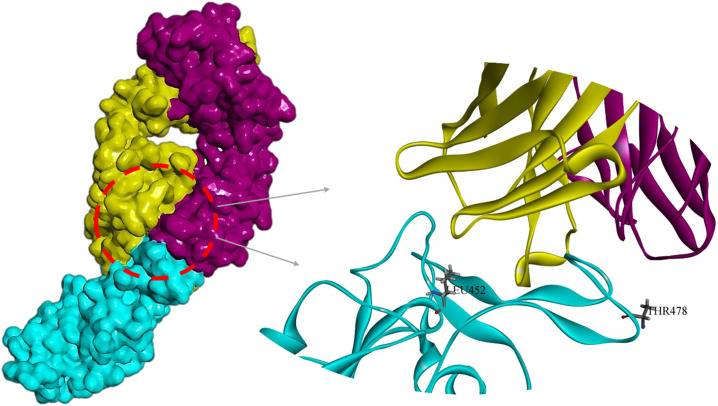
Cutting Spike protein in the S1-S2 position by Furin plays an important role in the infectivity and virus entry into the cell. Examination of the mutated region conformation disclosed that the P681R displacement causes the Furin target site to open more. Increasing the surface accessibility and reducing the fluctuations along with expanding the cavities and tunnels around the P681R (Fig. 12 ) can be a factor in improving the function and binding ability of the Furin to its target sequence.
Fig. 12.
A) Graphical representation of tunnels and cavities (shown in red) around Pro681 and Arg681, B) Representation of Furin target site in native (left panel) and mutant (right panel) Spikes. As shown in the right panel, the region near the Furin cleavage site is more open in the mutant structure than in the wild-type structure.
Compactness and fluctuation of Furin cleavage site and residues around this region to evaluate the effect of P681R mutation on S1–S2 cleavage were examined. Fluctuation of the Furin cleavage site following P681R was reduced. In addition, surface accessibility of residue around the cleavage site in the mutant Spike was increased (Table 8 ).
Table 8.
Compactness and fluctuation of the region around the Furin cleavage site.
| Spike | Fluctuation (nm) | Surface accessibility (nm2) |
|---|---|---|
| QTNSPro681RRARSVASQ | 0.213 | 19.1 |
| QTNSArg681RRARSVASQ | 0.156 | 21.4 |
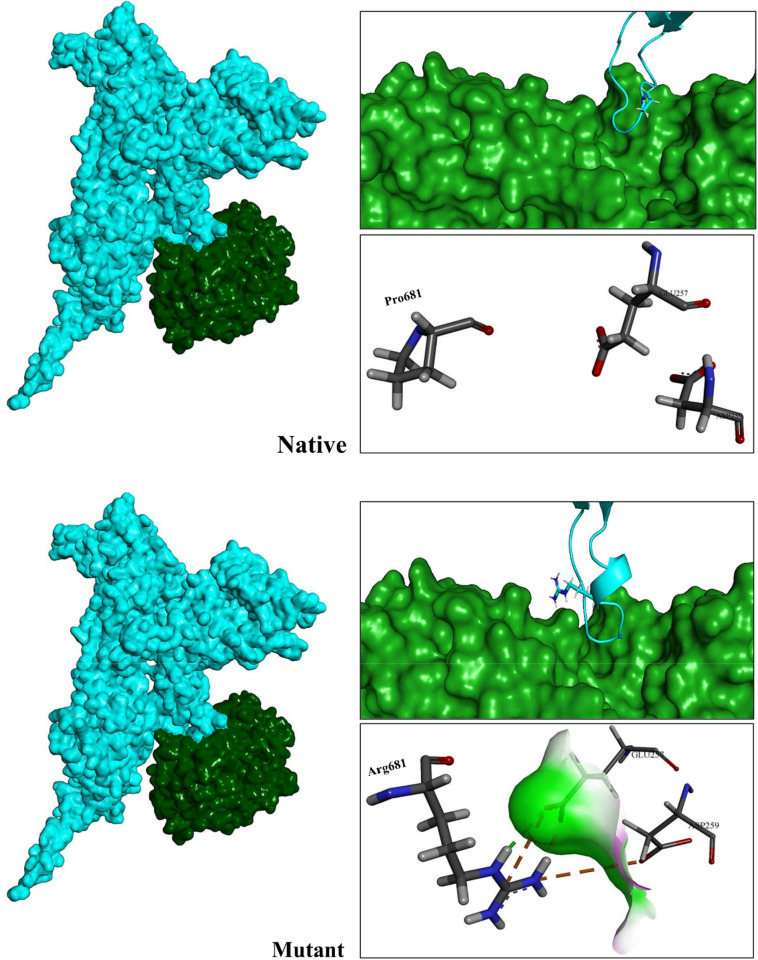
Previously conducted studies revealed displacement of proline by arginine at position 681 facilitates protein cleavage, enhances viral fusogenicity and improves virus replication [129], [130]. In our study, the results of the Rosetta docking protocol also showed that the Furin binds to the mutant Spike (−3825.14REU) with greater affinity than the native (−3561.68REU) structure. We found that longer side chain of arginine compared to proline allows the Spike to interact more strongly with the interface residues of Furin (Fig. 13 ). Cleavage of Spike by Furin is a critical step in the entry of the virus into the target cells. Increased Furin affinity for CS1 in the mutant Spike following the P681R mutation could promote the function of this enzyme and facilitate the entry of SARS-CoV-2 into the cell [122], [131], [132], [133], [134].
Fig. 13.
Representation of Furin interaction with native and mutant Spikes in monomeric forms. As can be seen, the active site of Furin interacts in a great manner with its cleavage region in the mutant structure.
