Abstract
The Gulf Coast tick, Amblyomma maculatum, is a vector of several tick-borne pathogens, including Rickettsia parkeri. The ability of R. parkeri to persist within the tick population through transovarial and transstadial transmission, without apparently harming the ticks, contributes to the pathogen’s perpetuation in the tick population. Previous studies have shown that the R. parkeri load in A. maculatum is regulated by the tick tissues’ oxidant/antioxidant balance and the non-pathogenic tick microbiome. To obtain further insights into the interaction between tick and pathogen, we performed a bulk RNA-Seq for differential transcriptomic analysis of ovaries and salivary glands from R. parkeri-infected and uninfected ticks over the feeding course on a host. The most differentially expressed functional category was of bacterial origin, exhibiting a massive overexpression of bacterial transcripts in response to the R. parkeri infection. Candidatus Midichloria mitochondrii and bacteria from the genus Rickettsia were mainly responsible for the overexpression of bacterial transcripts. Host genes were also modulated in R. parkeri-infected tick organs. A similar number of host transcripts from all analyzed functional categories was negatively and positively modulated, revealing a global alteration of the A. maculatum transcriptome in response to pathogen infection. R. parkeri infection led to an increase in salivary transcripts involved in blood feeding success as well as a decrease in ovarian immune transcripts. We hypothesize that these transcriptional alterations facilitate pathogen persistence and transmission within tick population.
Keywords: tick, salivary glands, transcriptome, ovary, Rickettsia parkeri, Amblyomma maculatum, hematophagy, Midichloria mitochondrii
Introduction
Ticks are obligate hematophagous parasites with a wide range of hosts capable of transmitting a variety of pathogens to humans and animals (de la Fuente et al., 2008). Ticks also harbor a diverse non-pathogenic microbiome, typically dominated by tick endosymbionts (Ponnusamy et al., 2014; Guizzo et al., 2020; Díaz-Sánchez et al., 2021; Lejal et al., 2021). Tick pathogens modulate their host’s metabolism to allow them to multiply and persist within the tick before being transmitted (Cabezas-Cruz et al., 2019; Kurokawa et al., 2020). In addition to evading tick defense mechanisms, pathogenic microorganisms must also compete with the non-pathogenic microbiome, which may impact pathogen multiplication and transmission (Narasimhan et al., 2014; Bonnet and Pollet, 2020). The Gulf Coast tick, Amblyomma maculatum is a vector of Rickettsia parkeri, which causes a febrile infection in humans (Sumner et al., 2007; Paddock et al., 2008; Cumbie et al., 2020), and also of Hepatozoon americanum, a pathogen of dogs (Mathew et al., 1998, 1999; Ewing et al., 2002). The distribution of A. maculatum extends from the southeastern states of the United States, bordering the Gulf of Mexico, into Mexico and several other Central and South American countries. In the past decades, it has extended Northwards and to the West in the United States, including the states of Arkansas, Oklahoma, Kansas, and southwestern Tennessee (Sonenshine, 2018).
Rickettsia parkeri infects A. maculatum ticks leading to an efficient transovarial and transstadial transmission without apparent harm to the ticks (Wright et al., 2015). R. parkeri is also transmitted to vertebrate hosts through tick saliva (Banajee et al., 2016). While infected, these mammalian hosts may transmit R. parkeri back to ticks, thus completing the horizontal transmission pathway of R. parkeri survival. Recent studies indicate that the rickettsial load in A. maculatum is regulated by the oxidant/antioxidant balance within tick tissues (Adamson et al., 2013; Budachetri et al., 2017a,b) and by the concurrent non-pathogenic tick microbiome (Budachetri et al., 2014, 2018).
The ability of R. parkeri to be transmitted transstadially and transovarially allows its persistence in the tick population. Therefore, the interaction between R. parkeri and tick tissues is critical for the pathogen’s continued vertical and horizontal transmission. In this study we examined the salivary and ovary transcriptomes of adult female A. maculatum ticks infected by R. parkeri to gain insights into the interactions of the pathogenic bacteria with its tick host and the endosymbiont microbiome. Our observations suggest that infection with R. parkeri leads to transcriptional changes in the host that favor its transmission and its perpetuation in the tick population.
Materials and methods
Ticks
Rickettsia parkeri infected (Rp+) and R. parkeri free (Rp-) Gulf coast tick (A. maculatum) colonies were maintained at the University of Southern Mississippi (United States) according to established methods (Patrick and Hair, 1975). Ticks were kept at room temperature under approximately 90% relative humidity and a 14 h light/10 h dark photoperiod before infestation on a sheep. Adult ticks were blood-fed on sheep and removed 2–7 days post-infestation (dpi) of their hosts, depending on the experimental protocol. The Institutional Animal Care and Use Committee at the University of Southern Mississippi approved all the protocols (#15101501.2 & 17191206.1) before the experiments commenced.
Immunolocalization assay
The infection status of R. parkeri infected (Rp+) ticks was confirmed by immunolocalization. This was done using unfed and partially fed salivary glands and ovarian tissues. Dissected tissues were fixed in 4% PFA and 4% sucrose diluted in 1X PBS and kept at 4°C until needed. Fixed samples were washed three times in 1X PBS prior to permeabilization. Samples were permeabilized in 0.25% Triton X-100 in PBS for 30 min followed by blocking in 2% BSA in PBS for an additional 1 h. Tissues were incubated overnight at 4°C with anti-Rickettsia antibody (M14-13, 1:500, kindly provided by Dr. Ted Hackstadt) that recognizes R. parkeri in 1X PBS containing 2% BSA. This was followed by incubation in the dark with Alexa-Fluor 568 goat anti-mouse secondary antibody (1,500, Invitrogen, Thermo Fisher Scientific, Eugene, Oregon, United States) in 1X PBS containing 2% BSA. Samples were washed three times to remove unbound antibodies and mounted on glass slides using VECTASHIELD antifade mounting medium with DAPI (Vector Laboratories Inc., Burlingame, CA, United States).
Image acquisition
A Leica STELLARIS STED confocal microscope was used to capture R. parkeri images in tick tissues. The 405 UV laser was used to acquire the DAPI channel while the tunable white light laser (WLL) was used to capture the Alexa-Fluor channel. For all images captured, a z-stack of the images consisting of 150–250 slices was compiled. The proprietary Leica built-in postprocessing plugin was used to deconvolute and carry out lightning processing. All images were exported as acquired and compiled in PowerPoint.
Tissue dissections
Salivary glands from R. parkeri-infected and non-infected adult female ticks were dissected from starving ticks and from ticks that fed on sheep for 2, 4, 5, or 7 days. For each time point RNA was isolated from a pool of three to five ticks. Three replicates were obtained for each data point. Ovaries from R. parkeri-infected and non-infected adult female ticks were dissected from ticks that fed on sheep for 2, 4, 5, or 7 days. A pool of three to five ticks was used for each individual time point.
Ticks were dissected within 4 h after removal from the sheep in ice-cold 100 mM 3-(N-Morpholino)-propanesulfonic acid (MOPS) buffer containing 20 mM ethylene glycol bis-(β-aminoethyl ether)-N, N, N′, N′-tetraacetic acid (EGTA), pH 6.8. After removal, each tissue was washed gently in the same ice-cold buffer. All other manipulations were carried out on the ice. The dissected tissues were preserved in RNAlater buffer and stored at −80°C for further RNA isolation.
RNA isolation
RNA was isolated from salivary glands and ovaries of unfed and partially fed ticks using an Illustra RNAspin Mini (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Briefly, the total RNA was eluted into nuclease-free water and the concentration of total RNA was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) and stored at −80°C for further analysis (Karim et al., 2011, 2012). All samples were DNase I treated before conducting RNAseq.
