Abstract
Pediatric SARS-CoV-2 vaccines are needed that elicit immunity directly in the airways as well as systemically. Building on pediatric parainfluenza virus vaccines in clinical development, we generated a live-attenuated parainfluenza-virus-vectored vaccine candidate expressing SARS-CoV-2 prefusion-stabilized spike (S) protein (B/HPIV3/S-6P) and evaluated its immunogenicity and protective efficacy in rhesus macaques. A single intranasal/intratracheal dose of B/HPIV3/S-6P induced strong S-specific airway mucosal immunoglobulin A (IgA) and IgG responses. High levels of S-specific antibodies were also induced in serum, which efficiently neutralized SARS-CoV-2 variants of concern of alpha, beta, and delta lineages, while their ability to neutralize Omicron sub-lineages was lower. Furthermore, B/HPIV3/S-6P induced robust systemic and pulmonary S-specific CD4+ and CD8+ T cell responses, including tissue-resident memory cells in the lungs. Following challenge, SARS-CoV-2 replication was undetectable in airways and lung tissues of immunized macaques. B/HPIV3/S-6P will be evaluated clinically as pediatric intranasal SARS-CoV-2/parainfluenza virus type 3 vaccine.
Keywords: intranasal vaccine, live-attenuated viral vector, parainfluenza virus, pediatric, SARS-CoV-2, nasal spray vaccine, mucosal immmunization, pediatric COVID vaccine, next-generation COVID vaccine
Graphical abstract
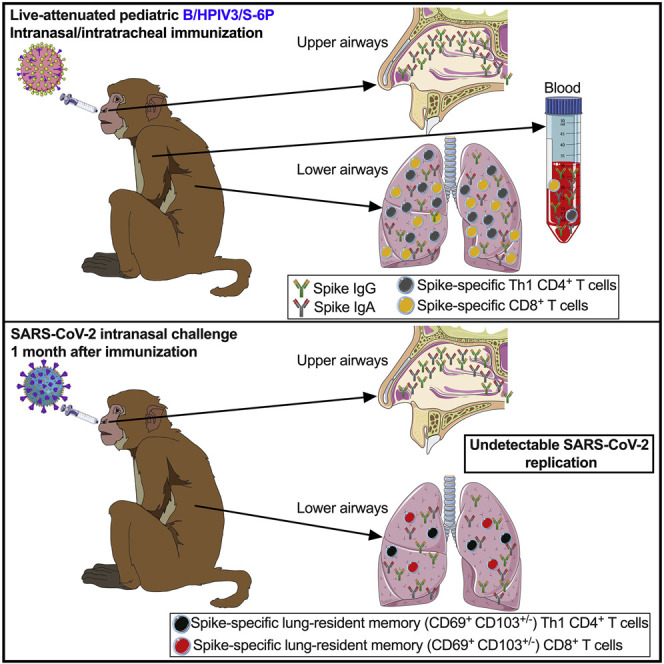
Use of an attenuated parainfluenza virus vector allows for effective vaccination against SARS-CoV-2 via the airways in young macaques, suggesting a promising approach for needle-free pediatric vaccination of humans against COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects and causes disease in all age groups. Although COVID-19 is generally milder in young children than in adults, thousands of children have been hospitalized due to COVID-19 in the United States alone, including about one-third without preexisting medical conditions.1 , 2 More than 900 children 0–11 years of age have died from COVID-19 in the United States (https://covid.cdc.gov/covid-data-tracker/#demographics, accessed on October 4, 2022), and during the fall/winter COVID-19 wave of 2021/2022, children accounted for over 25% of US cases.3 In rare cases (∼0.03% of infected children), COVID-19 can cause a multisystem inflammatory syndrome in children (MIS-C),4 , 5, arising within about 4 weeks after SARS-CoV-2 exposure. mRNA-based vaccines are available for children 6 months of age and older. One limitation of the current mRNA and other injectable SARS-CoV-2 vaccines is that they do not directly stimulate immunity in the respiratory tract, the major site of SARS-CoV-2 entry, replication, disease, and egress.6 Therefore, it is important to evaluate additional vaccine approaches that are suitable for pediatric use and stimulate both mucosal and systemic immunity. Ideally, a vaccine should be effective at a single dose and could be administered topically to the respiratory tract to induce robust systemic and respiratory mucosal immunity that restricts SARS-CoV-2 infection and shedding.
Here, we describe the preclinical evaluation of the safety, immunogenicity, and protective efficacy in rhesus macaques of a live intranasal SARS-CoV-2 vaccine candidate, bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the SARS-CoV-2 spike (S) protein trimer stabilized in its prefusion form with 6 proline substitutions (B/HPIV3/S-6P). The B/HPIV3 vector originally was developed as a pediatric vaccine candidate against HPIV3, a single-stranded negative-sense RNA virus, which is an important cause of respiratory illness, especially in infants and young children under 5 years of age.7 , 8 Previously, B/HPIV3 was well tolerated in a phase 1 study in infants and young children.9 B/HPIV3 also has been used to express the fusion (F) glycoprotein of another human respiratory pathogen, human respiratory syncytial virus (RSV), providing a bivalent HPIV3/RSV vaccine candidate, which was well tolerated in children >2 months of age10(Clinicaltrials.gov NCT00686075). We recently reported that B/HPIV3 expressing a stabilized prefusion form of S efficiently protected hamsters against infection with a vaccine antigen-matched SARS-CoV-2 isolate, preventing weight loss, lung inflammation, and efficiently reducing SARS-CoV-2 replication in the upper and lower respiratory tract.11 In this study, we evaluated the safety and efficacy of a single intranasal/intratracheal (IN/IT) dose of B/HPIV3/S-6P in rhesus macaques. Immunogenicity evaluations included S-specific mucosal and systemic antibody and T cell responses as well as neutralizing antibody responses to the vaccine-matched SARS-CoV-2 strain and major variants of concern (VoCs). In addition, we assessed the protective efficacy of B/HPIV3/S-6P against SARS-CoV-2 challenge prior to advancing this candidate to a phase 1 clinical study.
Results
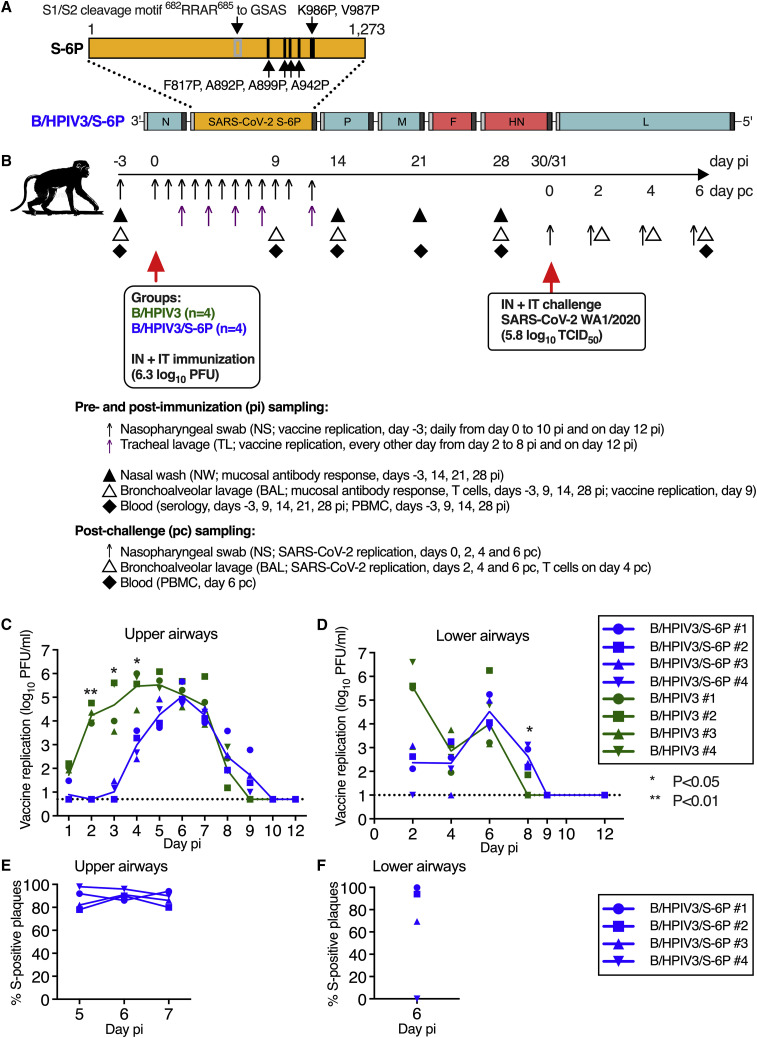
B/HPIV3/S-6P replicates in the upper and lower airways of rhesus macaques
We used B/HPIV3 to express the S-6P prefusion-stabilized full-length version of the SARS-CoV-2 S protein. B/HPIV3 is a cDNA-derived version of bovine PIV3 (BPIV3) strain Kansas in which the BPIV3 hemagglutinin-neuraminidase (HN) and fusion (F) glycoproteins (the two PIV3 neutralization antigens) have been replaced by those of the HPIV3 strain JS9 , 12 (Figure 1 A). The bovine PIV3 backbone provides stable attenuation due to host-range restriction in humans.9 , 10 The S-6P version of the S-protein contains 6 proline substitutions13 that stabilize S in its trimeric prefusion form and increase expression and immunogenicity. The S1/S2 polybasic furin cleavage motif (amino acids [aa] “RRAR”) was ablated by aa substitutions (RRAR-to-GSAS)14 (Figure 1A), rendering S-6P non-functional for virus entry, which eliminates the possibility of S altering the tissue tropism of the B/HPIV3 vector.
Figure 1.
Genome organization of B/HPIV3/S-6P; timeline of the rhesus macaque study, vaccine replication following intranasal/intratracheal immunization of rhesus macaques
(A) Diagram of the B/HPIV3/S-6P genome, with BPIV3 (N, P, M, and L; blue) and HPIV3 genes (F and HN; red). The full-length SARS-CoV-2 S ORF (codons 1–1,273) was codon optimized and inserted as additional gene (orange) between the N and P ORFs. The S sequence includes 6 stabilizing proline substitutions (S-6P) and RRAR-to-GSAS substitutions to ablate the S1/S2 cleavage site. Each gene begins and ends with PIV3 gene-start and gene-end transcription signals (light and dark bars).
(B) Experimental timeline for the immunization of groups of 4 macaques with the B/HPIV3/S-6P vaccine candidate or the empty B/HPIV3 vector used as a control. Challenge with the SARS-CoV-2 WA1/2020 isolate was performed on day 30 or 31 post-immunization. Pre- and post-challenge sampling schedules are summarized; pi, post-immunization; pc, post challenge. Details are described in STAR Methods.
(C and D) Replication of B/HPIV3/S-6P and B/HPIV3 in upper (C) and lower (D) airways of rhesus macaques. Two groups of 4 macaques were immunized intranasally and intratracheally with 6.3 log10 PFU of B/HPIV3/S-6P (blue) or B/HPIV3 (green). Nasopharyngeal swabs (upper airways; daily from days 0 to 10 and on day 12 pi), tracheal lavages (lower airways; every other day from days 2 to 8 and on day 12 pi), and bronchoalveolar lavages (lower airways; day 9 pi) were performed as described in (B). Vaccine virus titers were determined by immunoplaque assay (STAR Methods); expressed as log10 PFU/mL (limit of detection: 0.7 log10 PFU/mL for nasopharyngeal swabs; 1 log10 PFU/mL for tracheal lavages [dotted line]).
(E and F) The stability of S expression by B/HPIV3/S-6P in macaques was evaluated by dual-staining immunoplaque assay on Vero cells from nasopharyngeal swab (E) and tracheal lavage (F) samples collected at the peak of vaccine shedding (days 5 through 7). Plaques were immunostained with an HPIV3-specific rabbit hyperimmune serum to detect B/HPIV3 antigens, and a goat hyperimmune serum to the secreted SARS-CoV-2 S to detect co-expression of the S protein, followed by infrared-dye secondary antibodies. Fluorescent staining for PIV3 proteins and SARS-CoV-2 S was visualized in green and red, respectively, generating yellow plaques when merged. The percentage of yellow plaques expressing both HPIV3 and S proteins was determined. Each macaque is indicated by a symbol; lines represent medians (∗p 0.05, ∗∗p 0.01; two-way ANOVA, Sidak multiple comparison test).
To evaluate vaccine replication and immunogenicity, we immunized 2 groups of macaques (n = 4 per group) with a single dose of 6.3 log10 plaque-forming units (PFUs) of B/HPIV3/S-6P or the B/HPIV3 vector control, respectively, administered by the combined intranasal and intratracheal route (IN/IT) (Figure 1B). Replication of B/HPIV3/S-6P and the B/HPIV3 control was detectable through days 8 or 9 post-immunization (pi) in the upper and lower airways (UAs and LAs) (Figures 1C and 1D). In the UA, peak replication of B/HPIV3/S-6P and B/HPIV3 control was detected between study days 4 and 6 (medians independent of study day: 4.9 versus 5.9 log10 PFU/mL, respectively; p = 0.1429 by two-tailed Mann-Whitney test); replication of B/HPIV3/S-6P was delayed by 1–2 days compared with that of the empty vector (p < 0.01 on day 2; p < 0.05 on days 3 and 4) (Figure 1C). In the LA, B/HPIV3/S-6P replicated with similar kinetics to B/HPIV3, reaching median peak titers of 4.5 and 4.0 log10 PFU/mL, respectively, on day 6 pi (Figure 1D).
To evaluate the stability of S expression during vector replication, nasopharyngeal swab (NS) and tracheal lavage (TL) specimens positive for B/HPIV3/S-6P were evaluated by a dual-staining immunoplaque assay, which detects the expression of S and vector proteins. On average, 89% of the B/HPIV3/S-6P plaques recovered between days 5 and 7 from NS were positive for S expression (Figures 1E and 1F), suggesting stable S-6P expression in the UA. In TL specimens collected on day 6 pi, S expression was stable in 3 of 4 macaques, with on average 88% of the plaques positive for S expression. In TL samples from one B/HPIV3/S-6P-immunized macaque (B/HPIV3/S-6P #4), plaques were negative for S expression on day 6 pi. Sanger sequencing of the S gene revealed 13 cytidine-to-thymidine mutations in a 430-nucleotide region, suggestive of deaminase activity in the LA of this animal. Eleven were missense mutations resulting in amino acid substitutions, including 7 proline substitutions that might affect S protein folding.