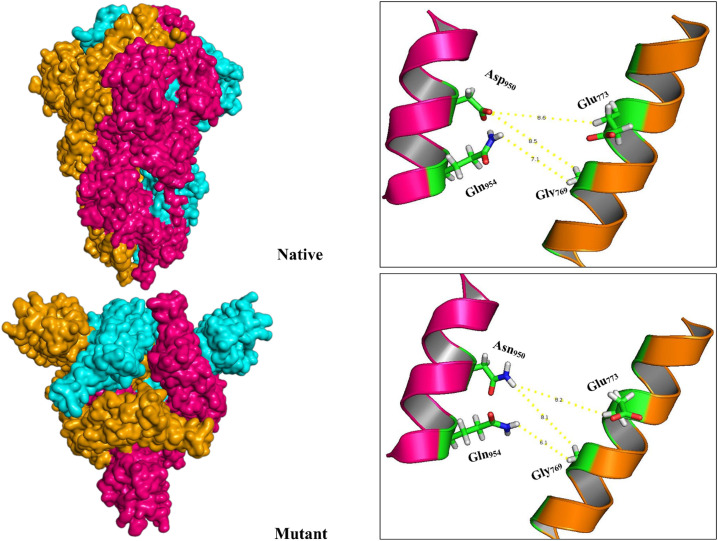
One of the unexplored facets of B.1.617.2 Spike is the effect of mutations on the capability of its trimeric form to oligomerize. Replacing Aspartate by Asparagine in D950N mutation seems to have a large effect on the Spike function due to its proximity to the oligomerization domain. Examination of the results of Rosetta symmetric docking showed that the structure of the mutant homotrimer (-3470REU) is more stable than the native structure (−3043.8REU). We found that D950N mutation, by reducing the distance between oligomers facilitated their interaction (Fig. 14 ).
Fig. 14.
Illustration of oligomerization state of native and mutant Spikes. Reducing the distance between the homomers of the mutant structure can improve Spike oligomerization in the Delta lineage of SARS-CoV-2.
3.6. Free energy and residue scanning analysis
The output trajectories of the MD simulation to calculate the stability of structures and binding energy of Spike-ACE2 complex were conducted to G_mmpbsa. Examination of the native and mutated Spike free energy showed that following the mutation protein becomes more stable. The energies of the native and mutant structures were −16,082.5 and −16,613.6 kJ/mol, respectively (Table 9 ). With the exception of apolar energy, other energy terms were improved in the mutant structure.
Table 9.
Different terms of structural free energy (kJ/mol) were calculated for target structures.
| Spike | Apolar solvation energy | polar solvation energy | Elec energy | VdW energy | Total energy |
|---|---|---|---|---|---|
| Native | 249.1 | −6174.5 | −7725.3 | −2431.7 | −16,082.5 |
| Mutant | 252.7 | −6668.8 | −7744.3 | −2453.1 | −16,613.6 |
Elec: Electrostatics; VdW: Van der Waals.
Similar to the G_mmpbsa result, the Score module of Rosetta revealed that mutated structure (−2052.4REU) achieved an optimal level of structural energy compared to the native Spike (−1879.9REU). In addition to estimate the energy for the whole structure, the results of residue scanning for mutated positions using Schrodinger (v2021.2) demonstrated that except for the T478K, other mutations cause target positions become more stable (Table 10 ).
Table 10.
Schrodinger residue scaning outputs.
| Position | Mutation | Score (kJ/mol) |
|---|---|---|
| 19 | T > R | −9.328 |
| 452 | L > R | −2.084 |
| 478 | T > K | 10.4 |
| 614 | D > G | −0.004 |
| 681 | P > R | −3.337 |
| 950 | D > N | −1.890 |
In addition to the free energy of Spike in single form (in the first phase), the binding free energy of the Spike-ACE2 complex was investigated. We found that the Spike affinity for ACE2 increased following mutation (Table 11 ). The observations of binding free energy confirm the results obtained throughout docking and MD, which show the positive effect of mutations in the B.1.617.2 on the ability of the virus to interact with its receptor for entering the target cells. This finding could justify the greater infectivity of the Delta lineage of SARS-CoV-2 compared to the Wuhan lineage.
Table 11.
Binding free energy (kJ/mol) calculated for Spike-ACE2 complexes.
| Spike | Apolar solvation energy | Polar solvation energy | Elec energy | VdW energy | Total energy |
|---|---|---|---|---|---|
| Native | −41.312 | 641.17 | −1321.58 | −313.115 | −1034.83 |
| Mutant | −34.058 | 611.6 | −2045.46 | −259.33 | −1727.25 |
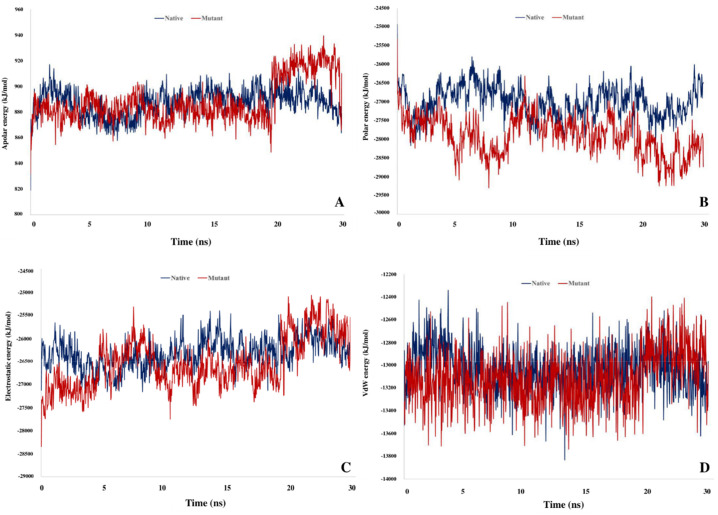
Apolar solvation and VdW energies in the native complex have reached more optimal levels, but polar solvation energy and electrostatic energy in the mutant complex significantly have improved the binding strength. Fig. 15 represents the binding free energy fluctuations of the studied complexes throughout the simulation. Major energy terms are equilibrated during the simulation.
Fig. 15.
Schematic representation of different terms of binding free energy throughout MD simulation. A) Apolar solvation energy B) Polar solvation energy C) Electrostatics energy D) VdW energy.
