Abstract
As the quality of image generation by deep learning increases, it is becoming difficult to discern its authenticity from the image alone. Currently, generative models represented by generative adversarial networks (GAN) are increasingly utilized in the research field of cardiology, and their potential risks are also being pointed out. In this context, we assessed whether expert cardiologists can detect synthesized myocardial perfusion images (MPI) generated by GAN as fake. A total of 1448 polar maps collected from consecutive patients who underwent MPI for known or suspected coronary artery disease from January 2020 to December 2021 were used for the analysis. A deep convolutional GAN was trained on the polar maps to synthesize realistic MPI. The realism of the generated images in terms of human perception was evaluated by the visual Turing test (VTT) on our original website. The average correct answer rate of the VTT was only 61.1% with a standard deviation of 21.5, but this improved to 80.0±15.8 (%) in the second trial when given the clue information. In the era of machine intelligence and virtual reality, digital literacy is becoming more necessary for healthcare professionals to identify deepfakes.
Keywords: digital literacy, deepfake, myocardial perfusion imaging, generative adversarial networks, visual turing test
Introduction
As the quality of image generation by deep learning increases, it is becoming difficult to discern its authenticity from the image alone. Currently, generative models represented by generative adversarial networks (GAN) are increasingly utilized in the medical domain [1], and their potential risks are also being pointed out [2,3]. One of the most important risks to be aware of is the possibility of misdiagnosis due to the attenuation of features in medical images [4]. Another risk includes the malicious use of artificial intelligence (AI)-based deepfake technology [5]. Recently, Thambawita and colleagues have reported that a realistic electrocardiogram could be synthesized by deepfake technologies [6]. Although the authors positively interpret their results as the end of privacy issues in medicine, the same result can be seen as the beginning of the confusion unless the generated data are distinguishable from the real objects. However, the importance of this issue does not seem to be fully recognized in the field of cardiology. In this context, the present study assessed whether expert cardiologists can detect synthesized myocardial perfusion images (MPI) generated by GAN as fake. Herein, we also introduce how we implemented a visual Turing test (VTT) as a web application.
Technical report
Image data collection
A total of 1448 polar maps collected from consecutive patients who underwent MPI for known or suspected coronary artery disease from January 2020 to December 2021 were used for the analysis. Stress/rest thallium-201 (Tl-201) myocardial perfusion scintigraphy was performed according to the American College of Cardiology/American Heart Association/American Society of Nuclear Cardiology clinical guidelines for cardiac radionuclide imaging [7]. The polar maps in stress and rest were extracted from the single-photon emission computed tomography (SPECT) studies as color images in JPEG format and rescaled to 64x64 respectively for the subsequent analysis. Details of the data collection are described in our previous literature [8].
Polar maps synthesized by generative adversarial networks
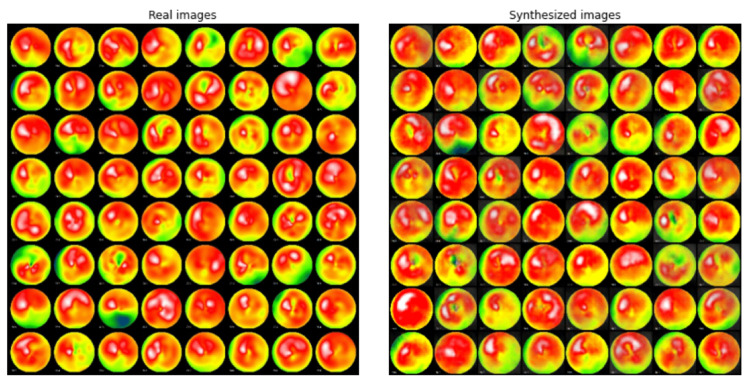
The original implementation of deep convolutional GAN (DCGAN) was trained on the collected image data [9]. In this study, both stress and rest images are used as training data. The DCGAN model was constructed referencing the “PyTorch DCGAN Tutorial (https://pytorch.org/tutorials/beginner/dcgan_faces_tutorial.html)”, but the batch normalization layers were removed from the discriminator network to regulate its performance. Figure 1 shows the example of real (true) and fake (synthesized) MPI polar maps.
Figure 1. Comparison of real and synthesized MPI polar maps.