Illumina sequencing
RNA isolated from tick salivary glands and ovaries as described above was submitted to Otogenetics Corporation (Norcross, GA, United States) for RNA-seq assays. Otogenetics checked the integrity and purity of the RNA samples using Agilent Bioanalyzer and OD 260/280. Total RNA was treated with the Ambion GLOBINclear-Human Kit to deplete globin mRNA that may have resulted from the bloodmeal; the Clontech SmartPCR cDNA kit (Clontech Laboratories, Inc., Mountain View, CA, United States) was used to generate 1–2 μg of cDNA from 100 ng of total RNA. Restriction digestion was used to remove adaptor sequences and the resulting cDNA was fragmented using Covaris (Covaris, Inc., Woburn, MA, United States), profiled using Agilent Bioanalyzer, and subjected to Illumina library preparation using NEBNext reagents (New England Biolabs, Ipswich, MA, United States). Agilent Bioanalzyer 2,100 was used to assess the quality, quantity, and size distribution of the Illumina libraries. The libraries were then submitted for Illumina HiSeq2000 sequencing according to the standard operation. Paired-end reads were generated and checked for data quality using FASTQC (Fuente, 2017). The number of reads for each of the 38 libraries ranged from 42 to 72 million, and the average read length was 128 nucleotides (nt) (Supplementary Spreadsheet S1).
Bioinformatic analysis
The resulting reads were trimmed of low quality reads and Illumina primer sequences using the program trim-galore1 that wraps the CutAdapt (Martin, 2011) and FastQC (Brown et al., 2017) programs. The clean reads were assembled in single-ended mode with the programs Trinity (Haas et al., 2013) and Abyss (Simpson et al., 2009) (with k values ranging from 25 to 95 in intervals of 10). The assemblies were compacted using the program CD-hit-est (Li and Godzik, 2006), using a cut-off limit of 98% identity. To obtain the coding sequences (CDS), the largest open reading frame (ORF) of each contig was extracted. CDS shorter than 201 bases were excluded. The putative starting methionine of each CDS was selected based on the identification of a signal peptide indicative of secretion (Nielsen, 2017) and/or a blast (Madden, 2013) similarity match to known proteins from the Swiss-prot (Bairoch, 2000) database and from the Arachnida protein sequences available on GenBank. If the starting methionine was not found, a non-methionine amino acid (aa) was chosen as the first aa of the truncated CDS (Benson, 2000). To identify the bacterial-derived transcripts, we compared by blastp the transcript translations to five strains of R. parkeri available in GenBank, as well as 513 proteomes from the genus Francisella, 24 from Coxiella, one of Candidatus Midichloria mitochondrii and 11 from Anaplasma phagocytophilum. The reads from each library were mapped to the CDS using the RSEM program using the Bowtie2 aligner with the sensitive setting, which reports the best score from a read seed of length 22, assigning negative score values for gaps and mismatches and taking in consideration the qual value of the reads. Supplementary Table S3 reports the number of reads obtained for each library and the number of mapped reads for each library according to the provenance of the transcript (Arachnida, Bacterial or Unknown) (Li and Dewey, 2011). Statistical analysis of differentially expressed genes (DEG) was done with EdgeR (Robinson et al., 2010). Heatmaps were done with the Gplots R package. The data is presented as a hyperlinked excel spreadsheet as previously described (Ribeiro et al., 2004).
Results and discussion
Rickettsia parkeri in ticks
We have previously demonstrated the presence of R. parkeri in A. maculatum salivary glands and ovaries by qPCR (Budachetri et al., 2018). In the current work, we confirmed this tissue distribution of R. parkeri using Rickettsia-specific antibodies. Confocal images showed intracellular localization of rod-shaped Rickettsiae in the unfed and partially fed salivary gland and ovarian tissues (Figures 1, 2). We next isolated RNA from infected and non-infected tissues to analyze the transcriptional changes in A. maculatum salivary glands and ovaries in response to the pathogen infection.
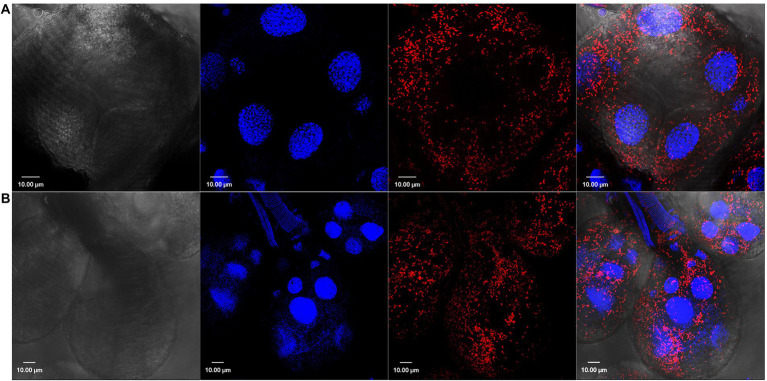
Figure 1.
Confocal images showing tissue localization of Rickettsia parkeri in (A) unfed and (B) partially blood-fed Amblyomma maculatum salivary glands. Rod-shaped rickettsiae are localized intracellularly and depicted by their location in each salivary acini. DAPI (blue) was used to stain nuclei; Rickettsiae-specific antibody (M14-13) followed by Alexa-Fluor 568 conjugated anti-mouse IgG (red) was used to stain R. parkeri and differential interference contrast was used to capture tissue outline. Scale bar = 10 μM.
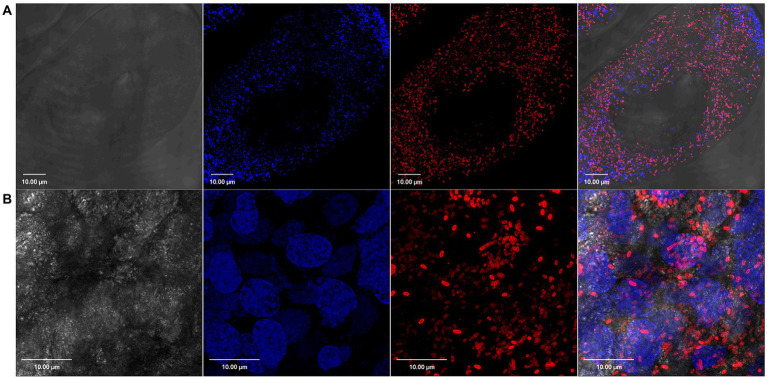
Figure 2.
Confocal images showing tissue localization of Rickettsia parkeri in (A) unfed and (B) partially blood-fed Amblyomma maculatum ovaries. Rod-shaped Rickettsiae are localized intracellularly as depicted by their location in each oocyte. DAPI (blue) was used to stain nuclei; Rickettsiae-specific antibody (M14-13) followed by Alexa-Fluor 568 conjugated anti-mouse IgG (red) was used to stain R. parkeri and differential interference contrast was used to capture tissue outline. Scale bar = 10 μM.
Transcriptomic analysis
Assembly of the 38 libraries from the salivary glands and ovaries of R. parkeri infected and non-infected adult female A. maculatum ticks under different blood engorgement regimens resulted in 66,578 CDS (coding sequences). After mapping the reads from each library to these transcripts, 45,459 had a minimum read value of 50 (Supplementary Spreadsheet S1). The following analyses were performed with this set of transcripts and will follow the statistical results from EdgeR which contrasted the several variables under study. Eight times or more underexpressed or overexpressed differentially expressed transcripts (DET) with a false discovery rate of 0.05 or less were further analyzed.
Differential gene expression in the Amblyomma maculatum microbiome in response to Rickettsia parkeri infection
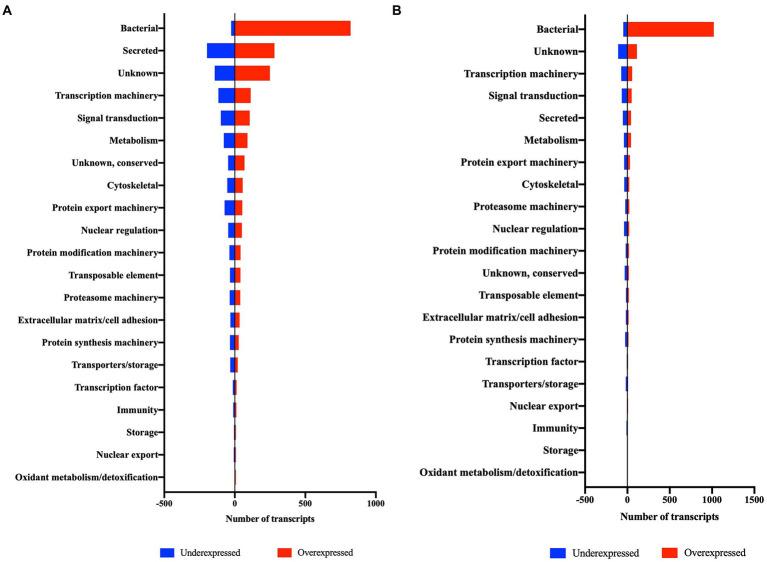
Infection by R. parkeri altered the expression of genes of several functional categories in A. maculatum salivary glands and ovaries over the feeding course on a host (Table 1). Intriguingly, the bacterial category, comprising of transcripts from bacterial origin, was the most differentially expressed between R. parkeri infected and non-infected salivary glands and ovaries, corresponding to 25.9 and 47.9% of the total, respectively (Table 1 and Figure 3). Of the bacterial transcripts, the vast majority were overexpressed (97% in salivary glands and 95.5% in ovaries), indicating a high metabolic activity consistent with bacterial multiplication.