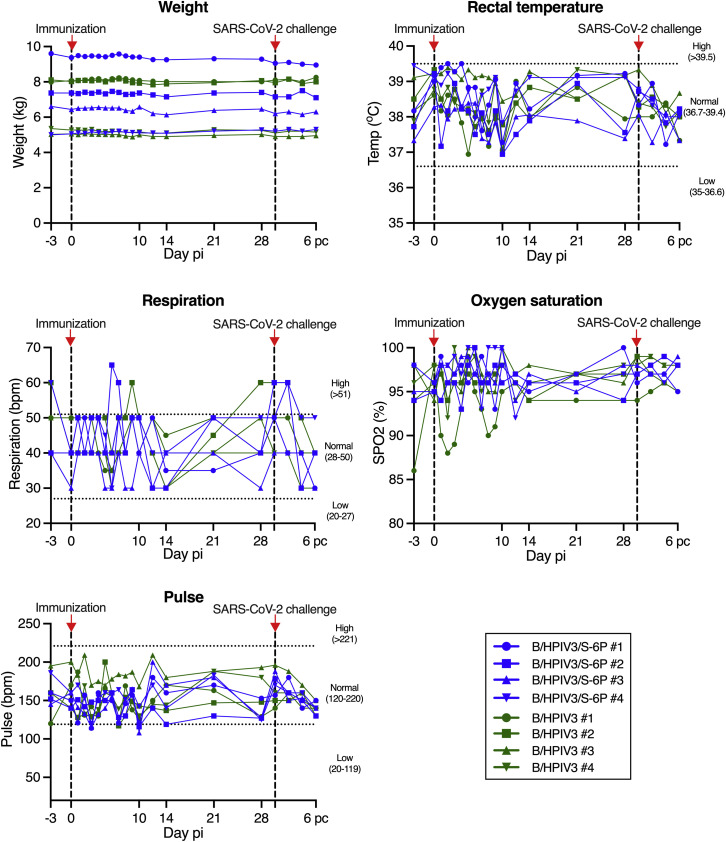
No changes in body weight, rectal temperature, respiration, oxygen saturation, or pulse were detected following immunization of macaques with B/HPIV3 or B/HPIV3/S-6P (Figure S1 ). Thus, B/HPIV3/S-6P replicates efficiently in the UA and LA of macaques, causes no apparent symptoms, and is cleared in approximately 10 days.
Figure S1.
Vital signs of macaques after immunization with B/HPIV3 or B/HPIV3/S-6P and SARS-CoV-2 challenge, related to Figures 1B–1D
Macaques in groups of 4 were immunized with B/HPIV3/S-6P or with B/HPIV3 (empty vector control). On day 30 post-immunization (pi), animals were challenged in a BSL3 facility with the SARS-CoV-2 WA1/2020 isolate. Animals were euthanized on day 36 pi (day 6 post challenge). The body weight, rectal temperature, respiration rate, oxygen saturation rate, and heart rate were monitored on the indicated day pi. Timing of immunization and SARS-CoV-2 challenge is indicated by dashed lines and red arrows. pi, post-immunization; pc, post challenge. B/HPIV3/S-6P-immunized animals are represented in blue, and B/HPIV3-immunized animals are represented in green. Each animal is represented by a unique symbol.
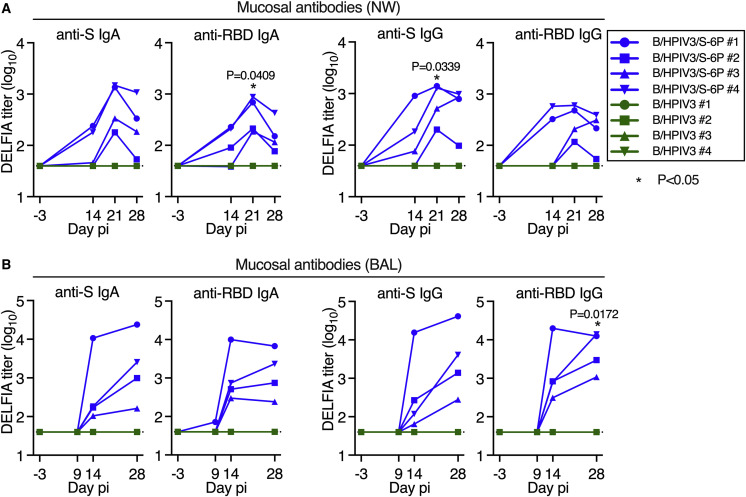
B/HPIV3/S-6P induces anti-SARS-CoV-2 S mucosal antibodies in the upper and lower airways
To assess the kinetics of airway mucosal antibody responses to the SARS-CoV-2 S protein in the UA and LA, we collected nasal washes (NWs) 3 days before immunization and on days 14, 21, and 28 after immunization and bronchoalveolar lavage (BAL) fluid on days 9, 14, and 28 pi (Figure 1B). In B/HPIV3/S-6P-immunized animals, we detected mucosal anti-S (2/4 animals) and anti-receptor binding domain (RBD) IgA (3/4 animals) in the UA as early as 14 days pi (Figure 2 A). By day 21 pi, all 4 B/HPIV3/S-6P immunized macaques exhibited anti-S and anti-RBD IgA (geometric mean titers [GMTs] between 2.3 and 3.2 log10, p = 0.0409 for anti-RBD IgA on day 21 pi). B/HPIV3/S-6P also induced mucosal anti-S and anti-RBD IgG responses in the UA on day 14 pi in 3/4 and 2/4 macaques, respectively, with responses observed in all animals by day 21 (anti-S titers between 2.1 and 3.1 log10, p = 0.0339).
Figure 2.
Intranasal/intratracheal immunization with B/HPIV3/S-6P induces mucosal antibody responses to SARS-CoV-2 S in the upper and lower airways
Rhesus macaques (n = 4 per group) were immunized with B/HPIV3/S-6P or B/HPIV3 (control) by the intranasal/intratracheal route (Figure 1B). To determine the mucosal antibody response in the upper airways, nasal washes (NWs) were performed before immunization and on days 14, 21, and 28. To analyze the antibody response in the lower airways, bronchoalveolar lavages (BALs) were collected before immunization and on days 9, 14, and 28 pi.
(A and B) S- and receptor binding domain (RBD)-specific mucosal IgA and IgG titers on indicated days pi in the upper (A) and lower (B) airways, determined by time-resolved dissociation-enhance lanthanide fluorescence immunoassay (DELFIA-TRF). Endpoint titers are expressed in log10 for mucosal IgA and IgG to a secreted prefusion-stabilized form (aa 1–1,208; S-2P14) of the S protein (left panels) or to a fragment of the S protein (aa 328–531) containing SARS-CoV-2 RBD (right panels). The limit of detection is 1.6 log10 (dotted line). B/HPIV3/S-6P-immunized macaques are shown in blue, while B/HPIV3-immunized macaques are in green, with each macaque represented by a symbol. ∗p 0.05 (two-way ANOVA, Sidak multiple comparison test).
B/HPIV3/S-6P also induced mucosal anti-S and anti-RBD IgA and IgG in the LA (Figure 2B). On day 14 pi, strong anti-S and anti-RBD IgA and IgG responses were detectable in the LA of all 4 B/HPIV3/S-6P-immunized macaques. Anti-S IgA and IgG titers continued to rise between days 14 and 28 pi. As expected, none of the macaques immunized with the empty B/HPIV3 vector had detectable anti-S or anti-RBD IgA or IgG antibodies.
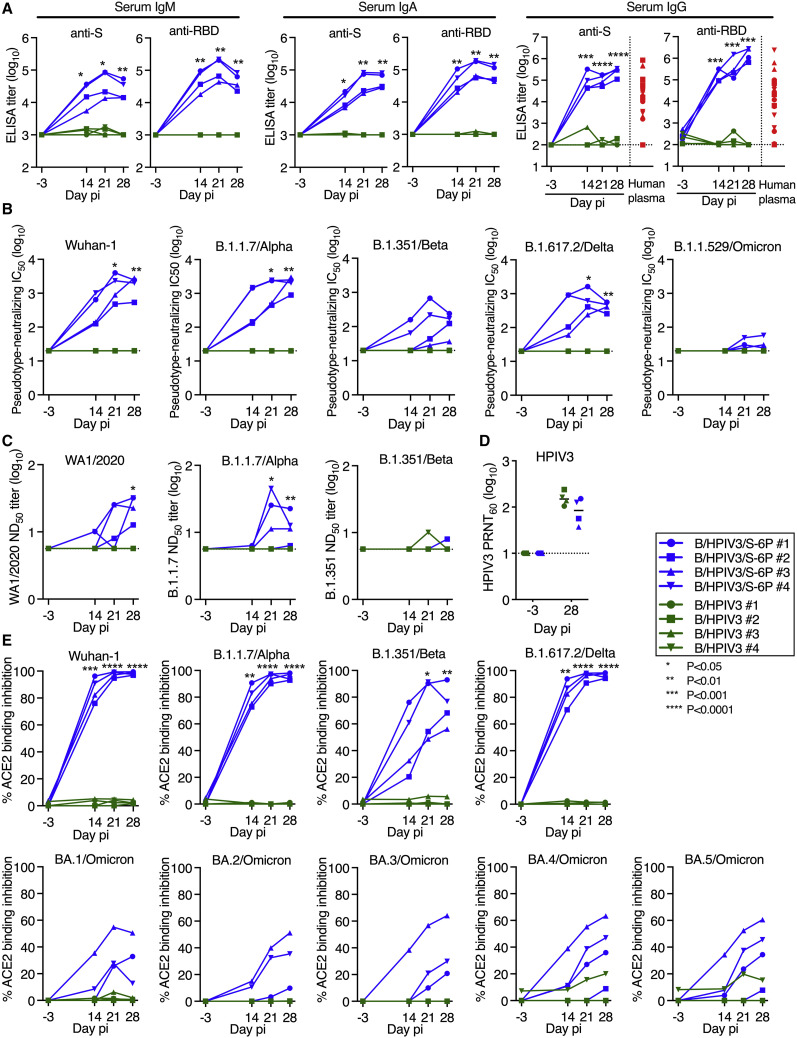
B/HPIV3/S-6P induces serum antibodies against SARS-CoV-2 S that neutralize SARS-CoV-2 WA1/2020 and VoCs
We next assessed the kinetics and breadth of the serum antibody response to B/HPIV3/S-6P (Figure 3 ). We detected robust serum IgM, IgA, and IgG binding antibody responses to the S protein and RBD by ELISAs in 4/4 B/HPIV3/S-6P-immunized macaques as early as 14 days pi (Figure 3A). Serum anti-S and anti-RBD IgM and IgA titers peaked on day 21 pi; in two macaques, anti-S IgA titers continued to rise until day 28 pi. High levels of serum anti-S and anti-RBD IgG were detected in all B/HPIV3/S-6P-immunized macaques as early as day 14 pi, continuing to rise in all macaques until day 28 pi. These levels of anti-S and anti-RBD IgG antibodies were 16- and 180-fold higher than the mean anti-S and anti-RBD IgG titers, respectively, detected in the plasma obtained from 23 SARS-CoV-2-convalescent humans. As expected, macaques immunized with empty B/HPIV3 control had no-serum anti-S or anti-RBD IgM, IgA, or IgG antibodies detectable at any time.
Figure 3.
B/HPIV3/S-6P induces serum binding antibody responses to SARS-CoV-2 S and neutralizing antibody responses to VoCs in macaques
Sera were collected from macaques before immunization and on days 14, 21, and 28 pi.
(A) Endpoint ELISA titers of serum IgM, IgA, and IgG to S-2P (left panels) or RBD (right panels), expressed in log10. Twenty-three plasma samples from COVID-19 convalescent individuals were evaluated in parallel for IgG to S-2P or the RBD (red symbols). The limits of detection are 3 log10 for IgM and IgA and 2.0 log10 for IgG.
(B) Serum neutralizing titers to pseudoviruses bearing spike proteins from SARS-CoV-2 Wuhan-1 (matching S-6P),14 B.1.1.7/Alpha, B.1.351/Beta, B.1.617.2/Delta, or B.1.1.529/Omicron. The 50% inhibitory concentration (IC50) titers of sera were determined. The detection limit is 1.3 log10.
(C) The 50% SARS-CoV-2 serum neutralizing titers (ND50) were determined on Vero E6 cells against vaccine-matched WA1/2020 or viruses from lineages B.1.1.7/Alpha or B.1.351/Beta. The limit of detection is 0.75 log10.
(D) Serum HPIV3 neutralizing antibody titers pi, determined by 60% plaque reduction neutralization test (PRNT60). The detection limit is 1 log10.
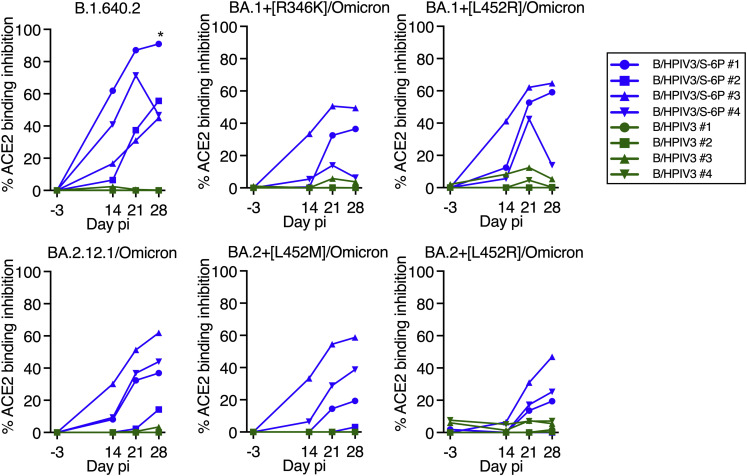
(E) Percentage of ACE2 binding inhibition to SARS-CoV-2 S proteins from Wuhan-1, B.1.1.7/Alpha, B.1.351/Beta, B.1.617.2/Delta, or indicated Omicron variants by serum antibodies from immunized macaques, relative to no-serum controls. ACE2 binding inhibition to additional S proteins is shown in Figure S2. Each macaque is represented by a symbol. ∗p 0.05, ∗∗p 0.01, ∗∗∗p 0.001, ∗∗∗∗p 0.0001, two-way ANOVA, Sidak multiple comparison test.
We also evaluated the kinetics and breadth of the serum neutralizing antibody response to vaccine-matched SARS-CoV-2 and to 4 VoCs (B.1.1.7/Alpha, B.1.351/Beta, B.1.617.2/Delta, and B.1.1.529/Omicron BA.1 sublineage) using a lentivirus-based pseudotype neutralization assay15 (Figure 3B). The sera efficiently and similarly neutralized lentivirus pseudotyped with vaccine-matched Wuhan-1 S protein (IC50 on day 28 between 2.7 and 3.5 log10) or with S from the B.1.1.7/Alpha lineage (IC50 between 3.0 and 3.5 log10). The sera also neutralized the B.1.351/Beta S-pseudotyped lentivirus although the titer was reduced compared with the vaccine match (IC50 between 1.6 and 2.4 log10). Day 14 sera from all 4 macaques efficiently neutralized the B.1.617.2/Delta S-pseudotyped lentivirus; titers further increased but, on day 28, were about 5-fold reduced compared with the vaccine match (IC50 between 2.4 and 2.8 log10). A low neutralizing activity against B.1.1.529/Omicron BA.1 was detected in day 28 sera from 3 of 4 macaques (IC50 between 1.4 and 1.8 log10) that was 59-fold reduced compared with the vaccine match.
The serum neutralizing antibody titers were also assessed by a live-virus SARS-CoV-2 neutralization assay using the vaccine-matched WA1/2020 isolate or an isolate of the Alpha or Beta lineages (Figure 3C). Results were overall comparable with those of the pseudotyped lentivirus neutralization assays although, as expected, the sensitivity and the dynamic range of the live-virus neutralization assays were lower than those of the pseudotype neutralization assays.