The left panel shows examples of real polar maps obtained from patients. The right panel shows examples of synthesized polar maps by GAN. The FID between the original and generated data was computed as 100.6. The BRISQUE score of the original images and generated images were 49.9±8.0 and 32.3±7.5, respectively.
GAN: generative adversarial networks; FID: Fréchet inception distance; BRISQUE score: Blind/Referenceless Image Spatial Quality Evaluator score
Evaluation of the image generation performance
Fréchet inception distance (FID) between the original and generated data was computed as 100.6. The Blind/Referenceless Image Spatial Quality Evaluator (BRISQUE) score was significantly higher in the original images than in generated images (49.9±8.0 vs 32.3±7.5, p<0.001 by Welch's t-test).
The realism of the generated images in terms of human perception was evaluated by the VTT [10]. Nine cardiologists (15.8±11.9 years of professional experience) certified by the Japanese Society of Cardiology participated in this test through a web application (https://visual-turing-test.glitch.me/). The participants were independently presented with a total of 10 bull’s eye images from the mixed dataset and asked to tell if the presented images were real or fake (Figure 2).
Figure 2. Screenshots of the web application of VTT.
A web application was implemented on Glitch with JavaScript + HTML. On the top page, participants are presented with examples of generated and real images, as well as an explanation of the VTT methodology (panel A). Once the test starts, participants are presented with the polar maps one by one and prompted to answer whether the image is real or fake, as shown in Panel B. Each time one of the buttons is clicked, the presented image is randomly selected from a database containing a mixture of original and generated images. After a total of 10 questions, the CAR is calculated and displayed as shown in Panel C.
CAR: correct answer rate
As for the initial evaluation, the average correct answer rate (CAR) of the VTT was 61.1% with a standard deviation of 21.5, which was not significantly higher than random guess (vs 50%, p=0.16 by Welch's t-test).
After a six-month interval, the same VTT was performed on the evaluators who were informed of the characteristics of the synthesized images; they were a bit blurred in color. When the evaluators were informed of the characteristics of the synthesized image, the CAR improved to 80.0±15.8 (%), which was significantly higher than random guess (vs 50%, p<0.01 by Welch's t-test). This improvement was statistically significant (p=0.01 by paired-samples t-test).
Table 1. Results of VTT by 9 cardiologists.
The CAR of the initial trial was not significantly higher than that of a random guess. Significant improvement in CAR was observed in the second trial after a six-month interval.
CAR: correct answer rate.
| Initial trial | Second trial | P value | |
| CAR (%) | 61.1 ± 21.5 | 80.0 ± 15.8 | 0.01 |
Data availability
The minimal dataset to reproduce our study results is accessible through a public repository (https://data.mendeley.com/datasets/mjhhw3zdwv/1).
Discussion
Risks in medical applications of deep generative models can be intentional or unintentional. The former is called malicious tampering and is increasingly recognized as a potential danger in radiology [11,12]. On the other hand, unintentional modification of images, such as the latter, is also a problem that cannot be overlooked. Therefore, we believe that healthcare professionals should recognize these issues and act accordingly.
This report assessed whether skilled cardiologists can distinguish synthesized MPI polar map images using VTT, which we implemented as a web application. As a result, the average CAR of the participants was not significantly higher than a random guess. This result is in line with the recent study by Skandarani et al. reporting that imaging experts’ CAR for identifying synthesized cardiac MRI or echocardiogram was no higher than 60% [13]. Thus, at the time of writing, the AI models are successfully fooling cardiologists. Because the physicians who participated in this experiment had no expertise in AI, they were not familiar with GAN-generated images, which may have contributed to the low CAR in this study. In fact, none of the nine participants knew about GAN at the beginning of the experiment. Interestingly, the synthesized polar maps shows a lower BRISQUE score than the real polar maps, meaning that humans perceive the synthetic image more naturally. Nevertheless, the fact that average CAR increased by about 20% on the second trial suggests that there is room for improvement through educational training in their discrimination skills.
There are several limitations to our study. First, we only generated polar maps and did not attempt to generate the original slices of MPI. Second, since we employed a classical DCGAN in this study, the CAR could have been lower if we had used a modern GAN or diffusion model, which can produce higher-resolution images [14]. Third, the relatively small number of images presented to the evaluator in the VTT may have affected the CAR. In any case, caution is still required when using the generated medical images in clinical practice.