Table 1.
Differentially expressed transcripts (DET– at least 8 fold over or underexpressed and with a false discovery rate of 0.05 or lower) from A. maculatum ovary and salivary glands in response to R. parkeri infection grouped into functional categories.
| Functional category | Number of transcripts DET | |||||||
|---|---|---|---|---|---|---|---|---|
| Salivary glands | Ovary | |||||||
| Over | Under | Over | Under | |||||
| Bacterial | 821 | 26 | 1,020 | 48 | ||||
| Cytoskeletal | 56 | 53 | 23 | 37 | ||||
| Oxidant metabolism/detoxification | 8 | 2 | 2 | 4 | ||||
| Extracellular matrix/cell adhesion | 34 | 31 | 15 | 18 | ||||
| Immunity | 11 | 11 | 6 | 12 | ||||
| Metabolism | 90 | 78 | 43 | 41 | ||||
| Nuclear export | 8 | 7 | 8 | 5 | ||||
| Nuclear regulation | 50 | 61 | 21 | 40 | ||||
| Protein export machinery | 53 | 72 | 30 | 38 | ||||
| Protein modification machinery | 41 | 38 | 19 | 21 | ||||
| Proteasome machinery | 39 | 36 | 22 | 26 | ||||
| Protein synthesis machinery | 28 | 34 | 13 | 27 | ||||
| Secreted | 281 | 197 | 43 | 53 | ||||
| Signal transduction | 106 | 99 | 50 | 66 | ||||
| Storage | 8 | 6 | 2 | 4 | ||||
| Transposable element | 40 | 34 | 17 | 17 | ||||
| Transcription factor | 13 | 14 | 9 | 8 | ||||
| Transcription machinery | 113 | 116 | 55 | 72 | ||||
| Transporters/storage | 21 | 32 | 8 | 19 | ||||
| Unknown, conserved | 69 | 47 | 17 | 32 | ||||
| Unknown | 249 | 142 | 112 | 109 | ||||
| Total | 2,139 | 1,136 | 1,535 | 697 | ||||
| Total over+under | 3,275 | 2,232 | ||||||
Figure 3.
Differentially expressed transcripts (DET) of salivary glands (A) and ovary (B) of R. parkeri-infected A. maculatum adult females in comparison to uninfected ticks grouped into functional categories.
The microorganism primarily responsible for the differential expression of bacterial genes in R. parkeri-infected ticks was Candidatus Midichloria mitochondrii (CMM) (Table 2). Transcripts from CMM corresponded to 56.1 and 60.6% of the bacterial transcripts in salivary glands and ovaries, respectively. All CMM differentially expressed transcripts were overexpressed in R. parkeri-infected ticks, showing a positive gene modulation in response to the pathogen. CMM is a maternally-inherited endosymbiont identified in several hard tick species (Epis et al., 2008), including A. maculatum (Budachetri et al., 2018). CMM prevalence is variable, depending on the tick species and the life-stage analyzed (Cafiso et al., 2016; Duron et al., 2017). Its unique lifestyle allows the bacterium to reside not only in the tick cell cytoplasm but also in the mitochondrial inter-membrane space (Sassera et al., 2006). Our previous work has shown that 5-day R. parkeri-infected A. maculatum adult females had an increase in CMM levels in ovaries and salivary glands (Budachetri et al., 2018), suggesting a synergistic relationship between R. parkeri and the endosymbiont. The transcriptomic analysis described in the current study corroborated these results, showing a significant increase in the number of transcripts from CMM in salivary glands and ovaries of R. parkeri-infected ticks over the feeding course (Table 2).
Table 2.
Bacterial differentially expressed transcripts (DET) in ovary and salivary glands in response to R. parkeri infection.
| Bacterial genera | Number of transcripts DET | |||||||
|---|---|---|---|---|---|---|---|---|
| Salivary glands | Ovary | |||||||
| Over | Under | Over | Under | |||||
| CMM | 475 | 0 | 647 | 0 | ||||
| Rickettsia parkeri | 207 | 3 | 183 | 3 | ||||
| Other Rickettsia | 135 | 17 | 151 | 18 | ||||
| Francisella sp. | 4 | 6 | 39 | 27 | ||||
| Total | 821 | 26 | 1,020 | 48 | ||||
| Total over+under | 847 | 1,068 | ||||||
CMM, Candidatus Midichloria mitochondrii; Over, overexpressed; Under, underexpressed.
In adult females of the species Ixodes ricinus, CMM has been found in all individuals examined (Sassera et al., 2006), being identified in high numbers as the dominant bacterium in the tick ovary (Guizzo et al., 2020). In contrast, CMM was present in low levels in A. maculatum ovary but multiplied in response to infection by R. parkeri (Budachetri et al., 2018). Low levels of CMM have been identified in I. ricinus salivary glands (Olivieri et al., 2019). In A. maculatum CMM was also found in low numbers, but expanded significantly in response to R. parkeri infection (Budachetri et al., 2018). CMM is suggested to act as a nutritional symbiont due to its ability to synthesize the B vitamins biotin (B7) and folic acid (B9) (Sassera et al., 2011). Obligate hematophagous arthropods are presumed to rely on symbionts to obtain micronutrients deficient in host blood that are essential to their development and fitness (Rio et al., 2016; Duron and Gottlieb, 2020). Our transcriptomic analysis showed that a CMM putative biotin synthase (transcript ID 7830) and a putative folate metabolism protein (transcript ID 1034) were overexpressed in R. parkeri-infected A. maculatum ticks in both salivary glands and ovaries. This indicates that CMM from A. maculatum as well as CMM identified in I. ricinus may encode genes for the biosynthetic pathways of B vitamins. Even though CMM has been identified as part of the indigenous microbiome of A. maculatum (Budachetri et al., 2018), the transcripts implicated in B vitamin synthesis were only expressed in R. parkeri-infected ticks. This suggests that CMM does not play a role as an essential B vitamin provider in this tick life-stage. Interestingly, a study showed that A. maculatum R. parkeri-free larva and nymph molting success was affected negatively in comparison to R. parkeri-infected ticks, indicating a positive effect of the pathogen on tick fitness (Wright et al., 2015). These observations led us to hypothesize that the increase in CMM numbers in infected ticks could confer a metabolic advantage, providing an extra support of vitamins to the tick, improving its fitness, and benefiting the pathogen indirectly. Biotin and folic acid are cofactors of several biological reactions that could be involved in pathogen persistence within the tick, transovarial and transstadial transmission between ticks, or the transmission to the vertebrate host. The physiological role of CMM in other A. maculatum life-stages and its interaction with R. parkeri and other pathogens remains to be elucidated and could reveal important insights into the non-pathogen-pathogen-tick interaction.