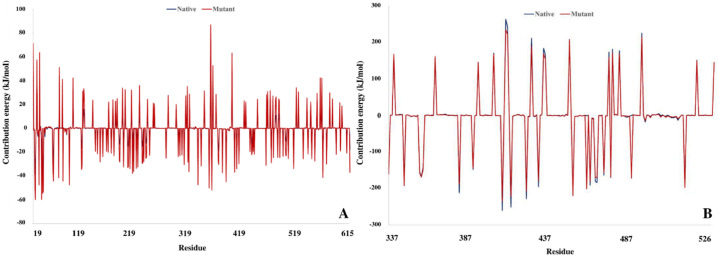
Decomposition of binding free energy throughout trajectory files confirmed the change in the affinity of Spike interface residue for the binding pocket of ACE2. As can be seen in Fig. 16 , the RBD mutated position along with their binding partner in the ACE2 have the optimal level of energy than the native complex.
Fig. 16.
Decomposition of binding free energy for Spike and ACE2 interface residues; A) ACE2, B) Spike.
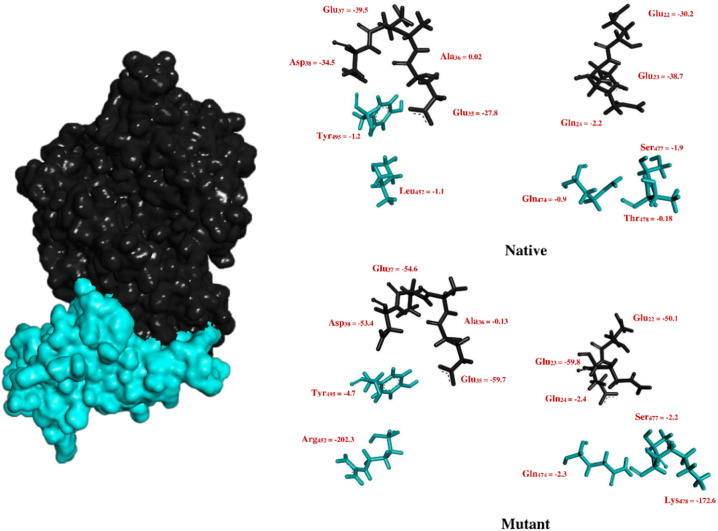
Binding potency improved dramatically following the L452R and T478K mutations. We found that the binding free energy of Arg452 and Lys478 were − 202.3 kJ/mol and − 172.6 kJ/mol, respectively. Fig. 17 shows the binding energy of the RBD mutated position and their adjacent residues.
Fig. 17.
Decomposition of binding free energy (kJ/mol) of mutated positions and residues around them along with related interface residues in the ACE2.
3.7. Antibody stability-affinity improvement
In order to improve the function of Bamlanivimab against Delta lineage of SARS-CoV-2, antibody complex with B.1.617.2 Spike (Fig. 18 ) was examined and all antibody residues at a distance of 7 Å from Spike were redesigned.
Fig. 18.
The complex of Spike-Bamlanivimab (PDB ID: 7KMG); Spike (cyan), Bamlanivimab heavy chain (yellow), Bamlanivimab light chain (purple).
At this stage, 27 residues from the heavy chain and 11 residues from the light chain of Bamlanivimab were chosen for redesigning. Also, interface residues of Spike with Bamlanivimab were repacked. During antibody improvement, >1000 structures were generated. We found that, among designed scaffolds, seven antibodies (Table 12 ) showed a better tendency to interact with Spike compared to native Bamlanivimab as a reference structure (score = −1304.2REU).
Table 12.
Affinity and stability matured antibodies designed by Rosetta based on Bamlanivimab scaffold.
| Chain | Target positions <0.7 (nm) | Model | Chain | Designed positions | Score (REU) |
|---|---|---|---|---|---|
| H | 28, 29, 30, 31, 32, 33, 47, 50, 52, 54, 55, 57, 58, 59, 60, 61, 65, 74, 100, 101, 102, 103, 104, 105, 106, 109, 110 | 1 | H | S30T, R50M, I54V, N59A, A103Y, Y110F | −1315.52 |
| L | Q27S, S28P, S30D, S91T, T94M, R96W | ||||
| 2 | H | S30T, R50M, I54V, N59A, K74Q, A103Y, Y110F | −1314.53 | ||
| L | Q27S, S28P, S30D, S91T, T94M, R96W | ||||
| 3 | H | S30T, R50M, I54V, N59A, A103Y, Y106W, Y110F | −1312.68 | ||
| L | Q27S, S28P, S30D, Y32F, S91T, T94M, R96W | ||||
| 4 | H | S30T, R50M, I54V, N59A, A103Y, Y110H | −1309.7 | ||
| L | 2, 27, 28, 29, 30, 32, 91, 92, 93, 94, 98 | L | Q27S, S28P, S30D, S91T, T94M, R96W | ||
| 5 | H | S30T, R50M, I54V, A58V, N59A, A103Y, Y110F | −1307.98 | ||
| L | Q27S, S28P, S30D, S91T, T94M, R96W | ||||
| 6 | H | S30T, N31D, Y32A, R50M, I54V, N59A, A103Y, R104K, H105N, Y110F | −1305.94 | ||
| L | Q27S, S28P, S30D, S91T, T94M, R96W | ||||
| 7 | H | S30T, R50M, I54V, N59A, A103Y, Y110F | −1305.49 | ||
| L | Q27T, S28E, S30G, S91T, T94M, R96W |
3.8. B.1.617.2 Spike targetability
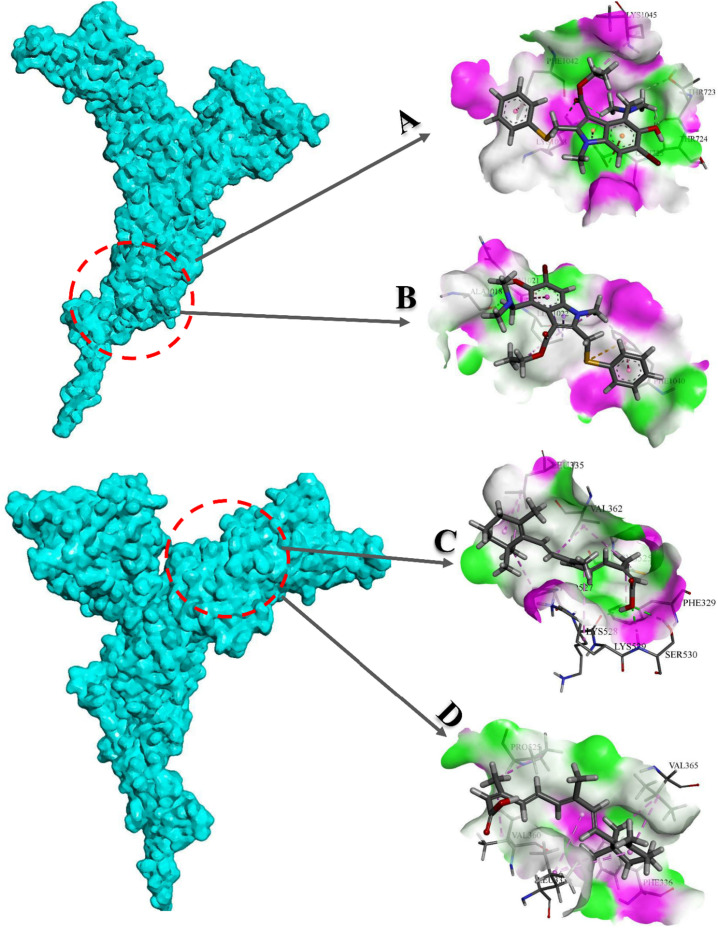
Targeting Spike protein as an important part of SARS-CoV-2 plays an essential role in the treatment of COVID-19. Inhibition of Spike function is possible by using different classes of therapeutics such as antibodies and peptides, but small molecules have attracted a lot of attention due to their ease of production and cost-effectiveness. Isotretinoin and Arbidol, which are in clinical phases III and VI, interfere with SARS-CoV-2 pathogenesis by targeting the Spike protein. In this part of the study, the capability of Arbidol and Isotretinoin to interact with B.1.617.2 Spike compared to the native structure was investigated using AutoDock Vina and MedusaDock. Due to the proximity of the L452R and T478K mutations to the Arbidol binding site and D950N mutation to the Isotretinoin binding pocket, it seems that the function of desired compounds has been affected in mutant lineage. The results of the molecular docking process showed that the mutant Spike has less tendency to interact with Arbidol and Isotretinoin (Table 13 ).