Finally, we evaluated the ability of serum antibodies to inhibit binding of soluble angiotensin-converting enzyme 2 (ACE2) to recombinant SARS-CoV-2 S proteins of several lineages (Figures 3E and S2 ). This sensitive ACE2 binding inhibition assay serves as an alternative to BSL3 SARS-CoV-2 neutralization assays. At a 1:20 dilution, serum antibodies from B/HPIV3/S-6P-immunized macaques completely inhibited ACE2 binding to the vaccine-matched S protein of the ancestral Wuhan-1 isolate and to S proteins of VoCs B.1.1.7/Alpha and B.1.617.2/Delta and less completely to VoC B.1.351/Beta and the B.1.640.2 variant (Figures 3E and S2), while binding inhibition to S proteins of Omicron BA.1, BA.2, BA.3, BA.4, and BA.5 derivatives was lower (Figures 3E and S2).
Figure S2.
ACE2 binding inhibition to SARS-CoV-2 spike proteins by the sera of immunized macaques, related to Figure 3E
The neutralizing activity of each serum to inhibit ACE2 binding to S from the variant under monitoring B.1.640.2 Omicron BA.2.12.1 or Omicron BA.1 and BA.2 subvariants that contain the indicated additional mutation is represented as percent inhibition relative to no-serum controls. pi, post-immunization. B/HPIV3/S-6P-immunized animals are represented in blue, and B/HPIV3-immunized animals are represented in green. Each animal is represented by a unique symbol.
As expected, neutralizing antibodies against the various SARS-CoV-2 lineages were undetectable in sera from B/HPIV3-control immunized macaques (except for the serum from one B/HPIV3-immunized macaque that induced background inhibition of ACE2 binding to BA.4 and BA.5/Omicron S; Figure 3E); additionally, all 8 macaques developed neutralizing serum antibodies against the HPIV3 vector (60% plaque reduction neutralization test [PRNT60] titers between 1.6 and 2.4 log10; Figure 3D).
B/HPIV3/S-6P immunization induces high frequencies of SARS-CoV-2 S-specific CD4+ and CD8+ T cells in the blood and the airways
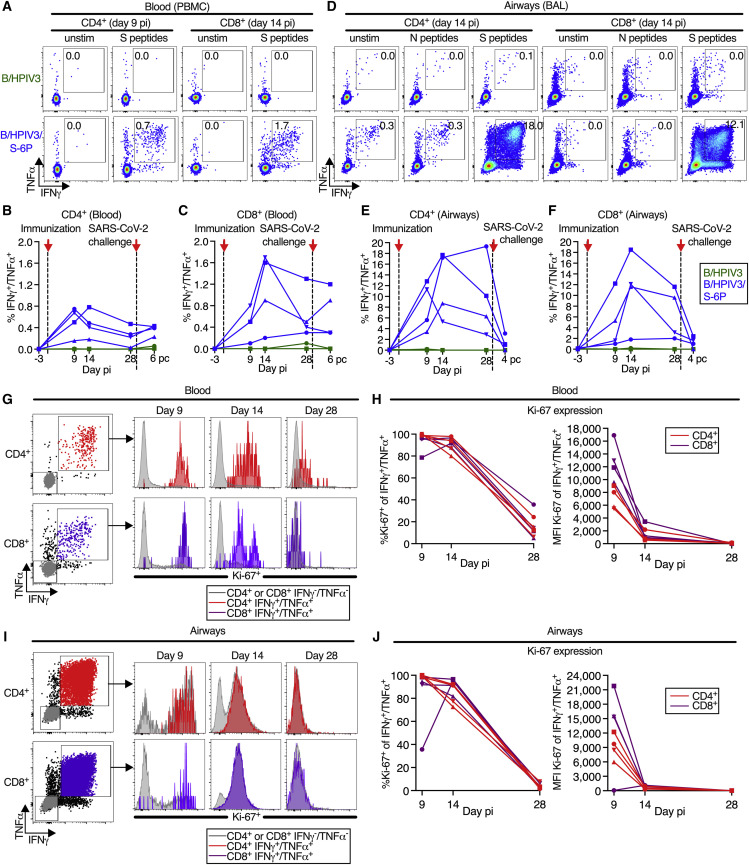
SARS-CoV-2 S-specific CD4+ and CD8+ T cell responses were evaluated using peripheral blood mononuclear cells (PBMCs) and cells recovered from the LA by BAL (see Figure S3 for the gating strategy) at the indicated time points following immunization with B/HPIV3/S-6P (Figures 4 , 5 , and Figure S4, Figure S5, Figure S6 ) and challenge with SARS-CoV-2 (Figures 4 and S7 ). SARS-CoV-2 S and N-specific CD4+ and CD8+ T cells were mostly identified as IFNγ+/TNFα+ double-positive cells after stimulation with pools of overlapping 15-mer peptides covering the entire length of these proteins. S-specific CD4+ T cells were present in the blood of all B/HPIV3/S-6P-immunized macaques by day 9 pi (Figure 4A, left panels; kinetics are shown in Figure 4B); frequencies peaked on day 9 (2 macaques) or day 14 pi (2 macaques; average peak of S-specific CD4+ T cells, irrespective of study day: 0.6%) and then steadily declined until day 28 pi. S-specific CD8+ T cells were also detectable in the blood of B/HPIV3/S-6P-immunized macaques on day 9 pi (Figures 4A, right panels and 4C), and their frequencies peaked on day 14 pi in 3 of 4 macaques (Figure 4C; average peak of S-specific CD8+ T cells irrespective of the study day: 1.1%).
Figure S3.
Gating strategy of the CD4+ and CD8+ T cells isolated from BAL of macaques, related to Figures 4 and 5
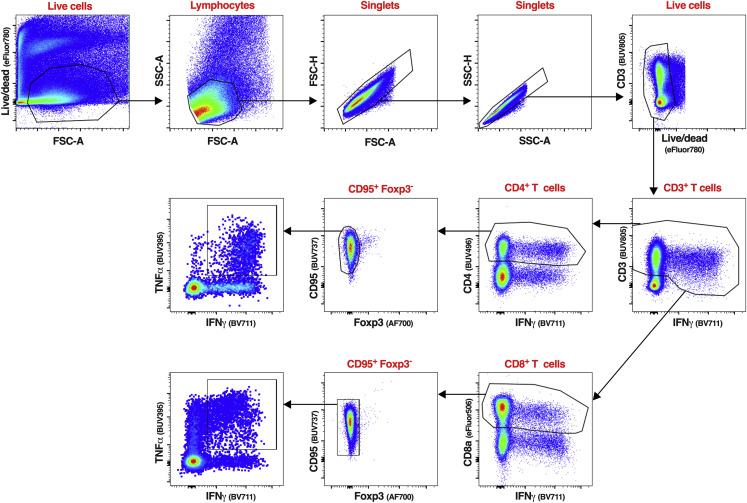
Representative flow cytometry dot plots of cells isolated from a BAL sample, visualizing the typical gating strategy used to identify the CD4+ and CD8+ T cell populations described in Figures 4, 5, S4, S5, and S7. The same gating strategy was applied to identify and analyze the CD4+ and CD8+ T cells from PBMC isolated from the blood (Figures 4, S6, and S7). Live cells were first gated based on a live/dead staining and forward scatter area. Live lymphocytes were identified based on forward and side scatter areas. Then, singlets were selected using a first gate based on forward scatter height and forward scatter area followed by a second gate based on side scatter height and side scatter area. An additional live/dead gating was performed to discard any remaining dead cells. The live single CD3+ IFNγ+ T cells were next gated using CD3 and IFNγ. As CD3 expression can be downregulated on activated T cells, a wide CD3 gate was applied. IFNγ+ CD4+ or CD8+ T cells were next identified using a CD4 or CD8 antibody. Non-naive, non-regulatory CD4+ or CD8+ T cells were finally gated using CD95 and Foxp3, respectively. The phenotypic analyses described in Figures 4, 5, and Figure S4, Figure S5, Figure S6, Figure S7 were performed on live single CD3+ CD4+ CD95+ Foxp3− or live single CD3+ CD8+ CD95+ Foxp3− T cells.
Figure 4.
Intranasal/intratracheal immunization with B/HPIV3/S-6P induces S-specific CD4+ and CD8+ T cell responses in blood and lower airways
(A–F) Frequencies of S-specific CD4+ and CD8+ T cells from the blood (A–C) or BAL (D–F). Mononuclear cells collected on indicated days pi were stimulated with overlapping SARS-CoV-2 S or (BAL only) N peptides or left unstimulated and processed for flow cytometry. Phenotypic analyses were performed on non-naive non-regulatory (CD95+/Foxp3−) CD4+ or CD8+ T cells (see Figure S3 for gating); frequencies are relative to that population.
(A and D) Dot plots showing IFNγ and TNFα expression by CD4+ or CD8+ T cells from blood (A) or BAL (D) of representative B/HPIV3 (top) or B/HPIV3/S-6P-immunized (bottom) macaques.
(B, C, E, and F) Background-corrected frequencies of S-specific IFNγ+/TNFα+ CD4+ (B and E) or CD8+ (C and F) T cells from blood (B and C) or BAL (E and F) on indicated days.
(G–J) Expression of proliferation marker Ki-67 by IFNγ+/TNFα+ CD4+ (red) or CD8+ (purple) T cells from the blood (G and H) or BAL (I and J) of B/HPIV3/S-6P-immunized macaques (n = 4, each represented by different symbols). IFNγ−/TNFα− cells in gray. (G and I) Gating and histograms showing Ki-67 expression. (H and J) Frequency and median fluorescence intensity (MFI) of Ki-67 in IFNγ+/TNFα+ T cells from the blood (H) and BAL (J). BAL, bronchoalveolar lavage.
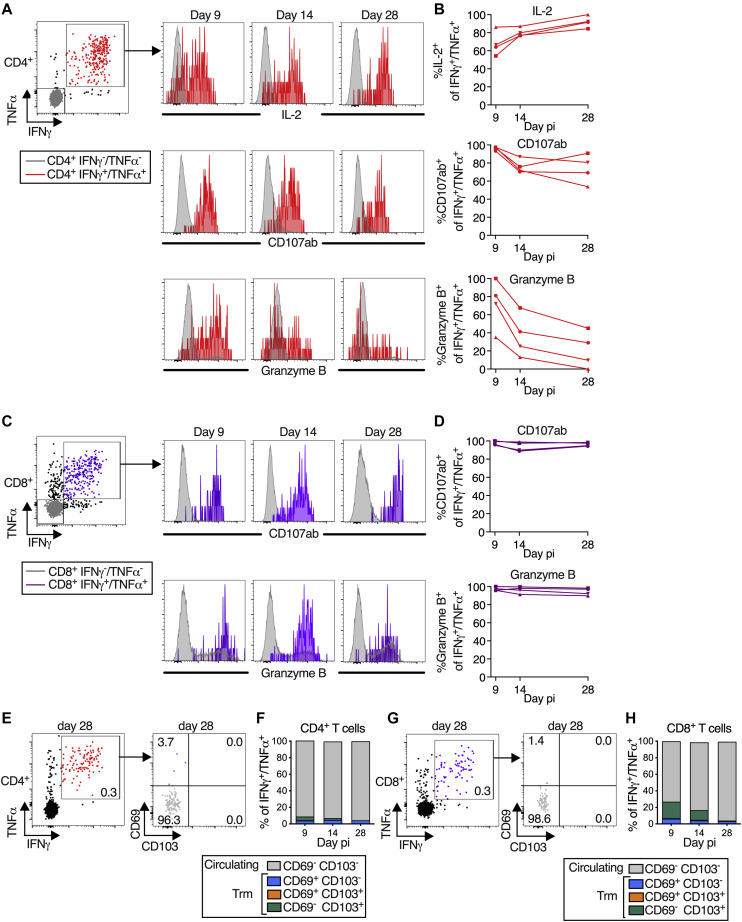
Figure 5.
B/HPIV3/S-6P-elicited S-specific CD4+ and CD8+ T cells in lower airways (LAs) transition to tissue-resident memory phenotype
T cells obtained by bronchoalveolar lavage (BAL) were stimulated with overlapping S peptides prior to flow cytometry analysis.
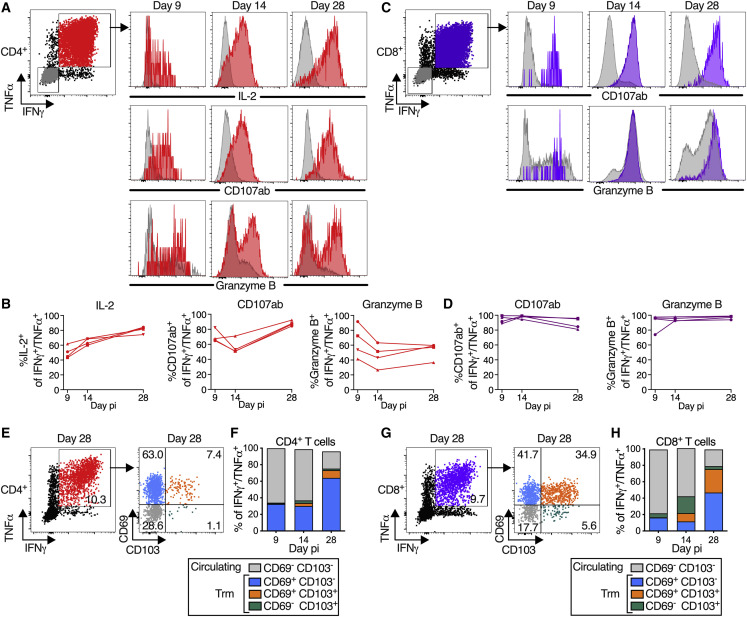
(A–D) S-specific T cells in LA are functional. (A and C) Representative dot plots showing gating on S-specific IFNγ+/TNFα+ CD4+ (red), CD8+ (purple), and IFNγ−/TNFα− (gray) T cells (for complete gating, see Figure S3); histograms showing expression of IL-2 (CD4+ T cells only), CD107ab and granzyme B by IFNγ+/TNFα+ T cells collected on indicated days pi. (B and D) Frequencies of IL-2+, CD107ab+, and granzyme B+ of IFNγ+/TNFα+ S-specific CD4+ (B) or CD8+ (D) T cells from 4 BHPIV3/S-6P-immunized macaques on indicated days.
(E–H) Transition to memory phenotype. (E and G) Representative dot plots showing gating on S-specific IFNγ+/TNFα+ T cells (left panels, % indicated). CD69 and CD103 were used to differentiate circulating (CD69−/CD103−, gray) and tissue-resident memory (Trm; CD69+/CD103− [blue], CD69+/CD103+ [orange], and CD69−/CD103+ [green]) S-specific IFNγ+/TNFα+ T cells from LA (right panels, % indicated). (F and H) The median % of circulating and each of the 3 Trm S-specific IFNγ+/TNFα+ CD4+ (F) or CD8+ (H) T cell subsets present on indicated days in BAL of 4 B/HPIV3/S-6P-immunized macaques are stacked.
Figure S4.