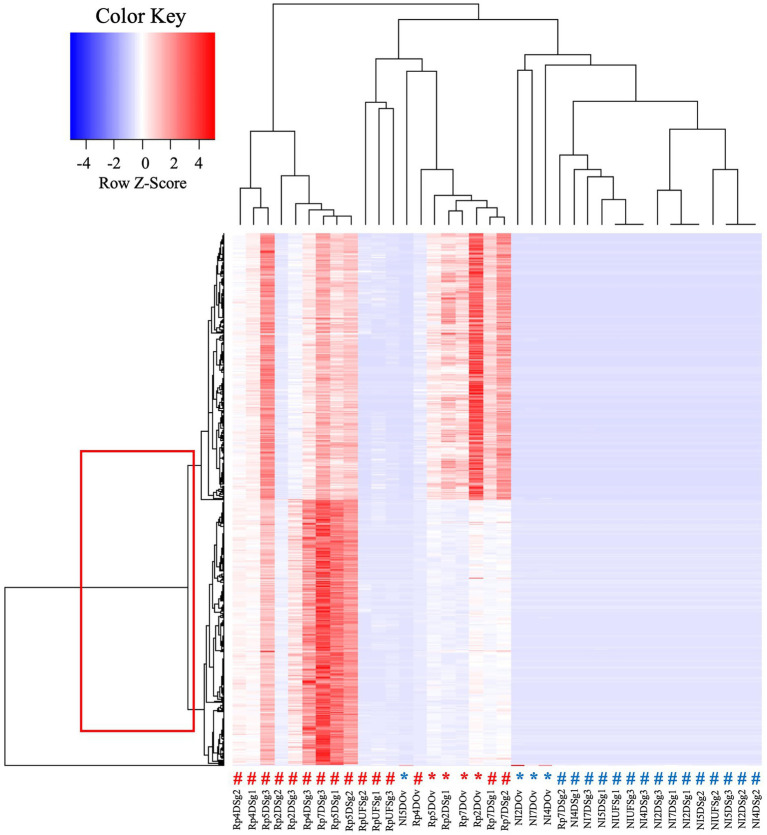
Intriguingly, the heat map analysis of transcripts from CMM in ovaries and salivary glands revealed two clusters, suggesting that there were two lineages of CMM in A. maculatum (Figure 4). Within the salivary glands of blood-fed R. parkeri-infected ticks the two strains of CMM were identified in most of the samples over the feeding course, while in the ovaries Cluster 2 was absent. Cluster 1, the most abundant lineage in the salivary glands, was 90% similar to CMM sequences deposited in GenBank while Cluster 2 was only 73% similar. Transcripts from both CMM clusters were overexpressed in R. parkeri infected ticks, suggesting a similar pattern of regulation of microbial growth regardless of the CMM strain. Three different lineages of CMM have already been described in association with ticks (Buysse and Duron, 2018). They present distinct evolutionary strategies: two of them are more generalists, being acquired horizontally between tick species, and the other one has a high degree of specificity to the Ixodes genus (Buysse and Duron, 2018). This variable degree of interdependency suggests that CMM might affect tick hosts differentially. Further studies are needed to elucidate the nature of the interactions between each CMM lineage and A. maculatum ticks. Of note, this study represents the first description of two potential lineages of CMM coexisting within the same tick species.
Figure 4.
Heat map analysis of transcripts from CMM in salivary glands and ovaries of R. parkeri-infected and non-infected A. maculatum ticks. The box indicates two clusters of transcripts. The # symbols indicate results for salivary gland transcripts, and the * indicate those for ovary transcripts, their red color indicating results for R. parkeri infected and the blue symbols for non-infected ticks. Rp, Rickettsia parkeri-infected; NI, non-infected; Sg, salivar glands; Ov:ovary; D, day of feeding. A more detailed description of the experimental condition of each sample can be found in the Supplementary Spreadsheet S3.
As expected, R. parkeri genes were identified as differentially overexpressed in infected ticks (Table 2). The pathogen infects the tick ovary and salivary glands contributing to both vertical and horizontal transmission (Budachetri et al., 2018). Transcripts from other Rickettsia species were also identified as largely overexpressed in ovary and salivary glands (Table 2). Bacteria from the genus Rickettsia have already been described either as tick pathogens or endosymbionts (Kurtti et al., 2016; Hensley et al., 2021). The non-pathogenic Candidatus Rickettsia andeanae has been identified in A. maculatum salivary glands and ovaries (Lee et al., 2019). The co-infection of Candidatus R. andeanae and R. parkeri resulted in increased levels of the pathogen, suggesting a synergic interaction (Lee et al., 2019). Rickettsia buchneri is the main symbiont of Ixodes scapularis, being vertically transmitted and capable of encoding genes for the biosynthetic pathway of the B vitamin biotin (Gillespie et al., 2012; Kurtti et al., 2015). In R. parkeri-infected ticks, transcripts from Rickettsias other than R. parkeri followed the same expression profile as CMM, being markedly overexpressed in response to the pathogen. A transcript for folate metabolism protein (Transcript ID 9558) expressed by a Rickettsia sp. was found overexpressed in ovary and salivary glands of R. parkeri-infected ticks, suggesting that it could act as a nutritional endosymbiont. However, was observed for CMM, the folate gene was only expressed in R. parkeri-infected ticks. This suggests that the bacterium does not play a nutritional role in this tick life-stage, while strengthening the hypothesis that there is a synergic interaction between non-pathogenic-pathogenic Rickettsiales.
Francisella constitutes an important fraction of the A. maculatum microbiome (Budachetri et al., 2014). Francisella species have been described as endosymbionts in several tick species (Binetruy et al., 2020; Buysse et al., 2021, 2022). In Ornithoros moubata, a Francisella bacterial species was shown to play a role as a nutritional symbiont, being essential for the tick development (Duron et al., 2018). Nevertheless, the role of other Francisellas in their tick hosts remain unexplored. Among the differentially expressed transcripts from bacteria, those from Francisella appeared to be the least altered in response to R. parkeri infection. This corroborated previous results showing that Francisella did not multiply in 5-day adult females salivary glands and ovaries in R. parkeri infected-ticks (Budachetri et al., 2018).
Together, these results showed that the A. maculatum non-pathogenic microbiome was differentially modulated in the presence of R. parkeri. While transcripts from CMM and of the genus Rickettsia were largely overexpressed in response to the pathogen, transcripts from Francisella were only slightly affected. It is interesting to point out that the infection by R. parkeri selectively led to an increase in the numbers of non-pathogenic Rickettsiales, such as CMM and other bacteria from the genus Rickettsia. We hypothesized that the synergic interaction between Rickettsiales could favor the pathogen’s transovarial and transstadial transmission through an improvement in tick fitness due to the B vitamins provided by CMM and Rickettsia sp. Alternatively, the expansion in non-pathogenic Rickettsiales numbers could benefit R. parkeri perpetuation by playing a role in masking the pathogen from tick immune recognition. Tick endosymbionts are assumed to be tolerated by the tick immune system due to the biological advantage they confer on host fitness. Therefore, the extra support of vitamins provided by the non-pathogenic Rickettsiales could confound the tick immune system allowing the multiplication of all Rickettsiales, including the pathogenic R. parkeri. More studies are needed to test this hypothesis and fully elucidate the interaction between the A. maculatum non-pathogenic microbiome and R. parkeri.
Differentially expressed tick genes in the salivary glands of Rickettsia parkeri-infected Amblyomma maculatum
Even though bacterial genes represented a large fraction of the differentially expressed genes in response to the R. parkeri infection, several functional categories of tick genes were also affected by the presence of the pathogen (Table 1; Figure 3). A similar number of transcripts were underexpressed and overexpressed in most functional categories, showing a global dysregulation of the tick metabolism in response to the pathogen. Intriguingly, while transcriptomic analysis of tick tissues in response to pathogen infection frequently display a marked overexpression of transcripts involved in tick host defense (Mulenga et al., 2003; Rudenko et al., 2005; Heekin et al., 2013; Kim et al., 2018; Martins et al., 2019), the A. maculatum infection by R. parkeri caused a phenomenon that we described as a “balanced dysregulation” of the tick metabolism.
Rickettsia parkeri infection resulted in the overexpression of a slight majority (54.3%) of diferentially-expressed tick transcripts detected in the salivary glands. The functional category comprising secreted transcripts was the main component responsible for the observed overexpression, representing 21.3% of the overexpressed transcripts. Tick infection by pathogens has previously been correlated with alterations in the expression of salivary secreted transcripts (Ribeiro et al., 2006; Mercado-Curiel et al., 2011; Martins et al., 2019; Paulino et al., 2021). Metalloproteases, evasins, Lipocalins, mucins, and protease inhibitors were some of the secreted proteins found in this study to be differentially modulated in response to R. parkeri infection (Table 3). Salivary transcripts have also been shown to be differentially expressed in A. americanum ticks infected by the Rickettsial pathogen Ehrlichia chaffeensis (Kim et al., 2018). Interestingly, while in R. parkeri-infected ticks most of the transcripts for lipocalins were overexpressed (Table 3), in E. chaffeensis-infected ticks they were underexpressed (Kim et al., 2018). Proteins from this family were also underexpressed in Amblyomma aureolatum ticks infected by R. rickettsii (Martins et al., 2019). Lipocalins counteract vertebrate host defenses by playing an anti-inflammatory role through the binding of small hydrophobic molecules, such as histamine, at the tick bite site (Valdés, 2014). In addition to lipocalins, metalloproteases were found to be underexpressed in E. chaffeensis-infected A. americanum ticks (Kim et al., 2018), but were overexpressed in R. parkeri-infected ticks (Table 3). Vaccination and gene silencing experiments have shown that salivary metalloproteases contribute to the blood feeding success in several tick species (Decrem et al., 2008; Imamura et al., 2009; Ali et al., 2015).