Table 13.
Vina and MedusaDock outputs for complex of native and mutant Spikes with Arbidol and Isotretinoin.
| Spike | Compund | MedusaDock score (kcal/mol) | Vina score (kcal/mol) | Inerface residues |
|---|---|---|---|---|
| Native | Arbidol | −12.3 | −4.9 | Thr723, Thr724, Glu725, Lys1028, Phe1042, Lys1045 |
| Isotretinoin | −14.6 | −6.5 | Phe329, Leu335, Val362, Cys525, Pro527, Lys528, Lys529, Ser530 | |
| Mutant | Arbidol | −10.9 | −4.6 | Ala1018, Asn1021, Leu1022, Phe1040 |
| Isotretinoin | −12.1 | −5.8 | Leu333, Phe336, Val360, Val365, Pro525 |
We observed that mentioned the mutations have a significant impact on the affinity and binding pattern of studied compounds (Fig. 19 ).
Fig. 19.
3D representation of native and mutant Spikes interaction with desired compounds. A) Arbidol (native), B) Arbidol (mutant), C) Isotretinoin (native), D) Isotretinoin (mutant).
4. Conclusions
Determining the molecular and biological attributes of pathogens at the beginning of emergence can help to prevent their spread. Due to the low speed and high cost of laboratory methods, computational biology approaches which have many advantages such as flexibility and affordability facilitated the fight against infectious diseases. There are many well-known methods in computational biology that are widely used for biological studies, such as molecular interaction analysis, MD, protein structure prediction, epitope mapping, and computational protein design. Using these methods, researchers have been able to study biological molecules on an atomic scale and diagnose and treat diseases earlier than ever. Recent advances in this field, such as the advent of Alphafold v2 and improvements in de novo protein design with tools such as Rosetta, have facilitated the study of biology in a way more than ever before. COVID-19, as a pandemic with a very high prevalence rate, has caused concern in the human health community. Due to the rapid evolution of SARS-CoV-2 and the high rate of mutations in its genome, early detection of the structural features of mutant lineages of this virus is important. In the present study, using computational biology approaches such as immunoinformatics, molecular dynamics, molecular docking and free energy calculation, the biological properties of B.1.617.2 Spike were investigated and we noticed extensive changes in the Spike structure and its response to existing drugs following mutation. The observed changes may justify the greater prevalence and infectivity of the Delta lineage. Also designed antibodies using the antibody design protocol of Rosetta based on Bamlanivimab scaffold, can be an excellent option against B.1.617.2 Spike.
CRediT authorship contribution statement
Mohammad Mahmoudi Gomari: Study design, Formal analysis, Methodology, Visualization, Investigation, Writing - Original Draft, Writing - Review & Editing.
Parastoo Tarighi: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Edris Choupani: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Shadi Abkhiz: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Masoud Mohamadzadeh: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Neda Rostami: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Esmaeil Sadroddiny: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Soukayna Baammi: Formal analysis, Investigation, Visualization, Writing - Review & Editing.
Vladimir N. Uversky: Study design, Formal analysis, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing.
Nikolay V. Dokholyan: Study design, Formal analysis, Methodology, Investigation, Visualization, Investigation, Writing - Original Draft, Writing - Review & Editing.
Declaration of competing interest
Mohammad Mahmoudi Gomari reports financial support was provided by Iran University of Medical Sciences.
Acknowledgment
This research was supported by grant No 22588 from Iran University of Medical Sciences.