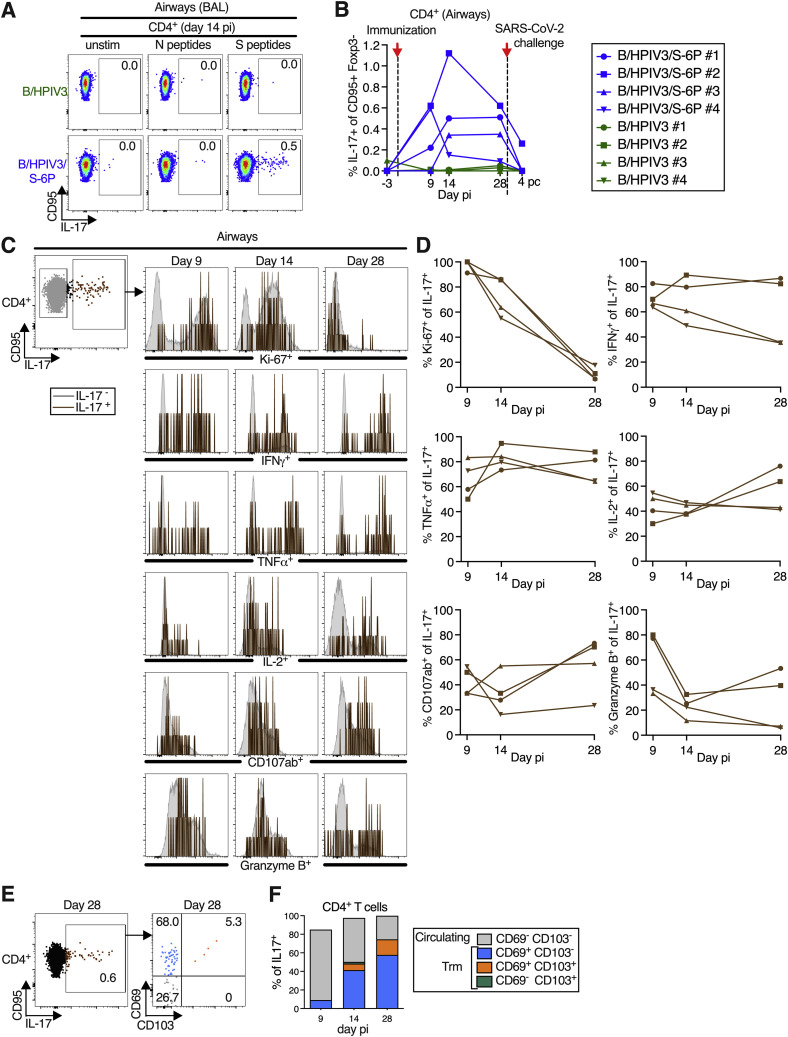
B/HPIV3/S-6P immunization induced a small population of S-specific IL-17+ CD4+ T cells in lower airways (LAs) that transitioned to tissue-resident memory phenotype, related to Figures 4 and 5
T cells obtained by bronchoalveolar lavage (BAL) were stimulated with overlapping N or S peptides and processed for flow cytometry. Phenotypic analyses were performed on non-naive non-regulatory (CD95+/Foxp3−) CD4+ T cells (see Figure S3 for gating) and frequencies are relative to that population.
(A and B) Frequencies of S-specific IL-17+ CD4+ T cells from BAL. (A) Dot plots showing IL-17 expression by CD95+ cells of representative B/HPIV3 (top) or B/HPIV3/S-6P-immunized (bottom) macaques. (B) Background-corrected frequencies of S-specific IL-17+ CD4+ on indicated days. Timing of immunization and SARS-CoV-2 challenge are indicated with red arrow and dotted line.
(C and D) Phenotype of the S-specific IL-17+ CD4+ T cells in LA. (C) Representative dot plots showing gating on S-specific CD95+/IL-17+ (brown), and CD95+/IL-17− (gray) CD4+ T cells. Histograms show expression of Ki-67, IFNγ, TNFα, IL-2, CD107ab, and granzyme B by IL-17+ CD4+ T cells on indicated days pi. (D) Frequencies of Ki-67+, IFNγ+, TNFα+, IL-2+, CD107ab+, and granzyme B+ of S-specific IL-17+ CD4+ T cells from 4 BHPIV3/S-6P-immunized macaques on indicated days.
(E and F) S-specific IL-17+ CD4+ T cells transition to memory phenotype. (E) Representative dot plots showing gating on S-specific CD95+/IL-17+ CD4 T+ cells (left panel, % of IL-17+ cells indicated). CD69 and CD103 were used to differentiate circulating (CD69−/CD103−, gray) and tissue-resident memory (Trm; CD69+/CD103− [blue], CD69+/CD103+ [orange], CD69−/CD103+ [green]) S-specific CD4+ T cells from LA (right panel, % indicated). (F) The median % of circulating and each of the 3 Trm S-specific IL-17+ CD4+ T cells present on indicated days in BAL of 4 B/HPIV3/S-6P-immunized macaques are stacked.
Figure S5.
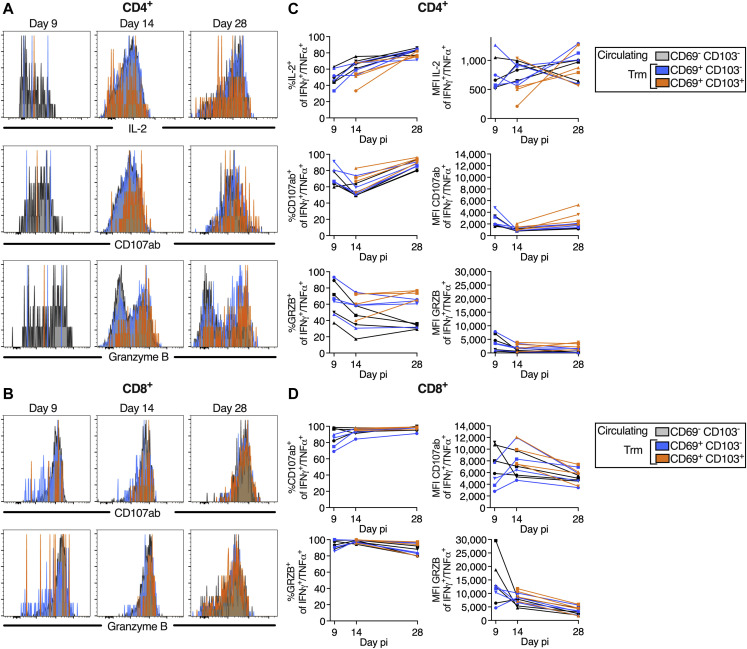
Comparable phenotype of circulating (CD69− CD103−) and tissue-resident memory (CD69+ CD103− and CD69+ CD103+) S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells in the airways, related to Figure 5
(A and B) Histograms representing IL-2 expression (A only), CD107ab, and granzyme B expression by S-specific circulating and tissue-resident memory (Trm) IFNγ+/TNFα+ CD4+ (A) or CD8+ (B) T cells obtained from BAL on indicated days pi.
(C and D) % and level of expression (MFI, median fluorescence intensity) of IL-2 (CD4+ T cells only), CD107ab and granzyme B by the S-specific circulating and Trm IFNγ+/TNFα+ CD4+ (C) and CD8+ T cells (D) in the 4 B/HPIV3/S-6P-immunized macaques. Due to the low frequency of CD69+ CD103+ T cells on day 9 pi, the frequencies and MFIs of IL-2, CD107ab and granzyme B by this subset are only indicated on days 14 and 28 pi. In (C) and (D), each macaque is indicated by a symbol.
Figure S6.
Phenotype of SARS-CoV-2 S-specific CD4+ and CD8+ T cells in the blood of the B/HPIV3/S-6P immunized macaques, related to Figures 4 and 5
(A) Dot blot of the CD4+ T cells from blood of a representative B/HPIV3/S-6P-immunized macaque describing the gating of S-specific IFNγ+/TNFα+ cells (red). The levels of expression of IL-2, CD107ab and granzyme B by the IFNγ+/TNFα+ CD4+ T cells from the same macaque are shown as histograms on the indicated day pi with the IFNγ−/TNFα− CD4+ T cells (gray) used for reference.
(B) % of IFNγ+/TNFα+ CD4+ T cells in the blood of the 4 B/HPIV3/S-6P-immunized macaques that expressed IL-2, CD107ab, or granzyme B on the indicated day pi.
(C) Dot blot of the CD8+ T cells in the blood of a representative B/HPIV3/S-6P-immunized macaque describing the gating of the S-specific IFNγ+/TNFα+ cells (purple). The level of expression of CD107ab and granzyme B by the IFNγ+/TNFα+ CD4+ T cells from the same macaque are shown as histograms on the indicated day pi with the IFNγ−/TNFα− CD4+ T cells (gray) used for reference.
(D) % CD107ab+ or granzyme B+ of IFNγ+/TNFα+ CD8+ T cells on the indicated days pi in the blood of the 4 B/HPIV3/S-6P-immunized macaques. Each macaque is represented by a different symbol.
(E and G) Representative dot plots showing gating on S-specific IFNγ+/TNFα+ T cells (left panels). CD69 and CD103 were used to differentiate circulating (CD69− CD103−, gray) and tissue-resident memory (Trm; CD69+ CD103− [blue], CD69+ CD103+ [orange], and CD69− CD103+ [green]) S-specific IFNγ+/TNFα+ T cells isolated from blood (right panels, % indicated).
(F and H) The median % of the circulating and each of the 3 Trm S-specific IFNγ+/TNFα+ CD4+ (F) or CD8+ (H) T cell subsets present in blood of 4 B/HPIV3/S-6P-immunized macaques on indicated days are stacked.
Figure S7.
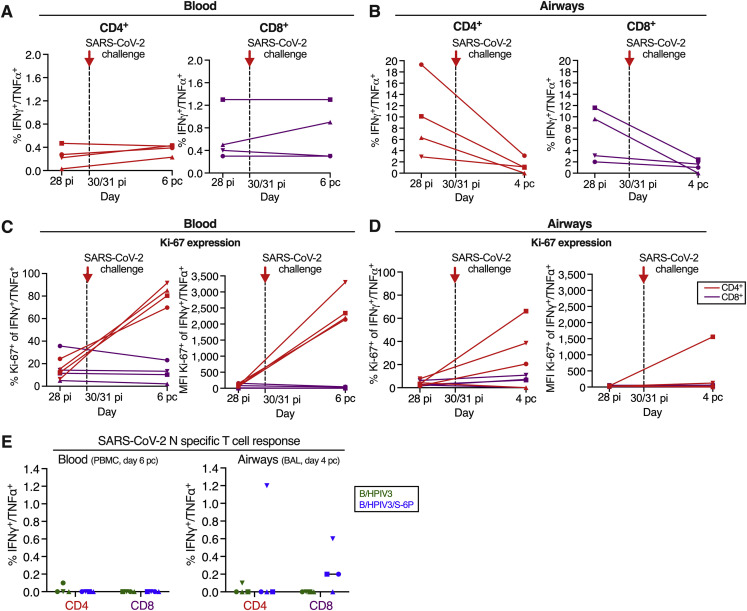
SARS-CoV-2 specific CD4+ and CD8+ T cell responses 4 days after SARS-CoV-2 challenge, related to Figure 4
(A and B) Background-corrected frequencies of S-specific IFNγ+/TNFα+ CD4+ or CD8+ T cells from blood (A) or airways (B) on days 28 post-infection and 4 post-challenge (airways) or 28 post-infection and 6 post-challenge (blood). These frequencies are similar to the frequencies shown in Figures 4B, 4C, 4E, and 4F for the blood and airways, respectively.
(C and D) % and median fluorescence intensity (MFI) of proliferation marker Ki-67 by IFNγ+/TNFα+ CD4+ (red) or CD8+ (purple) T cells from blood (C) or airways (D) of the 4 B/HPIV3/S-6P-immunized macaques, each represented by a different symbol; pi, post-immunization; pc, post-challenge.
(E) Background-corrected frequencies of N-specific IFNγ+/TNFα+ CD4+ or CD8+ T cells from blood (left panel) or airways (right panel) of B/HPIV3 (green)- and B/HPIV3/S-6P (blue)-immunized macaques on day 4 post challenge. Each macaque is represented by a different symbol. When appropriate, the median value from n = 4 macaques is indicated.
In the LA of B/HPIV3/S-6P-immunized animals, S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells were abundant by day 9 or 14 pi (Figures 4D–4F). Remarkably, the average peak percentage of S-specific CD4+ T cells recovered from BAL irrespective of day pi reached 14.3% (Figure 4E). In 3 of 4 animals, their frequency declined between days 14 and 28 pi. S-specific CD8+ T cells in BAL also peaked on day 14 pi in 3 of 4 macaques (Figure 4F; average peak of S-specific IFNγ+/TNFα+ CD8+ T cells irrespective of study day: 11.1%). No S-specific CD4+ or CD8+ T cells were detected in the blood or BAL of macaques immunized with B/HPIV3 (Figures 4A–4F). Finally, stimulation with SARS-CoV-2 N peptides of CD4+ or CD8+ T cells isolated from BAL, which was included as negative control, did not reveal IFNγ+/TNFα+-positive cells above the background present in unstimulated cells (Figure 4D).
On day 9 pi, close to 100% of the S-specific CD4+ and CD8+ T cells in the blood (Figures 4G and 4H) and BAL (Figures 4I and 4J) of the B/HPIV3/S-6P-immunized macaques expressed high levels of Ki-67, confirming active proliferation. While most of these cells still expressed Ki-67 on day 14, the level of expression was strongly reduced, and by day 28, the majority of cells were Ki-67− and had ceased to proliferate.
B/HPIV3/S-6P immunization induces highly functional SARS-CoV-2 S-specific Th1-biased CD4+ T cells and cytotoxic CD8+ T cells in the airways that transition to tissue-resident memory phenotypes
A more comprehensive functional analysis of the BAL-derived S-specific CD4+ T cells revealed that, in addition to expressing IFNγ and TNFα, a proportion of these cells (about 40% to 80% from days 9 to 28 pi) also expressed IL-2, consistent with Th1 bias (Figures 5A and 5B). A fraction of these S-specific IFNγ+/TNFα+ CD4+ T cells also expressed markers of cytotoxicity such as the degranulation markers CD107ab and granzyme B. Thus, most of the CD4+ T cells induced by this vaccine displayed a typical Th1-biased phenotype, similar to those generated after natural SARS-CoV-2 infection.6 , 16 , 17 In addition, a small population of S-specific CD4+ T cells in the BAL expressed IL-17, identifying them as Th17 cells (average peak % of S-specific IL-17+ CD4+ T cells irrespective of the peak day is 0.6%) (Figures S4A and S4B). Based on Ki-67 expression, these S-specific IL-17+ CD4+ T cells actively proliferated from days 9 to 14 pi (Figure S4C), with kinetics similar to IFNγ+/TNFα+ CD4+ T cells (Figure 4E); a fraction also expressed IFNγ, TNFα, IL-2, CD107ab, and granzyme B (Figures S4C and S4D).