Table 3.
Secreted differentially expressed transcripts (DET– at least 8 fold over or underexpressed and with a false discovery rate of 0.05 or lower) from A. maculatum salivary glands in response to R. parkeri infection grouped into functional categories.
| Functional category | Number of transcripts DET Salivary glands | |||
|---|---|---|---|---|
| Over | Under | |||
| Metalloproteases | 39 | 17 | ||
| Evasins | 26 | 18 | ||
| Lipocalins | 55 | 50 | ||
| Mucins | 27 | 22 | ||
| Protease inhibitors | 17 | 16 | ||
Over, overexpressed; Under, underexpressed.
Salivary mucins and evasins were also overexpressed in response to R. parkeri infection (Table 3). Proteins from these families were identified as components of the salivary gland and saliva of different tick species (Hayward et al., 2017; Ribeiro et al., 2017; Tirloni et al., 2020). Mucins are assumed to help to lubricate tick mouthparts facilitating the blood acquisition (Francischetti et al., 2008), while evasins bind host chemokines, inhibiting the inflammatory response of the host through the recruitment of leukocytes (Hayward et al., 2017). While several evasins were found to be differentially expressed in this study, with the majority being overexpressed, only one underexpressed transcript for evasin was found in the sialotranscriptome of Rhipicephalus microplus infected by the intracellular protozoan Theileria equi (Paulino et al., 2021). The overexpression of the salivary transcripts for lipocalins, metalloproteases, mucins and evasins induced by R. parkeri infection suggest that the pathogen can modulate the expression of tick salivary factors to facilitate its propagation by guaranteeing a successful host blood feeding. The second most overexpressed category was that of transcripts with unknown function, representing 18.9% of the total. However due to the lack of similarity with the analyzed databases, we cannot predict their function. Our results demonstrate that pathogen infection induced the expression of novel tick salivary transcripts, which may play a role in R. parkeri persistence and transmission.
Although pathogens have the ability to evade the immune system to survive within the tick host, immune transcripts are frequently overexpressed in tick salivary glands in response to pathogen infection (Kim et al., 2018; Martins et al., 2019; Paulino et al., 2021). It is assumed that the tick immune system is activated to limit pathogen numbers to levels that are not harmful to the host. Interestingly, in the salivary glands of R. parkeri-infected ticks, the same number of immune transcripts were underexpressed and overexpressed (Table 1), indicating a balanced dysregulated tick immune response to the infection. This suggests a modulation of the tick immunity, allowing the pathogen to survive without negatively impacting tick fitness. Other functional classes typically modulated positively or negatively in response to pathogen infection were balanced, in accordance with a dysregulated gene expression in R. parkeri-infected A. maculatum (Table 1). From the 33 differentially expressed transcripts grouped into the protease inhibitors functional class, a similar number was underexpressed and overexpressed (Table 3). Out of these, 24 were transcripts for Kunitz inhibitors, which also exhibited a balance of under-expressed and over-expressed (Table 3). An overall negative or positive modulation of Kunitz inhibitors, depending on the tick-pathogen interaction analyzed, has been shown in other reports (Kim et al., 2018; Martins et al., 2019). Protease inhibitors of Kunitz type are classically described as anti-coagulants, facilitating blood uptake (Corral-Rodríguez et al., 2009). Alternatively, a Kunitz inhibitor of Dermacentor variabilis was demonstrated to exert a bacteriostatic effect against Rickettsia montanensis (Ceraul et al., 2008). The balanced dysregulation of Kunitz inhibitors in response to R. parkeri infection could be part of a pathogen compensatory mechanism enabling A. maculatum successful blood feeding and/or pathogen survival without major alterations in tick metabolism. The remodeling of the tick cytoskeleton has been described as part of pathogens’ mechanisms to establish infection in both tick and vertebrate host (de la Fuente et al., 2016). In the salivary glands of adult female I. scapularis ticks infected by Anaplasma phagocytophilum, transcripts for components of the cytoskeleton were differentially expressed (Fuente, 2017). In our study a similar number of transcripts for tick cytoskeleton proteins were underexpressed and overexpressed (Table 1), showing a balanced dysregulated modulation of these tick genes in response to R. parkeri infection. This balanced dysregulation could facilitate tick cell invasion by the pathogen without considerably affecting the tick’s regular metabolism.
Differentially expressed tick genes in the ovaries of Rickettsia parkeri-infected Amblyomma maculatum
In the ovaries, several A. maculatum transcripts were differentially expressed in response to R. parkeri. In contrast to our findings in salivary glands, the pathogen caused a slight underexpression of the total transcripts (55.7%), across several functional categories (Table 1). It has been shown that tick pathogens that are transovarially transmitted, such as R. parkeri, cause an infection-related differential expression of tick proteins (Mulenga et al., 2003; Rachinsky et al., 2007; Heekin et al., 2013; Antunes et al., 2019). In R. microplus ovarian genes involved in the immune system, detoxification, and stress response were found to be differentially modulated in response to infection by Babesia bovis. (Heekin et al., 2013). The A. maculatum transcriptional response to R. parkeri infection, however, was distinct from those previously described. While immune-related transcripts for microplusin and defensins were overexpressed in the ovaries of B. bovis-infected R. microplus (Heekin et al., 2013), transcripts categorized as immune-related were found in our study to be mostly underexpressed in the ovaries of R. parkeri-infected ticks (Table 1). Immune transcripts were also overexpressed in the ovary of D. variabilis ticks infected by Ricketsia montanensis (Mulenga et al., 2003). We speculate that the downregulation of the A. maculatum ovarian immune response could facilitate vertical transmission of the pathogen, guaranteeing their successful persistence in tick population and reducing their dependence of reservoir hosts.
Moreover, Kunitz inhibitors, which are assumed to act as components of the ovarian immune response against pathogens, were overexpressed in the ovaries of R. microplus infected by B. bovis (Rachinsky et al., 2007; Heekin et al., 2013). In our analysis, two transcripts for Kunitz inhibitors were found differentially expressed in the ovaries in response to R. parkeri infection (Supplementary Spreadsheet S2). One of them was underexpressed and the other one overexpressed, corroborating to the suggested balanced dysregulated nature of the tick-pathogen interaction. A similar transcriptional modulatory profile was found for heat-shock proteins. While ovarian heat-shock proteins were induced in R. microplus and Rhipicephalus annulatus in response to B. bovis and B. bigemina, respectively (Heekin et al., 2013; Antunes et al., 2019), in R. parkeri-infected A. maculatum the same number of transcripts for these proteins were underexpressed and overexpressed (Supplementary Spreadsheet S2). Heat-shock proteins are stress response-related proteins induced as the result of protein misfolding caused by infection (Hernandez et al., 2019). Along with other tick molecules, heat-shock proteins play a role in limiting pathogen numbers (Sonenshine and Macaluso, 2017). The transcriptional regulatory profile for Kunitz inhibitors and heat-shock proteins falls into the suggested balanced dysregulation rule and could reflect compensatory mechanisms of the tick metabolism to the well-established R. parkeri infection.