Conclusions
This report demonstrated that GAN could synthesize realistic medical images that even skilled cardiologists could not detect as fakes. On the other hand, the results of repeated VTTs suggested that participants' knowledge of the AI may improve their discriminative ability. In the era of machine intelligence and virtual reality, digital literacy is becoming more necessary for healthcare professionals to identify deepfakes.
Acknowledgments
We would thank Shinsuke Kido, Kensho Matsuda, Saki Hosokawa, Tetsuya Kosaki, Go Kawamura, and Tatsuya Shigematsu for their kind participation in the VTT. We would also like to thank Prof. Osamu Yamaguchi and Prof. Hirofumi Ochi for their support in carrying out this project.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
The authors have declared that no competing interests exist.
Human Ethics
Consent was obtained or waived by all participants in this study. The ethics committee of Ehime Prefectural Central Hospital issued approval no. 03-51. All enrolled patients provided written informed consent. This study was approved by the ethics committee of Ehime Prefectural Central Hospital (no. 03-51).
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.Generative adversarial network in medical imaging: A review. Yi X, Walia E, Babyn P. Med Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 2.Concerns in the use of adversarial learning for image synthesis in cardiovascular intervention. Higaki A, Miyoshi T, Yamaguchi O. Eur Heart J Digit Health. 2021;2:556. doi: 10.1093/ehjdh/ztab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Translational challenges for synthetic imaging in cardiology. Olender ML, Nezami FR, Athanasiou LS, de la Torre Hernández JM, Edelman ER. Eur Heart J Digit Health. 2021;2:559–560. doi: 10.1093/ehjdh/ztab079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen JP, Luck M, Honari S. Medical Image Computing and Computer Assisted Intervention - MICCAI 2018. Cham: Springer; 2018. Distribution matching losses can hallucinate features in medical image translation; pp. 529–536. [Google Scholar]
- 5.Intelligent deep detection method for malicious tampering of cancer imagery. Khattab M, Alheeti A, Alzahrani A, Khoshnaw N, Al-Dosary D. 2022 7th International Conference on Data Science and Machine Learning Applications (CDMA) 2022:25–28. [Google Scholar]
- 6.DeepFake electrocardiograms using generative adversarial networks are the beginning of the end for privacy issues in medicine. Thambawita V, Isaksen JL, Hicks SA, et al. Sci Rep. 2021;11:21896. doi: 10.1038/s41598-021-01295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ACC/AHA/ASNC guidelines for the clinical use of cardiac radionuclide imaging--executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (ACC/AHA/ASNC Committee to Revise the 1995 Guidelines for the Clinical Use of Cardiac Radionuclide Imaging) Klocke FJ, Baird MG, Lorell BH, et al. J Am Coll Cardiol. 2003;42:1318–1333. doi: 10.1016/j.jacc.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Content-based image retrieval for the diagnosis of myocardial perfusion imaging using a deep convolutional autoencoder. Higaki A, Kawaguchi N, Kurokawa T, et al. J Nucl Cardiol. 2022 doi: 10.1007/s12350-022-03030-4. [DOI] [PubMed] [Google Scholar]
- 9.Unsupervised representation learning with deep convolutional generative adversarial networks, 4th International Conference on Learning Representations, ICLR 2016 - Conference Track Proceedings. Radford A, Metz L, Chintala S. arXiv. 2016 [Google Scholar]
- 10.Visual Turing test for computer vision systems. Geman D, Geman S, Hallonquist N, Younes L. Proc Natl Acad Sci U S A. 2015;112:3618–3623. doi: 10.1073/pnas.1422953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The potential dangers of artificial intelligence for radiology and radiologists. Chu LC, Anandkumar A, Shin HC, Fishman EK. J Am Coll Radiol. 2020;17:1309–1311. doi: 10.1016/j.jacr.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A review of malicious altering healthcare imagery using artificial intelligence. Hussain F, Ksantini R, Hammad M. 2021 International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT) 2021:646–651. [Google Scholar]
- 13.Generative adversarial networks in cardiology. Skandarani Y, Lalande A, Afilalo J, Jodoin PM. Can J Cardiol. 2022;38:196–203. doi: 10.1016/j.cjca.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Ho J, Jain A, Abbeel P. Advances in Neural Information Processing Systems 33 (NeurIPS 2020) NeurIPS 2020; 2020. Denoising diffusion probabilistic models; pp. 6840–6851. [Google Scholar]