Data availability
Data will be made available on request.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.COVID-19 Dashboard Coronavirus resource center. 2021. https://coronavirus.jhu.edu/map.html Available from:
- 3.Giovanetti M., Benedetti F., Campisi G., Ciccozzi A., Fabris S., Ceccarelli G., et al. Evolution patterns of SARS-CoV-2: snapshot on its genome variants. Biochem. Biophys. Res. Commun. 2021;538:88–91. doi: 10.1016/j.bbrc.2020.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mihaescu G., Chifiriuc M.C., Iliescu C., Vrancianu C.O., Ditu L.-M., Marutescu L.G., et al. SARS-CoV-2: from structure to pathology, host immune response and therapeutic management. Microorganisms. 2020;8(10):1468. doi: 10.3390/microorganisms8101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dound Y., Chaudhary S., Chaudhary S., Dound B. Plant based molecules for the management of Covid-19. J. Infect. Dis. Ther. S. 2020;2:2. [Google Scholar]
- 7.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder E., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo E., Escarmís C., Lázaro E., Manrubia S.C. Quasispecies dynamics and RNA virus extinction. Virus Res. 2005;107(2):129–139. doi: 10.1016/j.virusres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Smith E.C., Denison M.R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9(12) doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., Anderson D.E., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396(10251):603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18(1):1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callaway E. The coronavirus is mutating-does it matter? Nature. 2020;585(7824):174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 15.Mahmoudi Gomari M., Rostami N., Omidi-Ardali H., Arab S.S. Insight into molecular characteristics of SARS-CoV-2 spike protein following D614G point mutation, a molecular dynamics study. J. Biomol. Struct. Dyn. 2021:1–9. doi: 10.1080/07391102.2021.1872418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Control CfD . Prevention. SARS-CoV-2 Variant Classifications and Definitions. 2020. 16. Retrieved March. 2021. [Google Scholar]
- 17.Younes M., Hamze K., Nassar H., Makki M., Ghadar M., Nguewa P., et al. medRxiv; 2021. Emergence and Fast Spread of B. 1.1. 7 Lineage in Lebanon. [Google Scholar]
- 18.Tragni V., Preziusi F., Laera L., Onofrio A., Mercurio I., Todisco S., et al. Modeling SARS-CoV-2 spike/ACE2 protein-protein interactions for predicting the binding affinity of new spike variants for ACE2, and novel ACE2 structurally related human protein targets, for COVID-19 handling in the 3PM context. EPMA J. 2022;13(1):149–175. doi: 10.1007/s13167-021-00267-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quaglia F., Salladini E., Carraro M., Minervini G., Tosatto S.C.E., Le Mercier P. SARS-CoV-2 variants preferentially emerge at intrinsically disordered protein sites helping immune evasion. FEBS J. 2022;289(14):4240–4250. doi: 10.1111/febs.16379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rostami N, Choupani E, Hernandez Y, Arab SS, Jazayeri SM, Gomari MM. SARS-CoV-2 spike evolutionary behaviors; simulation of N501Y mutation outcomes in terms of immunogenicity and structural characteristic. Journal of Cellular Biochemistry.n/a(n/a). [DOI] [PMC free article] [PubMed]
- 21.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Assessing transmissibility of SARS-CoV-2 lineage B. 1.1. 7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 22.du Plessis L., McCrone J.T., Zarebski A.E., Hill V., Ruis C., Gutierrez B., et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371(6530):708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y.J., Jang U.S., Soh S.M., Lee J.-Y., Lee H.-R. The impact on infectivity and neutralization efficiency of SARS-CoV-2 lineage B. 1.351 pseudovirus. Viruses. 2021;13(4):633. doi: 10.3390/v13040633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dos Santos C.A., Bezerra G.V.B., de Azevedo A.R.R.A. SARS-CoV-2 genomic surveillance in Northeast Brazil: timing of emergence of the brazilian variant of concern P1. J. Travel Med. 2021;28:66–70. doi: 10.1093/jtm/taab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sallam M., Mahafzah A. Molecular analysis of SARS-CoV-2 genetic lineages in Jordan: tracking the introduction and spread of COVID-19 UK variant of concern at a country level. Pathogens. 2021;10(3):302. doi: 10.3390/pathogens10030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahimi F., Abadi A.T.B. Omicron: a highly transmissible SARS-CoV-2 variant. Gene Reports. 2022;27 doi: 10.1016/j.genrep.2022.101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakur V., Kanta Ratho R. OMICRON (B. 1.1. 529): a new SARS-CoV-2 variant of concern mounting worldwide fear. J. Med. Virol. 2021;94(5):1821–1824. doi: 10.1002/jmv.27541. [DOI] [PubMed] [Google Scholar]
- 28.Leung N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021;19(8):528–545. doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagpunar J.S. medRxiv; 2021. Interim Estimates of Increased Transmissibility, Growth Rate, and Reproduction Number of the Covid-19 B. 1.617. 2 Variant of Concern in the United Kingdom. [Google Scholar]
- 30.Yadav P.D., Mohandas S., Shete A.M., Nyayanit D.A., Gupta N., Patil D.Y., et al. bioRxiv; 2021. SARS CoV-2 Variant B. 1.617. 1 Is Highly Pathogenic in Hamsters Than B. 1 Variant. [Google Scholar]
- 31.Ferreira I., Datir R., Papa G., Kemp S., Meng B., Rakshit P., et al. bioRxiv; 2021. SARS-CoV-2 B. 1.617 Emergence and Sensitivity to Vaccine-elicited Antibodies. [Google Scholar]
- 32.Dougherty K., Mannell M., Naqvi O., Matson D., Stone J. 2021. SARS-CoV-2 B. 1.617. 2 (Delta) Variant COVID-19 Outbreak Associated With a Gymnastics Facility—Oklahoma, April–May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., et al. Effectiveness of Covid-19 vaccines against the B. 1.617. 2 (delta) variant. N. Engl. J. Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;1–7 doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 35.Muecksch F., Weisblum Y., Barnes C.O., Schmidt F., Schaefer-Babajew D., Lorenzi J.C.C., et al. bioRxiv; 2021. Development of Potency, Breadth and Resilience to Viral Escape Mutations in SARS-CoV-2 Neutralizing Antibodies. 2021.03.07.434227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahase E. Delta variant: What is happening with transmission, hospital admissions, and restrictions? British Medical Journal Publishing Group. 2021;37:1513–1521. doi: 10.1136/bmj.n1513. [DOI] [PubMed] [Google Scholar]
- 38.Wall E.C., Wu M., Harvey R., Kelly G., Warchal S., Sawyer C., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination. Lancet. 2021;397(10292):2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav P.D., Sapkal G.N., Ella R., Sahay R.R., Nyayanit D.A., Patil D.Y., et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J. Travel Med. 2021;28 doi: 10.1093/jtm/taab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malabadi RB, Kolkar KP, Meti NT, Chalannavar RK. Outbreak of Coronavirus (SARS-CoV-2) Delta variant (B. 1.617. 2) and Delta Plus (AY. 1) with fungal infections, Mucormycosis: Herbal medicine treatment.