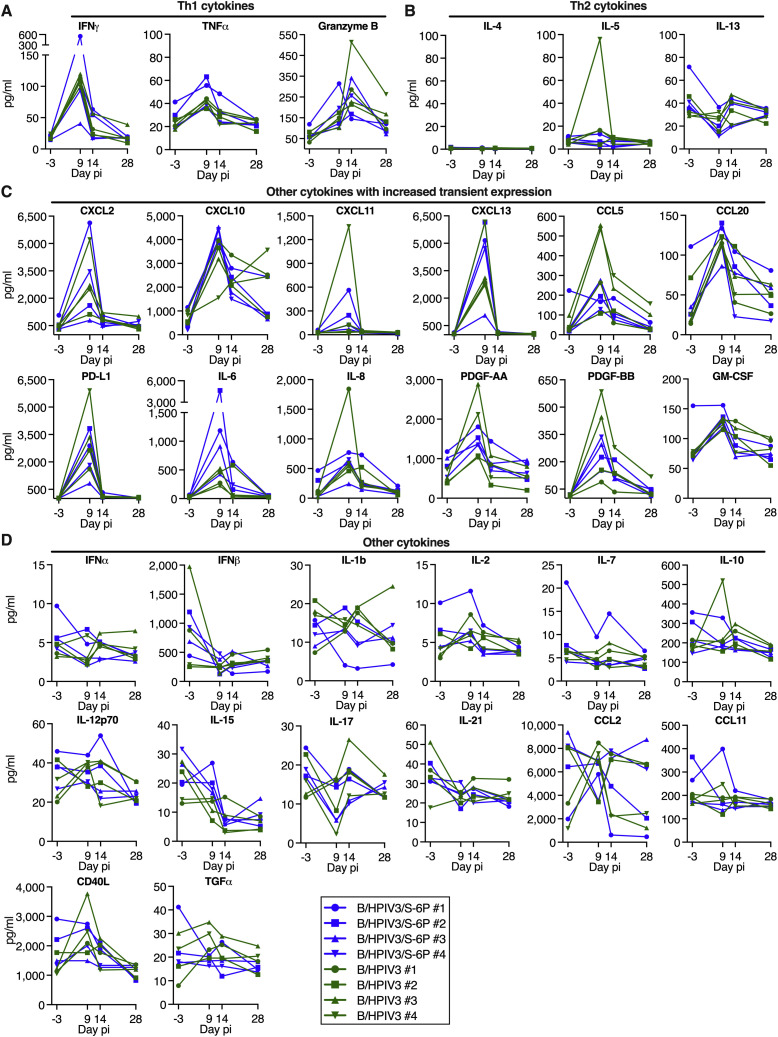
We also measured 36 cytokines in the BAL of the B/HPIV3- and B/HPIV3/S-6P-immunized macaques using multiplex-bead-based immunoassay (Figure S8 ). On days 9 or 14, we detected transient increases in the Th1 cytokines IFNγ, TNFα, and granzyme B in the BAL of all immunized macaques (Figure S8A) but not in Th2 cytokines (IL-4, IL-5, and IL-13, except for a transient increase in IL-5 in a single B/HPIV3-immunized macaque) (Figure S8B), consistent with a Th1-biased response. In addition, we detected a moderate and transient increase in some chemokines in BAL in both groups, including CXCL2, CXCL10, CXCL13, and CCL20 (Figure S8C). Eighteen of 36 cytokines were not increased in the BAL, including IFNα, IFNβ, IL-2, IL-10, and IL-21 (Figure S8D and not shown).
Figure S8.
Cytokines in BAL of B/HPIV3- and B/HPIV3/S-6P-immunized macaques, related to Figures 4 and 5
Macaques were immunized with B/HPIV3 or B/HPIV3/S-6P as described in Figure 1B. BAL collected before immunization (day −3) and on days 9, 14, and 28 after immunization with B/HPIV3 or B/HPIV3/S-6P were concentrated 10×, and the concentration of 36 cytokines from each BAL sample was determined using the NHP XL cytokine Luminex premixed kit. Th1-related cytokines (A), Th2-related cytokines (B), cytokines that showed transient increased expression (C), and cytokines that did not exhibit increased expression (D) are shown. Four additional cytokines (BDNF, VEGF, FGF basic, and G-CSF) that did not exhibit increased expression are not shown. Each macaque is represented by a different symbol. Cytokine concentration is expressed in pg/mL.
S-specific CD8+ T cells from the BAL, in addition to expressing IFNγ and TNFα, also expressed high levels of degranulation markers CD107ab and granzyme B from days 9 to 28 pi, suggesting that these cells were highly functional (Figures 5C and 5D).
Furthermore, S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells from BAL could be separated into circulating CD69− CD103− and tissue-resident memory (Trm) CD69+ CD103+/− subsets18 (Figures 5E and 5F for CD4+ T cells and Figures 5G and 5H for CD8+ T cells, respectively). An additional subset of presumably tissue-resident S-specific CD4+ and CD8+ T cells was identified as CD69− CD103+ and has been previously detected in SARS-CoV-2-infected macaques.19 Circulating CD69− CD103− S-specific CD4+ and CD8+ T cells were detectable in BAL on day 9 pi and were prominent until day 14, representing about 60% of the S-specific T cells on this day (Figures 5F and 5H). Lung-resident S-specific CD69+ CD103− CD4+ and CD8+ T cells were detectable from day 9 pi (Figures 5F and 5H), and their proportion increased through day 28 pi. On day 14, a fraction of these CD69+ S-specific T cells had acquired CD103, and the proportion of CD69+ CD103+ Trm CD4+ and CD8+ T cells further increased from day 14 to 28 pi. By day 28 pi, while the overall percentage of S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells decreased in the airways (Figures 4E and 4F), about 80% of the remaining S-specific T cells were positive for CD103 and/or CD69, indicating the contraction of the S-specific T cell populations and a transition to Trm phenotypes (Figures 5F and 5H). Similarly, a fraction of the S-specific IL-17+ CD4+ T cells in the airways also transitioned to Trm phenotypes (Figures S4E and S4F).
Following antigen stimulation, the S-specific circulating and tissue-resident IFNγ+/TNFα+ CD4+ or CD8+ T cells recovered from the airways on day 9, 14, or 28 were phenotypically comparable with respect to strong expression of CD107ab and granzyme B (Figure S5), suggesting that all S-specific CD69/CD103 subsets present in the airways were highly functional.
The phenotype of the blood-derived S-specific CD4+ and CD8+ T cells was overall comparable to that of the airway-derived S-specific T cells (Figures S6A–S6D) although no S-specific CD4+ or CD8+ T cells expressing IL-17 were identified. Unlike the BAL-derived cells, the S-specific (IFNγ+/TNFα+) CD4+ and CD8+ T cells in the blood mostly retained a circulating (CD69− CD103−) phenotype throughout day 28 pi (Figures S6E–S6H).
B/HPIV3/S-6P immunization protects macaques against SARS-CoV-2 challenge virus replication in the upper and lower airways
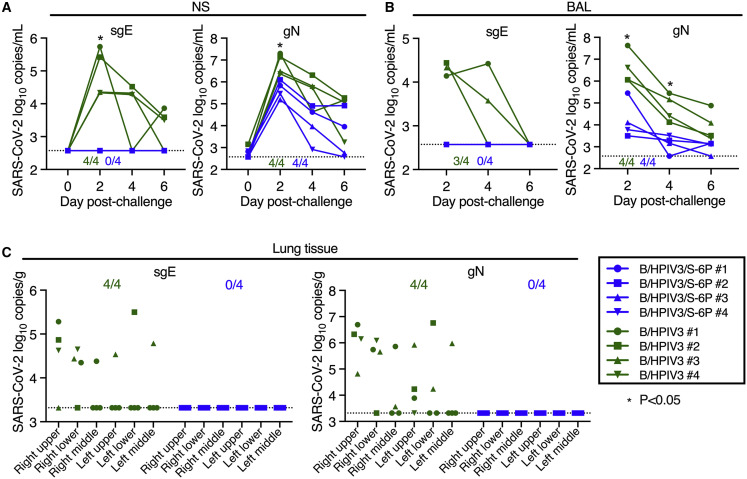
To assess protective efficacy of IN/IT immunization with B/HPIV3/S-6P, we challenged macaques from both groups intranasally and intratracheally with 5.8 log10 TCID50 of SARS-CoV-2 WA1/2020 on day 30 or 31 after immunization (Figure 1B). NS and BAL specimens were collected before challenge (NS only) and on days 2, 4, and 6 post challenge (pc). Viral RNA was extracted from these specimens, and the SARS-CoV-2 virus load was evaluated by RT-qPCR (Figures 6A and 6B). We quantified subgenomic E (sgE) mRNA, indicative of SARS-CoV-2 infection, mRNA synthesis, and challenge virus replication20 , 21 (Figures 6A and 6B, left panels). In B/HPIV3 empty-vector immunized macaques, sgE mRNA was detected in the UA of 4 of 4 and in the LA of 3 of 4 macaques, respectively, showing that all B/HPIV3 control immunized macaques were actively infected with SARS-CoV-2 challenge virus. Copy numbers were maximal on day 2 pc (mean 5.0 log10 copies/mL in the UA and 4.3 log10 copies/mL in the LA) and decreased until day 6 pc. In contrast, in all 4 B/HPIV3/S-6P-immunized macaques, sgE mRNA was undetectable in the UA and LA at all time points (p < 0.05), showing that IN/IT immunization with a single dose of B/HPIV3/S-6P induces robust protection against high levels of challenge virus replication.
Figure 6.
Absence of detectable SARS-CoV-2 challenge virus replication in the upper and lower airways and lung tissues of B/HPIV3/S-6P-immunized macaques
Rhesus macaques immunized with a single intranasal/intratracheal dose of B/HPIV3/S-6P or B/HPIV3 control (n = 4 per group) were challenged intranasally/intratracheally on day 30 pi with 5.8 TCID50 of SARS-CoV-2.
(A and B) Evaluation of challenge virus shedding by qRT-PCR. (A) Nasal swabs (NSs) and (B) bronchoalveolar lavage (BAL) fluid were collected on days 0 (NS only), 2, 4, and 6 post-challenge (pc) and SARS-CoV-2 subgenomic E mRNA (sgE, indicative of active SARS-CoV-2 replication) and genomic N RNA (gN, indicative of the presence of challenge virus) were quantified by RT-qPCR.
(C) Challenge virus detection in lung tissues. Animals were euthanized on day 6 pc, and RNA was extracted from indicated areas of lung tissue to quantify SARS-CoV-2 sgE mRNA and gN RNA by qRT-PCR.
(A–C) Copies/ml of SARS-CoV-2 sgE mRNA and gN RNA in NS (A) and BAL (B) and copies/g in lung tissues (C) are indicated for macaques immunized with B/HPIV3/S-6P (blue) or B/HPIV3 (green), each identified by a symbol. The numbers of animals in each group with detectable RNA are indicated above the x axis (A and B) or at the top of the graph (C). The limit of detection was 2.6 log10 copies/mL of NS or BAL fluid and 3.3 log10 copies/g of lung tissue. ∗p 0.05, two-way ANOVA, Sidak multiple comparison test.
From the same samples, we also quantified genomic N RNA (gN), indicative of the presence of challenge virus. gN RNA copies/mL were maximal on day 2 pi in the UA and LA of macaques, and then steadily decreased over time. The average of the gN RNA loads on day 2 in the UA and LA of macaques immunized with B/HPIV3/S-6P were 16- and 240-fold lower than those of macaques immunized with B/HPIV3 empty-vector control (UA: 6.8 versus 5.6 log10 copies/mL in the B/HPIV3- and the B/HPIV3/S-6P-immunized macaques, respectively, p < 0.05; LA: 6.6 versus 4.2 log10 copies/mL in the B/HPIV3- and the B/HPIV3/S-6P-immunized macaques, respectively, p < 0.05), which would be consistent with the presence of residual challenge virus, and absence of SARS-CoV-2 challenge virus replication, in B/HPIV3/S-6P immunized animals.
Quantification of sgE and gN RNA was also performed for lung tissues from different areas obtained on day 6 pc (Figure 6C). In all 4 B/HPIV3 empty-vector immunized macaques, gN RNA and sgE mRNA were detected, mostly in the right upper and right lower lobes of the lungs. Neither gN RNA nor sgE mRNA was detected in the lungs of the B/HPIV3/S-6P-immunized macaques, confirming robust protection against SARS-CoV-2 infection induced by B/HPIV3/S-6P. Furthermore, no active SARS-CoV-2 replication was detected from rectal swab samples.
We finally assessed the CD4+ and CD8+ T cell response in the blood (Figures 4B, 4C, and S7A) and LAs (Figures 4E, 4F, and S7B) of immunized macaques, 4 days (for LA) or 6 days (for blood) after challenge with SARS-CoV-2. In the blood, an increase of S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells was detected in 3 and 1 of 4 B/HPIV3/S-6P-immunized macaques, respectively, that correlated well with the increased expression of Ki-67 by the S-specific CD4+ T cells (Figure S7C). A modest increase of S-specific IFNγ+/TNFα+ CD4+ T cells was also detected in 1 B/HPIV3-immunized macaque (Figure 4B). However, in the LAs, a decrease rather than an increase of the S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells was detected in the B/HPIV3/S-6P-immunized macaques and no active T cell proliferation was detected (Figure S7D). As expected, no SARS-CoV-2 N-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells were detected in the blood of macaques following immunization with B/HPIV3- or B/HPIV3/S-6P (Figure S7E, left panel). On day 4 after SARS-CoV-2 challenge, only a modest N-specific IFNγ+/TNFα+ CD4+ and CD8+ T cell response was detected in the LA of a single B/HPIV3/S-6P macaque. This low N-specific primary response is consistent with the low recall response of S-specific IFNγ+/TNFα+ CD4+ and CD8+ T cells in the airways at this early time point after challenge (Figure S7E).
Discussion
SARS-CoV-2 vaccines for infants and young children are critically needed. Equally needed are topical SARS-CoV-2 vaccines that directly stimulate local respiratory tract immunity in addition to systemic immunity, which might effectively reduce infection and transmission. In this study, we evaluated a chimeric B/HPIV3 virus as a live topical viral vector to express the SARS-CoV-2 S protein, stabilized in its prefusion form, expected to be non-functional for virus entry.13 No effects of B/HPIV3 and B/HPIV3/S-6P on the general health of rhesus macaques were observed following immunization, indicating that this vector and the expressed S protein were safe in this nonhuman primate model.
The B/HPIV3 vector is in clinical development and is predicted to be safe. The HN and F surface glycoproteins of B/HPIV3 are derived from HPIV3. HN and F are the major protective antigens of HPIV3, a major respiratory pathogen in infants and children under 5 years of age. The BPIV3 genes encode for the nucleocapsid N, phosphoprotein P, interferon antagonist V, matrix M, and polymerase L that provide host-range restriction and represent the basis for the strong attenuation of this vector while the surface glycoproteins from HPIV3 mediate infection.9 , 10 The B/HPIV3 vector exhibits a natural tropism for the respiratory tract without any evidence of spread to other tissues. An important advantage of the B/HPIV3 vector is that it has been evaluated in young children both as the empty vector and expressing a foreign gene (RSV F protein), and in both cases it had an excellent clinical safety profile.9 , 10 This should expedite advancing this SARS-CoV-2 vaccine through clinical studies. A second important advantage of the B/HPIV3 vector is that it is administered intranasally and stimulates systemic and local mucosal immunity in the respiratory tract, the site of SARS-CoV-2 entry, replication, disease, and egress. Finally, another benefit associated with the use of B/HPIV3 as a vector for expressing a heterologous viral protein is that it also induces neutralizing antibody responses against HPIV3 itself,9 , 10 effectively serving as a dual HPIV3/SARS-CoV-2 vaccine.
A single IN/IT immunization with B/HPIV3/S-6P efficiently induced mucosal IgA and IgG in the UA and LA of all immunized macaques, as well as strong serum IgM, IgA, and IgG responses to SARS-CoV-2 S protein and its RBD. The anti-S and anti-RBD IgG responses were comparable to those detected in human convalescent plasma of individuals with high levels of anti-S and anti-RBD IgG antibodies. The serum antibodies efficiently neutralized the vaccine-matched SARS-CoV-2 WA1/2020 strain, as well as VoCs of B.1.1.7/Alpha and B.1.617.2/Delta lineages. However, the neutralizing activity of these sera against the B.1351/Beta and B.1.1.529/Omicron lineages was limited. This suggests that a second immunization with B/HPIV3/S-6P might be beneficial, as with perhaps any SARS-CoV-2 vaccine, to boost antibody titers and affinity maturation, providing for greater breadth of antigen recognition and protection against VoCs.