While several transcripts for cytochrome P450 were overexpressed in response to B. bovis infection in R. microplus (Heekin et al., 2013), in our study they were not differentially expressed. Other transcripts related to detoxification such as glutathione S-transferase, superoxidase dismutase and peroxinectin were induced by B. bovis infection (Heekin et al., 2013), but do not appear differentially expressed in our analysis. Similarly, a transcript for glutathione S-transferase was found overexpressed in the ovaries of D. variabilis infected by Ricketsia montanensis (Mulenga et al., 2003). Detoxification/oxidation molecules are suggested to be involved with the host protection against the oxidative stress caused by pathogen infection (Hernandez et al., 2019). In contrast to previous reports, our results show that infection by R. parkeri does not considerably affect the tick detoxification metabolism. Indeed, only six transcripts for the functional category of detoxification/oxidation were differentially expressed in R. parkeri-infected ticks, most of which were underexpressed (Table 1). We hypothesize that the underexpression of tick transcripts involved in detoxification and oxidation metabolism, in association with the underexpression of tick immune-related transcripts, may represent a mechanism by which the pathogen alters host transcription to favor perpetuation and vertical transmission. Nevertheless, it is important to point out that a small fraction of tick pathogens are vertically transmitted, and the number of studies focused on tick ovary transcriptional/proteomic modulation in response to vertically transmitted pathogens is still scarce. Further investigation of additional tick-pathogen associations could contribute to the understanding of the mechanisms by which specific pathogens manipulate tick metabolism to facilitate vertical transmission.
In both analyzed organs the presence of R. parkeri caused a balanced dysregulated expression of tick transcripts from most of the functional classes. While the majority of the reports describing transcriptomic analyses of ticks infected by pathogens have analyzed the tick metabolic responses to a novel infection (Kim et al., 2018), the ticks analyzed in this study were from an established R. parkeri-infected colony maintained in association with the pathogen over generations. As a consequence, the typical overexpression of transcripts involved with tick host defenses that occurs in response to the first encounter with a pathogen was not observed in this long-term tick-pathogen interaction. We speculate that the balanced dysregulation of the differentially expressed transcripts observed in this study could be the result of compensatory mechanisms of a well-adapted tick-pathogen interaction. This may explain how the pathogen is able to survive and continue transmission without significant deleterious effects to the development and fitness of the tick host.
Conclusion
In this study, we performed a transcriptomic differential analysis of A. maculatum ovaries and salivary glands in response to R. parkeri infection. The results revealed that bacterial genes were the most differentially expressed. This is the result of an expansion in non-pathogenic and pathogenic Rickettssiales numbers over the feeding course on the host. Tick genes were also modulated in response to the bacterial infection, being slightly overexpressed in the salivary glands and underexpressed in the ovaries. A balanced dysregulation of tick metabolism in response to R. parkeri-infection is suggested, reflecting a well-adapted tick-pathogen interaction.
While in the salivary glands a slight overexpression of transcripts involved in blood acquisition could facilitate pathogen propagation, in the ovaries the underexpression of immune transcripts may contribute to the pathogen’s vertical transmission. In addition, we speculate that there is a relationship of cooperation between tick and pathogen with mutual beneficial effects to both species, due to the significant increase in endosymbiont levels. The extra support of vitamins provided by the non-pathogenic Rickettssiales has the potential to improve tick fitness, benefiting R. parkeri perpetuation indirectly. This suggests an orchestrated synergic tripartite interaction between tick, pathogen, and endosymbionts. The generation of an A. maculatum germ-free tick line could provide further insights into this complex relationship, revealing intriguing aspects of the role played by endosymbionts in pathogen-infected and non-infected ticks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at the University of Southern Mississippi approved all the protocols (#15101501.2 and 17191206.1) before the experiments commenced.
Author contributions
JR and SK: conceptualization, formal analysis, project administration, resources, supervision, writing-review, and editing. KB, AA, JR, and SK: data curation, investigation, and methodology. SK: funding acquisition. AA and SK: validation. MG, JR, and SK: visualization. MG: writing original draft. All authors contributed to the article and approved the submitted version.
Funding
This research was principally supported by USDA NIFA award # 2017–67017-26171, the Mississippi INBRE (an Institutional Award (IdeA) from the National Institute of General Medical Sciences of the National Institutes of Health under award P20GM103476). JR and MG were supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (Vector-Borne Diseases: Biology of Vector Host Relationship, Z01AI000810-21. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The funders played no role in the study design, data collection, and analysis, decision to publish, or manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge to the NIH Fellows Editorial Board for the editorial assistance.
Footnotes
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1023980/full#supplementary-material
References
- Adamson S. W., Browning R. E., Budachetri K., Ribeiro J. M. C., Karim S. (2013). Knockdown of selenocysteine-specific elongation factor in Amblyomma maculatum alters the pathogen burden of rickettsia parkeri with epigenetic control by the Sin3 histone deacetylase corepressor complex. PLoS One 8:e82012. doi: 10.1371/journal.pone.0082012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A., Fernando Parizi L., Garcia Guizzo M., Tirloni L., Seixas A., Vaz S., et al. (2015). Immunoprotective potential of a rhipicephalus (boophilus) microplus metalloprotease. Vet. Parasitol. 207, 107–114. doi: 10.1016/j.vetpar.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Antunes S., Couto J., Ferrolho J., Sanches G. S., Merino Charrez J. O., De la Cruz Hernández N., et al. (2019). Transcriptome and proteome response of Rhipicephalus annulatus tick vector to Babesia bigemina infection. Front. Physiol. 10:318. doi: 10.3389/fphys.2019.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48. doi: 10.1093/nar/28.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banajee K. H., Verhoeve V. I., Harris E. K., Macaluso K. R. (2016). Effect of Amblyomma maculatum (Acari: Ixodidae) saliva on the acute cutaneous immune response to rickettsia parkeri infection in a murine model. J. Med. Entomol. 53, 1252–1260. doi: 10.1093/jme/tjw125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A. (2000). GenBank. Nucleic Acids Res. 28, 15–18. doi: 10.1093/nar/28.1.15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binetruy F., Buysse M., Lejarre Q., Barosi R., Villa M., Rahola N., et al. (2020). Microbial community structure reveals instability of nutritional symbiosis during the evolutionary radiation of Amblyomma ticks. Mol. Ecol. 29, 1016–1029. doi: 10.1111/mec.15373, PMID: [DOI] [PubMed] [Google Scholar]
- Bonnet S. I., Pollet T. (2020). Update on the intricate tango between tick microbiomes and tick-borne pathogens. Parasite Immunol. 43:e12813. doi: 10.1111/pim.12813, PMID: [DOI] [PubMed] [Google Scholar]