- 41.Gupta N., Kaur H., Yadav P., Mukhopadhyay L., Sahay R.R., Kumar A., et al. medRxiv; 2021. Clinical Characterization and Genomic Analysis of COVID-19 Breakthrough Infections During Second Wave in Different States of India. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., et al. medRxiv; 2021. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-induced Humoral Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alenquer M., Ferreira F., Lousa D., Valério M., Medina-Lopes M., Bergman M.L., et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021;17(8) doi: 10.1371/journal.ppat.1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramana J., Mehla K. Immunoinformatics and epitope prediction. Methods Mol. Biol. (Clifton, NJ) 2020;2131:155–171. doi: 10.1007/978-1-0716-0389-5_6. [DOI] [PubMed] [Google Scholar]
- 47.Raoufi E., Hemmati M., Eftekhari S., Khaksaran K., Mahmodi Z., Farajollahi M.M., et al. Epitope prediction by novel immunoinformatics approach: a state-of-the-art review. Int. J. Pept. Res. Ther. 2020;26(2):1155–1163. doi: 10.1007/s10989-019-09918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geoghegan J.L., Holmes E.C. The phylogenomics of evolving virus virulence. Nat. Rev. Genet. 2018;19(12):756–769. doi: 10.1038/s41576-018-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalsoom F., Sajjad Ur R., Mahmood M.S., Zahoor T. Association of Interleukin-1B gene polymorphism with H. Pylori infected dyspeptic gastric diseases and healthy population. Pakistan J. Med. Sci. 2020;36(4):825–830. doi: 10.12669/pjms.36.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54 doi: 10.1002/cpbi.3. 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janson G., Paiardini A. PyMod 3: a complete suite for structural bioinformatics in PyMOL. Bioinformatics (Oxford, England) 2021;37(10):1471–1472. doi: 10.1093/bioinformatics/btaa849. [DOI] [PubMed] [Google Scholar]
- 55.Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370(6513):203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercurio I., Tragni V., Busto F., De Grassi A., Pierri C.L. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell. Mol. Life Sci. 2021;78(4):1501–1522. doi: 10.1007/s00018-020-03580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierri C.L. SARS-CoV-2 spike protein: flexibility as a new target for fighting infection. Signal Transduct. Target. Ther. 2020;5(1):254. doi: 10.1038/s41392-020-00369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peters B., Nielsen M., Sette A. T cell epitope predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Odhar H.A., Ahjel S.W., Humadi S.S. Towards the design of epitope candidates for coronavirus 2. Bioinformation. 2020;16(5):375–386. doi: 10.6026/97320630016375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ayyagari V.S., TCV, KAP, Srirama K. Design of a multi-epitope-based vaccine targeting M-protein of SARS-CoV2: an immunoinformatics approach. Journal of Biomolecular Structure and Dynamics. 2020:1–15. doi: 10.1080/07391102.2020.1850357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8(12) doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rostami N, Davarnejad R. Characterization of folic acid-functionalized PLA–PEG nanomicelle to deliver Letrozole: A nanoinformatics study. IET Nanobiotechnology.n/a(n/a). [DOI] [PMC free article] [PubMed]
- 65.Bossis F., De Grassi A., Palese L.L., Pierri C.L. Prediction of high- and low-affinity quinol-analogue-binding sites in the aa3 and bo3 terminal oxidases from Bacillus subtilis and Escherichia coli1. Biochem. J. 2014;461(2):305–314. doi: 10.1042/BJ20140082. [DOI] [PubMed] [Google Scholar]
- 66.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 67.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nielsen S.O. Nested sampling in the canonical ensemble: direct calculation of the partition function from NVT trajectories. J. Chem. Phys. 2013;139(12) doi: 10.1063/1.4821761. [DOI] [PubMed] [Google Scholar]
- 69.Yamauchi M., Mori Y., Okumura H. Molecular simulations by generalized-ensemble algorithms in isothermal-isobaric ensemble. Biophys. Rev. 2019;11(3):457–469. doi: 10.1007/s12551-019-00537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rout A.K., Barnwal R.P., Agarwal G., Chary K.V. Root-mean-square-deviation-based rapid backbone resonance assignments in proteins. Magn. Reson. Chem. 2010;48(10):793–797. doi: 10.1002/mrc.2664. [DOI] [PubMed] [Google Scholar]
- 71.Tam B., Sinha S., Wang S.M. Combining Ramachandran plot and molecular dynamics simulation for structural-based variant classification: using TP53 variants as model. Comput. Struct. Biotechnol. J. 2020;18:4033–4039. doi: 10.1016/j.csbj.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuzmanic A., Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010;98(5):861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yanao T., Koon W.S., Marsden J.E., Kevrekidis I.G. Gyration-radius dynamics in structural transitions of atomic clusters. J. Chem. Phys. 2007;126(12) doi: 10.1063/1.2710272. [DOI] [PubMed] [Google Scholar]
- 74.Ali S.A., Hassan M.I., Islam A., Ahmad F. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci. 2014;15(5):456–476. doi: 10.2174/1389203715666140327114232. [DOI] [PubMed] [Google Scholar]
- 75.Benkovic S.J., Hammes G.G., Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47(11):3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 76.Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos. Transact. A Math. Phys. Eng. Sci. 2016;374(2065):20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pirovano W., Heringa J. Protein secondary structure prediction. Methods Mol. Biol. (Clifton, NJ) 2010;609:327–348. doi: 10.1007/978-1-60327-241-4_19. [DOI] [PubMed] [Google Scholar]