While not specifically investigated, B/HPIV3/S-6P immunization likely resulted in the generation of long-lived antigen-specific tissue-resident B and T cells in the mucosa. Antigen-specific B cells in the mucosa are sources of soluble dimeric IgA22 with increased neutralizing activity compared with serum-derived monomeric IgA.23 Injectable SARS-CoV-2 vaccines have been shown to result in the appearance of mucosal anti-S IgA and IgG in the UA and LA of immunized macaques, primarily by transudation of monomeric Igs from the blood, and in the absence of direct stimulation of mucosal immunity at the site of infection.24 To induce long-term mucosal humoral immunity in the airways, a homologous or heterologous prime/boost approach that includes an intranasal vaccine might be advantageous over a prime/boost approach based solely on immunizations with injectable vaccines. For adenovirus-vectored SARS-CoV-2 vaccines, the protective efficacy following administration by the intranasal versus the intramuscular route was compared in nonhuman primates. Overall, intranasal immunization with these vaccine candidates induced a modest systemic immune response, but generally provided better protection against SARS-CoV-2 challenge than intramuscular administration, substantially reducing challenge virus replication in the airways.25 , 26 , 27 The T cell responses elicited by these vaccines in the airways were not evaluated in detail.
B/HPIV3/S-6P also induced S-specific CD4+ and CD8+ T cells in the blood and the LA. Similar to immunization with injectable SARS-CoV-2 vaccines, IN/IT immunization with B/HPIV3/S-6P induced S-specific Th1-biased CD4+ T cells in the blood that expressed IFNγ, TNFα, and IL-2,24 , 28 , 29 , 30 and markers of cytotoxicity such as CD107ab and granzyme B, suggesting that they might also be directly involved in virus clearance. In addition, B/HPIV3/S-6P appeared to induce a stronger response of S-specific CD8+ T cells in the blood of macaques than injectable vaccines.29 , 30 , 31
To control respiratory virus infections, mucosal immune responses at the site of infection are essential.32 In longitudinal studies of SARS-CoV-2 infection in humans, T cells induced in airways exhibited activated and tissue-resident signatures and functionally protective profiles,33 and their frequencies in airways (but not in blood) correlated with younger age and survival.17 , 34 Depletion of CD8+ T cells partially abrogated the protective immunity against SARS-CoV-2 re-challenge in the macaque model.35 Intranasal immunization with B/HPIV3/S-6P induced strong T cell responses in the LA, with a very efficient induction of S-specific Th1-biased CD4+ T cells expressing IFNγ, TNFα, and IL-2 as well as S-specific CD8+ T cells that expressed IFNγ, TNFα, and granzyme B, while injectable mRNA or adenovirus-based vaccines do not appear to induce S-specific CD4+ and CD8+ T cells in the airways of immunized macaques.29 , 30 In our study, B/HPIV3/S-6P induced a small but detectable population of S-specific CD4+ Th17 cells in the LA. While the contribution of this cell population in mounting the immune response to B/HPIV3/S-6P is currently unknown, previous work suggested that Th17 cells that transitioned to tissue-resident T cells in the lungs facilitate the recruitment of T and B cells, resulting in an accelerated IgA response.36
A substantial fraction of the S-specific CD4+ and CD8+ T cells, including CD4+ Th17 cells, in the lungs expressed the Trm marker CD69, with a subpopulation also expressing CD103. The proportion of S-specific Trm T cells increased over time and represented the main S-specific T cell population 1 month after immunization. While the contribution of memory CD4+ and CD8+ T cells in controlling SARS-CoV-2 replication is still not fully understood,35 , 37 , 38 , 39 previous work suggested that Trm T cells contribute to long-term immunity and are associated with protection against respiratory disease.17 , 39 , 40 , 41 Furthermore, as T cell epitopes in the S protein are mostly conserved across SARS-CoV-2 lineages,42 , 43 the strong induction of S-specific Trm CD4+ and CD8+ T cells by B/HPIV3/S-6P should be beneficial to the protection against VoCs including Omicron.16 , 32 , 41 Indeed, in the rhesus model, the failure of injectable SARS-CoV-2 vaccines in controlling SARS-CoV-2 B.1.529/Omicron challenge virus replication in the upper respiratory tract was associated with negligible T cell responses but not with the level of anti-S antibody titers.44 In addition, even though the S expression by B/HPIV3/S-6P in the LA of one macaque was not detected (Figure 1F), this macaque still exhibited strong serum and mucosal anti-S IgA and IgG response as well as strong S-specific CD4+ and CD8+ T cells in the blood and LA. This suggested that even a low level of S expression in the airways was highly immunogenic.
We found that macaques were fully protected from SARS-CoV-2 challenge 1 month after immunization. No SARS-CoV-2 challenge virus replication was detectable in the UA or LA or in lung tissues of immunized macaques, suggesting sterilizing immunity under these experimental conditions. However, the duration of immunity is currently unknown and will be evaluated in a separate study. Following challenge, we detected a recall response of S-specific CD4+ T cells in the blood of B/HPIV3/S-6P-immunized macaques but not in the LA. The reasons for the apparent absence of a detectable recall response are still unknown. It is possible that the S-specific T cells were not present in the airways and inaccessible through BAL, or that the BAL cell collection on day 4 pc was too early to detect any recall response, or that in the absence of challenge virus replication, a recall response was not triggered in the LA. Later time points following challenge will be included in future studies. Even though we did not evaluate protection against HPIV3 challenge in this study, we showed that B/HPIV3 induced strong serum HPIV3 neutralizing antibodies, comparable to levels seen in previous studies;45 thus, B/HPIV3/S-6P represents a promising candidate for intranasal immunization against both SARS-CoV-2 and HPIV3, an important pediatric cause of lower respiratory tract disease.
In conclusion, a single mucosal immunization with B/HPIV3/S-6P was highly immunogenic and protective against SARS-CoV-2 in macaques. Our data support the further development of this vaccine candidate for potential use as a stand-alone vaccine and/or in a prime/boost combination with other SARS-CoV-2 vaccines for infants and young children. The B/HPIV3/S-6P vaccine candidate will be evaluated in a phase 1 study.
Limitations of the study
In our study, the sequence of the S-6P protein of B/HPIV3/S-6P was derived from the ancestral Wuhan-1 strain, and B/HPIV3/S-6P-immunized macaques were challenged with the SARS-CoV-2 strain WA1/2020, with an S amino acid sequence identical to that of the Wuhan-1 strain. While several functional domains and T cell epitopes are well conserved between the Wuhan-1 strain and currently circulating variants, antigenic differences between S-6P and Omicron variants are well documented. Thus, the efficacy of B/HPIV3/S-6P against Omicron sub-lineages will need to be evaluated in future studies. In another limitation, immunizations of macaques were performed by the combined IN/IT route, providing for direct vaccine delivery to the upper and lower respiratory tract, and optimal vaccine virus replication and immunogenicity in this semi-permissive animal model. IN/IT administration minimizes animal-to-animal variability, reducing the number of macaques required. In clinical studies, the B/HPIV3/S-6P vaccine candidate will be delivered by nasal spray. Finally, it is likely that anti-vector immunity induced by the first immunization with B/HPIV3/S-6P will restrict vaccine replication of a second dose, thereby limiting the immunogenicity of the second dose. This could pose a limitation if prime/boost approaches are needed but would be inconsequential in heterologous prime/boost combinations of intranasal and intramuscular vaccines, including mRNA-based or adenovirus-vectored vaccines or in combination with another vector vaccine for intranasal delivery.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD69-FITC, Clone FN50 | Biolegend | Cat #310903; RRID: AB_314838 |
| Anti-human Granzyme B-BV421, Clone GB11 | BD Biosciences | Cat #563389; RRID: AB_2738175 |
| Anti-human CD8a-eFluor 506, Clone RPA-T8 | Thermo Fisher | Cat #69-0088-42; RRID: AB_2637468 |
| Anti-human IL-2-BV605, Clone MQ1-17H12 | Biolegend | Cat #500332; RRID: AB_2563877 |
| Anti-human IFNg-BV711, Clone 4S.B3 | Biolegend | Cat #502540; RRID: AB_2563506 |
| Anti-human TNFa-BUV395, Clone Mab11 | BD Biosciences | Cat #563996; RRID: AB_2738533 |
| Anti-human CD4-BUV496, Clone SK3 | BD Biosciences | Cat #612937; RRID: AB_2916881 |
| Anti-human CD95-BUV737, Clone DX2 | BD Biosciences | Cat #612790; RRID: AB_2870117 |
| Anti-human CD3-BUV805, Clone SP34-2 | BD Biosciences | Cat #742053; RRID: AB_2871342 |
| Anti-human CD107a-AF647, Clone H4A3 | Biolegend | Cat #328612; RRID: AB_1227506 |
| Anti-human CD107b-AF647, Clone H4B4 | Biolegend | Cat #354312; RRID: AB_2721405 |
| Anti-human CD103-PE, Clone B-Ly7 | eBioscience | Cat #12-1038-42; RRID: AB_11150242 |
| Anti-human CD28-PE/Dazzle 594, Clone CD28.2 | Biolegend | Cat #302942; RRID: AB_2564235 |
| Anti-human Ki-67-PE-Cy7, Clone B56 | BD Biosciences | Cat #561283; RRID: AB_10716060 |
| Anti-human Foxp3-AF700, Clone PCH101 | Thermo Fisher | Cat #56-4776-41; RRID: AB_1582210 |
| Anti-monkey IgG(H+L)-HRP | Thermo Fisher | Cat #PA1-84631; RRID: AB_933605 |
| Anti-monkey IgA(alpha chain)-biotin | Alpha Diagnostic International | Cat #70049 |
| Anti-monkey IgM-biotin | Brookwood Biomedical | Cat #sab1152 |
| Rabbit hyperimmune serum against HPIV3 virions | Liang et al.46 | N/A |
| Goat hyperimmune serum against SARS-CoV-2 S-2P | Liu et al.11 | N/A |
| Anti-rabbit IRDye680RD IgG | Li-Cor | Cat #926-68073; RRID: AB_10954442 |
| Anti-goat IRDye800CW IgG | Li-Cor | Cat #926-32214; RRID: AB_621846 |
| Biological samples | ||
| SARS-CoV-2 USA-WA1/2020 isolate | Natalie Thornburg et al., CDC | GenBank MN985325; GISAID: EPI_ISL_404895 |
| SARS-CoV-2 lineage B.1.1.7/Alpha | CDC | USA/CA_CDC_5574/2020 isolate GISAID: EPI_ISL_751801 |
| SARS-CoV-2 lineage B.1.351/Beta | Dr. A Pekosz, Johns Hopkins University | USA/MD-HP01542/2021 isolate GISAID: EPI_ISL_890360 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 S-2P | Liu et al.11 | N/A |
| SARS-CoV-2 S RBD | Liu et al.11 | N/A |
| Viability Dye eFluor780 | Thermo Fisher | Cat #65-0865-14 |
| Ficoll-Paque density gradient | Cytivia | Cat #17144003 |
| Monensin solution | Thermo Fisher | Cat #00-4505-51 |
| Brefeldin A solution | Thermo Fisher | Cat #00-4506-51 |
| Peptivator SARS-CoV-2 Prot_S1 | Miltenyi Biotec | Cat #130-127-048 |
| Peptivator SARS-CoV-2 Prot_S+ | Miltenyi Biotec | Cat #130-127-312 |
| Peptivator SARS-CoV-2 Prot_S | Miltenyi Biotec | Cat #130-127-953 |
| Peptivator SARS-CoV-2 Prot_N | Miltenyi Biotec | Cat #130-126-699 |
| eBioscience intracellular fixation & permeabilization buffer set | Thermo Fisher | Cat #88-8824-00 |
| eBioscience permeabilization buffer | Thermo Fisher | Cat #00-83333-56 |
| Buffer AVL | Qiagen | Cat #19073 |
| Trizol LS Reagent | Thermo Fisher | Cat #10296028 |
| Critical commercial assays | ||
| DELFIA time resolved fluorescence immunoassay | Perkin Elmer | N/A |
| V-PLEX SARS-CoV-2 Panel 25 (ACE2) | MSD | Cat # K15578U |
| V-PLEX SARS-CoV-2 Panel 27 (ACE2) | MSD | Cat # K15609U |
| Amicon centrifugal filters | Millipore | Cat # UFC500396 |
| NHP XL Cyotkine Luminex Premixed kit | R&D Systems | Cat # FCSTM21 |
| QuickChange Lightning Multi Site-directed mutagenesis kit | Agilent | Cat # 210514 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat #52904 |
| PureLink RNA Mini Kit | Thermo Fisher | Cat #12183018A |
| TaqMan RNA-to-Ct 1-Step Kit | Thermo Fisher | Cat #4392938 |
| LiFect293 Transfection Reagent | LifeSct | Cat #M0002-01 |
| Bright-Glo Luciferase Assay System | Promega | Cat #E2620 |
| Experimental models: Cell lines | ||
| BSR T7/5 | Buchholz et al.47 | N/A |
| Vero | ATCC | Cat #CCL-81 |
| Vero E6 | ATCC | Cat #CRL-1586 |
| Vero E6 expressing human TMPRSS2 | Liu et al.11 | N/A |
| 293T/17 | Corbett et al.30 | ATCC Cat #CRL-11268 |
| 293T-hACE2.MF | Corbett et al.30 | N/A |
| Experimental models: Organisms/strains | ||
| Rhesus macaques (Macaca mulatta) | This paper | Young adult animals, 4-7 years of age, male. Animal study protocol approved by NIAID ACUC |
| Human plasma from SARS-CoV-2 convalescent donors, de-identified | Dr. Jeffrey I. Cohen, NIAID, NIH |
Exempt from IRB review |
| Oligonucleotides | ||
| 5’-CGATCTCTTGTAGATCTGTTCTC-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Subgenomic E Forward |
| 5’-ATATTGCAGCAGTACGCACACA-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Subgenomic E Reverse |
| 5’-ACACTAGCCATCCTTACTGCGCTTCG-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Subgenomic E Probe |
| 5’-GACCCCAAAATCAGCGAAAT-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Genomic N Forward |
| 5’-TCTGGTTACTGCCAGTTGAATCTG-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Genomic N Reverse |
| 5’-ACCCCGCATTACGTTTGGTGGACC-3’ | Chandrashekar et al.,20 Wölfel et al.,21 and Corman et al.48 | Genomic N Probe |
| Recombinant DNA | ||
| B/HPIV3 | Liu et al.11 | N/A |
| B/HPIV3/S-2P | Liu et al.11 | N/A |
| B/HPIV3/S-6P | This paper | N/A |
| BPIV3 N, P and L helper plasmids | Liu et al.11 and Schmidt et al.12 | N/A |
| SARS-CoV-2 S | Liu et al.11 | GenBank MN908947 |
| SARS-CoV-2 S-2P | Wrapp et al.14 | N/A |
| SARS-CoV-2 S RBD | Wrapp et al.14 | N/A |
| pHR’ CMV Luc | Corbett et al.30 | N/A |
| pCMV ΔR8.2 | Corbett et al.30 | N/A |
| TMPRSS2 | Corbett et al.30 | N/A |
| Software and algorithms | ||
| FlowJo v10 | BD | https://www.flowjo.com |
| Prism v9 | GraphPad Software, LLC | https://www.graphpad.com/scientificsoftware/prism/ |
| ImageStudioLite 5.2.5 | Li-Cor | https://www.licor.com/bio/image-studio-lite/resources#isl5 |
| Relative Quantification, Standard Curve | Thermo Fisher | https://apps.thermofisher.com |
| BioTek Gen 5 | BioTek | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/ |
Resource availability
Lead contact
Requests for resources, reagents and further information regarding this manuscript should be directed to and will be fulfilled by the lead contact, Ursula Buchholz (ubuchholz@niaid.nih.gov).