- Brown J., Pirrung M., McCue L. A. (2017). FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K., Browning R. E., Adamson S. W., Dowd S. E., Chao C.-C., Ching W.-M., et al. (2014). An insight into the microbiome of the Amblyomma maculatum (Acari: Ixodidae). J. Med. Entomol. 51, 119–129. doi: 10.1603/me12223, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K., Crispell G., Karim S. (2017a). Amblyomma maculatum SECIS binding protein 2 and putative selenoprotein P are indispensable for pathogen replication and tick fecundity. Insect Biochem. Mol. Biol. 88, 37–47. doi: 10.1016/j.ibmb.2017.07.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K., Kumar D., Crispell G., Beck C., Dasch G., Karim S. (2018). The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of rickettsia parkeri in the Gulf coast tick vector. Microbiome 6, 141–115. doi: 10.1186/s40168-018-0524-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K., Kumar D., Karim S. (2017b). Catalase is a determinant of the colonization and transovarial transmission of rickettsia parkeri in the Gulf coast tick Amblyomma maculatum. Insect Mol. Biol. 26, 414–419. doi: 10.1111/imb.12304, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysse M., Binetruy F., Leibson R., Gottlieb Y., Duron O. (2022). Ecological contacts and host specificity promote replacement of nutritional endosymbionts in ticks. Microb. Ecol. 83, 776–788. doi: 10.1007/s00248-021-01773-0, PMID: [DOI] [PubMed] [Google Scholar]
- Buysse M., Duron O. (2018). Multi-locus phylogenetics of the Midichloria endosymbionts reveals variable specificity of association with ticks. Parasitology 145, 1969–1978. doi: 10.1017/S0031182018000793, PMID: [DOI] [PubMed] [Google Scholar]
- Buysse M., Floriano A. M., Gottlieb Y., Nardi T., Comandatore F., Olivieri E., et al. (2021). A dual endosymbiosis supports nutritional adaptation to hematophagy in the invasive tick Hyalomma marginatum. elife 10:e72747. doi: 10.7554/eLife.72747, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Cruz A., Espinosa P., Alberdi P., de la Fuente J. (2019). Tick–pathogen interactions: the metabolic perspective. Trends Parasitol. 35, 316–328. doi: 10.1016/j.pt.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Cafiso A., Bazzocchi C., De Marco L., Opara M. N., Sassera D., Plantard O. (2016). Molecular screening for Midichloria in hard and soft ticks reveals variable prevalence levels and bacterial loads in different tick species. Ticks Tick. Borne. Dis. 7, 1186–1192. doi: 10.1016/j.ttbdis.2016.07.017, PMID: [DOI] [PubMed] [Google Scholar]
- Ceraul S. M., Dreher-Lesnick S. M., Mulenga A., Rahman M. S., Azad A. F. (2008). Functional characterization and novel rickettsiostatic effects of a Kunitz-type serine protease inhibitor from the tick Dermacentor variabilis. Infect. Immun. 76, 5429–5435. doi: 10.1128/IAI.00866-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Rodríguez M. Á., Macedo-Ribeiro S., Barbosa Pereira P. J., Fuentes-Prior P. (2009). Tick-derived Kunitz-type inhibitors as antihemostatic factors. Insect Biochem. Mol. Biol. 39, 579–595. doi: 10.1016/j.ibmb.2009.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Cumbie A. N., Espada C. D., Nadolny R. M., Rose R. K., Dueser R. D., Hynes W. L., et al. (2020). Survey of rickettsia parkeri and Amblyomma maculatum associated with small mammals in southeastern Virginia. Ticks Tick. Borne. Dis. 11:101550. doi: 10.1016/j.ttbdis.2020.101550, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J., Estrada-Peña A., Cabezas-Cruz A., Kocan K. M. (2016). Anaplasma phagocytophilum uses common strategies for infection of ticks and vertebrate hosts. Trends Microbiol. 24, 173–180. doi: 10.1016/j.tim.2015.12.001, PMID: [DOI] [PubMed] [Google Scholar]
- de la Fuente J., Estrada-Pena A., Venzal J. M., Kocan K. M., Sonenshine D. E. (2008). Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 13, 6938–6946. doi: 10.2741/3200 [DOI] [PubMed] [Google Scholar]
- Decrem Y., Mariller M., Lahaye K., Blasioli V., Beaufays J., Zouaoui Boudjeltia K., et al. (2008). The impact of gene knock-down and vaccination against salivary metalloproteases on blood feeding and egg laying by Ixodes ricinus. Int. J. Parasitol. 38, 549–560. doi: 10.1016/j.ijpara.2007.09.003, PMID: [DOI] [PubMed] [Google Scholar]
- Díaz-Sánchez S., Fernández A. M., Habela M. A., Calero-Bernal R., de Mera I. G. F., de la Fuente J. (2021). Microbial community of Hyalomma lusitanicum is dominated by Francisella-like endosymbiont. Ticks Tick. Borne. Dis. 12:101624. doi: 10.1016/j.ttbdis.2020.101624, PMID: [DOI] [PubMed] [Google Scholar]
- Duron O., Binetruy F., Noël V., Cremaschi J., McCoy K. D., Arnathau C., et al. (2017). Evolutionary changes in symbiont community structure in ticks. Mol. Ecol. 26, 2905–2921. doi: 10.1111/mec.14094, PMID: [DOI] [PubMed] [Google Scholar]
- Duron O., Gottlieb Y. (2020). Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol. 36, 816–825. doi: 10.1016/j.pt.2020.07.007, PMID: [DOI] [PubMed] [Google Scholar]
- Duron O., Morel O., Noël V., Buysse M., Binetruy F., Lancelot R., et al. (2018). Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr. Biol. 28, 1896–1902.e5. doi: 10.1016/j.cub.2018.04.038, PMID: [DOI] [PubMed] [Google Scholar]
- Epis S., Sassera D., Beninati T., Lo N., Beati L., Piesman J., et al. (2008). Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 135, 485–494. doi: 10.1017/S0031182007004052, PMID: [DOI] [PubMed] [Google Scholar]
- Ewing S. A., DuBois J. G., Mathew J. S., Panciera R. J. (2002). Larval Gulf Coast ticks (Amblyomma maculatum) [Acari: Ixodidae] as host for Hepatozoon americanum [Apicomplexa: Adeleorina]. Vet. Parasitol. 103, 43–51. doi: 10.1016/S0304-4017(01)00572-6, PMID: [DOI] [PubMed] [Google Scholar]
- Francischetti I. M. B., Mans B. J., Meng Z., Gudderra N., Veenstra T. D., Pham V. M., et al. (2008). An insight into the sialome of the soft tick, Ornithodorus parkeri. Insect Biochem. Mol. Biol. 38, 1–21. doi: 10.1016/j.ibmb.2007.09.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente J. (2017). Remodeling of tick cytoskeleton in response to infection with Anaplasma phagocytophilum. Front. Biosci. 22, 4574–1844. doi: 10.2741/4574, PMID: [DOI] [PubMed] [Google Scholar]
- Gillespie J. J., Joardar V., Williams K. P., Driscoll T., Hostetler J. B., Nordberg E., et al. (2012). A rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 194, 376–394. doi: 10.1128/JB.06244-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizzo M. G., Neupane S., Kucera M., Perner J., Frantová H., da Silva Vaz I., et al. (2020). Poor unstable midgut microbiome of hard ticks contrasts with abundant and stable monospecific microbiome in ovaries. Front. Cell. Infect. Microbiol. 10:211. doi: 10.3389/fcimb.2020.00211, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward J., Sanchez J., Perry A., Huang C., Rodriguez Valle M., Canals M., et al. (2017). Ticks from diverse genera encode chemokine-inhibitory evasin proteins. J. Biol. Chem. 292, 15670–15680. doi: 10.1074/jbc.M117.807255, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heekin A. M., Guerrero F. D., Bendele K. G., Saldivar L., Scoles G. A., Dowd S. E., et al. (2013). The ovarian transcriptome of the cattle tick, Rhipicephalus microplus (Boophilus), feeding upon a bovine host infected with Babesia bovis. Parasit. Vectors 6:276. doi: 10.1186/1756-3305-6-276, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley J. R., Zambrano M. L., Williams-Newkirk A. J., Dasch G. A. (2021). Detection of rickettsia species, and Coxiella -like and Francisella -like endosymbionts in Amblyomma americanum and Amblyomma maculatum from a shared field site in Georgia, United States of America. Vector-Borne Zoonotic Dis. 21, 509–516. doi: 10.1089/vbz.2020.2683, PMID: [DOI] [PubMed] [Google Scholar]
- Hernandez E. P., Talactac M. R., Fujisaki K., Tanaka T. (2019). The case for oxidative stress molecule involvement in the tick-pathogen interactions -an omics approach. Dev. Comp. Immunol. 100:103409. doi: 10.1016/j.dci.2019.103409, PMID: [DOI] [PubMed] [Google Scholar]
- Imamura S., Da Silva Vaz I., Konnai S., Yamada S., Nakajima C., Onuma M., et al. (2009). Effect of vaccination with a recombinant metalloprotease from Haemaphysalis longicornis. Exp. Appl. Acarol. 48, 345–358. doi: 10.1007/s10493-009-9245-3, PMID: [DOI] [PubMed] [Google Scholar]
- Karim S., Browning R., Ali L., Truhett R. (2012). Laboratory-infected Ehrlichia chaffeensis female adult Amblyomma americanum salivary glands reveal differential gene expression. J. Med. Entomol. 49, 547–554. doi: 10.1603/ME11214 [DOI] [PubMed] [Google Scholar]