- 78.Cao R., Cheng J. Protein single-model quality assessment by feature-based probability density functions. Sci. Rep. 2016;6:23990. doi: 10.1038/srep23990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bitencourt-Ferreira G., Veit-Acosta M., de Azevedo W.F., Jr. Hydrogen bonds in protein-ligand complexes. Methods Mol. Biol. (Clifton, NJ) 2019;2053:93–107. doi: 10.1007/978-1-4939-9752-7_7. [DOI] [PubMed] [Google Scholar]
- 80.Taghizadeh M., Goliaei B., Madadkar-Sobhani A. Variability of the cyclin-dependent kinase 2 flexibility without significant change in the initial conformation of the protein or its environment; a computational study. Iran. J. Biotechnol. 2016;14(2):1–12. doi: 10.15171/ijb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dass R., Mulder F.A.A., Nielsen J.T. ODiNPred: comprehensive prediction of protein order and disorder. Sci. Rep. 2020;10(1):14780. doi: 10.1038/s41598-020-71716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saikia S., Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr. Drug Targets. 2019;20(5):501–521. doi: 10.2174/1389450119666181022153016. [DOI] [PubMed] [Google Scholar]
- 83.Sulimov V.B., Kutov D.C., Sulimov A.V. Advances in docking. Curr. Med. Chem. 2019;26(42):7555–7580. doi: 10.2174/0929867325666180904115000. [DOI] [PubMed] [Google Scholar]
- 84.Leman J.K., Weitzner B.D., Lewis S.M., Adolf-Bryfogle J., Alam N., Alford R.F., et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods. 2020;17(7):665–680. doi: 10.1038/s41592-020-0848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khramushin A., Marcu O., Alam N., Shimony O., Padhorny D., Brini E., et al. Modeling beta-sheet peptide-protein interactions: Rosetta FlexPepDock in CAPRI rounds 38–45. Proteins. 2020;88(8):1037–1049. doi: 10.1002/prot.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Damborský J., Petřek M., Banáš P., Otyepka M. Wiley Online Library. 2007. Identification of tunnels in proteins, nucleic acids, inorganic materials and molecular ensembles. Report No.: 1860-6768. [DOI] [PubMed] [Google Scholar]
- 87.Marques S.M., Daniel L., Buryska T., Prokop Z., Brezovsky J., Damborsky J. Enzyme tunnels and gates as relevant targets in drug design. Med. Res. Rev. 2017;37(5):1095–1139. doi: 10.1002/med.21430. [DOI] [PubMed] [Google Scholar]
- 88.Biedermannová L., Prokop Z., Gora A., Chovancová E., Kovács M., Damborský J., et al. A single mutation in a tunnel to the active site changes the mechanism and kinetics of product release in haloalkane dehalogenase LinB. J. Biol. Chem. 2012;287(34):29062–29074. doi: 10.1074/jbc.M112.377853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B., et al. 2012. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy Burman S.S., Nance M.L., Jeliazkov J.R., Labonte J.W., Lubin J.H., Biswas N., et al. Novel sampling strategies and a coarse-grained score function for docking homomers, flexible heteromers, and oligosaccharides using Rosetta in CAPRI rounds 37–45. Proteins. 2020;88(8):973–985. doi: 10.1002/prot.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu T., Jin Z., Xiu Z., Li G. Binding free energy estimation for protein-ligand complex based on MM-PBSA with various partial charge models. Curr. Pharm. Des. 2013;19(12):2293–2307. doi: 10.2174/1381612811319120016. [DOI] [PubMed] [Google Scholar]
- 92.Kumari R., Kumar R., Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 93.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alford R.F., Leaver-Fay A., Jeliazkov J.R., O'Meara M.J., DiMaio F.P., Park H., et al. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 2017;13(6):3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hallen M.A., Martin J.W., Ojewole A., Jou J.D., Lowegard A.U., Frenkel M.S., et al. OSPREY 3.0: open-source protein redesign for you, with powerful new features. J. Comput. Chem. 2018;39(30):2494–2507. doi: 10.1002/jcc.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uversky V.N., Winter S., Löber G. Use of fluorescence decay times of 8-ANS-protein complexes to study the conformational transitions in proteins which unfold through the molten globule state. Biophys. Chem. 1996;60(3):79–88. doi: 10.1016/0301-4622(96)00009-9. [DOI] [PubMed] [Google Scholar]
- 97.Mahmoudi Gomari M., Saraygord-Afshari N., Farsimadan M., Rostami N., Aghamiri S., Farajollahi M.M. Opportunities and challenges of the tag-assisted protein purification techniques: applications in the pharmaceutical industry. Biotechnol. Adv. 2020;45 doi: 10.1016/j.biotechadv.2020.107653. [DOI] [PubMed] [Google Scholar]
- 98.Rajani H.F., Shahidi S., Gomari M.M. Protein and antibody engineering: suppressing degranulation of the mast cells and type I hypersensitivity reaction. Curr. Protein Pept. Sci. 2020;21(8):831–841. doi: 10.2174/1389203721666200511094717. [DOI] [PubMed] [Google Scholar]
- 99.Zhang Q., Xiang R., Huo S., Zhou Y., Jiang S., Wang Q., et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021;6(1):233. doi: 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Balamurugan Shanmugaraj K.S., Wangkanont Kittikhun, Phoolcharoen Waranyoo. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 101.Venkat Kumar G., Jeyanthi V., Ramakrishnan S. New Microbes and New Infections. Vol. 35. 2020. A short review on antibody therapy for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang P., Wang M., Yu J., Cerutti G., Nair M.S., Huang Y., et al. bioRxiv; 2021. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. 2021.03.01.433466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nivón L.G., Moretti R., Baker D. A pareto-optimal refinement method for protein design scaffolds. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Conway P., Tyka M.D., DiMaio F., Konerding D.E., Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23(1):47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., et al. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63(21):12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gomari M.M., Farsimadan M., Rostami N., Fadaie M., Farhani I., et al. Mahmoudi Z. CD44 polymorphisms and its variants, as an inconsistent marker in cancer investigations. Mutat. Res. Rev. Mutat. Res. 2021;787:108374. doi: 10.1016/j.mrrev.2021.108374. [DOI] [PubMed] [Google Scholar]