Materials availability
Plasmids and viruses newly generated in this study are available under material transfer request upon request to the lead contact.
Experimental model and subject details
Cell lines
Baby hamster kidney cells expressing T7 RNA polymerase (BSR T7/5) were grown in Glasgow minimum essential medium (MEM) (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS), 1% L-glutamine (Thermo Fisher Scientific), and 2% MEM Amino Acids (Thermo Fisher Scientific). African green monkey kidney Vero (ATCC CCL-81) and Vero E6 (ATCC CRL-1586) cells were cultured in Dulbecco’s MEM with GlutaMAX (Thermo Fisher Scientific) with 5% FBS and 1% L-glutamine. Vero E6 cells that express high levels of ACE249 , 50 and Vero E6 cells that stably express human TMPRSS211 were used to expand SARS-CoV-2.
SARS-CoV-2 virus stocks
The SARS-CoV-2 USA-WA1/2020 isolate (lineage A; GenBank MN985325; GISAID: EPI_ISL_404895; obtained from Dr. Natalie Thornburg et al., Centers for Disease Control and Prevention (CDC)) was passaged on Vero E6 cells. We used WA1/2020 as challenge virus in this study because its S sequence is homologous to the sequence from which S-6P was derived. The USA/CA_CDC_5574/2020 isolate [lineage B.1.1.7 (Alpha); GISAID: EPI_ISL_751801; obtained from CDC] and the USA/MD-HP01542/2021 isolate [lineage B.1.351 (Beta); GISAID: EPI_ISL_890360; obtained from Dr. Andrew Pekosz, Johns Hopkins University] were grown on TMPRSS2-expressing Vero E6 cells.11 The SARS-CoV-2 stocks were titrated in Vero E6 cells by determination of the 50% tissue culture infectious dose (TCID50).51 All experiments with SARS-CoV-2 were performed in biosafety level 3 (BSL3) containment laboratories approved by the USDA and CDC.
Animals and study design
All animal studies were approved by the NIAID Animal Care and Use Committee. The timeline of the experiment and sampling is summarized in Figure 1B. Eight juvenile to young adult male Indian-origin rhesus macaques (Macaca mulatta), 4-7 years of age, confirmed to be seronegative for HPIV3 and SARS-CoV-2, were randomly assigned to study groups, followed by adjustments for similar average weight of the animals in each group, and immunized intranasally (0.5 ml per nostril) and intratracheally (1 ml) with a total does of 6.3 log10 plaque-forming units (PFU) of B/HPIV3/S-6P or the empty vector control B/HPIV3. Animals were observed daily from day -3 until the end of the study. Each time they were sedated, animals were weighed, their rectal temperature was taken, as well as the pulse in beats per minute and the respiratory rate in breaths per minute. In addition, the blood oxygen levels were determined by pulse oximetry.
Blood for analysis of serum antibodies and peripheral blood mononuclear cells (PBMC) was collected on days -3, 9, 14, 21, and 28 pi. Nasopharyngeal swabs (NS) for vaccine virus quantification in the upper airways (UA) were performed on day -3 and daily from day 0 to day 10 pi and on day 12 pi using cotton-tipped applicators. Swabs were placed in 2 ml Leibovitz (L-15) medium with 1x sucrose phosphate (SP) used as stabilizer, and vortexed for 10 seconds. Aliquots were then snap frozen in dry ice and stored at -80°C. Nasal washes (NWs) for analysis of mucosal IgA and IgG were performed using 1 ml of Lactated Ringer’s solution per nostril (2 ml total) on days -3, 14, 21 and 28 pi and aliquots were snap frozen in dry ice and stored at -80°C until further analysis. Tracheal lavages (TL) for virus quantification were done every other day from day 2 to 8 pi and on day 12 pi using 3 ml PBS. The samples were mixed 1:1 with L-15 medium containing 2x SP and aliquots were snap frozen in dry ice and stored at -80°C for further analysis. Bronchoalveolar lavages (BALs) for analysis of mucosal IgA and IgG and airway immune cells from the lower airways (LA) were done on days -3, 9, 14 and 28 pi using 30 ml PBS (3 times 10 ml). For analysis of mononuclear cells, BAL was filtered through a 100 μm filter, and centrifuged at 544 x g for 15 min at 4°C. The cell pellet was resuspended at 2x107 cells/ml in X-VIVO 15 media supplemented with 10% FBS for subsequent analysis. The cell-free BAL was aliquoted, snap frozen in dry ice and stored at -80°C for further analysis. Rectal swabs were done on day -3 and then every other day from day 2 to 14 following the same procedure as NS.
Four weeks after immunization, animals were transferred to BSL3. On day 30 or 31 pi, animals were challenged intranasally and intratracheally with 105.8 TCID50 of SARS-CoV-2, USA-WA1/2020, that was entirely sequenced and free of any prominent adventitious mutations. Sample collections were done following the same procedures as during the immunization phase. Briefly, blood was collected on day 6 post-challenge (pc). NS were performed every other day from day 0 to day 6 pc. BAL were done on days 2, 4 and 6 pc, and rectal swabs on days 0, 2, 4 and 6 pc. Animals were necropsied on day 6 pc, and tissues were collected. Six samples per animal from individual lung lobes were collected, and snap frozen in dry ice for further analysis.
Method details
Generation of the B/HPIV3/S-6P vaccine
The B/HPIV3/S-6P vaccine candidate is an improved derivative of B/HPIV3/S-2P.11 To generate the B/HPIV3/S-2P cDNA,11 the ORF encoding the full-length 1,273 aa SARS-CoV-2 S protein from the first available sequence (GenBank MN908947, Wuhan-Hu-1; amino acid sequence of the S protein identical to that of WA1/2020) was codon-optimized for human expression and synthesized commercially (BioBasic). Two proline substitutions (aa positions 986 and 987) and four aa substitutions (RRAR to GSAS, aa 682-685) that stabilize S in the prefusion conformation and ablate the furin cleavage site between S1 and S214 were introduced to generate the S-2P cDNA.11 This S-2P ORF was then inserted into a cDNA clone encoding the B/HPIV3 antigenome between the N and P ORFs to create the B/HPIV3/S-2P cDNA.11 To create the B/HPIV3/S-6P cDNA, the B/HPIV3/S-2P cDNA was modified to introduce 4 additional proline substitutions in the S ORF (aa position 817, 892, 899, and 942 for a total of 6 proline substitutions). The 4 additional proline substitutions had been shown to confer increased stability to a soluble version of the prefusion-stabilized S protein.13 The B/HPIV3/S-6P cDNA was used to transfect BHK21 cells (clone BSR T7/5, stably expressing T7 RNA polymerase47), together with helper plasmids encoding the N, P and L proteins,11 , 52 to produce the B/HPIV3/S-6P recombinant virus. The empty vector control virus B/HPIV3 was recovered in parallel. Virus stocks were grown in Vero cells, and viral genomes of recovered viruses were completely sequenced by Sanger sequencing using overlapping RT-PCR fragments, confirming the absence of any adventitious mutations. In addition, stability of S expression was evaluated by dual-staining immunoplaque assay. In virus stocks originating from eight independent recoveries of B/HPIV3/S-6P, 96.3±1.7 of virus plaques were immunostaining-positive for PIV3 and SARS-CoV-2 S antigen, indicating that B/HPIV3/S-6P stably expressed the SARS-CoV-2 S protein.
Immunoplaque assay
Titers of B/HPIV3 and B/HPIV3/S-6P from NS and TLs were determined by dual-staining immunoplaque assay.11 Briefly, Vero cell monolayers in 24-well plates were infected in duplicate with 10-fold serially diluted samples. Infected monolayers were overlaid with culture medium containing 0.8% methylcellulose, and incubated at 32°C for 6 days, fixed with 80% methanol, and immunostained with a rabbit hyperimmune serum raised against purified HPIV3 virions to detect B/HPIV3 antigens, and a goat hyperimmune serum to the secreted SARS-CoV-2 S to detect co-expression of the S protein, followed by infrared-dye conjugated donkey anti-rabbit IRDye680 IgG and donkey anti-goat IRDye800 IgG secondary antibodies (LiCor). Plates were scanned with the Odyssey infrared imaging system (LiCor). Fluorescent staining for PIV3 proteins and SARS-CoV-2 S was visualized in green and red, respectively, providing for yellow plaque staining when merged.
DELFIA-TRF immunoassay, ELISA
Levels of anti-SARS-CoV-2 S antibodies elicited by B/HPIV3/S-6P were determined by dissociation-enhanced lanthanide fluorescent (DELFIA) time resolved fluorescence (TRF) immunoassay (Perkin Elmer) from NW or BAL following the supplier’s protocol and from serum samples by ELISA11 using the recombinantly-expressed secreted version of S-2P,14 or a fragment (aa 328-531) containing the receptor binding domain (RBD) of the SARS-CoV-2 S protein.53 The secondary antibodies used in both assays were goat anti-monkey IgG(H+L) horseradish peroxidase (HRP) (Thermo Fisher, Cat #PA1-84631), goat anti-monkey IgA (alpha chain)-biotin (Alpha Diagnostic International, Cat #70049), and goat anti-monkey IgM-biotin (Brookwood Biomedical, Cat #1152).
HPIV3 and SARS-CoV-2 neutralization assay
HPIV3-specific neutralizing antibody titers were measured by a 60% plaque reduction neutralization test (PRNT60) on Vero cells.11 The serum neutralizing antibody assays using live SARS-CoV-2 virus was performed in a BSL3 laboratory. Heat-inactivated sera were 2-fold serially diluted in Opti-MEM and mixed with an equal volume of SARS-CoV-2 (100 TCID50) and incubated at 37°C for 1 h. Mixtures were added to quadruplicate wells of Vero E6 cells in 96-well plates and incubated for four days. The 50% neutralizing dose (ND50) was defined as the highest dilution of serum that completely prevented cytopathic effect in 50% of the wells and was expressed as a log10 reciprocal value.11
Lentivirus pseudotype virus neutralization assay
The SARS-CoV-2 pseudovirus neutralization assays were performed as previously reported.30 Briefly, the single-round luciferase-expressing pseudoviruses were generated by co-transfection of plasmids encoding SARS-CoV-2 S of isolate Wuhan-1, GenBank accession number MN908947.3, or of lineages B.1.351/Beta, B.1.1.7/Alpha, B.1.617.2/Delta, B.1.1.529/Omicron), luciferase reporter (pHR’ CMV Luc), lentivirus backbone (pCMV ΔR8.2), and human transmembrane protease serine 2 (TMPRSS2) at a ratio of 1:20:20:0.3 into HEK293T/17 cells (ATCC) with transfection reagent LiFect293. The pseudoviruses were harvested at 72 h post transfection. The supernatants were collected after centrifugation at 478 x g for 10 minutes to remove cell debris, then filtered through a 0.45 μm filter, aliquoted and titrated before neutralization assay. For the antibody neutralization assay, 6-point, 5-fold dilution series were prepared in culture medium (DMEM medium with 10% FBS, 1 % Pen/Strep and 3 μg/ml puromycin). Fifty μl antibody dilution were mixed with 50 μl of diluted pseudoviruses in the 96-well plate and incubated for 30 min at 37°C. Ten thousand ACE2-expressing 293T-cells (293T-hACE2.MF stable cell line) were added in a final volume of 200 μl. Seventy two h later, after carefully removing all the supernatants, cells were lysed with Bright-Glo luciferase assay substrate (Promega), and luciferase activity (relative light units, RLU) was measured. Percent neutralization was normalized relative to uninfected cells as 100% neutralization and cells infected with only pseudoviruses as 0% neutralization. IC50 titers were determined using a log (agonist) vs. normalized response (variable slope) nonlinear function in Prism v8 (GraphPad).
ACE2 binding inhibition assay
As an alternative to a BSL3 SARS-CoV-2 neutralization assay, heat-inactivated macaque sera were evaluated for their ability to inhibit binding of soluble ACE2 to ectodomains of spike proteins [SARS-CoV-2 Panel 25 (K15578U) and 27 (K15609U), MesoScale Diagnostics (MSD)]. These panels contain 96-well, 10-spot plates coated with soluble ectodomains of spike proteins from the vaccine-matched SARS-CoV-2 (Wuhan-1, identical amino acid sequence as WA1/2020) and VoCs (B.1.1.7/Alpha, B.1.351/Beta, B.1.617.2/Delta, B.1.1.529/Omicron BA.1, BA.1+S[R346K], BA.1+S[L452R], B.1.640.2, BA.2.12.1, BA.2, BA.2+S[L452M], BA.2+S[L452R], BA.3, BA.4, and BA.5 sublineages. Briefly, spike-coated plates were blocked with MSD blocker A for 30 min and washed with MSD wash buffer. After a one-hour incubation with 1:20 diluted sera evaluated in duplicate, Sulfo-Tag labelled soluble ACE2 was added to each well. All incubations were performed on a plate shaker at room temperature. Following a one-hour incubation, plates were washed, MSD GOLD electrochemiluminescence read buffer B was added, and chemiluminescence of bound ACE2-Sulfo-Tag was detected using a Meso 1300 Quickplex reader. The average electrochemiluminescence signals in duplicate wells for each serum were determined, as well as the maximum electrochemiluminescence signals of ACE2/S protein binding in no-serum control wells. The ACE2 neutralizing activity of each serum is represented as percent inhibition relative to no-serum controls.
Evaluation of T cell responses
Blood and BAL collection procedures followed ACUC-approved standard operating procedures and limits. Blood that was collected in EDTA tubes was diluted 1:1 with PBS. Fifteen ml of Ficoll-Paque density gradient (GE Healthcare) was added to Leucosep PBMC isolation tubes (Greiner Bio-one) and centrifuged at 1,000 x g for 1 min at 22°C to collect Ficoll below the separation filter. The blood and PBS mixture was added to the Leucosep tubes with Ficoll-Paque and centrifuged at 863 x g for 10 min at 22°C. The upper layer was poured into a 50 ml conical tube and brought to 50 ml with PBS, and then centrifuged at 544 x g for 5 min at 4°C. The cell pellet was resuspended at 2x107 cells/ml in 90% FBS and 10% DMSO for storage at -80°C overnight before being transferred into liquid nitrogen.