- Karim S., Singh P., Ribeiro J. M. C. (2011). A deep insight into the Sialotranscriptome of the Gulf coast tick Amblyomma maculatum. PLoS One 6:e28525. doi: 10.1371/journal.pone.0028525, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Jaworski D. C., Cheng C., Nair A. D. S., Ganta R. R., Herndon N., et al. (2018). The transcriptome of the lone star tick, Amblyomma americanum, reveals molecular changes in response to infection with the pathogen, Ehrlichia chaffeensis. J. Asia Pac. Entomol. 21, 852–863. doi: 10.1016/j.aspen.2018.05.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa C., Lynn G. E., Pedra J. H. F., Pal U., Narasimhan S., Fikrig E. (2020). Interactions between Borrelia burgdorferi and ticks. Nat. Rev. Microbiol. 18, 587–600. doi: 10.1038/s41579-020-0400-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti T., Burkhardt N., Heu C., Munderloh U. (2016). Fluorescent protein expressing Rickettsia buchneri and Rickettsia peacockii for tracking symbiont-tick cell interactions. Vet. Sci. 3:34. doi: 10.3390/vetsci3040034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti T. J., Felsheim R. F., Burkhardt N. Y., Oliver J. D., Heu C. C., Munderloh U. G. (2015). Rickettsia buchneri sp. Nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int. J. Syst. Evol. Microbiol. 65, 965–970. doi: 10.1099/ijs.0.000047, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Moraru G. M., Stokes J. V., Benton A. N., Wills R. W., Nabors H. P., et al. (2019). Amblyomma maculatum-associated rickettsiae in vector tissues and vertebrate hosts during tick feeding. Exp. Appl. Acarol. 77, 187–205. doi: 10.1007/s10493-019-00343-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejal E., Chiquet J., Aubert J., Robin S., Estrada-Peña A., Rue O., et al. (2021). Temporal patterns in Ixodes ricinus microbial communities: an insight into tick-borne microbe interactions. Microbiome 9:153. doi: 10.1186/s40168-021-01051-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Madden T. (2013). “The BLAST Sequence Analysis Tool. 2013 Mar 15,” in The NCBI Handbook [Internet]. 2nd Edn. Bethesda (MD): National Center for Biotechnology Information (US). [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martins L. A., Malossi C. D., Galletti M. F. B. D. M., Ribeiro J. M., Fujita A., Esteves E., et al. (2019). The transcriptome of the salivary glands of Amblyomma aureolatum reveals the antimicrobial peptide Microplusin as an important factor for the tick protection against Rickettsia rickettsii infection. Front. Physiol. 10:529. doi: 10.3389/fphys.2019.00529, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew J. S., Ewing S. A., Panciera R. J., Kocan K. M. (1999). Sporogonic development of Hepatozoon americanum (Apicomplexa) in its definitive host, Amblyomma maculatum (Acarina). J. Parasitol. 85, 1023–1031. doi: 10.2307/3285663, PMID: [DOI] [PubMed] [Google Scholar]
- Mathew J., Ewing S., Panciera R., Woods J. (1998). Experimental transmission of Hepatozoon americanum Vincent-Johnson et al., 1997 to dogs by the Gulf coast tick Amblyomma maculatum. Koch. Vet. Parasitol. 80, 1–14. doi: 10.1016/S0304-4017(98)00189-7, PMID: [DOI] [PubMed] [Google Scholar]
- Mercado-Curiel R. F., Palmer G. H., Guerrero F. D., Brayton K. A. (2011). Temporal characterization of the organ-specific Rhipicephalus microplus transcriptional response to Anaplasma marginale infection. Int. J. Parasitol. 41, 851–860. doi: 10.1016/j.ijpara.2011.03.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A., Macaluso K. R., Simser J. A., Azad A. F. (2003). Dynamics of rickettsia-tick interactions: identification and characterization of differentially expressed mRNAs in uninfected and infected Dermacentor variabilis. Insect Mol. Biol. 12, 185–193. doi: 10.1046/j.1365-2583.2003.00400.x, PMID: [DOI] [PubMed] [Google Scholar]
- Narasimhan S., Rajeevan N., Liu L., Zhao Y. O., Heisig J., Pan J., et al. (2014). Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15, 58–71. doi: 10.1016/j.chom.2013.12.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. (2017). Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73. doi: 10.1007/978-1-4939-7015-5_6, PMID: [DOI] [PubMed] [Google Scholar]
- Olivieri E., Epis S., Castelli M., Varotto Boccazzi I., Romeo C., Desirò A., et al. (2019). Tissue tropism and metabolic pathways of Midichloria mitochondrii suggest tissue-specific functions in the symbiosis with Ixodes ricinus. Ticks Tick. Borne. Dis. 10, 1070–1077. doi: 10.1016/j.ttbdis.2019.05.019, PMID: [DOI] [PubMed] [Google Scholar]
- Paddock C. D., Finley R. W., Wright C. S., Robinson H. N., Schrodt B. J., Lane C. C., et al. (2008). Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin. Infect. Dis. 47, 1188–1196. doi: 10.1086/592254, PMID: [DOI] [PubMed] [Google Scholar]
- Patrick C. D., Hair J. A. (1975). Laboratory rearing procedures and equipment for multi: host ticks (Acarina: Ixodidae). J. Med. Entomol. 12, 389–390. doi: 10.1093/jmedent/12.3.389, PMID: [DOI] [PubMed] [Google Scholar]
- Paulino P., Vitari G., Rezende A., Couto J., Antunes S., Domingos A., et al. (2021). Characterization of the Rhipicephalus (Boophilus) microplus Sialotranscriptome profile in response to Theileria equi infection. Pathogens 10:167. doi: 10.3390/pathogens10020167, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy L., Gonzalez A., Van Treuren W., Weiss S., Parobek C. M., Juliano J. J., et al. (2014). Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl. Environ. Microbiol. 80, 354–359. doi: 10.1128/AEM.02987-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinsky A., Guerrero F. D., Scoles G. A. (2007). Differential protein expression in ovaries of uninfected and Babesia-infected southern cattle ticks, Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 37, 1291–1308. doi: 10.1016/j.ibmb.2007.08.001, PMID: [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M. C., Alarcon-Chaidez F., Ivo I. M., Mans B. J., Mather T. N., Valenzuela J. G., et al. (2006). An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 36, 111–129. doi: 10.1016/j.ibmb.2005.11.005, PMID: [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M. C., Slovák M., Francischetti I. M. B. (2017). An insight into the sialome of Hyalomma excavatum. Ticks Tick. Borne. Dis. 8, 201–207. doi: 10.1016/j.ttbdis.2016.08.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro J. M. C., Topalis P., Louis C. (2004). AnoXcel: an Anopheles gambiae protein database. Insect Mol. Biol. 13, 449–457. doi: 10.1111/j.0962-1075.2004.00503.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rio R. V. M., Attardo G. M., Weiss B. L. (2016). Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 32, 739–749. doi: 10.1016/j.pt.2016.05.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N., Golovchenko M., Edwards M. J., Grubhoffer L. (2005). Differential expression of Ixodes ricinus tick genes induced by blood feeding or Borrelia burgdorferi infection. J. Med. Entomol. 42, 36–41. doi: 10.1093/jmedent/42.1.36 [DOI] [PubMed] [Google Scholar]
- Sassera D., Beninati T., Bandi C., Bouman E. A. P., Sacchi L., Fabbi M., et al. (2006). Candidatus Midichloria mitochondrii’, an endosymbiont of the Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 56, 2535–2540. doi: 10.1099/ijs.0.64386-0, PMID: [DOI] [PubMed] [Google Scholar]
- Sassera D., Lo N., Epis S., D’Auria G., Montagna M., Comandatore F., et al. (2011). Phylogenomic evidence for the presence of a flagellum and cbb 3 oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 28, 3285–3296. doi: 10.1093/molbev/msr159, PMID: [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., Birol İ. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123. doi: 10.1101/gr.089532.108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine D. (2018). Range expansion of tick disease vectors in North America: implications for spread of tick-borne disease. Int. J. Environ. Res. Public Health 15:478. doi: 10.3390/ijerph15030478, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine D. E., Macaluso K. R. (2017). Microbial invasion vs. tick immune regulation. Front. Cell. Infect. Microbiol. 7:390. doi: 10.3389/fcimb.2017.00390, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner J. W., Durden L. A., Goddard J., Stromdahl E. Y., Clark K. L., Reeves W. K., et al. (2007). Gulf Coast ticks (Amblyomma maculatum) and rickettsia parkeri, United States. Emerg. Infect. Dis. 13, 751–753. doi: 10.3201/eid1305.061468, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni L., Braz G., Nunes R. D., Gandara A. C. P., Vieira L. R., Assumpcao T. C., et al. (2020). A physiologic overview of the organ-specific transcriptome of the cattle tick Rhipicephalus microplus. Sci. Rep. 10:18296. doi: 10.1038/s41598-020-75341-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés J. J. (2014). Antihistamine response: a dynamically refined function at the host-tick interface. Paras. Vect. 7:491. doi: 10.1186/s13071-014-0491-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. L., Gaff H. D., Sonenshine D. E., Hynes W. L. (2015). Experimental vertical transmission of rickettsia parkeri in the Gulf coast tick, Amblyomma maculatum. Ticks Tick. Borne. Dis. 6, 568–573. doi: 10.1016/j.ttbdis.2015.04.011, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.