- 107.Siahpoosh Z., Farsimadan M., Pazhohan M., Vaziri H., Mahmoudi Gomari M. KISS1R polymorphism rs587777844 (Tyr313His) is linked to female infertility. Br. J. Biomed. Sci. 2021;78(2):98–100. doi: 10.1080/09674845.2020.1856496. [DOI] [PubMed] [Google Scholar]
- 108.Mahmoudi Gomari M., Rostami N., Ghodrati A., Hernandez Y., Fadaie M., Sadegh Eslami S., et al. Implementation of docking, molecular dynamics and free energy to investigate drug potency of novel BCR-ABLT315I inhibitors as an alternative to ponatinib. Comput. Toxicol. 2021;20 [Google Scholar]
- 109.Wen W., Chen C., Tang J., Wang C., Zhou M., Cheng Y., et al. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19:a meta-analysis. Ann. Med. 2022;54(1):516–523. doi: 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tarighi P., Eftekhari S., Chizari M., Sabernavaei M., Jafari D., Mirzabeigi P. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur. J. Pharmacol. 2021;895 doi: 10.1016/j.ejphar.2021.173890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gundogdu M., Dere G. Is systemic isotretinoin use a risk factor for coronavirus disease 2019 (COVID-19)? J. Cosmet. Dermatol. 2021;20(6):1568–1570. doi: 10.1111/jocd.14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abdelmaksoud A., Vestita M., El-Amawy H.S., Ayhan E., An İ., öztürk M, et al. Systemic isotretinoin therapy in the era of COVID-19. Dermatol. Ther. 2020;33(4) doi: 10.1111/dth.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abdelmaksoud A., Patil A., Dursun R., Temiz S.A., Ayhan E., Goldust M., et al. Could isotretinoin be a protective agent against COVID-19?: a dermatologist perspective. J. Cosmet. Dermatol. 2021;20(8):2394–2395. doi: 10.1111/jocd.14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., et al. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81(1):e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang J., Dokholyan N.V. MedusaDock 2.0: efficient and accurate protein-ligand docking with constraints. J. Chem. Inf. Model. 2019;59(6):2509–2515. doi: 10.1021/acs.jcim.8b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220–4236. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen R.E., Gorman M.J., Zhu D.Y., Carreño J.M., Yuan D., VanBlargan L.A., et al. Reduced antibody activity against SARS-CoV-2 B.1.617.2 Delta virus in serum of mRNA-vaccinated patients receiving tumor necrosis factor-α inhibitors. Med (New York, NY) 2021;2:1327–1341.e4. doi: 10.1016/j.medj.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tani-Sassa C, Iwasaki Y, Ichimura N, Nagano K, Takatsuki Y, Yuasa S, et al. Viral loads and profile of the patients infected with SARS-CoV-2 Delta, Alpha, or R.1 variants in Tokyo. Journal of Medical Virology.n/a(n/a). [DOI] [PMC free article] [PubMed]
- 121.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124–1136. doi: 10.1016/j.chom.2021.06.006. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peacock T.P., Sheppard C.M., Brown J.C., Goonawardane N., Zhou J., Whiteley M., et al. bioRxiv; 2021. The SARS-CoV-2 Variants Associated With Infections in India, B.1.617, Show Enhanced Spike Cleavage by Furin. 2021.05.28.446163. [Google Scholar]
- 123.Chen J., Zaer S., Drori P., Zamel J., Joron K., Kalisman N., et al. The structural heterogeneity of α-synuclein is governed by several distinct subpopulations with interconversion times slower than milliseconds. Structure (London, England : 1993) 2021;29(9):1048–1064. doi: 10.1016/j.str.2021.05.002. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilhelm A., Toptan T., Pallas C., Wolf T., Goetsch U., Gottschalk R., et al. Antibody-mediated neutralization of authentic SARS-CoV-2 B.1.617 variants harboring L452R and T478K/E484Q. Viruses. 2021;13(9) doi: 10.3390/v13091693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goher S.S., Ali F., Amin M. The Delta variant mutations in the receptor binding domain of SARS-CoV-2 show enhanced electrostatic interactions with the ACE2. Med. Drug Discov. 2022;13 doi: 10.1016/j.medidd.2021.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gromiha M.M., Selvaraj S. Inter-residue interactions in protein folding and stability. Prog. Biophys. Mol. Biol. 2004;86(2):235–277. doi: 10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 128.Khazanov N.A., Carlson H.A. Exploring the composition of protein-ligand binding sites on a large scale. PLoS Comput. Biol. 2013;9(11) doi: 10.1371/journal.pcbi.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y., Liu J., Johnson B.A., Xia H., Ku Z., Schindewolf C., et al. bioRxiv; 2021. Delta Spike P681R Mutation Enhances SARS-CoV-2 Fitness Over Alpha Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mohammad A., Alshawaf E., Marafie S.K., Abu-Farha M., Abubaker J., Al-Mulla F. Higher binding affinity of furin for SARS-CoV-2 spike (S) protein D614G mutant could be associated with higher SARS-CoV-2 infectivity. Int. J. Infect. Dis. 2021;103:611–616. doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheng Y.W., Chao T.L., Li C.L., Wang S.H., Kao H.C., Tsai Y.M., et al. D614G substitution of SARS-CoV-2 spike protein increases syncytium formation and virus titer via enhanced furin-mediated spike cleavage. MBio. 2021;12(4) doi: 10.1128/mBio.00587-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen C., Boorla V.S., Banerjee D., Chowdhury R., Cavener V.S., Nissly R.H., et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. Proc. Natl. Acad. Sci. U. S. A. 2021;118(42) doi: 10.1073/pnas.2106480118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cheng M.H., Krieger J.M., Banerjee A., Xiang Y., Kaynak B., Shi Y., et al. Impact of new variants on SARS-CoV-2 infectivity and neutralization: a molecular assessment of the alterations in the spike-host protein interactions. Iscience. 2022;25(3) doi: 10.1016/j.isci.2022.103939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.