Single cell suspensions of PBMCs that had been rested overnight or freshly collected BAL cells were plated at 2x107 cells/ml in 200 μl in 96 well plates with X-VIVO 15 media, with 10% FBS, 1000x Brefeldin (Thermo Fisher Cat#00-4506-51) and 1000x Monensin (Thermo Fisher Cat #00-4505-51), CD107a APC, CD107b APC (both antibodies diluted 1:50), and the indicated peptide pools at 1μg/ml. Replica wells were not stimulated. Spike peptide pools consisted of Peptivator SARS-CoV-2 Prot_S1 (Miltenyi Cat #130-127-048), Peptivator SARS-CoV-2 Prot_S+ (Miltenyi Cat #130-127-312), and Peptivator SARS-CoV-2 Prot_S (Miltenyi Cat #130-127-953) covering the whole spike protein. Nucleocapsid peptide pool consisted of Peptivator SARS-CoV-2 Prot_N (Miltenyi Cat #130-126-699). Cells were stimulated for 6 h at 37°C with 5% CO2. After stimulation, cells were centrifuged at 544 x g for 5 min at 4°C, and further processed by surface staining.
Cells were resuspended in 50 μl surface stain antibodies diluted in PBS with 1% FBS and incubated for 20 min at 4°C. Cells were washed 3 times with PBS with 1% FBS before fixation with intracellular fixation & permeabilization buffer set (Thermo Fisher, Cat# 88-8824-00) for 16 h at 4°C. After fixation, cells were centrifuged at 1,028 x g for 5 min at 4°C without brake and washed once with permeabilization buffer. Cells were resuspended in 50 μl intracellular stains diluted in permeabilization buffer, and stained for 30 min at 4°C. The antibodies used for extracellular and intracellular staining were: CD69 (FITC, clone FN50, Biolegend), granzyme B (BV421, clone GB11, BD Biosciences), CD8a (eFluor 506, clone RPA-T8, Thermo Fisher), IL-2 (BV605, MQ-17H12, Biolegend), IFNγ (BV711, clone 4S.B3, Biolegend), IL-17 (BV785, clone BL168, Biolegend), TNFα (BUV395, clone Mab11, BD Biosciences), CD4 (BUV496, clone SK3, BD Biosciences), CD95 (BUV737, clone DX2, BD Biosciences), CD3 (BUV805, clone SP34-2, BD Biosciences), CD107a (AF647, clone H4A3, Biolegend), CD107b (AF647, clone H4B4, Biolegend), Viability Dye eFluor780 (Thermo Fisher), CD103 (PE, clone B-Ly7, eBioscience), CD28 (PE/Dazzle 594, clone CD28.2, Biolegend), Ki-67 (PE-Cy7, clone B56, BD Biosciences), Foxp3 (AF700, clone PCH101, Thermo Fisher). After staining, cells were washed twice with eBioscience permeabilization buffer and resuspended in PBS supplemented with 1% FBS and 0.05% sodium azide for flow cytometry analysis on the BD Symphony platform. Data were analyzed using FlowJo version 10.
Cytokine analysis
BAL collected on day 3 before immunization and on days 9, 14 and 21 after immunization with B/HPIV3 or B/HPIV3/S-6P were concentrated 10-fold using Amicon concentration tubes (3 kDa cutoff, Millipore). Then, the concentrations of 36 cytokines from each BAL sample were evaluated in duplicate by a bead-based multiplex assay (NHP XL cytokine Luminex premixed kit, R&D Systems) following the manufacturer’s instructions. Data were acquired and analyzed using Luminex xPONENT 4.3 software on Millipore MagPIX, and the concentration of each cytokine was expressed in pg/ml.
Quantification of SARS-CoV-2 RNA
Hundred μl each of NS and BAL fluid collected on day 2, 4 and 6 pc and rectal swabs collected on day 6 pc were inactivated in a BSL3 laboratory using 400 μl buffer AVL (Qiagen) and 500 μl ethanol, and RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s protocol. To extract total RNA from lung homogenates harvested on day 6 pc, 300 μl of each lung homogenate (at a concentration of 0.1 g of tissue/ml) was mixed with 900 μl TRIzol LS (Thermo Fisher) using Phasemaker Tubes (Thermo Fisher) and RNA was extracted using the PureLink RNA Mini Kit (Thermo Fisher) following the manufacturer’s instructions. Then, the SARS-CoV-2 genomic N RNA and subgenomic E mRNA were quantified in triplicate using the TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher) using previously reported TaqMan primers/probes20 , 21 , 48 on the QuantStudio 7 Pro (ThermoFisher). Standard curves were generated using serially diluted pcDNA3.1 plasmids encoding gN or sgE sequences. The limit of detection was 2.57 log10 copies per ml of NS, BAL fluid, or rectal swabs and 3.32 log10 copies per g of lung tissue.
Quantification and statistical analysis
Data sets were assessed for significance using a two-tailed Mann-Whitney test or two-way ANOVA with Sidak’s multiple comparison test in Prism 8 (GraphPad Software). Data were only considered significant at P 0.05. The statistical details of comparisons can be found in the figures, figure legends, and results. Values of n are listed in the text and figure legends, where n represents the number of macaques.
Acknowledgments
We thank Alicia Wojcik and Amanda Havenner and the staff of the National Institute of Allergy and Infectious Diseases (NIAID) Comparative Medicine Branch for animal study support, Jeffrey I. Cohen, Laboratory of Infectious Diseases, NIAID, NIH, for providing plasma from SARS-CoV-2 convalescent individuals, Melanie Cohen, Flow Cytometry Section, Research Technologies Branch, NIAID, NIH for help with the Luminex assay, Ruturaj R. Masvekar and Bibi Bielekova, Neuroimmunological Diseases Unit, NIAID, NIH, for help with the MSD reader, and Peter L. Collins for helpful discussions and for comments on the manuscript. This research was supported by the Intramural Research Program of the NIAID, NIH (project number ZIA AI001298-01).
Author contributions
Conceptualization, C.L.N., D.L.B., and U.J.B.; design of experiments, C.L.N., C.E.N., X.L., H.S.P., Y.M., C.L., R.H., A.C., R.M., P.L., S.M., D.L.B., and U.J.B.; investigation, C.L.N., C.E.N., X.L., H.S.P., Y.M., C.L., C.S., L.Y., I.N.M., T.W.K., A.W., R.M., P.Z., and L.E.V.; reagent development, R.F.J. and N.L.G.; data analysis and visualization, C.L.N., C.E.N., X.L., H.S.P., Y.M., C.L., I.N.M., P.Z., P.L., D.L.B., and U.J.B.; writing—original draft, C.L.N., D.L.B., and U.J.B.; writing—review and editing, C.L.N., C.E.N., X.L., H.S.P., Y.M., C.L., C.S., L.Y., R.H., A.C., I.N.M., T.W.K., A.W., R.M., P.Z., P.L., R.F.J., N.L.G., L.E.V., S.M., D.L.B., and U.J.B.
Declaration of interests
X.L., C.L., C.L.N., S.M., and U.J.B. are inventors on the provisional patent application number 63/180,534, entitled “recombinant chimeric B/HPIV3 expressing SARS-CoV-2 spike protein and its use,” filed by the United States Department of Health and Human Services.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 10, 2022
Data and code availability
There is no dataset or code associated with this paper.
References
- 1.Rankin D.A., Talj R., Howard L.M., Halasa N.B. Epidemiologic trends and characteristics of SARS-CoV-2 infections among children in the United States. Curr. Opin. Pediatr. 2021;33:114–121. doi: 10.1097/MOP.0000000000000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Funk A.L., Florin T.A., Kuppermann N., Tancredi D.J., Xie J., Kim K., Neuman M.I., Ambroggio L., Plint A.C., Mintegi S., et al. Outcomes of SARS-CoV-2-positive youths tested in emergency departments: the global PERN-COVID-19 study. JAMA Netw. Open. 2022;5:e2142322. doi: 10.1001/jamanetworkopen.2021.42322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber J.S., Offit P.A. COVID-19 vaccines for children. Science. 2021;374:913. doi: 10.1126/science.abn2566. [DOI] [PubMed] [Google Scholar]
- 4.Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D'Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiPiazza A.T., Graham B.S., Ruckwardt T.J. T cell immunity to SARS-CoV-2 following natural infection and vaccination. Biochem. Biophys. Res. Commun. 2021;538:211–217. doi: 10.1016/j.bbrc.2020.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard L.M., Edwards K.M., Zhu Y., Williams D.J., Self W.H., Jain S., Ampofo K., Pavia A.T., Arnold S.R., McCullers J.A., et al. Parainfluenza virus Types 1–3 infections among children and adults hospitalized with community-acquired pneumonia. Clin. Infect. Dis. 2021;73:e4433–e4443. doi: 10.1093/cid/ciaa973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGroote N.P., Haynes A.K., Taylor C., Killerby M.E., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human parainfluenza virus circulation, United States, 2011–2019. J. Clin. Virol. 2020;124:104261. doi: 10.1016/j.jcv.2020.104261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karron R.A., Thumar B., Schappell E., Surman S., Murphy B.R., Collins P.L., Schmidt A.C. Evaluation of two chimeric bovine-human parainfluenza virus type 3 vaccines in infants and young children. Vaccine. 2012;30:3975–3981. doi: 10.1016/j.vaccine.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein D.I., Malkin E., Abughali N., Falloon J., Yi T., Dubovsky F., Investigators M.-C. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr. Infect. Dis. J. 2012;31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 11.Liu X., Luongo C., Matsuoka Y., Park H.S., Santos C., Yang L., Moore I.N., Afroz S., Johnson R.F., Lafont B.A.P., et al. A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109744118. e2109744118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt A.C., McAuliffe J.M., Huang A., Surman S.R., Bailly J.E., Elkins W.R., Collins P.L., Murphy B.R., Skiadopoulos M.H. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. J. Virol. 2000;74:8922–8929. doi: 10.1128/jvi.74.19.8922-8929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarjour N.N., Masopust D., Jameson S.C. T cell memory: understanding COVID-19. Immunity. 2021;54:14–18. doi: 10.1016/j.immuni.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szabo P.A., Dogra P., Gray J.I., Wells S.B., Connors T.J., Weisberg S.P., Krupska I., Matsumoto R., Poon M.M.L., Idzikowski E., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814.e6. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng M.Z.M., Wakim L.M. Tissue resident memory T cells in the respiratory tract. Mucosal Immunol. 2022;15:379–388. doi: 10.1038/s41385-021-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson C.E., Namasivayam S., Foreman T.W., Kauffman K.D., Sakai S., Dorosky D.E., Lora N.E., Dagger N.D.T.I.P., Brooks K., Potter E.L., et al. Mild SARS-CoV-2 infection in rhesus macaques is associated with viral control prior to antigen-specific T cell responses in tissues. Sci Immunol. 2022;7:eabo0535. doi: 10.1126/sciimmunol.abo0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.Oh J.E., Song E., Moriyama M., Wong P., Zhang S., Jiang R., Strohmeier S., Kleinstein S.H., Krammer F., Iwasaki A. Intranasal priming induces local lung-resident B cell populations that secrete protective mucosal antiviral IgA. Sci. Immunol. 2021;6:eabj5129. doi: 10.1126/sciimmunol.abj5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbett K.S., Nason M.C., Flach B., Gagne M., O'Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Doremalen N., Purushotham J.N., Schulz J.E., Holbrook M.G., Bushmaker T., Carmody A., Port J.R., Yinda C.K., Okumura A., Saturday G., et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci. Transl. Med. 2021;13:eabh0755. doi: 10.1126/scitranslmed.abh0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan A.O., Feldmann F., Zhao H., Curiel D.T., Okumura A., Tang-Huau T.L., Case J.B., Meade-White K., Callison J., Chen R.E., et al. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. Cell Rep. Med. 2021;2:100230. doi: 10.1016/j.xcrm.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L., Wang Q., Shan C., Yang C., Feng Y., Wu J., Liu X., Zhou Y., Jiang R., Hu P., et al. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat. Commun. 2020;11:4207. doi: 10.1038/s41467-020-18077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce M.G., King H.A.D., Elakhal-Naouar I., Ahmed A., Peachman K.K., Macedo Cincotta C., Subra C., Chen R.E., Thomas P.V., Chen W.H., et al. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 2022;14:eabi5735. doi: 10.1126/scitranslmed.abi5735. [DOI] [PubMed] [Google Scholar]
- 29.Corbett K.S., Werner A.P., Connell S.O., Gagne M., Lai L., Moliva J.I., Flynn B., Choi A., Koch M., Foulds K.E., et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat. Immunol. 2021;22:1306–1315. doi: 10.1038/s41590-021-01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon M.M.L., Rybkina K., Kato Y., Kubota M., Matsumoto R., Bloom N.I., Zhang Z., Hastie K.M., Grifoni A., Weiskopf D., et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 2021;6:eabl9105. doi: 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen D., Mortensen R., Rosenkrands I., Dietrich J., Andersen P. Vaccine-induced Th17 cells are established as resident memory cells in the lung and promote local IgA responses. Mucosal Immunol. 2017;10:260–270. doi: 10.1038/mi.2016.28. [DOI] [PubMed] [Google Scholar]
- 37.Hasenkrug K.J., Feldmann F., Myers L., Santiago M.L., Guo K., Barrett B.S., Mickens K.L., Carmody A., Okumura A., Rao D., et al. Recovery from acute SARS-CoV-2 infection and development of anamnestic immune responses in T cell-depleted rhesus macaques. mBio. 2021;12:e0150321. doi: 10.1128/mBio.01503-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura T., Yamamoto H., Nishizawa M., Hau T.T.T., Harada S., Ishii H., Seki S., Nakamura-Hoshi M., Okazaki M., Daigen S., et al. Subacute SARS-CoV-2 replication can be controlled in the absence of CD8+ T cells in cynomolgus macaques. PLOS Pathog. 2021;17:e1009668. doi: 10.1371/journal.ppat.1009668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohlmeier J.E., Miller S.C., Woodland D.L. Cutting edge: antigen is not required for the activation and maintenance of virus-specific memory CD8+ T cells in the lung airways. J. Immunol. 2007;178:4721–4725. doi: 10.4049/jimmunol.178.8.4721. [DOI] [PubMed] [Google Scholar]
- 41.Auladell M., Jia X., Hensen L., Chua B., Fox A., Nguyen T.H.O., Doherty P.C., Kedzierska K. Recalling the future: immunological memory toward unpredictable influenza viruses. Front. Immunol. 2019;10:1400. doi: 10.3389/fimmu.2019.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell. Reprod. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandrashekar A., Yu J., McMahan K., Jacob-Dolan C., Liu J., He X., Hope D., Anioke T., Barrett J., Chung B., et al. Vaccine protection against the SARS-CoV-2 omicron variant in macaques. Cell. 2022;185:1549–1555.e11. doi: 10.1016/j.cell.2022.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt A.C., McAuliffe J.M., Murphy B.R., Collins P.L. Recombinant bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the respiratory syncytial virus (RSV) G and F proteins can be used to achieve simultaneous mucosal immunization against RSV and HPIV3. J. Virol. 2001;75:4594–4603. doi: 10.1128/JVI.75.10.4594-4603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang B., Munir S., Amaro-Carambot E., Surman S., Mackow N., Yang L., Buchholz U.J., Collins P.L., Schaap-Nutt A. Chimeric bovine/human parainfluenza virus type 3 expressing respiratory syncytial virus (RSV) F glycoprotein: effect of insert position on expression, replication, immunogenicity, stability, and protection against RSV infection. J. Virol. 2014;88:4237–4250. doi: 10.1128/JVI.03481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/JVI.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren X., Glende J., Al-Falah M., de Vries V., Schwegmann-Wessels C., Qu X., Tan L., Tschernig T., Deng H., Naim H.Y., Herrler G. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J. Gen. Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 51.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no dataset or code associated with this paper.